Abstract
The novel, anaerobic, Gram-positive, rod-shaped bacterial strain, ResAG-91T, was isolated from a faecal sample of a male human volunteer. Analysis of the 16S rRNA gene sequence revealed that strain ResAG-91T showed high similarity to the type strains of Adlercreutzia equolifaciens subsp. equolifaciens and Adlercreutzia equolifaciens subsp. celatus . Analysis of the whole draft genome sequences, i.e. digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI), of strain ResAG-91T and the type strains of Adlercreutzia species revealed that strain ResAG-91T represents a novel species of the genus Adlercreutzia . The genome size of strain ResAG-91T is 2.8 Mbp and the G+C content is 63.3 mol%. The major respiratory quinone of strain ResAG-91T was MMK-5 (methylmenaquinone). Major cellular fatty acids were C15 : 0 anteiso, C14 : 0 iso and C14 : 0 2-OH. Galactose and ribose were detected as major whole cell sugars. Furthermore, the peptidoglycan type of strain ResAG-91T was A1γ with meso-diaminopimelic acid. The polar lipids were phosphatidylglycerol, diphosphatidylglycerol, one unidentified lipid, three unidentified phospholipids and five unidentified glycolipids. Strain ResAG-91T was able to metabolize the stilbene resveratrol into dihydroresveratrol. On the basis of this polyphasic approach, including phenotypical, molecular (16S rRNA gene and whole genome sequencing) and biochemical (fatty acids, quinones, polar lipids, peptidoglycan, whole cell sugars, Rapid ID32A and API20A) analyses, we propose the novel species Adlercreutzia rubneri sp. nov. with the type and only strain ResAG-91T (=DSM 111416T=JCM 34176T=LMG 31897T).
Keywords: Adlercreutzia, anaerobic, Eggerthellaceae, faeces, taxonomy
In 2008, the genus Adlercreutzia was proposed in honour of Herman Adlercreutz for his contributions to research on the effects of phyto-oestrogens on human health [1]. The genus Adlercreutzia belongs to the family Eggerthellaceae within the class Coriobacteriia (phylum Actinobacteria ) [2]. In 2018, Nouioui et al. [3] investigated the phylum Actinobacteria using a genome-based taxonomic approach. At the time of the present writing, the genus Adlercreutzia includes the following species and subspecies: (I) Adlercreutzia caecimuris [4], (II) Adlercreutzia equolifaciens subsp. celatus [5], (III) Adlercreutzia equolifaciens subsp. equolifaciens (type species of the genus) [1], (IV) Adlercreutzia mucosicola [6] and (V) Adlercreutzia muris [7]. Adlercreutzia caecicola is not included in the analyses within this paper as the taxonomic position of the type strain of A. caecicola was recently proposed to need revision [8].
Members of the family Eggerthellaceae have mostly been isolated from the gastrointestinal tract or faecal samples of humans [9, 10], mice [4, 6, 7, 11], rats [5], dogs [12] and sheep [13]. Some strains of this family play an important role in the bioactivation and inactivation of secondary plant metabolites such as digoxin [14], daidzein [1, 5, 6], ellagic acid [15], resveratrol [16] and pyrrolizidine alkaloids [13]. In particular, strains of the genus Adlercreutzia (e.g. A. equolifaciens subsp. celatus do03T, A. equolifaciens subsp. equolifaciens FJC-B9T and A. mucosicola Mt1B8T) are described to metabolize the isoflavone daidzein to the more bioactive S-equol [1, 5, 6]. In addition, A. equolifaciens subsp. equolifaciens FJC-B9T was described to metabolize the stilbene resveratrol into dihydroresveratrol [16].
Strain ResAG-91T was isolated from a human faecal sample during a trial which aimed to isolate bacterial strains that are able to metabolize trans-resveratrol into dihydroresveratrol and/or lunularin. Within the same trial, the former newly described strains Rubneribacter badeniensis ResAG-85T and Enteroscipio rubneri ResAG-96T were isolated [10]. Therefore, the isolation conditions were the same as described previously [10]. Briefly, strain ResAG-91T was isolated from a fresh faecal sample of a healthy human, moderately obese male volunteer (30 years, body mass index 33.2 kg m−2). All dilutions and cultivation steps were performed at 37 °C under strictly anaerobic conditions, either in an A45 anaerobic workstation (Don Whitley Scientific) under atmospheric conditions of N2/CO2/H2 (80 : 10 : 10) or in Hungate tubes flushed with N2/CO2 (80 : 20). The faecal sample (7.5 g) was mixed with 22.5 ml modified brain heart infusion medium (MBHI). MBHI comprises BHI (Merck) supplemented with 0.5 % yeast extract, 0.05 % l-cysteine monohydrochloride (Roth), 1 mg l−1 resazurin sodium salt, 2.5 mg l−1 haem solution and 2 µg ml−1 vitamin K1 solution (Sigma-Aldrich). After a short period of incubation (10 min, 120 r.p.m., 37 °C), the faecal suspension was centrifuged (10 min, room temperature, 300 g ) and 1 ml of the faecal suspension supernatant was inoculated in MBHI (10 ml) supplemented with ampicillin (1 µg ml−1), colistin (5 µg ml−1), chloramphenicol (5 µg ml−1), cholic acid (18 µg ml−1) and trans-resveratrol (trans-3,5,4′-trihydroxystilbene; 80 µM). Every 48 h the above-mentioned supplements (antibiotics, cholic acid and trans-resveratrol) were added. Samples were serially diluted with PBS and plated onto MBHI agar plates with the same supplements as mentioned above. Strain ResAG-91T was isolated after 72 h, subcultured after 12 days of growth on MBHI agar plates at 37 °C and streaked repeatedly until purity.
After 48–72 h (strictly anaerobic conditions, 37 °C), strain ResAG-91T occurred as very small (Ø ≈ 1 mm), pale white, semi-translucent colonies. Optical density in liquid media was visually determined using McFarland standards (bioMérieux). Growth in MBHI was slow and similar to McFarland 0.5 at 48 h. Mid-exponential to stationary phase cells were visualized under a phase-contrast microscope (Leica) and occurred as non-motile, very small rods (singly or in short chains). Routine microbiological tests (i.e. Gram-staining, oxidase and catalase activity) using standard techniques showed that strain ResAG-91T was Gram-positive, oxidase-negative and catalase-negative.
For comparative analysis, the following type strains were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ): A. caecimuris DSM 21839T, A. equolifaciens subsp. celatus DSM 18785T, A. equolifaciens subsp. equolifaciens DSM 19450T, A. mucosicola DSM 19490T, A. muris DSM 29508T and Eggerthella lenta DSM 2243T.
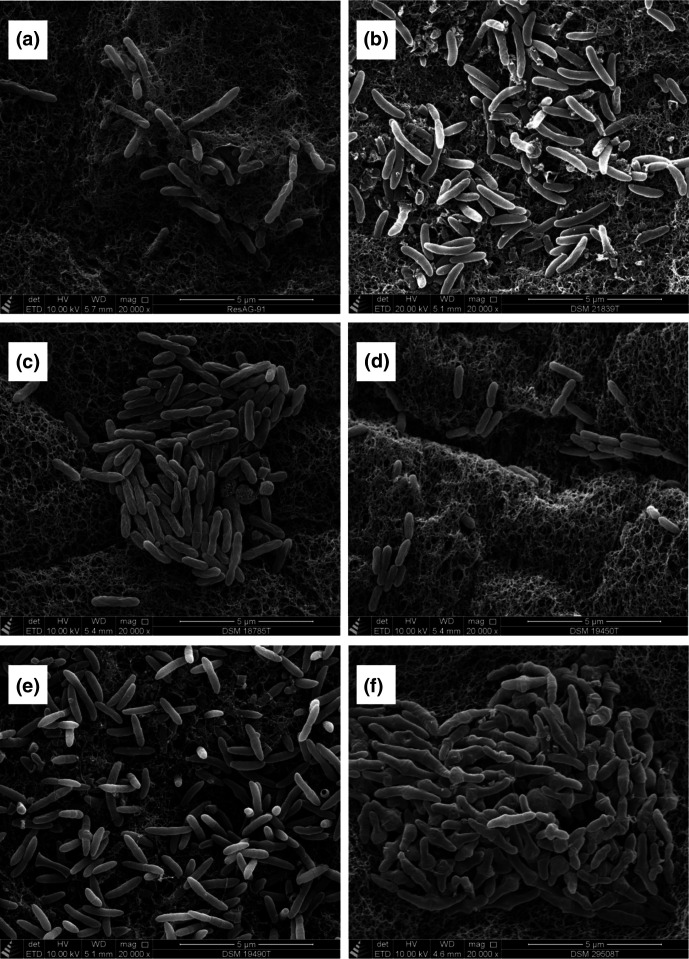
For characterization by scanning electron microscopy (SEM), all five type strains of the genus Adlercreutzia and strain ResAG-91T were grown on MBHI agar plates for 3 days (37 °C, strictly anaerobic conditions). Strains were fixed, dehydrated, dried and mounted on aluminium stubs as described previously [10]. SEM imaging was carried out by using a Quanta 250 FEG scanning electron microscope (FEI) with an Everhart-Thornley detector in high vacuum mode (0.0003 Pa) and an acceleration voltage of 10 kV. The rod-shaped morphology of strain ResAG-91T was confirmed by SEM (Fig. 1a). Furthermore, we confirmed the rod-shaped morphology of A. caecimuris DSM 21839T (Fig. 1b), A. equolifaciens subsp. celatus DSM 18785T (Fig. 1c), A. mucosicola DSM 19490T (Fig. 1e) and A. muris DSM 29508T (Fig. 1f). In the original strain description, the morphology of A. equolifaciens subsp. equolifaciens DSM 19450T was described as coccobacilli [1], but here we clearly could show the rod-shaped morphology (Fig. 1d). The cell lengths and diameters of 50 cells of strain ResAG-91T and the type strains of Adlercreutzia species were measured using ObjectJ in ImageJ 1.53c [17]. The mean length and mean diameter of strain cells of ResAG-91T were 1.29±0.33 and 0.31±0.04 µm, respectively (Table S1, available in the online version of this article). In addition, data for the cell length and diameter of the type strains of Adlercreutzia species are given in Table S1.
Fig. 1.
Scanning electron micrographs of cells of (a) strain ResAG-91T, (b) A. caecimuris DSM 21839T, (c) A. equolifaciens subsp. celatus DSM 18785T, (d) A. equolifaciens subsp. equolifaciens DSM 19450T, (e) A. mucosicola DSM 19490T and (f) A. muris DSM 29508T. Bars, 5 µm.
The biochemical characteristics of strain ResAG-91T were determined using Rapid ID 32A and API 20A test strips (bioMérieux) according to the manufacturer’s instructions. For comparative analysis the five type strains of the genus Adlercreutzia were also tested. After 48 h, the type strains of the genus Adlercreutzia (i.e. DSM 21839T, DSM 18785T, DSM 19450T, DSM 19490T, DSM 29508T) and strain ResAG-91T tested with API 20A showed no metabolic reaction. The results of Rapid ID 32A tests are listed in Table 1 if at least one strain gave a positive result for a specific substrate. All strains were negative for urease, α-galactosidase, β-galactosidase, β-galactosidase 6-phosphate, α-glucosidase, β-glucosidase, α-arabinosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, mannose fermentation, raffinose fermentation, α-fucosidase, nitrate reduction, indole production, alkaline phosphatase and glutamyl glutamic acid arylamidase. Strain ResAG-91T was capable of producing arginine dihydrolase and showed a weak reaction for the production of arginine arylamidase and leucine arylamidase. The production of arginine dihydrolase and production of arginine arylamidase seems to be a common characteristic of the genus Adlercreutzia .
Table 1.
Biochemical characteristics of strain ResAG-91T and type strains of species of the genus Adlercreutzia
Strains: 1, ResAG-91T; 2, A. caecimuris DSM 21839T; 3, A. equolifaciens subsp. celatus DSM 18785T; 4, A. equolifaciens subsp. equolifaciens DSM 19450T; 5, A. mucosicola DSM 19490T; 6, A. muris DSM 29508T. All strains were tested using the API 20A and Rapid ID 32A systems within this study. +, Positive; −, negative; w, weakly positive; (+), minor amounts; MK, menaquinone; MMK, methylmenaquinone; DMMK, dimethylmenaquinone; nd, not determined.
|
Characteristic |
1 |
2 |
3 |
4 |
5 |
6 |
|---|---|---|---|---|---|---|
|
Rapid ID 32A* |
||||||
|
Alanine arylamidase |
− |
− |
− |
− |
+ |
− |
|
Arginine arylamidase |
w |
w† |
+ |
+ |
+ |
w† |
|
Arginine dihydrolase |
+ |
+ |
+ |
+ |
+ |
+ |
|
Glutamic acid decarboxylase |
− |
+ |
− |
− |
− |
− |
|
Glycine arylamidase |
− |
− |
− |
− |
+ |
− |
|
Histidine arylamidase |
− |
− |
− |
w† |
+ |
− |
|
Leucine arylamidase |
w |
− |
− |
+ |
+ |
w† |
|
Leucyl glycine arylamidase |
− |
− |
− |
− |
+ |
− |
|
Phenylalanine arylamidase |
− |
− |
− |
− |
+ |
− |
|
Proline arylamidase |
− |
− |
− |
− |
+ |
− |
|
Pyroglutamic acid arylamidase |
− |
− |
− |
− |
w |
− |
|
Serine arylamidase |
− |
− |
− |
− |
+ |
− |
|
Tyrosine arylamidase |
− |
− |
− |
− |
+ |
− |
|
Ox-bile tolerant |
||||||
|
Growth on 2 % bile* |
+ |
+ |
+ |
+ |
+ |
+ |
|
Whole cell sugars |
||||||
|
Galactose |
+*‡ |
+§‡ |
+*‡ |
+‡ || |
+§‡ |
+*‡ |
|
Glucose |
(+)*‡ |
+§‡ |
+*‡ |
+‡ || |
−§‡ |
+*‡ |
|
Ribose |
+*‡ |
+§‡ |
+*‡ |
+‡ || |
+§‡ |
+*‡ |
|
Respiratory quinones |
MMK-5 (94.9%), MMK-6 (2.9%), DMMK-5 (0.9%), MK-6 (0.6%), DMMK-6 (0.5%), MK-5 (0.3%)*‡ |
MMK-6 (60%), DMMK-6 (40%)§‡ |
MMK-5 (83.0%), MMK-6 (8.2%), DMMK-6 (3.8%), DMMK-5 (3.1%), MK-5 (1.0%), MK-6 (0.9%)*‡ |
MMK-5 (68.9%), MMK-6 (16.4%), DMMK-5 (7.4%), DMMK-6 (6.1%), MK-5 (1.2%)‡ || |
MMK-6 (100%)§‡ |
MMK-5 (83.8%), MK-6 (9.3%), MMK-6 (4.5 %), MK-5 (1.9%), DMMK-6 (0.5%)*‡ |
|
Peptidoglycan |
||||||
|
Type |
A1γ*‡ |
A1γ or A4㧇 |
A1γ*‡ |
A1γ‡ || |
nd |
A1γ*‡ |
|
Diaminopimelic acid |
meso*‡ |
meso§‡ |
meso*‡ |
meso‡ || |
LL§‡ |
meso*‡ |
|
Polar lipids |
||||||
|
Aminolipid |
n=0*‡ |
n=0§‡ |
n=0*‡ |
n=0|| ‡ |
n=0§‡ |
n=1*‡ |
|
Diphosphatidylglycerol |
n=1*‡ |
n=1§‡ |
n=1*‡ |
n=1|| ‡ |
n=1§‡ |
n=1*‡ |
|
Glycolipids |
n=5*‡ |
n=2§‡ |
n=6*‡ |
n=3|| ‡ |
n=4§‡ |
n=4*‡ |
|
Lipid |
n=1*‡ |
n=1§‡ |
n=2*‡ |
n=3|| ‡ |
n=0§‡ |
n=2*‡ |
|
Phosphatidylglycerol |
n=1*‡ |
n=1§‡ |
n=1*‡ |
n=0|| ‡ |
n=1§‡ |
n=1*‡ |
|
Phospholipids |
n=3*‡ |
n=1§‡ |
n=2*‡ |
n=0|| ‡ |
n=3§‡ |
n=3*‡ |
*Results were obtained within this study.
†In original strain descriptions indicated as negative.
‡Analysed by the Identification Service of the DSMZ (Braunschweig, Germany).
§Data were obtained from the original strain descriptions.
||Data were obtained from the literature [8].
A bile-tolerant characteristic (growth with 20 % bile) was described for the type strain of A. equolifaciens subsp. celatus [5]. In addition, sensitivity against bile was described for the type strain of A. equolifaciens subsp. equolifaciens at a concentration of 20 % [1] whereas bile sensitivity at a concentration at 0.5 % bile was reported for the type strains of A. mucosicola [6] and A. caecimuris [4]. We investigated the bile tolerance of strain ResAG-91T and the type strains of the genus Adlercreutzia according to Nagai et al. [18] except that MBHI agar was used. Briefly, strains were plated on MBHI agar with and without ox-bile (Sigma-Aldrich) at a concentration of 2 % (w/v). Eggerthella lenta DSM 2243T was used as a bile-resistant positive control [19]. After 5 days of incubation at 37 °C under anaerobic conditions, well grown colonies of all tested strains including strain ResAG-91T were observed at concentrations of 2 % (w/v) ox-bile (Table 1).
Analyses of respiratory quinones, fatty acids, whole cell sugars, peptidoglycan structure and polar lipids of strains ResAG-91T, A. muris DSM 29508T and A. equolifaciens subsp. celatus DSM 18785T were carried out by the Identification Service of the DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). For this, the biomass of liquid cultures (MBHI, 37 °C, strictly anaerobic conditions) of strains ResAG-91T (24.3 L), A. muris DSM 29508T (36.9 L) and A. equolifaciens subsp. celatus DSM 18785T (27.0 L) were collected by centrifugation (9622 g ; 10 min; 4 °C) and the total moist biomass of strains ResAG-91T (6.45 g), A. muris DSM 29508T (3.94 g) and A. equolifaciens subsp. celatus DSM 18785T (4.50 g) was sent to the DSMZ. Ribose, galactose and minor amounts of glucose were detected as whole cell sugars in cells of strain ResAG-91T. The results of whole cell sugar analysis of strain ResAG-91T in comparison with the type strains of the genus Adlercreutzia are shown in Table 1. Polar lipids in strain ResAG-91T consisted of phosphatidylglycerol, diphosphatidylglycerol, one unidentified lipid, three unidentified phospholipids and five unidentified glycolipids (Fig. S1). The major fatty acids of strain ResAG-91T were C15 : 0 anteiso (40.59 %), C14 : 0 iso (14.95 %) and C14 : 0 2-OH (12.58 %). A list of the fatty acids of strain ResAG-91T, A. equolifaciens subsp. celatus DSM 18785T, A. equolifaciens subsp. equolifaciens DSM 19450T and A. muris DSM 29508T is given in Table S2 as these strains were cultivated and analysed under the same conditions. The following respiratory quinones were detected in strain ResAG-91T (Table 1): MMK-5 (methylmenaquinone; 94.9 %), MMK-6 (2.9 %) DMMK-5 (dimethylmenaquinone; 0.9 %), MK-6 (menaquinone; 0.6 %), DMMK-6 (0.5 %) and MK-5 (0.3 %). Table 1 shows the major menaquinones of strain ResAG-91T and the type strains of the genus Adlercreutzia . Predominant respiratory quinones were MMK-5 (menamethylquinone) or MMK-6. The peptidoglycan type of strain ResAG-91T was A1γ with meso-diaminopimelic acid. This peptidoglycan type was reported for the majority of the type strains of Adlercreutzia species (Table 1).
Strain ResAG-91T was investigated for its ability to metabolize trans-resveratrol into dihydroresveratrol: strain ResAG-91T was incubated in MBHI liquid medium with trans-resveratrol (78.125 µM) at 37 °C under strictly anaerobic conditions in Hungate tubes. At 0, 24, 48 and 72 h, an aliquot (500 µl) was taken and stored at −80 °C. For HPLC with diode array detection (HPLC-DAD) analysis, a 200 µl sample was extracted three times with 500 µl ethyl acetate/2-propanol/1-butanol (90:5:5, by vol.). The combined extracts were evaporated under a gentle stream of nitrogen and the residue was resolved in 200 µl of 0.1 % aqueous formic acid/methanol/acetonitrile (90:5:5, by vol.). The sample was filtrated using a syringe filter (PTFE, 0.2 µm, 4 mm; Phenomenex) and analysed by HPLC-DAD (Prominence system; Shimdazu) with LC conditions as described previously [16] and small alteration for the gradient (0–2 min isocratic with 15 % B, 2–11 min from 15 to 23 % B, 11–20.5 min isocratic with 23 % B, 20.5–31 min from 23 to 56 % B, 31–33 min from 56 to 100 % B, 33–36 min isocratic with 100 % B, 36–40 min from 100 to 15 % B, and 40–48 min isocratic at the initial conditions). To monitor the analytes, the trace at 250 nm was used. The identity of analytes was confirmed by the retention time and the UV–Vis spectra. To ensure proper analysis, negative controls (incubation of trans-resveratrol without strain ResAG-91T) as well as blanks (strain ResAG-91T incubated in MBHI liquid medium with DMSO) were measured together with the study samples. After 24 h, dihydroresveratrol was detected in samples previously inoculated with strain ResAG-91T and trans-resveratrol. This result was confirmed by an independent second experiment.
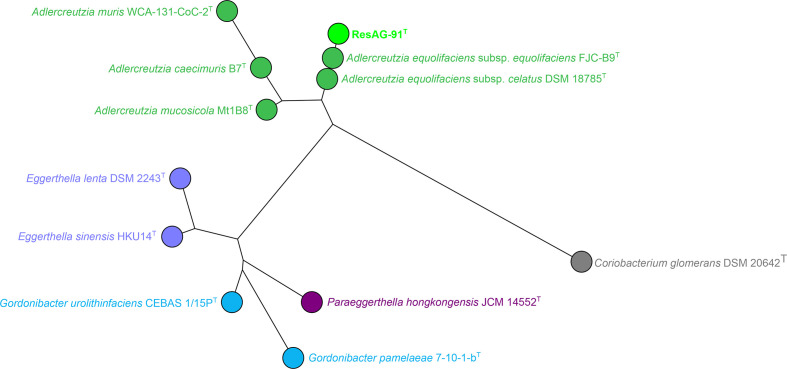
For 16S rRNA gene sequencing, isolation of bacterial DNA of strain ResAG-91T, 16S rRNA gene amplification and analysis were carried out as described previously [10]. Briefly, the bacterial DNA was isolated using the Blood and Tissue Kit (Qiagen) and the almost complete 16S rRNA gene was amplified using the primers 16Sseq fw (5′-ATAGTTTGATCMTGGCTCAG-3′) and 16Sseq rev (5′-GGNTACCTTGTTACGACTTC-3′). The 16S rRNA gene sequence (1394 bp, GenBank accession number MH553318) of strain ResAG-91T was used for blastn searches. Strain ResAG-91T was identified as a member of the genus Adlercreutzia and showed the highest similarity to A. equolifaciens subsp. equolifaciens DSM 19450T. Fig. 2 shows a maximum-likelihood tree (BioNumerics, version 7.6; Applied Maths) based on the 16S rRNA gene sequences of closely related validly published type strains of the family Eggerthellaceae . Comparable to the blastn analysis, the nearest neighbours of strain ResAG-91T were the type strains of A. equolifaciens subsp. equolifaciens and A. equolifaciens subsp. celatus (Fig. 2). The cluster analysis was repeated with two additional clustering methods, namely neighbour-joining and maximum-parsimony, and confirmed the taxonomic position of strain ResAG-91T (Figs S2 and S3).
Fig. 2.
Analysis of 16S rRNA gene sequences (length ca. 1500 bp) of strain ResAG-91T and the type strains of Adlercreutzia , Eggerthella, Paraeggerthella and Gordonibacter. Coriobacterium glomerans was used as an outgroup. The tree was built using maximum-likelihood with Jukes–Cantor as the evolutionary model (BioNumerics, version 7.6; Applied Maths). Type strains of the genera Adlercreutzia , Eggerthella , Gordonibacter , Paraeggerthella and Coriobacterium are labelled in green, purple, blue, plum and grey, respectively.
The whole draft genome sequences of strains ResAG-91T (GenBank accession no. WPOO00000000), A. equolifaciens subsp. celatus DSM 18785T (QICA00000000) and A. muris DSM 29508T (WAJS00000000) were sequenced on an Illumina MiSeq by our group and published previously [20–22]. Characteristics of the genome sequences of strain ResAG-91T and all type strains of Adlercreutzia (e.g. genome size, G+C content and number of proteins) were obtained by the Type (Strain) Genome Server (TYGS) [23] and are summarized in Table 2. The genome sizes, G+C content and number of proteins of Adlercreutzia species ranged from 2.76 to 3.01 Mbp, 63.1 to 65.1 mol% G+C and 2171 to 2455 proteins, respectively. The genome characteristics of strain ResAG-91T, with a length of 2.80 Mbp, 63.3 mol% G+C and 2369 proteins, were comparable to those of the type strains of Adlercreutzia species. In addition to basic genome characteristics, the results of digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) calculated as OrthoANIu values are included in Table 2. The dDDH values of strain ResAG-91T in comparison to the type strains of Adlercreutzia species were below the 70 % threshold for species delimitation [24, 25] (Table 2). The highest similarity was observed between strain ResAG-91T and the type strain of A. equolifaciens subsp. equolifaciens with a dDDH value of 55.4 %. The OrthoANIu of strain ResAG-91T compared to the type strains of Adlercreutzia species was calculated using the ANI calculator provided by EZBioCloud (https://www.ezbiocloud.net/tools/ani) [26, 27]. All OrthoANIu values of strain ResAG-91T compared to the type strains of Adlercreutzia species were below the 95–96% cut-off value for species delimitation [28].
Table 2.
Whole draft genome characteristics, values of digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) of strain ResAG-91T and type strains of species of the genus Adlercreutzia
Strains: 1, ResAG-91T (GCA_009755265.1); 2, A. caecimuris B7T (GCA_000403355.2); 3, A. equolifaciens subsp. celatus DSM 18785T (GCA_003726015.1); 4, A. equolifaciens subsp. equolifaciens DSM 19450T (GCA_000478885.1); 5, A. mucosicola DSM 19490T (GCA_000422625.1); 6, A. muris DSM 29508T (GCA_008831045.1).
|
Characteristic |
1 |
2 |
3 |
4 |
5 |
6 |
|---|---|---|---|---|---|---|
|
Genome size (Mbp)* |
2.80 |
2.94 |
2.88 |
2.86 |
3.01 |
2.76 |
|
G+C content (mol%)* |
63.3 |
64.1 |
63.1 |
63.5 |
64.3 |
65.1 |
|
Number of proteins* |
2369 |
2455 |
2386 |
2281 |
2437 |
2171 |
|
dDDH (%) versus ResAG-91T*: |
– |
26.2 |
55.3 |
55.4 |
26.0 |
25.9 |
|
OrthoANIu (%) versus ResAG-91T†: |
– |
81.6 |
93.7 |
93.6 |
81.4 |
81.4 |
*Values were obtained from TYGS [22]. For dDDH, the result for formula d4 is given.
†OrthoANIu was calculated using the ANI calculator provided by EZBioCloud (https://www.ezbiocloud.net/tools/ani) [26, 27].
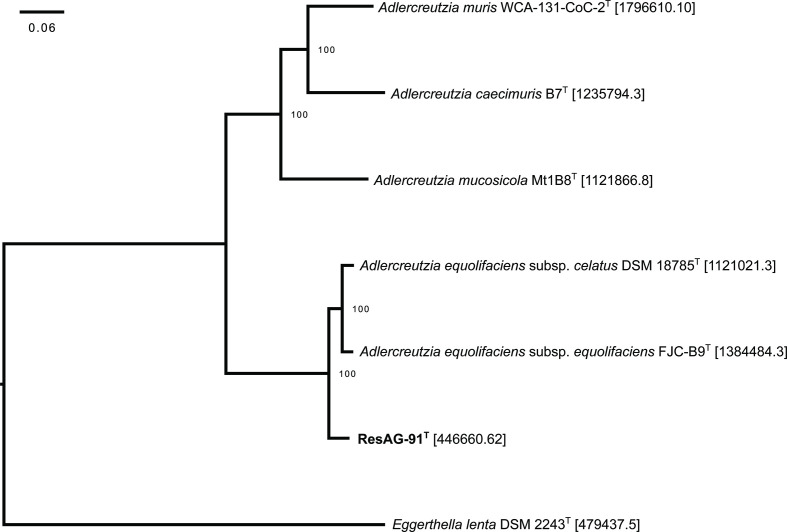
Fig. 3 shows a phylogenomic tree of strain ResAG-91T and the type strains of Adlercreutzia species. Eggerthella lenta DSM 2243T was used as an outgroup. The tree was calculated using the Pathosystems Resource Integration Center (PATRIC; version 3.6.9, www.patricbrc.org) [29] and the RAxML (Randomized Axelerated Maximum Likelihood) algorithm [30]. Bacterial species, strain number, GenBank accession and the PATRIC Genome ID of all genomes selected for phylogenetic comparison are given in Table S3 while the tree analysis statistics are listed in Table S4. For sufficient phylogenomic treeing, the involved minimum number of 31 genes is proposed [31]. To calculate the tree in Fig. 3, 378 coding gene sequences were used and the tree was built on protein alignments based on single-copy homology groups. The nearest neighbours of strain ResAG-91T were A. equolifaciens subsp. celatus DSM 18785T and A. equolifaciens subsp. equolifaciens DSM 19450T.
Fig. 3.
Cluster analysis based on ‘all shared proteins’ of strain ResAG-91T and the type strains of Adlercreutzia species. Eggerthella lenta DSM 2243T was used as an outgroup. The phylogenomic tree was built using PATRIC (version 3.6.9), the RAxML (Randomized Axelerated Maximum Likelihood) algorithm and fast bootstrapping. The tree was built on protein alignments based on single-copy homology groups (n=378 coding gene sequences). The numbers in square brackets indicate the respective PATRIC Genome ID.
In this study, we investigated the morphological, biochemical, chemo-taxonomic and genome-based characteristics of strain ResAG-91T. On the basis of our results, strain ResAG-91T represents the type strain of a novel species within the genus Adlercreutzia , for which we propose the name Adlercreutzia rubneri sp. nov.
Description of Adlercreutzia rubneri sp. nov.
A. rubneri (rub′ne.ri. N.L. gen. n. rubneri referring to Max Rubner, a German medical doctor after whom the Max Rubner-Institute was named, and where the type strain was isolated).
Cells occur as non-motile, Gram-positive and very small rods (singly or in short chains). Cells are on average 1.29 µm in length and 0.31 µm in width. Colonies are pale-white and semi-translucent (Ø ≈ 1 mm) after 48–72 h of incubation on MBHI agar at 37 °C under strictly anaerobic conditions. Optical density in liquid media is similar to McFarland 0.5. Oxidase-negative and catalase-negative. Cells are capable of producing arginine dihydrolase and show a weak reaction for the production of arginine arylamidase and leucine arylamidase. No other positive reactions are observed using API Rapid ID 32A and 20A strips. Growth occurs in the presence of 2 % (w/v) ox-bile. The major respiratory quinone is MMK-5 (methylmenaquinone). Ribose and galactose are detected as major whole cell sugars. The diamino acid in the peptidoglycan is meso-diaminopimelic acid. The polar lipids are phosphatidylglycerol, diphosphatidylglycerol, one unidentified lipid, three unidentified phospholipids and five unidentified glycolipids.
The type strain [5], ResAG-91T (=DSM 111416T=JCM 34176T=LMG 31897T), was isolated from a fresh faecal sample of a human, moderately obese male volunteer (30 years, body mass index 33.2 kg m−2) in Karlsruhe, Germany. The genome size of the type strain is 2.8 Mbp and the G+C content is 63.3 mol%.
Emended description of Adlercreutzia equolifaciens subsp. celatus (Minamida et al. 2008) Nouioui et al. 2018
The description is as given previously [5, 10] with the addition of the following characteristics: cell length is 1.19±0.22 µm and cell diameter is 0.32±0.04 µm. Whole cell sugars are galactose, glucose and ribose. The major respiratory quinone is MMK-5 (methylmenaquinone); minor amounts of MMK-6, DMMK-6 (dimethylmenaquinone), DMMK-5, MK-5 (menaquinone) and MK-6 may also be detected. Polar lipids consist of phosphatidylglycerol, diphosphatidylglycerol, unidentified lipids, unidentified phospholipids and unidentified glycolipids. The size of the genome is 2.88 Mbp.
Emended description of Adlercreutzia equolifaciens subsp. equolifaciens (Maruo et al. 2008) Nouioui et al. 2018
The description is as given previously [1] with the addition of the following characteristics: cell length is 0.98±0.18 µm and cell diameter is 0.31±0.03 µm. Whole cell sugars are galactose, glucose and ribose. The major respiratory quinone is MMK-5 (methylmenaquinone); minor amounts of MMK-6, DMMK-5 (dimethylmenaquinone), DMMK-6 and MK-5 (menaquinone) may also be detected. Polar lipids consist of diphosphatidylglycerol, unidentified lipids and unidentified glycolipids.
Emended description of Adlercreutzia muris (Lagkouvardos et al. 2016) Nouioui et al. 2018
The description is as given previously [7] with the addition of the following characteristics: cell length is 2.06±0.57 µm and cell diameter is 0.48±0.13 µm. Whole cell sugars are galactose, glucose and ribose. The major respiratory quinone is MMK-5 (methylmenaquinone); minor amounts of MK-6 (menaquinone), MMK-6, MK-5 and DMMK-6 (dimethylmenaquinone) may also be detected. The type of peptidoglycan is A1γ. Polar lipids consist of phosphatidylglycerol, diphosphatidylglycerol, an unidentified aminolipid, unidentified lipids, unidentified phospholipids and unidentified glycolipids.
Emended description of the genus Adlercreutzia (Maruo et al. 2008) Nouioui et al. 2018
The description is as given previously [1] with the addition of the following characteristics. Predominant respiratory quinones are MMK-5 (methylmenaquinone) or MMK-6. Whole cell sugars are ribose and galactose; glucose may be present. Production of arginine dihydrolase and production of arginine arylamidase are positive. Growth occurs on 2 % bile. Cell length and cell diameter range from ca. 1 to 2 µm and 0.3 to 0.5 µm, respectively. The genome sizes and the G+C contents range from 2.76 to 3.01 Mbp and 63.1 to 65.1 mol%, respectively.
Supplementary Data
Funding information
This work was part of the project ‘Importance and Bioactivity of the Microbial trans-Resveratrol Metabolites Dihydroresveratrol and Lunularin’ funded by the DFG (Deutsche Forschungsgemeinschaft; project number 274521263).
Acknowledgements
We thank Andrea Göbl for excellent technical assistance regarding the isolation of strain ResAG-91T. We thank Lilia Wiest, Stephanie Stricker and Jennifer Burke-Murphy for excellent technical assistance in the anaerobe laboratory as well as Bettina Schindler and Volker Müller for their excellent technical support regarding the HPLC-DAD analyses. Furthermore, we thank Simone Brümmer and Gunilla Breutmann for excellent support with the scanning electron microscope. Moreover, we thank the Microbe Division in RIKEN BioResource Centre (Japan Collection of Microorganisms, JCM), the Belgian Coordinated Collection of Microorganisms (BCCM) and the German Collection of Microorganisms and Cell Cultures (DSMZ) for strain deposition.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The faecal sample was obtained within the study entitled ‘Investigations into the bioavailability and metabolisation of Resveratrol in humans’, registered in the German Clinical Trials Register (DRKS00008788).
Footnotes
Abbreviations: ANI, average nucleotide identity; dDDH, digital DNA–DNA hybridization; DMMK, dimethylmenaquinone; MBHI, modified brain heart infusion; MK, menaquinone; MMK, methylmenaquinone; SEM, scanning electron microscopy.
Four supplementary tables and three supplementary figures are available with the online version of this article.
References
- 1.Maruo T, Sakamoto M, Ito C, Toda T, Benno Y. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella . Int J Syst Evol Microbiol. 2008;58:1221–1227. doi: 10.1099/ijs.0.65404-0. [DOI] [PubMed] [Google Scholar]
- 2.Gupta RS, Chen WJ, Adeolu M, Chai Y. Molecular signatures for the class Coriobacteriia and its different clades; proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int J Syst Evol Microbiol. 2013;63:3379–3397. doi: 10.1099/ijs.0.048371-0. [DOI] [PubMed] [Google Scholar]
- 3.Nouioui I, Carro L, Garcia-Lopez M, Meier-Kolthoff JP, Woyke T, et al. Genome-Based Taxonomic Classification of the Phylum Actinobacteria. Front Microbiol. 2018;9:2007. doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clavel T, Duck W, Charrier C, Wenning M, Elson C. Enterorhabdus caecimuris sp. nov., a member of the family Coriobacteriaceae isolated from a mouse model of spontaneous colitis, and emended description of the genus Enterorhabdus Clavel et al. 2009. Int J Syst Evol Microbiol. 2010;60:1527–1531. doi: 10.1099/ijs.0.015016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minamida K, Ota K, Nishimukai M, Tanaka M, Abe A, et al. Asaccharobacter celatus gen. nov., sp. nov., isolated from rat caecum. Int J Syst Evol Microbiol. 2008;58:1238–1240. doi: 10.1099/ijs.0.64894-0. [DOI] [PubMed] [Google Scholar]
- 6.Clavel T, Charrier C, Braune A, Wenning M, Blaut M, et al. Isolation of bacteria from the ileal mucosa of TNFdeltaARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. Int J Syst Evol Microbiol. 2009;59:1805–1812. doi: 10.1099/ijs.0.003087-0. [DOI] [PubMed] [Google Scholar]
- 7.Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 2016;1:16131. doi: 10.1038/nmicrobiol.2016.131. [DOI] [PubMed] [Google Scholar]
- 8.Stoll DA, Danylec N, Grimmler C, Kulling SE, Huch M. Genome analysis reveals that the correct name of type strain Adlercreutzia caecicola DSM 22242T is Parvibacter caecicola Clavel et al. 2013. Int J Syst Evol Microbiol. 2021;71 doi: 10.1099/ijsem.0.004814. [DOI] [PubMed] [Google Scholar]
- 9.Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, et al. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danylec N, Gobl A, Stoll DA, Hetzer B, Kulling SE, et al. Rubneribacter badeniensis gen. nov., sp. nov. and Enteroscipio rubneri gen. nov., sp. nov., new members of the Eggerthellaceae isolated from human faeces. Int J Syst Evol Microbiol. 2018;68:1533–1540. doi: 10.1099/ijsem.0.002705. [DOI] [PubMed] [Google Scholar]
- 11.Clavel T, Charrier C, Wenning M, Haller D. Parvibacter caecicola gen. nov., sp. nov., a bacterium of the family Coriobacteriaceae isolated from the caecum of a mouse. Int J Syst Evol Microbiol. 2013;63:2642–2648. doi: 10.1099/ijs.0.045344-0. [DOI] [PubMed] [Google Scholar]
- 12.Lawson PA, Greetham HL, Gibson GR, Giffard C, Falsen E, et al. Slackia faecicanis sp. nov., isolated from canine faeces. Int J Syst Evol Microbiol. 2005;55:1243–1246. doi: 10.1099/ijs.0.63531-0. [DOI] [PubMed] [Google Scholar]
- 13.Lanigan GW. Peptococcus heliotrinreducans, sp. nov., a cytochrome-producing anaerobe which metabolizes pyrrolizidine alkaloids. J Gen Microbiol. 1976;94:1–10. doi: 10.1099/00221287-94-1-1. [DOI] [PubMed] [Google Scholar]
- 14.Koppel N, Bisanz JE, Pandelia ME, Turnbaugh PJ, Balskus EP. Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. eLife. 2018;7 doi: 10.7554/eLife.33953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selma MV, Beltrán D, García-Villalba R, Espín JC, Tomás-Barberán FA. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food & function. 2014;5:1779–1784. doi: 10.1039/c4fo00092g. [DOI] [PubMed] [Google Scholar]
- 16.Bode LM, Bunzel D, Huch M, Cho GS, Ruhland D, et al. In vivo and in vitro metabolism of trans-resveratrol by human gut microbiota. Am J Clin Nutr. 2013;97:295–309. doi: 10.3945/ajcn.112.049379. [DOI] [PubMed] [Google Scholar]
- 17.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagai F, Watanabe Y, Morotomi M. Slackia piriformis sp. nov. and Collinsella tanakaei sp. nov., new members of the family Coriobacteriaceae, isolated from human faeces. Int J Syst Evol Microbiol. 2010;60:2639–2646. doi: 10.1099/ijs.0.017533-0. [DOI] [PubMed] [Google Scholar]
- 19.Wade WG, Downes J, Dymock D, Hiom SJ, Weightman AJ. The family Coriobacteriaceae: reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov., comb. nov. and Slackia heliotrinireducens gen. nov., comb. nov., and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov., comb. nov. Int J Syst Evol Microbiol. 1999;49:595–600. doi: 10.1099/00207713-49-2-595. [DOI] [PubMed] [Google Scholar]
- 20.Danylec N, Stoll DA, Gobl A, Huch M. Draft genome sequences of 13 isolates of Adlercreutzia equolifaciens, Eggerthella lenta, and Gordonibacter urolithinfaciens, isolated from Human Fecal Samples in Karlsruhe, Germany. Microbiol Resour Announc. 2020;9:e00017-00020. doi: 10.1128/MRA.00017-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danylec N, Stoll DA, Dotsch A, Huch M. Draft genome sequences of type strains of Gordonibacter faecihominis, Paraeggerthella hongkongensis, Parvibacter caecicola, Slackia equolifaciens, Slackia faecicanis, and Slackia isoflavoniconvertens . Microbiol Resour Announc. 2019;8:e01532-01518. doi: 10.1128/MRA.01532-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danylec N, Stoll DA, Huch M. Draft genome sequences of type strains of Adlercreutzia muris and Ellagibacter urolithinifaciens, belonging to the family Eggerthellaceae . Microbiol Resour Announc. 2019;8:e01306-01319. doi: 10.1128/MRA.01306-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 26.Yoon S-H, Ha S-M, Kwon S, Lim J, Kim Y, et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon S-H, Ha S-M, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 28.Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 29.Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.