Abstract
The dearth of new medicines effective against antibiotic-resistant bacteria presents a growing global public health concern1. For more than five decades, the search for new antibiotics has relied heavily on the chemical modification of natural products (semisynthesis), a method ill-equipped to combat rapidly evolving resistance threats. Semisynthetic modifications are typically of limited scope within polyfunctional antibiotics, usually increase molecular weight, and seldom permit modifications of the underlying scaffold. When properly designed, fully synthetic routes can easily address these shortcomings2. Here we report the structure-guided design and component-based synthesis of a rigid oxepanoproline scaffold which, when linked to the aminooctose residue of clindamycin, produces an antibiotic of exceptional potency and spectrum of activity, which we name iboxamycin. Iboxamycin is effective against ESKAPE pathogens including strains expressing Erm and Cfr ribosomal RNA methyltransferase enzymes, products of genes that confer resistance to all clinically relevant antibiotics targeting the large ribosomal subunit, namely macrolides, lincosamides, phenicols, oxazolidinones, pleuromutilins and streptogramins. X-ray crystallographic studies of iboxamycin in complex with the native bacterial ribosome, as well as with the Erm-methylated ribosome, uncover the structural basis for this enhanced activity, including a displacement of the nucleotide upon antibiotic binding. Iboxamycin is orally bioavailable, safe and effective in treating both Gram-positive and Gram-negative bacterial infections in mice, attesting to the capacity for chemical synthesis to provide new antibiotics in an era of increasing resistance.
Subject terms: Antibiotics, Antimicrobial resistance, X-ray crystallography, Chemical synthesis, Structure-based drug design
Structure-guided design and component-based synthesis are used to produce iboxamycin, a novel ribosome-binding antibiotic with potent activity against Gram-positive and Gram-negative bacteria.
Main
The rise and dissemination of bacteria resistant to most or all known antibiotics raises concerns of an impending global infectious-disease crisis1. Any plan to address the problem depends on the discovery of new antibiotics effective against modern bacterial pathogens, but opinions diverge on which strategies can best achieve these discoveries3. For decades, natural products have served as both conceptual and material starting points for antibiotics discovery, but making specific chemical modifications to structurally complex natural products (that is, semisynthesis) is inherently challenging, and the pace of drug discovery by this route has slowed markedly2. In an alternative approach, the development of fully synthetic platforms to construct tetracycline4, macrolide5,6, group A streptogramin7 and arylomycin8 antibiotics has enabled deep-seated structural modifications that are not achievable by semisynthesis. These technologies enable chemists to envision and reduce to practice an almost limitless array of design hypotheses. Here we use component-based chemical synthesis to achieve a substantive re-scaffolding of the lincosamide antibiotics, giving rise to a broad-spectrum agent that is effective against a wide range of multidrug-resistant bacterial pathogens.
The ribosome is a major antibiotic target in bacteria, and lincosamides are one of several ribosome-targeting classes that have proved essential to the modern pharmacopeia9,10. Lincomycin (1; Fig. 1a), the founding member of the class, was first isolated in 1963 from a Nebraskan soil streptomycete11 and quickly found use in the treatment of streptococcal, pneumococcal and staphylococcal infections. In an early semisynthetic modification of 1, stereoinvertive deoxychlorination at position C7 of the aminosugar residue resulted in the antibiotic now known as clindamycin (2; Fig. 1a), a molecule with improved pharmacokinetic properties and an enhanced spectrum of activity12 that has largely replaced lincomycin in human medicine13. Since the US Food and Drug Administration approved clindamycin in 1970, semisynthetic and fully synthetic approaches to lincosamide discovery have been explored, giving rise to candidates containing six- and seven-membered aminoacyl residues, each with an expanded spectrum of activity14,15 (for example, 3; Fig. 1a) More recently, modifications of the aminosugar residue by semisynthesis have been reported, expanding coverage to certain multidrug-resistant Gram-positive bacteria16
Fig. 1. Evolution of a novel antibiotic scaffold.
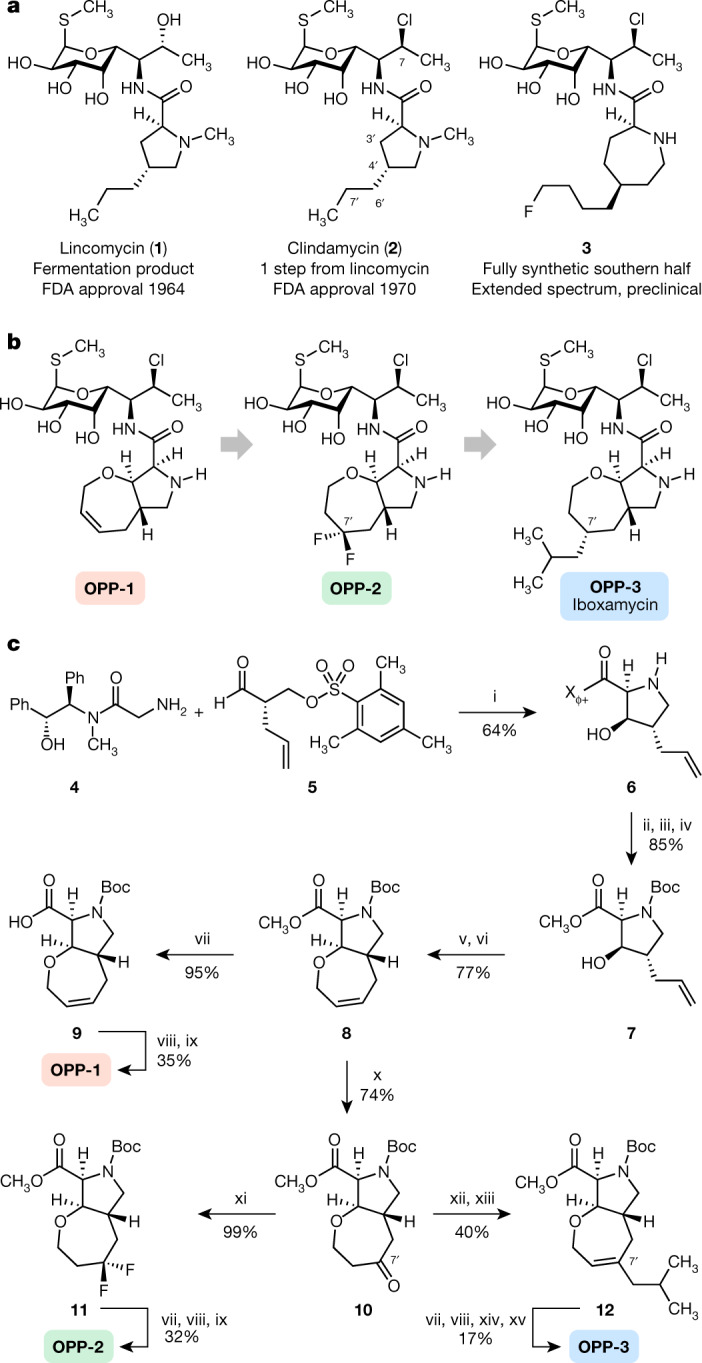
a, Chemical structures of lincomycin and clindamycin, and a preclinical antibiotic discovered by Vicuron scientists15. b, The design progression of oxepanoprolinamide antibiotics. Conformational constraint of the C4′ substituent within a bicyclic oxepanoprolyl scaffold, together with presentation of a lipophilic group at its 7′ position provided improved activity. c, Synthesis of oxepanoprolinamides OPP-1, OPP-2 and IBX (OPP-3). Reagents and conditions are as follows: (i) LiHMDS, LiCl; (ii) NaOH, >99% recovery of (R,R)-pseudoephenamine; (iii) Boc2O; (iv) MeI, Cs2CO3; (v) allyl ethyl carbonate, Pd(PPh3)4; (vi) Hoveyda–Grubbs catalyst I; (vii) LiOH; (viii) 7-Cl-MTL, HATU, EtiPr2N; (ix) TFA; (x) Pd(OAc)2, benzoquinone, HBF4 (aq.); (xi) DAST; (xii) LiHMDS, Comins’ reagent, 51% (plus, separately, 31% Δ6′ regioisomer); (xiii) iBuMgCl, Fe(acac)3; (xiv) H2, Pd(OH)2/C, 1:1 dr; (xv) BSTFA, TMSI, 17% over 2 steps (plus, separately, the 7′R epimer; 17% over 2 steps).
Clindamycin-resistant bacteria are globally distributed. Acquired resistance to clindamycin most commonly occurs through N6-dimethylation of 23S ribosomal RNA (rRNA) residue A2058 (nucleotide numbering in this Article refers to Escherichia coli) by erythromycin resistance rRNA methyltransferases17. These enzymes, which are encoded by erm genes, give rise to macrolide, lincosamide and streptogramin B (MLSB) cross-resistance; expression of these genes may be constitutive (c-erm), or inducible upon exposure to macrolide antibiotics18,19 (i-erm). The prevalence of erm genes among clinical streptococcal and staphylococcal isolates has led the US Centers for Disease Control and Prevention (CDC) to list MLSB-resistant pathogens among its most pressing threats in their 2019 report1. A distinct rRNA methyltransferase, Cfr (encoded by the horizontally transferrable gene cfr)20 methylates C8 of the 23S rRNA residue A2503 and confers resistance to phenicol, lincosamide, oxazolidinone, pleuromutilin and streptogramin A21,22 (PhLOPSA) antibiotics as well as 16-membered macrolides23. Moreover, recent studies have elucidated a third major resistance mechanism affecting the lincosamides, whereby target-protection proteins (for example, LsaA) bind to antibiotic-inhibited ribosomes, evict the drug and in so doing restore protein synthesis24,25. The prevalence of erm, cfr and target-protection resistance genotypes in staphylococcal, streptococcal and enterococcal clinical isolates, together with the risk of Clostridium difficile colitis with the use of clindamycin challenge the continued utility of this antibiotic in patients.
Discovery of oxepanoprolinamides
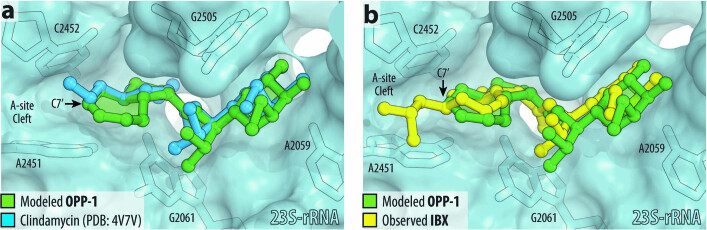
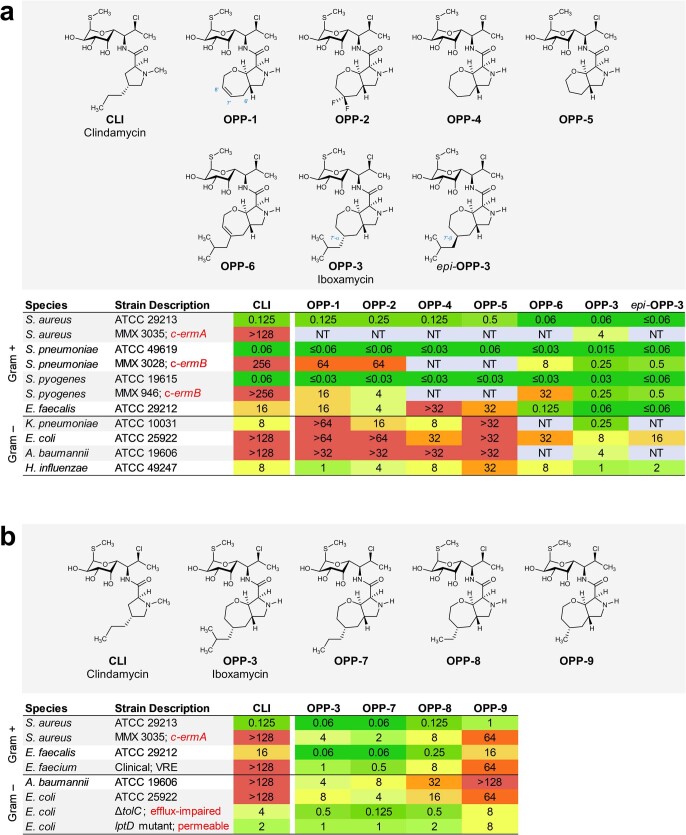
To enable full structural exploration of the lincosamides, we developed modular synthetic routes to both the aminooctose and aminoacyl residues, then linked these components by amide-bond formation to produce more than 500 antibacterial candidates26–29. Thus a wide array of structural changes were examined; we observed and report in this Article that bicyclic scaffold modifications of the aminoacyl hemisphere in combination with the aminooctose residue of clindamycin were especially advantageous with respect to antibacterial activity, notably among clindamycin-resistant strains. In brief, the bicyclic scaffold of oxepanoprolinamide 1 (OPP-1) was conceived to rigidify both the Cγ-endo proline ring pucker and the extended 4′-n-propyl chain of clindamycin as observed when bound to the ribosome30 (Extended Data Fig. 1a). These explorations were inspired and made feasible by a stereoselective aldol reaction developed for a key fragment coupling in the synthesis of macrolide antibiotics5. Thus, enolization of (R,R)-4, a chiral glycine equivalent31, followed by addition of the bis-electrophile 5 led to sequential syn-aldol addition, then spontaneous intramolecular N-alkylation, furnishing the β-hydroxy proline derivative 6 in a single operation (64% yield, 2.3 g scale) (Fig. 1c). A three-step sequence involving cleavage of the chiral auxiliary (with >99% recovery of pseudoephenamine), N-Boc protection, and esterification provided the suitably protected intermediate 7 in 85% overall yield. Construction of the oxepene ring was then achieved through palladium-catalysed O-allylation followed by ring-closing metathesis using the first-generation Hoveyda–Grubbs catalyst32. The target OPP-1 was assembled following an established sequence of saponification, amide coupling to the aminosugar component (7-chloro-1-methylthio-lincosamine, 7-Cl-MTL)33 and N-Boc removal15.
Extended Data Fig. 1. Structure-based design of 7′-substituted oxepanoprolinamides.
a, Superposition of the X-ray crystal structure of ribosome-bound lincosamide antibiotic clindamycin (2, blue, PDB entry 4V7V30) with the energy-minimized structure of OPP-1 (green). Note that in this configuration, the C7′-atom of OPP-1 contacts the lipophilic surface of the A-site cleft. b, The same structure, overlaid with the X-ray crystal structure of iboxamycin (IBX, yellow) bound to the bacterial ribosome.
Microbiological testing of OPP-1 showed it to be comparable in activity to clindamycin (Extended Data Fig. 2a). Substitution of position 7′ of the oxepene ring was prioritized next, as molecular modelling suggested that substituents there would project towards a hydrophobic cleft formed by 23S rRNA residues A2451 and C2452 (Extended Data Fig. 1a). Wacker oxidation of intermediate 8 using Grubbs’ protocol34 proceeded with complete regioselectivity to provide the 7′-keto product 10 with a yield of 74% (Fig. 1c); this selectivity is thought to arise from the σ-withdrawing effect of the oxygen atom embedded within the 7-membered ring. Deoxydifluorination of intermediate 8 with N,N-diethylaminosulfur trifluoride (DAST) followed by the same three-step sequence described earlier provided oxepanoprolinamide OPP-2, which exhibited substantially improved activity against multidrug-resistant Gram-positive strains (Streptococcus pneumoniae c-ermA; Streptococcus pyogenes c-ermB), as well as against Klebsiella pneumoniae (Extended Data Fig. 2a), lending support to our design strategy. Alkyl (and aryl) substitution of position 7′ was made possible by non-regioselective enolization of 10 with lithium hexamethyldisilazide (LiHMDS), followed by triflylation with Comins’ reagent. The resulting regioisomeric vinyl triflates both served as flexible intermediates for cross-coupling with aryl and alkyl nucleophiles. For example, iron-catalysed coupling of the Δ7′ vinyl triflate with isobutylmagnesium chloride35 provided intermediate 12, which upon saponification, amide coupling, non-diastereoselective olefin saturation (~1:1 7′-α:β diastereomeric ratio), N-Boc removal and purification by HPLC, provided separately OPP-3 (Fig. 1b) and its less active 7′-β stereoisomer epi-OPP-3. We found that, generally, 7′-α-alkyl analogues were more active than their unsaturated and 7′-β stereoisomeric congeners (Extended Data Fig. 2a). This sequence provided a viable discovery route to OPP-3—hereafter termed iboxamycin (IBX)—and other 7′-substituted analogues in amounts sufficient for minimum inhibitory concentration (MIC) analyses but could not provide sufficient material for in vivo evaluations in animal models of infection. For this purpose, we devised a different sequence to obtain IBX specifically, which produced the antibiotic in multigram quantities and could provide the basis for a manufacturing route36.
Extended Data Fig. 2. MICs (μg ml−1) of antibiotics containing a bicyclic aminoacyl residue.
a, Effects of of 7′ substitution, ring size, and saturation on antibacterial activity. b, Effects of 7′-alkyl substituent chain length on antibacterial activity, including against MLSB-resistant S. aureus and strains of E. coli engineered to lack key efflux or outer-membrane assembly machinery. c-ermA/B, constitutively expressed erythromycin ribosome methylase A/B gene.
IBX overcomes resistance and is effective in vivo
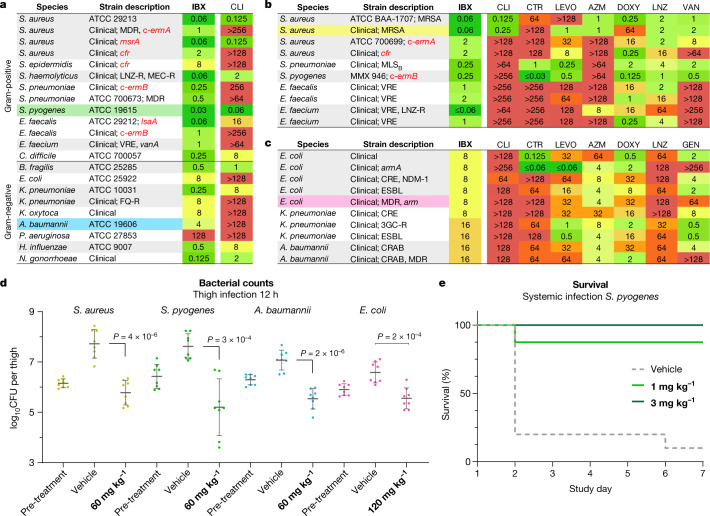
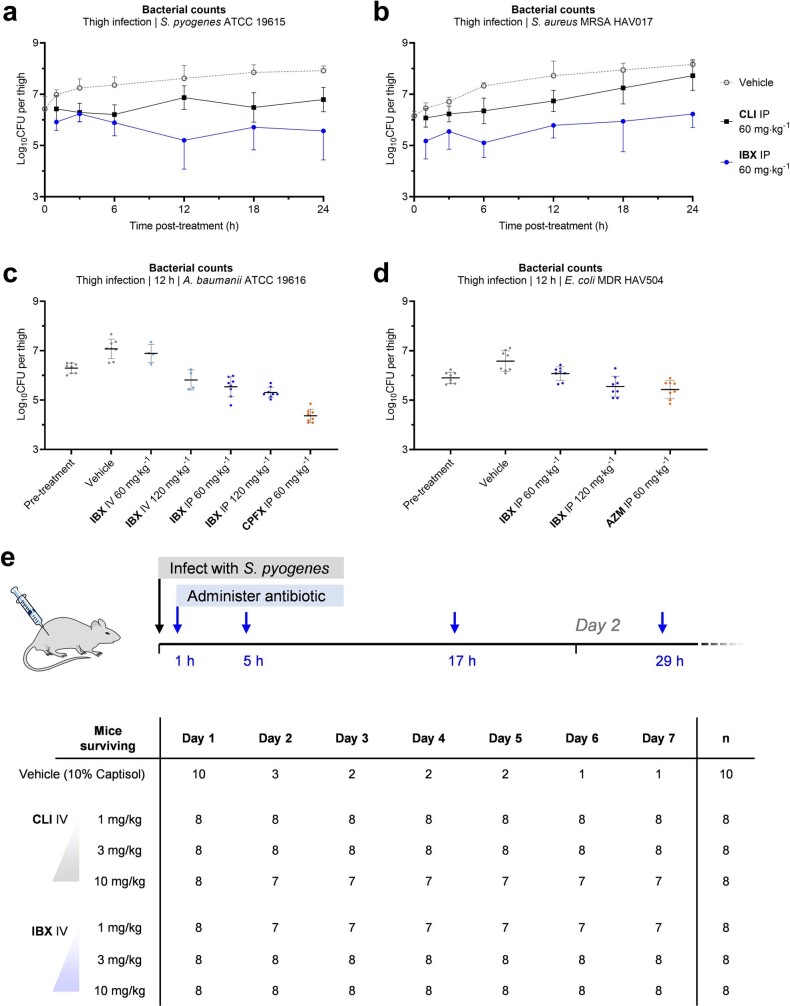
IBX exhibits a broad spectrum of activity when compared with clindamycin and displays an orthogonal resistance profile in side-by-side comparisons with widely prescribed antibiotics (Fig. 2, Supplementary Table 1). Broth culture susceptibility testing revealed that IBX is active against extensively drug-resistant bacterial strains, including ESKAPE pathogens and erm-, cfr- and lsaA-expressing strains that are resistant to lincosamide antibiotics (Fig. 2a). For instance, IBX overcame c-ermB-mediated resistance in streptococci, an increasingly widespread phenotype37, with MICs at least 1,000× lower than that of clindamycin against S. pneumoniae and S. pyogenes strains (Fig. 2a, b). Similarly, IBX displayed activity against methicillin-resistant Staphylococcus aureus isolates displaying MLSB and PhLOPSA cross-resistance phenotypes or expressing msrA, the gene encoding the MsrA target-protection protein (Fig. 2a, b). Enterococcal pathogens, which are responsible for widespread and life-threatening healthcare-associated infections38 and are invulnerable to lincosamides, also appear broadly susceptible to IBX: In Enterococcus faecalis, IBX overcame intrinsic resistance mediated by the target-protection gene lsaA39 (Fig. 2a), and displayed MICs ≤2 μg ml−1 against a panel of Enterococcus faecium and E. faecalis strains with varying resistance to vancomycin, linezolid, doxycycline, azithromycin and levofloxacin (Fig. 2b). IBX exhibited potent activity against C. difficile (listed as an urgent threat by the CDC)1, suggesting it may carry reduced risk of promoting C. difficile colitis, but validation of this hypothesis requires further study.
Fig. 2. In vitro and in vivo antibacterial activity of the broad-spectrum antibiotic IBX.
a–c, MICs in μg ml−1 of IBX in standard and multidrug-resistant bacterial strains. a, IBX displays a broad antibacterial spectrum of activity, and overcomes erm, cfr and lsaA resistance. B. fragilis, Bacteroides fragilis; H. influenzae, Haemophilus influenzae; K. oxytoca, Klebsiella oxytoca; S. epidermidis, Staphylococcus epidermis; S. haemolyticus, Staphylococcus haemolyticus. Comparisons of IBX with standard antibiotics against clinical Gram-positive (b) and Gram-negative (c) isolates illustrate the differentiated activity of the oxepanoprolinamide class. CLI, clindamycin; CTR, ceftriaxone; LEVO, levofloxacin; AZM, azithromycin; LNZ; linezolid; VAN, vancomycin; GEN, gentamicin. d, Efficacy of IBX in a neutropenic mouse thigh infection model; colours correspond to strains highlighted in a–c. Mice received vehicle or IBX intraperitoneally, and bacterial counts were determined 12 h after treatment. Data are mean ± s.d.; n = 8 thighs from 4 mice examined over 2 experiments; two-tailed unpaired Welch’s t-test. CFU, colony-forming units. e, Survival of mice receiving IBX or vehicle following systemic infection with S. pyogenes. 3GC-R, third-generation cephalosporin-resistant; CRAB, carbapenem-resistant A. baumannii; CRE, carbapenem-resistant Enterobacterales; ESBL, extended-spectrum beta-lactamase; FQ-R, fluoroquinolone-resistant; LNZ-R, linezolid-resistant; MDR, multidrug-resistant; MEC-R, methicillin-resistant; MRSA, methicillin-resistant S. aureus; VAN-R, vancomycin-resistant; VRE, vancomycin-resistant Enterococcus.
IBX showed activity against Gram-negative bacterial isolates, representing a further departure from approved lincosamides. High-priority ESKAPE pathogens such as carbapenem-resistant E. coli, K. pneumoniae and Acinetobacter baumannii, including strains expressing extended-spectrum β-lactamase or aminoglycoside resistance methylase (arm) genes, were largely susceptible (MIC ≤16 μg ml−1; Fig. 2c). In one example, IBX displayed activity against a strain of E. coli resistant to cephalosporin, fluoroquinolone, tetracycline and aminoglycoside antibiotics (MIC = 8 μg ml−1; Fig 2c). Clinical strains of Neisseria gonorrhoeae were also highly susceptible (MIC = 0.125 μg ml−1; Fig. 2a, Supplementary Table 1). To better understand the basis for this broad-spectrum activity, we determined the MICs of 7ʹ-substituted oxepanoprolinamides, including IBX, in laboratory strains engineered to illuminate the contributions of outer-membrane penetration and transporter-mediated efflux to Gram-negative activity6. Whereas clindamycin exhibited activity only against E. coli strains with loss-of-function mutations to major efflux transporter (tolC) or outer-membrane assembly (lptD) genes, we found that, as a class, oxepanoprolinamides were significantly less suceptible to these mechanisms of intrinsic antibiotic resistance (Extended Data Fig. 2b). Pseudomonas aeruginosa, while measurably inhibited by IBX (MIC = 128 μg ml−1; Fig. 2a), was comparatively resistant towards the antibiotic, reflecting the unique challenges facing the discovery of antibiotics effective against this pathogen40. It is notable that the expanded spectrum of activity of IBX relative to clindamycin is not readily explained by traditional physicochemical predictors of Gram-negative activity such as rotatable-bond count, molecular weight, relative polar surface area or lipophilicity41. Nor does it reinforce recently advanced rules for Gram-negative accumulation42 (see Supplementary Methods). Nevertheless, we believe that enhanced Gram-negative potency may be realized through optimization of the former metrics within the oxepanoprolinamide class.
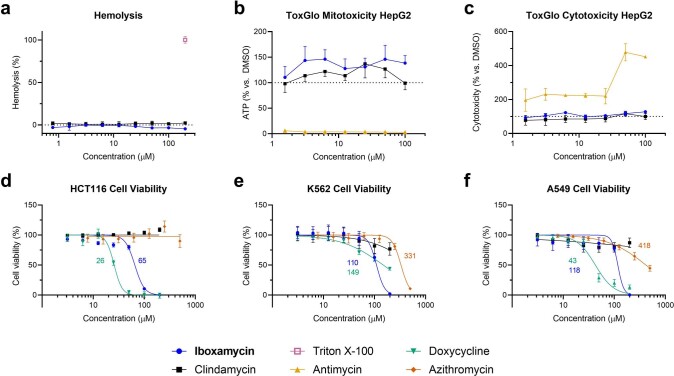
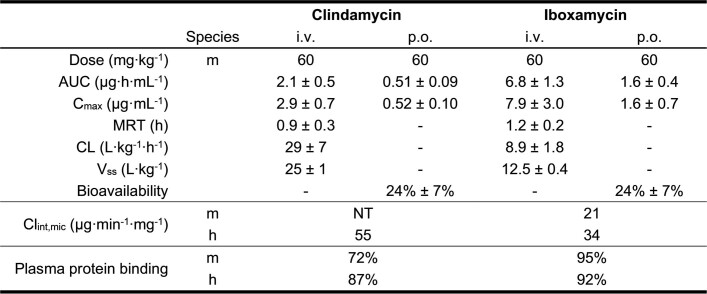
To assess the efficacy of oxepanoprolinamide antibiotics, we evaluated IBX in animal models of infection using both Gram-positive and Gram-negative bacteria. Cell culture safety profiling experiments showed that IBX is non-haemolytic and non-toxic towards mammalian cells (concentration causing 50% cell growth inhibition (GI50) > 50 μM, Extended Data Fig. 3a, d–f), and has no effect on membrane integrity or mitochondrial function (Extended Data Fig. 3b, c). In mice, IBX achieved greater exposure than clindamycin, exhibiting an intravenous mean residence time (MRT) of 1.2 h and 24% oral bioavailability (Extended Data Table 1). Next, we studied the efficacy of IBX in a murine neutropenic thigh infection model using standard strains of S. pyogenes and A. baumannii, a clinical strain of methicillin-resistant S. aureus, and a clinical strain of E. coli displaying aminoglycoside, tetracycline, fluoroquinolone and third-generation cephalosporin multi-resistance. In each experiment, when compared to vehicle, IBX achieved a statistically significant reduction in bacterial burden 12 h following treatment (Fig 2d, Extended Data Fig. 4a–d). Finally, in a model of systemic infection, we studied the ability of IBX to rescue mice from a lethal challenge of intraperitoneally administered S. pyogenes ATCC 19606. IBX was well-tolerated at all dose levels in this experiment, and resulted in the rescue of infected mice, with all mice treated with 3 or 10 mg kg−1 IBX surviving (Fig. 2e, Extended Data Fig. 4e). Together, these results illustrate the potential of the oxepanoprolinamide class to provide orally available, broad-spectrum antibiotics effective against a range of resistance mechanisms.
Extended Data Fig. 3. Effects of iboxamycin on mammalian cells.
a, Normalized hemolysis (mean ± s.d.) of human erythrocytes by IBX (n = 3 replicates) and clindamycin (n = 5 replicates) relative to Triton X-100 (n = 45 replicates) measured over one independent experiment. b, c, Mitochondrial ToxGlo data showing effects of IBX on HepG2 cellular membrane integrity and ATP production relative to vehicle-treated control (antimycin serves as a positive control for mitotoxicity). Data are mean ± s.d.; n = 3 technical replicates from one independent experiment. d–f, Comparison of effects of IBX, clindamycin, doxycycline, and azithromycin on cell viability (CellTiter-Blue). Data are the mean ± s.d. of n = 3 independent experiments performed in technical quintuplicate. Where applicable, GI50 values (µM) are reported beside the dose-response curves.
Extended Data Table 1.
Pharmacokinetics of clindamycin and iboxamycin
Parameters were calculated by a non-compartmental model (n = 3 mice per study arm), and are reported as their mean ± s.d., where applicable. i.v., intravenous; p.o., per os; AUC, area under the curve; MRT, mean retention time; CL, clearance; Vss, volume of distribution at steady state; Clint,mic, liver microsome intrinsic clearance; h, human; m, mouse.
Extended Data Fig. 4. Efficacy of iboxamycin (IBX) in mouse models of infection. Bacterial counts were quantified in the thighs of neutropenic mice infected with S. pyogenes ATCC 19615.
(a), S. aureus MRSA HAV017 (b), A. baumannii ATCC 19616 (c) or E. coli MDR HAV504 (d) treated with IBX, vehicle, or comparator antibiotic at the listed time points post-treatment. Data are mean ± s.d.; n = 8 thighs from 4 mice examined over 2 experiments, with the exception the sub-set of part c where mice received IBX via intravenous administration (n = 4 thighs from 2 mice examined over a single experiment). e, Mouse survival in an an S. pyogenes systemic infection model. Abbreviations: AZM, azithromycin; CLI, clindamycin; CPFX, ciprofloxacin; IP, intraperitoneal administration; IV, intravenous administration..
IBX targets wild-type and resistant ribosomes
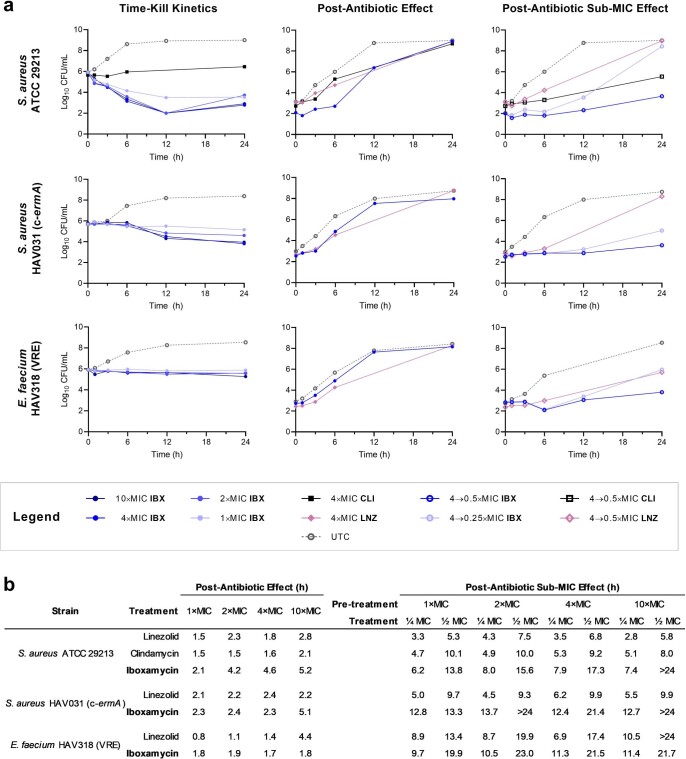
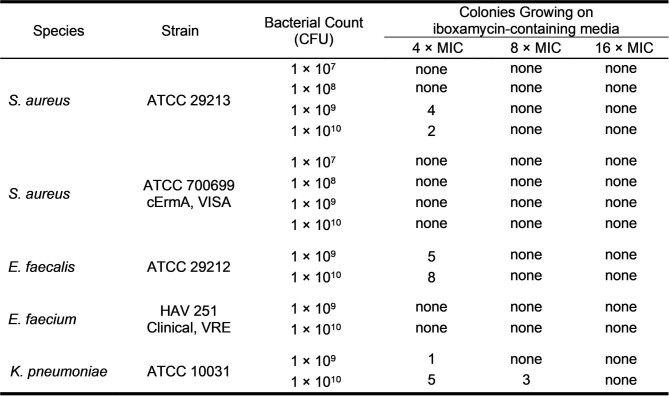
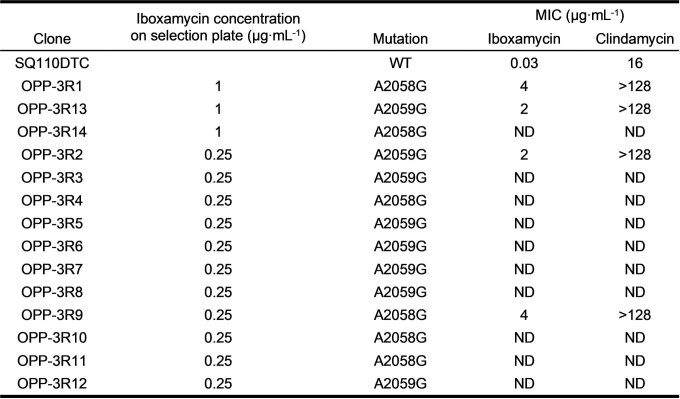
The remarkable antibacterial profile of IBX prompted us to study its mechanism of action, including the structural basis for its activity against Erm-methylated MLSB-resistant ribosomes. Broth culture profiling revealed that, like clindamycin, IBX is bacteriostatic but exhibits prolonged effects on bacterial growth following even brief exposure; concentration-dependent cidality was observed for certain highly susceptible strains (Extended Data Fig. 5). Spontaneous resistance was slow to develop in standard and clinical bacterial strains (approximate frequency of resistance ≤ 10–9 at 4×MIC; Extended Data Table 2). By contrast, IBX-resistant mutants did emerge in E. coli SQ110DTC, a strain specifically engineered for selecting mutations in rRNA that render cells resistant to ribosome-targeting inhibitors43. This strain lacks six out of seven rRNA alleles and the major multidrug efflux pump TolC. When SQ110DTC cells were plated on agar containing IBX, resistant clones with MICs in the range of 2–4 µg ml−1 appeared with a frequency of approximately 10–8. Of the 14 randomly selected clones we sequenced, all carried the single-nucleotide mutations A2058G or A2059G within the 23S rRNA (Extended Data Table 3), corresponding to base changes in the canonical lincosamide binding site known among clindamycin-resistant bacteria44. Notably, mammalian cytosolic and mitochondrial ribosomes carry G2058 residues45, explaining the non-toxicity of IBX in human cell culture experiments. These results demonstrated that, like clindamycin, IBX selectively targets the bacterial ribosome and thus has a low potential to select for resistance owing to the redundancy of rRNA genes in the genomes of most bacterial pathogens46.
Extended Data Fig. 5. Time-kill kinetics, post-antibiotic effect, and post-antibiotic sub-MIC effect data of iboxamycin (IBX) against susceptible strains.
a, Arrayed growth curves for three susceptible strains showing concentration effects on growth inhibition (time-kill), growth kinetics following exposure to antibiotic at 4×MIC (PAE), and growth kinetics under sub-MIC concentrations following exposure to antibiotic at 4×MIC (PA-SME). Points represent mean values from n = 2 biologically independent experiments. b, Tabulated PAE and PA-SME durations (determined as the difference in time required for bacterial counts to rise 10× between experimental and untreated control arms). Abbreviations: CLI, clindamycin; LNZ, linezolid.
Extended Data Table 2.
Frequency of resistance to iboxamycin in susceptible bacterial strains
Extended Data Table 3.
Mutations selected in E. coli strain SQ110DTC conferring resistance to iboxamycin
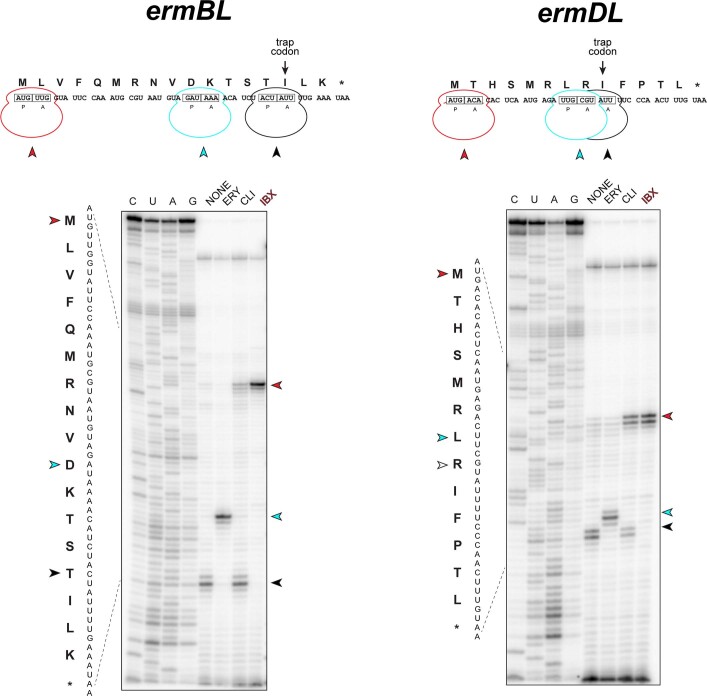
To understand the effect of IBX on ribosomal function, we performed primer extension inhibition analysis (‘toeprinting’), an in vitro technique that detects the position of drug-arrested ribosomes on mRNA47. Consistent with previous reports43, clindamycin arrested translation at the start codons of the model open reading frames (ORFs), yet allowed a fraction of ribosomes to translate ORFs up to the trap codon, a point at which ribosomes are arrested owing to the lack of isoleucyl tRNA in the in vitro translation system (Extended Data Fig. 6). By contrast, present at the same concentration (50 µM), IBX firmly locked the ribosome at the start codons, allowing virtually no escape, suggesting that IBX associates more strongly with the ribosome than does clindamycin.
Extended Data Fig. 6. Iboxamycin (IBX) efficiently arrests translation at the start codon.
Toeprinting analysis showing sites of IBX-induced translation arrest of ErmBL and ErmDL leader peptides. Because all reactions contained mupirocin, an inhibitor of Ile-tRNA synthetase, the ribosomes that escape inhibition by ribosome-targeting antibiotics are trapped at the codon preceding Ile (black arrowheads). Red arrowheads mark translation arrest at the start codon, while cyan arrowheads denote known erythromycin-induced arrest sites D10 (ermBL) and L7 (ermDL). Each gel is representative of two independent experiments, for source data see Supplementary Figure 1. ERY, erythromycin; CLI, clindamycin.
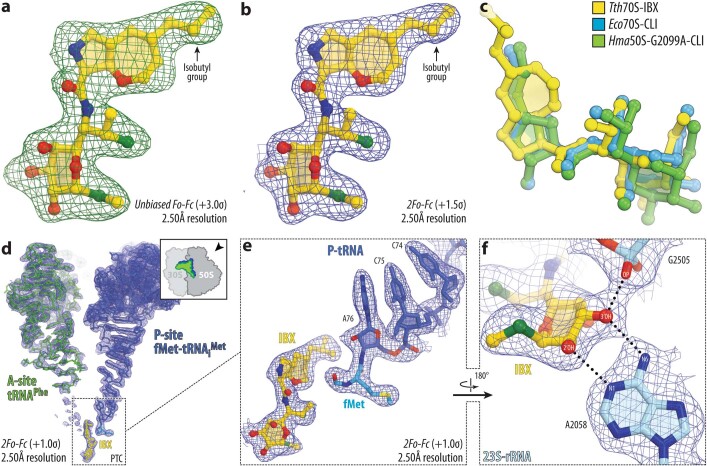
To illuminate the structural basis for this improved activity, we determined the structure of IBX bound to the bacterial ribosome using X-ray crystallography. 70S Ribosomes from the Gram-negative bacterium Thermus thermophilus were co-crystallized with IBX, mRNA, non-hydrolyzable aminoacyl-tRNA analogue fMet-NH-tRNAiMet (located within the P site), and deacylated tRNAPhe (within the A and E sites). The crystals we obtained diffracted to 2.50 Å resolution (Supplementary Table 2). The unbiased Fo − Fc difference Fourier map revealed positive electron density peaks resembling characteristic chemical features of IBX (Extended Data Fig. 7a, b), and confirmed that the antibiotic binds in the canonical binding pocket within the large ribosomal subunit (Fig. 3a), spanning the peptidyl transferase center (PTC) and extending into the nascent peptide exit tunnel (NPET) (Supplementary Video 1).
Extended Data Fig. 7. High-resolution electron density maps of iboxamycin (IBX) bound to the bacterial ribosome.
a, b, Unbiased Fo-Fc and 2Fo-Fc electron density maps of IBX in complex with the T. thermophilus 70S ribosome (green and blue mesh, respectively). The refined model of IBX is displayed in its electron density before (a) and after (b) the refinement contoured at 3.0σ and 1.5σ, respectively c, Superposition of ribosome-bound IBX (yellow) with prior structures of clindamycin bound to the tRNA-free 70S ribosome from eubacterium E. coli (blue, PDB entry 4V7V30) or to the 50S ribosomal subunit from archaeon H. marismortui harboring the 23S rRNA mutation G2099A (green, PDB entry 1YJN48). All structures were aligned based on domain V of the 23S rRNA. d, 2Fo-Fc electron density map (blue mesh) corresponding to ribosome-bound IBX (yellow), deacylated A-site tRNAPhe (green) and aminoacylated initiator P-site fMet-NH-tRNAiMet (dark blue). The refined models of tRNAs are displayed in their respective electron-density maps contoured at 1.0σ. In d, the entire bodies of the A- and P-site tRNAs are viewed from the back of the 50S subunit, as indicated by the inset. Ribosome subunits are omitted for clarity. Note that IBX binding to the ribosome prevents accommodation of the aminoacyl-bearing CCA-end of the A-site tRNA. e, Close-up view of the P-site tRNA CCA-end bearing a formyl-methionyl (cyan) residue. f, Detailed arrangement of the hydrogen bonds formed between the aminooctose component of IBX with 23S rRNA residues A2058 and G2505 (light blue).
Fig. 3. Structure of IBX in complex with the 70S ribosome, mRNA and tRNAs.
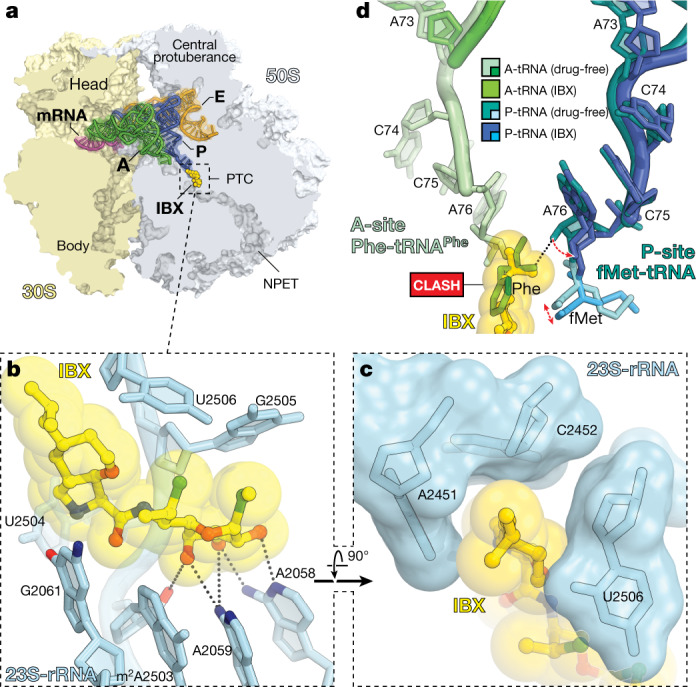
a, Overview of the IBX binding site in the T. thermophilus 70S ribosome viewed as a cross- section through the NPET. Indicated are the 30S subunit (light yellow), the 50S subunit (grey), mRNA (magenta), A-site tRNA (green), P-site tRNA (dark blue) and E-site tRNA (orange). b, c, Detail views of IBX bound in the PTC, highlighting hydrogen-bond interactions (dashed lines) and the positioning of the 7′-isobutyl substituent within the A-site cleft formed by 23S rRNA residues A2451 and C2452. d, Models of A- and P-site tRNAs from the IBX–ribosome complex (green and blue, respectively) superimposed with those found in the drug-free ribosome (PDB entry 6XHW51).
The binding of IBX to the functional T. thermophilus ribosome complex containing mRNA and tRNAs corresponds closely to the binding of clindamycin to the tRNA-free ribosome of E. coli30 or the large ribosomal subunit of the archaeon Haloarcula marismortui48 (Extended Data Fig. 7c). As with clindamycin and lincomycin, a network of hydrogen bonds anchors the aminooctose moiety of IBX to NPET nucleotides A2058, A2059, and A2503 (Fig. 3b). Similarly, the cationic aminoacyl residue of IBX occupies a hydrophilic pocket formed by the PTC residues G2061 and U2504 (Fig. 3b), displacing a divalent magnesium ion otherwise observed in the drug-free ribosome49,50. Consistent with our design hypothesis (Extended Data Fig. 1b), the oxepane ring of ribosome-bound IBX overlays closely with the n-propyl-group atoms of clindamycin and presents the 7′-isobutyl group for interaction with the A-site cleft formed by 23S rRNA residues A2451 and C2452 (Fig. 3c). This cleft, which normally accommodates the side-chains of incoming amino acids, has a key role in the positioning of aminoacylated 3′ ends of A-site tRNAs within the PTC during translation49,50. Electron density corresponding to the CCA terminus of the A-site tRNA is poorly resolved in our structure, suggesting that IBX blocks proper coordination of the aminoacylated 3ʹ end of A-site tRNA (Extended Data Fig. 7d). Notably, the 7′-isobutyl substituent of IBX extends deep enough within the A-site cleft to overlap not only with incoming amino acids (as the n-propyl group of clindamycin does, albeit to a lesser extent) but also with the P-site amino acid (Fig. 3d, Extended Data Fig. 7e). As tight coordination and proper placement of the P-site substrate are particularly important at the outset of protein synthesis, these observations, together with the toe-printing results described above, lead us to propose that compared to lincomycin and clindamycin, IBX more readily interferes with translation initiation.
The activity of IBX against bacteria harbouring erm resistance genes prompted us to determine the crystal structure of IBX bound to the -containing ribosome. Erm-mediated methylation of A2058 confers resistance to clindamycin by blocking the association of its 7-Cl-MTL residue with the NPET51, and because IBX bears the same aminooctose residue, its ability to overcome MLSB resistance was unexpected. Erm-modified 70S ribosomes with A2058 dimethylation levels of approximately 60% were isolated from T. thermophilus cells expressing the erm gene from Bifidobacterium thermophilum51. Co-crystallization of these ribosomes with IBX provided crystals diffracting to 2.60 Å resolution (Supplementary Table 2). Of note, the corresponding electron density map revealed that IBX binds to the methylated ribosome in a manner almost identical to its positioning within the wild-type ribosome, whereas undergoes a movement of about 2 Å relative to its canonical position to accommodate the antibiotic (Fig. 4). A2058-N6-dimethylation, together with this previously unknown displacement, disrupts the two hydrogen bonds typically formed between this residue and the aminooctose portion of lincosamides (Fig. 4d); this disruption appears sufficient to destabilize binding of clindamycin to the Erm-modified ribosome. However, new hydrophobic interactions between IBX and the A-site cleft appear to compensate for clashes with the modified nucleobase, providing sufficient affinity to defeat resistance. Indeed, oxepanoprolinamides’ activity against both MLSB-resistant S. aureus and Gram-negative species is governed rather precisely by the length of C7ʹ aliphatic substituents (Extended Data Fig. 2b), lending strong support to the hypothesis that enhanced target engagement underpins the improved activity of the class against multidrug-resistant pathogens. Thus, these results suggest that forming new interactions with the ribosomal A site may prove a general strategy in the design of expanded-spectrum antibiotics capable of defeating methylase-mediated resistance.
Fig. 4. Structure of iboxamycin (IBX) bound to the Erm-methylated 70S ribosome.
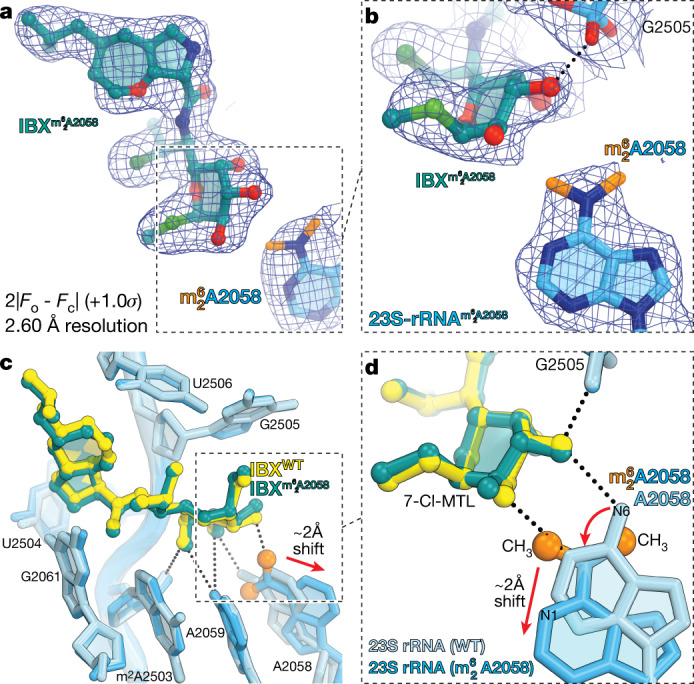
a, b, Electron density map (blue mesh), contoured at 1.0σ, of IBX (teal) in complex with the Erm-modified T. thermophilus 70S ribosome-containing N6-dimethylated A2058 residue in the 23S rRNA (a), highlighting the interaction with the methyl groups of (orange) (b). c, d, Superposition of IBX (yellow) in complex with the WT 70S ribosome containing an unmodified residue A2058 (light blue), and the structure of IBX (teal) in complex with the Erm-modified 70S ribosome containing an residue (medium blue) (c), highlighting hydrogen bonding (dashed lines) (d). Note that the position of IBX is almost identical in the two structures, whereas IBX binding to the Erm-modified ribosome causes substantial movement of from its canonical position (red arrow). WT, wild type.
Conclusion
Using a platform for antibiotics discovery rooted in component-based synthesis and structure-guided design, we identified the emergent oxepanoprolinamide antibiotic class. Departing radically from their lincosamide progenitors, the oxepanoprolinamides overcome Erm-, Cfr- and ABCF-mediated multidrug resistance and display activity against high-priority Gram-positive and Gram-negative pathogens, including multidrug-resistant strains of S. aureus, E. faecalis, E. coli, K. pneumoniae and A. baumannii. Methodologically, our findings attest to the continued role of empiricism in antibiotics discovery, as the activity of oxepanoprolinamides against posttranscriptionally methylated ribosomes could not have been predicted on the basis of existing antibiotic–ribosome co-crystal structures. Continued refinement of the scaffold, including modifications to the aminooctose residue whose unexpected capacity to displace we illustrate here, is likely to expand the oxepanoprolinamides’ spectrum of activity further. These findings, together with the safety, pharmacokinetic profile and efficacy of IBX in mouse models of Gram-positive and Gram-negative bacterial infection reaffirm the capacity for chemical synthesis to replenish our antibiotics armamentarium, delivering broad-spectrum agents capable of overcoming increasingly widespread resistance mechanisms.
Methods
In vitro susceptibility
MICs were determined by the broth microdilution method or the agar dilution method following the Clinical and Laboratory Standards Institute (CLSI) guidelines52–54. Standard bacterial strains were obtained from American Type Culture Collection (ATCC). Other bacterial strains were obtained from hospitals and commercial sources, as noted in Supplementary Table 1. Before the start of the MIC experiment, standard and test compound stock solutions were prepared in dimethyl sulfoxide (DMSO, Aldrich D2650) at a stock concentration of 5,120 µg ml−1. The compound concentration range typically employed for each experiment was 128–0.06 µg ml−1 (2.5% final DMSO concentration). Clindamycin was used as a comparator in all experiments and each experiment was performed in triplicate. Specialized procedures were employed for streptococci, anaerobic species and N. gonorrhoeae (see Supplementary Methods for details). All other bacterial strains were sub-cultured on blood agar plates (tryptic soy agar with 5% sheep blood, Hardy Diagnostics) and incubated overnight at 35 °C. Organisms were suspended in cation-adjusted Mueller–Hinton broth (CaMHB, BD 212322) and optical density was adjusted to 0.5 McFarland standard. Suspensions were further diluted to obtain a final inoculum of 5 × 105 CFU ml−1 for broth microdilution experiments. The minimum concentration of compound required to inhibit visible bacterial growth after 24 h of incubation was recorded as the MIC.
Time-kill studies
Time-kill studies were performed using four different concentrations of standard and test compounds (1, 2, 4 and 10×MIC). Experiments were performed in duplicate following CLSI guidelines. An inoculum was prepared in CaMHB containing 0.5 × 106–5 × 106 CFU ml−1 of test organism. Cultures were incubated at 37 °C in a shaker incubator at 110 rpm; a flask containing bacteria left unexposed to antibiotic was used as untreated control. At time points 0, 1, 3, 6 and 24 h following administration of antibiotic, bacterial counts were determined from each flask by serial dilution and plating on brain heart infusion agar (BD, Sparks, MD). Plates were incubated at 35 °C in incubators for 18–24 h to determine bacterial counts. Compounds exhibiting ≥3 log10CFU ml−1 reduction compared to initial counts are considered bactericidal. Compounds exhibiting ≤2 log10CFU reduction, or which maintain counts similar to the initial bacterial counts, are considered bacteriostatic.
Post-antibiotic and post-antibiotic sub-MIC effect
Measurements of post-antibiotic effect (PAE) and post-antibiotic sub-MIC effect (PA-SME) durations were performed according to the methods described by Odenholt-Tornqvist and colleagues55. Bacterial strains were cultured overnight on blood agar plates. Optical density was adjusted to 0.5 McFarland standard in CaMHB and suspensions were diluted further into 50-ml sterile Erlenmeyer flasks containing 20 ml CaMHB to achieve a bacterial load of 0.5–1 × 106 CFU ml−1. Cultures were then exposed to test and standard antibiotic at 1, 2, 4 and 10×MIC concentrations for 1 h at 37 °C in a shaker incubator at 110 rpm (0.5% final DMSO concentration). A flask containing the unexposed bacterial strain was left as untreated control. Post-exposure, 100 µl of bacterial culture was diluted in 900 µl of sterile CaMHB; 100 µl of this diluted sample was immediately diluted further in 19.9 ml of sterile CaMHB (1:2,000 final dilution). For PA-SME studies, post exposure and dilution, each flask (except those in the control group) was supplemented with 0.25 and 0.5×MIC per ml of specific antibiotic (0.5% final DMSO concentration). Flasks were incubated at 37 °C in a shaker incubator (110 rpm). Samples (100 µl from each flask per time point) were collected at 0, 1, 3, 6 and 24 h from each flask and were serially diluted. From each dilution, 100 µl and 10 µl from each dilution were plated in duplicate on tryptic soy agar plates. Plates were incubated overnight at 35 °C to determine the bacterial counts (limit of detection = 20 CFU per plate or 2 × 102 CFU ml−1). These counts were plotted against time with linear interpolation between time points to obtain the growth kinetic curves. PAE and PA-SME were determined from these curves using the formulae:
where T is the time required for bacteria previously exposed to antibiotic to multiply 1 log10 above counts immediately following dilution, and C is the corresponding time required for untreated culture to do the same; and
where Tpa is the time taken for bacteria previously exposed to antibiotic and then re-exposed to sub-MIC concentrations to increase 1 log10 above counts immediately following dilution.
Mammalian cell experiments
All cell lines were purchased from ATCC and used at a passage number of no greater than 8; cell lines were not authenticated or tested for mycoplasma contamination. All compounds were dissolved in sterile DMSO to a stock concentration of 20 µM and aliquoted prior to freezing at –20 °C. Aliquots were limited to a maximum of three freeze–thaw cycles. A549 (human pulmonary carcinoma; male) cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum. K562 (human chronic myelogenous leukaemia; female) cells were cultured in Iscove’s Modification of DMEM (IMDM) media supplemented with 10% fetal bovine serum. HCT116 (human colorectal carcinoma; male) cells were cultured in McCoy’s 5a (Iwakata and Grace Modification) media supplemented with 10% fetal bovine serum. HepG2 (human hepatocellular carcinoma; male) cells were cultured in Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% fetal bovine serum. All cell cultures were maintained in a 5% CO2 incubator at 35 °C.
Growth inhibition was studied using the Promega CellTiter-Blue cell viability assay pursuant to the manufacturer’s protocol. In brief, cells were harvested, diluted, and mixed with the desired concentration of test compound (or DMSO, used to define 0% growth inhibition). This suspension was then added to a 96-well plate (5 × 104 cells ml−1, 100 µl per well, 5 replicates per test compound), and the plates were incubated for 72 h at 35 °C in a 5% CO2 incubator. Next, 20 µl of CellTiter-Blue reagent was added to each well and the plates were incubated for 4 h. Plates were read on a SpectraMax i3 plate reader. GI50 values were determined by nonlinear regression using GraphPad Prism and represent the mean of at least three independent experiments.
Mitochondrial toxicity assays56,57 were conducted using the Promega Mitochondrial ToxGlo assay, according to the manufacturer’s protocol.
Animal studies
Animal experiments were performed at the Biology Research Infrastructure Laboratory of Harvard University following Institutional Animal Care and Use Committee (IACUC)-approved protocols. Animals were maintained in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Pathogen-free, 5- to 6-week-old, female CD-1 mice weighing 22–26 g were obtained from Charles River Laboratories, Inc. Animals were acclimated for a minimum of three days prior to start of the studies. Mice were caged as a group of four per cage, were housed at 21–23 °C with humidity ranging from 30–70%, were exposed to 12-h light and dark cycles, and were supplied with food and water ad libitum.
Mouse thigh infection studies
Mice were rendered neutropenic by administering cyclophosphamide intraperitoneally, 150 mg kg−1 4 days prior, and 100 mg kg−1 1 day prior to infection. Bacterial strains were cultured overnight on agar plates at 35 °C in 5% CO2. Bacterial inoculum was prepared in sterile brain heart infusion broth (BHIB) and OD was adjusted to 0.1 at 600 nm; the inoculum was further diluted to achieve a bacterial load of 0.5–1 × 107 CFU ml−1. Mice were infected by intramuscular injection of 0.1 ml of inoculum into each thigh (0.2 ml of inoculum was used per mouse). Mice were randomly divided into 4 treatment groups: IBX, comparator antibiotic, vehicle-treated control and untreated baseline. At 2 h post-infection (t = 0), the untreated mice (n = 2) were euthanized to obtain pre-treatment bacterial counts, while the others received a single dose of IBX (6 mg ml−1 in 10% Captisol, 250 µl, 60 mg kg−1), comparator antibiotic (clindamycin phosphate, ciprofloxacin, or azithromycin, 6 mg ml−1 in sterile saline, 250 µl−1, 60 mg kg−1), or sterile vehicle (10% Captisol, 250 µl) by intraperitoneal injection. Intramuscular bacterial counts were determined after euthanizing mice from each group (n = 2) by CO2 inhalation. Thighs (4 per time point) were aseptically removed and homogenized in sterile saline, and homogenates were serially diluted and plated (each sample was plated in duplicate). Bacterial counts (log10CFU per thigh) were determined after incubating the plates overnight at 35 °C in 5% CO2. Each experiment was repeated twice, and mean values were calculated (n = 16 counts across 4 biologically independent mice per time point). Because the outcome of the experiment (bacterial counts) was determined by serial dilution, a quantitative technique, blinding was not used. The results of these experiments are displayed in Extended Data Fig. 4.
Mouse systemic infection study
S. pyogenes ATCC 19615 was cultured overnight on blood agar plates in 5% CO2 at 35 °C before being suspended in BHIB. The optical density of the bacterial suspension was adjusted to 0.1 at 600 nm using sterile BHIB and was then further diluted 1:1 in 10% hog gastric mucin (HGM, type III) to prepare the infecting inoculum in 5% HGM. Mice were infected by intraperitoneal injection of 250 µl infecting inoculum, representing 1–2 × 106 CFU per mouse. Mice were randomly assigned to treatment groups; at time points 1, 5, 17 and 29 h post-infection, mice were administered intravenous infusions of IBX (1, 3 or 10 mg kg−1, 250 µl in 10% Captisol; 8 mice per dose level), clindamycin phosphate (1, 3 or 10 mg kg−1, 250 µl in normal saline; 8 mice per dose level), or vehicle as negative control (10% Captisol, 250 µl; 10 mice). Mice were monitored for survival for seven days following infection. Because the outcome of the experiment (survival) is objective, blinding was not used.
Frequency of resistance
Tripticase soy agar plates containing IBX at concentrations corresponding to 4×MIC, 8×MIC, and 16×MIC were prepared. Bacterial cultures (S. aureus ATCC 29213, S. aureus ATCC 700699, E. faecalis ATCC 29212, E. faecium HAV-251, and K. pneumoniae ATCC 10031) were prepared by overnight culture, and counts were determined by serial dilution and plating. Bacterial inocula (250 µl, containing approximately 1 × 107, 1 × 108, 1 × 109 or 1 × 1010 CFU) were spread onto the media plates. The plates were incubated at 37 °C for 72 h and were monitored for the appearance of colonies. The colony appearing after plating 1010 CFU of S. aureus ATCC 29213 on IBX-containing agar (4×MIC) was examined further to determine the fold change in antibiotic susceptibility; for details, see Supplementary Methods.
Selection of IBX-resistant mutants in SQ110DTC
Approximately 109 CFU (1.2 ml of the exponentially growing cell culture with optical density of A600 = 1.2), were plated on LB agar plates containing either 0.25 µg ml−1 or 1 µg ml−1 of IBX (~10× or ~30×MIC, respectively). Approximately 20 colonies appeared on both plates after incubation at 37 °C for 24 h. The segment of the 23S rRNA gene corresponding to domains V and VI of the 23S rRNA was PCR-amplified from 14 randomly selected colonies using the primers 2020R (CCCGAGACTCAGTGAAATTGAACTC) and L2904 (AAGGTTAAGCCTCACGG). PCR products were sequenced using the primer L2667 (GGTCCTCTCGTACTAGGAGCAG).
Toeprinting analysis
The ermBL DNA template for toeprinting was generated by a 4-primer cross-over PCR using primers T7, NV1, ermB3-F and ermB3-R. The ermDL template was generated in the same way using primers RLR-fwd, RLR-rev, T7-SD-fwd and T7-SD-rev. In vitro translation in the PURExpress system (New England Biolabs) and toeprinting analysis were carried out as described previously58. The antibiotics (mupirocin, clindamycin, erythromycin or IBX) were present in the reactions at a final concentration of 50 µg ml−1.
Primer sequences used for toeprinting are as follows: T7 (TAATACGACTCACTATAGGG); NV1 (GGTTATAATGAATTTTGCTTATTAAC); ermB3-F (TAATACGACTCACTATAGGGCTTAAGTATAAGGAGGAAAAAATATGTTGGTATTCCAAATGCGTAATGTAGATAAAACATCTAC); ermB3-R (GGTTATAATGAATTTTGCTTATTAACGATAGAATTCTATCACTTATTTCAAAATAGTAGATGTTTTATCTACATTACG); RLR-fwd (GGAGGAAAAAATATGACACACTCAATGAGACTTCGTATTTTCCC); RLR-rev (CTATCACTTACAAAGTTGGGAAAATACGAAGTCTCATTGAG); T7-SD-fwd (TAATACGACTCACTATAGGGCTTAAGTATAAGGAGGAAAAAATATGACACACTCAATG); T7-SD-rev (GGTTATAATGAATTTTGCTTATTAACGATAGAATTCTATCACTTACAAAGTTGGGAAAAT).
Crystallization of IBX in complex with wild-type and Erm-modified T. thermophilus 70S ribosomes
Wild-type 70S ribosomes from T. thermophilus (strain HB8) containing unmodified residue A2058 of the 23S rRNA were prepared as described previously50. Purification of the Erm-modified 70S ribosomes from T. thermophilus (strain HB27 expressing Erm-like enzyme from B. thermophilum) containing N6-dimethylated A2058 residue in the 23S rRNA was accomplished as optimized previously for the wild-type 70S ribosomes from T. thermophilus51. Deacylated tRNAPhe and non-hydrolyzable aminoacylated fMet-NH-tRNAiMet were prepared as described previously51. Complexes of the A2058-dimethylated or unmethylated T. thermophilus 70S ribosomes with mRNA, deacylated A-site tRNAPhe, and P-site hydrolysis-resistant aminoacylated fMet-NH-tRNAiMet were formed as described previously49–51. For T. thermophilus 70S ribosome complexes with IBX, the antibiotic was included in the crystallization mixture (250 µM each) and then later added to the stabilization buffers (250 µM each). Collection and processing of the X-ray diffraction data, model building, and structure refinement were performed as described in our previous reports49–51. Diffraction data were collected at beamlines 24ID-C and 24ID-E at the Advanced Photon Source (Argonne National Laboratory). A complete dataset for each complex was collected using 0.979 Å irradiation at 100 K from multiple regions of the same crystal, using 0.3-degree oscillations. Raw data were integrated and scaled using XDS software59 (5 Feb 2021). Molecular replacement was performed using PHASER from the CCP4 program suite60 (version 7.0). The search model was generated from the previously published structures of T. thermophilus 70S ribosome with bound mRNA and aminoacylated tRNAs (PDB entry 6XHW)51. Initial molecular replacement solutions were refined by rigid-body refinement with the ribosome split into multiple domains, followed by positional and individual B-factor refinement using PHENIX software61 (version 1.17). Non-crystallographic symmetry restraints were applied to four parts of the 30S ribosomal subunit (head, body, spur, and helix 44), and four parts of the 50S subunit (body, L1-stalk, L10-stalk, and C-terminus of the L9 protein). Structural models were built in Coot62 (version 0.8.2). Structural models and restraints for IBX were generated using PRODRG online software63 (http://prodrg1.dyndns.org). The statistics of data collection and refinement are compiled in Supplementary Table 2. All figures showing atomic models were rendered using PyMol software64.
Statistical analyses
Statistical analysis was performed using GraphPad Prism. Mouse thigh infection study data (12-h time points) were compared using Welch’s two-tailed unpaired t-test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-021-04045-6.
Supplementary information
Uncropped mRNA toeprinting gel image.
| .docx file | Antimicrobial susceptibility assays; susceptibility testing of a spontaneously resistant mutant; consideration of predictive rules for Gram-negative accumulation; structural basis for Cfr-mediated resistance to lincosamides; chemical synthesis of oxepanoprolinamide analogs.
In vitro antibacterial activity of iboxamycin.
X-ray crystallographic data for iboxamycin–ribosome complexes.
Illustration of iboxamycin binding to the wild-type ribosome, and iboxamycin-induced displacement of in the Erm-modified ribosome.
Source data
Acknowledgements
We thank C.-W. Chen for help with initial X-ray diffraction data collection; M. Svetlov for providing Erm-methylated ribosomes used in our structural studies; D. R. Andes, M. S. Gilmore, F. H. Lebreton, T. J. Dougherty, D. P. Nicolau, P. Courvalin, C. Grillot-Courvalin, H. E. Moser, S. Chiang, B. Wohl, C. Keith, S. Lahiri, R. Duggal and K. M. Otte for their invaluable input in the course of our research; the staff at NE-CAT beamlines 24ID-C and 24ID-E for their help with data collection, especially M. Capel, F. Murphy, I. Kourinov, A. Lynch, S. Banerjee, D. Neau, J. Schuermann, N. Sukumar, J. Withrow, K. Perry and C. Salbego; S.-L. Zheng for X-ray crystallographic data collection and structure determination of compounds 6 and OPP-3; D. L. Shinabarger and C. Pillar of Micromyx, L.L.C., W. J. Weiss of the UNT Health Science Center and M. D. Cameron of TSRI Florida for in vitro profiling of compounds. Ligand Pharmaceuticals kindly provided the Captisol used in our mouse experiments. M.J.M. was supported by the National Science Foundation Graduate Research Fellowship under grant no. DGE1144152. K.J.Y.W. was supported by a National Science Scholarship (PhD) by the Agency for Science, Technology and Research, Singapore. G.T. received postdoctoral support from the Deutsche Forschungsgemeinschaft (TE1311-1-1). This work is based upon research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30-GM124165). The Eiger 16M detector on 24ID-E beamline is funded by a NIH-ORIP HEI grant (S10-OD021527 to NE-CAT). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Extraordinary facility operations were supported in part by the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on the response to COVID-19, with funding provided by the Coronavirus CARES Act. This work was supported by Illinois State startup funds to Y.S.P. and National Institutes of Health R21-AI137584 (to A.S.M. and Y.S.P.), R01-GM132302 (to Y.S.P.), and R35GM127134 (to A.S.M.). A.G.M. gratefully acknowledges support from the LEO Foundation (grant LF18006) and the Blavatnik Biomedical Accelerator at Harvard University.
Extended data figures and tables
Author contributions
M.J.M. identified the oxepanoprolinamide scaffold and the synthetic routes used to prepare analogues OPP-1, OPP-2, OPP-4 and OPP-5. A.P. designed and performed MIC, TK, PAE, PA-SME and mouse infection experiments. E.A.S. and Y.S.P. designed and performed X-ray crystallography experiments. K.J.S. identified 7′-alkyl oxepanoprolinamides and the synthetic route used to prepare analogues OPP-6, epi-OPP-3 and IBX (OPP-3) from ketone 10. D.K. and A.S.M. selected and sequenced IBX-resistant SQ110DTC clones and conducted ribosome toeprinting analysis. M.J.M., K.J.S., J.D.M., D.W.T., G.T., A.R.P. and K.J.Y.W. synthesized and characterized oxepanoprolinamide analogues. A.P. and R.P.L. performed mammalian cell experiments. A.P. and K.C. designed and performed the pharmacokinetic experiments. A.G.M., A.S.M. and Y.S.P. supervised the experiments. All authors interpreted the results. M.J.M, A.G.M, A.S.M, and Y.S.P. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Data availability
Coordinates and structure factors are deposited in the RCSB Protein Data Bank with accession codes 7RQ8 for the T. thermophilus 70S ribosome in complex with IBX, mRNA, deacylated A-site tRNAPhe, aminoacylated P-site fMet-NH-tRNAiMet, and deacylated E-site tRNAPhe; and 7RQ9 for the T. thermophilus 70S ribosome in complex with IBX, mRNA, deacylated A-site tRNAPhe, aminoacylated P-site fMet-NH-tRNAiMet. Single-crystal X-ray crystallographic data for compound 6 are deposited at the Cambridge Crystallographic Data Centre under deposition number 2072277. All previously published structures that were used in this work for model building and structural comparisons were retrieved from the RCSB Protein Data Bank under accession codes 6XHW51, 4V7V30 and 1YJN48. Source data are provided with this paper.
Competing interests
A.G.M., M.J.M., K.J.S., J.D.M. and G.T are inventors in a provisional patent application submitted by the President and Fellows of Harvard College covering antibiotics of the type described in this work. A.G.M., M.J.M. and K.J.S. have filed an international patent application WO/2019/032936 ‘Lincosamide Antibiotics and Uses Thereof’. A.G.M. and M.J.M. have filed an international patent application WO/2019/032956 ‘Lincosamide Antibiotics and Uses Thereof’.
Footnotes
Peer review information Nature thanks Daniel Wilson, William Wuest and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Matthew J. Mitcheltree, Amarnath Pisipati, Egor A. Syroegin
Contributor Information
Yury S. Polikanov, Email: yuryp@uic.edu
Andrew G. Myers, Email: myers@chemistry.harvard.edu
Extended data
is available for this paper at 10.1038/s41586-021-04045-6.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-021-04045-6.
References
- 1.Antibiotic Resistance Threats in the United States, 2019 (Department of Health and Human Services, CDC, 2019); www.cdc.gov/DrugResistance/Biggest-Threats html.
- 2.Wright PM, Seiple IB, Myers AG. The evolving role of chemical synthesis in antibacterial drug discovery. Angew. Chem. Int. Ed. 2014;53:8840–8869. doi: 10.1002/anie.201310843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scientific Roadmap for Antibiotic Discovery (Pew Charitable Trusts, 2016); http://www.pewtrusts.org/antibiotic-discovery.
- 4.Charest MG, Lerner CD, Brubaker JD, Siegel DR, Myers AG. A convergent enantioselective route to structurally diverse 6-deoxytetracycline antibiotics. Science. 2005;308:395–398. doi: 10.1126/science.1109755. [DOI] [PubMed] [Google Scholar]
- 5.Seiple IB, et al. A platform for the discovery of new macrolide antibiotics. Nature. 2016;533:338–345. doi: 10.1038/nature17967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myers A, Clark RB. Discovery of macrolide antibiotics effective against multi-drug resistant Gram-negative pathogens. Acc. Chem. Res. 2021;54:1635–1645. doi: 10.1021/acs.accounts.1c00020. [DOI] [PubMed] [Google Scholar]
- 7.Li Q, et al. Synthetic group A streptogramin antibiotics that overcome Vat resistance. Nature. 2020;586:145–150. doi: 10.1038/s41586-020-2761-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith PA, et al. Optimized arylomycins are a new class of Gram-negative antibiotics. Nature. 2018;651:189–194. doi: 10.1038/s41586-018-0483-6. [DOI] [PubMed] [Google Scholar]
- 9.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Zhou D, Steitz TA, Polikanov YS, Gagnon MG. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 2018;87:451–478. doi: 10.1146/annurev-biochem-062917-011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason, D. J., Dietz, A. & De Boer, C. Lincomycin, a new antibiotic. I. Discovery and biological properties. Antimicrob. Agents Chemother. 554–559 (1962).
- 12.Birkenmeyer RD, Kagan F. Lincomycin. XI. Synthesis and structure of clindamycin. A potent antibacterial agent. J. Med. Chem. 1970;13:616–619. doi: 10.1021/jm00298a007. [DOI] [PubMed] [Google Scholar]
- 13.Phillips I. Past and current use of clindamycin and lincomycin. J. Antimicrob. Chemother. 1981;7:11–18. doi: 10.1093/jac/7.suppl_A.11. [DOI] [PubMed] [Google Scholar]
- 14.Birkenmeyer RD, Kroll SJ, Lewis C, Stern KF, Zurenko GE. Synthesis and antimicrobial activity of clindamycin analogs: pirlimycin, a potent antibacterial agent. J. Med. Chem. 1984;27:216–223. doi: 10.1021/jm00368a020. [DOI] [PubMed] [Google Scholar]
- 15.O’Dowd, H., Erwin, A. L. & Lewis, J. G. in Natural Products in Medicinal Chemistry (ed. Hanessian, S.) (Wiley-VCH, 2014).
- 16.Hirai Y, et al. Characterization of compound A, a novel lincosamide derivative active against methicillin-resistant Staphylococcus aureus. J. Antibiot. 2021;74:124–132. doi: 10.1038/s41429-020-00375-1. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq R, Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 1991;35:1267–1272. doi: 10.1128/AAC.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai CJ, Weisblum B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc. Natl Acad. Sci. USA. 1971;68:856–860. doi: 10.1073/pnas.68.4.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffith LJ, Ostrander WE, Mullins CG, Beswick DE. Drug antagonism between lincomycin and erythromycin. Science. 1965;147:746–747. doi: 10.1126/science.147.3659.746. [DOI] [PubMed] [Google Scholar]
- 20.Toh SM, et al. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 2007;64:1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giessing AM, et al. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith LK, Mankin AS. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 2008;52:1703–1712. doi: 10.1128/AAC.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowe-McAuliffe C, et al. Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens. Nat. Comm. 2021;12:3577. doi: 10.1038/s41467-021-23753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murina V, Kasari M, Hauryliuk V, Atkinson GC. Antibiotic resistance ABCF proteins reset the peptidyl transferase centre of the ribosome to counter translational arrest. Nucleic Acids Res. 2018;46:3753–3763. doi: 10.1093/nar/gky050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitcheltree MJ, Stevenson JW, Pisipati A, Myers AG. A practical, component-based synthetic route to methylthiolincosamine permitting facile northern-half diversification of lincosamide antibiotics. J. Am. Chem. Soc. 2021;143:6829–6835. doi: 10.1021/jacs.1c03536. [DOI] [PubMed] [Google Scholar]
- 27.Mitcheltree, M. J. A Platform for the Discovery of New Lincosamide Antibiotics. PhD Dissertation, Harvard University (2018).
- 28.Silvestre, K. J. Design, Synthesis, and Study of Lincosamide Antibiotics Containing a Bicyclic Amino Acid Moiety. PhD Dissertation, Harvard University (2019).
- 29.Moga, I. Novel Lincosamide Antibiotics Containing an Azepane Amino Acid Moiety. PhD Dissertation, Harvard University (2019).
- 30.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl Acad. Sci. USA. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seiple IB, Mercer JA, Sussman RJ, Zhang Z, Myers AG. Stereocontrolled synthesis of syn-β-hydroxy-α-amino acids by direct aldolization of pseudoephenamine glycinamide. Angew. Chem. Int. Ed. 2014;53:4642–4647. doi: 10.1002/anie.201400928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kingsbury JS, Harrity JPA, Bonitatebus PJ, Jr, Hoveyda AH. A recyclable ru-based metathesis catalyst. J. Am. Chem. Soc. 1999;121:791–799. doi: 10.1021/ja983222u. [DOI] [Google Scholar]
- 33.Magerlein BJ, Kagan F. Lincomycin. 8. 4′-alkyl-1′-demethyl-4′-depropylclindamycins, potent antibacterial and antimalarial agents. J. Med. Chem. 1969;12:780–784. doi: 10.1021/jm00305a013. [DOI] [PubMed] [Google Scholar]
- 34.Morandi B, Wickens ZK, Grubbs RH. Regioselective Wacker oxidation of internal alkenes: rapid access to functionalized ketones facilitated by cross-metathesis. Angew. Chem. Int. Ed. 2013;52:9751–9754. doi: 10.1002/anie.201303587. [DOI] [PubMed] [Google Scholar]
- 35.Scheiper B, Bonnekessel M, Krause H, Fürstner A. Selective iron-catalyzed cross-coupling reactions of Grignard reagents with enol triflates, acid chlorides, and dichloroarenes. J. Org. Chem. 2004;69:3943–3949. doi: 10.1021/jo0498866. [DOI] [PubMed] [Google Scholar]
- 36.Mason JD, Terwilliger DT, Pote AR, Myers AG. Practical gram-scale synthesis of iboxamycin, a potent antibiotic candidate. J. Am. Chem. Soc. 2021;143:11019–11025. doi: 10.1021/jacs.1c03529. [DOI] [PubMed] [Google Scholar]
- 37.Zhao C, et al. Investigation of antibiotic resistance, serotype distribution, and genetic characteristics of 164 invasive Streptococcus pneumoniae from north China between April 2016 and October 2017. Infect. Drug Resist. 2020;13:2117–2128. doi: 10.2147/IDR.S256663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner-Lastinger LM, et al. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect. Control Hosp. Epidemiol. 2020;41:1–18. doi: 10.1017/ice.2019.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh KV, Weinstock GM, Murray BE. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin–dalfopristin. Antimicrob. Agents Chemother. 2002;46:1845–1850. doi: 10.1128/AAC.46.6.1845-1850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice LB. Challenges in identifying new antimicrobial agents effective for treating infections with Acinetobacter baumannii and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006;43:S100–S105. doi: 10.1086/504487. [DOI] [PubMed] [Google Scholar]
- 41.O’Shea R, Moser HE. Physicochemical properties of antibacterial compounds: implications for drug discovery. J. Med. Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 42.Richter MF, et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. 2017;545:299–304. doi: 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orelle C, et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob. Agents Chemother. 2013;57:5994–6004. doi: 10.1128/AAC.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23s rRNA. Antimicrob. Agents Chemother. 2001;45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Böttger EC, Springer B, Prammananan T, Kidan Y, Sander P. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2001;2:318–323. doi: 10.1093/embo-reports/kve062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silver LL. Multi-targeting by monotherapeutic antibacterials. Nat. Rev. Drug Discov. 2007;6:41–55. doi: 10.1038/nrd2202. [DOI] [PubMed] [Google Scholar]
- 47.Orelle C, et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 2013;41:e144. doi: 10.1093/nar/gkt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Polikanov YS, Steitz TA, Innis CA. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014;21:787–793. doi: 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polikanov YS, Melnikov SV, Soll D, Steitz TA. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015;22:342–344. doi: 10.1038/nsmb.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svetlov, M. S. et al. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat. Chem. Biol. (2021). [DOI] [PMC free article] [PubMed]
- 52.Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, Approved Standard 11th edn (Clinical and Laboratory Standards Institute, 2018).
- 53.Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria;Approved Standard 8th edn (Clinical and Laboratory Standards Institute, 2012). [PubMed]
- 54.Performance Standards for Antimicrobial Susceptibility Testing 27th edn (Clinical and Laboratory Standards Institute, 2017).
- 55.Odenholt-Tornqvist I, Löwdin E, Cars O. Pharmacodynamic effects of subinhibitory concentrations of β-lactam antibiotics in vitro. Antimicrob. Agents Chemother. 1991;35:1834–1839. doi: 10.1128/AAC.35.9.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niles AL, et al. A homogeneous assay to measure live and dead cells in the same sample by detecting different protease markers. Anal. Biochem. 2007;366:197–206. doi: 10.1016/j.ab.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol. Sci. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- 58.Orelle C, et al. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 2013;41:e144. doi: 10.1093/nar/gkt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kabsch W. XDS. Acta Crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liebschner D, et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 64. The PyMOL Molecular Graphics System v.2.0 (Schrödinger, 2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Uncropped mRNA toeprinting gel image.
| .docx file | Antimicrobial susceptibility assays; susceptibility testing of a spontaneously resistant mutant; consideration of predictive rules for Gram-negative accumulation; structural basis for Cfr-mediated resistance to lincosamides; chemical synthesis of oxepanoprolinamide analogs.
In vitro antibacterial activity of iboxamycin.
X-ray crystallographic data for iboxamycin–ribosome complexes.
Illustration of iboxamycin binding to the wild-type ribosome, and iboxamycin-induced displacement of in the Erm-modified ribosome.
Data Availability Statement
Coordinates and structure factors are deposited in the RCSB Protein Data Bank with accession codes 7RQ8 for the T. thermophilus 70S ribosome in complex with IBX, mRNA, deacylated A-site tRNAPhe, aminoacylated P-site fMet-NH-tRNAiMet, and deacylated E-site tRNAPhe; and 7RQ9 for the T. thermophilus 70S ribosome in complex with IBX, mRNA, deacylated A-site tRNAPhe, aminoacylated P-site fMet-NH-tRNAiMet. Single-crystal X-ray crystallographic data for compound 6 are deposited at the Cambridge Crystallographic Data Centre under deposition number 2072277. All previously published structures that were used in this work for model building and structural comparisons were retrieved from the RCSB Protein Data Bank under accession codes 6XHW51, 4V7V30 and 1YJN48. Source data are provided with this paper.