Abstract
The status Candidatus was introduced to bacterial taxonomy in the 1990s to accommodate uncultured taxa defined by analyses of DNA sequences. Here I review the strengths, weaknesses, opportunities and threats (SWOT) associated with the status Candidatus in the light of a quarter century of use, twinned with recent developments in bacterial taxonomy and sequence-based taxonomic discovery. Despite ambiguities as to its scope, philosophical objections to its use and practical problems in implementation, the status Candidatus has now been applied to over 1000 taxa and has been widely adopted by journals and databases. Although lacking priority under the International Code for Nomenclature of Prokaryotes, many Candidatus names have already achieved de facto standing in the academic literature and in databases via description of a taxon in a peer-reviewed publication, alongside deposition of a genome sequence and there is a clear path to valid publication of such names on culture. Continued and increased use of Candidatus names provides an alternative to the potential upheaval that might accompany creation of a new additional code of nomenclature and provides a ready solution to the urgent challenge of naming many thousands of newly discovered but uncultured species.
Keywords: bacterial nomenclature, Candidatus, genome-based taxonomy, uncultured bacteria
Introduction
The International Code for Nomenclature of Prokaryotes (the ICNP or ‘the Code’) articulates principles, rules and recommendations for the naming of archaeal and bacterial taxa, including rules for the valid publication of names and establishing priority in the literature [1]. The current version of the Code is the descendent of earlier documents, stretching back to the 1860s [2, 3]. A major landmark in the development of the Code was the publication of the Approved List of Bacterial Names [4], which brought order out of chaos in the use of names for bacterial taxa, jettisoning over 20000 names, while giving priority to around 2300 names with effect from 1 January 1980 [5]. However, over 50 of these approved names were applied to species that had not, and still have not, been isolated in axenic culture, so that no type strain exists (Table 1), which means that these names could not be validly published if presented for the first time under today’s rules.
Table 1.
Species in the Approved List of Bacterial Names for which no cultured type strain is available
This list was compiled by downloading the genera, species and subspecies list from the List of Prokaryotic names with Standing in Nomenclature (https://lpsn.dsmz.de/downloads) and then sorting and selecting entries by nomenclatural type.
|
Species |
LPSN description of type strain |
|---|---|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No pure culture |
|
|
No culture available |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No culture available |
|
|
No pure culture |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture available |
|
|
No culture available |
|
|
Has not been cultivated |
|
|
None specified due to difficulties in cultivation |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture available |
|
|
No culture isolated |
|
|
Description from 1888 serves as type |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture available |
|
|
No culture isolated |
|
|
No pure culture |
|
|
No culture isolated |
|
|
No culture available |
|
|
No culture isolated |
|
|
No culture isolated |
|
|
No culture available; none designated |
|
|
No culture available |
|
|
No culture available; none designated |
|
|
No culture isolated |
|
|
No culture isolated |
In the 1980s and 1990s, there was a growing recognition that many, if not most, bacterial taxa could not be isolated or maintained in axenic culture, but could be identified and classified through analysis of macromolecular sequences [6–8]. To accommodate this new source of taxonomic information, in 1994 Murray and Schleifer proposed a new category of taxonomic name, which they called Candidatus, to ‘provide a proper record of sequence-based potential new taxa’ [9].
After consideration by the International Committee on Systematics of Bacteria, additional guidelines were published in 1995 [10]. Curiously, although the committee agreed that a ‘Candidatus name is by definition a preliminary name and therefore has no standing in prokaryote nomenclature’, they were happy to incorporate the Candidatus option within an appendix within the Code and suggested that a list of organisms with the status Candidatus should maintained and published in the International Journal of Systematic and Evolutionary Microbiology (IJSEM) at appropriate intervals [1, 11]. In parallel with these developments, the Committee discussed and then mandated a requirement that valid publication of names required deposition of viable pure cultures in strain repositories in two countries, which came into effect from 1 January 2001 [1, 11, 12].
Recent years have seen a dramatic growth in the discovery and classification of new uncultured taxa primarily through analysis of metagenomic sequences [13–15]. This has prompted calls for a change to the Code to give standing to names for uncultured taxa [16, 17]. A proposal to enable this was discussed and rejected by the International Committee on Systematics of Prokaryotes in 2020 [18]. This in turn has fuelled calls for the establishment of an additional or alternative code of nomenclature for uncultured microbes called the SeqCode [17, 19]. However, the Candidatus option has already been adopted for uncultured organisms [10, 20]. So, this begs the question: if the system works, do we actually need to change anything? To inform thinking on this issue, here I present a SWOT analysis, evaluating the strengths, weaknesses, opportunities and threats pertaining to the status Candidatus.
Strengths
Key strengths of the status Candidatus are that it has been in place for over a quarter of a century and that its use is already specified and permitted within the current version of the Code: ‘This category should be used for describing prokaryotic entities for which more than a mere nucleic acid sequence is available but for which characteristics required for description according to the Code are lacking’. Furthermore, the way in which Candidatus taxa are described has evolved over time. Thus, the examples provided in the original guidelines (copied over into Appendix 11 of the Code) failed to comply with the grammatical or orthographic norms of bacterial taxonomic nomenclature (e.g. adjectives and nouns agree in gender; connecting vowels are used consistently, binomials are specified). However, with the occasional exception—such as the orphaned species epithet Candidatus comitans [21]—Candidatus names have since then followed the conventions of Linnaean nomenclature, for example in specifying binomials for species or using relevant endings for proposed families (-aceae) or orders (-ales) [20].
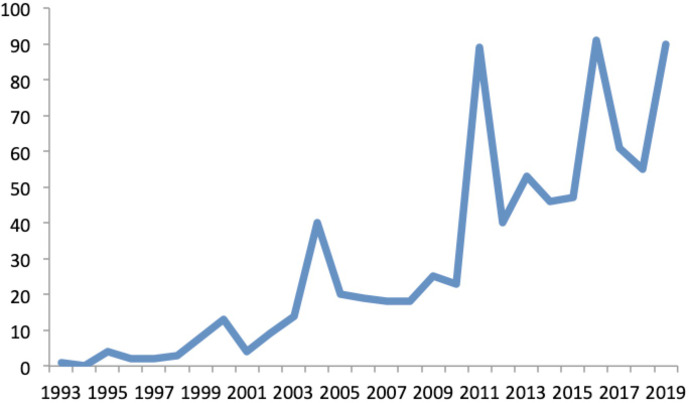
Candidatus designations have seen increasing usage over the last 25 years (Fig. 1). Lists of Candidatus names published from 1995 to 2019 document over 1000 names in total, including more than 700 species [20, 22]. By contrast, an alternative suggestion of using a superscripted ‘u’ to mark names of uncultured organisms has been adopted only a handful of times [16, 23, 24].
Fig. 1.
Number of Candidatus species names published each calendar year 1993–2019. Data extracted from published Candidatus lists [20, 22].
A PubMed search reveals that the term Candidatus has been used in over 500 journals, suggesting widespread acceptance among authors, reviewers and editors. In addition, the term Candidatus remains unambiguous, so that searches of the web or of the biomedical literature with this term return only appropriate hits. Crucially, Candidatus names are included in—and indeed often enforced by—the widely used NCBI taxonomy [25] and are documented in the List of Prokaryotic names with Standing in Nomenclature (LPSN) [26] and by the NamesforLife service [27]. However, it is worth noting the qualifier Candidatus is ignored by the widely used Genome Taxonomy Database (GTDB), which even goes so far as to strip the term off names imported from elsewhere [28].
Another potential strength is that the Candidatus approach provides a clear route to valid publication of names if taxa are subsequently cultured—in such circumstances, the term Candidatus can simply be deleted from the existing name. Over 30 Candidatus species have now been cultured and with existing names carried over, minus the term Candidatus and with occasional linguistic corrections [20, 22], with just one exception. And in that exceptional case, the re-designation of Candidatus Pectobacterium maceratum as Pectobacterium versatile was not based on a whim, but instead justified by the authors by reference to Recommendation 12c in the Code, aiming to avoid epithets based on a character common to all species within a genus [29].
Weaknesses
A key weakness is that the circumstances governing use of Candidatus status are poorly specified in the Code—rather than defined by Rules or Recommendations within the main body, the guidance is relegated to an Appendix—and so it remains unclear when use is allowed or denied, mandatory or merely optional. In fact, there has been confusion as to whether the status Candidatus can be used for taxa that have been cultured but not adequately characterized or is restricted to uncultured taxa. Thus, the term Candidatus has been applied to 20 or more taxa that had already been propagated in pure culture (or at least in culture free of other bacteria) at the time the description was published [20]. Although the original proposals for the use of Candidatus [10]—and recent expert opinion [20]—suggest it should apply only to uncultured taxa, Appendix 11 in the Code does not mention culture or lack of culture, but merely states: ‘This category should be used for describing prokaryotic entities … for which characteristics required for description according to the Code are lacking’. However, as we shall see later, this ambiguity might prove useful.
As the Code is also ambiguous on the issue of whether type strains have to be maintained in pure culture, it remains unclear whether the status Candidatus can be applied when viable but impure cultures of a bacterium or archaeon are available and could be deposited. Rule 18a states ‘The type strain is made up of living cultures of an organism, which … should have been maintained in pure culture’—note should rather than must as the modal verb. In addition, Rule 30 merely describes the need for ‘viable cultures’, while in Chapter 4, there is an advisory note that states that maintenance of type strains ‘may be by a variety of methods, e.g. in a medium, in a host by passage, in cells or exudates, or in the frozen or dried state’.
In the Approved Lists of Bacterial Names, there were over a dozen examples of species that can be propagated only in association with host cells, including Chlamydia psittaci and Chlamydia trachomatis , plus Rickettsia typhi and 11 other species of Rickettsia [4]. Since introduction of the requirement for deposition of type strains, dozens of additional names have been validly published for species of Chlamydia or Rickettsia , suggesting that most authorities take what Rule 18a describes as ‘pure culture’ to mean ‘viable culture’—an ambiguity that can be removed in subsequent editions of the Code.
However, such a move would exclude from Candidatus status any Archaea or Bacteria that can be stably propagated and deposited, but can be cultured only in association with another organism. Examples here include so-called Candidatus Nanosynbacter lyticus [30, 31], which can be cultured, but only in association with another bacterial species; Nanoarchaeum equitans, which can be cultured, but only in association with another archaeon [32]; and Mycobacterium lepromatosis, which can be cultured, but only in the footpads of nude mice [33, 34].
Similarly, there is no agreement on what counts as a satisfactory description of a Candidatus species. The examples provided in the initial proposal now appear dated, with their discussion of probes and primer sequences [10]. However, this has become less of an issue lately, as recent descriptions of Candidatus taxa have tended to converge on protologues similar in form to those used for cultured taxa, complete with etymological justifications for new names [35–38].
An initial weakness in the use of Candidatus names was the lack of any compilation of such names into published lists, despite a request for this in the original proposal and Appendix 11 of the Code [1, 10]. This meant that such names seldom underwent the linguistic quality control applied to validly published names of cultured taxa. However, after a first attempt at linguistic quality control for Candidatus names [39], this issue has now been addressed by annual publication of lists of Candidatus names within the IJSEM, together with suggested changes when names fail to comply with the rules and recommendations of the Code [20, 22].
However, it remains unclear how far suggested changes are actually taken up by authors, journals and databases. For example, when it was suggested that the genus name Candidatus Rohrkolberia should be changed to Candidatus Typhincola, the authors adopted an alternative name that they themselves crafted: Candidatus Symbiopectobacterium [40]. Similarly, the initially malformed Borkfalki was changed to Candidatus Borkfalkia, even though this clashed with Recommendation 6(10) that authors should not name taxa after themselves [36, 41]. However, similar issues also apply to the adoption of suggested changes to effectively published names, which may be ignored by authors if the original names merely flout recommendations, but do not break the rules of the Code. No more obvious example of a effectively published name that rides roughshod over the recommendations of the Code, but does not break the rules is Myxococcus llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogochensis [42].
A potential stylistic quibble is that adding the term Candidatus makes names longer than they have to be. However, for at least 20 years, Candidatus has been abbreviated in many publications to the simple two-letter moniker ‘Ca.’ [43]. Another criticism of the Candidatus option centres on its rather fussy and confusing orthographic requirements. Curiously, the Code does not mandate the use of italics for validly published Latin binomials or other taxonomic names, but in Appendix 11 does make clear: ‘the word Candidatus, but not the vernacular epithet is printed in italics’. However, a similar orthographic approach has been widely accepted in naming serovars of Salmonella enterica , where what was once considered a species epithet—but is now seen as merely a serovar—is presented in Roman type, for example, Salmonella enterica serovar Typhimurium or even S. Typhimurium [44].
Another source of uncertainty is whether a Candidatus name needs to be placed in quotation marks. Within the Code, some exemplar names are put quotation marks, whereas others are not. And among publications from 2021, some Candidatus names are published without quotation marks, e.g. Candidatus Sulfurimonas marisnigri [45], some sit within single quotation marks e.g. ‘Candidatus Liberibacter asiaticus’ [46] and some sit within double quotation marks, e.g. “Candidatus Laterigemmans baculatus” [47]. Simplicity suggests that no such orthographic encumbrance is needed.
One final and perhaps most important weakness is that Candidatus names lack priority under the Code’s rules on valid publication. This can be seen as a fundamental flaw, when priority is deemed to be of central importance to systems of nomenclature, including not just the ICNP, but also in the equivalent codes for zoology and botany. As noted, so far, there have been no disputes over the priority of such names in the literature or in databases. But it remains unclear how problematic this will be when there are many thousands of Candidatus species names. Will authors, reviewers or editors play ‘nice’ and respect the general principle of scientific priority in this context or will there be a free-for-all with endless disputes? A recent case where names for cultured taxa published in peer-reviewed journals, but never validated, were subsequently overturned by a new set of authors [48] suggests this remains a potential problem for any name that is in use but not validly published—whether Candidatus or not.
However, this brings to mind a deeper philosophical question: should the status of names depend on cultivability? This can be seen to conflict with the opening principle of the ICNP: ‘Nothing in this Code may be construed to restrict the freedom of taxonomic thought or action’ and with General Consideration 5: ‘This Code of Nomenclature of Prokaryotes applies to all Prokaryotes’ (my italics). In addition, the requirement to use the term Candidatus has not been applied retrospectively, so it does not apply to the names of uncultured species published in the Approved Lists of Bacterial Names or to the thirty or more validly published names of uncultured organisms approved between 1980 and 2001 [4, 49–67]. These glaring inconsistencies sit uneasily within what is supposed to be a precise rule-governed system of nomenclature.
However, there is also a serious operational issue at stake. According to the strictest interpretation, assigning a Candidatus name to a taxon depends on proving a negative—being confident that the taxon has never been cultured by anyone anywhere in the world. To take a lively example that applies at the time of writing, let’s say we wished to assign a new Candidatus name to the genus given the designation CAG-485 by GTDB (https://gtdb.ecogenomic.org/searches?s=gt&q=CAG-485). Almost all of the more than 100 genomes classified within this genus represent metagenome-associated genomes, so it might appear safe to propose a new Candidatus name. However, only after exhaustively working through the metadata associated with all of the BioSamples associated with these genome sequences in the NCBI databases does it become clear that at least one of these (NCBI BioSample SAMN10878315) is in fact derived from a cultured isolate, so that some might argue the status Candidatus cannot be applied. A similarly exhausting process awaits anyone attempting to prove that well-established Candidatus taxa do not yet contain cultured relatives. In all such cases, it is probably best if we fall back upon the looser definition of Candidatus, ‘used for describing prokaryotic entities … for which characteristics required for description according to the Code are lacking’ and deny Candidatus status only when someone provides proof of culture in a peer-reviewed publication.
Scrutiny of genome sequences assigned to CAG-485 reveals an additional problem. Several of these originate from a study of the mouse gut microbiota conducted in Germany and are tagged as derived from cultured isolates [68]. However, on reading the paper it becomes clear that these represent genomes from ‘strains that could be isolated but failed being maintained in culture’. Should taxa based on such criteria be allowed Candidatus status? If so—as complying with the requirements for effective and valid publication of names for cultured taxa is far more time-consuming than publishing Candidatus names—those interesting in cataloguing microbial diversity may well favour approaches that avoid stable culture altogether and simply assign Candidatus (or even SeqCode) names to new found organisms.
Opportunities
In the final chapter of the Origin of Species, Darwin prophesized: ‘Our classifications will come to be, as far as they can be so made, genealogies’ [69]. For plants and animals, this became a reality in the second half of the twentieth century with the adoption of cladistics, i.e. the scheme of phylogenetic taxonomy articulated by German entomologist Willi Hennig [70]. With the arrival of bacterial genome sequencing [71], terms and concepts from cladistics (e.g. clades and monophyletic groups) have permeated bacterial taxonomy [72], culminating in the near-universal adoption of sequence-based phylogenomic approaches [28]. Just as zoologists now accept that birds are dinosaurs, microbiologists now face similar challenges in say rejecting prokaryotes as a paraphyletic group [73–75] and accepting on cladistic grounds that mitochondria are bacteria and eukaryotes are archaea [76, 77].
This often unconscious appropriation of cladistics thinking has brought about a sea-change in microbial taxonomy—rather than an exhaustive description of phenotypic properties [78], the sine qua non in naming and describing a new species is now deposition of a genome sequence into the public domain [79], accompanied by a sequence-based circumscription built on a phylogenetic analysis of the type genome. In contrast to the chaos that predated the Approved List of Names, this means that it is now very easy to determine, through database searches, whether a bacterium characterized in say Europe does or does not belong to a taxon already defined in Asia or North America.
Such sequence-based comparisons provide a clear opportunity for ensuring that de facto priority of a name can be established by the deposition of a genome sequence into the public domain and a description of a taxon in a peer-reviewed publication. When twinned with Principle 1.1 of the Code ‘Aim at stability of names’, this effectively creates a default assumption that names of any taxa, including Candidatus taxa, should not be changed unless they conflict with any of the Code’s other Principles, Rules and Recommendations.
What this means is that although Candidatus names lack formal priority under the ICNP, there is in effect de facto establishment of priority through analyses of sequences. Thus, if I attempt to apply a new name to representatives of an existing Candidatus species, this will become obvious when I attempt classification using the NCBI or GTDB taxonomies and should, under the Code’s Principle 1.1, be blocked by reviewers, editors and the databases, unless I can provide a clear justification from the Code for overturning an existing name.
It is worth making a comparison here with names for bacterial and archaeal taxa published in peer-reviewed publications [80], but never validly published, which requires deposition of strains in two repositories and publication or listing in the IJSEM. Like Candidatus names, many of these have gained operational priority through sustained use—for example the name for the human pathogen Tropheryma whipplei [81], which has been used in over 600 publications listed in PubMed.
An alternative view is to see the absence of formal standing for Candidatus names not as a ‘bug’ but as a ‘feature’, as this means that they could be applied as provisional names to uncharacterized taxa, as an alternative to cumbersome alphanumeric labels, while leaving open the option that they could subsequently be changed after further characterization of taxa or changes in opinion.
The creation and curation of lists of Candidatus names published in the IJSEM provides a fresh opportunity for improving the linguistic quality of Candidatus names [20, 22]. From here, it is only a small step to encouraging authors, reviewers and editors to engage in linguistic quality control at the time such names are created, particularly as most errors are mundane problems (e.g. agreement in gender) that could be avoided through use of checklists—an approach that works even in high-risk contexts like medicine or aviation [82]—or through use of computer-based tools.
From my own experience, I can see that there is clearly scope for productive engagement with experts when creating Candidatus names en masse. Our recent progress with high-throughput name creation and quality control collaborating with an expert (Aharon Oren) provides proof-of-principle use of Candidatus names can be extended to cover huge numbers of new taxa discovered through metagenomic analyses. In an initial study of the chicken gut microbiome, we described names for 42 new Candidatus genera and 60 new Candidatus species [35]. Initially, this was done in a spreadsheet supplementary to the main manuscript. However, to ensure that names were propagated in lists and databases, conventional protologues were subsequently published in a corrigendum [83].
In a subsequent study, we described 657 Candidatus species names and 158 Candidatus genera names in a series of protologues that occupied over 100 pages of the paper [37]. These efforts primed a more ambitious study, which included software for generation of names and protologues, together with creation of over a million well-formed names for bacterial species [38]. Such efforts show that generating names for large numbers of taxa—whether cultured or uncultured—is indeed scalable in the age of the big-data metagenomics. The fact that a new version of the Code is now in preparation [84] also brings another opportunity to revisit and refine the Candidatus approach while clarifying ambiguities or contradictions in the current version of the Code (as detailed above). There would be an opportunity to solve the problem of priority, if the Candidatus status were brought within the main body of the Code, with Rules on its application, name formation, designation of type material etc., plus a Rule that if a Candidatus taxon is brought into culture then the name for the type species must be conserved.
Threats
A key challenge to continued use of the status Candidatus comes from the planned launch of a new code of nomenclature for bacteria and archaea termed the SeqCode [17, 19], which proposes to remove the need for deposition of type strains in culture collections, but instead to give de jure priority to names associated with a type sequence.
In preferring revolution to evolution, such an approach ignores the current status quo, in which adherence to the Code’s requirement for stability twinned with ease of database searches means that Candidatus names already have de facto priority in the literature. The success of this new venture will depend on whether authors, journals, editors and online resources (e.g. LPSN and NCBI taxonomy) will pay more attention to names issued under the new code than they already grant to Candidatus names. Similarly, uncertainties remain over whether the new code aims to govern nomenclature of all taxa of Bacteria and Archaea or only the uncultured. Clearly if the SeqCode is going to cover only uncultured organisms, this will require exhaustive checks to see which code should apply to which organisms and will require clarity on how conflicts with the ICNP will be managed.
Will the two codes run in parallel, with some authorities, e.g. NCBI, LPSN or the authors of a recent opinion piece [85], sticking with the Candidatus option, while others abandon it, just as GTDB ignores it already? Under this scenario, at least some microbiologists will continue to use the Candidatus option for years to come. More generally, only time will tell whether the SeqCode will see widespread acceptance or fall by the wayside, as has happened with similar efforts such as the PhyloCode and the BioCode [86, 87].
A potential operational challenge to the status quo comes from increasingly unsustainability in practice of the current approach to nomenclature. The requirement for deposition in culture collections in two countries has become more difficult under national and international rules on intellectual property, including the Nagoya protocol [88]—although if digital sequence information is seen as a genetic resource, the Nagoya protocol might also create problems for releasing genome sequences as type material.
Recent publications, in which names for novel species and genera have been published without deposition in two repositories, show that even those employed in culture collections do, on occasion, ignore the ICNP’s rules for effective publication [68, 89]. Furthermore, breakthroughs in culturing previously uncultured organisms, including co-culture and dilution-to-extinction culture [90] are probably not yet compatible with strain deposition as currently configured. All this suggests a re-think of the current rules may become necessary over the coming years, so that a subsequent vote on whether sequences can become type material may turn out differently from the most recent episode and sweep away the need for the Candidatus status.
Conclusions
The Candidatus option clearly works, as evidenced by a quarter century of use. What’s more, it has been shown to cope with modern high-throughput approaches to taxonomic discovery. Although lacking priority under the code, many Candidatus names have already achieved standing in the academic literature and in key databases and there is a clear path to valid publication of such names on culture. Continued use of Candidatus names provides an alternative to the upheaval that might accompany creation of a new additional code of nomenclature. As we have a solution that already works for naming uncultured organisms, faced with the calls for change, the pragmatist will probably want to invoke the Code’s opening call ‘Aim for stability’ and say if ‘it ain't broke, why fix it?’.
It is worth stressing that Candidatus names are not currently in competition with other well-formed names, but instead with an unpalatable alphanumerical spaghetti, epitomized in GTDB designations such as UBA6965 or sp000063525. We now face the urgent and exhilarating challenge of creating the many thousands, if not millions, of new well-formed Latin names for newly discovered species. So my own view is let’s get on with creating the names and let posterity decide whether they need to be prefaced with Candidatus. Carpe diem!
Funding information
Mark Pallen is supported by the Quadram Institute Bioscience BBSRC-funded Strategic Program: Microbes in the Food Chain (project no. BB/R012504/1) and its constituent project BBS/E/F/000PR10351 (Theme 3, Microbial Communities in the Food Chain) and by the Medical Research Council CLIMB-BIG-DATA grant MR/T030062/1.
Acknowledgements
I thank Aharon Oren and Iain Sutcliffe for comments on an early draft of this paper.
Conflicts of interest
The author declares that there are no conflicts of interest
References
- 1.Parker CT, Tindall BJ, Garrity GM. International Code of Nomenclature of Prokaryotes. Int J Syst Evol Microbiol. 2019;69:S1–S111. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 2.de Candolle A. Lois de la nomenclature botanique. Masson: 1867. [DOI] [Google Scholar]
- 3.Buchanan RE, St John-Brooks R, Breed RS. International Bacteriological Code of Nomenclature. J Bacteriol. 1948;55:287–306. doi: 10.1128/jb.55.3.287-306.1948. [DOI] [PubMed] [Google Scholar]
- 4.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Evol Microbiol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [Google Scholar]
- 5.Sneath PHA. The preparation of the approved lists of bacterial names. Int J Syst Evol Microbiol. 2005;55:2247–2249. doi: 10.1099/ijs.0.64137-0. [DOI] [PubMed] [Google Scholar]
- 6.Olsen GJ, Lane DJ, Giovannoni SJ, Pace NR, Stahl DA. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 7.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Relman DA. The identification of uncultured microbial pathogens. J Infect Dis. 1993;168:1–8. doi: 10.1093/infdis/168.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Murray RG, Schleifer KH. Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes. Int J Syst Bacteriol. 1994;44:174–176. doi: 10.1099/00207713-44-1-174. [DOI] [PubMed] [Google Scholar]
- 10.Murray RG, Stackebrandt E. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol. 1995;45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- 11.Labeda DP. Judicial Commission of the International Committee on Systematic Bacteriology VIIIth international Congress of microbiology and applied bacteriology: minutes of the meetings, 17 and 22 August 1996, Jerusalem, Israel. Int J Syst Bacteriol. 1997;47:240–241. doi: 10.1099/00207713-47-1-240. [DOI] [Google Scholar]
- 12.Labeda DP. Judicial Commission of the International Committee on Systematic Bacteriology IXth International (IUMS) Congress of Bacteriology and Applied Microbiology Minutes of the meetings, 14, 15 and 18 August 1999, Sydney, Australia. Int J Syst Bacteriol. 2000;50:2239–2244. [Google Scholar]
- 13.Nayfach S, Roux S, Seshadri R. A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 2020;39:499–509. doi: 10.1038/s41587-020-0718-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parks DH, Rinke C, Chuvochina M. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 15.Pasolli E, Asnicar F, Manara S. Extensive unexplored human microbiome diversity revealed by over 150000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray AE, Freudenstein J, Gribaldo S. Roadmap for naming uncultivated Archaea and Bacteria. Nat Microbiol. 2020;5:987–994. doi: 10.1038/s41564-020-0733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sutcliffe IC, Dijkshoorn L, Whitman WB. Minutes of the International Committee on Systematics of Prokaryotes online discussion on the proposed use of gene sequences as type for naming of prokaryotes, and outcome of vote. Int J Syst Evol Microbiol. 2020;70:4416–4417. doi: 10.1099/ijsem.0.004303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ISME SeqCode initiative: Path Forward for Naming the Uncultivated. 2021. https://www.isme-microbes.org/seqcode-initiative
- 20.Oren A, Garrity GM, Parker CT, Chuvochina M, Trujillo ME. Lists of names of prokaryotic Candidatus taxa. Int J Syst Evol Microbiol. 2020;70:3956–4042. doi: 10.1099/ijsem.0.003789. [DOI] [PubMed] [Google Scholar]
- 21.Jacobi CA, Reichenbach H, Tindall BJ, Stackebrandt E. “Candidatus comitans,” a bacterium living in coculture with Chondromyces crocatus (myxobacteria. Int J Syst Bacteriol. 1996;46:119–122. doi: 10.1099/00207713-46-1-119. [DOI] [PubMed] [Google Scholar]
- 22.Oren A, Garrity GM. Candidatus List No. 2. Lists of names of prokaryotic Candidatus taxa. Int J Syst Evol Microbiol. 2021;71 doi: 10.1099/ijsem.0.004671. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Song W, Wemheuer B, Reveillaud J, Webster N, et al. Comparative genomics reveals ecological and evolutionary insights into sponge-associated Thaumarchaeota . mSystems. 2019;4:e00288-19. doi: 10.1128/mSystems.00288-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hildebrand F, Moitinho-Silva L, Blasche S. Antibiotics-induced monodominance of a novel gut bacterial order. Gut. 2019;68:1781–1790. doi: 10.1136/gutjnl-2018-317715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoch CL, Ciufo S, Domrachev M. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database. 2020;2020 doi: 10.1093/database/baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parte AC, Sardà Carbasse J, Meier-Kolthoff JP, Reimer LC, Göker M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int J Syst Evol Microbiol. 2020;70:5607–5612. doi: 10.1099/ijsem.0.004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker C, Taylor D, Mannor K, Wigley S, Osier N, et al. NamesforLife semantic resolution services for the life sciences. Nature Precedings. 2010 doi: 10.1038/npre.2010.5137.1. [DOI] [Google Scholar]
- 28.Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 2020;38:1079–1086. doi: 10.1038/s41587-020-0501-8. [DOI] [PubMed] [Google Scholar]
- 29.Portier P, Pédron J, Taghouti G. Elevation of Pectobacterium carotovorum subsp. odoriferum to species level as Pectobacterium odoriferum sp. nov., proposal of Pectobacterium brasiliense sp. nov. and Pectobacterium actinidiae sp. nov., emended description of Pectobacterium carotovorum and description of Pectobacterium versatile sp. nov., isolated from streams and symptoms on diverse plants. Int J Syst Evol Microbiol. 2019;69:3207–3216. doi: 10.1099/ijsem.0.003611. [DOI] [PubMed] [Google Scholar]
- 30.He X, McLean JS, Edlund A. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA. 2015;112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedree JK, Bor B, Cen L. Quorum sensing modulates the epibiotic-parasitic relationship between Actinomyces odontolyticus and its Saccharibacteria epibiont, a Nanosynbacter lyticus strain, TM7x. Front Microbiol. 2018;9:2049. doi: 10.3389/fmicb.2018.02049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, et al. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 33.Han XY, Seo YH, Sizer KC. A new Mycobacterium species causing diffuse lepromatous leprosy. Am J Clin Pathol. 2008;130:856–864. doi: 10.1309/AJCPP72FJZZRRVMM. [DOI] [PubMed] [Google Scholar]
- 34.Sharma R, Singh P, McCoy RC. Isolation of Mycobacterium lepromatosis and development of molecular diagnostic assays to distinguish Mycobacterium leprae and M. lepromatosis . Clin Infect Dis. 2020;71:e262–e269. doi: 10.1093/cid/ciz1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glendinning L, Stewart RD, Pallen MJ, Watson KA, Watson M. Assembly of hundreds of novel bacterial genomes from the chicken caecum. Genome Biol. 2020;21:34. doi: 10.1186/s13059-020-1947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hildebrand F, Pallen MJ, Bork P. Towards standardisation of naming novel prokaryotic taxa in the age of high-throughput microbiology. Gut. 2020;69:1358–1359. doi: 10.1136/gutjnl-2019-319045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilroy R, Ravi A, Getino M. Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture. PeerJ. 2021;9:e10941. doi: 10.7717/peerj.10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pallen MJ, Telatin A, Oren A. The next million names for Archaea and Bacteria. Trends Microbiol. 2021;29:289–298. doi: 10.1016/j.tim.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Oren A. A plea for linguistic accuracy - also for Candidatus taxa. Int J Syst Evol Microbiol. 2017;67:1085–1094. doi: 10.1099/ijsem.0.001715. [DOI] [PubMed] [Google Scholar]
- 40.Martinson VG, Gawryluk RMR, Gowen BE, Curtis CI, Jaenike J, et al. Multiple origins of obligate nematode and insect symbionts by a clade of bacteria closely related to plant pathogens. Proc Natl Acad Sci USA. 2020;117:31979–31986. doi: 10.1073/pnas.2000860117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oren A. Naming novel prokaryotic taxa discovered in the human gut. Gut. 2020;69:969–970. doi: 10.1136/gutjnl-2019-318716. [DOI] [PubMed] [Google Scholar]
- 42.Chambers J, Sparks N, Sydney N, Livingstone PG, Cookson AR, et al. Comparative genomics and pan-genomics of the Myxococcaceae, including a description of five novel species: Myxococcus eversor sp. nov., Myxococcus llanfairpwllgwyngyllgogerychwyrndrobwllllantysiliogogogochensis sp. nov., Myxococcus vastator sp. nov., Pyxidicoccus caerfyrddinensis sp. nov., and Pyxidicoccus trucidator sp. nov. Genome Biol Evol. 2020;12:2289–2302. doi: 10.1093/gbe/evaa212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montano HG, Dally EL, Davis RE, Pimentel JP, Brioso PST. First report of natural infection by “Candidatus Phytoplasma brasiliense” in Catharanthus roseus . Plant Dis. 2001;85:1209. doi: 10.1094/PDIS.2001.85.11.1209C. [DOI] [PubMed] [Google Scholar]
- 44.Tindall BJ, Grimont PAD, Garrity GM, Euzéby JP. Nomenclature and taxonomy of the genus Salmonella . Int J Syst Evol Microbiol. 2005;55:521–524. doi: 10.1099/ijs.0.63580-0. [DOI] [PubMed] [Google Scholar]
- 45.Henkel JV, Vogts A, Werner J. Candidatus Sulfurimonas marisnigri sp. nov. and Candidatus Sulfurimonas baltica sp. nov., thiotrophic manganese oxide reducing chemolithoautotrophs of the class Campylobacteria isolated from the pelagic redoxclines of the Black Sea and the Baltic Sea. Syst Appl Microbiol. 2021;44:126155. doi: 10.1016/j.syapm.2020.126155. [DOI] [PubMed] [Google Scholar]
- 46.Silva PA, Huang J, Wulff NA, Zheng Z, Krugner R, et al. Genome sequence resource of ‘Candidatus Liberibacter asiaticus’ strain 9PA from Brazil. Plant Dis. 2021;105:199–201. doi: 10.1094/PDIS-05-20-1018-A. [DOI] [PubMed] [Google Scholar]
- 47.Kumar D, Kumar G, Jagadeeshwari U, Sasikala C, Ramana CV. “Candidatus Laterigemmans baculatus” gen. nov. sp. nov., the first representative of rod shaped planctomycetes with lateral budding in the family Pirellulaceae . Syst Appl Microbiol. 2021;44:126188. doi: 10.1016/j.syapm.2021.126188. [DOI] [PubMed] [Google Scholar]
- 48.Eisenberg T, Gronow S, Falgenhauer J. Sneathia vaginalis sp. nov. (Fusobacteriales, Leptotrichiaceae) as a replacement of the species ‘Sneathia amnii’ Harwich et al. 2012 and Leptotrichia amnionii Shukla et al. 2002, and emended description of Sneathia Collins et al. 2001. Int J Syst Evol Microbiol. 2019;71:ijsem.0.004663. doi: 10.1099/ijsem.0.004663. [DOI] [PubMed] [Google Scholar]
- 49.Aksoy S. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int J Syst Evol Microbiol. 1995;45:848–851. doi: 10.1099/00207713-45-4-848. [DOI] [PubMed] [Google Scholar]
- 50.Maier S. Description of Thioploca ingrica sp. nov., nom. rev. Int J Syst Evol Microbiol. 1984;34:344–345. [Google Scholar]
- 51.Maier S, Gallardo VA. Thioploca araucae sp. nov. and Thioploca chileae sp. nov. Int J Syst Evol Microbiol. 1984;34:414–418. [Google Scholar]
- 52.Schulz HN, Brinkhoff T, Ferdelman TG, Mariné MH, Teske A, et al. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science. 1999;284:493–495. doi: 10.1126/science.284.5413.493. [DOI] [PubMed] [Google Scholar]
- 53.Drozanski WJ. Sarcobium lyticum gen. nov., sp. nov., an obligate intracellular bacterial parasite of small free-living amoebae. Int J Syst Evol Microbiol. 1991;41:82–87. [Google Scholar]
- 54.Krumholz LR, Bryant MP, Brulla WJ, Vicini JL, Clark JH, et al. Proposal of Quinella ovalis gen. nov., sp. nov., based on phylogenetic analysis. Int J Syst Evol Microbiol. 1993;43:293–296. doi: 10.1099/00207713-43-2-293. [DOI] [PubMed] [Google Scholar]
- 55.Heckmann K, Schmidt HJ. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus . Int J Syst Evol Microbiol. 1987;37:456–457. [Google Scholar]
- 56.Brockman ER. In: Bergey’s Manual of Systematic Bacteriology. Staley J, Bryant M, Pfennig N, Holt J, editors. Baltimore: The Williams & Wilkins Co; 1989. Genus I. Polyangium link 1809, 42AL; pp. 2159–2162. Vol. 3. [Google Scholar]
- 57.Starr MP, Schmidt JM. Planctomyces stranskae (ex Wawrik 1952) sp. nov., nom. rev. and Planctomyces guttaeformis (ex Hortobágyi 1965) sp. nov., nom. rev. Int J Syst Evol Microbiol. 1984;34:470–477. [Google Scholar]
- 58.Hookey JV, Saunders NA, Fry NK, Birtles RJ, Harrison TG. Phylogeny of legionellaceae based on small-subunit ribosomal DNA sequences and proposal of Legionella lytica comb. nov. for legionella-like amoebal pathogens. Int J Syst Evol Microbiol. 1996;46:526–531. [Google Scholar]
- 59.Gromov BV, Ossipov DV. Holospora (ex Hafkine 1890) nom. rev., a genus of bacteria inhabiting the nuclei of Paramecia. Int J Syst Evol Microbiol. 1981;31:348–352. [Google Scholar]
- 60.Lewis GE, Huxsoll DL, Ristic M, Johnson AJ. Experimentally induced infection of dogs, cats, and nonhuman primates with Ehrlichia equi, etiologic agent of equine ehrlichiosis. Am J Vet Res. 1975;36:85–88. [PubMed] [Google Scholar]
- 61.Bermudes D, Chase D, Margulis L. Morphology as a basis for taxonomy of large spirochetes symbiotic in wood-eating cockroaches and termites: Pillotina gen. nov., nom. rev.; Pillotina calotermitidis sp. nov., nom. rev.; Diplocalyx gen. nov., nom. rev.; Diplocalyx calotermitidis sp. nov., nom. rev.; Hollandina gen. nov., nom. rev.; Hollandina pterotermitidis sp. nov., nom. rev.; and Clevelandina reticulitermitidis gen. nov., sp. nov. Int J Syst Bacteriol. 1988;38:291. doi: 10.1099/00207713-38-3-291. [DOI] [PubMed] [Google Scholar]
- 62.Starr MP, Sayre RM. Pasteuria thornei sp. nov. and Pasteuria penetrans sensu stricto emend., mycelial and endospore-forming bacteria parasitic, respectively, on plant-parasitic nematodes of the genera Pratylenchus and Meloidogyne . Ann Inst Pasteur Microbiol. 1988;139:11–31. doi: 10.1016/0769-2609(88)90094-4. [DOI] [PubMed] [Google Scholar]
- 63.Sayre RM, Starr MP. Pasteuria penetrans (ex Thorne, 1940) nom. rev., comb. n., sp. n., a mycelial and endospore-forming bacterium parasitic in plant-parasitic nematodes. Proc Helminthol Soc Wash. 1985;52:149–165. [Google Scholar]
- 64.Sayre RM, Wergin WP, Schmidt JM, Starr MP. Pasteuria nishizawae sp. nov., a mycelial and endospore-forming bacterium parasitic on cyst nematodes of genera Heterodera and Globodera. Res Microbiol. 1991;142:551–564. doi: 10.1016/0923-2508(91)90188-g. [DOI] [PubMed] [Google Scholar]
- 65.Gorlenko VM, Pivovarova TA. On the assignment of the blue-green alga Oscillatoria coerulescens Gicklhorn, 1921, to the new genus of Chlorobacteria oscillochloris nov. gen. Izv Akad Nauk SSSR Ser Biol. 1977;3:396–409. [Google Scholar]
- 66.Keppen OI, Tourova TP, Kuznetsov BB, Ivanovsky RN, Gorlenko VM. Proposal of Oscillochloridaceae fam. nov. on the basis of a phylogenetic analysis of the filamentous anoxygenic phototrophic bacteria, and emended description of Oscillochloris and Oscillochloris trichoides in comparison with further new isolates. Int J Syst Evol Microbiol. 2000;50:1529–1537. doi: 10.1099/00207713-50-4-1529. [DOI] [PubMed] [Google Scholar]
- 67.Duncan AJ, Carman RJ, Olsen GJ, Wilson KH. Assignment of the agent of Tyzzer’s disease to Clostridium piliforme comb. nov. on the basis of 16S rRNA sequence analysis. Int J Syst Bacteriol. 1993;43:314–318. doi: 10.1099/00207713-43-2-314. [DOI] [PubMed] [Google Scholar]
- 68.Lagkouvardos I, Lesker TR, Hitch TCA. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome. 2019;7:28. doi: 10.1186/s40168-019-0637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darwin C. On the Origin of Species. 1st edn. London: John Murray; 1859. [Google Scholar]
- 70.Hennig W. Grundzüge einer Theorie der phylogenetischen Systematik. Berlin: Deutscher Zentralverlag; 1950. [Google Scholar]
- 71.Loman NJ, Pallen MJ. Twenty years of bacterial genome sequencing. Nat Rev Microbiol. 2015;13:787–794. doi: 10.1038/nrmicro3565. [DOI] [PubMed] [Google Scholar]
- 72.Ho C-C, Lau SKP, Woo PCY. Romance of the three domains: How cladistics transformed the classification of cellular organisms. Protein Cell. 2013;4:664–676. doi: 10.1007/s13238-013-3050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pace NR. Time for a change. Nature. 2006;441:289. doi: 10.1038/441289a. [DOI] [PubMed] [Google Scholar]
- 74.Pace NR. Problems with “procaryote.”. J Bacteriol. 2009;191:2008–2010. doi: 10.1128/JB.01224-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pace NR. It’s time to retire the prokaryote. Microbiology Today. 2009;36:84. [Google Scholar]
- 76.Pallen MJ. Time to recognise that mitochondria are bacteria. Trends Microbiol. 2011;19:58–64. doi: 10.1016/j.tim.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 77.Castelle CJ, Banfield JF. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell. 2018;172:1181–1197. doi: 10.1016/j.cell.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 78.Sutcliffe IC. Challenging the anthropocentric emphasis on phenotypic testing in prokaryotic species descriptions: rip it up and start again. Front Genet. 2015;6:218. doi: 10.3389/fgene.2015.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chun J, Oren A, Ventosa A. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 80.Oren A, Garrity GM, Parte AC. Why are so many effectively published names of prokaryotic taxa never validated. Int J Syst Evol Microbiol. 2018;68:2125. doi: 10.1099/ijsem.0.002851. [DOI] [PubMed] [Google Scholar]
- 81.La Scola B, Fenollar F, Fournier PE, Altwegg M, Mallet MN, et al. Description of Tropheryma whipplei gen. nov., sp. nov., the Whipple’s disease bacillus. Int J Syst Evol Microbiol. 2001;51:1471–1479. doi: 10.1099/00207713-51-4-1471. [DOI] [PubMed] [Google Scholar]
- 82.Gawande A. The Checklist Manifesto: How To Get Things Right. New York: Metropolitan Books; 2010. [Google Scholar]
- 83.Glendinning L, Stewart RD, Pallen MJ, Watson KA, Watson M. Author Correction: Assembly of hundreds of novel bacterial genomes from the chicken caecum. Genome Biol. 2021;22:60. doi: 10.1186/s13059-021-02284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oren A, Arahal DR, Rosselló-Móra R, Sutcliffe IC, Moore ERB. Preparing a revision of the International Code of Nomenclature of Prokaryotes. Int J Syst Evol Microbiol. 2021;71:ijsem.0.004598. doi: 10.1099/ijsem.0.004598. [DOI] [PubMed] [Google Scholar]
- 85.Sanford RA, Lloyd KG, Konstantinidis KT, Löffler FE. Microbial Taxonomy Run Amok. Trends Microbiol. 2021;29:394–404. doi: 10.1016/j.tim.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 86.Brower AVZ. Dead on arrival: a postmortem assessment of ”phylogenetic nomenclature”, 20+ years on. Cladistics. 2020;36:627–637. doi: 10.1111/cla.12432. [DOI] [Google Scholar]
- 87.Greuter W, Garrity G, Hawksworth DL. Draft BioCode (2011): principles and rules regulating the naming of organisms. Taxon. 2011;60:201–212. doi: 10.1002/tax.601019. [DOI] [Google Scholar]
- 88.Smith D, da Silva M, Jackson J, Lyal C. Explanation of the Nagoya protocol on access and benefit sharing and its implication for microbiology. Microbiology. 2017;163:289–296. doi: 10.1099/mic.0.000425. [DOI] [PubMed] [Google Scholar]
- 89.Wylensek D, Hitch TCA, Riedel T. A collection of bacterial isolates from the pig intestine reveals functional and taxonomic diversity. Nat Commun. 2020;11:6389. doi: 10.1038/s41467-020-19929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lewis WH, Tahon G, Geesink P, Sousa DZ, Ettema TJG. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol. 2021;19:225–240. doi: 10.1038/s41579-020-00458-8. [DOI] [PubMed] [Google Scholar]