Abstract
Transcriptional activation requires both access to DNA assembled as chromatin and functional contact with components of the basal transcription machinery. Using the hormone-bound vitamin D3 receptor (VDR) ligand binding domain (LBD) as an affinity matrix, we previously identified a novel multisubunit coactivator complex, DRIP (VDR-interacting proteins), required for transcriptional activation by nuclear receptors and several other transcription factors. In this report, we characterize the nuclear receptor binding features of DRIP205, a key subunit of the DRIP complex, that interacts directly with VDR and thyroid hormone receptor in response to ligand and anchors the other DRIP subunits to the nuclear receptor LBD. In common with other nuclear receptor coactivators, DRIP205 interaction occurs through one of two LXXLL motifs and requires the receptor's AF-2 subdomain. Although the second motif of DRIP205 is required only for VDR binding in vitro, both motifs are used in the context of an retinoid X receptor-VDR heterodimer on DNA and in transactivation in vivo. We demonstrate that both endogenous p160 coactivators and DRIP complexes bind to the VDR LBD from nuclear extracts through similar sequence requirements, but they do so as distinct complexes. Moreover, in contrast to the p160 family of coactivators, the DRIP complex is devoid of any histone acetyltransferase activity. The results demonstrate that different coactivator complexes with distinct functions bind to the same transactivation region of nuclear receptors, suggesting that they are both required for transcription activation by nuclear receptors.
Nuclear receptors, including vitamin D3, thyroid hormone, and retinoic acid receptors (VDR, TR, and RAR, respectively), are intracellular factors that can transduce the signals of small lipophilic hormonal ligands by binding to target DNA sequences and regulating gene transcription in direct response to such ligands (13, 27). The VDR/TR/RAR subgroup typically acts in conjunction with a common partner, the retinoid X receptor (RXR), by recognizing and binding as heterodimers to specific DNA response elements composed of direct hexameric repeats of the (A/G)G(G/T)TCA consensus sequence separated by three, four, or five nucleotides (VDRE, TRE, or RARE, respectively). Nuclear receptors can be dissected into discrete functional regions, including a DNA binding domain and a ligand binding domain (LBD). The LBD contains at its C terminus a short α-helical motif called AF-2 (3, 8, 11) that is required for ligand-dependent transactivation, and it is a key determinant in interactions with other proteins, generally called coactivators, that mediate connections to the transcription machinery.
Transcriptional activation of genes regulated by nuclear receptors and other transcription factors involves both direct DNA binding of activators to their specific response elements and protein-protein interactions with components of the basal transcription machinery, ultimately targeting RNA polymerase II (RNA Pol II) or an RNA Pol II-associated factor (for reviews, see references 36 and 46). This process is mediated via connections with bridging factors that in some cases can remodel chromatin by various mechanisms, including the acetylation of histones, which presumably would result in greater promoter accessibility and in turn would lead to an enhancement of recruitment of the basal machinery.
Among the many nuclear receptor coactivators isolated and characterized over the last few years, the original enzymatic activity uncovered has been that of histone tail-modifying acetyltransferases (HATs). These proteins and protein complexes include pCAF and CREB-binding protein (CBP)/p300, as well as an emerging number of related factors, collectively called the p160 family, such as SRC-1/NCoA-1, GRIP1/TIF2/NCoA-2, and ACTR/pCIP/AIB1/RAC3 (for reviews, see references 15 and 39). The functional features that have been delineated within their sequences include the HAT domain (6, 33, 40) and nuclear receptor binding motifs, alternatively called signature motifs, NR boxes, or LXDs (18, 42). These short regions are composed of stretches of leucines and are defined by an LXXLL consensus sequence. Such motifs in SRC-1 and GRIP1/TIF2 are arranged as sets of three NR boxes required for interaction with the nuclear receptor AF-2 domain, and several combinations of these motifs appear to direct the specificity of interaction with nuclear receptor heterodimers (9, 28). For example, interaction of GRIP1 with RXR-TR and RXR-RAR heterodimers requires the second and third motifs (LXD2 and LXD3), whereas SRC-1 interaction with RXR/peroxisome proliferator-activated receptor (PPAR) is driven by LXD1 and LXD2. In contrast, estrogen receptor (ER) interaction requires mainly LXD2 (26, 28). Moreover, additional amino acids surrounding these motifs appear to be required for binding specificity (9, 28). Recent crystal structure analysis of a complex of liganded PPARγ with a peptide encompassing two LXXLL motifs of SRC-1 is consistent with a model where LXD2 and LXD3 each bind one receptor subunit of the heterodimer, yielding a stoichiometry of one SRC-1 molecule per receptor dimer (32). Finally, the SRC-1 family of coactivators appear to form complexes with the cointegrator CBP (6), whose HAT activity not only is required for histone modifications but also may regulate coactivator-nuclear receptor interactions, as recently demonstrated between ER and the coactivator ACTR (5).
Using liganded VDR LBD as a bait, we previously identified a novel multisubunit coactivator complex, DRIP (VDR-interacting proteins) (34, 35). The DRIP complex is identical to the ARC coactivator complex (30, 31) and very similar to the TRAP/SMCC complex (12, 16, 20). It is required for transcriptional activation by nuclear receptors and other transcription factors, as assayed in cell-free transcription systems. Moreover, the DRIP complex contains several subunits found within the NAT, CRSP, and mammalian Mediator complexes (21, 38, 41). In this report, we present the characterization of a key subunit of the DRIP complex, DRIP205, that interacts directly with nuclear receptors in response to ligand and anchors the other 14 DRIP subunits to the nuclear receptor LBD. We analyze DRIP205's nuclear receptor binding features and demonstrate that although both endogenous p160 coactivators and DRIP complex bind to the VDR LBD from nuclear extracts through similar sequence motifs, they do so as distinct complexes. In contrast to the p160 family of coactivators, neither DRIP205 nor the other DRIP subunits contain intrinsic HAT activity. The results suggest that distinct coactivator complexes with distinct functions bind to shared regions of nuclear receptors (i.e., the AF-2 domain), implying that they might act at discrete steps during the transcription preinitiation process.
MATERIALS AND METHODS
Materials.
Plasmids pSG5-VDR and pSG5-VDR-ΔAF2, pGSTag, and pCDNA3 were kindly provided by P. MacDonald, M. Garabedian, and R. Fisher, respectively. Anti-VDR and anti-Flag monoclonal antibodies (MAbs) were obtained from commercial sources (Affinity Bioreagent and Kodak IBI, respectively). Anti-hSRC-1 MAb GT12 2E9 was kindly provided by J. DiRenzo. Anti-DRIP205 was obtained by rabbit immunization (Covance, Denver, Pa.) with a glutathione S-transferase (GST)–DRIP205(527-970) fusion protein expressed in Escherichia coli. Anti-DRIP205 antibody was used as a crude antiserum. NR1 and NR2 peptides were synthesized by the Memorial Sloan-Kettering Cancer Center (MSKCC) Protein Core Facility. 1,25-Dihydroxyvitamin D3 [1,25(OH)2D3] (generously provided by M. Uskokovic [Hoffmann-LaRoche, Nutley, N.J.]) was used diluted in ethanol.
Identification and cloning of DRIP205.
Protein samples for microsequence analysis were prepared as previously described (35). Analysis was done by mass spectrometry as previously described (34). The cDNA encoding DRIP205 was obtained by reverse transcription-PCR (RT-PCR) using 1 to 2 μg of total RNA obtained from U-937 cells induced 8 or 12 h by 1,25(OH)2D3. cDNA was synthesized using Superscript reverse transcriptase II (Gibco-BRL); PCR was carried out using the Expand High Fidelity PCR system (Boehringer Mannheim) according to the manufacturer's protocol. Primer sequences were designed from the published sequence of the RB18A cDNA (10) in order to amplify the cDNA as three contiguous fragments, by use of the unique NheI and ApaI sites within the cDNA. The PCR products obtained were ligated directly into pGEM-T (Promega) and then subcloned into pGSTag and pCDNA3 vectors by the use of BamHI and XbaI restriction sites added at the 5′ and 3′ ends, respectively of the full-length cDNA. DRIP205 cDNA clones were verified by sequencing (Rockefeller University DNA Sequencing Facility). Point mutations within NR box motifs of DRIP205 (LXXLL to LXXAA) were created by a Quick Change site-directed mutagenesis kit (Stratagene). These clones were verified by sequencing (MSKCC DNA Sequencing Facility).
Overexpression and purification of recombinant proteins.
Recombinant full-length VDR and Flag-tagged RXR were overexpressed in a baculovirus system and purified as previously described (23). All GST fusion proteins were overexpressed in E. coli as previously described (14). Briefly, bacterial cultures were induced at room temperature by 0.1 mM isopropyl-β-d-thiogalactopyranoside for 3.5 h. Bacteria were then lysed by sonication in lysis buffer (phosphate-buffered saline [PBS] containing 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mg of leupeptin per ml, and 1 mM dithiothreitol [DTT]) and centrifuged. Soluble extracts were incubated with glutathione-Sepharose beads (Pharmacia) for 1 h at 4°C before washing three times in lysis buffer. The concentration of proteins immobilized on beads was quantitated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by comparison with a titration of bovine serum albumin (BSA; Sigma) after Coomassie blue staining.
GST affinity binding assays.
Binding assays were performed with either purified recombinant VDR and Flag-tagged RXRα or labeled proteins synthesized in vitro using the TnT coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine (Amersham). Aliquots of 2 to 10 μg of immobilized GST fusion proteins were preincubated for 1 h at 4°C with 10−6 M ligand or carrier in GST binding buffer (20 mM Tris-HCl [pH 7.9], 180 mM KCl, 0.2 mM EDTA, 0.05% Nonidet P-40 [NP-40], 0.5 mM phenylmethylsulfonyl fluoride, 1 mM DTT) containing 3 mg of BSA per ml (for VDR assays) or 0.5% nonfat dry milk (for TR assays) as described elsewhere (2). Immobilized proteins on beads were then incubated at 4°C for 2 h with 500 ng of purified receptors or 2 to 4 μl of labeled proteins. After three washes in GST binding buffer containing 0.1% NP-40, beads were boiled in SDS-sample buffer, resolved by SDS-PAGE, and analyzed by autoradiography or Western blotting.
Gel mobility shift assay.
GST fusion proteins used in gel shift assays were eluted from glutathione beads by incubation for 15 min at room temperature with 15 mM reduced glutathione in GST binding buffer. Aliquots of 10 ng of each recombinant purified VDR and Flag-RXR were incubated with 500 ng of GST or GST-DRIP205(527-970) for 1 h at 4°C with 10−6 M ligand or carrier in GST binding buffer. Protein-DNA complexes were then analyzed by gel mobility shift assays with 50,000 cpm of VDRE oligonucleotide as a probe as previously described (7) in binding buffer (20 mM Tris [pH 7.9], 100 mM KCl, 1 mM EDTA, 20% glycerol, 0.05% NP-40, 1 mM DTT) with 250 ng of BSA and 0.5 μg of poly(dI-C). Gel shift analysis with RXR-TR (45) was carried out as described above and previously (17).
Western blotting.
Protein samples were resolved by SDS-PAGE and then transferred onto a nitrocellulose membrane (Transblot 0.45; BioRad) in Towbin buffer (25 mM Tris-Cl, 192 mM glycine, 15% methanol). The membrane was blocked in PBS with 0.1% Tween 20 containing 5% nonfat dry milk and then probed with antibodies in a 1:5,000 dilution of MAb or in a 1:100 dilution of crude anti-DRIP205 antiserum. Immunoblots were developed with ECL (enhanced chemiluminescence) reagent (Amersham).
Transient transfection assay.
U-937 cells were maintained in RPMI 1640 medium supplemented with sodium pyruvate (300 μg/ml), 10% fetal bovine serum (Gibco-BRL), penicillin, and streptomycin. Cells were transfected by electroporation as described previously (37). Briefly, early-log-phase cells were electroporated with 5 μg of VDREx2-E1B-LUC reporter, 2 μg of cytomegalovirus (CMV)-based vector pCMVβ-gal as an internal control, 10 ng of pRc-CMV-VDR, and various amounts of pCDNA3-DRIP205, pCDNA3-205-Box, and its point mutants (as indicated in figure legends), balanced with pCDNA3 for a total of 6 μg of CMV expression vectors. Alternatively, cells were transfected with 5 μg of Gal4-UASx5-TK-LUC reporter, 2 μg of pCMVβ-gal, 50 ng of Gal-VP16, or 100 ng of Gal-E1A expression vector, together with DRIP205 constructs as described above. Two hours after electroporation, cells were treated for 20 h with 10−8 M 1,25(OH)2D3 or ethanol as a carrier. Cells were then harvested and resuspended in 50 μl of 250 mM Tris (pH 7.5); 5 to 10 μg of whole cell extracts, prepared by freeze-thaw lysis, were assayed for luciferase and β-galactosidase (β-Gal) activities as specified by the manufacturer (Promega). Luciferase activity was measured in a luminometer (Lumat 9501; Berthold), normalized to β-Gal activity, and expressed as relative luciferase units (RLU). Each value presented is the average of duplicate or triplicate samples and is representative of multiple independent experiments.
RESULTS
Identification and cloning of DRIP205.
We previously isolated a DRIP complex from human nuclear extracts by affinity purification using the VDR LBD (35). Mass spectrometric analysis of DRIP subunits revealed that DRIP205 (originally estimated as a 230-kDa protein by SDS-PAGE analysis [35]) is identical to RB18A, a human protein cloned in the context of p53 transactivation (10). DRIP205/RB18A is also highly homologous to mouse PPARγ binding protein (PBP) cloned in a yeast two-hybrid screen by virtue of its interaction with another nuclear receptor, PPARγ (47). The central portion of PBP is itself identical to TRIP2, a protein fragment cloned by the yeast two-hybrid method as a TR-interacting protein (22). Protein homology searches indicate that DRIP205 is also identical to TRAP220, ARC205, and CRSP200, components of the TRAP (12, 44), ARC (30, 31), and CRSP (38) coactivator complexes, respectively. These complexes together are highly homologous to the DRIP complex, based on the identity of a number of their subunits. We isolated DRIP205 cDNA from U-937 cells by RT-PCR. This clone matched the mass spectrometric data obtained from the cloned gene and the original protein used for its identification (34).
Ligand-dependent interaction of DRIP205 with VDR and TR.
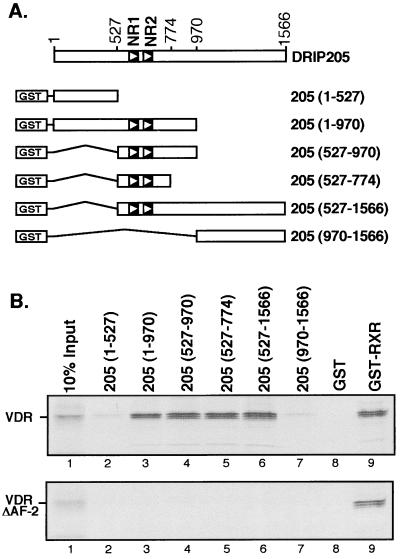
DRIP205 sequence contains two closely positioned consensus LXXLL nuclear receptor interaction motifs that we have termed NR1 and NR2 (Fig. 1 and 4A), suggesting that it directly interacts with VDR. The ligand-dependent effect that we previously observed for the interaction between the entire DRIP complex and nuclear receptors (35) was analyzed for several individual full-length DRIP subunits; only DRIP205 bound VDR in a ligand-dependent manner (34). Similar results have been reported for TRAP220 with several nuclear receptors (44). To more carefully map the region of DRIP205 required for nuclear receptor binding, we expressed a series of deletion mutants of DRIP205 (Fig. 1A) as GST fusion proteins in E. coli and tested their ability to bind in vitro-translated [35S]VDR in the presence of 1,25(OH)2D3 by GST pull-down assay. DRIP205(527-774) and all other DRIP205 fragments containing NR1 and NR2 motifs retained VDR binding activity in vitro (Fig. 1B, top, lanes 3 to 6); those derivatives lacking the NR motifs did not interact with VDR (Fig. 1B, top, lanes 2 and 7). This finding reinforces a potential role of NR1 and NR2 in nuclear receptor binding. VDR interaction with DRIP205 was abolished when VDR lacked its AF-2 domain (Fig. 1B, bottom). Thus, as with many other nuclear receptor coactivators, DRIP205 interaction with VDR is provided by contacts with the NR box region of DRIP205 and requires the AF-2 motif of liganded VDR.
FIG. 1.
Mapping of regions in DRIP205 required for VDR interaction. (A) Schematic representation of the various DRIP205 fragments fused to GST and used as baits in in vitro pull-down assays. The two potential nuclear receptor interaction motifs (NR1 and NR2) are indicated. (B) GST pull-down assay using GST-DRIP205 deletion mutants and [35S]VDR wild type (top) or [35S]VDR AF-2 deletion mutant (bottom). All incubations were carried out in the presence of 10−6 M 1,25(OH)2D3.
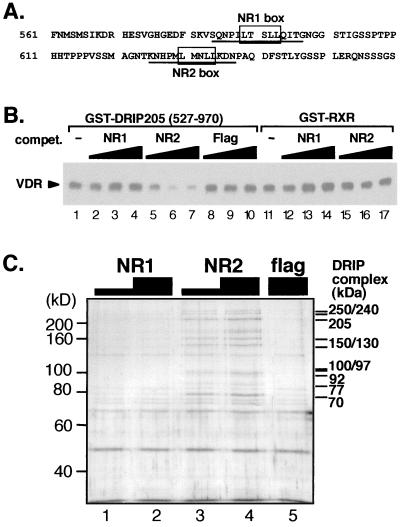
FIG. 4.
DRIP205-VDR interaction is selectively competed by the NR2 peptide. (A) Sequence of DRIP205 encompassing the two potential nuclear receptor-interacting motifs, NR1 and NR2. Underlined amino acids correspond to the sequences of NR1 and NR2 peptides used in subsequent experiments. (B) GST pull-down assay using 2 μg of GST-DRIP205 fragment (527 to 970) and purified VDR in the presence of 10−6 M 1,25(OH)2D3. NR1 (lanes 2 to 4) and NR2 (lanes 5 to 7) peptides (A) and an unrelated peptide (Flag; lanes 8 to 10) were used as competitors in 2-, 5-, and 20-fold molar excess over GST-DRIP205. The same amounts of NR peptides were coincubated with GST-RXR (lanes 11 to 17), here used as a control bait. (C) NR2 peptide is sufficient to elute the entire DRIP complex from VDR. GST-VDR LBD was used to pull down the DRIP complex from nuclear extracts as described previously (35). The complex immobilized on beads was then incubated with 5 or 30 μM NR1 or NR2 or 30 μM nonspecific (Flag) peptide. Bands matching subunits of the DRIP complex (34) are shown on the right.
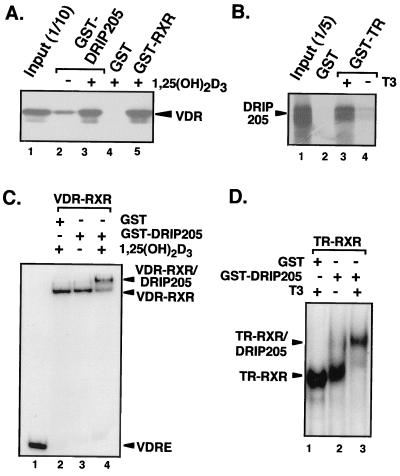
GST-DRIP205(527-970) was then used to assess ligand-dependent binding to both VDR and TR on and off DNA. First, the GST-DRIP205 fragment and 35S-labeled VDR (Fig. 2A), or GST-TR and 35S-labeled DRIP205 (Fig. 2B), were assayed for their direct interactions in vitro. DRIP205 binding to both nuclear receptors occurred in a strict ligand-dependent manner (Fig. 2A, lanes 2 and 3; Fig. 2B, lanes 3 and 4). We then investigated if the same effect is observed with DNA-bound receptor heterodimers. To do so, we examined the ability of DRIP205 to interact with RXR-VDR or RXR-TR heterodimers bound to a labeled VDRE or TRE by gel shift analysis. In both cases, we observed a strong ligand-dependent supershift of each heterodimer on DNA in the presence of both DRIP205 and the specific nuclear receptor ligand (Fig. 2C, lanes 3 and 4; Fig. 2D, lanes 2 and 3). These results suggest that the ligand-dependent effect previously observed on the binding of the 13-subunit DRIP complex to nuclear receptors occurs primarily through DRIP205's recruitment to the AF-2 in response to ligand binding.
FIG. 2.
DRIP205 ligand-dependent binding with VDR and TR. (A) GST pull-down assay using 5 μg of GST-DRIP205(527-970) together with purified VDR in the presence of 10−6 M 1,25(OH)2D3 (+) or ethanol (−). GST alone and GST-RXR were used as negative and positive controls, respectively. Detection was by immunoblotting with anti-VDR antibody. (B) GST pull-down assay using GST-TR and [35S]DRIP205 in the presence (+) or absence (−) of thyroid hormone (T3). Detection was by autoradiography. (C) Gel mobility shift assay using eluted GST-DRIP205(527-970) or GST alone to supershift a VDR-RXR heterodimer bound to a consensus VDRE in the presence of 10−6 M 1,25(OH)2D3 (+) or ethanol (−). (D) Gel mobility shift assay of GST-DRIP205 binding to a TR-RXR heterodimer bound to a TRE in the presence of 10−6 M T3 (+) or ethanol (−), as described in for panel C.
Two functionally distinct coactivator complexes bind to VDR.
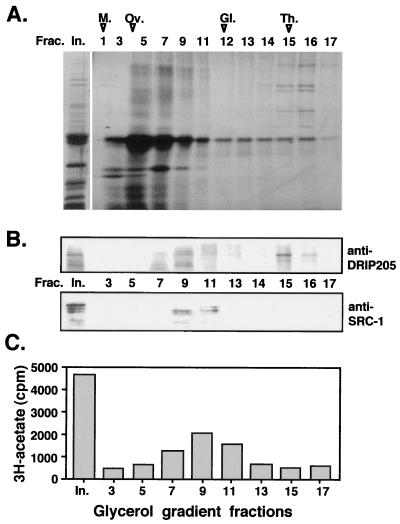
The similarity in the mode of interaction between VDR and DRIP205 relative to SRC-1/p160 family members raised the question of why none of the latter coactivators appear in the DRIP complex, or why they may not bind to VDR or TR on their own when transcriptionally active nuclear extracts are passed over immobilized LBD columns. Therefore, a closer analysis of the protein pattern obtained in the GST-VDR LBD pull-down assay was carried out by fractionation through glycerol gradients. This assay allowed us to distinguish between our large DRIP complex (Fig. 3A, fractions 15 to 17) and other smaller proteins or protein complexes (fractions 5 to 12) bound to VDR in apparent substoichiometric amounts. Western blot analysis was then performed on the gradient fractions, using antibodies directed against DRIP205 or other known coactivators. The signal observed for DRIP205 matched the sedimentation profile of the whole DRIP complex; it was detected most predominantly in fractions corresponding to the megadalton complex (Fig. 3, fractions 15 and 16 in panels A and B). However, immunoblotting with an anti-SRC-1 antibody detected the presence of SRC-1 only within smaller sedimenting fractions (Fig. 3B, fractions 9 to 11). We have also detected TIF-2 in these same fractions (C. Rachez, M. Gamble, and L. P. Freedman, unpublished data). Furthermore, a HAT activity assay performed on the same glycerol gradient fractions showed cosedimentation between the SRC-1 signal by Western blotting and HAT activity (Fig. 3C). No HAT activity, however, was detected with the DRIP complex fractions (compare Fig. 3B and C). These results demonstrate the absence of HAT activity within the DRIP complex itself, and importantly, they also demonstrate the ability of VDR to interact with different types of endogenous coactivators in nuclear extracts under the same biochemical conditions as distinct complexes. SRC-1/p160 is representative of one complex, and the DRIPs represent another, functionally distinct complex.
FIG. 3.
Two functionally distinct complexes bind VDR LBD. (A) Glycerol gradient fractionation of proteins immobilized on the GST-VDR LBD affinity column are shown by silver staining of a SDS–7.5% polyacrylamide gel. In., input; M., myoglobin; Ov., ovalbumin; Gl., gamma globulin; Th., thyroglobulin. (B) SRC-1 and DRIP205 exhibit distinct sedimentation profiles, as determined by Western blot analyses of glycerol gradient fractions in panel A for the presence of DRIP205 (probed with anti-DRIP205 serum) and SRC-1 (probed with MAb GT12 2E9). (C) HAT activity colocalizes with SRC-1 but not with the DRIP complex. Fractions were assayed for HAT activity in the presence of free histones by a filter binding assay as described previously (35). HAT activity was measured as the amount of [3H]acetate transferred from [3H]acetyl coenzyme A to histones.
DRIP205 interacts with VDR via only one of its two consensus NR box motifs.
To further analyze the contributions of the two NR box consensus sequences of DRIP205 in nuclear receptor binding, we performed competition assays in vitro using synthetic peptides encompassing each of two putative NR box sequences (i.e., NR1 and NR2 [Fig. 4A]). A 2- to 20-fold molar excess of NR2 peptide over DRIP205 was able to efficiently compete DRIP205 binding to VDR, but the same ratio of NR1 peptide or an unrelated nonspecific peptide was unable to compete for this interaction (Fig. 4B, compare lanes 5 to 7 to lanes 2 to 4 or 8 to 10). These results establish that the interaction of DRIP205 and VDR in solution requires only the NR2 box motif.
If DRIP205 anchors the entire DRIP complex to VDR, we would expect the NR2 peptide to compete the complex itself. The requirement for the NR2 motif of DRIP205 was therefore tested in the context of the whole DRIP complex. For this purpose, the DRIP complex from Namalwa nuclear extracts was immobilized on a VDR LBD affinity matrix as previously described (35). The NR1 and NR2 peptides were then incubated at different concentrations with the immobilized proteins. Analysis of the eluted material (Fig. 4C) demonstrated the ability of the NR2 peptide (5 μM) but not the NR1 peptide to compete off the entire immobilized DRIP complex (Fig. 4C). These data reinforce the key role of DRIP205 in the recruitment of the DRIP complex to VDR in response to ligand.
Differential requirement of NR1 and NR2 motifs of DRIP205 for binding of RXR-VDR heterodimers on DNA.
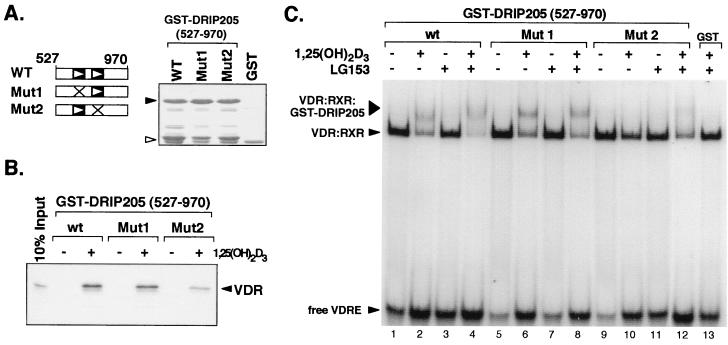
The requirement of the NR2 motif of DRIP205 for VDR binding was also tested in the context of VDR as a heterodimer with RXR bound to its response element VDRE. For this purpose, we created within the LXXLL motif in each NR box of DRIP205 (LXXAA mutations) point mutations that are known to attenuate binding to nuclear receptors (9, 28). GST pull-down assays between DRIP205 NR box mutants (amino acids 527 to 970) and in vitro-translated VDR (Fig. 5B) confirmed the essential role of the NR2 motif in nuclear receptor-coactivator interactions, as previously demonstrated by the peptide competition assay shown in Fig. 4. The ability of these mutants to bind to a VDR-RXR heterodimer on DNA was tested by gel shift analysis in response to VDR- and RXR-specific ligands [1,25(OH)2D3 and LG153, respectively]. Formation of a ternary complex between VDR, RXR, and GST-DRIP205 on the VDRE was strongly induced by 1,25(OH)2D3, but LG153 had no visible effect on DRIP205 binding (Fig. 5C, lanes 1 to 4). Point mutations in DRIP205's NR1 box (Mut1) did not significantly affect its binding to the heterodimer in the presence of 1,25(OH)2D3 or LG153 (lanes 5 to 8). However, mutations of the NR2 motif of DRIP205 (Mut2) abolished its ability to form a complex with the heterodimer in the presence of 1,25(OH)2D3 (lane 10). Interestingly, however, addition of LG153 together with 1,25(OH)2D3 was able to promote weak binding of DRIP205 (Mut2) to VDR-RXR (lane 12). These results reveal that, in addition to the strong requirement for the NR2 motif in DRIP205 binding to VDR-RXR, there is also a contribution of the NR1 motif to DRIP205 binding to VDR-RXR in the presence of both ligands, suggesting that each NR box contacts each subunit of the heterodimer.
FIG. 5.
DRIP205 binds a VDR-RXR heterodimer on DNA through contributions of both NR boxes. (A) Schematic representation of GST-DRIP205(527-970) wild-type protein (WT), or the same fragment containing point mutations in the NR1 or NR2 box that change each LXXLL motif to LXXAA (Mut1 or Mut2). The GST proteins used in the experiments depicted in panels B and C were quantitated by visualization on a Coomassie blue-stained SDS-polyacrylamide gel (right). Black arrowhead, GST-DRIP205(527-970) proteins; white arrowhead, GST alone. (B) GST pull-down assay of in vitro-translated, [35S]methionine-labeled VDR and GST-DRIP205(527-970) protein fragments. The pull-down assays were carried out in the presence (+) or absence (−) of 10−6 M 1,25(OH)2D3, as indicated, and VDR was visualized by autoradiography. (C) Association of DRIP205 to DNA-bound VDR-RXR heterodimers. Gel mobility shift analysis was performed in the presence of purified VDR, RXR, and GST-DRIP205(527-970), together with a VDRE oligonucleotide as a probe. GST-DRIP205 wild-type (wt), Mut1, and Mut2 proteins (A) were used in the presence (+) or absence (−) of 10−6 M of 1,25(OH)2D3 or LG153 ligands.
Functional requirement of DRIP205 NR box motifs in VDR transactivation.
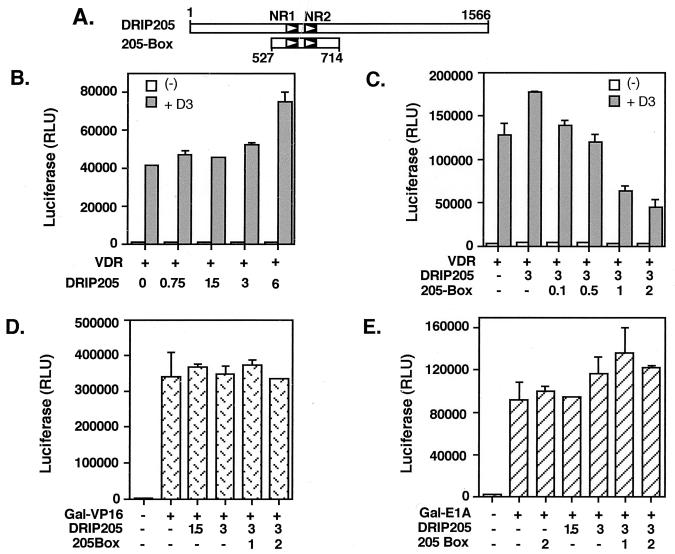
The results in Fig. 5 indicate that both NR1 and NR2 box motifs of DRIP205 are specifically required for RXR-VDR binding in vitro. We tested whether these motifs are also used in a functional context in vivo. For this purpose we analyzed the effect of DRIP205 on VDR transcription activity by transient transfection of U937 cells. Cotransfection of VDR and DRIP205 confered a modest dose-dependent costimulatory effect of DRIP205 on VDR activity (Fig. 6B). Interestingly, a 190-amino-acid fragment of DRIP205 (residues 527 to 714) that contains its two NR box motifs (205-Box [Fig. 6A]) had a strong dominant negative effect on VDR transactivation (Fig. 6C). Overexpression of the 205-Box also inhibited endogenous VDR function in this cell line (data not shown). This suggests that the region of DRIP205 that encompasses the NR box motifs is required for VDR transactivation by the DRIP complex. These data are also in agreement with the results of the in vitro binding assays (Fig. 5). The specificity of the NR box requirement for VDR transactivation was also tested in the context of other types of activators, such as VP16 and E1A. We expressed these activators as fusions with the Gal4 DNA binding domain and tested them on GAL4 upstream activation sequence promoters in transient transfection assays. DRIP205 was unable to potentiate the VP16 response, and the 205-Box did not confer a dominant negative effect over VP16 activation (Fig. 6D). DRIP205 and 205-Box also had no significant effect on E1A transactivation (Fig. 6E).
FIG. 6.
DRIP205 potentiates VDR transactivation, and a 190-amino-acid fragment containing both NR boxes (205-Box) acts as a dominant negative in vivo. (A) Schematic representation of the two NR box motifs (NR1 and NR2) in the full-length DRIP205 (amino acids 1 to 1566) and in 205-Box (527 to 714) used in the transient transfection experiments. (B) U-937 cells were transfected with a luciferase reporter plasmid containing a multimerized VDRE, together with 0.5 μg of VDR expression vector (+) and increasing amounts (in μg) of DRIP205 expression vector, in the absence (−) or presence (+D3) of 10−8 M 1,25(OH)2D3. Luciferase activity (expressed as RLU) was normalized relative to β-Gal activity. (C) Transient transfections were performed as for panel B, with the addition of increasing amounts (in micrograms) of 205-Box expression construct. (D and E) DRIP205 and 205-Box have no effect on transactivation by VP16 and E1A activation domains. Transient transfection assays were performed as for panels B and C except that U-937 cells were transfected with a luciferase reporter plasmid containing a multimerized GAL4 upstream activation sequence enhancer, together with increasing amounts (in micrograms) of DRIP205 and 205-Box expression vectors. Cells were also transfected with expression vectors for Gal4-VP16 (50 ng; D) or Gal4-E1A (100 ng; E).
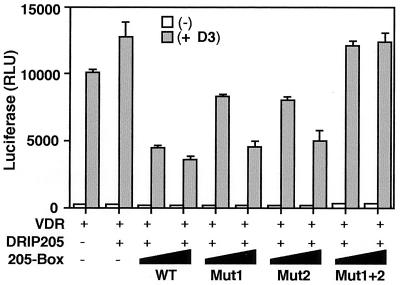
To examine more closely the requirements of both DRIP205 NR boxes for VDR transactivation, we tested the effect of point mutations in the context of the 205-Box that disrupt each NR box (i.e., Mut1 and Mut2 [Fig. 5]). Surprisingly, 205-Box Mut1 and Mut2 each had roughly the same dominant negative effect as the wild-type sequence (Fig. 7). Only a simultaneous mutation of both NR boxes in the context of the 205-Box construct (Mut1+2) relieved its dominant negative effect on VDR transactivation, suggesting that both NR boxes have equivalent functions in vivo, perhaps through their individual interactions with VDR and RXR.
FIG. 7.
Both NR boxes of DRIP205 are equally required for VDR signaling in vivo. A transient transfection assay was performed as for Fig. 6C, using 205-Box wild-type (WT) and mutant constructs.
In vitro interaction assays have shown that VP16 and E1A can recruit complexes related to the DRIP complex, such as TRAP/SMCC, ARC, and human Mediator, via direct contacts with subunits that are present in our complex, i.e., via TRAP80/DRIP77 and Sur-2/DRIP130, respectively (4, 20, 31); however, they do so independently of DRIP205. Taken together, these results suggest that distinct subunits are required for direct recruitment to different types of transcription activation motifs at a functional level. The NR boxes in DRIP205 therefore may be specific contact points for nuclear receptors and not other classes of transcription activators.
DISCUSSION
We have demonstrated that a single subunit of the multiprotein DRIP coactivator complex, DRIP205, interacts directly with the VDR LBD in an AF-2-dependent fashion. Moreover, a single, short motif within DRIP205, the NR2 box, is crucial to the DRIP complex's ligand-dependent interaction with VDR. A peptide corresponding to DRIP205 NR2 box was sufficient to compete the entire megadalton DRIP complex bound to VDR. However, in the context of a DNA-bound VDR-RXR heterodimer, both NR1 and NR2 in DRIP205 were required for binding, albeit to different extents. Furthermore, both NR boxes were equally required for the functional interaction of DRIP205 with VDR in vivo. Our results define for DRIP205 a mechanism of interaction with nuclear receptors that is similar to the one used by p160 coactivators such as SRC-1 and TIF-2/GRIP1. p160 coactivators use a combination of two NR boxes to achieve a degree of specificity for interactions with different nuclear receptor heterodimers, resulting in the binding of one coactivator per receptor dimer (9, 28).
Based on several AF-2 point mutants in VDR, we previously demonstrated that the DRIP complex shares similar sequence and perhaps structural requirements for receptor interaction with the SRC-1/p160 family (35). This type of interaction was shown to be competitive and mutually exclusive between TRAP220 and TIF2 for binding of TR-RXR heterodimers in vitro (43). Both coactivators interact with TR via the receptor's AF-2 motif with similar binding efficiencies. These results correlate with our own in vitro competition assays between VDR, DRIP205, and SRC-1, using peptides encompassing DRIP205 and SRC-1 NR boxes (data not shown). This raises the question of the functional relevance of receptor interactions with p160 coactivators and the DRIP complex in vivo. In a physiological context, transcription initiation requires promoter accessibility to transcription factors. This process involves chromatin remodeling via specific modifications of histone N-terminal tails, including acetylation by CBP/p300 and perhaps SRC-1-type coactivators. However, the prototypic model of coactivators acting as chromatin remodeling factors may be revised by several reports of HAT activity directed to modifications of nonhistone proteins. For example, acetylation of ACTR, a member of the p160 family, by CBP/p300 can regulate the association between ACTR and ER (5). These results fit with a model where CBP/p300 would act on TR transcription at a step subsequent to chromatin disruption and would not be essential for chromatin accessibility per se (25). Nucleosome remodeling by ATP-dependent activities has also been identified within large transcription complexes like SWI-SNF in mammals and yeast, ACF, CHRAC, NURF in Drosophila, RSC in yeast, and RSF in humans (reviewed in references 1 and 24). It still remains to be determined if the DRIP complex contains any remodeling activity, but we have established here that (i) it does not contain HAT activity and (ii) endogenous SRC-1 from nuclear extracts as well as several other related coactivators are not part of the DRIP complex, although they can clearly interact with VDR in the presence of ligand (Fig. 3).
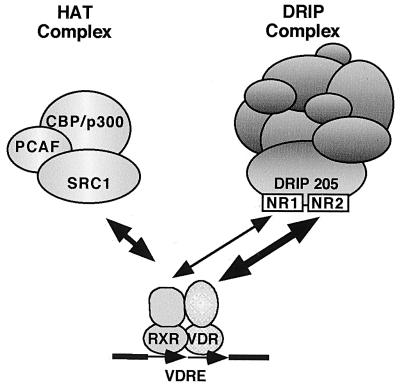
It should be pointed out that in an earlier publication (35), we reported that the LBD-bound material, which we identified as the DRIP complex, in fact contained HAT activity. Based on the results shown in Fig. 3, we must now reinterpret the data in light of the fact that the HAT activity does not cosediment with the DRIP complex on a glycerol gradient. We believe that the HAT activity derives from endogenous CBP/p300, which associates together with SRC-1 on other bound LBD moieties; these complexes appear to be distinct from DRIP since they sediment in distinct fractions on the gradient (Fig. 3). We cannot rule out, however, that both the DRIP and CBP/p160 complexes bind the LBD simultaneously but fall apart on the gradient, but this seems unlikely since they both require the AF-2 for ligand-dependent binding to the receptor. Importantly, we have carried out in vitro transcription assays using a minimal set of purified transcription components and have demonstrated an absolute requirement for the purified DRIP complex (e.g., gradient purified and devoid of HAT activity) in VDR-mediated transactivation, but only in the context of chromatin-assembled templates (34). This suggests that the DRIP complex may also require the cooperation of a remodeling activity, or itself contain a chromatin remodeling activity distinct from a HAT, at least when tested in vitro on chromatin templates. We propose that transcriptional activation by nuclear receptors requires the hormone-dependent recruitment of different complexes possessing distinct functions (Fig. 8). These would include a HAT activity-containing complex anchored on nuclear receptors via p160 coactivators (29) and a DRIP complex anchored via DRIP205. This model may correlate with our observation that the dominant negative 205-Box can in vivo compete equally well VDR transactivation potentiated by individual overexpression of DRIP205 or GRIP-1 (data not shown). Given the ability of NR box-containing peptides from each coactivator to cross-compete in vitro, this is not necessarily surprising. At this point, we have no idea if these proteins exchange with one another in equilibrium or interact with nuclear receptors in a stepwise fashion. The results of Chen et al. (5) demonstrating that the acetylation of ACTR by CBP results in the former's dissociation from ER suggests to us the possibility of a sequential mechanism, but this remains to be proven experimentally.
FIG. 8.
Model of VDR-coactivator interactions, as described in the text.
The potential homology of the DRIP complex with several other human complexes is based on the identity of a certain number of their subunits. For example, DRIP205 is identical to TRAP220, ARC205, and CRSP200, found in TRAP/SMCC, ARC, and CRSP complexes, respectively (31, 38, 44). Several DRIP subunits are also part of the NAT, mammalian Mediator, and human Mediator complexes (4, 21, 41). Functional assays performed on individual complexes, and a series of genetic experiments on Mediator, the apparent yeast counterpart of mammalian Mediator, provide some clues for functionality. Srb and Med subunits of yeast Mediator were identified for their role in modulating RNA Pol II activity. The DRIP and other mammalian complexes contain several Srb/Med subunits and therefore may also have a direct role in transcription initiation by RNA Pol II, since they contain phosphorylation activities targeting RNA Pol II C-terminal domain (21, 41) or regulating general transcription cofactors such as PC4 (16). The presence of Mediator subunits as components of the DRIP complex also suggests a role for the DRIPs in direct recruitment of RNA Pol II to the promoter (31, 34).
Recent studies have identified multiple binding sites for transcription factors within the DRIP complex. For example, the glucocorticoid receptor associates with the DRIP complex via both AF-1 and AF-2 motifs through direct interactions with DRIP150 and DRIP205, respectively (19). More generally, a number of transcription factors can functionally interact with several mammalian complexes homologous to the DRIP complex. The TRAP complex was originally purified by its ability to interact with TR (12). Other related complexes, such as ARC, SMCC, CRSP, and human Mediator, bind a number of targets that are distinct of nuclear receptors, such as SREBP-1a, NF-κB, VP16, E1A, p53, and SP-1, none of which have been found to interact with DRIP205 directly (4, 20, 31, 38). We believe that the association of some or most of the DRIP subunits with unique classes of activators suggests that transcriptional activation involves a limited, common set or subsets of general cofactors and that this type of complex provides a regulatory panel for targeting of RNA Pol II by multiple activators. Through this model, ligand-dependent transcription regulation by nuclear receptors would be supported by a strictly ligand-regulated association of a single subunit, DRIP205.
ACKNOWLEDGMENTS
We thank P. MacDonald, M. Stallcup, M. Garabedian, and R. Fisher for plasmids, and we thank J. DiRenzo and M. Brown for anti-hSRC1 MAb GT12 2E9. We also thank P. Tempst and H. Erdjument-Bromage for carrying out mass spectrometry. NR1 and NR2 peptides were synthesized by the MSKCC Protein Core Facility.
This work was supported by grants to L.P.F. from the NIH and the Human Frontiers Science Program.
REFERENCES
- 1.Armstrong J A, Emerson B M. Transcription of chromatin: these are complex times. Curr Opin Genet Dev. 1998;8:165–172. doi: 10.1016/s0959-437x(98)80137-8. [DOI] [PubMed] [Google Scholar]
- 2.Atkins G B, Guenther M G, Rachez C, Freedman L P, Lazar M A. Coactivators for the orphan nuclear receptor RORalpha. Mol Endocrinol. 1999;13:1550–1557. doi: 10.1210/mend.13.9.0343. [DOI] [PubMed] [Google Scholar]
- 3.Barettino D, Vivanco Ruiz M M, Stunnenberg H G. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer T G, Martin M E, Lees E, Ricciardi R P, Berk A J. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 6.Chen H W, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 7.Cheskis B, Freedman L P. Ligand modulates the conversion of DNA-bound vitamin D3 receptor (VDR) homodimers into VDR-retinoid X receptor homodimers. Mol Cell Biol. 1994;14:3329–3338. doi: 10.1128/mcb.14.5.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drane P, Barel M, Balbo M, Frade R. Identification of RB18A, a 205 kDa new p53 regulatory protein which shares antigenic and functional properties with p53. Oncogene. 1997;15:3013–3024. doi: 10.1038/sj.onc.1201492. [DOI] [PubMed] [Google Scholar]
- 11.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedman L P, editor. Molecular biology of steroid and nuclear hormone receptors. Boston, Mass: Birkhauser; 1997. [Google Scholar]
- 14.Freedman L P, Arce V, Perez Fernandez R. DNA sequences that act as high affinity targets for the vitamin D3 receptor in the absence of the retinoid-X receptor. Mol Endocrinol. 1994;8:265–273. doi: 10.1210/mend.8.3.8015545. [DOI] [PubMed] [Google Scholar]
- 15.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 16.Gu W, Malik S, Ito M, Yuan C X, Fondell J D, Zhang X, Martinez E, Qin J, Roeder R G. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 17.Harding H P, Atkins G B, Jaffe A B, Seo W J, Lazar M A. Transcriptional activation and repression by RORalpha, an orphan nuclear receptor required for cerebellar development. Mol Endocrinol. 1997;11:1737–1746. doi: 10.1210/mend.11.11.0002. [DOI] [PubMed] [Google Scholar]
- 18.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 19.Hittelman A B, Burakov D, Iñiguez-Lluhi J A, Freedman L P, Garabedian M J. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y W, Veschambre P, Erdjument-Bromage H, Tempst P, Conaway J W, Conaway R C, Kornberg R D. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J W, Choi H S, Gyuris J, Brent R, Moore D D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 23.Lemon B D, Fondell J D, Freedman L P. Retinoid X receptor:vitamin D3 receptor heterodimers promote stable preinitiation complex formation and direct 1,25-dihydroxyvitamin D3-dependent cell-free transcription. Mol Cell Biol. 1997;17:1923–1937. doi: 10.1128/mcb.17.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemon B D, Freedman L P. Nuclear receptor cofactors as chromatin remodelers. Curr Opin Genet Dev. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Imhof A, Collingwood T N, Urnov F D, Wolffe A P. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 1999;18:5634–5652. doi: 10.1093/emboj/18.20.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mak H Y, Hoare S, Henttu P M, Parker M G. Molecular determinants of the estrogen receptor-coactivator interface. Mol Cell Biol. 1999;19:3895–3903. doi: 10.1128/mcb.19.5.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 28.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna N J, Nawaz Z, Tsai S Y, Tsai M-J, O'Malley B W. Distinct steady-state nuclear receptor coregulator complexes exist in vivo. Proc Natl Acad Sci USA. 1998;95:11697–11702. doi: 10.1073/pnas.95.20.11697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Näär A M, Beaurang P A, Robinson K M, Oliner J D, Avizonis D, Scheek S, Zwicker J, Kadonaga J T, Tjian R. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 1998;12:3020–3031. doi: 10.1101/gad.12.19.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Näär A M, Beaurang P A, Zhou S, Abrahams A, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 32.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 33.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 34.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Näär A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 35.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 37.Rots N Y, Iavarone A, Bromleigh V, Freedman L P. Induced differentiation of U937 cells by 1,25-dihydroxyvitamin D3 involves cell cycle arrest in G1 that is preceded by a transient proliferative burst and an increase in cyclin expression. Blood. 1999;93:2721–2721. [PubMed] [Google Scholar]
- 38.Ryu S, Zhou S, Ladurner A G, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 39.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O'Malley B W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 40.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J X, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 41.Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- 42.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 43.Treuter E, Johansson L, Thomsen J S, Wärhmark A, Leers J, Pelto-Huikko M, Sjoberg M, Wright A P, Spyrou G, Gustafsson J. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of trap and p160 coactivator complexes. J Biol Chem. 1999;274:6667–6677. doi: 10.1074/jbc.274.10.6667. [DOI] [PubMed] [Google Scholar]
- 44.Yuan C X, Ito M, Fondell J D, Fu Z Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamir I, Zhang J, Lazar M A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 46.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Y, Qi C, Jain S, Rao M S, Reddy J K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]