Abstract
Myeloblastosis (MYB) genes are important transcriptional regulators of plant growth, development, and secondary metabolic biosynthesis pathways, such as capsaicinoid biosynthesis in Capsicum. Although MYB genes have been identified in Capsicum annuum, no comprehensive study has been conducted on other Capsicum species. We identified a total of 251 and 240 MYB encoding genes in Capsicum chinense MYBs (CcMYBs) and Capsicum baccatum MYBs (CbMYBs). The observation of twenty tandem and 41 segmental duplication events indicated expansion of the MYB gene family in the C. chinense genome. Five CcMYB genes, i.e., CcMYB101, CcMYB46, CcMYB6, CcPHR8, and CcRVE5, and two CaMYBs, i.e., CaMYB3 and CaHHO1, were found within the previously reported capsaicinoid biosynthesis quantitative trait loci. Based on phylogenetic analysis with tomato MYB proteins, the Capsicum MYBs were classified into 24 subgroups supported by conserved amino acid motifs and gene structures. Also, a total of 241 CcMYBs were homologous with 225 C. annuum, 213 C. baccatum, 125 potato, 79 tomato, and 23 Arabidopsis MYBs. Synteny analysis showed that all 251 CcMYBs were collinear with C. annuum, C. baccatum, tomato, potato, and Arabidopsis MYBs spanning over 717 conserved syntenic segments. Using transcriptome data from three fruit developmental stages, a total of 54 CcMYBs and 81 CaMYBs showed significant differential expression patterns. Furthermore, the expression of 24 CcMYBs from the transcriptome data was validated by quantitative real-time (qRT) PCR analysis. Eight out of the 24 CcMYBs validated by the qRT-PCR were highly expressed in fiery hot C. chinense than in the lowly pungent C. annuum. Furthermore, the co-expression analysis revealed several MYB genes clustered with genes from the capsaicinoid, anthocyanin, phenylpropanoid, carotenoid, and flavonoids biosynthesis pathways, and related to determining fruit shape and size. The homology modeling of 126 R2R3 CcMYBs showed high similarity with that of the Arabidopsis R2R3 MYB domain template, suggesting their potential functional similarity at the proteome level. Furthermore, we have identified simple sequence repeat (SSR) motifs in the CcMYB genes, which could be used in Capsicum breeding programs. The functional roles of the identified CcMYBs could be studied further so that they can be manipulated for Capsicum trait improvement.
Keywords: Capsicum chinense, baccatum, MYB, Solanaceae, fruit development, transcription factors
Introduction
The myeloblastosis (MYB) gene family is one of the largest transcription factor (TF) families in plants (Romero et al., 1998; Riechmann et al., 2000). MYB TFs have one or more imperfect repeats of the characteristic DNA-binding domain (DBD) in the basic region of a protein (Klempnauer and Sippel, 1987). Each repeat comprises about 53 amino acids with three regularly placed tryptophan residues forming a helix-turn-helix structure (Ogata et al., 1994; König et al., 1996). MYB TFs with only one repeat are called “MYB1R” or MYB-related, while those with two, three, or four repeats are called “R2R3-MYB,” “MYB3R,” or “MYB4R,” respectively. In plants, most MYBs belong to the R2R3 class, unlike in animals, and exhibit plant-specific responses (Martin and Paz-Ares, 1997). MYB TFs play key roles in the regulation of phenylpropanoid and flavonoid metabolism in plants; for instance, in Arabidopsis, MYB123 partly determines the accumulation of proanthocyanidin (PA) in the coat of seeds (Nesi et al., 2001), MYB11, MYB12, and MYB111 are involved in the transcriptional regulation of the chalcone synthase and flavonol synthase genes (Mehrtens et al., 2005), while MYB14 and MYB15, along with WRKY53, are reported to regulate stilbene synthesis in Chinese wild grapes (Wang et al., 2020b). MYB88 and MYB124 were reported to have diverse roles (Lei et al., 2015), such as in the regulation of mitotic divisions of the stomatal guard mother cell (Lai et al., 2005; Lee et al., 2013) and direct transcriptional regulation of auxin transporter PIN-FORMED proteins in roots of Arabidopsis thaliana (Xie et al., 2010; Wang et al., 2015; Geng et al., 2018). Reports also suggest their role in female reproductive development (Makkena et al., 2012) and conditional repression of non-stomatal epidermal cells in Arabidopsis cotyledons (Yang, 2016). MYB75 and MYB90 are known to activate phenylpropanoid biosynthetic genes and the accumulation of purple anthocyanins in Arabidopsis (Kranz et al., 1998; Borevitz et al., 2000). Furthermore, several MYBs have been reported to induce anthocyanin production in different organs, including fruits in tomatoes (Kiferle et al., 2015; Jian et al., 2019; Yan et al., 2020), potato (Li et al., 2021), and in other plants (Quattrocchio et al., 2006; Takos et al., 2006; Cutanda-Perez et al., 2009; Czemmel et al., 2009; Kortstee et al., 2011; Wang et al., 2017; Yan et al., 2019). In Capsicum, MYBA and CaAN2 control anthocyanin pigmentation in flower and fruit tissues (Aguilar-Barragán and Ochoa-Alejo, 2014; Jung et al., 2019). However, MYB TFs have been scarcely studied for their protein structures; for example, a MYB-related motif in Arabidopsis recognizes the major groove of target DNA via the amino acid residues present in three alpha helices while binding to the minor groove using an N-terminal arm (Hosoda et al., 2002). Another report characterized the crystal structure of the MYB domain from an Antirrhinum majus single MYB repeat RADIALIS (RAD) TF, which functions in the development of floral asymmetry (Stevenson et al., 2006).
The genus Capsicum comprises several species grown worldwide mostly for vegetables and spices, which are of high economic and nutritional value. The Capsicum fruit is known for its unique attribute of pungency owing to alkaloids, known as capsaicinoids complex, mainly capsaicin and dihydrocapsaicin (Antonious and Jarret, 2006), which have pharmaceutical applications (Fattori et al., 2016). There exists a wide variation in capsaicin content in Capsicum genotypes, with the highest being reported in Bhut jolokia (C. chinense; Sarpras et al., 2016; Chhapekar et al., 2020). R2R3-MYB31 was reported as a transcriptional regulator of capsaicinoid biosynthetic genes (CBGs; Arce-Rodríguez and Ochoa-Alejo, 2017). The same MYB was reported to be encoded by a pungency-controlling locus Pun3 (Han et al., 2019), and its gene promoter showed natural variations between high and low pungent C. annuum species (Zhu et al., 2019). CaMYB108 confers a pungent flavor to Capsicum genotypes and controls stamen development, and it is found to be induced by methyl jasmonate (Sun et al., 2019). Recently, CaMYB48 was discovered to directly control the expression of CBGs acyl transferase (AT3a) and ketoacyl-ACP synthase (KasIa) and the accumulation of capsaicinoids in C. annuum (Sun et al., 2020).
Myeloblastosis genes have been identified only in C. annuum (Wang et al., 2020c; Arce-Rodríguez et al., 2021), and no comprehensive study has been reported in other Capsicum species, such as C. chinense, C. baccatum, and C. frutescens. In this study, we identified MYB genes in the C. chinense, C. baccatum, and C. annuum genomes, and the analysis of their expression was performed using transcriptome data and validated by quantitative real-time (qRT) PCR in the early green (EG), mature green (MG), and breaker (Br) fruit developmental stages of highly pungent C. chinense and lowly pungent C. annuum. Seven MYB genes were found within the previously reported capsaicinoids quantitative trait loci (QTLs) (Han et al., 2018; Park et al., 2019). The co-expression analysis revealed several MYB genes that clustered with capsaicinoid, anthocyanin, phenylpropanoid, flavonoid, fruit shape and size, carotenoid, and vitamin C biosynthesis pathway genes. Furthermore, we analyzed duplications of MYB genes in C. chinense, and comparative analysis with C. baccatum, C. annuum, tomato, potato, eggplant, and Arabidopsis showed conserved syntenic segments (CSSs) and collinear MYB genes, and diversification among them. In addition, we performed homology modeling of R2R3 CcMYB proteins and developed simple sequence repeat (SSR) markers in genic and promoter regions of Capsicum MYBs, which can be used to manipulate pungency levels in future Capsicum breeding programs. The identified MYB genes in this study could be used in the future to understand their regulatory roles in diverse biological functions including the capsaicinoid biosynthetic pathway.
Materials and Methods
Identification of MYB Genes in Capsicum Spp
For the identification of MYB genes, a local blastp analysis (e-value <1e-05) was performed against 34,974, 35,853, and 31,353 full-length protein sequences of C. chinense (GCA_002271895.2; Kim et al., 2017), C. annuum (GCA_000710875.1; Qin et al., 2014), and C. baccatum (GCA_002271885.2; Kim et al., 2017), respectively, using the R2R3-MYB family protein sequences of Arabidopsis (Stracke et al., 2001), tomato (Li et al., 2016), potato (Li et al., 2021), and C. annuum (Arce-Rodríguez et al., 2021) as a query. For the Hidden Markov Model (HMM) analysis, the sequences of the Capsicum proteins were queried against the protein sequences of MYB DNA-binding domains (PF00249 and PF13921) using HMMER (v3.2.1; http://hmmer.janelia.org/). The duplicate sequences were removed manually, and the remaining sequences were cross-checked for the presence of the MYB domain using the Conserved Domains Database (CDD) of the National Centre for Biotechnology Information (NCBI) (Lu et al., 2020) and the Simple Modular Architecture Research Tool (SMART); http://smart.embl-heidelberg.de/) database. The resultant C. chinense and C. baccatum MYB encoding genes were given acronyms according to their amino acid sequence homology with C. annuum MYB genes (Arce-Rodríguez et al., 2021). Physicochemical features, such as molecular weight, theoretical isoelectric point (pI), instability index, aliphatic index, and grand average of hydrophobicity (GRAVY) of the MYB protein sequences were estimated using ProtParam (https://web.expasy.org/protparam/; Gasteiger et al., 2005). The subcellular localization of Capsicum MYBs was determined using Cello (v.2.5 http://cello.life.nctu.edu.tw/).
Chromosomal Distribution, Gene Duplication, and Co-localization With Capsaicinoid QTLs
The chromosomal positions of Capsicum MYB genes were obtained from the gene feature file (gff) of their genomes. The physical locations of Capsicum MYB genes, along with the capsaicin and dihydrocapsaicin QTLs as reported earlier (Han et al., 2018; Park et al., 2019), were represented across 12 Capsicum chromosomes using TBtools (v1.068; Chen et al., 2020). The duplication of MYB genes within the C. chinense genome was identified based on filter criteria of >75% identity and query coverage of above 75% of the gene length. Gene pairs with a <100-kb (kilobase) distance on the same chromosome were considered as tandem duplicates, while those with >100 kb were considered as segmental duplicates. The rate of non-synonymous (Ka) and synonymous substitutions (Ks) and their ratio (ω = Ka/Ks) for all duplicated gene pairs were estimated using KaKs_Calculator 2.0 (Wang et al., 2010). The value of ω ~ 0 indicates neutral selection, ω <1 indicates purifying selection, and ω > 1 indicates positive selection. The date of duplication (diversion time) was calculated using the formula T = Ks/2λ, assuming a clock-like rate (λ) of 6.96 synonymous substitutions per 10−9 years (Moniz de Sá and Drouin, 1996).
Gene Structure, Motifs, Cis-Elements Analysis, and Homology Modeling
The structure of the MYBs genes (exons and introns) was represented using Gene Structure Display Server (GSDS2.0; http://gsds.gao-lab.org/). The conserved motifs in the MYB protein sequences were identified using the Multiple Em for Motif Elicitation (MEME) suite (http://meme-suite.org/tools/meme). The maximum number of motif 40, the minimum width of each motif 6, and the maximum width of 120 were used as parameters. The identified conserved motifs were then confirmed with previously characterized Arabidopsis motifs (Stracke et al., 2001). The cis-regulatory elements and motifs in the 1,500 bp (base-pairs) upstream promoter region of Capsicum MYB genes were speculated using the PlantCARE website (Lescot et al., 2002). The 126 R2R3 CcMYBs were analyzed for protein tertiary (or 3D; three-dimensional) structure-based homology models using the Phyre2 server (Kelley et al., 2015). The models were predicted based on the alignment coverage, percent identity, and percent confidence score for the individual CcMYB protein sequences.
Phylogenetic Analysis
The multiple protein sequence alignment of Capsicum MYBs was performed using Clustal Omega (Madeira et al., 2019) with default parameters. The phylotree was constructed in MEGAX (v.10.1.8; Kumar et al., 2018) using maximum likelihood (ML) methods with a phylogeny test of 1,000 bootstrap replications and a Jones–Taylor–Thornton (JTT) model with uniform rates among sites applied to infer evolutionary history. The Nearest-Neighbor-Interchange (NNI), as an ML heuristic method, was used for phylogeny tree inference. The combined phylogenetic tree of C. chinense and C. baccatum R2R3 MYBs with already known Arabidopsis MYBs (AtMYBs) was generated using the above-mentioned parameters. The phylogenetic tree data with bootstrap values were visualized using the Interactive Tree of Life (iTOL) server (https://itol.embl.de/).
Plant Materials and Growth Conditions
Seeds of Capsicum genotypes belonging to highly pungent C. chinense (Bhut Jolokia; Acc-Cc74; 925084.8 Scoville Heat Unit; SHU) and lowly pungent C. annuum (Acc-Ca18; 7034.4 SHU) were sown in agro peat and vermiculite (in the proportion 3:1; Sarpras et al., 2019). The seedlings were grown in a glasshouse at 24–26°C temperature with 16-h light and 8-h dark photoperiod and 70% humidity. The 1-month-old plants were transferred into the soil and grown until fruit maturity in the glasshouse at Jawaharlal Nehru University, New Delhi. Fruit tissues of the early green (EG; 5–10 days postanthesis; DPA), mature green (MG; 20–25 DPA), and breaker (Br; 30–45 DPA) stages were harvested and immediately frozen in liquid nitrogen and stored in a deep freezer at −80°C until RNA extraction.
RNA Sequencing and Differential Gene Expression Analysis
Total RNA from the EG, MG, and Br fruit stages of C. chinense (Acc-Cc74) and C. annuum (Acc-Ca18) was extracted using an MN Nucleospin RNA Plant kit (Takara, Mountain View, CA, United States). The integrity of the RNA samples was checked using a bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). The RNA samples from three biological replicates of each fruit stage, with RNA integrity number (RIN) > 8, were used for library preparation using TruSeq RNA Sample Prep Kits (Illumina, San Diego, CA, United States) and sequenced using a HiSeq XTen (Illumina, San Diego, CA, United States) paired-end platform with an average read length of 150 bp. The quality of raw reads was evaluated with FastQC (v0.11.5), and adapter sequences along with low quality reads (phred score <20) were removed using TrimGalore (v0.4.4) as descrbied earlier (Chhapekar et al., 2020; Rawoof et al., 2020). The filtered good quality reads from C. chinense and C. annuum were mapped to their respective genomes (Qin et al., 2014; Kim et al., 2017) using the Hisat2 (Kim et al., 2019) program. The expression of all genes was estimated using StringTie (v2.0.6; Kim et al., 2019). Read counts of transcripts were quantified using the feature Counts (v1.5.1; Liao et al., 2014). The normalization of raw read counts was performed using TMM methods (Robinson and Oshlack, 2010), and differentially expressed genes (DEGs) between two tissues were identified using the glmQLFit and glmQLFTest functions in the edgeR package (Robinson et al., 2010). Genes with adjusted p < 0.01 and fold change (FC) >1.5 were considered as significantly expressed between the two tissues. The normalized expressions and differential expression pattern of Capsicum MYBs among fruit tissues were illustrated in the form of a heatmap using gplots (Warnes et al., 2020).
Expression Analysis of MYB Genes by Quantitative Real-Time (qRT) PCR
A total of 24 CcMYB genes showing DE among the fruit developmental stages of C. chinense and C. annuum, along with CBG (AT3, KasI, AMT, ACS1, BCAT, and COMT) genes, were validated by qRT-PCR. Gene-specific primers were designed from exonic sequences using standard criteria (Dieffenbach et al., 1993) (Supplementary Table 1). The total RNA was extracted as described above. The quality of RNA was checked on 1% agarose gel, and the quantity was measured using NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, United States). The total RNA (1 μg) was then converted into complementary DNA (cDNA) using PrimeScript IV 1st strand cDNA Synthesis Mix (Takara, United States) following the protocol of the manufacturer. The real-time PCR reaction was set up using SYBR Green Mix (Clontech, Mountain View, CA, United States) and run on the CFX96 Real-Time System (Bio-Rad Laboratories, Hercules, CA, United States). The thermal profile included the initial denaturation step (95°C for 2 min) and followed by a 40-cycle amplification step (95°C for 15 s and 60°C for 1 min). For qRT-PCR, three biological replicates of each fruit stage with three technical replicates were used. The relative expression of each gene was calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The actin gene was taken as an internal control. The student's t-test was performed for calculating significant differences in the expression of MYB genes (p < 0.05).
Co-expression Analysis of MYB Genes
The co-expression of Capsicum MYBs with genes involved in capsaicinoid biosynthesis (AT3: acyltransferase 3; Kas: ketoacyl-ACP synthase; pAMT: putative aminotransferase; BCKDH: branched-chain a-ketoacid dehydrogenase a-ketoacid decarboxylase; ACL: acyl carrier protein; FatA: acyl-ACP thioesterase), phenylpropanoid biosynthesis (PAL: phenylalanine ammonia-lyase; COMT: caffeic acid 3-O-methyltransferase; C4H: cinnamate 4-hydroxylase; 4CL: 4-coumaroyl-CoA ligase; BCAT: branched-chain amino acid aminotransferase), carotenoid biosynthesis (CCS: capsanthin-capsorubin synthase; BCH: b-carotene hydroxylase; PSY: phytoene synthase), anthocyanin biosynthesis genes (DFR: dihydroflavonol 4-reductase; F3′5′H: flavonoid 3′, 5′-hydroxylase; CHS: chalcone synthase), vitamin C biosynthesis genes (GLDH: L-galactono-1,4-lactone dehydrogenase; GalDH: L-galactose-1-dehydrogenase; GME: GDP-D-mannose-3′,5′-epimerase), and fruit shape and size genes (WD-40; CLAVATA1; auxin receptor; EAR1; SEC8: Arabidopsis Exocyst Complex; WUSCHEL; TTL3: TPR repeat-containing thioredoxin TTL3; OVATE; SUN) was represented using a heatmap. The expression of genes was shown as log2FPKM, and clustering was performed based on Pearson correlation analysis.
Synteny and Homologous MYB Gene Pairs
Genome-wide CSSs and the collinearity of C. chinense MYB genes with C. annuum, C. baccatum, tomato, potato, eggplant, and Arabidopsis genomes were identified using the MCScanX toolkit (Wang et al., 2012). Blastp search (e-value <1e-10) results among the total protein sequences of each genome against C. chinense total protein sequences were used as input. The whole genome sequences of C. chinense (GCA_002271895.2), C. annuum (GCA_000710875.1), and C. baccatum (GCA_002271885.2) were downloaded from NCBI, and the eggplant genome was downloaded from Solanaceae Genomics Network (https://solgenomics.net/; Barchi et al., 2019). The Arabidopsis, tomato, and potato genome sequences were retrieved from Phytozome v12 (https://phytozome.jgi.doe.gov/pz/portal.html). The CcMYB homologous proteins from the C. annuum, C. baccatum, tomato, potato, and Arabidopsis genomes were identified by blastp search with cut-off parameters: e-value <1e-03, percent identity > 75%, and coverage with > 80% of query length. The CSSs were represented using TBtools (v1.068; Chen et al., 2020), and collinear and homologous MYBs, along with physical positions on different chromosomes, were presented in a chord diagram using the circlize R package (Gu et al., 2014).
Simple Sequence Repeat (SSR) Prediction in Capsicum MYBs
Full length gene sequences and 1.5-kb upstream of promoter sequences from the Transcription Start Site (TSS) of 251 C. chinense MYBs were used to identify simple sequence repeats (SSRs) using the online WebSat tool (Martins et al., 2009) as described previously (Dubey et al., 2019; Jaiswal et al., 2020). The maximum size of an SSR motif was kept at 6 nucleotides, while the minimum number of repeats of the motif was kept at 6. Mononucleotide repeats were excluded from the analysis.
Results
Identification of MYB Genes in Capsicum Spp
After the duplicate sequences were removed from the blastp search results and Hidden Markov Model (HMM) analysis, a total of 301, 433, and 292 potential MYB-encoding genes were predicted in the C. chinense, C. annuum, and C. baccatum genomes, respectively. The remaining sequences were screened using CDD and the SMART database to ascertain the presence of the MYB domain. Ultimately, a total of 251, 245, and 240 MYB-encoding genes were identified in C. chinense, C. annuum, and C. baccatum genomes, respectively. In the C. chinense genome, out of the total CcMYBs, 126 were R2R3 type, while the remaining 99 were MYB1R type, 25 belonged to MYB with other domains type, and 1 was atypical MYB. A total of 128 C. annuum MYB (CaMYB) and 123 C. baccatum MYB (CbMYB) were of R2R3-type. The molecular weight of CcMYBs varied from 11.5 to 114.6 kDa (kilodalton), and that of MYB related from 8.8 to 183.5 kDa. Most of the CcMYBs were localized in the nucleus, seven in mitochondria, two in the chloroplast, two in the cytoplasm, and one in extracellular (Supplementary Table 2). The coding (CDS) and protein sequences of CcMYB genes are given in Supplementary File 1.
Genome-Wide Distribution, Duplication, and Co-mapping of Capsicum MYBs With Capsaicinoid QTLs
Of all the MYB genes identified, 232 (92.43%) were physically mapped on 12 chromosomes (chrs), and the remaining were mapped on scaffolds of the C. chinense genome (Figure 1). In C. annuum, 222 (90.61%) and in C. baccatum 221 (92.08%) MYBs were mapped on their respective chrs (Supplementary Figures 1A,B). Uneven distribution of MYB genes on the 12 Capsicum chrs (average 11–12/ chr) was observed. In C. chinense, the upper end of the arm on chr1 and the lower end of the arm on chrs 2 and 6, respectively, have a greater density of MYBs. A similar distribution of MYBs was observed for chrs 1 and 2 in both C. annuum and C. baccatum and for chr 6 in C. baccatum.
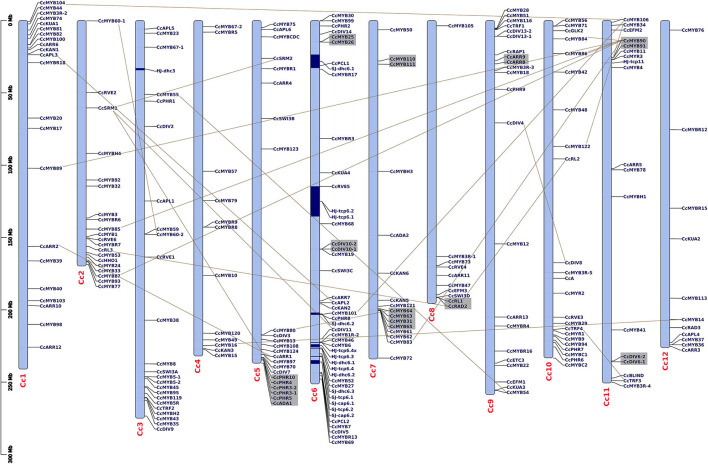
Figure 1.
Distribution of CcMYB genes on 12 Capsicum chromosomes along with the previously reported capsaicinoid QTLs (Han et al., 2018; Park et al., 2019). The QTLs are shown in blue blocks. The tandem duplicates are shown via gray blocks, while the segmental duplicates are shown via connecting lines. The scale is mentioned on the left of the chromosomes in million base pairs (Mbp).
In the C. chinense genome, nine clusters of 20 tandem and 41 segmental duplicated CcMYB gene pairs were observed with Ka/Ks ratios ranging from 0.001 to 1.0269 (Table 1). Among the duplicate pairs, CcMYB60-1 and CcMYB60-2 had the highest Ka/Ks ratio (1.0269) followed by CcPHR10 and CcPHR3-2 (0.8988), CcDIV6-2 and CcDIV6-1 (0.8396), and CcARR9 and CcARR8 (0.7781). Ka/Ks values of >1 indicate a positive selection, while a Ka/Ks ratio of <1 indicates purifying selection for these MYB gene pairs in the C. chinense genome. The minimum diversion time was 0.2414 MYA for the gene pair duplicated in tandem, CcGLK1-1 and CcGLK1-2, while it was highest between the CcMYB106 and CcMYB104 segmental duplicated pair, i.e., 309.78 MYA.
Table 1.
MYB gene duplicates in the Capsicum chinense genome.
| C. chinense MYB name | Duplicated MYB pair | Duplication type | Ka | Ks | Ka/Ks | Diversion time (T = Ks/2λ) MYA |
|---|---|---|---|---|---|---|
| CcAPL3 | CcAPL2 | Segmental | 0.131 | 0.526 | 0.248 | 37.77 |
| CcARR1 | CcARR2 | Segmental | 0.167 | 0.834 | 0.200 | 59.92 |
| CcARR9 | CcARR8 | Tandem | 0.092 | 0.118 | 0.778 | 8.50 |
| CcDIV10-1 | CcDIV10-2 | Tandem | 0.078 | 0.117 | 0.668 | 8.43 |
| CcDIV11 | CcSRM1 | Segmental | 0.427 | 3.501 | 0.122 | 251.48 |
| CcDIV4 | CcDIV8 | Segmental | 0.164 | 1.008 | 0.163 | 72.43 |
| CcDIV6-2 | CcDIV6-1 | Tandem | 0.091 | 0.108 | 0.840 | 7.77 |
| CcDIV7 | CcSRM1 | Segmental | 0.456 | 2.903 | 0.157 | 208.54 |
| CcGLK1-1 | CcGLK1-2 | Segmental | 0.000 | 0.003 | 0.001 | 0.24 |
| CcGLK1-3 | Segmental | 0.000 | 0.003 | 0.001 | 0.24 | |
| CcGLK1-4 | Segmental | 0.000 | 0.003 | 0.001 | 0.24 | |
| CcGLK1-2 | CcGLK1-3 | Segmental | NA | NA | NA | – |
| CcGLK1-4 | Segmental | NA | NA | NA | – | |
| CcGLK1-4 | CcGLK1-3 | Segmental | NA | NA | NA | – |
| CcMYB102 | CcMYB122 | Segmental | 0.235 | 1.185 | 0.198 | 85.10 |
| CcMYB106 | CcMYB102 | Segmental | 0.254 | 4.157 | 0.061 | 298.60 |
| CcMYB101 | Segmental | 0.346 | 3.527 | 0.098 | 253.35 | |
| CcMYB122 | Segmental | 0.334 | 4.018 | 0.083 | 288.64 | |
| CcMYB104 | Segmental | 0.336 | 4.312 | 0.078 | 309.78 | |
| CcMYB110 | CcMYB111 | Tandem | 0.014 | 0.083 | 0.164 | 5.94 |
| CcMYB115 | CcA | Segmental | 0.369 | 0.993 | 0.372 | 71.32 |
| CcMYB14 | CcMYB16 | Segmental | 0.396 | 2.037 | 0.195 | 146.36 |
| CcMYB25 | CcMYB26 | Tandem | 0.035 | 0.136 | 0.261 | 9.74 |
| CcMYB31 | CcMYB63 | Tandem | 0.137 | 0.456 | 0.300 | 32.76 |
| CcMYB65 | Tandem | 0.184 | 0.529 | 0.349 | 37.99 | |
| CcMYB32 | CcMYB33 | Segmental | 0.134 | 0.783 | 0.172 | 56.24 |
| CcMYB55 | CcMYB54 | Segmental | 0.233 | 2.843 | 0.082 | 204.22 |
| CcMYB60-1 | CcMYB60-2 | Segmental | 0.061 | 0.060 | 1.027 | 4.29 |
| CcMYB64 | CcMYB31 | Tandem | 0.090 | 0.334 | 0.269 | 23.98 |
| CcMYB63 | Tandem | 0.115 | 0.562 | 0.204 | 40.35 | |
| CcMYB80 | Segmental | 0.281 | 3.748 | 0.075 | 269.24 | |
| CcMYB90 | CcMYB91 | Tandem | 0.052 | 0.263 | 0.198 | 18.93 |
| CcMYB87 | Segmental | 0.172 | 3.742 | 0.046 | 268.83 | |
| CcMYB89 | Segmental | 0.156 | 0.985 | 0.159 | 70.74 | |
| CcMYB86 | Segmental | 0.347 | 2.739 | 0.127 | 196.74 | |
| CcMYB85 | Segmental | 0.329 | 3.449 | 0.096 | 247.77 | |
| CcMYB83 | Segmental | 0.359 | 3.568 | 0.101 | 256.29 | |
| CcMYB84 | Segmental | 0.381 | 2.282 | 0.167 | 163.91 | |
| CcMYBC2 | CcMYBC1 | Segmental | 0.158 | 1.221 | 0.130 | 87.75 |
| CcMYBH1 | CcMYBH5 | Segmental | 0.181 | 0.534 | 0.339 | 38.39 |
| CcMYBR10 | CcMYBR11 | Tandem | 0.046 | 0.070 | 0.655 | 4.99 |
| CcMYR1 | CcMYR2 | Segmental | 0.167 | 0.520 | 0.322 | 37.35 |
| CcPHR10 | CcPHR3-2 | Tandem | 0.196 | 0.218 | 0.899 | 15.69 |
| CcPHR4 | Tandem | 0.172 | 0.264 | 0.651 | 18.96 | |
| CcPHR10 | CcPHR3-1 | Tandem | 0.240 | 0.333 | 0.721 | 23.94 |
| CcPHR3-1 | CcPHR3-2 | Tandem | 0.052 | 0.098 | 0.528 | 7.03 |
| CcPHR4 | CcPHR3-2 | Tandem | NA | NA | NA | – |
| CcPHR3-1 | Tandem | 0.044 | 0.107 | 0.415 | 7.69 | |
| CcPHR5 | Tandem | 0.085 | 0.138 | 0.619 | 9.89 | |
| CcRAD3 | CcRAD6 | Segmental | 0.144 | 1.636 | 0.088 | 117.50 |
| CcRAD4 | Segmental | 0.288 | 3.758 | 0.077 | 269.94 | |
| CcRAD5 | Segmental | 0.213 | 2.091 | 0.102 | 150.25 | |
| CcRAD4 | CcRAD6 | Segmental | 0.188 | 3.621 | 0.052 | 260.12 |
| CcRAD3 | Segmental | 0.288 | 3.758 | 0.077 | 269.94 | |
| CcRAD5 | CcRAD6 | Tandem | 0.078 | 0.143 | 0.549 | 10.25 |
| CcRAD3 | Segmental | 0.213 | 2.091 | 0.102 | 150.25 | |
| CcRL1 | CcRAD2 | Tandem | 0.418 | 2.159 | 0.194 | 155.10 |
| CcRL2 | CcRL1 | Segmental | 0.210 | 1.744 | 0.120 | 125.30 |
| CcRL3 | CcRAD2 | Segmental | 0.225 | 4.043 | 0.056 | 290.42 |
| CcSRM1 | CcSRM2 | Segmental | 0.464 | 1.053 | 0.440 | 75.63 |
| CcTRF3 | CcTRF4 | Segmental | 0.066 | 0.107 | 0.617 | 7.70 |
| CcDIV7 | CcSRM1 | Segmental | 0.456 | 2.903 | 0.157 | 208.54 |
| CcGLK1-1 | CcGLK1-2 | Segmental | 0.000 | 0.003 | 0.001 | 0.24 |
| CcGLK1-3 | Segmental | 0.000 | 0.003 | 0.001 | 0.24 | |
| CcGLK1-4 | Segmental | 0.000 | 0.003 | 0.001 | 0.24 | |
| CcGLK1-2 | CcGLK1-3 | Segmental | NA | NA | NA | – |
| CcGLK1-4 | Segmental | NA | NA | NA | – | |
| CcGLK1-4 | CcGLK1-3 | Segmental | NA | NA | NA | – |
| CcMYB102 | CcMYB122 | Segmental | 0.235 | 1.185 | 0.198 | 85.10 |
| CcMYB106 | CcMYB102 | Segmental | 0.254 | 4.157 | 0.061 | 298.60 |
| CcMYB101 | Segmental | 0.346 | 3.527 | 0.098 | 253.35 | |
| CcMYB122 | Segmental | 0.334 | 4.018 | 0.083 | 288.64 | |
| CcMYB104 | Segmental | 0.336 | 4.312 | 0.078 | 309.78 | |
| CcMYB110 | CcMYB111 | Tandem | 0.014 | 0.083 | 0.164 | 5.94 |
| CcMYB115 | CcA | Segmental | 0.369 | 0.993 | 0.372 | 71.32 |
| CcMYB14 | CcMYB16 | Segmental | 0.396 | 2.037 | 0.195 | 146.36 |
| CcMYB25 | CcMYB26 | Tandem | 0.035 | 0.136 | 0.261 | 9.74 |
| CcMYB31 | CcMYB63 | Tandem | 0.137 | 0.456 | 0.300 | 32.76 |
| CcMYB65 | Tandem | 0.184 | 0.529 | 0.349 | 37.99 | |
| CcMYB32 | CcMYB33 | Segmental | 0.134 | 0.783 | 0.172 | 56.24 |
| CcMYB55 | CcMYB54 | Segmental | 0.233 | 2.843 | 0.082 | 204.22 |
| CcMYB60-1 | CcMYB60-2 | Segmental | 0.061 | 0.060 | 1.027 | 4.29 |
| CcMYB64 | CcMYB31 | Tandem | 0.090 | 0.334 | 0.269 | 23.98 |
| CcMYB63 | Tandem | 0.115 | 0.562 | 0.204 | 40.35 | |
| CcMYB80 | Segmental | 0.281 | 3.748 | 0.075 | 269.24 | |
| CcMYB90 | CcMYB91 | Tandem | 0.052 | 0.263 | 0.198 | 18.93 |
| CcMYB87 | Segmental | 0.172 | 3.742 | 0.046 | 268.83 | |
| CcMYB89 | Segmental | 0.156 | 0.985 | 0.159 | 70.74 | |
| CcMYB86 | Segmental | 0.347 | 2.739 | 0.127 | 196.74 | |
| CcMYB85 | Segmental | 0.329 | 3.449 | 0.096 | 247.77 | |
| CcMYB83 | Segmental | 0.359 | 3.568 | 0.101 | 256.29 | |
| CcMYB84 | Segmental | 0.381 | 2.282 | 0.167 | 163.91 | |
| CcMYBC2 | CcMYBC1 | Segmental | 0.158 | 1.221 | 0.130 | 87.75 |
| CcMYBH1 | CcMYBH5 | Segmental | 0.181 | 0.534 | 0.339 | 38.39 |
| CcMYBR10 | CcMYBR11 | Tandem | 0.046 | 0.070 | 0.655 | 4.99 |
| CcMYR1 | CcMYR2 | Segmental | 0.167 | 0.520 | 0.322 | 37.35 |
| CcPHR10 | CcPHR3-2 | Tandem | 0.196 | 0.218 | 0.899 | 15.69 |
| CcPHR4 | Tandem | 0.172 | 0.264 | 0.651 | 18.96 | |
| CcPHR10 | CcPHR3-1 | Tandem | 0.240 | 0.333 | 0.721 | 23.94 |
| CcPHR3-1 | CcPHR3-2 | Tandem | 0.052 | 0.098 | 0.528 | 7.03 |
| CcPHR4 | CcPHR3-2 | Tandem | NA | NA | NA | – |
| CcPHR3-1 | Tandem | 0.044 | 0.107 | 0.415 | 7.69 | |
| CcPHR5 | Tandem | 0.085 | 0.138 | 0.619 | 9.89 | |
| CcRAD3 | CcRAD6 | Segmental | 0.144 | 1.636 | 0.088 | 117.50 |
| CcRAD4 | Segmental | 0.288 | 3.758 | 0.077 | 269.94 | |
| CcRAD5 | Segmental | 0.213 | 2.091 | 0.102 | 150.25 | |
| CcRAD4 | CcRAD6 | Segmental | 0.188 | 3.621 | 0.052 | 260.12 |
| CcRAD3 | Segmental | 0.288 | 3.758 | 0.077 | 269.94 | |
| CcRAD5 | CcRAD6 | Tandem | 0.078 | 0.143 | 0.549 | 10.25 |
| CcRAD3 | Segmental | 0.213 | 2.091 | 0.102 | 150.25 | |
| CcRL1 | CcRAD2 | Tandem | 0.418 | 2.159 | 0.194 | 155.10 |
| CcRL2 | CcRL1 | Segmental | 0.210 | 1.744 | 0.120 | 125.30 |
| CcRL3 | CcRAD2 | Segmental | 0.225 | 4.043 | 0.056 | 290.42 |
| CcSRM1 | CcSRM2 | Segmental | 0.464 | 1.053 | 0.440 | 75.63 |
| CcTRF3 | CcTRF4 | Segmental | 0.066 | 0.107 | 0.617 | 7.70 |
| CcMYB104 | Segmental | 0.336 | 4.312 | 0.078 | 309.78 | |
| CcMYB110 | CcMYB111 | Tandem | 0.014 | 0.083 | 0.164 | 5.94 |
| CcMYB115 | CcA | Segmental | 0.369 | 0.993 | 0.372 | 71.32 |
| CcMYB14 | CcMYB16 | Segmental | 0.396 | 2.037 | 0.195 | 146.36 |
| CcMYB25 | CcMYB26 | Tandem | 0.035 | 0.136 | 0.261 | 9.74 |
| CcMYB31 | CcMYB63 | Tandem | 0.137 | 0.456 | 0.300 | 32.76 |
| CcMYB65 | Tandem | 0.184 | 0.529 | 0.349 | 37.99 | |
| CcMYB32 | CcMYB33 | Segmental | 0.134 | 0.783 | 0.172 | 56.24 |
| CcMYB55 | CcMYB54 | Segmental | 0.233 | 2.843 | 0.082 | 204.22 |
| CcMYB60-1 | CcMYB60-2 | Segmental | 0.061 | 0.060 | 1.027 | 4.29 |
| CcMYB64 | CcMYB31 | Tandem | 0.090 | 0.334 | 0.269 | 23.98 |
| CcMYB63 | Tandem | 0.115 | 0.562 | 0.204 | 40.35 | |
| CcMYB80 | Segmental | 0.281 | 3.748 | 0.075 | 269.24 | |
| CcMYB90 | CcMYB91 | Tandem | 0.052 | 0.263 | 0.198 | 18.93 |
| CcMYB87 | Segmental | 0.172 | 3.742 | 0.046 | 268.83 | |
| CcMYB89 | Segmental | 0.156 | 0.985 | 0.159 | 70.74 | |
| CcMYB86 | Segmental | 0.347 | 2.739 | 0.127 | 196.74 | |
| CcMYB85 | Segmental | 0.329 | 3.449 | 0.096 | 247.77 | |
| CcMYB83 | Segmental | 0.359 | 3.568 | 0.101 | 256.29 | |
| CcMYB84 | Segmental | 0.381 | 2.282 | 0.167 | 163.91 | |
| CcMYBC2 | CcMYBC1 | Segmental | 0.158 | 1.221 | 0.130 | 87.75 |
| CcMYBH1 | CcMYBH5 | Segmental | 0.181 | 0.534 | 0.339 | 38.39 |
| CcMYBR10 | CcMYBR11 | Tandem | 0.046 | 0.070 | 0.655 | 4.99 |
| CcMYR1 | CcMYR2 | Segmental | 0.167 | 0.520 | 0.322 | 37.35 |
| CcPHR10 | CcPHR3-2 | Tandem | 0.196 | 0.218 | 0.899 | 15.69 |
| CcPHR4 | Tandem | 0.172 | 0.264 | 0.651 | 18.96 | |
| CcPHR10 | CcPHR3-1 | Tandem | 0.240 | 0.333 | 0.721 | 23.94 |
| CcPHR3-1 | CcPHR3-2 | Tandem | 0.052 | 0.098 | 0.528 | 7.03 |
| CcPHR4 | CcPHR3-2 | Tandem | NA | NA | NA | – |
| CcPHR3-1 | Tandem | 0.044 | 0.107 | 0.415 | 7.69 | |
| CcPHR5 | Tandem | 0.085 | 0.138 | 0.619 | 9.89 | |
| CcRAD3 | CcRAD6 | Segmental | 0.144 | 1.636 | 0.088 | 117.50 |
| CcRAD4 | Segmental | 0.288 | 3.758 | 0.077 | 269.94 | |
| CcRAD5 | Segmental | 0.213 | 2.091 | 0.102 | 150.25 | |
| CcRAD4 | CcRAD6 | Segmental | 0.188 | 3.621 | 0.052 | 260.12 |
| CcRAD3 | Segmental | 0.288 | 3.758 | 0.077 | 269.94 | |
| CcRAD5 | CcRAD6 | Tandem | 0.078 | 0.143 | 0.549 | 10.25 |
| CcRAD3 | Segmental | 0.213 | 2.091 | 0.102 | 150.25 | |
| CcRL1 | CcRAD2 | Tandem | 0.418 | 2.159 | 0.194 | 155.10 |
| CcRL2 | CcRL1 | Segmental | 0.210 | 1.744 | 0.120 | 125.30 |
| CcRL3 | CcRAD2 | Segmental | 0.225 | 4.043 | 0.056 | 290.42 |
| CcSRM1 | CcSRM2 | Segmental | 0.464 | 1.053 | 0.440 | 75.63 |
| CcTRF3 | CcTRF4 | Segmental | 0.066 | 0.107 | 0.617 | 7.70 |
| CcPHR3-1 | Tandem | 0.044 | 0.107 | 0.415 | 7.69 | |
| CcPHR5 | Tandem | 0.085 | 0.138 | 0.619 | 9.89 | |
| CcRAD3 | CcRAD6 | Segmental | 0.144 | 1.636 | 0.088 | 117.50 |
| CcRAD4 | Segmental | 0.288 | 3.758 | 0.077 | 269.94 | |
| CcRAD5 | Segmental | 0.213 | 2.091 | 0.102 | 150.25 | |
| CcRAD4 | CcRAD6 | Segmental | 0.188 | 3.621 | 0.052 | 260.12 |
| CcRAD3 | Segmental | 0.288 | 3.758 | 0.077 | 269.94 | |
| CcRAD5 | CcRAD6 | Tandem | 0.078 | 0.143 | 0.549 | 10.25 |
| CcRAD3 | Segmental | 0.213 | 2.091 | 0.102 | 150.25 | |
| CcRL1 | CcRAD2 | Tandem | 0.418 | 2.159 | 0.194 | 155.10 |
| CcRL2 | CcRL1 | Segmental | 0.210 | 1.744 | 0.120 | 125.30 |
| CcRL3 | CcRAD2 | Segmental | 0.225 | 4.043 | 0.056 | 290.42 |
| CcSRM1 | CcSRM2 | Segmental | 0.464 | 1.053 | 0.440 | 75.63 |
| CcTRF3 | CcTRF4 | Segmental | 0.066 | 0.107 | 0.617 | 7.70 |
The type of duplication, Ka (rate of non-synonymous substitution per non-synonymous site), Ks (rate of synonymous substitution per synonymous site), Ka/Ks ratio, and the duplication time in million years ago (MYA) are mentioned (T = Ks/2λ; where λ = 6.96 × 10−9 years).
Our analysis showed at least seven MYB genes (five CcMYBs and two CaMYBs) within the previously reported capsaicinoid QTLs (Han et al., 2018; Park et al., 2019) (Figure 1). At chr 6 of C. chinense, CcMYB101 and CcPHR8 are located within the dihydrocapsaicin QTL SJ-dhc6.2; CcMYB46 and CcMYB6 within the capsaicinoid QTL HJ-tcp6.4; and CcRVE5 within capsaicinoid QTLs HJ-tcp6.1 and HJ-tcp6.2. At chr 2 of C. annuum, CaMYB3 and CaHHO1 are located within the two QTLs, i.e., a dihydrocapsaicin QTL PD-dicap2.2 and total capsaicinoid QTL PD-total2 (Supplementary Figure 1A).
Gene Structure, Motif, and Cis-Element Analysis
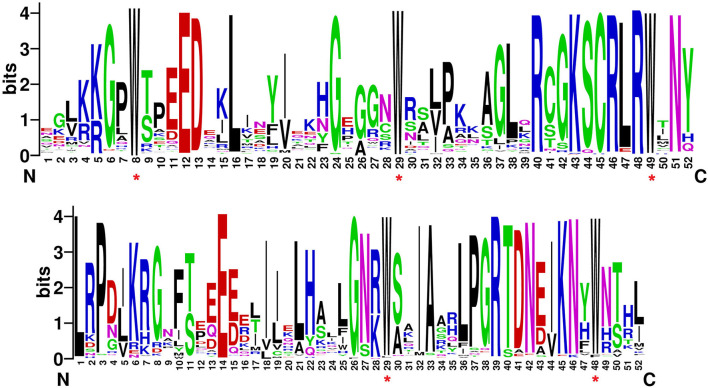
Most of the MYB genes, i.e., 60.95% in C. chinense, 61.22% in C. annuum, and 60.41% in C. baccatum, have two to three exons (Figure 2 and Supplementary Figure 2), and very few showed 11 or more exons. The motif analysis of CcMYB proteins revealed that in the R2 MYB domain, 18 (out of 52) amino acid (aa) positions were conserved in 80% of the R2R3-MYB protein sequences (Figure 3). However, 20 (out of 52) in the R3 domain were conserved, suggesting that R3 is relatively more conserved. The number and placement of tryptophan residues were found to be highly conserved; three tryptophan residues placed 20–21 residues apart in the R2 domain, and two tryptophan residues placed 19 residues apart in the R3 domain. The phenylalanine residue, which replaces the first tryptophan residue in the R3 repeat, is also found to be highly conserved. Apart from the MYB repeats, conserved motifs were observed on the C terminal of the MYB protein sequences (Supplementary Table 3). Previously identified motifs in AtMYBs were also identified in CcMYBs; for instance, motif-23 ([TY][SV]AN[LA][SR]HMA[QE]WESARLEAEARL[VS]R[EK]S[KQ]), which has previously been defined in Antirrhinum majus MIXTA MYB, was observed in CcMYB89, CcMYB90, CcMYB91, and CcMYB87 (Kranz et al., 1998) protein sequences. The putative cis-elements were also identified in C. chinense, C. annuum, and C. baccatum MYBs (Supplementary Table 4). Both elements binding to basic transcriptional machineries like the TATA and CAAT boxes (Forde et al., 1985) and cis-elements like hormone-responsive ABRE (ABA-Responsive Element; Yamaguchi-Shinozaki and Shinozaki, 1994); seed-specific like RY-element (Fujiwara and Beachy, 1994), AE-Box (Sevilla-Lecoq et al., 2003), AACA motif (Yoshihara et al., 1996), GCN4 motif (Müller and Knudsen, 1993) and Box II (Kim and Wu, 1990); light-responsive elements like MRE (Safrany et al., 2008), G-box (Schindler et al., 1992), GATA-motif (Reyes et al., 2004); low temperature-responsive LTRE (Dunn et al., 1994); and drought- and stress-responsive TC-rich repeats, were identified.
Figure 2.
Conserved amino acid motifs and structure of CcMYB genes. The motifs are differently colored with a specific number. The exons are shown in blue blocks, while the connecting lines in between represent intronic regions in kb (kilobase).
Figure 3.
Sequence logo of (A) R2 and (B) R3 MYB repeats in C. chinense.
Phylogenetic Analysis of Capsicum MYBs
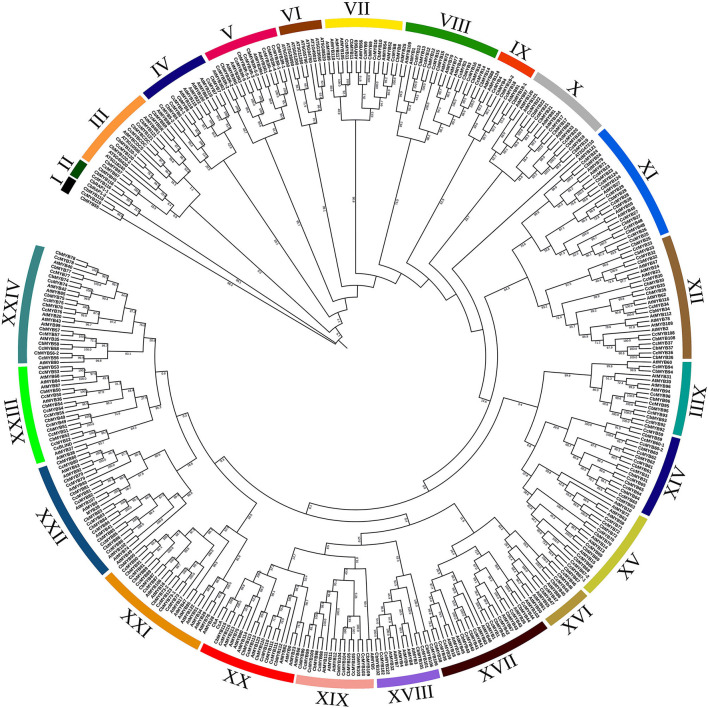
In the phylogenetic analysis, 126 CcMYBs, 123 CbMYBs, and 147 AtMYBs clustered into 24 subgroups (Figure 4) with >75% confidence in most of the branches, for instance, nearly 100% confidence between clades of CcMYB97 and CbMYB97 in subgroup III, CcMYB7 and CbMYB7 in subgroup IV, and AtMYB121, CcMYB24, and CbMYB24 in subgroup XI were observed. Similarly, subgroup VIII MYBs like CcMYB16, CcMYB13, and CcMYB14 share 99–100% confidence in their clades with CbMYB16, CbMYB13, and CbMYB14, respectively. However, CbMYB18-1 in subgroup IX shares a sister clade with AtMYB91 with only 51.6% confidence. MYB3Rs (subgroup V) formed a separate subgroup but share a distant common ancestor with R2R3MYBs, suggesting their common origin. Interestingly, Capsicum MYBs, such as MYB31, MYB59, MYB60, MYB62, MYB63, MYB64, and MYB65, were segregated separately in subgroup XIV. The phylogenetic tree constructed from a total of 734 MYB protein sequences from three Capsicum species resolved into 22 subgroups (Supplementary Figure 3). Several MYB genes, such as CcMYBR14, CcMYBR17, and CbLHY, clustered alone with no recent sister clade in the other two Capsicum species.
Figure 4.
Phylogenetic tree constructed using R2R3 MYBs from C. chinense, C. baccatum, and Arabidopsis. The tree was divided into 24 subgroups, and bootstrap values are shown on the branches of the tree.
Expression Analysis of MYB Genes
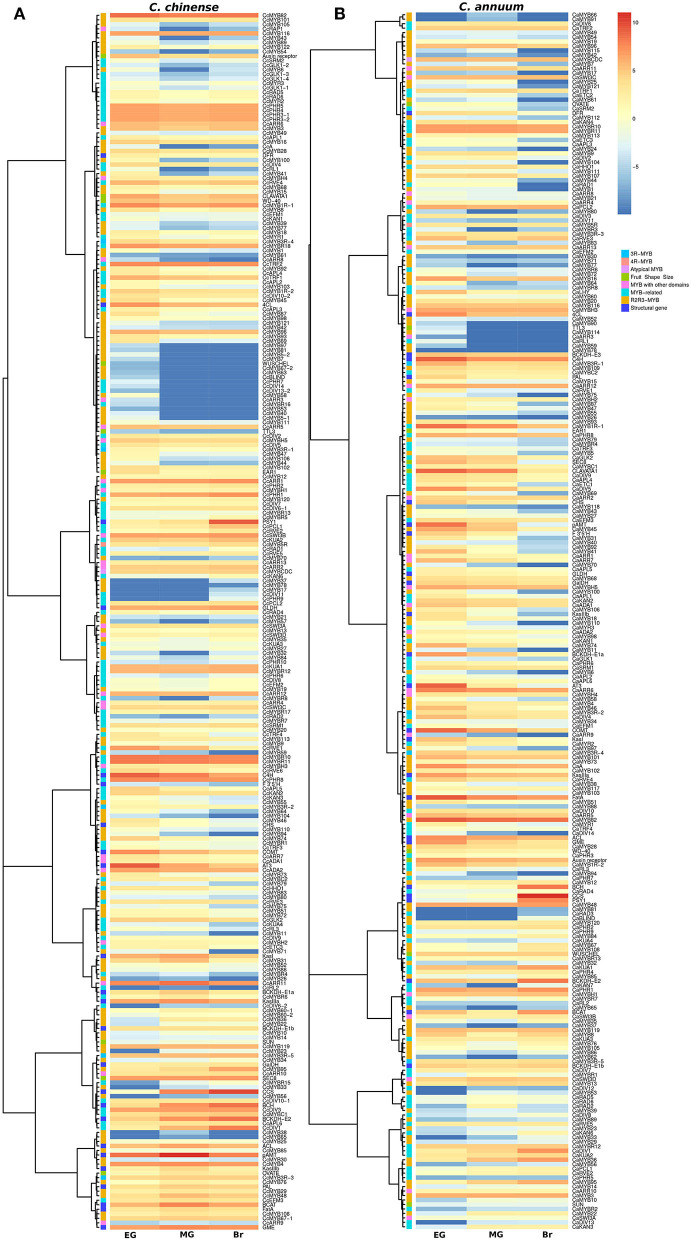
The transcriptome data of the EG, MG, and Br fruit development stages of C. chinense and C. annuum were used to determine the expression values of all the MYB genes based on the normalized FPKM (fragments per kilobase of transcript per million mapped reads). A total of ~458 million raw paired-end reads were generated from the fruit tissues of C. chinense and C. annuum. Around 196.8 and 200.9 million reads out of ~237.4 and 220.8 million clean reads were aligned with an average alignment rate of 82.9 and 91.02% against the C. chinense and C. annuum genomes (Qin et al., 2014; Kim et al., 2017), respectively (Supplementary Table 5). We observed a variation in the expression of MYB genes among the different fruit developmental stages in C. annuum and C. chinense. A total of 236 and 238 MYB genes were expressed in at least one tissue in C. chinense and C. annuum, respectively. Based on the expression patterns of these genes in the fruit developmental stages, the co-expression analysis identified 15 and 20 different clusters of MYB genes in C. chinense and C. annuum, respectively (Figure 5). Also, several MYB genes, such as CcMYB16, CcMYB28, CcMYB100, CcA, CcDIV4, CcMYB46, and CcMYB74, were co-expressed with DFR and CHS from the anthocyanin/flavonoid pathway, while CcDIV1, CcMYB4, CcMYB31, CcMYB52, CcMYB86, CcMYB108, CcMYBR6, and CcARR11 were co-expressed with Kas, FatA, and BCKDH from the capsaicinoid biosynthesis pathway. Moreover, the CcMYB10, CcMYB82, CcMYB102, CcMYB1R1, and CcRVE4 genes showed similar expression patterns with genes related to fruit shape and size (Figure 5). Further analysis revealed a total of 54 DE CcMYBs (adjusted p < 0.01) in C. chinense, 36 in MG compared with EG (12 upregulated and 24 downregulated), 66 in Br compared with EG (20 upregulated and 46 downregulated.), and 50 in MG compared with Br (34 upregulated and 16 downregulated; Figure 6). While a total of 81 CaMYBs were DEGs (adjusted p < 0.01) in C. annuum, 39 and 42 were DEs in MG and Br compared with the EG fruit stage and 13 were DEGs in MG with respect to the Br fruit stage (Figure 6). Furthermore, we analyzed the expression patterns of MYB genes in C. chinense and C. annuum using the transcriptome data and compared them using representative heatmaps (Figure 7A). Most of the MYB genes showed similar expression patterns, but several MYB genes showed contrasting expression levels in the two Capsicum spp. For instance, CcMYBR12 shows a higher expression in C. chinense and a moderate expression in C. annuum in all three fruit stages. CcPHR8 is highly expressed throughout the fruit developmental stages in C. chinense, while its expression decreases from high to low during fruit development in C. annuum. CcMYB31 is exclusively highly expressed in the MG stage in C. chinense, moderately expressed in EG, and lowly expressed in the Br fruit stage, while its homolog in C. annuum showed a low expression in the EG stage and a negligible expression in the rest of the two stages. MYB48 showed a higher expression throughout the fruit developmental stages in C. annuum but only slightly higher in MG in C. chinense than the rest of the fruit stages.
Figure 5.
Expression profiles of MYB genes in (A) C. chinense, (B) C. annuum at the three different fruit developmental stages. The heatmap color scale represents the log2 fragments per kilobase of transcript per Million mapped reads (FPKM) expression of MYB genes.
Figure 6.
Differentially expressed MYB genes in (A) C. chinense and (B) C. annuum. Significant DEG MYBs are enclosed within the yellow box.
Figure 7.
Ribonucleic acid sequencing (RNAseq) and quantitative real-time (qRT)-PCR expression data of Capsicum MYBs at the early green (EG), mature green (MG), and breaker (Br) fruit development stages. (A) Normalized FPKM expression of C. chinense MYBs and their C. annuum homologs by RNAseq, and (B) relative expression of 24 CcMYB genes and capsaicinoid biosynthesis pathway genes (AT3, KasI, pAMT, ACS1, BCAT, and COMT) between C. chinense (Bhut Jolokia; Acc-Cc74) and C. annuum (Acc-Ca18) by qRT-PCR in the three fruit developmental stages and correlation between RNAseq and qRT-PCR expression data. A Student t-test was performed to calculate the significant difference of expression. The significance level was represented as ***p < 0.001; **p < 0.01, and *p < 0.05).
The expression of 24 MYB genes showing DE in transcriptome data was validated by qRT-PCR analysis (Figure 7B). The MYB genes showed 74–83% similarity in their qRT-PCR expression profile with the RNA-seq data. Eight MYB genes- CcMYB100 (BC332_00785), CcMYB16 (BC332_11900), CcETC3 (BC332_24253), CcMYBR12 (BC332_30379), CcMYB106 (BC332_27082), CcMYB3R-1 (BC332_21354), CcMYB3 (BC332_04434), and CcMYB31 (BC332_19185) showed a significantly higher expression in one or more of the fruit stages of highly pungent C. chinense compared with lowly pungent C. annuum. The expression of CcMYBR12 increased from EG to the Br fruit stage in both the Capsicum spp., but the level of expression remains higher in C. chinense throughout. CcMYBR12 expression shows a high similarity between RNA seq and qRT-PCR data, except that it was highest in the Br stage in qRT-PCR and the MG stage in RNA-seq data. CcMYB16 and CcETC3 showed two to four times higher expression in C. chinense. The transcriptome data for CcMYB16 suggest the same for the MG and Br stages; however, it was the opposite for the EG stage. CcETC3 showed similar expression patterns in the transcriptome data. CcMYB106 and CcMYB100 showed expression only in the EG stage with a slightly higher level of expression in C. chinense as compared with that of C. annuum. CcMYB100 showed three times higher expression in the EG fruit of C. chinense as compared with that of C. annuum, and CcMYB31 showed approximately two times higher expression in the MG fruit stage in C. chinense compared with that of C. annuum. CcMYB3R-1 was 2-3 times highly expressed in all the three fruit stages of C. chinense, while CcMYB3 showed a lower expression in EG and MG, and a higher expression in the Br stage of C. chinense compared with those of C. annuum.
Protein Structure Prediction of C. chinense R2R3 MYB Genes
The 126 CcMYBs having the R2R3 MYB domain were analyzed for their secondary and tertiary structures using the Phyre2 server (Supplementary Figure 4). The best models for CcMYB proteins showed 20–68% identity and 98.6–100% query coverage with their template sequences. The majority of CcMYB (116) genes were observed to have high coverage and similarity with the c6kksA protein model/template of Arabidopsis R2R3 type MYB2 TF (WEREWOLF, WER). For instance, both CcMYB106 and CcMYB113 showed 68% similarity, while CcMYB67-2 was just 21% similar to this template sequence. The remaining seven CcMYBs were modeled with different protein templates, out of which two, namely, CcMYB3R-4 and CcMYB5R, were modeled with a template c1h88C of ternary protein-DNA complex1 of MYB TF, while CcMYB70, CcMYB5-1, and CcMYB100 were modeled with a template named d1mbja of c-MYB DNA-binding domain repeat 3. Two MYBs, i.e., CcMYB121 and CcMYB5-2, were modeled with templates d1gv2a2 and d1h8ac1, respectively, and were related to the Myb/SANT DNA-binding domain family and three MYBs, CcMYB116, CcMYB119, and CcMYB120, with c2yqkA, d2crga1, and c5ylzJ, respectively (Supplementary Table 6). The CcMYBs contained 21–61% α-helix in their secondary structure, while 1–6% β-strands were predicted in only 20 CcMYBs (Supplementary Table 6). Overall, the modeled 3D structures suggested the helix-turn-helix structure similarity of CcMYB proteins with already known Arabidopsis models and were highly reliable.
Synteny and Gene Duplication Analysis
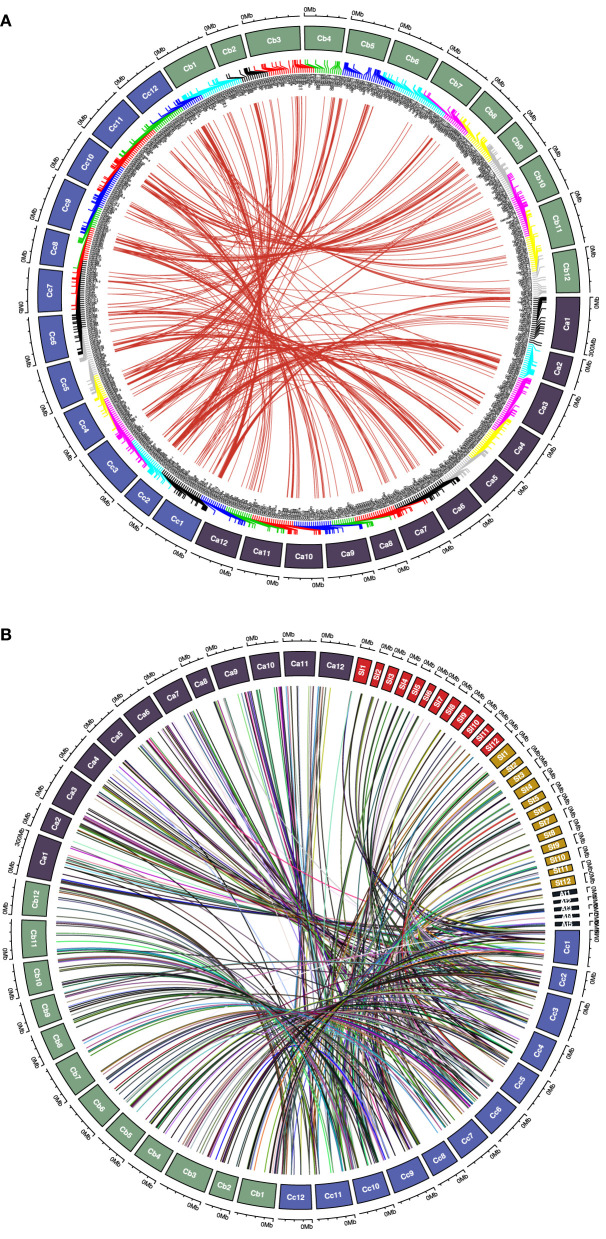
We analyzed the synteny and collinearity of five Solanaceae and Arabidopsis genomes with C. chinense (Figure 8) and identified a total of 717 conserved syntenic segments (CSSs) in all the species analyzed, ranging from 0.02 to 33.56 Mbp (million base pair) in size, which have at least one CcMYB gene in them along with other protein coding genes (Supplementary Table 7, Table 2). The highest number, i.e., 171 and 176 CSSs, was shared with C. annuum and C. baccatum, respectively. In these CSSs, 203 and 168 unique CcMYBs were homologous with MYBs of C. annuum and C. baccatum. Among the Capsicum species, most of the CSSs were present on the same chromosomes and had an order of genes similar to that of C. chinense, but few were found to be diverged (Table 2). For instance, 47 CSSs were present on different chromosomes, and 75 had a reversed gene order in C. annuum with respect to C. chinense. In particular, a CSS, harboring MYB44 along with other genes on chr 1, showed a reverse order of genes in C. annuum. The size of this CSS is 1.607 Mbp in C. chinense and 1.662 Mbp in C. annuum. Another CSS of 1.9 Mbp in C. chinense and of 3.17 Mbp in C. annuum were present on different chromosomes, chr 7 in C. chinense and on chr 9 in C. annuum and had a reverse order of genes. In C. baccatum, a CSS with CcMYB105 and CbMYB104 and other genes was 2.29 Mbp in size on chr 1, while it was 2.47 Mbp in C. chinense on chr 8 (Figure 9A, Tables 2, 3).
Figure 8.
Synteny and collinearity of the C. chinense (CcMYB) genes with the Capsicum annuum (Ca), C. baccatum (Cb), Solanum lycoperisucum (Sl), Solanum tuberosum (St), Solanum melongena (Sm), and Arabidopsis thaliana (At) genomes. Gray lines in the background show collinear segments, while red lines represent syntenic MYB gene pairs with respect to the C. chinense genome.
Table 2.
Number of conserved syntenic segments (CSSs), with at least one MYB gene in them, their size in million base pairs (Mbp), and their diversification in different Solanaceae genomes and Arabidopsis thaliana.
| Species | C. annuum | C. baccatum | S. lycopersicum | S. tuberosum | A. thaliana | C. chinense |
|---|---|---|---|---|---|---|
| No. of CSSs | 171 | 176 | 108 | 154 | 93 | 15 |
| Size (Mbp) | 0.06–28.79 | 0.07–33.56 | 0.05–2.75 | 0.06–3.07 | 0.02–0.89 | 0.29–10.95 |
| CSSs on diff chr | 47 | 71 | 55 | 74 | 86 | 12 |
| CSSs with reverse gene order | 75 | 76 | 42 | 65 | 37 | 5 |
| MYB genes in CSSs | 203 | 168 | 90 | 131 | 77 | 27 |
Figure 9.
Chord diagram representing (A) syntenic CcMYB gene pairs with C. annuum and C. baccatum MYBs and (B) CcMYB homologs in C. annuum, tomato (Sl), potato (St), and Arabidopsis (At) across their respective chromosomes.
Table 3.
Chromosome-wise distribution and the number of collinear CcMYB genes on the same chromosomes as C. chinense in Solanaceae members.
| chr No. | C. chinense | C. annuum | C. baccatum | S. lycopersicum | S. tuberosum | ||||
|---|---|---|---|---|---|---|---|---|---|
| MYB count | MYB Count | Collinear | MYB count | Collinear | MYB count | Collinear | MYB count | Collinear | |
| chr01 | 16 | 16 | 16 | 18 | 18 | 8 | 4 | 8 | 2 |
| chr02 | 18 | 15 | 15 | 12 | 12 | 10 | 10 | 14 | 14 |
| chr03 | 18 | 15 | 15 | 15 | 6 | 8 | 6 | 14 | 9 |
| chr04 | 10 | 12 | 9 | 7 | 6 | 9 | 5 | 13 | 7 |
| chr05 | 20 | 17 | 17 | 17 | 10 | 11 | 5 | 16 | 7 |
| chr06 | 28 | 20 | 19 | 2 | 2 | 11 | 10 | 17 | 16 |
| chr07 | 12 | 12 | 8 | 8 | 7 | 5 | 3 | 7 | 6 |
| chr08 | 9 | 8 | 6 | 8 | 7 | 5 | 2 | 7 | 2 |
| chr09 | 18 | 11 | 11 | 14 | 12 | 6 | 4 | 7 | 4 |
| chr10 | 21 | 14 | 14 | 12 | 12 | 8 | 7 | 14 | 14 |
| chr11 | 15 | 6 | 6 | 11 | 11 | 5 | 3 | 6 | 2 |
| chr12 | 10 | 9 | 6 | 9 | 8 | 3 | 1 | 8 | 3 |
| Scaffolds | 9 | 9 | – | 32 | 3 | 0 | – | 0 | – |
| Total | 204 | 164 | 142 | 165 | 114 | 89 | 60 | 131 | 86 |
As expected, the genomes of potato (154), tomato (108), and Arabidopsis (93) shared a lesser number of CSSs with the C. chinense genome. A number of CSSs were spread on different chromosomes compared with C. chinense, i.e., 55 in tomato, 74 in potato, and 86 in Arabidopsis, which was expected as they are more diverged compared with different species of Capsicum.
To study the effects of duplication events on the expansion of the MYB gene family, Capsicum MYB homolog proteins were identified in C. annuum, C. baccatum, tomato, potato, and Arabidopsis (Figure 9B). We found 758 pairs of MYB duplicates across the genomes of five species (Supplementary Table 8). Of these, our analysis showed 435 MYB duplicate pairs to be under purifying selection, 23 pairs under neutral selection, and 47 pairs under positive selection. Among these, 225, 213, 125, 79, and 23 CcMYB proteins were orthologous to C. annuum, C. baccatum, potato, tomato, and Arabidopsis MYBs, respectively. The value of Ka/Ks ranged from 0.001 to 50 in the homolog pairs. Homologous pairs CcMYB25 and CaMYB25, and CcMYB90 and CaMYB90 were under positive selection. Among others, CcMYB31 and CaMYB31, and CcMYB3 and SlMYB73 were under purifying selection. The average divergence time for CcMYB orthologs with tomato was 37.1 MYA, while that with potato was 33.56 MYA (Supplementary Table 8).
Identification of Simple Sequence Repeat (SSR) Motifs
We identified SSRs in the C. chinense MYB genes and their 1.5-kb upstream promoter, which can be used as molecular markers in future Capsicum breeding programs. A total of 169 SSRs were identified in the C. chinense MYBs. Out of these, 114 were gene-based from 77 C. chinense MYB genes. The remaining 55 SSRs were in the 1.5-Kb upstream regions from the TSS of 49 C. chinense MYB genes. Among all the SSRs, the dinucleotide repeats were the most common, i.e., 70.4% in C. chinense, followed by tri-, tetra-, penta-, and hexanucleotide repeats. The most common dinucleotide repeat was “AT,” with a frequency of 42. The maximum size of a SSR motif in C. chinense was seven repeats of hexanucleotide [ATTTTA] in CcMYBR11. The primer sequences and the expected amplicon length of all the SSR repeat motifs in the Capsicum species are given in Supplementary Table 9.
Discussion
MYB genes are important TFs involved in the regulation of several biological, developmental, and metabolic processes in plant species such as Capsicum (Liu et al., 2015; Xu et al., 2015b; Sun et al., 2019; Cao et al., 2020). Although previously reported in C. annuum; in C. chinense, however, being one of the most important species, including the naturally occurring highest pungency containing Capsicum genotype, Bhut jolokia/ghost chili, no report of MYB gene identification was reported. Additionally, in this study, we have identified MYB genes in C. baccatum (240). Previously, Arce-Rodríguez et al. (2021) reported 235 MYBs in C. annuum, but this study identified additional 10 new MYB genes (6 atypical R2R3 MYBs and 4 MYBs with other domains). The greater number of MYB genes in Capsicum might be due to genome expansion. Alternatively, it may also mean the deletion or loss of genes from other lineages. Despite a large number of MYB genes in Capsicum, there are few chances that their functions are redundant but more likely to overlap in their functionalities (Jin and Martin, 1999) or mask the functions of each other. The large size of the MYB gene family in plants can be attributed to the high rates of duplication and retention of duplicate copies compared with other TF families (Shiu et al., 2005). The retention of such large numbers of MYB genes, during evolution, in the members of Solanaceae indicates their positive selection and acquisition of new functions. R2R3-MYBs were the most common type of MYBs in other Capsicum like other plant species (Katiyar et al., 2012; Du et al., 2015; Li et al., 2019). The greater number of R2R3-MYBs in plants suggests their selective amplification and expansion after the loss of R1 repeat in ancestral three repeat MYBs (Lipsick, 1996) and can be involved in the plant speciation process. Whether an increase in genome size or the number of genes explains the huge number of MYB genes in phylogenetically related genomes or not remains to be explored further with broad sampling strategies.
CcMYBs Within Capsaicinoid QTLs Were Differentially Expressed
The chromosomal distribution of MYB genes was found to be random in the three Capsicum species. Seven Capsicum chromosomes had around 60% of the MYB genes (Figure 1 and Supplementary Figure 1). Five CcMYBs, CcMYB101, CcMYB46, CcMYB6, CcPHR8, and CcRVE5, on chr 6 of C. chinense and two CaMYBs, namely, CaMYB3 and CaHHO1, on chr 2 of C. annuum were found inside the previously reported capsaicinoid QTLs (Han et al., 2018; Park et al., 2019). Among these, two MYB genes, MYB3 and MYB46, also showed a significant differential expression in the fruit tissues of lowly pungent C. annuum and highly pungent C. chinense in the qRT-PCR analysis (Figure 7B). MYB46 was also found to be co-expressed with CHS in the co-expression analysis, suggesting its possible involvement in the transcriptional regulation of anthocyanin biosynthesis. Other two MYB genes in the QTLs, CcPHR8 and CaHHO1, were co-expressed with phenylpropanoid gene C4H and anthocyanin biosynthesis gene DFR (Figure 5), suggesting their potential roles in the respective pathways. The motif identification analysis revealed motifs that are important to impart functional significance to MYB proteins. For example, recently, motif-23 was reported to mediate the interaction between Cucumis sativus MYB6 and a MYB-related protein CsTRY (Yang et al., 2018). Although functions for most of these motifs are still unknown, they may be involved in protein-protein interactions or other biological roles and are subject to further exploration (Millard et al., 2019). MYB genes also respond to environmental and hormonal changes to regulate gene expression during abiotic stress responses in plants (Urao et al., 1996; Abe et al., 1997; Li et al., 2015). Here, the cis-element analysis also suggests environmental and hormonal regulatory mechanisms for the Capsicum MYB genes. MYB binding motifs (MBSI) and MYB recognition elements (MREs) observed in the promoter regions of Capsicum MYB genes may indicate their regulatory roles in flavonol biosynthesis (Solano et al., 1997; Mehrtens et al., 2005). Previously, MYB14 has been shown to contain MRE alongside AT-rich element, GATA-motif, ARE, Box 4, and circadian in its promoter region, and to be activated by UV-C light in order to activate stilbene synthesis in Vitis labrusca (Bai et al., 2019). Likewise, another MYB15 that controls basal immunity in V. quinquangularis was found to harbor cis-elements like GCN4-motif, MBS, and TCA-element in its promoter region (Luo et al., 2019). The identification of cis-elements may aid in deciphering regulatory networks and may further lead to the isolation and characterization of corresponding TFs.
Phylogenetic Analysis Showed Functional Importance of Capsicum MYBs
The phylogenetic relationships revealed in the MYB protein sequences can be utilized to relate their function. For instance, CcMYB3, CcMYB12, CcMYB14, CcMYB16, etc., along with their sister clades in C. baccatum and Arabidopsis, form subgroup VIII. AtMYB1 in the same subgroup is involved in pollen development, and AtMYB44, AtMYB73, and AtMYB77 are involved in lateral root growth and salinity response (Reňák, 2012; Kim et al., 2013; Zhao et al., 2014). Similar functions may be stipulated for Capsicum MYBs of the same subgroup. Subgroup XIII MYBs such as AtMYB30 positively regulate fatty acid biosynthesis as well as hypersensitive cell death response, while subgroup XXIV MYBs, such as AtMYB80, is, again, important for pollen development and tapetal growth (Raffaele et al., 2008; Phan et al., 2011). Subgroup XXII includes AtMYB49 and AtMYB74 that provide tolerance to abiotic stresses like salinity, and AtMYB102 provides resistance to biotic stresses like insect herbivores (De Vos et al., 2006; Xu et al., 2015a; Zhang et al., 2020). Subgroup X contains AtMYB33 and AtMYB101, which are targets of the miR159 family upon ABA accumulation during seed germination, and are shown to be involved in drought stress (Reyes and Chua, 2007). CcMYBs such as MYB31, MYB59, MYB60, MYB61, MYB63, MYB64, and MYB65 in subgroup XIV did not cluster with any AtMYBs (Figure 4). This was similar to the results of previous analysis on C. annuum, where clades 24 and 25 harbored Capsicum-specific MYBs (Arce-Rodríguez et al., 2021). Also, the same Capsicum MYBs clustered with the tomato and potato MYB homologs in a distinct subgroup, suggesting these MYBs to be Solanaceae-specific (Figure 4, Supplementary Figure 5). However, contrary to the findings of Arce-Rodríguez et al. (2021), other MYBs, such as MYB116 and MYB119, which were not reported earlier, clustered separately from AtMYBs and were present in subgroups I and II, respectively (Figure 4). CcMYB98, CcMYB99, and CcMYB100 lie in subgroup XIX with AtMYB11, AtMYB12, and AtMYB111 that control flavonol glycoside accumulation (Stracke et al., 2010). CcMYB115 and CcA in subgroup XX share a sister clade with AtMYB75 and AtMYB90. AtMYB75 has been extensively studied for its role in anthocyanin biosynthesis and AtMYB90 for phenylpropanoid biosynthesis (Borevitz et al., 2000; Teng et al., 2005). CcMYB46 lies along with AtMYB61 in subgroup XVI, which functions as a transcriptional control for the development of root, seed, and vascular tissues (Romano et al., 2012). MYB3Rs have been shown to regulate the cell cycle like c-MYB in animals by regulating cyclin genes via MYB recognition elements (MREs) in cyclin gene promoters via the R3 MYB repeat (Ito et al., 2001). One major challenge in the functional characterization of MYB genes in Capsicum could be the functional redundancy of duplicated MYBs (Dubos et al., 2010). The second phylogenetic tree among the MYBs in the three Capsicum species revealed C. chinense- and C. baccatum-specific lineages, leading to species-specific phenotypes. Similarly, when we analyzed the evolutionary relationship among MYBs between the Capsicum species and other Solanaceae family members along with Arabidopsis, C. chinense-specific clades having CcMYBR14 were observed (Supplementary Figures 3, 5). However, the phylogenetic tree has a low coverage sampling and may have omitted the evolutionary processes leading to the emergence of these clades and is less representative of the process of speciation in Capsicum species.
Expression Analysis Revealed Eight MYB Genes Were Highly Expressed in C. chinense
The expression analysis revealed the spatio-temporal expression pattern of CcMYB genes in the EG, MG, and Br fruit development stages of two Capsicum genotypes (Figures 6, 7). Of the 24 MYB genes showing DE in RNA-seq data and validated by qRT-PCR, eight showed a high expression in one or more of the fruit stages in C. chinense as compared with those of C. annuum. For instance, CcMYB106 and CcMYB100 showed a significantly higher expression in the EG fruit in C. chinense compared with that of C. annuum in both the transcriptome data and qRT analysis. CcMYB100 is a homolog of SlMYB12, which has been reported to regulate the flavonol biosynthesis pathway in tomatoes (Ballester et al., 2010). Moreover, our co-expression analysis also showed that CcMYB100 clustered with DFR, an anthocyanin pathway gene (Figure 5). CcMYB16 and CcETC3 show a maximum expression in the EG stage, which then decreases in the MG stage and then again increases in the Br stage in both the species. CcMYB16 is also clustered with DFR in the co-expression analysis, suggesting its role in anthocyanin biosynthesis. In the phylogenetic analysis, CcMYB16 had clustered with AtMYB44, AtMYB70, AtMYB77, etc., in subgroup VIII, which may suggest its role in plant growth and development, and abiotic and biotic stress responses (Figure 4; Jung et al., 2008; Shim et al., 2013; Zhao et al., 2014). CcETC3, on the other hand, was segregated with AtETC2, which plays a crucial role in trichome development and patterning (Supplementary Figure 3; Kirik et al., 2004; Hilscher et al., 2009). Here, the overall expression pattern of both the MYB genes indicates their possible functions in fruit ripening and development in Capsicum. CcMYB3R-1 showed a similar pattern of expression in both Capsicum species; however, the level of expression was higher in C. chinense. AtMYB3R-1, which lies in the same subgroup V as CcMYB3R-1, is known to regulate cell cycle and abiotic stress-responsive genes, suggesting similar functions for CcMYB3R-1 (Figure 4; Dai et al., 2007; Ma et al., 2009; Haga et al., 2011). CcMYB3 showed a similar pattern of increased expression from EG to Br in both the Capsicum species. CcMYB3 was found within the capsaicinoid QTL and clustered with AtMYB73 in subgroup VIII, suggesting its role in capsaicinoid biosynthesis and abiotic stress response (Figure 4, Supplementary Figure 1A; Kim et al., 2013). Another MYB gene, CcMYB46, however, present in capsaicinoid QTL, did not show a higher expression in C. chinense as well as C. annuum. In the co-expression analysis, CcMYB46 was also clustered with the anthocyanin pathway gene CHS (Figure 5). CcMYB31 showed a significantly higher expression in the MG fruit in C. chinense compared with that of C. annuum (Figure 7B). The qRT expression trends of CcMYB31 and its homolog CaMYB31 during fruit development were similar to those of their respective RNA-seq expression data (Figures 6, 7A). CcMYB31 and CcMYB48 were coexpressed with CBGs- KasI and PAL, respectively, in the co-expression analysis. Previously, the expression levels of their homologs, CaMYB31 and CaMYB48, have been reported to correlate with capsaicinoid levels in C. annuum (Arce-Rodríguez and Ochoa-Alejo, 2017; Han et al., 2019; Sun et al., 2020). Furthermore, other CcMYBs, such as CcDIV1, CcMYB4, CcMYB52, CcMYB86, CcMYB108, CcMYBR6, and CcARR11 also co-expressed with Kas, FatA, and BCKDH from the capsaicinoid biosynthesis pathway, suggesting their potential role in capsaicinoid biosynthesis regulation. However, the expression of CcMYB52 and CcMYB86 did not show a significant difference between C. chinense and C. annuum. Although pAMT, KasIIIb, ACL, BCAT, PAL, and FatA were grouped into the same major cluster in the co-expression analysis, other capsaicinoid pathway genes, such as AT3, clustered along with COMT and BCKDH with KasIIIa in their respective clusters (Figure 5). However, previously in C. annuum, AT3, pAMT, Kas, and BCKDH have been reported to be present in the same co-expression cluster (Arce-Rodríguez et al., 2021), which may be due to the selection of different species/genotypes and distinct fruit developmental stages. Sarpras et al. (2016) have previously reported that capsaicinoid pathway genes are highly expressed in C. chinense as compared with C. annuum and showed correlations with pungency levels of C. chinense, EG (315936 SHU), MG (763411.2 SHU), and Br (925084.8 SHU), and of C. annuum, EG (3478.4 SHU), MG (6656 SHU), and Br (7034.4 SHU) (Sarpras et al., 2016, 2019). In our study, several CcMYB genes also showed a significantly high expression in highly pungent C. chinense compared with lowly pungent C. annuum, while some of them co-expressed with capsaicinoid/phenylpropanoid biosynthesis pathway genes, which can be selected for further validation in correlation to pungency regulation (Figures 5–7). Additionally, we have also analyzed the expression of eight MYB genes in four other Capsicum accessions (two each from C. chinense and C. annuum) to further understand their expression pattern in high and low pungent Capsicum during fruit development (Supplementary Figure 6). Overall, the qRT-PCR expression and co-expression analysis suggested that CcMYBs potentially have a diverse role in the regulation of capsaicinoid, phenylpropanoid, and anthocyanin biosynthesis (Supplementary Figure 6). Furthermore, CcMYB10, CcMYB82, CcMYB1R-1, CcRVE4, and CcMYB102 were co-expressed with fruit shape and size genes like Auxin receptor, WD-40, SUN, and EAR1, suggesting their potential roles in fruit development (Figure 5).
Homology Modeling Suggested a High Structural Similarity of R2R3 CcMYBs With Arabidopsis R2R3 MYB Domain
With only sparse 3D structures characterized compared with increasingly known protein sequences, there is a massive need for the prediction of protein structures in order to bridge this ever-widening gap. In the absence of experimentally determined protein structures, computational tools for protein structure predictions provide a reliable prerequisite. For example, the 3D structure for MYB108-like involved in responses to drought and salt stresses in cotton was predicted using a Swiss model to better understand its mechanism of action (Ullah et al., 2020). We predicted the 3D models for the identified CcMYB protein sequences by sequence based homology modeling (Supplementary Figure 4). Most of the CcMYBs were modeled with a single chain model, c6kksA of MYB DNA-binding domain repeat with a unique helix-loop-helix structure in Arabidopsis R2R3 MYB2 TF WER (Supplementary Figure 4, Supplementary Table 6). The crystal structure of the WER complex with its target DNA was determined recently by X-Ray diffraction and showed that third recognition helices of both R2 and R3 MYB repeats bind to the major groove of DNA in a sequence-specific manner (Wang et al., 2020a). WER is a MYB-related protein that transcriptionally regulates the expression of GLABRA2 to control epidermal cell patterning in a position-dependent manner in Arabidopsis roots (Lee and Schiefelbein, 1999). The CcMYB R2R3 domains shared a moderate degree of sequence similarity (<68%) to the identified template sequence of the MYB domain, which may indicate similarity in the mechanism of binding to its target DNA. Provided there is still no method to determine protein structures solely based on sequence information without known reference structures, the computational based analysis of CcMYB protein sequences can be a stepping stone toward structural determination.
MYB Containing Conserved Syntenic Segments Showed Diversification Among Solanaceae Members
With an increasing number of sequenced plant genomes, little has been understood about the genomic divergence, chromosome evolution, and evolutionary relationships among them. Synteny and collinearity are one way to detect complex evolutionary relationships among plant genomes, especially in reference to multigene families. Synteny analysis among Solanaceae genomes displays the diversification and conservation of chromosomal segments containing MYB genes (Table 2). The Capsicum genomes shared more numbers of CSSs, most of them on similar chromosomes and with similar order of genes, as compared with Solanum genomes, which is expected. However, several CSSs were diversified in Capsicum spp. despite belonging to the same genus and being closely related. The least conservation in CSSs was observed with the Arabidopsis genome with only seven CSSs present on similar chromosomes as C. chinense. Only 142 (56.57%) and 114 (45.41%) C. chinense MYB genes were collinear with the C. annuum and C. baccatum genomes, respectively (Table 3). The lower level of collinearity suggests that Capsicum genomes have undergone large-scale chromosomal rearrangements during their evolution. Therefore, it may indicate that Capsicum genomes, and MYB genes, have diverged, and that there is a need to study specific genes and genomes. In the genomes of species belonging to the Solanum genus, even a lower level of conservation and collinearity of MYB genes was observed with respect to C. chinense as expected (Table 2).
Apart from the genome-wide duplications, tandem and segmental duplications within C. chinense highlight the duplication frequency of MYB genes within the Capsicum genome (Table 1). Genome-wide and tandem duplications have been implicated in the expansion of the R2R3-MYB gene family and are an important measure for the same (Du et al., 2015). In our study, we reported the average duplication time of Capsicum MYB homolog pairs as 28.56 MYA. The Ka/Ks ratios of 435 MYB duplicate pairs were <1, which indicates purifying selection. Forty-seven MYB pairs that had Ka/Ks >1 showed positive selection. We found 241 Capsicum MYBs with orthologs in tomato, potato, and Arabidopsis that were operating under purifying selection (Supplementary Table 8). The strong purifying selection of the Capsicum MYB gene family is similar to the tandem expansion and positive selection observed in the GRAS TF family and R-genes in Arabidopsis (Chen et al., 2010; Wu et al., 2014).
MYB-Specific SSR Repeats Can Serve as Potential Molecular Markers
In an earlier study, gene-specific SSRs related to Capsicum fruit ripening showed high polymorphism among Capsicum spp (Dubey et al., 2019). In this study, the SSRs predicted in the genic and non-genic regions of Capsicum MYBs have rendered them useful in Capsicum breeding and improvement programs. Compared with the 1.5-kb upstream regions, more SSRs were present in the genic regions, 67.45% (Supplementary Table 9). However, previous studies have suggested higher conservation and less variation in the genic regions of different species (Kim et al., 2008; Zhang et al., 2014; Chhapekar et al., 2020). This may be due to the less coverage of the upstream region of MYB genes for SSR prediction, which is 1.5 Kb from the TSS in our study. The di- and tri-nucleotide repeats have been found to be varying in number from species to species but are the most common SSRs in plants (Saha et al., 2017). We also found di-nucleotide repeats to be abundant among all the SSRs in Capsicum species. Both gene-based and non-genic SSRs in Capsicum MYBs can be used as potential markers for the selection of associated genes in fruit breeding programs.
Conclusion
A total of 251, 240, and 245 MYB genes were identified in the C. chinense, C. baccatum, and C. annuum genomes. Twenty tandem and 41 segmental duplication events may have led to the expansion of the MYB gene family in the C. chinense genome. Also, 225, 213, 125, 79, and 23 CcMYB proteins were orthologous to C. annuum, C. baccatum, potato, tomato, and Arabidopsis MYBs, respectively. The transcriptome analysis revealed that 54 CcMYB and 81 CaMYB genes were differentially expressed during fruit development in C. chinense and C. annuum, respectively. Eight CcMYB genes were highly expressed in highly pungent C. chinense compared with lowly pungent C. annuum in the qRT-PCR analysis. Additionally, our finding also suggests the CcMYBs, such as CcMYB16, CcMYB28, CcMYB100, CcDIV4, CcMYB46, and CcMYB74, as potential anthocyanin biosynthesis regulators in Capsicum. While along with already characterized MYB31 and MYB108 (Arce-Rodríguez and Ochoa-Alejo, 2017) other MYBs such as CcMYB4, CcDIV1, CcMYBR6, and CcARR11 may be used as potential targets for the regulation of capsaicinoid biosynthesis. On the other hand CcMYBs, such as CcMYB10, CcMYB82, CcMYB1R-1, CcDIV1, CcRVE4, and CcMYB102, may be investigated for their role in fruit development/shape-size regulation in fruits of Capsicum species. The MYB genes identified could be studied for their functional roles, so that they can be manipulated for Capsicum trait improvement.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: The RNA sequencing data related to this study were submitted on NCBI under BioProject (PRJNA679780). Sequence Read Archive (SRA) accessions for C. chinense samples includes SRR12963502, SRR12963513, and SRR12963514 for early green (EG), SRR12963488, SRR12963489, and SRR12963490 for mature green (MG), and SRR12963491, SRR12963492, and SRR12963493 for breaker (Br) fruit samples. SRA accessions for C. annuum samples are SRR12963501, SRR12963503 and SRR12963504 for EG, SRR12963495, SRR12963496, and SRR12963497 for MG and SRR12963498, SRR12963499, and SRR12963500 for Breaker fruit samples.
Author Contributions
NR conceived and designed the research. KI, AR, IA, MD, and JM conducted field and lab experiments. KI and AR performed in silico analysis and analyzed the data. KI and AR drafted and NR corrected and finalized the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Department of Biotechnology (DBT), Ministry of Science and Technology, Government of India in the form of Ramalingaswami Re-entry Fellowship cum Research Grant (BT/RLF/Re-entry/46/2011), and Science and Engineering Research Board (SERB) (SB/EMEQ-086/2014), Department of Science and Technology (DST), Government of India.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
KI acknowledges the University Grant Commission (UGC), Government of India, for the fellowship received in the form of a Senior Research Fellowship. AR, IA, and JM acknowledge the fellowships received from the Council of Scientific and Industrial Research (CSIR), Government of India, New Delhi in the form of Senior Research fellowships. The authors also acknowledge the DST, Govt. of India for DST-FIST-II given to School of Life Sciences, Jawaharlal Nehru University, New Delhi.
Glossary
Abbreviations
- CcMYB
Capsicum chinense MYB
- CbMYB
Capsicum baccatum MYB
- TF
transcription factor
- DBD
DNA-binding domain
- CSSs
conserved syntenic segments
- qRT-PCR
quantitative real-time PCR
- SSRs
simple sequence repeats
- HMM
Hidden Markov Model
- BLASTs
basic local alignment search tool sequence
- NCBI
National Centre for Biotechnology information
- CDD
Conserved Domains Database
- pI
isoelectric point
- GRAVY
grand average of hydropathicity
- SMART
simple modular architecture research tool
- CDS
coding sequences
- QTLs
quantitative trait loci
- GSDS
gene structure display server
- MEME
multiple em for motif elicitation
- NJ
neighbor-joining
- ML
maximum likelihood
- DPA
day post anthesis
- EG
early green
- MG
mature green
- Br
breaker
- RIN
RNA integrity number
- RNAseq
RNA sequencing
- FC
fold change
- DEG
differentially expressed gene
- Kb
kilobase
- TSS
transcription start site
- Chr
chromosome
- FPKM
fragments per kilobase of transcript per million mapped reads
- DE
differentially expressed
- Mbp
million base pair.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.721265/full#supplementary-material
Chromosomal distribution of MYB genes across 12 Capsicum chromosomes of (A) C. annuum and (B) C. baccatum. The capsaicinoid QTLs in C. annuum adapted from Han et al., 2018 are shown in shaded blocks.
Gene structure and phylogenetic tree of MYB genes in (A) C. annuum and (B) C. baccatum. The length of MYB genes are in a kb (kilobase) scale, and the exons are represented using purple and brown blocks, while the connecting lines represent the intronic regions.
Phylogenetic tree of MYB protein sequences from the three Capsicum species (C. chinense, C. annuum, and C. baccatum). The tree was constructed using the Maximum Likelihood method with 1,000 bootstrap replications.
Predicted 3D structural models of 126 C. chinense R2R3 MYB proteins. Models were shown in rainbow color from N to the C terminus.
The phylogenetic tree is drawn from MYB protein sequences of C. chinense, C. annuum, C. baccatum, tomato, potato, and Arabidopsis using the Maximum likelihood method with 1,000 bootstrap values.
Relative expression of eight CcMYB genes in six genotypes, three C. annuum (Kosom Moso, JH15 and JH7) and three C. chinense (Lota, CCGHY6, and BJS) by qRT-PCR in the three fruit developmental stages. A Student t-test was performed to calculate the significant difference of expression. The significance level was represented as ***p < 0.001; **p < 0.01, and *p < 0.05.
Primer sequences used for the qRT-PCR expression analysis of CcMYBs and capsaicinoids biosynthesis pathway genes.
The characteristic features of Capsicum MYBs. (A) C. chinense (CcMYBs) and (B) C. annuum (CaMYBs) genes with RNAseq expression data in the three fruit developmental stages. (C) C. baccatum (CbMYBs) along with their genomic coordinates.
Conserved amino acid motifs identified in CcMYB protein sequences using the MEME suite.
Cis-regulatory elements identified in the 1.5-kb upstream promoter region of CcMYBs using the PLANTCare database. (A) Cis-acting elements along with their sequences, function, and presence in the MYB genes and (B) their frequency in the upstream region of each MYB gene.
RNA sequencing alignment summary. Raw reads and aligned read summary with Hisat2 in the (A) C. chinense and (B) C. annuum fruit samples.
The 126 R2R3 CcMYBs and the probability and identity of their corresponding homologous protein model/template.
MYB containing conserved syntenic segments (CSSs) from the Capsicum chinense genome compared with the C. annuum, C. baccatum, tomato, potato, and Arabidopsis genomes [Note: Cc, Ca, Cb, St, Sl, and At represent the chromosomes from the C. chinense, C. annuum, C. baccatum, potato, tomato, and Arabidopsis genomes].
The C. chinense (CcMYBs) and their homologous gene pairs from C. annuum, C. baccatum, tomato, potato, and Arabidopsis along with the Ka/Ks ratios and duplication time (in MYA) for each homologous pair.
SSRs identified in the MYB genes and 1.5-kb promoter regions in C. chinense with the primer sequences and the amplicon size of the SSR motifs.
CDS and protein sequences of C. chinense MYB family members.
References
- Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. 10.1105/tpc.9.10.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Barragán A., Ochoa-Alejo N. (2014). Virus-induced silencing of MYB and WD40 transcription factor genes affects the accumulation of anthocyanins in chilli pepper fruit. Biol. Plant. 58, 567–574. 10.1007/s10535-014-0427-4 [DOI] [Google Scholar]
- Antonious G., Jarret R. (2006). Screening Capsicum accessions for capsaicinoids content. J. Environ. Sci. Heal. Part B Pestic. Food Contam. Agric. Wastes 41, 717–729. 10.1080/03601230600701908 [DOI] [PubMed] [Google Scholar]
- Arce-Rodríguez M. L., Martínez O., Ochoa-Alejo N. (2021). Genome-wide identification and analysis of the MYB transcription factor gene family in chili pepper (Capsicum spp.). Int. J. Mol. Sci. 22:2229. 10.3390/ijms22052229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce-Rodríguez M. L., Ochoa-Alejo N. (2017). An R2R3-MYB transcription factor regulates capsaicinoid biosynthesis. Plant Physiol. 174, 1359–1370. 10.1104/pp.17.00506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai R., Luo Y., Wang L., Li J., Wu K., Zhao G., et al. (2019). A specific allele of MYB14 in grapevine correlates with high stilbene inducibility triggered by Al 3+ and UV-C radiation. Plant Cell Rep. 38, 37–49. 10.1007/s00299-018-2347-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester A.-R., Molthoff J., de Vos R., Hekkert B., te L., Orzaez D., et al. (2010). Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152, 71–84. 10.1104/pp.109.147322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchi L., Pietrella M., Venturini L., Minio A., Toppino L., Acquadro A., et al. (2019). A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 9:11769. 10.1038/s41598-019-47985-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J. O., Xia Y., Blount J., Dixon R. A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2393. 10.1105/tpc.12.12.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Li K., Li Y., Zhao X., Wang L. (2020). MYB transcription factors as regulators of secondary metabolism in plants. Biology 9:61. 10.3390/biology9030061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen Q., Han Z., Jiang H., Tian D., Yang S. (2010). Strong positive selection drives rapid diversification of R-Genes in arabidopsis relatives. J. Mol. Evol. 70, 137–148. 10.1007/s00239-009-9316-4 [DOI] [PubMed] [Google Scholar]
- Chhapekar S. S., Brahma V., Rawoof A., Kumar N., Gaur R., Jaiswal V., et al. (2020). Transcriptome profiling, simple sequence repeat markers development and genetic diversity analysis of potential industrial crops Capsicum chinense and C. frutescens of Northeast India. Ind Crops Prod. 154:112687. [Google Scholar]
- Cutanda-Perez M.-C., Ageorges A., Gomez C., Vialet S., Terrier N., Romieu C., et al. (2009). Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 69, 633–648. 10.1007/s11103-008-9446-x [DOI] [PubMed] [Google Scholar]
- Czemmel S., Stracke R., Weisshaar B., Cordon N., Harris N. N., Walker A. R., et al. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530. 10.1104/pp.109.142059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Xu Y., Ma Q., Xu W., Wang T., Xue Y., et al. (2007). Overexpression of an R1R2R3 MYB Gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 143, 1739–1751. 10.1104/pp.106.094532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M., Denekamp M., Dicke M., Vuylsteke M., Van Loon L., Smeekens S. C., et al. (2006). The Arabidopsis thaliana transcription factor AtMYB102 functions in defense against the insect herbivore Pieris rapae. Plant Signal. Behav. 1, 305–311. 10.4161/psb.1.6.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieffenbach C. W., Lowe T. M. J., Dveksler G. S. (1993). General concepts for PCR primer design. Genome Res. 3, S30–S37. 10.1101/gr.3.3.S30 [DOI] [PubMed] [Google Scholar]
- Du H., Liang Z., Zhao S., Nan M. G., Tran L. S. P., Lu K., et al. (2015). The evolutionary history of R2R3-myb proteins across 50 eukaryotes: new insights into subfamily classification and expansion. Sci. Rep. 5: 11037. 10.1038/srep11037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey M., Jaiswal V., Rawoof A., Kumar A., Nitin M., Chhapekar S. S., et al. (2019). Identification of genes involved in fruit development/ripening in Capsicum and development of functional markers. Genomics 111, 1913–1922. 10.1016/j.ygeno.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Dunn M. A., Goddard N. J., Zhang L., Pearce R. S., Hughes M. A. (1994). Low-temperature-responsive barley genes have different control mechanisms. Plant Mol. Biol. 24, 879–888. 10.1007/BF00014442 [DOI] [PubMed] [Google Scholar]
- Fattori V., Hohmann M. S. N., Rossaneis A. C., Pinho-Ribeiro F. A., Verri W. A. (2016). Capsaicin: current understanding of its mechanisms and therapy of pain and other pre-clinical and clinical uses. Molecules 21:844. 10.3390/molecules21070844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B. G., Heyworth A., Pywell J., Kreis M. (1985). Nucleotide sequence of a B1 hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley, wheat and maize. Nucleic Acids Res. 13, 7327–7339. 10.1093/nar/13.20.7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Beachy R. N. (1994). Tissue-specific and temporal regulation of a β-conglycinin gene: roles of the RY repeat and other cis-acting elements. Plant Mol. Biol. 24, 261–272. 10.1007/BF00020166 [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., et al. (2005). ;Protein identification and analysis tools on the ExPASy server, in The Proteomics Protocols Handjournal, ed J. M. Walker (Hatfield: Humana Press; ), 571–607. [Google Scholar]
- Geng D., Chen P., Shen X., Zhang Y., Li X., Jiang L., et al. (2018). MdMYB88 and MdMYB124 enhance drought tolerance by modulating root vessels and cell walls in Apple. Plant Physiol. 178, 1296–1309. 10.1104/pp.18.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Gu L., Eils R., Schlesner M., Brors B. (2014). Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. 10.1093/bioinformatics/btu393 [DOI] [PubMed] [Google Scholar]
- Haga N., Kobayashi K., Suzuki T., Maeo K., Kubo M., Ohtani M., et al. (2011). Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 157, 706–717. 10.1104/pp.111.180836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Jang S., Lee J. H., Lee D. G., Kwon J. K., Kang B. C. (2019). A MYB transcription factor is a candidate to control pungency in Capsicum annuum. Theor. Appl. Genet. 132, 1235–1246. 10.1007/s00122-018-03275-z [DOI] [PubMed] [Google Scholar]
- Han K., Lee H. Y., Ro N. Y., Hur O. S., Lee J. H., Kwon J. K., et al. (2018). QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol. J. 16, 1546–1558. 10.1111/pbi.12894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilscher J., Schlötterer C., Hauser M.-T. (2009). A single amino acid replacement in ETC2 shapes trichome patterning in natural Arabidopsis populations. Curr. Biol. 19, 1747–1751. 10.1016/j.cub.2009.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K., Imamura A., Katoh E., Hatta T., Tachiki M., Yamada H., et al. (2002). Molecular structure of the GARP family of plant myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14, 2015–2029. 10.1105/tpc.002733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Araki S., Matsunaga S., Itoh T., Nishihama R., Machida Y., et al. (2001). G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13, 1891–1905. 10.1105/TPC.010102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal V., Rawoof A., Dubey M., Chhapekar S. S., Sharma V., Ramchiary N. (2020). Development and characterization of non-coding RNA based simple sequence repeat markers in Capsicum species. Genomics 112, 1554–1564. 10.1016/j.ygeno.2019.09.005 [DOI] [PubMed] [Google Scholar]
- Jian W., Cao H., Yuan S., Liu Y., Lu J., Lu W., et al. (2019). SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic. Res. 6, 1–15. 10.1038/s41438-018-0098-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Martin C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. 10.1023/A:1006319732410 [DOI] [PubMed] [Google Scholar]
- Jung C., Seo J. S., Han S. W., Koo Y. J., Kim C. H., Song S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. 10.1104/pp.107.110981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Venkatesh J., Kang M. Y., Kwon J. K., Kang B. C. (2019). A non-LTR retrotransposon activates anthocyanin biosynthesis by regulating a MYB transcription factor in Capsicum annuum. Plant Sci. 287:110181. 10.1016/j.plantsci.2019.110181 [DOI] [PubMed] [Google Scholar]
- Katiyar A., Smita S., Lenka S. K., Rajwanshi R., Chinnusamy V., Bansal K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. 10.1186/1471-2164-13-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiferle C., Fantini E., Bassolino L., Povero G., Spelt C., Buti S., et al. (2015). Tomato R2R3-MYB proteins SlANT1 and SlAN2: same protein activity, different roles. PLoS ONE 10:e0136365. 10.1371/journal.pone.0136365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Nguyen N. H., Jeong C. Y., Nguyen N. T., Hong S.-W., Lee H. (2013). Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 170, 1461–1465. 10.1016/j.jplph.2013.05.011 [DOI] [PubMed] [Google Scholar]
- Kim K. S., Ratcliffe S. T., French B. W., Liu L., Sappington T. W. (2008). Utility of EST-derived SSRs as population genetics markers in a Beetle. J. Hered. 99, 112–124. 10.1093/jhered/esm104 [DOI] [PubMed] [Google Scholar]
- Kim S., Park J., Yeom S. I., Kim Y. M., Seo E., Kim K. T., et al. (2017). New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18, 1–11. 10.1186/s13059-017-1341-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. Y., Wu R. (1990). Multiple protein factors bind to a rice glutelin promoter region. Nucleic Acids Res. 18, 6845–6852. 10.1093/nar/18.23.6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Simon M., Wester K., Schiefelbein J., Hulskamp M. (2004). ENHANCER of TRYand CPC 2(ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55, 389–398. 10.1007/s11103-004-0893-8 [DOI] [PubMed] [Google Scholar]
- Klempnauer K. H., Sippel A. E. (1987). The highly conserved amino-terminal region of the protein encoded by the v-myb oncogene functions as a DNA-binding domain. EMBO J. 6, 2719–2725. 10.1002/j.1460-2075.1987.tb02565.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- König P., Giraldo R., Chapman L., Rhodes D. (1996). The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85, 125–136. 10.1016/S0092-8674(00)81088-0 [DOI] [PubMed] [Google Scholar]
- Kortstee A. J., Khan S. A., Helderman C., Trindade L. M., Wu Y., Visser R. G. F., et al. (2011). Anthocyanin production as a potential visual selection marker during plant transformation. Transgenic Res. 20, 1253–1264. 10.1007/s11248-011-9490-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz H. D., Denekamp M., Greco R., Jin H., Leyva A., Meissner R. C., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. 10.1046/j.1365-313x.1998.00278.x [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L. B., Nadeau J. A., Lucas J., Lee E.-K., Nakagawa T., Zhao L., et al. (2005). The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. Plant Cell 17, 2754–2767. 10.1105/tpc.105.034116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Liu X., Eglit Y., Sack F. (2013). FOUR LIPS and MYB88 conditionally restrict the G1/S transition during stomatal formation. J. Exp. Bot. 64, 5207–5219. 10.1093/jxb/ert313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. 10.1016/S0092-8674(00)81536-6 [DOI] [PubMed] [Google Scholar]
- Lei Q., Lee E., Keerthisinghe S., Lai L., Li M., Lucas J. R., et al. (2015). The FOUR LIPS and MYB88 transcription factor genes are widely expressed in Arabidopsis thaliana during development. Am. J. Bot. 102, 1521–1528. 10.3732/ajb.1500056 [DOI] [PubMed] [Google Scholar]
- Lescot M., Déhais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. 10.1093/nar/30.1.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Ng C. K. Y., Fan L. M. (2015). MYB transcription factors; active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. 10.1016/j.envexpbot.2014.06.014 [DOI] [Google Scholar]
- Li W., Liu Y., Zhao J., Zhen X., Guo C., Shu Y. (2019). Genome-wide identification and characterization of r2r3-myb genes in medicago truncatula. Genet. Mol. Biol. 42, 611–623. 10.1590/1678-4685-gmb-2018-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liang J., Zeng X., Guo H., Luo Y., Kear P., et al. (2021). Genome-wide Analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis. Hortic. Plant J. 7, 129–141. 10.1016/j.hpj.2020.12.001 [DOI] [Google Scholar]
- Li Z., Peng R., Tian Y., Han H., Xu J., Yao Q. (2016). Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant cell Physiol. Physiol. 57, 1657–1677. 10.1093/pcp/pcw091 [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Lipsick J. S. (1996). One billion years of myb. Oncogene 13, 223–235. [PubMed] [Google Scholar]
- Liu J., Osbourn A., Ma P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. 10.1016/j.molp.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu S., Wang J., Chitsaz F., Derbyshire M. K., Geer R. C., Gonzales N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. 10.1093/nar/gkz991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Bai R., Li J., Yang W., Li R., Wang Q., et al. (2019). The transcription factor MYB15 is essential for basal immunity (PTI) in Chinese wild grape. Planta 249, 1889–1902. 10.1007/s00425-019-03130-5 [DOI] [PubMed] [Google Scholar]
- Ma Q., Dai X., Xu Y., Guo J., Liu Y., Chen N., et al. (2009). Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 150, 244–256. 10.1104/pp.108.133454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y. M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. 10.1093/nar/gkz268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkena S., Lee E., Sack F. D., Lamb R. S. (2012). The R2R3 MYB transcription factors FOUR LIPS and MYB88 regulate female reproductive development. J. Exp. Bot. 63, 5545–5558. 10.1093/jxb/ers209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Paz-Ares J. (1997). MYB transcription factors in plants. Trends Genet. 13, 67–73. 10.1016/S0168-9525(96)10049-4 [DOI] [PubMed] [Google Scholar]
- Martins W. S., César D., Lucas S., Fabricio K., Neves D. S., John D. (2009). Bioinformation WebSat - a web software for microsatellite marker development Bioinformation. Biomed. Informatics Publ. Gr. 3, 282–283. 10.6026/97320630003282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrtens F., Kranz H., Bednarek P., Weisshaar B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. 10.1104/pp.104.058032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard P. S., Kragelund B. B., Burow M. (2019). R2R3 MYB transcription factors – functions outside the DNA-binding domain. Trends Plant Sci. 24, 934–946. 10.1016/j.tplants.2019.07.003 [DOI] [PubMed] [Google Scholar]
- Moniz de Sá M., Drouin G. (1996). Phylogeny and substitution rates of angiosperm actin genes. Mol. Biol. Evol. 13, 1198–1212. 10.1093/oxfordjournals.molbev.a025685 [DOI] [PubMed] [Google Scholar]
- Müller M., Knudsen S. (1993). The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. Plant J. 4, 343–355. 10.1046/j.1365-313X.1993.04020343.x [DOI] [PubMed] [Google Scholar]
- Nesi N., Jond C., Debeaujon I., Caboche M., Lepiniec L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. 10.1105/TPC.010098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata K., Morikawa S., Nakamura H., Sekikawa A., Inoue T., Kanai H., et al. (1994). Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648. 10.1016/0092-8674(94)90549-5 [DOI] [PubMed] [Google Scholar]
- Park M., Lee J. H., Han K., Jang S., Han J., Lim J. H., et al. (2019). A major QTL and candidate genes for capsaicinoid biosynthesis in the pericarp of Capsicum chinense revealed using QTL-seq and RNA-seq. Theor. Appl. Genet. 132, 515–529. 10.1007/s00122-018-3238-8 [DOI] [PubMed] [Google Scholar]
- Phan H. A., Iacuone S., Li S. F., Parish R. W. (2011). The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23, 2209–2224. 10.1105/tpc.110.082651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Yu C., Shen Y., Fang X., Chen L., Min J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. 111, 5135–5140. 10.1073/pnas.1400975111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Verweij W., Kroon A., Spelt C., Mol J., Koes R. (2006). PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 18, 1274–1291. 10.1105/tpc.105.034041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., Vailleau F., Léger A., Joubès J., Miersch O., Huard C., et al. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20, 752–767. 10.1105/tpc.107.054858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawoof A., Chhapekar S. S., Jaiswal V., Brahma V., Kumar N., Ramchiary N. (2020). Single-base cytosine methylation analysis in fruits of three Capsicum species. Genomics, 112, 3342–3353. [DOI] [PubMed] [Google Scholar]
- Reňák D., Dupl'áková N., Honys D. (2012). Wide-scale screening of T-DNA lines for transcription factor genes affecting male gametophyte development in Arabidopsis. Sex Plant Reprod. 25, 39–60. 10.1007/s00497-011-0178-8 [DOI] [PubMed] [Google Scholar]
- Reyes J. C., Muro-Pastor M. I., Florencio F. J. (2004). The GATA family of transcription factors in arabidopsis and rice. Plant Physiol. 134, 1718–1732. 10.1104/pp.103.037788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J. L., Chua N.-H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. 10.1111/j.1365-313X.2006.02980.x [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C., Keddie J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. 10.1126/science.290.5499.2105 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano J. M., Dubos C., Prouse M. B., Wilkins O., Hong H., Poole M., et al. (2012). AtMYB61, an R2R3-MYB transcription factor, functions as a pleiotropic regulator via a small gene network. New Phytol. 195, 774–786. 10.1111/j.1469-8137.2012.04201.x [DOI] [PubMed] [Google Scholar]
- Romero I., Fuertes A., Benito M. J., Malpica J. M., Leyva A., Paz-Ares J. (1998). More than 80R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14, 273–284. 10.1046/j.1365-313X.1998.00113.x [DOI] [PubMed] [Google Scholar]
- Safrany J., Haasz V., Mate Z., Ciolfi A., Feher B., Oravecz A., et al. (2008). Identification of a novel cis-regulatory element for UV-B-induced transcription in Arabidopsis. Plant J. 54, 402–414. 10.1111/j.1365-313X.2008.03435.x [DOI] [PubMed] [Google Scholar]
- Saha D., Rana R. S., Chakraborty S., Datta S., Kumar A. A., Chakraborty A. K., et al. (2017). Development of a set of SSR markers for genetic polymorphism detection and interspecific hybrid jute breeding. Crop J. 5, 416–429. 10.1016/j.cj.2017.02.006 [DOI] [Google Scholar]
- Sarpras M., Ahmad I., Rawoof A., Ramchiary N. (2019). Comparative analysis of developmental changes of fruit metabolites, antioxidant activities and mineral elements content in Bhut jolokia and other Capsicum species. LWT 105, 363–370. 10.1016/j.lwt.2019.02.020 [DOI] [Google Scholar]
- Sarpras M., Gaur R., Sharma V., Chhapekar S. S., Das J., Kumar A., et al. (2016). Comparative analysis of fruit metabolites and pungency candidate genes expression between Bhut jolokia and other Capsicum species. PLoS ONE 11:e0167791. 10.1371/journal.pone.0167791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler U., Beckmann H., Cashmore A. R. (1992). TGA1 and G-Box binding factors: two distinct classes of arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell 4, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla-Lecoq S., Deguerry F., Matthys-Rochon E., Perez P., Dumas C., Rogowsky P. M. (2003). Analysis of ZmAE3 upstream sequences in maize endosperm and androgenic embryos. Sex. Plant Reprod. 16, 1–8. 10.1007/s00497-003-0176-6 [DOI] [Google Scholar]
- Shim J. S., Jung C., Lee S., Min K., Lee Y.-W., Choi Y., et al. (2013). AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 73, 483–495. 10.1111/tpj.12051 [DOI] [PubMed] [Google Scholar]
- Shiu S. H., Shih M. C., Li W. H. (2005). Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol. 139, 18–26. 10.1104/pp.105.065110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Fuertes A., Sánchez-Pulido L., Valencia A., Paz-Ares J. (1997). A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J. Biol. Chem. 272, 2889–2895. 10.1074/jbc.272.5.2889 [DOI] [PubMed] [Google Scholar]
- Stevenson C. E. M., Burton N., Costa M. M. R., Nath U., Dixon R. A., Coen E. S., et al. (2006). Crystal structure of the MYB domain of the RAD transcription factor from Antirrhinum majus. Proteins Struct. Funct. Genet. 65, 1041–1045. 10.1002/prot.21136 [DOI] [PubMed] [Google Scholar]
- Stracke R., Jahns O., Keck M., Tohge T., Niehaus K., Fernie A. R., et al. (2010). Analysis of production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 188, 985–1000. 10.1111/j.1469-8137.2010.03421.x [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Sun B., Zhou X., Chen C., Chen C., Chen K., Chen M., et al. (2020). Coexpression network analysis reveals an MYB transcriptional activator involved in capsaicinoid biosynthesis in hot peppers. Hortic. Res. 7:162. 10.1038/s41438-020-00381-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Zhu Z., Chen C., Chen G., Cao B., Chen C., et al. (2019). Jasmonate-inducible R2R3-MYB transcription factor regulates capsaicinoid biosynthesis and stamen development in Capsicum. J. Agric. Food Chem. 67, 10891–10903. 10.1021/acs.jafc.9b04978 [DOI] [PubMed] [Google Scholar]
- Takos A. M., Jaffé F. W., Jacob S. R., Bogs J., Robinson S. P., Walker A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142, 1216–1232. 10.1104/pp.106.088104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S., Keurentjes J., Bentsink L., Koornneef M., Smeekens S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. 10.1104/pp.105.066688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah A., Ul Qamar M. T., Nisar M., Hazrat A., Rahim G., Khan A. H., et al. (2020). Characterization of a novel cotton MYB gene, GhMYB108-like responsive to abiotic stresses. Mol. Biol. Rep. 47, 1573–1581. 10.1007/s11033-020-05244-6 [DOI] [PubMed] [Google Scholar]
- Urao T., Noji M., Yamaguchi-Shinozaki K., Shinozaki K. (1996). A transcriptional activation domain of ATMYB2, a drought-inducible Arabidopsis Myb-related protein. Plant J. 10, 1145–1148. 10.1046/j.1365-313X.1996.10061145.x [DOI] [PubMed] [Google Scholar]
- Wang B., Luo Q., Li Y., Yin L., Zhou N., Li X., et al. (2020a). Structural insights into target DNA recognition by R2R3-MYB transcription factors. Nucleic Acids Res. 48, 460–471. 10.1093/nar/gkz1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Jiang C., Liu W., Wang Y. (2020b). The WRKY53 transcription factor enhances stilbene synthesis and disease resistance by interacting with MYB14 and MYB15 in Chinese wild grape. J. Exp. Bot. 71, 3211–3226. 10.1093/jxb/eraa097 [DOI] [PubMed] [Google Scholar]
- Wang D., Zhang Y., Zhang Z., Zhu J., Yu J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics. Proteomics Bioinformatics 8, 77–80. 10.1016/S1672-0229(10)60008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-Z., Yang K.-Z., Zou J.-J., Zhu L.-L., Xie Z. D., Morita M. T., et al. (2015). Transcriptional regulation of PIN genes by FOUR LIPS and MYB88 during Arabidopsis root gravitropism. Nat. Commun. 6:8822. 10.1038/ncomms9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Liu Y., Tang B., Dai X., Xie L., Liu F., et al. (2020c). Genome-wide identification and capsaicinoid biosynthesis-related expression analysis of the R2R3-MYB gene family in Capsicum annuum L. Front. Genet. 11:598183. 10.3389/fgene.2020.598183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Hao J., Chen X., Zhang X. (2020d). SlMYB102 expression enhances low-temperature stress resistance in tomato plants. PeerJ 8:e10059. 10.7717/peerj.10059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Xu H., Jiang S., Zhang Z., Lu N., Qiu H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. 10.1111/tpj.13487 [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang H., DeBarry J. D., Tan X., Li J., Wang X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. 10.1093/nar/gkr1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes G. R., Bolker B., Bonebakker L., Gentleman R., Liaw W. H. A., Lum T. (2020). gplots: Various R Programming Tools for Plotting Data. R package version 3.1.1. Available online at: https://CRAN.R-project.org/package=gplots
- Wu N., Zhu Y., Song W., Li Y., Yan Y., Hu Y. (2014). Unusual tandem expansion and positive selection in subgroups of the plant GRAS transcription factor superfamily. BMC Plant Biol. 14:373. 10.1186/s12870-014-0373-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Li D., Wang L., Sack F. D., Grotewold E. (2010). Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J. 64, 731–739. 10.1111/j.1365-313X.2010.04364.x [DOI] [PubMed] [Google Scholar]
- Xu R., Wang Y., Zheng H., Lu W., Wu C., Huang J., et al. (2015a). Salt-induced transcription factor MYB74 is regulated by the RNA-directed DNA methylation pathway in Arabidopsis. J. Exp. Bot. 66, 5997–6008. 10.1093/jxb/erv312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Dubos C., Lepiniec L. (2015b). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. 10.1016/j.tplants.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. 10.1105/tpc.6.2.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., An G., Zhu T., Zhang W., Zhang L., Peng L., et al. (2019). Independent activation of the BoMYB2 gene leading to purple traits in Brassica oleracea. Theor. Appl. Genet. 132, 895–906. 10.1007/s00122-018-3245-9 [DOI] [PubMed] [Google Scholar]
- Yan S., Chen N., Huang Z., Li D., Zhi J., Yu B., et al. (2020). Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 225, 2048–2063. 10.1111/nph.16272 [DOI] [PubMed] [Google Scholar]
- Yang M. (2016). The FOUR LIPS (FLP) and MYB88 genes conditionally suppress the production of nonstomatal epidermal cells in Arabidopsis cotyledons. Am. J. Bot. 103, 1559–1566. 10.3732/ajb.1600238 [DOI] [PubMed] [Google Scholar]
- Yang S., Cai Y., Liu X., Dong M., Zhang Y., Chen S., et al. (2018). A CsMYB6-CsTRY module regulates fruit trichome initiation in cucumber. J. Exp. Bot. 69, 1887–1902. 10.1093/jxb/ery047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara T., Washida H., Takaiwa F. (1996). A 45-bp proximal region containing AACA and GCN4 motif is sufficient to confer endosperm-specific expression of the rice storage protein glutelin gene, GluA-3. FEBS Lett. 383, 213–218. 10.1016/0014-5793(96)00233-5 [DOI] [PubMed] [Google Scholar]
- Zhang M., Mao W., Zhang G., Wu F. (2014). Development and characterization of polymorphic ESTSSR and genomic SSR markers for tibetan annual wild barley. PLoS ONE 9:e94881. 10.1371/journal.pone.0094881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Wang R., Yang X., Ju Q., Li W., Lü S., et al. (2020). The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant. Cell Environ. 43, 1925–1943. 10.1111/pce.13784 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Xing L., Wang X., Hou Y.-J., Gao J., Wang P., et al. (2014). The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7:ra53. 10.1126/scisignal.2005051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Sun B., Cai W., Zhou X., Mao Y., Chen C., et al. (2019). Natural variations in the MYB transcription factor MYB31 determine the evolution of extremely pungent peppers. New Phytol. 223, 922–938. 10.1111/nph.15853 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chromosomal distribution of MYB genes across 12 Capsicum chromosomes of (A) C. annuum and (B) C. baccatum. The capsaicinoid QTLs in C. annuum adapted from Han et al., 2018 are shown in shaded blocks.
Gene structure and phylogenetic tree of MYB genes in (A) C. annuum and (B) C. baccatum. The length of MYB genes are in a kb (kilobase) scale, and the exons are represented using purple and brown blocks, while the connecting lines represent the intronic regions.
Phylogenetic tree of MYB protein sequences from the three Capsicum species (C. chinense, C. annuum, and C. baccatum). The tree was constructed using the Maximum Likelihood method with 1,000 bootstrap replications.
Predicted 3D structural models of 126 C. chinense R2R3 MYB proteins. Models were shown in rainbow color from N to the C terminus.
The phylogenetic tree is drawn from MYB protein sequences of C. chinense, C. annuum, C. baccatum, tomato, potato, and Arabidopsis using the Maximum likelihood method with 1,000 bootstrap values.
Relative expression of eight CcMYB genes in six genotypes, three C. annuum (Kosom Moso, JH15 and JH7) and three C. chinense (Lota, CCGHY6, and BJS) by qRT-PCR in the three fruit developmental stages. A Student t-test was performed to calculate the significant difference of expression. The significance level was represented as ***p < 0.001; **p < 0.01, and *p < 0.05.
Primer sequences used for the qRT-PCR expression analysis of CcMYBs and capsaicinoids biosynthesis pathway genes.
The characteristic features of Capsicum MYBs. (A) C. chinense (CcMYBs) and (B) C. annuum (CaMYBs) genes with RNAseq expression data in the three fruit developmental stages. (C) C. baccatum (CbMYBs) along with their genomic coordinates.
Conserved amino acid motifs identified in CcMYB protein sequences using the MEME suite.
Cis-regulatory elements identified in the 1.5-kb upstream promoter region of CcMYBs using the PLANTCare database. (A) Cis-acting elements along with their sequences, function, and presence in the MYB genes and (B) their frequency in the upstream region of each MYB gene.
RNA sequencing alignment summary. Raw reads and aligned read summary with Hisat2 in the (A) C. chinense and (B) C. annuum fruit samples.
The 126 R2R3 CcMYBs and the probability and identity of their corresponding homologous protein model/template.
MYB containing conserved syntenic segments (CSSs) from the Capsicum chinense genome compared with the C. annuum, C. baccatum, tomato, potato, and Arabidopsis genomes [Note: Cc, Ca, Cb, St, Sl, and At represent the chromosomes from the C. chinense, C. annuum, C. baccatum, potato, tomato, and Arabidopsis genomes].
The C. chinense (CcMYBs) and their homologous gene pairs from C. annuum, C. baccatum, tomato, potato, and Arabidopsis along with the Ka/Ks ratios and duplication time (in MYA) for each homologous pair.
SSRs identified in the MYB genes and 1.5-kb promoter regions in C. chinense with the primer sequences and the amplicon size of the SSR motifs.
CDS and protein sequences of C. chinense MYB family members.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: The RNA sequencing data related to this study were submitted on NCBI under BioProject (PRJNA679780). Sequence Read Archive (SRA) accessions for C. chinense samples includes SRR12963502, SRR12963513, and SRR12963514 for early green (EG), SRR12963488, SRR12963489, and SRR12963490 for mature green (MG), and SRR12963491, SRR12963492, and SRR12963493 for breaker (Br) fruit samples. SRA accessions for C. annuum samples are SRR12963501, SRR12963503 and SRR12963504 for EG, SRR12963495, SRR12963496, and SRR12963497 for MG and SRR12963498, SRR12963499, and SRR12963500 for Breaker fruit samples.