Abstract
The definition of a genus has wide-ranging implications both in terms of binomial species names and also evolutionary relationships. In recent years, the definition of the genus Mycobacterium has been debated due to the proposed split of this genus into five new genera ( Mycolicibacterium , Mycolicibacter , Mycolicibacillus , Mycobacteroides and an emended Mycobacterium ). Since this group of species contains many important obligate and opportunistic pathogens, it is important that any renaming of species does not cause confusion in clinical treatment as outlined by the nomen periculosum rule (56a) of the Prokaryotic Code. In this study, we evaluated the proposed and original genus boundaries for the mycobacteria, to determine if the split into five genera was warranted. By combining multiple approaches for defining genus boundaries (16S rRNA gene similarity, amino acid identity index, average nucleotide identity, alignment fraction and percentage of conserved proteins) we show that the original genus Mycobacterium is strongly supported over the proposed five-way split. Thus, we propose that the original genus label be reapplied to all species within this group, with the proposed five genera potentially used as sub-genus complex names.
Keywords: genus, genome, Mycobacterium
Introduction
The genus Mycobacterium was first named in 1896 by Lehmann and Neumann, based primarily on the features of the type strain Mycobacterium tuberculosis [1]. Such phenotypic characteristics include the presence of mycolic acids in the cell wall, aerobic growth and bacillary cell shape. Over the years, the taxonomic definition has been reinforced by 16S and phylogenomic analyses [2]. The genus contains over 190 named species, including major human pathogens such as M. tuberculosis and Mycobacterium leprae [2, 3]. It also encompasses the non-tuberculous mycobacteria (NTM), many of which are major opportunistic pathogens such as Mycobacterium abscessus and Mycobacterium avium . The vast majority of mycobacterial species are environmental and can be found in a wide array of niches.
Based on phenotypic and phylogenomic data, the genus is often split first into rapid and slow growers and then further split into specific complexes or groups (e.g. M. tuberculosis complex or M. avium complex) [4]. In 2018, the genus was split into four new genera: Mycolicibacterium , Mycolicibacter , Mycolicibacillus , Mycobacteroides and an emended Mycobacterium [5]. This was done first using a whole genome sequence-based phylogeny, which revealed five major groups within the original genus, corresponding loosely to many of the previously described complexes [4]. These groupings were then defined as genera based upon average amino acid identity (AAI), conserved signature indels (CSIs) and conserved signature proteins (CSPs).
Such taxonomic changes have far reaching consequences for genera that contain a considerable number of clinically important pathogens. Currently, the original genus name Mycobacterium serves as a synonym for these five genera, since validly published names are never withdrawn [6], adding to the confusion around species naming [7]. The recent splits at best add no benefit to clinical treatment [8] and at worst potentially cause much confusion for clinical treatment of mycobacterial diseases as some major opportunistic pathogens such as M. abscessus have been renamed. Indeed, the commonly used term non-tuberculous mycobacteria is called into question as this now includes many species no longer defined as mycobacteria. The renaming also means that the instructions for use for NTM diagnostics, such as the Hain GenoType system (Hain LifeSciences) or similar, may need to be changed as the species listed are not the current validly published species names, which could cause additional confusion for clinicians. Thus, there is a need to ensure this split is strongly supported before clinical guidelines and instructions for use are updated.
Despite being a widely used taxonomic rank, the definition of genus is somewhat elusive. Generally, it is defined as the taxonomic level above species and below family, without concrete methodology to circumscribe such a grouping. Phenotypic definitions usually include a combination of attributable features such as Gram staining or spore formation and biochemical test results such as catalase activity or amino acid degradation [9]. However, a unified genotypic approach has never been applied across several genera.
Early attempts to delimit genera included the use of the 16S rRNA gene identity cut-off of 94–95 % [10, 11], and AAI score of ≥65 % [12]. These options have now been expanded and include three genome-based methods. The relative evolutionary distance is the basis of the taxonomic structure underpinning the Genome Taxonomy Database (GTDB) [13]. This method uses a conserved set of proteins to build a phylogeny and assign taxonomic ranks based on normalized evolutionary distances. Within this approach, the original genus Mycobacterium is preserved as a single genus, with no evidence for sub-splitting above the level of species and is listed as such in the GTDB [14]. A second method, termed Percentage of Conserved Proteins (POCP) uses a blast-based approach to define homologs between species [15]. Two species belong to the same genus if 50 % of their proteins are shared. This allows for amino acid divergence and protein evolution over long time spans, as is common for members of the same genus, while still ensuring core functionality and characteristics are shared. This method has gained popularity for defining a genus and has been used many times before [16–18], but still has some issues as it relies on the 16S rRNA gene as a reference point. A third method uses a combination of the genome alignment fraction (AF) and the ANI to define genera boundaries [19]. This method is not directly dependent on the 16S rRNA gene, unlike POCP.
In this work, we apply the POCP and AF/ANI methods to the original Mycobacterium genus and the newly proposed five genera of Gupta et al. [5]. We show that the original genus fits the POCP definition of genus while the new genera have overlapping boundaries between genera, making their definitions unsupported. The AF/ANI method also shows that splitting the genus into five genera is unfounded, although the designation of Mycobacteroides as a separate genus warrants further investigation. We thus propose that the five newly created genera should be reconstituted into a single genus, named Mycobacterium .
Methods
The original designation of Mycobacterium before the genus was split into five genera [5] will be used here for clarity of the dataset under investigation. When referring to the new Mycobacterium genus this will be explicitly stated as Mycobacterium (Gupta).
Dataset
All genome sequences used in previous phylogenomic studies of the genus Mycobacterium [2, 4] were used here, covering the species re-designated in [5] and the corrigendum [20]. Subspecies names proposed by Tortoli et al. [2] are used here.
An outgroup was required to understand the boundary of genus delimitation for the proposed genus. To this end, all genomes in the GTDB [13, 21] assigned to Corynebacteriaceae , the same family as the genus Mycobacterium , were retrieved from NCBI on 31 January 2019.
A list of all genomes, their designation in the new genera, and their accession numbers are included in Table S1 (available in the online version of this article).
Genome quality control and annotation
Since annotation of the proteins for each species is vital for the proposed analyses, we wanted to ensure that a uniform approach was used for the detection and assigning of open reading frames. The program Prokka version 1.13.7 [22] was used to undertake genome annotation using the genomic sequence file retrieved from NCBI for each genome.
The program CheckM [23] was employed to ensure that only genomes of good quality were used for this study. The genomic sequence file (.fna) for each genome was input to CheckM. Those with an estimated genome completeness lower than 80 % (indicating incomplete sequencing [23]) were removed from further analysis.
16S rRNA gene similarity and AAI score estimation
The percentage similarity between the 16S sequences of each strain was calculated in a pairwise manner on the 16S sequences as annotated by Prokka using blastn, as implemented in blast+ version 2.5 [24].
The AAI between the proteomes of two strains is an estimate of their molecular relatedness using the amount of shared amino acids in their protein complements as a marker of divergence [12, 25]. Although this has been shown to be limited for delineating adjacent taxonomic ranks (e.g. species from genus and genus from family) it is still often used for this task. We used CompareM [26] to compute the AAI between all genomes in our dataset (i.e. the family Corynebacteriaceae ). The package ggplot2 [27] implemented in R version 3.5.1 [28] was used to construct histograms of these pairwise AAI scores.
POCP
Due to the drawbacks of the AAI method for delineating genera, Qin et al. implemented a method based on shared protein content (i.e. presence/absence of proteins in both strains) as a measure of relatedness [15]. Briefly, the POCP is calculated using the formula [(C1 +C2)/(T1 +T2)]*100 % where C1 is the number of proteins in strain 1 also present in strain 2 and T1 is the total protein count of strain 1. C2/T2 are the equivalent numbers for strain 2.
This method was applied to our data by first undertaking reciprocal blastp searches using blast+ version 2.5 with an e-value cut-off of 1e−5. A python 2.7 script was then used to filter these results, retaining only those with >40 % similarity. These similarity and e-value cut-offs were selected as they are the settings used by Qin et al. A second python script was used to calculate the POCP. These scripts can be found at https://githubcom/conmeehan/gentax. The resulting spread of POCP scores was visualised as a histogram using the ggplots2 [27] package in R.
AF and ANI
AF and average nucleotide identity (ANI) values were obtained by using ANIcalculator 2014–127, version 1.0 (https://ani.jgi.doe.gov/html/download.php?) [29]. A custom perl script was written to automate submission of sequence comparisons to ANIcalculator; this can be found at https://githubcom/eddieloh-usc/run_ANIcalculator. The genus demarcation approach that uses AF and ANI determines if a specific set of species constitutes a genus by using the type species of such genus as a stable reference for pairwise comparisons, which include genera within the same taxonomic order or family [19]. Since the genomic content of type strains is used to delineate genera, this makes the approach independent of a benchmarking ribosomal gene (e.g. 16S rRNA), which could introduce biases, as seen with other approaches. The spread of AF/ANI scores and the genus demarcation boundaries were visualized as a scatterplot using ggplots2.
Results
A total of 360 genomes were included in this study: 146 genomes from the original Mycobacterium genus with the remaining 214 genomes from other genera in the family Corynebacteriaceae (Table S1). Of these genomes, 13 were found to be incomplete by CheckM and were removed: nine Mycobacterium , two Gordonia and two Rhodococcus . This resulted in a dataset of 347 genomes, of which 137 were Mycobacterium , representing 123 Mycobacterium species based on the updated naming conventions set out by Tortoli et al. [3]. The remaining 16 ‘mycobacterial’ genomes represent subspecies that used to be designated as separate species [3], and four major M. tuberculosis lineages, besides lineage 4 (H37Rv reference strain) [30].
16S rRNA gene similarity and AAI support the original Mycobacterium designation
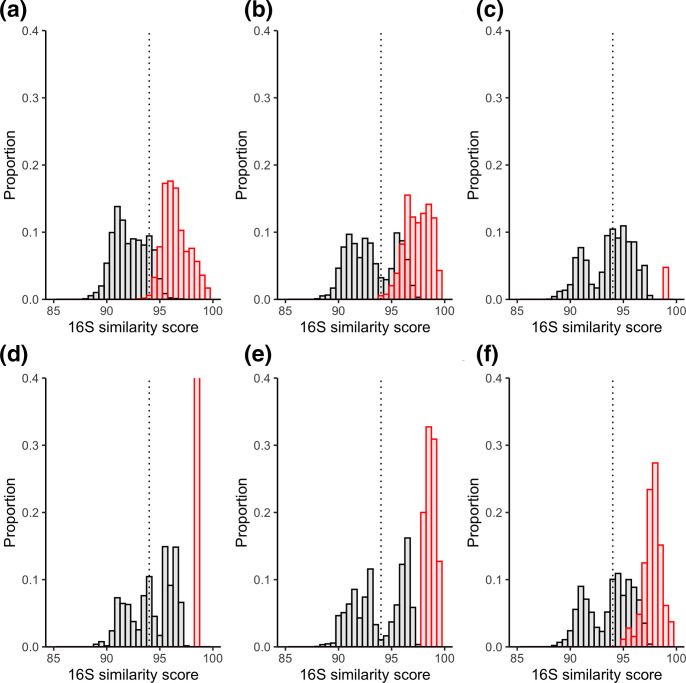
Although 16S rRNA gene similarity and AAI are not recommended for genus delineation [15, 25], they have been used for such in the past. Generally, a cut-off of >94 % is supportive of species belonging to the same genus. 16S similarity scores were compared within each genus (intra-genus) and between members of each specific genus and all other species in the dataset (inter-genus). The spread of scores are shown in Fig. 1 and the minimum intra-genus and maximum inter-genus scores are outlined in Table 1. If the 94.5 % boundary is used, we would expect no intra-genus score below 94.5 % and no inter-genus score above 94.5 % [11]. As expected, the spread of 16S intra-genus similarity scores were mostly above the 94.5 % cut-off; however, both the original and new Mycobacterium showed a very small percentage of organisms that cross this threshold (<1 %) (Table 1). Those scores below 94.5 % tended to be between species such as Mycobacterium xenopi , Mycobacterium heckeshornense , and species at the edges of the phylogenetic divergence of the genera, such as Mycobacterium intracellulare and Mycobacterium chelonae , suggesting those species closely related to M. xenopi have more highly divergent 16S sequences than the rest of the mycobacteria. Unexpectedly, inter-genus similarity scores were often well above the 94.5 % cut-off for all the genera, indicating general incongruences. Of all the genera, the original Mycobacterium had the lowest % of scores above 94 % for inter-genus comparisons (Table 1).
Fig. 1.
16S similarity scores between members of genera. Intra-genus scores are shown in red and inter-genus scores are shown in black. Histogram bin sizes are 0.5 %. The proposed genus boundary of 94.5 % is represented by the dashed line. Each genus is shown separately as follows: (a) Mycobacterium (original); (b) Mycobacterium (Gupta); (c) Mycobacteroides ; (d) Mycolicibacillus ; (e) Mycolicibacter ; (f) Mycolicibacterium . Note that the intra-genus proportion for Mycolicibacillus is 1.0 but the y-axis is cut at 0.4 for comparison between plots.
Table 1.
16S similarity scores
Inter-genus scores are those between members of the designated genus and all other species. Intra-genus scores are those only between members of the designated genus
|
Genus |
Max. inter-genus 16S similarity (i.e. ideally ≤94.5 %) |
% of inter-genus scores above 94.5 % |
Min. intra-genus 16S similarity (i.e. ideally >94.5 %) |
% intra-genus scores below 94.5 % |
|---|---|---|---|---|
|
Mycobacterium (original) |
97.57 |
16.61 |
92.73 |
0.5 |
|
Mycobacterium (Gupta) |
97.78 |
35.95 |
93.83 |
0.09 |
|
97.4 |
52.43 |
99.01 |
0 |
|
|
97.37 |
56.26 |
98.35 |
0 |
|
|
97.78 |
46.82 |
97.85 |
0 |
|
|
97.77 |
53.78 |
94.95 |
0 |
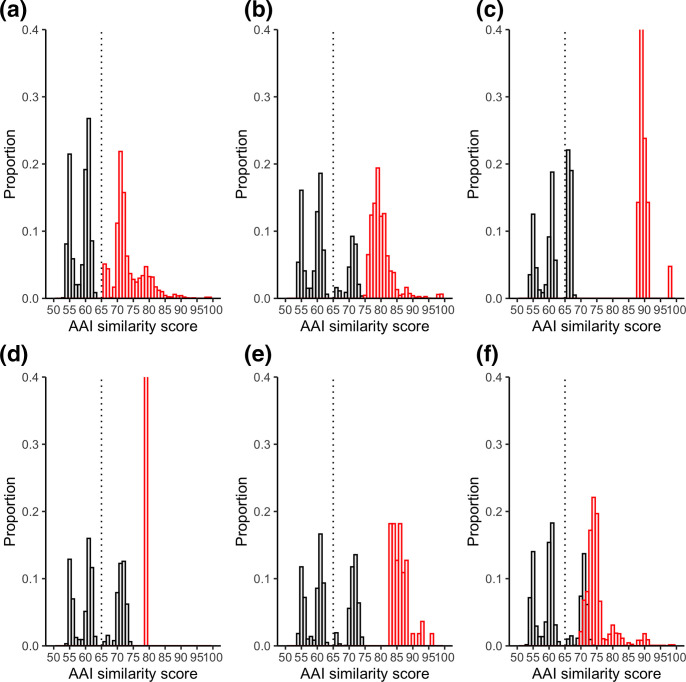
Designation of the five newly proposed genera was originally based on phylogenetic grouping and AAI similarity within those groups [5]. AAI scores above 65 % are often observed for members of the same genus [12]. Comparisons of inter- and intra-genus scores showed that for the original Mycobacterium genus, there is a clean split between these scores (Fig. 2, Table 2). Thus, all non-mycobacteria genera had an AAI of <65 % to any Mycobacterium (original), and all Mycobacterium (original) had an AAI score >65 % to any other Mycobacterium . The intra-genus scores for all five new genera were also consistently above 64 %. These combined results suggest that the split of Mycobacterium was not warranted.
Fig. 2.
AAI similarity scores between members of genera. Intra-genus scores are shown in red and inter-genus scores are shown in black. Histogram bin sizes are 1 %. The proposed genus AAI boundaries of 65 % is represented by a dashed line. Each genus is shown separately as follows: (a) Mycobacterium (original) (b) Mycobacterium (Gupta); (c) Mycobacteroides ; (d) Mycolicibacillus ; (e) Mycolicibacter ; (f) Mycolicibacterium . Note that the intra-genus proportion for Mycolicibacillus is 1.0 but the y-axis is cut at 0.4 for comparison between plots.
Table 2.
AAI similarity scores
Inter-genus scores are those between members of the designated genus and all other species. Intra-genus scores are those only between members of the designated genus
|
Genus |
Max inter-genus AAI similarity (i.e. ideally <65 %) |
% inter-genus scores above 65 % |
Min intra-genus AAI similarity (i.e. ideally >65 %) |
% intra-genus scores below 65 % |
|---|---|---|---|---|
|
Mycobacterium (original) |
63.75 |
0 |
65.48 |
0 |
|
Mycobacterium (Gupta) |
74.37 |
28.02 |
74.85 |
0 |
|
68.25 |
41.64 |
88.38 |
0 |
|
|
74.06 |
42.55 |
79.18 |
0 |
|
|
74.37 |
40.9 |
83.1 |
0 |
|
|
73.91 |
31.99 |
69.83 |
0 |
POCP supports original Mycobacterium genus over alternative sub-split
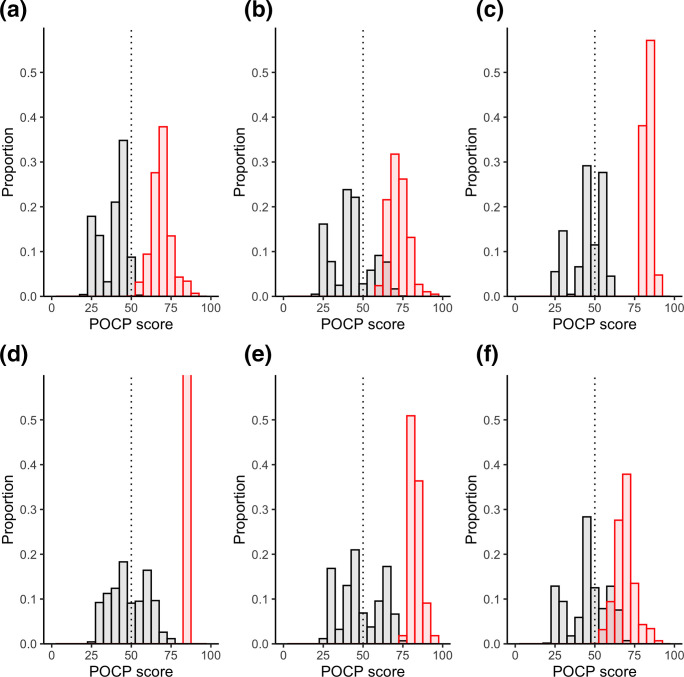
The POCP between members of the same genus (intra-genus) should be no less than 50 % and the POCP between members of the genus and other species (inter-genus) should be no more than 50 % [15]. The original Mycobacterium genus fits this designation almost exactly, with only 0.031 % of intra-genus POCP below 50 and 1.99 % of inter-genus POCP above 50 % (Fig. 3, Table 3). The intra-genus scores of the new genera also all fit this criterion well. The maximum inter-genus scores ranged from 55 to 88 %, with 2–43 % of comparisons above the 50 % cut-off. The original Mycobacterium genus has 2 % of the inter-genera comparisons >50 % while the new Mycobacterium genus has a much higher percentage (25 %) of the inter-genera comparisons above the 50 % cut-off. The rest of the other genera had even higher proportions of the inter-genera comparisons >50 % cut-off. This indicates that the newly designated splits contain numerous species with relatively high identity to species from other genera.
Fig. 3.
POCP scores between members of genera. Intra-genus scores are shown in red and inter-genus scores are shown in black. Histogram bin sizes are 5 %. The proposed genus POCP boundary of 50 % is represented by the dashed line. Each genus is shown separately as follows: (a) Mycobacterium (original); (b) Mycobacterium (Gupta); (c) Mycobacteroides ; (d) Mycolicibacillus ; (e) Mycolicibacter ; (f) Mycolicibacterium . Note that the intra-genus proportion for Mycolicibacillus is 1.0 but the y-axis is cut at 0.6 for comparison between plots.
Table 3.
POCP scores
Inter-genus scores are those between members of the designated genus and all other species. Intra-genus scores are those only between members of the designated genus.
|
Genus |
Max inter-genus POCP similarity (i.e. ideally <50 %) |
% inter-genus scores above 50% |
Min intra-genus POCP similarity (i.e. ideally >50 %) |
% intra-genus scores below 50 % |
|---|---|---|---|---|
|
Mycobacterium (original) |
55.29 |
1.99 |
48.85 |
0.03 |
|
Mycobacterium (Gupta) |
75.66 |
25.70 |
60.63 |
0 |
|
87.87 |
41.91 |
79.32 |
0 |
|
|
75.23 |
43.23 |
83.35 |
0 |
|
|
75.66 |
39.32 |
77.14 |
0 |
|
|
71.92 |
32.22 |
52.23 |
0 |
Genus demarcation boundary supports the reconstitution of Mycobacterium with weak support for Mycobacteroides
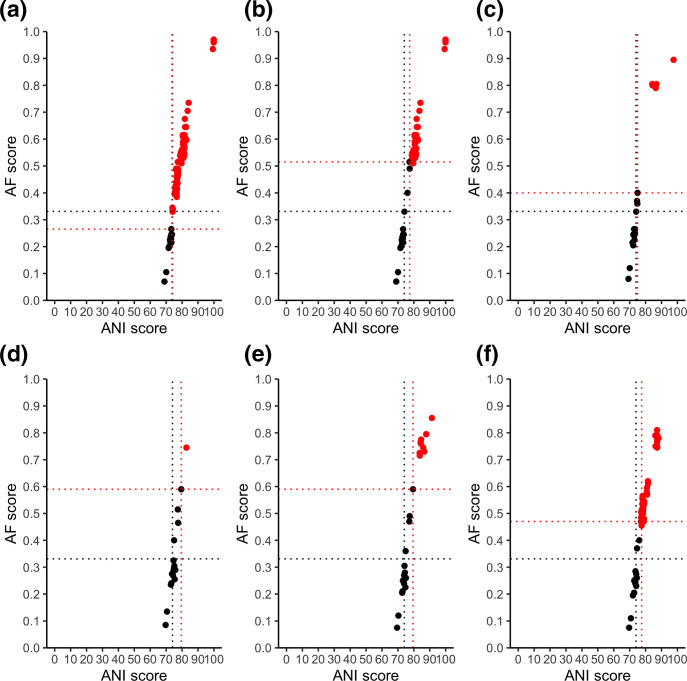
Pairwise comparisons against the type species of the proposed genus were made using all other species in that genus as well as other type species of closely related genera with available genomes. These comparisons were used to determine the genus demarcation boundary to support/refute that grouping. A study by Barco et al. of over 850 genera found the mean AF and ANI to be 0.331 (95 %; CI, 0.308–0.354) and 73.98 % (95 %; CI, 73.34–74.62 %) respectively which defined the genus demarcation boundary [19]. Thus, for a newly proposed genus, we would expect the AF and ANI of all species in that genus to be above these boundaries when compared to the proposed type species for that genus.
The comparison of all species within each proposed genus to the type species for that genus in terms of AF/ANI is outlined in Table 4 and Fig. 4. The five newly proposed genera all had AF and ANI genus demarcation boundaries well above the 95 % confidence interval of typical genera, indicating these new genera are too closely related to type species of other genera to be considered separate genera. Conversely, the original Mycobacterium genus had an ANI within the mean genus boundary confidence interval but an AF (0.265) considerably below the average genus demarcation boundary confidence interval (0.308–0.354). This supports its reconstitution as a genus over the proposed five-way split. However, the low AF value suggests that species at the genus boundary (i.e. those within the newly proposed Mycobacteroides ) could represent a separate genus, but would need additional species in this clade to be sequenced to better support this two-way split.
Table 4.
Genus demarcation boundary
The AF and ANI scores of each proposed genus along with associated type species used are outlined. These values combined demarcate each individual genus boundary.
|
Genus |
Type species |
AF boundary |
ANI boundary |
|---|---|---|---|
|
Mycobacterium (original) |
0.265 |
73.67 |
|
|
Mycobacterium (Gupta) |
0.515 |
77.4 |
|
|
0.4 |
74.98 |
||
|
0.59 |
79.5 |
||
|
0.59 |
79.49 |
||
|
0.47 |
77.53 |
Fig. 4.
Alignment fraction (AF) and average nucleotide identity (ANI) between genomes and the type species for each genus. Intra-genus scores are shown in red and inter-genus scores are shown in black. The proposed demarcation points for the genus is shown with a dashed red line. The mean demarcation points based on Barco et al. [19] are shown with dashed black lines. Each genus is shown separately as follows: (a) Mycobacterium (original); (b) Mycobacterium (Gupta); (c) Mycobacteroides ; (d) Mycolicibacillus ; (e) Mycolicibacter ; (f) Mycolicibacterium .
Discussion
The taxonomic labelling of clinically important pathogens can have wide-ranging implications in terms of correct, efficient identification and appropriate selection of treatment options. Of particular note, when renaming of species occurs, Rule 56a of the Prokaryotic Code states that nomen periculosum, ‘a name whose application is likely to lead to accidents endangering health or life or both or of serious economic consequences’ should be avoided [31]. The genus Mycobacterium falls within this rule as it contains several strict and opportunistic pathogens. Thus, any renaming of species within this genus should be strongly supported before implemented.
A variety of genetic analyses were employed here to examine the evidence supporting the split of the genus Mycobacterium into five new genera, as proposed by [5]. 16S rRNA similarity was found to be a very poor marker for delineation of the Mycobacterium genus, or new genera. The standard boundary of 94.5 % similarity did not create clear separation between any of the genera proposed by Gupta et al. (Table 1, Fig. 1). In each case, several species from one genus shared a 16S rRNA similarity above 94 % with species from other genera. In the case of Mycobacteroides , Mycolicibacillus and Mycolicibacterium , over 50 % of the comparisons between species inside these genera and species designated outside were above this cut-off. Conversely, when comparing species inside these genera classifications with each other (e.g. Mycobacteroides vs Mycobacteroides or Mycolicibacillus vs Mycolicibacillus ) the 94.5 % boundary did classify species correctly, suggesting that the 16S boundary could be useful as an inclusionary boundary, but not exclusionary. However, for both the original and new Mycobacterium genera, 0.5 and 0.09 % of comparisons were below this boundary, demonstrating that it is not completely clear cut.
Use of AAI scores was found to be better at demarcating genus boundaries. AAI clustering was used to define the new genera [5] and as expected, each genus formed groupings with high intra-genus AAI scores. However, as was seen with 16S, the inter-genera comparisons did not clearly separate species into the genera proposed by Gupta et al., with over 40 % of inter-genus comparisons being below the 65 % lower boundary cut-off for several of the newly proposed genera (Table 2, Fig. 2). Conversely, all inter-genus comparisons for the original Mycobacterium genus were above the 65 % boundary, strongly supporting this genus classification. Thus a sub-splitting is not warranted. The clustering and AAI scores more so suggest that the newly proposed genera are an expansion of the already defined complexes [4] and thus may represent sub-genus rankings instead.
The use of other genome-wide approaches for delineating genus boundaries further supports the re-amalgamation of the original Mycobacterium genus with the newly proposed genera forming sub-genus multi-species complexes. The POCP approach, using a score of shared protein coding genes across the genome, showed that the original Mycobacterium genus fell near the 50 % proposed demarcation line (Table 3, Fig. 3). Although the 50 % boundary is somewhat arbitrary, the redefining of several pathogenic species into new genera is not supported by this genus demarcation approach and conversely supports the retainment of the original designation.
While the POCP utilizes a non-standard approach, the [19] method uses AF and ANI, which is a standard genome relatedness index, to utilize genome similarity to look at genus boundary definitions. Comparing to the mean AF of 0.33 and ANI of 73.98 % genus demarcation boundaries set by a large group of genus comparisons [19], the original Mycobacterium genus most closely approximates this boundary, with the five newly proposed genera being well above these points. Interestingly, Mycobacteroides , which contains the most basal species such as M. abscessus and M. chelonae also sits close to this boundary. Thus, these species could perhaps constitute the basis of a new genus in the future, but careful analysis of more genome sequences of closely related species would be needed to confirm this, especially in light of issues brought up by the analyses based on 16S rRNA gene, AAI, POCP and ANI (e.g., inter-genera comparison results). Of note, due to the lack of Mycobacterium species with an ANI of 85–100 %, the exact genus inflection points could not be determined, hence the use of genus demarcation instead. With the constant sequencing and discovery of new species, hopefully future analysis of AF and ANI scores can confirm these findings and determine the exact genus inflection point.
Overall, the variety of methods used here strongly support the reconstitution of the original Mycobacterium genus, as has been previously suggested [7, 8]. Further study could support the redesignation of the newly proposed genera as sub-genus complexes. Such complexes are useful clinically as many species within the same complex tend to share similar clinical and biochemical features. The conserved signature indels (CSIs) and conserved signature proteins (CSPs) for each of these groups as outlined by [5] can serve as markers for these complexes, as CSIs and CSPs can be used to denote any taxonomic rank or phylogenetic clade, not just a genus [32, 33]. However, until they are confirmed as valid subgenera, they should be rejected as heterotrophic synonyms, as per Rule 56a of the Prokaryotic Code (nomina rejicienda) due to being nomen periculosum [34].
Emended description of the genus Mycobacterium Lehmann and Neuman 1896
Mycobacterium (My.co.bac.te´ri.um. Gr. n. mykes a fungus; N.L. neut. n. bacterium, a small rod; N.L. neut. n. Mycobacterium, a fungus rodlet).
The type species is Mycobacterium tuberculosis [35] Lehmann and Neumann 1896 (Approved Lists 1980) [36].
The characteristics of this genus match those of the emended description of the genus Mycobacterium as outlined in [5]. All species in the genera Mycobacterium , Mycobacteroides , Mycolicibacillus , Mycolicibacter and Mycolicibacterium are included in this genus. They can be separated from other species in the family Mycobacteriaceae using both the signature CSI and CSP for these groups outlined in [5] as well as having an AF above 0.265 and ANI above 73.76 % when compared to the type species, Mycobacterium tuberculosis .
Supplementary Data
Funding information
This work is supported by the Belgian Science Policy (Belspo).
Acknowledgements
The authors thank Oren Tzfadia for his useful comments on the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AAI, amino acid identity; AF, alignment fraction; ANI, average nucleotide identity; CSI, conserved signature indel; CSP, conserved signature protein; GTDB, Genome Taxonomy Database; NTM, non-tuberculous mycobacteria; POCP, Percentage of Conserved Proteins.
A supplementary table is available with the online version of this article.
References
- 1.Lehmann KB, Neumann R. Atlas und Grundriss der Bakeriologie und Lehrbuch der speziellen bakteriologischen Diagnositk. 1st edn. 1896. [Google Scholar]
- 2.Tortoli E, Fedrizzi T, Meehan CJ, Trovato A, Grottola A, et al. The new phylogeny of the genus Mycobacterium: The old and the new. Infect Genet Evol. 2017;56:19–25. doi: 10.1016/j.meegid.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 3.Tortoli E, Meehan CJ, Grottola A, Serpini GF, Fabio A, et al. Genome-based taxonomic revision detects a number of synonymous taxa in the genus Mycobacterium . Infect Genet Evol. 2019;103983 doi: 10.1016/j.meegid.2019.103983. [DOI] [PubMed] [Google Scholar]
- 4.Fedrizzi T, Meehan CJ, Grottola A, Giacobazzi E, Fregni Serpini G, et al. Genomic characterization of nontuberculous mycobacteria. Sci Rep. 2017;7:45258. doi: 10.1038/srep45258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta RS, Lo B, Son J. Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol. 2018;9:67. doi: 10.3389/fmicb.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tindall BJ. Misunderstanding the bacteriological code. Int J Syst Bacteriol. 1999;49 Pt 3:1313–1316. doi: 10.1099/00207713-49-3-1313. [DOI] [PubMed] [Google Scholar]
- 7.Tortoli E, Brown-Elliott BA, Chalmers JD, Cirillo DM, Daley CL, et al. Same meat, different gravy: ignore the new names of mycobacteria. Eur Respir J. 2019;54:1900795. doi: 10.1183/13993003.00795-2019. [DOI] [PubMed] [Google Scholar]
- 8.Armstrong DT, Parrish N. Current updates on mycobacterial taxonomy, 2018-2019. J Clin Microbiol. 2021;59:e0152820. doi: 10.1128/jcm.01528-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta AK. Classification. Springer Geology. Springer; 2015. pp. 69–87. [Google Scholar]
- 10.Yarza P, Richter M, Peplies J, Euzeby J, Amann R, et al. The All-Species Living Tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 12.Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME Journal. 2017;11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 14.GTDB Mycobacterium tree. 2021. https://gtdb.ecogenomic.org/tree?r=g__Mycobacterium
- 15.Qin QL, Bin XB, Zhang XY, Chen XL, Zhou BC, et al. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol. 2014;196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopes-Santos L, Castro DBA, Ferreira-Tonin M, Corrêa DBA, Weir BS, et al. Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie van Leeuwenhoek. 2017;110:727–736. doi: 10.1007/s10482-017-0842-6. [DOI] [PubMed] [Google Scholar]
- 17.Adeolu M, Alnajar S, Naushad S, Gupta RS. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol. 2016;66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 18.Nicholson AC, Gulvik CA, Whitney AM, Humrighouse BW, Graziano J, et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie van Leeuwenhoek, Int J Gen Mol Microbiol. 2018;111:55–72. doi: 10.1007/s10482-017-0926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barco RA, Garrity GM, Scott JJ, Amend JP, Nealson KH, et al. A genus definition for bacteria and archaea based on a standard genome relatedness index. mBio. 2020;11:e02475-19. doi: 10.1128/MBIO.02475-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta RS, Lo B, Son J. Corrigendum: Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol. 2019;10:714. doi: 10.3389/fmicb.2019.00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020;1–8 doi: 10.1038/s41587-020-0501-8. [DOI] [PubMed] [Google Scholar]
- 22.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 23.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. Checkm: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Konstantinidis KT, Tiedje JM. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parks DH. CompareM. 2014. https://github.com/dparks1134/CompareM
- 27.Warnes GR, Bolker B, Lumley T. gplots: Various R programming tools for plotting data. R package version 2.6.0. 2012. http://cran.r-project.org/web/packages/gplots
- 28.R Core Team R: A language and environment for statistical computing. 2017. http://www.r-project.org
- 29.Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, et al. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagneux S. Ecology and evolution of Mycobacterium tuberculosis . Nat Rev Microbiol. 2018;16:202–213. doi: 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- 31.Parker CT, Tindall BJ, Garrity GM. International Code of Nomenclature of Prokaryotes. Int J Syst Evol Microbiol. 2019;69 doi: 10.1099/ijsem.0.003239. [DOI] [PubMed] [Google Scholar]
- 32.Gupta RS. Commentary: Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol. 2019;10:206. doi: 10.3389/fmicb.2019.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao B, Gupta RS. Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol Mol Biol Rev. 2012;76:66–112. doi: 10.1128/MMBR.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oren A, Trujillo ME. On the valid publication of names of mycobacteria. Eur Respir J. 2019;54 doi: 10.1183/13993003.01483-2019. [DOI] [PubMed] [Google Scholar]
- 35.Zopf W. Die Spaltpilze: Nachdem neuesten Standpunkte. Breslau: Eduard Trewendt; 1883. [Google Scholar]
- 36.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.