Abstract
Background
Identification of non-human leukocyte antigen (HLA) genetic risk factors could improve survival after allogeneic blood or marrow transplant (BMT) through matching at additional loci or individualizing risk prediction. We hypothesized that non-HLA loci contributed significantly to 1-year overall survival (OS), disease related mortality (DRM) or transplant related mortality (TRM) after unrelated donor (URD)BMT.
Methods
We performed a genome-wide association study (GWAS) in 2,887 acute myeloid leukemia (AML), myelodysplastic syndrome (MDS) and acute lymphoblastic leukemia (ALL) patients and their ≥8/8 HLA-matched URDs comprising two independent cohorts treated from 2000–2011.
Findings
Using meta-analyses of both cohorts, genome-wide significant associations (p < 5 × 10−8) were identified in: recipient genomes with OS at MBNL1 (rs9990017, HR = 1.4, 95% CI 1.24–1.56, p = 3.3 × 10−8) and donor-recipient genotype mismatch with OS at LINC02774 (rs10927108, HR = 1.34, 95% CI 1.21–1.48, p = 2.0 × 10−8); donor genomes with DRM at PCNX4 (rs79076914, HR = 1.7, 95% CI 1.41–2.05, p = 3.15 × 10−8), LINC01194 (rs79498125, HR = 1.86, 95% CI 1.49–2.31, p = 2.84 × 10−8), ARID5B (rs2167710, HR = 1.5, 95% CI 1.31–1.73, p = 6.9 × 10−9) and CT49 (rs32250, HR = 1.44, 95% CI1.26–1.64, p = 2.6 × 10−8); recipient genomes at PILRB with TRM (rs141591562, HR = 2.33, 95% CI 1.74–3.12, p = 1.26 × 10−8) and donor-recipient genotype mismatch between EPGN and MTHF2DL with TRM (rs75868097, HR = 2.66, 95% CI 1.92–3.58, p = 4.6 × 10−9). Results publicly available at https://fuma.ctglab.nl/browse.
Interpretation
These data provide the first evidence that non-HLA common genetic variation at novel loci with biochemical function significantly impacts 1-year URD-BMT survival. Our findings have implications for donor selection, could guide treatment strategies and provide individualized risk prediction after future validation and functional studies.
Funding
This project was funded by grants from the National Institutes of Health, USA
Keywords: acute leukemia, myelodysplastic syndrome, allogeneic BMT, genome-wide association study, mortality
Research in Context.
Evidence before this study
Overall survival (OS) after blood or marrow transplantation (BMT) has improved dramatically; however, the 1-year all-cause mortality rate remains about 40% after human leukocyte antigen (HLA)-matched unrelated donor (URD) allogeneic BMT, leaving room for improvement. For over 20 years, inherited genetic susceptibility to BMT outcomes has been investigated with a candidate gene approach using small sample sizes and heterogeneous populations of disease, and the results could not be replicated or validated. Our prior exome-wide approach using DISCOVeRY-BMT showed that rare coding variation in donor and recipients significantly correlated with survival after BMT and identified new genes and pathways.
Added value of this study
Our DISCOVeRY-BMT multi-cohort genome-wide association study demonstrates that non-HLA common variants are associated with 1-year OS, transplant related mortality and disease related mortality. We identified several regions, many of which have clear biochemical function, in the donor genome, recipient genome, and the shared genome (defined as allele mismatch/difference between donor and recipient at a given loci) significantly associated with survival. Our findings of novel genes related to cellular metabolism, transcription factor binding, cytokine and chemokine receptors, and leukocyte immunoglobulin-like receptors, broadens our knowledge of post-allogeneic BMT survival outcomes and clearly show that genetic variation outside the HLA region impacts survival in the first year following BMT.
Implication of all the available evidence
Taken together, our findings to date provide evidence that both common and rare genetic variation outside the HLA region could improve donor selection and that better understanding of these loci may lead to improved knowledge of the pathogenesis of, and yield improvements in, specific causes of death after HLA-matched unrelated donor-BMT.
Alt-text: Unlabelled box
1. Introduction
Blood or Marrow Transplantation (BMT) is a curative therapy for acute leukemia and Myelodysplastic Syndrome (MDS); however, success is limited by death due to disease relapse/progression or transplant related causes, such as Graft versus Host Disease (GvHD), infection and/or organ failure which have the greatest impact on overall survival (OS) in the first year post-BMT [1]. OS after BMT has improved dramatically over the past two decades, owing mainly to improvements in reducing transplant-related mortality (TRM) through high resolution human leukocyte antigen (HLA) typing, patient selection, supportive care, and infection prophylaxis; nonetheless, the 1-year all-cause mortality rate remains about 40% after HLA-matched unrelated donor (URD) allogeneic BMT [2,3]. Several clinical variables, including disease status at transplant, stem cell source, graft-source, patient age and donor age are associated with survival outcomes [4], there are currently no established and validated genetic predictors of survival after BMT outside of HLA genes.
Prior studies have demonstrated strong effects on OS and TRM by genotypes at the HLA loci, however non-HLA candidate gene investigations of survival after transplant have been unsuccessful [5,6]. The identification of donor, recipient or a combination of donor-recipient genetic variation outside the Major Histocompatibility Complex (MHC) region associated with mortality outcomes in the first year after URD-BMT could provide new insight on the biology of these outcomes and/or ultimately improve BMT outcomes through matching at these additional loci. We hypothesized that non-HLA genetic variation in recipients and/or donors would significantly contribute to OS and its two competing causes: TRM and Disease Related Mortality (DRM). To inform our hypothesis we used two independent cohorts for our study, Determining the Influence of Susceptibility COnveying Variants Related to one-Year mortality after BMT (DISCOVeRY-BMT) composed of acute myeloid leukemia (AML), MDS and acute lymphoblastic leukemia (ALL) patients and their ≥8/8 HLA-matched unrelated donors. The use of a GWAS in the DISCOVeRY-BMT study population allowed us to identify SNPs and genes associated with TRM, DRM and OS at 1 year post URD-BMT.
2. Methods
2.1. DISCOVeRY-BMT Cohorts
The DISCOVeRY-BMT study population was contributed by 151 centers in the United States to the Center for International Blood and Marrow Transplant Research (CIBMTR) and consists of two cohorts of ALL, AML and MDS patients and their healthy ≥8/8 HLA-matched URDs described previously [5], [6], [7], [8], [9], [10], [11]. Cohort 1 received a first URD-BMT (10/10 HLA-matched) between 2000 and 2008; Cohort 2 received a first URD-BMT (10/10 HLA-matched) between 2009 and 2011 or an 8/8 (but <10/10) HLA-matched first URD-BMT between 2000 and 2011. Additional inclusion criteria were T-cell replete peripheral blood or marrow grafts, an available bio-repository sample from both recipient and donor, with no restrictions on conditioning regimen, age, sex or race/ethnicity.
Recipients and donors provided written informed consent for participation in the Center for International Blood and Marrow Transplant Research clinical outcome database and research repository. The bio-specimen and database protocols were approved by the National Marrow Donor Program (NMDP Institutional Review Board (IRB), as well as the individual transplant and donor centers’ IRBs. Recipients and donors were not compensated for their participation. This study was reviewed and approved by the Roswell Park Comprehensive Cancer Center and NMDP IRBs. LSC, JW, EK, AR, ACG, YW, AW, GB, QZ, LY, SL, SS, TH had access to all data and CH, DS, LP, XS, DV had access to genotyping data during the duration of the study.
2.2. Genotyping, imputation, and quality control
We previously described the DISCOVeRY-BMT cohorts and genotyping and quality control (QC) in detail. Supplemental Figures 1 and 2 show quality control and study workflow, respectively [5], [6], [7], [8], [9], [10], [11]. Briefly, samples were assigned to plates using the Optimal Sample Assignment Tool [12]. Genotyping was performed at the University of Southern California Genomics Facility using the Illumina Omni-Express BeadChip® containing approximately 733,000 single nucleotide polymorphisms (SNPs). Typed SNPs were removed if the minor allele frequency (MAF) < 1%, missing rate was > 2.0%, violation of Hardy Weinberg equilibrium proportions (p < 1.0 × 10−4). Population specific quality control was performed and samples were removed based on the SNP missing rate, reported-genotyped sex mismatch, abnormal heterozygosity, cryptic relatedness, and population outliers. The study is well powered to detect small effect sizes (HR<1.5) for common variants with minor allele frequency (MAF) of 30–40% and for clinically relevant effect sizes in the range of 1.5–2.5 for MAF of 5–10% [13]. Population stratification was assessed via principal components analysis using Eigenstrat software [14] and a genomic inflation factor (λ) was calculated for each cohort. Following quality control, the Omni Express Bead Chip in Cohorts 1 and 2 yielded 637,655 and 632, 823 SNPs, respectively, available for imputation. IMPUTE2 was used to perform SNP imputation via the Michigan Imputation server with the Haplotype Reference Consortium, hg19/b37 (http://www.haplotype-reference-consortium.org/home) as the reference genome [15,16]. After filtering to include SNPs with imputation quality scores >0.9 and MAF >0.005, 8,515,276 SNPs were available for analyses in both cohorts. Almost 95% of the total patient and donor population self-reported as Non-Hispanic White (European American (EA)), while individuals reporting as non-Hispanic African-Americans and Hispanic Whites each comprised approximately 1.5% of DISCOVeRY-BMT and the numbers were too small for separate analyses. The remainder of the racial and ethnic groups had fewer than 25 individuals each. After quality control, Cohorts 1 and 2 include 2111 and 779 non-Hispanic European Americans AML, MDS and ALL patients and 2219 and 808 non-Hispanic European American donors, respectively. Of these, there are 2053 and 763 complete donor-recipient pairs in Cohorts 1 and 2, respectively. (Supplemental Figure 1).
2.3. Statistical analysis
QC was implemented using QCTOOL-v2, R 3.5.1 (Feather Spray) and Plink-v1.9 [17]. OS analyses were performed using Cox proportional hazard models implemented in gwasurvivr [18]. To assess 1-year OS, surviving patients were censored at 1-year post BMT. Single SNP analyses were coded as 0–2 minor alleles of the recipients, their corresponding donors or the absolute value of the allele difference between recipients and donors. Causes of death were adjudicated as previously described [8]. Cox proportional hazard (OS) and competing risk models (TRM and DRM) were used to analyze single-variant association while controlling for the significant univariate risk factors of recipient age at BMT, disease (AML, ALL, MDS), disease status at BMT (complete remission (CR) /early, not in CR/advanced), cell source (peripheral blood, marrow), year of BMT and recipient Body Mass Index (TRM models only) [18]. The two cohorts were analyzed separately with meta-analyses performed by fitting random effects models with inverse variance weighting using the R package Metafor. Genome-wide statistical significance was set at p<5 × 10−8 for meta-analyses; experiment wide correction for multiple testing was not performed since donor and recipient (and D-R mismatch) are independent genomes and hypotheses. Disease specific mortality analyses were limited to myeloid diseases (AML, MDS) as were TRM, the competing risk of DRM; ALL specific mortality was not sufficiently powered to detect genome-wide associations across both cohorts. Death due to graft versus host disease (GvHD), organ failure and infection death comprise TRM [8], therefore TRM significant associations with MAF > 5% were further assessed for the association with GvHD-, infection- and organ failure-death to better understand the contribution of each cause specific mortality to the TRM association.
2.4. Co-localization analysis of the risk locus
Integrating GWAS and expression quantitative trait loci (eQTL) studies can elucidate mechanisms of non-coding variants on disease, however this is challenging due to the correlation (LD) between variants and because some loci could contain multiple causal variants [19]. We estimated the posterior probability that a genome wide significant variant was causal both in our GWAS and in genome-wide blood eQTL studies, while accounting for linkage disequilibrium (LD) and allelic heterogeneity, using the software program eCAVIAR [20]. To measure the degree of co-localization of GWAS and blood eQTL the co-localization posterior probability (CLPP) was calculated by estimating the probability that the same variant is causal in both eQTL and our GWAS studies. The CLPP was calculated with two separate inputs, blood eQTL and GWAS z-scores using the LD in the European (non-Hispanic white) population. A higher CLPP score indicates higher level of co-localization. A threshold of 1% for CLPP was used as recommended when selecting for candidate causal SNPs [20].
2.5. Functional annotation of genetic variation associated with OS, TRM and DRM
To better understand the potential function of the variants identified by GWAS, we derived additional information about significant SNPs and genomic regions and performed genome-wide gene-based testing using the web-based application Functional Mapping and Annotation (FUMA). For significant variants and those in LD, FUMA was used to: identify eQTLs across multiple tissues [21,22]; derive a measure of deleteriousness using the combined annotation dependent depletion (CADD) score computed by integrating 63 functional annotations (the higher the score, the more deleterious with a scaled score >10 for the top 10% and >20 for the top 1% variants in GRCh37v1.4) [23,24]; and retrieve Regulome DB scores and probabilities, the latter spanning 0–1 with 1 being most likely to be a regulatory variant [25] and show significant Hi-C data loops across 21 tissues from GSE87112 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87112 [26]. Additional analyses placing associations in genomic context of existing data are described in the Supplement material.
2.6. Role of the funding source
The funding sources had no role in the study design, data analysis, interpretation or decision to publish the results.
3. Results
Recipient and donor characteristics are shown in Table 1. Cohorts were comparable on all characteristics except for disease, disease status, graft source, performance status, and use of total body irradiation. Cohort 1 has a higher proportion of ALL and fewer MDS patients than Cohort 2, with similar proportions of AML in both cohorts. Similarly, there was a higher proportion of intermediate, and lower proportion of advanced disease in Cohort 1 versus Cohort 2. Despite these differences, there was no significant OS difference between Cohorts 1 and 2 (p = .8)
Table 1.
DISCOVeRY-BMT Recipient and Donor Characteristics by Cohort.
| Characteristic | Cohort 1, N = 2110* | Cohort 2, N = 777* | p-value# |
|---|---|---|---|
| Recipient Age, years | 0.057 | ||
| <20 | 240 (11%) | 79 (10%) | |
| 20–39 | 527 (25%) | 173 (22%) | |
| 40–59 | 931 (44%) | 340 (44%) | |
| ≥60 | 412 (20%) | 185 (24%) | |
| Recipient Age, years | <0.001 | ||
| Median (standard deviation) | 46.6 (17.5) | 50.0 (17.6) | |
| Donor Age, years | 0.010 | ||
| <20 | 11 (0.5%) | 12 (1.5%) | |
| 20–39 | 1487 (70%) | 569 (73%) | |
| 40–59 | 604 (29%) | 192 (25%) | |
| ≥60 | 8 (0.4%) | 4 (0.5%) | |
| Donor Age, years | <0.001 | ||
| Median (standard deviation) | 32.8 (9.3) | 30.6 (9.6) | <0.001 |
| Male Recipient | 1191 (56%) | 429 (55%) | 0.554 |
| Male Donor | 1437 (68%) | 563 (72%) | 0.025 |
| Disease | <0.001 | ||
| ALL | 483 (23%) | 94 (12%) | |
| AML | 1282 (61%) | 488 (63%) | |
| MDS | 345 (16%) | 195 (25%) | |
| Disease Status | <0.001 | ||
| Early | 1005 (48%) | 386 (50%) | |
| Intermediate | 528 (25%) | 127 (16%) | |
| Advanced | 577 (27%) | 264 (34%) | |
| Graft Source | <0.001 | ||
| Bone Marrow | 745 (35%) | 210 (27%) | |
| Peripheral blood | 1365 (65%) | 567 (73%) |
| Characteristic | Cohort 1, N = 2110 * | Cohort 2, N = 777 * | p-value # |
|---|---|---|---|
| KPS/LPS pre-BMT | <0.001 | ||
| <90 | 619 (29%) | 248 (32%) | |
| 90–100 | 1282 (61%) | 490 (63%) | |
| (Missing) | 209 (9.9%) | 39 (5.0%) | |
| Body Mass Index | 0.025 | ||
| Underweight | 41 (1.9%) | 20 (2.6%) | |
| Normal | 780 (37%) | 241 (31%) | |
| Overweight | 659 (31%) | 266 (34%) | |
| Obese | 630 (30%) | 250 (32%) | |
| Donor-Recipient Sex | 0.125 | ||
| F donor to F recipient | 341 (16%) | 113 (15%) | |
| F donor to M recipient | 332 (16%) | 101 (13%) | |
| M donor to F recipient | 578 (27%) | 235 (30%) | |
| M donor to M recipient | 859 (41%) | 328 (42%) | |
| Conditioning Regimen Intensity | 0.300 | ||
| Myeloablative | 1540 (73%) | 552 (71%) | |
| Reduced/NMA | 570 (27%) | 225 (29%) | |
| TBI-containing Regimen | <0.001 | ||
| Yes | 973 (46%) | 280 (36%) | |
| No | 1137 (54%) | 497 (64%) |
n (%).
Pearson's Chi-squared test; Fisher's exact test; Early=CR1 or MDS-RA/RARS, Intermediate=CR2+, Advanced=Not in Remission or MDS-RAEB, KPS: Karnofsky Performance Score, LPS: Lansky Performance Score, NMA: non-myeloablative, TBI: total body irradiation.
3.1. Associations with overall survival in AML, All and MDS patients
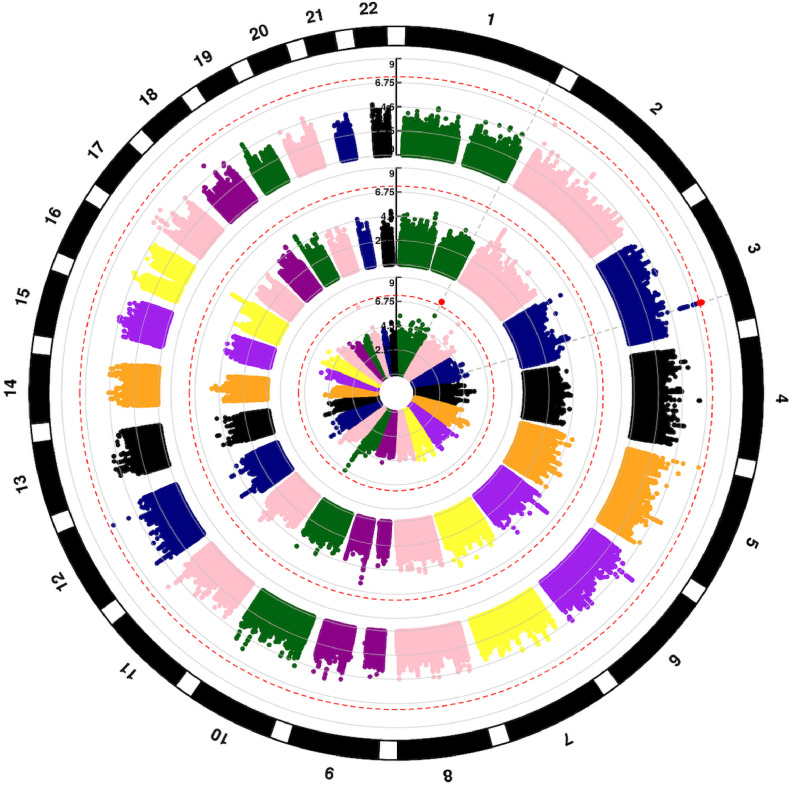
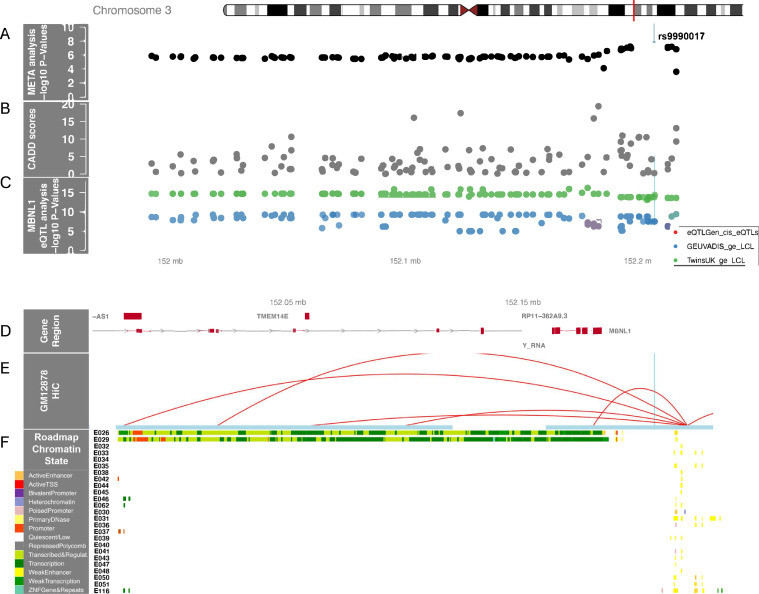
One region in recipient genomes, 3q25, contained five genome-wide significant variants associated with OS (Fig. 1, outer circle, Table 2, Supplemental Table 1). The quantile-quantile plot (QQplot) for all OS analyses shows low genomic inflation (λ<1.03) for Cohorts 1, 2 and meta-analyses (Supplemental Figure 3). The significant variants were strongly correlated (r2 > 0.95) with the lead variant, rs9990017 (typed), adjacent to Muscleblind Like Splicing Regulator 1 (MBNL1) a highly conserved gene whose protein is expressed in most blood and marrow hematopoietic cells. Supplemental Figure 4A-C shows the G allele at rs9990017 associates with increased risk of all-cause mortality (OS) in the first year after URD-BMT (HR = 1.4, 95% CI, 1.24, 1.56, p = 3.3 × 10−8), which is largely driven by TRM (HR = 1.48, 95% CI, 1.26, 1.75, p = 2.7 × 10−6) but is also associated with DRM (p = .002). Fig. 2 highlights the variants in the region significantly associated with gene expression of MBNL1 in blood (eQTLgen) and lymphoblastoid cell lines (GEUVADIS and TWINS_UK). Hi-C data show the region containing rs9990017 interacts with an active transcription start site (TSS) across multiple blood cell lines. The maximum CADD annotation in the region, rs79130661, approached 20 (CADD = 19.4) indicating the variant in the 3′ untranslated region (UTR), and in LD (r2 = 0.8) with rs9990017, is predicted to be near the top 1% of deleterious variants in the genome. MAGMA gene-based analyses show MBNL1 approaches (p = 5.3 × 10−6) but does not exceed genome-wide significance.
Fig. 1.
Circular Manhattan plot of SNP associations with overall survival. Chromosomes are numbered on the outside. p = 5 × 10−8 is marked with dashed red lines. Each SNP p value association with overall survival is a colored dot, with genome-wide significant SNPs as red dots. Results are shown for recipient (outer circle), donor (middle circle) and donor-recipient mismatches (inner circle), respectively.
Table 2.
Lead genome wide significant variants associated with 1-year survival outcomes in DISCOVeRY-BMT.
| BMT outcome | Rsid lead SNP | Chr | Position (hg 19) | Nearest Gene | Genome | Alleles | Effect Allele Freq | HR (95% CI) | P |
|---|---|---|---|---|---|---|---|---|---|
| OS | rs9990017 | 3 | 152,206,062 | MBNL1 | Recipient | T/G | 0.11 | 1.39 (1.24, 1.56) | 3.3 × 10−8 |
| rs10927108 | 1 | 244,184,670 | LINC02774 | Donor-Recipient allele mismatch | C/T | 0.27 | 1.34 (1.21, 1.48) | 2.0 × 10−8 | |
| DRM | rs79076914 | 14 | 60,559,239 | PCNX4 | Donor | C/A | 0.051 | 1.7 (1.41, 2.05) | 3.15 × 10−8 |
| rs79498125 | 10 | 63,695,702 | ARID5B | A/G | 0.06 | 1.86 (1.49, 2.31) | 2.8 × 10−8 | ||
| rs2167710 | 10 | 33,877,364 | LINC02628 | A/T | 0.65 | 1.5 (1.31, 1.73) | 6.9 × 10−9 | ||
| rs32250 | 5 | 12,465,740 | CT49 | T/G | 0.55 | 1.44 (1.26, 1.63) | 2.6 × 10−8 | ||
| TRM | rs75868097 | 4 | 75,148,954 | EPGN | Donor-Recipient allele mismatch | A/T | 0.027 | 2.65 (1.91,3.58) | 5.9 × 10−9 |
| rs141591562 | 7 | 99,947,974 | PILRB | Recipient | A/G | 0.03 | 2.33 (1.74, 3.12) | 1.26 × 10−8 |
Variants (SNPs) are within a gene or in LD (r2 > 0.8) with variants in a gene; bolded allele is the effect allele; for mismatch loci the effect allele frequency is the frequency of the associated mismatch across both cohorts
OS: Overall survival, DRM: Disease-related mortality, TRM: Transplant-related mortality, SNP: single nucleotide polymorphism, hg19: human genome 19 position, Freq: frequency, HR: hazard ratio, CI: confidence interval.
Fig. 2.
Chromosome 3 regional recipient association plot of overall survival. A) Regional meta-analysis SNP –log10(p-values) are plotted as solid black dots in the top panel. The higher the -log10 number, the lower the p-value. Rs9990017, the most significant genome-wide association is labeled and marked with a thin blue line that transverses all panels. The x-axis is the chromosome position in mega bases (Mb). B) Corresponding CADD scores in the region for the variants are shown as solid gray dots. C) Significant MBNL1 eQTLs identified in three separate cohorts: eQTLGen (solid red dots), GEUVADIA (solid blue dots) and TwinsUK (solid green dots). D) Genes in the region are plotted as red boxes on a gray line and annotated with E) HiC data from a lymphoblastoid cell line with the F) ENCODE epigenome roadmap for all blood, T-cell, HSC and B-cells; the cell line numbers shown down the left side (ie., E026, etc.) correspond to specific epigenome road map cell lines. The colors indicate the ChromHMM status as defined in the key on the bottom left of the figure.
Allele difference (mismatch) between donors and recipients was associated with a small region on Chromosome 1 containing long intergenic non-protein coding RNA 2774 (LINC02774) (Fig. 1, inner circle, Table 2, Supplemental Table 1). One variant in the region reached genome-wide significance, rs10927108 (HR = 1.34, 95% CI 1.21, 1.48, p = 2.0 × 10−8), with increasing copies of the T allele between the donor-recipient pairs showing increasing risk of all-cause mortality, driven by both TRM (HR=1.39, 95% CI,1.19, 1.61, p = 1.72 × 10−5) and DRM (HR = 1.31, 95% CI 1.14, 1.50, p = 1.68 × 10−4) (Supplemental Table 3, Supplemental Figure 5).
3.2. Associations with disease-related mortality in AML and MDS patients
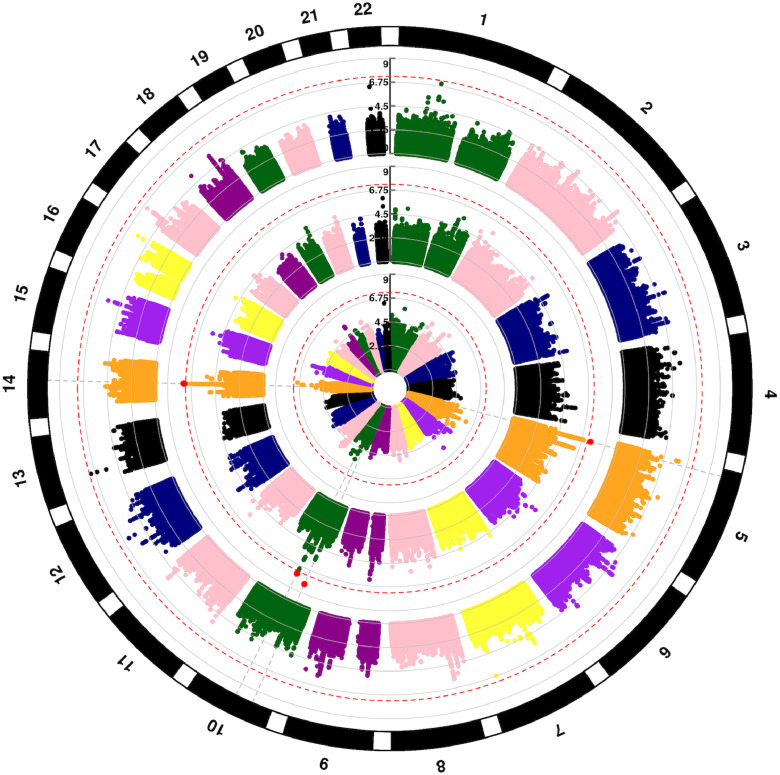
DISCOVeRY-BMT is comprised of lymphoid and myeloid diseases, thus all DRM analyses, and its competing risk, TRM, were done using myeloid diseases (AML and MDS) together. The DRM QQplots shows a low genomic inflation (λ<1.02) for both cohorts and the meta-analyses (Supplemental Figure 6). The donor genome has four genome-wide significant regions associated with death due to AML or MDS: at 14q23.1(rs79076914, HR = 1.7, 95% CI, 1.41–2.05, p = 3.15 × 10−8), 10q21.2 (rs79498125, HR = 1.86, 95% CI, 1.49, 2.31, p = 2.84 × 10−8), 10p11.22 (rs2167710, HR = 1.5, 95% CI, 1.31, 1.73, p = 6.9 × 10−9) and 5p15.2 (rs32250, HR = 1.44, 95% CI, 1.26, 1.64, p = 2.6 × 10−8) (Fig. 3, middle circle, Table 2, Supplemental Table 2). No genome-wide significant associations with death due to AML or MDS are seen in recipient genomes (Fig. 3, outer circle) or when comparing allele matching at each SNP in the recipient and donor (Fig. 3, inner circle). The chromosome 14 region (60,262,026–60,597,197 bp) contains several donor variants at Pmeta<5 × 10−8 that increase risk of DRM in AML and MDS patients. The most significant chromosome 14 donor association rs79076914, is located 610 bp from the canonical transcription start site of Pecanex 4 (PCNX4) increased risk of DRM 1.7-fold. This region contains numerous genome wide significant eQTLs in blood and LCLs for Dehydrogenase/Reductase 7 (DHRS7), a steroid and retinoid metabolizer gene, and PCNX4 (Supplemental Figure 7). The co-localization analyses done with eCAVIAR provides evidence that rs114514727 may be causal for the shared signal of the donor DRM GWAS association and PCNX4 expression (Supplemental Table 4). Rs79076914 also shows some association with worse OS (HR = 1.41, 95% CI, 1.21, 1.65, p = 1.06 × 10−5) but not at a genome-wide level. The Chromosome 10 donor genome associations with DRM reside in a well-known acute lymphoblastic leukemia-associated gene, ARID5B (Supplemental Figure 8) and is a molecular determinant of antimetabolite drug sensitivity [27]; rs79498125 confers an ∼1.9-fold increased risk of disease death (HR = 1.86, 95% CI 1.49, 2.31, p = 2.8 × 10−8) and resides in an area of strong transcription across multiple blood cell lines. We also identified a Chromosome 10 significant association independent of rs7949815 (Supplemental Figure 9), in LINC02628 rs2167710 (HR = 1.5, 95% CI 1.31, 1.73, p = 6.9 × 10−9). Rs32250, the most significant variant in the chromosome 5 region near CT49 does not show strong evidence of impacting transcription or gene expression (Supplemental Figure 10). These three variants show some association with OS but not at a genome-wide level: 10q21.2 (rs79498125, HR = 1.33, 95% CI, 1.12, 1.60, p=.001), 10p11.22 (rs2167710, HR = 1.24, 95% CI, 1.12, 1.37, p = 2.8 × 10−5), and 5p15.2 (rs32250, HR = 1.23, 95% CI, 1.12, 1.36, p = 1.08 × 10−5).
Fig. 3.
Circular Manhattan plot of SNP associations with disease related mortality. Chromosomes are numbered on the outside. p = 5 × 10−8 is marked with dashed red lines. Each SNP p value association with disease-related mortality is a colored dot, with genome-wide significant SNPs as red dots. Results are shown for recipient (outer circle), donor (middle circle) and donor-recipient mismatches (inner circle), respectively.
3.3. Associations with transplant related mortality in AML and MDS patients
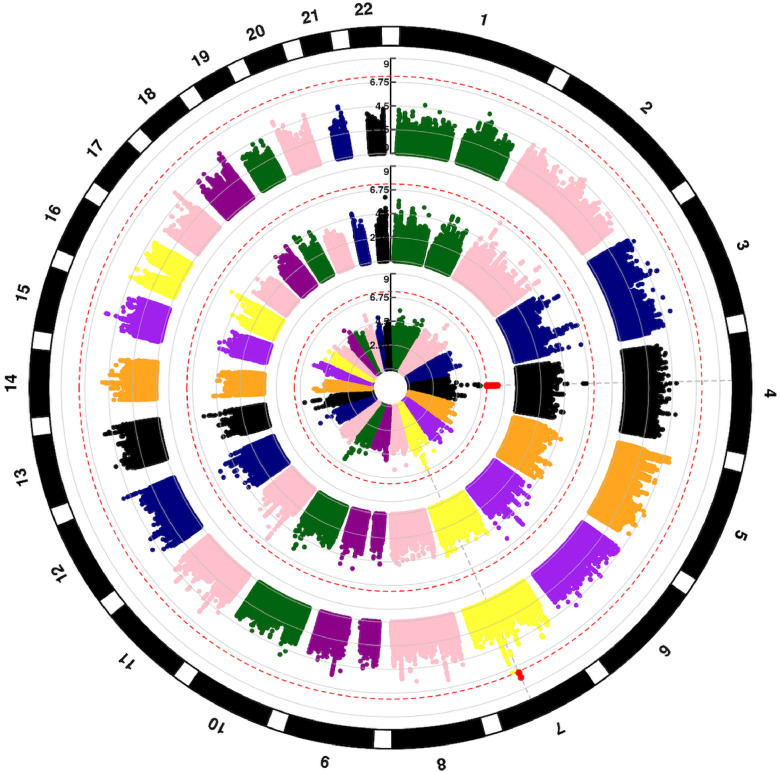
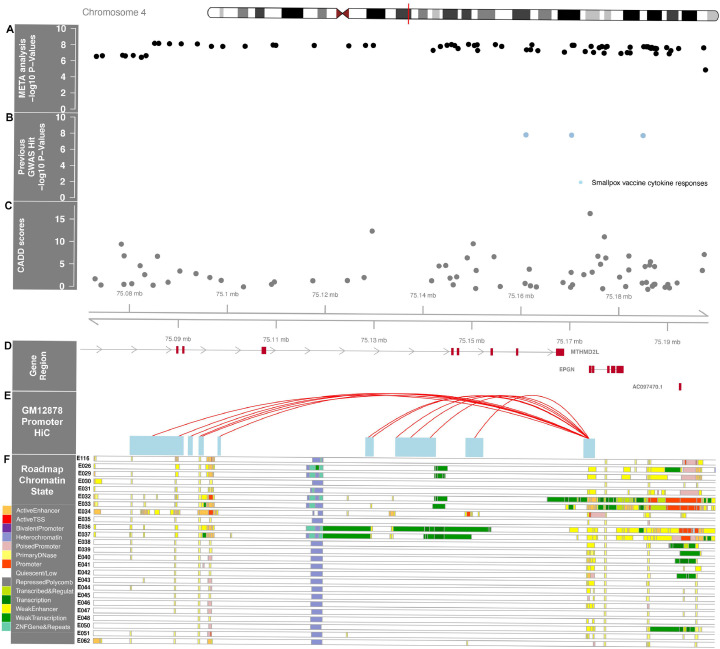
We identified SNP associations with TRM and donor-recipient genotype mismatch at 4q13.3 (Fig. 4, inner circle) and recipient variants at 7q22 (Fig. 4, outer circle, Table 2, Supplemental Table 2). The QQplot shows a low genomic inflation (λ<1.03) for both cohorts and the meta-analyses (Supplemental Figure 11). Genotype mismatches between the donor and recipient associate with a region spanning from 75,084,800–75,191,890 bp on Chromosome 4 (Fig. 5) at the genome-wide level. For rs75868097 (75,148,954 bp), the most significant variant in this region (HR = 2.66, 95% CI 1.92,3.58, p = 4.6 × 10−9), the increased risk of TRM was attributable to either donor or recipient having at least one A allele (A/A or A/T) when the corresponding match was TT; the frequency of this mismatch in cohorts 1 and 2 was ∼2.7% and 2% respectively (Supplemental Figure 12). Due to the low allele frequency, a dosage effect analysis was not possible as there are an inadequate number of donor-recipient pairs with 2, 3 or 4 of the A alleles between the pair. Given the association is due to a pair having 1 copy of the risk allele between them, unsurprisingly the risk allele (A) is associated with higher recipient TRM in donor genome only (HR = 2.98, 95% CI 1.92, 4.54, p = 7.4 × 10−7) and recipient genome only (HR = 2.19, 95% CI 1.36, 3.53, p = .001). This TRM association is also seen in OS (HR = 1.75, 95% CI 1.37, 2.26, p = 1.23 × 10−5) but not at the genome-wide level.
Fig. 4.
Circular Manhattan plot of SNP associations with transplant-related mortality. Chromosomes are numbered on the outside. p = 5 × 10−8 is marked with dashed red lines. Each SNP p value association with transplant-related mortality is a colored dot, with genome-wide significant SNPs as red dots. Results are shown for recipient (outer circle), donor (middle circle) and donor-recipient mismatches (inner circle), respectively.
Fig. 5.
Chromosome 4 regional association plot of transplant related mortality with donor-recipient allele difference. A) Regional meta-analysis SNP –log10(p-values) are plotted as solid black dots at the top of the figure. The higher the -log10 number, the lower the p-value. Rs75868097, the most significant genome-wide association, is labeled and marked with a thin blue line that transverses all panels. The x-axis is the chromosome position in mega bases (Mb). B) shows significant associations in previous GWAS as solid blue dots, including one study of smallpox vaccine cytokine responses. C) Corresponding CADD scores in the region for the variants are shown as solid gray dots. D) Genes in the region are plotted as red boxes on a gray line and annotated with E) HiC data from a lympho blastoid cell line with the F) ENCODE epigenome roadmap for all blood, T-cell, HSC and B-cells; the cell line numbers shown down the left side (ie., E026, etc.) correspond to specific epigenome road map cell lines. The colors indicate the Chrom HMM status as defined in the key on the bottom left of the figure.
Rs75868097 and the 57 associated variants in LD span two genes, methylenetetrahydrofolate dehydrogenase 2-like protein (MTHFD2L), responsible for carbon metabolism and conversion of folate to formate in the mitochondria [28] and Epithelial Mitogen (EPGN), an epidermal growth factor. Genome-wide gene-based association analyses using MAGMA show EPGN significantly associates with TRM (p = 5 × 10−8).
The most significant recipient SNP association with TRM is in 7q22 region at rs141591562 (Fig. 4, outer circle, Table 2, Supplemental Table 2) in Paired-immunoglobulin type 2-like receptor β (PILRB).The A allele confers a 2.3-fold increased risk of TRM (HR=2.33, 95% CI 1.74, 3.12, p = 1.26 × 10−8) (Supplemental Figure 13). Death due to GvHD, infection and organ failure comprise TRM and analyses of cause-specific mortality show an association with death due to GvHD (HR = 3.78, 95% CI 2.50, 5.70, p = 2.2 × 10−10), but not organ failure (p = .36) or infection (p = .33). This GvHD associations in recipient rs141591562 translates to a modest association with OS (HR = 1.32, 95% CI 1.04, 1.68, p = .02).
4. Discussion
We conducted the first GWAS of OS, DRM and TRM 1-year after BMT in two cohorts of recipients and their unrelated donors and identified multiple regions of association. Several of these associations have annotations implying biochemical function. Confirmation of these findings in additional cohorts and further studies of the functional consequences of these SNPs may contribute to donor selection and/or provide more individualized risk prediction/prognosis and the development of strategies to mitigate toxicity.
We demonstrated that genome-wide significant recipient SNPs in MBNL1 associate with OS (all-cause mortality), with contributions to both TRM and DRM. MBNL1 is an RNA-binding protein which can regulate alternative splicing with both repressor and activator functions, can auto-regulate MBNL1 mRNA and regulates the mRNA of hundreds of genes [29,30]. A recent study demonstrated that MBNL1 is required for the pathogenesis of AML or ALL with the mixed lineage leukemia (MLL) gene rearranged and that MBNL1 is frequently overexpressed in MLL-AML and MLL-ALL [31]. MLL-fusion proteins bind directly to the promoter region of MBNL1 which has been proposed as a potential therapeutic target for MLL-leukemias. In addition to its influence on leukemic cell survival, MBNL1 has a broad role in cellular RNA metabolism that is triggered in response to viral infections and stress [32,33]. GvHD is initiated via activation of antigen presenting cells (APCs) in response to stress from tissue damage which triggers a cascade of complex immune signaling pathways, expression of MHC class II, cell signaling and other immune-related genes requiring induction of RNA processing [34], [35], [36].
In donor genomes, DRM for AML and MDS patients was significantly associated with variants in PCNX4, with in silico analyses demonstrating rs114514727 is causal for this association and PCNX4 expression. PCNX4 is expressed in many organs and tissues including the blood and bone marrow, GI tract, liver and skin; these associations may warrant additional study to elucidate its association with death due to myeloid disease [37]. The other regions of association identified are less clear at this time and would benefit from a third cohort.
The Chromosome 4 region where donor and recipient allele mismatches significantly associate with TRM contains other genome-wide significant variants, including rs76183531 (p = 6.6 × 10−9), that are potentially deleterious (CADD = 17) and multiple loci that impact the binding of important transcription factors. Specifically, rs7694828 (p = 8.2 × 10−9) (RegulomeDB rank = 2b and probability score = 0.78) at 75,186,404 bp is located in a binding site of the transcriptional regulator CTCF (CCCTC-binding factor) across ∼150 cell lines including lymphoblastoid cell lines (LCL), lymph node, K562 (blast crisis cell line from chronic myeloid leukemia), bone, brain, lung, connective tissue and skin. CTCF functions as an insulator, preventing the influence of CIS-acting enhancers on gene activation and defines TAD boundaries. A conserved TAD boundary (75,100,000–75,260,000) encompasses this region [38] and is adjacent to a TAD containing CXCL (1,2,3,5,6,8,9,11). Interestingly, this region has been previously identified in a GWAS of vaccine response. Kennedy et al. identified four variants in strong LD where the homozygous minor allele associated with lower levels of IL-1β following smallpox vaccination in a population of almost 500 individuals [39]. All four variants were genome-wide significant in our GWAS, specifically if the donor-recipient pair had one or more minor alleles between them (eg, at least 1 out of 4 alleles), the recipient experienced significantly higher risk of TRM than those pairs who were both homozygous common. IL-1β is a pro-inflammatory cytokine secreted early in the inflammatory response and may be contributing to the risk of TRM through the combination of initiation and maintenance of host tissue inflammation and donor cell inflammatory response.
In recipient genomes, TRM, driven specifically by death due to GvHD, was associated with variants in the 7q22 region spanning the protein coding gene PILRB. The most significant variant, rs141591662 is in strong LD with rs7777462 (r2 = 0.78) and rs150702181 (r2 = 0.85), both of which impact transcription factor binding (RegulomeDB rank=2b, probability score ≥ .80). In LCLs, rs7777462 impacts binding of TBX21 (RegulomeDB rank = 2b, probability score = 0.79), which specifies Th1 lineage and represses alternative T cell fates [40]. Both variants reside in transcription start sites, an enhancer region or strong transcription region across all 27 T- and B-Cell, HSC and blood cell lines and are significant eQTL for PILRB and Paired-immunoglobulin type 2-like receptor α (PILRA) across blood and LCL [41].
Paired immunoglobulin-like receptors (PILRs) and killer immunoglobulin-like receptors (KIRs) are part of a family of leukocyte immunoglobulin-like receptors (LILRs) [42]. PILRB is an inhibitory receptor protein and its pair, PILRA, is an activating receptor with broad effects on immune response and neural function [43]. PILRA/B have opposing effects on development of acute GvHD in murine models [44,45]. High expression of PILRB and PD-L1 on regulatory dendritic cells has been shown to control murine GvHD, with younger mice exhibiting higher PILRB levels compared to older mice [46]. Recently, PILRA was shown to be required for MHC class I mediated alloantigen specific memory in monocytes and macrophages and promotes allograft rejection, with PILRB promoting tolerance [47].
While our study is the largest to date, it has some limitations. The cohorts are European American only and given the health disparities in access to and outcomes following transplant we are making a concerted effort to increase this dataset to better represent the US transplant population. While these data represent most unrelated allogeneic transplants performed in the US from 2000 to 2011, a third cohort would be a valuable addition for replication and for performing OS, TRM and DRM in the ALL recipient-donor pairs. Lastly, despite almost 3000 donor-recipient pairs, genome-wide analyses of cause-specific deaths remains underpowered for less common variants (MAF < 0.05), however we were able to examine genome-wide significant loci for cause specific mortality in more common variants (MAF > 0.05).
OS (all-cause mortality) is a composite phenotype of utmost importance to improving survivorship post URD-BMT. Detection of recipient and mismatch genetic variants associated with improved OS has implications for individualized patient risk/prognosis and advancing our understanding of contributors to all causes of death. Association of the donor variants to DRM and mismatches between donors and recipients to TRM clearly show that donor germline variation matters. Hence donor selection and matching on loci in addition to HLA may decrease specific causes of death and lead to improved understanding of the pathogenesis of those specific causes and yield improvements in OS after URD-BMT. Thus, we are pursuing additional replication and validation cohorts in European populations and a multi-ethnic cohort. Functional studies will be imperative to validate these findings if we are to successfully identify high-risk patients and alter approaches to treatment, prognosis and donor selection to improve survival after allogeneic BMT.
Data sharing statement
De-identified individual participant data that underlie the reported results are available at the Center for International Blood and Marrow Transplant (www.cibmtr.org). Both genotype and phenotype data will also be made available in dbGaP excluding 978 recipient-donor pairs for which the informed consent is not compliant with the NIH Genomic Data Sharing Policy. All GWAS summary data presented herein are publicly available on the FUMA website (https://fuma.ctglab.nl/browse), searchable by “TRM”, “DRM”, or “OS”.
Authorship contributions
L.S-C and TH designed the research and wrote the manuscript. L.S-C, L.M.P, J.W. E.K, A.R, A.C-G performed analyses and generated figures, C.A.H, D.V, D.S., X.S and L.P. performed the genotyping, A.C-G, Q.Z., X.Z., L.M.P, A.W and G.B performed imputation and quality control of genomic data, TH, KO, PM and MC adjudicated recipient causes of death. All authors reviewed and approved the manuscript.
Declaration of Competing Interest
PLM: Consulting: BlueBird Biotech, Bristol-Myers Squibb, Celgene, Fate Therapeutics, Janssen, Juno, Karyopharm, Magenta Therapeutics, Sanofi, Takeda; Honoraria: BlueBird Biotech, Bristol-Myers Squibb, Celgene, Fate Therapeutics, Janssen, Juno, Karyopharm, Magenta Therapeutics, Medscape, Takeda; Institutional Research Support: Celgene.
MCP: Institutional Research Support: Bristol-Myers Squibb, Novartis, Kite, GlaxoSmithKline; Consulting: Amgen (institution), Bristol-Myers Squibb (personal).
TH, LESC: report grants from NIH/NHLBI, grants from NIH/NCI, during the conduct of the study.
EK: reports a Graduate Student Fellowship from Pelotonia Foundation.
All other authors have nothing to declare.
Acknowledgments
Acknowledgements
The authors wish to acknowledge Dr. Mary Horowitz (MCW/CIBMTR), the late Dr. John Klein (MCW/CIBMTR), and the late Dr. John Hansen (FHCRC) for their support of this project. The authors also acknowledge the participation of all the patients and donors who consented to the bio repository and research database, as well as all transplant centers which participated in the CIBMTR Database and Bio repository studies.
Funding
This work was supported by grants from the National Institute of Health. LESC and TH were supported by NIH/NHLBI R01 HL102278 and NIH/NCI R03 CA188733 to perform this work. EK was supported by the Pelotonia Foundation Graduate Student Fellowship. Any opinions, findings, and conclusions expressed in this material are those of the author(s) and do not necessarily reflect those of the Pelotonia Fellowship Program or The Ohio State University.
The CIBMTR is supported primarily by Public Health Service U24CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI) and the National Institute of Allergy and Infectious Diseases (NIAID); U24HL138660 from NHLBI and NCI; OT3HL147741, and U01HL128568 from the NHLBI; HHSH250201700006C, and HHSH250201700007C from the Health Resources and Services Administration (HRSA); and N00014–20–1–2705 and N00014–20–1–2832 from the Office of Naval Research; Additional federal support is provided by P01CA111412, R01CA152108, R01CA215134, R01CA218285, R01CA231141, R01AI128775, R01HL126589, R01HL129472, R01HL130388, R01HL131731, U01AI069197, U01AI126612, UG1HL06924, and BARDA. Support is also provided by Be the Match Foundation, Boston Children's Hospital, Dana Farber, St. Baldrick's Foundation, Stanford University, the Medical College of Wisconsin the National Marrow Donor Program, and from the following commercial entities: Actinium Pharmaceuticals, Inc.; Adienne SA; Allovir, Inc.; Amgen, Inc.; Angiocrine Bioscience; Astellas Pharma US; bluebird bio, Inc.; Bristol Myers Squibb Co.; Celgene Corp.; CSL Behring; CytoSen Therapeutics, Inc.; Daiichi Sankyo Co., Ltd.; ExcellThera; Fate Therapeutics; Gamida-Cell, Ltd.; Genentech Inc; Incyte Corporation; Janssen/Johnson & Johnson; Jazz Pharmaceuticals, Inc.; Kiadis Pharma; Kite, a Gilead Company; Kyowa Kirin; Legend Biotech; Magenta Therapeutics; Merck Sharp & Dohme Corp.; Millennium, the Takeda Oncology Co.; Miltenyi Biotec, Inc.; Novartis Pharmaceuticals Corporation; Omeros Corporation; Oncoimmune, Inc.; Orca Biosystems, Inc.; Pfizer, Inc.; Pharmacyclics, LLC; Sanofi Genzyme; Stemcyte; Takeda Pharma; Vor Biopharma; Xenikos BV. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration (HRSA) or any other agency of the U.S. Government.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2021.101093.
Contributor Information
Theresa Hahn, Email: theresa.hahn@roswellpark.org.
Lara E. Sucheston-Campbell, Email: lara.sucheston-campbell@osumc.edu.
Appendix. Supplementary materials
References
- 1.Lee S.J. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110:4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed] [Google Scholar]
- 2.Hahn T. Significant improvement in survival after allogeneic hematopoietic cell transplantation during a period of significantly increased use, older recipient age, and use of unrelated donors. J Clin Oncol. 2013;31:2437–2449. doi: 10.1200/JCO.2012.46.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gooley T.A. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363:2091–2101. doi: 10.1056/NEJMoa1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majhail N.S. Significant improvement in survival after unrelated donor hematopoietic cell transplantation in the recent era. Biol Blood Marrow Transpl. 2015;21:142–150. doi: 10.1016/j.bbmt.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karaesmen E. Replication and validation of genetic polymorphisms associated with survival after allogeneic blood or marrow transplant. Blood. 2017;130:1585–1596. doi: 10.1182/blood-2017-05-784637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang H. Validation of genetic associations with acute GVHD and non-relapse mortality in DISCOVeRY-BMT. Blood Adv. 2019;3:2337–2341. doi: 10.1182/bloodadvances.2019000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clay-Gilmour A.I. Genetic association with B-cell acute lymphoblastic leukemia in allogeneic transplant patients differs by age and sex. Blood Adv. 2017;1:1717–1728. doi: 10.1182/bloodadvances.2017006023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hahn T. Establishment of definitions and review process for consistent adjudication of cause-specific mortality after allogeneic unrelated-donor hematopoietic cell transplantation. Biol Blood Marrow Transpl. 2015;21:1679–1686. doi: 10.1016/j.bbmt.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Q. Exome chip analyses identify genes affecting mortality after HLA-matched unrelated-donor blood and marrow transplantation. Blood. 2018;131:2490–2499. doi: 10.1182/blood-2017-11-817973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J. Genome-wide association analyses identify variants in <em>IRF4</em>associated with Acute Myeloid Leukemia and Myelodysplastic Syndrome susceptibility. bioRxiv. 2019 doi: 10.3389/fgene.2021.554948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y. Pre-HCT mosaicism increases relapse risk and lowers survival in acute lymphoblastic leukemia patients post-unrelated HCT. Blood Adv. 2021;5:66–70. doi: 10.1182/bloodadvances.2020003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan L. OSAT: a tool for sample-to-batch allocations in genomics experiments. BMC Genomics. 2012;13:689. doi: 10.1186/1471-2164-13-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sucheston-Campbell L.E. Identification and utilization of donor and recipient genetic variants to predict survival after HCT: are we ready for primetime? Curr Hematol Malig Rep. 2015;10:45. doi: 10.1007/s11899-014-0246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price A.L. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.Das S. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarthy S. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizvi A.A. gwasurvivr: an R package for genome wide survival analysis. Bioinformatics. 2018 doi: 10.1093/bioinformatics/bty920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamazon E.R. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hormozdiari F. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Võsa U. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv. 2018 [Google Scholar]
- 22.Carithers L.J. A novel approach to high-quality postmortem tissue procurement: the GTEx Project. Biopreserv Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong S., Boyle A.P. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum Mutat. 2019;40:1292–1298. doi: 10.1002/humu.23791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitt A.D. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H. ARID5B influences anti-metabolite drug sensitivity and prognosis of acute lymphoblastic leukemia. Clin Cancer Res. 2019 doi: 10.1158/1078-0432.CCR-19-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolusani S. Mammalian MTHFD2L encodes a mitochondrial methyl-enetetrahydrofolate dehydrogenase ISO-Zyme expressed in adult tissues. J Biol Chem. 2011;286:5166–5174. doi: 10.1074/jbc.M110.196840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teplova M., Patel D.J. Structural insights into RNA recognition by the alternative-splicing regulator muscle blind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konieczny P., Stepniak-Konieczna E., Sobczak K. MBNL expression in auto regulatory feedback loops. RNA Biol. 2018;15:1–8. doi: 10.1080/15476286.2017.1384119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itskovich S.S. MBNL1 regulates essential alternative RNA splicing patterns in MLL-rearranged leukemia. Nat Commun. 2020;11:2369. doi: 10.1038/s41467-020-15733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onishi H. MBNL1 associates with YB-1 in cytoplasmic stress granules. J Neurosci Res. 2008;86:1994–2002. doi: 10.1002/jnr.21655. [DOI] [PubMed] [Google Scholar]
- 33.White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Zeiser R., Blazar B.R. Pathophysiology of chronic graft-versus-host disease and therapeutic targets. N Engl J Med. 2017;377:2565–2579. doi: 10.1056/NEJMra1703472. [DOI] [PubMed] [Google Scholar]
- 35.Perkey E., Maillard I. New insights into graft-versus-host disease and graft rejection. Annu Rev Pathol. 2018;13:219–245. doi: 10.1146/annurev-pathol-020117-043720. [DOI] [PubMed] [Google Scholar]
- 36.Wolf D. Novel treatment concepts for graft-versus-host disease. Blood. 2012;119:16–25. doi: 10.1182/blood-2011-08-339465. [DOI] [PubMed] [Google Scholar]
- 37.Zahn-Zabal M. The neXtProt knowledgebase in 2020: data, tools and usability improvements. Nucleic Acids Res. 2020;48:D328–D334. doi: 10.1093/nar/gkz995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacArthur E., Capra J. 2020. Topologically Associating Domain (TAD) Boundaries Stable Across Diverse Cell Types are Evolutionarily Constrained and Enriched for Heritability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy R.B. Genome-wide analysis of polymorphisms associated with cytokine responses in smallpox vaccine recipients. Hum Genet. 2012;131:1403–1421. doi: 10.1007/s00439-012-1174-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soderquest K. Genetic variants alter T-bet binding and gene expression in mucosal inflammatory disease. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31:1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis Marffy A.L., McCarthy A.J. Leukocyte Immunoglobulin-Like Receptors (LILRs) on Human Neutrophils: modulators of Infection and Immunity. Front Immunol. 2020;11:857. doi: 10.3389/fimmu.2020.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda K., Nakamura A. Regulation of immune and neural function via leukocyte Ig-like receptors. J Biochem. 2017;162:73–80. doi: 10.1093/jb/mvx036. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J. Prevention of murine acute graft-versus-host disease by recipient-derived paired immunoglobulin-like receptor B lenti virus-transfected dendritic cells. Acta Haematol. 2010;124:134–140. doi: 10.1159/000315553. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura A., Kobayashi E., Takai T. Exacerbated graft-versus-host disease in Pirb-/- mice. Nat Immunol. 2004;5:623–629. doi: 10.1038/ni1074. [DOI] [PubMed] [Google Scholar]
- 46.Scroggins S.M., Olivier A.K., Meyerholz D.K., Schlueter A.J. Characterization of regulatory dendritic cells that mitigate acute graft-versus-host disease in older mice following allogeneic bone marrow transplantation. PLoS ONE. 2013;8:e75158. doi: 10.1371/journal.pone.0075158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai H. PIRs mediate innate myeloid cell memory to non-self MHC molecules. Science. 2020;368:1122–1127. doi: 10.1126/science.aax4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.