Abstract
Heme is essential for the survival of virtually all living systems—from bacteria, fungi, and yeast, through plants to animals. No eukaryote has been identified that can survive without heme. There are thousands of different proteins that require heme in order to function properly, and these are responsible for processes such as oxygen transport, electron transfer, oxidative stress response, respiration, and catalysis. Further to this, in the past few years, heme has been shown to have an important regulatory role in cells, in processes such as transcription, regulation of the circadian clock, and the gating of ion channels. To act in a regulatory capacity, heme needs to move from its place of synthesis (in mitochondria) to other locations in cells. But while there is detailed information on how the heme lifecycle begins (heme synthesis), and how it ends (heme degradation), what happens in between is largely a mystery. Here we summarize recent information on the quantification of heme in cells, and we present a discussion of a mechanistic framework that could meet the logistical challenge of heme distribution.
Keywords: heme, heme quantification, heme trafficking, heme homeostasis, heme sensors
Introduction
Heme is a macrocyclic complex of iron, with the metal ion coordinated equatorially to the four nitrogen atoms of an electronically delocalized protoporphyrin IX ring (Figure 1A) and to one or two axial ligands. Broadly following an IUPAC convention that defines heme as a complex of iron coordinated to a porphyrin and to one or two axial ligands,1 the term heme is used widely, and often interchangeably, to refer to different types of iron protoporphyrin IX where the precise heme structure (e.g., heme a, heme b, heme c, heme d, etc.) is not always elaborated. When one of these heme structures is bound to a protein, the two axial ligation positions are normally occupied by donor ligands provided by protein amino acids. Heme ligation to the side chains of His, Cys, Met, Tyr, Lys, Glu residues and even the N-terminus is known.2,3 In binding to a protein in this way, the heme becomes either 5- or 6-coordinated, Figure 1B. By convention, the proximal position, which is most commonly a histidine, is assigned as the fifth ligand and is normally shown underneath the heme when visualizing heme structures. The sixth ligand, if there is one, is then drawn above the heme on the so-called distal side. This proximal/distal terminology dates back at least as far as the crystallography studies of Kendrew and Perutz.4,5 The distal position can be occupied by a protein amino acid; by a diatomic gas (e.g., O2, NO, CO); by water, hydroxide, or another small molecule ligand (e.g., H2S, CN–); or it can be vacant giving a 5-coordinate heme species. The reactivity of the heme is controlled in part by the number and identity of these ligands at the proximal and distal positions, while stabilization of the heme molecule is further controlled by the heme-binding pocket via hydrophobic interactions with the porphyrin ring and vinyl groups, and hydrogen bonds between the propionate groups and the solvent.6
Figure 1.
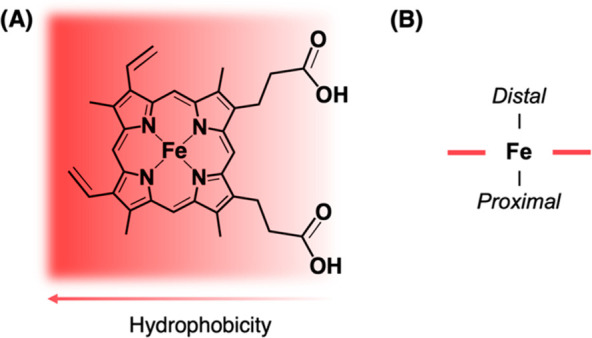
(A) The structure of iron protoporphyrin IX, also known as heme b. While heme is mostly hydrophobic, the carboxylate groups enable hydrogen-bonding interactions between the heme group and other molecules, including assisting the binding of heme to a protein. (B) The heme group is classified as containing distal and proximal sides, which are conventionally drawn above and below the plane of the ring, respectively. In heme proteins, the proximal side is usually bound to an amino acid residue provided by the protein; this helps to control its reactivity as a redox center190,191 and its properties as a gas-binding molecule for storage and signaling.11
The role of the protein can thus be envisaged as solubilizing the hydrophobic and thus poorly soluble heme molecule, and in doing so controlling the reactivity of the heme group for biological purposes. Over the past several decades, most of what has been learned about the role of heme in biology relates to binding of heme to individual proteins such as those involved in oxygen transport (e.g., myoglobin, hemoglobin) and sensing (e.g., cytoglobin, neuroglobin),7 bioenergetics (e.g., cytochrome c, cytochrome c oxidase),6 metabolism and catalysis driven by the insertion of oxygen (e.g., cytochrome P450, indoleamine 2,3-dioxygenase),6 peroxidase catalysis,8 formation of nitric oxide,9 and lipid metabolism (e.g., COX-2).10
In addition to these widely known roles for heme in biology, there is now a substantial body of evidence that identifies heme as a regulator of various cellular activities.11−16 The first report, to our knowledge, of heme as a regulator was as far back as 1975, when heme was observed as regulating the synthesis of δ-aminolevulinic acid synthase which catalyzes the first step of the 8-step heme biosynthesis pathway.17 We see chemical logic in the concept of regulatory mechanisms in cells that are linked to heme binding, but the idea of heme as a regulator laid dormant in the literature for many years. Despite its toxicity and lack of solubility, heme has a number of redeeming features that make it easier to understand why it could regulate cellular functions. The first is that heme is redox active and can respond to the redox status of the cell by modulating its oxidation state. The concentration of heme in cells is controlled by the balance of heme biosynthesis (to increase heme concentrations) and heme oxygenase activity (which degrades heme and produces CO as a byproduct). Cell signaling gases, such as O2, NO, and CO, can bind rapidly and sometimes reversibly to heme—this could be exploited alongside the association of heme with a protein to create a second layer of regulatory control. CO is itself produced via degradation of heme by the O2-dependent heme oxygenase enzyme,18−20 and NO is produced by the heme- and O2-dependent NO synthase enzyme;9 i.e., the production of cell signaling gases also depends on the presence of heme. We envisage that the redox state of the cell, the oxidation state of the heme, the balance between heme synthesis and degradation, and the relative concentrations of O2, NO, and CO may all be interconnected as outlined in Figure 2. This could be used as a versatile mechanism for cell signaling. Examples of heme-dependent regulation are beyond the scope of this review but have been summarized recently.11,12 They include, among others, transcriptional regulation and gas sensing,11,12,15 degradation of the p53 protein,21 regulation of the circadian clock,22−42 immune response,43,44 aging,45 and the gating of numerous ion channels.46−69
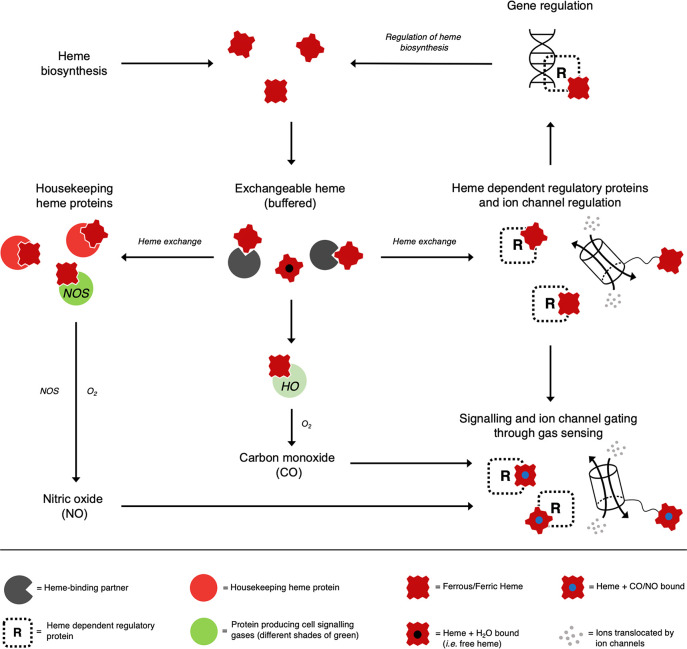
Figure 2.
The possible interconnected pathways for the movements of heme in cells, and the links to signaling gases such as CO and NO. From the total heme synthesized in the cell (top), a proportion is bound irreversibly to heme-binding housekeeping proteins (red circles) that are essential for cell survival. A body of exchangeable heme is envisaged as being mostly weakly bound, soon after heme biosynthesis, to heme-binding partners (dark gray pacmans in this and other figures, which could be heme proteins or non-heme proteins) and available for exchange to heme dependent regulatory proteins (R, right). These heme-binding partners constitute an exchangeable, buffered, reservoir that can provide a flexible supply of heme and protect against changes in heme concentration. Once formed, these heme-bound proteins can serve in regulatory roles by, for example, binding to DNA for transcriptional control (top right; including the regulation of heme biosynthesis17,21,28,37,192−194) or to ion channels (middle and bottom right54,195). In green circles are shown the proteins that produce cell signaling gases—nitric oxide synthase (NOS, left) and heme oxygenase (NOS, middle). The synthesis of NO by NOS, and the production of CO by the heme degrading HO enzyme, adds multiple layers of complexity by coupling the formation of cell signaling gases to the heme-binding process. This would allow both CO and NO to bind to any heme protein with a regulatory function (bottom right) but could equally well occur for other heme dependent regulatory processes. It is worth noting that, while the binding of π-acid ligands like CO is traditionally associated exclusively with heme in its ferrous form, ferric heme has also been shown capable of binding CO/NO.41 For the purposes of this review, movement of ferric/ferrous heme is presumed to mean heme b.
The Biological Need for Heme Distribution
The fundamental requirement for heme across virtually all organisms—both in catalytic and regulatory roles as outlined above—means that cells need mechanisms to manage heme supply and distribution. Supply of heme is regulated by the well-studied heme biosynthesis pathway.70 At the other end of the heme lifecycle, surplus heme (when it exists) is degraded by the O2-dependent heme oxygenase enzyme.18,19 But while the enzymatic machinery for making heme–heme synthesis—and removing heme–heme degradation—is well established, what happens in between is almost completely unknown. It has been recognized as far back as the 1970s71,72 that cells need a supply of heme to respond to cellular demands, but until quite recently tools to examine this question have been lacking. In this review, we outline recent advances in the development of methods for measuring heme in cells, and we present ideas on, and possible mechanisms for, heme mobilization consistent with quantitative determinations of heme concentration.
Terminology
To begin the discussion, we describe and define the terminology. For the purposes of this review, we take heme (in the context of heme transport) to mean heme b. Literature on the role and distributions of heme in cells has used a bewildering, and at times imprecise, range of terminologies. Phrases such as “labile heme”, “regulatory heme”, “free heme”, and “heme pool” are all used in the literature, often interchangeably.73 While the need to discriminate between these types of heme in cells is obvious, it is challenging to provide a precise description of each. What does “labile heme” mean? How can “free heme” exist in the cell, given that heme is cytotoxic and poorly soluble? Does the phrase “regulatory heme” describe a supply of heme dedicated exclusively for regulatory purposes, or might it be used for other purposes? Given its insolubility and cytotoxicity, does it actually make sense to talk about “pools” of heme when current estimates for heme concentration are in the nM to μM range?74−79 The lack of precise descriptions for the nature of different types of heme can lead to confusing or contradictory interpretations. Below we provide a summary and offer some suggestions, which we hope will be helpful in future discussions for both specialists and newcomers to the field.
Regulatory Heme
In disciplines that vary from blood disorder (e.g., porphyria),80,81 gene expression,71,72 and heme biosynthesis,82,83 a small portion of the total heme content of the cell has been referred to over many years as being present in a regulatory or intracellular heme pool. These terms originate from early papers71,72,83−86 where it was also suggested that the nature of heme was a free molecule (see below). The existence of a form of regulatory heme is conceptually useful, and because of this the nature of regulatory heme has been debated as long ago as 1975 when new roles for heme—above and beyond the traditional roles in oxygen binding, electron transfer, and catalysis—were identified by Granick.83 In Granick’s work, the regulation of heme biosynthesis was proposed as being controlled by fluctuations in the concentrations of heme in a dedicated heme pool where heme was envisaged as being weakly associated with an ensemble of cytosolic proteins.83,87 These weak binding interactions were considered important for heme to be readily exchangeable.83,86 The terms were then later adopted by scientists in multiple fields.46,80,88−98 Hence, since Granick’s initial studies, the phrases “regulatory heme” and “free heme” have been widely used although not precisely defined.
Free Heme
To the best of our knowledge, the term free heme was introduced in the 1970s72,85,99 and was interpreted as an intracellular population of free heme molecules either on their own (free) or all together (in a pool). These early papers noted the difficulties associated with the idea of a pool of free heme, but, nevertheless, the concept and the terms “free heme” and “pool of free heme” became ingrained in the literature.85,92,94,95,100,101 The term “free heme” has never been precisely defined—broadly speaking it has been taken to mean a heme molecule that is not bound to anything else. There are difficulties with that idea, however, and the concept of large amounts of free heme, because heme is cytotoxic through Fenton chemistry and radical formation.92,94,96,102−104 So if heme is free in the cells, it would cause problems. Heme is also a hydrophobic molecule and poorly soluble, so presumably cannot exist in cells without being solubilized by binding to a protein, cellular membranes,105 or even nucleic acids.45,106 Moreover, heme needs to be ligated at the fifth and sixth ligation position, otherwise, it will stack to form dimers, or higher multimers, in solution.107,108 This is problematic for cellular handing because, in a dimeric or multimeric form, heme could not be delivered to proteins that require only one molecule per binding site.
We envisage that a “free” molecule of heme can therefore only exist transiently in cells. In this review, we will use the term “free heme” to indicate molecules of iron protoporphyrin IX, in the ferrous or ferric oxidation state, existing in the absence of any protein binding partner but weakly associated with water molecules as ligands at the fifth and sixth coordination sites.
Labile Heme
In recent years the term “labile heme” has appeared.44,78,96,105,109−120 The use of this term allows a distinction to be made between the proportion of the total heme content that is available for mobilization, and the proportion that is unavailable (or, more precisely, inert for exchange) because it is bound with a high affinity, and therefore irreversibly, to proteins. Ideas on what the concepts of labile heme actually means mimic the early ideas on free heme.83,87 Labile heme is envisaged as being continuously engaged in transient binding to intracellular proteins that exist to actively buffer heme concentrations in the cell. Since some heme is unavailable for distribution in cells, labile heme is thus envisaged as a intermediary through which heme can move and be distributed through the cell.105,110−112 The term labile heme probably finds its origins as an adaptation of “labile iron pool” that predated it.121
Exchangeable Heme
The use of the term lability to distinguish a more mobile fraction of the total heme from that which is permanently (irreversibly) embedded into heme proteins is similar in concept to the kinetic lability of ligands in inorganic metal complexes.122−124 Labile ligands exchange very fast, and inert ligands exchange slowly. But the availability and distribution of heme will be defined not just by the kinetic lability of the ligands bound to the heme, but also by thermodynamics. This is because cascades of heme exchange events down a thermodynamic gradient (dictated by heme binding affinity) will control the overall distribution of heme. Hence, our opinion is that “exchangeable heme”—which has been adopted in a limited number of examples77,86,97,125—is a more precise term to convey the concept that both kinetic and thermodynamic processes are relevant when considering the bioavailability of heme.
Definitions Used in This Review
We propose here a working set of definitions.
Total Heme (Ht)
The total heme content of a cell. This includes the fraction of heme that is bound irreversibly to proteins and thus not available for other purposes (defined here as Hh), and the fraction of exchangeable heme (which includes small quantities of free heme, see below).
Exchangeable Heme (He)
A fraction of the total heme that is reversibly bound to proteins or small ligand molecules (e.g., free amino acids, H2O). Exchangeable heme can be considered as a reservoir that provides an accessible supply of heme to the cell.
Free Heme (Hf)
Molecules of heme that are not bound to a protein but weakly coordinated with water molecules. We expect free heme to exist in vanishingly small quantities. Long and co-workers126 have recently offered a definition for free heme which is similar to that proposed here.
In principle, Ht = Hh + He + Hf, where Hh is the amount of heme that is bound irreversibly to heme proteins (including membrane and cytosolic proteins).
The Logistics of Heme Supply and Demand
Given the need to move heme from its place of synthesis (in the mitochondria) to other regions of the cell, how can cells control the balance of supply and demand and what mechanisms are in place to do this?
Controlling Supply and Demand
The balance between heme biosynthesis and degradation is part of the regulatory control that the cell needs to maintain heme availability below the cytotoxic threshold (Figure 3A).94,102,127 Heme oxygenase, whose mechanism of degradation of heme into biliverdin and CO is well-known,18,19 plays a crucial role in maintaining this balance. Heme oxygenase-1 is transcriptionally regulated and induced by infection, inflammation, oxidative stress or increasing concentrations of heme.128,129 Interestingly, the post-transcriptional regulation of heme oxygenase-2 (abundant in the brain, testes, and neural tissue) has been shown to be controlled by heme occupancy in the active site.130 Specifically, heme oxygenase-2 appears to be destabilized and more likely to be degraded when heme is not bound, which is the likely situation under conditions of heme deficiency and when high heme oxygenase activity is undesirable. This regulation would help to keep heme concentrations within a suitable physiological range under conditions of cellular heme deficiency.130
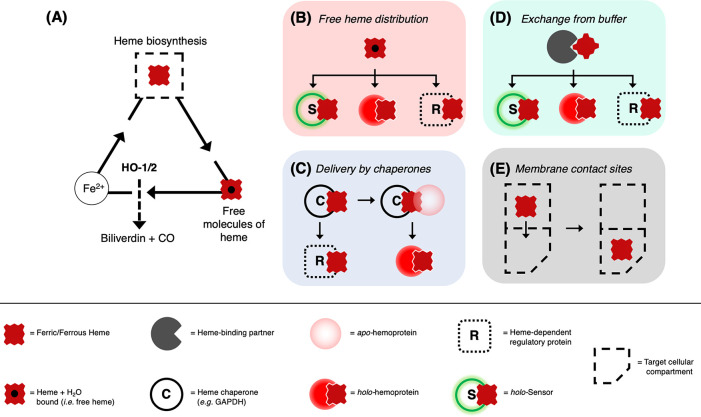
Figure 3.
Representation of possible mechanisms for distribution and delivery of heme across the cell. (A) The life cycle of heme in cells starts with its biosynthesis in the mitochondria (full dashed square) and ends with its degradation by heme oxygenase (HO-1/2). Heme oxygenase generates Fe2+ which can be recycled for the synthesis of new heme molecules. A balance between synthesis and degradation contributes to the controlling heme concentrations in cells. (B–E) The supply of heme to the locations where it is in demand is suggested as occurring via four possible mechanisms (colored panels). In (B), direct distribution of free heme into a heme protein (red circle), a heme-dependent regulatory function (R, white box), or a genetically encoded/synthetic heme sensor for quantification studies (S, green circle). Free heme (either ferrous or ferric) is envisaged as being present in minuscule concentrations but will still represent a mechanism for heme to be made available in cells.77 In (C), chaperone-mediated heme delivery to an apo-heme protein (pale red circle), for example by GAPDH.113−115 In (D), heme bound to heme-binding partners (dark gray pacman) constitutes a body of exchangeable heme readily available for downstream applications, in the same way as in (B). Possible candidates for heme-binding proteins are IDO, HBP22/23, SOUL, and albumin.111,163,196 In (E), distribution of heme via membrane contact sites between mitochondria—where heme is synthesized—to target cellular compartments which bypasses the need for transporters to mediate the delivery of heme.171
The Involvement of Heme Chaperones
The existence of biological chaperone systems is documented for other redox active transition metal ions (e.g., iron, copper, molybdenum, cobalt, nickel).131−134 Cells adopt strategies to tightly regulate the concentrations of different metals ions, ensuring suitable levels of bioavailability, effective physiological responses, and prevention of cytotoxicity.133−149 The concentration of transition metal ions in cells is kept at very low levels to avoid inappropriate activation of signaling pathways. For instance, the intracellular free copper concentration in S. cerevisiae is limited to less than one molecule per cell.133,137 Similarly, free zinc is estimated in the low femtomolar range in E. coli.133,136 Instead of being free to circulate around the cell, metal ions are stored, locked in an inaccessible state, and released to a network of chaperones or chelators that control their delivery, repurposing, and ensure fidelity in metalation when two competitive metals can bind to the same site.134,138−144 By comparison, it is reasonable to presume that cells use similar principles for the movement and distribution of free heme (Figure 3C,D).
The identification of heme chaperones with the specific role of transporting and selectively delivering heme to downstream proteins would thus clarify the picture considerably. But very few heme chaperones have been identified over the years. A notable example is the CcmE chaperone, part of the multi component Ccm membrane heme maturation machinery for heme translocation to cytochrome c in most proteobacteria, archaea, deinococcales, and plant mitochondria.150−153 However, this is a special case as it speaks only to the need for formation of covalent links to heme in the specialized c-type cytochromes. GAPDH is perhaps the most compelling example of a heme chaperone so far identified.113−115,154 The interaction between GAPDH and heme was initially thought to occur to protect heme from degradation during its transport (which at the same time reduces its cytotoxicity).155 But GAPDH has since been found to be responsible for the delivery of heme to the nuclear transcription factor Hap1156 in S. cerevisiae, to cytosolic iNOS in mouse macrophage cells,113 and to soluble guanylate cyclase in HEK293 cells.114
The involvement of GAPDH in heme delivery in both yeast and mammalian systems is indicative of a broadly conserved strategy for heme transport.113 GAPDH is primarily a glycolytic enzyme, and, therefore, its function as a heme chaperone looks to be opportunistic as far as the cell is concerned. This begs the question as to whether there are other as-yet unidentified heme chaperones lurking about the cell in disguise. Certainly, if heme chaperones are used at all then more than one could be required to create an effective supply chain. Arguably, chaperones of chaperones might even exist.115 But if heme binding can be opportunistic and does not need a specific heme-binding motif—as GAPDH seems to indicate—then identification of heme chaperones becomes an incredibly demanding task which would need to take into account noncanonical heme-binding motifs in addition to the large number of proteins with known heme-binding sites.3,157
All of this leads us to the conclusion that existing, well-known heme-binding proteins could be repurposed for the task of cellular heme delivery under certain conditions. For example, indoleamine 2,3-dioxygenase, whose mechanism and structure is well-documented,158−162 has recently been shown to lose its heme under certain conditions,163 which suggests the possibility that it could be used as a source of heme in specific regions of cells. In all likelihood, there may be other cellular proteins that can similarly masquerade as heme chaperones.
Possible Limitations on the Utility of Chaperones for the Distribution of Heme
The distribution of heme via chaperones can be used to explain how heme moves from the mitochondria to other locations, and then subsequently to the endoplasmic reticulum for degradation by heme oxygenase. But the chaperone mechanism is ill-equipped to deal with rapid changes in heme availability. If increases or decreases in heme concentration are required, then the rate-limiting steps are heme synthesis and degradation. Upregulation of heme synthesis, by expression of the ALA synthase enzyme, in response to low heme availability could take at least 30 min,164 while induction of heme oxygenase-1 for heme degradation could take hours.165 Both of these mechanisms are too slow by far if rapid control of heme concentration is required, for example in the case of ion channel regulation which is necessary for appropriate cardiac function.54 Hence, if the cell is solely reliant on heme chaperones, then it runs the risk of spikes in heme availability should there be a mismatch in timings between the delivery and the removal of heme. Taken together, and bearing in mind the failure (so far) to identify large numbers of heme chaperones, we suggest that complementary mechanisms will be required to regulate heme distribution. We elaborate on this below.
Buffering of Free Heme Concentration as a Mechanism of Control
An ability to store heme in cells would decrease the reliance of cells on heme synthesis and degradation and could be coupled to mechanisms to make heme more or less available, on-demand and at speed (Figure 3D). To date, there have been no proteins identified that have the dedicated function of storing heme, but there is evidence, from recent work,77 of a buffering capacity within cells that can accommodate changes in either the total concentration of heme (i.e., [Ht]), or the concentration of exchangeable heme (i.e., [He]), while maintain the capability to mobilize heme as and when necessary. As indicated above, it is entirely possible that known heme proteins (such as indoleamine 2,3-dioxygenase, and GAPDH) participate in the buffering of free heme concentration ([Hf]) and protect the cell against increases or decreases in total, or exchangeable, heme concentration. In this scenario for heme buffering, the ability of any given protein to acquire heme is based on how well it competes for heme (i.e., the heme affinity) against other proteins. Hence the availability of heme in the cell would be dependent entirely on the Kd, and abundance, of these heme-binding partners. In this way, the concentration of free heme is buffered—or, in other words, the availability of heme is kept within a limited range—by the many heme-binding proteins in the buffer and would be distributed among those proteins according to their relative heme affinities.
Crucially, in this buffering model the hierarchy of heme-binding affinity would be responsive to changes in the composition of the buffering proteins. As an example, upregulation of proteins in the buffer with high heme affinity (e.g., a globin, or a cytochrome) would deplete the capacity of the exchangeable heme reservoir to release heme, as heme was dragged to thermodynamically inaccessible sites. As well as providing a reservoir of exchangeable heme able to meet the localized and instantaneous demands of the cell, such a buffering system would, along with the heme degradation pathways, prevent heme-induced cytotoxicity by limiting the concentrations of free heme within the cell. The interplay between heme biosynthesis, degradation, transcriptional and post-transcriptional regulation of heme buffering proteins, are all envisaged as jointly contributing to the buffering of heme in cells.
Trafficking through Membranes
Evidence is emerging for the involvement of biological membranes as important factors in the trafficking of cellular heme. Heme has been shown to be capable of diffusing or even being retained by cellular lipid bilayers166−168 and examples of putative membrane heme transporters have been identified, most notably the mitochondrial exporter Flvcr1b, which have been reviewed elsewhere.94,169 But while heme may or may not necessitate membrane transporters for its movement across compartments, the contribution of lipid bilayers to heme trafficking is still not completely clarified. A recent study has proposed that the trafficking of heme through membranes does not rely on transporters and is, in contrast, a chaperone-less pathway made possible by interorganellar contact sites. This provides an elegant solution for the exchange of heme between compartments (Figure 3E).170,171
Quantifying Heme Concentrations in Cells
Early Estimation of Heme Concentration
Granick speculated in the 1970s83,87 that the concentration of exchangeable heme in the cytosol was in the range 10–100 nM. This value is, in fact, remarkably close to recent measurements (Table 1). In the past, the cellular heme concentration has had to be inferred from the amount of heme measured in soluble cell lysis extracts determined using either fluorometric approaches172 or enzymatic reconstitution techniques.74,89,97,173 These methods are likely to report a concentration that lies somewhere between that for exchangeable heme ([He]) and the total amount of heme present in the cell ([Ht]) because the denaturing methods lead to release of heme from many heme proteins. In order to selectively measure concentrations of exchangeable heme, it is necessary to use a method that is compatible with in vivo fluorescence imaging or spectroscopy of live cells.174 Such an approach is possible by the design and application of genetically encoded heme sensors which can be expressed recombinantly in different cell lines.
Table 1. Summary of Reported Cellular Heme Concentrations from Known Heme Reporters or Fluorescent Heme Sensors.
| reporter | heme probe | fluorescent tag(s) | Kd,heme (nM)a,b | measured heme concentration (nM) | quantitation method | tested in | ref. |
|---|---|---|---|---|---|---|---|
| CYSDY-9 | IsdX1, IsdC | ECFP, EYFP | 63.5 ± 14.3 | Cytosol 25.6 ± 5.5 | FRET | HeLa | (76) |
| Mitochondria 23.3 ± 4.9 | |||||||
| Nucleus 31.0 ± 7.0 | |||||||
| ER 5.4 ± 1.4c | |||||||
| HS1 | Cytochrome b562 | EGFP, mKATE2 | 3 (ferric), <1 (ferrous) | Cytosol 20–40 | FRET | S. cerevisiae | (79) |
| Nucleus <2.5 | |||||||
| Mitochondria <2.5 | |||||||
| CHY | P. falciparum Histidine rich protein | ECFP, EYFP | ∼250 | Cytosol 1600 | FRET | P. falciparum | (78) |
| apo-HRP | Horseradish peroxidase | – | – | 600 | peroxidase activityd | Human lung fibroblasts (IRM90) | (73,97) |
| apo-HRP | Horseradish peroxidase | – | – | 2100 ± 2 | Reconstitution of catalytic activityd | Human erythrocytes | (74) |
| apo-HRP | Horseradish peroxidase | – | 270 ± 40 | 433 ± 125 | Reconstitution of catalytic activityd | HEK293 | (75) |
| mAPXmEGFPe | Ascorbate peroxidase | mEGFP | 22 | Cytosol 4 | RET | HEK293 | (77) |
The heme affinity, or conversely the dissociation constant Kd,heme, is a key parameter in heme quantitation. Kd,heme values should be close to the physiological concentrations of heme (in the nM to μM range), to ensure prompt response of the sensor to changes in [heme] but also avoiding the sensor becoming either fully saturated or fully unsaturated (which is essential to study dynamic cellular processes). Kd,heme defines the fraction of total cellular heme which is available for donor–acceptor heme exchange (acceptor = apo-heme sensor). Heme sensors with the highest heme affinities will accept heme from a larger ensemble of heme donors, and vice versa. Caution is needed when comparing heme concentrations obtained with different sensors, as the sensors with lower Kd,heme will draw heme from a larger number of donors.
For ferric heme unless otherwise stated.
A relatively low concentration of heme was measured in the endoplasmic reticulum of HeLa cells compared to other cell lines (mouse melanoma B16, human HCT116 colon cancer cell, human PANC-1, hamster kidney fibroblast BHK-21, and CHO), which was interpreted as due to the high expression levels of heme oxygenase-1 in the endoplasmic reticulum surface of HeLa cells.
holo-HRP was formed by mixing apo-HRP with HEK293 lysates. The reconstitution of the catalytic activity of the HRP reporter by its binding to cellular heme was then used to measure heme concentration.
mAPX = monomeric APX; mEGFP = monomeric EGFP.
The Development of Cellular Heme Sensors
Genetically encoded sensors comprise a heme-binding protein conjugated to one or more fluorescent proteins. For the sensor to report on the presence and quantify the concentration of heme, the fluorescence output must be modulated by heme binding. There are different mechanisms that can lead to suitable modulation of the fluorescence output.
a. Förster Resonance-Energy Transfer between the Fluorescent Protein and a Bound Molecule of Heme
The heme-binding pocket of the sensor must be located in close proximity to the chromophore-forming amino acids in the fluorescent protein, and the emission of the fluorescent protein must have spectral overlap with either the Soret or Q absorption bands of heme.175 In this case, resonance-energy transfer provides a nonradiative decay pathway for the photoexcited state of the chromophore, via the heme moiety, that competes with the radiative-decay pathway responsible for the fluorescence signal (Figure 4A,C).
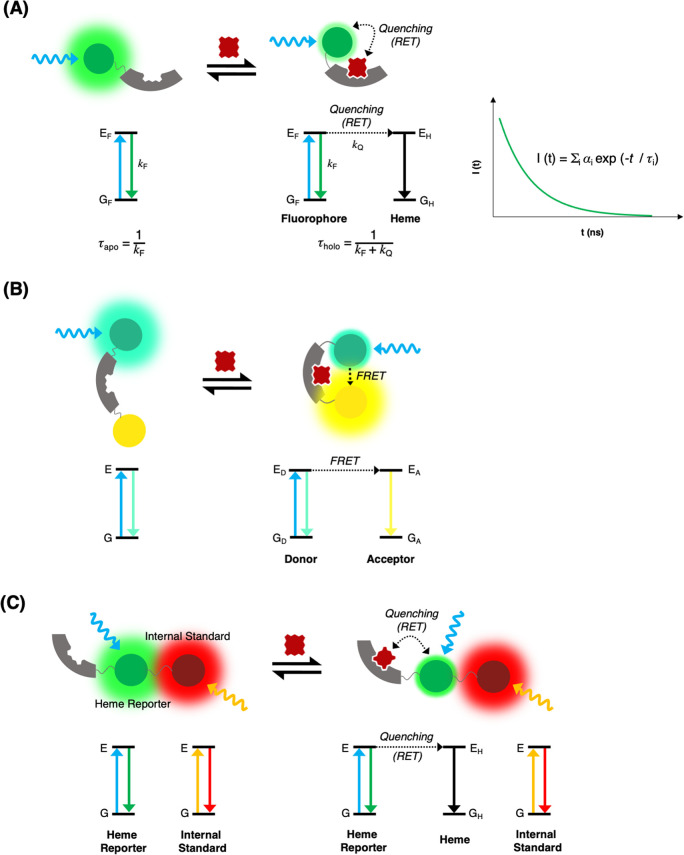
Figure 4.
Basic principles of heme sensor designs. (A) RET sensors: Heme-binding to this type of sensor introduces an additional relaxation pathway for the electronic excited state of the fluorophore. The mean fluorescence lifetime of the probe changes between the limiting values of τapo to τholo for the pure apo and holo forms of the sensor dependent on heme concentration.77 (B) FRET sensors: The heme-binding domain of the sensor undergoes a conformational change that brings two fluorophores into close proximity to one another. In this example, Förster energy transfer results in a decrease in the emission of a green fluorophore and an increase in emission of a yellow fluorophore. Hence changes in the relative emission intensities of the two fluorophores can be used to determine heme concentration.76 Multiple heme-binding sites may be present in the heme-binding domain.78 (C) A RET sensor (similar to that shown in (A)) that incorporates an additional fluorophore in order to measure a ratiometric intensity. The sensor is composed of two fluorophores, but heme-binding triggers the selective quenching of the fluorescence of only one of them. The unperturbed tag can then be used as an internal reference to monitor the changes in the intensity of emission for the quenched fluorophore, providing a method for precise heme quantitation.79,112
b. Förster Resonance-Energy Transfer between a Pair of Fluorescent Proteins
Unlike in (a), the heme-binding pocket must be distant from both fluorescent proteins, and, instead, the binding of heme to the sensor must lead to a conformational change that brings the chromophore-forming amino acids on each of the fluorescent proteins (i.e., the donor and acceptor sites) into close proximity; see Figure 4B. In this case, resonance-energy transfer between the donor and acceptor leads to subsequent radiative decay from the acceptor. Hence the binding of a molecule of heme is detected by the attenuation of the short wavelength emission band of one the fluorescent proteins (the donor), and the appearance of the long wavelength emission band of the other fluorescent protein (the acceptor).
In order to distinguish between the mode of operation of these two different designs for heme sensors, we will refer to the mechanism described in (a) as just resonance-energy transfer (RET) and the mechanism described in (b) as fluorescence resonance-energy transfer (FRET).
Further consideration is given here to the heme sensor design comprising a heme-binding domain and a single fluorescent protein (i.e., that shown in Figure 4A). If the distance between the chromophore-forming amino acids on the fluorescent protein and the heme-binding pocket, r, is much less than the Förster distance, R0 (calculated from the spectral overlap integral175), then the efficiency of RET in the holo-sensor will be near to 100%. The fluorescence quantum yield of the holo-sensor (i.e., the sensor with heme bound to it) will be near to 0%, and the presence of heme can be inferred from the quenching of the intensity of the fluorescence emission. Alternatively, if the distance r is close to the R0, then there will be an intermediate efficiency for RET in the holo-sensor (given by 100% × 1/(1 + (r/R0)6). The fluorescence quantum yield of the holo-sensor will be reduced (but will not equal zero), and the presence of heme can be inferred from either the reduction in the emission intensity or from a reduction in the fluorescence lifetime.
A change in the fluorescence lifetime can be observed by time-correlated single photon counting, where the proportions of the apo- and holo-forms of the sensor can also be deduced by determining the relative amplitudes of functions in the fitting of a multiexponential decay model, Figure 4A. For a genetically encoded heme sensor, a measurement of a fluorescence decay profile has significant advantages over a measurement of fluorescence intensity: (i) The fluorescence decay can be used to determine the ratio of holo- to apo-forms of the sensor, but the intensity can only be used to detect the apo-form of the sensor (because the holo-form of the sensor is quenched). Sensor designs that rely on measurements of fluorescence quenching require an additional tag (distant from the heme-binding pocket and the other chromophore site) to normalize the signal intensity. (ii) The quantitative accuracy of fluorescence lifetime imaging or spectroscopy is independent of the inner filter effect or partial photobleaching of the fluorescent protein.
Another property of the emission of a fluorescent protein is the anisotropy (polarization), which might be modulated by changes in the rotational diffusion between the apo- and holo-forms of the sensor. The sensitivity of a polarization measurement will be highly dependent on the local viscosity and, as yet, it has not been exploited in the context of heme sensing.
Maintaining Homeostasis in the Presence of Heme Sensors
The in vivo deployment of a heme sensor in cells introduces an additional heme-binding species into the cellular milieu. Hence the sensor will compete with other heme-binding proteins in the cell for molecules of exchangeable heme. While there would be no issue with the sensor competing for heme with either heme-binding proteins, or small molecules, whose roles are to buffer the concentration of free heme, it is critical that this competition does not extend to bona fide heme proteins that require heme either for their function or to adjust their properties. To accurately measure the availability of heme in cellular compartments, the expressed sensor must integrate into the network of buffering partners, and its presence must not result in a significant change either to the amount of exchangeable heme (He) associated with these heme-binding partners, or to the miniscule amount of free heme (Hf) in the cell. For a certain concentration of exchangeable heme, it is fairly straightforward to estimate an appropriate range of concentrations for the sensor that will preserve heme homeostasis in the cell.77 On the basis of measurements in the cytosol of HEK293 cells,77 a sensor concentration of up to 1 μM would not be expected to alter the availability of heme for exchangeable heme concentrations equal to, or in excess of, 3 μM. It is realistic to assume that, if a sensor is deployed in a localized area of the cell (e.g., directed specifically to the nucleus), then the sensor concentration may exceed 1 μM. Hence, we anticipate that, in future, controlling the concentration of expressed sensor will be increasingly important.
Summary of Known RET and FRET-Based Heme Sensors
There are a number of reports of different fluorescent heme sensors in the literature. We summarize them briefly below. Heme concentrations reported from the studies of these sensors are given in Table 1.
In the early 2000s, Takeda et al. provided the first proof-of-concept for a heme sensor. The sensor comprised an EGFP-cytochrome b562 fusion protein,176 and it was subsequently optimized for enhanced RET.177 Fluorescently labeled variants of heme oxygenase-1 (K18C178 and D140H179 variants) have also been used to detect heme in vitro. These sensors were a step forward in terms of heme quantification, but were not deployed in cells for real-time monitoring of heme concentrations.
The first genetically encoded heme sensor was CYSDY-9.76 CISDY-9 was designed to exploit the natural dimerization of a pair of bacterial chaperones (IsdX1 and IsdC). Tagging each chaperone with a fluorescent probe and linking them with a short peptide tether produced an intracellular FRET reporter which was used to analyze heme population in HeLa cells, Table 1. With the exception of the endoplasmic reticulum, heme concentrations were found to be evenly distributed across the cytosol, the mitochondria and the nucleus, which is different from the observations in yeast (see above and ref (79)). It may be the case that results from different sensor systems cannot be directly compared, see Table 1.
Cytochrome b562 was later engineered with EGFP and mKATE2 fluorescent tags. This created a sensor that could be deployed in cells79 in which the fluorescence intensity of EGFP was quenched (via RET) on heme-binding; the intensity of mKATE2 was unchanged providing a means of assessing heme concentration by ratiometric analysis. A somewhat heterogeneous distribution of heme throughout yeast cells was reported, Table 1, and the behavior of the sensor in cells112 supported the concept of a (buffered) reservoir of exchangeable heme that is readily available for regulatory roles and crucial in the overall physiology of yeast. The same sensor was then utilized to demonstrate that heme delivery from mitochondria—the place of heme biosynthesis—to the nucleus was 25% faster than to the cytosol and the mitochondrial inner matrix. The authors suggested direct membrane contacts between mitochondria and the ER as a possible pathway for heme delivery to the nucleus.170,171
A genetically encoded (FRET-based) heme sensor based on histidine rich protein 2 has been used to quantify heme concentrations in the parasite P. falciparum.78 In this case, heme concentrations are higher (ca. 1.6 μM), but this is not completely unexpected considering that 80% of the amount of heme contained in a human body is synthesized by red blood cells.73
Peroxidase-based fluorescent sensors, based on horseradish peroxidase and ascorbate peroxidase, were first used to measure dynamic heme fluxes in C. elegans.75 Ascorbate peroxidase has also been used recently to monitor heme concentrations and distributions in live cells77 using fluorescence lifetime imaging microscopy. In this work, miniscule concentrations (a few nM) of free heme were identified—corresponding to one molecule or less per cellular compartment. These observations are consistent with a system that sequesters free heme.
The Use of Heme Sensors to Study Heme Protein Maturation
Applications of heme sensors can extend beyond the determination of heme concentrations. A GFP-labeled version of cytochrome c peroxidase has been recently developed and used to study heme protein maturation by heme insertion.119 In this design, the percentage of the heme-bound sensor in yeast cells was revealed by the relative amplitudes of exponential-decay components in the emission measured by fluorescence lifetime imaging. This provides a forward-looking approach to establish in real-time the dynamic processes involved in the folding process that accompanies the formation of heme proteins.
Alternative Approaches
There are other technological approaches to heme quantification, including the use of the antimalarial 4-aminoquinoline probe180 and peptide-based fluorescent probes.181 The use of heme catabolites—biliverdin and bilirubin—as a way of tracking and studying heme biochemistry is also promising and is starting to provide interesting insights into the interplay between the biochemistry of heme and other biological pathways.182−189
Possible Mechanisms of Heme Transfer
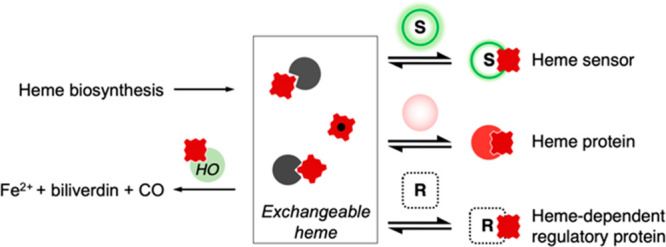
Detailed information on the mechanism to transfer heme from one place to another—for example between protein partners—is as yet poorly defined. It has been suggested77 that an exchange mechanism between protein partners might play a role, as shown in Figure 5. This is envisaged as heme exchange between a donor, which could be a buffering molecule or a chaperone, and an acceptor such as a heme protein or a heme sensor. This heme exchange could take place by a range of different pathways (corresponding to dissociative or associative mechanisms) for delivery of heme to specific acceptors. The existence of such a mechanism, as outlined in Figure 5, would account for the buffering of the concentration of free heme, changes in heme availability, and how cells are able to hoard supplies of heme despite the undesirable effects of heme. It would also provide a responsive capability for mobilization of heme when and where needed.
Figure 5.
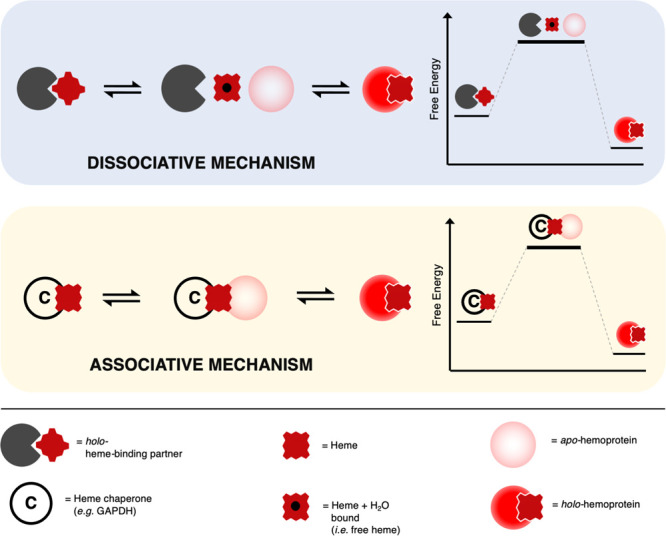
Possible mechanisms for exchange of heme. Heme-binding partners (dark gray pacmans) are envisaged as transferring heme to heme proteins (red circles) by dissociative (pale gray box) or associative (pale yellow box) pathways, resembling classical ligand exchange mechanisms in coordination chemistry. In a dissociative pathway, a free molecule of heme (assumed to be coordinated by a water molecule) is formed transiently following dissociation from a heme-binding partner, and is intercepted by an apo-heme protein (faded red circles). Alternatively, an associative exchange of heme is possible and is shown here for the example of heme delivery by chaperones (C, circles). This latter mechanism may provide better selectivity toward the target heme protein. However, we do not envisage this as being exclusive to chaperones, but a mechanism which, in principle, is available to be used by heme-binding partners as well in delivering heme to apo heme proteins. The different mechanisms of heme exchange may help to fine-tune the delivery of heme to specific acceptors.
Summary and Future Outlook
The logistics for the distribution of heme in cells have yet to be fully discovered. In practice, the dynamic requirements of cellular supply and demand may be complex, and perhaps there is no single mechanism for movement of heme within cells that dominates. Chaperones, heme-binding partners, membranes, and transporters may well work together in a concerted way and could provide contingencies for heme supply under conditions where the cell needs to react quickly to changes in heme demand. We see the role of a buffered reservoir of heme-binding proteins, potentially in large numbers, as being important. This could allow heme to travel from one place to another, where molecules of heme hitchhike across the cell by binding to whichever protein(s) they can find en route to their final destination.
Acknowledgments
The research was supported by the Leverhulme Trust (RPG-2016- 397) awarded to A.J.H. and E.L.R.; A.E.G. is grateful to the Engineering and Physical Sciences Research Council for a PhD studentship.
The authors declare no competing financial interest.
References
- Moss G. P.; Smith P. A. S.; Tavernier D. Glossary of class names of organic compounds and reactivity intermediates based on structure (IUPAC Recommendations 1995). Pure Appl. Chem. 1995, 67 (8–9), 1307–1375. 10.1351/pac199567081307. [DOI] [Google Scholar]
- Martinez S. E.; Huang D.; Szczepaniak A.; Cramer W. A.; Smith J. L. Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation. Structure 1994, 2 (2), 95–105. 10.1016/S0969-2126(00)00012-5. [DOI] [PubMed] [Google Scholar]
- Smith L. J.; Kahraman A.; Thornton J. M. Heme proteins--diversity in structural characteristics, function, and folding. Proteins: Struct., Funct., Genet. 2010, 78 (10), 2349–68. 10.1002/prot.22747. [DOI] [PubMed] [Google Scholar]
- Perutz M. F. Relation between structure and sequence of haemoglobin. Nature 1962, 194 (4832), 914–917. 10.1038/194914a0. [DOI] [PubMed] [Google Scholar]
- Kendrew J. C.; Watson H. C.; Strandberg B. E.; Dickerson R. E.; Phillips D. C.; Shore V. C. A Partial Determination by X-ray Methods, and its Correlation with Chemical Data. Nature 1961, 190 (4777), 666–670. 10.1038/190666a0. [DOI] [PubMed] [Google Scholar]
- Poulos T. L. Heme enzyme structure and function. Chem. Rev. 2014, 114 (7), 3919–62. 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi P.; Bellelli A.; Coletta M.; Colosimo A.; Falcioni G.; Giacometti G. M.; Ippoliti R.; Zolla L.; Giardina B. Multiple strategies for O2 transport: from simplicity to complexity. IUBMB Life 2007, 59 (8–9), 600–16. 10.1080/15216540701308424. [DOI] [PubMed] [Google Scholar]
- Moody P. C. E.; Raven E. L. The Nature and Reactivity of Ferryl Heme in Compounds I and II. Acc. Chem. Res. 2018, 51 (2), 427–435. 10.1021/acs.accounts.7b00463. [DOI] [PubMed] [Google Scholar]
- Stuehr D. J.; Haque M. M. Nitric oxide synthase enzymology in the 20 years after the Nobel Prize. Br. J. Pharmacol. 2019, 176 (2), 177–188. 10.1111/bph.14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzer C. A.; Marnett L. J. Cyclooxygenases: structural and functional insights. J. Lipid Res. 2009, 50, S29–S34. 10.1194/jlr.R800042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T.; Lengalova A.; Martinek V.; Martinkova M. Heme: emergent roles of heme in signal transduction, functional regulation and as catalytic centres. Chem. Soc. Rev. 2019, 48 (24), 5624–5657. 10.1039/C9CS00268E. [DOI] [PubMed] [Google Scholar]
- Shimizu T.; Huang D.; Yan F.; Stranava M.; Bartosova M.; Fojtikova V.; Martinkova M. Gaseous O2, NO, and CO in signal transduction: structure and function relationships of heme-based gas sensors and heme-redox sensors. Chem. Rev. 2015, 115 (13), 6491–533. 10.1021/acs.chemrev.5b00018. [DOI] [PubMed] [Google Scholar]
- Shimizu T. Binding of cysteine thiolate to the Fe(III) heme complex is critical for the function of heme sensor proteins. J. Inorg. Biochem. 2012, 108, 171–7. 10.1016/j.jinorgbio.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Sweeny E. A.; Schlanger S.; Stuehr D. J. Dynamic regulation of NADPH oxidase 5 by intracellular heme levels and cellular chaperones. Redox Biol. 2020, 36, 101656. 10.1016/j.redox.2020.101656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinaga M.; Sugimoto H.; Nishitani Y.; Nagai S.; Nagatoishi S.; Muraki N.; Tosha T.; Tsumoto K.; Aono S.; Shiro Y.; Sawai H. Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival. Commun. Biol. 2021, 4 (1), 467. 10.1038/s42003-021-01987-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.; Xie H.; Kang Y.; Lin Y.; Liu G.; Sakato-Antoku M.; Patel-King R. S.; Wang B.; Wan C.; King S. M.; Zhao C.; Huang K. Heme-binding protein CYB5D1 is a radial spoke component required for coordinated ciliary beating. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (17), e2015689118. 10.1073/pnas.2015689118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer G.; Reichelt J.; Jahn D.; Heinz D. W. Structure and function of enzymes in heme biosynthesis. Protein Sci. 2010, 19 (6), 1137–61. 10.1002/pro.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T.; Unno M.; Ikeda-Saito M. Heme oxygenase reveals its strategy for catalyzing three successive oxygenation reactions. Acc. Chem. Res. 2010, 43 (2), 240–7. 10.1021/ar9001685. [DOI] [PubMed] [Google Scholar]
- Unno M.; Matsui T.; Ikeda-Saito M. Structure and catalytic mechanism of heme oxygenase. Nat. Prod. Rep. 2007, 24 (3), 553–70. 10.1039/b604180a. [DOI] [PubMed] [Google Scholar]
- Siracusa R.; Schaufler A.; Calabrese V.; Fuller P. M.; Otterbein L. E. Carbon Monoxide: from Poison to Clinical Trials. Trends Pharmacol. Sci. 2021, 42 (5), 329–339. 10.1016/j.tips.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J.; Sheng X.; Chang Z.; Wu Q.; Wang S.; Xuan Z.; Li D.; Wu Y.; Shang Y.; Kong X.; Yu L.; Li L.; Ruan K.; Hu H.; Huang Y.; Hui L.; Xie D.; Wang F.; Hu R. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Rep. 2014, 7 (1), 180–93. 10.1016/j.celrep.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter E. L.; Ramirez Y.; Ragsdale S. W. The heme-regulatory motif of nuclear receptor Rev-erbbeta is a key mediator of heme and redox signaling in circadian rhythm maintenance and metabolism. J. Biol. Chem. 2017, 292 (27), 11280–11299. 10.1074/jbc.M117.783118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter E. L.; Gupta N.; Ragsdale S. W. High Affinity Heme Binding to a Heme Regulatory Motif on the Nuclear Receptor Rev-erbbeta Leads to Its Degradation and Indirectly Regulates Its Interaction with Nuclear Receptor Corepressor. J. Biol. Chem. 2016, 291 (5), 2196–222. 10.1074/jbc.M115.670281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.; Ragsdale S. W. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erb{beta}. J. Biol. Chem. 2011, 286 (6), 4392–403. 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.; Yin L.; Hanniman E. A.; Joshi S.; Lazar M. A. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha. Genes Dev. 2009, 23 (18), 2201–9. 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers P. M.; Ying L.; Burris T. P. Relationship between circadian oscillations of Rev-erbalpha expression and intracellular levels of its ligand, heme. Biochem. Biophys. Res. Commun. 2008, 368 (4), 955–8. 10.1016/j.bbrc.2008.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L.; Wu N.; Curtin J. C.; Qatanani M.; Szwergold N. R.; Reid R. A.; Waitt G. M.; Parks D. J.; Pearce K. H.; Wisely G. B.; Lazar M. A. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 2007, 318 (5857), 1786–9. 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Raghuram S.; Stayrook K. R.; Huang P.; Rogers P. M.; Nosie A. K.; McClure D. B.; Burris L. L.; Khorasanizadeh S.; Burris T. P.; Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat. Struct. Mol. Biol. 2007, 14 (12), 1207–13. 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanishi K.; Igarashi J.; Hayasaka K.; Hikage N.; Saiful I.; Yamauchi S.; Uchida T.; Ishimori K.; Shimizu T. Heme-binding characteristics of the isolated PAS-A domain of mouse Per2, a transcriptional regulatory factor associated with circadian rhythms. Biochemistry 2008, 47 (23), 6157–68. 10.1021/bi7023892. [DOI] [PubMed] [Google Scholar]
- Airola M. V.; Du J.; Dawson J. H.; Crane B. R. Heme binding to the Mammalian circadian clock protein period 2 is nonspecific. Biochemistry 2010, 49 (20), 4327–38. 10.1021/bi901945w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka K.; Kitanishi K.; Igarashi J.; Shimizu T. Heme-binding characteristics of the isolated PAS-B domain of mouse Per2, a transcriptional regulatory factor associated with circadian rhythms. Biochim. Biophys. Acta, Proteins Proteomics 2011, 1814 (2), 326–33. 10.1016/j.bbapap.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Mukaiyama Y.; Uchida T.; Sato E.; Sasaki A.; Sato Y.; Igarashi J.; Kurokawa H.; Sagami I.; Kitagawa T.; Shimizu T. Spectroscopic and DNA-binding characterization of the isolated heme-bound basic helix-loop-helix-PAS-A domain of neuronal PAS protein 2 (NPAS2), a transcription activator protein associated with circadian rhythms. FEBS J. 2006, 273 (11), 2528–39. 10.1111/j.1742-4658.2006.05259.x. [DOI] [PubMed] [Google Scholar]
- Dioum E. M.; Rutter J.; Tuckerman J. R.; Gonzalez G.; Gilles-Gonzalez M. A.; McKnight S. L. NPAS2: a gas-responsive transcription factor. Science 2002, 298 (5602), 2385–7. 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- Uchida T.; Sagami I.; Shimizu T.; Ishimori K.; Kitagawa T. Effects of the bHLH domain on axial coordination of heme in the PAS-A domain of neuronal PAS domain protein 2 (NPAS2): conversion from His119/Cys170 coordination to His119/His171 coordination. J. Inorg. Biochem. 2012, 108, 188–95. 10.1016/j.jinorgbio.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Koudo R.; Kurokawa H.; Sato E.; Igarashi J.; Uchida T.; Sagami I.; Kitagawa T.; Shimizu T. Spectroscopic characterization of the isolated heme-bound PAS-B domain of neuronal PAS domain protein 2 associated with circadian rhythms. FEBS J. 2005, 272 (16), 4153–62. 10.1111/j.1742-4658.2005.04828.x. [DOI] [PubMed] [Google Scholar]
- Ishida M.; Ueha T.; Sagami I. Effects of mutations in the heme domain on the transcriptional activity and DNA-binding activity of NPAS2. Biochem. Biophys. Res. Commun. 2008, 368 (2), 292–7. 10.1016/j.bbrc.2008.01.053. [DOI] [PubMed] [Google Scholar]
- Freeman S. L.; Kwon H.; Portolano N.; Parkin G.; Venkatraman Girija U.; Basran J.; Fielding A. J.; Fairall L.; Svistunenko D. A.; Moody P. C. E.; Schwabe J. W. R.; Kyriacou C. P.; Raven E. L. Heme binding to human CLOCK affects interactions with the E-box. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (40), 19911–19916. 10.1073/pnas.1905216116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat-Rodgers G. S.; Correia C.; Botuyan M. V.; Mer G.; Rodgers K. R. Heme-based sensing by the mammalian circadian protein CLOCK. Inorg. Chem. 2010, 49 (14), 6349–65. 10.1021/ic902388q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minegishi S.; Sagami I.; Negi S.; Kano K.; Kitagishi H. Circadian clock disruption by selective removal of endogenous carbon monoxide. Sci. Rep. 2018, 8 (1), 11996. 10.1038/s41598-018-30425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemz R.; Reischl S.; Wallach T.; Witte N.; Jurchott K.; Klemz S.; Lang V.; Lorenzen S.; Knauer M.; Heidenreich S.; Xu M.; Ripperger J. A.; Schupp M.; Stanewsky R.; Kramer A. Reciprocal regulation of carbon monoxide metabolism and the circadian clock. Nat. Struct. Mol. Biol. 2017, 24 (1), 15–22. 10.1038/nsmb.3331. [DOI] [PubMed] [Google Scholar]
- Sarkar A.; Carter E. L.; Harland J. B.; Speelman A. L.; Lehnert N.; Ragsdale S. W. Ferric heme as a CO/NO sensor in the nuclear receptor Rev-Erbss by coupling gas binding to electron transfer. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (3), e2016717118. 10.1073/pnas.2016717118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosure S. A.; Strutzenberg T. S.; Shang J.; Munoz-Tello P.; Solt L. A.; Griffin P. R.; Kojetin D. J. Structural basis for heme-dependent NCoR binding to the transcriptional repressor REV-ERBbeta. Sci. Adv. 2021, 7 (5), eabc6479. 10.1126/sciadv.abc6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janciauskiene S.; Vijayan V.; Immenschuh S. TLR4 Signaling by Heme and the Role of Heme-Binding Blood Proteins. Front. Immunol. 2020, 11, 1964. 10.3389/fimmu.2020.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudan K.; Vijayan V.; Madyaningrana K.; Gueler F.; Igarashi K.; Foresti R.; Motterlini R.; Immenschuh S. TLR4 activation alters labile heme levels to regulate BACH1 and heme oxygenase-1 expression in macrophages. Free Radical Biol. Med. 2019, 137, 131–142. 10.1016/j.freeradbiomed.2019.04.024. [DOI] [PubMed] [Google Scholar]
- van Wijk K.; Akabane T.; Kimura T.; Saitoh S.; Okano S.; Kelly V. P.; Takagi M.; Kodama K.; Takahashi K.; Tanaka T.; Nakajima M.; Nakajima O. Heterozygous disruption of ALAS1 in mice causes an accelerated age-dependent reduction in free heme, but not total heme, in skeletal muscle and liver. Arch. Biochem. Biophys. 2021, 697, 108721. 10.1016/j.abb.2020.108721. [DOI] [PubMed] [Google Scholar]
- Tang X. D.; Xu R.; Reynolds M. F.; Garcia M. L.; Heinemann S. H.; Hoshi T. Haem can bind to and inhibit mammalian calcium-dependent Slo1 BK channels. Nature 2003, 425 (6957), 531–5. 10.1038/nature02003. [DOI] [PubMed] [Google Scholar]
- Sahoo N.; Goradia N.; Ohlenschlager O.; Schonherr R.; Friedrich M.; Plass W.; Kappl R.; Hoshi T.; Heinemann S. H. Heme impairs the ball-and-chain inactivation of potassium channels. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (42), E4036–44. 10.1073/pnas.1313247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Publicover S.; Gu Y. An oxygen-sensitive mechanism in regulation of epithelial sodium channel. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (8), 2957–62. 10.1073/pnas.0809100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton M. J.; Kapetanaki S. M.; Chernova T.; Jamieson A. G.; Dorlet P.; Santolini J.; Moody P. C.; Mitcheson J. S.; Davies N. W.; Schmid R.; Raven E. L.; Storey N. M. A heme-binding domain controls regulation of ATP-dependent potassium channels. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (14), 3785–90. 10.1073/pnas.1600211113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustynek B.; Kudin A. P.; Bednarczyk P.; Szewczyk A.; Kunz W. S. Hemin inhibits the large conductance potassium channel in brain mitochondria: a putative novel mechanism of neurodegeneration. Exp. Neurol. 2014, 257, 70–5. 10.1016/j.expneurol.2014.04.022. [DOI] [PubMed] [Google Scholar]
- Horrigan F. T.; Heinemann S. H.; Hoshi T. Heme regulates allosteric activation of the Slo1 BK channel. J. Gen. Physiol. 2005, 126 (1), 7–21. 10.1085/jgp.200509262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton M. J.; Cresser-Brown J.; Thomas M.; Portolano N.; Basran J.; Freeman S. L.; Kwon H.; Bottrill A. R.; Llansola-Portoles M. J.; Pascal A. A.; Jukes-Jones R.; Chernova T.; Schmid R.; Davies N. W.; Storey N. M.; Dorlet P.; Moody P. C. E.; Mitcheson J. S.; Raven E. L. Discovery of a heme-binding domain in a neuronal voltage-gated potassium channel. J. Biol. Chem. 2020, 295 (38), 13277–13286. 10.1074/jbc.RA120.014150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson W. J.; Kemp P. J. Carbon monoxide: an emerging regulator of ion channels. J. Physiol. 2011, 589 (13), 3055–3062. 10.1113/jphysiol.2011.206706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanaki S. M.; Burton M. J.; Basran J.; Uragami C.; Moody P. C. E.; Mitcheson J. S.; Schmid R.; Davies N. W.; Dorlet P.; Vos M. H.; Storey N. M.; Raven E. A mechanism for CO regulation of ion channels. Nat. Commun. 2018, 9 (1), 907. 10.1038/s41467-018-03291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers C.; Boyle J. P.; Scragg J. L.; Dallas M. L.; Al-Owais M. M.; Hettiarachichi N. T.; Elies J.; Johnson E.; Gamper N.; Steele D. S. Diverse mechanisms underlying the regulation of ion channels by carbon monoxide. Br. J. Pharmacol. 2015, 172 (6), 1546–56. 10.1111/bph.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S.; Xu R.; Heinemann S. H.; Hoshi T. The RCK1 high-affinity Ca2+ sensor confers carbon monoxide sensitivity to Slo1 BK channels. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (10), 4039–43. 10.1073/pnas.0800304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L.; Morgan J. T.; Ragsdale S. W. Identification of a thiol/disulfide redox switch in the human BK channel that controls its affinity for heme and CO. J. Biol. Chem. 2010, 285 (26), 20117–27. 10.1074/jbc.M110.116483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl T.; Wissbrock A.; Goradia N.; Sahoo N.; Galler K.; Neugebauer U.; Popp J.; Heinemann S. H.; Ohlenschlager O.; Imhof D. Analysis of Fe(III) heme binding to cysteine-containing heme-regulatory motifs in proteins. ACS Chem. Biol. 2013, 8 (8), 1785–93. 10.1021/cb400317x. [DOI] [PubMed] [Google Scholar]
- Jaggar J. H.; Li A.; Parfenova H.; Liu J.; Umstot E. S.; Dopico A. M.; Leffler C. W. Heme is a carbon monoxide receptor for large-conductance Ca2+-activated K+ channels. Circ. Res. 2005, 97 (8), 805–12. 10.1161/01.RES.0000186180.47148.7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. E.; Brazier S. P.; Baban N.; Telezhkin V.; Muller C. T.; Riccardi D.; Kemp P. J. A structural motif in the C-terminal tail of slo1 confers carbon monoxide sensitivity to human BK Ca channels. Pfluegers Arch. 2008, 456 (3), 561–72. 10.1007/s00424-007-0439-4. [DOI] [PubMed] [Google Scholar]
- Kemp P. J. Hemeoxygenase-2 as an O2 sensor in K+ channel-dependent chemotransduction. Biochem. Biophys. Res. Commun. 2005, 338 (1), 648–52. 10.1016/j.bbrc.2005.08.110. [DOI] [PubMed] [Google Scholar]
- Williams S. E.; Wootton P.; Mason H. S.; Bould J.; Iles D. E.; Riccardi D.; Peers C.; Kemp P. J. Hemoxygenase-2 is an oxygen sensor for a calcium-sensitive potassium channel. Science 2004, 306 (5704), 2093–7. 10.1126/science.1105010. [DOI] [PubMed] [Google Scholar]
- Hou S.; Heinemann S. H.; Hoshi T. Modulation of BKCa channel gating by endogenous signaling molecules. Physiology 2009, 24, 26–35. 10.1152/physiol.00032.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Mechanistic insight into the heme-independent interplay between iron and carbon monoxide in CFTR and Slo1 BKCa channels. Metallomics 2017, 9 (6), 634–645. 10.1039/C7MT00065K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers C.; Dallas M. L.; Scragg J. L. Ion channels as effectors in carbon monoxide signaling. Commun. Integr. Biol. 2009, 2 (3), 241–2. 10.4161/cib.2.3.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motterlini R.; Otterbein L. E. The therapeutic potential of carbon monoxide. Nat. Rev. Drug Discovery 2010, 9 (9), 728–43. 10.1038/nrd3228. [DOI] [PubMed] [Google Scholar]
- Peers C. Modulation of ion channels and transporters by carbon monoxide: causes for concern?. Front. Physiol. 2012, 3, 477. 10.3389/fphys.2012.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. Y.; Qu H. Y.; Shang Z. L.; Tao S. T.; Xu G. H.; Wu J.; Wu H. Q.; Zhang S. L. Reciprocal regulation of Ca(2)+-activated outward K+ channels of Pyrus pyrifolia pollen by heme and carbon monoxide. New Phytol. 2011, 189 (4), 1060–1068. 10.1111/j.1469-8137.2010.03564.x. [DOI] [PubMed] [Google Scholar]
- Gessner G.; Ruhl P.; Westerhausen M.; Hoshi T.; Heinemann S. H. Fe(2+)-Mediated Activation of BKCa Channels by Rapid Photolysis of CORM-S1 Releasing CO and Fe(2). ACS Chem. Biol. 2020, 15 (8), 2098–2106. 10.1021/acschembio.0c00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey H. A.Biosynthesis of Heme and Cholorophylls; McGraw-Hill: New York, 1990. [Google Scholar]
- Hunt T.; Vanderhoff G.; London I. M. Control of globin synthesis: the role of heme. J. Mol. Biol. 1972, 66 (3), 471–81. 10.1016/0022-2836(72)90427-5. [DOI] [PubMed] [Google Scholar]
- Lodola A.; Jones O. T. G. Evidence for a rapidly turned over pool of haem in isolated hepatocytes. FEBS Lett. 1978, 90 (2), 250–254. 10.1016/0014-5793(78)80379-2. [DOI] [PubMed] [Google Scholar]
- Wissbrock A.; Imhof D. A Tough Nut to Crack: Intracellular Detection and Quantification of Heme in Malaria Parasites by a Genetically Encoded Protein Sensor. ChemBioChem 2017, 18 (16), 1561–1564. 10.1002/cbic.201700274. [DOI] [PubMed] [Google Scholar]
- Aich A.; Freundlich M.; Vekilov P. G. The free heme concentration in healthy human erythrocytes. Blood Cells, Mol., Dis. 2015, 55 (4), 402–9. 10.1016/j.bcmd.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X.; Rietzschel N.; Kwon H.; Walter Nuno A. B.; Hanna D. A.; Phillips J. D.; Raven E. L.; Reddi A. R.; Hamza I. Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (35), E5144–52. 10.1073/pnas.1609865113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.; Yang M.; Wegner S. V.; Zhao J.; Zhu R.; Wu Y.; He C.; Chen P. R. A Genetically Encoded FRET Sensor for Intracellular Heme. ACS Chem. Biol. 2015, 10 (7), 1610–5. 10.1021/cb5009734. [DOI] [PubMed] [Google Scholar]
- Leung G. C.; Fung S. S.; Gallio A. E.; Blore R.; Alibhai D.; Raven E. L.; Hudson A. J. Unravelling the mechanisms controlling heme supply and demand. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (22), E2104008118. 10.1073/pnas.2104008118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abshire J. R.; Rowlands C. J.; Ganesan S. M.; So P. T.; Niles J. C. Quantification of labile heme in live malaria parasites using a genetically encoded biosensor. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (11), E2068–E2076. 10.1073/pnas.1615195114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna D. A.; Harvey R. M.; Martinez-Guzman O.; Yuan X.; Chandrasekharan B.; Raju G.; Outten F. W.; Hamza I.; Reddi A. R. Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (27), 7539–44. 10.1073/pnas.1523802113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkovsky H. L.; Guo J. T.; Hou W.; Li T.; Narang T.; Thapar M. Porphyrin and heme metabolism and the porphyrias. Compr Physiol 2013, 3 (1), 365–401. 10.1002/cphy.c120006. [DOI] [PubMed] [Google Scholar]
- Bonkovsky H. L.; Healey J. F.; Lourie A. N.; Gerron G. G. Intravenous heme-albumin in acute intermittent porphyria: evidence for repletion of hepatic hemoproteins and regulatory heme pools. Am. J. Gastroenterol. 1991, 86 (8), 1050–1056. [PubMed] [Google Scholar]
- Woodard S. I.; Dailey H. A. Regulation of heme biosynthesis in Escherichia coli. Arch. Biochem. Biophys. 1995, 316 (1), 110–5. 10.1006/abbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- Granick S.; Sinclair P.; Sassa S.; Grieninger G. Effects by heme, insulin, and serum albumin on heme and protein synthesis in chick embryo liver cells cultured in a chemically defined medium, and a spectrofluorometric assay for porphyrin composition. J. Biol. Chem. 1975, 250 (24), 9215–25. 10.1016/S0021-9258(19)40633-9. [DOI] [PubMed] [Google Scholar]
- Eriksen L. So-called free erythrocyte protoporphyrin and its possible role in hemoglobin formation. Acta Physiol. Scand. 1961, 53, 288–99. 10.1111/j.1748-1716.1961.tb02287.x. [DOI] [PubMed] [Google Scholar]
- van Huystee R. B. Relationship of the Heme Moiety of Peroxidase and the Free Heme Pool in Cultured Peanut Cells. Z. Pflanzenphysiol. 1977, 84 (5), 427–433. 10.1016/S0044-328X(77)80234-1. [DOI] [Google Scholar]
- Tangeras A.; Flatmark T. In vitro binding of protoheme IX and protoporphyrin IX to components in the matrix of rat liver mitochondria. Biochim. Biophys. Acta, Gen. Subj. 1979, 588 (2), 201–10. 10.1016/0304-4165(79)90203-4. [DOI] [PubMed] [Google Scholar]
- Granick S.; Beale S. I. Hemes, chlorophylls, and related compounds: biosynthesis and metabolic regulation. Adv. Enzymol. Relat. Areas Mol. Biol. 2006, 46, 33–203. 10.1002/9780470122914.ch2. [DOI] [PubMed] [Google Scholar]
- Bonkowsky H. L.; Sinclair P. R.; Sinclair J. F. Hepatic heme metabolism and its control. Yale J. Biol. Med. 1979, 52 (1), 13–37. [PMC free article] [PubMed] [Google Scholar]
- Thomas J.; Weinstein J. D. Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol. 1990, 94 (3), 1414–23. 10.1104/pp.94.3.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celier C.; Cresteil T. Control of cytochromes P450 expression in Gunn rat liver: implication of the intracellular heme pool. Arch. Biochem. Biophys. 1991, 290 (2), 407–10. 10.1016/0003-9861(91)90559-2. [DOI] [PubMed] [Google Scholar]
- Atamna H. Heme, iron, and the mitochondrial decay of ageing. Ageing Res. Rev. 2004, 3 (3), 303–18. 10.1016/j.arr.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Bandyopadhyay U. Free heme toxicity and its detoxification systems in human. Toxicol. Lett. 2005, 157 (3), 175–88. 10.1016/j.toxlet.2005.03.004. [DOI] [PubMed] [Google Scholar]
- Correia M. A.; Sinclair P. R.; De Matteis F. Cytochrome P450 regulation: the interplay between its heme and apoprotein moieties in synthesis, assembly, repair, and disposal. Drug Metab. Rev. 2011, 43 (1), 1–26. 10.3109/03602532.2010.515222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. A.; Quigley J. G. Control of intracellular heme levels: heme transporters and heme oxygenases. Biochim. Biophys. Acta, Mol. Cell Res. 2011, 1813 (5), 668–82. 10.1016/j.bbamcr.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Santos D.; Schranzhofer M.; Horvathova M.; Jaberi M. M.; Bogo Chies J. A.; Sheftel A. D.; Ponka P. Heme oxygenase 1 is expressed in murine erythroid cells where it controls the level of regulatory heme. Blood 2014, 123 (14), 2269–77. 10.1182/blood-2013-04-496760. [DOI] [PubMed] [Google Scholar]
- Chiabrando D.; Vinchi F.; Fiorito V.; Mercurio S.; Tolosano E. Heme in pathophysiology: a matter of scavenging, metabolism and trafficking across cell membranes. Front. Pharmacol. 2014, 5, 61. 10.3389/fphar.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamna H.; Brahmbhatt M.; Atamna W.; Shanower G. A.; Dhahbi J. M. ApoHRP-based assay to measure intracellular regulatory heme. Metallomics 2015, 7 (2), 309–321. 10.1039/C4MT00246F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P.; Sheftel A. D.; English A. M.; Scott Bohle D.; Garcia-Santos D. Do Mammalian Cells Really Need to Export and Import Heme?. Trends Biochem. Sci. 2017, 42 (5), 395–406. 10.1016/j.tibs.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Israels L. G.; Yoda B.; Schacter B. A. Heme binding and its possible significance in heme movement and availability in the cell. Ann. N. Y. Acad. Sci. 1975, 244, 651–61. 10.1111/j.1749-6632.1975.tb41559.x. [DOI] [PubMed] [Google Scholar]
- Ponka P. Tissue-Specific Regulation of Iron Metabolism and Heme Synthesis: Distinct Control Mechanisms in Erythroid Cells. Blood 1997, 89 (1), 1–25. 10.1182/blood.V89.1.1. [DOI] [PubMed] [Google Scholar]
- Ryter S. W.; Tyrrell R. M. The heme synthesis and degradation pathways: role in oxidant sensitivity. Heme oxygenase has both pro- and antioxidant properties. Free Radical Biol. Med. 2000, 28 (2), 289–309. 10.1016/S0891-5849(99)00223-3. [DOI] [PubMed] [Google Scholar]
- Sassa S. Why heme needs to be degraded to iron, biliverdin IXalpha, and carbon monoxide?. Antioxid. Redox Signaling 2004, 6 (5), 819–824. 10.1089/1523086041798006. [DOI] [PubMed] [Google Scholar]
- Sachar M.; Anderson K. E.; Ma X. Protoporphyrin IX: the Good, the Bad, and the Ugly. J. Pharmacol. Exp. Ther. 2016, 356 (2), 267–75. 10.1124/jpet.115.228130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gbotosho O. T.; Kapetanaki M. G.; Kato G. J. The Worst Things in Life are Free: The Role of Free Heme in Sickle Cell Disease. Front. Immunol. 2021, 11, 561917. 10.3389/fimmu.2020.561917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi A. R.; Hamza I. Heme Mobilization in Animals: A Metallolipid’s Journey. Acc. Chem. Res. 2016, 49 (6), 1104–10. 10.1021/acs.accounts.5b00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda N.; Yabuki Y.; Yamaguchi K.; Onozato M.; Li Y.; Kurosawa K.; Tanabe H.; Okamoto N.; Era T.; Sugiyama H.; Wada T.; Fukunaga K. Targeting G-quadruplex DNA as cognitive function therapy for ATR-X syndrome. Nat. Med. 2018, 24 (6), 802–813. 10.1038/s41591-018-0018-6. [DOI] [PubMed] [Google Scholar]
- de Villiers K. A.; Kaschula C. H.; Egan T. J.; Marques H. M. Speciation and structure of ferriprotoporphyrin IX in aqueous solution: spectroscopic and diffusion measurements demonstrate dimerization, but not mu-oxo dimer formation. JBIC, J. Biol. Inorg. Chem. 2007, 12 (1), 101–17. 10.1007/s00775-006-0170-1. [DOI] [PubMed] [Google Scholar]
- Kuter D.; Streltsov V.; Davydova N.; Venter G. A.; Naidoo K. J.; Egan T. J. Molecular structures and solvation of free monomeric and dimeric ferriheme in aqueous solution: insights from molecular dynamics simulations and extended X-ray absorption fine structure spectroscopy. Inorg. Chem. 2014, 53 (20), 10811–24. 10.1021/ic500454d. [DOI] [PubMed] [Google Scholar]
- Soares M. P.; Bozza M. T. Red alert: labile heme is an alarmin. Curr. Opin. Immunol. 2016, 38, 94–100. 10.1016/j.coi.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Hanna D. A.; Martinez-Guzman O.; Reddi A. R. Heme Gazing: Illuminating Eukaryotic Heme Trafficking, Dynamics, and Signaling with Fluorescent Heme Sensors. Biochemistry 2017, 56 (13), 1815–1823. 10.1021/acs.biochem.7b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donegan R. K.; Moore C. M.; Hanna D. A.; Reddi A. R. Handling heme: The mechanisms underlying the movement of heme within and between cells. Free Radical Biol. Med. 2019, 133, 88–100. 10.1016/j.freeradbiomed.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna D. A.; Hu R.; Kim H.; Martinez-Guzman O.; Torres M. P.; Reddi A. R. Heme bioavailability and signaling in response to stress in yeast cells. J. Biol. Chem. 2018, 293 (32), 12378–12393. 10.1074/jbc.RA118.002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeny E. A.; Singh A. B.; Chakravarti R.; Martinez-Guzman O.; Saini A.; Haque M. M.; Garee G.; Dans P. D.; Hannibal L.; Reddi A. R.; Stuehr D. J. Glyceraldehyde-3-phosphate dehydrogenase is a chaperone that allocates labile heme in cells. J. Biol. Chem. 2018, 293 (37), 14557–14568. 10.1074/jbc.RA118.004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y.; Sweeny E. A.; Schlanger S.; Ghosh A.; Stuehr D. J. GAPDH Delivers Heme to Soluble Guanylyl Cyclase. J. Biol. Chem. 2020, 295 (24), 8145. 10.1074/jbc.RA120.013802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischhacker A. S.; Ragsdale S. W. An unlikely heme chaperone confirmed at last. J. Biol. Chem. 2018, 293 (37), 14569–14570. 10.1074/jbc.H118.005247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Vijayan V.; Jang M. S.; Thorenz A.; Greite R.; Rong S.; Chen R.; Shushakova N.; Tudorache I.; Derlin K.; Pradhan P.; Madyaningrana K.; Madrahimov N.; Brasen J. H.; Lichtinghagen R.; van Kooten C.; Huber-Lang M.; Haller H.; Immenschuh S.; Gueler F. Labile Heme Aggravates Renal Inflammation and Complement Activation After Ischemia Reperfusion Injury. Front. Immunol. 2019, 10, 2975. 10.3389/fimmu.2019.02975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.; Liu H. W.; Chen L.; Yuan J.; Liu Y.; Teng L.; Huan S. Y.; Yuan L.; Zhang X. B.; Tan W. Learning from Artemisinin: Bioinspired Design of a Reaction-Based Fluorescent Probe for the Selective Sensing of Labile Heme in Complex Biosystems. J. Am. Chem. Soc. 2020, 142 (5), 2129–2133. 10.1021/jacs.9b11245. [DOI] [PubMed] [Google Scholar]
- Fleischhacker A. S.; Gunawan A. L.; Kochert B. A.; Liu L.; Wales T. E.; Borowy M. C.; Engen J. R.; Ragsdale S. W. The heme regulatory motifs of heme oxygenase-2 contribute to the transfer of heme to the catalytic site for degradation. J. Biol. Chem. 2020, 295 (16), 5177–5191. 10.1074/jbc.RA120.012803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastpeyman S.; Godin R.; Cosa G.; English A. M. Quantifying Heme-Protein Maturation from Ratiometric Fluorescence Lifetime Measurements on the Single Fluorophore in Its GFP Fusion. J. Phys. Chem. A 2020, 124 (4), 746–754. 10.1021/acs.jpca.9b11901. [DOI] [PubMed] [Google Scholar]
- O’Keeffe R.; Latunde-Dada G. O.; Chen Y. L.; Kong X. L.; Cilibrizzi A.; Hider R. C. Glutathione and the intracellular labile heme pool. BioMetals 2021, 34 (2), 221–228. 10.1007/s10534-020-00274-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg G. R.; Wintrobe M. M. A labile iron pool. J. Biol. Chem. 1946, 165 (1), 397. 10.1016/S0021-9258(17)41250-6. [DOI] [PubMed] [Google Scholar]
- Chorkendorff I.; Niemantsverdriet J. W.. Concepts of Modern Catalysis and Kinetics; Wiley-VCH: Weinheim, Germany, 2003. [Google Scholar]
- Wilkins R. G.; Wilkins R. G.. Kinetics and Mechanism of Reactions of Transition Metal Complexes; VCH Publishers: Weinheim, New York, 1991. [Google Scholar]
- Langford C. H.; Gray H. B.. Ligand Substitution Processes; W.A. Benjamin: New York, 1966. [Google Scholar]
- de Villiers K. A.; Egan T. J. Heme Detoxification in the Malaria Parasite: A Target for Antimalarial Drug Development. Acc. Chem. Res. 2021, 54 (11), 2649–2659. 10.1021/acs.accounts.1c00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter E. R. H.; Ge Y.; Mason J. C.; Boyle J. J.; Long N. J. A Coumarin-Porphyrin FRET Break-Apart Probe for Heme Oxygenase-1. J. Am. Chem. Soc. 2021, 143 (17), 6460–6469. 10.1021/jacs.0c12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P.; Sheftel A. D.; English A. M.; Scott Bohle D.; Garcia-Santos D. Do Mammalian Cells Really Need to Export and Import Heme?. Trends Biochem. Sci. 2017, 42 (5), 395–406. 10.1016/j.tibs.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Maines M. D. The heme oxygenase system: a regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 1997, 37, 517–54. 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- Maines M. D. Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications. FASEB J. 1988, 2 (10), 2557–68. 10.1096/fasebj.2.10.3290025. [DOI] [PubMed] [Google Scholar]
- Liu L.; Dumbrepatil A. B.; Fleischhacker A. S.; Marsh E. N. G.; Ragsdale S. W. Heme oxygenase-2 is post-translationally regulated by heme occupancy in the catalytic site. J. Biol. Chem. 2020, 295 (50), 17227–17240. 10.1074/jbc.RA120.014919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C.; Bertini I.; Cavallaro G.; Holliday G. L.; Thornton J. M. Metal ions in biological catalysis: from enzyme databases to general principles. JBIC, J. Biol. Inorg. Chem. 2008, 13 (8), 1205–18. 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- Bleackley M. R.; Macgillivray R. T. Transition metal homeostasis: from yeast to human disease. BioMetals 2011, 24 (5), 785–809. 10.1007/s10534-011-9451-4. [DOI] [PubMed] [Google Scholar]
- Finney L. A.; O’Halloran T. V. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 2003, 300 (5621), 931–6. 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- Foster A. W.; Osman D.; Robinson N. J. Metal preferences and metallation. J. Biol. Chem. 2014, 289 (41), 28095–103. 10.1074/jbc.R114.588145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten C. E.; Albetel A. N. Iron sensing and regulation in Saccharomyces cerevisiae: Ironing out the mechanistic details. Curr. Opin. Microbiol. 2013, 16 (6), 662–8. 10.1016/j.mib.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten C. E.; O’Halloran T. V. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 2001, 292 (5526), 2488–92. 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- Rae T. D.; Schmidt P. J.; Pufahl R. A.; Culotta V. C.; O’Halloran T. V. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 1999, 284 (5415), 805–8. 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- Banci L.; Bertini I.; Cantini F.; Ciofi-Baffoni S. Cellular copper distribution: a mechanistic systems biology approach. Cell. Mol. Life Sci. 2010, 67 (15), 2563–89. 10.1007/s00018-010-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman D. L.; O’Halloran T. V. Function, structure, and mechanism of intracellular copper trafficking proteins. Annu. Rev. Biochem. 2001, 70, 677–701. 10.1146/annurev.biochem.70.1.677. [DOI] [PubMed] [Google Scholar]
- Rosenzweig A. C.; O’Halloran T. V. Structure and chemistry of the copper chaperone proteins. Curr. Opin. Chem. Biol. 2000, 4 (2), 140–7. 10.1016/S1367-5931(99)00066-6. [DOI] [PubMed] [Google Scholar]
- Osman D.; Martini M. A.; Foster A. W.; Chen J.; Scott A. J. P.; Morton R. J.; Steed J. W.; Lurie-Luke E.; Huggins T. G.; Lawrence A. D.; Deery E.; Warren M. J.; Chivers P. T.; Robinson N. J. Bacterial sensors define intracellular free energies for correct enzyme metalation. Nat. Chem. Biol. 2019, 15 (3), 241–249. 10.1038/s41589-018-0211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D.; Foster A. W.; Chen J.; Svedaite K.; Steed J. W.; Lurie-Luke E.; Huggins T. G.; Robinson N. J. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun. 2017, 8 (1), 1884. 10.1038/s41467-017-02085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S.; Harvie D. R.; Robinson N. J. Understanding how cells allocate metals using metal sensors and metallochaperones. Acc. Chem. Res. 2005, 38 (10), 775–83. 10.1021/ar0300118. [DOI] [PubMed] [Google Scholar]
- Waldron K. J.; Rutherford J. C.; Ford D.; Robinson N. J. Metalloproteins and metal sensing. Nature 2009, 460 (7257), 823–30. 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- Ba L. A.; Doering M.; Burkholz T.; Jacob C. Metal trafficking: from maintaining the metal homeostasis to future drug design. Metallomics 2009, 1 (4), 292–311. 10.1039/b904533c. [DOI] [PubMed] [Google Scholar]
- Ma Z.; Jacobsen F. E.; Giedroc D. P. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 2009, 109 (10), 4644–81. 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antelo G. T.; Vila A. J.; Giedroc D. P.; Capdevila D. A. Molecular Evolution of Transition Metal Bioavailability at the Host-Pathogen Interface. Trends Microbiol. 2021, 29 (5), 441–457. 10.1016/j.tim.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blindauer C. A. Bacterial metallothioneins: past, present, and questions for the future. JBIC, J. Biol. Inorg. Chem. 2011, 16 (7), 1011–24. 10.1007/s00775-011-0790-y. [DOI] [PubMed] [Google Scholar]
- Blindauer C. A.; Leszczyszyn O. I. Metallothioneins: unparalleled diversity in structures and functions for metal ion homeostasis and more. Nat. Prod. Rep. 2010, 27 (5), 720–41. 10.1039/b906685n. [DOI] [PubMed] [Google Scholar]
- Daltrop O.; Stevens J. M.; Higham C. W.; Ferguson S. J. The CcmE protein of the c-type cytochrome biogenesis system: unusual in vitro heme incorporation into apo-CcmE and transfer from holo-CcmE to apocytochrome. Proc. Natl. Acad. Sci. U. S. A. 2002, 99 (15), 9703–8. 10.1073/pnas.152120699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thony-Meyer L. A heme chaperone for cytochrome c biosynthesis. Biochemistry 2003, 42 (45), 13099–105. 10.1021/bi035598c. [DOI] [PubMed] [Google Scholar]
- Kranz R. G.; Richard-Fogal C.; Taylor J. S.; Frawley E. R. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 2009, 73 (3), 510–28. 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; Table of Contents.
- Brausemann A.; Zhang L.; Ilcu L.; Einsle O. Architecture of the membrane-bound cytochrome c heme lyase CcmF. Nat. Chem. Biol. 2021, 17, 800–805. 10.1038/s41589-021-00793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannibal L.; Collins D.; Brassard J.; Chakravarti R.; Vempati R.; Dorlet P.; Santolini J.; Dawson J. H.; Stuehr D. J. Heme binding properties of glyceraldehyde-3-phosphate dehydrogenase. Biochemistry 2012, 51 (43), 8514–29. 10.1021/bi300863a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.; Zhang P.; Yang Z.; Wang P.; Li H.; Gao Z. Interaction of glyceraldehyde-3-phosphate dehydrogenase and heme: The relevance of its biological function. Arch. Biochem. Biophys. 2017, 619, 54–61. 10.1016/j.abb.2017.03.005. [DOI] [PubMed] [Google Scholar]
- O’Halloran T. Transition metals in control of gene expression. Science 1993, 261 (5122), 715–725. 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- Handbook of Porphyrin Science; World Scientific Publishing Company, 2012; Vol. 30, p 512. [Google Scholar]
- Raven E. L. A short history of heme dioxygenases: rise, fall and rise again. JBIC, J. Biol. Inorg. Chem. 2017, 22 (2–3), 175–183. 10.1007/s00775-016-1412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov I.; Basran J.; Thackray S. J.; Handa S.; Mowat C. G.; Raven E. L. Structure and reaction mechanism in the heme dioxygenases. Biochemistry 2011, 50 (14), 2717–24. 10.1021/bi101732n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millett E. S.; Efimov I.; Basran J.; Handa S.; Mowat C. G.; Raven E. L. Heme-containing dioxygenases involved in tryptophan oxidation. Curr. Opin. Chem. Biol. 2012, 16 (1–2), 60–6. 10.1016/j.cbpa.2012.01.014. [DOI] [PubMed] [Google Scholar]
- Lewis-Ballester A.; Pham K. N.; Batabyal D.; Karkashon S.; Bonanno J. B.; Poulos T. L.; Yeh S. R. Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1. Nat. Commun. 2017, 8 (1), 1693. 10.1038/s41467-017-01725-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J.; Liu A. Heme-dependent dioxygenases in tryptophan oxidation. Arch. Biochem. Biophys. 2014, 544, 18–26. 10.1016/j.abb.2013.11.009. [DOI] [PubMed] [Google Scholar]
- Nelp M. T.; Kates P. A.; Hunt J. T.; Newitt J. A.; Balog A.; Maley D.; Zhu X.; Abell L.; Allentoff A.; Borzilleri R.; Lewis H. A.; Lin Z.; Seitz S. P.; Yan C.; Groves J. T. Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (13), 3249–3254. 10.1073/pnas.1719190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi G.; Hayashi N. Regulation by heme of synthesis and intracellular translocation of delta-aminolevulinate synthase in the liver. Mol. Cell. Biochem. 1981, 37 (1), 27–41. 10.1007/BF02355885. [DOI] [PubMed] [Google Scholar]
- Srivastava K. K.; Cable E. E.; Donohue S. E.; Bonkovsky H. L. Molecular basis for heme-dependent induction of heme oxygenase in primary cultures of chick embryo hepatocytes. Demonstration of acquired refractoriness to heme. Eur. J. Biochem. 1993, 213 (3), 909–17. 10.1111/j.1432-1033.1993.tb17835.x. [DOI] [PubMed] [Google Scholar]
- Light W. R. 3rd; Olson J. S. The effects of lipid composition on the rate and extent of heme binding to membranes. J. Biol. Chem. 1990, 265 (26), 15632–7. 10.1016/S0021-9258(18)55444-2. [DOI] [PubMed] [Google Scholar]
- Light W. R. 3rd; Olson J. S. Transmembrane movement of heme. J. Biol. Chem. 1990, 265 (26), 15623–31. 10.1016/S0021-9258(18)55443-0. [DOI] [PubMed] [Google Scholar]
- Rose M. Y.; Thompson R. A.; Light W. R.; Olson J. S. Heme Transfer between Phospholipid-Membranes and Uptake by Apohemoglobin. J. Biol. Chem. 1985, 260 (11), 6632–6640. 10.1016/S0021-9258(18)88828-7. [DOI] [PubMed] [Google Scholar]
- Chiabrando D.; Marro S.; Mercurio S.; Giorgi C.; Petrillo S.; Vinchi F.; Fiorito V.; Fagoonee S.; Camporeale A.; Turco E.; Merlo G. R.; Silengo L.; Altruda F.; Pinton P.; Tolosano E. The mitochondrial heme exporter FLVCR1b mediates erythroid differentiation. J. Clin. Invest. 2012, 122 (12), 4569–79. 10.1172/JCI62422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guzman O.; Willoughby M. M.; Saini A.; Dietz J. V.; Bohovych I.; Medlock A. E.; Khalimonchuk O.; Reddi A. R. Mitochondrial-nuclear heme trafficking in budding yeast is regulated by GTPases that control mitochondrial dynamics and ER contact sites. J. Cell Sci. 2020, 133 (10), jcs237917. 10.1242/jcs.237917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I. G.; Willoughby M. M.; Hamza I.; Reddi A. R. One ring to bring them all and in the darkness bind them: The trafficking of heme without deliverers. Biochim. Biophys. Acta, Mol. Cell Res. 2021, 1868 (1), 118881. 10.1016/j.bbamcr.2020.118881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison G. R. Fluorometric Microdetermination of Heme Protein. Anal. Chem. 1965, 37, 1124–6. 10.1021/ac60228a014. [DOI] [PubMed] [Google Scholar]
- Masuda T.; Takahashi S. Chemiluminescent-based method for heme determination by reconstitution with horseradish peroxidase apo-enzyme. Anal. Biochem. 2006, 355 (2), 307–9. 10.1016/j.ab.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Hopp M. T.; Schmalohr B. F.; Kuhl T.; Detzel M. S.; Wissbrock A.; Imhof D. Heme Determination and Quantification Methods and Their Suitability for Practical Applications and Everyday Use. Anal. Chem. 2020, 92 (14), 9429–9440. 10.1021/acs.analchem.0c00415. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R.Principles of Fluorescence Spectroscopy; Springer: New York, 2006. [Google Scholar]
- Takeda S.; Kamiya N.; Nagamune T. A novel protein-based heme sensor consisting of green fluorescent protein and apocytochrome b562. Anal. Biochem. 2003, 317 (1), 116–119. 10.1016/S0003-2697(03)00096-4. [DOI] [PubMed] [Google Scholar]
- Arpino J. A.; Czapinska H.; Piasecka A.; Edwards W. R.; Barker P.; Gajda M. J.; Bochtler M.; Jones D. D. Structural basis for efficient chromophore communication and energy transfer in a constructed didomain protein scaffold. J. Am. Chem. Soc. 2012, 134 (33), 13632–40. 10.1021/ja301987h. [DOI] [PubMed] [Google Scholar]
- Koga S.; Yoshihara S.; Bando H.; Yamasaki K.; Higashimoto Y.; Noguchi M.; Sueda S.; Komatsu H.; Sakamoto H. Development of a heme sensor using fluorescently labeled heme oxygenase-1. Anal. Biochem. 2013, 433 (1), 2–9. 10.1016/j.ab.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Taira J.; Nakashima Y.; Yoshihara S.; Koga S.; Sueda S.; Komatsu H.; Higashimoto Y.; Takahashi T.; Tanioka N.; Shimizu H.; Morimatsu H.; Sakamoto H. Improvement of heme oxygenase-1-based heme sensor for quantifying free heme in biological samples. Anal. Biochem. 2015, 489, 50–2. 10.1016/j.ab.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Hisamatsu Y.; Umezawa N.; Yagi H.; Kato K.; Higuchi T. Design and synthesis of a 4-aminoquinoline-based molecular tweezer that recognizes protoporphyrin IX and iron(iii) protoporphyrin IX and its application as a supramolecular photosensitizer. Chem. Sci. 2018, 9 (38), 7455–7467. 10.1039/C8SC02133C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton L. D.; Pascu S. I.; Tyrrell R. M.; Eggleston I. M. Development of a peptide-based fluorescent probe for biological heme monitoring. Org. Biomol. Chem. 2019, 17 (3), 467–471. 10.1039/C8OB02290A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T. A.; Mu A.; Tai T. T.; Kitajima S.; Taketani S. Continuous de novo biosynthesis of haem and its rapid turnover to bilirubin are necessary for cytoprotection against cell damage. Sci. Rep. 2015, 5, 10488. 10.1038/srep10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonagh A. F.; Palma L. A.; Lightner D. A. Blue light and bilirubin excretion. Science 1980, 208 (4440), 145–51. 10.1126/science.7361112. [DOI] [PubMed] [Google Scholar]
- Kumagai A.; Ando R.; Miyatake H.; Greimel P.; Kobayashi T.; Hirabayashi Y.; Shimogori T.; Miyawaki A. A bilirubin-inducible fluorescent protein from eel muscle. Cell 2013, 153 (7), 1602–11. 10.1016/j.cell.2013.05.038. [DOI] [PubMed] [Google Scholar]
- Nobles C. L.; Clark J. R.; Green S. I.; Maresso A. W. A dual component heme biosensor that integrates heme transport and synthesis in bacteria. J. Microbiol. Methods 2015, 118, 7–17. 10.1016/j.mimet.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville D. G.; Mullineaux-Sanders C.; Corcoran C. J.; Burger B. T.; Imam S.; Donohue T. J.; Ulijasz A. T. A High-Throughput Method for Identifying Novel Genes That Influence Metabolic Pathways Reveals New Iron and Heme Regulation in Pseudomonas aeruginosa. mSystems 2021, 6 (1), e00933-20. 10.1128/mSystems.00933-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourino S.; Giardina B. J.; Reyes-Caballero H.; Wilks A. Metabolite-driven Regulation of Heme Uptake by the Biliverdin IXbeta/delta-Selective Heme Oxygenase (HemO) of Pseudomonas aeruginosa. J. Biol. Chem. 2016, 291 (39), 20503–15. 10.1074/jbc.M116.728527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A.; Pye V. E.; Graham C.; Muir L.; Seow J.; Ng K. W.; Cook N. J.; Rees-Spear C.; Parker E.; Dos Santos M. S.; Rosadas C.; Susana A.; Rhys H.; Nans A.; Masino L.; Roustan C.; Christodoulou E.; Ulferts R.; Wrobel A. G.; Short C. E.; Fertleman M.; Sanders R. W.; Heaney J.; Spyer M.; Kjaer S.; Riddell A.; Malim M. H.; Beale R.; MacRae J. I.; Taylor G. P.; Nastouli E.; van Gils M. J.; Rosenthal P. B.; Pizzato M.; McClure M. O.; Tedder R. S.; Kassiotis G.; McCoy L. E.; Doores K. J.; Cherepanov P. SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity. Sci. Adv. 2021, 7 (22), eabg7607. 10.1126/sciadv.abg7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shum M.; Shintre C. A.; Althoff T.; Gutierrez V.; Segawa M.; Saxberg A. D.; Martinez M.; Adamson R.; Young M. R.; Faust B.; Gharakhanian R.; Su S.; Chella Krishnan K.; Mahdaviani K.; Veliova M.; Wolf D. M.; Ngo J.; Nocito L.; Stiles L.; Abramson J.; Lusis A. J.; Hevener A. L.; Zoghbi M. E.; Carpenter E. P.; Liesa M. ABCB10 exports mitochondrial biliverdin, driving metabolic maladaptation in obesity. Sci. Transl. Med. 2021, 13 (594), eabd1869. 10.1126/scitranslmed.abd1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson J. H. Probing structure-function relations in heme-containing oxygenases and peroxidases. Science 1988, 240 (4851), 433–9. 10.1126/science.3358128. [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano P. R. Control of the catalytic activity of prosthetic heme by the structure of hemoproteins. Acc. Chem. Res. 1987, 20 (8), 289–294. 10.1021/ar00140a004. [DOI] [Google Scholar]
- Ogawa K.; Sun J.; Taketani S.; Nakajima O.; Nishitani C.; Sassa S.; Hayashi N.; Yamamoto M.; Shibahara S.; Fujita H.; Igarashi K. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 2001, 20 (11), 2835–43. 10.1093/emboj/20.11.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–26. 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam J.; Killeen E.; Gong P.; Naquin R.; Hu B.; Stewart D.; Ingelfinger J. R.; Nath K. A. Heme activates the heme oxygenase-1 gene in renal epithelial cells by stabilizing Nrf2. Am. J. Physiol Renal Physiol 2003, 284 (4), F743–52. 10.1152/ajprenal.00376.2002. [DOI] [PubMed] [Google Scholar]
- Burton M. J.; Kapetanaki S. M.; Chernova T.; Jamieson A. G.; Dorlet P.; Santolini J.; Moody P. C. E.; Mitcheson J. S.; Davies N. W.; Schmid R.; Raven E. L.; Storey N. M. A heme-binding domain controls regulation of ATP-dependent potassium channels. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (14), 3785–3790. 10.1073/pnas.1600211113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I.; Dailey H. A. One ring to rule them all: trafficking of heme and heme synthesis intermediates in the metazoans. Biochim. Biophys. Acta, Mol. Cell Res. 2012, 1823 (9), 1617–32. 10.1016/j.bbamcr.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]