Abstract
Ras-GRF2 (GRF2) is a widely expressed, calcium-activated regulator of the small-type GTPases Ras and Rac. It is a multidomain protein composed of several recognizable sequence motifs in the following order (NH2 to COOH): pleckstrin homology (PH), coiled-coil, ilimaquinone (IQ), Dbl homology (DH), PH, REM (Ras exchanger motif), PEST/destruction box, Cdc25. The DH and Cdc25 domains possess guanine nucleotide exchange factor (GEF) activity and interact with Rac and Ras, respectively. The REM-Cdc25 region was found to be sufficient for maximal activation of Ras in vitro and in vivo caused Ras and extracellular signal-regulated kinase (ERK) activation independent of calcium signals, suggesting that, at least when expressed ectopically, it contains all of the determinants required to access and activate Ras signaling. Additional mutational analysis of GRF2 indicated that the carboxyl PH domain imparts a modest inhibitory effect on Ras GEF activity and probably normally participates in intermolecular interactions. A variant of GRF2 missing the Cdc25 domain did not activate Ras and functions as an inhibitor of wild-type GRF2, presumably by competing for interactions with molecules other than calmodulin, Ras, and ligands of the PH domain. The binding of calmodulin was found to require several amino-terminal domains of GRF2 in addition to the IQ sequence, and no correlation between calmodulin binding by GRF2 and its ability to directly activate Ras and indirectly stimulate the mitogen-activated protein (MAP) kinase ERK in response to calcium was found. The precise role of the GRF2-calmodulin association, therefore, remains to be determined. A GRF2 mutant missing the IQ sequence was competent for Ras activation but failed to couple this to stimulation of the ERK pathway. This demonstrates that Ras-GTP formation is not sufficient for MAP kinase signaling. We conclude that in addition to directly activating Ras, GRF2, and likely other GEFs, promote the assembly of a protein network able to couple the GTPase with particular effectors.
Ras-GRF2 (GRF2) is a widely expressed guanine nucleotide exchange factor (GEF) that stimulates the release of bound guanine nucleotide by the low-molecular-weight G protein Ras (6, 10). GRF2 stimulates the conversion of Ras from its GDP-bound state into a GTP-bound activated conformation. The Ras-binding domain of GRF2, which catalyzes the activation of Ras, is located in its carboxyl-terminal region and is approximately 40% identical to the Ras GEF domain of the Saccharomyces cerevisiae Cdc25 gene product (10). GRF2 is a bifunctional GEF. In addition to its activity on Ras, it binds to the small G protein Rac through its Dbl homology (DH) domain (11).
By virtue of its two distinct GEF activities, GRF2 is a potent activator of two different mitogen-activated protein kinases which function downstream of Rac and Ras (11). They are, respectively, the stress-activated protein kinase (SAPK) and the extracellular signal-regulated protein kinase (ERK). The brain-specific protein Ras-GRF1 (GRF1) has a domain structure similar to that of GRF2 (5, 17), and recently its DH domain was demonstrated to possess Rac GEF activity (14). The Son-of-sevenless gene product (Sos) also has been demonstrated to function as a Rac GEF (16). The frequent coupling of Ras and Rac GEF activities into a single polypeptide may reflect a strict requirement for the coordination of Ras and Rac effector pathways.
GRF2 has not been subjected to three-dimensional structural analysis, but inspection of its primary sequence and functional studies suggest that it is a modular protein composed of discrete functional domains (10, 11). It contains, in amino-to-carboxyl-terminal order, a pleckstrin homology (PH) domain, a coiled-coil, an ilimaquinone (IQ) motif, a DH domain, a second PH domain, a Ras exchanger motif (REM), a PEST region (rich in the amino acids proline, glutamate, serine, and threonine) that contains a candidate destruction box (DB), and, finally, the Cdc25 domain (10). Based on the solved structure of the REM and Cdc25 regions of the Son-of-sevenless (Sos) protein, it is likely the REM and Cdc25 regions of GRF2, and indeed of all Ras GEFs, interact to form a stable Ras-binding domain (3). DH domains, including that of GRF2, are flanked on their COOH sides by a PH domain. In Sos this neighboring PH domain may stabilize the Rac-binding region in the DH domain (18) and may be affected by the lipid products of phosphatidylinositol 3′-kinase (16). Hence, both the Cdc25 and DH classes of the GEF domain are augmented and perhaps regulated by a neighboring noncatalytic domain. However, the intra- and intermolecular interactions involving the various noncatalytic domains in GRF2 have not been determined.
Activation of the SAPK and ERK pathways by GRF2 is stimulated by calcium influx, and this requires the IQ motif of GRF2, which is required to maintain the calcium-dependent binding of calmodulin (CaM) to GRF2 (10, 11). GRF1-mediated activation of ERK is also regulated in a similar fashion (12). However, in contrast to what is shown by the signaling data, GRF1 Ras-specific GEF activity does not appear to be stimulated by the binding of calmodulin in vitro. In fact, some studies have shown that this association inhibits the Cdc25 domain (2). Therefore, the precise role of CaM in the regulation of GRF2 and GRF1 and their CaM-binding site(s) remain to be determined. Activated GRF1 is phosphorylated on serine/threonine (15) and tyrosine (14), suggesting that phosphorylation may play a direct role in the regulation of the GRFs. The recently described Ras-GRP protein is another Ras GEF. It contains calcium and diacylglycerol binding sites and like the other Ras GEFs appears to be regulated by translocation to the plasma membrane (9).
In this study, we addressed structure-function relationships in GRF2. Deleted and truncated versions of GRF2 were constructed and analyzed for their abilities to bind CaM, to activate Ras in vitro and in vivo, and to stimulate the ERK pathway. Our data indicate that CaM binding does not correlate with Ras-ERK activation, which is contrary to the previously held belief that CaM binding activates GRF-mediated signaling. This analysis indicates that GRF2 is a modular protein subject to complex regulatory mechanisms involving its noncatalytic domains.
MATERIALS AND METHODS
Construction of Flag-tagged GRF2 deletions.
The cloning of ΔIQ, ΔDH, and ΔCdc25 GRF2 mutants has been previously described (11). The GRF2 PH deletion mutants were generated by PCR. The ΔPHn construct was obtained by deleting GRF2 gene codons 23 to 135 using a 5′ primer containing an SpeI site and a 3′ primer containing a BamHI site. The PCR product was digested with SpeI and BamHI and subcloned into SpeI-BamHI-digested pcDNA3-Flag-GRF2 cDNA. GRF2 gene codons 460 to 590 were deleted to generate the ΔPHc mutant using a 5′ primer containing a BamHI site and a 3′ primer containing a KpnI site. The PCR product was digested with BamHI and ApaI and subcloned into BamHI-ApaI-digested pcDNA3-Flag-GRF2. ΔPHn+c, with both PH domains deleted, was obtained by digesting each of the ΔPHn and ΔPHc constructs with BamHI and KpnI and ligating together the 343-bp PHn insert and the 8,010-bp fragment containing pcDNA3-FlagΔPHc. The REM-Cdc25 construct was generated by PCR amplification of codons 591 to 1189 of the GRF2 gene using a 5′ primer containing a KpnI site, a Kozak sequence, and a Flag sequence and a 3′ primer containing a stop codon and a BamHI site. The PCR product was digested with KpnI and BamHI and subcloned into KpnI-BamHI-digested pcDNA3. The constructs were confirmed by automated sequencing or enzymatic restriction analysis. The ΔCdc25 construct was constructed by amplifying nucleotides (nt) 1770 to 2799 by PCR using full-length GRF2 DNA as a template. A stop codon was added after nt 2799 by the addition of the sequence TCA in the 3′ primer used for PCR. The PCR product was cloned into the pCR-BLunt II-TOPO PCR vector using the ZeroBlunt TOPO PCR cloning kit (Invitrogen), and the construct was verified by automated sequencing. The nt 1807 to 3570 of full-length pcDNA3-Flag-GRF2 were replaced by nt 1807 to 2799 (plus a stop codon) by digesting the PCR product in pCR-Blunt II-TOPO with EcoRI, and the 1.1-kb fragment was ligated to the 7-kb fragment produced by digestion of pcDNA3-Flag-GRF2 with EcoRI. A variety of restriction digests were performed to verify correct orientation of the 1.1-kb fragment.
Cell culture and transfections.
293T cells were maintained in Dulbecco's modified Eagle medium containing 10% fetal bovine serum, 4.5 g of l-glutamine/liter, 10 μM nonessential amino acids, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. 293T cells grown in 10-cm-diameter dishes were transiently transfected by calcium phosphate precipitation as described previously (19).
In vitro guanine nucleotide exchange assays.
293T cells expressing wild-type (WT) GRF2 or mutant proteins were immunoprecipitated with anti-Flag antibodies as previously described (10). These immunoprecipitates were then used in exchange assays with bacterially purified, recombinant H-Ras as described by Fan et al. (11). For exchange assays measuring the effect of bound CaM, the WT Flag immunoprecipitates were washed in buffer with or without EGTA (1 mM) (to remove CaM) as described previously (10) and then used in exchange assays as described above.
ERK1 in vitro kinase assays.
293T cells were transiently cotransfected with 4 μg of pJ3M-ERK1 (encoding myc epitope-tagged ERK1) and 4 μg of either pcDNA3 vector or pcDNA3-Flag-GRF2 constructs. After 48 h, the cells were starved for 18 h and then stimulated with 5 μM ionomycin for 5 min at 37°C, washed in phosphate-buffered saline (PBS), and then lysed in NP-40 lysis buffer (20 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1% NP-40, 50 mM NaF, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 10 μg of aprotinin/ml, 0.1 mM AEBSF, and 10 μg of leupeptin/ml. Clarified lysates were incubated with 1 μg of 9E10 myc monoclonal antibody precoupled to 20 μl of goat anti-mouse agarose beads (Sigma) for 2 h at 4°C with gentle rotation. The phosphorylation of myelin basic protein (MBP) by immunoprecipitated ERK1 was performed as previously described (10).
CaM binding.
293T cells were transiently transfected with 8 μg of pcDNA3 or pcDNA3-Flag-GRF2 constructs. Forty-eight hours after transfection, the cells were washed in PBS and lysed in NP-40 lysis buffer. The lysates were clarified, precleared with antimouse agarose beads, and then immunoprecipitated with 3 μg of anti-Flag (M2) monoclonal antibody (Kodak) in the presence of 20 μl of anti-mouse agarose beads for 2 h at 4°C with gentle rotation. The immunoprecipitates were washed three times in NP-40 lysis buffer and then resuspended and boiled in 30 μl of Laemmli loading buffer. The samples were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and immunoblotted with anti-Flag (M2) or anti-CaM antibodies (Upstate Biotechnology Inc.).
Activated Ras pulldown assay.
293T cells were transiently transfected with 5 μg of pcDNA3 or pcDNA3-Flag-GRF2 constructs. Forty-eight hours after transfection, the cells were washed in HEPES-buffered saline (HBS) and lysed in a solution containing 25 mM HEPES (pH 7.5), 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, 10 mM MgCl2, 25 mM NaF, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 1 mM sodium orthovanadate, and 0.1 mM AEBSF. Levels of activated Ras in the lysate were determined as described previously (8, 11, 19a, 20) by using a glutathione S-transferase (GST) fusion protein containing the Ras-GTP-binding domain of Raf (Upstate Biotechnology Inc.).
RESULTS
In vitro and in vivo activity of GRF2 mutants.
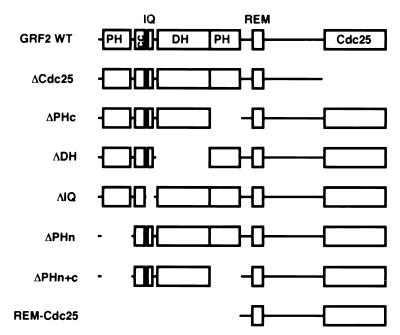
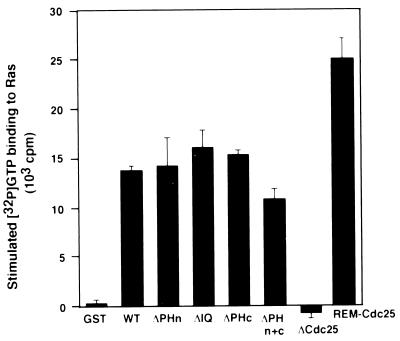
To investigate the function of the N-terminal region of GRF2, different deletion mutants, including one with a deletion of the entire N terminus, were made (Fig. 1). All these mutant proteins were expressed to similar levels in transiently transfected 293T cells (see below). To determine the effect of deleting different portions of the N terminus on Ras activation, WT GRF2 and the various mutants were compared for their abilities to catalyze in vitro nucleotide exchange of bound GDP for 32P-labeled GTP on purified, recombinant Ras. Different GRF2 constructs expressed in 293T cells were immunoprecipitated with anti-Flag antibodies. The extensively washed immunoprecipitates were then tested for guanine nucleotide exchange activity in vitro with Ras. Figure 2 shows that all the different deletion mutants retained Ras GEF activity approximately equivalent to that of WT GRF2 with the exception of REM-Cdc25, whose activity was increased approximately 1.5-fold. These results indicate that GRF2 is a modular protein; the amino-terminal domains of GRF2 are not required for in vitro nucleotide exchange on Ras by the Cdc25 domain and may in fact impart a modest inhibitory effect.
FIG. 1.
Schematic representation of the domain structures of GRF2 and of the constructs used in this report. Abbreviations are as defined in the text. cc, coiled-coil. The constructs were made using murine GRF2; the genetic sequences for the following mutants are missing the indicated codons: ΔCdc25, codons 934 to 1189; ΔPHc, codons 460 to 590; ΔPHn, codons 23 to 135; ΔDH, codons 245 to 504; ΔIQ, codons 205 to 229; ΔPHn+c, codons 23 to 135 and codons 460 to 590. The REM-Cdc25 genetic sequence contains codons 591 to 1189.
FIG. 2.
In vitro exchange activity of GRF2 and mutants. Assays were carried out 2 days after transfection of 293T cells with the indicated GRF2 construct. GRF2 was immunoprecipitated and used in an in vitro Ras exchange assay as described in Materials and Methods. Each bar shows the mean exchange activity and standard error of at least three experiments (duplicate readings in each experiment).
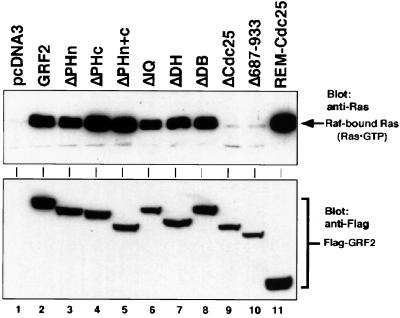
To test if the REM-Cdc25 protein was functional in vivo and to measure the activities of the various GRF2 deletion mutants, cell lysates expressing these proteins were analyzed for Ras-GTP levels by using the pulldown assay of Taylor and Shalloway (20). All of the Flag epitope-tagged proteins were comparably expressed in transfected 293T cells and migrated at their expected molecular masses relative to WT GRF2 (Fig. 3, lower panel). As demonstrated in Fig. 3 (upper panel), and consistent with Fig. 2, by this assay the N-terminal truncation mutant containing the REM and Cdc25 domains was approximately twice as efficient at activating Ras in vivo as WT GRF2. The Cdc25-deleted protein (ΔCdc25) and a negative-control mutant missing residues 687 to 933 and located immediately amino-terminal to, and perhaps infringing on, the Cdc25 domain, were inactive towards Ras in vivo. The deletion mutant missing the amino-terminal PH domain (ΔPHn) was as active as WT GRF2, whereas the mutants missing the carboxyl PH domain (ΔPHc) or both PH domains (ΔPHn+c) were more active (1.5-fold) than WT GRF2 (Fig. 3). Activated Ras was not detected in lysates from cells transfected with the empty expression vector (Fig. 3, lane 1). We showed previously that the mutant missing the DH domain (ΔDH) possesses a WT level of ERK-activating activity (11). As expected, this protein was able to cause activation of Ras in vivo that was similar to activation by WT GRF2 (Fig. 3, lane 7). Unexpectedly, the mutant missing the IQ motif (ΔIQ), previously found to be defective in basal and calcium-stimulated ERK activation (11), was still able to cause activation of Ras in vivo (Fig. 3, lane 6). These results confirm that expression of GRF2 in 293T cells causes activation of endogenous Ras proteins and that domains other than REM-Cdc25 are dispensable for this activity.
FIG. 3.
GTP-Ras in 293T cells expressing GRF and mutants. Assays were carried out 2 days after transfection of 293T cells with the indicated GRF2 construct. Lysate (1.5 mg) was incubated with 50 μg of GST–Ras-GTP binding domain beads for 30 min to pull down activated Ras. Upper panel, samples were Western blotted for Ras using the LAO45 pan-Ras monoclonal antibody, thereby allowing levels of GTP-bound Ras within the cells to be assessed; lower panel, the presence of GRF2 proteins was verified by Western blotting of lysates with Flag antibody. The data shown are representative of four experiments.
Effect of GRF2 deletion mutants on ERK1 signaling.
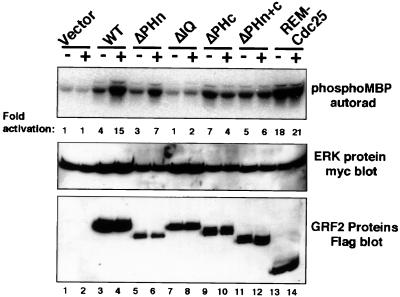
To investigate whether the N-terminal region of GRF2 played a role in ERK1 signaling, in vitro kinase assays were performed. 293T cells were cotransfected with the GRF2 constructs and myc-tagged ERK1 mitogen-activated protein kinase. After serum deprivation for approximately 18 h, the cells were treated with the calcium ionophore ionomycin or were left untreated. The cells were then lysed immediately, and ERK1 was immunoprecipitated with anti-myc antibodies. The immunoprecipitates were analyzed and quantified for kinase activity using MBP as a substrate (Fig. 4). WT GRF2 and the mutant missing the amino-terminal PH domain (ΔPHn) required stimulation with ionomycin to maximally activate ERK1 (Fig. 4, lanes 3 to 6), whereas the mutants missing the PH domain adjacent to the DH domain (ΔPHc and ΔPHn+c) were maximally activated under basal conditions (Fig. 4, lanes 9 and 10 and 11 and 12, respectively). Therefore, the PH domains of GRF2 are dispensable for ERK1 activation. The REM-Cdc25 protein was a potent activator of ERK1 and was more active (approximately 1.5-fold) than maximally stimulated WT GRF2 (Fig. 4, lanes 13 and 14). Activation of ERK1 by REM-Cdc25 was not stimulated by calcium and was maximal even under conditions of serum deprivation (Fig. 4). Despite its ability to efficiently activate Ras (Fig. 3) (10), the IQ-deleted GRF2 protein (ΔIQ) was defective for basal and calcium-stimulated ERK activation (Fig. 4, lanes 7 and 8).
FIG. 4.
ERK1 in vitro kinase assay in 293T cells. Assays were carried out 3 days after transfection of 293T cells with the indicated GRF2 construct and myc-ERK1. Cells were serum starved for 18 h and then either left untreated (−) or treated with 5 μM ionomycin for 5 min at 37°C (+). Upper panel, Erk activity was determined by precipitating lysates with 9E10 myc antibody, followed by a kinase assay of the immune complex with MBP as the substrate; middle panel, equal expression of myc-ERK1 was confirmed by Western blotting of the anti-myc immunoprecipitates; lower panel, GRF2 protein expression was verified by the Western blotting of the lysates. The data shown are representative of three experiments. Autorad, autoradiography.
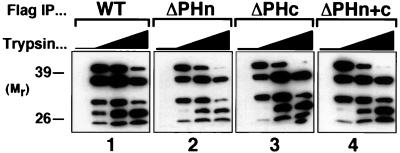
The three PH mutants (ΔPHn, ΔPHc, ΔPHn+c) were each active in the Ras and ERK assays, but the two mutants missing the PHc domain were more active than WT GRF2 for Ras activation (1.5-fold) and were fully active for ERK stimulation in the absence of Ca2+ treatment. One possible explanation for these differences was that the carboxyl Cdc25 domain might be directly affected by these mutations. To test this possibility, WT and PH-deleted GRF2 proteins were subjected to limited proteolysis with trypsin (Fig. 5). Specifically, the GRF2 proteins, in the form of anti-Flag immunoprecipitates, were incubated with a range of concentrations of trypsin, followed by separation of the digestion products by SDS-PAGE and analysis and imaging of the protease-resistant domains by using purified polyclonal antibodies directed against the Cdc25 domain (Fig. 5). The results for the WT and each of the PH-deleted proteins were equivalent. The proteins gave rise to similar series of trypsin-resistant domains derived from the Cdc25 regions of the proteins. This indicates that the PH domain deletions do not affect the intrinsic structure, and hence function, of their cognate Cdc25 domains and favors the interpretation that these deletions affect protein function through their effects on intermolecular interactions.
FIG. 5.
Partial trypsin proteolysis of GRF2 and the three PH deletion mutants. Assays were carried out 2 days after transfection of 293T cells with the indicated construct. GRF2 and the deletion mutants were isolated by anti-Flag immunoprecipitation (IP), and the immune complexes were treated with increasing amounts of trypsin (0, 1, 4, or 10 μg, from left to right) (Worthington Diagnostics) on ice for 15 min. The supernatant containing released fragments was separated by SDS-PAGE and blotted with purified anti-Cdc25 polyclonal antibodies. The data shown are representative of two experiments.
Role of CaM association in GRF2 activity.
We reported previously that the association of CaM with GRF2 is dependent on an intact IQ domain and is calcium dependent (10). Since deletion of this domain impaired GRF2-mediated activation of ERK1 but not Ras, we sought to further examine the correlation of the GRF2-CaM complex with GRF2 activity. First, the effect of CaM on the in vitro GEF activity of GRF2 was examined. Second, the association of CaM with the various GRF2 variants was measured.
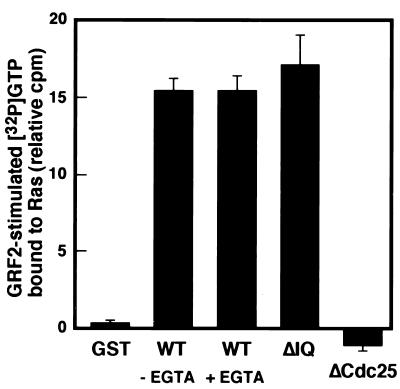
GRF2 isolated by immunoprecipitation was treated without or with EGTA to remove CaM as reported previously, and the presence or absence of CaM was verified by anti-CaM immunoblotting (data not shown) (10). GRF2 was then tested for Ras GEF activity. As shown in Fig. 6, there was no effect of associated CaM on GRF2 catalytic activity measured by the in vitro GEF assay. Therefore, loss of CaM association either by deletion of the IQ motif or by calcium chelation with EGTA does not affect the Ras GEF activity of GRF2.
FIG. 6.
In vitro exchange activity of GRF2 with and without CaM. Assays were carried out 2 days after transfection of 293T cells with the indicated GRF2 construct. WT GRF2 immunoprecipitates were washed in buffer without EGTA or with 1 mM EGTA (to remove bound CaM). The washed immune complexes were used in an in vitro Ras exchange assay as for Fig. 2. Each bar shows the mean exchange activity and standard error of two experiments (duplicate readings in each experiment).
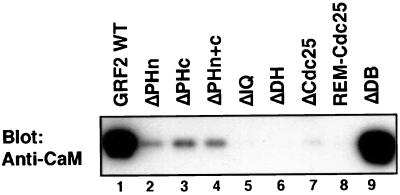
GRF2 immunoprecipitates containing approximately equivalent quantities of the indicated WT and mutant GRF2 proteins were analyzed for CaM by immunoblotting (Fig. 7). Surprisingly, all of the mutants tested were impaired to some extent in their association with CaM. In addition, the extent of CaM association did not change with ionomycin treatment (data not shown). GRF2 with either or both PH domains or the Cdc25 domain deleted still retained detectable CaM association, but at a level at least 10-fold reduced compared to WT GRF2. The REM-Cdc25 immunoprecipitate did not contain detectable CaM. As expected, the IQ-deleted GRF2 mutant did not bind CaM, whereas the mutant lacking the eight-residue candidate destruction motif (ΔDB) retained a WT level of associated CaM. Surprisingly, the DH-deleted protein (ΔDH) which causes calcium-induced ERK activation was devoid of associated CaM (Fig. 7).
FIG. 7.
CaM binding to GRF2 or mutants in 293T cells. Assays were carried out 2 days after transfection of 293T cells with the indicated GRF2 construct. GRF2 and mutants were immunoprecipitated from 293T cell lysates using M2 anti-Flag antibodies. The washed immune complexes were Western blotted for CaM.
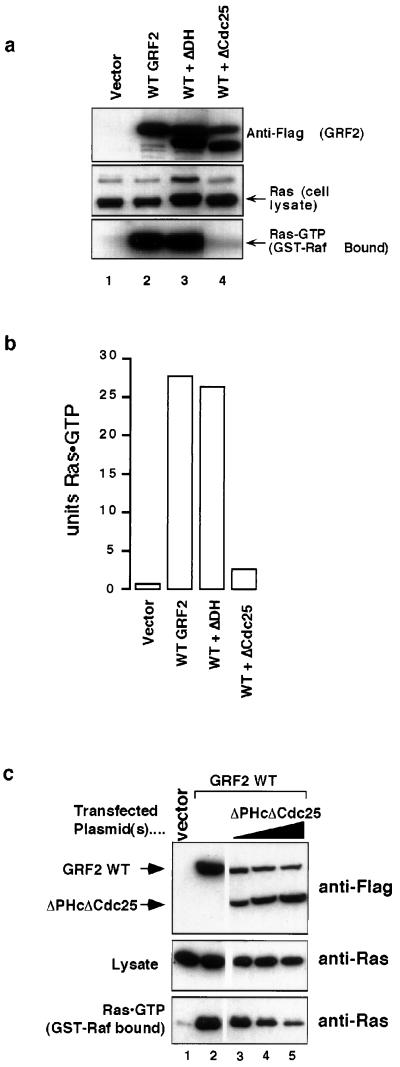
The above results indicate that interaction with CaM is not essential for calcium-induced signaling by GRF2. This suggests that the amino-terminal region of GRF2 interacts with targets other than CaM that are important for GRF2 regulation. In support of this postulate, expression of the ΔCdc25 protein inhibited Ras activation mediated by WT GRF2 (Fig. 8a). Lanes 1 and 2 of Fig. 8a are negative and positive controls, respectively, similar to the corresponding lanes in Fig. 3, and demonstrate the in vivo formation of Ras-GTP in cells expressing ectopic GRF2. Coexpression of ΔDH GRF2 and WT GRF2 caused Ras-GTP formation to the levels expected based on the ability of both these proteins to activate Ras, as shown in Fig. 3. Coexpression of ΔCdc25 and WT GRF2, however, interfered with Ras activation by WT GRF2 since Ras-GTP levels were only approximately 10% that expected based on the amount of WT GRF2 expression (Fig. 8b, right bar). Since the ΔCdc25 protein is unable to interact with Ras (11) and has only minimal interaction with CaM (Fig. 6), it does not inhibit GRF2 by competing with GRF2 for interactions with these proteins. Titrating the expression of a mutant GRF2 protein missing the Cdc25 domain revealed that inhibition of WT GRF2 became more efficient as the level of mutant protein expression exceeded that of the WT protein (Fig. 8c). This suggests that it functions in a codominant manner, commonly referred to as a dominant-negative inhibition. In this last experiment, the variant protein (ΔPHcΔCdc25) was missing both the PHc and Cdc25 domains, indicating that the carboxyl PH domain and any interactions it participates in are dispensable for the inhibitory activity (Fig. 8c).
FIG. 8.
ΔCdc25 protein acts as a codominant inhibitor. Assays were carried out 2 days after transfection of 293T cells with the indicated GRF2 constructs. (a) Ras-GTP levels in 293T cells. Lysate (1.5 mg) was incubated with 50 μg of GST–Ras-GTP binding domain beads for 30 min to pull down activated Ras. Top panel, the presence of GRF2 proteins was verified by Western blotting of lysates with Flag antibody; bottom panel, the samples were Western blotted for Ras, thereby allowing levels of GTP-bound Ras within the cells to be assessed; middle panel, lysates were blotted for Ras to ensure equivalent levels of endogenous Ras between samples. (b) Quantitation of Ras-GTP levels from panel a. The amount of activated Ras was quantitated using a phosphorimager. (c) Ras-GTP levels in 293T cells. Top panel, the presence of GRF2 proteins was verified by Western blotting of lysates with Flag antibody. Lysate (1.5 mg) was incubated with 50 μg of GST-RBD beads for 30 min to pull down activated Ras. Middle panel, lysates were blotted for Ras to ensure equivalent levels of endogenous Ras between samples. Bottom panel, the complexes were Western blotted for Ras, thereby allowing levels of GTP-bound Ras within the cells to be assessed.
DISCUSSION
The primary sequence of GRF2 is indicative of a modular, multidomain protein and even suggests some of the protein-protein interactions in which it may participate (6, 10). Indeed, the physical interaction of GRF2 with CaM and Ras is not surprising given its IQ and Cdc25 domains. Furthermore, since the GRF2-CaM interaction is calcium dependent, it has been tempting to conclude that it accounts for the calcium-mediated activation of the ERK pathway by the Ras-GRF proteins. However, the findings in this report indicate that the regulation of GRF2 and its function in the calcium-mediated stimulation of Ras and ERK are complex and are yet to be fully defined.
GRF2 is certainly a modular protein, and its carboxyl-terminal GEF region is remarkably tolerant of the various domain deletions analyzed in this report. The Ras-binding and Ras GEF activities of GRF2 reside in the carboxyl REM-Cdc25 region (10, 11). The PH domains, DH domain, IQ sequence, and associated CaM are not required for these activities and therefore do not obviously contribute to the stabilization of the functional Ras GEF domain. The REM-Cdc25 fragment was consistently up to twice as active as WT GRF2. While we cannot conclude that this difference in activity is biologically significant, it may indicate that GRF2 is capable of modest activation through relief of inhibition caused by intramolecular determinants that lie outside the REM-Cdc25 region. Similar conclusions have also been made for Sos (7).
The in vivo assay of GRF2-induced Ras-GTP formation gave results consistent with those obtained in vitro, indicating that the PH domains, IQ sequence, DB, and DH domain are dispensable for the constitutive activation of Ras which accompanies GRF2 expression in 293T cells. Activation of Ras by the ΔDH mutant was expected since this protein is not diminished in its ability to stimulate ERK (11). This indicates that an ability to interact with Rac through the DH domain is not necessary for Ras-ERK signaling by GRF2. The PH domains of GRF2 are also not essential for GRF2 to access Ras and ERK. Since the ΔPHc mutants were competent for Ras-ERK signaling and activated Ras to a greater extent than WT GRF2 in vivo, we conclude that the REM-Cdc25 portion of GRF2 may be repressed in a manner dependent on the PHc domain in unstimulated cells. The effects of deleting the DH and PHc domains in GRF2 contrast with those observed as a consequence of point mutations in the corresponding domains of GRF1 (13). In addition, deletion of the N-terminal PH domains in GRF2 and GRF1 has very different effects: GRF1 calcium-stimulated ERK activity is abolished, but there is little effect on GRF2 signaling (4) (Fig. 4). For GRF1, the effect of deleting the N-terminal PH domain results in a substantial redistribution of the protein from the particulate to the cytosolic fraction of cells (4). Therefore, this domain appears to be involved in targeting the protein to the membrane and appears to be required for maximal activation of Ras-ERK signaling. WT GRF2, on the other hand, is found predominantly localized in the cytosol of unstimulated cells, and further studies are aimed at identifying what factor(s) enable it to translocate to the cell periphery in response to calcium stimulation (10). Our present findings indicate that the REM-Cdc25 fragment contains the necessary localization determinants to access and activate the Ras-ERK pathway, at least when ectopically expressed in 293T cells.
Activation of Ras and activation of ERK are separable effects of GRF2. A similar resolution of these two activities has been observed for GRF1 (1). Two models, not mutually exclusive, are apparent. One, as proposed by Anborgh et al. (1) for GRF1, suggests that calcium-stimulated ERK activation by GRF1 is Ras independent. We suggest that for GRF2 it is Ras dependent. Both basal and calcium-stimulated modes of ERK activation by GRF2 are fully inhibited by N17 Ras, which targets the Cdc25 domains of Ras GEFs.
The effect of calcium on GRF2 may promote the assembly or proper orientation of a protein network able to couple with the ERK pathway. We have published other evidence in support of such a scaffolding or anchoring role for GRF2 in coupling Rac activation with stimulation of the SAPK pathway in 293T cells (11). In this model, GRF2 and perhaps other GEFs are not simply upstream activators of their target GTPases but may also determine effector interactions. For GRF2, the IQ sequence is required for the interaction of Ras-GTP with effectors ultimately required for activation of ERK. An alternative scheme, which does not exclude this kind of scaffolding model, is that in the absence of the IQ motif, GRF2 is able to bind and activate Ras in a futile manner such that GRF2 and the Ras-GTP produced are not properly localized, anchored, or oriented at the plasma membrane to physically couple to components of the ERK pathway.
Role of CaM in GRF2 regulation.
CaM binding to GRF2 is clearly not required for Ras GEF activity in vitro (Fig. 6) or in vivo (Fig. 3). Since both the IQ and DH deletion mutants no longer associate with CaM and because we have not generated a minimal CaM-binding fragment of GRF2, we cannot conclude that the IQ motif is the sole binding site for CaM. Indeed, since the PH mutants and the Cdc25-deleted protein were severely impaired in their association with CaM, it is clear that optimal CaM association with GRF2 requires an intact amino-terminal region of GRF2. In contrast, the association of CaM with GRF1 is not perturbed by similar deletions in the amino terminus (4). We did not detect any interaction of CaM with the REM-Cdc25 protein, but this does not eliminate the possibility that in the context of native GRF2 such an interaction may occur, as suggested for Ras-GRF1 (2).
There is no clear correlation between CaM association and Ras or ERK activation by GRF2. The ΔDH mutant remains responsive to calcium signals for ERK activation in the absence of CaM interaction. This indicates that calcium can affect GRF2 in a CaM-independent manner. For example, calcium may induce the phosphorylation of GRF2 and/or activate plasma membrane binding sites for GRF2 required for its activation. Furthermore, since the IQ motif is necessary for ERK activation by WT GRF2, we conclude that a function of the IQ motif, and possibly of the CaM interaction, is to overcome a constraint on GRF2-ERK signaling. The precise function of CaM association with GRF2 therefore remains to be determined.
The ability of Cdc25 domain-deleted GRF2 proteins, as described in this report, to function as negative inhibitors might be explained by their direct binding to and inhibition of GRF2 and/or by competition with GRF2 for interaction with other molecules required for GRF2 regulation. Self-association of GRF2 mediated by the DH domain has been reported (1), and we have also detected intermolecular interactions between GRF2 molecules in reciprocal coimmunoprecipitation experiments employing two differentially epitope-tagged GRF2 proteins (C. de Hoog, unpublished observations). However, given that the ΔDH mutant protein is fully functional for Ras and ERK activation, we cannot conclude that homo-oligomerization is critical for these effects.
The ability of the Ras-GRF proteins to respond to calcium signals distinguishes them from other Ras GEFs, but the physical association of GRF2 with CaM does not explain GRF2's ability to respond to calcium influx, as was previously thought. The ability of the ΔIQ mutant to activate Ras, but not to couple it to the ERK pathway, suggests that in addition to functioning as an upstream activator of Ras, GRF2, and interactions involving the IQ motif in particular, may serve to couple Ras with its effectors.
We conclude that GRF2 is a remarkably modular protein and that the interactions of its noncatalytic domains control its signaling functions. Ras, like the many other small GTPases, has several known effector proteins with which it can engage. We suggest a model wherein these interactions are determined by the GEF responsible for its activation. In this model, GEFs coordinate both the input and output signals of their target GTPase.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council (MRC) of Canada to M.F.M. C.A.K. is a Fellow of the Leukemia Research Society and C. H. Best Foundation, C.L.D.H. and W.-T.F. are MRC Students, M.D.G. is a Research Fellow of the National Cancer Institute of Canada supported with funds provided by the Terry Fox Run, and M.F.M. is an MRC Scientist.
We thank Hui Chen for expert technical assistance and L. McBroom, J. Koehler, and V. Simon for comments and suggestions.
REFERENCES
- 1.Anborgh P H, Qian X, Papageorge A G, Vass W C, DeClue J E, Lowy D R. Ras-specific exchange factor GRF: oligomerization through its dbl homology domain and calcium-dependent activation of Raf. Mol Cell Biol. 1999;19:4611–4622. doi: 10.1128/mcb.19.7.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baouz S, Jacquet E, Bernardi A, Parmeggiani A. The N-terminal moiety of CDC25(Mm), a GDP/GTP exchange factor of Ras proteins, controls the activity of the catalytic domain. Modulation by calmodulin and calpain. J Biol Chem. 1997;272:6671–6676. doi: 10.1074/jbc.272.10.6671. [DOI] [PubMed] [Google Scholar]
- 3.Boriak-Sjodin P A, Margarit S M, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 4.Buchsbaum R, Telliez J B, Goonesekera S, Feig L A. The N-terminal pleckstrin, coiled-coil, and IQ domains of the exchange factor Ras-GRF act cooperatively to facilitate activation by calcium. Mol Cell Biol. 1996;16:4888–4896. doi: 10.1128/mcb.16.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cen H, Papageorge A, Zippel R, Lowy D, Zhang K. Isolation of multiple mouse cDNAs with coding homology to Saccharomyces cerevisiae CDC25: identification of a region related to Bcr, Vav, Dbl and CDC24. EMBO J. 1992;11:4007–4015. doi: 10.1002/j.1460-2075.1992.tb05494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L, Zhang L-J, Greer P, Tung P S, Moran M F. A murine CDC25/ras-GRF-related protein implicated in Ras regulation. Dev Genet. 1993;14:339–346. doi: 10.1002/dvg.1020140503. [DOI] [PubMed] [Google Scholar]
- 7.Corbalan-Garcia S, Margarit S M, Galron D, Yang S S, Bar-Sagi D. Regulation of Sos activity by intramolecular interactions. Mol Cell Biol. 1998;18:880–886. doi: 10.1128/mcb.18.2.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Rooij J, Bos J L. Minimal Ras-binding domain of Raf1 can be used as an activation-specific probe for Ras. Oncogene. 1997;14:623–625. doi: 10.1038/sj.onc.1201005. [DOI] [PubMed] [Google Scholar]
- 9.Ebinu J O, Bottorff D A, Chan E Y, Stang S L, Dunn R J, Stone J C. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 10.Fam N, Fan W, Wang Z, Zhang L, Chen H, Moran M. Cloning and characterization of Ras-GRF2, a novel guanine nucleotide exchange factor for Ras. Mol Cell Biol. 1997;17:1396–1406. doi: 10.1128/mcb.17.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan W T, Koch C A, de Hoog C L, Fam N P, Moran M F. The exchange factor Ras-GRF2 activates Ras-dependent and Rac-dependent mitogen-activated protein kinase pathways. Curr Biol. 1998;8:935–938. doi: 10.1016/s0960-9822(07)00376-4. [DOI] [PubMed] [Google Scholar]
- 12.Farnsworth C L, Freshney N W, Rosen L B, Ghosh A, Greenberg M E, Feig L A. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 13.Freshney N W, Goonesekera S D, Feig L A. Activation of the exchange factor Ras-GRF by calcium requires an intact Dbl homology domain. FEBS Lett. 1997;407:111–115. doi: 10.1016/s0014-5793(97)00309-8. [DOI] [PubMed] [Google Scholar]
- 14.Kiyono M, Satoh T, Kaziro Y. G-protein beta-gamma subunit-dependent Rac-guanine nucleotide exchange activity of Ras-GRF1/CDC25(Mm) Proc Natl Acad Sci USA. 1999;96:4826–4831. doi: 10.1073/pnas.96.9.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattingly R, Macara I. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta-gamma subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- 16.Nimnual A S, Yatsula B A, Bar-Sagi D. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science. 1998;279:560–563. doi: 10.1126/science.279.5350.560. [DOI] [PubMed] [Google Scholar]
- 17.Shou C, Farnsworth C L, Neel B G, Feig L A. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature. 1992;358:351–354. doi: 10.1038/358351a0. [DOI] [PubMed] [Google Scholar]
- 18.Soisson S M, Nimnual A S, Uy M, Bar-Sagi D, Kuriyan J. Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell. 1998;95:259–268. doi: 10.1016/s0092-8674(00)81756-0. [DOI] [PubMed] [Google Scholar]
- 19.Stone J C, Moran M F, Pawson T. Construction and expression of linker insertion and site-directed mutants of the v-fps protein-tyrosine kinase. Methods Enzymol. 1991;200:673–692. doi: 10.1016/0076-6879(91)00180-5. [DOI] [PubMed] [Google Scholar]
- 19a.Taylor, S. J., R. J. Resnick, and D. Shalloway. Non-radioactive determination of Ras-GTP levels using ARIA. Methods Enzymol., in press. [DOI] [PubMed]
- 20.Taylor S J, Shalloway D. Cell cycle-dependent activation of Ras. Curr Biol. 1996;6:1621–1627. doi: 10.1016/s0960-9822(02)70785-9. [DOI] [PubMed] [Google Scholar]