Abstract
Non-small cell lung cancer (NSCLC) is the most common cause of cancer-associated mortality worldwide. Our previous study revealed that circular RNA (circRNA)-FOXO3 is highly expressed in lung cancer and inhibits cell proliferation. However, to the best of our knowledge, at present, no study has focused on the specific mechanism of circRNA-FOXO3 in drug resistance. Therefore, the present study aimed to provide novel perspectives on the role of circRNA-FOXO3 in cisplatin (DDP) resistance in NSCLC. A Cell Counting Kit-8 assay was used to determine the viability of cells overexpressed with circRNA-FOXO3 and under DDP treatment. Glycolysis was analyzed by measuring glucose consumption and lactate production. The interaction of circRNA-FOXO3, microRNA 543 (miR-543) and Foxo3 was confirmed using a dual-luciferase reporter assay. It was revealed that circRNA-FOXO3 improved cell sensitivity to DDP and repressed glycolysis in DDP-sensitive and DDP-resistant NSCLC cells. Bioinformatics analysis, luciferase reporter assays, quantitative PCR and RNA pull-down assays were employed to verify the binding of circRNA-FOXO3 to miR-543. Functionally, inhibition of miR-543 could sensitize NSCLC cells to DDP, and overexpression of miR-543 at least partially abolished the circRNA-FOXO3-induced decrease in chemoresistance. Furthermore, it was revealed that Foxo3 was a direct target of miR-543. Notably, the inhibitory action of miR-543 silencing on DDP resistance and glycolysis was reversed by overexpression of Foxo3 in DDP-sensitive and DDP-resistant NSCLC cells. In conclusion, the present study demonstrated that circRNA-FOXO3 promoted DDP sensitivity in NSCLC cells by regulating the miR-543/Foxo3 axis-mediated glycolysis balance. The present findings may provide novel perspectives for the treatment of patients with NSCLC resistant to DDP.
Keywords: NSCLC, DDP, resistance, circRNA, microRNA-543
Introduction
Non-small cell lung cancer (NSCLC) accounts for 85% of lung cancer cases worldwide, and the 5-year overall survival rate is only ~15% in 2019 (1,2). At present, supplementary therapy is used to treat patients with NSCLC following surgery, and cisplatin (DDP) is widely considered to be a first-line chemotherapeutic agent for such patients (3,4). Although DDP has broad-spectrum anticancer activity, its usefulness is limited by the high incidence of chemoresistance (5). Despite previous efforts, the mechanisms underlying DDP resistance in NSCLC are not well understood. Therefore, there is an urgent need to elucidate these mechanisms.
Circular RNAs (circRNAs) are covalently closed RNA molecules generated by the back-splicing process (6). They are abundant in the human body and have multiple biological functions (7), including acting as ‘sponges’ for microRNAs (miRNA/miRs), thus regulating gene expression at the post-transcriptional level (8). Aberrant expression of circRNAs is reportedly associated with cancer progression, and they have also been noted to act in the regulation of NSCLC pathogenesis (9,10). For example, hsa_circ_0076305 is highly expressed in NSCLC cells and tissues, and knockdown of circ_0076305 increases the sensitivity of NSCLC cells to DDP by modulating the miR-296-5p/STAT3 axis (11). In addition, Wu et al (12) identified that circ-ATP binding cassette subfamily B member 10 depletion suppresses lung cancer progression and sensitizes lung cancer cells to DDP. Our previous study revealed that circRNA-FOXO3 could act as a tumor suppressor in NSCLC (13). However, to the best of our knowledge, the role of circRNA-FOXO3 in DDP resistance remains unclear.
The regulation of tumor metabolism as a therapeutic strategy holds promise for all types of cancer, including NSCLC (14). Aberrant tumor glucose metabolism and the increased rates of glycolysis in tumors are associated with intrinsic/acquired resistance to routinely used anticancer drugs (15). The present study was designed to determine the effects of circRNA-FOXO3 on NSCLC glycolysis and DDP resistance. The results of the present study revealed the molecular mechanisms of circRNA-FOXO3 in NSCLC glycolysis via miR-543/Foxo3, suggesting the underlying pathogenesis for NSCLC and providing a therapeutic strategy for precise treatment.
Materials and methods
Tissue collection
NSCLC tissue samples and adjacent non-cancerous tissues were collected from 45 patients (25 men and 20 women; age range, 35–77 years; median age, 56 years) who received lobectomy at the Affiliated Zhongshan Hospital of Dalian University between January 2017 and September 2018. Paired adjacent normal tissues were also collected (2 cm from the lesion). All patients had pathologically confirmed disease, and the samples were collected before the initiation of chemotherapy or radiation therapy. The following patients were excluded: i) Patients with other pulmonary diseases, or severe hepatic or renal dysfunction; ii) patients with other comorbid malignant tumors; and iii) patients with immune system disorders. Tissue samples were obtained during surgery, snap-frozen with liquid nitrogen and stored at −80°C. The use of clinical samples was approved by the Research Scientific Ethics Committee of the Affiliated Zhongshan Hospital of Dalian University (Dalian, China) and written informed consent was obtained from all participants.
Cell culture
The NSCLC cell lines (NCI-H1650, NCI-H1299, A549 and SK-MES-1, (all previously obtained from the American Type Culture Collection) were maintained at the Affiliated Zhongshan Hospital of Dalian University as previously described (13). The DDP-resistant H1975 lung cancer cell line was established according to a previously described method (16). Specifically, H1975/DDP cells were obtained by exposing H1975 cells to stepwise-increasing concentrations of DDP (cat. no. 479306; Sigma-Aldrich; Merck KGaA). Cells were initially supplemented with DDP at 2 µM and after stable growth for 2 weeks, its concentration was increased to 4 µM for another 2 weeks of incubation. The dose of DDP was steadily increased until the cells reached a stable growth phase with the medium containing 50 µM DDP, thus generating a DDP-resistant cell line, H1975/DDP cells. The cells were maintained in RPMI medium (Sigma-Aldrich; Merck KGaA) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in an incubator at 37°C with 5% CO2.
Cell transduction
The circRNA-FOXO3 cDNA (5′-AAAAGAAGCTAGGACGTATCTAA−3′; 10 nM) was cloned into pcDNA3.1 plasmid (DNM3, V87020; Thermo Fisher Scientific, Inc.) and empty pcDNA3.1 plasmid (Thermo Fisher Scientific, Inc.) containing corresponding control RNAs (described as mock group, 5′-AGGCGATCTCCGTAGGACCTCTG-3′; 10 nM) were assembled by Shanghai GenePharma Co., Ltd. Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used for transfection according to the manufacturer's instructions. The has-miR-543 mimic, anti-miR-543 and negative controls (miR-NC and anti-miR-NC) were designed and constructed by Guangzhou RiboBio Co., Ltd. Cells were seeded into a 96-well plate, and then transfected with the miR-543 mimic, anti-miR-543 and negative controls at a concentration of 80 nmol/ml using Lipofectamine 3000® at 37°C for 24 h. Transfected cells were cultured in 6-well plates containing medium, which was refreshed every 12 h.
Microarray analysis A549 cells were transfected with circRNA-FOXO3 or control plasmid and cells were then incubated at 37°C for 24 h. Total RNA was extracted from cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and then purified using the RNasey Mini kit (Qiagen, Inc.), amplified and labeled using the Quick Amp Labeling kit, One-Color (Agilent Technologies, Inc.), according to the manufacturer's instructions. An equal amount of labeled cRNA from each sample was hybridized using the Agilent Gene Expression Hybridization kit (Agilent Technologies, Inc.), followed by image acquisition on the Agilent DNA Microarray Scanner (G5761A) and data analysis using the Feature Extraction software (version 12.0.3.1; Agilent Technologies, Inc.).
Cell Counting Kit-8 (CCK-8) cell viability assay
Cell viability was assessed using a CCK-8 assay. A549 cells or H1975/DDP cells were transferred into 96-well plates at a density of at 5×103 cells/well and maintained for 24 h. After transfection with indicated RNA oligonucleotides or vectors, cells were exposed to different doses of DDP (0–64 µM; Sigma-Aldrich; Merck KGaA) and kept for 24 h. CCK-8 (10 µl/well; MedChemExpress) was then added to the wells and cells were incubated in the dark at 37°C for 2 h. The optical density value at 450 nm was determined on a microplate reader (Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA from lung tissues or cells was extracted using TRIzol® reagent (Thermo Fisher Scientific, Inc.). The purity and concentration of the various RNA samples were measured using a NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.) by measuring the absorbance at 260 and 280 nm. Synthesis of cDNA was performed using a SuperScript VILO cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.). The conditions for RT were as follows: 95°C for 30 sec and 60°C for 30 min. SYBR-Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for PCR. The following thermocycling conditions were used for qPCR: 95°C for 10 min; followed by 40 cycles of 95°C for 10 sec, 60°C for 15 sec and 72°C for 10 sec. An All-in-One miR qRT-PCR detection kit (GeneCopoeia, Inc.) was used to detect the expression levels of miR-543. RNA quantification was performed using the 2−ΔΔCq method (17). The following primer sets were used: circRNA-FOXO3 forward, 5′-GTGGGGAACTTCACTGGTGCTAAG-3′ and reverse, 5′-GGGTTGATGATCCACCAAGAGCTCTT-3′; miR-543 forward, 5′-AAACATTCGCG-3′ and reverse, 5′-AAGAAGTGCAC-3′; Foxo3 forward, 5′-GCAAGAGCTCTTGGTGGATCATCAA-3′ and reverse, 5′-TGGGGCTGCCAGGCCACTTGGAGAG-3′; U6 forward, 5′-CCTGCGCAAGGATGAC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; and GAPDH forward, 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
Glucose and lactate measurements
After transfection for 48 h, the cultured cells were digested with 0.25% trypsin and centrifuged at 960 × g for 5 min at room temperature to obtain the supernatant. The glucose uptake and lactate production in the supernatant were measured using a glucose uptake colorimetric assay kit (cat. no. MAK083, Sigma-Aldrich; Merck KGaA) and a Lactic Acid assay kit (cat. no. BC2230; Nanjing KeyGen Biotech Co., Ltd.), respectively, according to the manufacturer's protocol.
Bioinformatics analysis
The StarBase 3.0 database (http://starbase.sysu.edu.cn) was used to determine the molecular mechanism by which circRNA-FOXO3 regulates miR-543.
Luciferase reporter assay
Luciferase reporter assay pGL3 Luciferase Reporter Vector (Promega BioVision; Promega Corporation) was used to construct the Foxo3 vector. A549 cells were transfected with Foxo3 vector (5′-TCACGCACCAATTCTAACGC-3′) and miR-543 mimic (miR-543 group: 5′-AAGUUGCCCGCGUGUUUUUCG-3′) or Foxo3 vector and NC miRNA (miR-NC: 5′-UUUGUACUACACAAAAGUACUG-3′). The wild-type (WT) Foxo3 sequence containing the miR-543-binding sequence and the mutant (MUT) Foxo3 sequence were cloned into the pmirGLO dual luciferase miRNA target expression vector in strict accordance with the provided instructions of the luciferase detection kit (Promega Corporation). Cells were harvested at 48 h post-transfection, and a Dual-Luciferase Reporter Assay System (Promega Corporation) was used to measure luciferase activity. Renilla luciferase activity was used for normalization.
RNA-pulldown assay
An RNA pull-down assay was conducted to verify the relationship between circRNA-FOXO3 and miR-543 using the Pierce Magnetic RNA-Protein Pull-Down kit (BersinBio; http://www.share-bio.com/product/943.html). Bio-circRNA-FOXO3 (or Bio-miR-543) and Bio-NC (Guangzhou RiboBio Co., Ltd.) were transcribed by the Biotin RNA Labeling Mix (Roche Diagnostics) and T7 RNA polymerase (Promega Corporation), and A549 cells were transfected with 50 nM of above Biotin-labeled RNA, using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were fixed with 1% formaldehyde and lysed with the RIPA lysis buffer (BersinBio). A total of 1 ml of lysates were subsequently centrifuged at 10,000 × g for 10 min at 4°C. The biotin-coupled RNA complex was pulled down using Streptavidin-Dyna beads M-280 (cat. no. 11206D; Invitrogen; Thermo Fisher Scientific, Inc.) and Proteinase K (Sigma-Aldrich; Merck KGaA) was added and incubated with the supernatants overnight at 4°C to isolate the RNA overnight at 4°C. The bead-probe-RNA mixture was then washed with washing buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 2 M NaCl and 0.1% Tween-20) followed by another centrifugation step (2,500 × g for 5 min at 4°C), and the RNA was extracted and measured by RT-PCR.
Western blotting
The cultured cells were collected and lysed in RIPA lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.) to obtain total protein. After protein quantification using a bicinchoninic acid kit (Beyotime Institute of Biotechnology), protein (30 µg) was subjected to 12% SDS-PAGE. Separated proteins were then transferred onto polyvinylidene fluoride membranes, which were blocked with 5% non-fat milk at room temperature for 1 h, followed by incubation with anti-human Foxo3 antibody (cat. no. sc-34894; 1:1,000; Santa Cruz Biotechnology, Inc.) or β-actin antibody (cat. no. sc-47778; 1:1,000; Santa Cruz Biotechnology, Inc.) at 4°C overnight. Subsequently, the membranes were incubated with horseradish peroxidase-labeled goat anti-rabbit secondary antibody (cat. no. A0208, 1:1,000; Beyotime Institute of Biotechnology) at room temperature for 1 h. Protein bands were visualized using SuperSignal™ West Pico PLUS (cat. no. 34580; Thermo Fisher Scientific, Inc.) and and protein quantification was performed using Quantity One software version 4.62 (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data were obtained from at least three independent experiments and are presented as the mean ± standard deviation. Data were analyzed using GraphPad Prism 8 software (GraphPad Software, Inc.). Differences between pairs of groups were evaluated using unpaired two-tailed Student's t-tests, with the exception that the comparison of circRNA-FOXO3, miR-543 or Foxo3 mRNA levels between lung cancer tissues and adjacent normal tissues was performed using paired Student's t-tests. One-way ANOVA with a subsequent Bonferroni test was performed to determine statistically significant differences among multiple groups. Spearman's correlation analysis was used to assess the correlation between miR-543 and circRNA-FOXO3 or Foxo3. P<0.05 was considered to indicate a statistically significant difference.
Results
Glycolysis inhibition and higher DDP sensitivity in NSCLC cells exposed to circRNA-FOXO3
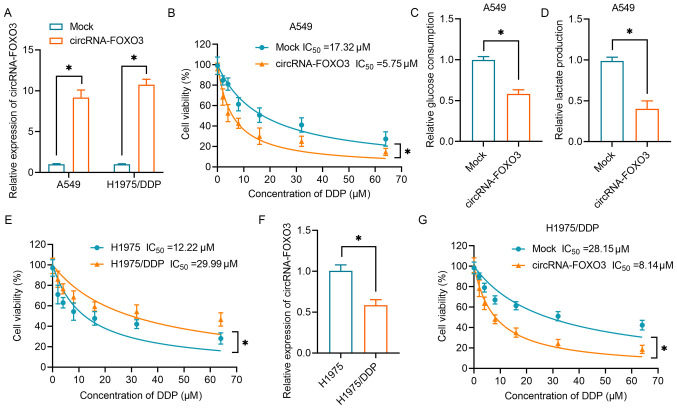
Our previous study revealed that circRNA-FOXO3 acts as a tumor suppressor in lung cancer via the Foxo3 axis (13). In the present study, circRNA-FOXO3 was overexpressed in A549 cells and H1975/DDP cells to investigate its effects on DDP resistance and its underlying mechanism (Fig. 1A). As shown in Fig. 1B, NSCLC cell viability gradually decreased as the concentration of DDP increased. The IC50 (17.32 µM) of mock-transfected A549 cells was higher than that of cells transfected with circRNA-FOXO3 (IC50, 5.75 µM). Subsequently, the present study investigated the effect of circRNA-FOXO3 on the glycolytic metabolism in NSCLC cells. The results demonstrated that overexpression of circRNA-FOXO3 was associated with significantly lower glucose consumption and lactate production (Fig. 1C and D). Furthermore, a DDP-resistant lung cancer cell line (H1975/DDP) was established by continuously exposing H1975 cells to a gradually increasing dose of DDP. The IC50 of H1975/DDP cells was significantly higher than that of H1975 parental cells (29.99 vs. 12.22 µM; Fig. 1E). Notably, circRNA-FOXO3 expression was lower in H1975/DDP cells than in H1975 parental cells (Fig. 1F). As was shown in normal lung cancer cells, H1975/DDP cells overexpressing circRNA-FOXO3 exhibited significantly higher sensitivity to DDP than the H1975/DDP cells in the mock group (Fig. 1G). Overall, these data suggested that circRNA-FOXO3 enhanced the sensitivity of lung cancer cells to DDP via regulation of the glycolytic metabolism.
Figure 1.
circRNA-FOXO3 enhances DDP sensitivity and inhibits glycolysis in NSCLC cells. (A) RT-qPCR was performed to detect the expression levels of circRNA-FOXO3 in A549 cells and H1975/DDP cells transfected with circRNA vectors. (B) A CCK-8 assay was used to determine proliferation following treatment with a series of concentrations of DDP (0–64 µM) in A549 cells transfected with circRNA vectors. (C) A glucose uptake colorimetric assay was performed to determine the glucose consumption of NSCLC cells transfected with circRNA vectors. (D) A lactic acid assay was performed to determine the lactate production of NSCLC cells transfected with circRNA vectors. (E) A CCK-8 assay was used to determine proliferation following treatment with a series of concentrations of DDP (0–64 µM) in H1975 and H1975/DDP cells. (F) RT-qPCR was used to detect circRNA-FOXO3 expression in H1975/DDP cells. (G) Proliferation of H1975/DDP cells transfected with circRNA vectors following treatment with different concentrations of DDP. *P<0.05. CCK-8, Cell Counting Kit-8; circRNA, circular RNA; DDP, cisplatin; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR.
Negative association between miR-543 and both lung cancer and DDP resistance
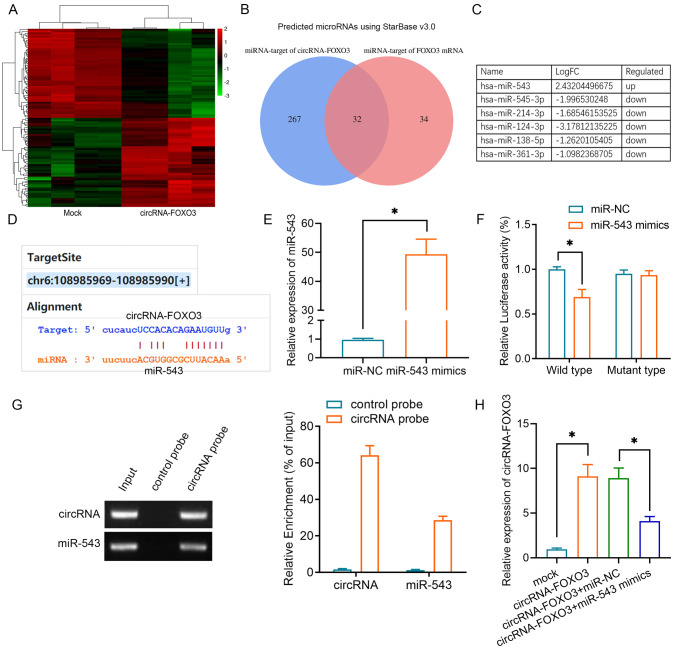
Given that circRNAs can function as competing endogenous RNAs (ceRNAs) in relation to miRNAs, a microRNA array was used to determine the miRNA expression profiles of A549 cells and the data were submitted to the Gene Expression Omnibus database (dataset accession no. GSE182576). In total, 87 upregulated miRNAs and 212 downregulated miRNAs were identified in A549 cells with or without circRNA-FOXO3 overexpression, and a heatmap of the top 50 upregulated and 50 downregulated miRNAs is shown in Fig. 2A. Putative binding sites of circRNA-FOXO3 with Foxo3 were predicted using StarBase 3.0. Among the miRNAs, 32 targeted one or more of these putative binding sites (Fig. 2B). Notably, six miRNAs, of which only one (miR-543) was upregulated, were identified in both the microarray and StarBase results (Fig. 2C). The 3′-untranslated region (UTR) of circRNA-FOXO3 contained a putative miR-543 seeding sequence binding region (Fig. 2D). To examine whether circRNA-FOXO3 was the target of miR-543, miR-543 mimics or miR-NC were transfected into A549 cells. The cells containing miR-543 mimics exhibited significantly higher miR-543 expression (Fig. 2E). Subsequently, dual-luciferase assays were performed in both of these cell lines to validate the regulation of circRNA-FOXO3 by miR-543, and following co-transfection with circRNA-FOXO3 and miR-543, the luciferase activity of the wild-type circRNA-FOXO3 reporter was attenuated (Fig. 2F). Furthermore, an RNA pull-down assay demonstrated that circRNA-FOXO3 interacted directly with miR-543. miR-543 was enriched in the circRNA-FOXO3 probe group compared with the levels in the control group (Fig. 2G). As expected, overexpression of miR-543 partly reversed the higher circRNA-FOXO3 expression (Fig. 2H).
Figure 2.
Identification of the regulatory effect of circRNA-FOXO3 on miR-543 in non-small cell lung cancer cells. (A) Top 50 upregulated and downregulated miRNAs between the mock and circRNA-FOXO3 groups are illustrated in a heatmap. Green represents downregulated, while red represents upregulated. (B) Venn diagram showing the mutual putative target miRNAs of circRNA-FOXO3 and Foxo3 mRNA. (C) Top 6 altered (log10 FC) miRNAs screened using a miRNA microarray and online prediction. (D) Putative miR-543 binding sites with respect to circRNA-FOXO3. (E) Reverse transcription-quantitative PCR was used to detect miR-543 expression in A549 cells transfected with miR-543 mimics or miR-NC. (F) Luciferase activity of A549 cells co-transfected with circRNA-FOXO3 vector and indicated miRNA mimics. (G) RNA pull-down assay indicating circRNA-FOXO3 binding to miR-543. (H) Relative expression levels of circRNA-FOXO3 in A549 cells transfected with circRNA vectors or miR-543 mimics. *P<0.05. circRNA, circular RNA; FC, fold-change; miR/miRNA, microRNA; NC, negative control.
Regulation of chemoresistance and glycolysis by circRNA-FOXO3/miR-543 in lung cancer cells
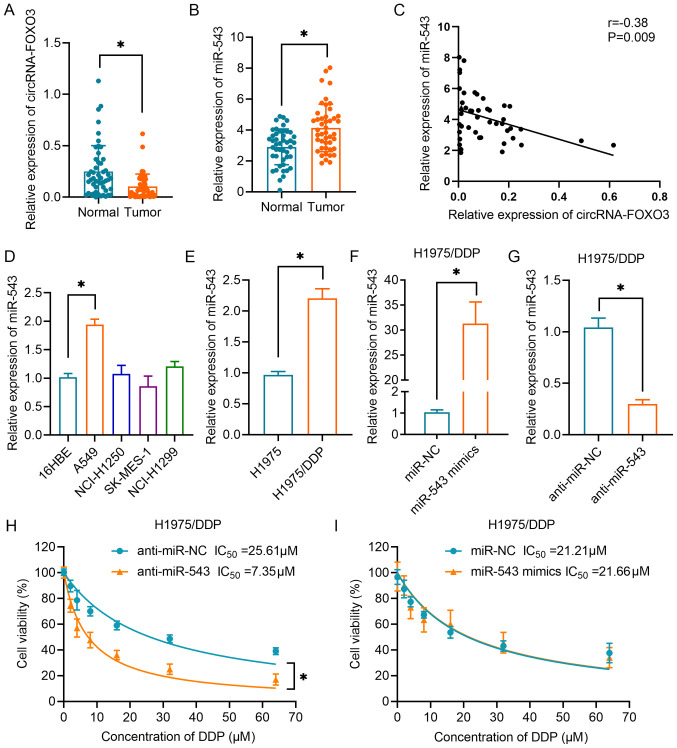
The results in Fig. 3A demonstrated that the expression levels of circRNA-FOXO3 were significantly downregulated in NSCLC tissues compared with in normal lung tissues. Additionally, the expression levels of miR-543 were markedly upregulated in NSCLC tissues compared with in the corresponding normal tissues (Fig. 3B). Correlation analysis confirmed that circRNA-FOXO3 expression was negatively correlated with miR-543 expression in NSCLC tissues (Fig. 3C). In addition, it was demonstrated that miR-543 expression levels were higher in A549 cells than in normal bronchial epithelial cells of the 16-HBE cell line (Fig. 3D). Furthermore, miR-543 expression was found to be upregulated in H1975/DDP cells compared with in H1975 parental cells (Fig. 3E). Subsequently, miR-543 was upregulated by transfection with miR-543 mimics (Fig. 3F) and downregulated by transfection with anti-miR-543 (Fig. 3G) in H1975/DDP cells. Next, the CCK-8 cell viability assay revealed that inhibition of miR-543 significantly sensitized lung cancer cells to DDP (Fig. 3H). However, overexpression of miR-543 had no effect on the resistance of H1975/DDP cells, as indicated by cell viability assay (Fig. 3I).
Figure 3.
Association between circRNA-FOXO3 and miR-543 in NSCLC. (A) RT-qPCR analysis of circRNA-FOXO3 in NSCLC tissues and corresponding normal tissues. (B) Relative expression levels of miR-543 in NSCLC tissues and non-tumor tissues. (C) A negative correlation between circRNA-FOXO3 and miR-543 expression in specimens from patients with NSCLC was identified using Spearman's correlation analysis. (D) Expression levels of miR-543 in 16HBE cells and NSCLC cell lines (NCI-H1250, NCI-H1299, A549 and SK-MES-1) were determined by RT-qPCR. (E) Relative expression levels of miR-543 in H1975 and H1975/DDP cells. (F) Relative expression levels of miR-543 in H1975/DDP cells transfected with miR-543 or miR-NC. (G) Relative expression of miR-543 in H1975/DDP cells transfected with anti-miR-543 mimics or anti-miR-NC. Cell proliferation following treatment with different concentrations of DDP in H1975/DDP cells transfected with (H) anti-miR-543 or anti-miR-NC, or (I) miR-543 mimics or miR-NC. *P<0.05. circRNA, circular RNA; DDP, cisplatin; miR/miRNA, microRNA; NC, negative control; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR.
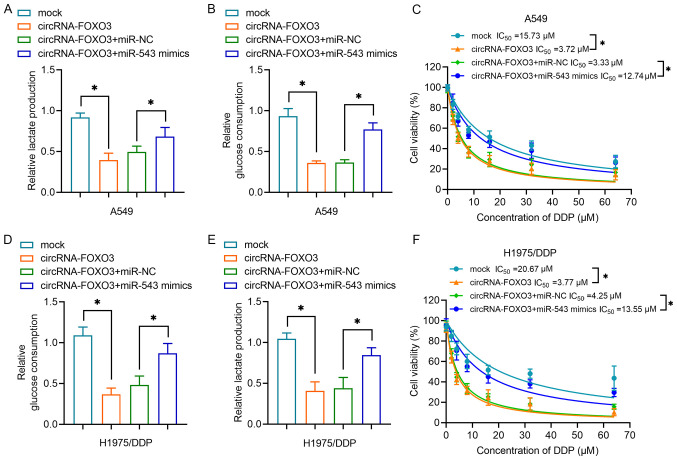
Subsequently, the present study investigated the role of the circRNA-FOXO3/miR-543 axis in chemoresistance in lung cancer cells. Overexpression of circRNA-FOXO3 reduced lactate production but overexpression of miR-543 reversed this effect (Fig. 4A). At the same time, circRNA-FOXO3 reduced glucose consumption, whereas miR-543 effectively reversed this effect in A549 cells (Fig. 4B). Furthermore, the results in Fig. 4C demonstrated that A549 cells co-transfected with circRNA-FOXO3 and miR-543 were more viable than the cells treated with circRNA-FOXO3 and miR-NC (IC50, 12.74 vs. 3.33 µM). In H1975/DDP cells, circRNA-FOXO3 caused lower glucose consumption (Fig. 4D) and lactate production (Fig. 4E), and higher sensitivity to DDP (Fig. 4F); however, miR-543 mimics reversed the effects of circRNA-FOXO3. Overall, these results indicated that circRNA-FOXO3 regulated chemoresistance and glycolysis in lung cancer cells by sponging miR-543.
Figure 4.
circRNA-FOXO3 regulates glycolysis and DDP sensitivity in non-small cell lung cancer cells by sponging miR-543. (A) A lactic acid assay was performed to examine lactate production of cells transfected with circRNA vectors and/or miR-543 mimics. (B) A glucose uptake colorimetric assay was performed to examine glucose consumption of A549 cells transfected with circRNA vectors and/or miR-543 mimics. (C) A CCK-8 assay was performed to measure A549 cell proliferation following treatment with a series of doses of DDP. (D) Relative glucose consumption and (E) lactate production of H1975/DDP cells transfected with circRNA vectors and/or miR-543 mimics. (F) A CCK-8 assay was used to determine H1975/DDP cell proliferation following treatment with a series of doses of DDP following transfection as indicated. *P<0.05. CCK-8, Cell Counting Kit-8; circRNA, circular RNA; DDP, cisplatin; miR/miRNA, microRNA; NC, negative control.
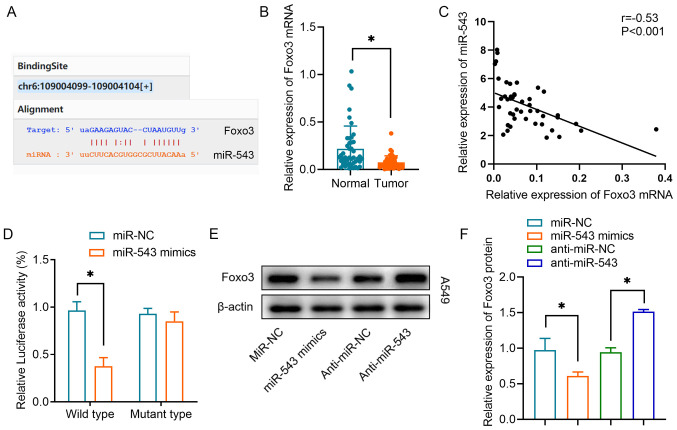
Foxo3 is a direct target of miR-543 in lung cancer
Our previous study demonstrated that circRNA-FOXO3 regulates Foxo3 expression via a ceRNA mechanism (13). The present study investigated whether miR-543 regulates Foxo3 expression. By searching the StarBase database, it was observed that binding sites between miR-543 and Foxo3 were proposed (Fig. 5A). It was revealed that Foxo3 mRNA expression was significantly downregulated in lung cancer tumor tissues compared with in the equivalent normal tissues (Fig. 5B). A negative correlation was also observed between miR-543 expression and Foxo3 mRNA expression in lung cancer tissues (Fig. 5C). Using a luciferase reporter assay, it was revealed that miR-543 could interact directly with the 3′-UTR of Foxo3 mRNA. The miR-543 mimics significantly inhibited luciferase activity in the wild-type reporter for Foxo3, whereas they did not inhibit the luciferase activity of the reporter vector containing the mutant binding sites for Foxo3 in A549 cells (Fig. 5D). Furthermore, western blot analysis demonstrated that overexpression of miR-543 significantly inhibited Foxo3 protein expression, whereas knockdown of miR-543 increased Foxo3 protein expression (Fig. 5E and F). These results indicated that Foxo3 was one of the targets of miR-543 in lung cancer.
Figure 5.
Foxo3 is a target of miR-543 in NSCLC cells. (A) Online bioinformatics prediction tools revealed the binding sequences of miR-543 and the Foxo3 mRNA 3′-untranslated region. (B) Relative expression levels of Foxo3 in NSCLC tissues and non-tumor tissues. (C) Spearman's correlation analysis was used to reveal the negative correlation between Foxo3 and miR-543 expression in NSCLC tissues. (D) Luciferase activity of the Foxo3 vector in A549 cells co-transfected with miR-543 mimics or miR-NC. (E and F) Western blot analysis revealed the relative protein expression levels of Foxo3 in A549 cells transfected with anti-miR-543, anti-miR-NC, miR-543 mimics or miR-NC. *P<0.05. miR/miRNA, microRNA; NC, negative control; NSCLC, non-small cell lung cancer.
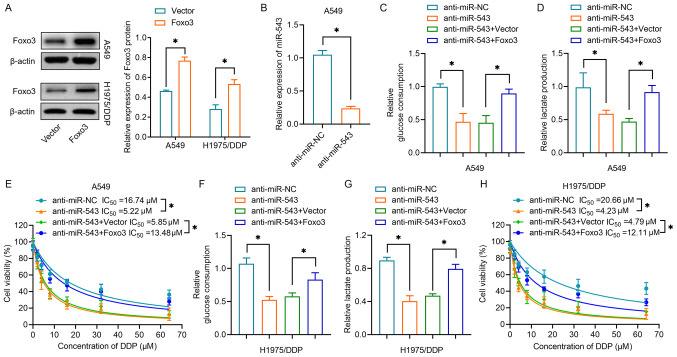
Restoration of Foxo3 reverses the effects of miR-543 silencing on glycolysis and DDP resistance
Subsequently, the present study investigated whether miR-543 affected drug resistance in lung cancer by regulating Foxo3. As shown in Fig. 6A, overexpression of Foxo3 was achieved by transfection with a Foxo3 plasmid, the efficiency of which was validated by western blot analysis in both A549 and H1975/DDP cells. miR-543 was then downregulated in A549 cells by transfection with anti-miR-543 (Fig. 6B). Co-transfection of anti-miR-543 and Foxo3 restored the glucose consumption suppressed by miR-543 in A549 cells (Fig. 6C). The present study also revealed that co-transfection of anti-miR-543 and Foxo3 overexpression plasmid increased lactate production compared with the levels in cells in which miR-543 had been silenced (Fig. 6D). The results of the subsequent CCK-8 assay demonstrated that cells overexpressing Foxo3 were less sensitive to DDP than cells transfected with anti-miR-543 in A549 cells (Fig. 6E). Notably, the present study further validated the effects of Foxo3 in H1975/DDP cells. Knocking down miR-543 reduced glucose consumption (Fig. 6F) and lactate production (Fig. 6G), whereas overexpression of Foxo3 reversed the effects of inhibition of miR-543. H1975/DDP cells displayed increased sensitivity to DDP following inhibition of miR-543; however, this sensitization was overridden by Foxo3 rescue (Fig. 6H). Collectively, the aforementioned results indicated that miR-543 suppressed glycolysis and DDP resistance by directly targeting Foxo3 in lung cancer cells.
Figure 6.
Foxo3 overexpression reverses the effects of miR-543 silencing on glycolysis and cisplatin resistance. (A) Western blot analysis revealed the relative protein expression levels of Foxo3 in A549 and H1975/DDP cells transfected with Foxo3 or negative control vector. (B) Relative expression levels of miR-543 in A549 cells transfected with anti-miR-543 or anti-miR-NC. (C) Glucose consumption was determined in A549 cells transfected with Foxo3 vectors and/or anti-miR-543. (D) Lactate production of A549 cells transfected with Foxo3 vectors and/or anti-miR-543. (E) A CCK-8 assay was performed to determine cell proliferation following DDP treatment of A549 cells transfected with Foxo3 vectors and/or anti-miR-543. (F) Relative glucose consumption and (G) lactate production of H1975/DDP cells transfected with Foxo3 vectors and/or anti-miR-543. (H) A CCK-8 assay was performed to determine cell proliferation following DDP treatment of H1975/DDP cells transfected with Foxo3 vectors and/or anti-miR-543. *P<0.05. CCK-8, Cell Counting Kit-8; DDP, cisplatin; miR/miRNA, microRNA; NC, negative control.
Discussion
Numerous studies have demonstrated that circRNAs are involved in NSCLC tumorigenesis. Yin et al (18) identified that downregulation of double homeobox A pseudogene 8 could suppress cell proliferation and glycolysis by regulating the miR-409-3p/hexokinase 2/lactate dehydrogenase A axis. Wang et al found hsa_circ_0008305 inhibited transforming growth factor-β-induced epithelial-mesenchymal transition and metastasis of NSCLC cells by controlling transcriptional intermediary factor 1 γ (19). However, our understanding of how circRNAs function in drug resistance in NSCLC is limited. Guo et al (20) found that circ_0011292 knockdown could suppress NSCLC progression and enhance paclitaxel sensitivity by regulating the miR-379-5p/tripartite motif containing 65 axis. Knocking down circ Rac GTPase activating protein 1 has also been reported to markedly inhibit tumor growth and enhance the sensitivity of NSCLC cells to gefitinib (21). In terms of DDP resistance, Song et al (22) were the first to report circRNA profiling in DDP-resistant NSCLC cells. Subsequently, Zhao et al (23) demonstrated a novel circRNA in the CDR1 antisense RNA/miR-641/homeobox protein Hox-A9 pathway, which regulates stemness and DDP chemoresistance in NSCLC. Consistent with this, the present results indicated that circRNA-FOXO3 could increase the sensitivity of lung cancer cells to DDP and inhibit glycolysis by regulating the miR-543-Foxo3 axis.
Glycolysis is an important process in tumor cells, providing the main source of energy for rapid tumor expansion (24). Previous studies have demonstrated that circRNAs can regulate glycolysis in cancer cells. For example, circ_0000140 has been found to inhibit the proliferation, metastasis and glycolysis metabolism of oral squamous cell carcinoma by targeting miR-182-5p (25). Notably, circRNA-AKT3 has been found to promote DDP resistance in NSCLC cells by regulating the miR-516b-5p/STAT3 axis-mediated glycolysis balance (26). Our previous study demonstrated that circRNA-FOXO3 inhibits the proliferation, migration and invasiveness of NSCLC cells (13). The present study further assessed the role of circRNA-FOXO3 in drug resistance. It was revealed that circRNA-FOXO3 could increase the sensitivity of NSCLC cells to DDP, as well as inhibit glycolysis. As circRNAs have ceRNA functionality, the miRNAs targeted by circRNA-FOXO3 were screened using a miRNA microarray and extracted from the Starbase datasets. Finally, miR-543 was predicted to target the circRNA-FOXO3 3′-UTR. Luciferase reporter, RNA-pulldown and RT-qPCR assays demonstrated the interaction between circRNA-FOXO3 and miR-543. At present, a role for miR-543 has been revealed in several types of malignant tumor. A review has reported a suppressive effect in various human cancer types (27), although its specific functions in NSCLC remain poorly understood (28). A recent study has suggested that miR-543 overexpression promotes cell proliferation and angiogenesis by targeting metastasis-associated protein 1 in NSCLC cells (29). In terms of drug resistance in cancer, miR-543 has been found to enhance resistance to 5-fluorouracil by regulating the PTEN/PI3K/AKT signaling pathway in colorectal cancer cells (30). In addition, a recent study revealed that prostate cancer-associated transcript 6 facilitates the proliferation and metastasis of cervical cancer cells, and their resistance to DDP, via the miR-543/zinc finger E-box binding homeobox 1 axis (31). To the best of our knowledge, the present study was the first to reveal that miR-543 is involved in DDP chemoresistance in NSCLC. Inhibition of miR-543 significantly increased the sensitivity of NSCLC cells to DDP.
It was revealed that circRNA-FOXO3 acted as a sponge for miR-543 and targeted the 3′-UTR of Foxo3 mRNA, thereby revealing the circRNA-FOXO3/miR-543/Foxo3 axis. Because the target effector of circRNA-FOXO3 is Foxo3, it was hypothesized that circRNA-FOXO3 regulates glycolysis by regulating Foxo3. A previous study has demonstrated that knockdown of Foxo3 is sufficient to induce apoptosis resistance in conjunction with elevated glycolysis (32). Activation of Foxo3 is compensated for by increased glycolysis and reduced apoptosis (33,34). Consistently, in the present study, overexpression of Foxo3 partially reversed the effects of miR-543 inhibition on glycolysis and DDP resistance in NSCLC cells. These data support our conclusion that circRNA-FOXO3 overexpression regulates glycolysis and thus sensitizes NSCLC cells to DDP by activating Foxo3.
The present study had some limitations that should be mentioned. Firstly, the number of samples from patients with NSCLC, which were used to analyze the association between the levels of circRNA-FOXO3 and miR-543, was small. Further studies involving more patients are required to further investigate the clinical significance of circRNA-FOXO3. Secondly, the mechanism study was carried out in two specific cell lines: DDP-sensitive A549 cells and DDP-resistant H1975/DDP cells. Further studies on other cell lines should be conducted. Finally, the methods were limited, in that the potential mechanism underlying the involvement of circRNA-FOXO3 in the chemoresistance of NSCLC cells was demonstrated by an in vitro assay only, and an in vivo xenograft tumor model will be required to fully explore the effect of the circRNA-FOXO3/miR-543/Foxo3 axis on the tumor growth and chemoresistance of NSCLC cells in vivo.
In summary, the present study demonstrated that circRNA-FOXO3 interacted with miR-543/Foxo3 to increase the sensitivity of NSCLC cells to DDP. The circRNA-FOXO3/miR-543/Foxo3 axis may provide a novel therapeutic approach for combating drug resistance during NSCLC treatment.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- NSCLC
non-small cell lung cancer
- circRNA
circular RNA
- DDP
cisplatin
- ceRNA
competing endogenous RNA
Funding Statement
The present study was supported by the National High Technology Research and Development Program (‘863’ Program) of China (grant no. 2015AA020409) and the Project of Liaoning Province Education Department (grant no. L2014358).
Funding
The present study was supported by the National High Technology Research and Development Program (‘863’ Program) of China (grant no. 2015AA020409) and the Project of Liaoning Province Education Department (grant no. L2014358).
Availability of data and materials
All data generated or analyzed during this study are included in this published article. The microarray dataset generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182576; accession no. GSE182576).
Authors' contributions
LB and PG designed the experiments, supervised the research and reviewed the manuscript. YZ, DZ and RX performed the experiments and analyzed data. YZ and LB confirmed the authenticity of all the raw data. YZ, DZ and LB wrote the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Research Scientific Ethics Committee of the Affiliated Zhongshan Hospital of Dalian University (Dalian, China) and written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Li C, Zhang L, Meng G, Wang Q, Lv X, Zhang J, Li J. Circular RNAs: Pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2019;145:2875–2889. doi: 10.1007/s00432-019-03045-4. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Xu C, Gao Y, Wang Y, Ding Z, Zhang Y, Shen W, Zheng Y, Wan Y. A novel long non-coding RNA, MSTRG.51053.2 regulates cisplatin resistance by sponging the miR-432-5p in non-small cell lung cancer cells. Front Oncol. 2020;10:215–215. doi: 10.3389/fonc.2020.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun M, He L, Fan Z, Tang R, Du J. Effective treatment of drug-resistant lung cancer via a nanogel capable of reactivating cisplatin and enhancing early apoptosis. Biomaterials. 2020;257:120252. doi: 10.1016/j.biomaterials.2020.120252. [DOI] [PubMed] [Google Scholar]
- 5.Schmidtova S, Kalavska K, Kucerova L. Molecular mechanisms of cisplatin chemoresistance and its circumventing in testicular germ cell tumors. Curr Oncol Rep. 2018;20:88. doi: 10.1007/s11912-018-0730-x. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, Xiong F, Xiang B, Zhou M, Liao Q, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 8.Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, Dai X, Zhou H, Zhu J, Zhang H, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:101–101. doi: 10.1186/s12943-020-1131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Tan S, Li J, Liu WR, Peng Y, Li W. CircRNAs in lung cancer - Biogenesis, function and clinical implication. Cancer Lett. 2020;492:106–115. doi: 10.1016/j.canlet.2020.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Zhao L, Wang Y. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by sponging miR-590-5p to regulate THBS2. Cancer Cell Int. 2020;20:387. doi: 10.1186/s12935-020-01444-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Y, Xu T, Zhong S, Wang B, Zhang H, Wang X, Wang P, Li G, Yang S. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239:116984. doi: 10.1016/j.lfs.2019.116984. [DOI] [PubMed] [Google Scholar]
- 12.Wu Z, Gong Q, Yu Y, Zhu J, Li W. Knockdown of circ-ABCB10 promotes sensitivity of lung cancer cells to cisplatin via miR-556-3p/AK4 axis. BMC Pulm Med. 2020;20:10. doi: 10.1186/s12890-019-1035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Zhao H, Zhang L. Identification of the tumor suppressive function of circular RNA FOXO3 in non small cell lung cancer through sponging miR 155. Mol Med Rep. 2018;17:7692–7700. doi: 10.3892/mmr.2018.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smolle E, Leko P, Stacher-Priehse E, Brcic L, El-Heliebi A, Hofmann L, Quehenberger F, Hrzenjak A, Popper HH, Olschewski H, et al. Distribution and prognostic significance of gluconeogenesis and glycolysis in lung cancer. Mol Oncol. 2020;14:2853–2867. doi: 10.1002/1878-0261.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haque MM, Desai KV. Pathways to endocrine therapy resistance in breast cancer. Front Endocrinol (Lausanne) 2019;10:573. doi: 10.3389/fendo.2019.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong Z, Fu Z, Shi J, Jiang X, Wan H. HtrA1 Down-regulation induces cisplatin resistance in colon cancer by increasing XIAP and Activating PI3K/Akt pathway. Ann Clin Lab Sci. 2017;47:264–270. [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Yin D, Hua L, Wang J, Liu Y, Li X. Long non-coding RNA DUXAP8 facilitates cell viability, migration, and glycolysis in non-small-cell lung cancer via regulating HK2 and LDHA by inhibition of miR-409-3p. OncoTargets Ther. 2020;13:7111–7123. doi: 10.2147/OTT.S243542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, Liu Z, Zeng Y, Li C, Zhao J, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-β-induced epithelial-mesenchymal transition and metastasis by controlling TIF1γ in non-small cell lung cancer. Mol Cancer. 2018;17:140. doi: 10.1186/s12943-018-0889-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo C, Wang H, Jiang H, Qiao L, Wang X. Circ_0011292 enhances paclitaxel resistance in non-small cell lung cancer by regulating miR-379-5p/TRIM65 axis. Cancer Biother Radiopharm. 2020 Aug 20; doi: 10.1089/cbr.2019.3546. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 21.Lu M, Xiong H, Xia ZK, Liu B, Wu F, Zhang HX, Hu CH, Liu P. circRACGAP1 promotes non small cell lung cancer proliferation by regulating miR 144 5p/CDKL1 signaling pathway. Cancer Gene Ther. 2021;28:197–211. doi: 10.1038/s41417-020-00209-0. [DOI] [PubMed] [Google Scholar]
- 22.Song L, Cui Z, Guo X. Comprehensive analysis of circular RNA expression profiles in cisplatin-resistant non-small cell lung cancer cell lines. Acta Biochim Biophys Sin (Shanghai) 2020;52:944–953. doi: 10.1093/abbs/gmaa085. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, Zheng R, Chen J, Ning D. circRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes to cisplatin resistance in non-small cell lung cancer (NSCLC) Cancer Cell Int. 2020;20:289. doi: 10.1186/s12935-020-01390-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akins NS, Nielson TC, Le HV. Inhibition of glycolysis and glutaminolysis: An emerging drug discovery approach to combat cancer. Curr Top Med Chem. 2018;18:494–504. doi: 10.2174/1568026618666180523111351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Su Y, Zhang M. Circ_0000140 restrains the proliferation, metastasis and glycolysis metabolism of oral squamous cell carcinoma through upregulating CDC73 via sponging miR-182-5p. Cancer Cell Int. 2020;20:407. doi: 10.1186/s12935-020-01501-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Jiang T, Wu C, Zhang Y. circAKT3 inhibits glycolysis balance in lung cancer cells by regulating miR-516b-5p/STAT3 to inhibit cisplatin sensitivity. Biotechnol Lett. 2020;42:1123–1135. doi: 10.1007/s10529-020-02846-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhou C, Zhao X, Duan S. The role of miR 543 in human cancerous and noncancerous diseases. J Cell Physiol. 2021;236:15–26. doi: 10.1002/jcp.29860. [DOI] [PubMed] [Google Scholar]
- 28.Hu J, Chen Y, Li X, Miao H, Li R, Chen D, Wen Z. THUMPD3-AS1 is correlated with non-small cell lung cancer and regulates self-renewal through miR-543 and ONECUT2. OncoTargets Ther. 2019;12:9849–9860. doi: 10.2147/OTT.S227995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang D, Cai L, Tian X, Li W. miR-543 promotes tumorigenesis and angiogenesis in non-small cell lung cancer via modulating metastasis associated protein 1. Mol Med. 2020;26:44. doi: 10.1186/s10020-020-00175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Zhou J, Dong M. Down regulation of miR 543 expression increases the sensitivity of colorectal cancer cells to 5 Fluorouracil through the PTEN/PI3K/AKT pathway. Biosci Rep. 2019;39:BSR20190249. doi: 10.1042/BSR20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Z, Gu G, Pan W, Chen X. LncRNA PCAT6 accelerates the progression and chemoresistance of cervical cancer through up-regulating ZEB1 by sponging miR-543. OncoTargets Ther. 2020;13:1159–1170. doi: 10.2147/OTT.S232354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khatri S, Yepiskoposyan H, Gallo CA, Tandon P, Plas DR. FOXO3a regulates glycolysis via transcriptional control of tumor suppressor TSC1. J Biol Chem. 2010;285:15960–15965. doi: 10.1074/jbc.M110.121871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bodur C, Karakas B, Timucin AC, Tezil T, Basaga H. AMP-activated protein kinase couples 3-bromopyruvate-induced energy depletion to apoptosis via activation of FoxO3a and upregulation of proapoptotic Bcl-2 proteins. Mol Carcinog. 2016;55:1584–1597. doi: 10.1002/mc.22411. [DOI] [PubMed] [Google Scholar]
- 34.Aldonza MB, Hong JY, Lee SK. Paclitaxel-resistant cancer cell-derived secretomes elicit ABCB1-associated docetaxel cross-resistance and escape from apoptosis through FOXO3a-driven glycolytic regulation. Exp Mol Med. 2017;49:e286. doi: 10.1038/emm.2016.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. The microarray dataset generated and/or analyzed during the current study are available in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182576; accession no. GSE182576).