Abstract
Several eukaryotic transcription factors such as Sp1 or Oct1 contain glutamine-rich domains that mediate transcriptional activation. In human cells, promoter-proximally bound glutamine-rich activation domains activate transcription poorly in the absence of acidic type activators bound at distal enhancers, but synergistically stimulate transcription with these remote activators. Glutamine-rich activation domains were previously reported to also function in the fission yeast Schizosaccharomyces pombe but not in the budding yeast Saccharomyces cerevisiae, suggesting that budding yeast lacks this pathway of transcriptional activation. The strong interaction of an Sp1 glutamine-rich domain with the general transcription factor TAFII110 (TAFII130), and the absence of any obvious TAFII110 homologue in the budding yeast genome, seemed to confirm this notion. We reinvestigated the phenomenon by reconstituting in the budding yeast an enhancer-promoter architecture that is prevalent in higher eukaryotes but less common in yeast. Under these conditions, we observed that glutamine-rich activation domains derived from both mammalian and yeast transcription factors activated only poorly on their own but strongly synergized with acidic activators bound at the remote enhancer position. The level of activation by the glutamine-rich activation domains of Sp1 and Oct1 in combination with a remote enhancer was similar in yeast and human cells. We also found that mutations in a glutamine-rich domain had similar phenotypes in budding yeast and human cells. Our results show that glutamine-rich activation domains behave very similarly in yeast and mammals and that their activity in budding yeast does not depend on the presence of a TAFII110 homologue.
The expression of protein-coding genes in eukaryotes depends on DNA-binding transcription factors that typically have at least two distinct domains: one domain responsible for specific DNA recognition and one responsible for transcriptional activation (5, 27). According to their predominant amino acid composition, activation domains have been classified mainly into acidic, proline-rich, and glutamine-rich domains (for reviews, see references 28 and 40). When tethered to DNA, these classes of activation domains possess different biological properties in their ability to influence gene expression (38). Acidic activation domains rich in acidic and hydrophobic amino acids, such as those found in VP16 or Gal4p proteins, are the most versatile activators, and they stimulate transcription when bound to DNA from proximal and distal enhancer positions in all eukaryotes. Proline-rich activation domains, e.g., those of AP-1 or CTF/NF1, generally activate from proximal and, albeit to a much reduced extent, from distal positions. Glutamine-rich activators like Sp1 or Oct1 on their own fail to activate transcription from a remote enhancer position, but they stimulate gene expression in response to remote enhancers when bound in close proximity to the TATA box in human cells (38). The same glutamine-rich activation domains were reported to be active in the fission yeast Schizosaccharomyces pombe when tethered to a proximal regulatory sequence (35). In contrast, with the budding yeast Saccharomyces cerevisiae, we and others have reported that glutamine-rich activation domains of the mammalian transcription factors Sp1, Oct1, and Oct2 or full-length Sp1, when tethered to DNA, are transcriptionally inactive at either proximal or remote position when tested on their own (24, 32).
In order to stimulate transcription, activation domains have to interact with components of the transcriptional complex (for reviews, see references 2 and 30). For Sp1, it has been shown that the two glutamine-rich activation domains directly interact with TAFII110, a component of the general transcription machinery (20). TAFII110 is present in higher eukaryotes (e.g., as dTAFII110 in Drosophila and as hTAFII130 in humans), but no homologue could be found in the genome of the budding yeast S. cerevisiae. This finding offered a straightforward explanation for the seeming inability of the Sp1 glutamine-rich domains to activate transcription in the budding yeast.
Another important observation is that the typical arrangement of regulatory sequences controlling gene expression in yeast differs from that of higher eukaryotes. In metazoans, binding sites for transcription factors are often found in close proximity to the transcriptional start site and also at a considerable distance. Proximal sites may contain binding sites for Sp1 and/or Oct1 (22). Distal enhancer elements can influence gene expression when positioned upstream, downstream, or even as part of an intron within the transcription unit (1; for a review, see reference 37). In the budding yeast, probably due to space constraints and the almost complete lack of introns, the majority of genes are controlled by a few binding sites for transcription factors, termed upstream activating sequences, which are located close to the TATA box.
To see whether enhancer or promoter structure could influence the activity of glutamine-rich domains in the budding yeast, we reconstituted a metazoan-like regulatory structure in the yeast chromosomal context by introducing transcription factor binding sites both in close proximity to the TATA box and at a remote upstream enhancer position. Under these conditions, we observed that the glutamine-rich activation domains of mammalian Sp1 or Oct1 and yeast Snf5p readily contributed to gene expression in yeast, in a manner similar to their behavior in mammalian cells tested in parallel. These results indicate that there is no functional difference for glutamine-rich activation domains in stimulating gene expression in yeast and mammalian cells, irrespective of the lack of TAFII110 in S. cerevisiae.
MATERIALS AND METHODS
Plasmids and yeast strains.
The yeast-integrating LacZ reporter plasmid pENH200 (see Fig. 2A) was derived via a combination of pJP158 (3), pDE200, and pDEYS2 (13). This resulted in a URA3-marked integrating vector that contains the LacZ gene under the control of two Lex, and three Gal4 binding sites 40 and 240 bp upstream of the GAL1 TATA box, respectively. The reporter plasmids pENH400 and pENH800 are derived from pENH200 and have 400- and 800-bp spacer sequences between the two proximal Lex binding sites and the three Gal4 binding sites, respectively. The integrating yeast plasmid pMG1, with switched Lex and Gal4 binding sites, was derived from pENH200 via replacement of the two proximal Lex (SacII-XbaI) and the three distal Gal4 (AflII-XmaI) sites with a double-stranded oligonucleotide that encodes two synthetic Gal4 and three Lex binding sites and possesses the corresponding SacII-XbaI and AflII-XmaI overhangs, respectively. The TATA-less TRP3 reporter plasmid pENH200TL was derived by subcloning the regulatory cassette that contains three Gal4 and two Lex binding sites (AflII [blunt ended]-XbaI) from pENH200 into the XhoI-filled-in XbaI sites immediately upstream of the TATA-less Trp promoter of pAB135, a URA3-integrating LacZ reporter vector.
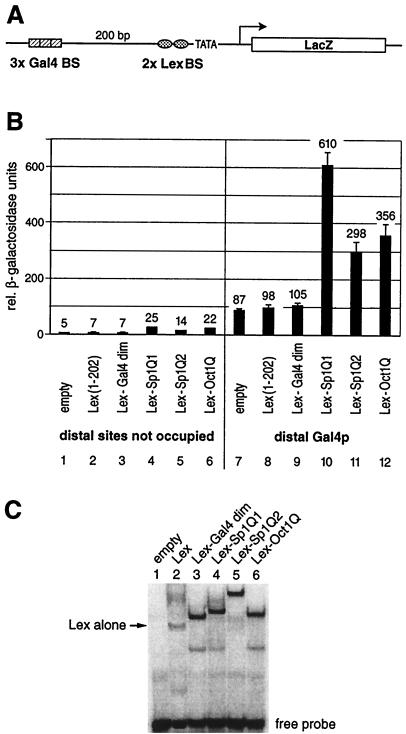
FIG. 2.
Glutamine-rich activation domains stimulate transcription from a chromosomally embedded reporter gene in yeast. (A) Schematic drawing of the integrated yeast reporter. The promoter architecture resembles that of a typical higher eukaryote. It contains distant (three Gal4 binding sites) as well as proximal (two Lex binding sites) regulatory elements upstream of the GAL1 TATA box. The reporter was integrated at the chromosomal URA3 locus of S. cerevisiae. (B) The reporter strain was transformed with yeast plasmids that express Lex fusion proteins to the Gal4 dimerization domain (amino acids 58 to 97) as a negative control and the same glutamine-rich activation domains of Sp1 (Q1 and Q2) and Oct1Q as tested in HeLa cells (Fig. 1). Their ability to stimulate transcription when bound to the proximal binding sites either alone or in combination with a distal Gal4p activator was monitored. The factors containing a glutamine-rich activation domain displayed a low intrinsic activation potential when tested on their own (lanes 4 to 6), which was above the background as determined by the Lex (lane 2) or Lex-Gal4 dimerization domain alone (lane 3). The influence on reporter gene activation of the distal Gal4p (lane 7) and the proximal glutamine-rich activation domains (lanes 4 to 6) was more than additive when tested in combination, i.e., they resulted in synergistic gene activation (lanes 10 to 12). As in the mammalian cells shown in Fig. 1, Sp1Q1 (lanes 4 and 10) was the strongest activation domain, followed by Oct1Q (lanes 6 and 12) and Sp1Q2 (lanes 5 and 11). (C) EMSAs were performed with total yeast protein extracts derived from reporter strains that expressed various Lex fusion proteins. A 32P-labeled oligonucleotide duplex with two Lex binding sites was used as a probe. Extracts from reporter strains without a Lex fusion protein did not reveal any protein interaction with the oligonucleotide (lane 1). Lex alone (lane 2) produced a weaker bandshift than the other fusion proteins. Fusion of the Gal4 dimerization domain to the Lex protein (lane 3) increased the stability of the protein to interact with the oligonucleotide and yielded a bandshift comparable to those of Lex fused to Sp1Q1 (lane 4), Sp1Q2 (lane 5), and Oct1Q (lane 6).
As a source strain for the integration of the yeast reporter plasmids, we used JPY9 (MATα ura3-52 his3Δ200 leu2Δ1 trp1Δ63 lys2Δ385 gal4Δ11) (3). All reporter plasmids (pENH200, pENH400, pENH800, pENH200TL, and pMG1) were linearized at the ApaI site in the URA3 gene and were integrated into the yeast genome, resulting in YENH200, YENH400, YENH800, YENHTL200, and YMG1, respectively. Correct, single integration was confirmed by genomic PCR analysis, and three independent yeast transformants from each integration were tested and compared in functional assays.
The yeast expression vectors encoding various Lex fusion proteins were derived from pDE101, a Trp1-marked ARS/CEN vector derived from pJP228 (3) coding for the Lex (amino acids 1 to 202)-Gal4 (amino acids 58 to 97) fusion protein under the control of the strong, constitutive yeast actin promoter. The Gal4 (amino acids 58 to 97) moiety was released by XbaI-SalI digestion and was replaced by different Sp1 or Oct1 fragments derived by PCR (Sp1Q1 amino acids 50 to 161, NRTVSGGQYVVAAAPNLQNQQVLTGLPGVMPNIQYQVIPQFQT VDGQQLQFAATGAQVQQDGSGQIQI I PGANQQI I T NRGSGG N I IAAMPNLLQQAVPLQGLANNVLSGQT; Sp1Q2 amino acids 257 to 403, SSGTNSQGQ T PQRVSGLQGSDALN IQQNQ TSGGS LQAGQQKEGEQNQQTQQQQI LIQPQLVQGGQALQALQAAPLSGQTF T TQAISQE T LQNLQLQAVPNSG P I I I RTPTVGPNGQVSWQTLQLQNLQVQNPQAQ TI T LAPMQGVS L; and Oct1Q amino acids 175-269, DLQQLQQLQQQNLNLQQFVLVHPTTNLQPAQF I I SQTPQGQQG L LQAQNLQTQLPQQSQAN L LQSQPS I T LTSQPATPTRTIAATPIQTLPQSQS). PCR of the different glutamine-rich activation domains was carried out on template as previously described (24). Three individual PCR-derived clones of each ligation were sequenced and then compared by functional assays. The section of the gene encoding the glutamine-rich domain of Snf5 was cloned by PCR by using yeast genomic DNA as a template and oligonucleotides containing an XbaI and SalI overhang, respectively, and annealed to SNF5 at nucleotide positions 550 and 859. The PCR product was cloned into XbaI/SalI-digested pDE101, resulting in pDESNF5#15 (Lex-Snf5 amino acids 185 to 286). The internal deletion (of Lex-Snf5 amino acids 211-260) in the glutamine-rich domain of Snf5 occurred fortuitously during cloning procedure, and is designated pDESNF5#9. The GAL4 full-length yeast expression vector is driven by the yeast ADH promoter on a HIS3-marked, high-copy-number (2μm) plasmid.
Gene expression.
Gene expression in yeast was monitored by using liquid β-galactosidase assays and were performed as previously described (13). For reliable measurement of the low signals obtained when the glutamine-rich activation domains were tested alone, we routinely extracted whole-cell proteins as described below. All assays were conducted with duplicate samples and were repeated at least once. For HeLa cell experiments, cells were transfected as previously described (14). As a reporter plasmid, we used the OVEC β-globin system (42). Subsequent S1 nuclease mapping was done as described (42).
EMSA and immunoblot analysis.
Yeast cultures were grown in 10 ml of selective media to an optical density at 600 nm of 1.5 and then were harvested by centrifugation and resuspended in 1 ml of buffer containing 20 mM HEPES (pH 7.5), 10% glycerol, and 0.45 M NaCl, then were reharvested and resuspended in 0.1 ml of the same buffer supplemented to 0.1 M phenylmethylsulfonyl fluoride and 1 M dithiothreitol. From this step on, the samples were kept on ice. After addition of an equal volume of glass beads, samples were vigorously vortexed (5 × 30 s), and 0.1 ml of the same buffer was added before an additional vortexing step (15 s). After a 10-min centrifugation step at 4°C, supernatants were collected and used for electrophoretic mobility shift assay (EMSA) experiments (approximately 2 to 6 μl) or for β-galactosidase assays according to standard protocol. As a probe, we used a double-stranded, 32P-labeled oligonucleotide (5′ GCAGTGCTGTATATAAAACGAGTGGTTATATGTACAGTAG 3′) that contains two Lex binding sites. EMSA was performed as described (38). Immunoblot analysis of the Lex-Sp1Q2 wild type and the different mutants was carried out by using an anti-LexA antibody (Clontech). The experimental procedure of the immunoblot was carried out as described (13).
RESULTS
Glutamine-rich activation domains have a low activation potential on their own, but strongly synergize with a distal enhancer in both human and yeast cells.
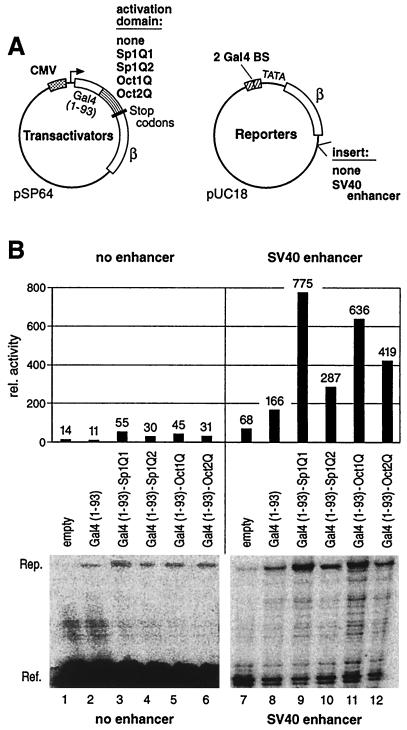
To clarify the transcriptional activation by glutamine-rich activation domains, we tested them in parallel in both mammalian and in yeast cells. In human cells, such domains were tethered to promoter-proximal binding sites, either on their own, or in combination with an enhancer. For this, we cloned the minimal glutamine-rich activation domains of Sp1 (Sp1Q1 and Sp1Q2, see Materials and Methods), Oct1 (Oct1Q), and Oct2 (Oct2Q) to the Gal4 DNA-binding domain (DBD) into a mammalian expression vector under the control of the strong cytomegalovirus promoter (Fig. 1A, Transactivators). As reporter, we used the β-globin gene, which was under the control of two Gal4 binding sites proximal to the TATA box, with or without a simian virus 40 (SV40) enhancer sequence in a downstream position (Fig. 1A, Reporters). The different transactivators and reporters were cotransfected into HeLa cells. Two days after transfection, we quantified the transcript levels of reporter and reference genes. In the absence of a distal enhancer, proximally bound glutamine-rich activation domains showed a very low intrinsic activation potential (Fig. 1B, lanes 2 to 6). The strongest activation domain (Sp1Q1, lane 3) was able to activate reporter gene transcription five- to sixfold above that of Gal4 DBD alone (lane 2). However, transactivation by Sp1Q1 was less than 5% of that mediated by the strong herpes simplex activator VP16 fused to the Gal4 DBD (data not shown). A remotely positioned SV40 enhancer by itself, without a proximally bound transcription activation domain, activated β-globin expression only weakly (lane 7). Binding of the Gal4 DBD (amino acids 1 to 93) in combination with the SV40 enhancer stimulated the reporter gene expression to some extent (lane 8), while a combination of proximal transactivator and distal SV40 enhancer led to strong synergistic gene activation (lanes 9 to 12). Quantification of reference and reporter β-globin expression revealed that the activity of a remote enhancer was six- to sevenfold higher in concert with Sp1Q1 (lane 9), compared to the background by the Gal4 DBD alone (lane 8). The other tested glutamine-rich activation domains, Sp1Q2, Oct1, and Oct2, also synergized with the remote SV40 enhancer to yield three-, six-, and fourfold-higher levels of activity (lanes 10 to 12), respectively. These results show that glutamine-rich activation domains in HeLa cells possess a very low intrinsic activation potential, but in combination with additional activators contribute to synergistic gene activation. These data, which are in agreement with previous results (38), were the basis for a direct comparison to the budding yeast, where the same glutamine-rich activation domains were tested. Unlike our previous study where we found no activity of glutamine-rich domains in yeast (24), this time we used a promoter architecture that more closely resembles that of higher eukaryotes by introducing two proximal Lex sites and three distal enhancer binding sites for Gal4p upstream of the TATA box (Fig. 2A). This LacZ reporter construct was integrated into the genomic URA3 locus. We fused the Lex DBD (amino acids 1 to 202) to the same glutamine-rich activation domains of Sp1 (Lex-Sp1Q1 and Lex-Sp1Q2) and Oct1 (Lex-Oct1Q) as used in the HeLa cell experiments. These effector plasmids were tested for their ability to activate LacZ gene expression when bound proximally. The activity of Lex fusion proteins, either alone or in combination with the remotely bound Gal4p activator, was measured by the β-galactosidase assay (Fig. 2B). All glutamine-rich activators when bound to the two proximal Lex binding sites were able to activate transcription to a low level only (Fig. 2B, lanes 4 to 6), although significantly above basal expression (lanes 1 to 3). This low expression was less than 5% of the value seen with acidic activators such as Lex-VP16 or Lex-Gal4p (data not shown).
FIG. 1.
Glutamine-rich activation domains are dependent on a distal enhancer in human cells. (A) HeLa cells were cotransfected with expression plasmids where the DBD of Gal4 (amino acids 1 to 93) was fused to glutamine-rich activation domains of Sp1, Oct1, or Oct2 (Transactivators), along with a reporter plasmid under the control of two proximal Gal4 binding sites, with or without a distal SV40 enhancer (Reporters). As an internal control, a reference plasmid expressing the 5′-truncated form of the β-globin gene was cotransfected. (B) Total RNA was harvested 2 days after transfection of HeLa cells, and S1 nuclease analysis was performed. Lanes 7 to 12, due to the stronger reporter signals, were exposed for a shorter time than lanes 1 to 6. Quantification of the reporter signal (Rep.) relative to the corresponding reference (Ref.) was carried out by using a PhosphorImager. Without a distal enhancer, the glutamine-rich activation domains stimulate reporter gene expression only slightly above the background level (lanes 3 to 6 versus lanes 1 and 2). In combination with the remote SV40 enhancer, the glutamine-rich activation domains synergistically activated gene expression (lanes 9 to 12) when compared to the influence of the SV40 enhancer alone (lanes 7 and 8).
To obtain a reliable quantification of β-galactosidase also at low expression levels, a highly efficient protein extraction method was employed (see Materials and Methods), and the experiment was repeated four times with independent yeast transformants. The yeast transactivator Gal4p, when bound to the remote enhancer position, activated the reporter gene to a similar extent when the proximal site was either not occupied or was bound by transcriptionally inert proteins like Lex (amino acids 1 to 202) or Lex fused to the Gal4 dimerization domain (Lex-Gal4 dim) (Fig. 2B, lanes 7 to 9). Combination of distal Gal4p with proximal glutamine-rich activators led to a strong reporter gene expression, in which the contributions of the remote Gal4p (lane 7) and the proximal glutamine-rich activators (lanes 4 to 6) were more than additive. Sp1Q1 synergized with the remote Gal4p activator approximately sixfold more efficiently than Gal4p alone (compare lanes 10 and 7). The weaker-stimulating activation domain Sp1Q2 and the glutamine-rich activation domain of Oct1 yielded three- and fourfold-higher activities, respectively, compared to the level of Gal4p alone (lanes 11, 12, and 7, respectively).
To control for protein stability and binding capacity of these Lex fusion proteins, whole-yeast protein extracts were subjected to gel mobility shift assays with labeled Lex site oligonucleotide (Fig. 2C). Lex fused to the Gal4 dimerization domain (amino acids 58 to 97) or fused to Sp1Q1, Sp1Q2, and Oct1Q showed comparable DNA-binding activities, while the signal with Lex (amino acids 1 to 202) was weaker. We therefore reasoned that the differential cooperation of the Lex fusion proteins with the Gal4p activator is due to their different transactivation potentials, rather than to differential expressions, bindings, or stabilities of these hybrid proteins.
In addition to the yeast Gal4p activator, we tested the artificial activator B42 (27) fused to the Gal4 DBD for its ability to influence gene expression from a remote enhancer position or in cooperation with proximal glutamine-rich activation domains. B42 synergized with the glutamine-rich activation domains of Sp1 and Oct1, comparable to the values observed with the Gal4p activator (data not shown). As an additional control, in order to see whether the observed synergistic gene activation was not a peculiarity of the Lex fusion proteins but was also evident with another heterologous DBD, we exchanged the positions of the cis-regulatory elements controlling the LacZ reporter gene. This resulted in a LacZ reporter gene driven by two proximal Gal4 and three distal Lex binding sites. The glutamine-rich activation domains Sp1Q1 and Oct1Q were fused to the Gal4 DBD (amino acids 1 to 147). With the distal amphipathic α-helical transcriptional activation domain (AH) (17) fused to Lex (amino acids 1 to 202), they synergized more than two- and threefold, respectively (data not shown).
In mammalian cells, Sp1 is also able to activate the class of promoters that lack a TATA box (TATA-less promoters) (4, 10, 21, 31, 33, 44). We were therefore interested to see whether the stronger glutamine-rich activation domain of Sp1 might also activate the yeast TATA-less TRP3 promoter. To this end, we replaced the core promoter of the reporter gene described in Fig. 2A with the TATA-less TRP3 core promoter. The distal Gal4p activator and the proximal Lex-Sp1Q1 activated the TATA-less driven reporter gene more than twofold (data not shown).
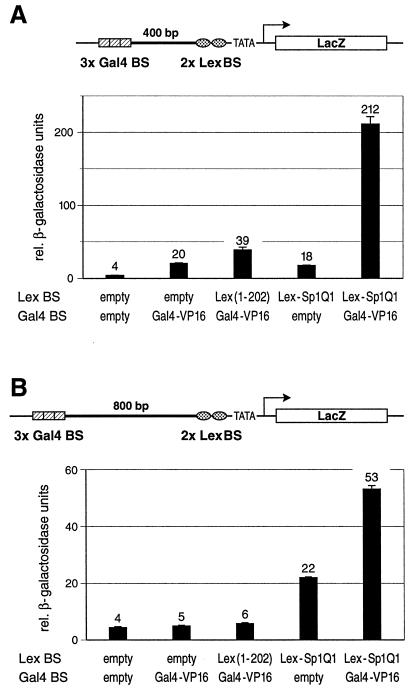
In addition to the remote activators Gal4p, α-helical transcriptional activation domain AH, and B42, we tested the acidic activation domain of the herpes simplex viral activator VP16 (41) fused to the Gal4 DBD (Gal4-VP16) for its effect to stimulate transcription either alone or in combination with proximally bound Sp1Q1. The reporter gene in the yeast strain YENH200, in which the remote enhancer was separated from the proximal binding sites by a 200-bp spacer, was activated by Gal4-VP16 by more than fourfold over that seen by the yeast activator Gal4p (data not shown). Combination of proximal glutamine-rich activation domains and Gal4-VP16 led only to a weak stimulation (less than a twofold increase). We considered whether Gal4-VP16 on its own could activate reporter gene expression to nearly the maximum extent under the tested promoter configuration. In such a scenario, a proximally bound glutamine-rich activator would only make minor contributions to gene activation. We attempted to weaken the influence of the strong activator by increasing the distance between its binding sites and the promoter. To this end, we replaced the 200-bp spacer sequence between the proximal and distal binding sites with 400- and 800-bp spacers, respectively. These reporters were also integrated into the genomic URA3 locus in yeast and were tested for relative β-galactosidase activity. The reporter containing the 400-bp spacer was only weakly activated by the remote Gal4-VP16 alone or by the proximally bound glutamine-rich Lex-Sp1Q1 alone (Fig. 3A). However, combination of distal Gal4-VP16 and proximal Lex-Sp1Q1 resulted in strong synergistic gene activation. The reporter containing the 800-bp spacer was not influenced by the remote Gal4-VP16 when tested alone or in combination with the proximal, transcriptionally inert Lex protein as compared to the background (Fig. 3B). Nevertheless, the combination of remote Gal4-VP16 and proximal Lex-Sp1Q1 did result in a significant increase in reporter gene expression compared to the control. Particularly striking is the fact that Gal4-VP16 activation over the very long distance of 800 bp entirely depends on the presence of a promoter-proximal glutamine-rich domain.
FIG. 3.
The herpes simplex viral activator VP16 synergizes with a proximal glutamine-rich activation domain of Sp1 over long distances in yeast. (A) The potent activation domain of the herpes simplex transactivator VP16 fused to the Gal4 DBD (Gal4-VP16) was tested for its ability to stimulate gene expression from remote Gal4 binding sites (Gal4 BS). These binding sites were separated from the proximal Lex binding sites (Lex BS) by a 400-bp spacer sequence. A schematic drawing of the chromosomal reporter construct is indicated above the graph. Distal Gal4-VP16 or proximal Sp1Q1 fused to LexA (Lex-Sp1Q1) alone activated to comparable low levels. Combination of both resulted in a more-than-tenfold increase of reporter gene transcription as compared to the effect of each activator tested on its own. (B) Over the very long distance of 800 bp, the Gal4-VP16 transactivator did not stimulate gene expression at all when tested either alone or with the proximal, transcriptionally inert LexA. However, in combination with the proximally bound glutamine-rich domain of Sp1 (Lex-Sp1Q1), Gal4-VP16 strongly contributed to gene expression, i.e., under these conditions, the enhancer-bound activator is strictly dependent on the presence of a promoter-proximal glutamine-rich activation domain.
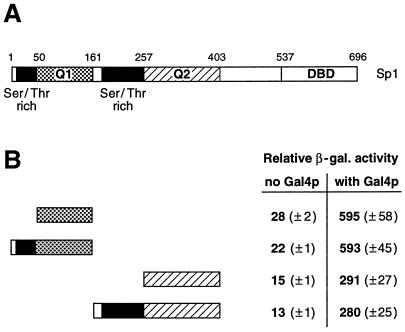
The serine/threonine-rich domains of Sp1 do not influence the adjacent glutamine-rich activation domains.
Several serine/threonine-rich domains have been reported to harbor transcriptional activity (18, 38, 39). The Sp1 transcription factor contains two serine/threonine-rich domains, each immediately N terminal to the two glutamine-rich activation domains (see Fig. 4A for a schematic drawing). We wanted to determine whether these two serine/threonine-rich domains of Sp1 could influence the activation potential of the adjacent glutamine-rich domains. For this, we fused fragments of Sp1 containing the serine/threonine-rich and Q1 domains (amino acids 1 to 161) as well as the serine/threonine-rich and Q2 domains (amino acids 161 to 403) to the Lex DBD. These fusion proteins were compared to their counterparts containing only the glutamine-rich activation domains Q1 and Q2 (Fig. 4B). We observed that the serine/threonine-domain-containing constructs behaved in a manner indistinguishable from that of the pure glutamine-rich activation domains. Therefore, the serine/threonine-rich domains apparently do not influence the transactivation potential of the adjacent Q1 or Q2 activation domain of Sp1 in yeast. These data are in agreement with previous results from Drosophila Schneider cells that indicated that the serine/threonine-rich domains of Sp1 do not contribute to transcriptional activation (9).
FIG. 4.
The serine/threonine-rich domains of Sp1 do not influence transactivation by the adjacent glutamine-rich activation domains. (A) Schematic drawing of full-length Sp1 transcription factor (696 amino acids). Sp1 harbors two glutamine-rich activation domains termed Q1 and Q2, two serine/threonine-rich domains, and a DBD at the C terminus. (B) Quantitative β-galactosidase assay. The activity of the glutamine-rich activation domains Q1 and Q2 were compared with N-terminal extensions that included the serine/threonine-rich domains. The activity of the different Lex DBD hybrids was assayed in yeast when bound to the proximal position, either alone (no Gal4p) or in combination with a distal Gal4p activator (with Gal4p). The serine/threonine-domain-containing constructs activated reporter gene expression indistinguishably from that of the respective activation domains Q1 or Q2.
Mutations in Sp1Q2 similarly affect its transactivation activity in yeast and in human cells.
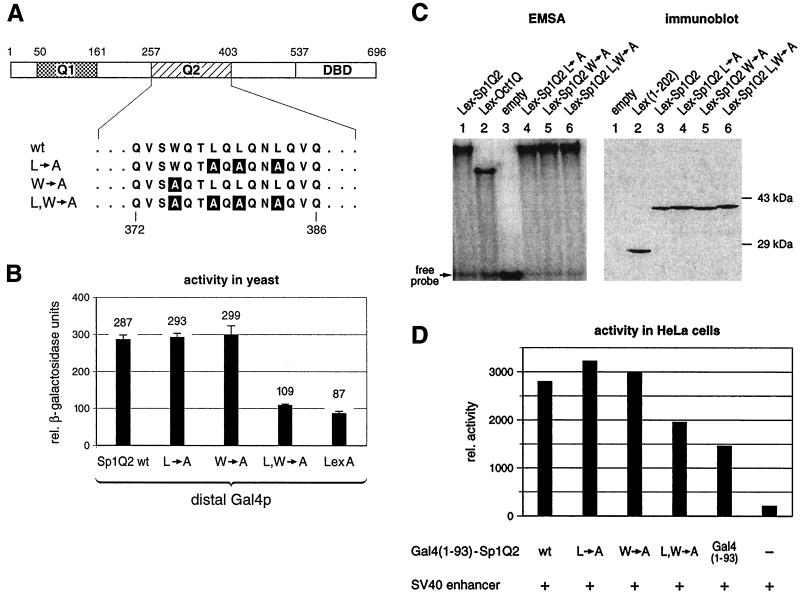
The Drosophila TATA-binding protein (TBP)-associated factor TAFII110 has been shown to interact with the human transcription factor Sp1 (20). The level of Sp1 transcriptional activity in Drosophila directly correlated with the strength of interaction between the glutamine-rich activation domains of Sp1 and TAFII110 (16). Substitutions in the weaker of the two glutamine-rich activation domains (Sp1Q2), changing three leucines to alanines (L→A) or one tryptophan to alanine (W→A), were reported to be defective in their ability to interact with the Drosophila TAFII110 in a yeast two-hybrid assay. When fused to the Zn finger DBD of Sp1, they also failed to activate transcription of a reporter gene dependent on six Sp1 binding sites from the SV40 promoter in Drosophila Schneider cells (16). We introduced the same mutations as described (L→A and W→A) and the combination of them (L, W→A) by site-directed mutagenesis (Fig. 5A). The resulting mutant Sp1Q2 domains fused to Lex (amino acids 1 to 202) were tested for their ability to synergize with the distal Gal4p activator in the yeast reporter strain described in Fig. 2A. Unexpectedly, we could not observe a significant difference of the two mutants (L→A and W→A) relative to transactivation by wild-type Sp1Q2 in yeast (Fig. 5B). Only a combination of the two mutants, with the four amino acids exchanged in the 147-amino-acid-long activation domain reduced activity to near background level.
FIG. 5.
Mutations in Sp1Q2 similarly affect its activation potential in both yeast and human cells. (A) Substitutions of three leucines to alanines (L→A) and a tryptophan to an alanine (W→A), which are known to affect the interaction with dTAFII110 and gene activation in Drosophila cells, were introduced in the Q2 activation domain of Sp1. In addition, both types of mutations were combined (L, W→A), resulting in a four-amino-acid exchange in the 147-amino-acid-long activation domain. (B) Yeast strains containing the reporter gene as described in Fig. 2A were transformed with plasmids encoding the Gal4 activator and different Lex-Sp1Q2 mutants or wild type. Quantitative β-galactosidase assays showed that the two mutants (L→A and W→A) still activated gene expression as much as the wild type. Only the combination mutant (L, W→A) diminished the transactivation potential of the Q2 domain. (C) Mutant and wild-type Sp1Q2 fusion proteins bind similarly to the Lex binding sites. EMSAs were performed by using total protein extracts from yeast cells and 32P-labeled oligonucleotide duplex containing two Lex binding sites. Equal amounts of protein were used for each EMSA. The Sp1Q2 mutants (lanes 4 to 6) bound to the Lex binding sites as well as the wild type (lane 1), indicating similar protein expression levels. Protein extracts from isogenic yeast cells containing an empty expression plasmid did not yield any detectable bandshifts (lane 3). The immunoblot using anti-LexA antibodies showed similar expression of the wild-type Sp1Q2 and the different mutants. (D) Transfection of HeLa cells with wild-type and mutant Sp1Q2. The mutants tested in yeast were subcloned as Gal4 (amino acids 1 to 93) hybrids into a mammalian expression vector. These transactivator plasmids were cotransfected into HeLa cells along with a reporter plasmid, under the control of two proximal Gal4 binding sites and a downstream SV40 enhancer (see Fig. 1A), and a reference plasmid. S1 nuclease analysis was performed, and the signals were quantified by using a PhosphorImager. As in yeast (Fig. 4B), the mutants L→A and W→A had the same transactivation potential as wild-type Sp1Q2. Only the combined mutations L, W→A showed a reduced ability to stimulate reporter gene expression, which was still above background.
We also addressed the possibility that the different stimulatory effects of the mutants observed were caused by different stabilities of the proteins in vivo. We therefore performed EMSAs with whole-yeast protein extracts and labeled oligonucleotides containing two Lex-binding sites (Fig. 5C). All the mutant Lex-Sp1Q2 proteins were apparently expressed at similar levels as judged from their DNA-binding signal, as compared to the wild-type Sp1Q2. This result was confirmed by immunoblot analysis by using anti-LexA antibodies (Fig. 5C).
We then tested these three mutants of Sp1Q2 in HeLa cells to see whether the seemingly greater permissivity of yeast in comparison to the published results with Drosophila (compare Fig. 5B to reference 16) also applied to human cells. For this purpose, the mutant domains were fused to the Gal4 DBD (amino acids 1 to 93) and were subcloned into a mammalian expression vector (same as described in Fig. 1A, Transactivators). The different expression vectors were cotransfected into HeLa cells with the β-globin reporter plasmid containing two proximal Gal4 binding sites and a downstream SV40 enhancer. As observed in yeast, the two mutants (L→A and W→A) did not affect the transactivation potential in HeLa cells. Only the combination of both mutants (L, W→A) displayed a reduced ability to synergize with the remote SV40 enhancer, again above the basal expression level mediated by Gal4 DBD alone (Fig. 5D).
The apparent difference between the results in Drosophila versus our results with yeast and human cells remains to be explained. For example, the reporter genes used in Drosophila, yeast, and humans were controlled by different core promoters that may respond differently to Sp1-mediated gene activation (11). In addition, we determined the influence of these mutants to synergize with a remote enhancer, whereas in Drosophila the transactivation potential of the mutants was determined in an isolated context (16).
The glutamine-rich domain of Snf5p activates transcription in yeast.
The above-mentioned results show that glutamine-rich activation domains from Sp1 and Oct1 on their own stimulate transcription in yeast and human cells only to a minor extent compared to acidic activators, but they readily synergize with a remote enhancer. Next, we wanted to test whether a glutamine-rich domain of an endogenous yeast protein could also activate gene expression. A search of the yeast genome database for open reading frames (ORFs) containing a stretch of at least 10 glutamine residues revealed that out of 64 hits, 48 are known ORFs. Of these 48, 18 (37%) are factors involved in transcriptional activation. They include the following genes: FLO8, DAT1, HAP2, MCM1, MED3, POP2, TAF61, NDD1, IXR1, CRZ1, CCR4, SNF5, DAL81, GAL11, YPR022C, GTS1, SRB9, and HAP1. The rest of the ORFs could be assigned to eight kinase or kinase-associated factors, five RNA-binding proteins, three factors involved in G-protein-coupled complexes, two chaperones, and eight other miscellaneous factors. This distribution indicates that the largest fraction of yeast glutamine-rich proteins are involved in transcriptional regulation and activation.
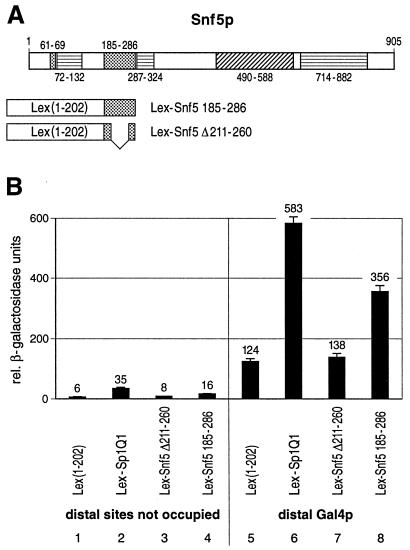
To see whether glutamine-rich domains derived from yeast transcription proteins can also activate gene expression, we chose Snf5, which contains a glutamine-rich domain of similar size to that of the glutamine-rich activation domains of Sp1 and Oct1, yet a higher glutamine content, including long polyglutamine tracts. Glutamine-rich domains and polyglutamine stretches were previously found to have similar properties in transcriptional activation in mammalian cells (15). Snf5p is a component of the SWI-SNF complex that is necessary for transcriptional activation of several genes, most probably by remodeling chromatin (6, 19, 25, 29, 34; for a review, see reference 7). Full-length Snf5p, when fused to a heterologous DBD, can activate transcription in yeast (26). The SNF5 gene codes for a 905-amino-acid protein (Fig. 6A) that contains an acidic domain (amino acids 490 to 588), three proline-rich domains (amino acids 72 to 132, 287 to 324, and 714 to 882) and two glutamine-rich domains (amino acids 61 to 69 and 185 to 286) (26). We fused the 102-amino-acid moiety of the long glutamine-rich domain (amino acids 185 to 286) of Snf5p to the DBD of Lex (Fig. 6A, Lex-Snf5 amino acids 185 to 286). This stretch contains 73 glutamine residues. In addition, we cloned an internal deletion of 50 amino acids that eliminates 42 glutamine residues (deletion of Lex-Snf5 amino acids 211 to 260). These constructs were tested for their abilities to activate transcription of the yeast reporter described in Fig. 2A when bound in a proximal position either alone or in combination with a distal Gal4p activator (Fig. 6B). For comparison, we used the stronger glutamine-rich activation domain of Sp1 (Lex-Sp1Q1). The glutamine-rich domain of Snf5p activated transcription significantly above the background control of Lex alone (lanes 4 and 1). This glutamine-rich domain also synergized with the distal Gal4p activator (lane 8); the reporter gene activity was more than threefold higher when the Lex-Snf5 amino acids 185 to 286 domain was bound at the proximal position in combination with distal Gal4p (lane 8) compared to binding of Gal4p and Lex only (lane 5). The deletion in the glutamine-rich domain of Snf5p (Lex-Snf5 amino acids 211 to 260) abolished the activation potential with a remaining activity comparable to Lex DBD alone (lanes 3 and 7).
FIG. 6.
The glutamine-rich domain of Snf5p activates transcription in yeast. (A) Schematic drawing of the 905-amino-acid-long Snf5p factor which contains an acidic region (amino acids 490 to 588), three proline-rich domains (amino acids 72 to 132, 287 to 324, and 714 to 882), and two glutamine-rich domains (amino acids 61 to 69 and 185 to 286). The boundary amino acid positions of the corresponding domains are indicated. The larger glutamine-rich domain of Snf5p (amino acids 185 to 286) (Lex-Snf5 185-286) as well as a deletion mutation (deletion of amino acids 211 to 260) of this glutamine-rich domain (Lex-Snf5 Δ211-260) were fused to the Lex DBD. (B) Quantitative β-galactosidase assay. The glutamine-rich domain of Snf5p stimulated LacZ expression when bound proximally to the TATA box (lane 4) and synergized with a distal Gal4p activator (lane 8). For comparison, the stronger glutamine-rich activation domain of Sp1 (Lex-Sp1Q1) was used (lanes 2 and 6). The 50-amino-acid deletion abolished the transactivation potential of the Snf5p glutamine-rich domain (lane 3) and hence the synergism with the distal Gal4p activator (lane 7).
DISCUSSION
Glutamine-rich domains and polyglutamine stretches are integral components of many proteins involved in transcriptional regulation, from yeast to human. We had previously shown that in mammalian cells, glutamine-rich domains poorly activate transcription on their own when tethered to DNA in a promoter-proximal position but strongly synergize with remotely bound transcriptional activators of the acidic type (38). While yeast cells readily respond to mammalian acidic activators, we and others had found them to be nonresponsive to glutamine-rich activation domains of mammalian factors (23, 24, 32), which at that time suggested that yeast lacks an important interaction partner for these latter domains. In Drosophila, which has no homologue of the mammalian Sp1 transcription factor and thus was suitable for testing the activity of ectopically expressed Sp1, its glutamine-rich activation domains were found to bind to TAFII110, a TBP-associated general transcription factor (8, 16, 20). The completion of the entire S. cerevisiae genome sequence revealed a number of homologues for mammalian TATA binding protein-associated factors, but no counterpart to Drosophila TAFII110/human TAFII130, which seemed to offer a straightforward explanation for the lack of activity of the glutamine-rich domains in yeast. More recently, the fission yeast S. pombe was found to be responsive to glutamine-rich domains of mammalian origin (35). In the sequence database of S. pombe we have found a 365-amino-acid ORF with considerable similarity to dTAFII110/hTAFII130. This prompted us to introduce this protein into the budding yeast and to study its effect on glutamine-rich activation domains. The S. pombe protein was expressed in S. cerevisiae, but failed to influence the expression of reporter genes driven by glutamine-rich activation domains in any of the enhancer/promoter constellations tested (D. Escher and W. Schaffner, unpublished results). Although we cannot exclude the possibility that an S. pombe-specific cofactor for TAFII110 is missing in the budding yeast, it has become clear by now that the latter can respond well to glutamine-rich domains in the absence of a TAFII110 homologue. Recently, it was reported that the glutamine-rich activation domain Sp1Q1 is able to activate transcription of a reporter gene when tethered to DNA in S. cerevisiae (43). This was, however, only observed when the reporter gene was present on a high-copy-number plasmid (2μm), while no activation was detected when it was integrated in a chromosomal locus. The present study clearly shows that glutamine-rich domains, both of mammalian and yeast origin, stimulate reporter gene expression in a yeast chromosomal context if the promoter region is structured in a manner that is prevalent in higher eukaryotes but less common in yeast, namely a promoter with proximal factor binding sites plus enhancer-type regulatory sequences further upstream. Such a configuration was not tested in previous experiments, including the ones from our lab; rather, a simpler promoter version with a few bindings sites in the immediate vicinity of the TATA box had been used. In the present study, the glutamine-rich activation domains of Sp1 and Oct1, when tethered to a proximal position, even synergized to comparable levels with remote acidic activators in yeast and human cells. Furthermore, mutations in the activation domain of Sp1Q2 had similar effects in yeast and HeLa cells, suggesting a common interaction pathway. Thus, it remains to be seen whether the interaction observed between TAFII110/TAFII130 and the human Sp1 glutamine-rich activation domains is of general significance. Whatever the role of the TAFII110-like protein in S. pombe, our results show that glutamine-rich domains behave very similarly in yeast and human and that their activity does not depend on the presence of a TAFII110 homologue in budding yeast. This again raises the question of the possible target(s) of glutamine-rich domains. The TBP itself was reported to interact with glutamine-rich activation domains. However, the good activity of glutamine-rich domains in yeast observed by us is difficult to reconcile with the report that TBPs from Drosophila and humans, but not from yeast, bind well to glutamine-rich domains (12, 32). Recently, a multiprotein complex termed “cofactor required for Sp1” was found to mediate Sp1 activity in extracts of human cells (36). Some of the characterized components are absent in yeast, whereas others do have a yeast homologue (36) and thus may be involved in mediating the activity of glutamine-rich domains both in human and yeast.
ACKNOWLEDGMENTS
We are indebted to Lee Martin and Werner Lutz for critical comments on the manuscript. We also thank Cristina Torres-de los Reyes for excellent technical assistance and Fritz Ochsenbein for excellent artwork.
This work was supported by the Kanton Zürich and the Schweizerischer Nationalfonds.
REFERENCES
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Barberis A, Gaudreau L. Recruitment of the RNA polymerase II holoenzyme and its implications in gene regulation. Biol Chem. 1998;379:1397–1405. doi: 10.1515/bchm.1998.379.12.1397. [DOI] [PubMed] [Google Scholar]
- 3.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 4.Boisclair Y R, Brown A L, Casola S, Rechler M M. Three clustered Sp1 sites are required for efficient transcription of the TATA-less promoter of the gene for insulin-like growth factor-binding protein-2 from the rat. J Biol Chem. 1993;268:24892–24901. [PubMed] [Google Scholar]
- 5.Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 6.Cairns B R, Kim Y J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson M, Laurent B C. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994;6:396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 8.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 9.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 10.Dusing M R, Wiginton D A. Sp1 is essential for both enhancer-mediated and basal activation of the TATA-less human adenosine deaminase promoter. Nucleic Acids Res. 1994;22:669–677. doi: 10.1093/nar/22.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emami K H, Navarre W W, Smale S T. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emili A, Greenblatt J, Ingles C J. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol Cell Biol. 1994;14:1582–1593. doi: 10.1128/mcb.14.3.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escher D, Schaffner W. Gene activation at a distance and telomeric silencing are not affected by yeast histone H1. Mol Gen Genet. 1997;256:456–461. doi: 10.1007/s004380050589. [DOI] [PubMed] [Google Scholar]
- 14.Escher D, Schaffner W. Improved “activator trap” method for the isolation of transcriptional activation domains from random DNA fragments. BioTechniques. 1996;21:848–854. doi: 10.2144/96215st02. [DOI] [PubMed] [Google Scholar]
- 15.Gerber H P, Seipel K, Georgiev O, Hofferer M, Hug M, Rusconi S, Schaffner W. Transcriptional activation modulated by homopolymeric glutamine and proline stretches. Science. 1994;263:808–811. doi: 10.1126/science.8303297. [DOI] [PubMed] [Google Scholar]
- 16.Gill G, Pascal E, Tseng Z H, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giniger E, Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 18.Gstaiger M, Schaffner W. Strong transcriptional activators isolated from viral DNA by the 'activator trap', a novel selection system in mammalian cells. Nucleic Acids Res. 1994;22:4031–4038. doi: 10.1093/nar/22.20.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 20.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 21.Huber R, Schlessinger D, Pilia G. Multiple Sp1 sites efficiently drive transcription of the TATA-less promoter of the human glypican 3 (GPC3) gene. Gene. 1998;214:35–44. doi: 10.1016/s0378-1119(98)00233-9. [DOI] [PubMed] [Google Scholar]
- 22.Jones K A, Tjian R. Sp1 binds to promoter sequences and activates herpes simplex virus 'immediate-early' gene transcription in vitro. Nature. 1985;317:179–182. doi: 10.1038/317179a0. [DOI] [PubMed] [Google Scholar]
- 23.Kim T K, Roeder R G. Transcriptional activation in yeast by the proline-rich activation domain of human CTF1. J Biol Chem. 1993;268:20866–20869. [PubMed] [Google Scholar]
- 24.Künzler M, Braus G H, Georgiev O, Seipel K, Schaffner W. Functional differences between mammalian transcription activation domains at the yeast GAL1 promoter. EMBO J. 1994;13:641–645. doi: 10.1002/j.1460-2075.1994.tb06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laurent B C, Treitel M A, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 29.Moreira J M, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 31.Parks C L, Shenk T. The serotonin 1a receptor gene contains a TATA-less promoter that responds to MAZ and Sp1. J Biol Chem. 1996;271:4417–4430. doi: 10.1074/jbc.271.8.4417. [DOI] [PubMed] [Google Scholar]
- 32.Ponticelli A S, Pardee T S, Struhl K. The glutamine-rich activation domains of human Sp1 do not stimulate transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:983–988. doi: 10.1128/mcb.15.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pugh B F, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 34.Recht J, Osley M A. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 1999;18:229–240. doi: 10.1093/emboj/18.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Remacle J E, Albrecht G, Brys R, Braus G H, Huylebroeck D. Three classes of mammalian transcription activation domain stimulate transcription in Schizosaccharomyces pombe. EMBO J. 1997;16:5722–5729. doi: 10.1093/emboj/16.18.5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ryu S, Zhou S, Ladurner A G, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- 37.Schaffner W. Enhancer. In: Creighton T, editor. The encyclopedia of molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1999. pp. 823–828. [Google Scholar]
- 38.Seipel K, Georgiev O, Schaffner W. Different activation domains stimulate transcription from remote ('enhancer') and proximal ('promoter') positions. EMBO J. 1992;11:4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Serra E, Zemzoumi K, di Silvio A, Mantovani R, Lardans V, Dissous C. Conservation and divergence of NF-Y transcriptional activation function. Nucleic Acids Res. 1998;26:3800–3805. doi: 10.1093/nar/26.16.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Triezenberg S J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 41.Triezenberg S J, Kingsbury R C, McKnight S L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 42.Westin G, Gerster T, Muller M M, Schaffner G, Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987;15:6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao H, Jeang K T. Glutamine-rich domains activate transcription in yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:22873–22876. doi: 10.1074/jbc.273.36.22873. [DOI] [PubMed] [Google Scholar]
- 44.Yang M, Nomura H, Hu Y, Kaneko S, Kaneko H, Tanaka M, Nakashima K. Prolactin-induced expression of TATA-less cyclin D3 gene is mediated by Sp1 and AP2. Biochem Mol Biol Int. 1998;44:51–58. doi: 10.1080/15216549800201052. [DOI] [PubMed] [Google Scholar]