ABSTRACT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged at the end of 2019 and has been responsible for the still ongoing coronavirus disease 2019 (COVID-19) pandemic. Prophylactic vaccines have been authorized by the U.S. Food and Drug Administration (FDA) for the prevention of COVID-19. Identification of SARS-CoV-2-neutralizing antibodies (NAbs) is important to assess vaccine protection efficacy, including their ability to protect against emerging SARS-CoV-2 variants of concern (VoC). Here, we report the generation and use of a recombinant (r)SARS-CoV-2 USA/WA1/2020 (WA-1) strain expressing Venus and an rSARS-CoV-2 strain expressing mCherry and containing mutations K417N, E484K, and N501Y found in the receptor binding domain (RBD) of the spike (S) glycoprotein of the South African (SA) B.1.351 (beta [β]) VoC in bifluorescent-based assays to rapidly and accurately identify human monoclonal antibodies (hMAbs) able to neutralize both viral infections in vitro and in vivo. Importantly, our bifluorescent-based system accurately recapitulated findings observed using individual viruses. Moreover, fluorescent-expressing rSARS-CoV-2 strain and the parental wild-type (WT) rSARS-CoV-2 WA-1 strain had similar viral fitness in vitro, as well as similar virulence and pathogenicity in vivo in the K18 human angiotensin-converting enzyme 2 (hACE2) transgenic mouse model of SARS-CoV-2 infection. We demonstrate that these new fluorescent-expressing rSARS-CoV-2 can be used in vitro and in vivo to easily identify hMAbs that simultaneously neutralize different SARS-CoV-2 strains, including VoC, for the rapid assessment of vaccine efficacy or the identification of prophylactic and/or therapeutic broadly NAbs for the treatment of SARS-CoV-2 infection.
IMPORTANCE SARS-CoV-2 is responsible of the COVID-19 pandemic that has warped daily routines and socioeconomics. There is still an urgent need for prophylactics and therapeutics to treat SARS-CoV-2 infections. In this study, we demonstrate the feasibility of using bifluorescent-based assays for the rapid identification of hMAbs with neutralizing activity against SARS-CoV-2, including VoC in vitro and in vivo. Importantly, results obtained with these bifluorescent-based assays recapitulate those observed with individual viruses, demonstrating their feasibility to rapidly advance our understanding of vaccine efficacy and to identify broadly protective human NAbs for the therapeutic treatment of SARS-CoV-2.
KEYWORDS: SARS-CoV-2, coronavirus, in vitro, in vivo, monoclonal antibodies, neutralizing antibodies, recombinant virus, reporter genes, reverse genetics
INTRODUCTION
The emergence of SARS-CoV-2 at the end of 2019 has been responsible for the COVID-19 pandemic (1). Despite numerous efforts to contain viral spread, SARS-CoV-2 disseminated worldwide, and as of today, it has been linked to over 175 million infections and more than 3.8 million deaths around the world (2). To date, one antiviral drug (remdesivir) and three human monoclonal antibodies (hMAbs) (casirivimab/imdevimab, bamlanivimab/etesevimab, and sotrovimab) have been approved by the U.S. Food and Drug Administration (FDA) for the treatment of COVID-19 (3–5). As of June 2021, six prophylactic vaccines against SARS-CoV-2 have been authorized by the U.S. FDA to prevent SARS-CoV-2 infection (6–8). However, recent evidence suggests that newly identified SARS-CoV-2 variants of concern (VoC) are not efficiently neutralized by sera from naturally infected or vaccinated individuals (9), raising concerns about the protective efficacy of current vaccines against emerging SARS-CoV-2 VoC (10–12).
To investigate SARS-CoV-2 infection in vitro and in vivo, including tissue and cell tropism and pathogenesis, recombinant viruses expressing a variety of reporter genes have been generated. We and others have documented the generation of recombinant (r)SARS-CoV-2 expressing fluorescent (Venus, mCherry, mNeonGreen, and green fluorescent protein [GFP]) or luciferase (Nluc) reporter genes (13–16) and their use for the identification of neutralizing antibodies (NAbs) or antivirals (14–19). Importantly, these reporter-expressing rSARS-CoV-2 strains have been shown to have growth kinetics and plaque phenotypes in cultured cells like those of their parental rSARS-CoV-2 wild type (WT). Current rSARS-CoV-2 strains have been genetically engineered to express the reporter gene by replacing the open reading frame (ORF) encoding the 7a viral protein, an approach similar to that used with SARS-CoV (16, 20).
Recently, we described the generation of rSARS-CoV-2 expressing reporter genes where the porcine teschovirus 1 (PTV-1) 2A autoproteolytic cleavage site was placed between the reporter gene of choice and the viral nucleocapsid (N) protein (20). Three major advantages of this new approach are (i) all viral proteins are expressed (e.g., the insertion of the reporter does not replace or remove a viral protein) (20), (ii) high levels of reporter gene expression from the N locus in the viral genome (20), and (iii) high genetic stability of the viral genome in vitro and in vivo because of the need of the viral N protein for genome replication and gene transcription (20). Importantly, this new approach allowed the visualization of infected cells in vitro and supported tracking SARS-CoV-2 infection in vivo (20). Notably, these new reporter-expressing rSARS-CoV-2 exhibited WT-like plaque size phenotype and viral growth kinetics in vitro, as well as pathogenicity in K18 human angiotensin-converting enzyme 2 (hACE2) transgenic mice.
Using this strategy, we have successfully rescued Venus-and mCherry-expressing rSARS-CoV-2 USA/WA1/2020 (WA-1) and a new rSARS-CoV-2 strain expressing mCherry and containing mutations K417N, E484K, and N501Y present in the receptor binding domain (RBD) of the viral spike (S) glycoprotein of the South Africa (SA) B.1.351 (beta [β]) VoC (12). Using rSARS-CoV-2 WA-1 expressing Venus and rSARS-CoV-2 SA expressing mCherry, we developed a novel bifluorescent-based assay to readily and accurately evaluate hMAbs able to specifically neutralize one or both viral variants. Importantly, the 50% neutralizing titers (NT50) obtained with this new bifluorescent-based assay correlated well with those obtained using individual viruses in separated wells. Moreover, we also demonstrated the feasibility of using rSARS-CoV-2 expressing different S and fluorescent proteins (FP) to rapidly identify hMAbs able to neutralize in vivo both SARS-CoV-2 strains using an in vivo imaging system (IVIS). These new tools will help advance our understanding of the efficacy of current and future SARS-CoV-2 vaccines, as well as contribute to the identification of hMAbs with broadly neutralizing activity against SARS-CoV-2 strains, including VoC, for the therapeutic or prophylactic treatment of SARS-CoV-2 infection.
RESULTS
Generation and characterization of rSARS-CoV-2 expressing FPs.
The pBeloBAC11 plasmid encoding the full-length viral genome of SARS-CoV-2 WA-1 was used as backbone to generate the different rSARS-CoV-2 (16, 20, 21). We constructed new rSARS-CoV-2 reporter viruses that retained all viral genes by cloning the Venus or mCherry FP upstream of the viral N gene using the PTV-1 2A autocleavage sequence (Fig. 1A) (20). Recombinant viruses expressing FPs using this experimental approach based on the use of the 2A cleavage site from the N locus do not require removing any viral genes (20), express higher levels of reporter gene expression compared to those previously described from the locus of the ORF7a (20), and are genetically more stable (20).
FIG 1.
Generation and characterization of Venus and mCherry-expressing rSARS-CoV-2. (A) Schematic representation of Venus and mCherry rSARS-CoV-2. Reporter genes Venus (green) or mCherry (red) were inserted upstream of the N protein (dark blue), flanked by the PTV-1 2A autocleavage sequence (light blue). (B and C) Venus and mCherry expression from rSARS-CoV-2. (B) Vero E6 cells (6-well plate format, 106 cells/well, triplicates) were mock infected or infected (MOI of 0.01) with rSARS-CoV-2 Venus or rSARS-CoV-2 mCherry. (C) At 24 hpi, cells were fixed in 10% neutral buffered formalin and visualized under a fluorescence microscope for Venus or mCherry expression. A cross-reactive mMAb against SARS-CoV N protein (1C7C7) was used for staining of infected cells. DAPI was used for nuclear staining. FL, fluorescent field. (D to F) Multistep growth kinetics. (D)Vero E6 cells (6-well plate format, 106 cells/well, triplicates) were mock infected or infected (MOI of 0.01) with rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry, alone or together, or rSARS-CoV-2 WT, and tissue-cultured supernatants were collected at the indicated times postinfection to assess viral titers using standard plaque assay. (E) The number of Venus- and/or mCherry-positive rSARS-CoV-2 strains at the same times postinfection in cells infected with both viruses was also determined using plaque assay. (F) Images of infected cells under a fluorescence microscope at the same times postinfection are shown. (G) Plaque assays. Vero E6 cells (6-well plate format, 106 cells/well, triplicates) were mock infected or infected with ∼20 PFU of rSARS-CoV-2, rSARS-CoV-2 Venus, rSARS-CoV-2 mCherry, or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry. At 72 hpi, fluorescent plaques were assessed using a ChemiDoc instrument. Viral plaques were also immunostained with the SARS-CoV N protein 1C7C7 cross-reactive mMAb. Fluorescent green, red, and merged images are shown. Representative images are shown for panels B, C, F, and G. Scale bars, 300 μm. Statistical significance was determined by one-way ANOVA analysis on GraphPad Prism. *, P < 0.05; ns, no significance.
To characterize the newly generated FP-expressing rSARS-CoV-2, we first assessed Venus and mCherry expression levels. Confluent monolayers of Vero E6 cells were infected (multiplicity of infection [MOI] of 0.01 PFU/cell) with either rSARS-CoV-2 wild type (WT), rSARS-CoV-2 Venus, or rSARS-CoV-2 mCherry, or mock infected, and then examined by fluorescence microscopy (Fig. 1B). As expected, only cells infected with rSARS-CoV-2 Venus or rSARS-CoV-2 mCherry were detected under a fluorescence microscope (Fig. 1B). Cells infected with rSARS-CoV-2 WT, rSARS-CoV-2 Venus, and rSARS-CoV-2 mCherry showed comparable levels of N protein expression (Fig. 1C).
We next determined the multistep growth kinetics of the newly generated rSARS-CoV-2. Vero E6 cells were infected (MOI of 0.01 PFU/cell) with rSARS-CoV-2 Venus or rSARS-CoV-2 mCherry, individually or together, and tissue culture supernatants collected over a course of 96 h to determine viral titers (Fig. 1D). Wild-type rSARS-CoV-2 was included as a control. Kinetics of production and peak titers of infectious progeny were similar for rSARS-CoV-2 expressing Venus or mCherry and WT. Results from coinfection experiments using Venus- and mCherry-expressing rSARS-CoV-2 indicated that both viruses had similar fitness under the experimental conditions used (Fig. 1E). This conclusion was further validated by assessing FP expression in cells infected with rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry, alone or in combination (Fig. 1F). Moreover, both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry exhibited similar plaque formation efficiency and plaque size phenotype as the parental rSARS-CoV-2 WT (Fig. 1G).
A bifluorescent-based assay for the identification of SARS-CoV-2 NAbs.
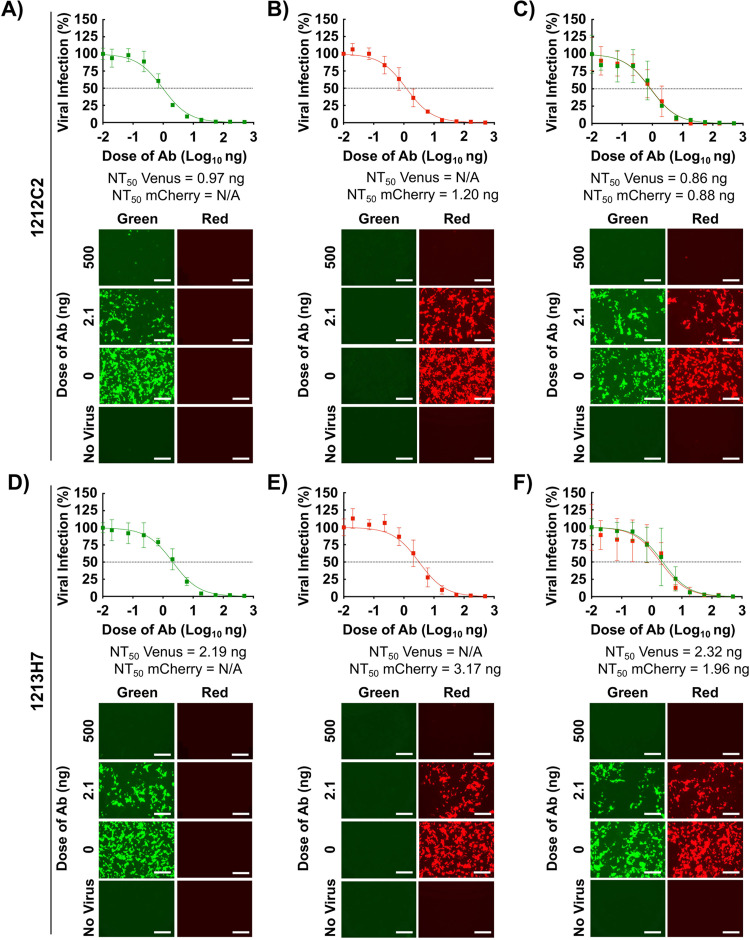
We next assessed the feasibility of using these two FP-expressing rSARS-CoV-2, alone and in combination, to identify NAbs against SARS-CoV-2. For proof of concept, we used hMAbs 1212C2 and 1213H7, both previously shown to potently neutralize rSARS-CoV-2 (22, 23). The NT50 values of 1212C2 against rSARS-CoV-2 Venus (0.97 ng) (Fig. 2A), rSARS-CoV-2 mCherry (1.20 ng) (Fig. 2B), as well as rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry together (0.86 ng and 0.88 ng, respectively) (Fig. 2C) were similar to those reported using a natural SARS-CoV-2 WA-1 isolate (16, 22). The NT50 of 1213H7 against rSARS-CoV-2 Venus (2.19 ng) (Fig. 2D), rSARS-CoV-2 mCherry (3.17 ng) (Fig. 2E), and both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry together (2.32 ng and 1.96 ng, respectively) (Fig. 2F) were similar to those obtained with the natural SARS-CoV-2 WA-1 isolate (16). These results demonstrated the feasibility of using rSARS-CoV-2 expressing Venus and mCherry reporter genes in a new bifluorescent-based assay to identify SARS-CoV-2 NAbs.
FIG 2.
Bifluorescent-based assay to identify Nabs. Confluent monolayers of Vero E6 cells (4 × 104 cells/well, 96-plate well format, quadruplicates) were infected (MOI of 0.1) with rSARS-CoV-2 Venus (A and D), rSARS-CoV-2 mCherry (B and E), or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry (C and F). After 1 h infection, postinfection medium containing 3-fold serial dilutions of 1212C2 (A to C) or 1213H7 (D to F) hMAbs (starting concentration, 500 ng) was added to the cells. At 48 hpi, cells were fixed with 10% neutral buffered formalin, and levels of fluorescence expression were quantified in a fluorescent plate reader and analyzed using Gen5 data analysis software (BioTek). The NT50 values of 1212C2 and 1213H7 hMAbs for each virus, alone or in combination, were determined using GraphPad Prism. Dashed lines indicate 50% viral neutralization. Data are means and SD from quadruplicate wells. Representative images are shown. Scale bars, 300 μm.
Generation and characterization of rSARS-CoV-2 mCherry SA.
The emergence of new SARS-CoV-2 VoC, including the SA B.1.351 (beta [β]) (12), is a major health threat since the efficacy of current vaccines against recently identified VoC may be diminished. We sought to develop an assay that would allow us to evaluate the protective efficacy of hMAbs against WA-1 and SA VoC within the same well. Towards this objective, we generated an rSARS-CoV-2 containing the K417N, E484K, and N501Y mutations found in the S RBD of the SA strain of SARS-CoV-2 and expressing also mCherry, referred to as rSARS-CoV-2 mCherry SA (Fig. 3A). The genetic identity of the rescued rSARS-CoV-2 mCherry SA was confirmed by Sanger sequencing (Fig. 3B).
FIG 3.
Generation and characterization of rSARS-CoV-2 mCherry SA. (A) Schematic representation of rSARS-CoV-2 mCherry SA. The genome of a rSARS-CoV-2 Venus strain (top) and the rSARS-CoV-2 strain with the three mutations (K417N, E484K, and N501Y) present in the S RBD of the SA B.1.351 (β) VoC expressing mCherry (bottom) is shown. (B) Sequencing of rSARS-CoV-2 mCherry SA. Sanger sequencing results of the rSARS-CoV-2 Venus strain (top) and the rSARS-CoV-2 mCherry SA with the K417N, E484K, and N501Y substitutions in the RBD of the S glycoprotein (bottom) are indicated. (C) Reporter gene expression. Vero E6 cells (6-well plate format, 106 cells/well, triplicates) were mock infected or infected (MOI of 0.01) with rSARS-CoV-2, rSARS-CoV-2 Venus, or rSARS-CoV-2 mCherry SA. Infected cells were fixed in 10% neutral buffered formalin at 24 hpi and visualized under a fluorescence microscope for Venus or mCherry expression. (D to F) Multicycle growth kinetics. Vero E6 cells (6-well plate format, 106 cells/well, triplicates) were mock infected or infected (MOI 0.01) with rSARS-CoV-2 Venus, rSARS-CoV-2 mCherry SA, or both rSARS-CoV-2 Venus and rSARS-COV-2 mCherry SA. (D) Tissue-cultured supernatants were collected at the indicated times postinfection to assess viral titers using standard plaque assay. (E) The number of Venus- and/or mCherry-positive plaques at the same times postinfection was determined using fluorescence microscopy. (F) Images of infected cells under a fluorescence microscope at the same times postinfection are shown. (G) Plaque assays. Vero E6 cells (6-well plate format, 106 cells/well, triplicates) were mock infected or infected with ∼20 PFU of rSARS-CoV-2, rSARS-CoV-2 Venus, rSARS-CoV-2 mCherry SA, or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA. At 72 hpi, fluorescent plaques were assessed using a ChemiDoc instrument. Viral plaques were also immunostained with the SARS-CoV N protein 1C7C7 cross-reactive mMAb. Fluorescent green, red, and merged images are shown. Representative images are shown for panels C, F, and G. Scale bars, 300 μm. Statistical significance was determined by one-way ANOVA analysis on GraphPad Prism. **, P < 0.01; ***, P < 0.001; ns, no significance.
Next, we aimed to characterize the rSARS-CoV-2 mCherry SA by assessing reporter expression levels using rSARS-CoV-2 and rSARS-CoV-2 Venus as controls. Vero E6 cells were infected (MOI, 0.01 PFU/cell) with rSARS-CoV-2 WT, rSARS-CoV-2 Venus, or rSARS-CoV-2 mCherry SA, and expression of Venus and mCherry assessed by epifluorescence microscopy (Fig. 3C). Only cells infected with rSARS-CoV-2 Venus or rSARS-CoV-2 mCherry SA were fluorescent. However, immunostaining with the SARS-CoV cross-reactive N protein mouse MAb (mMAb) (1C7C7) detected cells infected with rSARS-CoV-2 WT, rSARS-CoV-2 Venus, and rSARS-CoV-2 mCherry SA (Fig. 3C). Next, we compared the growth kinetics of rSARS-CoV-2 mCherry SA and rSARS-CoV-2 Venus in Vero E6 cells (Fig. 3D to F). Interestingly, at all hours postinfection (hpi) tested, tissue culture supernatants from rSARS-CoV-2 mCherry SA-infected cells had higher viral titers than those from rSARS-CoV-2 Venus-infected cells (Fig. 3D), which correlated with a higher number of mCherry- than Venus-positive cells in cells coinfected with rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA (Fig. 3E and F). These results were further confirmed when we assessed multiplication of rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA by plaque assay (Fig. 3G). Larger plaque foci were observed in cells infected with rSARS-CoV-2 mCherry SA than those infected with rSARS-CoV-2 Venus (Fig. 3G). We have also observed a similar fitness advantage of a natural SARS-CoV-2 SA natural isolate over SARS-CoV-2 WA-1 (24).
A bifluorescent-based assay to identify SARS-CoV-2 broadly NAbs.
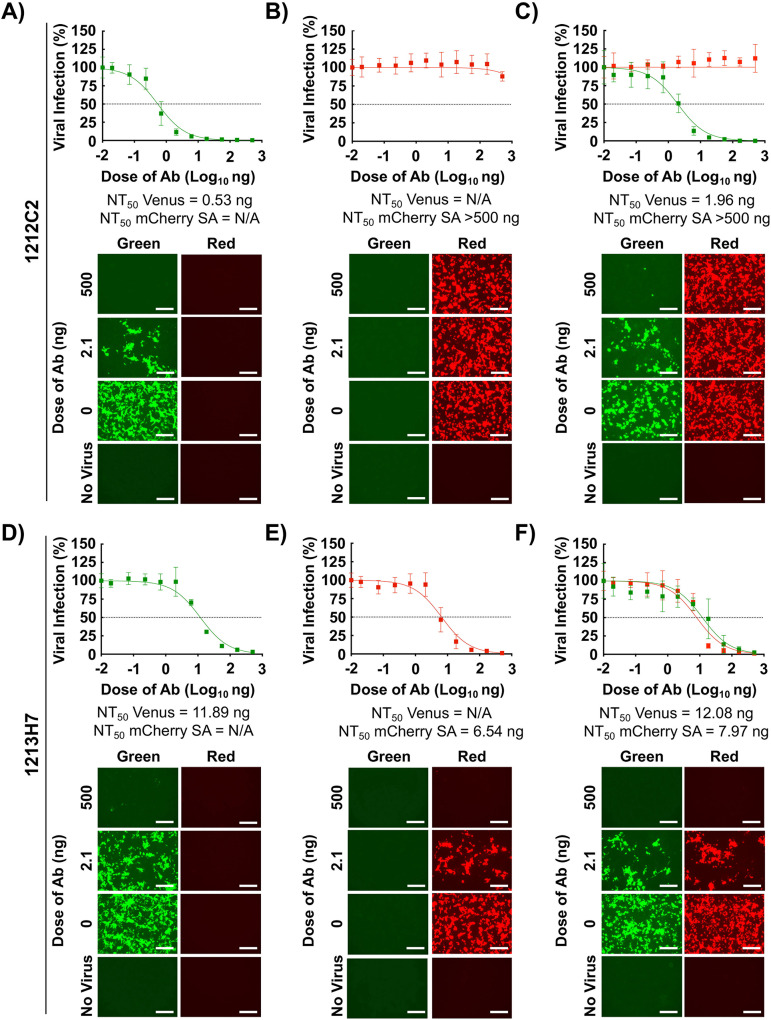
We next evaluated whether the rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA could be used in a bifluorescent-based assay to identify broadly NAbs, using the 1212C2 and 1213H7 hMAbs (Fig. 2). Preliminary data using natural SARS-CoV-2 WA-1 and SA isolates showed that 1212C2 neutralized SARS-CoV-2 WA-1 but not SARS-CoV-2 SA VoC, while 1213H7 neutralized both viral isolates (22, 23). As expected, 1212C2 was able to efficiently neutralize rSARS-CoV-2 Venus (NT50, 0.53 ng) (Fig. 4A) but not rSARS-CoV-2 mCherry SA (NT50 > 500 ng) (Fig. 4B), alone or in combination (NT50 values of 1.96 ng and >500 ng, respectively) (Fig. 4C). In contrast, 1213H7 was able to efficiently neutralize both rSARS-CoV-2 Venus (NT50, 11.89 ng) (Fig. 4D) and rSARS-CoV-2 mCherry SA (NT50, 6.54 ng) (Fig. 4E), alone or in combination (NT50 values of 12.08 and 7.97 ng, respectively) (Fig. 4F). These results demonstrated the feasibility of using this novel bifluorescent-based assay to readily and reliably identify hMAbs with neutralizing activity against both SARS-CoV-2 strains within the same assay and that these results recapitulated those of experiments following individual viral infections and classical neutralization assays using natural viral isolates.
FIG 4.
A bifluorescent-based assay to identify SARS-CoV-2 broadly Nabs. Confluent monolayers of Vero E6 cells (4 × 104 cells/well, 96-plate well format, quadruplicates) were infected (MOI of 0.1) with rSARS-CoV-2 Venus (A and D), rSARS-CoV-2 mCherry SA (MOI of 0.01) (B and E), or both rSARS-CoV-2 Venus (MOI of 0.1) and rSARS-CoV-2 mCherry SA (MOI of 0.01) (C and F). After 1 h infection, postinfection media containing 3-fold serial dilutions of 12C2C2 (A to C) or 1213H7 (D to F) hMAbs (starting concentration, 500 ng) were added to the cells. At 48 hpi, cells were fixed with 10% neutral buffered formalin, and levels of fluorescence expression were quantified in a fluorescent plate reader and analyzed using Gen5 data analysis software (BioTek). The NT50 values of 1212C2 and 1213H7 hMAbs for each virus, alone or in combination, were determined using GraphPad Prism. Dashed lines indicate 50% viral neutralization. Data are means and SD from quadruplicate wells. Representative images are shown. Scale bars, 300 μm.
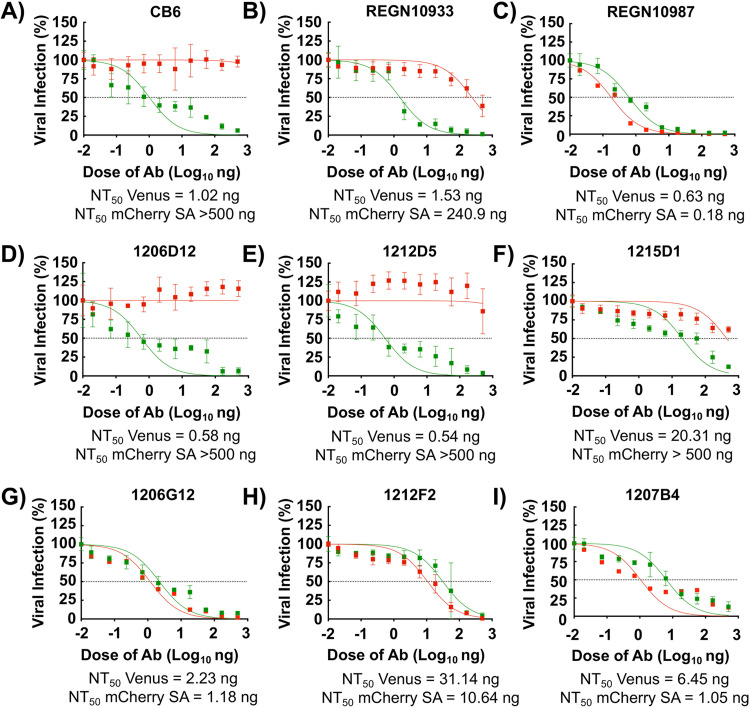
To further demonstrate the feasibility of this new bifluorescent-based assay to identify hMAbs able to neutralize different SARS-CoV-2 strains present in the same sample, we assessed the neutralizing activity of a selected set of previously described hMAbs (22). CB6, REGN10933, and REGN10987 hMAbs were used as internal controls in the assay (25, 26). CB6 (Fig. 5A) and REGN10933 (Fig. 5B) neutralized rSARS-CoV-2 Venus (NT50 values of 1.02 and 1.53 ng, respectively) but exhibited limited (REGN10933, NT50 > 240.9 ng) or no (CB6, NT50 > 500 ng) neutralization activity against rSARS-CoV-2 mCherry SA. On the other hand, REGN10987 (Fig. 5C) efficiently neutralized both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA (NT50 values of 0.63 and 0.18 ng, respectively) (Fig. 5C). Some of the tested hMAbs were also able to specifically neutralize rSARS-CoV-2 Venus but not rSARS-CoV-2 mCherry SA, including 1206D12 (NT50 values of 0.58 and >500 ng, respectively) (Fig. 5D), 1212D5 (NT50 values of 0.54 and >500 ng, respectively) (Fig. 5E), and 1215D1 (NT50 values of 20.31 and >500 ng, respectively) (Fig. 5F). We identified hMAbs with broadly neutralizing activity against both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA, including 1206G12 (NT50 values of 2.23 and 1.18 ng, respectively) (Fig. 5G), 1212F2 (NT50 values of 31.14 and 10.64 ng, respectively) (Fig. 5H), and 1207B4 (NT50 values of 6.45 and 1.05 ng, respectively) (Fig. 5I). These results support the feasibility of this novel bifluorescent-based assay to identify broad neutralizing hMAbs against different SARS-CoV-2 strains within the same assay.
FIG 5.
Identification of SARS-CoV-2 broadly NAbs using the bifluorescent-based assay. Confluent monolayers of Vero E6 cells (4 × 104 cells/well, 96-plate well format, quadruplicates) were coinfected with rSARS-CoV-2 Venus (MOI of 0.1) and rSARS-CoV-2 mCherry SA (MOI of 0.01). After 1 h infection, postinfection media containing 3-fold serial dilutions (starting concentration, 500 ng) of CB6 (A), REGN10933 (B), REGN10987 (C), 1206D12 (D), 1212D5 (E), 1215D1 (F), 1206G12 (H), 1212F2 (H), or 1207B4 (I) hMAbs were added to the cells. At 48 hpi, cells were fixed with 10% neutral buffered formalin, and levels of fluorescence were quantified using a fluorescent plate reader and analyzed using Gen5 data analysis software (BioTek). The NT50 value of each of the hMAbs was determined using GraphPad Prism. Dashed lines indicate 50% viral neutralization. Data are means and SD from quadruplicate wells. Representative images are shown. Scale bars, 300 μm.
An in vivo bifluorescent-based assay to identify SARS-CoV-2 broadly NAbs.
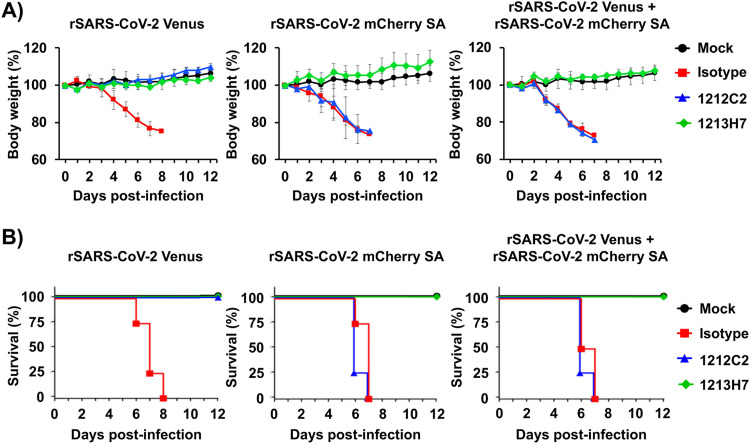
Based on our in vitro results, we hypothesized that our novel bifluorescent-based assay to identify NAbs against different SARS-CoV-2 strains could be adapted to assess the neutralizing activity of hMAbs in vivo. To test this hypothesis, we assessed the ability of 1212C2 and 1213H7 hMAbs to neutralize rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA, alone or in combination, in the K18 hACE2 transgenic mouse model of SARS-CoV-2 infection (Fig. 6) (27). Mice were treated intraperitoneally (i.p.) with 25 mg/kg of 1212C2, 1213H7, or an IgG isotype control 24 h prior to challenge with 104 PFU of rSARS-CoV-2 Venus, rSARS-CoV-2 mCherry SA, or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA together. Body weight (Fig. 6A) and survival (Fig. 6B) were evaluated for 12 days postinfection (dpi). IgG isotype control-treated mice infected with rSARS-CoV-2 Venus, rSARS-CoV-2 mCherry SA, or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA together exhibited weight loss starting on 4 dpi (Fig. 6A) and succumbed to viral infection between 6 and 8 dpi (Fig. 6B). However, all mice treated with 1212C2 or 1213H7 survived challenge with rSARS-CoV-2 Venus, consistent with efficient neutralization of SARS-CoV-2 WA-1 in vitro by these two hMAbs (Fig. 2 and 4). In contrast, only 1213H7, but not 1212C2, was able to protect mice infected with rSARS-CoV-2 mCherry SA (Fig. 6A and B), consistent with the inability of 1212C2 to neutralize rSARS-CoV-2 mCherry SA in vitro (Fig. 4). When mice were coinfected with both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA, only mice treated with 1213H7 retained their initial body weight and survived infection (Fig. 6A and B, right panels), similar to results obtained using individual infections.
FIG 6.
Prophylactic activity of 1212C2 and 1213H7 against rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA, alone or in combination, in K18 hACE2 transgenic mice. Six- to 8-week-old female K18 hACE2 transgenic mice (n = 5) were treated (i.p.) with 25 mg/kg of IgG isotype control, hMAb 1212C2, or hMAb 1213H7 and infected with 104 PFU of rSARS-CoV-2 Venus (left), rSARS-CoV-2 mCherry SA (middle), or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA (right). Mice were monitored for 12 days for changes in body weight (A) and survival (B). Data represent the means and SD of the results determined for individual mice.
Use of FP expression to assess kinetics of SARS-CoV-2 multiplication in the lungs of infected K18 hACE2 transgenic mice.
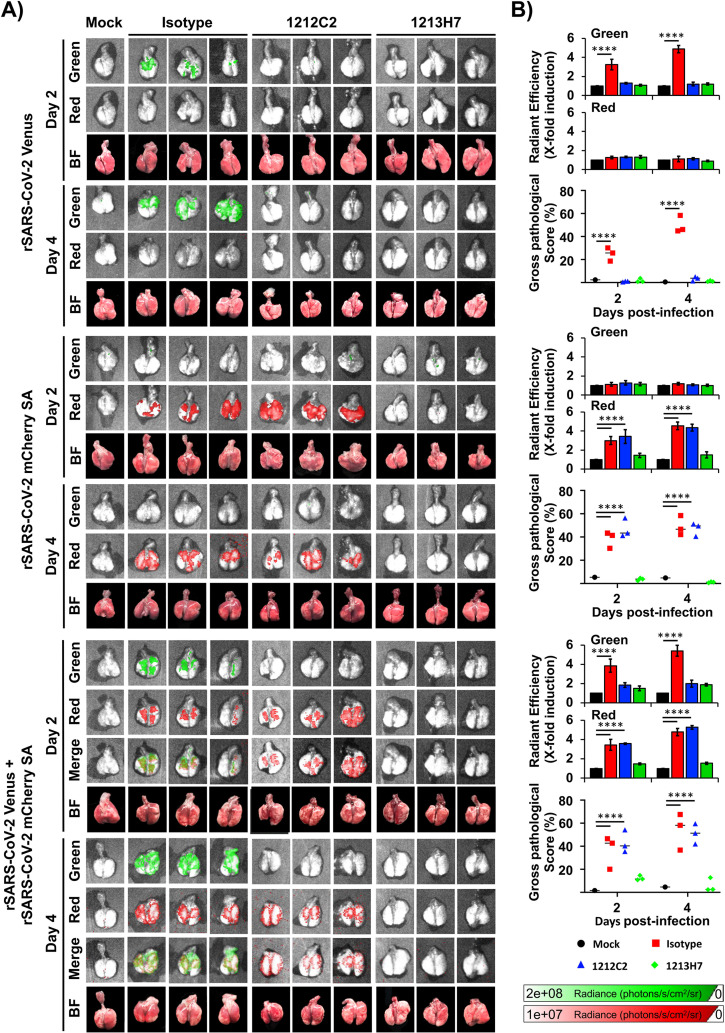
We next examined whether FP expression could be used as a surrogate of SARS-CoV-2 multiplication in the lungs of infected mice, providing a readout to assess the in vivo protective activity of 1212C2 and 1213H7 hMAbs through IVIS (Fig. 7). K18 hACE2 transgenic mice were treated (i.p., 25 mg/kg) with IgG isotype control, 1212C2, or 1213H7 hMAbs 24 h before infection (104 PFU/mouse) with rSARS-CoV-2 Venus and/or rSARS-CoV-2 mCherry SA, singly or in combination. Mock-infected mice were included as control. At 2 and 4 dpi, Venus and mCherry expression in the lungs was evaluated using IVIS (Fig. 7A) and quantified using Aura imaging software (Fig. 7B). Excised lungs were also evaluated in a blinded manner by a certified pathologist to provide gross pathological scoring (Fig. 7A). Both Venus and mCherry expression was detected in the lungs of mice treated with the IgG isotype control and infected with rSARS-CoV-2 Venus and/or rSARS-CoV-2 mCherry SA, respectively (Fig. 7A), alone or in combination. Fluorescent signal increased from 2 to 4 dpi in the lungs of all IgG isotype control-treated infected mice (Fig. 7B). Mice treated with 1212C2 and infected with rSARS-CoV-2 Venus showed no detectable Venus signal, indicating that 1212C2 protects against rSARS-CoV-2 Venus infection (Fig. 7A, top). In contrast, 1212C2-treated mice infected with rSARS-CoV-2 mCherry SA expressed mCherry in the lungs (Fig. 7A, middle). In mice treated with 1212C2 and coinfected with both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA, we observed only mCherry expression, consistent with the ability of 1212C2 to neutralize rSARS-CoV-2 Venus but not rSARS-CoV-2 mCherry SA (Fig. 7A, bottom). Corroborating our previous in vitro and in vivo results (Fig. 4 and 6, respectively), mice treated with 1213H7 were protected against infection with both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA, when administered alone or in combination, and presented no detectable fluorescence in the lungs (Fig. 7A). These data were further supported by quantification of the average radiant efficiency of fluorescence signals, which was high in the lungs of IgG isotype control-treated mice infected with rSARS-CoV-2 Venus or rSARS-CoV-2 mCherry SA, and in the lungs of 1212C2-treated mice infected with rSARS-CoV-2 mCherry SA (Fig. 7B). Importantly, gross pathological scoring correlated with levels of FP expression in the lungs of infected mice.
FIG 7.
Kinetics of fluorescent expression in the lungs of K18 hACE2 transgenic mice treated with 1212C2 or 1213H7 hMAbs and infected with rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA. Six- to 8-week-old female K18 hACE2 transgenic mice (n = 3) were injected (i.p.) with 25 mg/kg of an IgG isotype control, hMAb 1212C2, or hMAb 1213H7 and infected with 104 PFU of rSARS-CoV-2 Venus (top), rSARS-CoV-2 mCherry SA (middle), or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA (bottom). (A) At 2 and 4 dpi, lungs were collected to determine Venus and mCherry fluorescence expression using an Ami HT imaging system. BF, bright field. (B) Venus and mCherry radiance values were quantified based on the mean values for the regions of interest in mouse lungs. Mean values were normalized to the autofluorescence in mock-infected mice at each time point and were used to calculate fold induction. Gross pathological scores in the lungs of mock-infected and rSARS-CoV-2-infected K18 hACE2 transgenic mice were calculated based on the percentage of area of the lungs affected by infection. Statistical significance was determined by one-way ANOVA analysis on GraphPad Prism. ****, P < 0.0001.
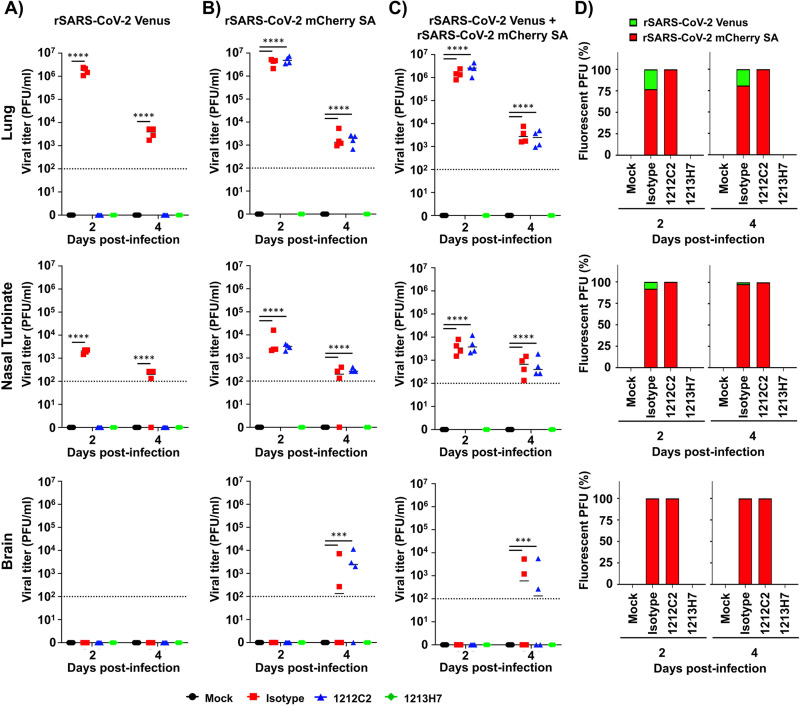
As predicted, IgG isotype control-treated K18 hACE2 transgenic mice infected with rSARS-CoV-2 Venus (Fig. 8A), rSARS-CoV-2 mCherry SA (Fig. 8B), or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA (Fig. 8C) presented high viral titers. In contrast, lungs of 1212C2-treated and infected mice had undetectable levels of rSARS-CoV-2 Venus (Fig. 8A) but high titers of rSARS-CoV-2 mCherry SA when mice were individually infected (Fig. 8B) or coinfected with both viruses (Fig. 8C). In 1213H7-treated and infected mice, we did not detect rSARS-CoV-2 Venus (Fig. 8A) or rSARS-CoV-2 mCherry SA (Fig. 8B), including double-infected mice (Fig. 8C), consistent with the ability of 1213H7 to potently neutralize both viruses in vitro and in vivo (Fig. 4 and 6, respectively).
FIG 8.
Viral titers in the lungs, nasal turbinate, and brain of K18 hACE2 transgenic mice treated with 1212C2 or 1213H7 hMAbs and infected with rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA. Six-to-8-week-old female K18 hACE2 transgenic mice (n = 3) injected (i.p.) with 25 mg/kg of an IgG isotype control, hMAb 1212C2, or hMAb 1213H7, and infected with 104 PFU of rSARS-CoV-2 Venus (A), rSARS-CoV-2 mCherry SA (B), or both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA (C). Viral titers in the lungs (top), nasal turbinate (middle) and brain (bottom) at 2 and 4 dpi were determined by plaque assay in Vero E6 cells. Bars indicate the mean and SD of lung virus titers. Dotted lines indicate the limit of detection. (D) Quantification of rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA in the lungs (top), nasal turbinate (middle), and brain (bottom) from mice coinfected with both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA at 2 and 4 dpi. Statistical significance was determined by one-way ANOVA analysis on GraphPad Prism. ***, P < 0.001; ****, P < 0.0001.
Lung homogenates from IgG isotype control-treated mice infected with both reporter viruses contained ∼25% and ∼75% of Venus- and mCherry-positive plaques, respectively, by 2 dpi. This finding suggested that rSARS-CoV-2 mCherry SA had a higher fitness than rSARS-CoV-2 Venus in vivo (Fig. 8D), which was similar to our in vitro studies (Fig. 3). Notably, by 4 dpi, all viral plaques were mCherry-positive, further supporting a higher fitness of rSARS-CoV-2 mCherry SA than rSARS-CoV-2 Venus in vivo (Fig. 8D). Lung homogenates from 1212C2-treated mice contained rSARS-CoV-2 mCherry SA, reflecting the ability of 1212C2 to efficiently neutralize rSARS-CoV-2 Venus but not rSARS-CoV-2 mCherry SA. In contrast, no viral plaques were detected in lung homogenates from mice treated with 1213H7, as this hMAb efficiently neutralizes both viruses. We obtained similar results in the nasal turbinate (Fig. 8, middle) and brain (Fig. 8, bottom) of hMAb-treated and infected K18 hACE2 transgenic mice.
DISCUSSION
The COVID-19 pandemic caused by SARS-CoV-2 started at the end of 2019 (1). Despite U.S. FDA-authorized prophylactic vaccines (6–8) and some, although still limited, available therapeutic antiviral drugs (remdesivir) and neutralizing hMAbs (casirivimab/imdevimab, bamlanivimab/etesevimab, and sotrovimab) interventions (3–5), over 175 million infections and more than 3.8 million deaths have been attributed to the COVID-19 pandemic globally (2). As with other viruses, SARS-CoV-2 is continuously evolving, resulting in viral variants (e.g., VoC) that are becoming dominant within the human population due to increased fitness, transmission, and/or resilience against natural or vaccine-induced immune responses. To date, several VoC have been identified, including the United Kingdom B.1.1.7 (alpha [α])) (28), SA B.1.351 (beta [β]) (12), Brazil P.1 (gamma [γ]) (30), India B.1.617.2 (delta [δ]) (31), and California B.1.427 (epsilon [ε]) (32). There is limited information on the ability of current vaccines to protect against these newly identified SARS-CoV-2 VoC (9, 10, 33). Moreover, it is likely that additional VoC will emerge in the future.
Reporter-expressing recombinant viruses can circumvent limitations imposed by the need for secondary methods to detect the presence of viruses in infected cells. These reporter viruses have been used to evaluate viral infections, identify therapeutics, and study viral virulence in vivo. Here, we have documented the generation of novel rSARS-CoV-2 strains to facilitate tracking infection of two different SARS-CoV-2 strains (WA-1 and SA) in vitro and in vivo based on the use of two different FPs (Venus and mCherry, respectively). The FP-expressing rSARS-CoV-2 strains encode the fluorescent Venus or mCherry proteins from the locus of the N protein without the need for deletion of any viral protein (20). Notably, the use of this approach to generate FP-expressing rSARS-CoV-2 strains resulted in higher FP expression levels than those afforded by rSARS-CoV-2 expressing FPs from the locus of the viral ORF7a protein (20). Moreover, rSARS-CoV-2 expressing reporter genes from the N locus are more genetically stable than those expressing reporter genes from the ORF7a locus of the SARS-CoV-2 genome (20).
We showed that rSARS-CoV-2 expressing Venus or mCherry from the N locus exhibited similar growth kinetics, peak titers, and plaque phenotype as the parental WT rSARS-CoV-2 WA-1 strain. Importantly, we were able to use these novel reporter rSARS-CoV-2 strains in bifluorescent-based assays to determine the neutralization efficacy of hMAbs based on FP expression levels. We also generated rSARS-CoV-2 mCherry SA, an mCherry-expressing rSARS-CoV-2 strain containing the K417N, E484K, and N501Y mutations in the RBD of the S glycoprotein of the SA VoC. Notably, rSARS-CoV-2 mCherry SA had higher fitness than rSARS-CoV-2 Venus in cultured cells, as evidenced by higher viral titers reached and a bigger plaque size phenotype. Interestingly, when used in the bifluorescent-based assay, hMAb 1212C2 was unable to neutralize rSARS-CoV-2 mCherry SA, but efficiently neutralized rSARS-CoV-2 Venus. In contrast, hMAb 1213H7 displayed efficient neutralization of both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA. Importantly, these in vitro results correlated with in vivo studies in which K18 hACE2 transgenic mice pretreated with 1212C2 were protected against challenge with rSARS-CoV-2 Venus but not rSARS-CoV-2 mCherry SA, alone or in combination. In contrast, mice treated with 1213H7 were protected against lethal challenge with both reporter-expressing rSARS-CoV-2 strains, alone or in combination. These protection results were corroborated through IVIS studies, in which fluorescence and viral titers demonstrated the neutralizing protective efficacy of 1212C2 against rSARS-CoV-2 Venus but not rSARS-CoV-2 mCherry SA, while 1213H7 efficiently protected mice against challenge with both viruses, alone or in combination. These results prove the feasibility of using both rSARS-CoV-2 Venus and rSARS-CoV-2 mCherry SA to accurately assess the ability of hMAbs to efficiently neutralize one or both SARS-CoV-2 strains, alone or in combination, in vitro and/or in vivo, and establish that the readouts of the bifluorescent-based assays correlate well with those of individual viral infections.
rSARS-CoV-2 strains expressing FP or luciferase reporter genes have been described by us and others (13–16), but in this study, we have documented, for the first time, the use of two rSARS-CoV-2 strains expressing different FP and S glycoproteins in a bifluorescent-based assay to identify NAbs exhibiting differences in their neutralizing activity against different SARS-CoV-2 strains present in the same biological sample in vitro and in vivo. Notably, this approach can be extended to identify broadly NAbs against different SARS-CoV-2 VoC by generating rSARS-CoV-2 strains expressing additional FP and containing the S glycoproteins of different VoC in multiplex-based fluorescent assays in vitro and/or in vivo. These reporter rSARS-CoV-2 strains expressing the S glycoprotein of VoC also represent an excellent option to investigate viral infection, dissemination, pathogenesis, and therapeutic interventions, including protective efficacy of vaccines or antivirals, for the treatment of SARS-CoV-2 infection in cultured cells and/or in validated animal models of SARS-CoV-2 infection.
MATERIALS AND METHODS
Biosafety.
Experiments involving the use of infectious SARS-CoV-2 were performed at biosafety level 3 (BSL3) containment laboratories at Texas Biomedical Research Institute. All experiments using SARS-CoV-2 were approved by the Institutional Biosafety Committee (IBC) at Texas Biomedical Research Institute.
Cells.
African green monkey kidney epithelial cells (Vero E6; CRL-1586) were grown and maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1× PSG (100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine), and incubated at 37°C in an 5% CO2 atmosphere.
Generation of pBeloBAC11-SARS-CoV-2 encoding fluorescent proteins.
The pBeloBAC11 plasmid (NEB) containing the entire viral genome of SARS-CoV-2 USA/WA1/2020 (WA-1) isolate (GenBank accession no. MN985325) has been described (20, 21). The rSARS-CoV-2 strains expressing Venus or mCherry from the locus of the viral N protein using the PTV-1 2A autocleavage sequence were generated as previously described (20, 21). The rSARS-CoV-2 strain containing mutations K417N, E484K, and N501Y present in the receptor binding domain (RBD) within the spike (S) gene of the South African (SA) B.1.351 (beta[β]) VoC (12) and expressing mCherry was generated using standard molecular biology techniques. Plasmids containing the full-length genome of the different rSARS-CoV-2 strains were analyzed by digestion using specific restriction enzymes and validated by deep sequencing. Oligonucleotides for cloning the Venus or mCherry FP or K417N, E484K, and N501Y mutations are available upon request.
Generation of rSARS-CoV-2 strains expressing FP.
Wild-type (WT, WA-1), Venus (Venus WA-1), and mCherry (mCherry WA-1) reporter-expressing rSARS-CoV-2 strains, as well as rSARS-CoV-2 strain encoding the SA B.1.351 (β) mutations K417N, E484K, and N501Y in the S RBD expressing mCherry (mCherry SA) were rescued as previously described (21, 34). Briefly, confluent monolayers of Vero E6 cells (1.2 × 106 cells/well, 6-well plate format, triplicates) were transfected with 4 μg/well of pBeloBAC11-SARS-CoV-2 (WA-1), -2A/Venus, -2A/mCherry, or -2A/mCherry-SA-RBD plasmids using Lipofectamine 2000 (Thermo Fisher). After 24 h posttransfection, medium was exchanged with postinfection medium (DMEM containing 2% FBS), and 24 h later, cells were scaled up to T75 flasks and incubated for 72 h at 37°C. Viral rescues were first confirmed under a bright-field microscope by assessing cytopathic effect (CPE) before supernatants were collected, aliquoted, and stored at −80°C. To confirm the rescue of rSARS-CoV-2, Vero E6 cells (1.2 × 106 cells/well, 6-well plates, triplicates) were infected with virus-containing tissue culture supernatants and incubated at 37°C in a 5% CO2 incubator for 48 h. Viruses were detected by fluorescence or immunostaining with a SARS-CoV N protein cross-reactive mouse (m)MAb (1C7C7). Plaque assays were used to determine viral titers (plaque-forming units [PFU])/ml). Viral stocks were generated by infecting fresh monolayers of Vero E6 cells at low multiplicity of infection ([MOI] of 0.0001) for 72 h before being aliquoted and stored at −80°C.
Sequencing.
To confirm the identity of the rescued rSARS-CoV-2 mCherry SA, total RNA from infected (MOI of 0.01) Vero E6 cells (1.2 × 106 cells/well, 6-well format, triplicates) was extracted using TRIzol reagent (Thermo Fisher Scientific) and used in reverse transcription-quantitative PCRs (RT-PCRs) to amplify a fragment of 1,174 bp around the RBD of the S gene. RT-PCR was done using SuperScript II reverse transcriptase (Thermo Fisher Scientific) and Expand high-fidelity PCR system (Sigma-Aldrich). RT-PCR products were purified on 0.7% agarose gel and subjected to Sanger sequencing (ACGT). All primer sequences are available upon request.
Immunofluorescence assays.
Confluent monolayers of Vero E6 cells (1.2 × 106 cells/well, 6-well format, triplicates) were mock infected or infected (MOI, 0.01) with WT, Venus-, or mCherry-expressing rSARS-CoV-2 WA-1 (WA-1, Venus WA-1, or mCherry WA-1, respectively) or rSARS-CoV-2 mCherry SA. At 48 hours postinfection (hpi), cells were submerged in 10% neutral buffered formalin at 4°C overnight for fixation and viral inactivation and then permeabilized with 0.5% Triton X-100 phosphate-buffered saline (PBS) at room temperature for 10 min. Thereafter, cells were washed with PBS before blocking with 2.5% bovine albumin serum (BSA)-PBS for 1 h. Cells were then incubated with 1 μg/ml of SARS-CoV anti-N mMAb 1C7C7 in 1% BSA at 37°C for 1 h. Reporter-expressing rSARS-CoV-2 strains were detected directly by epifluorescence and using either Alexa Fluor 594 goat anti-mouse IgG (Invitrogen; 1:1,000) or fluorescein isothiocynate (FITC)-conjugated goat anti-mouse IgG (Dako; 1:200), depending on whether the viruses express Venus or mCherry, respectively. Cell nuclei were detected with 4′,6-diamidino-2-phenylindole (DAPI; Research Organics). An EVOS M5000 imaging system was used to acquire representative images (×10 magnification).
Viral growth kinetics.
Vero E6 cells (1.2 × 106 cells/well, 6-well plate format, triplicates) were infected (MOI, 0.01) with the indicated viruses at 37°C for 1 h. After viral adsorption, cells were washed with PBS and incubated at 37°C in postinfection media. At 24, 48, 72, and 96 hpi, fluorescence-positive cells were imaged with an EVOS M5000 fluorescence microscope for rSARS-CoV-2 expressing Venus or mCherry FP, and viral titers in the tissue culture supernatants were determined by plaque assay and immunostaining using the anti-SARS-CoV N mMAb 1C7C7. Mean values and standard deviation (SD) were calculated with Microsoft Excel software.
Plaque assays and immunostaining.
Confluent monolayers of Vero E6 cells (2 × 105 cells/well, 24-well plate format, triplicates) were infected with WT or reporter-expressing rSARS-CoV-2 for 1 h before being overlaid with postinfection media containing 1% agar (Oxoid) and incubated at 37°C in a 5% CO2 incubator. After 72 h, cells were fixed in 10% neutral buffered formalin overnight at 4°C. Next, overlays were removed, PBS was added to each well, and fluorescent plaques were detected and quantified using a ChemiDoc MP imaging system (Bio-Rad). Cells were then permeabilized with 0.5% Triton X-100 in PBS for 5 min, blocked with 2.5% BSA in PBS for 1 h, and incubated with the SARS-CoV N mMAb 1C7C7, and plaques were detected using a Vectastain ABC kit and 3,3'-diaminobenzidine horseradish peroxidase (DAB HRP) substrate kit (Vector Laboratories) following the manufacturer’s instructions.
A bifluorescent-based neutralization assay.
The hMAbs used in this study were generated and purified as described (22). CB6, REGN10987, and REGN10933 hMAbs were included as controls (25, 26). To test the neutralizing activity of hMAbs, confluent monolayers of Vero E6 cells (4 × 104 cells/well, 96-plate well format, quadruplicates) were infected (MOI of 0.01 or 0.1) with the indicated rSARS-CoV-2 for 1 h at 37°C. After viral absorption, postinfection media containing 3-fold dilutions of the indicated hMAbs (starting concentration of 500 ng/well) were added to the cells and incubated at 37°C for 48 h. Cells were then fixed in 10% neutral buffered formalin overnight and washed with PBS before fluorescence signal was measured and quantified using a Synergy LX microplate reader and Gen5 data analysis software (Bio-Tek). The mean and SD of viral infections were calculated from individual wells of three independent experiments conducted in quadruplicates with Microsoft Excel software. Nonlinear regression curves and NT50 values were determined using GraphPad Prism Software (San Diego, CA, USA; version 8.2.1). Representative images were captured with an EVOS M5000 imaging system (Thermo Fisher) at ×10 magnification.
Mouse experiments.
All animal protocols were approved by Texas Biomed IACUC (1718MU). Six- to 8-week-old female K18 human angiotensin-converting enzyme 2 (hACE2) transgenic mice were purchased from The Jackson Laboratory and maintained in the animal biosafety laboratory level 3 (ABSL-3) at Texas Biomedical Research Institute. All mouse procedures were approved by Texas Biomedical Research Institute IACUC. To assess the in vivo efficacy of hMAbs, K18 hACE2 transgenic mice (n = 5/group) were anesthetized with isoflurane and injected (i.p.) with hMAbs IgG isotype control, 1212C2, or 1213H7 (25 mg/kg) using a 1-ml syringe 23- to 2- gauge, 5/8-in. needle 24 h prior to challenge with rSARS-CoV-2. For viral challenges, mice were anesthetized and inoculated intranasally (i.n.) with 104 PFU of the indicated rSARS-CoV-2 and monitored daily for morbidity as determined by changes in body weight and survival. Mice that lost greater than 25% of their initial weight were considered to have reached their experimental endpoint and were humanely euthanized. In parallel, K18 hACE2 transgenic mice (n = 3/group) were treated (i.p.) with 1212C2 or 1213H7 hMAbs and challenged i.n. with 104 PFU of the indicated rSARS-CoV-2 for viral titer determination. Viral titers in the lungs of infected mice at 2 and 4 dpi were determined by plaque assay. In vivo fluorescence imaging of mouse lungs was conducted using an Ami HT in vivo imaging system (IVIS; Spectral Instruments). Mice were euthanized with a lethal dose of Fatal-Plus solution, and lungs were surgically extracted and washed in PBS before imaging in the Ami HT. Images were analyzed with Aura software to determine radiance with the region of interest (ROI), and fluorescence signal was normalized to background signal of lungs from mock-infected mice. Bright-field images of lungs were captured using an iPhone X camera. After imaging, lungs were homogenized using a Precellys tissue homogenizer (Bertin Instruments) in 1 ml of PBS and centrifuged at 21,500 × g for 10 min to pellet cell debris. Clarified supernatants were collected and used to determine viral titers by plaque assay. Macroscopic pathological scoring was determined from the percentage of total surface area affected by congestion, consolidation, and atelectasis of excised lungs, using NIH ImageJ software as previously described (21, 35).
Statistical analysis.
All data are presented as mean values and SD for each group and were analyzed using Microsoft Excel software. One-way analysis of variance (ANOVA) or Student's t test were used for statistical analysis on GraphPad Prism or Microsoft Word software, respectively. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, no significance.
Data availability.
All of the data supporting the findings of this work can be found within the paper. The raw data are available from the corresponding authors upon request. The plasmids described in this study are available at the following website: https://www.txbiomed.org/services-2/reverse-genetics-plasmids.
ACKNOWLEDGMENTS
We are grateful to Thomas Moran at the Icahn School of Medicine at Mount Sinai for providing the SARS-CoV cross-reactive 1C7C7 N protein mMAb.
J.-G.P., M.S.P., M.R.W., J.K.K., and L.M.-S. are coinventors on a patent that includes claims related to some of the hMAbs described in this paper.
Contributor Information
Chengjin Ye, Email: cye@txbiomed.org.
Luis Martinez-Sobrido, Email: lmartinez@txbiomed.org.
Colin R. Parrish, Cornell University
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H, Gardner L. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20:533–534. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G. 2020. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh MD, Ruiz-Palacios GM, Benfield T, Fatkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC, Members A-SG. 2020. Remdesivir for the treatment of Covid-19 - final report. N Engl J Med 383:1813–1826. 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones BE, Brown-Augsburger PL, Corbett KS, Westendorf K, Davies J, Cujec TP, Wiethoff CM, Blackbourne JL, Heinz BA, Foster D, Higgs RE, Balasubramaniam D, Wang L, Zhang Y, Yang ES, Bidshahri R, Kraft L, Hwang Y, Žentelis S, Jepson KR, Goya R, Smith MA, Collins DW, Hinshaw SJ, Tycho SA, Pellacani D, Xiang P, Muthuraman K, Sobhanifar S, Piper MH, Triana FJ, Hendle J, Pustilnik A, Adams AC, Berens SJ, Baric RS, Martinez DR, Cross RW, Geisbert TW, Borisevich V, Abiona O, Belli HM, de Vries M, Mohamed A, Dittmann M, Samanovic MI, Mulligan MJ, Goldsmith JA, Hsieh CL, Johnson NV, et al. 2021. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci Transl Med 13:eabf1906. 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, C4591001 Clinical Trial Group . 2020. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliver SE, Gargano JW, Marin M, Wallace M, Curran KG, Chamberland M, McClung N, Campos-Outcalt D, Morgan RL, Mbaeyi S, Romero JR, Talbot HK, Lee GM, Bell BP, Dooling K. 2021. The Advisory Committee on Immunization Practices' interim recommendation for use of Moderna COVID-19 vaccine - United States, December 2020. MMWR Morb Mortal Wkly Rep 69:1653–1656. 10.15585/mmwr.mm695152e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livingston EH, Malani PN, Creech CB. 2021. The Johnson & Johnson vaccine for COVID-19. JAMA 325:1575. 10.1001/jama.2021.2927. [DOI] [PubMed] [Google Scholar]
- 9.Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, Wang B, Paesen GC, Lopez-Camacho C, Slon-Campos J, Hallis B, Coombes N, Bewley K, Charlton S, Walter TS, Skelly D, Lumley SF, Dold C, Levin R, Dong T, Pollard AJ, Knight JC, Crook D, Lambe T, Clutterbuck E, Bibi S, Flaxman A, Bittaye M, Belij-Rammerstorfer S, Gilbert S, James W, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Fry EE, Mongkolsapaya J, Ren J, Stuart DI, Screaton GR. 2021. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 184:2348–2361.e6. 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madhi SA, Baillie V, Cutland CL, Voysey M, Koen AL, Fairlie L, Padayachee SD, Dheda K, Barnabas SL, Bhorat QE, Briner C, Kwatra G, Ahmed K, Aley P, Bhikha S, Bhiman JN, Bhorat AE, Du Plessis J, Esmail A, Groenewald M, Horne E, Hwa SH, Jose A, Lambe T, Laubscher M, Malahleha M, Masenya M, Masilela M, McKenzie S, Molapo K, Moultrie A, Oelofse S, Patel F, Pillay S, Rhead S, Rodel H, Rossouw L, Taoushanis C, Tegally H, Thombrayil A, van Eck S, Wibmer CK, Durham NM, Kelly EJ, Villafana TL, Gilbert S, Pollard AJ, de Oliveira T, Moore PL, Sigal A, et al. 2021. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N Engl J Med 384:1885–1898. 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irfan N, Chagla Z. 2021. In South Africa, a 2-dose Oxford/AZ vaccine did not prevent mild to moderate COVID-19 (cases mainly B.1.351 variant). Ann Intern Med 174:JC50. 10.7326/ACPJ202105180-050. [DOI] [PubMed] [Google Scholar]
- 12.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao M, Korsman S, Davies M-A, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Pond SLK, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. 2020. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 10.1101/2020.12.21.20248640:2020.12.21.20248640. [DOI] [Google Scholar]
- 13.Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, III, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G, Olivier KN, Ghio A, Tse LV, Leist SR, Gralinski LE, Schafer A, Dang H, Gilmore R, Nakano S, Sun L, Fulcher ML, Livraghi-Butrico A, Nicely NI, Cameron M, Cameron C, Kelvin DJ, de Silva A, Margolis DM, Markmann A, Bartelt L, Zumwalt R, Martinez FJ, Salvatore SP, Borczuk A, Tata PR, Sontake V, Kimple A, Jaspers I, O'Neal WK, Randell SH, Boucher RC, Baric RS. 2020. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182:429–446.e14. 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, Plante KS, Weaver SC, Makino S, LeDuc JW, Menachery VD, Shi PY. 2020. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe 27:841–848.e3. 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie X, Muruato AE, Zhang X, Lokugamage KG, Fontes-Garfias CR, Zou J, Liu J, Ren P, Balakrishnan M, Cihlar T, Tseng C-TK, Makino S, Menachery VD, Bilello JP, Shi P-Y. 2020. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat Commun 11:5214. 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiem K, Morales Vasquez D, Park JG, Platt RN, Anderson T, Walter MR, Kobie JJ, Ye C, Martinez-Sobrido L. 2021. Generation and characterization of recombinant SARS-CoV-2 expressing reporter genes. J Virol 95:e2209-20. 10.1128/JVI.02209-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nogales A, Ávila-Pérez G, Rangel-Moreno J, Chiem K, DeDiego ML, Martínez-Sobrido L. 2019. A novel fluorescent and bioluminescent bireporter influenza A virus (BIRFLU) to evaluate viral infections. J Virol 95:e00032-19. 10.1128/JVI.00032-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Creanga A, Gillespie RA, Fisher BE, Andrews SF, Lederhofer J, Yap C, Hatch L, Stephens T, Tsybovsky Y, Crank MC, Ledgerwood JE, McDermott AB, Mascola JR, Graham BS, Kanekiyo M. 2021. A comprehensive influenza reporter virus panel for high-throughput deep profiling of neutralizing antibodies. Nat Commun 12:1722. 10.1038/s41467-021-21954-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park JG, Oladunni FS, Chiem K, Ye C, Pipenbrink M, Moran T, Walter MR, Kobie J, Martinez-Sobrido L. 2021. Rapid in vitro assays for screening neutralizing antibodies and antivirals against SARS-CoV-2. J Virol Methods 287:113995. 10.1016/j.jviromet.2020.113995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye C, Chiem K, Park J-G, Silvas JA, Vasquez DM, Torrelles JB, Kobie JJ, Walter MR, de la Torre JC, Martinez-Sobrido L. 2021. Visualization of SARS-CoV-2 infection dynamic. bioRxiv 10.1101/2021.06.03.446942:2021.06.03.446942. [DOI]
- 21.Ye C, Chiem K, Park JG, Oladunni F, Platt RN, II, Anderson T, Almazan F, de la Torre JC, Martinez-Sobrido L. 2020. Rescue of SARS-CoV-2 from a single bacterial artificial chromosome. mBio 11:e02168-20. 10.1128/mBio.02168-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piepenbrink MS, Park J-G, Oladunni FS, Deshpande A, Basu M, Sarkar S, Loos A, Woo J, Lovalenti P, Sloan D, Ye C, Chiem K, Erdmann NB, Goepfert PA, Truong VL, Walter MR, Martinez-Sobrido L, Kobie JJ. 2021. Therapeutic activity of an inhaled potent SARS-CoV-2 neutralizing human monoclonal antibody in hamsters. Cell Rep Med 2:100218. 10.1016/j.xcrm.2021.100218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deshpande A, Harris BD, Martinez-Sobrido L, Kobie JJ, Walter MR. 2021. Epitope classification and RBD binding properties of neutralizing antibodies against SARS-CoV-2 variants of concern. Front Immunol 12:691715. 10.3389/fimmu.2021.691715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horspool AM, Ye C, Wong TY, Russ BP, Lee KS, Winters MT, Bevere JR, Kieffer T, Martinez I, Sourimant J, Greninger A, Plemper RK, Denvir J, Cyphert HA, Torrelles J, Martinez-Sobrido L, Damron FH. 2021. SARS-CoV-2 B.1.1.7 and B.1.351 variants of concern induce lethal disease in K18-hACE2 transgenic mice despite convalescent plasma therapy. bioRxiv 10.1101/2021.05.05.442784. [DOI]
- 25.Shi R, Shan C, Duan X, Chen Z, Liu P, Song J, Song T, Bi X, Han C, Wu L, Gao G, Hu X, Zhang Y, Tong Z, Huang W, Liu WJ, Wu G, Zhang B, Wang L, Qi J, Feng H, Wang FS, Wang Q, Gao GF, Yuan Z, Yan J. 2020. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584:120–124. 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 26.Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, Fulton BO, Yan Y, Koon K, Patel K, Chung KM, Hermann A, Ullman E, Cruz J, Rafique A, Huang T, Fairhurst J, Libertiny C, Malbec M, Lee WY, Welsh R, Farr G, Pennington S, Deshpande D, Cheng J, Watty A, Bouffard P, Babb R, Levenkova N, Chen C, Zhang B, Romero Hernandez A, Saotome K, Zhou Y, Franklin M, Sivapalasingam S, Lye DC, Weston S, Logue J, Haupt R, Frieman M, Chen G, Olson W, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. 2020. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369:1010–1014. 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oladunni FS, Park JG, Pino PA, Gonzalez O, Akhter A, Allué-Guardia A, Olmo-Fontánez A, Gautam S, Garcia-Vilanova A, Ye C, Chiem K, Headley C, Dwivedi V, Parodi LM, Alfson KJ, Staples HM, Schami A, Garcia JI, Whigham A, Platt RN, Gazi M, Martinez J, Chuba C, Earley S, Rodriguez OH, Mdaki SD, Kavelish KN, Escalona R, Hallam CRA, Christie C, Patterson JL, Anderson TJC, Carrion R, Dick EJ, Hall-Ursone S, Schlesinger LS, Alvarez X, Kaushal D, Giavedoni LD, Turner J, Martinez-Sobrido L, Torrelles JB. 2020. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat Commun 11:6122. 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambaut A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, Robertson DL, Volz E. 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Genom Epidemiol. [Google Scholar]
- 29.Reference deleted.
- 30.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, Crispim MAE, Sales FCS, Hawryluk I, McCrone JT, Hulswit RJG, Franco LAM, Ramundo MS, de Jesus JG, Andrade PS, Coletti TM, Ferreira GM, Silva CAM, Manuli ER, Pereira RHM, Peixoto PS, Kraemer MUG, Gaburo N, Camilo CDC, Hoeltgebaum H, Souza WM, Rocha EC, de Souza LM, de Pinho MC, Araujo LJT, Malta FSV, de Lima AB, Silva JDP, Zauli DAG, Ferreira ACS, Schnekenberg RP, Laydon DJ, Walker PGT, Schlüter HM, Dos Santos ALP, Vidal MS, Del Caro VS, Filho RMF, Dos Santos HM, Aguiar RS, Proença-Modena JL, Nelson B, Hay JA, Monod M, Miscouridou X, et al. 2021. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372:815–821. 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yadav PD, Sapkal GN, Abraham P, Ella R, Deshpande G, Patil DY, Nyayanit DA, Gupta N, Sahay RR, Shete AM, Panda S, Bhargava B, Mohan VK. 2021. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin Infect Dis 10.1093/cid/ciab411. [DOI] [PubMed] [Google Scholar]
- 32.Martin Webb L, Matzinger S, Grano C, Kawasaki B, Stringer G, Bankers L, Herlihy R. 2021. Identification of and surveillance for the SARS-CoV-2 variants B.1.427 and B.1.429 - Colorado, January-March 2021. MMWR Morb Mortal Wkly Rep 70:717–718. 10.15585/mmwr.mm7019e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL, COVID-19 Genomics UK (COG-UK) Consortium . 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avila-Perez G, Park JG, Nogales A, Almazan F, Martinez-Sobrido L. 2019. Rescue of recombinant Zika virus from a bacterial artificial chromosome cDNA clone. J Vis Exp e59537. 10.3791/59537. [DOI] [PubMed] [Google Scholar]
- 35.Jensen EC. 2013. Quantitative analysis of histological staining and fluorescence using ImageJ. Anat Rec (Hoboken) 296:378–381. 10.1002/ar.22641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data supporting the findings of this work can be found within the paper. The raw data are available from the corresponding authors upon request. The plasmids described in this study are available at the following website: https://www.txbiomed.org/services-2/reverse-genetics-plasmids.