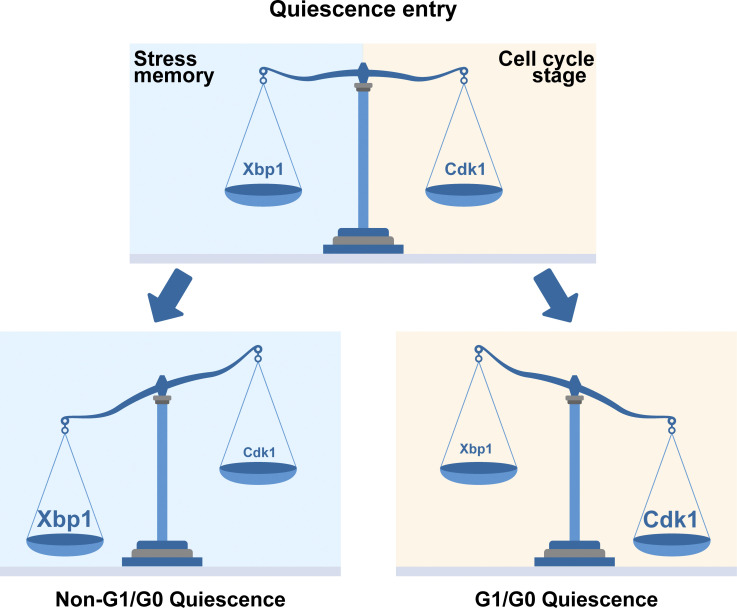
Argüello-Miranda et al. use machine learning and spectral imaging to show how the stress-activated transcriptional repressor Xbp1 can drive cellular quiescence—a reversible arrest of proliferation crucial for stress survival and development—even outside the G1 cell cycle stage.
Abstract
Cellular quiescence is a nonproliferative state required for cell survival under stress and during development. In most quiescent cells, proliferation is stopped in a reversible state of low Cdk1 kinase activity; in many organisms, however, quiescent states with high-Cdk1 activity can also be established through still uncharacterized stress or developmental mechanisms. Here, we used a microfluidics approach coupled to phenotypic classification by machine learning to identify stress pathways associated with starvation-triggered high-Cdk1 quiescent states in Saccharomyces cerevisiae. We found that low- and high-Cdk1 quiescent states shared a core of stress-associated processes, such as autophagy, protein aggregation, and mitochondrial up-regulation, but differed in the nuclear accumulation of the stress transcription factors Xbp1, Gln3, and Sfp1. The decision between low- or high-Cdk1 quiescence was controlled by cell cycle–independent accumulation of Xbp1, which acted as a time-delayed integrator of the duration of stress stimuli. Our results show how cell cycle–independent stress-activated factors promote cellular quiescence outside G1/G0.
Graphical Abstract
Introduction
Quiescence is a nonproliferative cellular state regulated by stimuli including starvation, DNA damage, and developmental signals (Cheung and Rando, 2013; van Velthoven and Rando, 2019). In unicellular organisms, quiescent states are essential for surviving adverse environmental conditions ranging from starvation to antibiotic exposure (Rittershaus et al., 2013). In multicellular organisms, quiescence is essential for tissue homeostasis and the maintenance of adult stem cells (Sun and Buttitta, 2017; Bainbridge, 2013; Apte et al., 2015; Nakamura-Ishizu et al., 2014; Li and Clevers, 2010).
Quiescence in eukaryotes commonly occurs after cells halt proliferation in a state where the activity of the proliferation-promoting kinase Cdk1 is low (low-Cdk1 quiescence). In many organisms, however, cells can enter quiescence in states of high-Cdk1 activity (high-Cdk1 quiescence), which are essential for development and single-cell survival (Wei et al., 1993; Hajeri et al., 2005; Velappan et al., 2017; Baisch, 1988). High-Cdk1 quiescence occurs in mammalian oocyte development (Nixon et al., 2002), embryonic development of invertebrates (Hajeri et al., 2005; Nystul et al., 2003; Otsuki and Brand, 2018), plant meristems (Velappan et al., 2017), microorganisms (Laporte et al., 2011; Klosinska et al., 2011), and metastasis-initiating cancer cells (Wang et al., 2015). Despite its ubiquitous nature, mechanisms for high-Cdk1 quiescence remain largely uncharacterized (Sun and Gresham, 2021).
Several conserved mechanisms maintain low-Cdk1 quiescence including (1) the accumulation of Cdk1 transcriptional repressors, such as Whi5/Rb; (2) the accumulation of Cdk1 inhibitors, such as Sic1/p27/p21; and (3) the destruction of Cdk1 activators by anaphase promoting complex/cyclosome (APC/C)-Cdh1–mediated proteolysis (Moser et al., 2018; Cappell et al., 2016; Hopkins et al., 2017). By contrast, only a few factors promoting high-Cdk1 quiescence have been identified, such as Caenorhabditis elegans spindle assembly components SAN-1 and MDF-2 (Nystul et al., 2003) or the pseudokinase Tribbles in Drosophila melanogaster neural stem cells (Otsuki and Brand, 2018).
Stress-activated pathways are good candidates to promote quiescence regardless of Cdk1 activity due to their capacity to decrease overall translation and transcription and modulate cell cycle protein networks (Miles et al., 2013; Marion et al., 2004; Escoté et al., 2004; Wysocki et al., 2006). However, quiescence-promoting stimuli, such as starvation, often trigger multiple stress signaling pathways that are experimentally challenging to simultaneously measure in single cells (De Virgilio, 2012), and high-Cdk1 quiescent states are particularly difficult to characterize because they are less common than low-Cdk1 quiescence or occur in complex tissues (Velappan et al., 2017; Otsuki and Brand, 2018). These challenges can be overcome by studying quiescence in a tractable unicellular model organism such as Saccharomyces cerevisiae; however, most protocols to induce quiescence require long-term batch cultures, where interactions between cells and changing chemical and physical medium parameters confound the assessment of whether stress factors are associated or causative of quiescent states (Klosinska et al., 2011). Furthermore, because single cells are not tracked in batch cultures, it remains controversial whether high-Cdk1 quiescent states in S. cerevisiae correspond to a failure of arresting cell cycle in response to starvation, a population of slowly dividing cells, or a proper cellular strategy to survive acute starvation stress after G1 exit.
In this work, we used a microfluidics assay with controlled environmental conditions to study S. cerevisiae’s transition from proliferation into quiescence after acute starvation. To characterize quiescent states with high-Cdk1 activity, we used machine learning algorithms to automatically distinguish proper high-Cdk1 quiescent states from slowly dividing or senescent cells. Quiescent states were classified based on established markers of the high-Cdk1 activity state, such as destruction of the Cdk1 inhibitor Sic1, inactivation of APC/C-Cdh1–mediated proteolysis, and assembly of the septin ring at the budding site after G1 exit (Zhang et al., 2011).
We found that high-Cdk1 quiescent states reproducibly occurred when a proliferating population was challenged by acute starvation. The frequency of high-Cdk1 quiescent cells (Q-cells) depended on the stress status of the population at starvation onset. Although all quiescent states shared a core of stress-associated processes such as the up-regulation of autophagy, protein aggregation, and mitochondrial biomass, high-Cdk1 quiescence was distinguished by increased nuclear accumulation of the general transcriptional repressor Xbp1, the nitrogen stress transcription factor Gln3, and the ribosome biogenesis factor Sfp1. The establishment of high-Cdk1 quiescence was controlled by cell cycle–independent nuclear accumulation of Xbp1, which acted as a time-delayed integrator of stress stimuli. Our results show that cell cycle–independent integration of stress stimuli by transcriptional repressors is a viable cellular response to establish quiescence outside a G1/G0 state.
Results
Acute starvation induces heterogeneous quiescent states
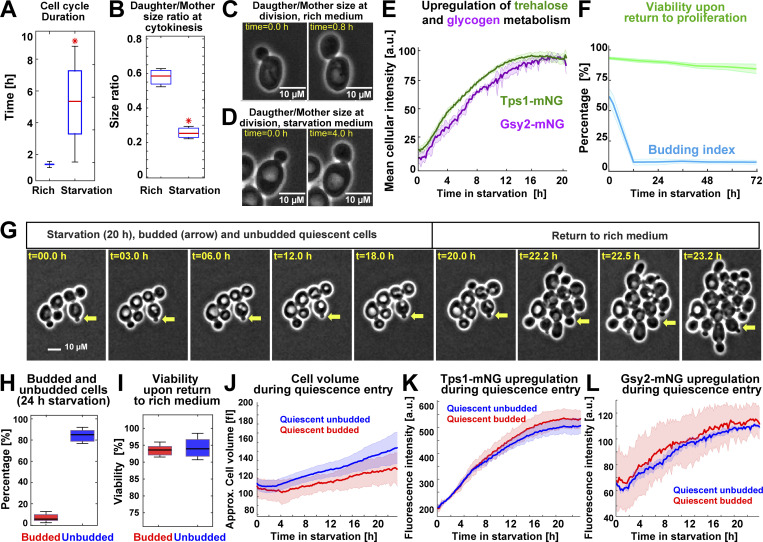
To study the establishment of quiescence in single cells, we modified a previously published microfluidics assay for quiescence induction in S. cerevisiae (Argüello-Miranda et al., 2018). Briefly, proliferating W303 diploid cells were cultured in a flow cell with rich medium (synthetic complete defined [SCD]) before being exposed to 20 h of acute starvation medium (0.6% potassium acetate, pH 7.2, 0.6 psi, which is a modified version of meiosis-inducing medium; Buonomo et al., 2000). Upon acute starvation, cells recapitulated quiescence properties previously observed in batch cultures (Miles et al., 2019). Proliferation was stopped after a slow and asymmetric final cell division, which required 5 ± 3 h under starvation conditions instead of 93 ± 7 min in SCD (Fig. 1 A). The daughter/mother cell size ratio at cytokinesis decreased from 0.58 ± 0.5 in rich medium to 0.25 ± 0.04 for cells born after starvation (Fig. 1, B–D). As reported in quiescent batch cultures (Shi et al., 2010), cells up-regulated trehalose and glycogen metabolism during quiescence entry as measured by tracking the level of the mNeonGreen-tagged trehalose synthase catalytic subunit, Tps1, and the mNeonGreen-tagged glycogen synthase, Gsy2 (Fig. 1 E). Q-cells maintained high viability as judged by return to proliferation (Fig. 1 F; 72 h is the limit of the microfluidics device) and acquired a stress-resistant state after 20 h of starvation as judged by 94 ± 3% survival (n = 7; 443 cells) after exposure to 4 M NaCl for 4 h (Video 1). Strikingly, although most cells arrested in G1, as judged by maintaining an unbudded state, a reproducible minority of 7 ± 3% cells remained arrested in a budded state (Fig. 1, G and H), which was highly viable upon return to rich medium (Fig. 1 I) and was reminiscent of budded quiescent states previously observed in stationary cultures in carbon-limited medium (Laporte et al., 2011). Single-cell tracking of budded and unbudded Q-cells showed no significant differences in cell volume (Fig. 1 J), Tps1 or Gsy2 up-regulation (Fig. 1, K and L), or cell morphology (Fig. S1 A), as assessed by the overlap of the 95% confidence intervals. We concluded that a fraction of proliferating S. cerevisiae cells entered viable quiescent states outside G1/G0 when challenged by acute starvation.
Figure 1.
S. cerevisiae enters heterogeneous quiescent states under acute starvation. (A) WT cell cycle duration (strain OAM128) measured by the time between two consecutive cytokinesis events in rich medium (n = 13; 400 cells) or after exposure to acute starvation (n = 9; 300 cells). (B) Daughter/Mother cell size ratio at cytokinesis, measured as cell area in pixels, in rich medium (n = 14; 400 cells) or for cells that completed cell cycle under starvation (n = 9; 300 cells). (C and D) Representative phase-contrast (PC) micrographs for cell division in rich (C) and starvation (D) medium. (E) Scaled mean cellular fluorescence intensity of the trehalose metabolism marker Tps1 tagged with mNeonGreen (mNG; n = 9; 499 cells, OAM394) and the glycogen metabolism marker Gsy2-mNeonGreen (n = 9; 300 cells, OAM396) during starvation. (F) Population budding index during starvation (blue) and viability (green) of Q-cells upon return to proliferation (n = 3; 237 cells). (G) Representative PC micrographs of cells during the starvation–proliferation transition showing a budded Q-cell (yellow arrow). (H and I) Percentage of budded and unbudded Q-cells (H) after 24-h starvation (n = 6; 400 cells) and their viability measured as proliferation resumption upon return to rich medium (I). (J) Average cell volume of quiescent budded and unbudded cells during starvation. (K) Average Tps1-mNeonGreen intensity in budded and unbudded Q-cells. (L) Average Gsy2-mNeonGreen intensity in budded and unbudded Q-cells. All data from biological replicates. Red star = P < 0.05, K-S test. Solid lines with shaded area = average plus 95% confidence interval. Box plots: central mark, median; box bottom and top limit, 25th and 75th percentiles; whiskers, most extreme nonoutlier values. Approx., approximate.
Video 1.
A microfluidics assay to study the transition from proliferation into quiescence. After 90 min in rich medium, proliferating WT cells (OAM128) are exposed to starvation medium for 20 h and reached a stress-resistant quiescent state that survives exposure to 4 M NaCl for 4 h before resumption of proliferation by exposure to rich medium. Sampling rate = 12 min. Square side = 90 µM. Playback = 22 frames per second.
Figure S1.
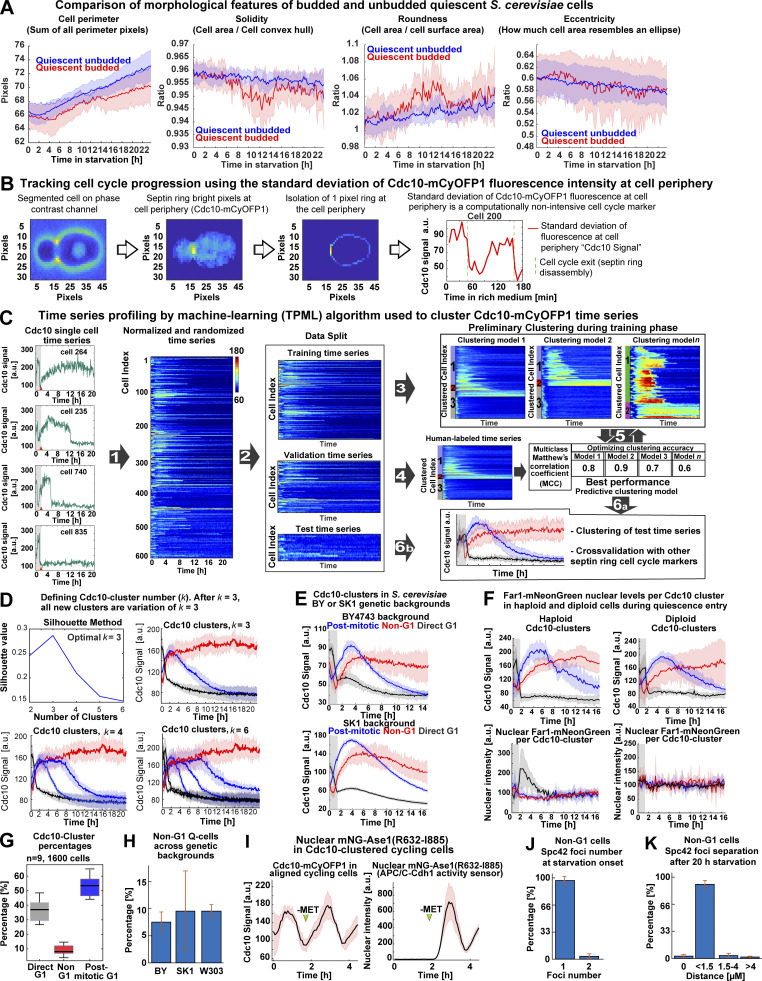
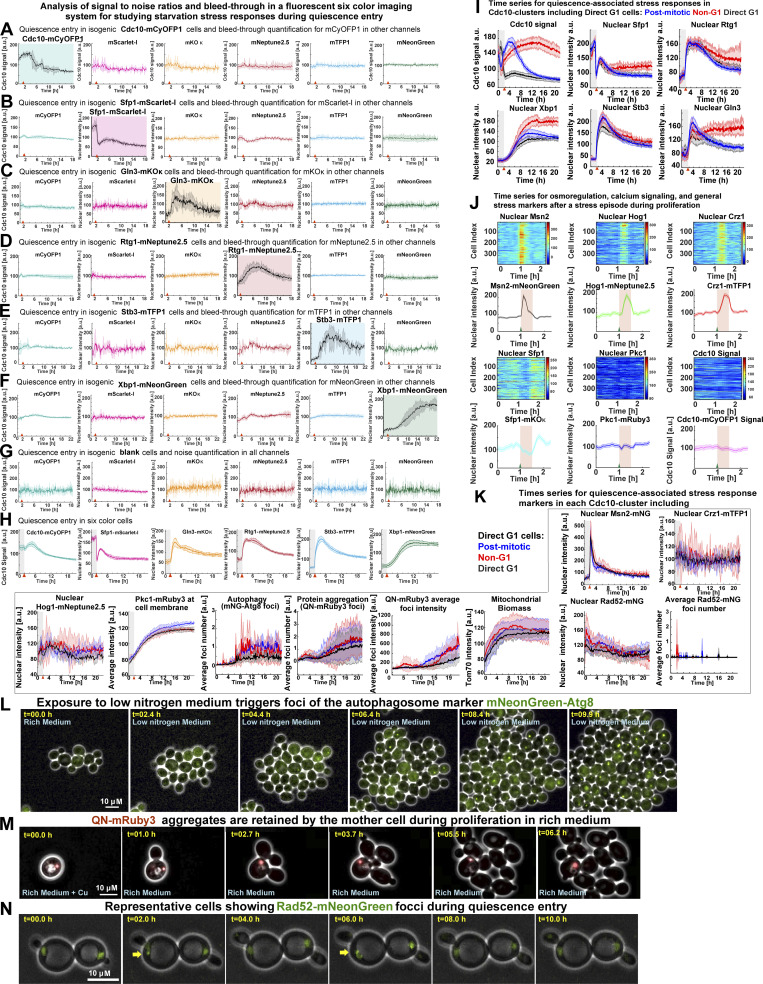
Analysis of single-cell morphological features during quiescence entry, Cdc10 signal quantification algorithm workflow, TPML algorithm workflow, cluster number analysis, mitotic induction of the APC/C-Cdh1 activity sensor MET3p-mNeonGreen (mNG)-Ase1(R632-I885), and extended Spc42 foci quantification. (A) Comparison of morphological parameters between budded and unbudded Q-cells. (B) Algorithm to detect cell cycle progression based on the SD of Cdc10-mCyOFP1 fluorescence at the cell periphery or “Cdc10 Signal a.u.” (C) Generation of a machine learning algorithm to sort cells into clusters depending on the pattern of cell cycle arrest indicated by Cdc10. Left: Representative single-cell time series for Cdc10-mCyOFP1 signal (OAM421). (1) Time series are normalized per biological replicate, and their cell index is randomized. (2) Time series data are split into 50% training time series, 30% validation time series, and 20% test time series. (3) Training time series are used to establish different clustering models depending on parameters such as clustering method (k-means, k-medoids), time window (number of time points in time series), number of clusters, and distance metric. (4) Validation time series are labeled by experienced cell biologists who assign different cluster affiliations to each time series manually. (5) The preliminary clustering models are optimized by comparing the unsupervised clustering solutions to the human clustering solution using a multiclass Matthews’s coefficient. (6a) The best-performing clustering model is tested using the (6b) test time series. The best-performing algorithm is used to classify cells in future experiments. (D) Evaluation of the number of clusters using the silhouette method or by inspecting increasing number of clusters (k values). k = 3 is the minimum number of clusters that recapitulate the major cell cycle arrest patterns in the population. (E) Average time series for Cdc10 clusters in different S. cerevisiae strain backgrounds (Up, BY4743, OAM857; down, SK1, OAM856) bearing Cdc10-mCyOFP1. (F) Average time series in haploid (left, OAM846) and diploid (right, OAM840) strains bearing Cdc10-mCyOFP1 and Far1-mNeonGreen (n = 3; at least 400 cells per strain). (G) Percentage of each Cdc10 cluster in a representative population undergoing quiescence upon acute stress (n = 9; 1,600 cells, OAM425). (H) Percentage of Non-G1 Q-cells in three S. cerevisiae strain backgrounds (n = at least 4 biological replicates). (I) Induction of the APC/C-Cdh1 activity sensor, mNeonGreen-Ase1(R632-I885), upon depletion of methionine (MET) in rich medium (green arrowhead); OAM487 cells were aligned in silico according to their Cdc10 signal, and the average times series was plotted, Cdc10 (left) and APC/C-Cdh1 sensor (right). (J) Percentage of Non-G1 Q-cells displaying one or two Spc42-mTFP1 foci at starvation onset. (K) Percentage of Non-G1 Q-cells displaying a distance separation between Spc42-mTFP1 foci of 0 (single foci), <1.5 (S-G2), 1.5–4 (metaphase), or >4 (anaphase) µM. Solid lines with shaded area = average ± 95% confidence intervals. Box plots display data from biological replicates: central mark, median; box bottom and top limit, 25th and 75th percentiles; whiskers, most extreme nonoutlier values. Bar plots = mean + SD.
Acute starvation induces quiescence in states of low- or high-Cdk1 activity
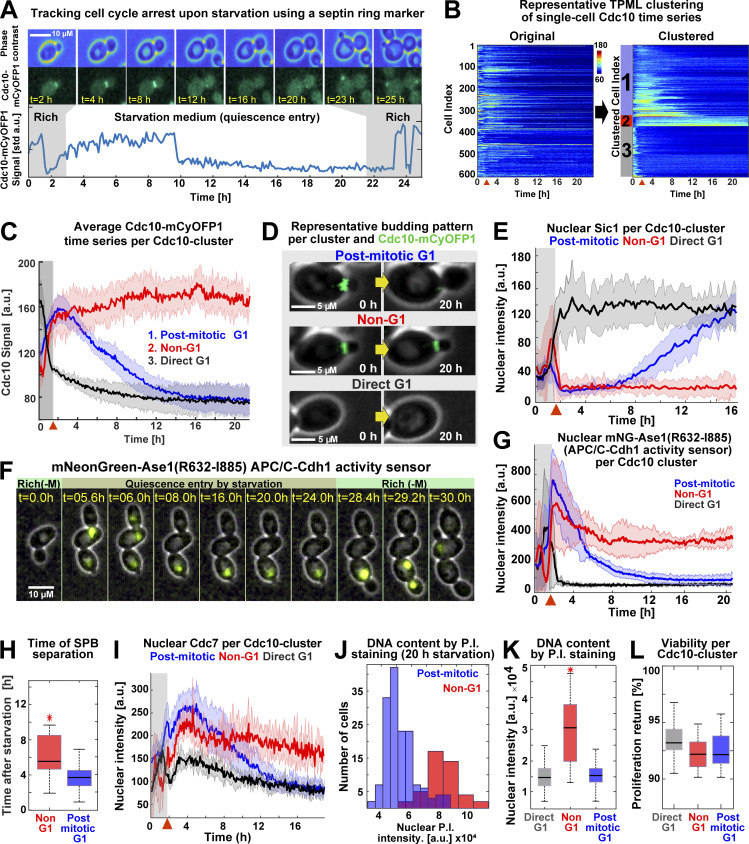
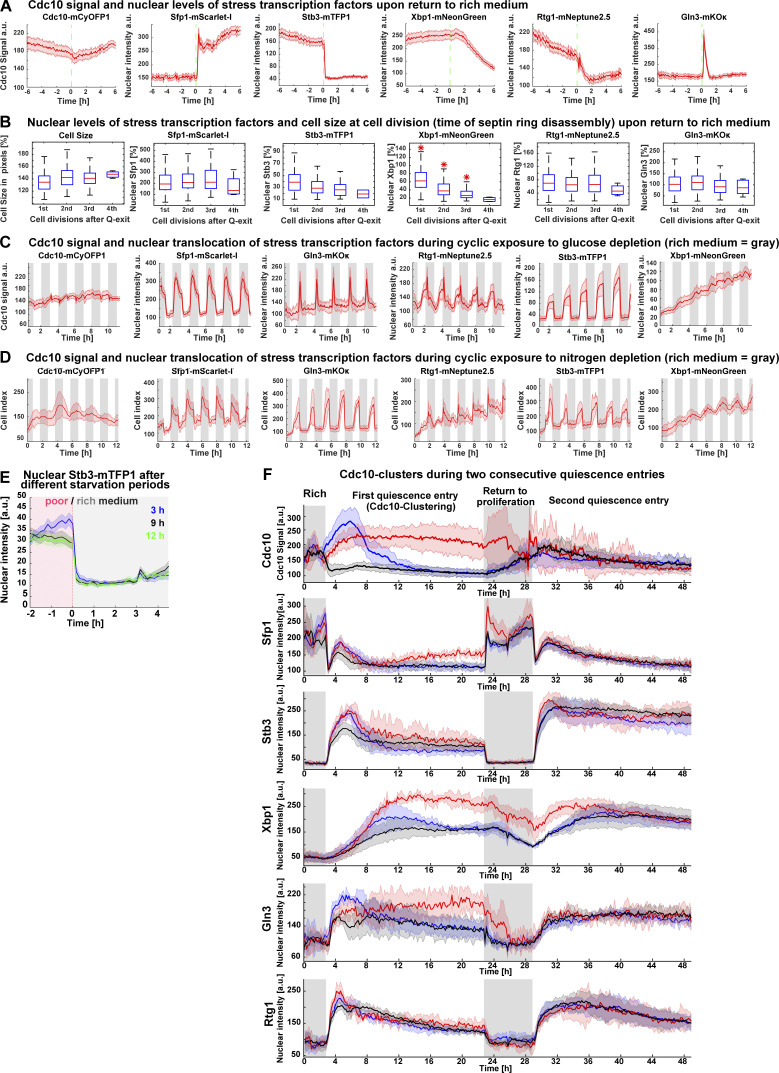
To analyze the Cdk1 activity state of budded and unbudded Q-cells, we tracked the formation of the septin ring at the bud site—which is absent during the low-Cdk1 state of G1 but present during the high-Cdk1 state of S-M phase—by C-terminally tagging the septin Cdc10 with the fluorophore mCyOFP1 (McMurray and Thorner, 2009). The presence of the septin ring was measured by the SD of Cdc10 fluorescence at the cell periphery (Cdc10 signal; Fig. 2 A and Fig. S1 B). To analyze the Cdc10 signal of single cells during the proliferation–quiescence transition, we developed the algorithm “Time series Profiling by Machine Learning” (TPML), which groups single-cell time series according to similarity of cell cycle arrest into hierarchically sorted heatmaps (Fig. 2 B; Fig. S1, C and D; and Video 2). TPML analysis of Cdc10-mCyOFP1 time series established three major groups or “Cdc10 clusters” named according to whether cells reached a G1 cell cycle arrest during the proliferation–quiescence transition. Cells in cluster one—or “post-mitotic G1”—were in a high-Cdk1 state at starvation onset, as judged by the presence of the septin ring, and finished one last division before arresting in G1 (Fig. 2, C and D, blue). Cells in cluster two—or “Non-G1”—were also in a high-Cdk1 state at starvation onset but remained arrested in this state without reaching G1 conditions, as judged by a persistent septin ring (Fig. 2, C and D, red). Cells in cluster three—or “direct G1”—were in the low-Cdk1 state of G1 at starvation onset and remained arrested in this state (Fig. 2, C and D, black). Cdc10-cluster classification was independent of ploidy or strain genetic background (Fig. S1, E–H).
Figure 2.
Acute starvation induces low- and high-Cdk1 quiescent states. (A) Schematic of cell cycle tracking during starvation using the septin Cdc10-mCyOFP1 signal (OAM421). (B) Representative heatmaps showing TPML arrangement of single-cell Cdc10 time series into Cdc10 clusters corresponding to the main patterns of cell cycle arrest upon starvation. (C) Average Cdc10 time series for each Cdc10 cluster (n = 6; 633 cells). (D) Representative micrographs of budding patterns per Cdc10 cluster during starvation. Maximum intensity projection (MIP) mCyOFP1 image superimposed on phase contrast. Yellow arrows represent passage of time. (E) Average nuclear Sic1 intensity per Cdc10 cluster (n = 3; 341 cells, OAM672). (F) Representative micrographs of the APC/C-Cdh1 activity sensor mNeonGreen-Ase1(R632-I885) during starvation in post-mitotic (up) and Non-G1 (down) Q-cells (OAM487). MIP mNeonGreen (mNG) image superimposed on phase contrast. Rich(−M), rich medium minus methionine. (G) Average mNeonGreen-Ase1(R632-I885) nuclear intensity per Cdc10 cluster (n = 3; 255 cells). (H) Time of SPB separation in Non-G1 and post-mitotic G1 Q-cells upon starvation (n = 3; 353 cells, OAM423). (I) Average Cdc7–mScarlet-I nuclear intensity per Cdc10 cluster (n = 3; 345 cells, OAM422). (J and K) Representative nuclear intensity histograms (J) and comparison of nuclear fluorescence intensity per Cdc10 cluster (K) after propidium iodide (P.I.) staining (n = 3; 304 cells, OAM421). (L) Average viability of cells in each Cdc10 cluster upon return to rich medium after 24 h of starvation (n = 3; 565 cells, OAM421). All data from biological replicates. Red arrowhead = onset of starvation. Solid lines with shaded area = average ± 95% confidence intervals. Red star = P < 0.05, K-S test. Box plots display data from biological replicates: central mark, median; box bottom and top limits, 25th and 75th percentiles; whiskers, most extreme nonoutlier values. std, SD.
Video 2.
TPML training on single-cell time series (Cdc10-mCyOFP1 data). In this example, TPML finds the best interval (number of time points) for clustering cells according to cell cycle arrest pattern. Each frame corresponds to a potential clustering solution evaluated against a human-labeled validation dataset using the MCC. Clustering solutions with the highest MCC are further optimized and eventually converge in a clustering solution (see Fig. S1, B and C). tp, time point. Playback = 8 frames per second.
To assess the Cdk1 activity state of Cdc10 clusters, we imaged quiescence entry in cells bearing Cdc10-mCyOFP1 as a marker for the high-Cdk1 state of S-M phase and mNeonGreen C–terminally tagged Sic1 as a marker for the low-Cdk1 state of G1 (Hopkins et al., 2017). After starvation onset, nuclear Sic1 accumulated in post-mitotic G1 and direct G1 Q-cells according to their cell cycle progression (Fig. 2 E, black, blue). In contrast, nuclear Sic1 did not accumulate in Non-G1 Q-cells (Fig. 2 E, red), suggesting persistent Cdk1 activity.
The high-Cdk1 kinase state of S-M phase is characterized by the inactivation of APC/C-Cdh1–mediated proteolysis (Zachariae et al., 1998). To measure the activity of APC/C-Cdh1 in Q-cells, we tracked the nuclear levels of the APC/C-Cdh1 activity sensor mNeonGreen-Ase1(R632-I885), expressed from the MET3 promoter (Ondracka et al., 2016). During proliferation, the accumulation–degradation of the sensor followed the Cdc10 signal as expected for APC/C-Cdh1 substrates (Fig. S1 I). After starvation, the APC/C-Cdh1 sensor was degraded in post-mitotic G1 and direct G1 Q-cells (Fig. 2, F and G, black, blue) but persisted in Non-G1 Q-cells (Fig. 2, F and G, red), indicating APC/C-Cdh1 inactivation. Consistent with this observation, spindle pole body (SPB) separation, a process that requires APC/C-Cdh1 inactivation (Crasta et al., 2006), occurred, although with a delay, in Non-G1 Q-cells as judged by tracking of the SPB component Spc42 tagged with mTFP1 (Fig. 2 H; and Fig. S1, J and K).
To assess whether Non-G1 Q-cells entered S-phase, we tracked the DNA replication kinase Cdc7 C-terminally tagged with mScarlet-I. During starvation, nuclear Cdc7 persisted at higher levels in Non-G1 Q-cells (Fig. 2 I, red) in comparison to the other clusters (Fig. 2 I, blue, black). To measure DNA content in Q-cells, Cdc10-mCyOFP1 cells were microfluidically stained with propidium iodide after 20 h in starvation medium. On average, Non-G1 Q-cells had duplicated DNA content (Fig. 2, J and K).
Together, these results indicated that Non-G1 Q-cells persisted in a state of high-Cdk1 activity as judged by APC/C-Cdh1 inactivation, SPBs separation, DNA replication, and lack of Sic1 accumulation. High-Cdk1 quiescent states were equally viable to low-Cdk1 quiescence, as judged by comparing proliferation resumption among Cdc10 clusters (Fig. 2 L). We concluded that proliferating S. cerevisiae cells challenged by acute starvation after exiting G1 faced a decision-making process between remaining arrested in a high-Cdk1 state or completing a final cell cycle to reach a low-Cdk1 quiescent state in G1. In the rest of this study, we focused on cells experiencing starvation after G1 exit (post-mitotic and Non-G1 Cdc10 clusters) and used the TPML algorithm to classify them into “low-Cdk1 quiescence” if they finished one last division or “high-Cdk1 quiescence” if they remained arrested.
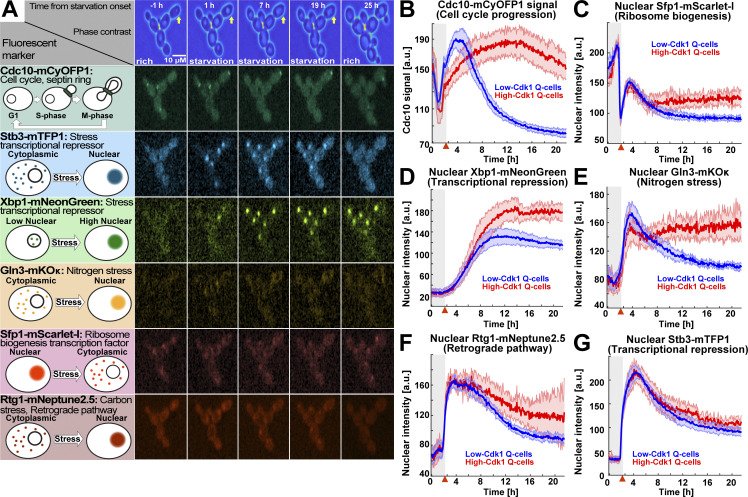
Low- and high-Cdk1 quiescent states share a core of stress responses but differ in the nuclear accumulation of Sfp1, Gln3, and Xbp1
To study how single cells challenged by starvation after G1 exit subsequently enter low- or high-Cdk1 quiescent states, we used a six-color fluorescent imaging system to assess to what extent differences in cell cycle progression or stress-activated factors determined high-Cdk1 quiescence entry (Argüello-Miranda et al., 2018). Imaging was optimized for using Cdc10-mCyOFP1 as cell cycle marker while simultaneously tracking five reporters for the major stress responses associated with the establishment of quiescence (Fig. 3 A; Fig. S2 A–H; and Video 3). Metabolic stress was measured by the nuclear translocation of the nitrogen stress transcriptional regulator Gln3 (Crespo et al., 2002) and the carbon stress/retrograde pathway transcriptional regulator Rtg1 (Komeili et al., 2000). The biosynthetic capacity of the cell was tracked using the nuclear translocation of the ribosome biogenesis factor Sfp1 (Marion et al., 2004). Transcriptional repression was tracked using the histone deacetylase regulators Stb3 and Xbp1, which are key transcriptional repressors during quiescence (Mai and Breeden, 1997; McKnight et al., 2015).
Figure 3.
The stress transcription factors Sfp1, Gln3, and Xbp1 are selectively up-regulated during high-Cdk1 quiescence entry. (A) Left: Schematic nuclear accumulation of fluorescently tagged transcription factors used as sensors for stress responses. Right: Representative MIP micrographs of the six fluorescent channels imaged in 6C1 cells upon starvation. Yellow arrow = high-Cdk1 Q-cell. (B–G) Quantification of cell cycle progression and stress responses in low-Cdk1 (blue) or high-Cdk1 (red) Q-cells upon starvation (n = 9; 825 cells, OAM425). (B) Average Cdc10-mCyOFP1 time series per cluster. (C) Average nuclear intensity of the ribosome biogenesis factor Sfp1–mScarlet-I per cluster. (D) Average nuclear intensity of the transcriptional repressor Xbp1-mNeonGreen per cluster. (E) Average nuclear intensity of the nitrogen stress response regulator Gln3-mKOκ per cluster. (F) Average nuclear intensity of the carbon/retrograde pathway regulator Rtg1-mNeptune2.5 per cluster. (G) Average nuclear intensity of the transcriptional repressor Stb3-mTFP1 per cluster. Red arrowheads = onset of starvation. Solid lines with shaded area = average ± 95% confidence intervals.
Figure S2.
Analysis of signal-to-noise ratios and bleed-through in the six-color imaging system, analysis of osmoregulatory responses in 6C2 cells, and detection of mNeonGreen (mNG)-Atg8, QN-mRuby3, or Rad52-mNeonGreen foci. (A–H) Assessment of signal-to-noise ratios and bleed-through in a six-color fluorescent microscopy system by imaging quiescence entry in isogenic single fluorophore–tagged strains for Cdc10-mCyOFP1 (OAM421; A) and the stress markers Sfp1–mScarlet-I (OAM407; B), Gln3-mKOκ (OAM507; C), Rtg1-mNeptune2.5 (OAM510; D), Stb3-mTFP1 (OAM395; E), Xbp1-mNeonGreen (OAM397; F), and isogenic blank cells (OAM128; G) or an isogenic six-color strain (OAM425; H); at least 75 cells per strain were analyzed from three different biological replicates; each channel is normalized to a mean of 100 a.u. Plot with colored background = channel containing a fluorescent signal. Plot with white plot background = channel without a fluorescent signal. Red arrowhead = onset of starvation. (I) Average time series for Cdc10 clusters, including direct G1 cells, for the cell cycle marker Cdc10-mCyOFP1 and the stress response factors Sfp1–mScarlet-1, Rtg1-mNeptune2.5, Xbp1-mNeonGreen, Stb3-mTFP1, and Gln3-mKOκ. (J) Up-regulation of osmotic/mechano-sensitive stress responses by 200 mM CaCl2 in unsynchronized 6C2 cells (OAM667) bearing the stress markers Crz1-mTFP1, Pkc1-mRuby3, Msn2-mNeonGreen, Sfp1-mKOκ, Hog1-mNeptune2.5, and Cdc10-mCyOFP1. Heatmaps display unsynchronized single-cell time series. The plots display average time series (n = 4; 364 cells). Red area, period in 200 mM CaCl2-containing rich medium. Green triangle, onset of CaCl2 exposure. (K) Average time series for quiescence-associated stress responses in Cdc10 clusters, including the nuclear accumulation of Msn2, Crz1, Hog1, and Rad52; the cell membrane association of Pkc1; up-regulation of mNeonGreen-Atg8; and QN-mRuby3 foci or Tom70-mTFP1 total intensity. (L) Representative micrographs showing the accumulation of the autophagy reporter mNeonGreen-Atg8 upon proliferation in low-nitrogen medium (SCD without ammonium sulfate, OAM654). (M) Representative time-lapse micrographs of a mother cell retaining a fluorescent QN-mRuby3 aggregate during proliferation in rich medium after 2 h in 25 µM CuSO4-containing SCD (OAM671). (N) Representative time-lapse micrographs of cells (OAM670) with recurring Rad52-mNeonGreen foci (yellow arrows) during starvation. Solid lines with shaded area = average ± 95% confidence intervals. Red arrowhead = onset of starvation.
Video 3.
Six-color imaging of the proliferation–quiescence transition in 6C1 cells (OAM425) carrying the cell cycle marker Cdc10-mCyOFP1 and the stress response markers Sfp1–mScarlet-I, Gln3-mKOκ, Rtg1-mNeptune2.5, Stb3-mTFP1, and Xbp1-mNeonGreen. Each peripheral hexagon displays a median-filtered fluorescent (Fl.) channel. Each subplot corresponds to a time series obtained from the representative high-Cdk1 Q-cell highlighted in yellow. Sampling rate = 12 min. Hexagon side = 65 µM. Playback = 60 frames per second.
Sfp1-Stb3-Rtg1-Gln3-Xbp1-Cdc10 (6C1) cells modulated the nuclear levels of stress markers upon starvation (Fig. S2 H). Low- and high-Cdk1 quiescent states were identified by TPML, and the differences in the average time series of each group were assessed using the overlap in the 95% confidence interval. Strikingly, the most significant difference between low- and high-Cdk1 Q-cells was not cell cycle progression at starvation onset (Fig. 3 B), but the accumulation of nuclear Sfp1 (Fig. 3 C), Xbp1 (Fig. 3 D), and Gln3 (Fig. 3 E). In contrast, nuclear Rtg1 (Fig. 3 F) and Stb3 (Fig. 3 G) were accumulated to the same extent in low- and high-Cdk1 Q-cells (Fig. S2 I).
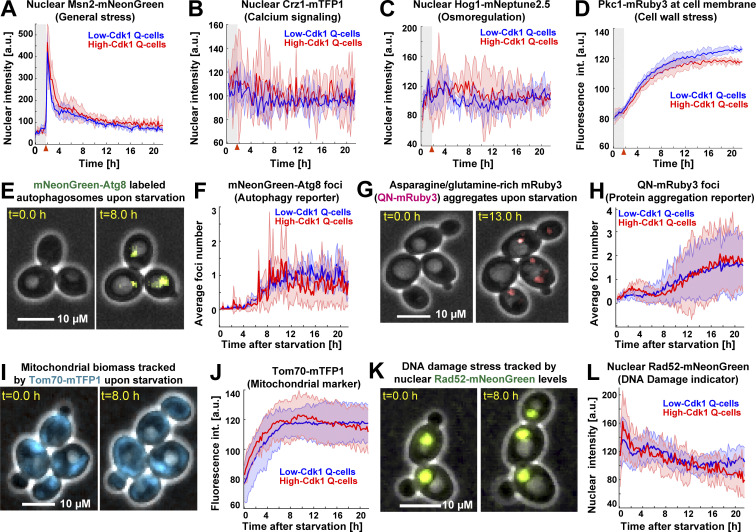
To assess whether low- and high-Cdk1 Q-cells presented differences in stress responses not directly related to starvation, we imaged a second six-color (6C2) strain bearing fluorescent reporters for calcium signaling (Crz1-mTFP1; Cyert, 2003), cell wall stress (Pkc1-mRuby3; Mishra et al., 2017), osmoregulation (Hog1-mNeptune2.5; Westfall et al., 2004), general stress (Msn2-mNeonGreen, Sfp1-mKOκ; Granados et al., 2018), and cell cycle (Cdc10-mCyOFP1). 6C2 cells readily reported stress stimuli if perturbed during proliferation (Fig. S2 J). Upon starvation, the nuclear levels of Msn2 (Fig. 4 A), Crz1 (Fig. 4 B), and Hog1 (Fig. 4 C) and the cell membrane-associated levels of Pkc1 (Fig. 4 D) were similar in low- and high-Cdk1 Q-cells.
Figure 4.
Low- and high-Cdk1 quiescent states share a core of stress-associated processes. (A–D) Quantification of nonmetabolic stress responses in low-Cdk1 (blue) or high-Cdk1 (red) Q-cells upon starvation (n = 3; 255 cells, OAM667). (A) Average nuclear intensity of the general stress transcription factor Msn2-mNeonGreen per cluster. (B) Average nuclear intensity of the calcium stress transcription factor Crz1-mTFP1 per cluster. (C) Average nuclear intensity of the osmotic stress kinase Hog1-mNeptune2.5 per cluster. (D) Average intensity (int.) of the cell wall stress kinase Pkc1-mRuby3 at cell membrane per cluster. (E) Representative micrographs of cells bearing the autophagy reporter mNeonGreen-Atg8 upon starvation. MIP mNeonGreen image superimposed on phase contrast. (F) Average number of mNeonGreen-Atg8 foci per cluster upon starvation (n = 3; 450 cells, OAM654). (G) Representative micrographs of cells bearing the protein aggregation sensor QN-mRuby3 upon starvation. MIP fluorescent mRuby3 image superimposed on phase contrast. (H) Average number of QN-mRuby3 foci per cluster upon starvation (n = 3; 333 cells, OAM671). (I) Representative micrographs of cells bearing the mitochondrial marker Tom70-mTFP1 upon starvation. MIP mTFP1 image superimposed on phase contrast. (J) Average total intensity of Tom70-mTFP1 per cluster upon starvation (n = 3; 225 cells, OAM673). (K) Representative micrographs of cells bearing the DNA damage marker Rad52-mNeonGreen upon starvation. MIP mNeonGreen image superimposed on phase contrast. (L) Average Rad52-mNeonGreen nuclear intensity per cluster upon starvation (n = 3; 240 cells, OAM670). Solid lines with shaded area = average ± 95% confidence intervals. Red arrowheads = onset of starvation.
To investigate the status of quiescence-associated processes (Sagot and Laporte, 2019; Laporte et al., 2018) in low- and high-Cdk1 Q-cells, we induced starvation in Cdc10-mCyOFP1 strains containing fluorescent sensors for autophagy, protein aggregation, mitochondrial biomass, and DNA damage. Autophagy was measured by the foci number of an autophagosome marker, mNeonGreen-Atg8, expressed from its own promoter (Fig. S2 L; Nair et al., 2011). Upon starvation, Atg8 foci increased with no difference between quiescent states (Fig. 4, E and F). Protein aggregation was measured using a glutamine-asparagine–rich peptide (Antonets et al., 2016) bound to an mRuby3 fluorophore (QN-mRuby3) under the control of a copper-inducible promoter (CUP1-1). CUP1 promoter (CUP1p)expression of QN-mRuby3 during proliferation produced few foci that remained within mother cells, as expected from protein aggregates in S. cerevisiae (Fig. S2 M; Saarikangas et al., 2017). By contrast, expression of QN-mRuby3 during quiescence led to several foci per cell independent of cell cycle progression (Fig. 4, G and H). Total mitochondrial biomass, as measured by the mean fluorescent intensity of the translocase Tom70 (Hughes et al., 2016), showed no difference between quiescent states (Fig. 4, I and J). Rad52 nuclear levels and foci, which increase upon DNA damage (Alvaro et al., 2007), were not significantly different between quiescent states (Fig. 4, K and L).
These results showed that although quiescent states shared a core of stress-associated processes (Fig. S2 K), high-Cdk1 quiescence entry was characterized by increased nuclear accumulation of Sfp1, Gln3, and Xbp1.
High-Cdk1 quiescence entry is determined by the stress status of the cell
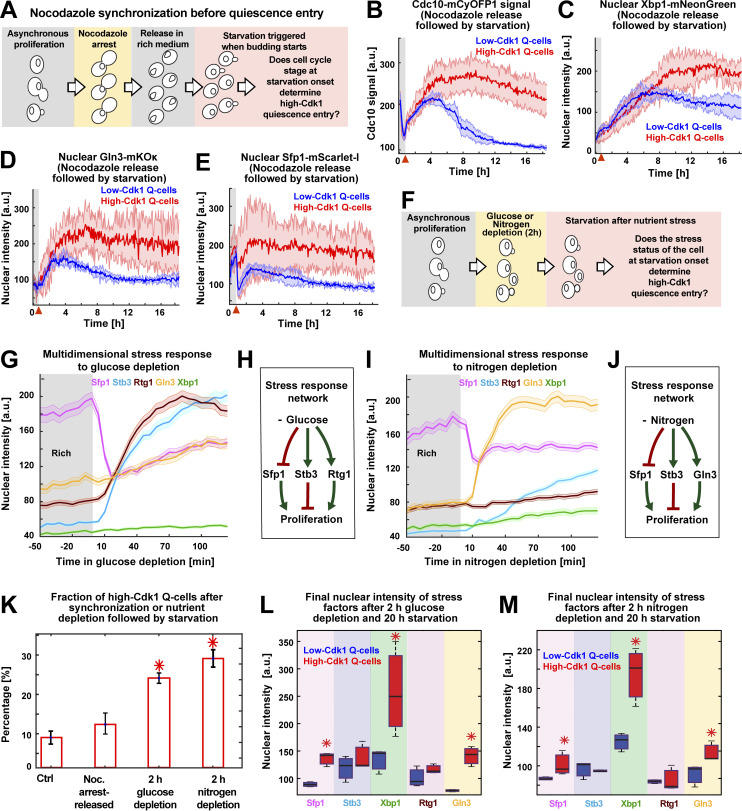
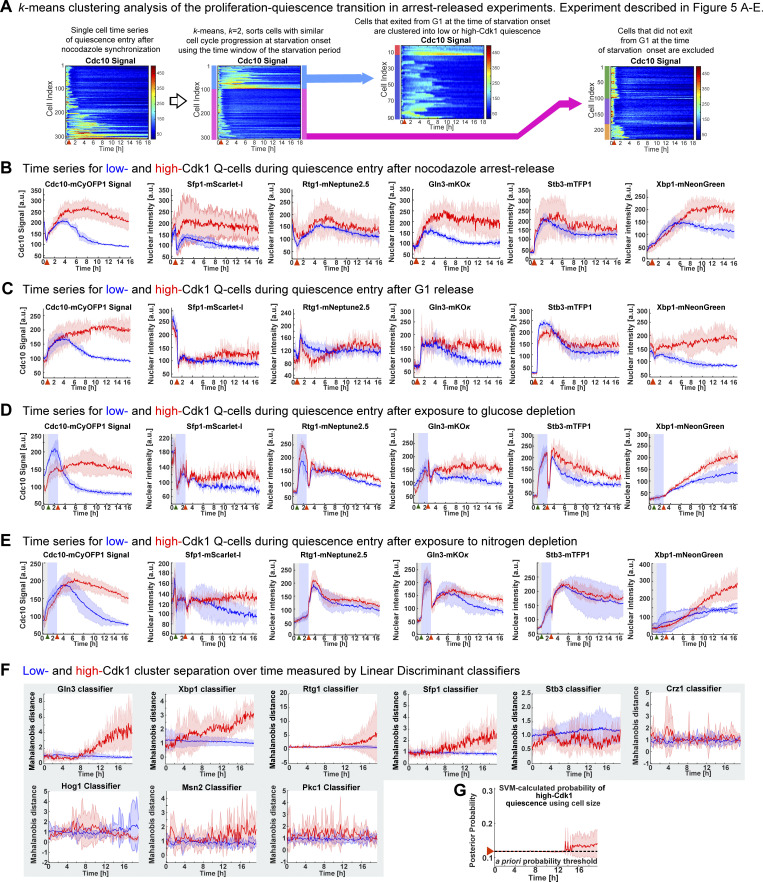
The increased nuclear accumulation of Xbp1, Gln3, and Sfp1 in high-Cdk1 Q-cells suggested that the stress status of the cell, rather than differences in cell cycle progression, promoted high-Cdk1 quiescence entry upon starvation. To test this hypothesis, 6C1 cells were cell cycle synchronized with a nocodazole arrest-release protocol before exposure to starvation (Fig. 5 A) and then clustered using TPML to identify cells with the same cell cycle synchrony around starvation onset (Fig. S3 A). This revealed that cells with identical synchrony, as judged by the Cdc10 signal overlap, could diverge into low- or high-Cdk1 quiescent states (Fig. 5 B and Fig. S3 B). A G1 synchronization protocol confirmed this result (Fig. S3 C). As in unsynchronized cultures, high-Cdk1 Q-cells accumulated higher nuclear levels of Xbp1 (Fig. 5 C), Gln3 (Fig. 5 D), and Sfp1 (Fig. 5 E), but not Rtg1 or Stb3 (Fig. S3 B). This showed that high-Cdk1 quiescence was not defined by cell cycle stage at starvation onset.
Figure 5.
High-Cdk1 quiescence entry is determined by the stress status of the cell during starvation. (A) Schematic of nocodazole cell cycle synchronization before starvation onset. (B–E) Average time series for low-Cdk1 (blue) and high-Cdk1 (red) Q-cells with the same cell cycle progression around starvation onset after nocodazole arrest-release. Average time series for Cdc10 (B) and the nuclear intensity of Xbp1 (C), Gln3 (D), and Sfp1 (E) in nocodazole arrest-released cells with same cell cycle progression at starvation onset (n = 3; 96 cells). (F) Schematic of stress induction before starvation onset. (G) Nuclear intensity of stress factors as population average upon glucose depletion before starvation (504 cells). (H) Schematic of glucose depletion–induced stress response network. (I) Nuclear intensity of stress factors as population average upon nitrogen depletion before starvation (222 cells). (J) Schematic of nitrogen depletion–induced stress response network. (K) Percentage of high-Cdk1 Q-cells upon 20-h starvation in 6C1 cells with the following treatments before starvation: unsynchronized and unstressed control (Ctrl); nocodazole-synchronized (Noc.); 2-h glucose depletion; 2-h nitrogen depletion (n = 7 per treatment). (L and M) Quantification of the final nuclear intensity of stress markers after 20-h starvation in low-Cdk1 (blue) and high-Cdk1 (red) Q-cells pretreated with glucose (L) or nitrogen (M) depletion for 2 h before starvation (n = 7; >233 cells). Solid lines with shaded area = average ± 95% confidence intervals. Red arrowheads = onset of starvation. Red star = P < 0.05, K-S test. Bar plots = mean ± SD. Box plots display data from biological replicates: central mark, median; box bottom and top limit, 25th and 75th percentiles; whiskers, most extreme nonoutlier values.
Figure S3.
Analysis of quiescence entry after cell cycle synchronization or nutrient depletion and linear discriminant analysis of cluster separation by stress factors. (A) A two-step k means–based algorithm to sort time series before TPML. Heatmaps = single-cell time series derived from 6C1 cells that were M-phase (nocodazole) synchronized before starvation. Clustering identifies cells that exited from G1 shortly after starvation onset. (B) Average time series for cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, Stb3, and Xbp1) in M-phase arrest-released 6C1 cells that exited G1 shortly before starvation onset (n = 3; 96 cells, OAM425). (C) Average time series for cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, Stb3, and Xbp1) in G1-released 6C1 cells that exited G1 shortly before starvation onset (n = 3; 86 cells, OAM425). (D) Average time series of cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, Stb3, and Xbp1) in 6C1 cells that were exposed to glucose depletion for 2 h before quiescence entry (n = 3; 504 cells, OAM425). (E) Average time series of cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, Stb3, and Xbp1) in 6C1 cells that were exposed to nitrogen depletion for 2 h before quiescence entry (n = 3; 222 cells, OAM425). (F) Average cluster separation at each time point between low- and high-Cdk1 Q-cells, as measured by the Mahalanobis distance calculated using LDCs based on the time series for each stress marker derived from OAM425 (Fig. 3) and OAM667 (Fig. 4) cells. (G) Average probability of high-Cdk1 quiescence assigned to high-Cdk1 Q-cells by SVMs based on final cell size after rich-starvation medium transition. Solid lines with shaded area = average ± 95% confidence intervals from biological replicates. Green arrowhead = onset of nutrient depletion. X-axis red arrowheads = onset of starvation. Y-axis red arrowhead with dotted line = a priori probability.
To assess whether high-Cdk1 quiescence was predisposed by the stress status of the cell at starvation onset, 6C1 cells were exposed to medium lacking glucose or nitrogen for 2 h before starvation (Fig. 5 F). Glucose depletion generated a multidimensional stress response characterized by down-regulation of nuclear Sfp1 and up-regulation of Stb3 and Rtg1 (Fig. 5, G and H). Nitrogen depletion produced a different multidimensional stress response with nuclear translocations for Gln3, Sfp1, and Stb3 (Fig. 5, I and J). Triggering acute starvation from these two different stress states led to a significant increase of high-Cdk1 Q-cells (P < 0.05; Fig. 5 K) with high final levels of nuclear Xbp1, Gln3, and Sfp1 (Fig. 5, L and M; and Fig. S4, D and E).
Figure S4.
Sorting of time series according to the presence/absence of a fluorescent signal, analysis of quiescence entry in cells CUP1p-expressing transcriptional repressors, and in xbp1Δ cells. (A) Sorting of mixed time series from cells expressing mTFP1-tagged transcription factors and control cells without the mTFP1 construct that were loaded in the same microfluidics device (see Fig. 7). (B–E) Analysis of quiescence entry in strains bearing Gln3-mKOκ, Rtg1-mNeptune2.5, Sfp1–mScarlet-I, Xbp1-mNeonGreen, and Cdc10-mCyOFP1 after CUP1p-expression of mTFP1 C-terminally tagged WT Xbp1 (n = 6; OAM458), the DNA-binding domain mutant xbp1-EE (n = 7; OAM466), the Xbp1-related transcriptional repressor Stb3 (n = 4; OAM497), or the cell cycle repressor Whi5 (n = 7; OAM455) at starvation onset. (B) Average time series per cluster for cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, and Xbp1) in cells expressing CUP1p-XBP1-mTFP1 during quiescence entry. Cells whose Xbp1-mTFP1 levels exceeded two SDs from the mean Xbp1-mTFP1 levels were excluded. (C) Average time series per cluster for cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, and Xbp1) in cells expressing CUP1p-xbp1-EE-mTFP1 during quiescence entry. (D) Average time series per cluster for cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, and Xbp1) in cells expressing CUP1p-STB3-mTFP1 during quiescence entry. (E) Average time series per cluster for cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, and Xbp1) in cells expressing CUP1p-WHI5-mTFP1 during quiescence entry. (F) Average time series per cluster for cell cycle (Cdc10 signal) and stress markers (Sfp1–mScarlet-I, Gln3-mKOκ, Rtg1-mNeptune2.5, Msn2-mNeonGreen, and Stb3-mTFP1) in six-color xbp1Δ cells (n = 4; 500 cells, OAM500). Unlike XBP1 strains, quiescence entry in xbp1Δ cells could not be described as three main clusters; instead, TPML sorted xbp1Δ cells into four clusters (four-color bar) and cells undergoing ectopic divisions. (G) Percentage of cells entering ectopic proliferation during starvation in control (Ctrl; OAM502) and xbp1Δ (OAM500) cells. (H) Schematic quantification of quiescence entry in control (OAM502) and xbp1Δ (OAM500) cells, including the percentage of cells with ectopic divisions in each cluster. Red arrows indicate paths with <75% viability upon return to rich medium (n > 3). (I) Viability of direct G1 and post-mitotic Q-cells in WT (left) and xbp1Δ cells (right) upon return to rich medium after 20-h starvation. Gray area on plot = period in rich medium. Red arrowhead = onset of starvation. Red star = P < 0.05, K-S test. Solid lines with shaded area = average ± 95% confidence intervals. Bar plots = mean + SD.
Together, these experiments showed that high-Cdk1 quiescence entry was determined by the stress status of the cell and not by cell cycle stage at starvation onset.
Nuclear levels of stress transcription factors predict high-Cdk1 quiescence
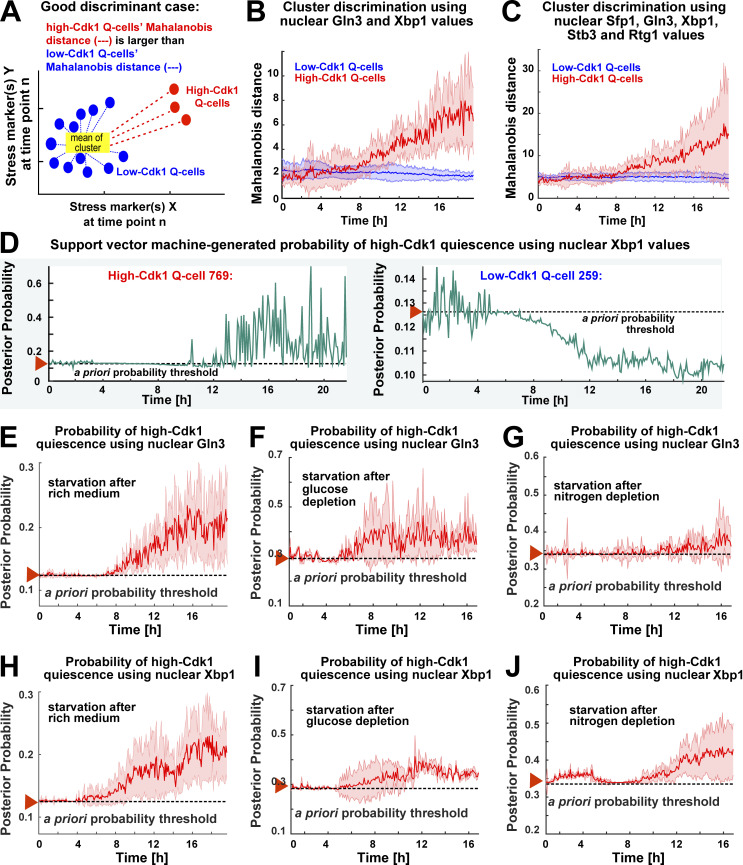
To assess the contribution of stress responses to the establishment of high-Cdk1 quiescence, we used linear discriminant classifiers (LDCs) to find the best combinations of stress markers that discriminated between low- and high-Cdk1 quiescent states. To quantify the discrimination between quiescent states, the single-cell Mahalanobis distance to the mean of the low-Cdk1 Q-cells cluster was used. If a stress marker discriminates well between clusters, this metric should be small for low-Cdk1 Q-cells and large for high-Cdk1 Q-cells (Fig. 6 A). Time series obtained during quiescence entry in 6C1 (Fig. 3) and 6C2 (Fig. 4) cells were used as input for LDCs, and the average separation between clusters using single or combined parameters at each time point was calculated. LDCs based on Xbp1 and Gln3 could significantly discriminate between low- and high-Cdk1 quiescent states after 10 h into starvation (Fig. 6 B), whereas all other stress markers or their combinations did not (Fig. 6 C and Fig. S3 F).
Figure 6.
Nuclear accumulation of stress-activated transcription factors predicts high-Cdk1 quiescence entry depending on starvation conditions. (A) Schematic quantification of cluster discrimination by an LDC based on the Mahalanobis distance. (B and C) Average cluster separation between low-Cdk1 (blue) and high-Cdk1 (red) Q-cells, as measured by the Mahalanobis distance, calculated using LDCs based on the combined nuclear intensity values of Xbp1 and Gln3 (B) or all stress markers (C). (D) Probabilities of high-Cdk1 quiescence during starvation in a representative high-Cdk1 (left) and low-Cdk1 (right) Q-cell as measured by an SVM approach. (E–G) Average probability of high-Cdk1 quiescence assigned to high-Cdk1 Q-cells by SVMs based on the final nuclear intensity of Gln3 after exposure to rich medium (E), glucose depletion (F), or nitrogen depletion (G) before starvation. (H–J) Average probability of high-Cdk1 quiescence assigned to high-Cdk1 Q-cells by SVMs based on the final nuclear Xbp1 intensity after exposure to rich medium (H), glucose depletion (I), or nitrogen depletion (J) before starvation. Red arrowhead with horizontal dotted line = a priori probability threshold corresponding to the fraction of high-Cdk1 Q-cells in each experiment. Solid lines with shaded area = average ± 95% confidence intervals based on biological replicates (n > 3).
To evaluate the predictive power of Xbp1 and Gln3, support vector machine classifiers (SVMs) were used to calculate probabilities of high-Cdk1 quiescence using data from assays in which the 6C1 strain was exposed to rich medium (Fig. 3), glucose depletion (Fig. S3 D), or nitrogen depletion (Fig. S3 E) before starvation. SVMs were trained using the final nuclear values of Xbp1 or Gln3, and single-cell probabilities of high-Cdk1 quiescence upon starvation were computed for all time points (Fig. 6 D and Fig. S3 G). Factors were considered predictive when correctly assigning probabilities of high-Cdk1 quiescence over the a priori probability threshold given by the fraction of high-Cdk1 Q-cells in the experiment.
Nuclear Gln3 produced predictions for high-Cdk1 quiescence over the a priori threshold only when cells were transferred from rich medium into starvation (Fig. 6 E) but not when cells were pretreated with glucose (Fig. 6 F) or nitrogen depletion (Fig. 6 G) before starvation. In contrast, nuclear Xbp1 produced predictions for high-Cdk1 quiescence over the a priori threshold regardless of quiescence entry conditions (Fig. 6, H and I) and as early as 1 h into starvation for cells pretreated with nitrogen depletion (Fig. 6 J). The average predictive power of Xbp1 across experiments, however, was significant after 8–9 h into starvation, suggesting that Xbp1 was correlated to high-Cdk1 quiescence when accumulated to high levels.
These results indicated that although different stress responses could contribute to high-Cdk1 quiescence depending on starvation conditions, increased nuclear Xbp1 was a general and potentially causative factor of high-Cdk1 quiescence.
Nuclear accumulation of the transcriptional repressor Xbp1 promotes high-Cdk1 quiescence entry during starvation
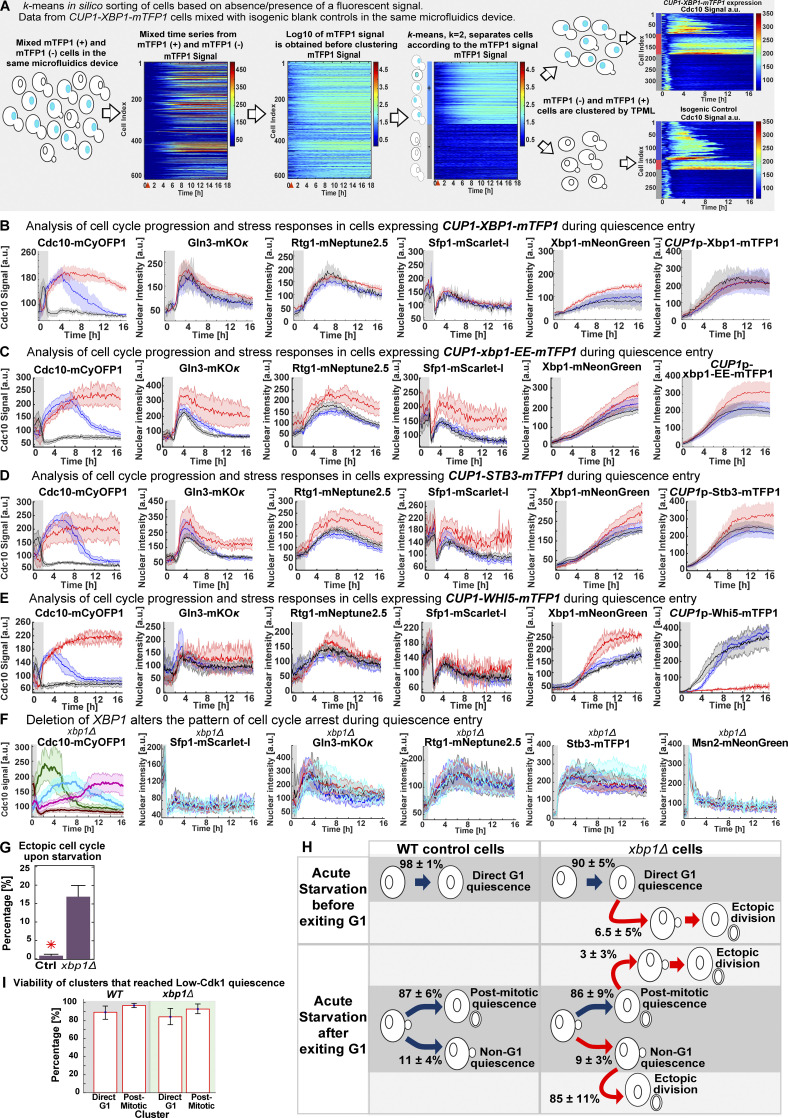
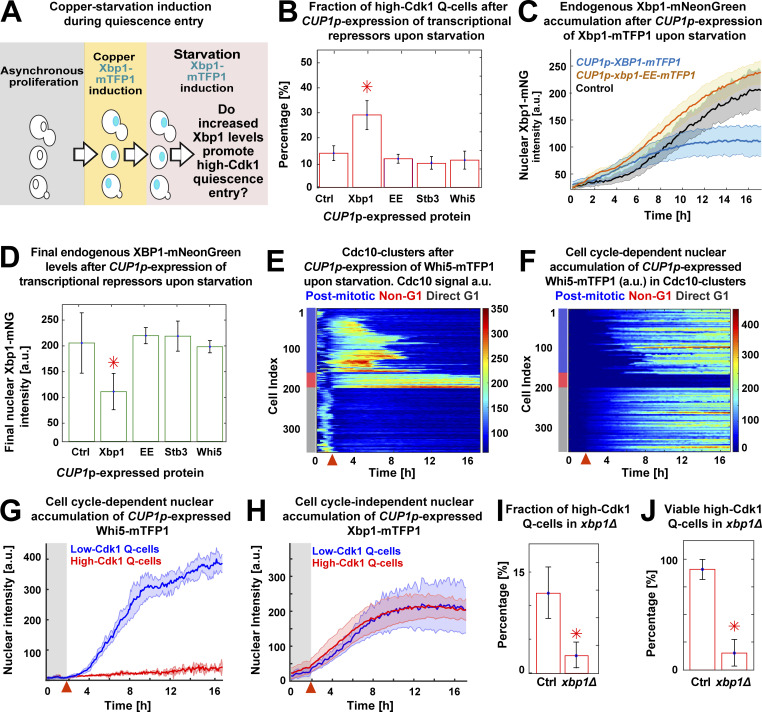
To evaluate whether increased nuclear Xbp1 levels were causative of high-Cdk1 quiescence while simultaneously measuring the stress status of the cell, we tracked quiescence entry in a Cdc10-mCyOFP1, Gln3-mKOκ, Rtg1-mNeptune2.5, Sfp1–mScarlet-I strain endogenously expressing Xbp1-mNeonGreen and also expressing Xbp1-mTFP1 from a CUP1 promoter that is mildly induced by starvation (Fig. 7 A; Peng et al., 2015). As an internal control for copper induction experiments, cultures were spiked with isogenic cells lacking CUP1p-XBP1-mTFP1, and both populations were algorithmically sorted using the mTFP1 signal before TPML analysis (Fig. S4 A).
Figure 7.
Nuclear accumulation of the transcriptional repressor Xbp1 promotes high-Cdk1 quiescence. (A) Schematic copper/starvation–induction protocol to assess the effect of increased Xbp1 levels during starvation. (B) Percentage of high-Cdk1 Q-cells after CUP1p-expression of mTFP1 C-terminally tagged WT Xbp1 (n = 6; OAM458), the DNA-binding domain mutant xbp1-EE (n = 7; OAM466), the Xbp1-related transcriptional repressor Stb3 (n = 4; OAM497), or the cell cycle repressor Whi5 (n = 7; OAM455) during starvation. Ctrl, isogenic control strain (OAM454). (C) Nuclear accumulation of endogenously expressed Xbp1-mNeonGreen in cells expressing mTFP1 C-terminally tagged wild-type Xbp1 (n = 6) or the DNA-binding domain mutant xbp1-EE (n = 7). (D) Final nuclear intensity of endogenously expressed Xbp1-mNeonGreen in cells expressing mTFP1–C-terminally tagged Xbp1 (n = 6), xbp1-EE (n = 7), Stb3 (n = 4), or Whi5 (n = 7) from the CUP1 promoter during starvation. (E and F) Single-cell time series heatmaps contrasting the opposing pattern of Cdc10-mCyOFP1 (E) and nuclear Whi5-mTFP1 (F) after CUP1p-expression of Whi5-mTFP1 during starvation. (G and H) Average nuclear intensity of CUP1p-expressed Whi5-mTFP1 (G; n = 7) or Xbp1-mTFP1 (H; n = 4) in low-Cdk1 (blue) and high-Cdk1 (red) Q-cells. (I and J) Percentage of high-Cdk1 Q-cells (I) and their viability (J), as measured by proliferation upon return to rich medium after 20 h of starvation, in xbp1Δ (n = 4; 500 cells, OAM500) and control XBP1 cells (Ctrl; n = 4; 311 cells, OAM502). Red arrowheads = onset of starvation. Solid lines with shaded area = average ± 95% confidence intervals. Red star = P < 0.05, K-S test. Bar plots = mean ± SD.
CUP1p-expression of Xbp1-mTFP1 during starvation (Video 4) significantly increased the fraction of high-Cdk1 Q-cells (P < 0.05; Fig. 7 B) without altering stress responses (Fig. S4 B). Interestingly, CUP1p-expressed Xbp1-mTFP1 reduced the levels of endogenously expressed Xbp1-mNeonGreen (Fig. 7, C and D), suggesting autoinhibition.
Video 4.
Six-color imaging of cells CUP1p-expressing Xbp1-mTFP1 (OAM458) and control cells (without Xbp1-mTFP1, OAM454) during quiescence entry. Unlike control cells, CUP1p-XBP1-mTFP1 cells underwent high-Cdk1 quiescence entry with high frequency. Notice the brighter Xbp1-mNeonGreen nuclear signal in mTFP1(−) control cells. Cells also carry the cell cycle marker Cdc10-mCyOFP1 and the stress response markers Sfp1–mScarlet-I, Gln3-mKOκ, Rtg1-mNeptune2.5, and Xbp1-mNeonGreen. Each peripheral hexagon displays a median-filtered fluorescent channel. Sampling rate = 12 min. Hexagon side = 65 µM. Playback = 60 frames per second.
To test whether the effect of CUP1p-XBP1-mTFP1 required its transcriptional repressor activity, we generated a DNA-binding domain mutant of XBP1 by introducing the mutations R349E and Q356E, which prevent DNA binding in related transcription factors (Mai and Breeden, 1997; Liu et al., 2015; Nair et al., 2003). CUP1p-expression of xbp1-EE-mTFP1 during starvation had no impact on the fraction of high-Cdk1 Q-cells (Fig. 7 B), the endogenous Xbp1-mNeonGreen levels (Fig. 7, C and D), or stress responses (Fig. S4 C).
To determine the specificity of Xbp1’s effect in promoting high-Cdk1 quiescence, we used the CUP1 promoter to express Stb3, which is a transcriptional repressor functionally related to Xbp1 (Liko et al., 2010), and Whi5, a major transcriptional repressor of cell cycle (Schmoller et al., 2015). CUP1p-expression of Stb3-mTFP1 or Whi5-mTFP1 did not alter the fraction of high-Cdk1 Q-cells (Fig. 7 B) or endogenous Xbp1 levels (Fig. 7 D) and had negligible effects on stress responses (Fig. S4, D and E).
The nuclear accumulation of CUP1p-expressed Whi5-mTFP1 strictly depended on cell cycle progression, as indicated by the opposing patterns of Cdc10 (Fig. 7 E) and Whi5 (Fig. 7 F) in Cdc10 clusters, remaining excluded from the nucleus of high-Cdk1 Q-cells (Fig. 7 G and Video 5). In stark contrast, nuclear accumulation of CUP1p-expressed Xbp1-mTFP1 was entirely independent of Cdk1 activity state (Fig. 7 H) or cell cycle progression (Fig. S4 B).
Video 5.
Six-color imaging of cells CUP1p-expressing Whi5-mTFP1 (OAM455) during quiescence entry. Notice that high-Cdk1 Q-cells expressing CUP1p-WHI5-mTFP1 keep Whi5-mTFP1 in the cytoplasm during starvation (budded cell in the middle). Cells also carry the cell cycle marker Cdc10-mCyOFP1 and the stress response markers Sfp1–mScarlet-I, Gln3-mKOκ, Rtg1-mNeptune2.5, and Xbp1-mNeonGreen. Each peripheral hexagon displays a median-filtered fluorescent channel. Sampling rate = 12 min. Hexagon side = 40 µM. Playback = 60 frames per second.
TPML analysis of xbp1Δ cells bearing markers for stress (Msn2-mNeonGreen, Gln3-mKOκ, Rtg1-mNeptune2.5, Sfp1–mScarlet-I, Stb2-mTFP1) and cell cycle progression (Cdc10-mCyOFP1) showed that deletion of XBP1 led to altered patterns of cell cycle arrest during starvation with 16 ± 4% of cells entering an ectopic, less viable cell cycle under stress (Fig. S4, G and H; and Video 6). Comparison of cells that arrested for >6 h under starvation in WT and xbp1Δ showed that 40 ± 10% of WT cells remained arrested, leading to high-Cdk1 quiescence, whereas most xbp1Δ cells bypassed entry into high-Cdk1 quiescence by completing a last division, with only 7 ± 5% remaining arrested. XBP1 deletion reduced the proportion (Fig. 7 I) and viability (Fig. 7 J) of high-Cdk1 Q-cells, whereas the onset of stress responses (Fig. S4 F) or the viability of cells with stable cell cycle arrest (Fig. S4 I) remained unaffected.
Video 6.
Six-color imaging of quiescence entry in xbp1Δ cells (OAM500). Left: Cells carry Cdc10-mCyOFP1 as cell cycle marker and Sfp1–mScarlet-I, Gln3-mKOκ, Rtg1-mNeptune2.5, Stb3-mTFP1, and Msn2-mNeonGreen as stress response markers. Right: Detail of the phase-contrast image with labeled example cells that failed to maintain a cell cycle arrest during starvation. Each peripheral hexagon displays a median-filtered fluorescent channel. Sampling rate = 12 min. Hexagon side = 38 µM. Playback = 60 frames per second.
We concluded that Xbp1 was a causative factor of high-Cdk1 quiescence entry, which was sufficient to increase the frequency of high-Cdk1 Q-cells when accumulated at high levels.
Nuclear Xbp1 acts as a time-delayed integrator to record single-cell history of stress stimuli
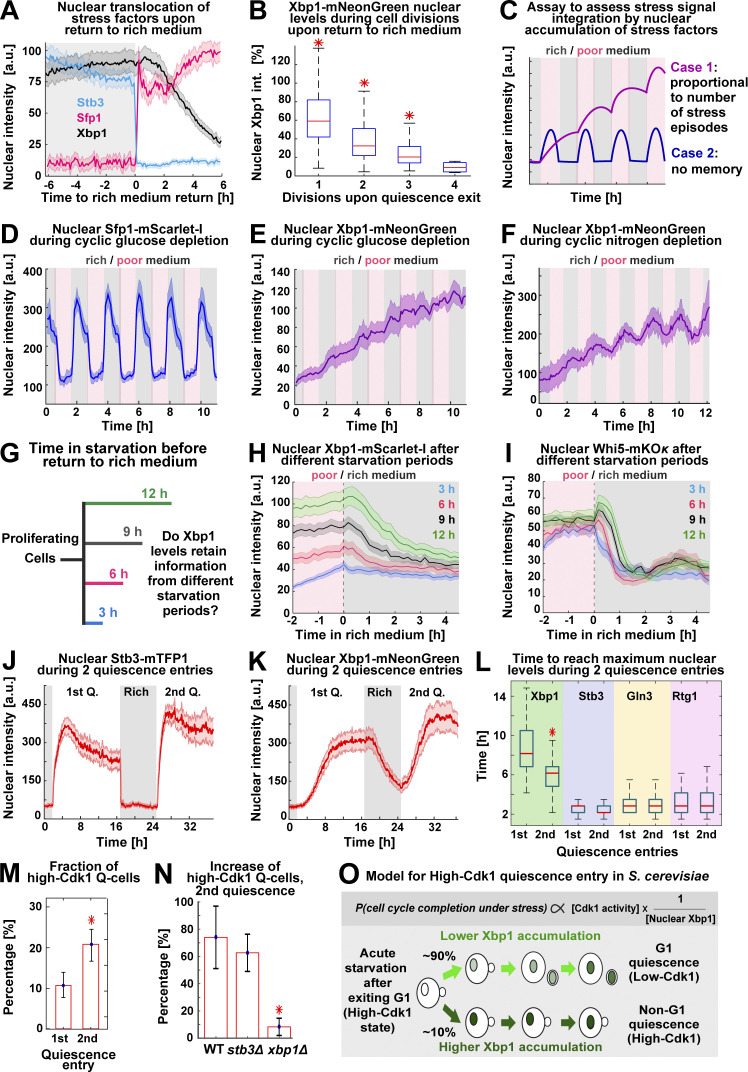
The capacity of Xbp1 to promote quiescent states suggested that it must be rapidly down-regulated upon return to proliferation. To test this hypothesis, we analyzed the nuclear levels of stress transcription factors in 6C1 cells that were returned to rich medium after 20 h of starvation. Surprisingly, we found that whereas stress factors such as Sfp1 or Stb3 readily translocated upon exposure to rich medium, the nuclear levels of Xbp1 remained invariable for 2 h ± 20 min after exposure to rich medium (Fig. 8 A and Fig. S5 A) and after that decreased by dilution through cell division (Fig. 8 B and Fig. S5 B). This indicated that Xbp1 was slowly down-regulated after return to rich medium, suggesting that information from past quiescent states persisted in the nuclear levels of Xbp1.
Figure 8.
Xbp1 acts as a time-delayed integrator of stress stimuli. (A) Average nuclear intensity of Sfp1–mScarlet-I, Stb3-mTFP1, and Xbp1-mNeonGreen as percentage of their maximum nuclear accumulation ±6 h around transfer to rich medium (n = 6; 210 cells, OAM425). (B) Box plots of nuclear Xbp1 intensity as percentage of nuclear Xbp1 intensity (int.) at 20-h starvation, during cell divisions after return to proliferation (n = 6; >101 cells, OAM425). (C) Schematic assay to assess stress stimuli integration by nuclear accumulation of stress factors. (D and E) Average nuclear Sfp1 (D) and Xbp1 (E) intensity during hourly episodes of glucose depletion (n = 5; 335 cells). (F) Average nuclear Xbp1 intensity during hourly episodes of nitrogen depletion (n = 5; 235 cells, OAM425). (G) Schematic assay to assess the integration of starvation duration by nuclear accumulation of stress factors. (H) Nuclear Xbp1–mScarlet-I average intensity in cells transferred to rich medium after different starvation periods (n = 3; >89 cells, OAM849) (I) Nuclear Whi5-mKOκ average intensity in cells transferred to rich medium after different starvation periods (n = 3; >64 cells, OAM850). (J and K) Average nuclear intensity of Stb3 (J) and Xbp1 (K) during two consecutive quiescence (Q) entries (n = 4; 310 cells, OAM425). (L) Box plots of time to reach maximum nuclear intensity by stress factors during two quiescence entries (n = 4; 310 cells, OAM425). (M) Percentage of high-Cdk1 Q-cells during two consecutive quiescence entries (n = 4; 310 cells, OAM425). (N) Percentage increase of high-Cdk1 Q-cells during two consecutive quiescence entries in WT (OAM421), stb3Δ (OAM851), and xbp1Δ cells (OAM852; n > 3; at least 350 cells). (O) Schematic model of high-Cdk1 quiescence entry. Top: Probability of cell cycle completion under starvation (P) is proportional to Cdk1 activity (given by cell cycle stage at starvation onset) and inversely proportional to nuclear Xbp1 levels (given by single-cell history). Bottom: Depending on Xbp1 accumulation, cells with identical cell cycle progression at starvation onset enter different quiescence states. Solid lines with shaded area = average ± 95% confidence intervals. Red star = P < 0.05, K-S test. Bar plots = mean ± SD. Box plots display data from biological replicates: central mark, median; box bottom and top limit, 25th and 75th percentiles; whiskers, most extreme nonoutlier values.
Figure S5.
Analysis of nuclear translocation of stress transcription factors upon return to rich medium and during cyclic episodes of glucose/nitrogen depletion or two consecutive quiescence entries. (A) Average time series for cell cycle (Cdc10 signal) and stress markers (Sfp1, Rtg1, Gln3, Stb3, and Xbp1) during return to proliferation in 6C1 cells (n = 6; OAM425); green dotted line, transfer from starvation to rich medium. (B) Nuclear intensity of Sfp1, Stb3, Xbp1, Rtg1, and Gln3 and cell size expressed as percentage of their values at 20 h of starvation, during cell divisions upon return to rich medium (n = 4; >101 cells, OAM425). (C) Average times series for cell cycle (Cdc10-mCyOFP1) or nuclear stress markers (Sfp1, Gln3, Rtg1, Stb3, and Xbp1) during hourly cycles of glucose depletion (n = 5; 335 cells, OAM425). (D) Average times series for cell cycle (Cdc10-mCyOFP1) or nuclear stress markers (Sfp1, Gln3, Rtg1, Stb3, and Xbp1) during hourly cycles of nitrogen depletion (n = 5; 235 cells, OAM425). (E) Average nuclear Stb3-mTFP1 intensity in cells transferred to rich medium after different starvation periods (n = 3; >71 cells, OAM395). (F) Average of Cdc10 clusters for cell cycle (Cdc10-mCyOFP1) and stress markers (Sfp1, Stb3, Xbp1, Gln3, and Rtg1) during two consecutive quiescence entries (n = 3; 354 cells). Cdc10 clusters were established according to the first quiescence entry; only cells present during the first quiescence entry are included. Gray rectangles = period in rich medium. Solid lines with shaded area = average ± 95% confidence intervals. Red star = P < 0.05, K-S test. Box plots display data from biological replicates: central mark, median; box bottom and top limit, 25th and 75th percentiles; whiskers, most extreme nonoutlier values. Q-exit, quiescence exit.
To test whether information from past stress stimuli (Morimoto and Koshland, 1991) was recorded in the nuclear levels of Xbp1, we exposed 6C1 cells to hourly changes between rich medium (SCD) and either glucose-depleted medium or nitrogen-depleted medium (Fig. 8 C). Under these conditions, stress responses oscillated (Fig. 8 D; Fig. S5, C and D; and Video 7), but only the nuclear levels of Xbp1 increased in proportion to the number of previous stress episodes (Fig. 8, E and F; and Fig. S5, C and D).
Video 7.
Six-color imaging of 6C1 cells (OAM425) during cyclic exposure to stress episodes triggered by hourly changes between rich (SCD) and glucose-deficient (synthetic complete glycerol) medium. Cells carry Cdc10-mCyOFP1 as cell cycle marker and Sfp1–mScarlet-I, Gln3-mKOκ, Rtg1-mNeptune2.5, Stb3-mTFP1, and Xbp1-mNeonGreen as stress response markers. Each peripheral hexagon displays a median-filtered fluorescent channel. Sampling rate = 12 min. Hexagon side = 45 µM. Playback = 60 frames per second.
To test whether nuclear Xbp1 levels retained information from past starvation episodes, we tracked Xbp1 in cells exposed to starvation for 3, 6, 9, or 12 h before return to rich medium (Fig. 8 G). Starvation periods of increasing duration produced populations that maintained significantly different Xbp1 levels after transfer to rich medium, with the 3-h and 12-h starvation-exposed populations maintaining different Xbp1 levels beyond 4 h after return to rich medium (P < 0.05; Fig. 8 H; cells aligned to the time of return to rich medium). This behavior was analogous to a time-delayed integrator in control theory (the output signal—nuclear Xbp1 levels—corresponds to the time integral, or duration in this case, of the input signal—starvation period; Hao et al., 2013). To test whether other histone deacetylase regulators could act as temporal integrators of starvation signals, we tracked nuclear Whi5 and Stb3 levels in cells exposed to different starvation periods before return to rich medium; however, Whi5 (Fig. 8 I) and Stb3 (Fig. S5 E) nuclear levels did not reflect starvation duration.
A time-delayed integrator role for Xbp1 predicted that previous quiescence entries could increase the frequency of high-Cdk1 Q-cells in future quiescence entries by producing populations with higher Xbp1 levels. To test this hypothesis, we imaged 6C1 cells during two quiescence entries separated by a period in rich medium (Video 8). Although Stb3 accumulated to higher levels during the second quiescence entry (Fig. 8 J), only Xbp1 persisted in rich medium after the first quiescence exit (Fig. 8 K) and accumulated to maximum levels 2 h ± 29 min faster during the second quiescence entry (Fig. 8 L). TPML Cdc10 clustering according to the first or second quiescence entry (Fig. S5 F) showed that high-Cdk1 Q-cells were indeed enriched during the second quiescence entry in WT (Fig. 8 M) and stb3Δ cells, but not in xbp1Δ cells (Fig. 8 N).
Video 8.
Six-color imaging of 6C1 cells (OAM425) during two consecutive quiescence entries separated by a period in rich medium. Cells carry Cdc10-mCyOFP1 as cell cycle marker and the stress response markers Sfp1–mScarlet-I, Gln3-mKOκ, Rtg1-mNeptune2.5, Stb3-mTFP1, and Xbp1-mNeonGreen. Each peripheral hexagon displays a median-filtered fluorescent channel. Sampling rate = 12 min. Hexagon side = 85 µM. Playback = 60 frames per second.
These results showed that Xbp1 acted as an integrator of previous stress stimuli, which rendered the propensity of high-Cdk1 quiescence dependent on single-cell history.
Discussion
In this study, we showed how proliferating S. cerevisiae populations challenged by acute starvation enter quiescent states with low- or high-Cdk1 kinase activity. Our results obtained under controlled microfluidic conditions confirm previous batch studies that described quiescence states in cell cycle stages other than G1 after depletion of nutrients in a carbon-limited batch culture (Laporte et al., 2011) or by inducing cell cycle arrest before starvation (Wei et al., 1993). The halt of proliferation upon acute starvation was accompanied by increased storage carbohydrate metabolism, mitochondrial biomass, protein aggregation, autophagy, stress resistance, and size asymmetry during cell division. This indicates that the general processes of quiescence entry observed in batch cultures can also be triggered by starvation under constant environmental conditions.
The use of machine learning classifiers allowed us to in silico isolate and characterize cells entering quiescence in a high-Cdk1 state. By turning the values of single or combined stress markers into probabilities of cell fate, this approach distinguished correlated (Sfp1 and Gln3) from causative (Xbp1) factors of high-Cdk1 quiescence. Our results show that stress responses are remarkably homogeneous during the initial stages of quiescence entry and that cells can diverge into different quiescent states after starvation stimuli are integrated by the histone deacetylase regulator Xbp1. Although Xbp1 represses crucial cell cycle genes involved in proliferation-promoting positive feedback loops, such as cyclins (CLN3, CLB2), mitotic regulators (CDC5, CDC20), and mitotic transcriptional activators (NDD1; Miles et al., 2013), it has been considered dispensable for quiescence because xbp1Δ cells still access G1/G0 quiescence in batch cultures (Mai and Breeden, 1997); however, we find that Xbp1 is essential for viable high-Cdk1 quiescence.
The role of Xbp1 as a cell cycle–independent integrator of past stress signals shows that quiescence entry in S. cerevisiae depends on single-cell history. Remarkably, this implies that each cell can have a different point of commitment to finish cell cycle under stress or restriction point (Johnson and Skotheim, 2013; Zetterberg et al., 1995), determined not only by Cdk1 activity (given by cell cycle stage at starvation onset) but also by nuclear Xbp1 levels (given by single-cell history; Fig. 8 O).
The mechanism to control Xbp1 activity is unknown but likely involves auto-inhibition when accumulated to high levels (Fig. 7 C). Interestingly, quiescence in hair follicles relies on an inhibitory network motif whereby a Cdk1 inhibitor, p21, transcriptionally represses another Cdk1 inhibitor, p15 (Lee et al., 2013). Auto-inhibition or mutual inhibition of quiescence-promoting factors might be a universal mechanism to ensure the reversibility of the quiescent state.
How can cells persist in high-Cdk1 quiescence for a long time? Due to the bistable nature of cell cycle networks, the low-Cdk1 state of G1 and the high-Cdk1 state of S-M phase are stable steady states, which can be, by definition, sustained for prolonged periods (Novak et al., 2007). We envision that under persistent starvation, high levels of Xbp1 force cells to settle in the available stable steady state by inhibiting genes required for proliferation-promoting positive feedback loops through transcriptional repression. Consistent with this, high-Cdk1 quiescence is promoted by high levels of Xbp1 and abolished by XBP1 deletion. Increased nuclear Xbp1, however, did not stop all cells from completing cell cycle under acute starvation (Fig. 7 B), indicating that other events, such as completion of DNA replication under stress or accumulation of mitotic cyclins, could also regulate high-Cdk1 quiescence entry.
The control of quiescence entry by cell cycle–independent integrators of stress stimuli could provide an adaptive advantage in environments with strong fluctuations of nutrients, as is the case for parasites with complex life cycles and cancer cells attempting metastasis (Wang et al., 2015). In mammalian cells, transcriptional repressors that could play an analogous role to Xbp1 are HES1 (Sueda et al., 2019; Coller, 2011), the Dimerization partner, RB-like, E2F and Multi-vulval class B complex (Bainor et al., 2018; Miles and Breeden, 2017; Schade et al., 2019), and developmentally regulated histone deacetylase regulators (Wang et al., 2019); however, it is unknown whether they integrate stimuli during quiescence entry.
In this study, we have characterized, using microfluidics and machine learning algorithms, the emergence of low- and high-Cdk1 quiescent states in proliferating S. cerevisiae cells challenged by acute starvation. Our results outline the temporal integration of stress stimuli through cell cycle–independent transcriptional repressors as a mechanism to establish cellular quiescence outside of G1/G0.
Materials and methods
Media
Rich medium was SCD (1% succinic acid, 0.6% sodium hydroxide, 0.5% ammonium sulfate, 0.17% yeast nitrogen base without amino acids or ammonium sulfate, 0.13% amino acid dropout powder [complete], and 2% glucose). Glucose-depleted medium was synthetic complete (SC) glycerol medium (SC with 2% glycerol instead of glucose). Nitrogen-depleted medium was SCD minus ammonium sulfate and amino acid dropout powder. Starvation medium was 0.6% potassium acetate, delivered at 0.6-psi flow rate, pH 7.2, adjusted with 0.125 M Na2CO3. For copper expression of proteins, cells were maintained in SCD plus 50 µM bathocuproine disulfonate (Gross et al., 2000) until loading in the microfluidic device. CUP1p-expression was triggered with SCD containing 25 µM CuSO4, added as a 1:1,000 dilution from a 25-mM stock solution (Labbé and Thiele, 1999). MET3p-expression was triggered with SCD lacking methionine. 200 mM CaCl2-containing SCD had NH4SO4 replaced by NHCl3.
Plasmid construction
All plasmids were confirmed by sequencing and restriction analysis. Table S1 lists the used primers and Table S2 lists the used plasmids. The MET3p-mNeonGreen-Ase1(R632-I885) plasmid resulted from a quadruple ligation of the following fragments: (1) a SacI-PacI–flanked MET3 promoter sequence (493–1 to −1); (2) a PacI-AscI–flanked mNeonGreen (yeast optimized) fluorophore released from pYLB10 (Argüello-Miranda et al., 2018); (3) an AscI-NotI–flanked C-terminus of ASE1 (R632-I885); and (4) a pRS305 backbone cut with NotI and SacI. The MET3 promoter and ASE1 fragment were amplified from WT W303 genomic DNA. Linearization with AgeI allowed integration at LEU2. pRS304 carrying ATG8p-mNeonGreen-ATG8-ATG8ter was created by replacing the PacI-BamHI–flanked yeGFP sequence in pRS304-ATG8p-2xyeGFP-ATG8-ATG8ter (a gift from the Henne Lab, University of Texas Southwestern Medical Center) by a PacI-BamHI–flanked mNeonGreen fluorophore amplified from pYLB10 (Argüello-Miranda et al., 2018). The pRS306 plasmid carrying CUP1p–QN-mRUBY3 was created by triple ligation of the following fragments: (1) a SalI-HindIII–flanked CUP1-1 promoter sequence (434–16 bp upstream of start codon); (2) a HindIII-PacI–flanked QN-rich sequence of GLN3 (aa 166–242); and (3) a PacI-SalI pLondon266 backbone containing the mRuby3 fluorophore. CUP1p and GLN3 (aa 166–242) fragments were amplified from W303 genomic DNA. Linearization with StuI allowed integration at URA3. CUP1 expression plasmids were created by replacing the PacI-AscI–flanked mCherry fluorophore in pLondon266 (Argüello-Miranda et al., 2018) with a PacI-AscI–flanked mTFP1 fluorophore. The resulting pLondon266-mTFP1 was cut with SalI-PacI and triple-ligated to a SalI-HindIII–flanked CUP1 promoter and the HindIII-PacI–flanked open reading frame of XBP1, STB3, or WHI5, which were amplified from W303 genomic DNA. Linearization with StuI allowed integration at the URA3. CUP1p-xbp1-EE-mTFP1 was created by swapping the DNA-binding domain of XBP1, which is naturally flanked by ClaI-BsaBI sites in S. cerevisiae W303, by a ClaI-BsaBI fragment with arginine 349 and glutamine 356 changed to glutamic acid by PCR mutagenesis with the appropriate primers.
Strain construction
Strains were diploid S. cerevisiae W303 (leu2-3,112 his3-11,15 ura3-1 trp1-1 can1-100 ade2) unless otherwise noted (Table S3). Diploids were created by mating appropriate haploids obtained by tetrad dissection or by transformation with PCR tagging cassettes, deletion cassettes, or plasmids using the polyethylene glycol/lithium acetate protocol (Longtine et al., 1998). xbp1Δ::KANMX4 and stb3Δ::KANMX4 deletion cassette was amplified from BY4741 genomic DNA. C-terminal PCR tagging cassettes were amplified from Longtine (pFA6a) plasmids (Table S2). Tagged strains were confirmed by DNA sequencing, PCR, or functional assays. Tagged fluorescent proteins were heterozygous unless otherwise stated.
Microfluidic cell culture
A Y04C CellASIC microfluidic device (http://www.cellasic.com/) operated at 25°C, 0.6-psi flow rate, was used for culturing cells in all experiments. 50 µl of sonicated (4–6 s at 3 W) liquid SCD culture, OD600 = 0.3, were loaded with 1–2 pulses of 8 psi for 5 s. All medium changes were isobaric. To trigger quiescence, cells grew for 2 or 3 h in SCD before exposure to starvation medium for 20 h. For MET3p protein induction, cells grew 1 h in SCD before exposure to SCD minus methionine for 2 h. For CUP1p induction, cells were maintained in 50 µM of bathocuproine disulfonate–containing SCD before exposure to 25 µM CuSO4-containing SCD for 2 h. For osmoregulatory tests, cells grew 2 h in SCD before exposure to 200 mM CaCl2-containing SCD for 30 min.
Microfluidic staining of DNA
After 20 h in starvation, cells in the Y04C microfluidic device were treated at 25°C, 0.6-psi flow rate with (1) a solution of 4 M LiCl containing 50 mM Nystatin (from a stock solution of 3 mg/ml Nystatin in DMSO) for 4 h; (2) a solution of 50 mM Tris/HCl, pH 7.8, containing 0.3 mg/ml RNase A (from a 10-mg/ml RNase stock solution in 10 mM sodium-acetate, pH 5.2) for 4 h; and (3) FACS buffer (180 mM Tris/HCl, 190 mM NaCl, 70 mM MgCl2) containing 1.85 µM propidium iodide (added before use from a 0.5-mg/ml solution) for 2 h. Nuclear fluorescent intensity was measured in segmented cells as describe below.
Statistical analysis
All statistical analyses were done in MATLAB and considered each lane of the microfluidic device as a biological replicate. Statistical significance was assessed using the Kolmogorov-Smirnov test (K-S test) using a 0.05 P value. Bar plots display mean and SD. Box plots display the median as a central mark, the 25th and 75th percentiles are the bottom and top limits of the box, and the whiskers are the most extreme nonoutlier values. Outliers were excluded using the function isoutlier(). Average times series were plotted as a solid line surrounded by a shaded area corresponding to 95% confidence intervals defined by biological replicates.
Time-lapse microscopy
A motorized, temperature-controlled, ZEN software–operated Zeiss Observer Z1 microscope with Definite Focus 2.0 was used. A 40X Zeiss EC Plan-Neofluar 40X 1.3 NA oil immersion objective and an AxioCam HR Rev 3 camera were used for image acquisition. At least five fields of view were imaged per biological replicate using a 6- or 12-min acquisition rate. A set of custom dichroic mirrors and bandpass filters was used to simultaneously image six different fluorophores (Table S4; Argüello-Miranda et al., 2018). Fluorophores and exposure times were cyan-excitable orange fluorescent protein (mCyOFP1) 35 ms (Chu et al., 2016), teal-cyan fluorescent protein (mTFP1) 400 ms (Ai et al., 2006), green-yellow fluorescent protein (mNeonGreen) 100 ms (Shaner et al., 2013), mKusabira-Orange-κ fluorescent protein (mKOκ) 400 ms (Tsutsui et al., 2008), red fluorescent protein variant (mScarlet-I) 120 ms (Bindels et al., 2017), eqFP611 red fluorescent protein variant (mRuby3) 120 ms (Bajar et al., 2016), mKate far-red fluorescent protein variant (mNeptune2.5) 400 ms (Chu et al., 2014), and phase-contrast 20 ms. CUP1p-expressed fluorescent proteins were imaged using 20 ms. Light-emitting diode (LED) light sources were kept at 12.5% intensity (25% for mNeptune2.5). Correction for crosstalk between fluorescent channels was assessed using single-fluorophore–tagged strains by calculating the cross-correlation between pixels in the signal-containing channel and the empty channels; the code for crosstalk detection is available at GitHub: https://github.com/alejandrolvido/Spectral-Imaging. Crosstalk corrections were for strains OAM421 and OAM425, the corrected mKOκ image equals original mKOκ − 0.23 mCyOFP1, and for strains OAM455/458/466/497, the corrected mCyOFP1 image equals original mCyOFP1 − 0.33 mTFP1.
Image processing and quantification of cellular features
The code for image analysis, single-cell quantifications, and time series visualization is available at https://github.com/alejandrolvido/Quiescence-Entry. The code is organized in subfolders corresponding to each main figure of this work. Images were collected using ZEN pro software (Zeiss) with 2 × 2 binning, exported in noncompressed TIFF format, and converted to double format before quantification. Shot noise was reduced using a medfilt2() filter with a 3 × 3 structuring element. After segmenting cells with published algorithms (Doncic et al., 2013; Wood and Doncic, 2019), image background was calculated as the median intensity of the space not occupied by cells. A 2D Gaussian fit to the brightest pixel of nuclear proteins was used to calculate nuclear parameters (Doncic et al., 2013), where nuclear intensity is defined as the mean intensity of nuclear pixels minus the mean intensity of cytoplasmic pixels. The septin ring was detected by measuring the SD Cdc10-mCyOFP1 at cell periphery “Cdc10 Signal” (Fig. S1 B).
TPML algorithm
This algorithm aims to use a small number of single-cell time series classified by experimentalists to construct a clustering strategy to classify cells from future experiments automatically. Single-cell time series for the Cdc10-mCyOFP1 signal were normalized per biological replicate before being pulled together into matrices where each row represented a cell and each column corresponded to a time point. The order of the rows was then randomized, and the data were divided into 50% training data, 30% validation data, and 20% test data (Chicco, 2017). The silhouette and the Calinski-Harabasz methods defined the minimum number of clusters in the data, which were confirmed by assessing higher cluster numbers using the function kmeans(). Unsupervised clustering algorithms were used to classify cells in the training dataset into three clusters according to their Cdc10-mCyOFP1 signal. The algorithms differed in the clustering method (k-means, k-medoids, hierarchical clustering), the number of time points, the interval of time points, and the cluster centroid distance metric (sqeuclidean, correlation, cityblock, cosine). Clustering algorithms’ performance was evaluated using the validation data, which were manually labeled by three cell biologists tasked with classifying 300 cells from movies as “direct G1 arrest after starvation onset,” “completion of one final cell division after starvation onset,” and “persistent arrest as a budded cell after starvation onset.” Clustering algorithms were then asked to classify the human-labeled validation data using the function pdist2(), and the coherence of the cluster assignments was scored by a multiclass Matthews correlation coefficient (MCC) (Gorodkin, 2004). The best-performing classifier algorithms were optimized by adjusting clustering parameters until their clustering closely recapitulated the human cluster assignments as judged by MCC. The centroids of the best clustering solutions were used to write the function fcentroids(), which uses the function pdist2() and the saved cluster centroids to classify new time series from new experiments into Cdc10 clusters automatically. In strains carrying multiple fluorescent markers, clustering according to Cdc10 was used to arrange all fluorescent channels.
Clustering analysis of synchronized cultures
For M-phase synchronization, exponentially growing cells, OD600 = 0.3, were spun down and resuspended in 20 µg/ml nocodazole-containing SCD and cultured for 2 h (∼90% of dumbbell-shaped cells) before loading in the microfluidics device and being released by exposure to nocodazole-free SCD. For G1 synchronization, an early stationary culture OD600 = 2 was centrifuged at 500 rpm for 2 min on a discontinuous PEG 5000: SC gradient (1:3, 1:27, 1:81), and cells in the top 200 μl, which were mostly unbudded, were washed and resuspended in SCD before loading in the flow cell. As soon as 15% of the synchronized population started to bud, cells were transferred to starvation medium.
Linear discriminant classifier (LDC) analysis
Affiliation to “low-Cdk1” or “high-Cdk1” clusters was used as a categorical variable to establish LDCs that measured how well stress and cell cycle markers discriminated between the two clusters at each time point. LDC hyperparameters were optimized using the function fitcdiscr(), holding out 30% of the cross-validation data. Discrimination between clusters at a given time point was measured by the Mahalanobis distance calculated with the function mahal().
Support vector machine (SVM)–based calculation of single-cell probabilities
Single-cell probabilities of high-Cdk1 quiescence were calculated using an SVM given by the function fitcsvm() with radial basis kernel. SVMs were trained using the last time point of the starvation period, assuming that stress and cell cycle markers relevant for cluster determination displayed their maximum divergence at this stage. A priori probabilities equal to the average ratio of high-Cdk1 cells to the total number of cells were assumed. The trained SVMs were used to classify every cell at every time point according to each stress or cell cycle marker’s values. The function fitPosterior() was used to obtain the posterior probability that a cell was correctly assigned to its cluster at a given time point.
In silico cell sorting according to cellular fluorescence
In copper induction experiments, mTFP1(+) and control mTFP1(−) strains were mixed in the same microfluidic space and sorted in silico using the following algorithm (Fig. S4 A): (1) single-cell time series for mTFP1 were transformed into log10 values; (2) the mean of the log10 time series was calculated; and (3) the mean log10 values derived from each single-cell time series were clustered using kmeans(), distance metric cityblock, which separated mTFP1(−) from mTFP1(+) cells before further analysis.
Semi-automatic quantifications of single cells and intracellular features
The quantification of the time of SPB separation, time point of cell division, time of maximum nuclear accumulation, and foci detection was made on a previously published pipeline for single-cell analysis with a semi-automated user interface for scoring of single-cell data (Argüello-Miranda et al., 2018), available at https://github.com/alejandrolvido/Quiescence-Entry.
Online supplemental material
Fig. S1 shows morphological quantifications of Q-cells under starvation, the algorithm for Cdc10 signal quantification, the workflow of the TPML algorithm, cluster number analysis, and the mitotic induction of the APC/C-Cdh1 activity sensor MET3p-mNeonGreen-Ase1(R632-I885). Fig. S2 shows the assessment of signal-to-noise ratios and crosstalk between fluorescent channels in the six-color imaging setup, the osmoregulatory response in the 6C2 strain, and representative micrographs of cells expressing mNeonGreen-Atg8, QN-mRuby3, or Rad52-mNeonGreen. Fig. S3 shows the clustering analysis of cells treated with nocodazole and nutrient depletion before starvation, as well as the linear discriminant analysis for low- and high-Cdk1 Q-cells. Fig. S4 shows the algorithm to sort cells according to the presence/absence of a fluorescent signal, the extended analysis of the CUP1p-expression of Xbp1, xbp1-EE, Stb3, and Whi5 during quiescence entry, and the TPML analysis of xbp1Δ cells. Fig. S5 shows the analysis of nuclear translocation of stress transcription factors during return to rich medium, cyclic episodes of nutrient depletion, or two consecutive quiescence entries. Video 1 shows the assay to study the proliferation–quiescence transition, including a stress-resistance test by exposure to hyperosmotic medium. Video 2 shows the TPML algorithm being optimized to classify cells in three main clusters. Video 3 shows six-color imaging of the proliferation–quiescence transition in 6C1 cells with single-cell tracking of a representative high-Cdk1 Q-cell. Video 4 shows six-color imaging of quiescence entry in 6C1 cells CUP1p-expressing Xbp1-mTFP1. Video 5 shows six-color imaging of quiescence entry in 6C1 cells CUP1p-expressing Whi5-mTFP1. Video 6 shows six-color imaging of quiescence entry in xbp1Δ cells bearing cell cycle and stress markers. Video 7 shows six-color imaging of 6C1 cells during cyclic exposure to stress. Video 8 shows six-color imaging of 6C1 cells during two consecutive quiescence entries. Table S1 lists primers used. Table S2 lists plasmids used. Table S3 lists strains used. Table S4 lists the optics set up for six-color imaging.
Supplementary Material
lists the primers used for strain construction.
lists the plasmids used for strain construction.
shows the strain list.
shows the optics setup for a six-color imaging system to study the proliferation–quiescence transition.
Acknowledgments
We thank Gaudenz Danuser, Mike Henne, Sandra Schmid, Jan Skotheim, Jennifer Ewald, and Peter Michaely. We thank Andreas Doncic posthumously.
This work was supported by the Cancer Prevention and Research Institute of Texas (RP150596 and RR150058), the Welch Foundation (I-1919-20170325), and the National Institute of General Medical Sciences of the National Institutes of Health (K99GM135487).
The authors declare no competing financial interests.
Author contributions: O. Argüello-Miranda: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Software, Validation, Writing - original draft, and Writing - review and editing. J. Noh: Conceptualization, Data curation, Formal analysis, Software, Supervision, Validation, Writing - original draft, and Writing - review and editing. A.J. Marchand, T. Kennedy, and M.A.X. Russo: Investigation, Methodology, Resources, and Validation.
Footnotes
A preprint of this paper was posted in bioRxiv on March 24, 2021.
References
- Ai, H.W., Henderson J.N., Remington S.J., and Campbell R.E.. 2006. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem. J. 400:531–540. 10.1042/BJ20060874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvaro, D., Lisby M., and Rothstein R.. 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3:e228. 10.1371/journal.pgen.0030228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonets, K.S., Sargsyan H.M., and Nizhnikov A.A.. 2016. A Glutamine/Asparagine-Rich Fragment of Gln3, but not the Full-Length Protein, Aggregates in Saccharomyces cerevisiae. Biochemistry (Mosc.). 81:407–413. 10.1134/S0006297916040118 [DOI] [PubMed] [Google Scholar]
- Apte, M., Pirola R.C., and Wilson J.S.. 2015. Pancreatic stellate cell: physiologic role, role in fibrosis and cancer. Curr. Opin. Gastroenterol. 31:416–423. 10.1097/MOG.0000000000000196 [DOI] [PubMed] [Google Scholar]
- Argüello-Miranda, O., Liu Y., Wood N.E., Kositangool P., and Doncic A.. 2018. Integration of Multiple Metabolic Signals Determines Cell Fate Prior to Commitment. Mol. Cell. 71:733–744.e11. 10.1016/j.molcel.2018.07.041 [DOI] [PubMed] [Google Scholar]
- Bainbridge, P. 2013. Wound healing and the role of fibroblasts. J. Wound Care. 22:407–408: 410–412. 10.12968/jowc.2013.22.8.407 [DOI] [PubMed] [Google Scholar]
- Bainor, A.J., Saini S., Calderon A., Casado-Polanco R., Giner-Ramirez B., Moncada C., Cantor D.J., Ernlund A., Litovchick L., and David G.. 2018. The HDAC-Associated Sin3B Protein Represses DREAM Complex Targets and Cooperates with APC/C to Promote Quiescence. Cell Rep. 25:2797–2807.e8. 10.1016/j.celrep.2018.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baisch, H. 1988. Different quiescence states of three culture cell lines detected by acridine orange staining of cellular RNA. Cytometry. 9:325–331. 10.1002/cyto.990090409 [DOI] [PubMed] [Google Scholar]
- Bajar, B.T., Wang E.S., Lam A.J., Kim B.B., Jacobs C.L., Howe E.S., Davidson M.W., Lin M.Z., and Chu J.. 2016. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 6:20889. 10.1038/srep20889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels, D.S., Haarbosch L., van Weeren L., Postma M., Wiese K.E., Mastop M., Aumonier S., Gotthard G., Royant A., Hink M.A., and Gadella T.W. Jr. 2017. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat. Methods. 14:53–56. 10.1038/nmeth.4074 [DOI] [PubMed] [Google Scholar]
- Buonomo, S.B., Clyne R.K., Fuchs J., Loidl J., Uhlmann F., and Nasmyth K.. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 103:387–398. 10.1016/S0092-8674(00)00131-8 [DOI] [PubMed] [Google Scholar]
- Cappell, S.D., Chung M., Jaimovich A., Spencer S.L., and Meyer T.. 2016. Irreversible APC(Cdh1) Inactivation Underlies the Point of No Return for Cell-Cycle Entry. Cell. 166:167–180. 10.1016/j.cell.2016.05.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, T.H., and Rando T.A.. 2013. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14:329–340. 10.1038/nrm3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicco, D. 2017. Ten quick tips for machine learning in computational biology. BioData Min. 10:35. 10.1186/s13040-017-0155-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, J., Haynes R.D., Corbel S.Y., Li P., González-González E., Burg J.S., Ataie N.J., Lam A.J., Cranfill P.J., Baird M.A., et al. 2014. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat. Methods. 11:572–578. 10.1038/nmeth.2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, J., Oh Y., Sens A., Ataie N., Dana H., Macklin J.J., Laviv T., Welf E.S., Dean K.M., Zhang F., et al. 2016. A bright cyan-excitable orange fluorescent protein facilitates dual-emission microscopy and enhances bioluminescence imaging in vivo. Nat. Biotechnol. 34:760–767. 10.1038/nbt.3550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, H.A. 2011. Cell biology. The essence of quiescence. Science. 334:1074–1075. 10.1126/science.1216242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta, K., Huang P., Morgan G., Winey M., and Surana U.. 2006. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. EMBO J. 25:2551–2563. 10.1038/sj.emboj.7601136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo, J.L., Powers T., Fowler B., and Hall M.N.. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. USA. 99:6784–6789. 10.1073/pnas.102687599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert, M.S. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143–1150. 10.1016/S0006-291X(03)01552-3 [DOI] [PubMed] [Google Scholar]
- De Virgilio, C. 2012. The essence of yeast quiescence. FEMS Microbiol. Rev. 36:306–339. 10.1111/j.1574-6976.2011.00287.x [DOI] [PubMed] [Google Scholar]
- Doncic, A., Eser U., Atay O., and Skotheim J.M.. 2013. An algorithm to automate yeast segmentation and tracking. PLoS One. 8:e57970. 10.1371/journal.pone.0057970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoté, X., Zapater M., Clotet J., and Posas F.. 2004. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell Biol. 6:997–1002. 10.1038/ncb1174 [DOI] [PubMed] [Google Scholar]
- Gorodkin, J. 2004. Comparing two K-category assignments by a K-category correlation coefficient. Comput. Biol. Chem. 28:367–374. 10.1016/j.compbiolchem.2004.09.006 [DOI] [PubMed] [Google Scholar]
- Granados, A.A., Pietsch J.M.J., Cepeda-Humerez S.A., Farquhar I.L., Tkačik G., and Swain P.S.. 2018. Distributed and dynamic intracellular organization of extracellular information. Proc. Natl. Acad. Sci. USA. 115:6088–6093. 10.1073/pnas.1716659115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, C., Kelleher M., Iyer V.R., Brown P.O., and Winge D.R.. 2000. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 275:32310–32316. 10.1074/jbc.M005946200 [DOI] [PubMed] [Google Scholar]
- Hajeri, V.A., Trejo J., and Padilla P.A.. 2005. Characterization of sub-nuclear changes in Caenorhabditis elegans embryos exposed to brief, intermediate and long-term anoxia to analyze anoxia-induced cell cycle arrest. BMC Cell Biol. 6:47. 10.1186/1471-2121-6-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, N., Budnik B.A., Gunawardena J., and O’Shea E.K.. 2013. Tunable signal processing through modular control of transcription factor translocation. Science. 339:460–464. 10.1126/science.1227299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, M., Tyson J.J., and Novák B.. 2017. Cell-cycle transitions: a common role for stoichiometric inhibitors. Mol. Biol. Cell. 28:3437–3446. 10.1091/mbc.e17-06-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L., Hughes C.E., Henderson K.A., Yazvenko N., and Gottschling D.E.. 2016. Selective sorting and destruction of mitochondrial membrane proteins in aged yeast. eLife. 5:e13943. 10.7554/eLife.13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A., and Skotheim J.M.. 2013. Start and the restriction point. Curr. Opin. Cell Biol. 25:717–723. 10.1016/j.ceb.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska, M.M., Crutchfield C.A., Bradley P.H., Rabinowitz J.D., and Broach J.R.. 2011. Yeast cells can access distinct quiescent states. Genes Dev. 25:336–349. 10.1101/gad.2011311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili, A., Wedaman K.P., O’Shea E.K., and Powers T.. 2000. Mechanism of metabolic control. Target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151:863–878. 10.1083/jcb.151.4.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé, S., and Thiele D.J.. 1999. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 306:145–153. 10.1016/S0076-6879(99)06010-3 [DOI] [PubMed] [Google Scholar]
- Laporte, D., Lebaudy A., Sahin A., Pinson B., Ceschin J., Daignan-Fornier B., and Sagot I.. 2011. Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 192:949–957. 10.1083/jcb.201009028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte, D., Gouleme L., Jimenez L., Khemiri I., and Sagot I.. 2018. Mitochondria reorganization upon proliferation arrest predicts individual yeast cell fate. eLife. 7:e35685. 10.7554/eLife.35685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Hoi C.S., Lilja K.C., White B.S., Lee S.E., Shalloway D., and Tumbar T.. 2013. Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo. Proc. Natl. Acad. Sci. USA. 110:4634–4639. 10.1073/pnas.1213015110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., and Clevers H.. 2010. Coexistence of quiescent and active adult stem cells in mammals. Science. 327:542–545. 10.1126/science.1180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liko, D., Conway M.K., Grunwald D.S., and Heideman W.. 2010. Stb3 plays a role in the glucose-induced transition from quiescence to growth in Saccharomyces cerevisiae. Genetics. 185:797–810. 10.1534/genetics.110.116665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Huang J., Zhao Y., Liu H., Wang D., Yang J., Zhao W., Taylor I.A., and Peng Y.L.. 2015. Structural basis of DNA recognition by PCG2 reveals a novel DNA binding mode for winged helix-turn-helix domains. Nucleic Acids Res. 43:1231–1240. 10.1093/nar/gku1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Mai, B., and Breeden L.. 1997. Xbp1, a stress-induced transcriptional repressor of the Saccharomyces cerevisiae Swi4/Mbp1 family. Mol. Cell. Biol. 17:6491–6501. 10.1128/MCB.17.11.6491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion, R.M., Regev A., Segal E., Barash Y., Koller D., Friedman N., and O’Shea E.K.. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA. 101:14315–14322. 10.1073/pnas.0405353101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight, J.N., Boerma J.W., Breeden L.L., and Tsukiyama T.. 2015. Global Promoter Targeting of a Conserved Lysine Deacetylase for Transcriptional Shutoff during Quiescence Entry. Mol. Cell. 59:732–743. 10.1016/j.molcel.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, M.A., and Thorner J.. 2009. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 4:18. 10.1186/1747-1028-4-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, S., and Breeden L.. 2017. A common strategy for initiating the transition from proliferation to quiescence. Curr. Genet. 63:179–186. 10.1007/s00294-016-0640-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, S., Li L., Davison J., and Breeden L.L.. 2013. Xbp1 directs global repression of budding yeast transcription during the transition to quiescence and is important for the longevity and reversibility of the quiescent state. PLoS Genet. 9:e1003854. 10.1371/journal.pgen.1003854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles, S., Li L.H., Melville Z., and Breeden L.L.. 2019. Ssd1 and the cell wall integrity pathway promote entry, maintenance, and recovery from quiescence in budding yeast. Mol. Biol. Cell. 30:2205–2217. 10.1091/mbc.E19-04-0190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, R., van Drogen F., Dechant R., Oh S., Jeon N.L., Lee S.S., and Peter M.. 2017. Protein kinase C and calcineurin cooperatively mediate cell survival under compressive mechanical stress. Proc. Natl. Acad. Sci. USA. 114:13471–13476. 10.1073/pnas.1709079114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, B.H., and Koshland D.E. Jr. 1991. Short-term and long-term memory in single cells. FASEB J. 5:2061–2067. 10.1096/fasebj.5.7.2010059 [DOI] [PubMed] [Google Scholar]
- Moser, J., Miller I., Carter D., and Spencer S.L.. 2018. Control of the Restriction Point by Rb and p21. Proc. Natl. Acad. Sci. USA. 115:E8219–E8227. 10.1073/pnas.1722446115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair, M., McIntosh P.B., Frenkiel T.A., Kelly G., Taylor I.A., Smerdon S.J., and Lane A.N.. 2003. NMR structure of the DNA-binding domain of the cell cycle protein Mbp1 from Saccharomyces cerevisiae. Biochemistry. 42:1266–1273. 10.1021/bi0205247 [DOI] [PubMed] [Google Scholar]
- Nair, U., Thumm M., Klionsky D.J., and Krick R.. 2011. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy. 7:1546–1550. 10.4161/auto.7.12.18424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-Ishizu, A., Takizawa H., and Suda T.. 2014. The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development. 141:4656–4666. 10.1242/dev.106575 [DOI] [PubMed] [Google Scholar]
- Nixon, V.L., Levasseur M., McDougall A., and Jones K.T.. 2002. Ca(2+) oscillations promote APC/C-dependent cyclin B1 degradation during metaphase arrest and completion of meiosis in fertilizing mouse eggs. Curr. Biol. 12:746–750. 10.1016/S0960-9822(02)00811-4 [DOI] [PubMed] [Google Scholar]
- Novak, B., Tyson J.J., Gyorffy B., and Csikasz-Nagy A.. 2007. Irreversible cell-cycle transitions are due to systems-level feedback. Nat. Cell Biol. 9:724–728. 10.1038/ncb0707-724 [DOI] [PubMed] [Google Scholar]
- Nystul, T.G., Goldmark J.P., Padilla P.A., and Roth M.B.. 2003. Suspended animation in C. elegans requires the spindle checkpoint. Science. 302:1038–1041. 10.1126/science.1089705 [DOI] [PubMed] [Google Scholar]
- Ondracka, A., Robbins J.A., and Cross F.R.. 2016. An APC/C-Cdh1 Biosensor Reveals the Dynamics of Cdh1 Inactivation at the G1/S Transition. PLoS One. 11:e0159166. 10.1371/journal.pone.0159166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki, L., and Brand A.H.. 2018. Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science. 360:99–102. 10.1126/science.aan8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, B., Williams T.C., Henry M., Nielsen L.K., and Vickers C.E.. 2015. Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microb. Cell Fact. 14:91. 10.1186/s12934-015-0278-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittershaus, E.S., Baek S.H., and Sassetti C.M.. 2013. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe. 13:643–651. 10.1016/j.chom.2013.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas, J., Caudron F., Prasad R., Moreno D.F., Bolognesi A., Aldea M., and Barral Y.. 2017. Compartmentalization of ER-Bound Chaperone Confines Protein Deposit Formation to the Aging Yeast Cell. Curr. Biol. 27:773–783. 10.1016/j.cub.2017.01.069 [DOI] [PubMed] [Google Scholar]
- Sagot, I., and Laporte D.. 2019. The cell biology of quiescent yeast - a diversity of individual scenarios. J. Cell Sci. 132:jcs213025. 10.1242/jcs.213025 [DOI] [PubMed] [Google Scholar]
- Schade, A.E., Oser M.G., Nicholson H.E., and DeCaprio J.A.. 2019. Cyclin D-CDK4 relieves cooperative repression of proliferation and cell cycle gene expression by DREAM and RB. Oncogene. 38:4962–4976. 10.1038/s41388-019-0767-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoller, K.M., Turner J.J., Kõivomägi M., and Skotheim J.M.. 2015. Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size. Nature. 526:268–272. 10.1038/nature14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner, N.C., Lambert G.G., Chammas A., Ni Y., Cranfill P.J., Baird M.A., Sell B.R., Allen J.R., Day R.N., Israelsson M., et al. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods. 10:407–409. 10.1038/nmeth.2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, L., Sutter B.M., Ye X., and Tu B.P.. 2010. Trehalose is a key determinant of the quiescent metabolic state that fuels cell cycle progression upon return to growth. Mol. Biol. Cell. 21:1982–1990. 10.1091/mbc.e10-01-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueda, R., Imayoshi I., Harima Y., and Kageyama R.. 2019. High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 33:511–523. 10.1101/gad.323196.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D., and Buttitta L.. 2017. States of G0 and the proliferation-quiescence decision in cells, tissues and during development. Int. J. Dev. Biol. 61:357–366. 10.1387/ijdb.160343LB [DOI] [PubMed] [Google Scholar]
- Sun, S., and Gresham D.. 2021. Cellular quiescence in budding yeast. Yeast. 38:12–29. 10.1002/yea.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui, H., Karasawa S., Okamura Y., and Miyawaki A.. 2008. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat. Methods. 5:683–685. 10.1038/nmeth.1235 [DOI] [PubMed] [Google Scholar]
- van Velthoven, C.T.J., and Rando T.A.. 2019. Stem Cell Quiescence: Dynamism, Restraint, and Cellular Idling. Cell Stem Cell. 24:213–225. 10.1016/j.stem.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velappan, Y., Signorelli S., and Considine M.J.. 2017. Cell cycle arrest in plants: what distinguishes quiescence, dormancy and differentiated G1? Ann. Bot. 120:495–509. 10.1093/aob/mcx082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N., Docherty F., Brown H.K., Reeves K., Fowles A., Lawson M., Ottewell P.D., Holen I., Croucher P.I., and Eaton C.L.. 2015. Mitotic quiescence, but not unique “stemness,” marks the phenotype of bone metastasis-initiating cells in prostate cancer. FASEB J. 29:3141–3150. 10.1096/fj.14-266379 [DOI] [PubMed] [Google Scholar]
- Wang, H., Cai H., Wang X., Zhang M., Liu B., Chen Z., Yang T., Fang J., Zhang Y., Liu W., et al. 2019. HDAC3 maintains oocyte meiosis arrest by repressing amphiregulin expression before the LH surge. Nat. Commun. 10:5719. 10.1038/s41467-019-13671-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W., Nurse P., and Broek D.. 1993. Yeast cells can enter a quiescent state through G1, S, G2, or M phase of the cell cycle. Cancer Res. 53:1867–1870. [PubMed] [Google Scholar]
- Westfall, P.J., Ballon D.R., and Thorner J.. 2004. When the stress of your environment makes you go HOG wild. Science. 306:1511–1512. 10.1126/science.1104879 [DOI] [PubMed] [Google Scholar]
- Wood, N.E., and Doncic A.. 2019. A fully-automated, robust, and versatile algorithm for long-term budding yeast segmentation and tracking. PLoS One. 14:e0206395. 10.1371/journal.pone.0206395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki, R., Javaheri A., Kristjansdottir K., Sha F., and Kron S.J.. 2006. CDK Pho85 targets CDK inhibitor Sic1 to relieve yeast G1 checkpoint arrest after DNA damage. Nat. Struct. Mol. Biol. 13:908–914. 10.1038/nsmb1139 [DOI] [PubMed] [Google Scholar]
- Zachariae, W., Schwab M., Nasmyth K., and Seufert W.. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 282:1721–1724. 10.1126/science.282.5394.1721 [DOI] [PubMed] [Google Scholar]
- Zetterberg, A., Larsson O., and Wiman K.G.. 1995. What is the restriction point? Curr. Opin. Cell Biol. 7:835–842. 10.1016/0955-0674(95)80067-0 [DOI] [PubMed] [Google Scholar]
- Zhang, T., Schmierer B., and Novák B.. 2011. Cell cycle commitment in budding yeast emerges from the cooperation of multiple bistable switches. Open Biol. 1:110009. 10.1098/rsob.110009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists the primers used for strain construction.
lists the plasmids used for strain construction.
shows the strain list.
shows the optics setup for a six-color imaging system to study the proliferation–quiescence transition.