Abstract
Background and Objective
To test the hypothesis that a multicenter-validated computer deep learning algorithm detects MRI-negative focal cortical dysplasia (FCD).
Methods
We used clinically acquired 3-dimensional (3D) T1-weighted and 3D fluid-attenuated inversion recovery MRI of 148 patients (median age 23 years [range 2–55 years]; 47% female) with histologically verified FCD at 9 centers to train a deep convolutional neural network (CNN) classifier. Images were initially deemed MRI-negative in 51% of patients, in whom intracranial EEG determined the focus. For risk stratification, the CNN incorporated bayesian uncertainty estimation as a measure of confidence. To evaluate performance, detection maps were compared to expert FCD manual labels. Sensitivity was tested in an independent cohort of 23 cases with FCD (13 ± 10 years). Applying the algorithm to 42 healthy controls and 89 controls with temporal lobe epilepsy disease tested specificity.
Results
Overall sensitivity was 93% (137 of 148 FCD detected) using a leave-one-site-out cross-validation, with an average of 6 false positives per patient. Sensitivity in MRI-negative FCD was 85%. In 73% of patients, the FCD was among the clusters with the highest confidence; in half, it ranked the highest. Sensitivity in the independent cohort was 83% (19 of 23; average of 5 false positives per patient). Specificity was 89% in healthy and disease controls.
Discussion
This first multicenter-validated deep learning detection algorithm yields the highest sensitivity to date in MRI-negative FCD. By pairing predictions with risk stratification, this classifier may assist clinicians in adjusting hypotheses relative to other tests, increasing diagnostic confidence. Moreover, generalizability across age and MRI hardware makes this approach ideal for presurgical evaluation of MRI-negative epilepsy.
Classification of Evidence
This study provides Class III evidence that deep learning on multimodal MRI accurately identifies FCD in patients with epilepsy initially diagnosed as MRI negative.
Focal cortical dysplasia (FCD), a surgery-amenable developmental epileptogenic brain malformation, presents with cortical thickening on T1-weighted MRI, as well as hyperintensity and blurring of the gray matter (GM)–white matter (WM) interface on fluid-attenuated inversion recovery (FLAIR) images. While these features are often visible to the naked eye, FCD may be overlooked and found only at surgery.1 MRI-negative patients represent a major diagnostic challenge.2
Currently, benchmark automated detection methods fail in 20% to 40% of patients,3-6 particularly those with subtle FCD, and suffer from high false-positive (FP) rates.7 Conversely, deep neural networks outperform state-of-the-art methods at disease detection (see elsewhere8,9 for review). Specifically, convolutional neural networks (CNNs) learn abstract concepts from high-dimensional data, alleviating the challenging task of hand-crafting features.10 The integration of convolutional operators that implicitly encode spatial covariance of neighboring voxels (rather than treating each voxel independently) with nonlinearity capturing complex patterns and variability is expected to optimize the detection of the full FCD spectrum. With regard to diagnostic performance, the deterministic nature of conventional algorithms does not permit risk assessment of the automated decisions, a requirement to be integrated into clinical diagnostic systems. Alternatively, bayesian CNNs provide a distribution of predictions from which the mean and variance can be computed, the latter being interpreted as a measure of uncertainty.11
Here, we tested the hypothesis that a multicenter-validated computer deep learning algorithm operating directly on T1-weighted and FLAIR MRI voxel detects MRI-negative FCD.
Methods
The primary question of this study was whether a deep learning algorithm operating on multimodal MRI has significant diagnostic value, including in MRI-negative patients. Our automated algorithm was trained and validated on a multicenter dataset of patients with histologically confirmed FCD. We ruled out sources of spectrum bias12 by evaluating specificity against healthy individuals and a disease control cohort of patients with temporal lobe epilepsy (TLE) and histologically confirmed hippocampal sclerosis (HS). To minimize incorporation bias,12 the classifier was iteratively trained and tested using a leave-one-site-out scheme; that is, the classifier was trained iteratively on all sites minus the one held out for testing, which guaranteed the out-of-fold validation in which tested cohorts were never part of the training. Moreover, the classifier trained on the full dataset was tested on an independent cohort of patients who were never part of training. According to the Classification of Evidence schemes of the American Academy of Neurology,13 this study satisfies the rating for Class III evidence for diagnostic accuracy, demonstrating that deep learning operating on multimodal MRI has significant diagnostic value, including in MRI-negative patients, with 85% sensitivity.
Participants
We studied consecutive retrospective cohorts from 9 tertiary epilepsy centers worldwide with histologically validated FCD lesions collected from October 2012 to January 2018 and in whom both 3-dimensional (3D) T1-weighted MRI and 3D FLAIR were acquired as part of the clinical presurgical investigation.14 The TLE cohort included both patients with MRI-visible HS (n = 49, comparable to MRI-positive FCD) and those in whom the MRI was unremarkable but the histologic examination of the surgical specimen revealed the presence of subtle HS (n = 40, comparable to MRI-negative, histology-positive FCD). Patients had been investigated for drug-resistant epilepsy with a standard presurgical workup that included neurologic examination, assessment of seizure history, neuroimaging, and video-EEG recordings.
On histologic examination of the surgical specimen,15 FCD type II was defined as disrupted cortical lamination with dysmorphic neurons in isolation (IIA, n = 70) or together with balloon cells (IIB, n = 78). At a mean ± SD postoperative follow-up of 31.2 ± 14.4 months (range 12–78 months), 103 patients (70%) became seizure-free (Engel class I), 33 (22%) had rare disabling seizures (Engel class II), 9 (6%) had worthwhile improvement (Engel class III), and 3 (2%) had no improvement (Engel class IV). In patients with Engel classes III and IV, the resection was incomplete because the FCD encroached eloquent areas in primary cortices (7 in sensorimotor, 2 in primary visual, and 3 in language areas); the residual lesion and extent of resection were evaluated on postoperative MRI.
Standard Protocol Approvals, Registrations, and Patient Consents
The ethics committees and institutional review boards at all participating sites (S1–S9) approved the study, and written informed consent was obtained from all participants.
MRI Acquisition and Image Processing
High-resolution 3D T1-weighted and 3D FLAIR MRIs were acquired in all individuals.14 All images were obtained on 3T scanners; 1 site provided additional cases with 1.5T MRI. Imaging parameters are listed in eTable 1 (available from Dryad: doi.org/10.5061/dryad.h70rxwdgm). MRI data were deidentified; files were converted from Digital Imaging and Communications in Medicine to Neuroimaging Informatics Technology Initiative with header anonymization. T1-weighted images were linearly registered to the Montreal Neurological Institute (MNI) 152 symmetric template. FLAIR images were linearly mapped to T1-weighted MRI in MNI space. T1-weighted and FLAIR underwent intensity nonuniformity correction16 followed by intensity standardization with scaling of values between 0 and 100. Finally, images were skull-stripped with an in-house deep learning method (version 1.0.0) trained on manually corrected brain masks from patients with cortical malformations. Two experts manually segmented lesions on coregistered T1-weighted and FLAIR images; interrater Dice agreement was 0.92 ± 0.10, calculated as 2|M1∩M2|/(|M1| + |M2|), where M1 is label 1, M2 is label 2, and M1 ∩ M2 is the intersection of M1 and M2.
Classifier Design
Figure 1 and eFigure 1 (available from Dryad: doi.org/10.5061/dryad.h70rxwdgm) illustrate the design. The full methodology is described in additional Methods (available from Dryad).
Figure 1. Classifier Design.
Training and inference (or testing) workflow. In the cascaded system, the output of convolutional neural network (CNN) -1 serves as an input for CNN-2. CNN-1 maximizes the detection of lesional voxels; CNN-2 reduces the number of misclassified voxels, removing false positives (FPs) while maintaining optimal sensitivity. The training procedure (indicated by dashed arrows) operating on T1-weighted (T1w) and fluid-attenuated inversion recovery (FLAIR) MRI extracts 3-dimensional patches from lesional and nonlesional tissue to yield tCNN-1 (trained model 1) and tCNN-2 (trained model 2) models with optimized weights (indicated by vertical dashed-dotted arrows). These models are then used for participant-level inference. For each unseen participant, the inference pipeline (solid arrows) uses tCNN-1 and generates a mean (μdroupout) of 20 predictions (forward passes); the mean map is then thresholded voxel-wise to discard improbable lesion candidates (μdroupout > 0.1). The resulting binary mask serves to sample the input patches for the tCNN-2. Mean probability and uncertainty maps are obtained by collating 50 predictions; uncertainty is transformed into confidence. The sampling strategy (identical for training and inference) is illustrated only for testing.
Data Sampling and Network Architecture
In each individual, we thresholded FLAIR images by z-normalizing intensities and discarding the bottom 10 percentile intensities. This internal thresholding resulted in a mask containing voxels within the GM and its interface with the WM. This mask was then used to extract 3D patches (i.e., regions of interest centered around a given voxel) from coregistered 3D T1-weighted and FLAIR images, which served as input to the network. The 3D patches seamlessly sampled the FCD across orthogonal planes and tissue types. We designed a cascaded system in which the output of the first CNN (CNN-1) served as input to the second (CNN-2). CNN-1 aimed at maximizing the detection of lesional voxels; CNN-2 reduced the number of misclassified voxels, removing FPs while maintaining optimal sensitivity. In brief, for each test participant, 3D T1-weighted and FLAIR patches were fed to CNN-1. To discard improbable lesional candidates, the mean of 20 forward passes (or predictions) was thresholded at >0.1 (equivalent to rejecting bottom 10 percentile probabilities); voxels surviving this threshold served as the input to sample patches for CNN-2.
Estimation of Prediction Uncertainty
Bayesian inference in deep CNNs with a large number of parameters is computationally intensive.17 By probabilistically excluding neurons (or units) after every convolutional layer during training, the Monte Carlo dropout method18 simulates an ensemble of neural networks with diverse architectures, thus preventing overfitting without compromising on accuracy. This procedure provides a distribution of posterior probabilities at each voxel resulting from multiple stochastic forward passes through the classifier; their variance provides a measure of uncertainty. Here, we used the mean and variance of 50 voxel-wise forward passes to generate probability and uncertainty maps. The mean probability map was binarized by thresholding at >0.7 (empirically determined by setting the cluster-level FP rate to <6) and underwent a postprocessing routine entailing morphologic erosion, dilation, and extraction of connected components (>75 voxels) to remove flat blobs and noise, a procedure that resulted in nonoverlapping clusters. To evaluate performance, this detection map was compared to the manual expert annotation.
Transforming Uncertainty Into Confidence and Ranking
For each cluster of the detection map, we estimated confidence by computing the median uncertainty across its voxels; we then aggregated uncertainties across all clusters and normalized values between 0 and 1 to obtain a measure of confidence. All clusters were then ranked according to their confidence estimates, with the highest-confidence cluster as rank 1, second-highest-confidence cluster rank 2, and so on until all clusters surviving the threshold (probability >0.7 and spatial extent >75 voxels) had been ranked. Confidence maps were evaluated together with a diagram plotting lesion probability against lesion ranking, with rank 1 signifying highest confidence to be lesional, regardless of cluster size.
Performance Evaluation
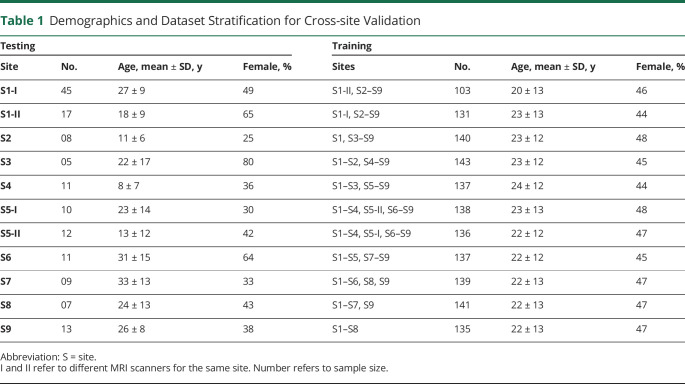
To assess performance, we use a leave-one-site-out cross-validation by which the classifier trained on 8 sites was tested iteratively on the held-out site until all sites had served as testing set. A minimum of 1 voxel colocalizing with the manually segmented FCD (ground truth label) was deemed true positive (TP); any detection not colocalizing was deemed FP. Consistent with previous FCD detection literature,3,19-21 we deemed partial overlap to be sufficient without requiring the detection to be completely within the expert label. Demographics and dataset stratification are shown in Table 1. In addition, we evaluated the algorithm trained on the complete dataset of the 148 patients with FCD on an independent cohort of 23 cases with FCD (11 female patients, age 13 ± 10 years, 70% MRI negative) from S1 and S2.
Table 1.
Demographics and Dataset Stratification for Cross-site Validation
Patient-level (i.e., lesion-level) evaluation metrics included sensitivity (P, L) = |P1∩L1|/|L1| and specificity (P, L) = |P0∩L0|/|L0|, where P is the model prediction and L is the ground truth label; L1 and L0 signify voxels predicted as positive (lesional) and negative (not lesional); and P1 and P0 represent the same for model predictions. We evaluated specificity as the absence of any findings by applying the algorithm trained on the complete dataset of patients with FCD to healthy controls and TLE disease controls; in other words, specificity was calculated as the proportion of healthy or disease controls in whom no FCD lesion cluster was falsely identified. Site-wise area under the receiver operating characteristic curve evaluated voxel-wise classification performance (i.e., the TP vs FP rate) stratified by sites.
We evaluated the spatial relation between lesional clusters and FPs in patients as well as healthy and disease controls. To this end, we generated a lesional probability map by overlaying all manually segmented FCD labels; the Dice coefficient quantified the overlap between the FCD probability map and both the group-wise probability and uncertainty maps of FPs.
The Pearson correlation quantified associations between probability and uncertainty and between age and the number of FPs. Biserial correlation evaluated the association between MRI-negative status and the number of FPs. The Spearman correlation quantified the association between lesion rank and probability. Nonparametric permutations (10,000 iterations with replacement) tested group differences at p < 0.05 (2 tailed), with Bonferroni correction for multiple comparisons.
Data Availability
These datasets are not publicly available because they contain information that could compromise the privacy of research participants. The source code and pretrained model weights are available for download online (version 1.0.0 at GitHub). In addition, a derivative dataset composed of lesional and nonlesional patches from 148 patients with FCD is available as a Hierarchical Data Format dataset (available from Zenodo).
Results
Demographics
The primary site (S1) comprised 62 patients with FCD (33 female patients, mean ± SD age 25 ± 10 years) and control groups consisting of age- and sex-matched healthy individuals (n = 42, 22 female controls, age 30 ± 7 years) and patients with TLE and histologically verified HS (n = 89, 47 female controls, age 31 ± 8 years). Across the remaining 8 sites (S2–S9), the cohort comprised 86 patients with FCD (36 female patients, age 20 ± 14 years). In 75 patients (51%) in whom routine MRI evaluation was initially reported as unremarkable in the initial readings of the neuroradiologists at each participating center, the location of the seizure focus was established through intracranial EEG.
Patient-Level Performance
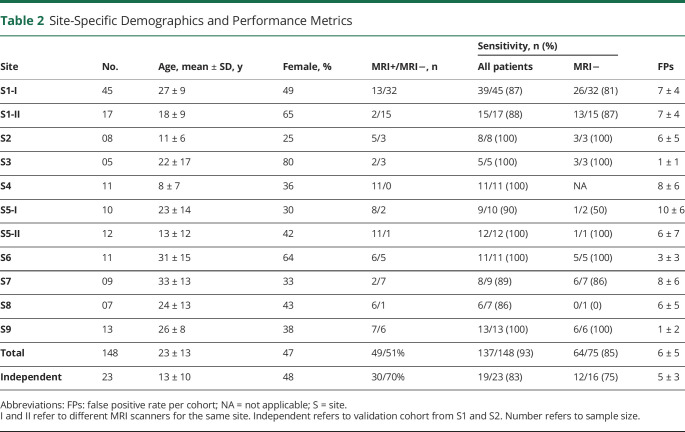
The overall sensitivity of the classifier based on leave-one-site-out cross-validation was 93% (137 of 148 FCD lesions detected), with 6 ± 5 FP clusters per patient. Stratifying children and adults yielded sensitivity of 98% for the former (52 of 53, 7 ± 5 FP clusters) and 89% (85 of 95, 5 ± 5 FP) for the latter. Eight-five percent of MRI-negative and 100% of MRI-positive lesions were detected. When the classifier was tested on the independent cohort (using the model trained on the complete dataset of the 148 patients with FCD), overall sensitivity was 83% (19 of 23 FCD lesions detected, 5 ± 3 FP clusters per patient), with 100% of MRI-positive and 75% of MRI-negative lesions detected. Specificity was 90% in healthy controls (4 of 42 with 2 ± 1 FP clusters) and 89% in TLE disease controls (10 of 89, 1 ± 0 FP cluster). With respect to the latter, specificity was similar between MRI-positive HS (92%, 5 of 49, 1 ± 0 FP cluster) and MRI-negative HS (88%, 5 of 40, 1 ± 0 FP cluster) controls. Per-site sensitivity and FP rates are shown in Table 2.
Table 2.
Site-Specific Demographics and Performance Metrics
Voxel-Wise Performance
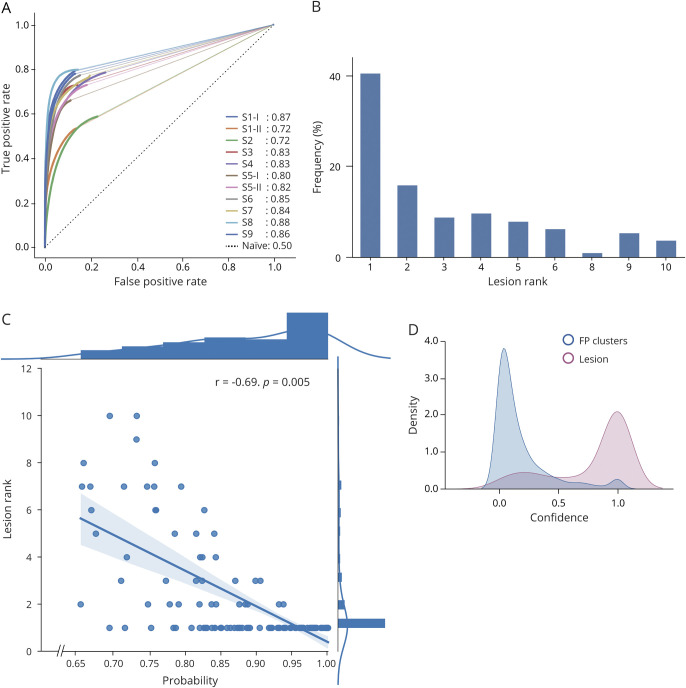
The median area under the receiver operating characteristic curve was 0.83 (range 0.72–0.87), indicative of high sensitivity (high TP rates) and specificity (low FP rates), with comparable performance across sites (Figure 2A).
Figure 2. Performance Evaluation.
(A) Site (S)-wise area under the receiver operating characteristic curve using the leave-one-site-out cross-validation (solid colored lines with values; black dotted line represents a naive classifier). (B) Frequency of lesions according to their rank. Rank 1 signifies highest confidence to be lesional. Seventy-three percent of lesions were distributed across ranks 1 through 5. (C) Lesion rank plotted against probability of being lesional shows a significant correlation with focal cortical dysplasia voxels having low rank values (high confidence) and high probability. (D) Distribution (kernel density estimation) of confidence for lesional and false-positive (FP) clusters; lesions exhibit high confidence values, while FP clusters show low confidence.
Analysis of Confidence
In 73% of patients, the FCD lesion was among the 5 clusters with the highest confidence; in half of them, it ranked the highest, with a mean probability of 72% (95% confidence interval 69%–76%; Figure 2B). Lesion rank negatively correlated with probability, i.e., the lower the rank, the higher the probability of being lesional (r = −0.69, p = 0.005, Figure 2C). Moreover, confidence for a cluster to be lesional centered around 1 (i.e., 100% confidence), while for FPs it centered around zero (Figure 2D). Representative MRI-negative cases are shown in Figures 3 and 4.
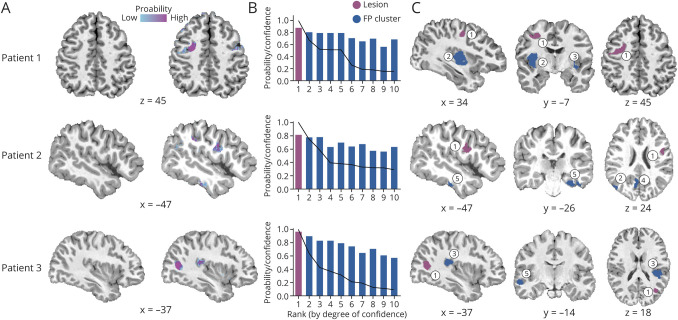
Figure 3. Automated Detection of MRI-Negative FCD.
(A) T1-weighted MRI and prediction probability maps with the lesion circled. (B) Probability of the lesion and false-positive (FP) clusters sorted by their rank; superimposed line indicates the degree of confidence for each cluster. (C) Location of the focal cortical dysplasia (FCD) lesion (rank 1, highest confidence; purple) and FP clusters (ranks 2–5; blue). In these cases, the lesion has both the highest confidence (rank 1) and high probability (>0.8). S = site.
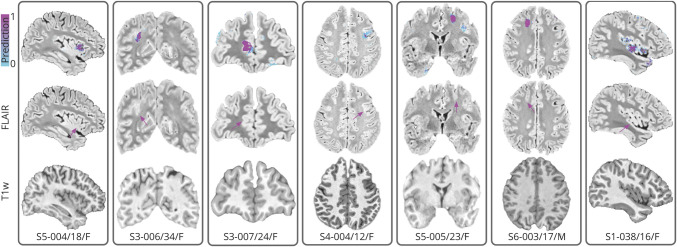
Figure 4. Representative FCD Detection Examples.
Seven representative MRI-negative focal cortical dysplasia (FCD) lesions across sites (Ss) are shown (top row: prediction overlaid on the fluid-attenuated inversion recovery [FLAIR]; flame scale indicates the probability strength). Bottom labels are interpreted as site-patient-identification/age/sex. Arrows indicate the ground-truth lesion location. T1w = T1 weighted.
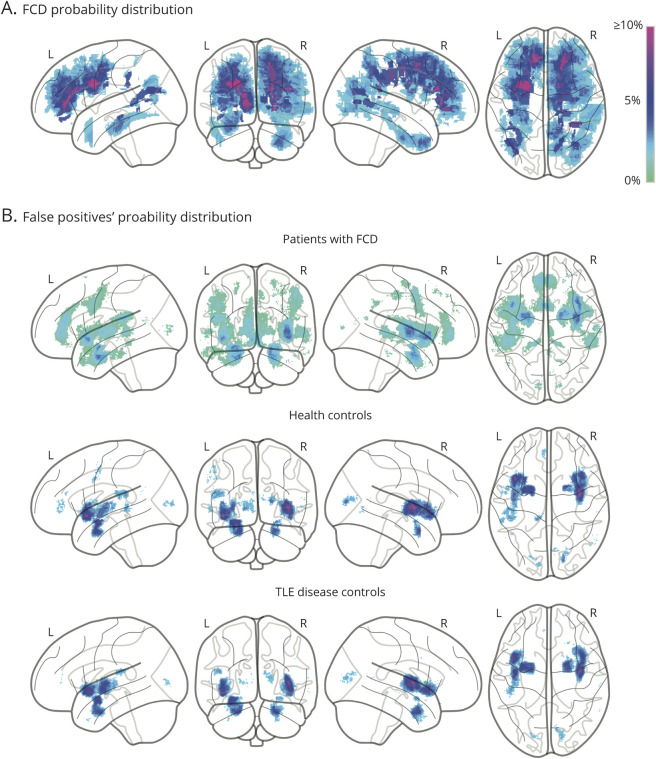
Spatial Distribution of FCD and FPs
The majority of FCD lesions were located within the frontal lobes (Figure 5A). Overall, FPs in patients and healthy and disease controls (Figure 5B) were found in the insula and the parahippocampus (Dice overlap with FCD: 21%, 22%, and 34%, respectively). FPs in healthy and disease controls overlapped to a greater extent (Dice 52%) and exhibited low confidence to be lesional (i.e., high uncertainty). Conversely, FPs in patients with FCD tended to display high confidence to be lesional (p = 0.013). Coordinates for the lesion and FPs are listed in eTables 2 and 3 (available from Dryad: doi.org/10.5061/dryad.h70rxwdgm). The incidence of FP clusters was negatively correlated with age (r = −0.23, p = 0.004): the younger the patients, the higher the number of FPs. The number of FPs was not significantly different between MRI-positive and MRI-negative patients.
Figure 5. Probability Distributions of FCD and False Positives.
(A) Lesional probability maps of manually labelled focal cortical dysplasia (FCD) lesions superimposed on glass brains. (B) Probability maps of confidence of false-positive (FP) clusters across cohorts. Colors indicate proportions (in percent) of (A) lesional and (B) FP voxels. TLE = temporal lobe epilepsy.
Discussion
MRI-negative FCD represents a major diagnostic challenge. To define the epileptogenic area, patients undergo long and costly hospitalizations for EEG monitoring with intracerebral electrodes, a procedure that carries risks similar to those of surgery itself.22,23 Moreover, patients without MRI evidence for FCD are less likely to undergo surgery and consistently show worse seizure control compared to those with visible lesions.24,25 Here, we present a deep learning method for automated FCD detection trained and validated on histologically verified data from multiple centers worldwide. The classifier uses T1- and T2-weighted FLAIR, contrasts available on most recent magnetic resonance scanners14; operates in 3D voxel space without laborious preprocessing and feature extraction; and pairs predictions with confidence. It yields the highest performance to date with a sensitivity of 93% using a leave-one-site-out cross-validation and 83% when tested on an independent cohort while maintaining a high specificity of 89% in both healthy and disease controls. Deep learning detected MRI-negative FCD with 85% sensitivity, thus offering a considerable gain over standard radiologic assessment. Results were generalizable across cohorts with variable age, hardware, and sequence parameters. Taken together, such characteristics and performance promise potential for broad clinical translation. These advantages notwithstanding, good-quality scans are essential to guarantee valid results because motion can mimic lesions14; we thus advise against analyzing low-quality motion-corrupted scans. While a Classification III for the level of evidence was assigned to our study, the current American Academy of Neurology scheme for diagnostic accuracy does not specify criteria for designs based on machine learning algorithms. A revision of these guidelines, ideally disease specific, would likely better reflect the level of evidence for studies relying on artificial intelligence.
Over the past decade, several automated FCD detection algorithms have been developed, the most recent relying on surface-based representations.3,6,19,20 While the majority operate on T1-weighted MRI, recent methods have combined T1-weighted and T2-weighted MRI for improved performance.6,21 A few have used shallow (single-layer) artificial neural networks.4,21 Notably, all require arduous preprocessing, including manual corrections of tissue segmentation and surface extraction, thus precluding integration into clinical workflow. They rely on domain knowledge to engineer features. These procedures generally fail to detect subtle lesions.7 In comparison, our approach offers several advantages. First, to optimize lesion detection across the FCD spectrum, we leveraged the power of CNNs that recursively learn complex properties from the data themselves. Second, contrary to previous medical imaging applications relying on 2-dimensional orthogonal sampling, we extracted 3D patches to model the spatial extent of FCD across multiple slices and tissue types. Operating in true volumetric domain allowed assessment of the spatial neighborhood of the lesion, whereas prior surface-based methods have considered each vertex location independently. Third, restricting training to the GM reduced the nearly infinite dataset to a manageable finite set. Last, by relying on participant-wise feature normalization rather than group-wise, our implementation obviates the need for a matched normative dataset, an expensive and time-intensive undertaking. Compared to previous deep learning methods26-28 in which clinical description was scarce to absent and information on the FCD expert labels and histologic validation of lesions was not provided, our study relied on best-practice multimodal MRI, histologically validated lesions, and a large dataset. Moreover, in previous work, FLAIR images in presumably MRI-positive patients were acquired with interslice gaps ranging from 0.5 to 1.0 mm,26,27 and the acquisition parameters for the 3D T1-weighted images were different from those in healthy and disease controls.28
The practical advantages of our method notwithstanding, a general limitation of deep learning is the reduced transparency of the process leading to the predictions, a consequence of the high dimensionality of learned features. The tradeoff is a richer encoding and learning of complex spatial covariances of intensity and morphology that is beyond the ability of human eye. To maximize transparency and validity, we trained our algorithm on manual expert labels of histologically validated FCD lesions. In addition to a rigorous cross-validation design, including application of the classifier to a totally independent cohort of patients with FCD, our predictions were stratified according to confidence to be lesional. These precautions notwithstanding, as for many diagnostic tests, the convergence of findings with independent tests is essential to increase confidence even further.
Estimation of generalizability is key to any diagnostic method. To guarantee unbiased evaluation, training and testing datasets should remain distinct. We thus devised a strategy in which the model was iteratively trained on patient data from all sites, except the 1 held-out. this guaranteed out-of-distribution validation in which tested cohorts were never part of the training. This leave-one-site-out cross-validation simulated a real-world scenario with optimal bias-variance trade-off compared to conventional train-test split of k-folds; it also exploited the full richness of data during training and the out-of-distribution samples from a single site during testing. Moreover, the classifier trained on the full dataset was tested on a totally independent cohort of patients that were never part of training. Consistent high performance across cohorts, as well as modest FPs in healthy and disease controls, demonstrates that our cascaded CNN classifier learns and optimizes parameters specific to FCD, a fact validated by histologic confirmation.
In machine learning, human in the loop refers to the need for human interaction with the learner to improve human performance, machine performance, or both. Human involvement expedites the efficient labeling of difficult or novel cases that the machine has previously not encountered, reducing the potential for errors, a requirement of utmost importance in health care. In FCD, the outcome of surgery depends heavily on the identification of the lesion; it is thus crucial to decide which putative lesional clusters are significant. In this context, thresholding the final probabilistic mean map is essential to evaluate the balance between TPs and FPs. Notably, to guarantee an objective assessment of sensitivity and specificity across cohorts, in this study, we defined an empirical threshold. However, in clinical practice, a judicious approach would imply adaptive thresholding of the maps at single-patient level, taking into account independent tests. Indeed, in 5 of 11 of the undetected MRI-negative cases, the lesion could be resolved when modulating the threshold in light of seizure semiology and electrophysiology. Besides thresholding, confidence is pivotal in any diagnostic assessment, an aspect so far neglected. To fill this gap, we incorporated a bayesian uncertainty estimation that enables risk stratification. In practical terms, we ranked putative lesional clusters in a given patient according to confidence, thus assisting the examiner in gauging the significance of all findings. In 73% of cases, the FCD was among the top 5 clusters with the highest confidence to be lesional; in half of them, it ranked the highest. In the remaining 27%, lesions manifested with low confidence. In a real-world scenario, when location is unknown (i.e., no FCD label is available), a concerted evaluation including electroclinical and other imaging tests is likely to increase diagnostic certainty.29 While the good performance of our classifier is also attributable to the richness of the training set, including a large spectrum of anatomic locations, 11 MRI-negative FCDs remained unresolved, with 6 located in the orbitofrontal cortex, an area for which limited data were available for training. The prospective use of our classifier trained on the entire cohort would likely reclaim these lesions.
The analysis of the spatial distribution of FPs was moderately comparable across patients with FCD and healthy and disease controls, involving mainly the insula and parahippocampal region bilaterally. A possible explanation may lie in the similarity of the cytoarchitectonic signature of these cortices with FCD histopathologic traits. The 3-layered cortex of the hippocampal formation, the transitional mesocortex of the parahippocampus, and the mesocortex-like insula present with indistinct boundaries between laminas compared to the typical 6-layered neocortex30,31; these cortices may thus mimic dyslamination and blurring. Notably, however, our algorithm detected 3 of 3 FCD lesions in the insula with a high degree of confidence. Because these lesions were provided by different sites, the leave-one-site-out strategy guaranteed that each training set had at least 1 lesion. Nonetheless, adding more lesions to the training set would increase the ability of the classifier to learn better discriminative features in the insular region. Alternatively, an impact of developmental trajectory32 on FPs is suggested by the high prevalence in younger patients, possibly in relation to age-varying tissue contrast, cortical myelination, and maturation, which may also manifest as lesion-like on MRI. Conversely, registration errors are less likely in our voxel-based method compared to surface-based algorithms. For the latter, to align a participant’s brain into a standardized stereotaxic space, registration strongly depends on the accuracy of GM/WM segmentation, while our method does not require tissue segmentation. Notably, some FPs were seen only in cases with FCD, particularly in frontocentral regions, and tended to gather around the lesion, suggesting subthreshold perilesional anomalies not included in the manually segmented label.3,33 Given the favorable surgical outcome, a biological explanation for FPs in our FCD cohort may thus include a combination of normal cytoarchitectural nuances and nonepileptogenic peri-lesional developmental anomalies. In a previous study,3 we found FPs to manifest as abnormal sulcal depth, while the FCD lesions had higher cortical thickness relative to controls. Sulcal abnormalities in cortical malformations have been described in the proximity and at a distance of MRI-visible lesions and are thought to result from disruptions of neuronal connectivity and WM organization.3,34 Finally, it is also plausible that some FP clusters my represent dysplastic tissue, an entity so far reported in only 5 cases.35
While our algorithm was trained on histologically verified FCD-II lesions and is aimed mainly at identifying MRI-negative FCD, it is possible that it could identify difficult-to-detect low-grade tumors that may resemble dysplastic lesions, a rare occurrence because these tumors are generally easy to see on routine MRI. Regardless, the dilemma of differentiation of FCD from low-grade tumors uniquely based on MRI features may arise; the differential diagnosis is then evaluated with additional tests, including magnetic resonance spectroscopy. On the other hand, our algorithm may be useful in identifying associated often occult dysplastic lesions in the peritumoral area.36
Traditional machine learning adopts a centralized approach that requires training datasets to be aggregated in a single center. A significant obstacle to clinical adoption of such strategy is privacy and ethical concerns. Federated learning,37 on the other hand, is a distributed approach that enables multi-institutional collaboration without sharing of patient data. Our proposed approach of patch-based data augmentation preserves privacy because only a portion of each patient's data are collated and randomized before exposure to the neural network, an implementation that can be flexibly reconfigured to support federated learning. As the data corpus diversifies and expands to include more edge cases, performance and confidence of future classifiers will inevitably improve.
Glossary
- CNN
convolutional neural network
- FCD
focal cortical dysplasia
- FLAIR
fluid-attenuated inversion recovery
- FP
false positive
- GM
gray matter
- HS
hippocampal sclerosis
- MNI
Montreal Neurological Institute
- 3D
3-dimensional
- TLE
temporal lobe epilepsy
- TP
true positive
- WM
white matter
Appendix. Authors
Footnotes
Editorial, page 754
Class of Evidence: NPub.org/coe
Study Funding
This work was supported by the Canadian Institutes of Health Research to A.B. and N.B. (MOP-57840 and 123,520), Natural Sciences and Engineering Research Council of Canada (Discovery-243141 to AB and 24779 to N.B.), Epilepsy Canada (Jay & Aiden Barker Breakthrough Grant in Clinical & Basic Sciences to A.B.), and Brain Canada. Salary supports were provided by Fonds de Recherche Santé – Quebec, Savoy Foundation for Epilepsy (H.-M.L.), and Lloyd Carr-Harris Foundation (B.C.).
Disclosure
R.S. Gill, H.-M. Lee, B. Caldairou, S.J. Hong, C. Barba, F. Deleo, L. D'Incerti, V.C. Mendes Coelho, M. Lenge, M. Semmelroch, D. Schrader, F. Bartolomei, M. Guye, A. Schulze-Bonhage, H. Urbach, K.H. Cho, F. Cendes, R. Guerrini, G. Jackson, R.E. Hogan, N. Bernasconi, and A. Bernasconi report no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Bernasconi A, Bernasconi N, Bernhardt BC, Schrader D. Advances in MRI for “cryptogenic” epilepsies. Nat Rev Neurol. 2011;7(2):99-108. [DOI] [PubMed] [Google Scholar]
- 2.So EL, Lee RW. Epilepsy surgery in MRI-negative epilepsies. Curr Opin Neurol. 2014;27(2):206-212. [DOI] [PubMed] [Google Scholar]
- 3.Hong S-J, Kim H, Schrader D, Bernasconi N, Bernhardt BC, Bernasconi A. Automated detection of cortical dysplasia type II in MRI-negative epilepsy. Neurology. 2014;83(1):48-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, Ahmed B, Thesen T, et al. A Non-Parametric Approach to Detect Epileptogenic Lesions Using Restricted Boltzmann MachinesKDD '16. ACM Press; 2016:373-382. [Google Scholar]
- 5.Snyder K, Whitehead EP, Theodore WH, Zaghloul KA, Inati SJ, Inati SK. Distinguishing type II focal cortical dysplasias from normal cortex: a novel normative modeling approach. Neuroimage Clin. 2021;15:102565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill RS, Hong S-J, Fadaie F, et al. Automated detection of epileptogenic cortical malformations using multimodal MRI. In: Cardoso MJ, Arbel T, Carneiro G, et al., editors. Medical Image Computing and Computer-Assisted Intervention. Springer International Publishing; 2017:349-356. [Google Scholar]
- 7.Kini LG, Gee JC, Litt B. Computational analysis in epilepsy neuroimaging: a survey of features and methods. Neuroimage Clin. 2016;11:515-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88. [DOI] [PubMed] [Google Scholar]
- 9.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44-56. [DOI] [PubMed] [Google Scholar]
- 10.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436-444. [DOI] [PubMed] [Google Scholar]
- 11.Leibig C, Allken V, Ayhan MS, Berens P, Wahl S. Leveraging uncertainty information from deep neural networks for disease detection. Sci Rep. 2017;7(1):17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sica GT. Bias in research studies. Radiology. 2006;238(3):780-789. [DOI] [PubMed] [Google Scholar]
- 13.Ashman EJ, Gronseth GS. Level of evidence reviews: three years of progress. Neurology. 2012;79(1):13-14. [DOI] [PubMed] [Google Scholar]
- 14.Bernasconi A, Cendes F, Theodore WH, et al. , Recommendations for the use of structural magnetic resonance imaging in the care of patients with epilepsy: a consensus report from the International League Against Epilepsy Neuroimaging Task Force. Epilepsia. 2019;60(6):1054-1068. [DOI] [PubMed] [Google Scholar]
- 15.Blümcke I, Thom M, Aronica E, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc task force of the ILAE Diagnostic Methods Commission. Epilepsia. 2011;52(1):158-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17(1):87-97. [DOI] [PubMed] [Google Scholar]
- 17.Gal Y, Ghahramani Z. Bayesian convolutional neural networks with bernoulli approximate variational inference. arXiv preprint. 2015;arXiv:1506.02158.
- 18.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Machine Learn Res. 2014;15(56):1929-1958. [Google Scholar]
- 19.Thesen T, Quinn BT, Carlson C, et al. Detection of epileptogenic cortical malformations with surface-based MRI morphometry. PLoS One, 2011;6(2):e16430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmed B, Brodley CE, Blackmon KE, et al. Cortical feature analysis and machine learning improves detection of “MRI-negative” focal cortical dysplasia. Epilepsy Behav. 2015;48:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler S, Wagstyl K, Gunny R, et al. Novel surface features for automated detection of focal cortical dysplasias in paediatric epilepsy. Neuroimage Clin. 2017;14:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hader WJ, Tellez-Zenteno J, Metcalfe A, et al. Complications of epilepsy surgery: a systematic review of focal surgical resections and invasive EEG monitoring. Epilepsia. 2013;54(5):840-847. [DOI] [PubMed] [Google Scholar]
- 23.Hedegärd E, Bjellvi J, Edelvik A, Rydenhag B, Flink R, Malmgren K. Complications to invasive epilepsy surgery workup with subdural and depth electrodes: a prospective population-based observational study. J Neurol Neurosurg Psychiatry. 2014;85(7):716-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Téllez-Zenteno JF, Hernandez-Ronquillo L, Moien-Afshari F, Wiebe S. Surgical outcomes in lesional and non-lesional epilepsy: a systematic review and meta-analysis. Epilepsy Res. 2010;89(2-3):310-318. [DOI] [PubMed] [Google Scholar]
- 25.Noe K, Sulc V, Wong-Kisiel L, et al. Long-term outcomes after nonlesional extratemporal lobe epilepsy surgery. JAMA Neurol. 2013;70(8):1003-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bijay Dev KM, Jogi PS, Niyas S, Vinayagamani S, Kesavadas C, Rajan J. Automatic detection and localization of focal cortical dysplasia lesions in MRI using fully convolutional neural network. Biomed Signal Process Control. 2019;52:218-225. [Google Scholar]
- 27.Thomas E, Pawan SJ, Kumar S, et al. Multi-res-attention UNet: a CNN model for the segmentation of focal cortical dysplasia lesions from magnetic resonance images. IEEE J Biomed Health Inform. 2020;25(5):1724-1734. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Ahmed SN, Mandal M. Automated detection of focal cortical dysplasia using a deep convolutional neural network. Comput Med Imaging Graph. 2020;79:101662. [DOI] [PubMed] [Google Scholar]
- 29.Guerrini R, Duchowny M, Jayakar P, et al. Diagnostic methods and treatment options for focal cortical dysplasia. Epilepsia. 2015;56(11):1669-1686. [DOI] [PubMed] [Google Scholar]
- 30.Nieuwenhuys R. The insular cortex: a review. Prog Brain Res. 2012;195:123-163. [DOI] [PubMed] [Google Scholar]
- 31.Gogolla N. The insular cortex. Curr Biol. 2017;27(12):R580-R586. [DOI] [PubMed] [Google Scholar]
- 32.Gilmore JH, Knickmeyer RC, Gao W. Imaging structural and functional brain development in early childhood. Nat Rev Neurosci. 2018;19(3):123-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong S-J, Bernhardt BC, Caldairou B, et al. Multimodal MRI profiling of focal cortical dysplasia type II. Neurology. 2017;88(8):734-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Besson P, Andermann F, Dubeau F, Bernasconi A. Small focal cortical dysplasia lesions are located at the bottom of a deep sulcus. Brain, 2008;131(pt 12):3246-3255. [DOI] [PubMed] [Google Scholar]
- 35.Fauser S, Sisodiya SM, Martinian L, et al. Multi-focal occurrence of cortical dysplasia in epilepsy patients. Brain. 2009;132(pt 8):2079-2090. [DOI] [PubMed] [Google Scholar]
- 36.Slegers RJ, Blümcke I. Low-grade developmental and epilepsy associated brain tumors: a critical update 2020. Acta Neuropathol Commun. 2020;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaissis GA, Makowski MR, Rückert D, Braren RF. Secure, privacy-preserving and federated machine learning in medical imaging. Nat Mach Intell. 2020;2:305-311. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
These datasets are not publicly available because they contain information that could compromise the privacy of research participants. The source code and pretrained model weights are available for download online (version 1.0.0 at GitHub). In addition, a derivative dataset composed of lesional and nonlesional patches from 148 patients with FCD is available as a Hierarchical Data Format dataset (available from Zenodo).