Abstract
Coronaviruses (CoVs) have very large RNA viral genomes with a distinct genomic architecture of core and accessory open reading frames (ORFs). It is of utmost importance to understand their patterns and limits of homologous and nonhomologous recombination, because such events may affect the emergence of novel CoV strains, alter their host range, infection rate, tissue tropism pathogenicity, and their ability to escape vaccination programs. Intratypic recombination among closely related CoVs of the same subgenus has often been reported; however, the patterns and limits of genomic exchange between more distantly related CoV lineages (intertypic recombination) need further investigation. Here, we report computational/evolutionary analyses that clearly demonstrate a substantial ability for CoVs of different subgenera to recombine. Furthermore, we show that CoVs can obtain—through nonhomologous recombination—accessory ORFs from core ORFs, exchange accessory ORFs with different CoV genera, with other viruses (i.e., toroviruses, influenza C/D, reoviruses, rotaviruses, astroviruses) and even with hosts. Intriguingly, most of these radical events result from double crossovers surrounding the Spike ORF, thus highlighting both the instability and mobile nature of this genomic region. Although many such events have often occurred during the evolution of various CoVs, the genomic architecture of the relatively young SARS-CoV/SARS-CoV-2 lineage so far appears to be stable.
Keywords: coronavirus, recombination, genome evolution, horizontal gene transfer, bioinformatics, molecular evolution
Introduction
Genomic analyses of single-stranded RNA viruses, including coronaviruses (CoVs), have repeatedly demonstrated how recombination affects their emergence, host range, and pathogenicity (Decaro et al. 2009; Simon-Loriere and Holmes 2011; Terada et al. 2014; Tian et al. 2014; Su et al. 2016; Lau et al. 2018). Given the current pandemic of SARS-CoV-2 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses 2020; Wu et al. 2020), it is of utmost importance to fully understand the patterns and limits of homologous and nonhomologous genomic exchange of the entire CoV subfamily. This knowledge will allow us to better evaluate any risks from cross-species transmission and recombination with other closely or distantly related viruses. It may also guide the development of future vaccines, by allowing the selection of stable antigenic regions and avoiding reversion (via recombination) of any future live-attenuated vaccine strains (Guillot et al. 2000; Racaniello 2006; Pliaka et al. 2012; Burns et al. 2013; Graham et al. 2018; Nikolaidis et al. 2019).
According to the ICTV 2020 release, the CoV subfamily (Orthocoronavirinae) harbors significant genomic diversity, comprising 4 genera (α−δ), further subdivided into 25 subgenera (Lauber and Gorbalenya 2012; Lauber et al. 2012; ICTV Coronaviridae Study Group 2020). Various CoVs are found in a wide range of animal species, causing respiratory, enteric, hepatic, and nervous system disorders with mild to severe symptoms (Rota et al. 2003; Weiss and Navas-Martin 2005; Woo et al. 2007; Bermingham et al. 2012; Wheeler et al. 2018; Chen et al. 2020; Wu et al. 2020). Bats are reservoirs for the α- and β-CoVs, whereas wild birds are reservoirs for the γ- and δ-CoVs (Woo et al. 2009, 2012; Wong et al. 2019; Latinne et al. 2020; Wille and Holmes 2020). Human CoVs are found in the α- and β-genera and have a zoonotic origin, with bats as the key reservoir, but intermediate hosts may also be involved in the cross-species transmission (Song et al. 2005; Reusken et al. 2013; Fan et al. 2019).
CoVs possess very large genomes among RNA viruses (25–32 kb) and contain at least 6 core open reading frames (ORFs; 1a, 1b, Spike, Envelope, Membrane, and Nucleocapsid; Gorbalenya et al. 2006; Cui et al. 2019; Chen et al. 2020). Lineage-specific accessory ORFs are also present and may be involved in host adaptation, including the modulation of interferon signaling and the production of proinflammatory cytokines (Gorbalenya et al. 2006; Liu et al. 2014; Cui et al. 2019; Hartenian et al. 2020). This large genome size and complex architecture allow division of labor and flexibility for cross-species adaptation (Lauber et al. 2013). Importantly, the Spike protein facilitates binding to host receptors and so determines host range, cell tropism, and even the transition from a mild toward a highly pathogenic phenotype, via point mutations and recombination (Sánchez et al. 1999; Kuo et al. 2000; Casais et al. 2003; Rottier et al. 2005; Menachery et al. 2015).
Recombination events among closely related CoV strains/genotypes/species of the same subgenus have been reported frequently (Keck et al. 1988; Kottier et al. 1995; Herrewegh et al. 1998; Decaro et al. 2009; Tian et al. 2014; Dudas and Rambaut 2016; Forni et al. 2017; Bobay et al. 2020; Boni et al. 2020; Saeng-Chuto et al. 2020; Goldstein et al. 2021; Yang et al. 2021); we denote this category of events as intratypic recombination. The corresponding recombination junctions are scattered across the genome, although enrichment around transcriptional regulatory sequences (TRS-B) has been reported (Yang et al. 2021). These TRS are needed for template switching during the transcription of the CoV ORFs (Sawicki et al. 2007; Sola et al. 2015), but they may also facilitate recombination via template switching among different CoVs (Graham et al. 2018; Yang et al. 2021). The genomes of several CoVs are mosaic, but many of their donors have yet to be sequenced (Goldstein et al. 2021). Furthermore, recombination events among more distantly related CoVs have also been observed. Such radical evolutionary events probably result from the presence of highly conserved TRS-B sequences (shared between the recombining CoVs) at the beginning of the various ORFs (Sawicki et al. 2007; Sola et al. 2015; Boniotti et al. 2016; Graham et al. 2018; Banerjee et al. 2020). Nevertheless, very disparate TRS-B sequences between two CoVs cause incompatibility and thus may also present barriers to such recombination events (Yount et al. 2006). In this study, we define as intertypic any recombination event among members of different CoV subgenera. In addition, nonhomologous recombination events may occur with other viruses or taxa, leading to the acquisition of new genomic regions that appear as lineage-specific accessory ORFs (Zeng et al. 2008; Woo et al. 2014; Forni et al. 2017). The goal of this study is to understand the patterns and limits of radical (intertypic) genomic exchange of CoVs and to see whether any genomic regions emerge as hotspots of recombination. The first part of this analysis focuses on homologous recombination of core ORFs among different CoV subgenera, whereas the second part deals with nonhomologous recombination of accessory ORFs among CoV subgenera/genera and even with other taxa.
Results
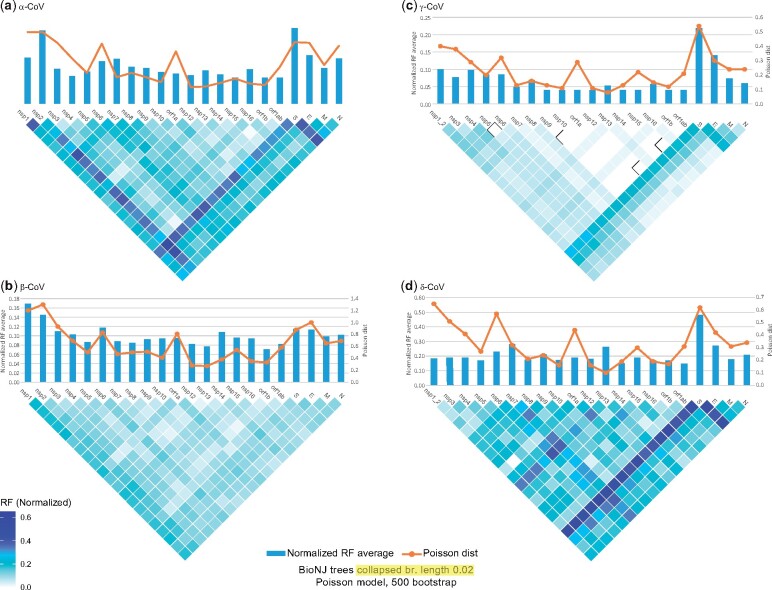
Several computational methods exist for detecting and analyzing recombination events among closely related viruses (Posada et al. 2002; Pond et al. 2005; Martin et al. 2011). In this study, we have implemented phylogenetic tree incongruence methods, which are best suited for macroevolutionary analyses, as well as similarity plots (see Materials and Methods). BioNJ, PhyMl, and Bayesian protein phylogenetic trees and tanglegrams (or “cophylo plots,” a way of graphically representing correspondence between two phylogenies with the same tip labels) were generated for the nonstructural peptides (nsps) of ORFs 1a/1b and the other core ORFs. This was done both for all four genera together and for each of the four genera individually. In addition, phylogenetic trees (BioNJ and PhyML) of the various regions were compared against each other for incongruence, using the normalized Robinson–Foulds (RF) method for unrooted trees (see Materials and Methods). We further validated the statistical significance of detected incongruities with CONSEL, to ensure the robustness of our conclusions. In this study, we only consider highly confident phylogenetic incongruence events that are supported by high bootstrap, aLRT, and posterior probability values for all three tree methods and are also statistically supported by the corresponding CONSEL analyses. In all analyses, the neighborhood of the Spike ORF emerges as an intertypic recombination hotspot.
The Spike ORF Displays Elevated Phylogenetic Tree Incongruence
Phylogenetic trees based on the Spike ORF consistently display the highest or next-highest phylogenetic incongruence compared with all other analyzed regions, in α-, γ-, and δ-CoVs (fig. 1; supplementary file 1 and figs. 32, 33, 38, 39, 50, 51, 57, and 58, Supplementary Material online). In contrast, the corresponding regions of β-CoVs display relatively low phylogenetic incongruence. The Spike sequence is one of the most variable core genomic regions. However, other core regions also have similar sequence variability but do not display such high levels of phylogenetic incongruence. Therefore, this pattern (confirmed by subsequent phylogenetic tree tanglegram analyses) does not result from badly aligned regions, rather, it may be attributed to divergence combined with cassette-like intertypic recombination. If the majority of intertypic recombination events involved single crossovers, then there should be high phylogenetic incongruence among the regions flanking the Spike ORF, but this is not the case. Furthermore, if most of the intertypic recombination (in various regions) involved single crossovers, then the incongruence among the 5′ terminal nsps and the 3′ terminal ORFs, such as Membrane and Nucleocapsid, should also be high, resembling linkage disequilibrium decay (Dudas and Rambaut 2016), but it is not.
Fig. 1.
Matrices of incongruence among the core genomic regions of the four CoV genera (A–D) based on the normalized RF method, for unrooted trees (calculated with the TreeCMP server). BioNJ phylogenetic trees were generated with the Poisson model of evolution and 500 bootstrap replicates. In addition, branch lengths <0.02 were collapsed. The orange line above each matrix displays the average Poisson distance among sequences of the same genomic region (calculated with the MegaX software). Blue bars above each matrix display the average RF value for that particular region (against all other regions).
Tanglegram-Based Detection of Intertypic Recombination Events in the Common Ancestors of CoV Genera and Subgenera
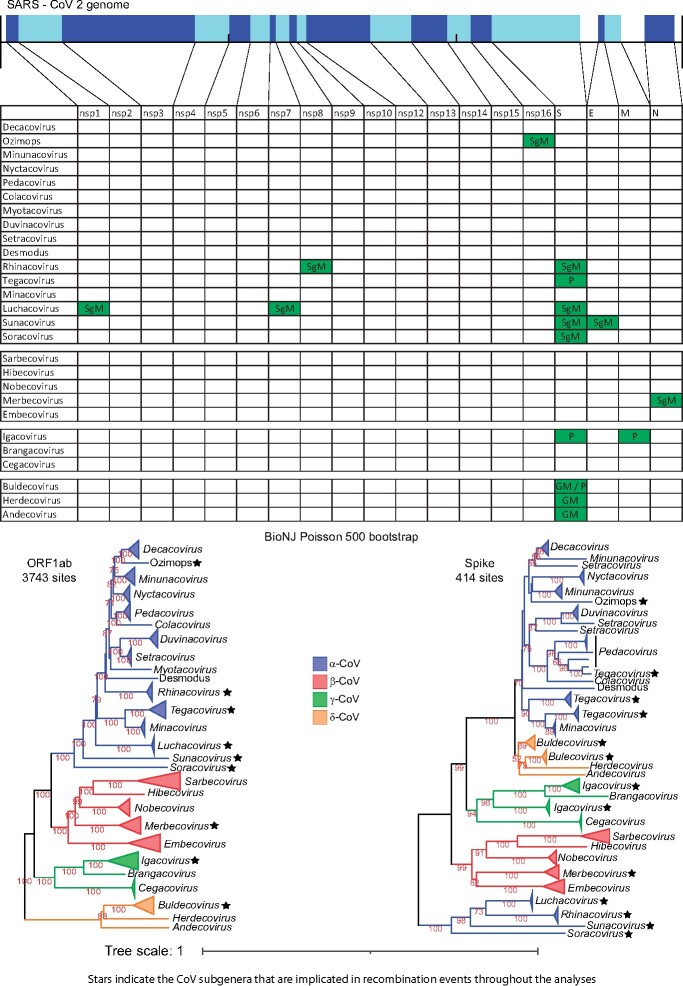
α- and β-CoVs consistently cluster together as a major clade for all core genomic regions except for Spike, for which most of the α- and all the δ-CoVs form a single group (fig. 2 and supplementary fig. 1, Supplementary Material online, recombination event 21). Moreover, cryo-electron microscopy has demonstrated that the Spike proteins of α- and δ-CoVs are structurally more similar to each other (Shang et al. 2018). Thus, at least one recombination event occurred in which the common ancestor of all δ-CoVs obtained a Spike ORF from an α-CoV ancestor.
Fig. 2.
The genomic organization of the core ORFs and peptides of the SARS-CoV-2 genome are displayed on the top of the figure. The table/matrix below it shows which genomic regions of the various subgenera are involved in intertypic recombination events. “GM” represents events that occurred at the common ancestor of the genus. “SgM” represents events that occurred at the common ancestor of the subgenus. “P” represents more recent events that occurred for one or few members of the subgenus and have resulted in a polyphyletic tree pattern (for that region and subgenus). All incongruence events in the matrix are supported by the three phylogenetic tree methods (NJ, PhyML, and Bayesian) and are also statistically significant, based on the AU test of CONSEL. Two phylogenetic trees (of ORF1ab and Spike) for all four genera are also included below the matrix, to visualize the recombination events of the Spike region. In these trees, we use stars to denote subgenera that have been involved in intertypic homologous recombination events, in any genomic region (not only the Spike).
We also observed several cases of phylogenetic incongruence involving entire subgenera (mostly in α-CoVs); they displayed a major shift in their phylogenetic position (for a certain genomic region), as a monophyletic group. We interpret this as a major event that occurred in the common ancestor of the representative sequences of that subgenus. Here, we only report cases well supported by BioNJ, PhyML, and Bayesian tree tanglegrams and also statistically supported (for their incongruence) by CONSEL. The regions that are involved in such events are shown in figure 2 and are designated as SgM (Subgenus Movement).
More specifically, in α-CoVs, there exist 14 well-established subgenera, with the Ozimops and Desmodus genomes possibly forming two extra subgenera. The first 9 subgenera (Decacovirus, Pedacovirus, Colacovirus, Nyctacovirus, Minunacovirus, Duvinacovirus, Setracovirus, Myotacovirus, and Rhinacovirus) together with Ozimops and Desmodus constitute a major clade that we designate A1. Another two subgenera (Tegacovirus and Minacovirus) constitute a major clade that we designate A2 and is a sister group to A1. Luchacovirus (found in rodents), Sunacovirus, and Soracovirus (both found in shrews) constitute three very diverse additional clades that we designate A3, A4, and A5, respectively. The tanglegrams reveal that Ozimops is a sister group to Decacovirus, but for nsp16 it pairs with Minunacovirus (recombination event 4, supplementary figs. 16–19, Supplementary Material online). The Rhinacovirus (A1 clade) nsp8 is no longer part of the A1 clade, but clusters with the A3 Luchacovirus (recombination event 3, supplementary figs. 12–15, Supplementary Material online). Luchacovirus (A3 clade) moves within the A1 clade for both nsp1 (recombination event 1, supplementary figs. 4–7, Supplementary Material online) and nsp7 (recombination event 2, supplementary figs. 8–11, Supplementary Material online). Similarly, Sunacovirus (A4 clade) moves within the A1 clade for Envelope (recombination event 11, supplementary figs. 28–31, Supplementary Material online). We observed many other incongruities for most of the subgenera in various genomic regions, but their new positions (in the trees) were not supported by both high bootstrap/aLRT values and different trees, thus they may actually represent cases of rapid divergence.
Although α-CoVs form five distinct lineages, their Spike ORFs are organized into two major evolutionary clusters. The smaller cluster comprises Rhinacovirus (a member of clade A1), Luchacovirus (clade A3), Sunacovirus (clade A4), Soracovirus (clade A5), whereas the major cluster comprises all the other members of clades A1 and A2 (see Spike tree in fig. 2, recombination events 5, 8, 9, 10 in supplementary figs. 1, 2, and 20–23, Supplementary Material online). The Spike ORF of this smaller cluster has been suggested to originate from β-CoVs via an ancient recombination event (Tsoleridis et al. 2019).
Phylogenetic incongruence was also observed for the Nucleocapsid region of β-CoV Merbecovirus (fig. 2 and recombination event 12 in supplementary figs. 34–37, Supplementary Material online). By taking Sarbecovirus as the reference point, Hibecovirus is their closest subgenus, followed by Nobecovirus, Merbecovirus, and finally Embecovirus (most distant). The only exception to this pattern is observed in the Nucleocapsid region, where Merbecovirus seems to be the closest subgenus to the Sarbecovirus–Hibecovirus group. An alternative explanation is that the ancestral Nobecovirus Nucleocapsids underwent recombination or significant sequence divergence. However, manual inspection of the trees, their branch lengths, and the Poisson distances leads us to favor the first explanation, whilst acknowledging that the second cannot be excluded at present.
Tanglegram-Based Detection of Intertypic Recombination between Some Members of Different Subgenera
We investigated instances where certain genomic regions of the members of a particular subgenus did not form a monophyletic group. These observations could be attributed to rapid divergence or intertypic recombination events in some, but not all, members. These events are more recent than the ones (described above) that occurred in the common ancestor of a subgenus. Such regions are shown in figure 2 (designated as “P”: polyphyletic). We checked whether these candidate recombinant sequences clustered within or next to other subgenera with high bootstrap/aLRT/posterior probability values and also performed similarity plot and bootscan analyses with RDP4 (Martin et al. 2015; see Materials and Methods), whenever possible. We detected several events; two in α-CoVs, five in γ-CoVs, and three in δ-CoVs. Interestingly, nine of these ten events are located at the Spike ORF.
The most striking and recent event has been documented for Swine Enteric CoV (Boniotti et al. 2016), which is essentially a swine Tegacovirus (A2 lineage) that obtained the Spike ORF of a swine Pedacovirus (A1 lineage; recombination event 6, supplementary figs. 20–22 and 24–25, Supplementary Material online). A second case (again in the Spike ORF) concerns 5 of the 13 analyzed Tegacovirus sequences that form a monophyletic sister group to Minacoviruses (recombination event 7, supplementary figs. 20–22, 26, and 27, Supplementary Material online). An alternative sequence of events is that the other seven Tegacovirus (from cats and dogs) that form the second Spike monophyletic group recombined with an as yet unknown donor from the A2 lineage. Inspection of the phylogenetic trees and their branches leads us to favor the first option, whereas the host range of the second group favors the second option. Yet another instance concerns four γ-CoV Igacovirus Spike sequences (from birds) that form a monophyletic cluster outside of the Igacovirus (recombination events 13–16, supplementary figs. 40–44, Supplementary Material online). This is a case of three or most probably four independent events where members from an as yet unknown γ-CoV subgenus repeatedly served as Spike donors to several Igacoviruses. A further case involves a duck IgacovirusMembrane sequence that clusters with the γ−CoV Brangacovirus (recombination event 17, supplementary figs. 45–49, Supplementary Material online). A final example concerns five δ-CoV Buldecovirus Spike sequences forming a monophyletic cluster (that is outside of Buldecoviruses) and is a sister group to Herdecovirus (recombination events 18–20, supplementary figs. 52–56, Supplementary Material online). Our interpretation is that this is a case of three independent events, where members from an, as yet unknown, δ-CoV subgenus (a close relative of Herdecoviruses) repeatedly served as Spike donors to these Buldecoviruses.
In addition, we detected several low-confidence intertypic recombination events for α-CoV subgenera, where the incongruent sequences cluster with other subgenera, but with low bootstrap/aLRT/posterior probability support. Here, either the donor is unknown or the incongruence is due to rapid divergence; they were not considered further in our study. Finally, we also observed previously reported intratypic recombination events, that is, within Sarbecovirus (supplementary figs. 60–68, Supplementary Material online). Although such events are not the focus of this study, it should be mentioned that, at the beginning of the COVID-19 pandemic, several studies analyzed the available genomic data for evidence of recombination that could have led to the emergence of SARS-CoV-2 (Boni et al. 2020; Lam et al. 2020; Paraskevis et al. 2020; Yang et al. 2021). Although the data show that SARS-CoV-2 did not emerge via a recent recombination event, recombinant sequences (from other species) among the SARS-CoV and SARS-CoV-2 lineages have been detected and were also confirmed by our study.
Accessory ORF Evolution: Nonhomologous Recombination of Accessory ORFs between Different CoV Subgenera and Genera
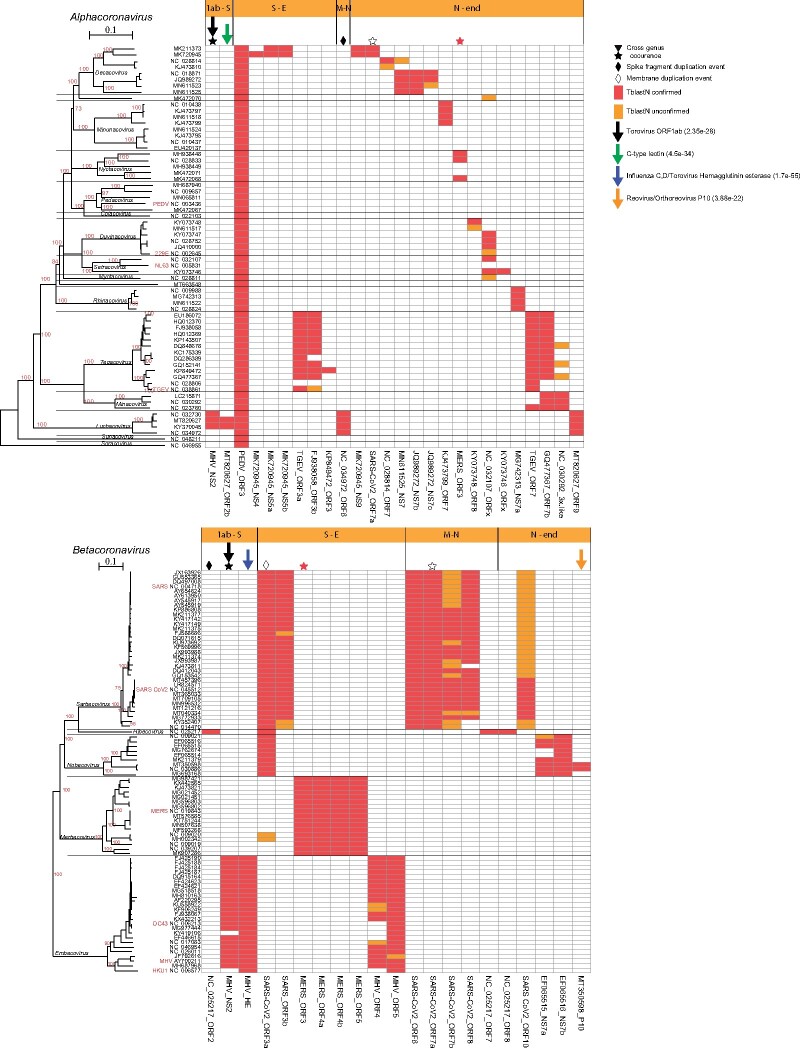
Based on PSI-BLAST, we built position-specific scoring matrices (PSSMs) for the various annotated accessory ORFs and thus identified 73 nonredundant Accessory ORF Families (AOFs; see Materials and Methods). The PSSMs allowed for a very sensitive homology search and revealed very distinct distributions in the various genera and subgenera (figs. 3 and 4 and supplementary file 2, Supplementary Material online). Although no AOF was present in all four genera, three AOFs were present in some subgenera of both α- and β-CoVs and three AOFs were present in subgenera of both γ- and δ-CoVs. Interestingly, three of these intergenus AOFs are localized in the neighborhood of the Spike ORF. Possibly, some AOFs with restricted distributions may actually be distant homologs of other AOFs that significantly diverged (Ouzounis 2020; Neches et al. 2021) and lost their homology signal.
Fig. 3.
Presence and distribution of AOFs in the α- and β-CoVs. Each column in the matrix represents a certain AOF. Red color (within the matrix cells) denotes the (TblastN) presence of an AOF that is also verified by a predicted ORF with length ≥30 aa, whereas if the length of the predicted ORF is <30 aa, then it is denoted with orange color. Stars denote AOFs that are present in both α- and β-CoV members, whereas diamonds denote an AOF that resulted from duplication of a core ORF. Downward arrows denote AOFs that have homologs in non-CoV genomes, together with their best PSI-BLAST hit e-value. Horizontal orange bars (above the matrices) denote the genomic region where the AOF is located, that is, S-E denotes the region between the Spike and Envelope ORFs.
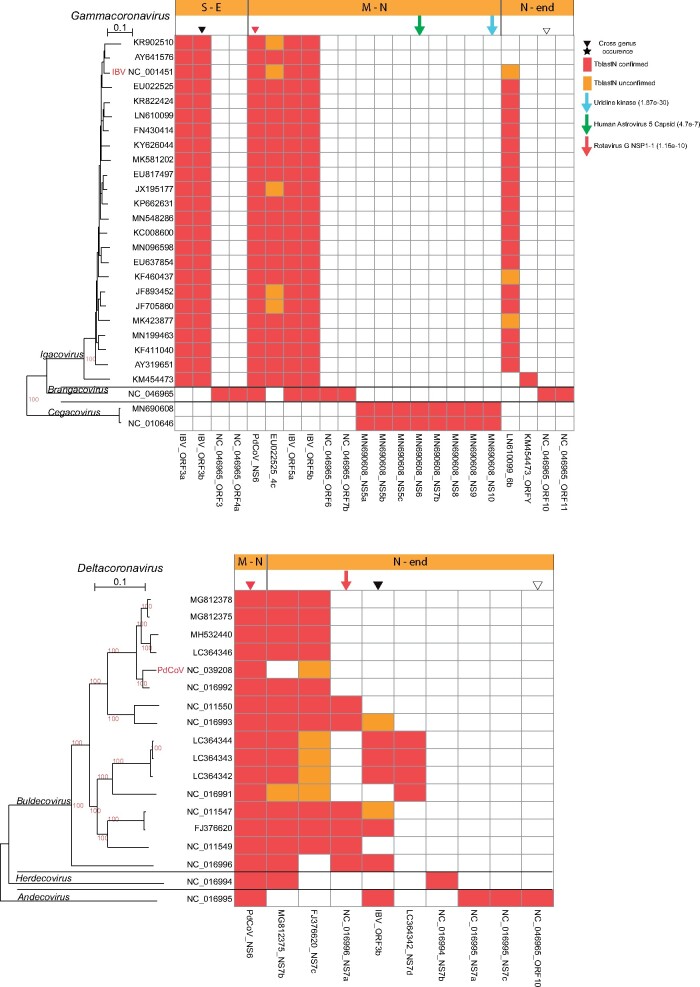
Fig. 4.
Presence and distribution of AOFs in the γ- and δ-CoVs. Each column in the matrix represents a certain AOF. Red color (within the matrix cells) denotes the (TblastN) presence of AOFs that is also verified by a predicted ORF with length ≥30 aa, whereas if the length of the predicted ORF is <30 aa, then it is denoted with orange color. Inverted triangles denote AOFs that are present in both γ- and δ-CoV members. Downward arrows denote AOFs that have homologs in non-CoV genomes, together with their best PSI-BLAST hit e-value. Horizontal orange bars (above the matrices) denote the genomic region where the AOF is located, that is, M-N denotes the region between the Membrane and Nucleocapsid ORFs.
Intriguingly, we detected two AOFs with very restricted distributions that originated either from gene duplication or horizontal gene transfer (HGT) of a Spike ORF fragment. The first instance concerns a bat β-CoV Hibecovirus ORF2 that is situated between ORF1ab and Spike, that is distantly homologous to the N-terminal region of its Spike (supplementary file 2, Supplementary Material online: PSSM_TBlastN: 4e−39; 27% identity). This is either a case of nonhomologous recombination/gene-fragment duplication within the same genome (followed by rapid divergence) or horizontal transfer from another related HibecovirusSpike N-terminal region. The second instance concerns a similar Spike gene-fragment duplication event for ORF6 of some Luchacoviruses (supplementary file 2, Supplementary Material online: PSSM_TBlastN: 7e−63; 25% identity).
We also detected distant homology between the ORF3a of β-CoV Sarbecovirus/Hibecovirus/Nobecovirus and the Membrane ORF of α-CoV A2 Tegacovirus and A4 Sunacovirus (supplementary file 2, Supplementary Material online: PSSM_TBlastN: 2.4e−4 and 3.9e−4, respectively). Accordingly, a bioinformatics analysis (Ouzounis, 2020) recently reported a very distant homology among the SARS-CoV-2 ORF3a and Membrane ORFs. Based on our extended genome sampling and the observed e-values of the ORF3a PSSM against α−CoVs (best PSSM_TBlastN: 2.4e−4) and β-CoVs (best PSSM_TBlastN: 2e−3), possibly a Membrane region from α-CoVs jumped via nonhomologous recombination to the common ancestor of Sarbecovirus/Hibecovirus/Nobecovirus and rapidly diverged to an accessory ORF.
Nonhomologous Recombination of Accessory ORFs between CoVs and Other Taxa
We detected seven AOFs that had homologs in other taxa, outside of the Coronavirinae (supplementary file 2, Supplementary Material online), with three of them situated in the neighborhood of Spike. The most striking and well-studied example is a hemagglutinin-esterase (MHV_HE) that is present in all the members of β-CoV Embecovirus, situated just before the Spike. It has homologs in toroviruses (porcine torovirus PSI-BLAST e-value: 1.7e−55) and influenza C/D. Most probably, it was acquired either indirectly (via a torovirus intermediate step) or directly from an influenza C/D-like virus, and subsequently adapted and coevolved with the Spike (Snijder et al. 1991; Zeng et al. 2008; Caprari et al. 2015; Lang et al. 2020).
Another case is the β-CoV NS2 Embecovirus AOF (MHV_NS2) that belongs to the 2H phosphoesterase superfamily (Mazumder et al. 2002). This AOF is observed in most Embecoviruses, like HCoV-OC43, and is situated between ORF1ab and the hemagglutinin-esterase (HE). Interestingly, close homologs (NCBI-BlastP e-value: 6e−61) of this AOF (from β-CoVs) are consistently found in several rodent α-CoV Luchacoviruses as well (Tsoleridis et al. 2019), at the same genomic location, but they do not have the neighboring HE ORF. This AOF is also homologous to a region within the central part of polyprotein 1ab of several toroviruses, including porcine torovirus (PSI-BLAST e-value: 2e−28). Apparently, nonhomologous genomic exchange among CoVs and toroviruses has happened more than once.
Next to the α-CoV LuchacovirusORF2/NS2, there exists another accessory ORF (instead of HE in Embecoviruses), designated ORF2b. It is present in some, but not all α-CoV Luchacoviruses. It is homologous to rodent C-type lectins (PSI-BLAST e-value: 4e−34) found in natural killer cell receptors as well as in many poxviruses and some herpesviruses. This AOF probably originated from its hosts (Wang et al. 2020). Furthermore, both ORF2a and ORF2b are missing from another closely related Luchacovirus genome (MT820625.1), thus highlighting the dynamic nature of this genomic region (Wang et al. 2020) and the potential for gene loss (Forni et al. 2017).
We also identified four more interesting AOFs. p10, situated just after the nucleocapsid region of some β-CoV Nobecoviruses in bats, is homologous (PSI-BLAST e-value: 3.9e−22) to p10 proteins from reoviruses (Huang et al. 2016). The BuldecovirusNS7a AOF (situated after the Nucleocapsid) of several avian δ-CoV Buldecoviruses is homologous (PSI-BLAST e-value: 1e−10) to NSP1-1 from avian rotavirus-g. An uridine kinase (closest PSI-BLAST hit: fungi; e-value: 2e−30) is found only in γ-CoV Cegacoviruses (Mihindukulasuriya et al. 2008). Finally, the same γ-CoV Cegacoviruses contain ORF6 that is distantly homologous to the capsid protein of human astrovirus 5 (PSI-BLAST e-value: 4.7e−7).
Discussion
The integration of our extensive phylogenetic and genome architecture analyses has revealed intertypic homologous and nonhomologous recombination events among the genomes of different CoV subgenera/genera, and even with other taxa. Intriguingly, many of these events are localized around the Spike ORF and occur as double crossovers, where an entire region is exchanged as a cassette/module and the rest of the genome stays intact. It is unlikely that these observed and statistically supported phylogenetic incongruities (especially for Spike) are artifacts of rapid divergence or convergent evolution, because the “incongruent” regions actually cluster with regions from other genera/subgenera with high bootstrap/aLRT/posterior probability support (among other evidence, like site-wise likelihood of alternative hypotheses—results not shown). The Spike recombination of Swine Enteric CoV is the most recent and clear example. We have applied stringent analysis criteria involving the phylogeny of entire regions and it is possible that many genuine intertypic recombination events may not have passed our filters, especially if they involved small segments of an ORF (Forni et al. 2017). Another major problem is genomic sampling, where the donor has yet to be sequenced (Goldstein et al. 2021).
Our interpretation for the frequently observed modular recombination events around the Spike ORF is that long-range genetic interactions of various genomic regions may actually block radical (intertypic) single-crossover recombination events (Sola et al. 2011, 2015) but allow for double-crossover events in certain genomic islands. This conclusion is supported by various independent experimental observations. Nucleocapsid proteins (N-proteins) from different members of the same genus may only be partially compatible, whereas N-proteins from different genera are completely incompatible (Schelle et al. 2005; Sungsuwan et al. 2020) and may even have a suppressive effect (Masters 2019; Sungsuwan et al. 2020). N-proteins are also involved in the circularization of the genome (Lo et al. 2019). CoV RNA secondary structures have been shown to form long-range interactions within a CoV genome (Ziv et al. 2020) and to interact with cellular components, to initiate transcription and replication (Sola et al. 2011). Genetic interactions have been observed between the nsp8, nsp9 peptide regions (from ORF1a), and the pseudoknot at the 3′ end of the genome (Züst et al. 2008). Thus, single-crossover recombination events among different subgenera may break such long-range interactions, whereas double-crossover/modular events may allow their retention.
We also observed distinct subgenus-specific accessory ORF genomic architectures. These may function as an additional barrier to single-crossover intertypic recombination events that would otherwise disrupt certain coevolved combinations of ORFs. Several of these AOFs have been introduced from other genera/subgenera. However, some of these AOFs do not have homologs in any other subgenera and may have emerged via 1) de novo gene birth, 2) rapid divergence of existing ORFs and loss of the homology signal, or 3) via nonhomologous recombination with ORFs (followed by rapid divergence) from other CoVs, other viruses, or even hosts (Elhaik et al. 2006; McLysaght and Hurst 2016; Moyers and Zhang 2016; Schmitz and Bornberg-Bauer 2017; Ouzounis 2020).
We observed the exchange of genomic regions between CoVs and toroviruses, influenza C/D (directly or indirectly), reoviruses, rotaviruses, astroviruses, and even with hosts. Such events were frequent in the neighborhood of the Spike ORF. Toroviruses are of particular interest, because they belong to the same order (Nidovirales) as CoVs and can also act as gene donors in other viral orders, for example, porcine Enterovirus-G (Shang et al. 2017; Hu et al. 2019). Worryingly, porcine toroviruses have both a worldwide distribution and a high infection rate (Hu et al. 2019). Thus, future genomic sampling of yet undiscovered CoVs may reveal an even more extensive exchange between CoVs and toroviruses. Moreover, genomic exchange between viruses (Flaviviridae, Hepeviridae, Dicistroviridae, and Potyviridae) and their hosts has been observed repeatedly (Gilbert and Cordaux 2017). It is conceivable that some of the abovementioned CoV AOFs did not move from one virus to the other, but independently from similar hosts; however, the PSI-BLAST results show other viral sequences, and not cellular proteins, to be the closest hits.
Importantly, members of the relatively young (Boni et al. 2020) SARS-CoV/SARS-CoV-2 lineages (within Sarbecoviruses) do not yet appear to act as recipients in radical intertypic recombination events. They also display a very distinct AOF architecture. Thus, current evolutionary data do not favor a scenario where SARS-CoV-2 may (homologously) recombine with other currently circulating human CoVs of other subgenera/genera. Furthermore, SARS-CoV/SARS-CoV-2 do not seem to exchange accessory ORFs with other CoV subgenera or other viruses/hosts, with the exceptions of ORF3a that is an old and unresolved event and ORF7a (with some Decacoviruses). It should be noted that their closest relatives, Hibecoviruses, have a divergent Spike-like accessory ORF that resulted from either gene duplication or horizontal transfer event. Nevertheless, SARS-like viruses can recombine with SARS2-like viruses, as our and other analyses have shown (Boni et al. 2020; Lam et al. 2020; Yang et al. 2021). This finding has very important implications, because, combined with the ability of Sarbecoviruses to easily move from one host to another, it demonstrates a potential for a future intratypic recombination event (within Sarbecoviruses), where a highly infectious SARS-CoV-2 variant (e.g., the Delta variant) could recombine with a SARS-like sequence in another host species and give rise to a recombinant that combines the high infectivity of SARS-CoV-2 with the much higher mortality rate of SARS itself.
Many of the events that we have observed are very old; nevertheless, our results suggest that researchers and those responsible for public health should be vigilant. Certain key taxa like bats and/or farmed animals (especially pigs) have the potential to play a key role in any future emergence of a recombinant SARS-CoV-2 strain or some other CoV epidemic (from another genus/subgenus). SARS-CoV-2 spill-back from humans to other animals (domesticated or wild) that also harbor many and diverse CoVs has been reported (de Morais et al. 2020; Olival et al. 2020; Sit et al. 2020). Ferrets, cats, and dogs are susceptible to the currently circulating SARS-CoV-2 strains, whereas pigs, chicken, and ducks appear to have lesser, or no susceptibility (Meekins et al. 2020; Shi et al. 2020; Sit et al. 2020; Pickering et al. 2021). CoVs demonstrate a high capacity for cross-species infection, even from birds to mammals, either directly or via a few evolutionary steps (Li et al. 2006; Graham and Baric 2010; Menachery et al. 2015, 2016; Li et al. 2018; Boley et al. 2020). Furthermore, pigs are carriers of very diverse α-, β-, as well as δ-CoVs and have been shown to function as “recombination bioreactors,” with the notable example of Swine Enteric CoV (Boniotti et al. 2016). In addition, intensively farmed pigs are hosts for many other viruses, such as toroviruses or influenza A (Hu et al. 2019; Henritzi et al. 2020; Sun et al. 2020). Fortunately, genomics is a valuable new tool for monitoring the emergence, spread, and ongoing adaptations of SARS-CoV-2 (Boni et al. 2020; Neches et al. 2020; Worobey et al. 2020; Kemp et al. 2021; Volz et al. 2021). It is conceivable that what we have observed is only the “tip of the iceberg”; that past unknown recombination events of various CoVs may have led to many unnoticed (or, perhaps, readily contained) localized small-scale epidemics that died out. However, given the observed genomic diversity and inherent genomic instability of CoVs, in this new era of urbanization, global transport, intensive farming, and habitat destruction (Beyer et al. 2021), intratypic and intertypic recombination events may lead to new epidemic strains that may prove much more difficult to contain (Bedford et al. 2019). As a final note, these results highlight the need to further investigate the inclusion of other, and much more stable, genomic regions (in addition to Spike) in the design and development of the next generation of CoV vaccines.
Materials and Methods
Phylogenetic Analyses
We obtained the taxonomy IDs for α-, β-, γ-, and δ-CoVs from NCBI Taxonomy in order to search for available nucleotide sequences in GenBank (Benson et al. 2013), using (as two extra criteria) the keyword “complete” and nucleotide length higher than 24,000. We obtained 1,102, 14,769, 435, and 154 genomic sequences from α-, β-, γ-, and δ-CoVs, respectively, in August 2020. Redundancy with the set of retrieved sequences was removed with the UCLUST software (Edgar 2010), using 90% nucleotide identity and 98% query coverage at the whole-genome level, in order to filter out the thousands of available genomes from the same virus that have been involved in large outbreaks, like SARS-CoV-2, Porcine Epidemic Diarrhea Virus (PEDV), and Infectious Bronchitis Virus (IBV). From each nonredundant group, we retained one representative sequence, or more if they were obtained from different hosts. We designate these groups as NRG90 (Nonredundant Group—90% nucleotide sequence identity). In addition, within each NRG90 group, we ensured that we retained the representative RefSeq sequences for each species that were obtained from ICTV taxonomy (ICTV Coronaviridae Study Group 2020). Sequences were aligned with Muscle (Edgar 2004) and MAFTT (parameters: –auto; Nakamura et al. 2018). Multiple alignment views and manual editing were performed with the Seaview4 software (Gouy et al. 2010). The boundaries of nsps within ORF1ab, as well as those of Spike, Envelope,Membrane,Nucleocapsid, and the accessory ORFs, were determined based on GenBank annotation and from manual inspection of the multiple alignments. Filtering of poorly aligned regions was performed with the g-blocks software (Castresana 2000), where we retained sites with less than 50% gaps and blocks of two consecutive sites. Model selection for maximum likelihood (ML) and Bayesian trees was performed with Prottest3 (Darriba et al. 2011). Subsequent ML tree reconstruction was performed with PhyML (Guindon and Gascuel 2003; applying Shimodaira-Hasegawa-like (SH-like) approximate likelihood ratio test, Subtree-Prunning-Regrafting (SPR) algorithm for tree search). Neighbor-Joining (BioNJ) trees were generated with Seaview4 (Gouy et al. 2010), using the Kimura two-parameter and Poisson models with 500 bootstraps, for nucleotide and protein sequences, respectively. Bayesian phylogenetic trees were calculated using the BEAST software v.1.10.4 (Drummond et al. 2012; Suchard et al. 2018) with Markov chain Monte Carlo length of 1 million and a burn-in value of 10,000 (all the other operators and priors were set to default). Phylogenetic trees were visualized with Treedyn (Chevenet et al. 2006), iTOL (Letunic and Bork 2019), and Dendroscope (Huson and Scornavacca 2012). Phylogenetic trees were generated for all regions (nsps, ORF1ab, Spike, Envelope, Membrane, and Nucleocapsid) of each CoV genus independently. In addition, phylogenetic trees that included all sequences of all four CoV genera together were generated for those regions (nsps 3–10, 12–16, ORF1ab, Spike, Membrane, and Nucleocapsid) whose multiple alignments had a sufficient number of columns, after g-blocks filtering.
Phylogenetic tree incongruence was estimated/quantified with the RF method (Robinson and Foulds 1981) for unrooted trees, within the Visual Treecmp server (Goluch et al. 2020). A certain genomic region is considered incongruent when its phylogeny is not in agreement with the phylogeny of the other regions (from the same genome). Visualization of the triangular matrix of RF normalized values among the various trees was performed with Python and R pheatmap packages. This RF-matrix resembles the linkage disequilibrium matrix, at the macroevolutionary level, but for specified genomic regions. Since the goal was to investigate incongruence at the macroevolutionary level and not within the virus species level, for these type of analyses, branches with length less than 0.02 were collapsed with the TreeGraph v2 software (Stöver and Müller 2010). Otherwise, the incongruence of strains of the same virus species would artificially inflate the RF values. This would especially be the case for γ-CoVs, where many divergent strains of IBV (Igacovirus) were available. Phylogenetic tree tanglegrams were visualized with Dendroscope (Huson and Scornavacca 2012), using the ML, BioNJ, and Bayesian tree of ORF1ab as the reference tree against each of the ML, BioNJ, and Bayesian trees of the individual nsps and ORFs S, E, M, N, for each of the four CoV genera separately. Estimation of evolutionary distance among homologous aligned sequence regions (for visualization in the RF matrices) was performed with the Poisson distance method within the MEGA X software (Kumar et al. 2018; parameters—gap missing data: pairwise deletion; rates among sites: uniform). The statistical significance of phylogenetic incongruence of specific suspected recombination events was further assessed with the approximately unbiased (AU) test, using CONSEL (Shimodaira and Hasegawa 2001). For a certain set of sequences, the reference PhyML tree was obtained from the suspected recombined region and it was compared against the PhyML tree of the corresponding ORF1ab regions.
Accessory ORF Analysis
In the first step of this analysis, all annotated accessory ORFs from our nonredundant set of 196 CoV genomes were retrieved from GenBank. We only retained accessory ORFs with a length of ≥50 amino acids, with the exception of human CoVs (length ≥ 30) that were situated in the regions among the 6 core ORFs and not any accessory ORFs that were entirely overlapping with any of the core ORFs. The selected annotated accessory ORFs in all analyzed genomes were further clustered in 88 homologous groups, using as cutoff, pairwise BLASTP e-values of 1e−10, followed by grouping with mcl-clustering (Enright et al. 2002). Afterwards, a representative peptide sequence from each cluster was used to build a corresponding PSSM with locally installed PSI-BLAST, against the Coronaviridae proteins of the (locally installed) NCBI nonredundant protein database, with an e-value cutoff 1e−3 and as many iterations as needed, until convergence was achieved. Next, 15 redundant PSSMs were removed and we ended up with 73 annotated accessory ORF PSSMs. Accordingly, each nonredundant PSSM corresponded to one homologous AOF. All 73 PSSMs are available in supplementary file 3, Supplementary Material online.
Afterwards, each AOF PSSM was used to scan all the analyzed CoV representative genomes for the presence of the corresponding family with TBlastN (cutoff: 1e−3). Each TBlastN hit was inspected to determine whether it encoded a peptide of at least 30 amino acids, otherwise, it was considered to be pseudogenized (represented with orange color in the matrices of figs. 3 and 4). The coordinates of the detected homologous regions were visualized in each genome with Biopython and the genomic architectures were manually inspected. Genomic regions from the representative CoV genomes containing a certain AOF were aligned with Muscle. Each multiple alignment is available within the zipped supplementary file 4, Supplementary Material online. Next, the annotated ORF PSSMs were used as queries to scan the entire NCBI nonredundant protein database, in order to detect AOF homologs in taxa outside of Coronavirinae and thus detect potential nonhomologous recombination events (HGTs). Intriguingly, bacterial draft genomes were found to include CoV AOFs with very high sequence identity. These draft genomes were reassembled with Spades (Bankevich et al. 2012) and the relevant contigs were manually investigated for copresence of CoV and bacterial genes, but they eventually appeared to be contaminations and were not further investigated.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
M.N. would like to thank the Bodossakis foundation (studentship: BDA-394) and the University of Thessaly (studentship: DEKA-UTH-259) for financial support. We thank Stephane Rombauts for useful discussions concerning bacterial genome assembly artifacts.
Author Contributions
M.N and G.D.A. analyzed the data; M.N, P.M., Y.V.d.P., S.G.O., and G.D.A designed the analyses, wrote and edited the manuscript; G.D.A. supervised M.N.
Data Availability
All necessary data are incorporated into the article and its online supplementary material. Any further data are available on request.
References
- Banerjee A, Doxey AC, Tremblay BJ-M, Mansfield MJ, Subudhi S, Hirota JA, Miller MS, McArthur AG, Mubareka S, Mossman K.. 2020. Predicting the recombination potential of severe acute respiratory syndrome coronavirus 2 and Middle East respiratory syndrome coronavirus. J Gen Virol. 101(12):1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford J, Farrar J, Ihekweazu C, Kang G, Koopmans M, Nkengasong J.. 2019. A new twenty-first century science for effective epidemic response. Nature 575(7781):130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW.. 2013. GenBank. Nucleic Acids Res. 41(Database issue):D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, et al. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East. Euro Surveill. 17(20290):2012. [PubMed] [Google Scholar]
- Beyer RM, Manica A, Mora C.. 2021. Shifts in global bat diversity suggest a possible role of climate change in the emergence of SARS-CoV-1 and SARS-CoV-2. Sci Total Environ. 767:145413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay L-M, O’Donnell AC, Ochman H.. 2020. Recombination events are concentrated in the spike protein region of Betacoronaviruses. PLoS Genet. 16(12):e1009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boley PA, Alhamo MA, Lossie G, Yadav KK, Vasquez-Lee M, Saif LJ, Kenney SP.. 2020. Porcine deltacoronavirus infection and transmission in poultry, United States. Emerg Infect Dis. 26(2):255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni MF, Lemey P, Jiang X, Lam TT-Y, Perry BW, Castoe TA, Rambaut A, Robertson DL.. 2020. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol. 5(11):1408–1417. [DOI] [PubMed] [Google Scholar]
- Boniotti MB, Papetti A, Lavazza A, Alborali G, Sozzi E, Chiapponi C, Faccini S, Bonilauri P, Cordioli P, Marthaler D.. 2016. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. 22(1):83–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CC, Shaw J, Jorba J, Bukbuk D, Adu F, Gumede N, Pate MA, Abanida EA, Gasasira A, Iber J, et al. 2013. Multiple independent emergences of type 2 vaccine-derived polioviruses during a large outbreak in northern Nigeria. J Virol. 87(9):4907–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprari S, Metzler S, Lengauer T, Kalinina OV.. 2015. Sequence and structure analysis of distantly-related viruses reveals extensive gene transfer between viruses and hosts and among viruses. Viruses 7(10):5388–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R, Dove B, Cavanagh D, Britton P.. 2003. Recombinant avian infectious bronchitis virus expressing a heterologous spike gene demonstrates that the spike protein is a determinant of cell tropism. J Virol. 77(16):9084–9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu Q, Guo D.. 2020. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 92(4):418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R.. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 5:536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Li F, Shi Z-L.. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 17(3):181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Morais HA, Dos Santos AP, do Nascimento NC, Kmetiuk LB, Barbosa DS, Brandão PE, Guimarães AMS, Pettan-Brewer C, Biondo AW.. 2020. Natural infection by SARS-CoV-2 in companion animals: a review of case reports and current evidence of their role in the epidemiology of COVID-19. Front Vet Sci. 7:591216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N, Mari V, Campolo M, Lorusso A, Camero M, Elia G, Martella V, Cordioli P, Enjuanes L, Buonavoglia C.. 2009. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of Swine are circulating in dogs. J Virol. 83(3):1532–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29(8):1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G, Rambaut A.. 2016. MERS-CoV recombination: implications about the reservoir and potential for adaptation. Virus Evol. 2(1):vev023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Elhaik E, Sabath N, Graur D.. 2006. The “inverse relationship between evolutionary rate and age of mammalian genes” is an artifact of increased genetic distance with rate of evolution and time of divergence. Mol Biol Evol. 23(1):1–3. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA.. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30(7):1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Zhao K, Shi Z-L, Zhou P.. 2019. Bat coronaviruses in China. Viruses 11(3):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D, Cagliani R, Clerici M, Sironi M.. 2017. Molecular evolution of human coronavirus genomes. Trends Microbiol. 25(1):35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Cordaux R.. 2017. Viruses as vectors of horizontal transfer of genetic material in eukaryotes. Curr Opin Virol. 25:16–22. [DOI] [PubMed] [Google Scholar]
- Goldstein SA, Brown J, Pedersen BS, Quinlan AR, Elde NC.. 2021. Extensive recombination-driven coronavirus diversification expands the pool of potential pandemic pathogens. bioRxiv. 2021.02.03.429646. doi: 10.1101/2021.02.03.429646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goluch T, Bogdanowicz D, Giaro K.. 2020. Visual TreeCmp: comprehensive comparison of phylogenetic trees on the Web. Methods Ecol Evol. 11(4):494–499. [Google Scholar]
- Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ.. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117(1):17–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O.. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27(2):221–224. [DOI] [PubMed] [Google Scholar]
- Graham RL, Baric RS.. 2010. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 84(7):3134–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Deming DJ, Deming ME, Yount BL, Baric RS.. 2018. Evaluation of a recombination-resistant coronavirus as a broadly applicable, rapidly implementable vaccine platform. Commun Biol. 1:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot S, Caro V, Cuervo N, Korotkova E, Combiescu M, Persu A, Aubert-Combiescu A, Delpeyroux F, Crainic R.. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J Virol. 74(18):8434–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA.. 2020. The molecular virology of coronaviruses. J Biol Chem. 295(37):12910–12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henritzi D, Petric PP, Lewis NS, Graaf A, Pessia A, Starick E, Breithaupt A, Strebelow G, Luttermann C, Parker LMK, et al. 2020. Surveillance of European domestic pig populations identifies an emerging reservoir of potentially zoonotic swine influenza a viruses. Cell Host Microbe. 28(4):614–627.e6. [DOI] [PubMed] [Google Scholar]
- Herrewegh AA, Smeenk I, Horzinek MC, Rottier PJ, de Groot RJ.. 1998. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. 72(5):4508–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z-M, Yang Y-L, Xu L-D, Wang B, Qin P, Huang Y-W.. 2019. Porcine Torovirus (PToV)—a brief review of etiology, diagnostic assays and current epidemiology. Front Vet Sci. 6:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liu WJ, Xu W, Jin T, Zhao Y, Song J, Shi Y, Ji W, Jia H, Zhou Y, et al. 2016. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathog. 12(9):e1005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C.. 2012. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 61(6):1061–1067. [DOI] [PubMed] [Google Scholar]
- ICTV Coronaviridae Study Group. 2020. International Committee on Taxonomy of Viruses (ICTV). Available from: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/223/coronaviridae-figures. Accessed October 5, 2021.
- Keck JG, Matsushima GK, Makino S, Fleming JO, Vannier DM, Stohlman SA, Lai MM.. 1988. In vivo RNA-RNA recombination of coronavirus in mouse brain. J Virol. 62(5):1810–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, Hosmillo M, Rees-Spear C, Mlcochova P, Lumb IU, et al. ; The CITIID-NIHR BioResource COVID-19 Collaboration. 2021. SARS-CoV-2 evolution during treatment of chronic infection. Nature 592(7853):277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottier SA, Cavanagh D, Britton P.. 1995. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology 213(2):569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L, Godeke GJ, Raamsman MJ, Masters PS, Rottier PJ.. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol. 74(3):1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT-Y, Jia N, Zhang Y-W, Shum MH-H, Jiang J-F, Zhu H-C, Tong Y-G, Shi Y-X, Ni X-B, Liao Y-S, et al. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583(7815):282–285. [DOI] [PubMed] [Google Scholar]
- Lang Y, Li W, Li Z, Koerhuis D, van den Burg ACS, Rozemuller E, Bosch B-J, van Kuppeveld FJM, Boons G-J, Huizinga EG, et al. 2020. Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity. Proc Natl Acad Sci U S A. 117(41):25759–25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latinne A, Hu B, Olival KJ, Zhu G, Zhang L, Li H, Chmura AA, Field HE, Zambrana-Torrelio C, Epstein JH, et al. 2020. Origin and cross-species transmission of bat coronaviruses in China. Nat Commun. 11(1):4235. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lau SKP, Wong EYM, Tsang C-C, Ahmed SS, Au-Yeung RKH, Yuen K-Y, Wernery U, Woo PCY.. 2018. Discovery and sequence analysis of four deltacoronaviruses from birds in the Middle East reveal interspecies jumping with recombination as a potential mechanism for avian-to-avian and avian-to-mammalian transmission. J Virol. 92(15):e00265-18. doi: 10.1128/JVI.00265-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C, Goeman JJ, Parquet M del C, Thi Nga P, Snijder EJ, Morita K, Gorbalenya AE.. 2013. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. 9(7):e1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C, Gorbalenya AE.. 2012. Partitioning the genetic diversity of a virus family: approach and evaluation through a case study of picornaviruses. J Virol. 86(7):3890–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C, Ziebuhr J, Junglen S, Drosten C, Zirkel F, Nga PT, Morita K, Snijder EJ, Gorbalenya AE.. 2012. Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch Virol. 157(8):1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47(W1):W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch B-J.. 2018. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci U S A. 115(22):E5135–E5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wong S-K, Li F, Kuhn JH, Huang I-C, Choe H, Farzan M.. 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol. 80(9):4211–4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DX, Fung TS, Chong KK-L, Shukla A, Hilgenfeld R.. 2014. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 109:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C-Y, Tsai T-L, Lin C-N, Lin C-H, Wu H-Y.. 2019. Interaction of coronavirus nucleocapsid protein with the 5’- and 3’-ends of the coronavirus genome is involved in genome circularization and negative-strand RNA synthesis. FEBS J. 286(16):3222–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Posada D.. 2011. Analysing recombination in nucleotide sequences. Mol Ecol Resour. 11(6):943–955. [DOI] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B.. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 1(1):vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS. 2019. Coronavirus genomic RNA packaging. Virology 537:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R, Iyer LM, Vasudevan S, Aravind L.. 2002. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 30(23):5229–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Hurst LD.. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet. 17(9):567–578. [DOI] [PubMed] [Google Scholar]
- Meekins DA, Morozov I, Trujillo JD, Gaudreault NN, Bold D, Carossino M, Artiaga BL, Indran SV, Kwon T, Balaraman V, et al. 2020. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg Microbes Infect. 9(1):2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, Yount BL, Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge X-Y, Donaldson EF, et al. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 21(12):1508–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, Yount BL, Sims AC, Debbink K, Agnihothram SS, Gralinski LE, Graham RL, Scobey T, Plante JA, Royal SR, et al. 2016. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A. 113(11):3048–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihindukulasuriya KA, Wu G, St Leger J, Nordhausen RW, Wang D.. 2008. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J Virol. 82(10):5084–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyers BA, Zhang J.. 2016. Evaluating phylostratigraphic evidence for widespread de novo gene birth in genome evolution. Mol Biol Evol. 33(5):1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Yamada KD, Tomii K, Katoh K.. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34(14):2490–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neches RY, Kyrpides NC, Ouzounis CA.. 2021. Atypical divergence of SARS-CoV-2 Orf8 from Orf7a within the coronavirus lineage suggests potential stealthy viral strategies in immune evasion. mBio. 12(1):e03014-20. doi: 10.1128/mBio.03014-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neches RY, McGee MD, Kyrpides NC.. 2020. Recombination should not be an afterthought. Nat Rev Microbiol. 18(11):606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaidis M, Mimouli K, Kyriakopoulou Z, Tsimpidis M, Tsakogiannis D, Markoulatos P, Amoutzias GD.. 2019. Large-scale genomic analysis reveals recurrent patterns of intertypic recombination in human enteroviruses. Virology 526:72–80. [DOI] [PubMed] [Google Scholar]
- Olival KJ, Cryan PM, Amman BR, Baric RS, Blehert DS, Brook CE, Calisher CH, Castle KT, Coleman JTH, Daszak P, et al. 2020. Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: a case study of bats. PLoS Pathog. 16(9):e1008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounis CA. 2020. A recent origin of Orf3a from M protein across the coronavirus lineage arising by sharp divergence. Comput Struct Biotechnol J. 18:4093–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S.. 2020. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 79:104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering BS, Smith G, Pinette MM, Embury-Hyatt C, Moffat E, Marszal P, Lewis CE.. 2021. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 27(1):104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliaka V, Kyriakopoulou Z, Markoulatos P.. 2012. Risks associated with the use of live-attenuated vaccine poliovirus strains and the strategies for control and eradication of paralytic poliomyelitis. Expert Rev Vaccines. 11(5):609–628. [DOI] [PubMed] [Google Scholar]
- Pond SLK, Frost SDW, Muse SV.. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21(5):676–679. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA, Holmes EC.. 2002. Recombination in evolutionary genomics. Annu Rev Genet. 36:75–97. [DOI] [PubMed] [Google Scholar]
- Racaniello VR. 2006. One hundred years of poliovirus pathogenesis. Virology 344(1):9–16. [DOI] [PubMed] [Google Scholar]
- Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke G-J, Meyer B, Muth D, Raj VS, Vries LS-D, Corman VM, et al. 2013. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 13(10):859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR.. 1981. Comparison of phylogenetic trees. Math Biosci. 53(1–2):131–147. [Google Scholar]
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen M-H, et al. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300(5624):1394–1399. [DOI] [PubMed] [Google Scholar]
- Rottier PJM, Nakamura K, Schellen P, Volders H, Haijema BJ.. 2005. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J Virol. 79(22):14122–14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeng-Chuto K, Jermsutjarit P, Stott CJ, Vui DT, Tantituvanont A, Nilubol D.. 2020. Retrospective study, full-length genome characterization and evaluation of viral infectivity and pathogenicity of chimeric porcine deltacoronavirus detected in Vietnam. Transbound Emerg Dis. 67(1):183–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez CM, Izeta A, Sánchez-Morgado JM, Alonso S, Sola I, Balasch M, Plana-Durán J, Enjuanes L.. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J Virol. 73(9):7607–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki SG, Sawicki DL, Siddell SG.. 2007. A contemporary view of coronavirus transcription. J Virol. 81(1):20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelle B, Karl N, Ludewig B, Siddell SG, Thiel V.. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J Virol. 79(11):6620–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JF, Bornberg-Bauer E.. 2017. Fact or fiction: updates on how protein-coding genes might emerge de novo from previously non-coding DNA. F1000Res 6:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Zheng Y, Yang Y, Liu C, Geng Q, Tai W, Du L, Zhou Y, Zhang W, Li F.. 2018. Cryo-electron microscopy structure of porcine deltacoronavirus spike protein in the prefusion state. J Virol. 92(4):e01556-17. doi: 10.1128/JVI.01556-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang P, Misra S, Hause B, Fang Y.. 2017. A naturally occurring recombinant enterovirus expresses a torovirus deubiquitinase. J Virol. 91(14):e00450-17. doi: 10.1128/JVI.00450-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, et al. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 368(6494):1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M.. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17(12):1246–1247. [DOI] [PubMed] [Google Scholar]
- Simon-Loriere E, Holmes EC.. 2011. Why do RNA viruses recombine? Nat Rev Microbiol. 9(8):617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW, Yu VYT, Sims LD, Tsang DNC, Chu DKW, et al. 2020. Infection of dogs with SARS-CoV-2. Nature 586(7831):776–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, den Boon JA, Horzinek MC, Spaan WJ.. 1991. Comparison of the genome organization of toro- and coronaviruses: evidence for two nonhomologous RNA recombination events during Berne virus evolution. Virology 180(1):448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Almazán F, Zúñiga S, Enjuanes L.. 2015. Continuous and discontinuous RNA synthesis in coronaviruses. Annu Rev Virol. 2(1):265–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I, Mateos-Gomez PA, Almazan F, Zuñiga S, Enjuanes L.. 2011. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 8(2):237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H-D, Tu C-C, Zhang G-W, Wang S-Y, Zheng K, Lei L-C, Chen Q-X, Gao Y-W, Zhou H-Q, Xiang H, et al. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A. 102(7):2430–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöver BC, Müller KF.. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF.. 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24(6):490–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A.. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4(1):vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Xiao Y, Liu J, Wang D, Li F, Wang C, Li C, Zhu J, Song J, Sun H, et al. 2020. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci U S A. 117(29):17204–17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungsuwan S, Jongkaewwattana A, Jaru-Ampornpan P.. 2020. Nucleocapsid proteins from other swine enteric coronaviruses differentially modulate PEDV replication. Virology 540:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y, Matsui N, Noguchi K, Kuwata R, Shimoda H, Soma T, Mochizuki M, Maeda K.. 2014. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS One 9(9):e106534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P-F, Jin Y-L, Xing G, Qv L-L, Huang Y-W, Zhou J-Y.. 2014. Evidence of recombinant strains of porcine epidemic diarrhea virus, United States, 2013. Emerg Infect Dis. 20(10):1735–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoleridis T, Chappell JG, Onianwa O, Marston DA, Fooks AR, Monchatre-Leroy E, Umhang G, Müller MA, Drexler JF, Drosten C, et al. 2019. Shared common ancestry of rodent alphacoronaviruses sampled globally. Viruses 11(2):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF, et al. ; COG-UK Consortium. 2021. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184(1):64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lin X-D, Zhang H-L, Wang M-R, Guan X-Q, Holmes EC, Zhang Y-Z.. 2020. Extensive genetic diversity and host range of rodent-borne coronaviruses. Virus Evol. 6(2):veaa078. doi: 10.1093/ve/veaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SR, Navas-Martin S.. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol Mol Biol Rev. 69(4):635–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Sariol A, Meyerholz DK, Perlman S.. 2018. Microglia are required for protection against lethal coronavirus encephalitis in mice. J Clin Invest. 128(3):931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille M, Holmes EC.. 2020. Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses. FEMS Microbiol Rev. 44(5):631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ACP, Li X, Lau SKP, Woo PCY.. 2019. Global epidemiology of bat coronaviruses. Viruses 11(2):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Lau SKP, Huang Y, Yuen K-Y.. 2009. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med (Maywood). 234(10):1117–1127. [DOI] [PubMed] [Google Scholar]
- Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, Bai R, Teng JLL, Tsang CCC, Wang M, et al. 2012. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 86(7):3995–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Lau SKP, Lam CSF, Tsang AKL, Hui S-W, Fan RYY, Martelli P, Yuen K-Y.. 2014. Discovery of a novel bottlenose dolphin coronavirus reveals a distinct species of marine mammal coronavirus in Gammacoronavirus. J Virol. 88(2):1318–1331. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Woo PCY, Wang M, Lau SKP, Xu H, Poon RWS, Guo R, Wong BHL, Gao K, Tsoi H-W, Huang Y, et al. 2007. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 81(4):1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Pekar J, Larsen BB, Nelson MI, Hill V, Joy JB, Rambaut A, Suchard MA, Wertheim JO, Lemey P.. 2020. The emergence of SARS-CoV-2 in Europe and North America. Science 370(6516):564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, et al. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yan W, Hall AB, Jiang X.. 2021. Characterizing transcriptional regulatory sequences in coronaviruses and their role in recombination. Mol Biol Evol. 38(4):1241–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount B, Roberts RS, Lindesmith L, Baric RS.. 2006. Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome. Proc Natl Acad Sci U S A. 103(33):12546–12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Langereis MA, van Vliet ALW, Huizinga EG, de Groot RJ.. 2008. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci U S A. 105(26):9065–9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O, Price J, Shalamova L, Kamenova T, Goodfellow I, Weber F, Miska EA.. 2020. The short- and long-range RNA-RNA interactome of SARS-CoV-2. Mol Cell. 80(6):1067–1077.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst R, Miller TB, Goebel SJ, Thiel V, Masters PS.. 2008. Genetic interactions between an essential 3’ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3’ end of the mouse coronavirus genome. J Virol. 82(3):1214–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All necessary data are incorporated into the article and its online supplementary material. Any further data are available on request.