Abstract
A continual rise in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection causing coronavirus disease (COVID-19) has become a global threat. The main problem comes when SARS-CoV-2 gets mutated with the rising infection and becomes more lethal for humankind than ever. Mutations in the structural proteins of SARS-CoV-2, i.e., the spike surface glycoprotein (S), envelope (E), membrane (M) and nucleocapsid (N), and replication machinery enzymes, i.e., main protease (Mpro) and RNA-dependent RNA polymerase (RdRp) creating more complexities towards pathogenesis and the available COVID-19 therapeutic strategies. This study analyzes how a minimal variation in these enzymes, especially in S protein at the genomic/proteomic level, affects pathogenesis. The structural variations are discussed in light of the failure of small molecule development in COVID-19 therapeutic strategies. We have performed in-depth sequence- and structure-based analyses of these proteins to get deeper insights into the mechanism of pathogenesis, structure-function relationships, and development of modern therapeutic approaches. Structural and functional consequences of the selected mutations on these proteins and their association with SARS-CoV-2 virulency and human health are discussed in detail in the light of our comparative genomics analysis.
Keywords: severe acute respiratory syndrome coronavirus-2, coronavirus disease 2019, single amino acid substitutions, SARS-CoV-2 mutations, SARS-CoV-2 pathogenesis
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a seventh strain and the third member of the coronavirus family, has rapidly spread all across the globe since 2019 and has been a leading cause of death worldwide (Seo et al., 2020). The urgency and health crisis forced the World Health Organization (WHO) to enforce a state of health emergency and declare it a pandemic (Jebril, 2020). People with existing comorbidity and those belonging to the elderly were more prone to this infection earlier. Still, now many young individuals are losing the battle to Coronavirus disease 2019 (COVID-19) (Ruan, 2020; Brookman et al., 2021). Such changes in the patterns of SARS-CoV-2 infection as compared to the previous strains of coronaviruses and among different variants of SARS-CoV-2 has been attributed to the mutations of the virus in the Spike (S) protein, a part of the structural component which allows it to enter into the host cells (Wan et al., 2020). Currently, the total number of confirmed cases across the world as of now stands at > 219 million, whereas 4.5 million people have died (assessed on 11th September 2021) (Organization, 2020). Many countries have witnessed this expeditious spread of infection in the form of different waves occurring after a certain time interval (Asrani et al., 2021c; Boroujeni et al., 2021; Iftimie et al., 2021). Many scientists have warned against the upcoming peaks of the current waves and the arrival of new waves, which are yet to come in several countries in the future (Lai and Cheong, 2020). The incubation period of this virus is said to be between 10-14 days. Therefore, immediate diagnosis post-viral entry is not possible, putting undue pressure on the healthcare infrastructure and medical facilities. Diagnosis is mostly performed through reverse transcription real-time – polymerase chain reaction (rRT-PCR) approach, but it takes time to provide results (Singh et al., 2020a; Singh et al., 2020b; Vo et al., 2020; Asrani et al., 2021a). Rapid antigen test is also being used in certain parts; however, rRT-PCR is a more accurate procedure to be followed for the diagnosis (Asrani et al., 2021b). The serious complications of this virus have led to the complete lockdown in major parts of the world, leading to physical and psychological effects on their citizens (Ju et al., 2021; Kim et al., 2021; Van Vo et al., 2021).
SARS-CoV-2 mediates its entry into the host via the S protein of the virus, which interacts with the ACE2 receptors on the host cells (Lan et al., 2020). In this positive sense, a single-stranded RNA virus escapes the host’s innate and adaptive immune response, causing overproduction of cytokines leading to the formation of cytokine storm (Song et al., 2020). Patients in serious conditions have shown an alleviated expression of IL-2, IL-7, IL-10, IP10, MIP1A, MCP1, G-CSF and TNFα cytokines (Huang C. et al., 2020). The death is mainly observed to be caused by pneumonia affecting the patient’s respiratory system (Xu et al., 2020). Along with acute respiratory distress syndrome, COVID-19 causes the manifestation of acute heart injuries, heart failures, inflammation leading to sepsis and multi-organ dysfunction in individuals in chronic cases (Wang D. et al., 2020). The virus was initially thought to spread through droplets of infected individuals via sneezing or coughing; however, recent reports claim their airborne transmission (Zou et al., 2020; Tang et al., 2021).
The virus possesses four structural proteins- spike (S) protein that helps in attachment of the virus to the host cells ACE-2 receptors (Kandeel et al., 2018); membrane (M) protein typically involved in the formation of viral membrane for enclosing the mature virus particles (Neuman et al., 2011); nucleocapsid (N) protein involved in the formation of a viral protein coat, i.e., N which surrounds the genetic material of the virus (Risco et al., 1996); and envelope (E) protein which is involved in the formation of the envelope that assembles the virion particles (Ruch and Machamer, 2012). The following gene arrangement has been observed in SARS-CoV-2 structural analysis: 5' untranslated region (UTR) [non-structural genes (ORF 1a/ORF1b replicase gene), structural genes (S, M, E, and N) and accessory genes (ORF 3, ORF 6, ORF 7a, ORF 7b, ORF 8, ORF 9b)] 3' UTR (Song et al., 2019; Asrani et al., 2020). Replicase genes account for the synthesis of non-structural proteins (NSPs). Sixteen NSPs assist in the replication and packaging of the virus (Naqvi et al., 2020). Accessory proteins usually differ among the different strains of Coronaviruses (Li et al., 2020). SARS-CoV-2 shares more than 80% genomic similarity to the previous SARS-CoV strain that caused an outbreak in 2003 (Asrani et al., 2020; Malik, 2020). Thus, it is known to exhibit a similar replication process as observed in the previous cases.
Now, different mutant strains of this virus have been identified from different parts of the world, such as B.1.1.7 variant of SARS-CoV-2 was originally acknowledged in United Kingdom (UK), B.1.351 variant from South Africa, B.1.1.28 variant from Brazil, B.1.36 variant, N440K and E484Q mutations from India however; all these variants have now been identified and cultured following their spread to different parts of the world (Tang J. W. et al., 2020; Islam et al., 2021; Planas et al., 2021). Apart from these single-site mutations, few variants have been reported to have double and triple mutations. B.1.617, a double mutant variant that originated from a combination of previously identified Coronavirus variants L452R and E484Q, has been found to cause major deaths in certain parts of India (Cherian et al., 2021). A triple mutant (B.1.618) strain was recently found to cause major outbreaks and deaths in the Bengal region in India, leading to the worst COVID-19 outbreak (Huh et al., 2021).
Since the mutation rate of SARS-CoV-2 is very high, it is important to identify the major sites in its genome that show potential in mutating further and posing a risk to humankind (Chen J. et al., 2020). It is also necessary to identify the mutation types that have occurred predominately to understand the selection pressure on this novel coronavirus strain (Presti et al., 2020). In this article, we have performed mutational analysis on different proteins specific to SARS-CoV-2. We have explored the structural and functional consequences of the selected mutations on the protein structures and their interaction with respective binding partners. The association between SARS-CoV-2 virulence properties and its effect on human health has also been discussed subsequently so that different mutations that may happen in the future and their implications on humanity could be assessed.
The presence of the SARS-CoV-2 S protein assists in attaching the virus to the host cell membrane (Letko et al., 2020; Walls et al., 2020). It belongs to transmembrane (TM) glycoprotein class I and is trimeric in structure (Hoffmann et al., 2020). The activation of S protein occurs by TM protease serine 2, which is present on the host cell membrane. Post-viral entry, the release of viral RNA, translation of polyproteins and assembly of replicase-transcriptase complex for replication and transcription of viral genome occurs. This results in the synthesis of structural proteins assembled, packed and released from the host cells (Fehr and Perlman, 2015). S protein plays an important role in recognizing the ACE2 receptor, attaching virion to the host cell, and their subsequent coronavirus entry induced pathogenesis (Lan et al., 2020; Wang Q. et al., 2020). The S proteins are common to many coronaviruses and other members of the influenza family, including HIV, Ebola virus, influenza virus, paramyxovirus etc. (Hoffmann et al., 2020; Huang Y. et al., 2020).
The size and shape of the virus are maintained by the action of the most abundant structural protein in a virus referred to as M proteins (Mahtarin et al., 2020). These are membrane glycoproteins that are conserved among the β-coronaviruses (Bianchi et al., 2020). They have embedded inside the lipid bilayer and consist of an amino-terminal (NH2) domain at the extracellular region and a cytoplasmic domain (COOH) within the viral cell (Mousavizadeh and Ghasemi, 2020). M proteins have 222 amino acid residues in length, and they exhibit a conserved sequence suggesting a common structure of these proteins among different variants (Tang T. et al., 2020). Although higher conservation in the M protein sequence was observed among BAT-CoV, SARS-CoV and SARS-CoV-2 through multiple sequence alignment (MSA) studies, despite this, great variability was observed within the sequence of MERS-CoV suggesting their divergence from the traits shown by other members of coronaviruses (Naqvi et al., 2020).
M proteins usually interact with other (structural, non-structural, and accessory) proteins of the virus to mediate several functions. One of the main features of M protein is that it helps assemble structural proteins (S, E, and N) required for virus budding (Neuman et al., 2011; Schoeman and Fielding, 2019). These interactions between M, N, and E proteins help form virus-like particles (VLP), their intracellular trafficking, and subsequent release from the host cells (Siu et al., 2008). The stabilization of the viral RNA-N protein complex is maintained by the interaction of M with N structural proteins (Astuti, 2020). Similarly, they also interact with S protein and help in their incorporation into the virion. Their interaction is also observed during viral attachment to the host cells and in the regulation of entry processes (Naskalska et al., 2019). M proteins show self-association behavior, and their protein-protein interactions account for their ability in processing, modification and trafficking of structural proteins for assembling virus particles before release (Li et al., 2021).
Experiments involving the deletion of E protein from a highly pathogenic strain of SARS-CoV showed attenuated properties, which could be a basis of an effective vaccine against the virus (Dediego et al., 2008; Netland et al., 2010; Fett et al., 2013); however, reversions into the virulent form were reported when similar experiments were performed in cell cultures and in-vivo (Jimenez-Guardeño et al., 2015). Soon after, stable vaccine candidates in mice were identified by introducing deletion mutations in the C-terminal region without interrupting the PDZ binding motif (PDM) (Jimenez-Guardeño et al., 2015). Therefore, E protein serves as an excellent candidate for vaccine development in comparison to the other structural proteins (Mandala et al., 2020).
Among all structural proteins, N protein is a potent immunogen whose expression increases during infection (Shang et al., 2005; Liu et al., 2006). Most of the serological assays for the coronavirus diagnosis rely on identifying N proteins during the diagnostic procedures (Ahmed et al., 2020). More N protein-specific antibodies were detected in the serum of patients infected with SARS-CoV (Tan et al., 2004). These antibodies were more persistent and highly sensitive than other structural proteins within serum (Shi et al., 2003). Post-viral infection, these proteins bind to the viral RNA genome and play a major role in forming a ribonucleoprotein core and assist in their replication, assembly, and subsequent release from the cells to infect the new host (Ji et al., 2020). In complex with genomic RNA of the virus, N proteins provide stability and improve viral transcription and assembly (Mcbride et al., 2014). In addition to this, they also assert their role in mediating the viral life cycle (Chang et al., 2014).
To get deeper insights into the mechanism of pathogenesis, we have performed extensive sequence and structure analysis of structural and enzymatic proteins of SARS-CoV-2. The emergence of new variants and their harmful impact on human health concern healthcare experts and drug/vaccine development. In such context, our findings establish gene to disease relationships and provide the molecular basis of pathogenesis.
Materials and Methods
Mutational and Structural Data
The mutational data for the SARS-CoV-2 proteins, i.e., S, E, N, and Mpro was fetched out from the NCBI Virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/scov2_snp). The structural coordinates of all four proteins were taken from the Protein Data Bank (PDB). The sequence information was taken from the UniProt database.
Mutational Analysis
To study the impact of the reported mutations on the S, E, N, and Mpro proteins structure, we have performed a structure-based analysis using various bioinformatics tools, such as DynaMut2 (Rodrigues et al., 2021), mCSM (Pires et al., 2014), CUPSAT (Ham, 2020), MAESTROweb (Laimer et al., 2016), SDM (Ju et al., 2021). MAESTROweb, mCSM, CUPSAT. SDM provides Gibbs free energy values (ΔΔG); The change in free energy during the unfolding of a kinetically stable protein is described by this ΔΔG value. Sometimes the mutation in proteins differentiates the free energy landscape between the mutant and the native protein. This variance in the free energy landscape is why the mutation affects the stability of a protein. DynaMut2 is based on vibrational entropy (VE); VE describes how a protein residue in an energy landscape is likely to be occupied based on average configurational entropy. A decrease in VE would increase the rigidity of a protein. If a mutation is shown destabilizing by four out of the five tools, we have considered that as a destabilizing mutation. A detailed protocol of structure-based mutational analyses can be found in our previous reports (Amir et al., 2019; Mohammad et al., 2020; Choudhury et al., 2021; Habib et al., 2021; Umair et al., 2021).
Results
Stabilizing and Destabilizing Mutations in SARS-CoV-2 Proteins
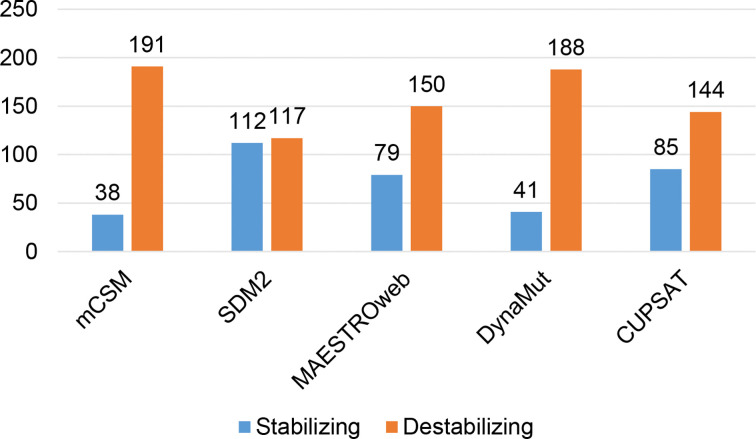
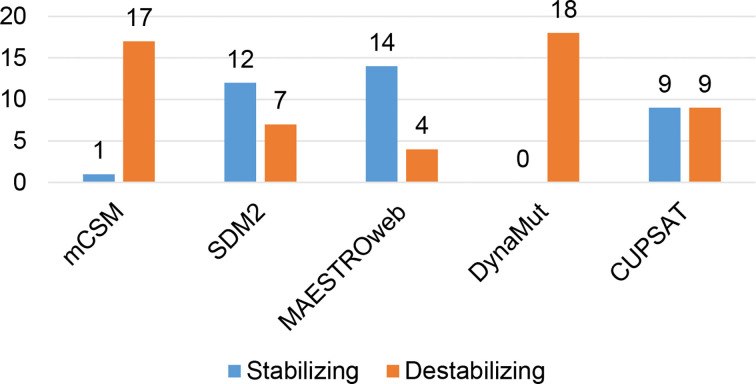
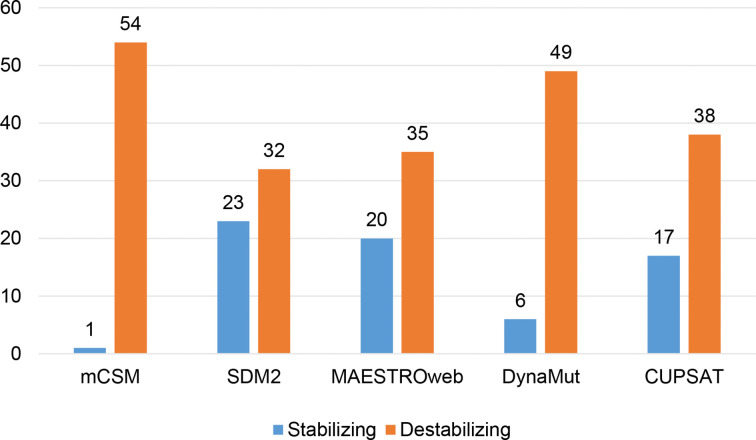
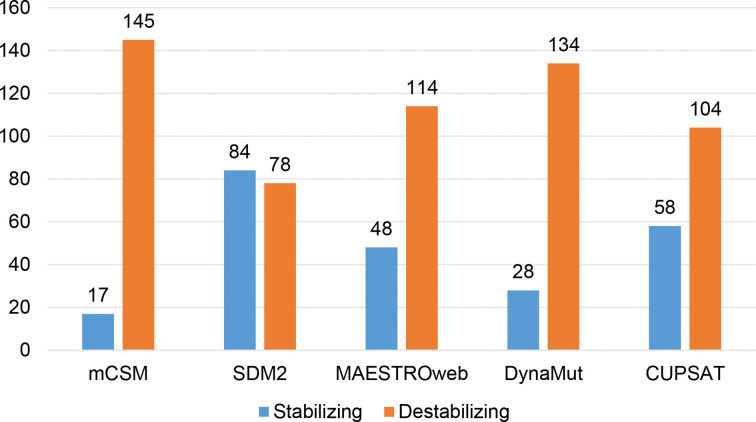
Different sets of reported mutations in the SARS-CoV-2 proteins were extracted from the NCBI Virus database. For S protein, 229 mutations were analyzed, where 123 mutations were found to be destabilizing ( Figure 1 ). In contrast, 6 mutations were destabilizing out of 18 mutations in E protein ( Figure 2 ). At the same time, out of 57 mutations in Mpro, 36 mutations have a destabilizing effect ( Figure 3 ). Here, 25 mutations are present in domain I and 11 mutations in domain II. While two mutations lie in the loop region, and 19 mutations are in the C-terminal domain III. The analysis of 162 N protein mutations showed 85 mutations as destabilizing ( Figure 4 ). The analysis revealed that the SARS-CoV-2 structural proteins, i.e., S, E, and N, and replication machinery enzyme, i.e., Mpro have several mutations found in the concerning variants ( Table 1 ).
Figure 1.
Distribution of stabilizing and destabilizing mutations in SARS-CoV-2 Spike protein predicted by structure-based tools.
Figure 2.
Distribution of stabilizing and destabilizing mutations in SARS-CoV-2 Envelope protein predicted by structure-based tools.
Figure 3.
Distribution of stabilizing and destabilizing mutations in SARS-CoV-2 Main Protease predicted by structure-based tools.
Figure 4.
Distribution of stabilizing and destabilizing mutations in SARS-CoV-2 Nucleocapsid protein predicted by structure-based tools.
Table 1.
Selected mutations predicted to be destabilizing and stabilizing through different structure-based approaches.
| Protein | Destabilizing Mutations | Stabilizing Mutations |
|---|---|---|
| Spike Protein | L5F, Q14H, T20I, T22N, P26S, P26L, P26T, T29I, T29P, N30K, S31T, F32L, R34P, R34S, L54F, V62L, V70I, S71F, G75V, T76I, D80Y, T95I, D111N, L141F, G142S, G142D, Y144N, Y145H, W152C, R158T, L176F, M177I, L189F, L212F, D215N, D215E, L216I, L216F, P217A, P217H, G219C, S221L, W258L, G261D, A262S, A262T, F318Y, F318I, V320F, V320I, V367F, V382L, I402V, R403K, E406D, K417N, K417E, L452R, L455F, R466G, R466I, G485R, Q498H, P499T, N501Y, N501T, L518Q, A520S, A520E, P521T, A522V, A522S, A522G, E583D, D614G, V622F, A626V, T632N, S640F, E654K, Q675H, S691F, M731I, T732A, A771S, Q779H, I788M, I794F, I794M, S810P, L822F, I834M, A845D, A845S, A846S, E868D, A879S, I882T, I882V, S884A, A899S, A930V, S939T, S940T, A942S, L945F, L1004F, Y1047N, S1055A, V1068F, A1070S, K1073N, A1078T, A1078S, V1104L, P1112L, V1122L, G1124V, N1125S, V1137I, Y1138H, P1140A, L1145F | P9L, S12F, S13I, L18F, R21I, A27V, G35C, G35R, H49Y, S50L, A67V, H69Y, D88H, S98F, D138Y, H146Y, N149Y, M153I, S155I, Q173K, E180V, G181R, V213L, R214L, R214S, D215V, D215Y, Q218L, A222V, H245Y, S247R, D253G, S254F, S255F, T307I, Q321R, Q321H, K417M, N439K, G446V, S477N, S477I, Q493K, S494P, S514C, L517P, L518I, H519Y, H519Q, H519N, K558N, Q613H, N641K, A653V, H655Y, Q677H, Q677P, P681H, P681L, R682W, R682L, A688V, S698L, A701V, S704L, T716I, T719I, G769V, V772I, T778I, Q779R, P793T, D796Y, D796H, S810L, P812S, P812L, S813T, D839E, D839Y, L841I, L841R, A845T, R847T, T859I, L894F, S929N, S929R, G932S, K933Q, D936Y, D936E, S939F, S943T, T1027I, A1078V, D1084E, D1084Y, S1097L, H1101Y, T1117I, V1133I, D1139V, L1141M, Q1142E, E1144Q |
| Envelope Protein | L37H, V49L, L51F, F56V, R69K, V75L | T9I, T30I, S55T, S55F, S68F, R69I, R69G, P71S, P71L, D72G, L73F, L74M |
| MPro | F8L, G15S, L30F, D34G, V35A, I43V, T45I, L50F, K61R, G71S, L75F, R76S, V86L, K88R, L89F, A94V, P96L, P108S, P108L, P132S, F134L, F134Y, V157L, L167F, P168S, N180K, V202F, V204A, L227F, M235L, M235I, E270A, L282F, L286F, P293S, R298K, | S10A, S10Y, L32F, V35I, S46A, L67F, K90R, A94S, T135I, N180T, A191V, T196M, A234V, S254F, A266V, S267A, R279C, A285T, L286I |
| Nucleocapsid | N11S, P13L, R14C, P20S, T24I, G25D, S26N, E31G, R32H, G34W, A35V, K65R, P67S, V72I, P80Q, D81Y, I94V, D103Y, A119S, G120A, I131V, L139F, D144Y, T148S, P151L, A152T, A152S, A156S, I157T, P168S, P168A, G178V, S180I, S183Y, S188P, R191L, N192D, S194L, P199S, R203K, R203S, G204R, T205I, T205N, P207L, R209S, R209K, G212S, G212V, N213K, N213D, G215D, D216E, A217T, Q229H, M234V, S235F, G238C, A251V, T265I, E290D, I292T, A308S, S327L, S327W, T334K, T334P, N345K, N345H, D348H, Q349K, A359S, T366A, E367G, P368S, A376T, E378Q, T379A, A381P, P383L, Q384H, T391I, L407F, Q409K, Q409H | Q9H, S21T, D22G, S33N, S37P, N48I, D63E, D63Y, S79G, S79T, P80T, D103E, D128E, D128Y, T135I, A152D, A173V, A182S, R185C, S187L, S188L, S190I, N192S, S193N, S193I, S194T, R195I, S197L, P199L, S202C, S202N, S202T, G204V, S206F, A208G, A208S, R209I, M210I, M210K, A211V, N213S, A220V, L230F, M234I, G243C, T247I, A267E, A267P, V270L, T271I, Q289H, H300Y, A305V, T325I, T329M, T334I, P344S, Q349H, T362I, T362K, P364L, T366I, D371Y, D377Y, T379I, P383S, Q389L, Q390P, Q390H, T393I, A398V, L400M, D401Y, D402Y, K405R, S413R, A414S |
Discussion
The SARS-CoV-2 proteins have several mutations found in the different variants emerging day by day. These mutations have various adverse impacts on the structure functions of the SARS-CoV-2 proteins making COVID-19 complex to administrate. Here we have discussed such mutations and their roles in the SARS-CoV-2 virulence.
Spike Protein Mutations
S protein consists of 1273 amino acids and is approximately 180-200 kDa (Hoffmann et al., 2020). Several polysaccharide molecules are coated onto the surface of S protein to help escape the host’s immune response (Watanabe et al., 2020). Like other typical proteins, S protein also has an extracellular N-terminal domain (NTD), transmembrane domain and a cytoplasmic C-terminal domain (CTD) (Hwang and Yu, 2021). It has a signal peptide ranging from 1 to 13 amino acid (aa) residues followed by two different subunits, namely S1 (14-685 aa residues) and S2 (686-1273 aa residues), each one of which plays an essential role in adherence of SARS-CoV-2 to the host cells for their entry (Xia et al., 2020). In the native state, S protein is inactive. The presence of TMPRSS2 on the target cell membrane and other cellular proteases cleaves the S protein into its S1 and S2 subunits required for the fusion of viral-host membrane after the viral invasion (Bertram et al., 2013; Hoffmann et al., 2020).
S1 subunit further consists of an N-terminal region and a receptor-binding domain (RBD) (Xia et al., 2020). It initiates the viral infection cycle by binding of trimeric S protein via RBD of S1 subunit to ACE2 receptors on the host cell (Wrapp et al., 2020; Yan et al., 2020). This interaction induces a conformational change that directs them to form endosomes to trigger viral fusion with the host cell under the influence of low pH (Shang et al., 2020). Understanding such conformational changes provides a base for the development of drugs that disrupt the entry mechanisms (Walls et al., 2020; Shamsi et al., 2021). The atomic-level studies by cryo-electron microscopy have revealed different open and closed conformations of the RBD. These domains account for the variability of SARS-CoV-2 (Walls et al., 2020; Wrapp et al., 2020). The amino acid composition of the RBD reflects the evolutionary relatedness of SARS-CoV-2 with other members of the Coronavirus family, and it is the least conserved unit. However, the RBD of MERS-CoV was distinct, indicating the divergence from the previous strains (Andersen et al., 2020). The intermediate hosts of SARS-CoV-2 can be identified by analyzing binding affinities and the RBD domain of the S1 subunit with the ACE2 receptors (Chen Y. et al., 2020; Huang Y. et al., 2020). Mutations in the S1 region are associated with changed antigenicity, and thus, it accounts for some strains to be more infectious than others. The affinity of the receptor binding region of S1 subunit with ACE-2 receptors might change because of mutations, but their interaction is preserved among humans, cats and swine (Chan et al., 2020; Wrapp et al., 2020; Yan et al., 2020).
S2, another subunit of S protein, mediates the fusion of the virus to the host cell membrane. It consists of a fusion peptide (FP), heptapeptide repeat sequence 1 (HR1), HR2, TM domain and a cytoplasm domain at the last (Xia et al., 2020). Various large-scale structural rearrangements allow the virus to fuse with the host cell membrane (Watanabe et al., 2020). The main basis of viral-host fusion lies in the cleavage of S1 and S2 subunits, releasing the viral genome into the host cell (Tortorici et al., 2019; Rabaan et al., 2020). Multiple furin cleavage sites have been found in SARS-CoV-2, which are susceptible to attack by furin-like proteases (Millet and Whittaker, 2015; Hasan et al., 2020). These sites were absent from previously known SARS-CoV and, thus, might have increased the infectivity of SARS-CoV-2 (Coutard et al., 2020; Rabaan et al., 2020). Besides, furin-like proteases TMPRSS2 and trypsin have been found to exhibit a profound role in activating the membrane fusion domain (Heurich et al., 2014; Limburg et al., 2019; Ou et al., 2020). The fusion peptide, rich in hydrophobic residues like glycine and alanine, anchors to the host cell membrane and disrupts their lipid bilayer (Millet and Whittaker, 2018). The exposure of FP following the cleavage of S protein into two subunits and the two HR domains of the S2 subunit is important for mediating the viral fusion (Kawase et al., 2019). Certain receptor (FP) and ligand (on host cell) interactions trigger a conformational change which shortens the distance between the two membranes for fusion (Harrison, 2015). As a result, the HR1 domain becomes closer to the host cell, whereas the proximity of the HR2 domain increases with the viral membrane. The folding of HR1 and HR2 together creates a six-helical bundle structure aligned in an antiparallel form to the fusion core. In this way, viral and host cell membrane pulls each other close for bringing out the necessary fusion (Eckert and Kim, 2001). The potential mutations in the SARS-CoV-2 S protein found in different variants are discussed here:
N501Y: The N501Y mutation was initially found in the B.1.1.7 UK variant (Leung et al., 2021), and later it appeared in South African and Brazilian variants, which were B.1.128.1 and B.1.351, respectively (Weisblum et al., 2020). It was also detected in the Theta (P.3 or GR/1092K.V1) variant in Japan and the Philippines in February 2021. This amino acid substitution is located in the RBD of the S protein. It was found that the N501Y substitution increases the affinity of RBD to bind to the ACE2 receptor and thus enhances the transmission rate (Starr et al., 2020; Makowski et al., 2021). Our study also shows that the N501Y mutation destabilizes the SARS-CoV-2 S protein ( Table 1 ).
K417N: This mutation is first reported in the B.1.351 variant (Beta variant or GH501Y.V2) in South Africa (Tegally et al., 2021). Later it was also detected in the P.1 variant (GR/501Y.V3, Gamma variant) in Brazil in December 2020 (Faria NR, 2021). The K417N substitution is also present in the RBD. It was found that this substitution significantly increases the binding affinity of SARS-CoV-2 RBD to the ACE2 receptor. Also, the mutation causes a huge decrease in the binding affinity STE90-C11 antibody to RBD (Fratev, 2020).
E484K: The E484K mutation was first reported in the South African variant B.1.351 variant (GH501Y.V2, Beta variant) (Tegally et al., 2021) and later in the P.1 variant (Gamma variant or GR/501Y.V3) (Faria NR, 2021). This mutation was also found in the Zeta (P.2), Eta (B.1.525) and Iota (B.1.526) variants (Huh et al., 2021). This mutation significantly alters the electrostatic complementarity of antibody binding to the RBD (Andreano et al., 2020). The impact of E484K mutation in the RBD was also seen in the binding of serum polyclonal neutralizing antibodies to SARS-CoV-2 (Jangra et al., 2021).
L18F: This mutation occurred in the South Africa B.1.351 variant (Beta variant or GH501Y.V2) [5]. Later it was also found in the P.1 variant (Gamma variant or GR/501Y.V3) and Zeta (P.2) variants in Brazil. This substitution has been found to affect the binding of neutralizing antibodies negatively [16]. Studies have shown that mutants with L18F substitution are highly sequenced variants, escaping S2L28-mediated neutralization (Mccallum et al., 2021).
A570D: The A570D substitution was first identified in the B.1.1.7 UK variant (Alpha or GRY GR/501Y.V1) in December 2020. Along with D614G, this mutation induces significant conformational destabilization (Socher et al., 2021).
P681H: This mutation was detected first in the UK B.1.1.7 UK variant. Later it was identified in P.3 (Theta or GR/1092K.V1) variant found in Japan and the Philippines in February 2021. The P681H mutation is present at a proteolytic cleavage site for furin or furin-like proteases at the junction of the fusion domain and S protein RBD (Jaimes et al., 2020). It has been shown that P681H increases cleavability at the S1/S2 junction but does not surely indicate increased membrane fusion and infectivity (Lubinski et al., 2021).
S982A: The S982A substitution was first identified in the B.1.1.7 UK variant (Alpha or GRY GR/501Y.V1). The S982A mutation of B.1.1.7 lineage is present on the S2 subunit of the S protein. This substitution in UK variant B.1.1.7 does not have intermolecular hydrogen bonding potential between S protein subunits (Ostrov, 2021).
D1118H: This mutation was detected first in the B.1.1.7 UK variant. The D1118H substitution is also present in the S2 subunit of the S protein (Chrysostomou et al., 2021). It has been suggested that this mutation can alter the stability and dynamics of trimer assembly (Zhao et al., 2021).
A701V: The A701V substitution was first detected in the B.1.351 variant (Beta variant or GH501Y.V2) in South Africa and later found in the Iota (B.1.526) (Annavajhala et al., 2021) variant in New York. This non-synonymous substitution is located in the cleavage site of the neighboring promoter of the S2 subunit (West et al., 2021).
D614G: The D614G is the most widespread mutation of the S protein; it has been found to increase the infectivity of the SARS-CoV-2 virus (Korber et al., 2020). This non-synonymous substitution was identified in the South Africa B.1.351 variant (Beta variant or GH501Y.V2), B.1.617.2 variant (Delta variant), Epsilon (B.1.427 and B.1.429) variants, Zeta (P.2), Kappa (B.1.617.1), Eta (B.1.525) and Iota (B.1.526) variants. The D614G mutation increases infectivity and enhances the replication of the SARS-CoV-2 virus in the upper respiratory tract (Plante et al., 2021). This substitution is away from the RBD. It decreases the binding affinity of S protein to the ACE2 receptor and introduces a conformational change in the S1 subunit of the S protein (Yurkovetskiy et al., 2020). D614G is present at the SD2 domain, enhancing the furin cleavage at the S1/S2 domain junction (Gobeil et al., 2021).
R158G: The R158G substitution was first found in the B.1.617.2 variant (Delta variant) in December 2020 in India. The mutation is absent from the RBD of the S protein (Baral et al., 2021).
L452R: This mutation was reported in the B.1.617.2 variant (Delta variant) and Kappa (B.1.617.1) variant in India as well as in the Epsilon (B.1.427 and B.1.429) variants in the US (Huh et al., 2021). This mutation is present in the RBD of SARS-COV-2 S protein and was found to reduce neutralizing activities in many monoclonal antibodies (Baral et al., 2021).
T478K: This mutation was first identified in the B.1.617.2 variant (Delta variant) in India. The mutation is also present in the RBD of the S protein (Baral et al., 2021).
P681R: This mutation was first detected in the B.1.617.2 variant (Delta variant) in December 2020 in India and later in the Kappa (B.1.617.1) variant. The mutation is absent from the RBD of the S protein.
Envelope Protein Mutations
The E protein with an 8-12 kDa size is one of the smallest structural proteins in SARS-CoV-2 (Schoeman and Fielding, 2019). The sequence of E protein is highly conserved among different members of Coronaviruses. The major function of E protein involves activation of host inflammasome, viral progeny budding and release from the host cells (Nieto-Torres et al., 2015; Schoeman and Fielding, 2019). Like other structural proteins, it also possesses three distinct domains- the extracellular domain at N-terminus consisting of hydrophilic (7-12 amino acid) residues followed by a transmembrane domain of 25 hydrophobic amino acid residues and a cytosolic or C-terminus domain-containing hydrophilic amino acid sequences (Corse and Machamer, 2000; Torres et al., 2007; Surya et al., 2018). The characteristic feature of E protein is viroporin, a pentameric ion channel with no or low selectivity of ions formed from the oligomerization of the transmembrane domain (Verdiá-Báguena et al., 2012; Nieto-Torres et al., 2014). Viroporins are small M proteins that get incorporated into the host membrane and assists in the maturation and release of the viral particles (Nieva et al., 2012). Therefore, these pentameric structures with an ion-conducting pore mediate the host-pathogen interactions (Torres et al., 2006; Parthasarathy et al., 2008; Pervushin et al., 2009). Besides regulating the assembly and release of virions, they have been found to possess a significant role in the pathogenesis of the virus (Nieto-Torres et al., 2014), where pathogenesis is directly proportional to the ion channeling (IC) activity (Chellasamy et al., 2020). For example, no effect on the replication of the virus was observed after E gene knockdown, but reduced edema accumulation was witnessed. This might be because the loss of ion channeling activity of E protein resulted in the correct localization of Na+/K+ ATPase, which is probably involved in decreased edema accumulation and an increase in edema resolution. Often, the accumulation of edema is one of the reasons contributing to ARDS. Also, studies on animal models infected with IC activity lacking viruses exhibited reduced levels of IL-1β, which further reduced the production of TNF and IL-6 in the lung airways. Therefore, it was estimated that IC activity of E protein is essential in the development and progression of cytokine storm leading to permanent lung damage and ARDS in the later stages (Nieto-Torres et al., 2014).
In addition to this, the selective cation ion channel formed from viroporin is localized towards the ERGIC membrane (Wilson et al., 2004; Verdiá-Báguena et al., 2012). The C-terminal region within the E protein contains a β motif with a conserved proline amino acid residue that is important for localization into the ER-Golgi complex (Li et al., 2014; Chellasamy et al., 2020). A small part of E protein inside the host cell membrane is transferred to the virion when a virus replicates. In contrast, the larger section of this protein remains at the location of intracellular trafficking within the mammalian cells, i.e., ER-Golgi network and the ERGIC (Nieto-Torres et al., 2011; Venkatagopalan et al., 2015). Such localization of E proteins assists in viral structural assembling and budding from the host cell (Nieto-Torres et al., 2011).
In all the variants of SARS-COV-2 except the Beta (B.1.351) variant, there are no reported mutations in the E protein. The Beta variant was first discovered in the Eastern Cape province of South Africa in October 2020 (Tegally et al., 2021). India has reported more than 200 cases of the Beta variant from the time of its discovery. The potential mutation in the SARS-CoV-2 E protein found in the Beta (B.1.351) variant are discussed here:
P71L: It is the amino acid substitution found in the E protein of the Beta variant. Statistically, the mutation P71L was associated with disease severity and death rate. The mutation was present in deceased patients’ datasets and virus isolates of patients from high case-fatality-ratio countries (Rizwan et al., 2021).
Mpro Mutations
The SARS-CoV-2 Mpro has several mutations reportedly found in different variants are discussed here:
T45I: This mutation in the domain I region of SARS-CoV-2 Mpro is reported in variants B.1.1.7, B.1.351, P.1, B1.617, B.1.429+ B.1.427 (Philot et al., 2021). It presented a polar to non-polar substitution due to which there is a reduction in its hydrogen bonding potential. SDM predicted the free energy change as stabilizing w.r.t. WT SARS-CoV-2 Mpro. Also, Dynamut2 indicates no substantial change in the flexibility of the protein compared to the WT SARS-CoV-2 Mpro.
K90R: This mutant lies in the domain-I region; it has shown relevant modifications in its potential energy concerning the WT SARS-CoV-2 Mpro (Philot et al., 2021). SDM has indicated this substitution as destabilizing, and the Dynamut2 score suggests the substitution will increase the flexibility of the protein. K90R mutant is reported in the variants B.1.1.7, B.1.351, P.1, B1.617, which are mainly are our variant of concern. Furthermore, a lower energy configuration and a more extensive dimeric interface have resulted from the mutant K90R (Philot et al., 2021).
P99L: It is reported in the B.1.1.7 variant only. The Dynamut2 score suggests the mutant will induce stability to the structure, whereas SDM shows destabilizing; a higher probability of the formation of dimeric interfaces was reported than WT SARS-CoV-2 Mpro (Sheik Amamuddy et al., 2020; Philot et al., 2021).
P108S: This mutant is reported only in the B.1.1.7 variant. The substitution showed a significant variation in potential energy (Philot et al., 2021); nevertheless, the SDM and Dynamut2 scores suggest no substantial change in the structure.
T135I: This substitution brought polar to non-polar replacement. It is reported in variant B.1.1.7 and P.1. SDM has predicted this as stabilizing, whereas there may be a potential increase in the protein flexibility as per Dynamut2 w.r.t. WT SARS-CoV-2 Mpro.
N151D: This substitution is reported in variants B.1.1.7, B.1.351, P.1, B1.617. Compared to WT SARS-CoV-2 Mpro, this mutant shows behavior that may induce catalysis and create distinct dimeric interfaces (Philot et al., 2021). In addition, a significant variation in potential energy was also detected (Sheik Amamuddy et al., 2020). SDM predicts destabilizing, whereas, according to Dynamut2, there will be no change in the structure.
A234V: The substitution A234V in the Domain III region is reported in variants B.1.1.7, B.1.351, P.1, and B.1.617. This mutant is associated with the protein’s mobility. However, it might also affect the flexibility since it lies in the highly flexible region of the protein (Sheik Amamuddy et al., 2020).
A266V: This mutation was found in B.1.1.7, B.1.351, P.1, and B.1.617 varients. The mutation occurred in a highly flexible region involved in protein mobility and might affect protein flexibility (Sheik Amamuddy et al., 2020). The variation might induce rigidity as per the Dynamut2 result and is also destabilizing according to SDM.
R279C: This mutant is also related to protein’s mobility and lies in the Domain III region. It is reported in variants B.1.1.7, B.1.351, P.1, and B.1.617. The substitution R279C increases potential energy more than WT SARS-CoV-2 Mpro (Philot et al., 2021). As per SDM, the mutation may destabilize the protein. However, there may be an increase in protein’s flexibility, as per Dynamut2, which might benefit the protein.
Nucleocapsid Mutations
N proteins play a major role in packaging the viral genome into a ribonucleoprotein complex known as a capsid. This packaging ensures the proper replication and self-assembly of the virus (Chang et al., 2014). N protein contains N-terminal (NTD) and C-terminal domains (CTD) connected by a linkage region (LKR) having serine/arginine-rich (SR-rich) domain within their structural sequence (Huang et al., 2004; Hurst et al., 2009). The presence of these positive amino acid residues favors the binding of the viral genome to both the domains, i.e., NTD and CTD (Chen et al., 2007; Saikatendu et al., 2007). LKR is mostly associated with the oligomerization process (He et al., 2004a; Chang et al., 2013). This binding occurs through a long stretch of RNA binding domain in N protein consisting of approximately 140 amino acid residues (Fehr and Perlman, 2015). Structural analysis of the N protein of SARS-CoV-2 has revealed a disordered region in high content that is not in the bound state to the genomic DNA (Zeng et al., 2020). The linker region is also disordered (Chang et al., 2006; Yu et al., 2006), suggesting its ability to bind to several other partners to maintain appropriate N protein conformation (He et al., 2004a; He et al., 2004b; Luo et al., 2005). Likewise, other structural proteins, N protein of SARS-CoV-2 is also conserved among coronavirus family and shares ~ 90% sequence similarity with SARS-CoV N protein sequence (Naqvi et al., 2020). The potential mutations in the SARS-CoV-2 N protein found in different variants are discussed here:
R203K: Mutation R203K was observed in the B.1.1.7 variant or the alpha variant and the P.1 variant or the gamma variant, initially found in the United Kingdom and Japan/Brazil. It is found in the Ser/Arg-rick linker region (LKR), one of the protein’s most phosphorylated regions. It is one of the most found mutants in N protein. Variant R203L and G204R were aroused by homologous recombination in the SARS-CoV-2 genome (Leary et al., 2021).
G204R: The mutation G204R was also observed in the B.1.1.7 variant (Alpha variant) and the P.1 variant (Gamma variant), initially found in the United Kingdom and Japan/Brazil. It is also located in the LKR region. G204R, along with R203K, is one of the most found mutations in the N protein. Their presence was associated with the increase in N protein and sub-genomic RNA expression from the other ORFs (Leary et al., 2021). N protein shows the high protein intrinsic disorder, and 203/204 residue sites showed increasing entropy and their neighborhoods aligned with areas of the high disorder (Tomaszewski et al., 2020).
T205I: The mutation T205I was first observed in the B.1.351 variant or the beta variant of the Coronavirus, which was first observed in South Africa in October 2020. It is also found in the LKR region. T205I mutant was a common mutation at around 43% [35] since it is highly phosphorylated. Hence, the mutation disrupts the activation of N protein and thus interferes with the virus life cycle.
S235F: The mutation S235F was observed in the B.1.1.7 variant, I.e., the alpha variant, first observed in the United Kingdom in November 2020. It is also found in the LKR region. This mutation was seen to alter the corresponding epitopes, which can cause changes in the specificity of certain antibodies and alter the vaccine-induced protection against the disease.
Altogether, studies suggest that mutations affect pathogenesis by changing the phenotype of a protein, disrupting its stability, structure, macromolecular binding, ablation of posttranslational modification sites, etc. (Jubb et al., 2017; Joo and Liu, 2021; Padhi et al., 2021a). Some mutations increase/decrease the binding affinity of the protein towards its receptor (Lee et al., 2020). The increased binding affinity in viral proteins results in a higher infection probability (Lee et al., 2020). The N501Y mutation, located in the receptor-binding domain of the S protein, increases its binding affinity towards the ACE2 receptor (Shi et al., 2021). Various mutations in the S protein reportedly affect vaccine development, efficacy, and neutralization. The D614G mutation of S protein enhances the viral replication rate and is the most prevalent mutation, predominantly reported in B.1.617.2, B.1.427, B.1.429, P.2, B.1.617.1., B.1.525, and B.1.526 variants. It was also found that the D614G mutation may decrease binding affinity and could also change the predicted MHC binding (Akkiz, 2021; Daniloski et al., 2021). The spike protein mutations, N501Y, E484K, P681H, and K417N, found in variant B.1.1.7, B.1.351, and the B.1.128.1 could decrease the virus’s ability to attach to antibodies (Mahase, 2021). Hence, it becomes a necessity to consider the variant during the development of vaccines. However, according to some recent findings, despite the N501Y and P681H mutants in B.1.1.7, vaccine efficacy would not be affected (Seo et al., 2020; Mahase, 2021; Shen et al., 2021). The Spike mutations N501Y (B.1.1.7, B.1.351 and the B.1.128.), L18F, K417T (B.1.351), E484K (B.1.1.7, B.1.351, B.1.128.1, B.1.525 and B.1.526), and D614G (B.1.617.2, B.1.427, B.1.429, P.2, B.1.617.1., B.1.525, and B.1.526) are reported in most concerning variants and has significant inferences for evading antibody-mediated immunity (Altmann et al., 2021; Gupta, 2021; Mohammadi et al., 2021).
The T190I mutation in Mpro has brought a polar to non-polar substitution near the active site cavity, which might have a significant role in enzymatic activity, particularly when coupled with mutations in neighboring areas (Sheik Amamuddy et al., 2020). Another mutation adjacent to T190I is A191V; both residues belong to the most flexible regions, as substrate recognition sites often require structural flexibility to recognize the binding sites precisely (Sheik Amamuddy et al., 2020). Such mutations would alter the native conformation and activity of the SARS-CoV-2 Mpro and RdRp, which might affect the binding of therapeutic molecules (Padhi et al., 2021b; Padhi and Tripathi, 2021).
Conclusion
SARS-CoV-2 is more potent than the previous strains of coronaviruses; this reflects their enormous ability to mutate into new strains. When a virus enters a host cell, it uses its machinery to replicate and synthesize viral particles. Mutations lead to the evolution of the viral genome, allowing them to better adapt and survive in the human host for their active reproduction. Such mutations are achieved by modifications in the epitopes of viral genes, making them more infective, transmissible and helps in escaping the immune responses of the host. Most of the structural proteins of SARS-CoV-2 conserved among coronavirus family and shares ~ 90% sequence similarity. However, a slight change in sequence causes a great impact on the structure and pathogenesis of SARS-CoV-2. Despite major mutations do not affect vaccine efficacy, however, sometimes become more pathogenic.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Author Contributions
Conceptualization, TM, FA, and MIH. Methodology, AC, IH, DY, TM, and YM. Software, AC and MU. Validation, TM and MIH. Formal analysis, AC, AS, and MU. Investigation, MA, AS, and TM. Resources; data curation AC, and IH. Writing—original draft preparation, TM, PA, DY, AS, and AC. Writing—review and editing, TM, FA, and MH. Visualization, FA, AC, and IH. Supervision, MIH. Project administration, DY and MIH. Funding acquisition, FA and MIH. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Taif University Researchers Supporting Project Number (TURSP-2020/131), Taif University, Taif, Saudi Arabia and the Indian Council of Medical Research (Grant No. ECD/Adhoc/2/2021-22).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This work was supported by Taif University Researchers Supporting Project Number (TURSP-2020/131), Taif University, Taif, Saudi Arabia. MIH acknowledges the Council of Scientific and Industrial Research for financial support [Project No. 27(0368)/20/EMR-II]. The authors thank the Department of Science and Technology, Government of India, for the FIST support (FIST program No. SR/FST/LSII/2020/782).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.765039/full#supplementary-material
References
- Ahmed S. F., Quadeer A. A., Mckay M. R. (2020). Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 12, 254. doi: 10.3390/v12030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkiz H. (2021). Implications of the Novel Mutations in the SARS-CoV-2 Genome for Transmission, Disease Severity, and the Vaccine Development. Front. Med. (Lausanne) 8, 636532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann D. M., Boyton R. J., Beale R. (2021). Immunity to SARS-CoV-2 Variants of Concern. Science 371, 1103–1104. doi: 10.1126/science.abg7404 [DOI] [PubMed] [Google Scholar]
- Amir M., Ahmad S., Ahamad S., Kumar V., Mohammad T., Dohare R., et al. (2019). Impact of Gln94Glu Mutation on the Structure and Function of Protection of Telomere 1, a Cause of Cutaneous Familial Melanoma. J. Biomol. Struct. Dynamics. doi: 10.1080/07391102.2019.1610500 [DOI] [PubMed] [Google Scholar]
- Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F. (2020). The Proximal Origin of SARS-CoV-2. Nat. Med. 26, 450–452. doi: 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N. V., Paciello I., et al. (2020). SARS-CoV-2 Escape In Vitro From a Highly Neutralizing COVID-19 Convalescent Plasma. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annavajhala M. K., Mohri H., Wang P., Nair M., Zucker J. E., Sheng Z., et al. (2021). A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv. doi: 10.1101/2021.02.23.21252259 [DOI] [Google Scholar]
- Asrani P., Afzal Hussain K. N., Alajmi M. F., Amir S., Sohal S. S., Hassan M. I. (2021. a). Guidelines and Safety Considerations in the Laboratory Diagnosis of SARS-CoV-2 Infection: A Prerequisite Study for Health Professionals. Risk Manage. Healthc. Policy 14, 379. doi: 10.2147/RMHP.S284473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrani P., Eapen M. S., Chia C., Haug G., Weber H. C., Hassan M. I., et al. (2021. b). Diagnostic Approaches in COVID-19: Clinical Updates. Expert Rev. Respir. Med. 15, 197–212. doi: 10.1080/17476348.2021.1823833 [DOI] [PubMed] [Google Scholar]
- Asrani P., Eapen M. S., Hassan M. I., Sohal S. S. (2021. c). Implications of the Second Wave of COVID-19 in India. Lancet Respir. Med. 9(9), e93–e94. doi: 10.1016/S2213-2600(21)00312-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrani P., Hasan G. M., Sohal S. S., Hassan M. I. (2020). Molecular Basis of Pathogenesis of Coronaviruses: A Comparative Genomics Approach to Planetary Health to Prevent Zoonotic Outbreaks in the 21st Century. Omics: J. Integr. Biol. 24, 634–644. doi: 10.1089/omi.2020.0131 [DOI] [PubMed] [Google Scholar]
- Astuti I. (2020). Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response. Diabetes Metab. Syndrome: Clin. Res. Rev. 14, 407–412. doi: 10.1016/j.dsx.2020.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral P., Bhattarai N., Hossen M. L., Stebliankin V., Gerstman B., Narasimhan G., et al. (2021). Mutation-Induced Changes in the Receptor-Binding Interface of the SARS-CoV-2 Delta Variant B. 1.617. 2 and Implications for Immune Evasion. Biochem. Biophysic. Res. Comm. 574, 14–19. doi: 10.1016/j.bbrc.2021.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Dijkman R., Habjan M., Heurich A., Gierer S., Glowacka I., et al. (2013). TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium. J. Virol. 87, 6150–6160. doi: 10.1128/JVI.03372-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. (2020). Sars-CoV-2 Envelope and Membrane Proteins: Structural Differences Linked to Virus Characteristics? BioMed. Res. Int. 2020. doi: 10.1155/2020/4389089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroujeni M., Saberian M., Li J. (2021). Environmental Impacts of COVID-19 on Victoria, Australia, Witnessed Two Waves of Coronavirus. Environ. Sci. Pollution Res. 28, 14182–14191. doi: 10.1007/s11356-021-12556-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookman S., Cook J., Zucherman M., Broughton S., Harman K., Gupta A. (2021). Effect of the New SARS-CoV-2 Variant B. 1.1. 7 on Children and Young People. Lancet Child Adolesc. Health 5, e9–e10. doi: 10.1016/S2352-4642(21)00030-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-K., Chen C.-M. M., Chiang M.-H., Hsu Y.-L., Huang T.-H. (2013). Transient Oligomerization of the SARS-CoV N Protein–Implication for Virus Ribonucleoprotein Packaging. PloS One 8, e65045. doi: 10.1371/journal.pone.0065045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-K., Hou M.-H., Chang C.-F., Hsiao C.-D., Huang T.-H. (2014). The SARS Coronavirus Nucleocapsid Protein–Forms and Functions. Antiviral Res. 103, 39–50. doi: 10.1016/j.antiviral.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-K., Sue S.-C., Yu T.-H., Hsieh C.-M., Tsai C.-K., Chiang Y.-C., et al. (2006). Modular Organization of SARS Coronavirus Nucleocapsid Protein. J. Biomed. Sci. 13, 59–72. doi: 10.1007/s11373-005-9035-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. F.-W., Yip C. C.-Y., To K. K.-W., Tang T. H.-C., Wong S. C.-Y., Leung K.-H., et al. (2020). Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and With Clinical Specimens. J. Clin. Microbiol. 58, e00310–20. doi: 10.1128/JCM.00310-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellasamy G., Arumugasamy S. K., Govindaraju S., Yun K. (2020). Analytical Insights of COVID-19 Pandemic. Trends Analyt. Chem. 133, 116072. doi: 10.1016/j.trac.2020.116072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-Y., Chang C.-K., Chang Y.-W., Sue S.-C., Bai H.-I., Riang L., et al. (2007). Structure of the SARS Coronavirus Nucleocapsid Protein RNA-Binding Dimerization Domain Suggests a Mechanism for Helical Packaging of Viral RNA. J. Mol. Biol. 368, 1075–1086. doi: 10.1016/j.jmb.2007.02.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Guo Y., Pan Y., Zhao Z. J. (2020). Structure Analysis of the Receptor Binding of 2019-Ncov. Biochem. Biophys. Res. Commun. 525, 135–140. doi: 10.1016/j.bbrc.2020.02.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang R., Wang M., Wei G.-W. (2020). Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 432, 5212–5226. doi: 10.1016/j.jmb.2020.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., et al. (2021). Convergent Evolution of SARS-CoV-2 Spike Mutations, L452R, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 9(7). doi: 10.3390/microorganisms9071542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A., Mohammad T., Samarth N., Hussain A., Rehman M. T., Islam A., et al. (2021). Structural Genomics Approach to Investigate Deleterious Impact of nsSNPs in Conserved Telomere Maintenance Component 1. Sci. Rep. 11, 1–13. doi: 10.1038/s41598-021-89450-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysostomou A. C., Vrancken B., Koumbaris G., Themistokleous G., Aristokleous A., Masia C., et al. (2021). A Comprehensive Molecular Epidemiological Analysis of SARS-CoV-2 Infection in Cyprus From April 2020 to January 2021: Evidence of a Highly Polyphyletic and Evolving Epidemic. Viruses 13, 1098. doi: 10.3390/v13061098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corse E., Machamer C. E. (2000). Infectious Bronchitis Virus E Protein Is Targeted to the Golgi Complex and Directs Release of Virus-Like Particles. J. Virol. 74, 4319–4326. doi: 10.1128/JVI.74.9.4319-4326.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., De Lamballerie X., Canard B., Seidah N., Decroly E. (2020). The Spike Glycoprotein of the New Coronavirus 2019-Ncov Contains a Furin-Like Cleavage Site Absent in CoV of the Same Clade. Antiviral Res. 176, 104742. doi: 10.1016/j.antiviral.2020.104742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloski Z., Jordan T. X., Ilmain J. K., Guo X., Bhabha G., Tenoever B. R., et al. (2021). The Spike D614G Mutation Increases SARS-CoV-2 Infection of Multiple Human Cell Types. Elife 10, e65365. doi: 10.7554/eLife.65365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dediego M. L., Pewe L., Alvarez E., Rejas M. T., Perlman S., Enjuanes L. (2008). Pathogenicity of Severe Acute Respiratory Coronavirus Deletion Mutants in hACE-2 Transgenic Mice. Virology 376, 379–389. doi: 10.1016/j.virol.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert D. M., Kim P. S. (2001). Mechanisms of Viral Membrane Fusion and its Inhibition. Annu. Rev. Biochem. 70, 777–810. doi: 10.1146/annurev.biochem.70.1.777 [DOI] [PubMed] [Google Scholar]
- Faria Nr C. I., Candido D., Franco L., Andrade P. S., Coletti T. M., Silva C., et al. (2021) Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings. Available at: https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586.
- Fehr A. R., Perlman S. (2015). Coronaviruses: An Overview of Their Replication and Pathogenesis. Coronaviruses 1–23. doi: 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett C., Dediego M. L., Regla-Nava J. A., Enjuanes L., Perlman S. (2013). Complete Protection Against Severe Acute Respiratory Syndrome Coronavirus-Mediated Lethal Respiratory Disease in Aged Mice by Immunization With a Mouse-Adapted Virus Lacking E Protein. J. Virol. 87, 6551–6559. doi: 10.1128/JVI.00087-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratev F. J. B. (2020). The N501Y and K417N Mutations in the Spike Protein of SARS-CoV-2 Alter the Interactions With Both Hace2 and Human Derived Antibody: A Free Energy of Perturbation Study. BioRxiv. doi: 10.1101/2020.12.23.424283 [DOI] [PubMed] [Google Scholar]
- Gobeil S. M., Janowska K., Mcdowell S., Mansouri K., Parks R., Manne K., et al. (2021). D614G Mutation Alters SARS-CoV-2 Spike Conformation and Enhances Protease Cleavage at the S1/S2 Junction. Cell Rep. 34, 108630. doi: 10.1016/j.celrep.2020.108630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. K. (2021). Will SARS-CoV-2 Variants of Concern Affect the Promise of Vaccines? Nat. Rev. Immunol. 21, 340–341. doi: 10.1038/s41577-021-00556-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib I., Khan S., Mohammad T., Hussain A., Alajmi M. F., Rehman T., et al. (2021). Impact of non-Synonymous Mutations on the Structure and Function of Telomeric Repeat Binding Factor 1. J. Biomol. Struct. Dynamics 1–14. doi: 10.1080/07391102.2021.1922313 [DOI] [PubMed] [Google Scholar]
- Ham S. (2020). Prevention of Exposure to and Spread of COVID-19 Using Air Purifiers: Challenges and Concerns. Epidemiol Health 42, e2020027. doi: 10.4178/epih.e2020027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C. (2015). Viral Membrane Fusion. Virology 479, 498–507. doi: 10.1016/j.virol.2015.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan A., Paray B. A., Hussain A., Qadir F. A., Attar F., Aziz F. M., et al. (2020). A Review on the Cleavage Priming of the Spike Protein on Coronavirus by Angiotensin-Converting Enzyme-2 and Furin. J. Biomol. Struct. Dynamics 1–9. doi: 10.1080/07391102.2020.1754293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Dobie F., Ballantine M., Leeson A., Li Y., Bastien N., et al. (2004. a). Analysis of Multimerization of the SARS Coronavirus Nucleocapsid Protein. Biochem. Biophys. Res. Commun. 316, 476–483. doi: 10.1016/j.bbrc.2004.02.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R., Leeson A., Ballantine M., Andonov A., Baker L., Dobie F., et al. (2004. b). Characterization of Protein–Protein Interactions Between the Nucleocapsid Protein and Membrane Protein of the SARS Coronavirus. Virus Res. 105, 121–125. doi: 10.1016/j.virusres.2004.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pöhlmann S. (2014). TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 88, 1293–1307. doi: 10.1128/JVI.02202-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280. e278. doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. (2020). Clinical Features of Patients Infected With 2019 Novel Coronavirus in Wuhan, China. Lancet 395, 497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. (2020). Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacologica Sin. 41, 1141–1149. doi: 10.1038/s41401-020-0485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Yu L., Petros A. M., Gunasekera A., Liu Z., Xu N., et al. (2004). Structure of the N-Terminal RNA-Binding Domain of the SARS CoV Nucleocapsid Protein. Biochemistry 43, 6059–6063. doi: 10.1021/bi036155b [DOI] [PubMed] [Google Scholar]
- Huh K., Jung J., Hong J., Kim M., Ahn J. G., Kim J. H. (2021). Impact of Nonpharmaceutical Interventions on the Incidence of Respiratory Infections During the Coronavirus Disease 2019 (COVID-19) Outbreak in Korea: A Nationwide Surveillance Study. Clin. Infect. Dis. 72 (7), e184–e191. doi: 10.1093/cid/ciaa1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst K. R., Koetzner C. A., Masters P. S. (2009). Identification of In Vivo-Interacting Domains of the Murine Coronavirus Nucleocapsid Protein. J. Virol. 83, 7221–7234. doi: 10.1128/JVI.00440-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E., Yu S. (2021). Modeling and Forecasting the COVID-19 Pandemic With Heterogeneous Autoregression Approaches: South Korea. Results Phys. 29, 104631. doi: 10.1016/j.rinp.2021.104631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftimie S., López-Azcona A. F., Vallverdú I., Hernández-Flix S., De Febrer G., Parra S., et al. (2021). First and Second Waves of Coronavirus Disease-19: A Comparative Study in Hospitalized Patients in Reus, Spain. PloS One 16, e0248029. doi: 10.1371/journal.pone.0248029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam O. K., Al-Emran H. M., Hasan M. S., Anwar A., Jahid M. I. K., Hossain M. A. (2021). Emergence of European and North American Mutant Variants of SARS-CoV-2 in South-East Asia. Transboundary Emerg. Dis. 68, 824–832. doi: 10.1111/tbed.13748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J. A., André N. M., Chappie J. S., Millet J. K., Whittaker G. R. (2020). Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 432, 3309–3325. doi: 10.1016/j.jmb.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Krammer F., Simon V., et al. (2021). SARS-CoV-2 Spike E484K Mutation Reduces Antibody Neutralisation. Lancet Microbe 2, e283–e284. doi: 10.1016/S2666-5247(21)00068-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebril N. (2020). World Health Organization Declared a Pandemic Public Health Menace: A Systematic Review of the Coronavirus Disease 2019 “COVID-19”, Up to 26th March 2020. Available at SSRN 3566298. doi: 10.2139/ssrn.3566298 [DOI] [Google Scholar]
- Ji W., Huh K., Kang M., Hong J., Bae G. H., Lee R., et al. (2020). Effect of Underlying Comorbidities on the Infection and Severity of COVID-19 in Korea: A Nationwide Case-Control Study. J Korean Med Sci. 35(25), e237. doi: 10.3346/jkms.2020.35.e237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeño J. M., Regla-Nava J. A., Nieto-Torres J. L., Dediego M. L., Castaño-Rodriguez C., Fernandez-Delgado R., et al. (2015). Identification of the Mechanisms Causing Reversion to Virulence in an Attenuated SARS-CoV for the Design of a Genetically Stable Vaccine. PloS Pathog. 11, e1005215. doi: 10.1371/journal.ppat.1005215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo J. Y., Liu M. F. (2021). Nurses' Barriers to Caring for Patients with COVID-19: A Qualitative Systematic Review. Int. Nurs. Rev. 68(2), 202–213. doi: 10.1111/inr.12648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju M. J., Oh J., Choi Y. H. (2021). Changes in Air Pollution Levels After COVID-19 Outbreak in Korea. Sci Total Environ. 750, 141521. doi: 10.1016/j.amsu.2020.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubb H. C., Pandurangan A. P., Turner M. A., Ochoa-Montaño B., Blundell T. L., Ascher D. B. (2017). Mutations at Protein-Protein Interfaces: Small Changes Over Big Surfaces Have Large Impacts on Human Health. Prog. biophysics Mol. Biol. 128, 3–13. doi: 10.1016/j.pbiomolbio.2016.10.002 [DOI] [PubMed] [Google Scholar]
- Kandeel M., Al-Taher A., Li H., Schwingenschlogl U., Al-Nazawi M. (2018). Molecular Dynamics of Middle East Respiratory Syndrome Coronavirus (MERS CoV) Fusion Heptad Repeat Trimers. Comput. Biol. Chem. 75, 205–212. doi: 10.1016/j.compbiolchem.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Kataoka M., Shirato K., Matsuyama S. (2019). Biochemical Analysis of Coronavirus Spike Glycoprotein Conformational Intermediates During Membrane Fusion. J. Virol. 93, e00785–19. doi: 10.1128/JVI.00785-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. E., Kim D. K., Song Y. J. (2021). SARS-CoV-2 Nonstructural Proteins 1 and 13 Suppress Caspase-1 and the NLRP3 Inflammasome Activation. Microorganisms 9, 494. doi: 10.3390/microorganisms9030494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. (2020). Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 812–827.e819. doi: 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai J. W., Cheong K. H. (2020). Superposition of COVID-19 Waves, Anticipating a Sustained Wave, and Lessons for the Future. Bioessays 42, 2000178. doi: 10.1002/bies.202000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimer J., Hiebl-Flach J., Lengauer D., Lackner P. (2016). MAESTROweb: A Web Server for Structure-Based Protein Stability Prediction. Bioinformatics 32, 1414–1416. doi: 10.1093/bioinformatics/btv769 [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. (2020). Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 581, 215–220. doi: 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- Leary S., Gaudieri S., Parker M. D., Chopra A., James I., Pakala S., et al. (2021). Generation of a Novel SARS-CoV-2 Sub-Genomic RNA Due to the R203K/G204R Variant in Nucleocapsid. BioRxiv 2020.04.10.029454. doi: 10.1101/2020.04.10.029454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Yum H. K., Si H. J., Han S. H., Park S. Y., Peck K. R., et al. (2020). Guidelines for Surgery of Confirmed or Suspected COVID-19 Patients. Infect Chemother 52 (3), 453–459. doi: 10.3947/ic.2020.52.3.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. (2020). Functional Assessment of Cell Entry and Receptor Usage for SARS-CoV-2 and Other Lineage B Betacoronaviruses. Nat. Microbiol. 5, 562–569. doi: 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K., Shum M. H., Leung G. M., Lam T. T., Wu J. T. (2021). Early Transmissibility Assessment of the N501Y Mutant Strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill 26, 2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Guo M., Tian X., Wang X., Yang X., Wu P., et al. (2021). Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis. Med 2, 99–112.e117. doi: 10.1016/j.medj.2020.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Liao C.-H., Wang Q., Tan Y.-J., Luo R., Qiu Y., et al. (2020). The ORF6, ORF8 and Nucleocapsid Proteins of SARS-CoV-2 Inhibit Type I Interferon Signaling Pathway. Virus Res. 286, 198074. doi: 10.1016/j.virusres.2020.198074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limburg H., Harbig A., Bestle D., Stein D. A., Moulton H. M., Jaeger J., et al. (2019). TMPRSS2 Is the Major Activating Protease of Influenza A Virus in Primary Human Airway Cells and Influenza B Virus in Human Type II Pneumocytes. J. Virol. 93, e00649–19. doi: 10.1128/JVI.00649-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Surya W., Claudine S., Torres J. (2014). Structure of a Conserved Golgi Complex-Targeting Signal in Coronavirus Envelope Proteins. J. Biol. Chem. 289, 12535–12549. doi: 10.1074/jbc.M114.560094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-J., Leng C.-H., Lien S.-P., Chi H.-Y., Huang C.-Y., Lin C.-L., et al. (2006). Immunological Characterizations of the Nucleocapsid Protein Based SARS Vaccine Candidates. Vaccine 24, 3100–3108. doi: 10.1016/j.vaccine.2006.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubinski B., Tang T., Daniel S., Jaimes J. A., Whittaker G. R. (2021). Functional Evaluation of Proteolytic Activation for the SARS-CoV-2 Variant B.1.1.7: Role of the P681H Mutation. bioRxiv. doi: 10.1101/2021.04.06.438731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Chen Q., Chen J., Chen K., Shen X., Jiang H. (2005). The Nucleocapsid Protein of SARS Coronavirus has a High Binding Affinity to the Human Cellular Heterogeneous Nuclear Ribonucleoprotein A1. FEBS Lett. 579, 2623–2628. doi: 10.1016/j.febslet.2005.03.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. (2021). Covid-19: Novavax Vaccine Efficacy Is 86% Against UK Variant and 60% Against South African Variant. BMJ 372, n296. doi: 10.1136/bmj.n296 [DOI] [PubMed] [Google Scholar]
- Mahtarin R., Islam S., Islam M. J., Ullah M. O., Ali M. A., Halim M. A. (2020). Structure and Dynamics of Membrane Protein in SARS-CoV-2. J. Biomol. Struct. Dynamics, 1–14. doi: 10.1080/07391102.2020.1861983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Olson-Sidford W., J W. W. (2021). Biological and Clinical Consequences of Integrin Binding via a Rogue RGD Motif in the SARS CoV-2 Spike Protein. Viruses 13, 146. doi: 10.3390/v13020146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Y. A. (2020). Properties of Coronavirus and SARS-CoV-2. Malaysian J. Pathol. 42, 3–11. [PubMed] [Google Scholar]
- Mandala V. S., Mckay M. J., Shcherbakov A. A., Dregni A. J., Kolocouris A., Hong M. (2020). Structure and Drug Binding of the SARS-CoV-2 Envelope Protein Transmembrane Domain in Lipid Bilayers. Nat. Struct. Mol. Biol. 27, 1202–1208. doi: 10.1038/s41594-020-00536-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcbride R., Van Zyl M., Fielding B. C. (2014). The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 6, 2991–3018. doi: 10.3390/v6082991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccallum M., De Marco A., Lempp F. A., Tortorici M. A., Pinto D., Walls A. C., et al. (2021). N-Terminal Domain Antigenic Mapping Reveals a Site of Vulnerability for SARS-CoV-2. Cell 184, 2332–2347.e2316. doi: 10.1016/j.cell.2021.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J. K., Whittaker G. R. (2015). Host Cell Proteases: Critical Determinants of Coronavirus Tropism and Pathogenesis. Virus Res. 202, 120–134. doi: 10.1016/j.virusres.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J. K., Whittaker G. R. (2018). Physiological and Molecular Triggers for SARS-CoV Membrane Fusion and Entry Into Host Cells. Virology 517, 3–8. doi: 10.1016/j.virol.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad T., Amir M., Prasad K., Batra S., Kumar V., Hussain A., et al. (2020). Impact of Amino Acid Substitution in the Kinase Domain of Bruton Tyrosine Kinase and its Association With X-Linked Agammaglobulinemia. Int. J. Biol. Macromolecules 164, 2399–2408. doi: 10.1016/j.ijbiomac.2020.08.057 [DOI] [PubMed] [Google Scholar]
- Mousavizadeh L., Ghasemi S. (2020). Genotype and Phenotype of COVID-19: Their Roles in Pathogenesis. J. Microbiol. Immunol. Infect. 54(2), 159–163. doi: 10.1016/j.jmii.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A., Fatima K., Mohammad T., Fatima U., Singh I. K., Singh A., et al. (2020). Insights Into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Biophys. Acta (BBA) Molecular Basis Dis. 1866(10), 165878. doi: 10.1016/j.bbadis.2020.165878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naskalska A., Dabrowska A., Szczepanski A., Milewska A., Jasik K. P., Pyrc K. (2019). Membrane Protein of Human Coronavirus NL63 Is Responsible for Interaction With the Adhesion Receptor. J. Virol. 93, e00355–19. doi: 10.1128/JVI.00355-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netland J., Dediego M. L., Zhao J., Fett C., Álvarez E., Nieto-Torres J. L., et al. (2010). Immunization With an Attenuated Severe Acute Respiratory Syndrome Coronavirus Deleted in E Protein Protects Against Lethal Respiratory Disease. Virology 399, 120–128. doi: 10.1016/j.virol.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B. W., Kiss G., Kunding A. H., Bhella D., Baksh M. F., Connelly S., et al. (2011). A Structural Analysis of M Protein in Coronavirus Assembly and Morphology. J. Struct. Biol. 174, 11–22. doi: 10.1016/j.jsb.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J. L., Dediego M. L., Álvarez E., Jiménez-Guardeño J. M., Regla-Nava J. A., Llorente M., et al. (2011). Subcellular Location and Topology of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. Virology 415, 69–82. doi: 10.1016/j.virol.2011.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J. L., Dediego M. L., Verdiá-Báguena C., Jimenez-Guardeño J. M., Regla-Nava J. A., Fernandez-Delgado R., et al. (2014). Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Ion Channel Activity Promotes Virus Fitness and Pathogenesis. PloS Pathog. 10, e1004077. doi: 10.1371/journal.ppat.1004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J. L., Verdiá-Báguena C., Jimenez-Guardeño J. M., Regla-Nava J. A., Castaño-Rodriguez C., Fernandez-Delgado R., et al. (2015). Severe Acute Respiratory Syndrome Coronavirus E Protein Transports Calcium Ions and Activates the NLRP3 Inflammasome. Virology 485, 330–339. doi: 10.1016/j.virol.2015.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva J. L., Madan V., Carrasco L. (2012). Viroporins: Structure and Biological Functions. Nat. Rev. Microbiol. 10, 563–574. doi: 10.1038/nrmicro2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization W. H. (2020). WHO Coronavirus Disease (COVID-19) Dashboard. 2020. [Google Scholar]
- Ostrov D. A. (2021). Structural Consequences of Variation in SARS-CoV-2 B.1.1.7. J. Cell Immunol. 3, 103–108. doi: 10.33696/immunology.3.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. (2020). Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and its Immune Cross-Reactivity With SARS-CoV. Nat. Commun. 11, 1–12. doi: 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi A. K., Dandapat J., Saudagar P., Uversky V. N., Tripathi T. (2021. a). Interface-Based Design of the Favipiravir-Binding Site in SARS-CoV-2 RNA-Dependent RNA Polymerase Reveals Mutations Conferring Resistance to Chain Termination. FEBS Lett. doi: 10.1002/1873-3468.14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi A. K., Shukla R., Saudagar P., Tripathi T. (2021. b). High-Throughput Rational Design of the Remdesivir Binding Site in the RdRp of SARS-CoV-2: Implications for Potential Resistance. Iscience 24, 101992. doi: 10.1016/j.isci.2020.101992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi A. K., Tripathi T. (2021). Targeted Design of Drug Binding Sites in the Main Protease of SARS-CoV-2 Reveals Potential Signatures of Adaptation. Biochem. Biophys. Res. Commun. 555, 147–153. doi: 10.1016/j.bbrc.2021.03.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy K., Ng L., Lin X., Liu D. X., Pervushin K., Gong X., et al. (2008). Structural Flexibility of the Pentameric SARS Coronavirus Envelope Protein Ion Channel. Biophys. J. 95, L39–L41. doi: 10.1529/biophysj.108.133041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K., Tan E., Parthasarathy K., Lin X., Jiang F. L., Yu D., et al. (2009). Structure and Inhibition of the SARS Coronavirus Envelope Protein Ion Channel. PloS Pathog. 5, e1000511. doi: 10.1371/journal.ppat.1000511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philot E. A., Gasparini P., De Mattos J. C., Magro A., Torres-Bonfim E., Quiroz R. C. N., et al. (2021). Unveiling Mutation Effects on the Structural Dynamics of the Main Protease From SARS-CoV-2 With Hybrid Simulation Methods. BioRxiv. doi: 10.1101/2021.07.17.452787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. E., Ascher D. B., Blundell T. L. (2014). mCSM: Predicting the Effects of Mutations in Proteins Using Graph-Based Signatures. Bioinformatics 30, 335–342. doi: 10.1093/bioinformatics/btt691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., et al. (2021). Sensitivity of Infectious SARS-CoV-2 B. 1.1. 7 and B. 1.351 Variants to Neutralizing Antibodies. Nat. Med. 27, 917–924. doi: 10.1038/s41591-021-01318-5 [DOI] [PubMed] [Google Scholar]
- Plante J. A., Liu Y., Liu J., Xia H., Johnson B. A., Lokugamage K. G., et al. (2021). Spike Mutation D614G Alters SARS-CoV-2 Fitness. Nature 592, 116–121. doi: 10.1038/s41586-020-2895-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presti A. L., Rezza G., Stefanelli P. (2020). Selective Pressure on SARS-CoV-2 Protein Coding Genes and Glycosylation Site Prediction. Heliyon 6, e05001. doi: 10.1016/j.heliyon.2020.e05001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan A. A., Al-Ahmed S. H., Haque S., Sah R., Tiwari R., Malik Y. S., et al. (2020). SARS-CoV-2, SARS-CoV, and MERS-COV: A Comparative Overview. Infez Med. 28, 174–184. [PubMed] [Google Scholar]
- Risco C., Antón I. M., Enjuanes L., Carrascosa J. L. (1996). The Transmissible Gastroenteritis Coronavirus Contains a Spherical Core Shell Consisting of M and N Proteins. J. Virol. 70, 4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizwan T., Kothidar A., Meghwani H., Sharma V., Shobhawat R., Saini R., et al. (2021). Comparative Analysis of SARS-CoV-2 Envelope Viroporin Mutations From COVID-19 Deceased and Surviving Patients Revealed Implications on its Ion-Channel Activities and Correlation With Patient Mortality. J. Biomol Struct. Dyn, 1–16. doi: 10.1080/07391102.2021.1944319 [DOI] [PubMed] [Google Scholar]
- Rodrigues C. H. M., Pires D. E. V., Ascher D. B. (2021). DynaMut2: Assessing Changes in Stability and Flexibility Upon Single and Multiple Point Missense Mutations. Protein Sci. 30, 60–69. doi: 10.1002/pro.3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan S. (2020). Likelihood of Survival of Coronavirus Disease 2019. Lancet Infect. Dis. 20, 630–631. doi: 10.1016/S1473-3099(20)30257-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruch T. R., Machamer C. E. (2012). The Coronavirus E Protein: Assembly and Beyond. Viruses 4, 363–382. doi: 10.3390/v4030363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikatendu K. S., Joseph J. S., Subramanian V., Neuman B. W., Buchmeier M. J., Stevens R. C., et al. (2007). Ribonucleocapsid Formation of Severe Acute Respiratory Syndrome Coronavirus Through Molecular Action of the N-Terminal Domain of N Protein. J. Virol. 81, 3913–3921. doi: 10.1128/JVI.02236-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B. C. (2019). Coronavirus Envelope Protein: Current Knowledge. Virol. J. 16, 1–22. doi: 10.1186/s12985-019-1182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M. R., Kim J. W., Park E. J., Jung S. M., Sung Y. K., Kim H., et al. (2020). Recommendations for the Management of Patients with Systemic Rheumatic Diseases During the Coronavirus Disease Pandemic. Korean J Intern Med 35(6), 1317–1332. doi: 10.3904/kjim.2020.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi A., Mohammad T., Anwar S., Amani S., Khan M. S., Husain F. M., et al. (2021). Potential Drug Targets of SARS-CoV-2: From Genomics to Therapeutics. Int. J. Biol. Macromolecules. doi: 10.1016/j.ijbiomac.2021.02.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B., Wang X.-Y., Yuan J.-W., Vabret A., Wu X.-D., Yang R.-F., et al. (2005). Characterization and Application of Monoclonal Antibodies Against N Protein of SARS-Coronavirus. Biochem. Biophys. Res. Commun. 336, 110–117. doi: 10.1016/j.bbrc.2005.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., et al. (2020). Cell Entry Mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. 117, 11727–11734. doi: 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheik Amamuddy O., Verkhivker G. M., Tastan Bishop O. Z. (2020). Impact of Early Pandemic Stage Mutations on Molecular Dynamics of SARS-CoV-2 Mpro. J. Chem. Inf. Modeling 60, 5080–5102. doi: 10.1021/acs.jcim.0c00634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Tang H., Mcdanal C., Wagh K., Fischer W., Theiler J., et al. (2021). SARS-CoV-2 Variant B. 1.1. 7 Is Susceptible to Neutralizing Antibodies Elicited by Ancestral Spike Vaccines. Cell Host Microbe 29, 529–539.e523. doi: 10.1016/j.chom.2021.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Yi Y., Li P., Kuang T., Li L., Dong M., et al. (2003). Diagnosis of Severe Acute Respiratory Syndrome (SARS) by Detection of SARS Coronavirus Nucleocapsid Antibodies in an Antigen-Capturing Enzyme-Linked Immunosorbent Assay. J. Clin. Microbiol. 41, 5781–5782. doi: 10.1128/JCM.41.12.5781-5782.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H. J., Lee J. B., Choi M. K., Jang Y. R., Cho Y. K., Eom J. S. (2021). Protection and Response of a Tertiary Hospital in South Korea to the COVID-19 Outbreak Disaster Med Public Health Prep. 15(1), e1–e5. doi: 10.1017/dmp.2020.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D. D., Han I., Choi E. H., Yadav D. K. (2020. a). Immunopathology, Host-Virus Genome Interactions, and Effective Vaccine Development in SARS-CoV-2. Comput. Struct. Biotechnol. J. 18, 3774–3787. doi: 10.1016/j.csbj.2020.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D. D., Han I., Choi E. H., Yadav D. K. (2020. b). Recent Advances in Pathophysiology, Drug Development and Future Perspectives of SARS-CoV-2. Front. Cell Dev. Biol. 8, 580202. doi: 10.3389/fcell.2020.580202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y., Teoh K., Lo J., Chan C., Kien F., Escriou N., et al. (2008). The M, E, and N Structural Proteins of the Severe Acute Respiratory Syndrome Coronavirus Are Required for Efficient Assembly, Trafficking, and Release of Virus-Like Particles. J. Virol. 82, 11318–11330. doi: 10.1128/JVI.01052-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socher E., Conrad M., Heger L., Paulsen F., Sticht H., Zunke F., et al. (2021). Mutations in the B.1.1.7 SARS-CoV-2 Spike Protein Reduce Receptor-Binding Affinity and Induce a Flexible Link to the Fusion Peptide. Biomedicines 9, 525. doi: 10.3390/biomedicines9050525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song P., Li W., Xie J., Hou Y., You C. (2020). Cytokine Storm Induced by SARS-CoV-2. Clinica Chimica Acta 509, 280–287. doi: 10.1016/j.cca.2020.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., et al. (2019). From SARS to MERS, Thrusting Coronaviruses Into the Spotlight. Viruses 11, 59. doi: 10.3390/v11010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T. N., Greaney A. J., Hilton S. K., Ellis D., Crawford K. H. D., Dingens A. S., et al. (2020). Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310.e1220. doi: 10.1016/j.cell.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya W., Li Y., Torres J. (2018). Structural Model of the SARS Coronavirus E Channel in LMPG Micelles. Biochim. Biophys. Acta (BBA) Biomembranes 1860, 1309–1317. doi: 10.1016/j.bbamem.2018.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Bidon M., Jaimes J. A., Whittaker G. R., Daniel S. (2020). Coronavirus Membrane Fusion Mechanism Offers a Potential Target for Antiviral Development. Antiviral Res. 178, 104792. doi: 10.1016/j.antiviral.2020.104792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. W., Marr L. C., Li Y., Dancer S. J. (2021). Covid-19 has Redefined Airborne Transmission. (United Kingdom: British Medical Journal Publishing Group; ). doi: 10.1136/bmj.n913 [DOI] [PubMed] [Google Scholar]
- Tan Y.-J., Goh P.-Y., Fielding B. C., Shen S., Chou C.-F., Fu J.-L., et al. (2004). Profiles of Antibody Responses Against Severe Acute Respiratory Syndrome Coronavirus Recombinant Proteins and Their Potential Use as Diagnostic Markers. Clin. Diagn. Lab. Immunol. 11, 362–371. doi: 10.1128/CDLI.11.2.362-371.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. W., Tambyah P. A., Hui D. S. (2020). Emergence of a New SARS-CoV-2 Variant in the UK. J. Infect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. (2021). Detection of a SARS-CoV-2 Variant of Concern in South Africa. Nature 592, 438–443. doi: 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- Tomaszewski T., Devries R. S., Dong M., Bhatia G., Norsworthy M. D., Zheng X., et al. (2020). New Pathways of Mutational Change in SARS-CoV-2 Proteomes Involve Regions of Intrinsic Disorder Important for Virus Replication and Release. Evol. Bioinf. 16, 1176934320965149. doi: 10.1177/1176934320965149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Maheswari U., Parthasarathy K., Ng L., Liu D. X., Gong X. (2007). Conductance and Amantadine Binding of a Pore Formed by a Lysine-Flanked Transmembrane Domain of SARS Coronavirus Envelope Protein. Protein Sci. 16, 2065–2071. doi: 10.1110/ps.062730007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J., Parthasarathy K., Lin X., Saravanan R., Kukol A., Liu D. X. (2006). Model of a Putative Pore: The Pentameric α-Helical Bundle of SARS Coronavirus E Protein in Lipid Bilayers. Biophys. J. 91, 938–947. doi: 10.1529/biophysj.105.080119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M. A., Walls A. C., Lang Y., Wang C., Li Z., Koerhuis D., et al. (2019). Structural Basis for Human Coronavirus Attachment to Sialic Acid Receptors. Nat. Struct. Mol. Biol. 26, 481–489. doi: 10.1038/s41594-019-0233-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umair M., Khan S., Mohammad T., Shafie A., Anjum F., Islam A., et al. (2021). Impact of Single Amino Acid Substitution on the Structure and Function of TANK-Binding Kinase-1. J. Cell. Biochem. doi: 10.1002/jcb.30070 [DOI] [PubMed] [Google Scholar]
- Van Vo G., Bagyinszky E., Park Y. S., Hulme J., An S. S. A. (2021). SARS-CoV-2 (COVID-19): Beginning to Understand a New Virus. Adv. Exp. Med. Biol. 1321, 3–19. doi: 10.1007/978-3-030-59261-5_1 [DOI] [PubMed] [Google Scholar]
- Venkatagopalan P., Daskalova S. M., Lopez L. A., Dolezal K. A., Hogue B. G. (2015). Coronavirus Envelope (E) Protein Remains at the Site of Assembly. Virology 478, 75–85. doi: 10.1016/j.virol.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdiá-Báguena C., Nieto-Torres J. L., Alcaraz A., Dediego M. L., Torres J., Aguilella V. M., et al. (2012). Coronavirus E Protein Forms Ion Channels With Functionally and Structurally-Involved Membrane Lipids. Virology 432, 485–494. doi: 10.1016/j.virol.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo V. G., Bagyinszky E., Shim K., Park Y. S., An S. S. A. (2020). Additional Diagnostic Testing of the 2019 Novel Coronavirus (SARS-CoV-2). Mol. Cell Toxicol., 1–3. doi: 10.1007/s13273-020-00096-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C., Park Y.-J., Tortorici M. A., Wall A., Mcguire A. T., Veesler D. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181, 281–292.e286. doi: 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. (2020). Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. Jama 323, 1061–1069. doi: 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., et al. (2020). Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 181, 894–904. e899. doi: 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R. S., Li F. (2020). Receptor Recognition by the Novel Coronavirus From Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 94, e00127–20. doi: 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J. D., Wrapp D., Mclellan J. S., Crispin M. (2020). Site-Specific Glycan Analysis of the SARS-CoV-2 Spike. Science 369, 330–333. doi: 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., Dasilva J., Poston D., Lorenzi J. C., et al. (2020). Escape From Neutralizing Antibodies by SARS-CoV-2 Spike Protein Variants. Elife 9, e61312. doi: 10.7554/eLife.61312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A. P., Wertheim J. O., Wang J. C., Vasylyeva T. I., Havens J. L., Chowdhury M. A., et al. (2021). Detection and Characterization of the SARS-CoV-2 Lineage B.1.526 in New York. bioRxiv. doi: 10.1101/2021.02.14.431043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L., Mckinlay C., Gage P., Ewart G. (2004). SARS Coronavirus E Protein Forms Cation-Selective Ion Channels. Virology 330, 322–331. doi: 10.1016/j.virol.2004.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., et al. (2020). Cryo-EM Structure of the 2019-Ncov Spike in the Prefusion Conformation. Science 367, 1260–1263. doi: 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., et al. (2020). Fusion Mechanism of 2019-Ncov and Fusion Inhibitors Targeting HR1 Domain in Spike Protein. Cell. Mol. Immunol. 1–3. doi: 10.1038/s41423-020-0374-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. (2020). Pathological Findings of COVID-19 Associated With Acute Respiratory Distress Syndrome. Lancet Respir. Med. 8, 420–422. doi: 10.1016/S2213-2600(20)30076-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. (2020). Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 367, 1444–1448. doi: 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu I.-M., Oldham M. L., Zhang J., Chen J. (2006). Crystal Structure of the Severe Acute Respiratory Syndrome (SARS) Coronavirus Nucleocapsid Protein Dimerization Domain Reveals Evolutionary Linkage Between Corona-and Arteriviridae. J. Biol. Chem. 281, 17134–17139. doi: 10.1074/jbc.M602107200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K. E., Tomkins-Tinch C., Nyalile T. P., Wang Y., et al. (2020). Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 183, 739–751.e738. doi: 10.1016/j.cell.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Liu G., Ma H., Zhao D., Yang Y., Liu M., et al. (2020). Biochemical Characterization of SARS-CoV-2 Nucleocapsid Protein. Biochem. Biophys. Res. Commun. 527, 618–623. doi: 10.1016/j.bbrc.2020.04.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. P., Lybrand T. P., Gilbert P. B., Hawn T. R., Schiffer J. T., Stamatatos L., et al. (2021). Tracking SARS-CoV-2 Spike Protein Mutations in the United State/01 - 2021/03) Using a Statistical Learning Strategy. bioRxiv. doi: 10.2139/ssrn.3844900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., et al. (2020). SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl. J. Med. 382, 1177–1179. doi: 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.