Abstract
Dental pulp stem cells (DPSCs) are one promising cell source of mesenchymal stem cells in bone tissue engineering. However, it remains unknown that the molecules and signaling pathways involved in osteogenesis of DPSCs. Hence, this study investigated the functional roles and underlying mechanisms of circRFWD2 during osteogenesis of DPSCs. Knockdown of circRFWD2 suppressed osteogenesis of DPSCs significantly. Mechanistically, circRFWD2 could crosstalk with miR-6817-5p, which was an inhibitor of DPSCs osteogenesis. MiR-6817-5p functioned as a sponge of BMPR2, which regulated the phosphorylation of Smad5 and p38 to impact osteogenesis activity of DPSCs. Collectively, circRFWD2/miR-6817-5p/BMPR2 axis could regulate DPSCs osteogenesis via BMP-Smad and p38 MAPK pathway, which are novel mechanisms in the osteogenic differentiation of DPSCs and suggest potential therapeutic methods for bone defects regeneration.
Keywords: circRFWD2, miR-6817-5p, BMPR2, BMP-Smad pathway, p38 MAPK pathway, bone tissue engineering
Introduction
Bone defects, which could be caused by traumas, tumors, and congenital malformations, are one kind of most prevalent skeletal diseases 1–2 . They reduce the quality of life seriously and pose heavy physical, emotional, and economic burden to the individuals and society 3 . Cell-based bone tissue engineering, which utilizes a combination of mesenchymal stem cells (MSCs), scaffolds, and growth factors, is a promising strategy for bone defects regeneration 4–5 . The best approach to acquiring MSCs should meet the requirements of sufficient quantity, painless and straightforward sampling, and low risk of complications 6 .
Dental pulp stem cells (DPSCs), neural crest-derived cells with an outstanding ability to differentiate into multiple lineages, have become a popular cell source of MSCs in bone tissue engineering 7 –9 . As DPSCs can be isolated from extracted teeth after routine surgical practice (e.g., third molars, orthodontic teeth), they are more easily accessible than bone marrow or adipose tissue derived MSCs which require invasive biopsies 10 . Some studies reported that simple in vitro protocols and/or considerable interactions with certain biomaterials could induce DPSCs into mature osteoblasts to produce a mineralized matrix 11,12 . However, it remains unknown that the molecules and signaling pathways involved in osteogenesis of DPSCs which induce DPSCs osteogenesis effectively and safely in bone defects treatment. Therefore, understanding the functions and characteristics of DPSCs at the genetic and molecular levels are of great importance.
Circular RNAs (circRNAs) are novel noncoding RNAs. Unlike linear RNAs, circRNAs have unique closed loop structures with 3’ heads and 5’ tails bound together, which makes them resistant to digestion by RNAse R 13 . Additionally, a growing body of studies suggest that circRNAs express in a spatial- and temporal-specific manner, and participate in embryonic development, cellular activities, and many human diseases 14,15 . It is showed that circRNAs orchestrate a variety of molecules and signaling pathways to regulate osteogenesis 16,17 . Our previous study performed RNA-sequencing to detect the expression pattern of circRNAs in recombinant NELL-1-induced osteogenic differentiation 18 . The results showed circRFWD2 were up-regulated during recombinant NELL-1-induced osteogenesis of adipose tissue derived stem cells (ASCs) and circRFWD2 could regulate the expression of miR-6817-5p to influence the recombinant NELL-1-induced osteoblast differentiation of ASCs, indicating circRFWD2 plays key roles in the osteogenic differentiation process of MSCs. However, it remains to be studied that the roles of circRFWD2 during DPSCs osteogenesis.
In this study, we investigated the functional roles and underlying mechanisms of circRFWD2 during osteogenesis of DPSCs for the first time. Knockdown of circRFWD2 significantly retarded osteoblast differentiation of DPSCs, while overexpression of miR-6817-5p could induced a similar change. Mechanistically, miR-6817-5p served as a sponge of BMPR2, which regulated the phosphorylation of Smad5 and p38 to impact DPSCs osteogenesis activity. Therefore, this study reveals novel mechanisms in the osteogenic differentiation of DPSCs and suggests potential therapeutic methods for bone defects regeneration.
Material and Methods
hDPSCs Isolation and Identification
As described previously, hDPSCs were extracted and cultured from premolars extracted for orthodontic treatment which were collected from 3 adolescents (12–14 years old) with informed consent 19,20 . Briefly, type I collagenase (Gibco, Grand Island, NY, USA) was used to digest the dental pulp tissues for 1 hour at 37°C. The digested pulp tissue was then maintained in the culture medium composed of low glucose Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Grand Island, NY, USA), penicillin-streptomycin (Gibco, Grand Island, NY, USA), as well as 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA). The 4th passage hDPSCs were used for subsequent experiments.
To identify MSCs surface markers, hDPSCs were collected in phosphate-buffered saline (PBS) and incubated with FITC-conjugated anti-human CD29, FITC-conjugated anti-human CD31, FITC-conjugated anti-human CD44, FITC-conjugated anti-human CD45, FITC-conjugated anti-human HLA-ABC (all from Thermo Fisher Scientific, Waltham, MA, USA). After incubation for 30 min, the hDPSCs samples were identified in the flow cytometer (Beckman Coulter, FC500, FL, USA), and FlowJo software (Tree Star, San Carlos, CA, USA) were used for data analysis.
Trilineage differentiation that consisting of osteogenic, chondrogenic, and adipogenic differentiation was designed to evaluate the differentiation potential of hDPSCs. Briefly, hDPSCs were seeded into 24-well plates (2.0 × 104 cells/well) with normal medium and transferred into trilineage differentiation induction medium after cell confluence reached 60%. These plates were cultured for 21 days in the osteogenic, chondrogenic, and adipogenic induction medium (Cyagen, Guangzhou, China). The ability of osteogenic differentiation was determined by Alizarin Red S staining (ARS, Solarbio, Beijing, China). Alcian Blue (Sigma-Aldrich, St. Louis, MO, USA) staining was utilized to evaluate chondrogenic differentiation potential. Oil Red O (Sigma-Aldrich, St. Louis, MO, USA) staining was performed for visualizing adipogenic differentiation.
Osteogenic Induction
The hDPSCs were seeded in a 24-well plate, and osteogenic differentiation was initiated after 80% cell confluence. The osteogenic induction medium consisted of general culture medium, 10 nM dexamethasone, 50 mg/mL vitamin C, and 10 mM β-glycerophosphate (all from Sigma-Aldrich, St. Louis, MO, USA). All the plates were cultured in an incubator under 5% CO2 at 37°C, and induction medium was replaced every 3 days.
RNA Oligoribonucleotides and Cell Transfection
For alteration expression of circRFWD2, miR-6817-5p, and BMPR2, cell transfection was conducted by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions after cell reached 70% confluence. The RNA oligoribonucleotides were obtained from GenePharma Co. (Shanghai, China) which were showed in Table S1. The si-circRFWD2, si-BMPR2, miR-6817-5p mimic, miR-6817-5p inhibitor, and negative controls underwent cell transfection with 100 nM concentration. The transfected cells were collected after 2 and 3 days for qRT-PCR and Western blot, respectively.
RNA Isolation and Real-Time PCR Analysis
Total RNA from cultured cells were extracted by the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s protocol. The concentration and quality of RNAs were evaluated by the spectrophotometer (NanoDrop 2000, ThermoFisher, Carlsbad, CA, USA). Real-time PCR analysis was performed on an Applied Biosystems 7500 Fast Systems (Applied Biosystems, Shanghai, China). The primers of circRNAs and mRNAs were synthesized by GenePharma Co. (Shanghai, China) which were showed in Table S2. The miRNAs qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA) was used for detecting miRNAs expression, and U6 was used for expression normalization. The relative expression in this study was calculated by the formula 2−ΔΔCt.
Western Blot
Total cell protein was extracted by using radio immunoprecipitation assay (RIPA) lysis buffer. Protein concentration was measured by the BCA Protein Assay Kit (Beyotime Institute of Biotechnology, Nanjing, China). Equal amount of protein samples was used for 10% SDS-PAGE electrophoresis. After that, the protein was transferred onto the PVDF membranes blocked with 5% BSA. These PVDF membranes were added with primary antibodies and incubated overnight at 4°C. After washed with TBST, the secondary antibodies were added to the membranes. The antibodies were as follows: RUNX2 (ab76956, Abcam), BMPR2 (EM1709-92, Huabio), p-SMAD5 (ET1605-5, Huabio), SMAD5 (ET1606-26, Huabio), p-p38 (ER1903-01, Huabio), p38 (ET1602-26, Huabio), and GAPDH (ab181602, Abcam).
Alkaline Phosphatase (ALP) Staining and Activity
Alkaline phosphatase staining was performed by utilizing the Leukocyte Alkaline Phosphatase Kit 86C (Sigma-Aldrich, St. Louis, MO, USA). Briefly, after washed with PBS, the cell samples were fixed in citrate solution for 30 seconds. After fixation, samples were stained with a mixed solution composed of naphthol AS-BI alkaline, FRV alkaline, and sodium nitrite for 15 minutes. To measure the ALP activity, the Alkaline Phosphatase Assay Kit (Beyotime Institute of Biotechnology, Nanjing, China) was used to analyze the ALP activity. In brief, the cell samples were cleaved with RIPA lysis buffer and scraped into distilled water, the supernatant was added into a 96-well plate and incubated at 37°C for 30 minutes. The p-nitrophenol and reaction substrates were used for detection of ALP activity. Absorbance was calculated at 405 nm.
Alizarin red S (ARS) Staining and Quantification
Cell samples were fixed in 4% para-formaldehyde solution for 30 minutes at room temperature, and stained with 1% Alizarin Red S (pH 4.2; Solarbio, Beijing, China) for 5 minutes. The Alizarin Red staining results were quantified by a spectrophotometer at 595 nm. To determine the quantity of mineralized nodules, the ARS stain was dissolved in 10% cetylpyridinium chloride (Sigma-Aldrich, St. Louis, MO, USA) for 60 minutes, and the absorbance was measured at 570 nm.
Dual Luciferase Reporter Assay
Dual luciferase reporter assay was conducted to confirm the binding relationship between miR-6817-5p and BMPR2 as well as circRFWD2 and miR-6817-5p. In brief, HEK293 cells were seeded in the 24-well plate with 50% density. The wild-type 3’UTR sequences of BMPR2 and circRFWD2 mRNA contained putative binding sites for miR-6817-5p, and the mutant 3’UTR of BMPR2 and circRFWD2 were constructed with the Mut Express MultiS Fast Mutagenesis Kit (Vazyme Biotech, Nanjing, China). 40 ng BMPR2 luciferase reporter plasmids with 100 nM miR-NC or miR-6817-5p mimic were co-transfected into HEK 293 T cells by using Lipofectamine 3000. After transfected for 48 h, luciferase activity was measured using the Dual-Luciferase Reporter System (Promega, Madison, WI, USA), while Renilla luciferase activity was used for normalizing firefly luciferase activity. All experiments were conducted in triplicate.
Statistical Analyses
Statistical analyses in this study were performed by using SPSS version 16.0 (SPSS, Chicago, IL, USA). All the data were shown as the means ± standard deviation (SD) of at least three independent experiments. The differences between the two groups were analyzed using Student’s t test. One-way analysis of variance (ANOVA) was used for measuring differences among three or more groups. P-value < 0.05 was regarded as statistical significance. In all cases, *P < 0.05, **P < 0.01.
Results
hDPSCs Showed Multidirectional Differentiation Ability and Expressed MSCs Markers
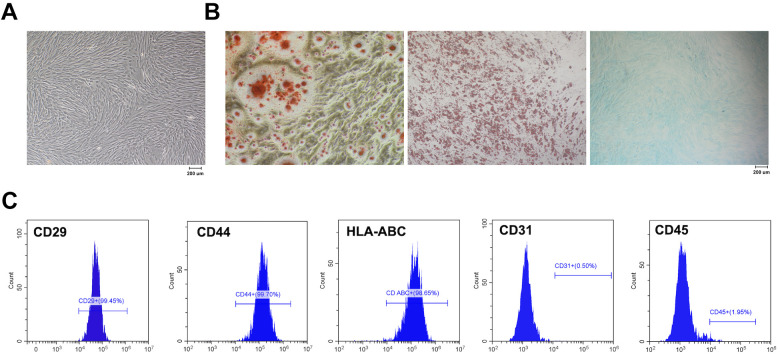
hDPSCs were extracted from dental pulp, and subsequently cultured with medium for proliferation. hDPSCs at passage 4 appeared to be spindle-shaped, indicating the typical MSCs morphology (Fig. 1A). To determine the potential of multidirectional differentiation of hDPSCs, the multidirectional induction was performed with osteogenesis, adipogenesis, and chondrogenesis inducing medium, respectively. ARS staining showed apparent calcium mineral deposits (Fig. 1B), while Oil Red O staining showed numerous cytoplasmic lipid droplets (Fig. 1B), and Alcian Blue staining exhibited more glycosaminoglycans after chondrogenesis induction (Fig. 1B). Flow cytometry was performed to identify MSCs surface markers, indicating that hDPSCs were positive for CD29 (99.45%), CD44 (99.70%), and HLA-ABC (98.65%), but negative for CD31 (0.50%) and CD45 (1.95%) (Fig. 1C). These results demonstrated that hDPSCs in this study showed MSCs morphology, conserved multidirectional differentiation ability, and exhibited MSCs markers expression.
Figure 1.
hDPSCs showed multidirectional differentiation ability and expressed MSCs markers. (A) The typical MSCs morphology of hDPSCs. (B) The multidirectional induction was performed with osteogenesis, adipogenesis, and chondrogenesis inducing medium. (C) hDPSCs were positive for CD29, CD44, and HLA-ABC, while negative for CD31 and CD45. hDPSCs: human dental pulp stem cells; MSCs: mesenchymal stem cells.
Knockdown of circRFWD2 Inhibited Osteogenic Differentiation of hDPSCs
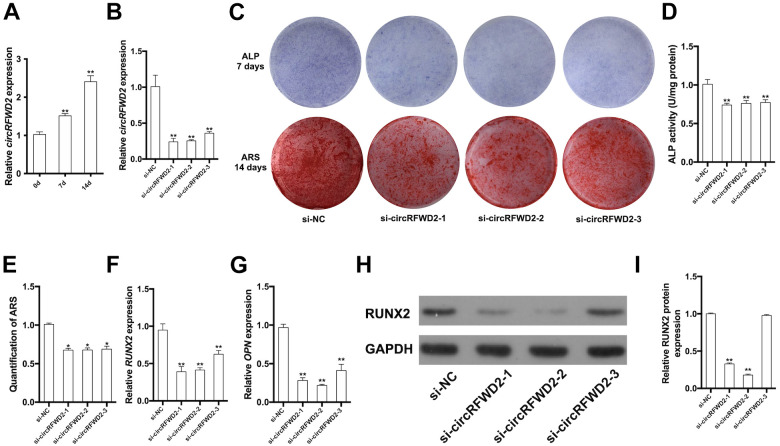
It was showed that circRFWD2 was up-regulated after 7-day and 14-day osteogenic induction (Fig. 2A). Cell transfection was performed to knock down the expression of circRFWD2 for determining its biological effect on osteogenesis of hDPSCs. Three si-RNA plasmids were constructed to avoid the off-targets effect during transfection. We used qRT-PCR analysis to detect the expression of circRFWD2, finding that circRFWD2 was down-regulated by approximately 76%, 74%, and 60% in three si-circRFWD2 groups (Fig. 2B). After osteogenic induction for 7 days, ALP staining and activity were both attenuated in si-circRFWD2 groups (Fig. 2C, D). Similarly, the intensity and quantification of ARS staining were decreased in si-circRFWD2 groups, as a result of impaired matrix mineralization (Fig. 2C, E). qRT-PCR results demonstrated that the relative mRNA expressions of RUNX2 and OPN declined significantly in si-circRFWD2 groups (Fig. 2F–G). Meanwhile, the results of Western blot indicated that RUNX2 protein level was down-regulated when circRFWD2 was knocked down (Fig. 2H–I).
Figure 2.
Knockdown of circRFWD2 inhibited osteogenic differentiation of hDPSCs. (A) Relative expression of circRFWD2 after osteogenic induction for 0, 7, and 14 days. (B) qRT-PCR was performed to determine the expression of circRFWD2 in si-NC and si-circRFWD2 groups. (C) ALP staining and ARS staining in si-NC and si-circRFWD2 groups. (D) ALP activity was measured by spectrophotometry and normalized to the si-NC group. (E) The quantity of ARS staining was detected by densitometry at 562 nm. (F) Relative expression of RUNX2 mRNA in si-NC and si-circRFWD2 groups. (G) Relative expression of OPN mRNA in si-NC and si-circRFWD2 groups. (H) Western blot analysis of RUNX2 in si-NC and si-circRFWD2 groups. (I) Quantity of western blot analysis of RUNX2 in si-NC and si-circRFWD2 groups. GAPDH is a protein-loading control. All experiments were repeated at least three times. The data are shown as the mean ± SD (n = 3). *P < 0.05, **P < 0.01. ALP: Alkaline phosphatase; ARS: Alizarin Red S; si-circRFWD2: small interfering RNAs targeting circRFWD2; si-NC: small interfering RNA negative control.
miR-6817-5p Suppressed Osteogenic Differentiation of hDPSCs
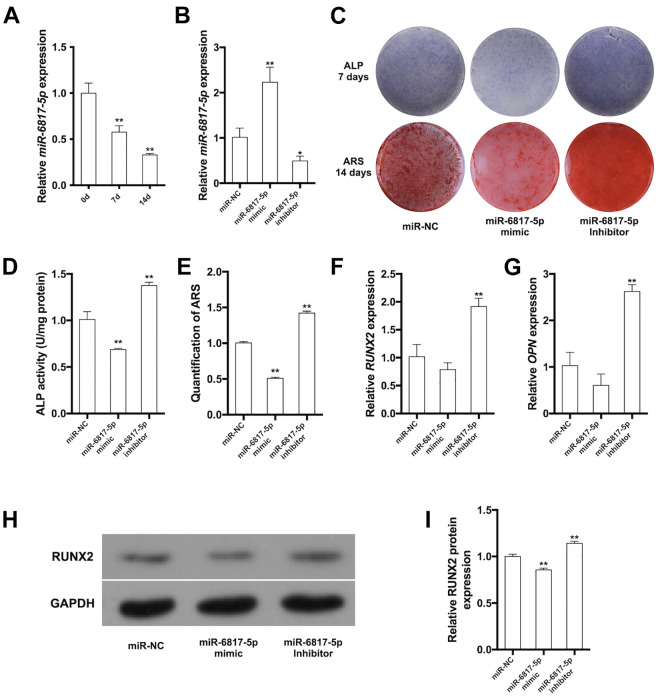
The expression levels of miR-6817-5p were decreased from day 0 to day 14 after osteogenic induction (Fig. 3A). The hDPSCs were incubated with miR-6817-5p mimic or miR-6817-5p inhibitor for 2 days to over- or low-express miR-6817-5p, respectively. The results of qRT-PCR showed that miR-6817-5p increased to more than 2-fold in miR-6817-5p mimic group, while it was declined by approximately 50% in miR-6817-5p inhibitor group (Fig. 3B). After 7 days of osteogenic induction, ALP staining and activity showed decreased osteogenesis ability in the miR-6817-5p mimic group, while osteogenesis ability was increased in the miR-6817-5p inhibitor group (Fig. 3C, D). A similar trend was observed in the intensity and quantification of ARS staining after a 14-day induction (Fig. 3C, E). qRT-PCR analysis was performed to detect the mRNA expression levels of osteogenic markers, showing that the mRNA expression levels of RUNX2 and OPN were decreased in the miR-6817-5p mimic group, while they were increased in the miR-6817-5p inhibitor group (Fig. 3F, G). In accordance, miR-6817-5p overexpression could inhibit RUNX2 protein expression, while miR-6817-5p knockdown promoted the protein level of RUNX2(Fig. 3H, I). These results supported that miR-6817-5p could suppress osteogenic differentiation of hDPSCs.
Figure 3.
miR-6817-5p suppressed osteogenic differentiation of hDPSCs. (A) Relative expression of miR-6817-5p after osteogenic induction for 0, 7, and 14 days. (B) qRT-PCR was performed to determine the expression of miR-6817-5p in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (C) ALP staining and ARS staining in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (D) ALP activity in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (E) The quantity of ARS staining at 562 nm in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (F) Relative expression of RUNX2 mRNA in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (G) Relative expression of OPN mRNA in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (H) Western blot analysis of RUNX2 in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (I) Quantity of western blot analysis of RUNX2 in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. GAPDH is a protein-loading control. All experiments were repeated at least three times. The data are shown as the mean ± SD (n = 3). *P < 0.05, **P < 0.01. ALP: Alkaline phosphatase; ARS: Alizarin Red S; NC: negative control.
circRFWD2 Could Talk with miR-6817-5p Through BMPR2
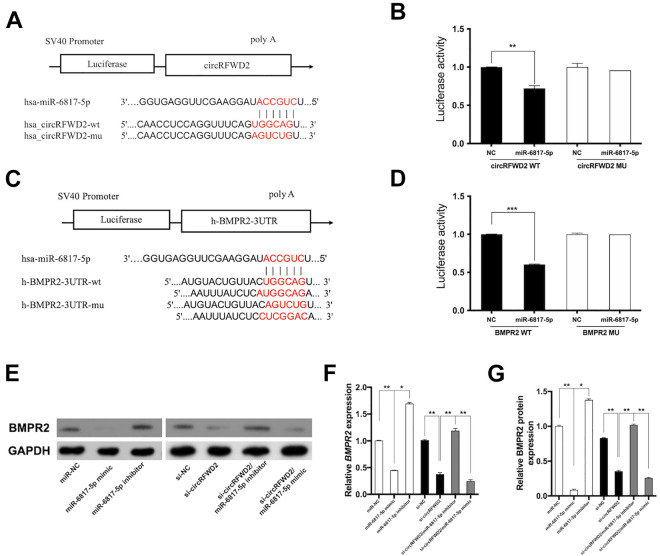
The binding sites between circRFWD2 and miR-6817-5p were confirmed by dual luciferase reporter assay. The mutated sequences of the miR-6817-5p binding site were designed in the circRFWD2 mutant-type (circRFWD2 MU) reporter. The relative luciferase activity in circRFWD2 wild type declined significantly, while the relative luciferase activity in circRFWD2 MU remained stable (Fig. 4A, B). These results suggested circRFWD2 could bind to miR-6817-5p. Similarly, TargetScanhuman 7.2 database was used to predict the potential target genes of miR-6817-5p, which showed that the 3’ untranslated region (UTR) of BMPR2 may bind to miR-6817-5p. Therefore, we constructed two luciferase reporter plasmids to confirm the binding sites between miR-6817-5p and BMPR2. We designed mutated sequences of the miR-6817-5p binding site in the mutant-type (MU) reporter, where the relative luciferase activity of BMPR2 remained stable (Fig. 4C, D). These results confirmed miR-6817-5p could bind to BMPR2 directly.
Figure 4.
circRFWD2 could talk with miR-6817-5p through BMPR2. (A) The putative binding sites between miR-6817-5p and circRFWD2 was displayed. (B) 40 ng circRFWD2 luciferase reporter plasmids with 100 nM miR-NC or miR-6817-5p mimic were co-transfected into HEK 293 T cells, luciferase assays were conducted 48 h after transfection. (C) The putative binding sites of miR-6817-5p to the 3’ untranslated region of BMPR2 predicted by TargetScanhuman 7.2 database, and the mutation site in the 3’ UTR of BMPR2 was shown. (D) 40 ng BMPR2 luciferase reporter plasmids with 100 nM miR-NC or miR-6817-5p mimic were co-transfected into HEK 293 T cells, luciferase assays were conducted 48 h after transfection. (E) Western blot analysis of BMPR2 in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor, si-NC, si-circRFWD2, si-circRFWD2/miR-6817-5p inhibitor, and si-circRFWD2/miR-6817-5p mimic groups. (F) The mRNA levels of BMPR2 in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor, si-NC, si-circRFWD2, si-circRFWD2/miR-6817-5p inhibitor, and si-circRFWD2/miR-6817-5p mimic groups. (G) The protein levels of BMPR2 in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor, si-NC, si-circRFWD2, si-circRFWD2/miR-6817-5p inhibitor, and si-circRFWD2/miR-6817-5p mimic groups. GAPDH is a protein-loading control. All experiments were repeated at least three times. The data are shown as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001. si-circRFWD2: small interfering RNAs targeting circRFWD2; si-NC: small interfering RNA negative control; NC: negative control.
To further confirm whether BMPR2 could be regulated by circRFWD2 and miR-6817-5p, cell transfection was conducted to control the expression of miR-6817-5p and circRFWD2. The results of qRT-PCR and Western blot showed that knockdown of circRFWD2 could suppress expression of BMPR2 both at mRNA and protein levels (Fig. 4E–G). Consistently, miR-6817-5p mimic decreased the mRNA and protein levels of BMPR2 (Fig. 4E–G), while miR-6817-5p inhibitor could promote the expression of BMPR2 (Fig. 4E–G).
Cell co-transfection was used to explore how the crosstalk of circRFWD2 and miR-6817-5p affected the expression of BMPR2. The results showed that the mRNA and protein levels of BMPR2 declined significantly in si-circRFWD2 and miR-6817-5p mimic group, when compared with those in si-circRFWD2 group (Fig. 4E–G). In contrast, the mRNA and protein levels of BMPR2 were increased significantly in si-circRFWD2 and miR-6817-5p inhibitor group (Fig. 4E–G). Taken together, BMPR2 may serve as the target of miR-6817-5p, and the cross-talk between circRFWD2 and miR-6817-5p could function via BMPR2.
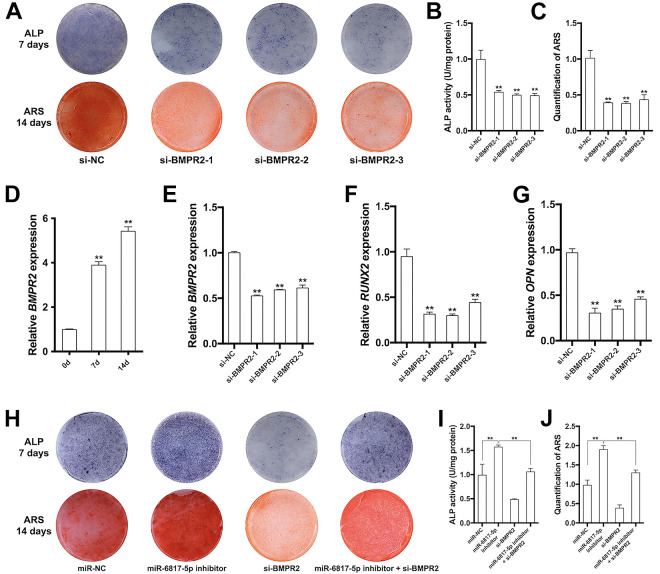
Silence of BMPR2 inhibited osteogenic differentiation of hDPSCs and miR-6817-5p inhibitor partially reversed the inhibition effect of si-BMPR2 on osteogenesis
After 7-day and 14-day osteogenic induction, the mRNA expression levels of BMPR2 were up-regulated significantly (Fig. 5D). hDPSCs transfected with si-BMPR2 were further used to investigate the effect of BMPR2 on osteogenic differentiation of hDPSCs. The mRNA expression of BMPR2 decreased by approximately 50% in si-BMPR2 groups when compared to negative control, indicating the gene knock-down was successful (Fig. 5E). ALP and ARS staining on both quality and quantity levels also showed the weaken osteogenesis ability in si-BMPR2 groups (Fig. 5A–C). The relative mRNA expression of osteogenic markers, including RUNX2 and OPN, declined significantly in si-BMPR2 groups (Fig. 5F–G).
Figure 5.
Silence of BMPR2 inhibited osteogenic differentiation of hDPSCs and miR-6817-5p inhibitor partially reversed the inhibition effect of si-BMPR2 on osteogenesis. (A) ALP staining and ARS staining in si-NC and si-BMPR2 groups. (B) ALP activity in si-NC and si-BMPR2 groups. (C) The quantity of ARS staining at 562 nm in si-NC and si-BMPR2 groups. (D) Relative expression of BMPR2 mRNA after osteogenic induction for 0, 7, and 14 days. (E) Relative expression of BMPR2 mRNA in si-NC and si-BMPR2 groups. (F) Relative expression of RUNX2 mRNA in si-NC and si-BMPR2 groups. (G) Relative expression of OPN mRNA in si-NC and si-BMPR2 groups. (H) ALP staining and ARS staining in si-NC, miR-6817-5p inhibitor, si-BMPR2, and miR-6817-5p inhibitor/si-BMPR2 groups. (I) ALP activity in si-NC, miR-6817-5p inhibitor, si-BMPR2, and miR-6817-5p inhibitor/si-BMPR2 groups. (J) The quantity of ARS staining at 562 nm in si-NC, miR-6817-5p inhibitor, si-BMPR2, and miR-6817-5p inhibitor/si-BMPR2 groups. All experiments were repeated at least three times. The data are shown as the mean ± SD (n = 3). *P < 0.05, **P < 0.01. ALP: Alkaline phosphatase; ARS: Alizarin Red S; NC: negative control.
To investigate whether miR-6817-5p affect osteogenesis of hDPSCs via BMPR2, we utilized cell co-transfection including miR-6817-5p inhibitor and si-BMPR2. After osteogenesis induction for 7 and 14 days, ALP and ARS staining suggested that miR-6817 inhibitor partially reversed the inhibition effect of si-BMPR2 on osteogenesis (Fig. 5H–J).
circRFWD2/miR-6817-5p/BMPR2 Axis Could Regulate the Osteogenesis of hDPSCs Through Smad5 and p38 Phosphorylation
Smad5 served as the essential transcript factor and p38 is an important kinase of BMPs family. To investigate the specific molecular signaling pathways involved in this study, we selected Smad5 and p38 as the potential candidates. Compared to control group, p-Smad5 was decreased significantly in si-circRFWD2 (Fig. 6A, B), miR-6817-5p mimic groups (Fig. 6C, D), and si-BMPR2 (Fig. 6E, F), whereas increased in the miR-6817-5p inhibitor group (Fig. 6C, D). Similarly, phosphorylation of p38 (p-p38) was showed to decline significantly in si-circRFWD2 (Fig. 6A, B), miR-6817-5p mimic groups (Fig. 6C, D), and si-BMPR2 (Fig. 6E, F), while it was increased in miR-6817-5p inhibitor group (Fig. 6C, D). These results demonstrated that circRFWD2/miR-6817-5p/BMPR2 axis may function by activating p-Smad5 and p-p38. The Schematic model showed that miR-6817-5p, targeted by circRFWD2, suppressed osteogenic differentiation through the BMPR2-mediated activation of pSmad5 and p38 MAPK pathway (Fig. 6G).
Figure 6.
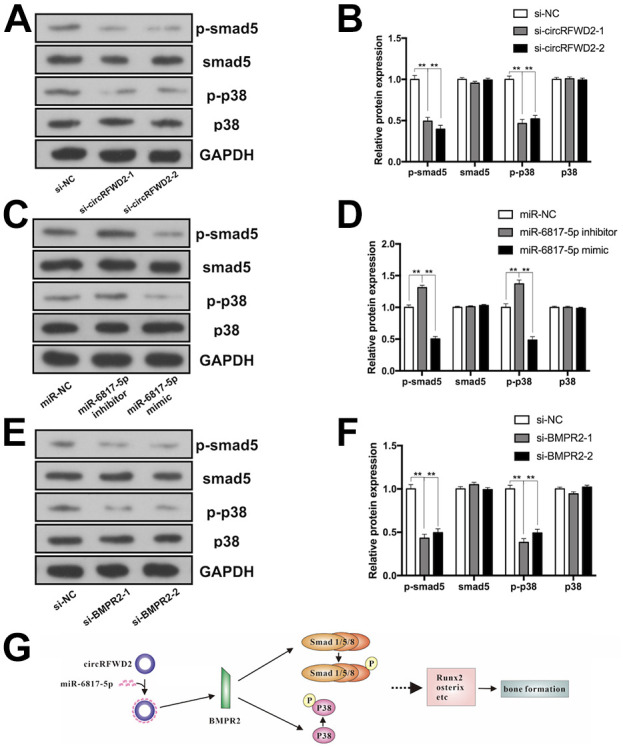
circRFWD2/miR-6817-5p/BMPR2 axis could regulate the osteogenesis of hDPSCs through Smad5 and p38 phosphorylation. (A) Western blot analyses of phosphorylated Smad5 (p-Smad5), Smad5, phosphorylated p-38 (p-p38), p38, and the internal control GAPDH in si-NC and si-circRFWD2 groups. (B) The protein levels of p-Smad5, Smad5, p-p38, p38, and GAPDH in si-NC and si-circRFWD2 groups. (C) Western blot analyses of p-Smad5, Smad5, p-p38, p38, and GAPDH in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (D) The protein levels of p-Smad5, Smad5, p-p38, p38, and GAPDH in miR-NC, miR-6817-5p mimic, and miR-6817-5p inhibitor groups. (E) Western blot analyses of p-Smad5, Smad5, p-p38, p38, and GAPDH in si-NC and si-BMPR2 groups. (F) The protein levels of p-Smad5, Smad5, p-p38, p38, and GAPDH in si-NC and si-BMPR2 groups. (G) Schematic model showed that miR-6817-5p, targeted by circRFWD2, suppressed osteogenic differentiation through the BMPR2-mediated activation of pSmad5 and p38 MAPK pathway. All experiments were repeated at least three times. The data are shown as the mean ± SD (n = 3). **P < 0.01. NC: negative control; p-Smad5: phosphorylated Smad5; p-p38: phosphorylated p-38.
Discussion
The treatment of bone defects by bone tissue engineering requires numerous stem cells. Although MSCs can be expanded in vitro, their ability of self-renewing and proliferation is decreased during passaging 21 . Additionally, bone defects are not terminal diseases typically and treatment should balance risks and benefits simultaneously. Therefore, the best approach to acquiring MSCs should meet the requirements of sufficient quantity, painless and straightforward sampling, and low risk of complications. DPSCs are multipotent MSCs which are can differentiate into a variety of cell lineages, such as osteoblasts, chondrocytes, and adipocytes, in response to stimulation by multiple environmental factors 22,23 . It is showed that BMSCs have a longer population doubling time (PDT) than DPSCs during a 10-day period and that DPSCs have greater proliferative ability than BMSCs 24 . Moreover, DPSCs could be easily isolated from extracted teeth such as orthodontic teeth and third molars, which are usually discarded 25 . Therefore, DPSCs are one kind of promising cell sources in bone tissue engineering and further exploration are required to understand more deeply the specific regulation of DPSCs osteogenesis.
In this study, we investigated the functional roles and underlying mechanisms of circRFWD2 during osteogenesis of DPSCs for the first time. The results showed that both of ALP staining and activity, as well as the intensity and quantification of ARS staining, were decreased in si-circRFWD2 groups. Similarly, mRNA and protein expression levels of osteogenic markers were down-regulated when circRFWD2 was knocked down. These results suggested that knockdown of circRFWD2 significantly retarded osteoblast differentiation of DPSCs.
Several circRNAs have been showed to prevent miRNAs efficiently to regulate the expression levels of mRNAs, and function as the critical orchestrator of signaling pathway during the osteogenesis 26 . It is reported that circSIPA1L1 functions as the sponge of miR-617 to phosphorylate Smad3, which is responsible for stimulating DPSC osteogenesis 27 . CircAKT3 sponges miR-206 and arrests the negative effect of miR-206 on CX43 expression, and thus serves as a positive regulator during osteogenic differentiation of DPSCs 19 . CircLPAR1 could eliminate the inhibitive effect of hsa-miR-31 on osteogenesis by competently bind to hsa-miR-31, which promotes osteogenesis of the recipient homotypic DPSCs 28 . Our study showed that circRFWD2 could talk with miR-6817-5p through BMPR2. MiR-6817-5p could bind with BMPR2 and decrease expression of BMPR2 both at mRNA and protein levels, while knockdown of circRFWD2 could down-regulate BMPR2. Cell co-transfection was used to explore how the crosstalk of circRFWD2 and miR-6817-5p affected the expression of BMPR2. The results showed that the mRNA and protein levels of BMPR2 declined significantly in si-circRFWD2 and miR-6817-5p mimic group, while the expression levels of BMPR2 were increased significantly in si-circRFWD2 and miR-6817-5p inhibitor group. Our study further showed that miR-6817-5p could suppress osteogenic differentiation of hDPSCs. Silence of BMPR2 inhibited osteogenic differentiation of hDPSCs and miR-6817-5p inhibitor partially reversed the inhibition effect of si-BMPR2 on osteogenesis.
BMPR2 is a transmembrane serine/threonine kinase receptor which forms a heterodimeric complex with BMP receptor type 1 29 . BMPs bind to the preformed heterocomplexes to phosphorylate R-Smad proteins, including Smad1, Smad5, and Smad8. Subsequently, the phosphorylated Smad1/5/8 have a high affinity for the common-mediator Smad (Co-Smad), Smad4. The resulting complex serves as a transcription activator, which translocates into the nuclei and activates the expression of corresponding target genes to regulate osteogenic gene expression 30 . BMP–Smad signaling could enhance almost every step during osteogenesis 31 . In addition to this Smad-dependent signaling pathway, p38-mitogen activated protein kinase (p38MAPK) signaling cascade could be activated by BMP-induced signaling complexes to affect transcriptional responses 32 . MAPK pathway is showed to phosphorylate Runx2 and Osterix to promote their transcriptional activity and regulate MSC differentiation positively 33 . It is reported that p38 could perform a negative feedback on canonical BMP-Smad signaling, which contributes to a tight regulated network of osteogenesis 34 . In our study, the results showed that circRFWD2 and BMPR2 could up-regulate the expression level of p-Smad5 and p-p38, while miR-6817-5p suppressed the phosphorylation of p38 and Smad5. Therefore, it is suggested that circRFWD2/miR-6817-5p/BMPR2 axis regulated the osteogenesis of hDPSCs through BMP-Smad and p38 MAPK pathway.
Conclusions
In conclusion, silencing of circRFWD2 was showed to suppress osteogenesis of DPSCs. Mechanistically, miR-6817-5p suppressed osteogenic differentiation of DPSCs, and circRFWD2 could talk with miR-6817-5p through BMPR2. MiR-6817-5p served as a sponge of BMPR2, which regulated BMP-Smad and p38 MAPK pathway to impact DPSCs osteogenesis activity. Thus, this study reveals novel mechanisms of osteogenic differentiation of DPSCs and suggests potential therapeutic methods for bone defects regeneration.
Supplemental Material
Supplemental Material, sj-doc-1-cll-10.1177_09636897211052959 for CircRFWD2 Promotes Osteogenic Differentiation of human Dental Pulp Stem Cells by Targeting miR-6817-5p Through BMP-Smad and p38 MAPK Pathway by Xinqi Huang, Xuefeng Pan, Bo Zhang, Wei Huang, Xiao Cen, Jun Liu and Zhihe Zhao in Cell Transplantation
Footnotes
Authors’ contributions: JL, ZZ, and XC designed the research; XH, XP, WH, and BZ conducted the research; XH and XC contributed to figures and linguistic revision; XH and XC wrote the paper. All authors have approved the final manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval: This study was approved by the Ethics Committee at West China School of Stomatology, Sichuan University (Chengdu, China).
Statement of Human and Animal Rights: Human tissues involved in the study were harvested in accordance with the Committee on Human Research of the West China Hospital of Stomatology of Sichuan University (Approval Number: WCHSIRB-D- 2017-277) approved protocols. The animal studies were approved by the Panel on Laboratory Animal Care of the State Key Laboratory of Oral Diseases, Sichuan University (WCHSIRB-D-2017-201).
Statement of Informed Consent: Written informed consent was obtained from the patients for their anonymized information to be published in this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Natural Science Foundation of China (81771048;81900981;81870743), China Postdoctoral Science Foundation (2019M663530), Sichuan Science and Technology Program (2020YFS0170;2021YJ0149), Research and Development Foundation of West China Hospital of Stomatology (RD-02-201904), and Research Funding from West China School/Hospital of Stomatology Sichuan University (RCDWJS2020-18).
ORCID iDs: Xinqi Huang  https://orcid.org/0000-0002-2762-2438
https://orcid.org/0000-0002-2762-2438
Supplemental Material: Supplemental material for this article is available online.
References
- 1. García-Gareta E, Coathup MJ, Blunn GW. Osteoinduction of bone grafting materials for bone repair and regeneration. Bone. 2015;81:112–121. [DOI] [PubMed] [Google Scholar]
- 2. Kumar BP, Venkatesh V, Kumar KA, Yadav BY, Mohan SR. Mandibular reconstruction: overview. J Maxillofac Oral Surg. 2016;15(4):425–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cowan CM, Shi YY, Aalami OO, Chou YF, Mari C, Thomas R, Quarto N, Contag CH, Wu B, Longaker MT. Adipose-derived adult stromal cells heal critical-size mouse calvarial defects. Nat Biotechnol. 2004;22(5):560–567. [DOI] [PubMed] [Google Scholar]
- 4. Stegen S, van Gastel N, Eelen G, Ghesquière B, D’Anna F, Thienpont B, Goveia J, Torrekens S, Van Looveren R, Luyten FP. HIF-1α Promotes glutamine-mediated redox homeostasis and glycogen-dependent bioenergetics to support postimplantation bone cell survival. Cell Metab 2016;23(2):265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Narita T, Shintani Y, Ikebe C, Kaneko M, Campbell NG, Coppen SR, Uppal R, Sawa Y, Yashiro K, Suzuki K. The use of scaffold-free cell sheet technique to refine mesenchymal stromal cell-based therapy for heart failure. Mol Ther. 2013;21(4):860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi Y, Wang Y, Li Q, Liu K, Hou J, Shao C, Wang Y. Immunoregulatory mechanisms of mesenchymal stem and stromal cells in inflammatory diseases. Nat Rev Nephrol. 2018;14(8):493–507. [DOI] [PubMed] [Google Scholar]
- 7. Graziano A, d’Aquino R, Laino G, Papaccio G. Dental pulp stem cells: a promising tool for bone regeneration. Stem Cell Rev. 2008;4(1):21–26. [DOI] [PubMed] [Google Scholar]
- 8. Shen WC, Lai YC, Li LH, Liao K, Lai HC, Kao SY, Wang J, Chuong CM, Hung SC. Methylation and PTEN activation in dental pulp mesenchymal stem cells promotes osteogenesis and reduces oncogenesis. Nat Commun. 2019;10(1):2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ai T, Zhang J, Wang X, Zheng X, Qin X, Zhang Q, Li W, Hu W, Lin J, Chen F. DNA methylation profile is associated with the osteogenic potential of three distinct human odontogenic stem cells. Signal Transduct Target Ther. 2018;3(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aurrekoetxea M, Garcia-Gallastegui P, Irastorza I, Luzuriaga J, Uribe-Etxebarria V, Unda F, Ibarretxe G. Dental pulp stem cells as a multifaceted tool for bioengineering and the regeneration of craniomaxillofacial tissues. Front Physiol. 2015;6:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. De Colli M, Radunovic M, Zizzari VL, Di Giacomo V, Di Nisio C, Piattelli A, Guirado JL, Zavan B, Cataldi A, Zara S. Osteoblastic differentiating potential of dental pulp stem cells in vitro cultured on a chemically modified microrough titanium surface. Dent Mater J. 2018;37(2):197–205. [DOI] [PubMed] [Google Scholar]
- 12. Rapino M, Di Valerio V, Zara S, Gallorini M, Marconi GD, Sancilio S, Marsich E, Ghinassi B, di Giacomo V, Cataldi A. Chitlac-coated thermosets enhance osteogenesis and angiogenesis in a co-culture of dental pulp stem cells and endothelial cells. Nanomaterials (Basel). 2019;9(7):928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. [DOI] [PubMed] [Google Scholar]
- 15. Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;166(4):1055–1056. [DOI] [PubMed] [Google Scholar]
- 16. Chia W, Liu J, Huang YG, Zhang C. A circular RNA derived from DAB1 promotes cell proliferation and osteogenic differentiation of BMSCs via RBPJ/DAB1 axis. Cell Death Dis. 2020;11(5):372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang X, Cen X, Zhang B, Liao Y, Zhu G, Liu J, Zhao Z. Prospect of circular RNA in osteogenesis: a novel orchestrator of signaling pathways. J Cell Physiol. 2019;234(12):21450–21459. [DOI] [PubMed] [Google Scholar]
- 18. Huang X, Cen X, Zhang B, Liao Y, Zhao Z, Zhu G, Zhao Z, Liu J. The roles of circRFWD2 and circINO80 during NELL-1-induced osteogenesis. J Cell Mol Med. 2019;23(12):8432–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang B, Huo S, Cen X, Pan X, Huang X, Zhao Z. circAKT3 positively regulates osteogenic differentiation of human dental pulp stromal cells via miR-206/CX43 axis. Stem Cell Res Ther. 2020;11(1):531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun Y, Luo T, Shen Y, Haapasalo M, Zou L, Liu J. Effect of iroot fast set root repair material on the proliferation, migration and differentiation of human dental pulp stem cells in vitro. PLoS One. 2017;12(10):e0186848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan XZ, van den Beucken JJ, Both SK, Yang PS, Jansen JA, Yang F. Biomaterial strategies for stem cell maintenance during in vitro expansion. Tissue Eng Part B Rev. 2014;20(4):340–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu J, He H, Tang C, Zhang G, Li Y, Wang R, Shi J, Jin Y. Differentiation potential of STRO-1+ dental pulp stem cells changes during cell passaging. BMC Cell Biol. 2010;11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ferrarotti F, Romano F, Gamba MN, Quirico A, Giraudi M, Audagna M, Aimetti M. Human intrabony defect regeneration with micrografts containing dental pulp stem cells: a randomized controlled clinical trial. J Clin Periodontol. 2018;45(7):841–850. [DOI] [PubMed] [Google Scholar]
- 24. Kunimatsu R, Nakajima K, Awada T, Tsuka Y, Abe T, Ando K, Hiraki T, Kimura A, Tanimoto K. Comparative characterization of stem cells from human exfoliated deciduous teeth, dental pulp, and bone marrow-derived mesenchymal stem cells. Biochem Biophys Res Commun. 2018;501(1):193–198. [DOI] [PubMed] [Google Scholar]
- 25. Ge X, Li Z, Jing S, Wang Y, Li N, Lu J, Yu J. Parathyroid hormone enhances the osteo/odontogenic differentiation of dental pulp stem cells via ERK and P38 MAPK pathways. J Cell Physiol. 2020;235(2):1209–1221. [DOI] [PubMed] [Google Scholar]
- 26. Panda AC. Circular RNAs Act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- 27. Ge X, Li Z, Zhou Z, Xia Y, Bian M, Yu J. Circular RNA SIPA1L1 promotes osteogenesis via regulating the miR-617/Smad3 axis in dental pulp stem cells. Stem Cell Res Ther. 2020;11(1):364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie L, Guan Z, Zhang M, Lyu S, Thuaksuban N, Kamolmattayakul S, Nuntanaranont T. Exosomal circLPAR1 promoted osteogenic differentiation of homotypic dental pulp stem cells by competitively binding to hsa-miR-31. Biomed Res Int. 2020;2020:6319395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heining E, Bhushan R, Paarmann P, Henis YI, Knaus P. Spatial segregation of BMP/Smad signaling affects osteoblast differentiation in C2C12 cells. PLoS One. 2011;6(10):e25163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guzman A, Zelman-Femiak M, Boergermann JH, Paschkowsky S, Kreuzaler PA, Fratzl P, Harms GS, Knaus P. SMAD versus non-SMAD signaling is determined by lateral mobility of bone morphogenetic protein (BMP) receptors. J Biol Chem. 2012;287(47):39492–39504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu M, Chen G, Li YP. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sánchez-Duffhues G, Hiepen C, Knaus P, Ten Dijke P. Bone morphogenetic protein signaling in bone homeostasis. Bone 2015;80:43–59. [DOI] [PubMed] [Google Scholar]
- 33. Lee KS, Hong SH, Bae SC. Both the Smad and p38 MAPK pathways play a crucial role in Runx2 expression following induction by transforming growth factor-beta and bone morphogenetic protein. Oncogene. 2002;21(47):7156–7163. [DOI] [PubMed] [Google Scholar]
- 34. Ortuño MJ, Ruiz-Gaspà S, Rodríguez-Carballo E, Susperregui AR, Bartrons R, Rosa JL, Ventura F. p38 regulates expression of osteoblast-specific genes by phosphorylation of osterix. J Biol Chem. 2010;285(42):31985–31994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-doc-1-cll-10.1177_09636897211052959 for CircRFWD2 Promotes Osteogenic Differentiation of human Dental Pulp Stem Cells by Targeting miR-6817-5p Through BMP-Smad and p38 MAPK Pathway by Xinqi Huang, Xuefeng Pan, Bo Zhang, Wei Huang, Xiao Cen, Jun Liu and Zhihe Zhao in Cell Transplantation