Abstract
Natural products that contain distinctive chemical functionality can serve as useful starting points to develop Nature’s compounds into viable therapeutics. Peptide natural products, an under-represented class of medicines, such as ribosomally synthesized and post-translationally modified peptides (RiPPs), often contain noncanonical amino acids and structural motifs that give rise to potent biological activity. However, these motifs can be difficult to obtain synthetically, thereby limiting the transition of RiPPs to the clinic. Aminovinyl cysteine containing peptides, which display potent antimicrobial or anticancer activity, possess an intricate C-terminal ring that is critical for bioactivity. To date, successful methods for the total chemical synthesis of such peptides are yet to be realized, although several advancements have been achieved. In this perspective, we review this burgeoning class of aminovinyl cysteine peptides and critically evaluate the chemical strategies to install the distinct aminovinyl cysteine motif.
Keywords: Lanthipeptides: aminovinyl cysteine, antimicrobials, cyclic peptides, biosynthesis, chemical synthesis
1. Introduction
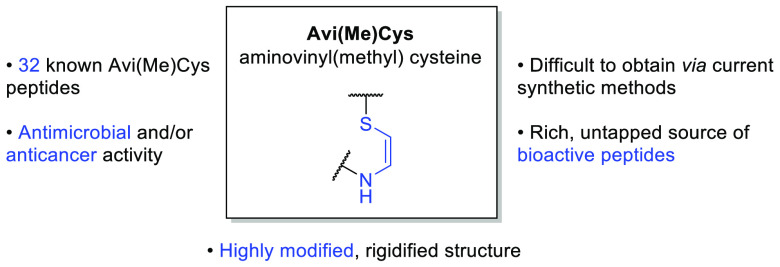
Ribosomally synthesized and post-translationally modified peptides (RiPPs) are a major class of diverse natural products that cover a broad range of bioactivities.1,2 They are genetically encoded and expressed as large precursor peptides which are enzymatically processed into smaller modified bioactive peptides.1 These enzymatic modifications are known as post-translational modifications (PTMs) and may include cyclization, halogenation, dehydration, and incorporation of d-amino acids.3 These PTMs act to improve the peptides’ bioactivity and increase their stability against proteolytic degradation.3 One such PTM is the rigidifying C-terminal S-[(Z)-2-aminovinyl]-d-cysteine (AviCys) or (2S,3S)-S-[(Z)-2-aminovinyl]-3-methyl-d-cysteine (AviMeCys) unit, herein jointly referred to as Avi(Me)Cys units (Figure 1).
Figure 1.
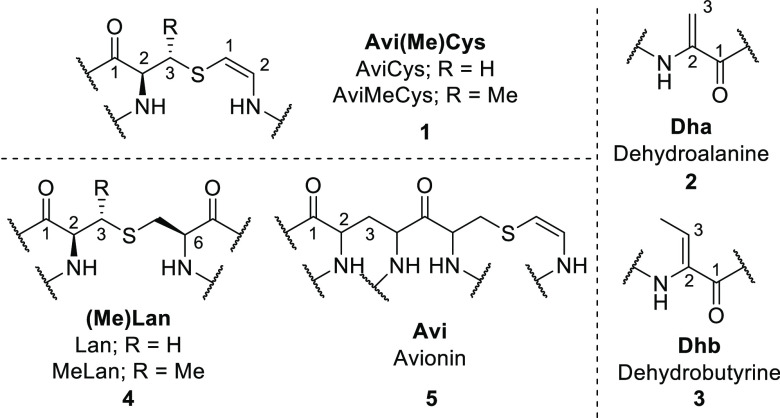
Chemical structures of the noncanonical amino acids commonly found in Avi(Me)Cys-containing peptides. (Me)Lan = (methyl)lanthionine (4), Avi = avionin (5), AviCys = S-[(Z)-2-aminovinyl]-d-cysteine (1), AviMeCys = (2S,3S)-S-[(Z)-2-aminovinyl]-3-methyl-d-cysteine, jointly referred to as Avi(Me)Cys (1).
To date, there are 32 peptides that contain this moiety. These are found among five major peptide families: lanthipeptides (Figures 2, 3, and 4), linaridins (Figure 5), thioamitides (Figure 6), lanthidins (Figure 7), and the lipolanthines (Figure 7). Gram-positive bacteria produce these compounds as a defense mechanism against competing bacterial species; therefore, many RiPPs have potent antibacterial activity against Staphylococcus, Streptococcus, Enterococcus, and Clostridia species (Table 1).4 Some Avi(Me)Cys-containing peptides, such as the thioamitides, exhibit cytotoxicity toward human cancer cell lines.5
Figure 2.
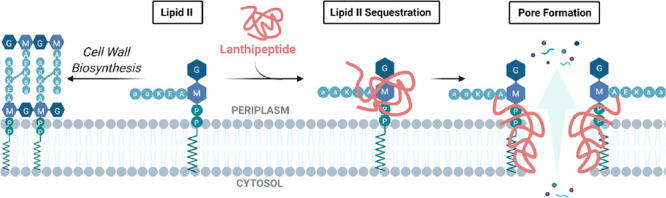
Sequestration of lipid II and concomitant pore formation by the lanthipeptide–lipid II complex, leading to bacterial cell lysis and death. This dual mechanism of action has been described for several lanthipeptides including mutacin-1140 and epidermin.28,31 However, other lanthipeptides such as mersacidin are currently believed to only partake in sequestration of lipid II without pore formation.32 Further investigation is required to identify other pore-forming lanthipeptides. A = l-alanine, a = d-alanine, K = lysine, E = glutamic acid, G = GlcNAc (N-acetylglucosamine), M = MurNAc (N-acetylmuramic acid), P = phospho group. Created with BioRender.com.
Figure 3.
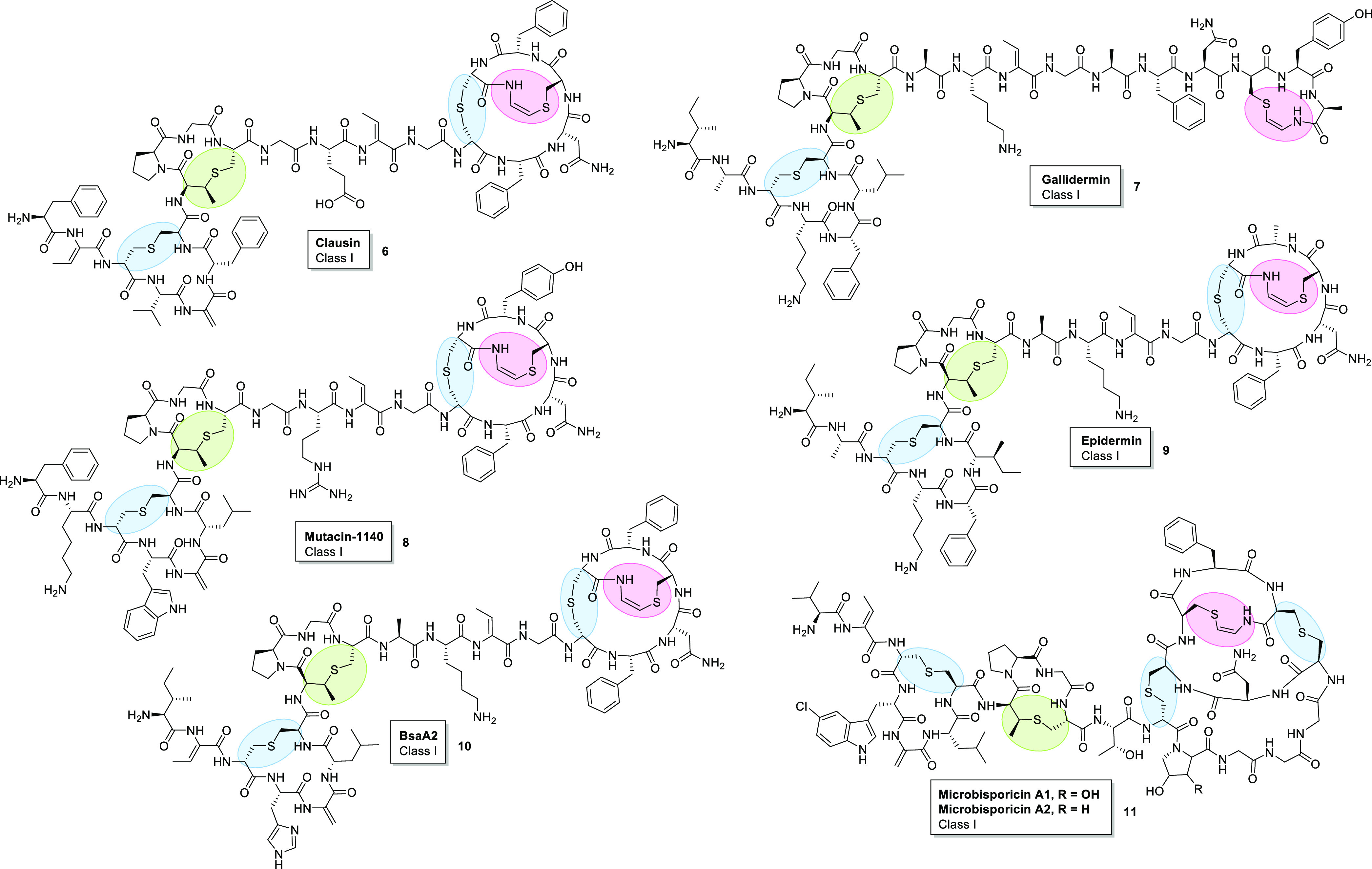
Structures of Avi(Me)Cys-containing class I lanthipeptides; clausin (6),21 gallidermin (7),19 mutacin-1140 (8),53 epidermin (9),20 BsaA2 (10),37 and microbisporicins A1 and A2 (11).18 Pink = Avi(Me)Cys. Blue = Lan. Green = MeLan.
Figure 4.
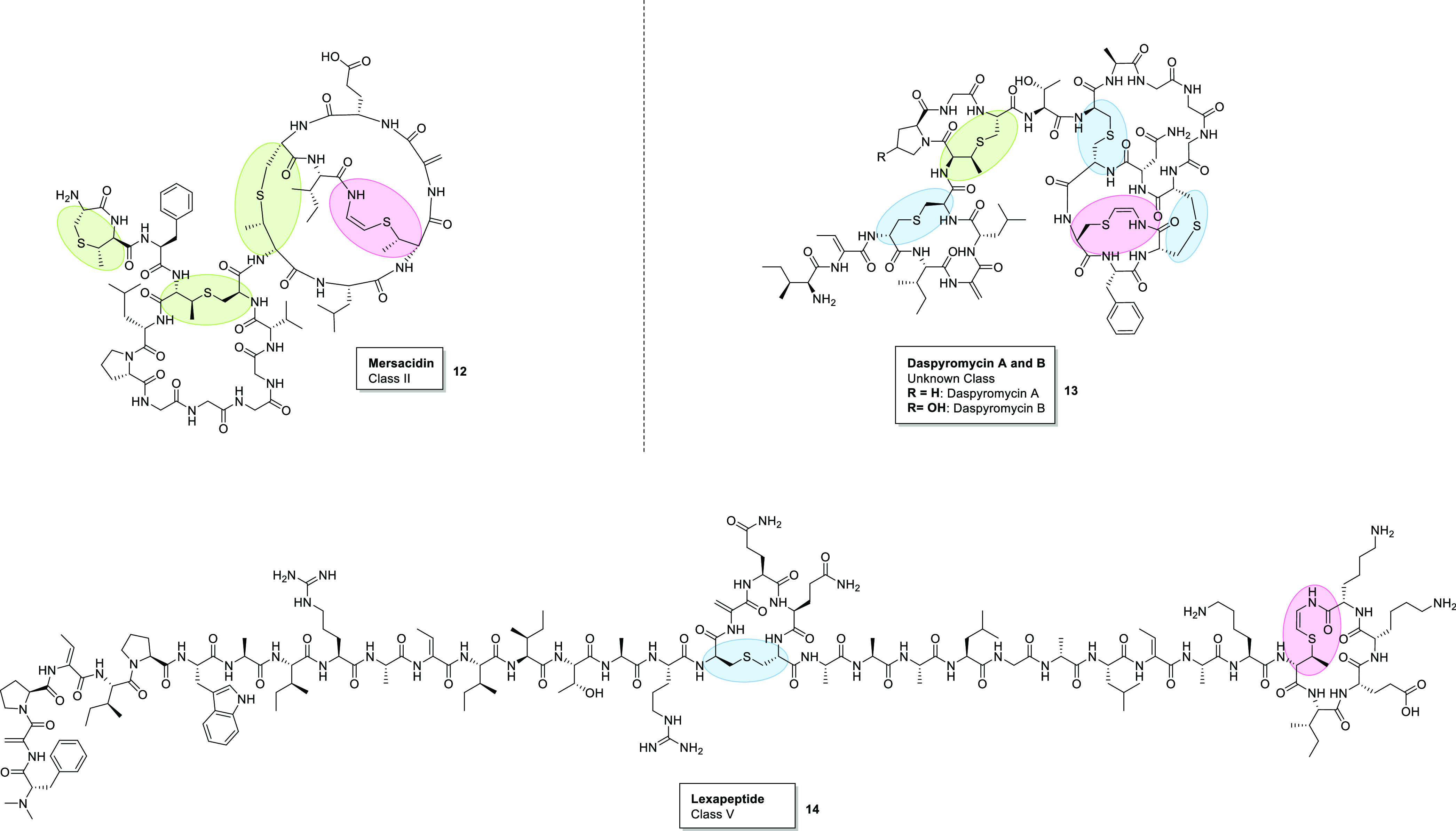
Structures of Avi(Me)Cys-containing class II and class V lanthipeptides; mersacidin (12) (class II),22 daspyromycins A and B (13) (unknown class),24 and lexapeptide (13) (class V).23 Pink = Avi(Me)Cys. Green = MeLan. Blue = Lan.
Figure 5.
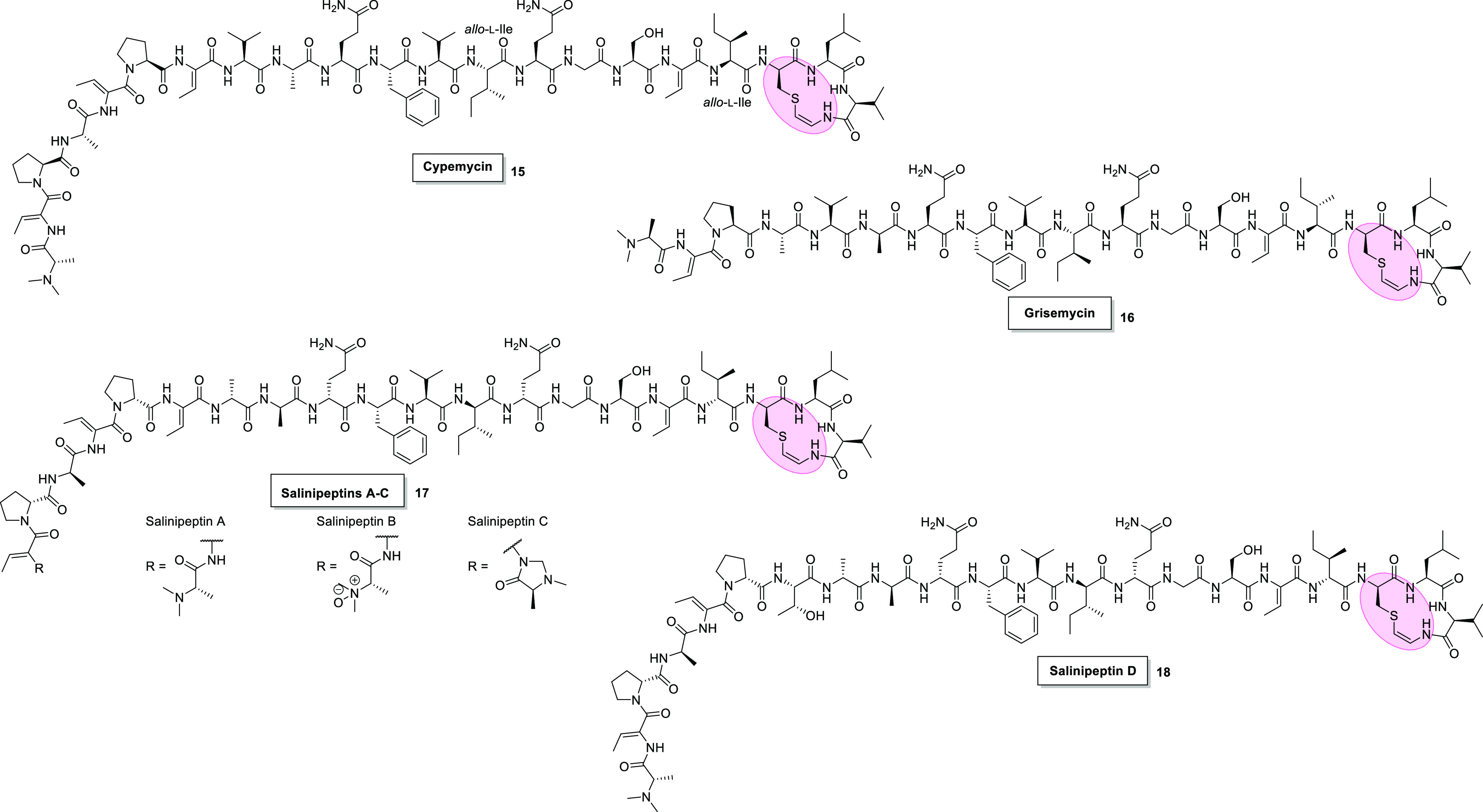
Chemical structures of Avi(Me)Cys-containing peptides of the linaridin family; cypemycin (15),40 grisemycin (16),41 and salinipeptins A–D (17 and 18).42 Pink = AviCys.
Figure 6.
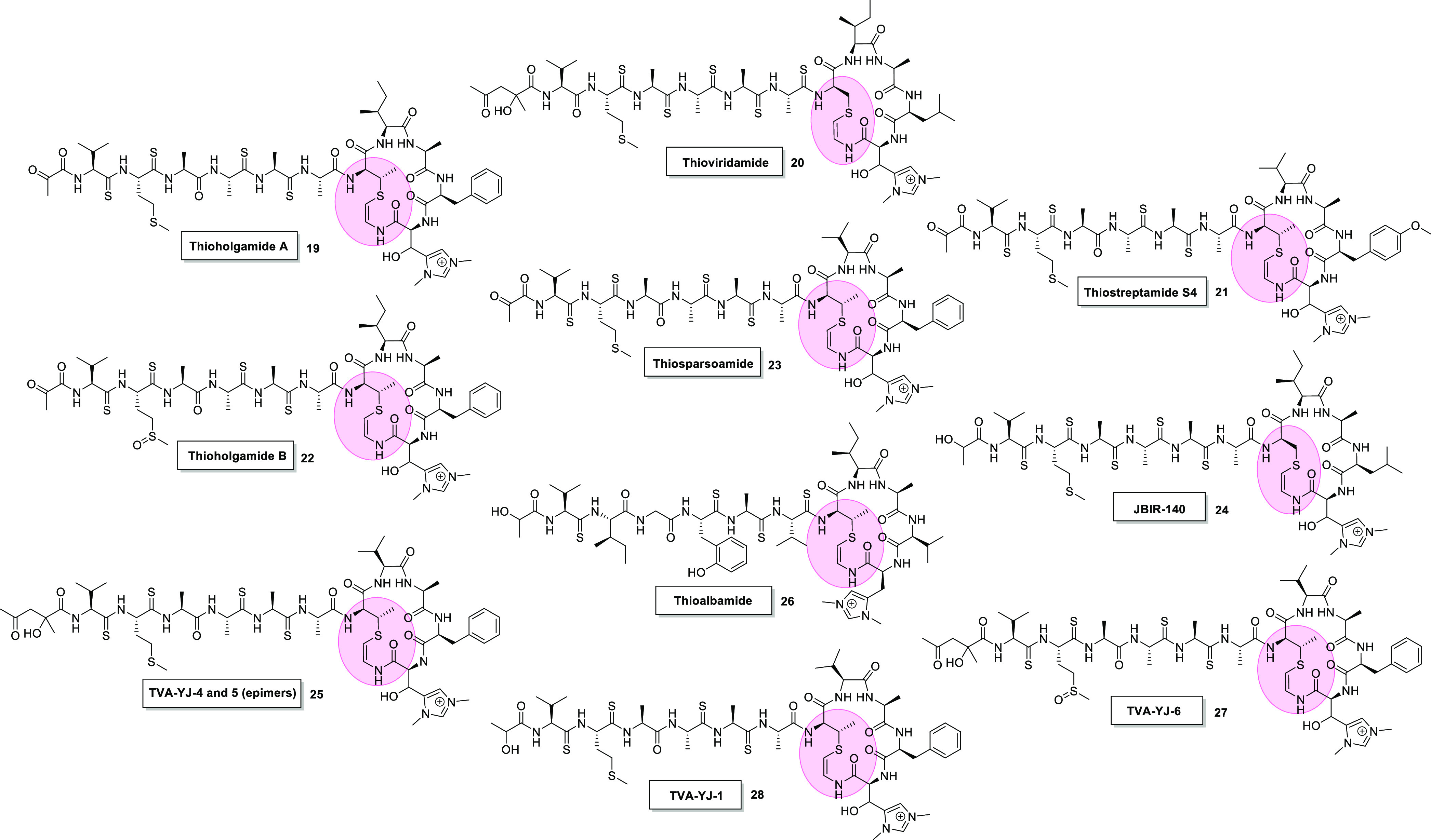
Chemical structures of Avi(Me)Cys-containing peptides of the thioamitide family; thioholgamide A (19),44 thioviridamide (20),5 thiostreptamide S4 (21),45 thioholgamide B (22),44 thiosparsoamide (23),46 JBIR-140 (24),48 TVA-YJ-4 and 5 (epimers at α-hydroxyamide) (25),43 thioalbamide (26),47 TVA-YJ-6 (27),43 and TVA-YJ-1 (28).43 Pink = Avi(Me)Cys.
Figure 7.
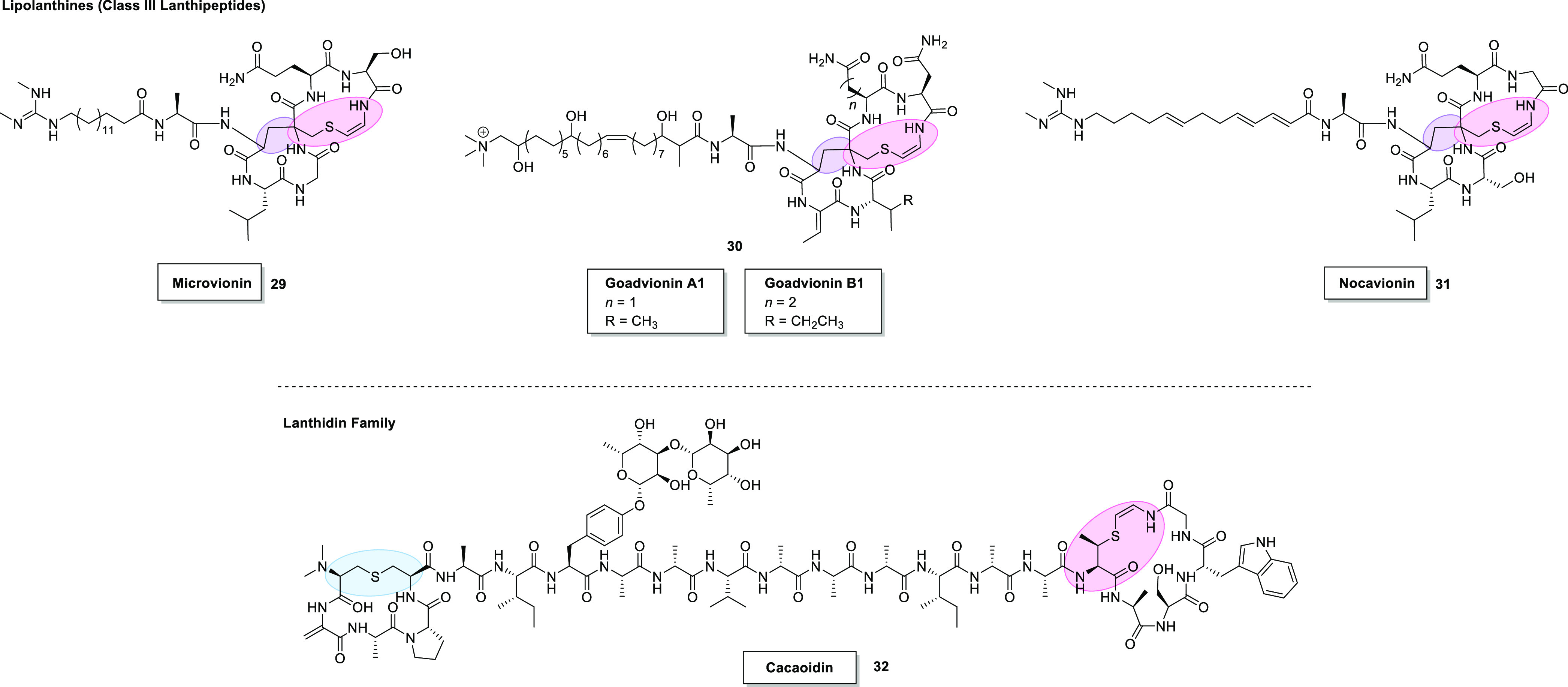
Chemical structures of Avi(Me)Cys-containing peptides of the lipolanthine family (since reclassified as class III lanthipeptides); microvionin (29),50 goadvionins A1/B1 (30)51 and nocavionin (31),50 and the lanthidin family; cacaoidin (32).52 Pink = Avi(Me)Cys. Purple = avionin (Avi). Blue = Lan.
Table 1. Producing Bacterial Strain and Bioactivity of Known Avi(Me)Cys-Containing Peptides (G+ = Gram-Positive, G– = Gram-Negative).
| family | peptide | producing strain | notable bioactivity |
|---|---|---|---|
| lanthipeptide | mutacin-114017 (class I) | Streptococcus mutans | Neisseria gonorrhea (G−), Enterococcus faecalis (G+), Staphylococcus epidermidis (G+), Helicobacter pylori (G−) |
| microbisporicins18 (class I) | Microbispora corallina | Staphylococcus aureus (G+), Streptococcus pneumoniae (G+) | |
| gallidermin19 (class I) | Staphylococcus gallinarum | Mariniluteicoccus flavus (G+), Staphylococcus simulans (G+) | |
| epidermin20 (class I) | Staphylococcus epidermidis | Mariniluteicoccus flavus (G+), Staphylococcus simulans (G+) | |
| clausin21 (class I) | Bacillus clausii O/C | Clostridium difficile (G+) | |
| BsaA237 (class I) | Staphylococcus aureus | antibacterial (G+) | |
| mersacidin22 (class II) | Bacillus amyloliquefaciens | Staphylococcus aureus (G+), MRSA (G+) | |
| lexapeptide23 (class V) | Streptomyces rochei | MRSA (G+), MRSE (G+), Micrococcus luteus (G+), Bacillus subtilis (G+) | |
| daspyromycin A and B38 (unknown class) | Actinokineospora diospyrosa NBRC 15665 | MRSA (G+), VRE (G+) | |
| linaridin | cypemycin39,40 | Streptomyces sp. OH-4156 | cytotoxic against P388 leukemia cells, antibacterial against Micrococcus luteus |
| grisemycin41 | Streptomyces griseus | not reported | |
| salinipeptins A–D42 | Streptomyces sp. strain GSL-6C | group A Streptococcus pyogenes M1T1 | |
| thioamitide | thioviridamide5 | Streptomyces olivoviridis | apoptosis inducer (anticancer) |
| TVA-YJ-4, 5, and 643 | Streptomyces sp. NRRL S-87 | cytotoxic (anticancer) | |
| thioholgamides A and B44 | Streptomyces malayseiense | antiproliferative and cytotoxic (anticancer) | |
| thiostreptamide S445 | Streptomyces sp. NRRL S-4 | anticancer (specific activity yet to be reported) | |
| thiosparsoamide46 | Streptomyces sparsogenes | ||
| thioalbamide47 | Amycolatopsis alba | ||
| JBIR-14048 | Streptomyces avermitilis | ||
| TVA-YJ-149 | Streptomyces laurentii | ||
| lipolanthine | microvionin50 | Microbacterium arborescens | MRSA (G+), Streptococcus pneumoniae (G+) |
| nocavionin50 | Nocardia terpenica | not reported | |
| goadvionins51 | Streptomyces sp. TP-A0584 | Staphylococcus aureus (G+), Bacillus subtilis (G+), Micrococcus luteus (G+) | |
| lanthidin | cacaoidin52 | Streptomyces cacaoi | MRSA (G+), Clostridium difficile (G+), Staphylococcus simulans (G+) |
These Avi(Me)Cys-containing peptides possess highly modified structures that give rise to desirable drug-like properties such as heat and pH stability, high target specificity, and resistance toward proteases.6 These natural products also commonly contain other noncanonical amino acids, including 2,3-dehydroalanine (Dha, 2), (Z)-2,3-dehydrobutyrine (Dhb, 3), d-amino acids, and lanthionine (Lan, 4), β-methyllanthionine ((Me)Lan (4), and avionin (Avi, 5) (Figure 1).3,4,7 These PTMs restrict the conformational freedom of the peptide, providing a dual function of (a) locking the peptide in an active conformation for target binding and (b) inhibiting proteolytic degradation.6 In particular, unsaturated Dha, Dhb, and Avi(Me)Cys moieties increase structural rigidity by including sp2 α-carbons in the peptide backbone.8
Despite the “drug-like” potential of Avi(Me)Cys-containing peptides, lack of viable synthetic routes to access the Avi(Me)Cys moiety limits their transition to the clinic. To date, there is no total synthesis of an Avi(Me)Cys-containing natural product peptide reported, although several routes have been investigated.9−13 Therefore, these natural products present an intriguing synthetic target with important therapeutic applications and require the development of novel methodology. This perspective discusses in detail the different families of Avi(Me)Cys-containing peptides and the current attempts to install the Avi(Me)Cys unit by chemical methods.
2.0. Lanthipeptides
Over 100 lanthipeptides have been discovered and characterized since Rogers and Whittier discovered the lanthipeptide nisin in 1928.14 This family encompasses RiPPs that contain one or more (Me)Lan thioether linkages and are further categorized into five classes (I–V) determined by their genetically encoded biosynthetic enzymes responsible for installing the Lan and (Me)Lan motifs.15,16 The seven known Avi(Me)Cys-containing lanthipeptides are found only in classes I, II, and V. Class I comprises mutacin-1140,17 microbisporicin,18 gallidermin,19 epidermin,20 and clausin (Figure 3).21 Class II and V contain the peptides mersacidin22 and lexapeptide,23 respectively (Figure 4). Daspyromycin (Figure 4), the most recently discovered Avi(Me)Cys-containing lanthipeptide, is yet to be categorized.24
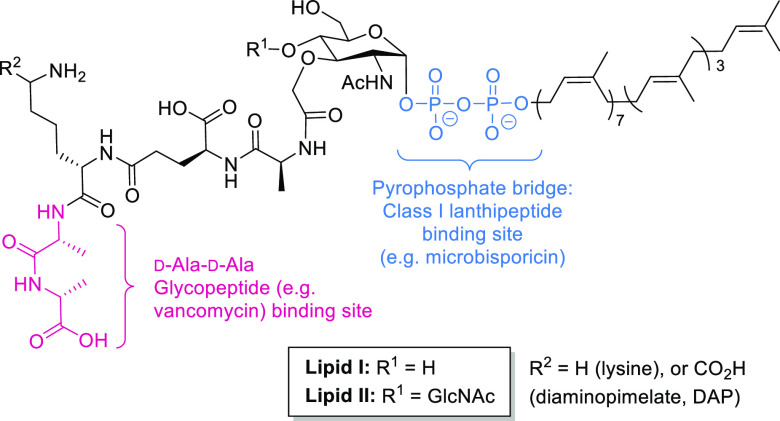
The Avi(Me)Cys-containing peptides within the lanthipeptide family are the most well-known and studied of the Avi(Me)Cys natural products.7 All display antimicrobial activity against Gram-positive bacterial species including multi-drug-resistant Staphylococcus epidermidis and Staphylococcus aureus.25 The antibacterial activity of Avi(Me)Cys-containing lanthipeptides arises from targeting lipid II, an essential precursor for the cell wall of Gram-positive bacteria (Figure 2).4,26 Lipid II consists of a bactoprenol “carrier lipid” coupled to a peptidoglycan building block (N-acetylmuramyl pentapeptide-N-acetylglucosamine) via a pyrophosphate bridge.27 The lanthipeptide nisin and the Avi(Me)Cys-containing microbisporicin sequester lipid II by forming lanthipeptide–lipid II complexes.28,29 Sequestration of lipid II blocks cell wall biosynthesis, resulting in poor structural integrity and lysis due to high intracellular osmotic pressure (Figure 2).27 Additionally, some lanthipeptide–lipid complexes can form pores in the bacterial cell membrane by insertion of the complexed lanthipeptide into the phospholipid bilayer (Figure 2).27,28,30 These pores provide a permeable channel through which cellular components may leak through into the periplasm, causing loss of osmotic pressure and cell death.28 This pore-forming mode of action remains poorly understood, with only a handful of lanthipeptides known to exhibit this behavior, notably the Avi(Me)Cys-containing peptides epidermin and mutacin-1140.28,31
Glycopeptide antibiotics such as vancomycin typically target the peptidyl side chain (d-Ala-d-Ala) of lipid II.33 In vancomycin-resistant bacteria, the d-Ala-d-Ala sequence is mutated to prevent vancomycin from binding, therefore reducing its efficacy.34,35 Another mechanism of resistance is a “false target” wherein vancomycin-resistant bacteria produce excess peptidoglycan containing the d-Ala-d-Ala unit, which trap and sequester vancomycin.35 Lanthipeptides typically do not target the d-Ala-d-Ala sequence of lipid II and instead bind via the lipid II pyrophosphate bridge (Figure 8).30 Microbisporicin, an Avi(Me)Cys-containing lanthipeptide, interacts electrostatically with the negatively charged lipid II pyrophosphate bridge, an interaction that is also favored by the association of the hydrophobic ring system in microbisporicin with the cell membrane.36 The pyrophosphate bridge of lipid II is less susceptible to mutation than the d-Ala-d-Ala sequence, thereby reducing the probability of antibiotic resistance toward microbisporicin developing compared to vancomycin.36 The Avi(Me)Cys-containing lanthipeptides lexapeptide and microbisporicin exhibit potent activity against vancomycin-resistant bacteria, indicating their potential for therapeutic use to combat antimicrobial resistance.18,23
Figure 8.
Generic chemical structure of lipid I and lipid II, precursors for the biosynthesis of the peptidoglycan cell wall.27 GlcNAc = N-acetylglucosamine, postulated binding site of mersacidin. Pink = d-Ala-d-Ala vancomycin binding site;33 blue = pyrophosphate bridge of lipid II, the postulated microbisporicin binding site.18
Microbisporicins A1 and A2 (Figure 3), produced in a 60:40 ratio by fermentation of the actinomycete Microbispora corallina, were previously being developed by Naicons SRL and Sentinella Pharmaceuticals for the treatment of multi-drug-resistant bacterial infections.54,55 Known as NAI-107,56 both peptides exhibit potent antibacterial activity against both Gram-positive and Gram-negative bacteria, including against vancomycin-intermediate S. aureus (VISA) and vancomycin-resistant Enterococcus faecalis.54,57 Microbisporicins A1 and A2 are both 24-mer pentacyclic lanthipeptides, identical except at Pro14, wherein A1 contains 3,4-dihydroxyproline, and A2 contains 4-hydroxyproline.56 With promising activity in preliminary in vivo studies,58,59 NAI-107 was in preclinical development by Naicons SRL and Sentinella Pharmaceuticals for treatment of multi-drug-resistant Gram-positive infections; however, the current status of NAI-107 development is unknown.54,55
In contrast to lexapeptide and microbisporicin, the class II Avi(Me)Cys-containing lanthipeptide mersacidin (Figure 4) does not target the pyrophosphate bridge of lipid II.22,60 Instead, mersacidin exhibits affinity for the lipid II GlcNAc (N-acetylglucosamine) unit, which is absent in lipid I (Figure 8).61 As such, mersacidin does not show affinity for lipid I, unlike class I lanthipeptides which can bind to lipid I via the pyrophosphate bridge.30 The mersacidin–lipid II complex does not lead to the formation of pores in the bacterial membrane, and therefore, the primary mechanism of action of mersacidin is by the sequestration of lipid II and inhibition of cell wall biosynthesis.60
Mersacidin is also unusual in that it does not possess any positively charged amino acids. Typically, antimicrobial peptides (AMPs) have a net positive charge, which facilities association of the peptide with the negatively charged bacterial membrane.62 Mersacidin, however, contains a negatively charged glutamate at position 17 (Figure 4). Mutation of this residue by amidation or substitution for an alanine greatly reduces the activity of mersacidin, which indicates that Glu17 is essential for activity.63 It has been postulated that Glu17 interacts with calcium ions (Ca2+) to form a salt bridge with negatively charged moieties on the bacterial cell membrane.32
Mersacidin has previously displayed moderate in vitro antimicrobial activity; however, preliminary in vivo antibacterial activity studies have shown promise.64,65 Antimicrobial assays have shown low to moderate activity against some Gram-positive bacteria and negligible activity against Gram-negative strains, with mersacidin having significantly less activity compared to that of the glycopeptide class overall.66 However, work by Kruszewska et al.65 found that mersacidin eliminated methicillin-resistant Staphylococcus aureus (MRSA, strain 99308) from the nasal cavity in mouse rhinitis models. A twice daily treatment of mersacidin (1.66 mg/kg per dose) over 3 days effectively cured mice infected with MRSA in the nasal cavity (n = 12) compared to mice that did not receive mersacidin.65 Additionally, cytotoxic effects such as morphological changes, mucosal lesions, and cytokines related to MRSA infection were not detected in mouse models when treated with mersacidin intranasally.65
2.1. Biosynthesis of Avi(Me)Cys in Lanthipeptides
Within the lanthipeptide family, the incorporation of Avi(Me)Cys units across the relevant lanthipeptides is catalyzed by structurally homologous flavoprotein decarboxylase (LanD) enzymes.67 These flavoproteins require a redox-active flavin cofactor, such as flavin mononucleotide (FMN) or flavin adenine dinucleotide (FAD), for catalytic activity.67
LanD enzymes are part of the homo-oligomeric flavoprotein cysteine decarboxylase (HFCD) family.68 These proteins form complex homo-oligomeric quaternary structures with active site(s) at the central interface of the complex.69 LanD enzymes form dodecameric oligomers with a substrate binding clamp that is highly disordered in the absence of a peptide substrate. The flavin cofactor (FMN or FAD) is anchored in a structurally conserved region of the active site.67,69,70
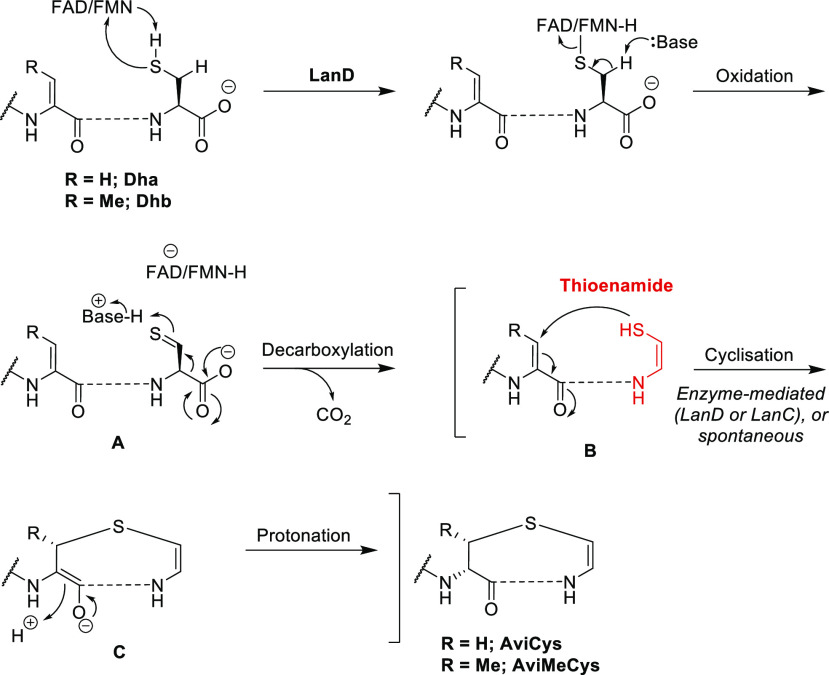
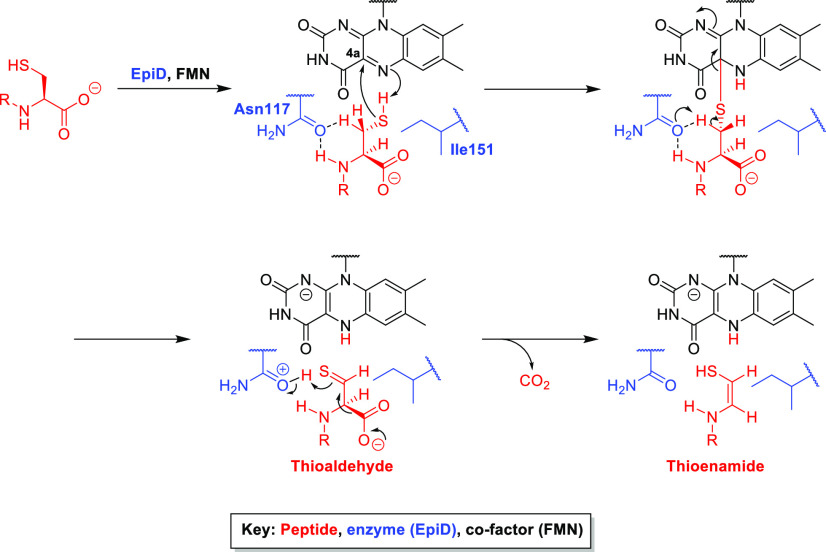
For the biosynthesis of Avi(Me)Cys-containing peptides, the C-terminal Cys-sulfhydryl of the LanA prepeptide undergoes flavin-mediated oxidation to give a corresponding thioaldehyde A (Scheme 1).67,69,70 Spontaneous decarboxylation of the C-terminal carboxylate then occurs, driven by tautomerization of the thioaldehyde to enethiol B to install the Cα–Cβ double bond of Avi(Me)Cys (Scheme 1).67,69,70 Thioether cyclization then occurs by nucleophilic attack of the sulfhydryl upon the β-carbon of an α,β-unsaturated amino acid (Dha or Dhb), and the resulting enolate C is protonated to afford the (2S)-AviCys or (2S,3S)-AviMeCys moiety (Scheme 1).15,67 To date, this thioether cyclization step during Avi(Me)Cys formation is not fully understood and may be enzyme-catalyzed (e.g., by LanC or LanD) or a spontaneous process.15,23
Scheme 1. Oxidation and Decarboxylation of a C-Terminal Cysteine-Bearing Peptide, Catalyzed by a Decarboxylase (LanD)68 (Reactive Thioenamide (Red) Then Undergoes Thiol-Michael Cyclization with an Unsaturated Amino Acid (Dha/Dhb) to Provide the Avi(Me)Cys Unit Following Protonation).
Oxidative decarboxylation of C-terminal cysteine peptides by the epidermin decarboxylase, EpiD, has been reported to occur in the absence of a Michael acceptor residue (Dha/Dhb), giving the linear thioenamide-containing products.71 Kupke et al.70 demonstrated the decarboxylase activity of EpiD on C-terminal Cys-sulfhydryl substrates and the absence of activity on S-alkylated peptides, confirming that the decarboxylation step occurs prior to cyclization.67 Furthermore, EpiD has been successfully applied to the formation of Avi(Me)Cys linkages on non-epidermin peptides from 4 to 52 amino acids in length with the C-terminal tripeptide sequence AA1-AA2-Cys, where AA1 = Val, Ile, Leu, Met, Phe, Tyr, or Trp and AA2 = Ala, Ser, Val, Thr, Cys, Ile, or Leu.67,71 However, the leader peptide sequence is seemingly nonessential for the enzymatic activity of EpiD.71 Combined with its broad substrate scope, the use of EpiD for the decarboxylation of synthetic peptides may be a viable strategy for the efficient production of bioactive compounds.67
The X-ray crystal structures of the LanD enzymes EpiD70 and MrsD (mersacidin decarboxylase)69 have been determined. The X-ray crystal structure of EpiD complexed with a C-terminal cysteine containing a pentapeptide substrate revealed a contact between the FMN cofactor and the Cys-sulfhydryl but not the Cys-carboxylate, supporting the idea that decarboxylation occurs indirectly as a result of sulfhydryl modification.70 Obtaining this X-ray crystal structure was achieved by mutation of EpiD-His67 to an asparagine (Asn) residue, which abolished decarboxylase activity without preventing substrate binding.70 Therefore, His67 of EpiD plays an essential role in the oxidative decarboxylation of C-terminal cysteines, proposed to occur by increasing acidity of the Cys-sulfhydryl proton.68−70
In the proposed mechanism (Scheme 2), nucleophilic attack of the Cys-sulfhydryl on the C4a position of flavin initiates the oxidative decarboxylation reaction of a C-terminal cysteine.67,70 H-bonding interactions between the side-chain carbonyl of Asn117 in EpiD and the peptide Cys-Cβ protons facilitate Cβ-deprotonation to afford the thioaldehyde intermediate.70 The subsequent decarboxylation step is favored over Cα-deprotonation as the Cα-proton is blocked by Ile151 of EpiD (Scheme 2), and decarboxylation to give the C–C double bond of Avi(Me)Cys is further promoted through exposure of the Cys-carboxylate to solvent, allowing carbon dioxide to exit the active site.70
Scheme 2. Postulated Mechanism of Oxidation and Subsequent Decarboxylation Catalyzed by EpiD70 (Peptide Substrate Is Indicated in Red, Enzyme (EpiD) in Blue, and Enzyme Cofactor (FMN) in Black).
The observed interaction of Asn117 with the Cys-Cβ protons provides insight to the selective formation of the (Z)-alkene in Avi(Me)Cys, as these contacts enable rotation of the Cys-sulfhydryl toward a syn conformation with respect to the Cys-Nα (Scheme 2).70 When the peptide substrate binds in the enzyme active site, the C-terminal Cys sits above the flavin re face, with the sulfhydryl orientated downward toward flavin-C4a and -N5 (Scheme 2).70 Formation of thioaldehyde then positions the sulfur atom toward the protonated Asn117 oxygen, and decarboxylation generates the (Z)-alkene (Scheme 2).70 The role of EpiD Asn117 in Avi(Me)Cys formation is further supported by the presence of Asn residues at structurally similar sites across other LanD enzymes, such as Asn125 in MrsD.69
2.2. Lipolanthines
In 2018, Wiebach et al. isolated microvionin (Figure 7) from a culture of Microbacterium arborescens. This peptide natural product exhibits an unusual triaminodicarboxylic acid moiety named avionin, as well as an Avi(Me)Cys macrocycle and N-terminal guanidino fatty acid.50,72 It displayed potent antibacterial activity against both MRSA and Streptococcus pneumoniae, which makes it a promising candidate for the treatment of antibiotic-resistant bacteria.50 Nocavionin (Figure 7), from Nocardia terpenica, was also isolated by Wiebach et al. in 2018 and contains the same defining structures as microvionin yet with a shortened and unsaturated N-terminal fatty acid.50 In 2020, Kozakai et al. characterized eight new lipolanthines and coined the name goadvionins.51 Bioactivity assays established antibacterial activity of the goadvionins against Gram-positive bacteria, the exact mechanism of which is currently unknown.51
Previously understood to be a family of their own, lipolanthines have since been classified as class III lanthipeptides, owing to the utilization of class-III-type lanthipeptide synthetases in biosynthesis. Formation of the Avi(Me)Cys unit in microvionin is catalyzed by FAD-dependent cysteine decarboxylases, MicD and MicKC, which resemble class III lanthipeptide-modifying enzymes.50,73,74 Mechanistic studies show both enzymes function in a mutual regulatory manner, wherein the activity of MicKC is enhanced by MicD through substrate binding.73
3.0. Linaridins
In 1993, Komiyama et al. isolated a new peptide natural product from Streptomyces sp. OH-4156 called cypemycin, displaying antibacterial activity against Micrococcus luteus and cytocidal activity against mouse P388 leukemia cells.39 Cypemycin possesses four Dhb residues, two l-allo-isoleucines, an Nα-dimethylated N-terminal alanine, and an AviCys ring at the C-terminus (Figure 5).39 Due to the presence of the Dhb and AviCys moieties, cypemycin was initially classified as a lanthipeptide until in 2010, when Claesen and Bibb identified the cypemycin biosynthetic gene cluster which revealed that there was no lanthipeptide dehydratase-like enzyme encoded. Therefore, cypemycin was reclassified as a linaridin.40,75
Linaridin natural products are a small but growing family of linear dehydrated peptides, and to date, there are 10 characterized members: cypemycin, grisemycin, legonaridin, salinipeptin A–D, mononaridin, and pegvadin A and B.39,41,42,76−78 Of these 10, six contain the Avi(Me)Cys moiety (Figure 5); however, only two of them have shown antimicrobial and anticancer properties, cypemycin and salinipeptin A, with the latter displaying modest levels of activity against Streptococcus pyogenes, U87 glioblastoma and HCT-116 colon carcinoma cells.39,42 The mechanism of their antimicrobial activity is unclear, but for cypemycin, it is postulated that it inserts itself into the bacterial membranes and forms pores, thereby causing cell lysis.41
Biochemically, incorporation of the Avi(Me)Cys moiety into these natural products is achieved via a decarboxylation using a homo-oligomeric flavin-containing Cys decarboxylase known as LinD followed by Michael-type addition of the thioenol to a Dha residue formed prior to decarboxylation.79 Unlike most other Avi(Me)Cys-containing peptides, the Dha intermediate is derived from a Cys rather than a Ser.80 The cyclization step to form Avi(Me)Cys is still unclear for linaridins as their biosynthetic gene clusters (BGCs) do not encode any cyclase enzymes like those found in lanthipeptide and lipolanthines (LanC).80 It is thought that a LinD enzyme (e.g., CypD) catalyzes both decarboxylation and cyclization, although further investigation is required.80
4.0. Thioamitides
Thioviridamide (Figure 6), the first thioamitide, was characterized by Hayakawa et al. in 2005.5 This unusual natural peptide was isolated from Streptomyces olivoviridis and was revealed to contain a 2-hydroxy-2-methyl-4-oxopentanoyl (HMOP) group, a β-hydroxy-N1,N3-dimethylhistidinium, and an AviCys ring.5 However, the most unique part of the structure was that it contained thioamide bonds in the peptide backbone.2 This natural product displayed potent cytotoxic properties against rat fibroblasts transformed with adenovirus oncogenes with an IC50 = 3.9 ng/mL.5 Since then, nine other Avi(Me)Cys-containing thioamitides have been discovered, with all of them displaying anticancer activity across multiple cell lines.5,45,46,49,81 Thioviridamide acts as an apoptosis inducer by targeting the F1F0-ATP synthase and inducing the integrated stress response, a cellular process that downregulates protein synthesis in response to internal or environmental stresses such as amino acid deprivation or endoplasmic reticulum stress.82 Thioholgamide A, which was discovered in 2020 by Kiemer et al., also targets the ATP synthase which causes antiproliferative and cytotoxic effects on cancer cells.44 The antiproliferative nature of these compounds is due to the initiation of the integrated stress response which causes the cancer cells to enter a resting state, thereby preventing proliferation before they undergo apoptosis.44
Much like the other Avi(Me)Cys-containing peptide families, formation of the Avi(Me)Cys motif involves the dehydration of serine/threonine residues, cysteine decarboxylation, and Avi(Me)Cys macrocyclization. Apart from that, little is known about the biosynthetic pathway, although it has been postulated that enzymes similar to those found in the class III lanthipeptides (LanKCt) are responsible for forming the Avi(Me)Cys structure. These enzymes dehydrate the precursor peptide and form a complex with the cysteine decarboxylase to produce the Avi(Me)Cys macrocycle.46
5.0. Lanthidins
Lanthidins are the most recently discovered family of Avi(Me)Cys-containing peptides, currently only encompassing one member, cacaoidin (Figure 7).52 Cacaoidin was characterized in 2020 by Ortiz-López et al. and found to contain a dimethylated N-terminus, a lanthionine, an Avi(Me)Cys ring, and a rare O-glycosylated tyrosine residue.52 It was isolated from Streptomyces cacaoi CA-170360 and demonstrated potent antibacterial activity against MRSA and moderate activity against Clostridium difficile.52 Cacaoidin was observed to trigger the induction of the lipid II cycle interfering with the antibiotic response regulator and sensor (LiaRS) bioreceptor, indicating interference with the lipid II biosynthesis cycle.52 This suggests that cacaoidin targets lipid II in the bacterial membrane, resulting in poor structural integrity and lysis. Formation of the Avi(Me)Cys moiety is believed to be facilitated by a protein homologous to the cypemycin decarboxylase CypD.52
6.0. Chemical Synthesis of Avi(Me)Cys Units
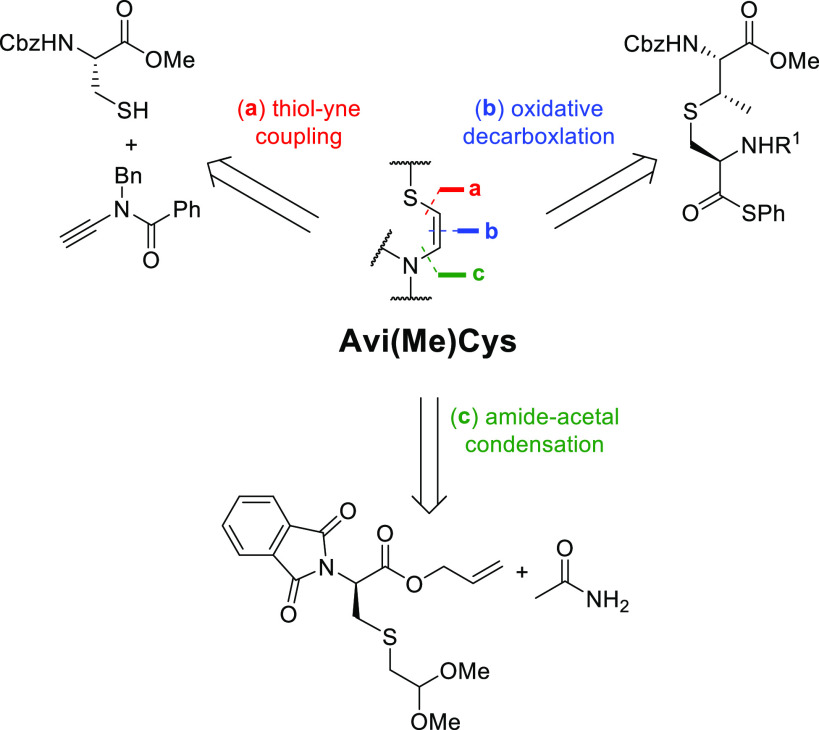
The preparation of multicyclic peptides containing dehydrated residues (Dha, Dhb) and (Me)Lan rings has been researched extensively;83,84 however, the total chemical synthesis of Avi(Me)Cys-containing natural product peptides has remained elusive. Several research groups have investigated different chemical methods for forming the Avi(Me)Cys moiety on building-block-type compounds and peptide fragments including thiol-yne conjugation by Castle et al.,9 oxidative decarbonylation by VanNieuwenhze et al.,10 and condensation of amines with acetals by Taylor et al. (Scheme 3).11,12 However, each route only generated a low to moderate yield of the Avi(Me)Cys and in E/Z mixtures. Further modification of these peptides is yet to be reported, perhaps due to the chemical instability of the Avi(Me)Cys unit. Therefore, a total chemical synthesis of any bioactive Avi(Me)Cys-containing natural product has yet to be reported.
Scheme 3. Retrosynthetic Scheme Depicting the Different Avi(Me)Cys Synthesis Pathways Reported to Date9−13.
6.1. Radical Thiol-Yne Couplings
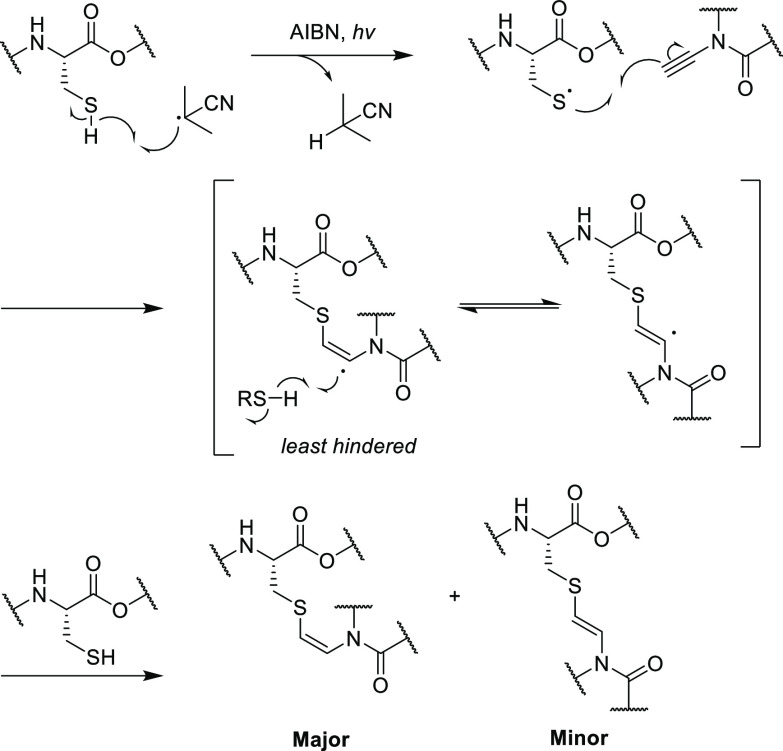
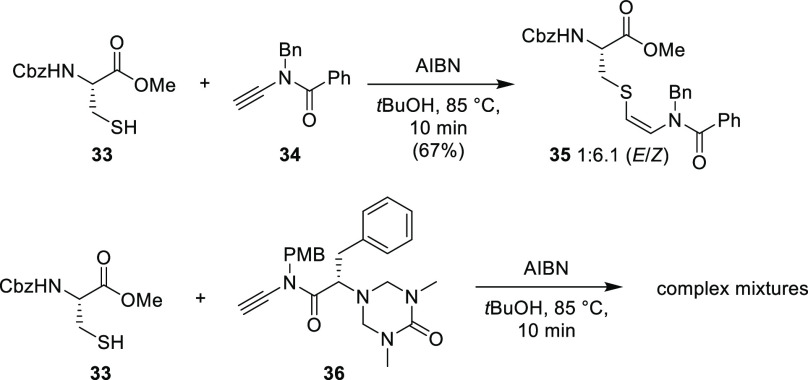
In a study toward the synthesis of thioviridamide, Castle et al. prepared building block analogues of AviCys using a radical thiol-yne reaction, where backbone-protected cysteine derivatives were coupled with various ynamides.9 In this synthesis, the radical initiator, 2,2′-azobis(2-methylproionitrile) (AIBN), first abstracts the sulfhydryl proton to yield a thiyl radical, which propagates across the electron-rich alkyne (Scheme 4). The resulting carbon-centered alkene radical is then quenched by hydrogen atom transfer (HAT) from a second equivalent of the sulfhydryl. Preliminary studies suggested the (Z)-alkene dominates as the kinetically favored product, due to rapid quenching of the alkene radical by HAT on the least hindered side (Scheme 4). Under these conditions, the reaction proceeded well with small substrates, successfully affording 35 as the desired isomer in good yield (Scheme 5).9 However, when these methods were applied to more complex and potentially useful ynamide building blocks, such as 36, no desired product was obtained. Therefore, although this procedure proved to be effective for small molecules, it may not be practical for the installation of AviCys into more complex substrates.
Scheme 4. Postulated Mechanism for Radical Thiol-Yne Reaction for the Synthesis of an AviCys Derivative by Castle et al.9 (AIBN = 2,2′-Azobis(2-methylpropionitrile)).
Scheme 5. Radical Thiol-Yne Reaction for the Synthesis of an AviCys Derivative and Attempted Radical Thiol-Yne Coupling of Cysteine Derivative with Ynamides by Castle et al.9 (AIBN = 2,2′-Azobis(2-methylpropionitrile), Cbz = Carboxybenzyl, PMB = para-Methoxybenzyl).
6.2. Oxidative Decarboxylation/Decarbonylation
In 2012, VanNieuwenhze et al.10,13 published two back-to-back research articles investigating decarbonylation/decarboxylation strategies toward the synthesis of mersacidin. Their work focused on mimicking the biosynthetic route to Avi(Me)Cys-containing peptides to create building blocks and peptide fragments.
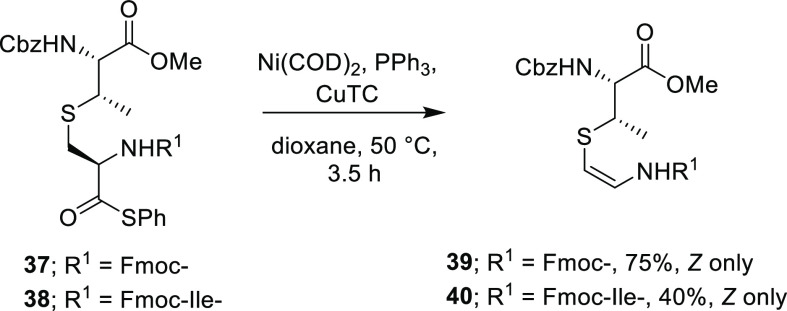
Decarbonylation of model cysteine-containing thioester derivatives afforded the corresponding Avi(Me)Cys derivatives selectively using a Ni(COD)4 and copper(I) thiophene-2-carboxylate (CuTC) catalyst system (Scheme 6). Upon application of the optimized method to building block 37, complete Z-selectivity successfully afforded 39 in 75% yield. However, they found that the yield decreased with increasing substrate complexity, illustrated by the low to moderate yield of 40 from 38.10 Furthermore, incorporation of Avi(Me)Cys-containing amino acids such as 39 or 40 into a peptide has not yet been reported, suggesting that a building block approach may not be viable for the synthesis of Avi(Me)Cys-containing peptides.
Scheme 6. Decarbonylation of Thioesters to Give AviMeCys Derivatives and Building Blocks10 (Ni(COD)2 = Bis(1,5-cyclooctadiene)nickel(0), CuTC = Copper(I) Thiophene-2-carboxylate, Cbz = Carboxybenzyl).
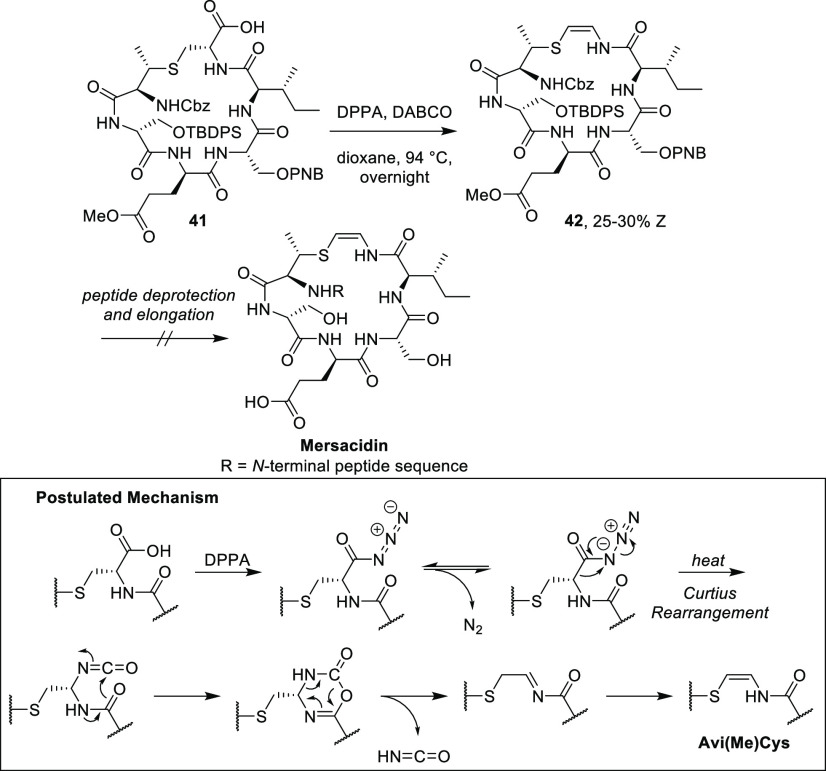
VanNieuwenhze et al.13 then reported a similar approach to form a protected fragment of the C-terminal D-ring fragment of mersacidin. The cyclic substrate 41 was prepared by solution-phase peptide synthesis utilizing protecting groups that are orthogonal to TFA-mediated deprotection of the C-terminal carboxylic acid. The MeLan building block was prepared by literature procedures and incorporated into the growing peptide chain, followed by lactamization to yield 41 (Scheme 7). Preliminary attempts at decarboxlation of 41 to afford 42 using diphenylphosphoryl azide (DPPA) with Et3N in toluene gave promising but low-yielding results.13 Optimization of this route afforded Z-isomer 42 in 25–30% yield using DPPA and 1,4-diazabicyclo[2.2.2]octane (DABCO) in dioxane at reflux overnight (Scheme 7).13
Scheme 7. Oxidative Decarboxylation/Decarbonylation of 41 to Afford 42, the C-Terminal Ring of Mersacidin by VanNieuwenhze et al.13.
Inset: Curtius rearrangement and postulated mechanism of intramolecular isocyanate trapping and decomposition to give the AviMeCys unit. To date, there has been no further publication utilizing this strategy toward the total synthesis of mersacidin or other Avi(Me)Cys-containing peptides. Cbz = carboxybenzyl, TBDPS = tert-butyldiphenylsilyl, PNB = para-nitrobenzyl, DPPA = diphenylphosphoryl azide, DABCO = 1,4-diazabicyclo[2.2.2]octane.
This method employed by VanNieuwenhze et al.13 is presumed to undergo a Curtius rearrangement followed by intramolecular trapping of the isocyanate and collapse of the resulting six-membered ring (Scheme 7, inset). The resulting protected analogue 42 remains the largest Avi(Me)Cys-containing substrate synthesized to date; however, deprotection and/or ligation of this fragment for the total or partial synthesis of mersacidin is yet to be reported.
6.3. Condensation of Amides with Acetals
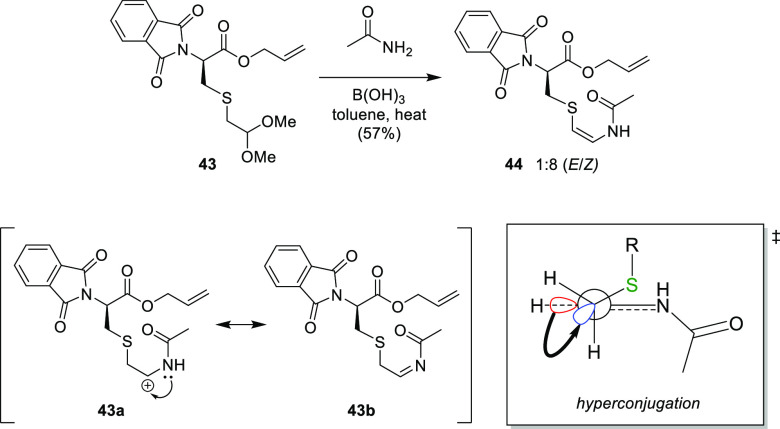
Taylor et al.11 published a unique strategy toward (Z)-thioenamide moieties and AviCys derivatives based on condensation of amides with acetals, followed by β-hydride elimination. The optimized reaction used the mild Lewis acid, B(OH)3, in toluene at reflux to catalyze the condensation of cysteine derivative 43 with acetamide, affording (Z)-AviCys derivative 44 in moderate yield with high stereoselectivity.11
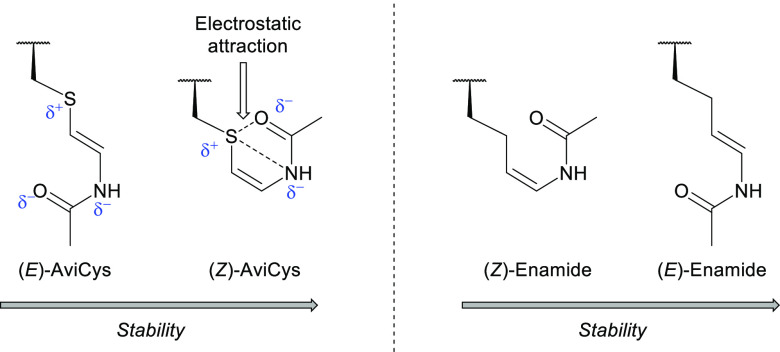
The presence of the sulfur atom directed the E/Z selectivity of the reaction.11 Electronic structure calculations were performed for the reaction of butanal dimethyl acetal with acetamide, with a methylene unit replacing the sulfur atom. The calculations were performed using the Gaussian 09 suite of programs, and it was found that the (E)-enamide was the favored product. Conversely, the Z-isomer was favored in all reactions with sulfur-containing substrates. This strongly supported the idea that the sulfur atom was favoring the formation of the desired Z-isomer; hence computational studies were carried out. By determining the relative energies of each reagent, transition state, and product, it was found that C–C bond rotation occurs, leading to a defined transition state exhibiting an s-cis orientation of the heteroatom substituents (43a and 43b, Scheme 8, inset).11 This transition state has a stabilization energy of 4.04 kcal mol–1 associated with the hyperconjugative donation from the σ-orbital of the adjacent C–H bond (red) into the σ*-orbital of the C–S bond (blue) (Scheme 8).11 This does not occur with substrates lacking sulfur, as the intermediate cation loses a proton resulting in the rapid formation of the (E)-enamide.11 This was consistent with their original hypothesis that a rotational barrier to elimination exists, which favors the arrangement of the electronegative substituents to maximize hyperconjugative stabilization.11 Additionally, their calculations found that the sulfur atom of the final product possesses a partial positive charge enabling electrostatic attraction with both the oxygen and the nitrogen, thus stabilizing the Z- over the E-isomer by 1.8 kcal mol–1 (Figure 9).11
Scheme 8. Synthesis of AviCys Derivatives via Condensation of Acetamide upon Acetal 43 in the Presence of a Mild Lewis Acid11.
Inset: (left) transition state 43a and 43b occurring with sulfur-containing substrates; (right) illustration of hyperconjugation occurring with sulfur-containing substrates leading to the Z-isomer 44.
Figure 9.
E- and Z-isomers of AviCys derivatives and enamides prepared by Taylor et al.11 Left: Due to an electrostatic interaction between the sulfur and both the nitrogen and oxygen atoms, the (Z)-AviCys is thermodynamically more stable than the corresponding (E)-AviCys.11 Right: Synthesis of enamides (i.e., substrates lacking a sulfur atom) using the same methods as used for AviCys formation favor the E-isomer.11 Without an electrostatic interaction stabilizing the Z-configuration, the (E)-enamide predominates as it is less sterically hindered than the (Z)-enamide.
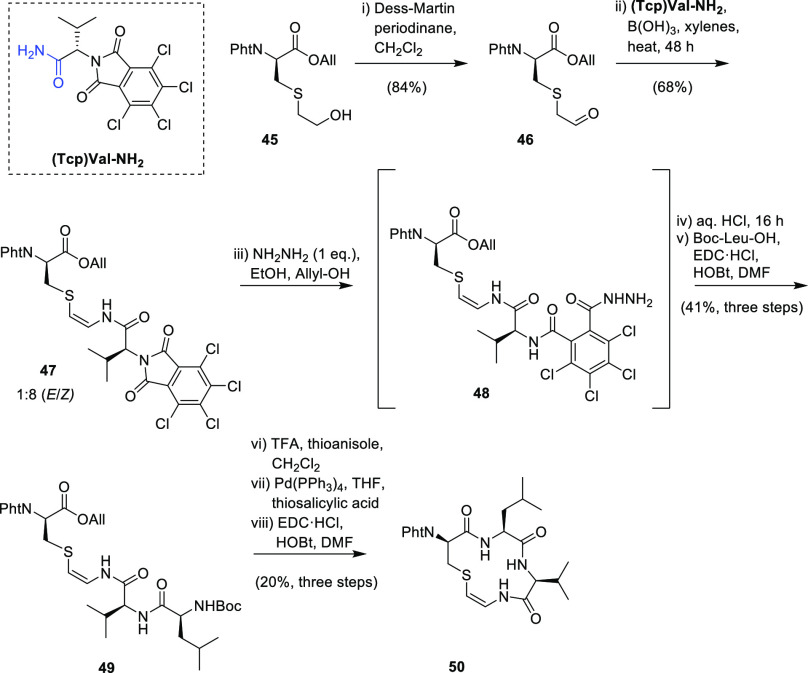
Recently, Taylor et al. successfully applied this methodology to the synthesis of the AviCys C-terminal macrocycle of all known members of the linaridin family (Scheme 9).12 First, the primary alcohol of 45 was oxidized to an aldehyde with Dess–Martin periodinane to afford aldehyde 46 in high yield (84%).12 Condensation of 46 with (Tcp)Val-NH2 (Tcp = 3,4,5,6-tetrachlorophthalimide) in the presence of the Lewis acid B(OH)3 afforded a 1:8 E/Z mixture of isomers of the AviCys-containing compound 47 in good yield (68%).12 Previous work had demonstrated that the Tcp group in 47 could be removed in the presence of a Pht (Pht = phthalimide) group using ethylenediamine; however, for Tcp removal of 47, this led to complex mixtures.12 It was then found that treatment of 47 with 1 equiv of hydrazine led to a single product: the ring-opened acyl hydrazide intermediate 48, which was hydrolyzed in aqueous HCl to afford a free amine.12 Boc-Leu-OH was then coupled using EDC·HCl and HOBt to afford 49 (Scheme 9).
Scheme 9. Synthesis of the AviCys-Containing Ring of Cypemycin via Condensation of Aldehyde 46 with Amide (Tcp)Val-NH2, Followed by Elongation of the Peptide Chain and Lactamization to Give 50 in 4.6% Yield from 46(12) (Tcp = 3,4,5,6-Tetrachlorophthalimide, Pht = Phthalimide).
The final step was deprotection of the N- and C-termini with required modifications to the standard reaction conditions due to the sensitivity of the thioenamide.12 The N-terminal deprotection of 49 was achieved using TFA and thioanisole. Thioanisole was used instead of the more common triethylsilane due to partial reduction of the thioenamide double bond when triethylsilane was used.12 For deprotection of the C-terminus, initial deallylation conditions using palladium(0) with barbituric acid as an allyl acceptor were unsuccessful and resulted in oxidation of the thioenamide sulfur to the corresponding sulfoxide.12 Deallylation of the C-terminus was then successfully achieved using thiosalicylic acid as both the allyl acceptor and sacrificial reductant.12 Finally, lactamization with EDC/HOBt afforded the cyclic AviCys-containing peptide 50 as a single isomer (4.6% overall yield from 45) (Scheme 9).12
7.0. Conclusion
RiPPs are a large class of diverse natural peptides that contain a number of PTMs, including the structurally distinct Avi(Me)Cys macrocycle. Nearly all peptides that contain this unusual motif exhibit antimicrobial or anticancer activity, suggesting a biological importance of this scaffold. This is further highlighted by the fact that the biosynthesis of Avi(Me)Cys is conserved across all families, involving the dehydration of serine/threonine or cysteine residues, cysteine decarboxylation, and final macrocyclization to fashion the Avi(Me)Cys unit.
Despite the clinical potential these of natural products, difficulties in synthesizing this important structural moiety hinder drug development. Several synthetic strategies toward Avi(Me)Cys moieties have been attempted; however, the application of these methods to the total synthesis of an Avi(Me)Cys-containing peptide is yet to be reported. Nevertheless, the successes of VanNieuwenhze et al.13 and Taylor et al.12 in the syntheses of the Avi(Me)Cys-containing rings of mersacidin and the linaridins, respectively, offer encouragement.
Glossary
Abbreviations
- AIBN
2,2′-azobis(2-methylproionitrile)
- AMP
antimicrobial peptide
- AviCys
S-[(Z)-2-aminovinyl]-d-cysteine
- AviMeCys
(2S,3S)-S-[(Z)-2-aminovinyl]-3-methyl-d-cysteine
- Cbz
carboxybenzyl
- CuMeSal
copper(I) 3-methylsalicylate
- CuTC
copper(I) thiophene-2-carboxylate
- DABCO
1,4 diazabicyclo[2.2.2]octane
- Dha
2,3-didehydroalanine
- Dhb
(Z)-2,3-didehydrobutyrine
- DPPA
diphenylphosphoryl azide
- FAD
flavin adenine dinucleotide
- FMN
flavin mononucleotide
- GlcNAc
N-acetylglucosamine
- HAT
hydrogen atom transfer
- HFCD
homo-oligomeric flavoprotein cysteine decarboxylase
- HMOP
2-hydroxy-2-methyl-4-oxopentanoyl
- Lan
lanthionine
- LiaRS
lipid II cycle interfering antibiotic response regulator and sensor
- (Me)Lan
β-methyllanthionine
- MRSA
methicillin-resistant Staphylococcus aureus
- Ni(COD)2
bis(1,5-cyclooctadiene)nickel(0)
- PMB
para-methoxybenzyl
- PNB
para-nitrobenzyl
- RiPP
post-translationally modified peptide
- TBDPS
tert-butyldiphenylsilyl
- Tcp
3,4,5,6-tetrachlorophthalimide
- VISA
vancomycin-intermediate Staphylococcus aureus
- VRE
vancomycin-resistant Enterococcus faecalis
Author Present Address
∥ Department of Chemistry, University of Zurich, Winterthurerstrasse 190, 8057 Zürich, Switzerland
Author Contributions
⊥ E.S.G.-M. and E.T.W. contributed equally to this work.
The authors wish to acknowledge the Ministry of Business, Innovation and Employment (MBIE Endeavor Grant No. UOAX2010) for generous financial support and the Maurice Wilkins Centre for Molecular Biodiscovery.
The authors declare no competing financial interest.
References
- Cao L.; Do T.; Link A. J. Mechanisms of Action of Ribosomally Synthesized and Posttranslationally Modified Peptides (RiPPs). J. Ind. Microbiol. Biotechnol. 2021, 10.1093/jimb/kuab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalban-Lopez M.; Scott T. A.; Ramesh S.; Rahman I. R.; van Heel A. J.; Viel J. H.; Bandarian V.; Dittmann E.; Genilloud O.; Goto Y.; Grande Burgos M. J.; Hill C.; Kim S.; Koehnke J.; Latham J. A.; Link A. J.; Martinez B.; Nair S. K.; Nicolet Y.; Rebuffat S.; Sahl H.-G.; Sareen D.; Schmidt E. W.; Schmitt L.; Severinov K.; Sussmuth R. D.; Truman A. W.; Wang H.; Weng J.-K.; van Wezel G. P.; Zhang Q.; Zhong J.; Piel J.; Mitchell D. A.; Kuipers O. P.; van der Donk W. A. New Developments in RiPP Discovery, Enzymology and Engineering. Nat. Prod. Rep. 2021, 38 (1), 130–239. 10.1039/D0NP00027B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidler C. M.; Kick L. M.; Schneider S. Ribosomal Peptides and Small Proteins on the Rise. ChemBioChem 2019, 20 (12), 1479–1486. 10.1002/cbic.201800715. [DOI] [PubMed] [Google Scholar]
- Dischinger J.; Basi Chipalu S.; Bierbaum G. Lantibiotics: Promising Candidates for Future Applications in Health Care. Int. J. Med. Microbiol. 2014, 304 (1), 51–62. 10.1016/j.ijmm.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Hayakawa Y.; Sasaki K.; Adachi H.; Furihata K.; Nagai K.; Shin-ya K. Thioviridamide, a Novel Apoptosis Inducer in Transformed Cells from Streptomyces Olivoviridis. J. Antibiot. 2006, 59 (1), 1–5. 10.1038/ja.2006.1. [DOI] [PubMed] [Google Scholar]
- Ongey E. L.; Yassi H.; Pflugmacher S.; Neubauer P. Pharmacological and Pharmacokinetic Properties of Lanthipeptides Undergoing Clinical Studies. Biotechnol. Lett. 2017, 39 (4), 473–482. 10.1007/s10529-016-2279-9. [DOI] [PubMed] [Google Scholar]
- Ongey E. L.; Neubauer P. Lanthipeptides: Chemical Synthesis versus in Vivo Biosynthesis as Tools for Pharmaceutical Production. Microb. Cell Fact. 2016, 15 (1), 97. 10.1186/s12934-016-0502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon Rodriguez L. M.; Williams E. T.; Brimble M. A. Chemical Synthesis of Bioactive Naturally Derived Cyclic Peptides Containing Ene-Like Rigidifying Motifs. Chem. - Eur. J. 2018, 24 (68), 17869–17880. 10.1002/chem.201802533. [DOI] [PubMed] [Google Scholar]
- Banerjee B.; Litvinov D. N.; Kang J.; Bettale J. D.; Castle S. L. Stereoselective Additions of Thiyl Radicals to Terminal Ynamides. Org. Lett. 2010, 12 (11), 2650–2652. 10.1021/ol1008679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Reynaga P.; Carrillo A. K.; VanNieuwenhze M. S. Decarbonylative Approach to the Synthesis of Enamides from Amino Acids: Stereoselective Synthesis of the (Z)-Aminovinyl-d-Cysteine Unit of Mersacidin. Org. Lett. 2012, 14 (4), 1030–1033. 10.1021/ol203399x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz J. A.; Subasinghege Don V.; Kumar R.; Taylor C. M. Influence of Sulfur on Acid-Mediated Enamide Formation. Org. Lett. 2017, 19 (19), 5146–5149. 10.1021/acs.orglett.7b02432. [DOI] [PubMed] [Google Scholar]
- Lutz J. A.; Taylor C. M. Synthesis of the Aminovinylcysteine-Containing C-Terminal Macrocycle of the Linaridins. Org. Lett. 2020, 22 (5), 1874–1877. 10.1021/acs.orglett.0c00218. [DOI] [PubMed] [Google Scholar]
- Carrillo A. K.; VanNieuwenhze M. S. Synthesis of the AviMeCys-Containing D-Ring of Mersacidin. Org. Lett. 2012, 14 (4), 1034–1037. 10.1021/ol2034806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers L. A.; Whittier E. O. Limiting Factors in the Lactic Fermentation. J. Bacteriol. 1928, 16 (4), 211–229. 10.1128/jb.16.4.211-229.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repka L. M.; Chekan J. R.; Nair S. K.; van der Donk W. A. Mechanistic Understanding of Lanthipeptide Biosynthetic Enzymes. Chem. Rev. 2017, 117 (8), 5457–5520. 10.1021/acs.chemrev.6b00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K.-D.; Fischbach M. A.; Garavelli J. S.; Göransson U.; et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013, 30 (1), 108–160. 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman J. D.; Novák J.; Sagura E.; Gutierrez J. A.; Brooks T. A.; Crowley P. J.; Hess M.; Azizi A.; Leung K.-P.; Cvitkovitch D.; Bleiweis A. S. Genetic and Biochemical Analysis of Mutacin 1140, a Lantibiotic from Streptococcus Mutans. Infect. Immun. 1998, 66 (6), 2743–2749. 10.1128/IAI.66.6.2743-2749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglione F.; Lazzarini A.; Carrano L.; Corti E.; Ciciliato I.; Gastaldo L.; Candiani P.; Losi D.; Marinelli F.; Selva E.; Parenti F. Determining the Structure and Mode of Action of Microbisporicin, a Potent Lantibiotic Active Against Multiresistant Pathogens. Chem. Biol. 2008, 15 (1), 22–31. 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Kellner R.; Jung G.; Hörner T.; Zähner H.; Schnell N.; Entian K.-D.; Götz F. Gallidermin: A New Lanthionine-Containing Polypeptide Antibiotic. Eur. J. Biochem. 1988, 177 (1), 53–59. 10.1111/j.1432-1033.1988.tb14344.x-i2. [DOI] [PubMed] [Google Scholar]
- Allgaier H.; Jung G.; Werner R. G.; Schneider U.; Zähner H. Epidermin: Sequencing of a Heterodet Tetracyclic 21-Peptide Amide Antibiotic. Eur. J. Biochem. 1986, 160 (1), 9–22. 10.1111/j.1432-1033.1986.tb09933.x. [DOI] [PubMed] [Google Scholar]
- Barbosa J.; Caetano T.; Mendo S. Class I and Class II Lanthipeptides Produced by Bacillus Spp. J. Nat. Prod. 2015, 78 (11), 2850–2866. 10.1021/np500424y. [DOI] [PubMed] [Google Scholar]
- Chatterjee S.; Chatterjee D. K.; Jani R. H.; Blumbach J.; Ganguli B. N.; Klesel N.; Limbert M.; Seibert G. Mersacidin, a New Antibiotic from Bacillius. J. Antibiot. 1992, 45 (6), 839–845. 10.7164/antibiotics.45.839. [DOI] [PubMed] [Google Scholar]
- Xu M.; Zhang F.; Cheng Z.; Bashiri G.; Wang J.; Hong J.; Wang Y.; Xu L.; Chen X.; Huang S.-X.; Lin S.; Deng Z.; Tao M. Functional Genome Mining Reveals a Class V Lanthipeptide Containing a D-Amino Acid Introduced by an F420H2-Dependent Reductase. Angew. Chem., Int. Ed. 2020, 59 (41), 18029–18035. 10.1002/anie.202008035. [DOI] [PubMed] [Google Scholar]
- Shi J.; Ma J.-Q.; Wang Y.-C.; Xu Z.-F.; Zhang B.; Jiao R.-H.; Tan R.-X.; Ge H.-M. Discovery of Daspyromycins A and B, 2-Aminovinyl-Cysteine Containing Lanthipeptides, through a Genomics-Based Approach. Chin. Chem. Lett. 2021, 10.1016/j.cclet.2021.06.010. [DOI] [Google Scholar]
- Geng M.; Smith L. Improving the Attrition Rate of Lanthipeptide Discovery for Commercial Applications. Expert Opin. Drug Discovery 2018, 13 (2), 155–167. 10.1080/17460441.2018.1410137. [DOI] [PubMed] [Google Scholar]
- Bakhtiary A.; Cochrane S. A.; Mercier P.; McKay R. T.; Miskolzie M.; Sit C. S.; Vederas J. C. Insights into the Mechanism of Action of the Two-Peptide Lantibiotic Lacticin 3147. J. Am. Chem. Soc. 2017, 139 (49), 17803–17810. 10.1021/jacs.7b04728. [DOI] [PubMed] [Google Scholar]
- Breukink E.; de Kruijff B. Lipid II as a Target for Antibiotics. Nat. Rev. Drug Discovery 2006, 5 (4), 321–323. 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- Brötz H.; Josten M.; Wiedemann I.; Schneider U.; Götz F.; Bierbaum G.; Sahl H.-G. Role of Lipid-Bound Peptidoglycan Precursors in the Formation of Pores by Nisin, Epidermin and Other Lantibiotics. Mol. Microbiol. 1998, 30 (2), 317–327. 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- Hsu S.-T. D.; Breukink E.; Tischenko E.; Lutters M. A. G.; de Kruijff B.; Kaptein R.; Bonvin A. M. J. J.; van Nuland N. A. J. The Nisin–Lipid II Complex Reveals a Pyrophosphate Cage That Provides a Blueprint for Novel Antibiotics. Nat. Struct. Mol. Biol. 2004, 11 (10), 963–967. 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- Dickman R.; Mitchell S. A.; Figueiredo A. M.; Hansen D. F.; Tabor A. B. Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B. J. Org. Chem. 2019, 84 (18), 11493–11512. 10.1021/acs.joc.9b01253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhrel R.; Bhattarai N.; Baral P.; Gerstman B. S.; Park J. H.; Handfield M.; Chapagain P. Molecular Mechanisms of Pore Formation and Membrane Disruption by the Antimicrobial Lantibiotic Peptide Mutacin 1140. Phys. Chem. Chem. Phys. 2019, 21 (23), 12530–12539. 10.1039/C9CP01558B. [DOI] [PubMed] [Google Scholar]
- Böttiger T.; Schneider T.; Martínez B.; Sahl H.-G.; Wiedemann I. Influence of Ca2+ Ions on the Activity of Lantibiotics Containing a Mersacidin-Like Lipid II Binding Motif. Appl. Environ. Microbiol. 2009, 75 (13), 4427–4434. 10.1128/AEM.00262-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna J. C. J.; Williams D. H. The Structure and Mode of Action of Glycopeptide Antibiotics of the Vancomycin Group. Annu. Rev. Microbiol. 1984, 38 (1), 339–357. 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- Courvalin P. Vancomycin Resistance in Gram-Positive Cocci. Clin. Infect. Dis. 2006, 42 (Supplement_1), S25–S34. 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- Gardete S.; Tomasz A. Mechanisms of Vancomycin Resistance in Staphylococcus Aureus. J. Clin. Invest. 2014, 124 (7), 2836–2840. 10.1172/JCI68834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch D.; Müller A.; Schneider T.; Kohl B.; Wenzel M.; Bandow J. E.; Maffioli S.; Sosio M.; Donadio S.; Wimmer R.; Sahl H.-G. The Lantibiotic NAI-107 Binds to Bactoprenol-Bound Cell Wall Precursors and Impairs Membrane Functions*. J. Biol. Chem. 2014, 289 (17), 12063–12076. 10.1074/jbc.M113.537449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhalili R. N.; Canbäck B. Identification of Putative Novel Class-I Lanthipeptides in Firmicutes: A Combinatorial In Silico Analysis Approach Performed on Genome Sequenced Bacteria and a Close Inspection of Z-Geobacillin Lanthipeptide Biosynthesis Gene Cluster of the Thermophilic Geobacillus Sp. Strain ZGt-1. Int. J. Mol. Sci. 2018, 19 (9), 2650. 10.3390/ijms19092650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Ma J.-Q.; Wang Y.-C.; Xu Z.-F.; Zhang B.; Jiao R.-H.; Tan R.-X.; Ge H.-M. Discovery of Daspyromycins A and B, 2-Aminovinyl-Cysteine Containing Lanthipeptides, through a Genomics-Based Approach. Chin. Chem. Lett. 2021, 10.1016/j.cclet.2021.06.010. [DOI] [Google Scholar]
- Komiyama K.; Otoguro K.; Segawa T.; Shiomi K.; Yang H.; Takahashi Y.; Hayashi M.; Oxani T.; Omura S. A New Antibiotic, Cypemycin Taxonomy, Fermentation, Isolation and Biological Characteristics. J. Antibiot. 1993, 46 (11), 1666–1671. 10.7164/antibiotics.46.1666. [DOI] [PubMed] [Google Scholar]
- Minami Y.; Yoshida K.; Azuma R.; Urakawa A.; Kawauchi T.; Otani T.; Komiyama K.; O̅mura S. Structure of Cypemycin, a New Peptide Antibiotic. Tetrahedron Lett. 1994, 35 (43), 8001–8004. 10.1016/0040-4039(94)80033-2. [DOI] [Google Scholar]
- Claesen J.; Bibb M. J. Biosynthesis and Regulation of Grisemycin, a New Member of the Linaridin Family of Ribosomally Synthesized Peptides Produced by Streptomyces Griseus IFO 13350. J. Bacteriol. 2011, 193 (10), 2510–2516. 10.1128/JB.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Z.; Winter J. M.; Kauffman C. A.; Yang I.; Fenical W. Salinipeptins: Integrated Genomic and Chemical Approaches Reveal Unusual d-Amino Acid-Containing Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs) from a Great Salt Lake Streptomyces Sp. ACS Chem. Biol. 2019, 14 (3), 415–425. 10.1021/acschembio.8b01058. [DOI] [PubMed] [Google Scholar]
- Li Y.; Liu J.; Tang H.; Qiu Y.; Chen D.; Liu W. Discovery of New Thioviridamide-Like Compounds with Antitumor Activities. Chin. J. Chem. 2019, 37 (10), 1015–1020. 10.1002/cjoc.201900235. [DOI] [Google Scholar]
- Dahlem C.; Siow W. X.; Lopatniuk M.; Tse W. K. F.; Kessler S. M.; Kirsch S. H.; Hoppstädter J.; Vollmar A. M.; Müller R.; Luzhetskyy A.; Bartel K.; Kiemer A. K. Thioholgamide A, a New Anti-Proliferative Anti-Tumor Agent, Modulates Macrophage Polarization and Metabolism. Cancers 2020, 12 (5), 1288. 10.3390/cancers12051288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattaruolo L.; Lacret R.; Cappello A. R.; Truman A. W. A Genomics-Based Approach Identifies a Thioviridamide-Like Compound with Selective Anticancer Activity. ACS Chem. Biol. 2017, 12 (11), 2815–2822. 10.1021/acschembio.7b00677. [DOI] [PubMed] [Google Scholar]
- Lu J.; Wu Y.; Li J.; Li Y.; Zhang Y.; Bai Z.; Zheng J.; Zhu J.; Wang H.. Lanthipeptide Synthetases Participate the Biosynthesis of 2-Aminovinyl-Cysteine Motifs in Thioamitides bioRxiv 2020; https://doi.org/10.1101/2020.08.21.260323 [Google Scholar]
- Frattaruolo L.; Fiorillo M.; Brindisi M.; Curcio R.; Dolce V.; Lacret R.; Truman A. W.; Sotgia F.; Lisanti M. P.; Cappello A. R. Thioalbamide, A Thioamidated Peptide from Amycolatopsis Alba, Affects Tumor Growth and Stemness by Inducing Metabolic Dysfunction and Oxidative Stress. Cells 2019, 8 (11), 1408. 10.3390/cells8111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumikawa M.; Kozone I.; Hashimoto J.; Kagaya N.; Takagi M.; Koiwai H.; Komatsu M.; Fujie M.; Satoh N.; Ikeda H.; Shin-ya K. Novel Thioviridamide Derivative—JBIR-140: Heterologous Expression of the Gene Cluster for Thioviridamide Biosynthesis. J. Antibiot. 2015, 68 (8), 533–536. 10.1038/ja.2015.20. [DOI] [PubMed] [Google Scholar]
- Qiu Y.; Liu J.; Li Y.; Xue Y.; Liu W. Formation of an Aminovinyl-Cysteine Residue in Thioviridamides Occurs through a Path Independent of Known Lanthionine Synthetase Activity. Cell Chem. Biol. 2021, 28 (5), 675–685. 10.1016/j.chembiol.2020.12.016. [DOI] [PubMed] [Google Scholar]
- Wiebach V.; Mainz A.; Siegert M.-A. J.; Jungmann N. A.; Lesquame G.; Tirat S.; Dreux-Zigha A.; Aszodi J.; Le Beller D.; Süssmuth R. D. The Anti-Staphylococcal Lipolanthines Are Ribosomally Synthesized Lipopeptides. Nat. Chem. Biol. 2018, 14 (7), 652–654. 10.1038/s41589-018-0068-6. [DOI] [PubMed] [Google Scholar]
- Kozakai R.; Ono T.; Hoshino S.; Takahashi H.; Katsuyama Y.; Sugai Y.; Ozaki T.; Teramoto K.; Teramoto K.; Tanaka K.; Abe I.; Asamizu S.; Onaka H. Acyltransferase That Catalyses the Condensation of Polyketide and Peptide Moieties of Goadvionin Hybrid Lipopeptides. Nat. Chem. 2020, 12 (9), 869–877. 10.1038/s41557-020-0508-2. [DOI] [PubMed] [Google Scholar]
- Ortiz-López F. J.; Carretero-Molina D.; Sánchez-Hidalgo M.; Martín J.; González I.; Román-Hurtado F.; Cruz M. de la; García-Fernández S.; Reyes F.; Deisinger J. P.; Müller A.; Schneider T.; Genilloud O. Cacaoidin, First Member of the New Lanthidin RiPP Family. Angew. Chem. 2020, 132 (31), 12754–12758. 10.1002/ange.202005187. [DOI] [PubMed] [Google Scholar]
- Kers J. A.; Sharp R. E.; Defusco A. W.; Park J. H.; Xu J.; Pulse M. E.; Weiss W. J.; Handfield M. Mutacin 1140 Lantibiotic Variants Are Efficacious Against Clostridium Difficile Infection. Front. Microbiol. 2018, 9, 415. 10.3389/fmicb.2018.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunati C.; Thomsen T. T.; Gaspari E.; Maffioli S.; Sosio M.; Jabes D.; Løbner-Olesen A.; Donadio S. Expanding the Potential of NAI-107 for Treating Serious ESKAPE Pathogens: Synergistic Combinations against Gram-Negatives and Bactericidal Activity against Non-Dividing Cells. J. Antimicrob. Chemother. 2018, 73 (2), 414–424. 10.1093/jac/dkx395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabés D.; Brunati C.; Candiani G.; Riva S.; Romanó G.; Donadio S. Efficacy of the New Lantibiotic NAI-107 in Experimental Infections Induced by Multidrug-Resistant Gram-Positive Pathogens. Antimicrob. Agents Chemother. 2011, 55 (4), 1671–1676. 10.1128/AAC.01288-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarini A.; Gastaldo L.; Candiani G.; Ciciliato I.; Losi D.; Marinelli F.; Selva E.; Parenti F.. Antibiotic 107891, Its Factors A1 and A2, Pharmaceutically Acceptable Salts and Compositions, and Use Thereof. U.S. Patent Appl. US7319088B2, January 15, 2008.
- Cruz J. C. S.; Iorio M.; Monciardini P.; Simone M.; Brunati C.; Gaspari E.; Maffioli S. I.; Wellington E.; Sosio M.; Donadio S. Brominated Variant of the Lantibiotic NAI-107 with Enhanced Antibacterial Potency. J. Nat. Prod. 2015, 78 (11), 2642–2647. 10.1021/acs.jnatprod.5b00576. [DOI] [PubMed] [Google Scholar]
- Lepak A. J.; Marchillo K.; Craig W. A.; Andes D. R. In Vivo Pharmacokinetics and Pharmacodynamics of the Lantibiotic NAI-107 in a Neutropenic Murine Thigh Infection Model. Antimicrob. Agents Chemother. 2015, 59 (2), 1258–1264. 10.1128/AAC.04444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen T. T.; Mojsoska B.; Cruz J. C. S.; Donadio S.; Jenssen H.; Løbner-Olesen A.; Rewitz K. The Lantibiotic NAI-107 Efficiently Rescues Drosophila Melanogaster from Infection with Methicillin-Resistant Staphylococcus Aureus USA300. Antimicrob. Agents Chemother. 2016, 60 (9), 5427–5436. 10.1128/AAC.02965-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.-T. D.; Breukink E.; Bierbaum G.; Sahl H.-G.; de Kruijff B.; Kaptein R.; van Nuland N. A. J.; Bonvin A. M. J. J. NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles: Conformational Changes Are a Key to Antimicrobial Activity*. J. Biol. Chem. 2003, 278 (15), 13110–13117. 10.1074/jbc.M211144200. [DOI] [PubMed] [Google Scholar]
- Brötz H.; Bierbaum G.; Leopold K.; Reynolds P. E.; Sahl H.-G. The Lantibiotic Mersacidin Inhibits Peptidoglycan Synthesis by Targeting Lipid II. Antimicrob. Agents Chemother. 1998, 42 (1), 154–160. 10.1128/AAC.42.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell C. D.; Hiss J. A.; Hancock R. E. W.; Schneider G. Designing Antimicrobial Peptides: Form Follows Function. Nat. Rev. Drug Discovery 2012, 11 (1), 37–51. 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- Bashiruddin N. K.; Suga H. Construction and Screening of Vast Libraries of Natural Product-like Macrocyclic Peptides Using in Vitro Display Technologies. Curr. Opin. Chem. Biol. 2015, 24, 131–138. 10.1016/j.cbpa.2014.11.011. [DOI] [PubMed] [Google Scholar]
- Niu W. W.; Neu H. C. Activity of Mersacidin, a Novel Peptide, Compared with That of Vancomycin, Teicoplanin, and Daptomycin. Antimicrob. Antimicrob. Agents Chemother. 1991, 35 (5), 998–1000. 10.1128/AAC.35.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszewska D.; Sahl H.-G.; Bierbaum G.; Pag U.; Hynes S. O.; Ljungh Å. Mersacidin Eradicates Methicillin-Resistant Staphylococcus Aureus (MRSA) in a Mouse Rhinitis Model. J. Antimicrob. Chemother. 2004, 54 (3), 648–653. 10.1093/jac/dkh387. [DOI] [PubMed] [Google Scholar]
- Niu W. W.; Neu H. C. Activity of Mersacidin, a Novel Peptide, Compared with That of Vancomycin, Teicoplanin, and Daptomycin. Antimicrob. Antimicrob. Agents Chemother. 1991, 35 (5), 998–1000. 10.1128/AAC.35.5.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit C. S.; Yoganathan S.; Vederas J. C. Biosynthesis of Aminovinyl-Cysteine-Containing Peptides and Its Application in the Production of Potential Drug Candidates. Acc. Chem. Res. 2011, 44 (4), 261–268. 10.1021/ar1001395. [DOI] [PubMed] [Google Scholar]
- Mo T.; Yuan H.; Wang F.; Ma S.; Wang J.; Li T.; Liu G.; Yu S.; Tan X.; Ding W.; Zhang Q. Convergent Evolution of the Cys Decarboxylases Involved in Aminovinyl-Cysteine (AviCys) Biosynthesis. FEBS Lett. 2019, 593 (6), 573–580. 10.1002/1873-3468.13341. [DOI] [PubMed] [Google Scholar]
- Blaesse M.; Kupke T.; Huber R.; Steinbacher S. Structure of MrsD, an FAD-Binding Protein of the HFCD Family. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2003, 59 (8), 1414–1421. 10.1107/S0907444903011831. [DOI] [PubMed] [Google Scholar]
- Blaesse M.; Kupke T.; Huber R.; Steinbacher S. Crystal Structure of the Peptidyl-Cysteine Decarboxylase EpiD Complexed with a Pentapeptide Substrate. EMBO J. 2000, 19 (23), 6299–6310. 10.1093/emboj/19.23.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupke T.; Kempter C.; Jung G.; Götz F. Oxidative Decarboxylation of Peptides Catalyzed by Flavoprotein EpiD: Determination of Substrate Specificity Using Peptide Libraries and Neutral Loss Mass Spectrometry*. J. Biol. Chem. 1995, 270 (19), 11282–11289. 10.1074/jbc.270.19.11282. [DOI] [PubMed] [Google Scholar]
- Hegemann J. D.; Süssmuth R. D. Matters of Class: Coming of Age of Class III and IV Lanthipeptides. RSC Chem. Biol. 2020, 1, 110–127. 10.1039/D0CB00073F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J.; Li Y.; Bai Z.; Lv H.; Wang H. Enzymatic Macrocyclization of Ribosomally Synthesized and Posttranslational Modified Peptides via C–S and C–C Bond Formation. Nat. Prod. Rep. 2021, 38, 981. 10.1039/D0NP00044B. [DOI] [PubMed] [Google Scholar]
- Wiebach V.; Mainz A.; Schnegotzki R.; Siegert M.-A. J.; Hügelland M.; Pliszka N.; Süssmuth R. D. An Amphipathic Alpha-Helix Guides Maturation of the Ribosomally-Synthesized Lipolanthines. Angew. Chem., Int. Ed. 2020, 59 (38), 16777–16785. 10.1002/anie.202003804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesen J.; Bibb M. Genome Mining and Genetic Analysis of Cypemycin Biosynthesis Reveal an Unusual Class of Posttranslationally Modified Peptides. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (37), 16297–16302. 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E. Rateb M.; Zhai Y.; Ehrner E.; M. Rath C.; Wang X.; Tabudravu J.; Ebel R.; Bibb M.; Kyeremeh K.; C. Dorrestein P.; Hong K.; Jaspars M.; Deng H. Legonaridin, a New Member of Linaridin RiPP from a Ghanaian Streptomyces Isolate. Org. Biomol. Chem. 2015, 13 (37), 9585–9592. 10.1039/C5OB01269D. [DOI] [PubMed] [Google Scholar]
- Wang F.; Wei W.; Zhao J.; Mo T.; Wang X.; Huang X.; Ma S.; Wang S.; Deng Z.; Ding W.; Liang Y.; Zhang Q. Genome Mining and Biosynthesis Study of a Type B Linaridin Reveals a Highly Versatile α-N-Methyltransferase. CCS Chem. 2021, 3 (3), 1049–1057. 10.31635/ccschem.020.202000247. [DOI] [Google Scholar]
- Georgiou M. A.; Dommaraju S. R.; Guo X.; Mast D. H.; Mitchell D. A. Bioinformatic and Reactivity-Based Discovery of Linaridins. ACS Chem. Biol. 2020, 15 (11), 2976–2985. 10.1021/acschembio.0c00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W.; Yuan N.; Mandalapu D.; Mo T.; Dong S.; Zhang Q. Cypemycin Decarboxylase CypD Is Not Responsible for Aminovinyl–Cysteine (AviCys) Ring Formation. Org. Lett. 2018, 20 (23), 7670–7673. 10.1021/acs.orglett.8b03380. [DOI] [PubMed] [Google Scholar]
- Ma S.; Zhang Q. Linaridin Natural Products. Nat. Prod. Rep. 2020, 37 (9), 1152–1163. 10.1039/C9NP00074G. [DOI] [PubMed] [Google Scholar]
- Tang J.; Lu J.; Luo Q.; Wang H. Discovery and Biosynthesis of Thioviridamide-like Compounds. Chin. Chem. Lett. 2018, 29 (7), 1022–1028. 10.1016/j.cclet.2018.05.004. [DOI] [Google Scholar]
- Takase S.; Kurokawa R.; Kondoh Y.; Honda K.; Suzuki T.; Kawahara T.; Ikeda H.; Dohmae N.; Osada H.; Shin-ya K.; Kushiro T.; Yoshida M.; Matsumoto K. Mechanism of Action of Prethioviridamide, an Anticancer Ribosomally Synthesized and Post-Translationally Modified Peptide with a Polythioamide Structure. ACS Chem. Biol. 2019, 14 (8), 1819–1828. 10.1021/acschembio.9b00410. [DOI] [PubMed] [Google Scholar]
- Jimenez J. C.; Bayo N.; Chavarria B.; Lopez-Macra A.; Royo M.; Nicolas E.; Giralt E.; Albericio F. Synthesis of Peptides Containing α or, β-Didehydroamino Acids. Scope and Limitations. Lett. Pept. Sci. 2002, 9 (2–3), 135–141. 10.1007/BF02576876. [DOI] [Google Scholar]
- Denoël T.; Lemaire C.; Luxen A. Progress in Lanthionine and Protected Lanthionine Synthesis. Chem. - Eur. J. 2018, 24 (58), 15421–15441. 10.1002/chem.201801115. [DOI] [PubMed] [Google Scholar]