ABSTRACT
Zika virus (ZIKV) is a mosquito-borne pathogen classified by the World Health Organization (WHO) as a public health emergency of international concern in 2016, and it is still identified as a priority disease. Although most infected individuals are asymptomatic or show mild symptoms, a risk of neurologic complications is associated with infection in adults. Additionally, infection during pregnancy is directly linked to microcephaly and other congenital malformations. Since there are no currently available vaccines or approved therapeutics for this virus, there is a critical unmet need in developing treatments to prevent future ZIKV outbreaks. Toward this end, we performed a large-scale cell-based high-content screen of 51,520 chemical compounds to identify potential antiviral drug candidates. The compound (2E)-N-benzyl-3-(4-butoxyphenyl)prop-2-enamide (SBI-0090799) was found to inhibit replication of multiple ZIKV strains and in different cell systems. SBI-0090799 did not affect viral entry or RNA translation but suppressed RNA replication by preventing the formation of the membranous replication compartment. Selection of drug-resistant viruses identified single-amino-acid substitutions in the N-terminal region of nonstructural protein NS4A, arguing this is the likely drug target. These resistance mutations rescued viral RNA replication and restored the formation of the membranous replication compartment. This mechanism of action is similar to clinically approved NS5A inhibitors for hepatitis C virus (HCV). Taken together, SBI-0090799 represents a promising lead candidate for the development of an antiviral treatment against ZIKV infection for the mitigation of severe complications and potential resurgent outbreaks of the virus.
IMPORTANCE This study describes the elucidation of (2E)-N-benzyl-3-(4-butoxyphenyl)prop-2-enamide (SBI-0090799) as a selective and potent inhibitor of Zika virus (ZIKV) replication using a high-throughput screening approach. Mapping and resistance studies, supported by electron microscopy observations, indicate that the small molecule is functioning through inhibition of NS4A-mediated formation of ZIKV replication compartments in the endoplasmic reticulum (ER). Intriguingly, this defines a novel nonenzymatic target and chemical matter for the development of a new class of ZIKV antivirals. Moreover, chemical modulation affecting this nonstructural protein mirrors the identification and development of hepatitis C virus (HCV) NS5A inhibitor daclatasvir and its derivatives, similarly interfering with the formation of the viral replication compartment and also targeting a protein with no enzymatic activity, which have been part of a curative strategy for HCV.
KEYWORDS: antiviral, NS4A, replication compartment, Zika virus, drug discovery
INTRODUCTION
The ongoing SARS-CoV-2 pandemic has highlighted the global threat represented by zoonotic viruses. Emergent and reemergent viruses can significantly impact the health and health care infrastructure of the countries affected and can also cause economic devastation across the globe. Zika virus (ZIKV) is a human mosquito-borne virus responsible for major epidemics in 2015 to 2016, when 87 countries and territories confirmed evidence of ZIKV transmission (1, 2). Although symptoms are generally mild, a risk of neurologic complications (e.g., Guillain-Barré syndrome) is associated with infection in adults (3), and infection during pregnancy can lead to microcephaly and other congenital malformations, referred to as congenital Zika syndrome (4). The World Health Organization (WHO) classified ZIKV and ZIKV-associated diseases as a public health emergency of international concern in 2016 and still identifies Zika as a priority disease (5). Unfortunately, no antiviral treatments or vaccines have been developed and approved to prevent or treat disease caused by this pathogen.
ZIKV is a positive-sense single-stranded RNA virus that belongs to the Flavivirus genus in the Flaviviridae family and is related to dengue virus (DENV), yellow fever virus (YFV), Japanese encephalitis virus (JEV), Kunjin virus (KUNV), and West Nile virus (WNV) (6, 7). The ∼11-kb genome codes for a polyprotein that is proteolytically cleaved by cellular and viral proteases into three structural (capsid protein, premembrane/membrane protein, and envelope glycoprotein) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins (8). The structural proteins constitute the components of the virus particle, protecting the viral genome and representing a major target for antibody-mediated immune responses, while most of the nonstructural proteins are involved in other functions, including restriction of immune response and viral RNA replication (8, 9). RNA replication of flaviviruses occurs in virus-induced vesicle-like membrane rearrangements corresponding to endoplasmic reticulum (ER) invaginations and designated vesicle packets (VPs) (10, 11). These structures contain viral nonstructural proteins and intermediates of viral genome replication (double-stranded RNA) and are the sites where viral RNA replication most likely takes place (10, 11). Nonstructural protein 4A (NS4A) has been described to induce membrane curvatures and, thus, is considered a crucial initiator of membrane rearrangements required for assembly of the viral replicase complexes (12–16).
In this report, using a large-scale high-content screening approach, we identified (2E)-N-benzyl-3-(4-butoxyphenyl)prop-2-enamide (SBI-0090799) as a specific inhibitor of ZIKV replication. It was found to exert antiviral activity through blocking the formation of the vesicle packets. Single point mutations in the N-terminal region of NS4A conferred resistance to the drug and restored formation of vesicle packets, suggesting a mechanism of action mediated by the role of NS4A as an initiator of the de novo formation of the membranous ZIKV replication compartment. Thus, blocking the biogenesis of these membranous replication factories represents a promising antiviral approach against ZIKV.
RESULTS
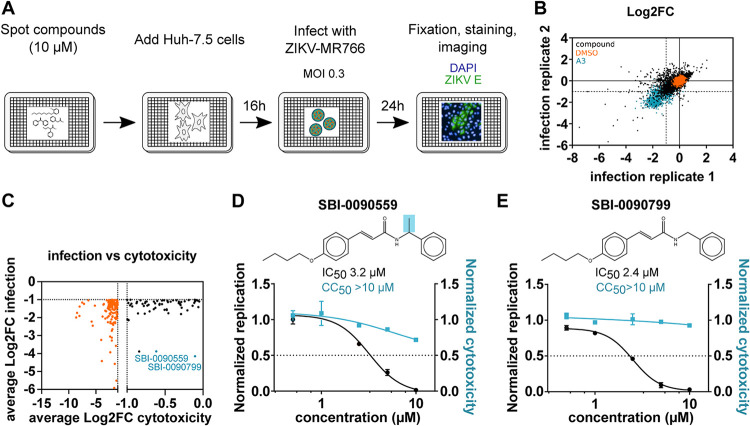
To identify antivirals targeting ZIKV infection, we performed a high-content phenotypic screen of 51,520 chemical compounds covering diverse chemical structures and chemotypes in Huh-7.5 cells infected with the ZIKV strain MR766, as depicted in Fig. 1A. The percentage of infected cells in each well was quantified 24 h postinfection (h.p.i.) after staining using an antibody targeting the envelope protein of the virus. Total cells were identified by 4′,6-diamidino-2-phenylindole (DAPI) staining. The screen was performed in duplicate, and the values corresponding to each well were normalized to the median of each plate. The average correlation coefficient between the two replicates was 0.54 and the average Z′ factor was 0.48, suggesting acceptable sensitivity and reproducibility for such a large chemical compound screen (Fig. 1B; see also Fig. S1A in the supplemental material). Aiming to select only strongly active compounds showing reliable and reproducible antiviral activity as well as limited cytotoxicity, stringent selection criteria were applied. One hundred ninety-eight compounds were identified that reduce the percentage of infection by at least 50% in the average of the replicate screens (Fig. 1B, lower left-hand quadrant). Among these, only 63 did not induce cytotoxicity (<50% compared to the vehicle control) according to the total cell numbers (Fig. 1C, black dots). These selected hits were cherry picked and retested for validation over a range of five concentrations spanning from 625 nM to 10 μM. Antiviral activity was confirmed with 38 of the 63 compounds at the same concentration as the primary screen (10 μM), with only 3 compounds exerting cytotoxicity (Fig. S1B and C). Based on their preliminary dose-response curves, 16 compounds were retested across a broader range of concentrations (Table S1). Compounds (2E)-N-benzyl-3-(4-butoxyphenyl)prop-2-enamide (indicated as SBI-0090799) and (2E)-3-(4-butoxyphenyl)-N-(1-phenylethyl)prop-2-enamide (indicated as SBI-0090559) were selected as the top two candidates, showing the highest ratio of 50% cytotoxic concentration (CC50)/half-maximal inhibitory concentration (IC50) (>3) at the concentrations tested (Fig. 1D and E and 2A, Fig. S2A). Interestingly, they share the same core structure, with SBI-0090559 harboring an additional methyl group (Fig. 1D and E).
FIG 1.
Identification of SBI-0090799 as inhibitor of ZIKV infection through a high-throughput small chemical compound screen. (A) Schematic of the screening strategy used for the identification of chemical compounds showing antiviral activity directed against ZIKV. Huh-7.5 cells were pretreated for 16 h with the compounds (10 μM final concentration) and then infected with ZIKV MR766 (MOI, 0.3). Twenty-four hours after infection, cells were fixed and analyzed by immunofluorescence imaging. For each condition, the percentage of infection was calculated as the ratio of the number of infected cells stained for ZIKV envelope protein to the number of cells stained with DAPI. (B) Correlation plot indicating the log2 fold change (Log2FC) of each compound in the two replicate screens, after normalization to the median of each plate for ZIKV percentage of infection, across all screening plates (black dots). Values corresponding to DMSO (orange dots)- and A3 (cyan dots)-treated wells are also represented. (C) Dot plot indicating the average log2 fold change (Log2FC) of each compound in the two replicate screens, after normalization to the median of each plate, for cell count (cytotox, x axis) versus ZIKV percent infection (infection, y axis) for compounds reaching the criteria of selection of at least 50% inhibition of infection and less than 50% decrease in cell count (black dots). Compounds not meeting the cytotoxicity criteria are indicated by orange dots. The top 2 compounds selected for follow-up studies are indicated in cyan. (D and E) Huh-7.5 cells were pretreated for 16 h with increasing concentrations of SBI-0090559 (D) or SBI-0090799 (E) and then infected with ZIKV MR766 at an MOI of 0.3. Twenty-four hours after infection, cells were fixed and analyzed by immunofluorescence imaging. For each condition, the percentage of infection was calculated as described for panel A. Dose-response curves for infectivity (black) and cell number (cyan) are shown. Data are normalized to the mean for DMSO-treated wells and represent means ± SEM from n = 3 independent experiments. IC50 and CC50 for each compound were calculated using a four-parameter logistic nonlinear regression model and are indicated. Highlighted in blue is the structural difference between compound SBI-0090559 and SBI-0090799.
To identify the key substitutions around the scaffold, 19 analogues were selected in addition to SBI-0090799 (Fig. 2A) and SBI-0090559 (Fig. S2A). The antiviral activity of these molecules was assessed in dose-response infection assays in parallel to cytotoxicity studies (Fig. S2B to T). These structure-activity relationship studies allowed us to identify the active core of the molecule, highlighted in blue in Fig. S2U. Among the 20 chemical analogues tested, compound SBI-0090799 appeared to be the optimal candidate for follow-up studies, showing a selectivity index (SI; CC50/IC50) of >38 (Fig. 2A), an IC50 of 2.1 μM, and an IC90 of 6 μM.
FIG 2.
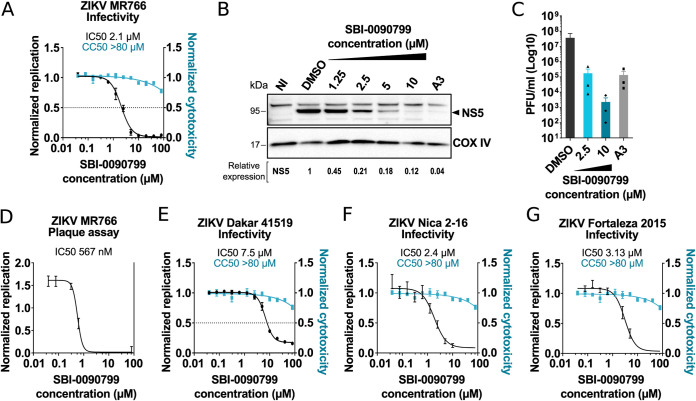
SBI-0090799 inhibits ZIKV strains from both African and Asian lineages. Huh-7.5 cells were pretreated for 16 h with increasing concentrations of the indicated compounds and then infected with ZIKV MR766 (A), ZIKV Dakar 41519 (E), ZIKV Nica 2–16 (F), or ZIKV Fortaleza 2015 (G), at MOIs of 0.3, 0.5, 0.9, or 0.6, respectively. Twenty-four hours after infection, cells were fixed and analyzed by immunofluorescence imaging. For each condition, the percentage of infection was calculated as the ratio of the number of infected cells staining for ZIKV envelope protein to the number of cells stained with DAPI (left axis, black line). In parallel, Huh-7.5 cells were treated with increasing concentrations of compound, and 30 h later cell viability was assessed by measurement of the ATP content (right axis, cyan line). Data represent the means ± SEM from n = 3, normalized to mean values for DMSO-treated wells. The anti-ZIKV IC50 values and the CC50 values were determined by nonlinear regression analysis. (B) Huh-7.5 cells were treated with increasing concentrations of the indicated compounds. Sixteen hours later, cells were infected for 1 h at 37°C with ZIKV MR766 at an MOI of 0.3 in the presence of the compounds. After removal of the inoculum, fresh medium supplemented with the indicated compounds was added to each well. Twenty-four hours later, cells were harvested, and a Western blot was performed. Infection was quantified by immunoblotting of NS5. COXIV was used as a loading control. Relative quantification of NS5 is indicated. (C and D) Huh-7.5 cells were pretreated for 16 h with A3 (10 μM)] or the indicated concentration of SBI-0090799 (C), or 12 increasing concentrations of SBI-0090799 ranging from 39 nM to 80 μM (D), and then infected with ZIKV MR766 at an MOI of 0.3 for 24 h. The released infectious virions were quantified by plaque assay in Vero cells. (C) Data are presented as log10 ± SEM of the concentration of infectious particles (PFU/ml) for n = 3 independent experiments. (D) Data represent the means ± SEM from n = 2 of infectious virions, normalized to mean values for DMSO-treated wells. The anti-ZIKV IC50 value was determined by nonlinear regression analysis.
The antiviral activity of SBI-0090799 in Huh-7.5 cells infected with ZIKV MR766 was then confirmed using orthogonal assays. Quantification of NS5 by Western blotting showed a reduction of this viral protein of ∼80% at a concentration of 2.5 μM (Fig. 2B), consistent with the IC50 calculated by immunostaining. Quantification by plaque assay of infectious viral particles released revealed a >2-log10 reduction in cells treated with SBI-0090799 at 2.5 μM and a >4.5-log10 reduction in cells treated with 10 μM SBI-0090799 (Fig. 2C). The efficacy of SBI-0090799 was then measured by plaque assay over a 12-point dose-response covering a concentration range from 40 nM to 80 μM, yielding an IC50 of 567 nM (SI > 114) (Fig. 2D). This result shows a larger impact of the drug on the release of infectious viral particles than measurement of intracellular accumulation of viral proteins. However, it is worth noting that the design of this experiment captures several cycles of replication, and the dynamic range measurable by plaque assay is wider than that of immunostaining or Western blotting.
The antiviral activity of SBI-0090799 was also assessed using other ZIKV strains belonging to both African (Dakar 41519) (Fig. 2E) and Asian/American lineages (Nica 2–16 and Fortaleza 2015 strains) (Fig. 2F and G), resulting in an SI above 10 for all the strains tested. SBI-0090799 did not show any antiviral activities against other human flaviviruses tested, including DENV-2, JEV, YFV, and KUNV (Fig. S3), suggesting ZIKV-specific inhibition of infection.
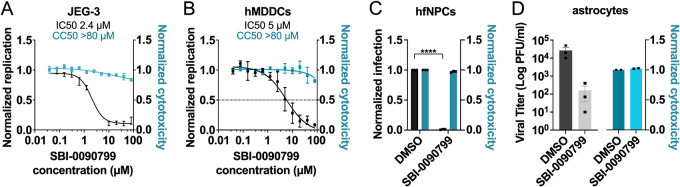
Antiviral activity against ZIKV and cytotoxicity of SBI-0090799 were then assessed in other cell types, including natural cellular targets of the virus (Fig. 3). The compound was shown to inhibit ZIKV with similar efficacy in all cell models tested, including placenta-derived JEG-3 cells (Fig. 3A), human monocyte-derived dendritic cells (hMDDCs) (Fig. 3B), human fetal neural progenitor cells (hfNPCs) (Fig. 3C), and primary human astrocytes (Fig. 3D).
FIG 3.
Antiviral activity of SBI-0090799 in relevant cell lines and primary cells. JEG-3 cells (A), hMDDCs (B), hfNPC (C), or astrocytes (D) were pretreated for 16 h with various concentrations (A and B) or 10 μM (C and D) SBI-0090799 and then infected with ZIKV MR766 (A and B), ZIKV Fortaleza 2015 (C), or ZIKV Nica 2–16 (D) at an MOI of 0.5 (A), 2.5 (B), 1 (C), and 5 (D), respectively. The infection was quantified 24 h (A and B) or 72 h (C) later by immunostaining for the ZIKV envelope protein and nuclear staining with DAPI (A and B; left axes, black line) or 24 h later by plaque assay (D). In parallel, JEG-3 cells (A), hMDDCs (B), or astrocytes (D) were treated with increasing concentrations of SBI-0090799, and 30 h later cell viability was assessed by measurement of the ATP content (A and B, right axis, cyan line) or LDH release (D). (C) Cytotoxicity in hNPCs was evaluated by cell count. Data represent the mean ± SEM from n = 3, normalized to mean values for DMSO-treated wells. ****, P ≤ 0.001. The anti-ZIKV IC50 values and the CC50 values in panels A and B were determined by nonlinear regression analysis.
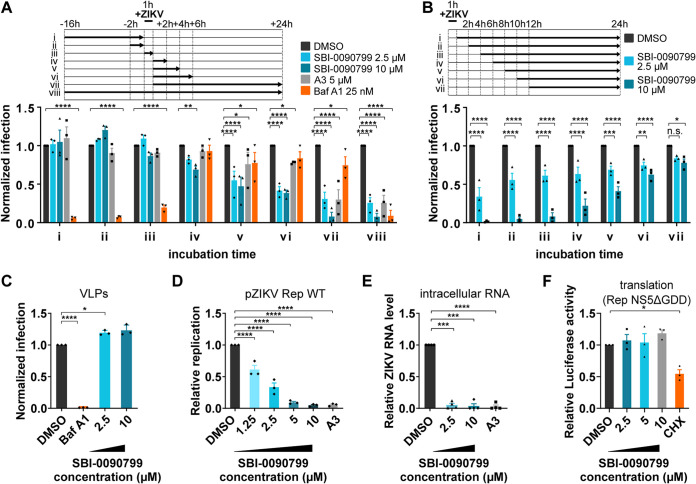
Time-of-addition experiments showed that SBI-0090799 inhibits the infection when added after the viral inoculation (Fig. 4A, iv to viii), with earlier kinetics than the de novo pyrimidine biosynthesis inhibitor A3 (Fig. 4A). Antiviral activity was virtually lost when SB-0090799 was added 12 h after infection, consistent with antiviral impact of the compound on post-entry steps (Fig. 4B). This is also corroborated by the lack of inhibition of virus-like particles (VLPs) bearing ZIKV structural proteins and mimicking virus entry (Fig. 4C).
FIG 4.
Mapping the step of the ZIKV replication cycle targeted by SBI-0090799. (A and B) Time-of-addition assays. To synchronize infection, Huh-7.5 cells were infected for 1 h with ZIKV MR766 (MOI of 0.6), and the inoculum was then removed. Cells were also incubated with the indicated compound at the indicated time points. Infection was quantified 24 h postinoculation after fixation and staining for ZIKV envelope protein. Data are normalized to the mean of DMSO-treated wells for each corresponding time point and are presented as mean ± SEM from n = 3 independent experiments. Bafilomycin A1 and compound A3 were used as positive controls at the indicated concentrations. (C) Reporter virus-like particle (VLP) assay. Huh-7.5 cells were infected for 2 h with ZIKV pseudotyped particles harboring a WNV replicon expressing GFP in the presence of the indicated compounds. Inoculum was removed after 2 h, and fresh medium containing the same compounds was added to the wells. GFP-positive cells were quantified 24 h postinoculation. The percentage of infected cells was calculated as the ratio of the number of GFP-positive cells to number of cells stained with DAPI. (D) Huh-7.5 cells were electroporated with RNA encoding the replicon pZIKV Rep WT. The antiviral activity of SBI-0090799 was assessed 24 h postelectroporation by measuring the Renilla luciferase activity as a readout of viral replication. The graph shows the level of replication (mean ± SEM) relative to the DMSO control. Renilla counts were normalized to 4 h posttransfection values, reflecting electroporation efficiency. (E) Huh-7.5 cells were infected with ZIKV MR766 at an MOI of 0.3 in the presence of the indicated compounds. Inoculum was removed after 1 h, and fresh medium supplemented with the compounds was added to the wells. Cells were harvested 24 h postinfection, and the intracellular RNA was isolated and quantified by RT-qPCR. For each condition, ZIKV RNA levels relative to the DMSO-treated wells are shown for n = 3 biological replicates. RPLP0 was used as the housekeeping gene to normalize for input RNA levels. (F) Huh-7.5 cells were electroporated with RNA encoding the ZIKV replicon Rep-NS5ΔGDD. The antiviral activity of SBI-0090799 was assessed 4 h postelectroporation by measuring the Renilla luciferase activity as readout of viral RNA translation. The graph shows the level of luciferase (mean ± SEM) relative to the DMSO control. Cycloheximide (50 μg/ml) is used as a positive control. Results are presented as means ± SEM (n = 3). Two-way (A and B) or one-way (C to F) analysis of variance followed by Tukey’s or Dunnett’s posttest was performed for statistical analysis. Baf A1, bafilomycin A1; CHX, cycloheximide; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.001; n.s., not significant.
To better assess which post-entry step was targeted by SBI-0090799, Huh-7.5 cells were electroporated with RNA encoding a ZIKV replicon (pZIKV Rep WT, Cambodia strain) lacking the structural region of the virus genome, which is replaced by a Renilla luciferase reporter gene (17), and then incubated either with dimethyl sulfoxide (DMSO), A3, or SBI-0090799. SBI-0090799 inhibited viral replication with an IC50 of 1.64 μM, based upon measurement of luciferase activity 24 h postelectroporation (h.p.e.) (Fig. 4D). Similar results were obtained using a replicon of the H/PF/2013 strain (Fig. S4A and B). In agreement with this observation, quantification of intracellular viral RNA confirmed that SBI-0090799 blocks the accumulation of viral RNA at 24 h.p.i. (Fig. 4E). Attenuation of ZIKV replication observed using ZIKV replicons can be explained either by inhibition of the RNA translation step or by abrogation of RNA replication. To assess if the observed phenotype could be caused by an inhibition of RNA translation, a replicon containing a GDD mutation in the NS5 polymerase active site, making it deficient for replication, was used (17), and the luciferase activity was measured 4 h postelectroporation. No inhibition was observed in cells treated with SBI-0090799 compared to DMSO. Conversely, the inhibitor of translation cycloheximide resulted in a reduction of luciferase activity (Fig. 4F). These data suggest SBI-0090799 inhibits ZIKV RNA replication.
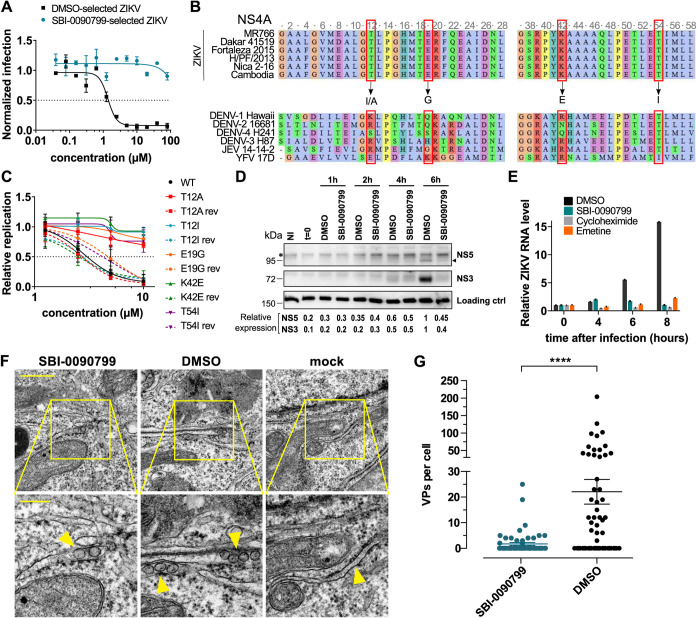
To evaluate the potential mechanism of action of SBI-0090799, we passaged the virus in the presence of DMSO or SBI-0090799 at a concentration corresponding to 5-fold the 50% effective concentration (EC50) of the compound in Huh-7.5 cells. Virus was collected when ∼80% cytopathic effect (CPE) was observed, and the selection was carried out for 10 sequential passages. We observed that as early as passage 2, the virus started developing resistance, and at passage 5, the level of CPE was comparable to the DMSO condition. The replication of the virus selected in the presence of SBI-0090799 at passage 5 was then compared to the virus passaged in the presence of DMSO, confirming its resistance to SBI-0090799 (Fig. 5A, IC50 > 80 μM). The viral RNA from both DMSO- and SBI-0090799-passaged virus was then sequenced in parallel. This analysis identified 33 mutations present only in the SBI-0090799-selected virus (Table S2), of which 4 present with a frequency above 10%. Interestingly, these 4 mutations all appeared to be localized in NS4A, two of which involved the T12 amino acids T12A (19.3%), T12I (10.5%), E19G (19.8%), and K42E (11.1%). Specifically, the mutations targeted 3 amino acid residues localized in the cytoplasmic N-terminal region of NS4A (18) that are conserved among the different ZIKV strains (T12, E19, and K42) but not among the other flaviviruses (Fig. 5B, Fig. S4C). An additional mutation in NS4A with a frequency of 5.7% was identified, affecting amino acid residue T54 (T54A), which localizes in the N-terminal region of the predicted first transmembrane domain (18) (Fig. 5B, Fig. S4C). Interestingly, this amino acid residue was previously reported to be important for NS4A-NS4B interaction in DENV, and a mutation at this position reduced DENV replication (19). The insertion of these 4 mutations in the ZIKV replicon allowed us to confirm that each mutation was sufficient to confer resistance to SBI-0090799, restoring the susceptibility to the drug by reverting the mutation to the wild-type (WT) sequence (rev) without affecting viral fitness (Fig. 5C, Fig. S4D and E). The mutations did not affect polyprotein processing or stability of the viral proteins NS1, NS3, NS4B, and NS5, as determined by using an expression construct (Fig. S4F to I). This system, designated pIRO-Z (Fig. S5A) (20), was used to avoid confounding effects arising from replication inhibition and, thus, reduced protein levels when conducting such analyses with a replicon or viral genome. This observation suggests a mechanism of action of SBI-0090799 related to the role of NS4A in RNA replication. Given the multifunctional nature of NS4A, this could be (i) formation of the replication organelle through induction of membrane rearrangements (12), (ii) induction of autophagy (21), and (iii) repression of RIG-I-like receptor (RLR)-mediated innate immune responses (22). An additional role as a cofactor for NS3 helicase activity has also been recently suggested (23).
FIG 5.
Identification of mutations in NS4A conferring resistance to SBI-0090799 and inhibition of ZIKV replication organelle formation by this compound. (A) Huh-7.5 cells were pretreated for 16 h with increasing concentrations of SBI-0090799 or DMSO and then infected with ZIKV MR766 selected for 5 passages in the presence of either DMSO (black line) or SBI-0090799 (10 μM) (cyan line). Twenty-four hours after infection, cells were fixed and analyzed by immunofluorescence imaging. For each condition, the percentage of infection was calculated as the ratio of the number of infected cells stained for ZIKV envelope protein to the number of cells stained with DAPI. Data represent the means ± SEM from n = 3, normalized to mean values for DMSO-treated wells. Curves were fitted by nonlinear regression analysis. (B) Sequence alignment for NS4A N-terminal region. Red squares indicate the amino acid residues replaced after 5 passages of viral selection in the presence of SBI-0090799. Arrows indicate the replacing residues. (C) Huh-7.5 cells were electroporated with RNA encoding the replicon pZIKV Rep, bearing the indicated mutation in NS4A, or the corresponding sequence revertant to wild type (rev). The antiviral activity of SBI-0090799 was assessed 24 h postelectroporation by measuring the Renilla luciferase activity as a readout of viral replication. Renilla counts at 24 h were normalized to the corresponding values at 4 h posttransfection, reflecting electroporation efficiency. Data represent the means ± SEM from n = 3, with each replicon normalized to its mean values for DMSO-treated wells. Curves were fitted by nonlinear regression analysis. (D) Huh-7.5 cells were infected for 1 h at 37°C with ZIKV MR766 at an MOI of 1. After removal of the inoculum and two washes with PBS, fresh medium supplemented with either DMSO or SBI-0090799 (10 μM) was added to each well. At the indicated time postinfection, cells were harvested and a Western blot was performed. Replication was quantified by immunoblotting of NS3 and NS5, and relative quantification for these two proteins is indicated. Asterisk indicates a nonspecific band recognized by the NS5A antiserum. An additional nonspecific band recognized by the NS5 antiserum is shown as a loading control. (E) Huh-7.5 cells were infected for 1 h at 37°C with ZIKV MR766 at an MOI of 1. After removal of the inoculum, fresh medium supplemented with either DMSO, SBI-0090799 (10 μM), or the RNA translation inhibitors cycloheximide (50 μg/ml) or emetine (2 μM) was added to each well. At the indicated time postinfection, cells were harvested, and intracellular RNA was isolated and quantified by RT-qPCR. For each condition, ZIKV RNA levels relative to the DMSO-treated cells are shown for n = 3 biological replicates. RPLP0 was used as a housekeeping gene. (F and G) SBI-0090799 inhibits ZIKV replication organelle formation. (F) Transmission electron microscopy images of 70-nm sections of resin-embedded Huh7/Lunet-T7 cells transfected for 18 h with pIRO-Z in the presence of either SBI-0090799 (12.5 μM) or DMSO (0.125%, vol/vol). Lower panels are magnifications of yellow-squared areas in the upper panels. Scale bars, 500 nm and 200 nm in the upper and lower panels, respectively. Yellow arrows indicate VPs or ER membrane in the case of mock-infected cells. (G) For determining the number of pIRO-Z-induced vesicle packets (VPs) per cell, 20 cell profiles were randomly selected, and VPs were manually counted. Means ± SEM are shown for n = 3 independent experiments. Differences in numbers of vesicle packets per cell were analyzed using the two-tailed unpaired t test with Welch’s correction. ****, P < 0.0001.
Considering SBI-0090799 is active in Huh-7.5 cells, which has been described as having a deficiency in the RIG-I pathway (24), we exclude a mechanism of action through the modulation of NS4A in the RLR-mediated immune signaling. A role for SBI-0090799 through the modulation of the role of NS4A as a regulator of Akt-mTOR signaling and induction of the autophagy pathway (21) was also considered unlikely, since the kinetics of SBI-0090799 activity (Fig. 4A and B) are inconsistent with the induction of autophagy in response to NS4A accumulation and, importantly, no difference in LC3 II conversion was observed in ZIKV-infected Huh-7.5 cells upon treatment with SBI-0090799 (data not shown). Thus, we focused on the potential mechanism of action of SBI-0090799 as an inhibitor of the formation of viral replication compartments (10, 25, 26). Consistent with this hypothesis, kinetic studies revealed an inhibition of both protein and RNA synthesis at early time points after infection (Fig. 5D and E), consistent with the time-of-addition experiments (Fig. 4A and B).
Flaviviral replication compartments appear as invaginations of the endoplasmic reticulum membranes, known as vesicle packets (VPs), which can be observed by electron microscopy in Huh7/Lunet-T7 cells (20, 26) transfected with the pIRO-Z polyprotein expression construct (10, 25, 26). Using this approach, the number of VPs was counted in DMSO- or SBI-0090799-treated Huh7/Lunet-T7 cells transfected with the ZIKV polyprotein expression construct (Fig. S5A) (25, 27). This showed a significant decrease in the number of VPs in the presence of SBI-0090799 (Fig. 5F and G). Of note, SBI-0090799 affected neither the expression of NS1, NS3, NS4B, and NS5 nor the transfection efficiency (Fig. S5B to E). These results indicate that SBI-0090799 inhibits NS4A’s capability to induce membrane rearrangements (12), which most likely is required for the formation of VPs, the sites of viral RNA replication. Interestingly, the ability of NS4A to multimerize was not affected by the drug in 293T cells transfected with NS4A. As shown in Fig. 5F, under non-reducing conditions (no dithiothreitol [DTT]) NS4A monomers can be observed at 14 kDa and NS4A dimers at 28 kDa. When DTT was added, only NS4A monomers were detected. Thus, treatment with SBI-0090799 did not affect the formation of NS4A dimers, suggesting NS4A’s role in membrane rearrangement is independent from its multimerization.
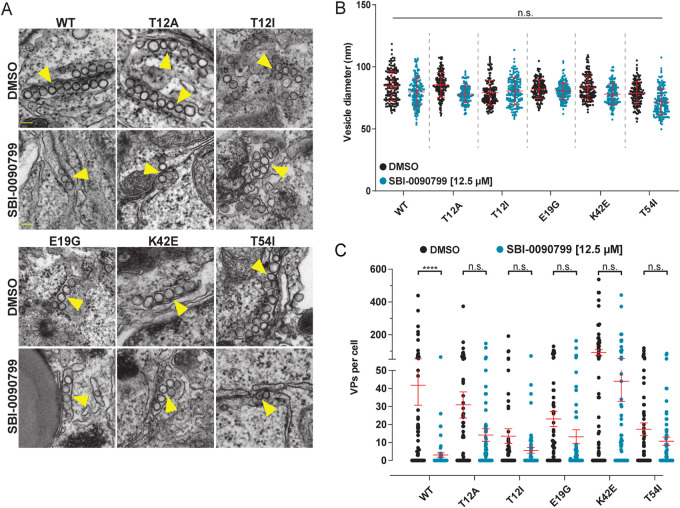
To further investigate whether SBI-0090799 suppresses ZIKV replication via NS4A, the mutations conferring resistance to SBI-0090799 were inserted in the ZIKV H/PF/2013 polyprotein-encoding plasmid, and VPs detected in transfected cells treated with SBI-0090799 were quantified for each of these mutants. As expected, each of the mutants induced VPs that were morphologically comparable to those generated by the wild type (Fig. 6A and B). Of note, each of the mutations conferring replication resistance to SBI-0090799 also conferred resistance against disruption of VP formation (Fig. 6A and C). Taken together, these results provide compelling evidence that the antiviral activity of SBI-0090799 is based on the modulation of the role of NS4A in the formation of the membranous replication compartments.
FIG 6.
Mutations in NS4A conferring replication resistance to SBI-0090799 also confer resistance to inhibition of ZIKV replication organelle formation. (A) Transmission electron microscopy images of 70-nm sections of resin-embedded Huh7/Lunet-T7 cells transfected with respective pIRO-Z constructs in the presence of either SBI-0090799 (12.5 μM) or DMSO (0.125%, vol/vol). Scale bars, 200 nm. (B) Vesicle diameter within VPs is not affected by SBI-0090799. Using the FIJI software package, vesicle diameters were measured. Means ± standard deviations are shown from 3 different experiments. Within each experiment, 50 VPs were analyzed for each condition. n.s., not significant. (C) For determining the number of pIRO-Z-induced VPs per cell, 20 cell profiles were randomly selected, and VPs were manually counted. Means ± SEM are shown for n = 3 independent experiments (****, P < 0.0001). Yellow arrows indicate VPs.
DISCUSSION
Zoonotic viruses are now recognized as a major threat to global health. The ongoing SARS-CoV-2 pandemic has highlighted the urgency of developing antivirals and vaccines against emergent and reemergent viruses to contain viral spread. ZIKV is a mosquito-borne virus responsible for an outbreak in 2015 to 2016, mainly affecting the Americas (1, 28). The main vectors transmitting ZIKV are Aedes mosquitoes (A. aegypti and A. albopictus). These vectors are also shared between dengue and chikungunya viruses and are mainly distributed in tropical and subtropical areas, although they have recently spread into more temperate regions (29) as climate change is currently altering the areas mosquitoes species inhabit (30, 31). This change is likely to have an impact on the transmission of mosquito-borne diseases, potentially enlarging the viral target population (30) and influencing the efficacy of the vector to transmit the disease (31). Future outbreaks of ZIKV may impact a larger population in expanded geographic zones compared to the 2015–2016 pandemic. Therefore, the need for developing antivirals targeting this virus remains a priority for the medical community. To identify promising chemical candidates for the development of antivirals targeting ZIKV, we employed a large-scale high-content screening approach. Previous drug repurposing screens had identified several ZIKV antiviral candidates (32–34), which, unfortunately, did not show success in the clinic (35). Therefore, instead of screening approved or known bioactive compounds as already described (32–34), we profiled a library that encompassed diverse chemical structures and chemotypes, aiming to identify novel chemical scaffolds that harbor antiviral activity against ZIKV.
This strategy enabled the identification of (2E)-N-benzyl-3-(4-butoxyphenyl)prop-2-enamide (SBI-0090799) as an inhibitor of ZIKV replication. The compound was found to be active specifically against ZIKV and not other flaviviruses. We find that it functions through inhibiting the formation of viral replication compartments, likely through blocking the membrane-altering activity of NS4A, as suggested by the appearance of drug-resistant viruses accumulating mutations in the N-terminal region of this protein. By analogy to dengue virus, the first 48 amino acid residues of ZIKV NS4A are assumed to be part of a cytosolic disordered region, described to fold into a helix upon interaction with lipids, supporting a role in membrane rearrangements, although the in-membrane structure of NS4A has not been solved (15). We hypothesize that SBI-0090799 interferes with NS4A, promoting the formation of the replication compartments, possibly by preventing its ability to induce ER membrane curvature (13, 14) or precluding NS4A from binding viral (19) or cellular partners (36, 37) independently from its multimerization. Of note, this region of NS4A was recently reported to also increase the helicase activity of NS3 in vitro, suggesting a role as a cofactor of the viral replicase complex (23). Further investigations will be required to elucidate both the structural and functional impact of SBI-0090799 on NS4A.
Structure-activity relationship studies (SAR) have been performed here, allowing the identification of the active core of the molecule. Further medicinal chemistry optimization may result in enhanced potency, enabling the molecule to be more suitable for in vivo application and clinical translation. For example, the first generation of hepatitis C virus (HCV) NS5A inhibitors (38, 39) was identified in 2009 through a large-scale chemical compound screen that showed total inhibition of HCV at 15 μM (38). After extensive SAR, compounds >10,000-fold more potent were generated (39), ultimately resulting in the development of clinically approved drugs (e.g., daclatasvir and ledipasvir) (40). Moreover, in the course of these studies, drugs with increased resistance barrier and broad-spectrum activity, targeting most, if not all, HCV genotypes have been generated (reviewed in reference 41). With respect to SBI-0090799, one current limitation is the rapid development of resistance, which is due to the lack of proofreading activity of the ZIKV RNA-dependent RNA polymerase (42), giving rise to a mutation frequency in the range of 1.4 × 10−4 (43). Several strategies can be employed to overcome antiviral drug resistance, including optimization of the molecule to improve the barrier for resistance as well as combinatorial administration of an antiviral cocktail for the treatment of ZIKV. Thus, medicinal chemistry around SBI-0090799 could also improve the barrier for resistance. Importantly, it will be critical to also understand if the mutations induced by SBI-0090799 in NS4A impact the fitness of the virus in vivo and its transmission via the mosquito vector.
Interestingly, a number of parallels exist between SBI-0090799 and HCV NS5A inhibitors. Similar to ZIKV NS4A, HCV NS5A has no enzymatic activity but is essential for HCV replication. NS5A is sufficient to induce the formation of the membranous replication compartment, which, for HCV, is composed of double membrane vesicles (10, 25, 26). In addition, NS5A is part of the viral replicase complex catalyzing viral genome amplification and required for the assembly of the virus particles (44). Notably, daclatasvir and derivatives thereof have also been shown to block the formation of double membrane vesicles, the site where HCV RNA replication occurs (45, 46), in analogy to SBI-0090799 activity on ZIKV. Similarly, the development of HCV variants resistant to daclatasvir has been broadly documented in vitro and in vivo (41). However, although preferentially used in combinatorial therapies, second and third generations of NS5A inhibitors showed improved resistance profiles and efficacy against the resistant variants (41, 47).
Taken together, this work identified SBI-0090799 as an inhibitor of ZIKV replication, acting by blocking the de novo formation of viral replication compartments, likely through direct or indirect modulation of NS4A’s activity to rearrange ER-derived membranes. SBI-0090799 represents a promising lead candidate for further development and optimization that can enable the realization of efficacious antiviral therapies for the treatment of ZIKV.
MATERIALS AND METHODS
Cells.
Huh-7.5 cells (24), JEG-3 (ATCC HTB-36), Vero cells (ATCC CCL-81), and 293T/17 cells (ATCC CRL-11268) were cultivated at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Gibco, Life Technologies) and 1× penicillin-streptomycin-glutamine (Gibco, Life Technologies). Huh7/Lunet-T7 cells (20) were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM l-glutamine, and 1% nonessential amino acids (all purchased from Gibco, Life Technologies). For maintaining stable expression of the T7 RNA polymerase, zeocin was added to the medium at a final concentration of 5 μg/ml.
Viruses.
The following viruses were used in this study: ZIKV MR766 (ATCC VR84), ZIKV Dakar 41519 (GenBank accession no. HQ234501), ZIKV Nica 2–16 (GenBank accession no. KX421194), ZIKV Fortaleza 2015 (GenBank accession no. KX811222.1), DENV-2 16681 (GenBank accession no. NC_001474.2), YFV-17D (GenBank accession no. MG051217.1), JEV SA-14-14-2 (GenBank accession no. JN604986.1), and KUNV (GenBank accession no. MF289571.1). All the viral stocks were amplified in Vero cells. After harvesting, the supernatants were clarified by centrifugation for 15 min at 4°C at 2,000 × g, aliquoted, and stored at −80°C until use.
hMDDC preparation and infection.
Human primary monocyte-derived dendritic cells (hMDDCs) were prepared from fresh, healthy donor blood from the San Diego Blood Bank. Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient sedimentation with Ficoll-Paque (GE Healthcare). CD14-positive monocytes were positively selected using anti-CD14 microbeads according to the manufacturer’s instructions (Miltenyi Biotec). The isolated CD14+ monocytes were incubated in RPMI 1640 (Life Technology) supplemented with 10% FBS (Gibco), 1,000 U/ml of human interleukin-4 (IL-4) (Peprotech), and 500 U/ml of human granulocyte macrophage colony-stimulating factor (GM-CSF) (Peprotech) and allowed to differentiate for 5 days. Fresh medium with cytokines was added on day 3. On the 5th day, the cells were reseeded in 384-well plates in the presence of DMSO, A3, or increasing concentrations of SBI-0090799 at a density of 9,000 cells/well. After a 16-h incubation, cells were infected by addition of ZIKV MR766 at a multiplicity of infection (MOI) of 2.5. Cells were fixed at 24 h.p.i. with 4% paraformaldehyde (PFA), and the infection was quantified as described in the immunostaining section. This study was approved by the Sanford Burnham Prebys Medical Discovery Institute Biosafety Committee (IBC).
Infection of hfNPC culture and compound treatment against ZIKV infection.
Human fetal neural progenitor cells (hfNPCs) were cultured as previously described (48). Ten thousand hfNPCs at passage 11 cultured overnight on a Matrigel-coated 96-well plate (Perkin Elmer) were infected with ZIKV (Fortaleza 2015 strain) at an MOI of 1. DMSO, A3 (2 μM), or SBI-0090799 (10 μM) was added to the cells at the same time as the virus. At 24 h.p.i., cells were fixed with 4% PFA and stained as previously reported (48). Briefly, mouse antiflavivirus group antigen antibody clone D1-4G2-4-15 was used to visualize the ZIKV-infected cells. Nuclei were stained with DAPI to quantify the total cell number in each well. Twenty-one images per well of a 96-well plate were acquired automatically via an IC200-KIC microscope (Vala Sciences) and analyzed using Acapella 2.6 software (Perkin Elmer). The nuclei and cell soma were segmented using the DAPI channel. A ZIKV-positive cell was determined if the intensity of flavivirus group antigen in the soma was above the empirically determined threshold. The percentage of ZIKV-infected cells was calculated per well, with triplicate wells per condition.
Preparation and infection of primary human astrocytes.
Normal human astrocytes (NHA) were purchased from Lonza (CC-2565) and cultured with the astrocyte growth medium (AGM) bullet kit (CC-3186; Lonza) per the manufacturer’s instructions. For ZIKV infections, 1 × 105 NHA were seeded in 24-well plates to achieve a confluence of ∼70%. The day after seeding, cells were infected with ZIKV (Nica 2–16) at an MOI of 5 for 30 min at 37°C. The viral inoculum was then removed, cells were washed twice in 1× phosphate-buffered saline (PBS), and 1 ml of standard AGM medium was added back. Viral titers were assessed by plaque assay in Vero cells (1:10 dilutions) from total cell lysates (1 round of freeze-thaw) at 24 h.p.i. Cell viability was assessed using the CytoTox96 cytotoxicity assay (G1781; Promega), measuring the lactate dehydrogenase (LDH) release as a measurement of cell lysis. The percent cytotoxicity was normalized to maximum LHD release from a lysis buffer, and results were then normalized to the DMSO-treated cells.
Infection of JEG-3 cells.
Two thousand five hundred JEG-3 cells were seeded per well of a 384-well plate in the presence of DMSO, A3, or increasing concentrations of SBI-0090799. After a 16-h incubation, cells were infected by addition of ZIKV MR766 at an MOI of 0.5. Twenty-four h.p.i., cells were fixed with 4% PFA, and the infection was quantified as described in the immunostaining section.
Compounds, antibodies, and reagents.
The following compounds were used in this study: (2E)-N-benzyl-3-(4-butoxyphenyl)prop-2-enamide (indicated as SBI-0090799; CSC032625200; ChemSpace; and MolPort-002-049-778; MolPort), (2E)-3-(4-butoxyphenyl)-N-(1-phenylethyl)prop-2-enamide (indicated as SBI-0090559; MolPort-002-049-758), A3 (49), bafilomycin A1 (Selleckchem). All the analogs of SBI-0090799 were purchased from Molport: MolPort-000-214-439, MolPort-000-666-063, MolPort-000-667-334, MolPort-001-540-126, MolPort-001-632-769, MolPort-001-837-976, MolPort-001-838-962, MolPort-002-048-412, MolPort-002-233-296, MolPort-002-885-075, MolPort-009-663-306, MolPort-019-745-256, MolPort-027-892-391, MolPort-044-257-513, MolPort-002-866-259, MolPort-002-860-297, MolPort-002-863-114, MolPort-002-049-740, and MolPort-002-049-562. The source of additional compounds purchased for validation of the screen hits are indicated in Table S1. The following primary antibodies were used in this study: mouse anti-ZIKV ZV-2 monoclonal antibody (MAb) (50), mouse anti-flavivirus group antigen MAb clone D1-4G2-4-15 (MAB10216; Millipore), mouse MAb E60 (51), human MAb hE16 (52), mouse MAb JEV-31 (53), rabbit NS3 and NS5 antisera (54), rabbit anti-CoxIV MAb (4850; Cell Signaling), mouse MAb anti-ZIKV NS1 (GTX634158; GeneTex), rabbit polyclonal anti-ZIKV NS3 (GTX133320; GeneTex), anti-ZIKV NS4A (GTX133704; GeneTex), anti-ZIKV NS4B (GTX133321; GeneTex), anti-ZIKV NS5 (GTX133327; GeneTex) antibodies, mouse anti-GAPDH MAb (sc-47724/0411; Santa Cruz), anti-mouse IgG-horseradish peroxidase (HRP) antibody (A4416; Sigma-Aldrich), anti-rabbit IgG-HRP antibody (A6154; Sigma-Aldrich), Alexa Fluor 568-conjugated donkey anti-rabbit IgG (A10042; Thermofisher). Alexa Fluor 488-conjugated goat anti-mouse secondary antibody (A-11001; Invitrogen), Alexa Fluor 488-conjugated goat anti-human secondary antibody (A-11013; Invitrogen), HRP-conjugated goat anti-rabbit IgG (H+L) secondary antibody (1706515; Bio-Rad), and HRP-conjugated goat anti-mouse IgG (H+L) secondary antibody (1706516; Bio-Rad). Additional reagents used were DMSO (Sigma-Aldrich), DAPI (BioLegend), paraformaldehyde (Fisher Scientific), bovine serum albumin (BSA) (Sigma-Aldrich), methyl cellulose (Sigma-Aldrich), and phenol-chloroform (Sigma-Aldrich).
High-content screen.
A total of 51,520 small chemical compounds from the SBP 320K library (DIVERSet Diverse Screening Library; Chembridge), at 10 mM in pure DMSO, were dispensed into 384-well clear-bottom black plates (Greiner Bio-One) using an Echo 550 acoustic liquid handler (LabCyte), adding 50 nl per well to reach a final concentration of 10 μM during the assay. The same volumes of DMSO and DHODH inhibitor A3 (49) (10 mM stock) were added to each plate as negative and positive controls. Two thousand Huh-7.5 cells resuspended in 30 μl were added to each well. Sixteen hours later, cells were infected by adding 20 μl of ZIKV MR766 at an MOI of 0.3 and incubated at 37°C and 5% CO2 for 24 additional hours. Cells were then fixed by addition of 50 μl of 8% PFA and stained as described in the immunofluorescence staining section. Four fields per well were acquired with an IC200 high-content imager (Vala Sciences) using a 20× objective. Images were analyzed with Columbus software, allowing automated quantification of the number of cells and the percentage of infected cells per well. Compounds decreasing the percentage of infection by at least 50% and reducing the cell number by less than 50% compared to the median of each plate were selected for validation. These compounds were cherry picked from the same library, and the validation was performed by following the same protocol used for the primary screen.
Immunofluorescence staining.
At several time points throughout experimentation, infected Huh-7.5 cells, JEG-3 cells, and hMDDCs were subjected to an immunofluorescence-based imaging assay, labeling the viral envelope in ZIKV-infected cells. In each assay detailed below, cells were fixed at the indicated time with 4% PFA for 30 min and permeabilized with 0.5% Triton X-100 for 10 min. After blocking with 3% BSA for 30 min, cells were incubated for 1 h at room temperature with mouse anti-ZIKV ZV-2 MAb (50). After two washes with phosphate-buffered saline (PBS), the cells were incubated with Alexa Fluor 488-conjugated goat-anti-mouse secondary antibody supplemented with 0.1 μg/ml DAPI for 1 h at room temperature. After three additional washes with PBS, plates were imaged using an IC200 high-content imager (Vala Sciences) or Celigo image cytometer (Nexcelom). The assay results and data analyses enabled us to determine infectivity and viability or cytotoxicity. From the infectivity and cytotoxicity values, a four-parameter logistic nonlinear regression model was used to calculate IC50 and 50% cytotoxic concentration (CC50) values whenever required. For DENV-2 16881, JEV-SA14-14-2, YFV-17D, and KUNV, the same protocol was used, with the following antibodies: mouse anti-flavivirus group antigen MAb clone D1-4G2-4-15, mouse MAb E60 (51), human MAb hE16 (52), and mouse MAb JEV-31 (53) were used as primary antibodies to detect DENV, JEV, YFV and KUNV, respectively. Alexa Fluor 488-conjugated goat anti-mouse secondary antibody was used for DENV, YFV, and JEV, while Alexa Fluor 488-conjugated goat anti-human secondary antibody was used for KUNV.
Cell viability assay.
To determine cell viability and cytotoxicity of the chemical compounds, the commercially available kit CellTiter-Glo 2.0 assay (Promega) was used. Huh-7.5 cells, JEG-3 cells, hMDDCs, or astrocytes were seeded at 30 μl/well in 384-well plates at the same density indicated for the antiviral assays and incubated with 2-fold serial dilutions of compound SBI-0090799, diluted from a 20 μM stock solution. After 16 h, 20 μl of fresh medium was added to each well. Cell viability was quantified 24 h later by measuring the ATP levels in live cells through CellTiter-Glo (Promega) luminescent cell viability assay according to the manufacturer’s protocol. The signal was quantified using an EnSpire multilabel plate reader (Perkin Elmer). To determine cell viability and cytotoxicity of SBI-0090799, Huh7/Lunet-T7 cells were seeded at a density of 10,000 cells/well in a total volume of 100 μl DMEM complete (without zeocin) in a 96-well plate. Cells were treated with 2-fold serial dilutions of the compound SBI-0090799, of which an 80 μM stock solution was prepared. Each concentration was evaluated in triplicate. After treating the cells for 24 h, cell viability was determined by measuring ATP levels using the protocol provided. Once the substrate was added, plates were placed in a Mithras LB940 plate reader (Berthold Technologies), and luminescence was measured at 560 nm. As a control, cells were treated with equal concentrations of DMSO only.
Time-of-addition assays.
For time-of-addition assays, 9,000 Huh-7.5 cells were seeded per well of a 96-well plate. The following day, cells were infected with ZIKV MR766 (MOI, 0.6). After 1 h, the viral inoculum was removed, and cells were washed with ice-cold PBS before addition of fresh medium. DMSO or the indicated compounds were added at the indicated concentrations at different time points, according to the timelines illustrated in Fig. 4A and B. Cells were fixed at 24 h.p.i., and an immunofluorescence assay was performed as described in the immunofluorescence staining section.
VLPs.
VLPs were produced as previously described (55). Briefly, 500,000 293T/17 cells were seeded per well of a 6-well plate. The following day, cells in each well were transfected with 1 μg of plasmid expressing WNV green fluorescent protein (GFP) replicon and 1 μg ZIKA MR766 prM-C-E plasmid, using 4 μl of Lipofectamine 2000 (Life Technologies). The supernatant containing reporter viral particles was collected 48 h posttransfection, centrifuged for 5 min at 2,000 × g, aliquoted, and stored at −80°C until use. Ten microliters of stock was used to infect Huh-7.5 cells seeded in 96-well plates (10,000 cells/well) in the presence of DMSO or the indicated compound. Cells were fixed at 24 h.p.i. with 4% PFA for 30 min and stained with 0.1 μg/ml DAPI for 1 h at room temperature. After three washes with PBS, plates were imaged using a Celigo image cytometer (Nexcelom).
ZIKA replicons: in vitro transcription, RNA transfection, and measurement of luciferase activity.
To study ZIKV replication in the presence of chemical compounds, the subgenomic pZIKV RepWT, Rep NS5ΔGDD, and H/PF/2013 replicons, encoding the Renilla luciferase, were used (17, 56). pZIKV RepWT and plasmid Rep NS5ΔGDD were linearized using the restriction enzyme ClaI (New England Biolabs). The digested plasmids were extracted with phenol-chloroform and precipitated with ethanol. The pellet was resuspended with RNase-free water, and DNA was in vitro transcribed using the mMESSAGE mMACHINE kit (Ambion), as previously described (17). The RNA was introduced in Huh-7.5 cells by electroporation. Briefly, 500,000 cells were resuspended in 100 μl resuspension buffer R (Life Technologies) and electroporated with 500 ng of RNA, using the Neon transfection system (Life Technologies) with the following conditions: 1,600 V, 20 ms, 1 pulse (57). Directly after electroporation, cells were resuspended in fresh DMEM supplemented with 10% FBS and seeded in a 96-well plate at a density of 20,000 cells/well in the presence of the indicated compound. Four and 24 h postelectroporation, supernatant was removed and cells were lysed with 30 μl of Glo lysis buffer (Promega). The lysates were then mixed with the same volume of Renilla Glo substrate (Promega), and the luciferase signal was quantified using an EnSpire multilabel plate reader (Perkin Elmer). The viral RNA for the subgenomic H/PF/2013 replicon was generated by linearizing 10 μg of DNA plasmid with XhoI (New England Biolabs), followed by DNA purification using the Nucleo-Spin extraction kit (Macherey-Nagel). Upon elution of the linearized DNA with RNase-free water, the in vitro transcription (IVT) reaction was carried out in a total volume of 100 μl containing 20 μl 5× RRL buffer (400 mM HEPES [pH 7.5], 60 mM MgCl2, 10 mM spermidine, and 200 mM DTT), 12.5 μl rNTP-mix (3.125 mM ATP, CTP, and UTP, 1.56 mM GTP), 2.5 μl RNasin (1 U/μl; Promega), 4 μl T7 RNA polymerase (2 U/μl; New England Biolabs), and 20 μl anti-reverse cap analogue (ARCA; 3′-O-Me-m7G(5′)ppp(5′)G; stock, 1 mM; England Biolabs). IVT was conducted for 2.5 h at 37°C, followed by addition of 2 μl of T7 RNA polymerase and an additional incubation of 16 h at 37°C. The DNA template was digested by adding 20 μl RNase-free DNase I and incubating at 37°C for 1 h. In vitro transcripts were purified via acidic phenol-chloroform extraction and isopropanol precipitation. Purified RNA was resuspended in RNase-free water at a final concentration of ∼1 μg/μl, and quality and size were validated by agarose gel electrophoresis. Aliquots of 10 μg were stored at −80°C until needed. In vitro transcripts were introduced into Huh7/Lunet-T7 cells via electroporation. Subconfluent Huh7/Lunet-T7 cells were trypsinized and collected in complete DMEM followed by centrifugation at 700 × g for 5 min. Subsequently, cells were washed once with 20 ml sterile PBS and counted using the TC20 automated cell counter (Bio-Rad). After an additional centrifugation step at 700 × g for 5 min, cells were resuspended in cytomix buffer (120 mM KCl, 0.15 mM CaCl2, 10 mM potassium phosphate, 25 mM HEPES [pH 7.6], 2 mM EGTA, 5 mM MgCl2, freshly supplemented with 2 mM ATP and 5 mM glutathione) at a concentration of 1 × 107 cells/ml. For electroporation, 10 μg of in vitro-transcribed ZIKV RNA was mixed with 400 μl cell suspension and transferred into an electroporation cuvette with a gap width of 0.4 cm. By using the Gene Pulser II system (Bio-Rad), a pulse of 270 V and 975 μF was applied, resulting in a time constant of 17 to 20 ms. Cells were immediately resuspended in 10 ml prewarmed complete DMEM and seeded into 12-well plates at a density of 200,000 cells/well. Prior to seeding of transfected cells, 12-well plates were prepared by adding SBI-0090799 (concentration range, 160 μM to 0.08 μM) or the respective DMSO control. Renilla luciferase activity was measured 24 h.p.e. by lysing the cells in 100 μl luciferase buffer (10% [vol/vol] glycerol, 0.1% Triton X-100, 25 mM glycine-glycine [pH 7.8], 15 mM MgSO4, 4 mM EGTA, and freshly added 1 mM DTT). Lysates were stored at –20°C for at least 1 h before further processing. For each concentration of SBI-0090799 or DMSO (control), 20 μl of lysate was mixed with 100 μl freshly prepared luciferase assay buffer (25 mM glycine-glycine [pH 7.8], 15 mM K4PO4 buffer [pH 7.8], 15 mM MgSO4, 4 mM EGTA, and 1.42 μM coelenterazine). To quantify luciferase activity, samples were measured for 10 s in a tube luminometer (Lumat LB9507; Berthold Technologies). Each sample was analyzed in duplicate. For normalization, values obtained with samples harvested 4 h.p.e. were used.
Plaque assay.
Huh-7.5 cells were infected for 1 h at 37°C with ZIKV MR766 at an MOI of 0.6. After one wash with cold PBS, fresh medium was added to the cells. Whenever specified, either DMSO or chemical compounds were added to the cells, according to the indicated kinetics. Supernatant was harvested at 24 h.p.i., and a plaque assay to quantify the infectious virus was performed on Vero cells, similar to previous descriptions (58). Briefly, Vero cells were seeded in 12-well plates at a density of 150,000 cells/well. The following day, 10× dilutions of ZIKV MR766 were added to each well and 0.8% methyl cellulose was added as overlay after 2 h of infection. Cells were fixed at 72 h.p.i. and stained with crystal violet. Plaques were counted and the viral titer was determined as PFU/ml.
NS4A multimerization.
Fifty thousand 293T/17 cells were seeded per well of a 24-well plate. The following day, cells in each well were transfected with 250 ng of plasmid (pCDNA3.1, pCDNA ZIKV NS4A Puerto Rico, pCDNA ZIKV NS4A-2K Puerto Rico) (36) using 0.5 μl of Lipofectamine 2000 (Life Technologies). Twenty-four hours later, cells were treated with DMSO or SB-0090799 (10 μM). Samples were harvested 48 h posttransfection and analyzed by Western blotting comparing reducing (DTT-treated) and nonreducing (no DTT) conditions.
Western blot.
Huh-7.5 cells were infected for 1 h at 37°C with ZIKV MR766 at an MOI of 1. After one wash with cold PBS, fresh medium was added to the cells. Whenever specified, either DMSO or the specified chemical compounds were added to the cells according to the indicated kinetics. Cells were harvested at the indicated time point and lysed with lysis buffer (50 mM Tris-HCl [pH 8], 5 mM EDTA, 150 mM NaCl, 10 mM MgCl2, 1% Triton X-100), and the proteins were subjected to Western blotting. Protein samples were mixed with 4× NuPAGE LDS sample buffer (Thermo Fisher) supplemented with 50 mM DTT or not, whenever specified, and heated at 95°C for 10 min. Proteins were then separated by SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). All the membranes were blocked with PBS-Tween containing 3% BSA and then incubated with the indicated antibodies. The proteins of interest were detected using the West Pico plus chemiluminescent substrate (GE Healthcare) and the Chemidoc XRS+ imaging system (Bio-Rad). Proteins were quantified based on the intensities of the corresponding bands, using FIJI software (59). Huh7/Lunet-T7 cells seeded in a 6-well plate were subjected to Western blot analysis at 18 h posttransfection by adding 100 μl of sample buffer (120 mM Tris-HCl [pH 6.8], 60 mM SDS, 100 mM DTT, 1.75% glycerol, 0.1% bromophenol blue) supplied with 1 μl of Benzonase (70746-3; Millipore) to digest nucleic acids. After a 5-min incubation period at room temperature, samples were transferred into 1.5-ml reaction tubes and denatured for 5 min at 95°C. Protein samples (10 μl/well) were separated by electrophoresis into a 12% polyacrylamide gel. Proteins were transferred onto a PVDF membrane (0.45-μm pore size; Millipore) at 350 mA for 1 h at 4°C using the wet blotting system (transfer buffer, containing 20% methanol, 150 mM glycine, and 20 mM Tris [pH 8]). Membranes were blocked in PBS supplemented with 5% nonfat dry milk powder for 1 h at room temperature. Primary antibodies were diluted in 1% blocking buffer, and membranes were incubated at 4°C overnight. The next day, membranes were washed three times with PBS-T (0.1% Tween 20), 10 min each, before being incubated with secondary HRP-conjugated antibodies diluted in 1% blocking buffer. After an incubation period of 1 h at room temperature, membranes were washed three times with PBS-T, 10 min each, before being developed using the Western Lightning Plus-ECL reagent (NEL105001EA; Perkin Elmer). For imaging, the Intas ECL Chemocam imager and the corresponding ChemoStar imager software package were used. Whenever indicated, the bands have been indicated by densitometry using the Fiji software (59).
RNA extraction and RT-qPCR.
RNA was extracted from Huh-7.5 cells by use of a NucleoSpin total RNA isolation kit (Macherey-Nagel) and reverse transcribed to cDNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Quantitative PCR (qPCR) was performed using SYBR select master mix (Applied Biosystems). The ZIKV-specific primers ZIKV_Fw (5′-CCGCTGCCCAACACAAG-3′) and ZIKV_Rv (5′-CCACTAACGTTCTTTTGCAGACAT-3′) were used to quantify viral RNA (60). RPLP0 (Hs00420895_gH) was used as a housekeeping gene (RPLP0_Fw, 5′-CCCATGTGAAGTCACTGTGC-3′; RPLP0_Rv, 5′-GGTTGTAGATGCTGCCATTG-3′). Change in cycle threshold (ΔCT) values were calculated as ΔCT = CT(ZIKV) − CT(RPLP0). Relative gene expression was calculated using the 2−ΔΔCT method (61).
Selection of drug-resistant viruses.
Huh-7.5 cells were infected for 1 h at 37°C with ZIKV MR766 at an MOI of 0.3. After one wash with cold PBS, fresh medium containing either DMSO or SBI-0090799 (10 μM) was added to the cells. Supernatant was collected after 3 days and used to reinfect new Huh-7.5 cells for 9 additional passages. Viruses from passages 2, 5, and 10 were tested in 12-point dose-response assays in the presence of DMSO, A3, or SBI-0090799 to assess their resistance. The IC50 was calculated as previously described. RNA was isolated from the virus at passage 5 using the NucleoSpin viral RNA isolation kit (Macherey-Nagel). ZIKV sequencing libraries were generated using the NEBNext Ultra II directional RNA library prep kit for Illumina (NEB, Ipswich MA). Libraries were sequenced on the Illumina NextSeq 500 and data were processed in BaseSpace (basespace.illumina.com). Reads were aligned to the ZIKV genome (NC_012532.1) and aligned with BWA using default settings. Variants were called with the Pisces Variant Caller (https://github.com/Illumina/Pisces).
Cloning of drug-resistant mutants in SGR and revertant.
Single-point mutations and sequences reverted to wild type were inserted in the pZIKV Rep WT construct (17) by two-step overlap PCR cloning. Standard overlap PCR was performed with Pfu Turbo DNA polymerase (Agilent Technologies) to amplify the DNA fragment between unique restriction enzyme sites NaeI and EcoRI. A first PCR was performed using the primers NaeI_F, 5′-ACAGTGTGCCGGCAGA-3′; EcoRI_R, 5′-CTCCTGAATTCTCTCTCCCCAT-3′, each coupled with one of the following: NS4A_T12A_F, 5′-CTGGGAGCACTGCCAGGA-3′; NS4A_T12A_R, 5′-TGGCAGTGCTCCCAGGG-3′; NS4A_T12I_F, 5′-CTGGGAATACTGCCAGGA-3′; NS4A_T12I_R, 5′-TGGCAGTATTCCCAGGG-3′; NS4A_T12V_F, 5′-CTGGGAGTACTGCCAGGA-3′; NS4A_T12V_R, 5′-TCCTGGCAGTACTCCCAG-3′; NS4A_E19G_F, 5′-ATGACAGGGAGATTCCAG-3′; NS4A_E19G_SGR_R, 5′-TGGAATCTCCCTGTCATATG-3′; NS4A_K42E_SGR_F, 5′-CTACGAAGCCGCGGCG-3′; NS4A_K42E_SGR_R, 5′-CGGCTTCGTAGGGCCT-3′; NS4A_T54I_SGR_F, 5′-CCCTAGAGATTATCATGCTT-3′; NS4A_T54I_SGR_R, 5′-CATGATAATCTCTAGGGTCTC-3′; NS4A_T12rev_F, 5′-CTGGGAACACTGCCAGGA-3′; NS4A_T12REV_R, 5′-TGGCAGTGTTCCCAGGG-3′; NS4A_E19REV_F, 5′-ATGACAGAGAGATTCCAG-3′; NS4A_E19rev_SGR_R, 5′-TGGAATCTCTCTGTCATATG-3′; NS4A_K42rev_SGR_F, 5′-CTACAAAGCCGCGGCG-3′; NS4A_K42rev_SGR_R, 5′-CGGCTTtGTAGGGCCT-3′; NS4A_T54rev_SGR_F, 5′-CCCTAGAGACTATCATGCTT-3′; NS4A_T54rev_SGR_R, 5′-CATGATAGTCTCTAGGGTCTC-3′. Overlap PCR on the purified products was then performed using the NaeI_F and EcoRI_R primers, resulting in a DNA fragment spanning restriction enzyme sites NaeI and EcoRI. The purified PCR fragment was inserted into pZIKV Rep WT, previously digested with these same enzymes (New England Biolabs), and purified using NucleoSpin Gel and a PCR clean-up kit (Macherey-Nagel). All plasmids were validated by DNA sequencing.
Transient transfection of mammalian cell lines.
One day prior to transfection, Huh7/Lunet-T7 cells were seeded in 24-well plates on glass coverslips (for immunofluorescence microscopy and electron microscopy) or in a 6-well plate (for Western blotting) at a density of 30,000 cells/well or 200,000 cells/well, respectively. On the day of transfection, medium was replaced by fresh DMEM supplemented with either 12.5 μM SBI-0090799 (5× IC90) or DMSO (0.125%, vol/vol). For cells seeded on coverslips, 500 ng/well plasmid DNA was mixed with 100 μl/well Opti-MEM reduced serum medium (31985070; Thermo Fisher) and 1.5 μl/well of Mirus TransIT-LT1 transfection reagent (MIR2304; Mirus). The mixture was incubated for 20 min at room temperature and added to the cells in a dropwise manner. After a 4-h incubation period, medium was exchanged by fresh DMEM supplemented with either 12.5 μM SBI-0090799 or DMSO (0.125%, vol/vol). Cells seeded in 6-well plates were transfected by mixing 2 μg/well plasmid DNA with 400 μl/well Opti-MEM medium and 6 μl/well Mirus TransIT-LT1.
Immunofluorescence microscopy of transfected Huh7/Lunet-T7 cells.
For determining transfection efficiency, cells grown on glass coverslips were fixed at 18 h posttransfection. Cells were washed three times with sterile PBS before being fixed with 4% PFA in PBS for 10 min at room temperature. Fixed cells were washed three times with PBS, permeabilized with 0.2% Triton X-100 in PBS for 7 min at room temperature, and blocked for 1 h at room temperature in blocking buffer (10% FBS in PBS). Subsequently, samples were incubated with ZIKV NS4B antibody (diluted 1:250 in 3% BSA in PBS) for 2 h at room temperature or at 4°C overnight. After washing the samples three times with PBS-T (0.01% Tween 20), samples were incubated for 1 h with Alexa Fluor 568-conjugated secondary antibodies (1:1,000 diluted in 3% BSA in PBS) in the dark. Next, samples were washed three times with PBS-T, 10 min each, in the dark. Coverslips were mounted on microscopy slides using Fluoromount G (SouthernBiotech) supplemented with DAPI to stain nuclear DNA. Fluorescence images were acquired with a Nikon Ti Eclipse microscope and analyzed with the NIS-Element AR software package. Overview images (5 by 5) were taken with the ×20 magnification objective.
Transmission electron microscopy.
To investigate whether SBI-0090799 affects the formation of ZIKV replication organelles, Huh7/Lunet-T7 cells were fixed with 2.5% glutaraldehyde and 1% paraformaldehyde (in PBS) in cacodylate buffer (50 mM cacodylate, 50 mM KCl, 2.6 mM CaCl2, 2.6 mM MgCl2, and 2% sucrose) for at least 30 min at room temperature. Samples were rinsed 5× with cacodylate buffer (50 mM) and incubated with 2% osmium tetroxide in 50 mM cacodylate buffer for at least 40 min on ice. Samples were washed three times with electron microscopy (EM)-grade water and incubated for 16 h in 0.5% uranyl acetate in water at 4°C. The next day, samples were rinsed three times with EM-grade water, dehydrated in a graded ethanol series (from 50% to 100%) at room temperature before being embedded in epoxy resin, and polymerized for at least 48 h at 60°C. Ultrathin sections of 70 nm were obtained by sectioning with a Leica EM UC6 ultramicrotome (Leica Microsystems) and a diamond knife. Sections were collected on EM grids, counterstained using lead citrate and uranyl acetate, and examined with a JEOL electron microscope (JEM 1400). For each experiment, a total of 20 cells were analyzed and the number of VPs per complete cell section was manually counted at ×6,000 magnification. Representative images of VPs were acquired at ×12,000 magnification and processed using FIJI software (59).
Quantification and statistical analysis.
Data are presented as means ± standard errors of the means (SEM) except where specified differently. Statistical parameters and analyses are reported in the figure legends, with n representing the number of independent experiments. Statistical analyses were performed using GraphPad Prism 7.
ACKNOWLEDGMENTS
We thank Michael Diamond for sharing the viruses ZIKV MR766, ZIKV-Dakar 41519, YFV-17D, JEV-SA14-14-2, and KUNV and the antibodies E60, hE16, JEV-31, and ZV-2; Ana Sesma for providing DENV-2 16681; Megan Shaw for supplying compound A3; Ricardo Rajsbaum for sharing JEG-3 cells; Andres Merits for providing the ZIKV NS3 and NS5 antisera; Jose Luis Slon Campos and Oscar Burrone for sharing the ZIKA MR766 prM-C-E plasmid; Priya Shah for pCDNA NS4A and NS4A-2K Puerto Rico plasmids; Sunnie Yoh, Naoko Matsunaga, Vikram Shende, and Mike Gardner for scientific input; Sylvie Blondelle and Larry Adelman for facilities and biosafety support; Ian Pass, Luis Orozco, Fu-Yue Zeng, and Breda Walsh at the SBP High-Throughput Screening Core for their support with compounds management and help with the coordination of the primary large-scale screen and validation; Susanne Heynen-Genel and Debbie Chen at the SBP High Content Imaging Platform for their technical assistance with plate imaging and analysis of the primary screen; Brian James at the SBP Genomic Core for his support with RNA-seq analyses; and Marisol Chacon for administrative assistance. S.G., U.H., and R.B. are grateful to the Electron Microscopy Core Facility at Heidelberg University, headed by Stefan Hillmer, and the Infectious Diseases Imaging Platform (IDIP) at the Center for Integrative Infectious Disease Research in Heidelberg, headed by Vibor Laketa, for expert support and access to their equipment.
This work was supported by National Institutes of Health grants R01 AI104972 and U19 AI118610. S.B.B. was supported in part as an Open Philanthropy awardee of the Life Sciences Research Foundation (LSRF). R.B. was supported by the Deutsche Forschungsgemeinschaft (DFG), grants Ba1505/8-1 and 240245660-SFB 1129, and the German Center for Infection Research (DZIF).
L.R., S.G., S.B.B., H.C.-T., A.R., U.H., C.M.W., and R.B. designed and/or performed experiments. L.R., S.G., S.B.B., H.C.-T., and P.D.D.J. analyzed data. A.V.T., E.H., A.B.P., R.B., and S.K.C. oversaw the conception and design of the experiments. L.R. and S.K.C. wrote the manuscript. All authors reviewed the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Sumit K. Chanda, Email: schanda@sbpdiscovery.org.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.World Health Organization. https://www.who.int/docs/default-source/documents/emergencies/zika/zika-countries-with-zika-and-vectors-table-july2019.pdf?sfvrsn=591689c1_2.
- 2.World Health Organization. https://www.who.int/emergencies/zika-virus/classification-tables/en/.
- 3.Carod-Artal FJ. 2018. Neurological complications of Zika virus infection. Expert Rev Anti Infect Ther 16:399–410. 10.1080/14787210.2018.1466702. [DOI] [PubMed] [Google Scholar]
- 4.Freitas DA, Souza-Santos R, Carvalho LMA, Barros WB, Neves LM, Brasil P, Wakimoto MD. 2020. Congenital Zika syndrome: a systematic review. PLoS One 15:e0242367. 10.1371/journal.pone.0242367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts.
- 6.Pierson TC, Diamond MS. 2020. The continued threat of emerging flaviviruses. Nat Microbiol 5:796–812. 10.1038/s41564-020-0714-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Institute of Allergy and Infectious Diseases. https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens.
- 8.Acosta EG, Kumar A, Bartenschlager R. 2014. Revisiting Dengue virus–host cell interaction. Adv Virus Res 88:1–109. 10.1016/B978-0-12-800098-4.00001-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Wu Z, Wang M, Cheng A. 2017. Innate immune evasion mediated by flaviviridae non-structural proteins. Viruses 9:291. 10.3390/v9100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CKE, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P, Schorb M, Pruunsild P, Schwab Y, Chatel-Chaix L, Ruggieri A, Bartenschlager R. 2017. Ultrastructural characterization of Zika virus replication factories. Cell Rep 18:2113–2123. 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. 2007. The non-structural protein 4A of Dengue virus is an integral membrane protein inducing membrane alterations in a 2k-regulated manner. J Biol Chem 282:8873–8882. 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 13.Hung Y-F, Schwarten M, Hoffmann S, Willbold D, Sklan E, Koenig B. 2015. Amino terminal region of dengue virus NS4A cytosolic domain binds to highly curved liposomes. Viruses 7:4119–4130. 10.3390/v7072812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung Y-F, Schwarten M, Schünke S, Thiagarajan-Rosenkranz P, Hoffmann S, Sklan EH, Willbold D, Koenig BW. 2015. Dengue virus NS4A cytoplasmic domain binding to liposomes is sensitive to membrane curvature. Biochim Biophys Acta 1848:1119–1126. 10.1016/j.bbamem.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Kumar P, Giri R. 2020. Zika virus NS4A cytosolic region (residues 1–48) is an intrinsically disordered domain and folds upon binding to lipids. Virology 550:27–36. 10.1016/j.virol.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Roosendaal J, Westaway EG, Khromykh A, Mackenzie JM. 2006. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol 80:4623–4632. 10.1128/JVI.80.9.4623-4632.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie X, Zou J, Shan C, Yang Y, Kum DB, Dallmeier K, Neyts J, Shi P-Y. 2016. Zika virus replicons for drug discovery. EBioMedicine 12:156–160. 10.1016/j.ebiom.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CM, Xie X, Zou J, Li S-H, Lee MYQ, Dong H, Qin C-F, Kang C, Shi P-Y. 2015. Determinants of Dengue virus NS4A protein oligomerization. J Virol 89:6171–6183. 10.1128/JVI.00546-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou J, Xie X, Wang Q-Y, Dong H, Lee MY, Kang C, Yuan Z, Shi P-Y. 2015. Characterization of Dengue Virus NS4A and NS4B Protein Interaction. J Virol 89:3455–3470. 10.1128/JVI.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appel N, Pietschmann T, Bartenschlager R. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol 79:3187–3194. 10.1128/JVI.79.5.3187-3194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Q, Luo Z, Zeng J, Chen W, Foo S-S, Lee S-A, Ge J, Wang S, Goldman SA, Zlokovic BV, Zhao Z, Jung JU. 2016. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 19:663–671. 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma J, Ketkar H, Geng T, Lo E, Wang L, Xi J, Sun Q, Zhu Z, Cui Y, Yang L, Wang P. 2018. Zika virus non-structural protein 4A blocks the RLR-MAVS signaling. Front Microbiol 9:1350. 10.3389/fmicb.2018.01350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar D, Kumar A, Bhardwaj T, Giri R. 2020. Zika virus NS4A N-terminal region (1-48) acts as a cofactor for inducing NTPase activity of NS3 helicase but not NS3 protease. Arch Biochem Biophys 695:108631. 10.1016/j.abb.2020.108631. [DOI] [PubMed] [Google Scholar]
- 24.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. 10.1128/jvi.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerikan B, Goellner S, Neufeldt CJ, Haselmann U, Mulder K, Chatel-Chaix L, Cortese M, Bartenschlager R. 2020. A non-replicative role of the 3′ terminal sequence of the dengue virus genome in membranous replication organelle formation. Cell Rep 32:107859. 10.1016/j.celrep.2020.107859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romero-Brey I, Merz A, Chiramel A, Lee J-Y, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8:e1003056. 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goellner S, Cerikan B, Cortese M, Neufeldt CJ, Haselmann U, Bartenschlager R. 2020. Replication-independent generation and morphological analysis of flavivirus replication organelles. STAR Protoc 1:100173. 10.1016/j.xpro.2020.100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan Am Health Organization/World Health Organization. 2016. Zika cumulative cases. Pan Am Health Organization/World Health Organization, Geneva, Switzerland. [Google Scholar]
- 29.Kraemer MU, Sinka ME, Duda KA, Mylne AQ, Shearer FM, Barker CM, Moore CG, Carvalho RG, Coelho GE, Van Bortel W, Hendrickx G, Schaffner F, Elyazar IR, Teng H-J, Brady OJ, Messina JP, Pigott DM, Scott TW, Smith DL, Wint GW, Golding N, Hay SI. 2015. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 4:e08347. 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellone R, Failloux A-B. 2020. The role of temperature in shaping mosquito-borne viruses transmission. Front Microbiol 11:584846. 10.3389/fmicb.2020.584846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dohm DJ, O'Guinn ML, Turell MJ. 2002. Effect of environmental temperature on the ability of Culex pipiens (Diptera: Culicidae) to transmit West Nile virus. J Med Entomol 39:221–225. 10.1603/0022-2585-39.1.221. [DOI] [PubMed] [Google Scholar]
- 32.Barrows NJ, Campos RK, Powell ST, Prasanth KR, Schott-Lerner G, Soto-Acosta R, Galarza-Muñoz G, McGrath EL, Urrabaz-Garza R, Gao J, Wu P, Menon R, Saade G, Fernandez-Salas I, Rossi SL, Vasilakis N, Routh A, Bradrick SS, Garcia-Blanco MA. 2016. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe 20:259–270. 10.1016/j.chom.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rausch K, Hackett BA, Weinbren NL, Reeder SM, Sadovsky Y, Hunter CA, Schultz DC, Coyne CB, Cherry S. 2017. Screening bioactives reveals nanchangmycin as a broad spectrum antiviral active against Zika virus. Cell Rep 18:804–815. 10.1016/j.celrep.2016.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adcock RS, Chu Y-K, Golden JE, Chung D-H. 2017. Evaluation of anti-Zika virus activities of broad-spectrum antivirals and NIH clinical collection compounds using a cell-based, high-throughput screen assay. Antiviral Res 138:47–56. 10.1016/j.antiviral.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 35.Han Y, Mesplède T. 2018. Investigational drugs for the treatment of Zika virus infection: a preclinical and clinical update. Expert Opin Invest Drugs 27:951–962. 10.1080/13543784.2018.1548609. [DOI] [PubMed] [Google Scholar]
- 36.Shah PS, Link N, Jang GM, Sharp PP, Zhu T, Swaney DL, Johnson JR, Von Dollen J, Ramage HR, Satkamp L, Newton B, Hüttenhain R, Petit MJ, Baum T, Everitt A, Laufman O, Tassetto M, Shales M, Stevenson E, Iglesias GN, Shokat L, Tripathi S, Balasubramaniam V, Webb LG, Aguirre S, Willsey AJ, Garcia-Sastre A, Pollard KS, Cherry S, Gamarnik AV, Marazzi I, Taunton J, Fernandez-Sesma A, Bellen HJ, Andino R, Krogan NJ. 2018. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and Zika virus pathogenesis. Cell 175:1931–1945. 10.1016/j.cell.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teo CSH, Chu JJH. 2014. Cellular vimentin regulates construction of dengue virus replication complexes through interaction with NS4A protein. J Virol 88:1897–1913. 10.1128/JVI.01249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemm JA, O'Boyle D, Liu M, Nower PT, Colonno R, Deshpande MS, Snyder LB, Martin SW, St Laurent DR, Serrano-Wu MH, Romine JL, Meanwell NA, Gao M. 2010. Identification of hepatitis C virus NS5A inhibitors. J Virol 84:482–491. 10.1128/JVI.01360-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romine JL, St Laurent DR, Leet JE, Martin SW, Serrano-Wu MH, Yang F, Gao M, O'Boyle DR, Lemm JA, Sun J-H, Nower PT, Huang XS, Deshpande MS, Meanwell NA, Snyder LB. 2011. Inhibitors of HCV NS5A: from iminothiazolidinones to symmetrical stilbenes. ACS Med Chem Lett 2:224–229. 10.1021/ml1002647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta SP. 2018. The medicinal chemistry of antihepatitis agents III, p 133–210. In Studies on hepatitis viruses. Elsevier, New York, NY. [Google Scholar]
- 41.Nakamoto S, Kanda T, Wu S, Shirasawa H, Yokosuka O. 2014. Hepatitis C virus NS5A inhibitors and drug resistance mutations. World J Gastroenterol 20:2902–2912. 10.3748/wjg.v20.i11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drake JW, Holland JJ. 1999. Mutation rates among RNA viruses. Proc Natl Acad Sci USA 96:13910–13913. 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Boheemen S, Tas A, Anvar SY, van Grootveld R, Albulescu IC, Bauer MP, Feltkamp MC, Bredenbeek PJ, van Hemert MJ. 2017. Quasispecies composition and evolution of a typical Zika virus clinical isolate from Suriname. Sci Rep 7:2368. 10.1038/s41598-017-02652-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross-Thriepland D, Harris M. 2015. Hepatitis C virus NS5A: enigmatic but still promiscuous 10 years on! J Gen Virol 96:727–738. 10.1099/jgv.0.000009. [DOI] [PubMed] [Google Scholar]
- 45.Berger C, Romero-Brey I, Radujkovic D, Terreux R, Zayas M, Paul D, Harak C, Hoppe S, Gao M, Penin F, Lohmann V, Bartenschlager R. 2014. Daclatasvir-like inhibitors of NS5A block early biogenesis of hepatitis C virus–induced membranous replication factories, independent of RNA replication. Gastroenterology 147:1094–1105. 10.1053/j.gastro.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 46.McGivern DR, Masaki T, Williford S, Ingravallo P, Feng Z, Lahser F, Asante-Appiah E, Neddermann P, De Francesco R, Howe AY, Lemon SM. 2014. Kinetic analyses reveal potent and early blockade of hepatitis C virus assembly by NS5A inhibitors. Gastroenterology 147:453–462. 10.1053/j.gastro.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sofia MJ (ed). 2019. HCV: the journey from discovery to a cure: volume II. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 48.Shiryaev SA, Farhy C, Pinto A, Huang C-T, Simonetti N, Ngono AE, Dewing A, Shresta S, Pinkerton AB, Cieplak P, Strongin AY, Terskikh AV. 2017. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antiviral Res 143:218–229. 10.1016/j.antiviral.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffmann H-H, Kunz A, Simon VA, Palese P, Shaw ML. 2011. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci USA 108:5777–5782. 10.1073/pnas.1101143108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, Fremont DH. 2016. Structural basis of Zika virus-specific antibody protection. Cell 166:1016–1027. 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi B-E, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J Virol 80:12149–12159. 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat Med 11:522–530. 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez E, Kose N, Edeling MA, Adhikari J, Sapparapu G, Lazarte SM, Nelson CA, Govero J, Gross ML, Fremont DH, Crowe JE, Diamond MS. 2018. Mouse and human monoclonal antibodies protect against infection by multiple genotypes of Japanese encephalitis virus. mBio 9:e00008-18. 10.1128/mBio.00008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mutso M, Saul S, Rausalu K, Susova O, Žusinaite E, Mahalingam S, Merits A. 2017. Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on a Brazilian Zika virus isolate. J Gen Virol 98:2712–2724. 10.1099/jgv.0.000938. [DOI] [PubMed] [Google Scholar]
- 55.Mossenta M, Marchese S, Poggianella M, Slon Campos JL, Burrone OR. 2017. Role of N-glycosylation on Zika virus E protein secretion, viral assembly and infectivity. Biochem Biophys Res Commun 492:579–586. 10.1016/j.bbrc.2017.01.022. [DOI] [PubMed] [Google Scholar]
- 56.Münster M, Płaszczyca A, Cortese M, Neufeldt C, Goellner S, Long G, Bartenschlager R. 2018. A reverse genetics system for Zika virus based on a simple molecular cloning strategy. Viruses 10:368. 10.3390/v10070368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flint M, McMullan LK, Dodd KA, Bird BH, Khristova ML, Nichol ST, Spiropoulou CF. 2014. Inhibitors of the tick-borne, hemorrhagic fever-associated flaviviruses. Antimicrob Agents Chemother 58:3206–3216. 10.1128/AAC.02393-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tripathi S, Balasubramaniam VRMT, Brown JA, Mena I, Grant A, Bardina SV, Maringer K, Schwarz MC, Maestre AM, Sourisseau M, Albrecht RA, Krammer F, Evans MJ, Fernandez-Sesma A, Lim JK, García-Sastre A. 2017. A novel Zika virus mouse model reveals strain specific differences in virus pathogenesis and host inflammatory immune responses. PLoS Pathog 13:e1006258. 10.1371/journal.ppat.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barnard T, Rajah M, Sagan S. 2018. Contemporary Zika virus isolates induce more dsRNA and produce more negative-strand intermediate in human astrocytoma cells. Viruses 10:728. 10.3390/v10120728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (Dose response confirmations.)<br>. Download JVI.00996-21-s0001.xlsx, XLSX file, 1.0 MB (1MB, xlsx)
Table S2 (Resistance mutations.)<br>. Download JVI.00996-21-s0002.xlsx, XLSX file, 0.01 MB (13KB, xlsx)
Fig. S1 to S5<br>. Download JVI.00996-21-s0003.pdf, PDF file, 1.5 MB (1.5MB, pdf)
Legend to Table S1<br>. Download JVI.00996-21-s0004.pdf, PDF file, 0.2 MB (166.8KB, pdf)
Legend to Table S2<br>. Download JVI.00996-21-s0005.pdf, PDF file, 0.2 MB (188.1KB, pdf)