Abstract
The spread of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged suddenly at the end of 2019 and the disease came to be known as coronavirus disease 2019 (COVID-19). To date, there is no specific therapy established to treat COVID-19. Identifying effective treatments is urgently required to treat patients and stop the transmission of SARS-CoV-2 in humans. For the present review, >100 publications on therapeutic agents for COVID-19, including in vitro and in vivo animal studies, case reports, retrospective analyses and meta-analyses were retrieved from PubMed and analyzed, and promising therapeutic agents that may be used to combat SARS-CoV-2 infection were highlighted. Since the outbreak of COVID-19, different drugs have been repurposed for its treatment. Existing drugs, including chloroquine (CQ), its derivative hydroxychloroquine (HCQ), remdesivir and nucleoside analogues, monoclonal antibodies, convalescent plasma, Chinese herbal medicine and natural compounds for treating COVID-19 evaluated in experimental and clinical studies were discussed. Although early clinical studies suggested that CQ/HCQ produces antiviral action, later research indicated certain controversy regarding their use for treating COVID-19. The molecular mechanisms of these therapeutic agents against SARS-CoV2 have been investigated, including inhibition of viral interactions with angiotensin-converting enzyme 2 receptors in human cells, viral RNA-dependent RNA polymerase, RNA replication and the packaging of viral particles. Potent therapeutic options were reviewed and future challenges to accelerate the development of novel therapeutic agents to treat and prevent COVID-19 were acknowledged.
Keywords: coronavirus disease 2019, severe acute respiratory syndrome coronavirus 2, RNA-dependent RNA polymerase, nucleoside analogues, antibodies, Chinese herbal medicine
1. Introduction: Identification of the COVID-19 pandemic
In December 2019, the novel coronavirus (2019-nCoV) emerged, eventually causing a global pandemic of severe acute respiratory syndrome (SARS). Soon after the outbreak, the virus was identified in January of 2020 and was temporally named 2019-nCoV, and then labelled SARS-CoV-2 by the World Health Organization (WHO). Later on, the disease was re-named as coronavirus disease 2019 (COVID-19) to differentiate it from other coronavirus infections of SARS or Middle Eastern respiratory syndrome (MERS). The acute clinical symptoms of patients with COVID-19 include fever, dry cough and dyspnea, which are similar to those of SARS and MERS. By now, more than several million confirmed COVID-19 cases have been reported in >200 countries, with an estimated mortality risk of 2–6% worldwide (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen).
The entire genome of novel SARS-CoV-2 (MN908947.3) was soon sequenced and determined to have 29,903 nucleotides. It was revealed to have 79.5% nucleotide similarity with SARS-CoV, which appeared in Guangdong Province in China in 2002, >50% similarity with MERS-CoV, which emerged in Middle Eastern countries in 2012, and 96% similarity to the bat coronavirus bat-CoV-RaTG13 (1). The coronavirus genome structures of SARS-CoV, MERS-CoV and SARS-CoV-2 are similar in that they contain a single strand of RNA, which is organized by the 5′-leader-UTR-RNA-dependent RNA polymerase (RdRp)-spike (S)-envelope (E)-Membrane (M)-nucleocapsid (N)-3′UTR poly(A) tail, with accessory non-structural genes interspersed throughout the structural genes, as presented in Fig. 1A-D (2).
Figure 1.
Genomic structure and potential drug targets for SARS-Cov-2, SARS-CoV and MERS-CoV. (A-C) Genomic organization of (A) SARS-Cov-2 (GenBank ID, MN908947.3), (B) SARS-CoV and (C) MERS-CoV, indicating the coding regions for proteins that are potential drug targets. (D) The main structural proteins of SARS-Cov-2 were indicated on viral particles. SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle Eastern respiratory syndrome coronavirus; RdRp, RNA-dependent RNA polymerase; S, spike; M, membrane; N, nucleocapsid; E, envelope.
In the history of human coronavirus-caused infections, at the international level, there have been six types of coronavirus disease: 229E, NL63, HKU1, OC43, MERS and SARS. The first human coronavirus was identified in the 1960s and SARS-CoV and MERS-CoV caused epidemics in 2002 and 2012, respectively. SARS-CoV2 is the seventh coronavirus, which triggered the most severe global spread of COVID-19 in humans ever recorded. The virus particle is composed of a single-stranded, positive-sense RNA enveloped by glycoproteins and has a typical coronavirus appearance under an electron microscope. As that of SARS-CoV or MERS-CoV, the genomic RNA of SARS-CoV2 encodes for non-structural proteins, such as RdRps, other viral enzymes and at least four structural proteins: The S protein, the E protein, the M protein and the N protein, as visualized in Fig. 1D. The N protein of SARS-CoV2 has nearly 90% similarity with the amino acid sequence of SARS-CoV (1).
Along with SARS-CoV and MERS-CoV, SARS-CoV2 belongs to the subgenus Sarbecovirus of the genus β-coronavirus, which is closely related to bat-SL-CoVZC45 or bat-SL-CoVZXC21. Although several animals are potential carriers of SARS-CoV2, none has been confirmed to be an intermediate host for SARS-CoV2. Further studies have indicated that SARS-CoV2 uses its S protein to bind to the angiotensin-converting enzyme 2 (ACE2) receptors in human cells, similar to how SARS-CoV used to infect humans. Other studies have demonstrated that both SARS-CoV and SARS-CoV-2 use S proteins to bind to the ACE2 receptors of host cells for viral infection (2–6).
The pathogenesis of SARS-CoV-2 infection is composed of four stages including at least two consecutive stages: The viral replication phase and the immunological response phase (7–9). The viral phase is the initial one during which ~80% of patients with COVID-19 may be asymptomatic or have mild symptoms. Whereas, ~20% of patients enter the second immunological phase in which their condition becomes more severe and their immune system becomes hyper-responsive. Certain studies have reported that the circulating natural killer (NK) cells of patients with COVID-19 have been reduced, whereas the expression of inhibitory receptor NKG2A in NK cells is increased to lower the cytolytic activity of NK cells. Furthermore, dendritic cells, activated neutrophils and inflammatory monocytes increase in patients with COVID-19, with higher percentages of patients experiencing severe damage to their respiratory tract and lungs (10,11). At times, a third phase of hypercoagulability among patients is followed again by the second immunological phase. Finally, in the fourth stage, organ injury and failure occur in the most severe COVID-19 cases.
Although no drugs have been approved to treat coronavirus infections, certain existing antiviral drugs for SARS, MERS or other viral infections have been used as emerging treatments for COVID-19 and demonstrated beneficial effects in patients. In order to provide solid evidence to select therapeutics to immediately treat COVID-19, the PubMed database was searched for the present review using the key words ‘therapeutics’ and ‘COVID-19’ to identify articles published since December 2019. Recent advances in the use of chloroquine (CQ; 4-aminoquinoline), remdesivir, ribavirin, favipiravir, galidesivir, monoclonal antibodies (mAbs), patient-derived plasma, natural products and Chinese herbal medicine (CHM) for the treatment of SARS or MERS and COVID-19 were reviewed in order to outline treatment options for health professionals and medical researchers (12–14).
2. Chloroquine and hydroxychloroquine
In vitro and in vivo studies suggested that CQ and hydroxychloroquine (HCQ) are effective against SARS and MERS. CQ was initially approved as an immuno-modulating drug to treat malaria (15). Later on, the less toxic HCQ was synthesized and approved by the US Food and Drug Administration (FDA) to treat malaria (16). Both CQ and HCQ appear to generate antiviral actions either by inhibiting the virus from binding to the ACE2 receptors on human cells or through their anti-inflammatory effect. In cultured primate cells treated with CQ either prior to or after SARS-CoV infection, CQ had strong antiviral effects, indicating that CQ may be used to treat SARS-CoV infections, including SARS-CoV-2 (17). By detecting viral RNA copy numbers and the N protein expression of SARS-CoV-2 in the supernatant of treated Vero E6 cells, it was revealed that CQ strongly inhibited the RNA replication of SARS-CoV-2 (18).
In several clinical trials in China, CQ and HCQ were tested as emerging treatments for COVID-19 and improved patients' recovery. CQ was effective against SARS-CoV2 infections in >100 patients by shortening their hospital stay and alleviating their clinical symptoms (19). In a clinical study involving 22 patients with COVID-19, the percentage of patients who became SARS-CoV2-negative in the CQ group was slightly higher than that in the lopinavir/ritonavir group at days 7, 10 and 14 of treatment with CQ (20). Another study reported that HCQ had stronger antiviral activity against COVID-19 than CQ. A similar therapeutic effect was indicated in the use of HCQ sulfate for patients with COVID-19 compared to the use of CQ phosphate (21). In addition, a group of French patients with COVID-19 were treated with a combination of HCQ and azithromycin, which significantly reduced the viral load in the patients after 6 days of treatment. This aforementioned study suggests that azithromycin has a synergistic, antiviral effect with HCQ. To validate this outcome, the SARS-CoV-2 IHUMI-3 strain from the same group of patients was used to infect the cultured Vero E6 cells in order to observe the combined antiviral effects of HCQ and azithromycin. The viral replication copy numbers from treated cell lysates decreased significantly through a combined treatment involving HCQ and azithromycin (22,23). A multi-center retrospective study was performed to include patients with a COVID-19-related admission in six hospitals in Detroit (USA) from March 10 to May 2, 2020. In total, 2,541 patients were enrolled and followed up for 28.5 days; the patients had a median hospitalization time of 6 days. HCQ reduced the mortality of infected patients by 66% and HCQ combined with azithromycin decreased the mortality of infected patients by 71% compared to that of patients who did not receive any HCQ or azithromycin. This clinical study implied that treatment with HCQ alone or in combination with azithromycin is associated with significantly reduced mortality among patients with COVID-19 (24).
However, certain studies have indicated that CQ or HCQ does not improve the clinical outcomes of patients with COVID-19. A systematic review using PubMed, EMBASE, the Cochrane Library, MedRxiv and International Clinical Trials Registry Platform suggested that five of seven completed clinical trials indicated positive results using CQ or HCQ to treat COVID-19, whereas two of seven trials revealed no improvement in patients with COVID-19 treated with CQ or HCQ. To further learn about the potential benefits and harms of CQ or HCQ for treating COVID-19, four randomized controlled trials (RCTs), 10 cohort studies and 9 case series published between December 2019 and May 2020 were collected and analyzed in order to determine possible treatment effects of CQ or HCQ. The efficacy of CQ or HCQ treatment for COVID-19 is relatively low and some results are positive, whereas some results are negative (25). To systematically review the available data on the efficacy and safety of CQ and HCQ for treating COVID-19, 12 observational and 3 RCTs to include 10,659 patients were reviewed. The efficacy of CQ/HCQ for COVID-19 was analyzed for 5,713 patients who received CQ/HCQ in comparison with 4,966 patients who only received standard care. Meta-analysis indicated no significant reduction in the mortality of COVID-19 patients by treatment with HCQ. Hence, CQ or HCQ may not improve clinical outcomes for COVID-19 (26,27). A recent study also searched PubMed, Embase and Web of Science for RCTs and assessed the efficacy and safety of HCQ/CQ therapy alone for SARS-CoV-2 infection. HCQ did not reduce the requirement for hospitalization among outpatients, but led to a significantly higher rate of any adverse event compared to the control group (28).
Overall, the antiviral activity of CQ and HCQ is evident in both experimental and clinical studies, even though certain research has reported negative outcomes of CQ and HCQ for treating COVID-19 in clinical trials. This discrepancy may be due to several reasons. First, CQ and HCQ may have been used in patients with different time phases of the disease in different clinical trials. Since CQ and HCQ inhibit the entry and replication of the virus into lung cells, the therapeutic effect may be decreased if patients are in the late stage of the disease progression. Furthermore, the severity of a patient may impact the therapeutic effects. The therapeutic effects of CQ and HCQ may be weaker or have no significant impact on patients with a severe status. Finally, the therapeutic effects of CQ and HCQ are influenced by treatment time and dosage. It usually takes 5–7 continuous days to apply CQ or HCQ to patients with COVID-19 to achieve clinical improvements.
3. Remdesivir and other nucleoside analogues
As non-structural proteins, such as RdRp and helicase, have a critical role in the replication, transcription and translation processes of the coronavirus, nucleoside analogues (NAs), which have similar structures to adenosine or guanine, are able to target RdRp and be integrated into RNA synthesis to inhibit viral infection (12). However, it is far more difficult to develop NAs against coronaviruses such as SARS-CoV, MERS-CoV and SARS-CoV-2 due to the unique proofreading of 3′-5′ exoribonuclease in coronaviruses (29). The NAs and remdesivir (GS-5734), which was originally approved as a reverse transcriptase inhibitor to treat HIV, produced potential antiviral actions against coronaviruses. In 2016, Warren et al (30) reported on the antiviral activity of remdesivir against Ebola virus (EBOV) by inhibiting viral replication, decreasing pathological changes and reducing clinical symptoms in non-human primate models with EBOV infections. Later, in 2017, remdesivir strongly inhibited viral replication in primary human airway cultures infected with coronavirus and effectively decreased the viral load in a mouse model infected with SARS-CoV. In 2018, remdesivir was reported to significantly inhibit SARS-CoV and MERS-CoV infections in in vitro and in vivo models. Other studies also suggested that remdesivir has significant antiviral activity against several CoVs, including SARS-CoV, MERS-CoV, the highly pathogenic SARS-CoV-2 and emergent Bat CoVs in cultured cells through in vitro experiments and mouse models in vivo (31,32). Recently, a combination of remdesivir and GC376, an inhibitor of the 3C-like protease (3CLpro) for treating feline infectious peritonitis (coronavirus infection), had a synergistic antiviral effect against SARS-CoV-2 in Vero cells, implying that these compounds inhibit the replication of SARS-CoV-2 through different drug targets (33).
Several clinical trials have been performed to evaluate the efficacy and safety of intravenous remdesivir for patients with SARS-CoV-2 infection (NCT04292899, NCT04292730, NCT04257656, NCT04252664 and NCT04280705) (34). In 2020, a clinical trial to use remdesivir to treat patients with severe COVID-19 suggested that 36 out of 53 patients (68%) exhibited significant improvement in clinical symptoms. The 53 patients were given remdesivir for a period of 10 days at a dose of 200 mg on day 1, followed by 100 mg daily for the remaining 9 days of treatment (35). An RCT involving 596 patients with SARS-CoV-2 infection at 105 hospitals in the US, Europe and Asia was performed from March 15 through to April 18, 2020. Patients were randomized into three groups with a 10-day treatment with remdesivir (n=197), a 5-day treatment with remdesivir (n=199) or standard care (n=200). Patients in the 5-day remdesivir group were significantly improved regarding their clinical symptoms compared to those who received standard care (36). Another large-scale, double-blinded, randomized, placebo-controlled trial enrolled 1,062 patients; 541 were treated with remdesivir and 521 were treated with placebo. The patients with COVID-19 treated with remdesivir recovered more rapidly than the patients treated with placebo. Compared to the placebo, remdesivir shortened the recovery time by 5 days (37). Recently, a retrospective comparative study was performed in 5 hospitals in the US. Out of 2,483 patients with confirmed COVID-19, 342 were treated with remdesivir. 184 patients received remdesivir and corticosteroids, while 158 patients were given remdesivir alone. Remdesivir-treated patients recovered more rapidly than the matched control patients who did not receive remdesivir (38). In 2020, the FDA approved remdesivir for the emerging treatment of hospitalized patients with COVID-19 and it was recommended for use in adult and pediatric patients requiring hospitalization aged >12 years with a bodyweight of at least 40 kg (39).
In addition to remdesivir, other NAs are able to target RdRp and inhibit the RNA replication of SARS-CoV-2. A synthetic RdRp model with 801 amino acid residues was generated to have >97% sequence identity to the RdRp of SARS-CoV-2 and the FDA-approved drugs of ribavirin, galidesivir and remdesivir were tested in this model. These compounds use different hydrogen bonds to interact with the residues of RdRp of SARS-CoV-2 (40). Ribavirin was originally approved to treat the hepatitis C virus (HCV), respiratory syncytial virus and EBOV infection. As a guanine derivative, ribavirin targets RdRp and interferes with RNA synthesis and the mRNA capping of viruses. Recent studies have also indicated that ribavirin has antiviral activity against SARS-COV-2 (3,40). Importantly, ribavirin, combined with interferon, significantly increased antiviral activity against SARS-CoV and MERS-CoV via in vitro and in vivo studies, including in primate models (41,42).
Favipiravir also effectively inhibited the RdRp of RNA viruses, such as EBOV and HCV (42,43). In a recent study, favipiravir inhibited the replication of SARS-COV-2 in cultured Vero E6 cells (44). In clinical studies, the symptoms of patients with COVID-19 improved significantly after they were treated with favipiravir for 2.5 days, compared to 4.2 days in control patients at hospitals in Wuhan and Shenzhen (China) (45). Galidesivir (BCX4430) is an adenosine analogue that was developed to treat RNA viral infections, including HCV. Galidesivir significantly decreased the viral load and improved the survival of animal models with MERS-CoV and SARS-CoV infections (46). In the recently validated RdRp model, created to examine the antiviral activity of drugs against COVID-19, ribavirin, remdesivir and galidesivir effectively inhibited the replication of SARS-COV-2, further suggesting their potential use in clinical practice (40).
4. Antibody-based therapy for COVID-19
Antibodies (Abs) have long been used to neutralize viral antigens to treat viral diseases. One adaptive immune response in patients with SARS-CoV-2 infection is the production of specific Abs. The Abs from patients with COVID-19 are able to bind to the S protein or RBD of SARS-CoV-2 to block it from interacting with ACE2 receptors in order to prevent viral replication. A subset of the Abs may be able to inhibit authentic SARS-CoV-2 infection. Although the titers of neutralizing Abs against SARS-CoV-2 in human plasma eventually decrease over time, these Abs may remain in the human body for at least three months (47,48). mAbs are able to specifically bind to epitope proteins on virions to inhibit virus packaging. The neutralizing mAbs are generated to target the S protein of SARS-CoV and MERS-CoV to inhibit them from entering the human body (49,50). CR3022 is a neutralizing mAb used to combat SARS-CoV; it binds to the receptor-binding domain (RBD) of the S protein to inhibit the virus from entering. Thus, it may be used to treat COVID-19 (51).
Since the viruses tend to develop mutations in the targeted epitopes to escape Ab-mediated neutralization, a cocktail of mAbs that targets different epitopes in the viruses to neutralize mutant viruses is required in experimental and clinical therapy. Previously, a more effective mAb against SARS-CoV was obtained by combing several mAbs to target conserved epitopes of the coronavirus (52). In January 2020, Distributed Bio developed a panel of ultra-high affinity mAbs to recognize and neutralize a panel of isotopes on SARS-CoV-2 to block COVID-19 infection. Certain mAbs against COVID-19 have been developed and proceeded to clinical trials (53). An RCT against EBOV indicated that single or triple mAbs significantly reduced the mortality of patients with Ebola virus disease, providing clinical evidence that mAbs have potential utility for the treatment of COVID-19 (54).
To date, >30 Abs have demonstrated a potentially neutralizing effect against SARS-CoV-2 infection. Most Abs have been generated from B cells or fresh peripheral blood mononuclear cells, depending on the neutralizing targets on SARS-CoV-2. The antiviral efficacy of most of these Abs has been demonstrated in preclinical studies and certain Abs, including CT-P59, ADG20, CB6LALA, AZD8895, AZD1061 and DXP-593, are in phase 2 or 3 clinical trials against COVID-19 (55,56). Most recently, a clinical trial included 452 COVID-19 patients to test the efficacy of a neutralizing Ab, LY-CoV555, based on three doses (700, 2,800 or 7,000 mg) or a placebo; the clinical outcomes were evaluated at the time of the interim analysis of the phase 2 trial. From days 2 to 6, patients who had received LY-CoV555 had slightly lower severity of symptoms than those who had received the placebo. The LY-CoV555 appeared to accelerate the natural decline of viral load over time (ClinicalTrials.gov; no. NCT04427501) (57). Since the immune response factors are usually upregulated in the pathogenesis of SARS-CoV-2 infection, a human mAb of canakinumab, targeting interleukin (IL)-1β, was used to treat 17 patients with mild or severe non-intensive care COVID-19. Another 17 patients with similar symptoms were treated with standard HCQ plus lopinavir/ritonavir. Canakinumab therapy caused rapid, long-lasting improvement in oxygenation levels in 60.3% of the patients with COVID-19, which is better than that of standard therapy (58).
To improve patient survival, another Ab-based treatment that has been developed involves using convalescent plasma (CP) or immunoglobulins isolated from patients who have recovered from SARS, MERS or COVID-19. Over the past two decades, CP immunotherapy has been indicated to be effective and safe to treat patients with SARS, MERS and H1N1. Clinical studies have reported that patients with SARS treated with CP had lower mortality and shorter hospital stays than those not treated with CP (59–61). Since the patients' viral peak usually appears in the first week of infection in most viral diseases and a primary immune response of the host normally develops 10–14 days after infection (62), the CP should be collected earlier during the second or third week of the SARS-CoV-2 infection. During the recent outbreak of COVID-19, one study used CP transfusion to treat 5 critical patients with laboratory-confirmed COVID-19 in Shenzhen (China). The donor patients had been previously diagnosed with confirmed COVID-19 and were subsequently screened to exclude patients who were positive for SARS-CoV-2, hepatitis B virus, HCV, HIV and other respiratory viruses at the time of blood donation. Prior to and after CP transfusion, the patients were evaluated and had an increased pressure of arterial oxygen/fraction of inspired oxygen ratio, from 172–276 to 206–290 for 4 out of 5 patients within 7 days after transfusion, indicating therapeutic efficacy of CP for COVID-19 (63). A Korean group reported that two patients with severe and acute pneumonia caused by SARS-CoV-2 improved after the infusion of CP from donors (64). Hence, the clinical symptoms of transfused patients were improved and the patients had a significant increase in oxyhemoglobin saturation and lymphocyte counts. The average time from disease onset to transfusion was 16.5 days among these patients and the neutralizing Ab was maintained at a high level (above 1:640) after transfusion. In addition, the viral particles were undetectable and no severe adverse effects were observed in patients after transfusion, thus supporting the efficacy and safety of CP for treating SARS-CoV-2 infection (65).
5. Chinese herbal medicine for treating SARS-COV-2 infection
CHM is a potential treatment option for SARS-CoV-2 infection. Natural compounds from plants or CHM may have antiviral effects against SARS-CoV in certain aspects. Due to its safety and lower toxicity, combined with other conventional drugs, CHM has beneficial effects in terms of decreasing mortality and relieving the symptoms of patients with SARS-CoV in clinical trials (66,67).
In further studies, the clinical antiviral impact of CHM on SARS-CoV infection was supported through the identification of the active components in Chinese herbs. As a major component of the Chinese herb liquorice root (Glycyrrhiza uralensis), glycyrrhizin inhibited the replication of clinically isolated SARS-CoV (68). Wang et al (69) indicated that a compound derived from CHM, MOL376, had potential antiviral efficacy against SARS. Certain Chinese herbal components, such as emodin from the genus Rheumand polygonum, baicalin from Scutellaria baicalensis, scutellarin from Erigeron breviscapus, tetra-O-galloyl-β-D-glucose from Galla chinensis and luteolin from Veronicalina riifolia markedly inhibited the interaction of the S-protein of SARS-CoV with ACE2 receptors in human cells, implying that CHM exerts its antiviral activity by inhibiting viral entry into human cells. However, the antiviral activity of these compounds in relation to SARS-CoV-2 requires to be validated in future studies (70).
During the outbreak of COVID-19, common Chinese herb formulae have been used to treat SARS-CoV-2 infection. Based on the pathological mechanism of viral infection, CHM, with a matched structure to target ACE2 receptors in human cells, holds the antiviral potential to prevent SARS-CoV-2 infection. A Chinese group reported that more than 40 active ingredients of seven Chinese herbs, including Lonicerae Japonicae Flos and Mori Folium, displayed an antiviral effect against SARS-CoV by screening the CHM drug library acting on the S-protein-binding site of human ACE2 receptors for SARS-CoV (71). Lianhuaqingwen capsule is a Traditional Chinese Medicine (TCM) formula that is commonly used to treat influenza and other viral diseases. In a recent study, it was demonstrated to inhibit the replication of SARS-CoV-2 and its related inflammatory activity in vitro (72). As a widely used antiviral herb, Scutellariae radix the root of Scutellaria baicalensis, was studied for its antiviral action against SARS-CoV2. The ethanolic extract of Scutellaria baicalensis inhibited the replication of SARS-CoV2 by interfering with a key protease, 3CLpro enzyme of SARS-CoV2, in Vero cells. Further investigation identified that the major antiviral component of Scutellaria baicalensis is baicalein, which may effectively inhibit the 3CLpro activity of SARS-CoV2 with an IC50 of 0.39 µM (73). Liquiritin is one of the main flavonoids in Glycyrrhiza uralensis and has an antiviral function by mimicking type I interferon. In a transcriptional analysis to screen for potentially antiviral compounds, liquiritin significantly inhibited the replication of SARS-CoV2 in Vero E6 cells; hence, liquiritin has therapeutic effects on SARS-CoV2 infection (74). Other studies have extensively examined >100 herbal formulae in Chinese government-issued guidelines and Korean guidelines for treating COVID-19 with traditional medicine and support the idea that the formula of Qinfei Paidu Tang is recommended in both Chinese and Korean guidelines. Qinfei Paidu Tang is mainly composed of >20 formula, including Ephedrae herba, Glycyrrhizae radix and Rhizoma (75–77).
As CHM contains millions of natural compounds and it is tedious to screen for herbs effective against SARS-CoV-2, a recently developed computational prediction was employed as a rapid and efficient method to identify potentially active compounds of CHM against SARS-CoV-2, based on the interactions between the herb in question and the protein structures of the S-/N-proteins or the RdRp of SARS-CoV-2. The publicly available TCMSP database, a common antiviral and gene database, has provided invaluable as a source to predict the molecular mechanisms of CHM in relation to the target proteins of SARS-CoV2 (http://sm.nwsuaf.edu.cn/lsp/tcmsp.php). Certain novel components of CHM have been discovered to have antiviral activity against SARS-CoV-2 (78,79). Most recently, a study confirmed natural compounds against SARS-CoV and MERS-CoV in published papers, cross-checked these compounds in Chinese herbal databases to determine the commonly shared compounds and then used computer-based prediction to identify 13 compounds in CHM with potential antiviral activity in relation to SARS-CoV-2. In addition, >125 Chinese herbs contain 2 or more of these 13 compounds (80). These predicted herbs still require to be validated for their antiviral effects against COVID-19 through experimental and clinical studies.
6. Other options for treating COVID-19
In addition to the above-mentioned therapeutics, certain other drugs have demonstrated antiviral effects on SARS-CoV-2 and in patients with COVID-19. Tocilizumab is a recombinant, anti-human mAb of IgG1, which improved a patient's condition by inhibiting the IL-6 receptors. A clinical trial in China used tocilizumab to treat 20 patients with acute COVID-19 and indicated that 95% of patients (19) were cured and discharged from the hospital after two weeks (81). Tocilizumab has been approved to treat severe complications related to SARS-CoV-2 in China. Recent clinical trials have demonstrated the therapeutic efficacy of tocilizumab in severe COVID-19 cases. In a clinical trial, tocilizumab was used to treat 100 patients with COVID-19. After 10 days, 77 (77%) patients had improved in terms of respiratory conditions. Of the 77 patients, 61 achieved significant improvement as indicated by chest X-ray. As such, the therapeutic efficacy of tocilizumab for COVID-19 is rapid, sustained and associated with significant clinical improvement (82,83).
Ivermectin is an FDA-approved anti-parasitic drug that was effective against the SARS-CoV-2 virus in an in vitro study (84). Cultured Vero/hSLAM cells were infected with SARS-CoV-2 isolate and 5 µM ivermectin was then added to treat the cells for 24–72 h. After 24 h, there was a 93% reduction of viral RNA in the supernatant of samples treated with ivermectin compared to the control, DMSO. By 48 h, a 99.8% reduction in cell-associated viral RNA was observed with ivermectin treatment. Another study indicated that the antiviral activity of ivermectin targets and inhibits the host's importin α/β1 nuclear transport proteins of viral proteins, as also indicated for other types of RNA (85).
Based on the pathological immune regulation of COVID-19, blocking the IL signaling pathway may control the cytokine release from SARS-CoV-2 infection. Thus, a potential therapeutic option is to use the IL-1 receptor antagonist anakinra. Since anakinra blocks the activity of the pro-inflammatory cytokines IL-1α and IL-1β, and is commonly used to treat rheumatologic ailments (such as systemic-onset juvenile idiopathic arthritis), anakinra reduced mortality in pediatric and adult patients with secondary hemophagocytic lymphohistiocytosis triggered by a virus (86). A clinical study involving 112 patients with COVID-19 (56 treated with anakinra and 56 controls) was performed to evaluate the therapeutic effect of anakinra. The survival rate at day 28 was obtained for 69 patients (61.6%) and was significantly higher in anakinra-treated patients than in the controls (75.0 vs. 48.2%, P=0.007). Thus, anakinra significantly improved overall survival of COVID-19 patients (87).
Anti-inflammatory treatment is another option for preventing COVID-19. Several clinical trials have used anti-inflammatory drugs, including corticosteroids, cytokines and drugs that interfere with cytokine activities (such as tocilizumab and sarilumab) to block IL-6 activity, or infliximab and adalimumab to block TNF-α, and/or baricitinib and ruxolitinib as JAK1/2 signaling pathway inhibitors. The efficacy of corticosteroids was supported for the treatment of patients with severe COVID-19, although certain studies suggested that treatment with corticosteroid may not have an effect in hospitalized patients. A systematic analysis of several clinical trials that included 1,703 critically ill patients determined that corticosteroid treatment resulted in lower mortality among COVID-19 cases compared to standard care or a placebo (the WHO Rapid Evidence Appraisal for COVID-19 Therapies 2020) (88–90). In a recent clinical trial, 2,014 patients with COVID-19 were treated with dexamethasone; 22.9% of patients in the dexamethasone group and 25.7% of patients in the usual care group died within 28 days after treatment, suggesting antiviral efficacy of dexamethasone. Thus, dexamethasone is strongly recommended for hospitalized COVID-19 cases who require oxygen delivery (ClinicalTrials.gov no. NCT04381936; ISRCTN no. 50189673) (91).
The therapeutic efficacy of other drugs, such as pidotimod and flavonoids, has also been investigated for COVID-19. One clinical study enrolled 20 patients with COVID-19 and divided them into two groups: The pidotimod group and the control group. These patients were treated for 14 days and pidotimod significantly reduced their symptoms (92). Flavonoids are abundant in plants, fruits and vegetables and have antiviral activities that may offer protection against COVID-19. The flavonoids exhibit potential inhibitory activity against SARS-CoV-2 by binding to essential viral targets required for viral entry and/or replication. Flavonoids also exert marked immune-modulatory and anti-inflammatory effects, including the inhibition of various inflammatory cytokines. Further studies suggested that flavonoids are able to reduce the severity of COVID-19 by promoting lipids metabolism. Two flavonoids, namely quercetin and luteolin, have demonstrated promising multi-target activity against SARS-CoV-2 infection. Therefore, flavonoid-rich plants may be recommended as a supplementary treatment for SARS-CoV-2 infection (14). The mechanism of the antiviral activity of quercetin against SARS-CoV-2 including effects on 3CLpro, peptidase, the S glycoprotein, RNA replicase, RNA binding protein and the papain-like protease of SARS-CoV-2, to regulate microRNA genes involved in viral pathogenesis (92). To accelerate the identification of therapeutic drugs to treat COVID-19 effectively, a recent study used an in silico analysis of the immune protein network, single-cell RNA sequencing and neural networks to search for potential therapeutic drug targets against COVID-19. After screening for 1,584 immune proteins in cells to co-express the receptors of ACE2 and TMPRSS2, 25 potential therapeutic targets were determined to be significantly overexpressed in nasal goblet secretory cells, lung type II pneumocytes and ileal absorptive enterocytes in patients with several immunopathologies. Overall, 10,672 drugs with potential antiviral activity were predicted, which may be used to treat COVID-19. These drugs require to be further validated in experiments and approved for clinical trials to treat patients with COVID-19. (https://github.com/muntisa/immuno-drug-repurposing-COVID-19) (93,94).
7. Antiviral mechanisms of therapeutic agents against SARS-CoV2
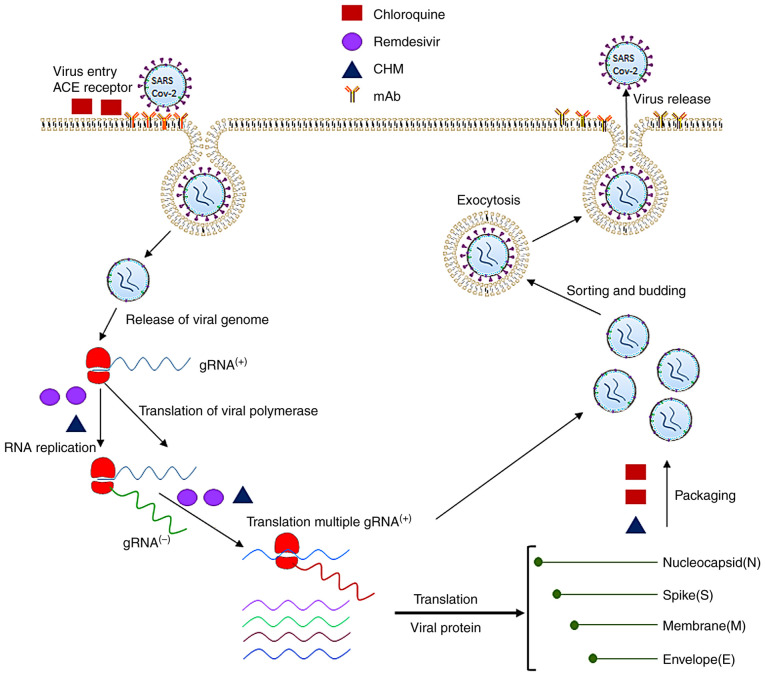
As discussed above, certain therapeutic drugs or Abs are able to inhibit SARS-CoV2 infection by targeting the different pathological stages of the disease involved in the virus' entry, RNA replication, synthesis of the polymerases, proteases, the non-structural proteins and structural proteins of SARS-CoV2, as well as the adaptive immune response to SARS-CoV2 infection. Fig. 2 displays the molecular pathogenesis of the viral infection and antiviral mechanisms by which therapeutic agents block SARS-CoV2 infection. Based on the therapeutic targets of SARS-CoV2, the antiviral agents may be classified into four major categories.
Figure 2.
Pathogenesis of SARS-CoV-2 infection and molecular targets of antiviral therapeutics. The SARS-CoV-2 virus uses its spike proteins to bind to ACE2 receptors to enter human cells. Its RNA is released into the cytoplasm and translated to non-structural proteins such as RNA-dependent RNA polymerase. The positive strand of viral RNA is replicated to produce the negative strand of viral RNA, which is then transcribed to multiple positive-strand subgenomic RNA. The viral structural proteins, including the spike, envelope, membrane and nucleocapsid proteins, are translated and inserted into the endoplasmic reticulum. The subgenomic RNAs then combine with structural proteins and are packaged into the virus, which enters the Golgi vesicle to form the mature virion. Finally, the virion-containing vesicle fuses with the membrane of the host's cell to release the virus particles. The molecular targets of the antiviral therapeutics are denoted in green (chloroquine), purple (remdesivir), orange (mAbs) and black (CHM). SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; mAb, monoclonal antibodies; CHM, Chinese herbal medicine; ACE, angiotensin-converting enzyme; gRNA, guide RNA.
Prevention of binding of coronavirus proteins to human cell receptors and inhibition of viral self-assembly
SARS-CoV-2 uses human ACE2 receptors for the virus' S protein to bind to host cells so that the virus is able to invade the host cells. Thus, the application of drugs that block the S proteins of SARS-CoV-2 from binding to ACE2 receptors in human cells is an effective antiviral approach (95–98). ACE2 receptors have a high similarity of amino acid sequences to the RBD of SARS-CoVs. CQ and HCQ exert antiviral effects against SARS-CoV-2 by inhibiting the coronavirus binding to the ACE2 receptors on a cell's surface. Due to their alkaline nature, CQ and HCQ may also increase the pH within a cell's cytoplasm to block the virus from entering and facilitate transport to inhibit SARS-CoV-2 infection both in vitro and in vivo. In addition, CQ and HCQ may interfere with the packaging process of viral genomic positive RNAs and structural proteins to block or delay the formation of viral particles and their release from infected cells.
Certain Chinese herbs and natural flavonoids (e.g., licoflavonol from Glycyrrhiza uralensis) blocked the binding of the viral S protein to ACE2 receptors to exert an antiviral effect (80). Pudilan (PDL) is a four-herb formula that includes Ban Lan Gen (Isatis indigotica Fort., indigowoad root) and Huang Qin (Scutellaria baicalensis Georgi., Baikal skullcap). PDL has been recommended to treat H1N1 infection. An experimental study revealed that PDL has a therapeutic effect against SARS-CoV-2 by blocking the virus from binding to the ACE2 receptors in cultured Vero E6 cells and in hACE2-transgenic mice (99).
Inhibition of the replication of coronavirus RNA by action on polymerases
Coronaviral non-structural proteins (nsps), such as RdRp, are involved in the virus' RNA replication, transcription and protein translation, as well as the modification and processing of proteins. RdRp (nsp12) is the critical enzyme of the viral replication and transcriptional complex, which is conserved in coronaviruses. Therefore, RdRp has been used as an important antiviral target to develop drugs against SARS, MERS and COVID-19 (44,100). As a small molecule NA, remdesivir was developed to mimic the structure of adenosine and to target RdRp for integration into the replication synthesis of RNA to inhibit coronavirus replication. After remdesivir enters a cell's cytoplasm, it is metabolized into the active triphosphate metabolite NTP, which interferes with the RdRp of SARS-CoV-2. Incorporation of remdesivir at position i causes the termination of RNA synthesis at position i+3. Of note, the same antiviral effects are obtained by interfering with the RdRps of SARS-CoV, MERS-CoV and SARS-CoV-2 after the cells are treated with remdesivir and other NAs (101). Besides remdesivir, other NAs, such as ribavirin, favipiravir and galidesivir have potential antiviral actions against SARS-CoV-2 by inhibiting the activity of RdRps. One study indicated that favipiravir exerted an antiviral effect through the lethal mutagenesis of RdRp of SARS-CoV-2. The SARS-CoV RdRp complex is far more active than any other viral RdRps. During the replication of viral RNA, favipiravir was inserted into viral RNA, inducing C-to-U and G-to-A transitions in the SARS-CoV-2 genome, leading to the mutated RNA to block viral replication (102). The latter study further supported the concept of NAs as promising treatments for COVID-19.
Certain Chinese herbs or natural products have also demonstrated antiviral activity by binding to RdRp; betulonal (from Cassine xylocarpa), gnidicin and gniditrin (from Gnidia lamprantha) bind to and inhibit the RdRp of coronaviruses. The active theaflavin of CHM markedly inhibited RdRp to block SARS-CoV-2 replication. Through structural prediction and screening for a total of 6,842 natural drugs from natural resources in South Africa, four natural products (diosmetin-7-O-β-d-apiofuranoside, 3-O-α-l-arabinopyranosyl-echinocystic acid, 3′-epi-afroside and genkwanin-8-C-β-glucopyranoside) were identified to have a similar binding domain to that of remdesivir, allowing them to block the RdRp-mediated RNA replication of SARS-CoV-2. Each of these products had similar antiviral activity to remdesivir based on computational screening and molecular docking predictions, with docking scores between −7.1 and −10. 4 kcal/mol (103).
Inhibition of synthesis of viral protein and the enzymes of the coronavirus
The coronavirus main proteinase 3CLpro, papain-like proteinase (PLpro) and helicase are potential molecular targets for antiviral small molecules to inhibit the viral RNA replication of SARS-CoV-2. 3CLpro is responsible for the processing and maturation of the viral polyproteins; thus, it is a promising target for developing drugs against SARS-CoV-2 infection. Recently, treatment combining GC376 and remdesivir significantly inhibited viral replication by binding to the catalytically active site of 3CLpro of SARS-CoV-2 as a major antiviral mechanism (33). The Chinese herb, the root extract of Isatis indigotica (Ban Lan Gen), dose-dependently inhibited the cleavage activity of 3CLpro of SARS-coronavirus to exert its antiviral action (104). The active component of the natural compound, resveratrol, which is widely present in different plants, including Vitis vinifera, Polygonum cuspidatum and Vaccinium macrocarpon, has antiviral effects on MERS-CoV by targeting the N protein to block the packaging of viral particles.
Ab-mediated antiviral mechanisms for blocking SARS-CoV-2 infection
Neutralizing Abs are useful for treating coronavirus infections such as COVID-19. These Abs may be obtained either by isolation from the CP of a patient with SARS-CoV-2 or produced by a biotech company (43). In most viral infections, the titer of the antiviral serum usually reaches a higher concentration after the first week of viral infection. Thus, the early transfusion of CP is probably more effective to improve the survival rate of critical COVD-19 patients in the early stage of the disease (52,54). It should be noted that CP transfusion may not be useful for patients with end-stage disease, given the severity of the disease. On the other hand, certain patients with mild disease may self-recover; hence, CP transfusion may not be required (59).
mAbs may be designed to target the specific viral antigens in different steps of the pathogenesis of SARS-CoV-2 infection. Usually, mAbs that combat SARS-CoV-2 are able to neutralize the virus by targeting the virus' structural proteins or host receptors to inhibit the virus from attaching and entering. Certain mAbs are able to inhibit the virus' replication and transcription or increase the immune response of patients with SARS-CoV-2 infection. Since SARS-CoV-2 enters human cells through their RBD in the S1 subunit of the S protein to bind to ACE2 receptors, mAbs (targeting S proteins), are the most effective Abs against SARS-CoV-2 infection (105). In addition to the Abs that inhibit the virus from entering, other Abs may neutralize the specific enzymes or proteins of SARS-CoV-2 by preventing its replication. These Abs are required to cross the cell membrane of infected cells to neutralize the RdRp or other enzymes to inhibit viral replication. The critical viral enzymes, such as PLpro, 3CLpro and certain nsps, may be targets to develop Abs to block the RNA replication cycle of SARS-CoV-2 (106,107).
Anti-inflammatory mechanism of drugs for inhibiting the pathogenesis of SARS-CoV-2
In addition to inhibiting the entry and replication of the SARS-CoV-2, certain drugs exert antiviral effects by decreasing the pathological inflammatory response induced by SARS-CoV-2. CQ and HCQ may also have an immunomodulatory effect to inhibit pathogenesis by decreasing cytokine production and inhibiting cellular autophagy in infected cells. HCQ was reported to inhibit the release of IL-6, IL-1β and TNF-α (108–110). It was also reported that the combination of azithromycin with HCQ increased viral clearance in certain patients with COVID-19. Various anti-inflammatory drugs, such as corticosteroids, cytokines and drugs that interfere with cytokine activities (such as tocilizumab and sarilumab) to block IL-6 activity for decreasing the pathological inflammatory reaction of SARS-CoV-2, were described in the above sections.
8. Conclusions and future perspectives
Although experimental and clinical studies indicated that certain therapeutic agents may be used to treat or prevent SARS-CoV-2 infection, major challenges remain to develop effective drugs and vaccines for SARS-CoV-2 infection.
First, the novel NAs and single compounds of Chinese herbs should be further identified and modified to validate their antiviral efficacy against COVID-19. Certain NAs such as remdesivir cannot be administered to the human body directly in the active triphosphate form. NAs are usually given as a prodrug that requires to be phosphorylated into the triphosphate form to be recognized by the RdRp once it enters a cell. To overcome this problem, the nucleosides of NAs require to be modified to allow active NAs to cross the cell membrane (40,44). Numerous natural products and Chinese herbs inhibit the entry and packaging of SARS-CoV-2, but the molecular targets of the compounds in Chinese herbs require to be extensively characterized to increase their antiviral activity. Furthermore, Ab-based therapy appears to be the most validated treatment for COVID-19. Since mAbs have the advantages of specifically neutralizing viral antigens and may be produced on a large scale within a short period, a standard process of development and production is required to accelerate the approval of mAbs to treat COVID-19. Due to the diversity of the epitopes on SARS-CoV-2, a combination of several mAbs or CP should be used to achieve therapeutic efficacy. In addition, it must be decided how many injections of the Abs are required to maintain a sufficient concentration and efficacy of mAbs in treated patients. Secondly, certain natural plant compounds and TCMs have strong antiviral properties and no toxicity and are thus recommended as candidate drugs for immediate treatment of SARS-CoV-2 infection (111,112). Finally, as the genomic RNA, structural proteins and non-structural proteins of SARS-CoV-2 have been characterized, the effective DNA or mRNA, or inactivated-virus vaccines, have been developed for immunization of non-affected populations to prevent the further spread of SARS-CoV-2. Certain mRNA vaccines or inactivated virus vaccines for SARS-CoV2 have been approved and have been used to immunize normal populations in numerous countries, including the USA, Canada, China, Japan and the UK. Therefore, with fast advances in drug discovery and vaccine development, the risk of SARS-CoV2 infection in different populations around the world will largely decline in the near future.
Acknowledgements
Not applicable.
Funding Statement
This study was funded by the Start-up Research fund of Shandong University of Traditional Chinese Medicine (grant no. 2020-220259). Furthermore, the Quancheng Talent Scholar Fund of Jinan City (grant no. 5150) provided financial support.
Funding
This study was funded by the Start-up Research fund of Shandong University of Traditional Chinese Medicine (grant no. 2020-220259). Furthermore, the Quancheng Talent Scholar Fund of Jinan City (grant no. 5150) provided financial support.
Availability of data and materials
Not applicable.
Authors' contributions
FH conceived and designed the review. FH and JW were major contributors to writing the manuscript. YY, YL, MM, CW and JC wrote parts of the manuscript and prepared the figures. FH, YL and JC performed the literature search and selection, and were responsible for editing the references. All authors read and approved the final version of the manuscript. All authors are responsible for all aspects of the work and approve the submission in its current form. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn JH, Radoshitzky SR, Li W, Wong SK, Choe H, Farzan M. The SARS Coronavirus receptor ACE 2 A potential target for antiviral therapy. In: Holzenburg A, Bogner E, editors. New Concepts of Antiviral Therapy. Springer US; Boston, MA: 2006. pp. 397–418. [DOI] [Google Scholar]
- 6.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez Y, Novelli L, Rojas M, De Santis M, Acosta-Ampudia Y, Monsalve DM, Ramírez-Santana C, Costanzo A, Ridgway WM, Ansari AA, et al. Autoinflammatory and autoimmune conditions at the crossroad of COVID-19. J Autoimmun. 2020;114:102506. doi: 10.1016/j.jaut.2020.102506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: Immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fricke-Galindo I, Falfán-Valencia R. Genetics insight for COVID-19 susceptibility and severity: A review. Front Immunol. 2021;12:622176. doi: 10.3389/fimmu.2021.622176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maggi E, Canonica GW, Moretta L. COVID-19: Unanswered questions on immune response and pathogenesis. J Allergy Clin Immunol. 2020;146:18–22. doi: 10.1016/j.jaci.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmed-Hassan H, Sisson B, Shukla RK, Wijewantha Y, Funderburg NT, Li Z, Hayes D, Jr, Demberg T, Liyanage NPM. Innate immune responses to highly pathogenic coronaviruses and other significant respiratory viral infections. Front Immunol. 2020;11:1979. doi: 10.3389/fimmu.2020.01979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 13.Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses-drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alzaabi MM, Hamdy R, Ashmawy NS, Hamoda AM, Alkhayat F, Khademi NN, Al Joud SMA, El-Keblawy AA, Soliman SSM. Flavonoids are promising safe therapy against COVID-19. Phytochem Rev. 2021 May 22; doi: 10.1007/s11101-021-09759-z. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savarino A, Di Trani L, Donatelli I, Cauda R, Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect Dis. 2006;6:67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Bari MA. Chloroquine analogues in drug discovery: New directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. J Antimicrob Chemother. 2015;70:1608–1621. doi: 10.1093/jac/dkv018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keyaerts E, Vijgen L, Maes P, Neyts J, Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem Biophys Res Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 20.Huang M, Tang T, Pang P, Li M, Ma R, Lu J, Shu J, You Y, Chen B, Liang J, et al. Treating COVID-19 with chloroquine. J Mol Cell Biol. 2020;12:322–325. doi: 10.1093/jmcb/mjaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, Liu X, Zhao L, Dong E, Song C, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreani J, Le Bideau M, Duflot I, Jardot P, Rolland C, Boxberger M, Wurtz N, Rolain JM, Colson P, La Scola B, Raoult D. In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb Pathog. 2020;145:104228. doi: 10.1016/j.micpath.2020.104228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Arshad S, Kilgore P, Chaudhry ZS, Jacobsen G, Wang DD, Huitsing K, Brar I, Alangaden GJ, Ramesh MS, McKinnon JE, et al. Treatment with hydroxychloroquine, azithromycin, and combination in patients hospitalized with COVID-19. Int J Infect Dis. 2020;97:396–403. doi: 10.1016/j.ijid.2020.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez AV, Roman YM, Pasupuleti V, Barboza JJ, White CM. Hydroxychloroquine or chloroquine for treatment or prophylaxis of COVID-19: A living systematic review. Ann Intern Med. 2020;173:287–296. doi: 10.7326/M20-2496. [DOI] [PubMed] [Google Scholar]
- 26.Chowdhury MS, Rathod J, Gernsheimer J. A rapid systematic review of clinical trials utilizing chloroquine and hydroxychloroquine as a treatment for COVID-19. Acad Emerg Med. 2020;27:493–504. doi: 10.1111/acem.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elavarasi A, Prasad M, Seth T, Sahoo RK, Madan K, Nischal N, Soneja M, Sharma A, Maulik SK, Shalimar, Garg P. Chloroquine and hydroxychloroquine for the treatment of COVID-19: A systematic review and meta-analysis. J Gen Intern Med. 2020;35:3308–3314. doi: 10.1007/s11606-020-06146-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar J, Jain S, Meena J, Yadav A. Efficacy and safety of hydroxychloroquine/chloroquine against SARS-CoV-2 infection: A systematic review and meta-analysis. J Infect Chemother. 2021;27:882–889. doi: 10.1016/j.jiac.2021.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, Siegel D, Perron M, Bannister R, Hui HC, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 32.Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, Smith EC, Case JB, Feng JY, Jordan R, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9:e00221–e00228. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu L, Ye F, Feng Y, Yu F, Wang Q, Wu Y, Zhao C, Sun H, Huang B, Niu P, et al. Both boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat Commun. 2020;11:4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, et al. First case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grein J, Ohmagari N, Shin D, Diaz G, Asperges E, Castagna A, Feldt T, Green G, Green ML, Lescure FX, et al. Compassionate use of remdesivir for patients with severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spinner CD, Gottlieb RL, Criner GJ, Arribas López JR, Cattelan AM, Soriano Viladomiu A, Ogbuagu O, Malhotra P, Mullane KM, Castagna A, et al. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: A randomized clinical trial. JAMA. 2020;324:1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, et al. Remdesivir for the treatment of Covid-19-final report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garibaldi BT, Wang K, Robinson ML, Zeger SL, Bandeen-Roche K, Wang MC, Alexander GC, Gupta A, Bollinger R, Xu Y. Comparison of time to clinical improvement with vs without remdesivir treatment in hospitalized patients with COVID-19. JAMA Netw Open. 2021;4:e213071. doi: 10.1001/jamanetworkopen.2021.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aleissa MM, Silverman EA, Paredes Acosta LM, Nutt CT, Richterman A, Marty FM. New perspectives on antimicrobial agents: Remdesivir treatment for COVID-19. Antimicrob Agents Chemother. 2020;65:e01814–20. doi: 10.1128/AAC.01814-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falzarano D, de Wit E, Martellaro C, Callison J, Munster VJ, Feldmann H. Inhibition of novel β coronavirus replication by a combination of interferon-α2b and ribavirin. Sci Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP, Brining D, Bushmaker T, Martellaro C, Baseler L, et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Clercq E. New nucleoside analogues for the treatment of hemorrhagic fever virus infections. Chem Asian J. 2019;14:3962–3968. doi: 10.1002/asia.201900841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choy KT, Wong AY, Kaewpreedee P, Sia SF, Chen D, Hui KPY, Chu DKW, Chan MCW, Cheung PP, Huang X, et al. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C, Zhang Y, Huang J, Yin P, Cheng Z, Wu J, Chen S, Zhang Y, Chen B, Lu M, et al. Favipiravir versus Arbidol for COVID-19: A randomized clinical trial. 2020 doi: 10.1101/2020.03.17.20037432. medRxiv: 2020.03.17.20037432. [DOI] [Google Scholar]
- 46.Taylor R, Kotian P, Warren T, Panchal R, Bavari S, Julander J, Dobo S, Rose A, El-Kattan Y, Taubenheim B, et al. BCX4430-A broad-spectrum antiviral adenosine nucleoside analog under development for the treatment of Ebola virus disease. J Infect Public Health. 2016;9:220–226. doi: 10.1016/j.jiph.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu H, Wang S, Ruan S, Wang Y, Zhang J, Yuan Y, Liu H, Wu Y, Li R, Pan S, et al. Dynamic changes of antibodies to SARS-CoV-2 in COVID-19 patients at early stage of outbreak. Virol Sin. 2020;35:744–751. doi: 10.1007/s12250-020-00268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA, Claireaux M, Kerster G, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan KH, Chan JF, Tse H, Chen H, Lau CC, Cai JP, Tsang AK, Xiao X, To KK, Lau SK, et al. Cross-reactive antibodies in convalescent SARS patients' sera against the emerging novel human coronavirus EMC (2012) by both immunofluorescent and neutralizing antibody tests. J Infect. 2013;67:130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mair-Jenkins J, Saavedra-Campos M, Baillie JK, Cleary P, Khaw FM, Lim WS, Makki S, Rooney KD, Nguyen-Van-Tam JS, Beck CR, Convalescent Plasma Study Group The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: A systematic review and exploratory meta-analysis. J Infect Dis. 2015;211:80–90. doi: 10.1093/infdis/jiu396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, Lu L, Jiang S, Yang Z, Wu Y, Ying T. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sui J, Deming M, Rockx B, Liddington RC, Zhu QK, Baric RS, Marasco WA. Effects of human anti-spike protein receptor binding domain antibodies on severe acute respiratory syndrome coronavirus neutralization escape and fitness. J Virol. 2014;88:13769–13780. doi: 10.1128/JVI.02232-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses: (Trends in Immunology 41, 355–359; 2020) Trends Immunol. 2020;41:545. doi: 10.1016/j.it.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulangu S, Dodd LE, Davey RT, Jr, Tshiani Mbaya O, Proschan M, Mukadi D, Lusakibanza Manzo M, Nzolo D, Tshomba Oloma A, Ibanda A, et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 56.Valdez-Cruz NA, García-Hernández E, Espitia C, Cobos-Marín L, Altamirano C, Bando-Campos CG, Cofas-Vargas LF, Coronado-Aceves EW, González-Hernández RA, Hernández-Peralta P, et al. Integrative overview of antibodies against SARS-CoV-2 and their possible applications in COVID-19 prophylaxis and treatment. Microb Cell Fact. 2021;20:88. doi: 10.1186/s12934-021-01576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katia F, Myriam DP, Ucciferri C, Auricchio A, Di Nicola M, Marchioni M, Eleonora C, Emanuela S, Cipollone F, Vecchiet J. Efficacy of canakinumab in mild or severe COVID-19 pneumonia. Immun Inflamm Dis. 2021;9:399–405. doi: 10.1002/iid3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hung IF, To KK, Lee CK, Lee KL, Chan K, Yan WW, Liu R, Watt CL, Chan WM, Lai KY, et al. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin Infect Dis. 2011;52:447–456. doi: 10.1093/cid/ciq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ko JH, Seok H, Cho SY, Ha YE, Baek JY, Kim SH, Kim YJ, Park JK, Chung CR, Kang ES, et al. Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: A single centre experience. Antivir Ther. 2018;23:617–622. doi: 10.3851/IMP3243. [DOI] [PubMed] [Google Scholar]
- 61.Soo YO, Cheng Y, Wong R, Hui DS, Lee CK, Tsang KK, Ng MH, Chan P, Cheng G, Sung JJ. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin Microbiol Infect. 2004;10:676–678. doi: 10.1111/j.1469-0691.2004.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casadevall A, Grossman BJ, Henderson JP, Joyner MJ, Shoham S, Pirofski LA, Paneth N. The assessment of convalescent plasma efficacy against COVID-19. Med (N Y) 2020;1:66–77. doi: 10.1016/j.medj.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arabi Y, Balkhy H, Hajeer AH, Bouchama A, Hayden FG, Al-Omari A, Al-Hameed FM, Taha Y, Shindo N, Whitehead J, et al. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: A study protocol. Springerplus. 2015;4:709. doi: 10.1186/s40064-015-1490-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahn JY, Sohn Y, Lee SH, Cho Y, Hyun JH, Baek YJ, Jeong SJ, Kim JH, Ku NS, Yeom JS, et al. Use of convalescent plasma therapy in two COVID-19 patients with acute respiratory distress syndrome in Korea. J Korean Med Sci. 2020;35:e149. doi: 10.3346/jkms.2020.35.e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu S, Shergis J, Chen X, Yu X, Guo X, Zhang AL, Lu C, Xue CC. Chinese herbal medicine (weijing decoction) combined with pharmacotherapy for the treatment of acute exacerbations of chronic obstructive pulmonary disease. Evid Based Complement Alternat Med. 2014;2014:257012. doi: 10.1155/2014/257012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leung PC. The efficacy of Chinese medicine for SARS: A review of Chinese publications after the crisis. Am J Chin Med. 2007;35:575–581. doi: 10.1142/S0192415X07005077. [DOI] [PubMed] [Google Scholar]
- 68.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang SQ, Du QS, Zhao K, Li AX, Wei DQ, Chou KC. Virtual screening for finding natural inhibitor against cathepsin-L for SARS therapy. Amino Acids. 2007;33:129–135. doi: 10.1007/s00726-006-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho TY, Wu SL, Chen JC, Li CC, Hsiang CY. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. 2007;74:92–101. doi: 10.1016/j.antiviral.2006.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Niu M, Wang RL, Wang ZX, Zhang P, Bai ZF, Jing J, Guo YM, Zhao X, Zhan XY, Zhang ZT, et al. Rapid establishment of traditional Chinese medicine prevention and treatment of 2019-nCoV based on clinical experience and molecular docking. Zhongguo Zhong Yao Za Zhi. 2020;45:1213–1218. doi: 10.19540/j.cnki.cjcmm.20200206.501. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 72.Runfeng L, Yunlong H, Jicheng H, Weiqi P, Qinhai M, Yongxia S, Chufang L, Jin Z, Zhenhua J, Haiming J, et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu H, Ye F, Sun Q, Liang H, Li C, Li S, Lu R, Huang B, Tan W, Lai L. Scutellaria baicalensis extract and baicalein inhibit replication of SARS-CoV-2 and its 3C-like protease in vitro. 2020 doi: 10.1080/14756366.2021.1873977. bioRxiv: 2020.04.10.035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu J, Deng YQ, Wang X, Li XF, Zhang NN, Liu Z, Zhang B, Qin CF, Xie Z. An artificial intelligence system reveals liquiritin inhibits SARS-CoV-2 by mimicking type I interferon. doi: 10.1101/2020.05.02.074021. bioRxiv: doi: https://doi.org/10.1101/2020.05.02.074021. [DOI] [Google Scholar]
- 75.Chen Z, Nakamura T. Statistical evidence for the usefulness of Chinese medicine in the treatment of SARS. Phytother Res. 2004;18:592–594. doi: 10.1002/ptr.1485. [DOI] [PubMed] [Google Scholar]
- 76.Luo H, Tang QL, Shang YX, Liang SB, Yang M, Robinson N, Liu JP. Can Chinese medicine be used for prevention of corona virus disease 2019 (COVID-19)? A review of historical classics, research evidence and current prevention programs. Chin J Integr Med. 2020;26:243–250. doi: 10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ang L, Lee HW, Choi JY, Zhang J, Soo Lee M. Herbal medicine and pattern identification for treating COVID-19: A rapid review of guidelines. Integr Med Res. 2020;9:100407. doi: 10.1016/j.imr.2020.100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu S, Wang J, Shen H. Network pharmacology-based analysis of the role of traditional Chinese herbal medicines in the treatment of COVID-19. Ann Palliat Med. 2020;9:437–446. doi: 10.21037/apm.2020.03.27. [DOI] [PubMed] [Google Scholar]
- 79.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang DH, Wu KL, Zhang X, Deng SQ, Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med. 2020;18:152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Q, Wang Y, Qi C, Shen L, Li J. Clinical trial analysis of 2019-nCoV therapy registered in China. J Med Virol. 2020;92:540–545. doi: 10.1002/jmv.25733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kulanthaivel S, Kaliberdenko VB, Balasundaram K, Shterenshis MV, Scarpellini E, Abenavoli L. Tocilizumab in SARS-CoV-2 patients with the syndrome of cytokine storm: A narrative review. Rev Recent Clin Trials. 2021;16:138–145. doi: 10.2174/1574887115666200917110954. [DOI] [PubMed] [Google Scholar]
- 83.Toniati P, Piva S, Cattalini M, Garrafa E, Regola F, Castelli F, Franceschini F, Airò P, Bazzani C, Beindorf EA, et al. Tocilizumab for the treatment of severe COVID-19 pneumonia with hyperinflammatory syndrome and acute respiratory failure: A single center study of 100 patients in Brescia, Italy. Autoimmun Rev. 2020;19:102568. doi: 10.1016/j.autrev.2020.102568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang SNY, Atkinson SC, Wang C, Lee A, Bogoyevitch MA, Borg NA, Jans DA. The broad spectrum antiviral ivermectin targets the host nuclear transport importin α/β1 heterodimer. Antiviral Res. 2020;177:104760. doi: 10.1016/j.antiviral.2020.104760. [DOI] [PubMed] [Google Scholar]
- 86.Monteagudo LA, Boothby A, Gertner E. Continuous intravenous anakinra infusion to calm the cytokine storm in macrophage activation syndrome. ACR Open Rheumatol. 2020;2:276–282. doi: 10.1002/acr2.11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Franzetti M, Forastieri A, Borsa N, Pandolfo A, Molteni C, Borghesi L, Pontiggia S, Evasi G, Guiotto L, Erba M, et al. IL-1 receptor antagonist anakinra in the treatment of COVID-19 acute respiratory distress syndrome: A retrospective, observational study. J Immunol. 2021;206:1569–1575. doi: 10.4049/jimmunol.2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Romani L, Tomino C, Puccetti P, Garaci E. Off-label therapy targeting pathogenic inflammation in COVID-19. Cell Death Discov. 2020;6:49. doi: 10.1038/s41420-020-0283-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Puccetti M, Costantini C, Ricci M, Giovagnoli S. Tackling immune pathogenesis of COVID-19 through molecular pharmaceutics. Pharmaceutics. 2021;13:494. doi: 10.3390/pharmaceutics13040494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.D'Ardes D, Pontolillo M, Esposito L, Masciarelli M, Boccatonda A, Rossi I, Bucci M, Guagnano MT, Ucciferri C, Santilli F, et al. Duration of COVID-19: Data from an Italian cohort and potential role for steroids. Microorganisms. 2020;8:1327. doi: 10.3390/microorganisms8091327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Horby P, White NJ, Landray MJ. Hydroxychloroquine in hospitalized patients with Covid-19. Reply. N Engl J Med. 2021;384:882. doi: 10.1056/NEJMc2035374. [DOI] [PubMed] [Google Scholar]
- 92.Ucciferri C, Barone M, Vecchiet J, Falasca K. Pidotimod in paucisymptomatic SARS-CoV2 infected patients. Mediterr J Hematol Infect Dis. 2020;12:e2020048. doi: 10.4084/mjhid.2020.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saakre M, Mathew D, Ravisankar V. Perspectives on plant flavonoid quercetin-based drugs for novel SARS-CoV-2. Beni Suef Univ J Basic Appl Sci. 2021;10:21. doi: 10.1186/s43088-021-00107-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.López-Cortés A, Guevara-Ramírez P, Kyriakidis NC, Barba-Ostria C, León Cáceres Á, Guerrero S, Ortiz-Prado E, Munteanu CR, Tejera E, Cevallos-Robalino D, et al. In silico analyses of immune system protein interactome network, single-cell rna sequencing of human tissues, and artificial neural networks reveal potential therapeutic targets for drug repurposing against COVID-19. Front Pharmacol. 2021;12:598925. doi: 10.3389/fphar.2021.598925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hong W. Combating COVID-19 with chloroquine. J Mol Cell Biol. 2020;12:249–250. doi: 10.1093/jmcb/mjaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 97.Liu J, Cao R, Xu M, Wang X, Zhang H, Hu H, Li Y, Hu Z, Zhong W, Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kong Q, Wu Y, Gu Y, Lv Q, Qi F, Gong S, Chen X. Analysis of the molecular mechanism of Pudilan (PDL) treatment for COVID-19 by network pharmacology tools. Biomed Pharmacother. 2020;128:110316. doi: 10.1016/j.biopha.2020.110316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chu CK, Gadthula S, Chen X, Choo H, Olgen S, Barnard DL, Sidwell RW. Antiviral activity of nucleoside analogues against SARS-coronavirus (SARS-coV) Antivir Chem Chemother. 2006;17:285–289. doi: 10.1177/095632020601700506. [DOI] [PubMed] [Google Scholar]
- 101.Gordon CJ, Tchesnokov EP, Woolner E, Perry JK, Feng JY, Porter DP, Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J Biol Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shannon A, Selisko B, Le NT, Huchting J, Touret F, Piorkowski G, Fattorini V, Ferron F, Decroly E, Meier C, et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat Commun. 2020;11:4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Khan A, Khan M, Saleem S, Babar Z, Ali A, Khan AA, Sardar Z, Hamayun F, Ali SS, Wei DQ. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscip Sci. 2020;12:335–348. doi: 10.1007/s12539-020-00381-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lin CW, Tsai FJ, Tsai CH, Lai CC, Wan L, Ho TY, Hsieh CC, Chao PD. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res. 2005;68:36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park T, Lee SY, Kim S, Kim MJ, Kim HG, Jun S, Kim SI, Kim BT, Park EC, Park D. Spike protein binding prediction with neutralizing antibodies of SARS-CoV-2. 2020 doi: 10.1101/2020.02.22.951178. bioRxiv: doi: 2020.02.22.951178. [DOI] [Google Scholar]
- 106.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: Basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 107.Zhou G, Zhao Q. Perspectives on therapeutic neutralizing antibodies against the novel coronavirus SARS-CoV-2. Int J Biol Sci. 2020;16:1718–1723. doi: 10.7150/ijbs.45123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Quiros Roldan E, Biasiotto G, Magro P, Zanella I. The possible mechanisms of action of 4-aminoquinolines (chloroquine/hydroxychloroquine) against Sars-Cov-2 infection (COVID-19): A role for iron homeostasis? Pharmacol Res. 2020;158:104904. doi: 10.1016/j.phrs.2020.104904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhou D, Dai SM, Tong Q. COVID-19: A recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J Antimicrob Chemother. 2020;75:1667–1670. doi: 10.1093/jac/dkaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Devaux CA, Rolain JM, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: What to expect for COVID-19? Int J Antimicrob Agents. 2020;55:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adhikari B, Marasini BP, Rayamajhee B, Bhattarai BR, Lamichhane G, Khadayat K, Adhikari A, Khanal S, Parajuli N. Potential roles of medicinal plants for the treatment of viral diseases focusing on COVID-19: A review. Phytother Res. 2021;35:1298–1312. doi: 10.1002/ptr.6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee DYW, Li QY, Liu J, Efferth T. Traditional Chinese herbal medicine at the forefront battle against COVID-19: Clinical experience and scientific basis. Phytomedicine. 2021;80:153337. doi: 10.1016/j.phymed.2020.153337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.