ABSTRACT
Human norovirus is the leading cause of gastroenteritis worldwide, with no approved vaccine or antiviral treatment to mitigate infection. These plus-strand RNA viruses have T = 3 icosahedral protein capsids with 90 pronounced protruding (P) domain dimers, to which antibodies and cellular receptors bind. We previously demonstrated that bile binding to the capsid of mouse norovirus (MNV) causes several major conformational changes; the entire P domain rotates by ∼90° and contracts onto the shell, the P domain dimers rotate about each other, and the structural equilibrium of the epitopes at the top of the P domain shifts toward the closed conformation, which favors receptor binding while blocking antibody binding. Here, we demonstrate that MNV undergoes reversible conformational changes at pH 5.0 that are nearly identical to those observed when bile binds. Notably, at low pH or when metals bind, a cluster of acidic resides in the G′-H′ loop interact and distort the G′-H′ loop, and this may drive C′-D′ loop movement toward the closed conformation. Enzyme-linked immunosorbent assays with infectious virus particles at low pH or in the presence of metals demonstrated that all tested antibodies do not bind to this contracted form, akin to what was observed with the MNV-bile complex. Therefore, low pH, cationic metals, and bile salts are physiological triggers in the gut for P domain contraction and structural rearrangement, which synergistically prime the virus for receptor binding while blocking antibody binding.
IMPORTANCE The protruding domains on the calicivirus capsids are recognized by cell receptors and antibodies. We demonstrated that MNV P domains are highly mobile, and bile causes contraction onto the shell surface while allosterically blocking antibody binding. We present the near-atomic cryo-electron microscopy structures of infectious MNV at pH 5.0 and pH 7.5. Surprisingly, low pH is sufficient to cause the same conformational changes as when bile binds. A cluster of acidic residues on the G′-H′ loop were most likely involved in the pH effects. These residues also bound divalent cations and had the same conformation as observed here at pH 5. Binding assays demonstrated that low pH and metals block antibody binding, and thus the G′-H′ loop might be driving the conformational changes. Therefore, low pH, cationic metals, and bile salts in the gut synergistically prime the virus for receptor binding while blocking antibody binding.
KEYWORDS: cryo-EM, monoclonal antibodies, neutralizing antibodies, norovirus, structure
INTRODUCTION
Human norovirus infections are responsible for a majority of acute, nonbacterial gastroenteritis cases worldwide, and the economic impact in the United States is ∼$10 billion per year. There are ∼200 million cases per year in children less than 5 years of age, with an estimated mortality rate of 50,000 deaths per year, mainly in countries with underdeveloped or underfunded health care systems (1). Currently, no vaccines or directed antiviral treatment options are approved (2), and controlling infection has proved difficult, given the highly infectious nature of the virus and antigenic differences between strains. Further complicating matters, culture systems for producing infectious human norovirus have proved difficult to maintain and are limited in viral yields (3). Mouse norovirus (MNV) is an ideal model system to study norovirus infection because mice are the native host and represent a widely used animal model for infection, a reverse genetics system has been developed, and the virus grows well in macrophage-like cell lines (4). MNV is an enteric virus and causes both asymptomatic and symptomatic infections in mice (5, 6).
Both human norovirus and MNV belong to the Caliciviridae family and share similar structural features. The major capsid protein (VP1) (∼58 kDa) comprises the T = 3 icosahedral viral capsid. VP1 is subdivided into the N-terminal, shell (S), and protruding (P) domains (7, 8). Viral plus-stranded RNA is contained within the shell domain. The P domain is further subdivided into the P1 and P2 domains; the P1 domain lies at the base, and the P2 domain is at the apical tip. Most neutralizing antibodies (9, 10) and viral receptors/cofactors (11, 12) bind to the P2 domain.
The calicivirus capsid is not a static structure that merely transports viral RNA. In the original X-ray structure of Norwalk virus, the P domain sat directly on the shell (7, 13). Subsequently, our cryo-electron microscopy (cryo-EM) work showed that, in physiological buffers, the MNV P domains appeared to “float” >15 Å above the shell (8). This flexible P domain feature was later also found in rabbit hemorrhagic fever virus-like particles (VLPs) and human norovirus VLPs of genotype GII.10 (14, 15). Not only is the entire P domain flexible, but there is also motility within the P domain itself. The X-ray structure of the isolated P domain showed that the two most apical loops of the P2 domain (A′-B′ and E′-F′) can adopt two different conformations, in which these two loops are splayed apart (open) or tightly associated (closed). (16).
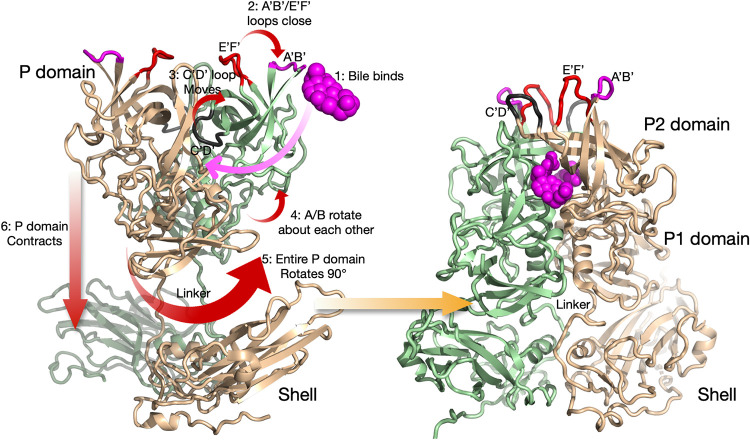
We recently showed that the MNV viral capsid undergoes dramatic structural changes upon exposure to bile salts (Fig. 1), such as glycochenodeoxycholic acid (GCDCA) (17). Although Fig. 1 shows the conformational changes in particular sequence, there may be multiple pathways that reach the same end. In the absence of added ligands, the P domain A′-B′ and E′-F′ loops (Fig. 1, mauve and red, respectively) are in equilibrium between the open and closed conformations. Two molecules of bile bind to the P domain dimer interface (step 1) to push the equilibrium toward the closed conformation (step 2). Similarly, the C′-D′ loop (Fig. 1, highlighted in black) is in equilibrium between the down position, which points toward the shell, and the up position, which associates with the A′-B′ and E′-F′ loops. When bile binds, the position of the C′-D′ loop greatly favors the up orientation (step 3). We know that the conformations of all of these loops are in equilibrium in solution, since the A′-B′ and E′-F′ loops were in both the open and closed conformations in the apo X-ray structure, while the C′-D′ loop was always in the down position (16). The C′-D′ loop also appears to be correlated with a rotation of the two P domains in the dimer relative to each other (step 4). When bile is bound, the C′-D′ loop moves to the up position and the P domains rotate counterclockwise to each other (18). This rotation creates a surface complementary to the shell and may be necessary for the P domains to rotate by 90° (step 5) and contract onto the shell (step 6). Evidence for a role of the C′-D′ loop in the contraction process was that the apo P domain X-ray structure in the closed conformation did not show the dimer proteins rotated about each other (16). However, it was not clear how the other conformational changes either cause or are a result of this contraction. In this context, the structure of MNV at low pH starts to detail the connections and correlations between these conformational changes.
FIG 1.
Summary of the conformational changes in MNV upon bile binding (18). Shown here is a capsid dimer, with the two subunits colored blue and tan. The P domain is highly flexible and in structural equilibrium in solution. The binding of bile likely drives the structural equilibrium in the following manner. Bile binds between the P domain dimers (step 1), and this is likely facilitated by the flexibility of the C′-D′ and A′-B′/E′-F′ loops. As bile binds, the equilibrium is pushed toward a closed conformation for the A′-B′ and E′-F′ loops (step 2), and the C′-D′ loop moves up to stabilize that conformation (step 3). Likely related to the C′-D′ loop movement, the P domains in the dimer rotate slightly about each other (step 4), and this new surface at the base is now compatible with that of the top of the shell. This leads to an ∼90° rotation of the entire P domain (step 5) and contraction of the P domain onto the shell (step 6). The contracted structure with the P domain in the closed conformation, after all of the conformational changes have occurred, is shown on the right.
Interestingly, antibody escape mutations for some broadly neutralizing MNV antibodies are found near the C′-D′ loop (10), far removed from the epitopes. Since bile enhances receptor binding (12), the receptor favors the contracted capsid with the P domain A′-B′ and E′-F′ loops in the closed conformation. However, from our recent high-resolution cryo-EM structure of the isolated P domain complexed with neutralizing Fab A6.2 (18), antibodies cannot bind to the closed conformation favored by the receptor. Therefore, we proposed that noroviruses (in particular, MNV) can evade host immunity by presenting one conformation to B cells while presenting a completely different structure in the gut. We also proposed that some antibody escape mutants may block antibody binding in a manner akin to bile, by pushing the C′-D′ loop into the up conformation, thereby closing the A′-B′ and E′-F′ loops.
In the work presented here, we demonstrate that MNV at pH 5.0 has a structure nearly identical to that of the MNV-bile complex. The A′-B′ and E′-F′ loops adopt the closed conformation, the C′-D′ loop moves upward toward the A′-B′ and E′-F′ loops, and the entire P domain rotates down onto the capsid shell. This may be driven at least in part by the protonation of a cluster of acidic residues on the G′-H′ loop that are immediately adjacent to the C′-D′ loop on the other P domain in the dimer. This distorted conformation of the G′-H′ loop was also found at neutral pH in the presence of cationic metal ions, with these acidic residues chelating the metal ions (12). Another major change in charge interactions is in the linker region, which is fully extended at neutral pH, with acidic and basic residues distributed along its length. At pH 5.0, these residues form a tight bundle at the P domain-shell interface, which may also be facilitated by the acidic conditions. We previously demonstrated that antibody binding is abrogated by the bile-induced conformational changes (18). Similarly, we show here that antibody binding is inhibited by low-pH conditions and the presence of cationic metals. These results further support our model in which conditions in the gut convert the structure of MNV to a contracted P domain conformation with a closed A′-B′/E′-F′ loop. This structure is primed for receptor binding while at the same time burying epitopes that are exposed outside the gut environment. These results suggest that vaccine development may need to consider the possibility that host factors may alter the structure of epitopes, as recently shown with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (19).
RESULTS
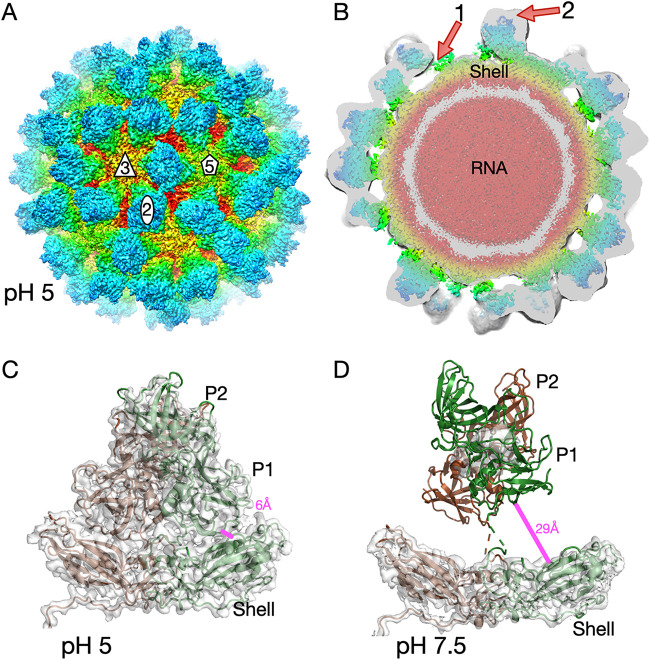
The structure of MNV is markedly flexible, with various environmental conditions causing a range of conformational changes (Fig. 1 and Table 1). This low-pH structure offers another opportunity to look at possible correlations and causations of specific structural changes. Since noroviruses are enteric viruses, they must pass through a wide range of pH conditions while traveling down the alimentary tract. Since we previously showed that the MNV capsid has marked flexibility, it was of interest to see what conformation it would adopt under acidic conditions. To that end, a single preparation of MNV was divided in two equal fractions, pelleted via ultracentrifugation, and resuspended in 25 mM sodium phosphate, 25 mM sodium citrate,100 mM sodium chloride, at either pH 5.0, the mean intestinal pH of mice (20), or pH 7.5, the pH used in previous structural studies (8, 14, 17). For both data sets, ∼200 particles were manually selected, and ab initio models were generated in cryoSPARC. Automatic particle picking was then performed with this volume and resulted in the final data sets of 244,657 and 145,347 particles for the pH 5.0 and pH 7.5 data sets, respectively. The final reconstructions had gold standard cryo-EM resolutions of ∼3.2 Å at pH 5.0 and ∼3.0 Å at pH 7.5 (Fig. 2). However, as we observed previously (17), resolution calculation at pH 7.5 did not include the entire P domain, due to its marked flexibility leading to weak density (Fig. 2D). As shown in Fig. 2, MNV at pH 5.0 is in the contracted state, in which the P domain has rotated by ∼90° and lies directly on the shell surface. As was observed in the presence of bile (17), this stabilizes the position of the P domain, and nearly all of the capsid structure could be built (Fig. 2A and C). The shell domain density was the most well defined, while the apical loops of the P domain showed some disorder. Alignment of this pH 5.0 structure with our recent structure of the MNV-bile complex (Protein Data Bank [PDB] code 6P4J) (17) yielded a root mean square deviation (RMSD) of ∼1.0 Å using all backbone atoms in PyMOL (21).
TABLE 1.
Data collection and refinement statistics for MNV at pH 5.0 and pH 7.5
| Parameter | Data for: |
|
|---|---|---|
| MNV at pH 5 | MNV at pH 7.5 | |
| PDB code | 7N6Y | 7N7F |
| EMDB accession no. | 24211 | 24226 |
| Instrument used | Titan Krios G3i | Titan Krios G3i |
| No. of images | 25,794 | 12,829 |
| No. of particles used | 244,657 | 145,347 |
| Software | cryoSPARC v2 | cryoSPARC v2 |
| Resolution (Å) | 3.28 | 3.02 |
| No. of atoms | 23,524 | 9,402 |
| Rwork (%) | 36.9 | 32 |
| Correlation coefficient, masked | 0.85 | 0.87 |
| Ramachandran plot (%) | ||
| Outliers | 0.07 | 0.16 |
| Allowed | 14.0 | 7.08 |
| Favored | 85.9 | 92.75 |
| RMSD | ||
| Bond length (Å) | 0.021 | 0.007 |
| Bond angle (°) | 1.3 | 0.72 |
| B values (Å2) | ||
| Minimum | 33 | 44 |
| Maximum | 183 | 197 |
| Mean | 92 | 105 |
FIG 2.
Cryo-EM image reconstructions of MNV at pH 7.5 and 5.0. (A) Exterior view of MNV at pH 5.0. The surface is colored from red to blue, going from the interior to the exterior surface. Icosahedral 5-, 3-, and 2-fold axes are noted on the virion. The P domains are blue and green, while the shell surface varies from yellow to red in color. (B) Central section of MNV at pH 5 (colored), with the corresponding section of the MNV pH 7.5 structure shown in translucent gray. Note that the contour level for the pH 7.5 structure was far lower than that for the pH 5.0 structure, to visualize the flexible P domain. Red arrow 1 shows the gap between the P and shell domains in the pH 7.5 structure, and arrow 2 shows that the floating P domain in the pH 7.5 structure extends much further than that in the pH 5.0 structure, while the shells in the two structures follow the same contours. (C) Structure of a P domain dimer in the pH 5.0 structure, showing that the P domain has contracted onto the shell surface. This stabilizes the position of the P domain such that the entire chain can be traced. (D) Electron density of the pH 7.5 structure at a higher (3 σ) contouring threshold. The model for the shell represents the refined structure of that domain. The P domain model from the pH 5 model was placed in the P domain density at much lower contours than those shown in panel B.
In contrast, the structure at pH 7.5 is the same as when MNV was frozen in phosphate-buffered saline (PBS) in the absence of bile (8, 14, 17). Although the cryo-EM resolution of MNV at pH 7.5 is estimated to be ∼3.0 Å, the P domain was only visible at very low contouring levels (Fig. 2B), while the shell density was clear and unambiguous (Fig. 2D). Note that, at higher contouring levels, the shell is clear at pH 7.5 but the P domain is barely visible (Fig. 2D). To highlight the difference between the floating P domain structure at pH 7.5 and the contracted structure at pH 5.0, the density of the pH 7.5 structure at a low contouring level is shown in translucent gray in Fig. 2B and superimposed on the pH 5.0 density. Arrow 1 in Fig. 2B indicates the gap between the P and shell domains in the pH 7.5 structure, and arrow 2 notes the difference in the heights of the P domains above the shell in the two structures. To further highlight the movement, the distance between residue 64 of the shell and residue 522 at the base of the P domain was measured under the two conditions. In the contracted form at pH 5.0, the distance between the two residues is ∼6.2 Å; this increases to 29.4 Å as the P domain lifts and rotates by ∼90° in the expanded conformation.
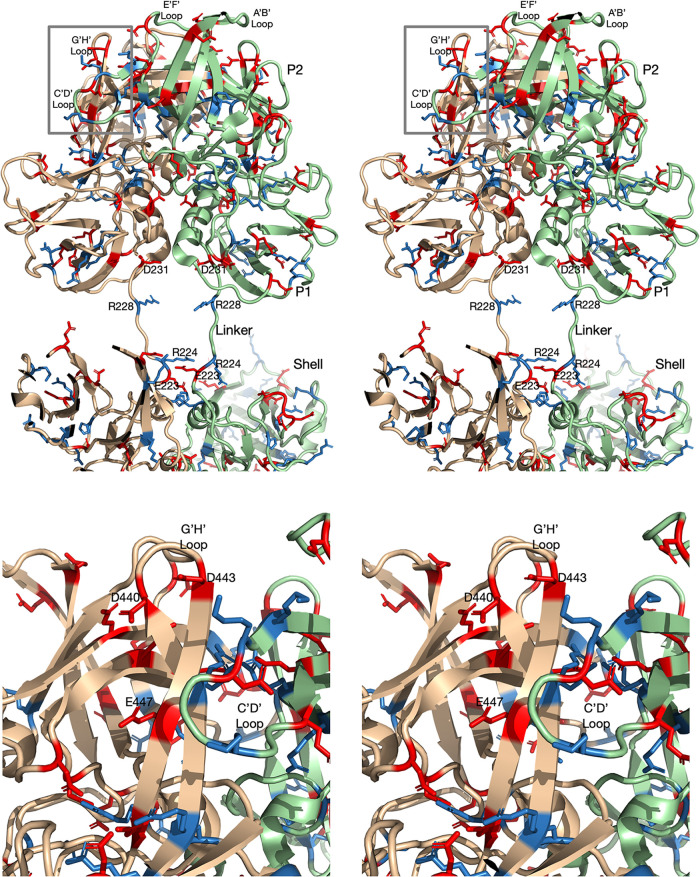
Changing the pH from 7.5 to 5.0 caused the same conformational changes as adding bile (17). As noted in Fig. 1, one of these conformational changes involves the rotation of the P domains in the dimer with respect to each other. Using PDBePISA (22), the interfaces between the A and B subunits at pH 5.0 and pH 7.5 (the P domain from the P domain-Fab A6.2 structure [18]) were compared. These contact surfaces are shown in Fig. 3, in which the acidic and basic residues are highlighted in red and blue, respectively. As is clear in Fig. 3, the contact area between the subunits increases when the pH is lowered to 5.0. From the PDBePISA calculations, the buried surface area at 7.5 is ∼1,789 Å2 and that at pH 5.0 is ∼2,174 Å2. However, as shown in Fig. 4 and 5, the linker region is extended at pH 7.5 but is tightly coiled at pH 5.0. This accounts for an additional ∼280 Å2 of P domain dimer interactions found in the contracted pH 5.0 structure that are not observed in the pH 7.5 structure, making for a total of ∼2,460 Å2 of contact between the P domain dimers at low pH.
FIG 3.
Surface contact between the A and B P domain dimers at pH 7.5 and pH 5.0. (A) for this stereo figure, the open conformation of the P domain from the P domain-Fab A6.2 structure (18) was used. The contact residues were identified using PDBePISA (22), and the surface was rendered using PyMOL (21). The acidic and basic residues are highlighted in red and blue, respectively. (B) Shown here is the contact area between the P domain dimers in the pH 5.0 EM structure. Note that the contact surface is larger than that at pH 7.5.
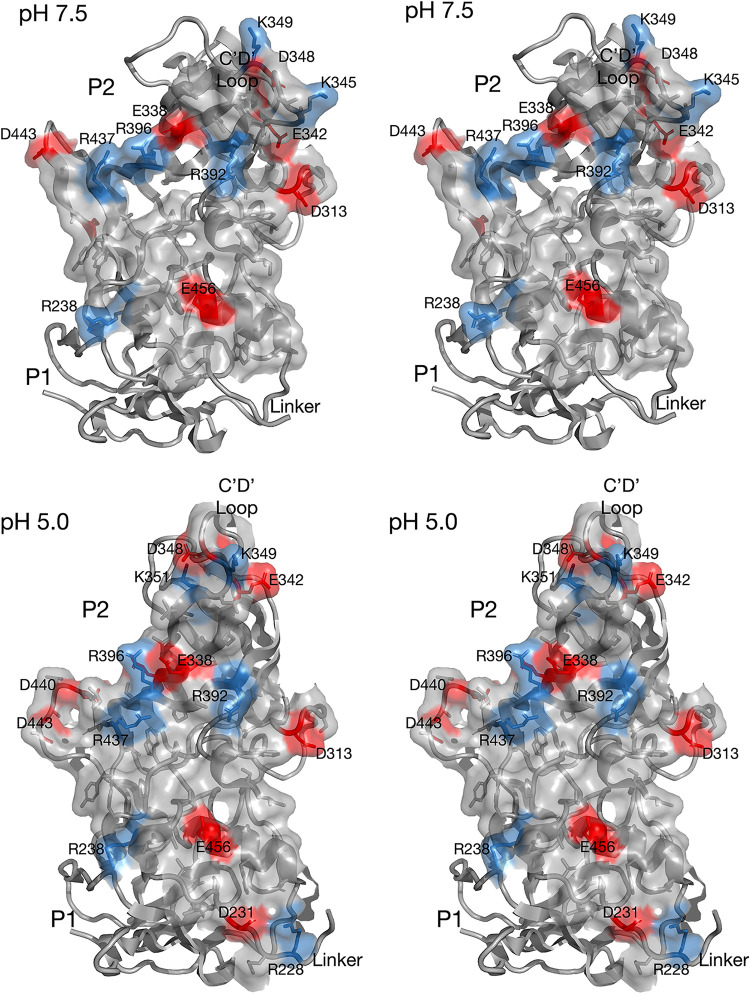
FIG 4.
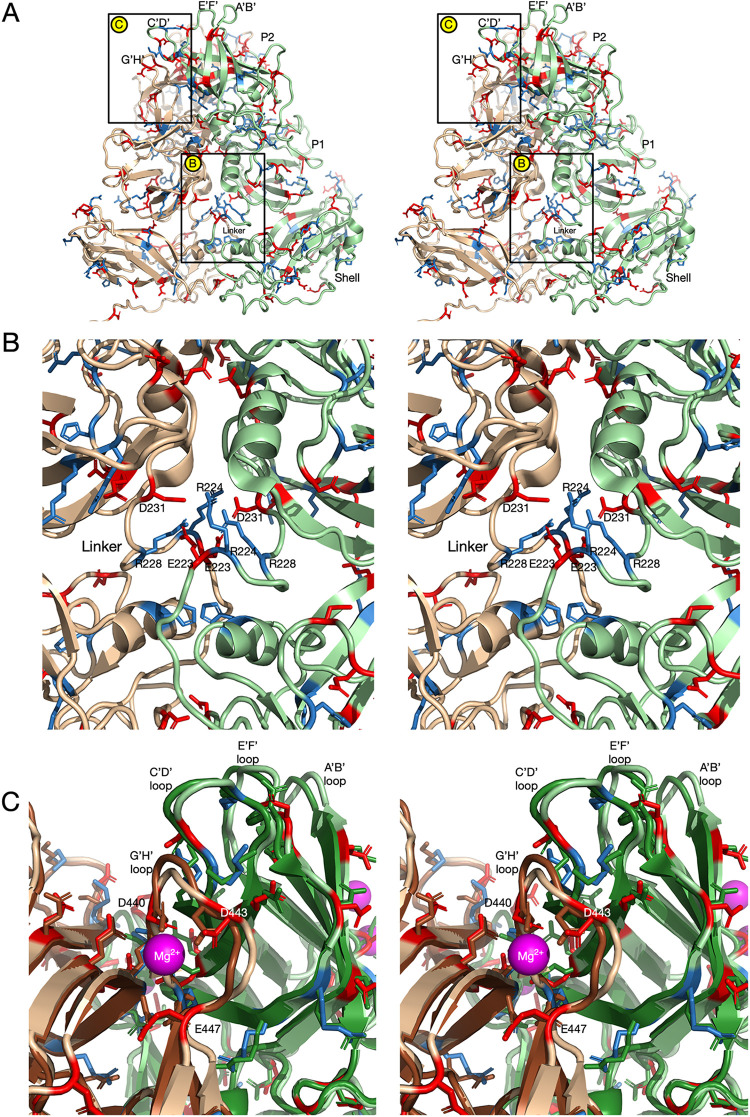
(A) Pseudo-atomic model of the MNV structure at pH 7.5. For this model, the MNV pH 7.5 shell structure was used. The structure of the P domain dimer was taken from the pH 7.5 P domain-Fab A6.2 structure and fit as a rigid body into the EM envelope of the pH 7.5 structure at low contour. The connection between the shell and P domains is visible at lower contour and forms a nearly straight tube connecting the two domains. Therefore, the backbone for the linker region is defined, but the positions of the side chains are not. To account for the conformational changes at pH 5.0, the acidic and basic residues are shown in red and blue, respectively. Note that the linker region is fully extended and that charged residues are evenly distributed along its length. (B) Close-up view of the G′-H′ and C′-D′ loops in the pH 7.5 P domain structure indicated by the box in panel A. The C′-D′ loop is pointed down and away from the A′-B′ and E′-F′ loops, and the G′-H′ loop is in a nearly vertical position. Noted are the locations of D440, D443, and E447, which cluster together at pH 5.0, as shown in Fig. 5.
FIG 5.
Structure and distribution of charged residues in MNV at pH 5.0. (A) At pH 5.0, the structure of MNV is nearly identical to the MNV-bile complex (17); the P domain has rotated and contracted onto the shell surface, the C′-D′ loop is in the up position, and the A′-B′ and E′-F′ loops are in the closed conformation. The magnified sections for panels B and C are highlighted by labeled boxes. (B) Show here is the linker region coiled up in this contracted conformation. Notably, the charged residues that are evenly distributed along the length of the linker in the expanded pH 7.5 structure are all compressed into this small volume in the contracted state. (C) Shown here are the A′-B′, E′-F′, C′-D′, and G′-H′ loops in the pH 5.0 structure (tan and light green), compared to their conformations in the X-ray structure of the isolated P domain dimer in the presence of Mg2+ (12) (brown and dark green). In both structures, the G′-H′ loop contracts onto itself and moves into the area previously occupied by the C′-D′ loop at pH 7.5. The G′-H′ loops are nearly identical between the two structures, and D440, D443, and E447 move to chelate Mg2+ in the X-ray structure. It may be that protonation at low pH has a similar effect on these acidic residues in the pH 5.0 structure.
Since the P domain dimer contact area increased at pH 5.0, this raised the question of whether this change could be due to pH effects on hydrogen bonds or salt bridges between the subunits. A summary of dimer contacts at the two pH values is shown in Table 2. Interestingly, there are only three salt bridges between the dimers, with only one uniquely found at pH 7.5. The larger changes are in the hydrogen bonds (both side chain and main chain). There are 8 hydrogen bonds at pH 7.0, and this increases to 12 at pH 5.0, with only 1 hydrogen bond being shared between the two structures. The lack of commonality between hydrogen bonds in the two structures is due to the conformational changes in the loops at the tip of the P2 domain and the rotation of the P domains relative to each other.
TABLE 2.
List of P domain dimer contacts found in MNV structures at pH 7.5, pH 5.0, or both
| First residue in bond |
Second residue in bond |
||
|---|---|---|---|
| Amino acid | Residue no. | Amino acid | Residue no. |
| Hydrogen bonds | |||
| Pa | 246 | Ra | 392 |
| Pa | 248 | Sa | 284 |
| Ya | 250 | Qa | 312 |
| Sa | 282 | Ta | 241 |
| Gb | 283 | Tb | 241 |
| Sa | 284 | Ta | 241 |
| T | 344 | Y | 435 |
| T | 346 | A | 446 |
| T | 346 | E | 447 |
| Ga | 347 | Ta | 441 |
| G | 347 | A | 446 |
| D | 348 | A | 446 |
| Ka | 349 | Ta | 441 |
| Ka | 349 | Aa | 442 |
| La | 350 | Da | 440 |
| Ra | 396 | Ta | 353 |
| Ra | 396 | Ta | 354 |
| Y | 435 | T | 346 |
| S | 459 | E | 456 |
| Salt bridges | |||
| Rb | 238 | Db | 313 |
| Rb | 396 | Eb | 338 |
| R | 437 | E | 338 |
At pH 5.0.
At both pH 7.5 and pH 5.0.
The pH-induced conformational change in the P domain is likely due to structural changes at multiple locations. To find possible key locations involved in these conformational changes, the locations of titratable charged residues were examined. Figure 4 shows a pseudo-atomic model of the expanded conformation of MNV at pH 7.5. For the P domain structure, the near-atomic resolution cryo-EM structure of the P domain of our recent P domain-Fab A6.2 complex (18) was used. The shell shown here is the ∼3-Å structure determined in this study. The linker region was modeled using the tube of density observed in this pH 7.5 density. The resolution of the linker was insufficient to accurately model side chains, but its highly extended nature constrained the linear placement of the residues. This linker region is particularly interesting since it has acidic and basic residues along its length, but the linkers in the P domain dimers are too distal to interact. This is completely different from what is found at pH 5.0 (Fig. 5), at which the linkers of the P domains in the dimer interact extensively and form numerous hydrogen bonds and salt bridges. The other region that undergoes significant pH-dependent conformational changes is at the C′-D′ and G′-H′ loops (boxed region in Fig. 4A). As discussed previously (18), there is evidence that the conformation of the C′-D′ loop may be a control point for both P domain rotation and the open/closed state of the A′-B′ and E′-F′ loops. At pH 7.5, the C′-D′ loop points downward and away from the A′-B′ and E′-F′ loops, and the β-strands leading to and from the G′-H′ loop are extended, giving the loop a rigid and vertical conformation (16, 18). In this conformation, the G′-H′ loop has three acidic residues that are too far apart to interact. Technically, E447 is not part of the G′-H′ loop at this pH, but this changes at pH 5.0 when the amino end of the H′ loop loses its β-strand conformation and becomes part of the enlarged G′-H′ loop. At pH 7.5, these acidic residues are expected to be deprotonated and repulsive due to their negative charges. It is possible that the extended conformation of the G′-H′ loop is due, at least in part, to this repulsion.
The pH 5.0 structure shows significant differences that may be driving the large conformational changes observed in the virion (Fig. 5). As shown in Fig. 2, decreasing the pH from 7.5 to 5.0 is sufficient to contract the P domain onto the shell and cause the structural changes within the P domain, as reviewed in Fig. 1. As shown in Fig. 5B, a major difference from the pH 7.5 structure is that the linker forms a tightly coiled structure with extensive interactions between the P domain dimers. While the interactions between the P domain linkers are too complex to interpret without extensive computational modeling, it is interesting to note that D231 of one subunit is close to E223 of the other. It may be that protonation of the acids at pH 5.0 partially facilitates the extensive coiling of the two linker regions.
The other region that may be affected by the acidic pH is at the C′-D′ and G′-H′ loops. At pH 5.0 (tan and light green in Fig. 5), the G′-H′ loop changes from the extended vertical conformation to a shorter bulging loop that spreads into the region previously occupied by the C′-D′ loop at pH 7.5. In this conformation, D440, D443, and E447 are clustered together, and the pKa values for the side chains likely increase to resist deprotonation. It is thus not entirely surprising that the conformation of this loop is sensitive to changes in pH. The other way to neutralize the charges of these side chains is to bind cationic metals. The previous X-ray structure of the P domain was crystallized in the presence of Mg2+ (12), and one of the locations of bound metal is at this cluster of G′-H′ acidic groups (mauve sphere and brown/light green models in Fig. 5). Notably, in the presence of metal ions, the A′-B′ and E′-F′ loops are in the closed conformation, the C′-D′ loop points up toward the A′-B′ and E′-F′ loops, and the G′-H′ loop has moved into the location previously occupied by the C′-D′ loop. This is thus consistent with the hypothesis that the G′-H′ loop conformation is at least partially dictated by the charges of these acidic residues. Interestingly, the cryo-EM structure of MNV was determined in buffer that contained magnesium and calcium and yielded a structure nearly identical to the low-pH structure presented here, although it was not clear at the time that the metals were responsible for the conformational differences from our structures (23). Therefore, we now have essentially identical structures when bile salts, metals, or pH 5.0 buffer is added to the virus, and all three conditions block antibody binding. For an animation showing the conformational changes in these loops switching from pH 7.4 to pH 5.0 and back, see Movie S1 in the supplemental material.
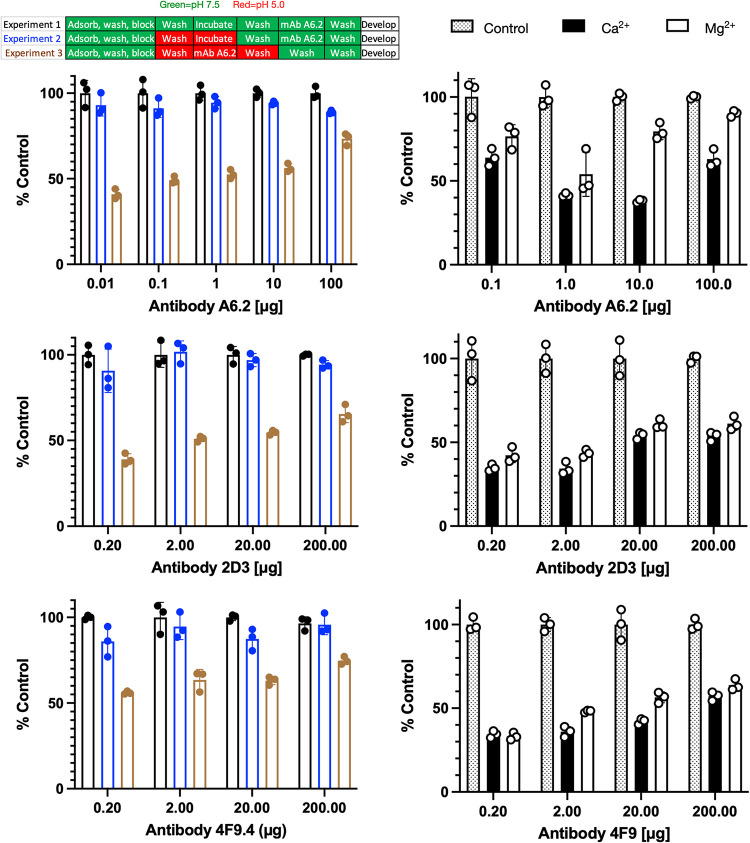
These results have a possible biological implication. As we demonstrated recently (17), bile pushes the A′-B′/E′-F′ structural equilibrium to the closed conformation, which is no longer recognized by several neutralizing monoclonal antibodies (MAbs) (18). Since acidic conditions also lead to the closed conformation, it follows that antibody binding would be inhibited at low pH (Fig. 6, left side). Since cell culture is not conducive to low-pH conditions, enzyme-linked immunosorbent assays (ELISAs) were performed using infectious virus particles. In experimental design 1 (black bars), a standard ELISA was performed using partially purified MNV. The virus was adsorbed to the plastic well and MAb A6.2 was added using the same pH 7.5 citrate-phosphate buffer as used for the structural studies. After allowing the virus and antibody to interact for 1.5 h, the excess MAb was washed away with the same buffer and the plate was developed using horseradish peroxidase (HRP)-labeled anti-mouse IgG secondary antibody. Since there was concern that the long exposure to acidic conditions might affect the virus, a viral stability control was performed (experimental design 2, blue bars). Here, the virus was treated with the citrate-phosphate buffer at pH 5.0 for 1.5 h. The acidic buffer was washed away with the pH 7.5 buffer, and then MAb A6.2 was incubated at pH 7.5. If the virus suffered irreversible damage with the low-pH treatment, then a significant drop in signal would be expected despite MAb A6.2 being added at pH 7.5. At pH 5.0, a slight drop in signal was observed. While this could be due to denaturation of the virus, the low-pH buffers might be affecting the virus adhering to the plates. In the final experiment (experimental design 3, brown bars), the adsorbed virus was washed with pH 5.0 buffer and incubated with MAb A6.2 in the same buffer. After incubation with MAb A6.2, the unbound antibody was washed away with pH 5 buffer since neutral-pH buffer could allow some antibody to bind during the washing process. The wells were then washed with pH 7.5 buffer and developed as described above. These results clearly show that MAb A6.2 does not bind as well to the whole virus at low pH, compared to neutral pH. In addition, the results from experimental design 2 show that the conformational changes in the capsid caused by low pH are reversible. Abrogation of antibody binding is consistent with the fact that the low-pH MNV structure is nearly identical to the MNV-bile complex (17), which does not bind antibody (18). These experiments were repeated with two additional neutralizing antibodies, 2D3 and 4F9, and the same reduction in binding at low pH was observed. It is important to note that the low-pH conditions, as with bile, alter the structural equilibrium and do not completely lock MNV into a particular conformation. Therefore, the effect of pH on antibody is dependent on the balance of energetics between antibody binding and pH changes, which is related to factors such as MAb concentration, MAb avidity, and pH. Therefore, as the concentration of antibody is increased, there is a notable increase in the ELISA signal.
FIG 6.
Effects of pH and metals on MAb A6.2, 2D3, and 4F9 binding to infectious particles of MNV. Shown here is the effect of low pH on A6.2 binding using the same citrate/phosphate buffers used for the structural work. The design of the experiment is outlined in the bar figure shown at the top. For experiment 1, the pH 7.5 buffer was used throughout the experiment. For experiment 2, the virus was adsorbed to the plate, treated with pH 5.0 buffer for 1 h, and washed with pH 7.5 buffer, and the experiment was completed in the same buffer. Loss of signal during this process would imply irreversible denaturation of MNV at low pH. Finally, in experiment 3, MNV was washed and incubated with pH 5.0 buffer immediately before, during, and after incubation with antibody. Loss of signal, compared to experiment 2, demonstrates that all three antibodies do not bind as well to the pH 5 conformation. Since the pH 5.0 structure is nearly identical to the X-ray structure in the presence of Mg2+ (12), ELISAs were performed with metals added during the incubation with MAbs A6.2, 2D3, and 4F9. As shown here, both calcium and magnesium significantly blocked antibody binding. Therefore, bile (18), low pH, and metals all decrease the affinity of all three antibodies to the virion by pushing the structural equilibrium to the closed conformation.
Since the conformation at pH 5.0 was nearly identical to the X-ray structure in the presence of cationic metals, a similar ELISA was performed at different Mg2+ and Ca2+ concentrations. Here, the ELISA was performed at pH 7.5 using Tris buffer, because of the interaction between the citrate-phosphate buffer and the metals. During incubation with MAbs A6.2, 2D3, and 4F9, 100 mM calcium chloride or magnesium chloride was added. As shown in Fig. 6, right side, both metals significantly blocked antibody binding. This is particularly important since calcium and magnesium, as well as bile, were found to enhance receptor binding (12). This is consistent with our previously proposed model (17, 18, 24), in which the open and closed P domain conformations are exclusively recognized by either antibodies or receptor, respectively. It is also important to note that all metal-binding locations in the X-ray structure (12) are distal to antibody contact sites (18). Therefore, metal abrogation of antibody binding is via allosteric conformational changes, as with bile, rather than through direct competition with antibody binding. It is interesting to speculate that the metal enhancement of receptor binding could be due to both direct interactions at the CD300lf receptor-P domain interface and the distortion of the G′-H′ loop that would shift the structural equilibrium to the receptor-binding closed conformation. It should be noted that the IgA antibodies (MAbs 2D3 and 4F9) are not as pure as the IgG MAb (MAb 6A), since IgA antibodies cannot be affinity purified. Therefore, it is not clear whether the small differences in metal sensitivity among the antibodies are statistically significant.
DISCUSSION
As shown in this study, there are multiple physiologically relevant triggers (bile salts, low pH, and divalent metal ions) that convert MNV from a highly flexible structure recognized by antibodies in the serum to the contracted conformation in the gut that blocks antibody binding while enhancing the affinity for receptor. As reviewed in Fig. 1, the MNV transition from the antibody-binding conformation, with open A′-B′ and E′-F′ loops and a floating P domain, to the receptor-binding, contracted capsid with a closed P domain, requires movement at several locations in the capsid. The order of these changes is likely interchangeable, with multiple possibilities, but this study has identified areas that are likely key to the transition. In this case, antibody binding acts as a sensor for the structural transition by binding only to the open A′-B′/E′-F′ conformation. The effects of low pH and metals on antibody binding are more than likely allosteric in nature. In the case of acidic pH conditions, most of the residues at the epitope-paratope interface are hydrophobic and unlikely to be greatly affected by pH changes. Similarly, three magnesium-binding sites were observed in the crystal structure of the P domain-CD300lf receptor complex (12), i.e., one at the G′-H′ loop (Fig. 5C) and two at the P domain-CD300lf interface. While the latter two metal locations could account for the enhanced receptor binding, it is also possible that the binding site at the G′-H′ loop may play an indirect role. The common feature among the bile, metal, and low-pH complexes is that the A′-B′ and E′-F′ loops are in the closed conformation, the C′-D′ loop is in the up orientation, and the G′-H′ loop fills the space formerly occupied by the C′-D′ loop. Further, in all of these structures, the A and B subunits are rotated about each other, compared to the pH 7.5 apo structure. It is fascinating that these three rather disparate conditions yield the same collection of conformational changes. As we suggested previously, perhaps the C′-D′ loop is the key to these transitions (18). For bile to bind, the C′-D′ loop is necessarily in the up position. That, in turn, requires the A′-B′ and E′-F′ loops to be in the closed conformation, and the G′-H′ loop becomes distorted to fill in the space left after the G′-H′ movement. The effects of acidic pH and metals may indirectly affect the C′-D′ conformation by distorting the G′-H′ loop structure. Since receptor binding favors the closed conformation, these conditions may directly and/or indirectly promote receptor binding.
Perhaps one of the more puzzling changes in the MNV capsid structure under all of these conditions is the contraction of the P domain onto the shell. We have demonstrated that the rotation of the P domains within the spike dimers is necessary to create a surface complementary to the shell, and this may be related to the C′-D′ loop movement (18). What we observed here is that the linker region in the floating conformation has charged residues evenly dispersed along its length, which become a dense bundle of charges in the contracted state. Perhaps these interactions help to decrease the energy barrier for the capsid to transition from the expanded state to the contracted state. The fact that such disparate environmental conditions can lead to the same conformation suggests that the capsid is unstable at neutral pH and there are multiple trigger sites throughout the P domain that can lead to the contracted state.
It is important to note that these results show that the rotation of the P domains about each other in the dimer and the conformational change to the closed conformation drives the contraction onto the shell, rather than the other way around. The original crystal structure of the P domain at neutral pH, without metals, and without bile has the C′-D′ loop in the down position (16), the P domains are not rotated about each other (in contrast to the contracted structure), and the A′-B′ and E′-F′ loops are observed in both the open and closed conformations. Even in the isolated P domain, the presence of bile (12) moves the C′-D′ loop up and the P domains rotate about each other, which creates a surface at the P domain base that complements the shell surface (18). At pH 7.5, in the absence of bile and metals (18), the isolated P domain appears to be in the open conformation, the C′-D′ loop is in the down position, and the P domains have not rotated about each other. Finally, in structures of the infectious particles in the presence of bile (17), at pH 5.0, or in the presence of metals (23), the C′-D′ loop is in the up position, the P domains are rotated about each other, the A′-B′ and E′-F′ loops are in the closed conformation, and the P domain contracts onto the surface. All of these results strongly suggest that the P domain does not need the shell to adopt the closed, contracted conformation. Further, these results show that the P domain is not driven to the closed conformation by the binding of the receptor; rather, these conditions drive the P domain to the closed conformation to which the receptor binds. What is not clear, however, is the biological significance of the contraction of the P domain onto the surface. While we have presented models suggesting that this contraction might increase receptor binding valency, bury the P1 and shell domain epitopes (24), or protect the virion during transit through the gut, the true biological function remains to be determined.
These results demonstrate the remarkably plastic nature of MNV and the biological importance of this flexibility. Viruses exist on the precipice of disaster; they need to be stable enough to survive the movement between cells and hosts, while being sufficiently unstable to respond to environmental stimuli required for infection and genome release. For example, a number of plant viruses just need a relatively slight push with neutral-pH conditions and low metal concentrations to start uncoating (e.g., references 25 and 26). In the case of human rhinovirus, the virus transiently exposes its myristylated capsid N termini in a “breathing” process while waiting for the receptor to bind (27). This delicate state is disrupted if the capsid is stabilized by the addition of hydrophobic antiviral compounds (27) or destabilized by acidic conditions.
MNV differs from these viruses in that it uses at least three cues, i.e., pH, bile, and cationic metals, to adapt its structure to the changing environment. Since MNV is an enteric virus, it needs to adapt to wildly changing conditions in the gastrointestinal tract. From these results, MNV is likely in the contracted state as it passes from the stomach through to the jejunum, where the pH values start at ∼2 and eventually reach 7 in the later jejunum. Interestingly, there are several possible trypsin cleavage sites (i.e., lysine and arginine residues) in the extended, flexible linker (Fig. 4). Therefore, it is tempting to speculate that MNV, much like a turtle, contracts at low pH to protect itself from denaturation and digestion. MNV then enters the duodenum, where it is exposed to all three compounds that cause contraction and enhanced receptor binding. The acidic conditions enhance metal solubility (e.g., reference 28), which both enhances receptor binding (12) and causes contraction (Fig. 2). Finally, the common bile duct connects at the duodenum, and bile concentrations in the early gut are 2.5 to 45 mM (29). Therefore, there are multiple environmental cues in the gut that push the MNV structural equilibrium to favor receptor binding while distorting the epitopes at the tip of the P domain and occluding those at the base of the P domain and shell (18, 24). Finally, once the virus spreads from the gut lumen and enters cells or serum, the pH is no longer acidic and the bile and metal concentrations drop to negligible levels. Under these conditions, the P domain rises off the shell and the P domain changes to the open conformation. In this way, MNV uses all of the environmental cues in the gut to protect itself while optimizing cell binding. Once the infection spreads outside the gut, it may present a completely different mask to confuse the immune response.
It is interesting to consider the details of what structures are presented in the serum as the virus spreads and the immune system responds. All of the MAbs raised from infected mice that we have studied to date (8–10, 14, 16, 18, 30) clearly recognize the open conformation observed under low-metal, neutral-pH, and low-bile conditions. Importantly, the conditions used in the ELISAs when screening the hybridomas also favored the open conformation. Recognizing that the MNV structure is in a structural equilibrium between the open and closed conformations, it is possible that there were hybridomas that were able to bind to the closed conformation but were not selected in the screening. Therefore, we cannot say that the mice did not produce any antibodies to the closed conformation. Regarding the fact that the receptor apparently binds to the closed conformation, it is incorrect to suggest that MNV cannot bind to receptors and infect tissue outside the gut and, indeed, MNV causes systemic infection. Bile and metals enhance receptor binding to MNV but are not absolutely required for virus-receptor interactions. Surface plasmon resonance studies using isolated CD300lf and P domains showed weak but measurable interactions, with a binding constant of ∼220 μM. Calcium improved that ∼10-fold to ∼25 μM, and the combination of bile and calcium acted synergistically to yield a dissociation constant (Kd) of ∼12 μM (12). Therefore, it is more accurate to say that the conditions in the gut enhance receptor binding while decreasing the ability of antibodies to bind. Certainly, these levels of enhanced cell attachment and antibody blockade are sufficient to drive the evolution of these structural switches.
The capsid plasticity observed here is quite different from that observed in viruses that mainly use large conformational changes to initiate uncoating, such as the picornaviruses and influenza virus. In this regard, recent studies on SARS-CoV-2 suggest that MNV may not be alone in using metabolites to thwart antibody binding (19). Those authors demonstrated that heme metabolites bind to the N-terminal domain and allosterically abrogate neutralizing antibody binding. Clearly, identification of hijacked metabolites or changes in physiological conditions that viruses employ to evade the immune response will be critical for the development of more efficacious vaccines for a potentially wide range of viruses.
MATERIALS AND METHODS
MAb A6.2, 4F9, and 2D3 production, purification, and digestion.
MAbs were produced as described previously (9). Briefly, hybridoma cells were grown in Bioreactor 1000 flasks, the cells were removed by centrifugation, and the antibody was precipitated with a 50% (final concentration) saturated solution of ammonia sulfate. A6.2, an IgG, was then purified using a protein G column according to the manufacturer’s recommendation. 4F9 and 2D3 are IgA antibodies and were purified via size exclusion chromatography, as described previously (10).
ELISAs.
An indirect ELISA was used to measure the effect of pH on antibody binding. In the control experiment, 1 × 105 PFU/ml MNV in pH 7.5 buffer was used as the antigen; 100 μl of this diluted virus was added to the wells and allowed to attach overnight at 4°C. Wells were blocked with 5% milk in pH 7.5 buffer for 4 h at room temperature. The plate was washed extensively with pH 7.5 buffer before the addition of the intact IgG A6.2 antibody at neutral pH; 0.1 μg, 1 μg, 10 μg, or 100 μg antibody was added to the wells and allowed to attach for 1.5 h before extensive washing at neutral pH. A secondary HRP-conjugated anti-mouse IgG antibody (1:5,000 dilution) was added at neutral pH and incubated for 1.5 h. Plates were developed using the HRP substrate o-phenylenediamine dihydrochloride (OPD), according to the manufacturer’s protocol, and the reaction was quenched with 3 M HCl after ∼30 min. A PHERAstar plate reader equipped with a 490-nm filter was used for plate reading, and Prism was used for analysis.
A second control experiment (experiment 2) tested the effects of low pH on the virus. Virus was adsorbed to the plates and blocked as described above. Wells were then washed with acidic buffers. Wells were incubated at low pH for 1 h at room temperature and then extensively washed at neutral pH. Secondary antibody was added, and the plate was developed as described above.
Finally, the effects of low pH on antibody binding were determined (experiment 3). Virus was adsorbed, and the plates were blocked as described above and then washed extensively with low-pH buffer. Antibody that had been dialyzed in low-pH buffer was added in various amounts and incubated for 1.5 h at room temperature. Excess antibody was removed by washing with low-pH buffer, and then the wells were washed with neutral-pH buffer to prepare for secondary antibody binding. Secondary antibody was added at neutral pH, and the plates were developed as described above.
Mg2+ and Ca2+ were shown to enhance receptor binding (12), the atomic structure of the P domain in the presence of receptor and metal had the closed conformation (12), and a previous cryo-EM structure showed a contracted structure at neutral pH in the presence of these metals (23). Together, these results suggested that these metals push the P domain to the closed conformation and would block antibody binding. To that end, the effects of Mg2+ and Ca2+ on antibody binding were determined. MNV1 adsorption to the ELISA wells and blocking with milk were performed as in the low-pH experiments, and 100 mM CaCl2 or MgCl2 was added to the antibody dilutions prior to antibody addition to the wells. Primary antibody was allowed to attach for 1.5 h, with all subsequent steps and data processing as described for the low-pH conditions. The buffer used was 50 mM Tris (pH 7.6) instead of the citrate-phosphate buffer, to prevent metal precipitation. The plates were washed, developed, and analyzed as described above.
Virus production and purification.
MNV-1 was produced and purified essentially as described previously (17). In brief, when BV-2 (RRID: CVCL_0182) cells reached a density of ∼0.5 × 106 to 1.0 × 106 cells/ml, ∼6 liters of cells were harvested by centrifugation at 4,000 × g for 10 min. The cells were suspended in ∼200 ml of AH medium and placed into a 4-liter flask, and ∼1 × 109 PFU of MNV was added. AH media, per liter, is composed of Hyclone DMEM/Low Glucose (Cat. SH30002.04), 10 ml of a 1× solution of non-essential amino acids (NEAA), 0.062 g Penicillin G, 0.4 g streptomycin sulfate, 2.2 g NaHCO3, and 5.9 g HEPES. This media is adjusted to pH 7.2 and sterilized by filtration. The suspension was slowly shaken for 1 h at room temperature, ∼800 ml of fresh AH medium was added, and the suspension was then transferred to a 37°C incubator without CO2 and shaken at ∼70 rpm in the dark for 24 to 32 h. The infected cell suspension was centrifuged for 30 min at 5,000 × g, and the supernatant was collected. To the supernatant, dry NaCl and polyethylene glycol (PEG) 8000 were added to yield 0.3 M and 10%, respectively. The solution was then mixed at 4°C overnight. The solution was centrifuged for 30 min at 10,000 × g, and the pellet was resuspended in 50 to 80 ml of PBS. After incubation at 4°C for several hours, the debris was removed by centrifugation at 10,000 × g for 30 min. Glycerol was added to the supernatant to a final concentration of 10% (vol/vol) as a cryoprotectant, and the mixture was divided into 1.5-ml aliquots and stored at −80°C.
Immediately before the cryo-EM experiments, 50 to 100 ml of this material was thawed and centrifuged at 45,000 rpm for 2 h with a 1-ml, 30% solution (wt/vol) of sucrose as a cushion at the bottom. The pellets were resuspended in less than 3 ml of PBS and allowed to incubate for several hours at 4°C. Debris was removed by centrifugation, and the supernatant was then layered into Beckman SW41 tubes containing 7.5 to 45% linear sucrose gradients, using PBS as the buffer. After centrifugation for 1.5 to 2.0 h at 35,000 rpm at 4°C, MNV forms a band approximately two-thirds of the way down the tube. The virus was collected by puncturing the side of the tube with a syringe. The pooled bands were divided into two equal parts and pelleted as described above. The pellets were resuspended in ∼100 μl of 100 mM NaCl, 25 mM citrate, 25 mM phosphate, at either pH 5.0 or pH 7.5, and immediately frozen for cryo-EM studies. In this way, the virus samples for both conditions came from the same preparation, were purified at the same time, and were resuspended in the same buffer but at different pH values.
Cryo-EM.
Purified MNV at pH 5 and pH 7.4 was at concentrations of ∼1 mg/ml. The virus was vitrified as described previously (31), on carbon holey film (R2x1 Quantifoil; Micro Tools GmbH, Jena, Germany) grids. Briefly, grids were cleaned with Gatan 950 Solarus plasma cleaner for 40 s in a hydrogen-oxygen gas mixture, and 4-μl aliquots of the two virus solutions were applied to the holey films; the films were blotted with filter paper and plunged into liquid ethane. The EM-GP2 (Leica) automated plunger was used for vitrification.
The grids were screened for ice and sample quality and imaged with a Titan Krios G3i microscope (Thermo Fisher Scientific). The microscope was equipped with a Gatan K3 BioQuantum filter (Ametek, Inc.) and operated at 300 keV. A slit width of 20 eV was used for data collection. Images were acquired with EPU software (Thermo Fisher Scientific) using fast acquisition mode, with beam-image shift used for hole centering instead of stage movement. The direct detector camera (K3; Ametek) was operated in superresolution counted mode, images were recorded with an overall electron dose of 48 electrons/Å2, and the defocus range was −1.5 to −2.5 μm. The data collection statistics are summarized in Table 1.
Image processing.
For the sample of MNV at pH 5.0, 14,244 micrographs were recorded, and a final total of 244,657 particles were picked using the cryoSPARC v2 (32) template picker and culled using two-dimensional (2D) classification. The initial model was generated using the ab initio routine in cryoSPARC v2 with icosahedral symmetry. This initial model was used as an initial volume in the nonuniform refinement algorithm and after several cycles yielded a cryo-EM density with an effective resolution of the MNV structure at pH 5.0 of 3.3 Å, according to the 0.143 gold standard Fourier shell correlation (FSC) criterion.
For the sample of MNV at pH 7.5, 12,829 micrographs were collected, and 145,347 particles were extracted using the cryoSPARC v2 template picker and used for 2D classification. Initial model generation and refinement in cryoSPARC used the same parameters as for pH 5.0. After several rounds of nonuniform refinement, MNV at pH 7.5 was found to have an effective resolution of 3.0 Å, according to the 0.143 FSC criterion. It should be noted that, while the shell domain had a calculated resolution of ∼3 Å, the highly flexible P domain was significantly disordered and could be interpreted only by rigid body fitting of the known P domain structure into the envelope.
Structure refinement.
All structure refinement was performed using PHENIX (33). From the density of the pH 5.0 complex, it was clear that the virion had the contracted conformation, as observed for the MNV-bile complex (17); therefore, that structure was used as an initial model for building and refinement in PHENIX. The model was manually fitted into the EM envelope, and several cycles of real space refinement (rigid body, global minimization, and simulated annealing) in PHENIX (33) and rebuilding in COOT (34) were performed. To ensure that the model of the icosahedral asymmetric unit did not move into the density of adjacent subunits, the model used for real space refinement included neighboring subunits. The final refinement statistics are shown in Table 1.
In the case of virus at pH 7.5, it was evident that the capsid was in an expanded conformation with the P domain floating >10 Å above the shell, identical to the apo state described previously (17). Just as with previous structures (8, 16, 17), the density of the P domain was too diffuse to allow for proper building. To create an approximate model for the figures, the P domain structure from the MNV P domain-Fab A6.2 complex at pH 7.5 (18) was manually fitted into the envelop and then refined as a rigid body in Chimera (35). Given the weak density of the P domain at pH 7.5, only the shell domain was used for refinement. The apo MNV shell domain served as an initial model. Just as for pH 5.0, multiple rounds of refinement and rebuilding were performed using PHENIX (33) and COOT (34), respectively. The final refinement statistics are shown in Table 1.
Data availability.
The coordinates have been deposited in the PDB with accession codes 7N6Y and 7N7F for the pH 5.0 and pH 7.5 structures, respectively, and the cryo-EM maps have been deposited in the EMDB with accession numbers 24211 and 24226 for the pH 5.0 and pH 7.5 structures, respectively.
ACKNOWLEDGMENTS
This work was supported by NIH grants 1R01-AI141465 (T.J.S. and B.M.P.) and R21-AI154647 (C.E.W.). A.N.W. was supported by the James W. McLaughlin Fellowship Fund. We acknowledge the support of the Sealy Center for Structural Biology at the University of Texas Medical Branch at Galveston. B.M.P. thanks the Robert A. Welch Foundation, H-0013.
Footnotes
Supplemental material is available online only.
Contributor Information
Thomas J. Smith, Email: thosmith@utmb.edu.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCES
- 1.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. 2014. Global prevalence of norovirus in cases of gastroenteritis; a systemic review and meta-analysis. Lancet Infect Dis 14:725–730. 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debbink K, Lindesmith LC, Baric RS. 2014. The state of norovirus vaccines. Clin Infect Dis 58:1746–1752. 10.1093/cid/ciu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Estes MK, Ettayebi K, Tenge VR, Murakami K, Karandikar U, Lin S-C, Ayyar BV, Cortes-Penfield NW, Haga K, Neill FH, Opekun AR, Broughman JR, Zeng X-L, Blutt SE, Crawford SE, Ramani S, Graham DY, Atmar RL. 2019. Human norovirus cultivation in nontransformed stem cell-derived human intestinal enteroid cultures: success and challenges. Viruses 11:638. 10.3390/v11070638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hwang S, Alhatlani B, Arias A, Caddy SL, Christodoulou C, Cunha JB, Emmott E, Gonzalez-Hernandez M, Kolawole A, Lu J, Rippinger C, Sorgeloos F, Thorne L, Vashist S, Goodfellow I, Wobus CE. 2014. Murine norovirus: propagation, quantification, and genetic manipulation. Curr Protoc Microbiol 33:15K.2.1–15K.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wobus CE, Thackray LB, Virgin HWI. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol 80:5104–5112. 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wobus CE. 2018. The dual tropism of noroviruses. J Virol 92:e01010-17. 10.1128/JVI.01010-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasad BVV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290. 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 8.Katpally U, Wobus CE, Dryden K, Virgin HWI, Smith TJ. 2008. Structure of antibody-neutralized murine norovirus and unexpected differences from viruslike particles. J Virol 82:2079–2088. 10.1128/JVI.02200-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolawole AO, Li M, Xia C, Fischer AE, Giacobbi NS, Rippinger CM, Proescher JB, Wu SK, Bessling SL, Gamez M, Yu C, Zhang R, Mehoke TS, Pipas JM, Wolfe JT, Lin JS, Feldman AB, Smith TJ, Wobus CE. 2014. Flexibility in surface-exposed loops in a virus capsid mediates escape from antibody neutralization. J Virol 88:4543–4557. 10.1128/JVI.03685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolawole AO, Smith HQ, Svoboda SA, Lewis MS, Sherman MB, Lynch GC, Pettitt BM, Smith TJ, Wobus CE. 2017. Norovirus escape from broadly neutralizing antibodies is limited to allosteric-like mechanisms. mSphere 2:e00334-17. 10.1128/mSphere.00334-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilic T, Koromyslova A, Malak V, Hansman GS. 2018. Atomic structure of the murine norovirus protruding domain and soluble CD300lf receptor complex. J Virol 92:e00413-18. 10.1128/JVI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson CA, Wilen CB, Dai Y-N, Orchard RC, Kim AS, Stegeman RA, Hsieh LL, Smith TJ, Virgin HW, Fremont DH. 2018. Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. Proc Natl Acad Sci USA 115:E9201–E9210. 10.1073/pnas.1805797115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasad BVV, Hardy ME, Estes MK. 2000. Structural studies of recombinant Norwalk capsids. J Infect Dis 181(Suppl 2):S317–S321. 10.1086/315576. [DOI] [PubMed] [Google Scholar]
- 14.Katpally U, Voss NR, Cavazza T, Taube S, Rubin JR, Young VL, Stuckey J, Ward VK, Virgin HW, Wobus CE, Smith TJ. 2010. High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. J Virol 84:5836–5841. 10.1128/JVI.00314-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansman GS, Taylor DW, McLellan JS, Smith TJ, Georgiev I, Tame JRH, Park S-Y, Yamazaki M, Gondaira F, Miki M, Katayama K, Murata K, Kwong PD. 2012. Structural basis for broad detection of genogroup II noroviruses by a monoclonal antibody that binds to a site occluded in the viral particle. J Virol 86:3635–3646. 10.1128/JVI.06868-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taube S, Rubin JR, Katpally U, Smith TJ, Kendall A, Stuckey JA, Wobus CE. 2010. High-resolution X-ray structure and functional analysis of the murine norovirus 1 capsid protein protruding domain. J Virol 84:5695–5705. 10.1128/JVI.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman MB, Williams AN, Smith HQ, Nelson C, Wilen CB, Fremont DH, Virgin HW, Smith TJ. 2019. Bile salts alter the mouse norovirus capsid conformation: possible implications for cell attachment and immune evasion. J Virol 12:e00970-19. 10.1128/JVI.00970-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams AN, Sherman MB, Smith HQ, Taube S, Pettitt BM, Wobus CE, Smith TJ. 2021. A norovirus uses bile salts to escape antibody recognition while enhancing receptor binding. J Virol 95:e00176-21. 10.1128/JVI.00176-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosa A, Pye VE, Graham C, Muir L, Seow J, Ng KW, Cook NJ, Rees-Spear C, Parker E, dos Santos MS, Rosadas C, Susana A, Rhys H, Nans A, Masino L, Roustan C, Christodoulou E, Ulferts R, Wrobel A, Short C-E, Fertleman M, Sanders RW, Heaney J, Spyer M, Kjær S, Riddell A, Malim MH, Beale R, MacRae JI, Taylor GP, Nastouli E, van Gils MJ, Rosenthal PB, Pizzato M, McClure MO, Tedder RS, Kassiotis G, McCoy LE, Doores KJ, Cherepanov P. 2021. SARS-CoV-2 recruits a haem metabolite to evade antibody immunity. medRxiv 2021.01.21.21249203. 10.1101/2021.01.21.21249203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McConnell EL, Basit AW, Murdan S. 2010. Measurements of rat and mouse gastrointestinal pH, fluid and lymphoid tissue, and implications for in-vivo experiments. J Pharm Pharmacol 60:63–70. 10.1211/jpp.60.1.0008. [DOI] [PubMed] [Google Scholar]
- 21.DeLano WL. 2008. The PyMOL molecular graphics system. DeLano Scientific LLC, Palo Alto, CA. [Google Scholar]
- 22.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Snowden JS, Hurdiss DL, Adeyemi OO, Ranson NA, Herod MR, Stonehouse NJ. 2020. Dynamics in the murine norovirus capsid revealed by high-resolution cryo-EM. PLoS Biol 18:e3000649. 10.1371/journal.pbio.3000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith HQ, Smith TJ. 2019. The dynamic capsid structures of the noroviruses. Viruses 11:235. 10.3390/v11030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherman MB, Kakani K, Rochon DA, Jiang W, Voss NR, Smith TJ. 2017. Stability of cucumber necrosis virus at the quasi-6-fold axis affects zoospore transmission. J Virol 91:e01030-17. 10.1128/JVI.01030-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman MB, Guenther R, Reade R, Rochon DA, Sit T, Smith TJ. 2020. Near atomic resolution cryo-electron microscopy structures of cucumber leaf spot virus and red clover necrotic mosaic virus: evolutionary divergence at the icosahedral three-fold axes. J Virol 94:e01439-19. 10.1128/JVI.01439-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis JK, Bothner B, Smith TJ, Siuzdak G. 1998. Antiviral agent blocks breathing of the common cold virus. Proc Natl Acad Sci USA 95:6774–6778. 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goss SL, Lemons KA, Kerstetter JE, Bogner RH. 2010. Determination of calcium salt solubility with changes in pH and PCO2, simulating varying gastrointestinal environments. J Pharm Pharmacol 59:1485–1492. 10.1211/jpp.59.11.0004. [DOI] [PubMed] [Google Scholar]
- 29.Sjövall J. 1959. On the concentration of bile acids in the human intestine during absorption. bile acids and steroids 74. Acta Physiol Scand 46:339–345. 10.1111/j.1748-1716.1959.tb01763.x. [DOI] [PubMed] [Google Scholar]
- 30.Kolawole AO, Xia C, Li M, Gamez M, Yu C, Rippinger CM, Yucha RE, Smith TJ, Wobus CE. 2014. Newly isolated mAbs broaden the neutralizing epitope in murine norovirus. J Gen Virol 95:1958–1968. 10.1099/vir.0.066753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman MB, Guenther RH, Tama F, Sit TL, Brooks CL, Mikhailov AM, Orlova EV, Baker TS, Lommel SA. 2006. Removal of divalent cations induces structural transitions in red clover necrotic mosaic virus revealing a potential mechanism for RNA release. J Virol 80:10395–10406. 10.1128/JVI.01137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Punjani A, Rubinstein JA-O, Fleet DJ, Brubaker MA-O. 2017. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14:290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 33.Afonine PV, Grosse-Kunstleve RW, Adams PD. 2005. The Phenix refinement framework. CCP4 Newsl 42:contribution 8. [Google Scholar]
- 34.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Goddard TD, Huang CC, Ferrin TE. 2007. Visualizing density maps with UCSF Chimera. J Struct Biol 157:281–287. 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1<br>. Download JVI.01471-21-s0001.mov, MOV file, 2.4 MB (2.4MB, mov)
Supplemental Movie Legend<br>. Download JVI.01471-21-s0002.pdf, PDF file, 0.01 MB (15.9KB, pdf)
Data Availability Statement
The coordinates have been deposited in the PDB with accession codes 7N6Y and 7N7F for the pH 5.0 and pH 7.5 structures, respectively, and the cryo-EM maps have been deposited in the EMDB with accession numbers 24211 and 24226 for the pH 5.0 and pH 7.5 structures, respectively.