ABSTRACT
The high pathogenicity of SARS-CoV-2 requires it to be handled under biosafety level 3 conditions. Consequently, Spike protein-pseudotyped vectors are a useful tool to study viral entry and its inhibition, with retroviral, lentiviral (LV), and vesicular stomatitis virus (VSV) vectors the most commonly used systems. Methods to increase the titer of such vectors commonly include concentration by ultracentrifugation and truncation of the Spike protein cytoplasmic tail. However, limited studies have examined whether such a modification also impacts the protein’s function. Here, we optimized concentration methods for SARS-CoV-2 Spike-pseudotyped VSV vectors, finding that tangential flow filtration produced vectors with more consistent titers than ultracentrifugation. We also examined the impact of Spike tail truncation on transduction of various cell types and sensitivity to convalescent serum neutralization. We found that tail truncation increased Spike incorporation into both LV and VSV vectors and resulted in enhanced titers but had no impact on sensitivity to convalescent serum. In addition, we analyzed the effect of the D614G mutation, which became a dominant SARS-CoV-2 variant early in the pandemic. Our studies revealed that, similar to the tail truncation, D614G independently increases Spike incorporation and vector titers, but this effect is masked by also including the cytoplasmic tail truncation. Therefore, the use of full-length Spike protein, combined with tangential flow filtration, is recommended as a method to generate high titer pseudotyped vectors that retain native Spike protein functions.
IMPORTANCE Pseudotyped viral vectors are useful tools to study the properties of viral fusion proteins, especially those from highly pathogenic viruses. The Spike protein of SARS-CoV-2 has been investigated using pseudotyped lentiviral and VSV vector systems, where truncation of its cytoplasmic tail is commonly used to enhance Spike incorporation into vectors and to increase the titers of the resulting vectors. However, our studies have shown that such effects can also mask the phenotype of the D614G mutation in the ectodomain of the protein, which was a dominant variant arising early in the COVID-19 pandemic. To better ensure the authenticity of Spike protein phenotypes when using pseudotyped vectors, we recommend using full-length Spike proteins, combined with tangential flow filtration methods of concentration if higher-titer vectors are required.
KEYWORDS: D614G mutation, SARS-CoV-2, Spike protein, cytoplasmic tail truncation, pseudotyped vectors
INTRODUCTION
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and was first reported in Wuhan, China, in December 2019 (1). The disease rapidly spread worldwide, causing over 150 million confirmed cases and more than 3 million reported deaths by May 2021 (2). The accompanying worldwide research effort has resulted in a large number of vaccine candidates and both national and international clinical trials to assess novel and repurposed drug regimens (3). In the United States, the SARS-CoV-2 Spike glycoprotein has been a primary target of such efforts. Spike is a major viral antigen that induces protective immune responses in COVID-19 patients (4–6) and mediates cell entry by binding to angiotensin-converting enzyme 2 (ACE2) (1, 7, 8) or other receptors (9, 10) and employs cellular proteases such as TMPRSS2 to prime the Spike protein for membrane fusion (11, 12). ACE2 is expressed in the human respiratory system (13), especially on type II pneumocytes (14), which are the main target cell for SARS-CoV-2 infection. Expression of ACE2 in other organs also allows infection outside the lung (13).
Due to the high pathogenicity of SARS-CoV-2, biosafety level 3 (BSL3) labs are required for studies that involve replication-competent virus. Therefore, investigators often use Spike protein pseudovirus vector systems, based on replication-incompetent vector particles and attenuated or conditional viruses. Identification of an optimal pseudovirus system for any particular viral entry glycoprotein typically involves comparing the most commonly used systems: replication-incompetent lentiviral (LV) or retroviral (RV) vectors or conditional vesicular stomatitis viruses (VSV) that are deleted for the VSV glycoprotein (G) (15, 16). SARS-CoV-2 Spike protein can pseudotype all three vector systems, which have been used to investigate viral entry (11, 17–19), neutralization by monoclonal antibodies or convalescent plasma (4, 5, 17, 20–28), and entry inhibitors (17, 24, 29, 30) and to characterize surging viral variants (31–45). In approximately one-third of these studies, deletions of 18 to 21 amino acids from the cytoplasmic tail of Spike were used to enhance vector titers and thereby facilitate the study.
Cytoplasmic tail truncation of viral glycoproteins is a common strategy to enhance pseudovirus formation since this can remove steric interference that may occur between the heterologous viral glycoproteins and the vector matrix or capsid proteins (46–51). Also employed are cytoplasmic tail swaps, whereby the tail from the natural viral glycoprotein is used to create a chimeric glycoprotein with enhanced incorporation properties (47, 52). However, we and others have shown that tail modifications can also have functional consequences, for example, removing endocytosis signals that lead to increased cell surface levels and enhanced incorporation into vector particles (53, 54), alterations of the ectodomain conformation (53, 55), changes to fusogenicity (47, 55–57), and altered antigenic characteristics (58, 59).
SARS-CoV-2 Spike contains a putative endoplasmic reticulum (ER) retention signal (KLHYT) at its C terminus, which is removed by the tail truncations of 13 amino acids (60) or 18 to 21 amino acids that are frequently employed (23, 25, 61–63). Compared to the full-length Spike, such truncations were reported to generate ∼10- to 20-fold higher titers of both LV vectors (23, 60, 61, 63) and VSV vectors (25, 61, 62). Truncated Spike also enhances RV vector titers, albeit with a smaller effect compared with LV and VSV vectors (61). Havranek et al. (62) and Yu et al. (60) investigated the mechanism for such an effect for VSV and LV pseudoviruses and found that tail truncation enhanced both Spike incorporation into the viral particles and cell-cell fusion for Spike-expressing cells without altering cell surface expression levels. However, the impact of Spike tail truncations on any other ectodomain functions remains unclear.
In this report, we compared the practicality and functionality of using Spike pseudotyped vectors based on LV and VSV and pseudotyped with either full-length or tail truncated proteins. We compared methods to prepare such vectors and identified tangential flow filtration as a facile method that is superior to ultracentrifugation and allows efficient production at a larger scale. An optimized system based on VSV vectors was used to assess the impact of the Spike mutation D614G (35) and to assess neutralizing activity in convalescent serum. Our studies determined that although Spike tail truncation boosts incorporation into vectors and enhances the titers achieved for unconcentrated supernatants, it also blunted the ability to observe differences caused by this specific Spike mutation. We therefore recommend that studies using Spike pseudotyped vectors retain the natural full-length cytoplasmic tail and use other strategies, such as concentration method and vector system choice, to achieve the required vector titers.
RESULTS
Spike cytoplasmic tail truncation facilitates vector incorporation and enhances titer.
Cytoplasmic tail truncation of SARS-CoV-2 Spike has been reported to enhance the transduction efficiency of pseudotyped LV and VSV vectors (23, 60, 61, 63), with the effect suggested to be the result of enhanced incorporation and/or fusogenicity of Spike (60, 62). Using both the full-length Spike (S) and an 18-amino-acid cytoplasmic tail truncation (SΔ18), we generated pseudotyped LV and VSV vectors carrying reporter green fluorescent protein (GFP) or luciferase genes, respectively. We compared the ability of the two Spike proteins to be incorporated into the vectors and to transduce HeLa cells expressing human ACE2 (HeLa-ACE2). Transduction by LV-GFP vectors was analyzed at 48 h postransduction, while VSV-Luc vectors were analyzed as early as 16 to 24 h postransduction.
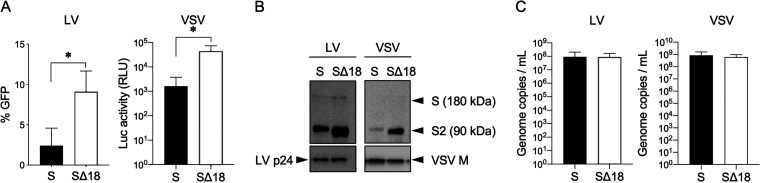
Consistent with previous findings, we found that the cytoplasmic tail truncation increased vector transduction efficiency on HeLa-ACE2 cells by approximately 4- and 30-fold for the LV-GFP and VSV-Luc vectors, respectively (Fig. 1A). We also observed a significant increase in incorporation for the truncated Spike protein in both vector systems (Fig. 1B) while having no impact on other viral particle proteins (Fig. 1B) or vector genome copy number (Fig. 1C). Together, these results suggest that cytoplasmic tail truncation increases Spike incorporation into both LV and VSV particles, and this results in higher infectivity per particle. Since the VSV-Luc vectors have a faster readout time, we chose this pseudovirus system for the rest of our studies.
FIG 1.
Impact of Spike protein cytoplasmic tail truncation on LV and VSV vectors. (A) Transduction of HeLa-ACE2 cells by equal volumes of unconcentrated vector supernatants of LV-GFP or VSV-Luc vectors, pseudotyped with full-length (S) or truncated (SΔ18) Spike proteins. Shown are means and standard deviations from 3 independent vector stocks. *, P < 0.05, unpaired t test, one-tail. (B) Spike protein incorporation into vector particles, analyzed by Western blotting using antibodies against the Spike S2 subunit and vector particle components p24 (LV) and M (VSV). Full-length Spike (S) and S2 subunit are indicated. (C) Genomic copy number for indicated vectors. Shown are means and standard deviations from 3 independent vector stocks.
Susceptibility of different cell lines and lung organoids to Spike protein pseudovectors.
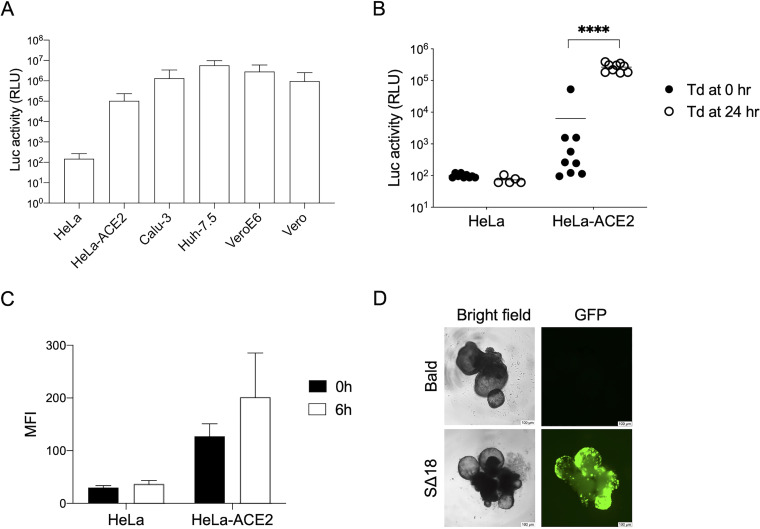
Next, we tested the permissivity of different cell lines and a lung organoid model to SΔ18 pseudotyped VSV vectors. In agreement with previous findings, several ACE2-expressing cells were found to be susceptible to the vectors (1, 11), while ACE2 overexpression was required to support transduction of HeLa cells (Fig. 2A). We also evaluated an alternative transduction protocol with a shortened timeline, whereby trypsinized HeLa and HeLa-ACE2 cells were incubated with vectors simultaneously with seeding onto plates instead of transduction occurring 24 h after seeding (64). However, this protocol significantly reduced the transduction efficiency (Fig. 2B), which we hypothesize is a result of reduced cell surface ACE2 after trypsinization (Fig. 2C).
FIG 2.
Transduction of cells by Spike VSV pseudovectors. (A) Indicated cell lines were transduced with equal amounts of SΔ18 VSV-Luc vectors and luciferase activity in cell lysates analyzed 24 h later. Shown are means and standard deviations from 3 independent vector stocks. (B) HeLa and HeLa-ACE2 cells were detached from culture flasks by trypsin, seeded into 96-well plates, and transduced (Td) with equal amounts of SΔ18 VSV-Luc vectors, either immediately (0 h) or 24 h after seeding, and luciferase was measured 24 h later. Data from 9 different wells in a single experiment are shown. ****, P < 0.001, multiple t test. (C) ACE2 expression levels on cell surface measured by flow cytometry. Cells were stained with anti-ACE2 antibody at 0 or 6 h after trypsinization. Mean and standard deviation MFI from four (HeLa) or five (HeLa-ACE2) independent experiments are shown. (D) Lung bud organoids were transduced with equal amounts of VSV-GFP vectors pseudotyped with SΔ18 or control (bald) vectors with no Spike protein. GFP expression was visualized 24 h later. Scale bars represent 100 μm.
Finally, we tested the susceptibility of a three-dimensional lung bud organoid model to SΔ18 VSV pseudovectors carrying a GFP reporter. Compared to cell lines, lung organoids provide more physiologically relevant models of virus infection and have been used to identify candidate COVID-19 therapeutics (30). SΔ18-pseudotyped VSV-GFP vectors were able to efficiently transduce the cells, with GFP expression observed throughout the organoid by 24 h (Fig. 2D).
TFF facilitates scale-up of vector production and concentration.
To identify an optimal method for concentration of Spike protein pseudovectors suitable for a research laboratory, we compared ultracentrifugation through a 20% (wt/vol) sucrose cushion with tangential flow filtration (TFF). Ultracentrifugation is limited by the capacity of a rotor, for example, SW28 rotors have a maximum capacity of ∼230 ml of vector supernatant per 2-h run. In contrast, TFF can process much larger volume (65, 66), and a single TFF filter with 1,000-cm2 surface area can process up to 3,000 ml in 2 h. Larger filter systems with capacities of up to 15 liters are also available. In addition, VSV-G-pseudotyped LV vectors produced by TFF are reported to have a higher recovery rate than when prepared by ultracentrifugation (66).
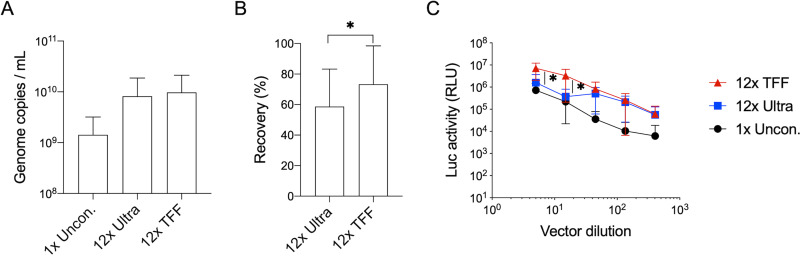
To compare these approaches, 100 ml of SΔ18-pseudotyped VSV-Luc vector supernatants was subjected to either ultracentrifugation (Ultra) or TFF and concentrated into an 8-ml final stock (12×, vol/vol). Vector genome copies in the unconcentrated and 12× concentrated vector stocks were measured by droplet digital PCR (ddPCR), which revealed slightly better recovery rates following TFF (∼70%) compared to ultracentrifugation (∼60%) (Fig. 3A and B). At the same time, the transduction efficiencies of the three vector stocks were measured on HeLa-ACE2 cells, using serially diluted vectors (1:5 to 1:450 dilutions) (Fig. 3C). At the higher dilution points, both 12× Ultra and 12× TFF vector preparations produced about a 10-fold higher luciferase signal than the unconcentrated vectors. Interestingly, at the lower dilutions (1:5 and 1:15), the 12× Ultra vector stock showed no enhancement over unconcentrated vectors, while the 12× TFF vector stocks retained their 10-fold higher transduction rates. This observation is suggestive of the presence of an inhibitory factor that is concentrated during ultracentrifugation but was not retained following TFF.
FIG 3.
Concentration methods for Spike VSV pseudovectors. (A) Genome copy numbers of VSV-Luc vectors pseudotyped with SΔ18 Spike protein, from unconcentrated supernatants (1× Uncon.) or following 12× concentration (vol/vol) by either ultracentrifugation (Ultra) or tangential flow filtration (TFF). Shown are means and standard deviations from 3 independent vector stocks. (B) Vector recovery, calculated by comparing genome copies in concentrated versus unconcentrated vector stocks. Shown are means and standard deviations from 3 independent vector concentrations for each method. *, P < 0.05, one-tailed paired t test. (C) Transduction of HeLa-ACE2 cells by serial dilutions (1:5 to 1:405) of indicated vectors. Shown are mean and standard deviation from three independent vector stocks. *, P < 0.01, two-tail paired t test for comparison between 12× Ultra and 12× TFF at the same dilutions.
In summary, we found that TFF facilitates large-scale processing of vector stocks, with genomic copy number recovery rates similar to those of the more typical ultracentrifugation method. More importantly, TFF results in vector stocks that retain a more consistent titer throughout a broader range of different dilutions than those produced by ultracentrifugation.
Cytoplasmic tail truncation alters Spike protein functional properties.
We used the VSV pseudovirus system to examine the impact of the D614G mutation of Spike protein. This mutation was first detected in China and Germany in late January and became the dominant circulating variant of SARS-CoV-2 globally by April 2020 (35). It has functional consequences for the virus, resulting in higher viral loads in the upper respiratory tract of patients (35, 67). In parallel, in vitro studies have shown that D614G enhances replication of the virus on human lung epithelial cells and primary airway tissue (42) and increases replication or transmissibility in human ACE2 transgenic mice and hamster models (42, 68, 69). Effects were also observed using Spike protein pseudoviruses, where the D614G mutation increased Spike incorporation into vector particles, despite minimal or no effect on Spike expression in vector-producing cells (44, 70), and increased transduction rates on various cell lines (31, 33, 35, 38, 44, 63, 71). Of interest, residue 164 is not located within the receptor binding domain (RBD) of Spike, and it has been suggested instead that it impacts Spike protein structure and stability, leading to downstream effects on cell entry (35, 70, 72).
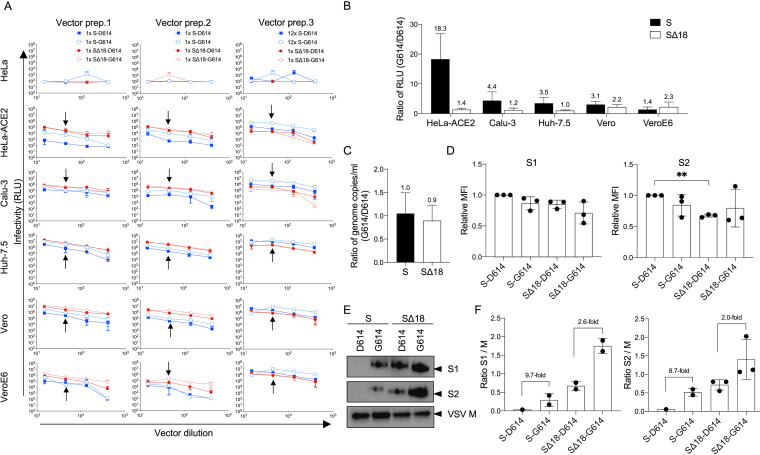
Since we had noted that cytoplasmic tail truncation of Spike protein had effects similar to those of the D614G mutation and increased incorporation rates and transduction efficiencies (Fig. 1), we next examined the impact of the D614G mutation in the context of both full-length and truncated Spike proteins. For the full-length Spike protein, we observed up to 18-fold higher transduction rates for the G614 variant on HeLa-ACE2 cells, with less striking effects on the other cell lines we tested. In contrast, transduction rates for the variants in the SΔ18 backbone showed minimal to no differences across the range of cell types tested (Fig. 4A and B).
FIG 4.
Impact of D614G mutation is only observed with full-length Spike protein. (A) Indicated cell lines were transduced with G614 or D614 variants of VSV-Luc vectors for both full-length and truncated Spike protein versions. Three preparations of all 4 vectors were produced and tested independently (vector prep. 1 to 3), using the same serial dilutions and volumes of vectors applied to the cells for transduction. For the full-length Spike variants, both unconcentrated (1×) and 12× concentrated vectors were tested. Luciferase activity was measured after 24 h. Means and standard deviations for 3 technical repeats on the same plate are shown. Arrows indicate the luciferase activity (RLU) data points used to compare G614 and D614 variants of specific Spike protein and cell line combinations. (B) Ratio of luciferase activity for the G614:D614 pair combinations indicated, on different cell lines, calculated using the RLU data points arrowed in panel A. Each pair of compared vectors was produced in the same way, and equal volumes were applied in transduction. Means and standard deviations for ratios calculated from each of the three independent vector preparations are shown. (C) Ratio of genomic titers of G614 versus D614 vectors, for both full-length and truncated Spike proteins. Shown are means and standard deviations from equal volumes of 3 independent vector stocks, produced in the same way for each pairwise comparison. (D) Cell surface expression levels of different Spike protein variants on 293T vector-producing cells, measured by flow cytometry using anti-S1 or anti-S2 antibodies at the time of vector harvest. Expression levels are reported as mean fluorescence intensity (MFI) and normalized to the level of S-D614 in each independent experiment. Means and standard deviations from 3 independent experiments are shown. **, P < 0.01, one-way analysis of variance with Tukey’s multiple comparisons. (E) Representative Western blot showing incorporation of Spike proteins into VSV particles, from equal volumes of 100× concentrated vector supernatants, using antibodies against S1 or S2 subunits of Spike or VSV M protein. (F) Comparison of Spike subunit incorporation into vectors, normalized to VSV M. Data are from 2 to 3 independent vector stocks, indicated by individual dots. S1 and S2 subunits were only detected in one stock of S-D614 vectors.
The discrepancy between the behavior of the full-length S and SΔ18 vectors occurred despite similar genome copy numbers (Fig. 4C), ruling out an effect on vector production. We also observed minimal effects on cell surface expression levels of Spike when comparing the different variants in vector-producing cells (Fig. 4D). Instead, in agreement with previous studies using full-length Spike-pseudotyped RV and LV vectors, we found that the D614G mutation enhanced Spike incorporation, albeit with a much larger effect for the full-length Spike versus the truncated protein (∼9-fold versus ∼2-fold effect) (Fig. 4E and F).
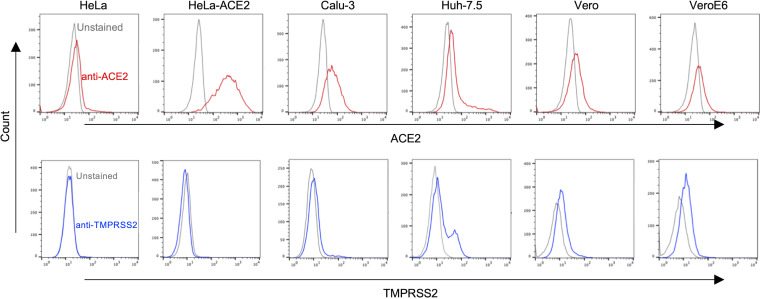
To further investigate the observation that the magnitude of titer enhancement caused by the G614 mutation in full-length Spike protein varied between different cell types, we also measured the levels of the cellular receptor ACE2 and the protease TMPRSS2 (Fig. 5). This revealed that the largest impact of the mutation occurred in the cells with the highest levels of ACE2, namely, HeLa-ACE2 and, to a lesser extent, Calu-3 cells. We hypothesize that higher ACE2 expression renders such cells more susceptible to vectors with higher Spike densities due to enhanced avidity between the Spike protein and receptors. In sum, these observations suggest that a primary effect of both the tail truncation and the D614G mutation is on Spike protein incorporation, which in turn leads to enhanced titers, at least on cells with high enough levels of ACE2. Furthermore, it is likely that an upper limit for these effects reduces the impact of the D614G mutation when combined with a tail truncation.
FIG 5.
ACE2 and TMPRSS2 expression on different cell lines. Cell surface levels of ACE2 and TMPRSS2 on indicated cell lines, measured by flow cytometry using anti-ACE2 or anti-TMPRSS2 antibodies. Unstained cells were used as negative controls.
D614G mutation or cytoplasmic tail truncation does not alter Spike protein sensitivity to convalescent serum.
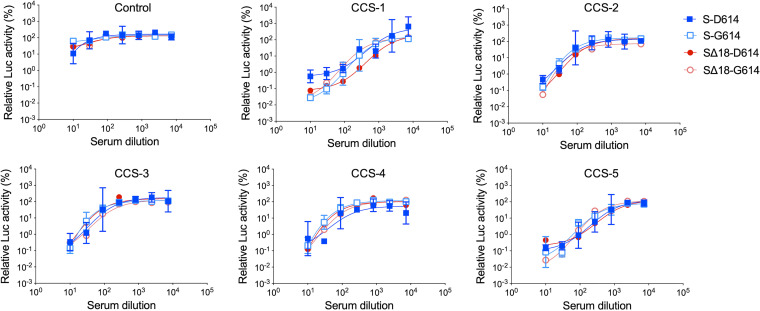
Spike protein pseudovectors are a useful tool to measure antibody neutralizing activity in COVID-19 patient or convalescent sera (4, 5, 17, 20, 21, 23–25, 27, 28). We examined whether the D614G mutation or the cytoplasmic tail truncation altered sensitivity to neutralization by a panel of convalescent sera. VSV-Luc vectors displaying the four different Spike proteins were incubated with serially diluted sera for 30 min before being applied to HeLa-ACE2 cells. After normalizing values to the luciferase signals obtained from cells transduced in the absence of sera, we observed that all four Spike proteins exhibited similar sensitivities to each serum (Fig. 6). This suggests that neither the D614G mutation nor the cytoplasmic tail truncation alters the sensitivity of the Spike protein to neutralization.
FIG 6.
Sensitivity of different Spike proteins to convalescent serum neutralization. Indicated VSV-Luc pseudoviruses were incubated with serially diluted sera (1:10- to 1:7,290-fold) from control or convalescent COVID-19 patients (CCS) for 30 min before addition to HeLa-ACE2 cells. Luciferase activity was determined 24 h later. All values were normalized to the luciferase signal from cells transduced with the same pseudovirus without serum. Means and standard deviations from 3 technical replicates of single vector stocks are shown.
DISCUSSION
Pseudotyped vectors are a useful system to study the entry glycoproteins from highly pathogenic viruses such as SARS-CoV-2, as they remove the need for BSL3 laboratory conditions. We confirmed that the SARS-CoV-2 Spike protein was able to pseudotype both LV and VSV vectors and determined that the combination of using a conditional VSV vector and a luciferase reporter gene had the advantage of allowing titers to be read at 16 h postransduction. Such Spike-pseudotyped VSV vectors supported entry into a variety of mammalian cell types, including lung organoid systems, making them a useful system with which to study SARS-CoV-2 entry under standard laboratory conditions.
Optimization of pseudotyped vectors includes selection of an appropriate concentration method, such as centrifugation, polyethylene glycol (PEG) precipitation, or ultrafiltration. For VSV pseudovectors, we found that concentration by TFF produced vector stocks with higher recovery rates and more consistent titers throughout a dilution series than those produced by ultracentrifugation. TFF also has the advantage of providing a partial purification due to the selective loss of potential contaminants below the cutoff value of the filter and provides a larger processing capacity than ultracentrifugation. As a result, TFF is frequently used to facilitate large-scale vector production, including for clinical use (65, 66, 73). In our own experience, 3 liters of supernatant can be concentrated down to 50 ml in 2 h.
Since pseudovector titers can be impacted by incompatibilities between a viral fusion protein and the heterologous viral particle (46–51), an additional strategy to enhance vector titers has been to truncate the fusion protein’s cytoplasmic tail. We found that this approach increased the titers of Spike protein pseudovectors based on both LV and VSV, in agreement with previous reports (23, 25, 60–63). Furthermore, as others have also noted (60, 62), the enhanced vector titers correlated with increased levels of Spike protein incorporation that were not simply the result of higher levels of cell surface expression following tail truncation, and tail truncation has also been reported to enhance the fusogenicity of Spike (60, 62). Together, this suggests that truncation of the cytoplasmic tail could also alter the conformation or function of the protein’s ectodomain, as has been reported for viral fusion proteins in HIV (58), measles virus (57), simian immunodeficiency virus (56), and gibbon ape leukemia virus (47), where truncation of the cytoplasmic tail impacted ectodomain conformation or functions such as receptor binding or fusogenicity.
Our comparison of techniques to enhance vector titers also identified an area for caution. Although cytoplasmic tail truncation enhanced pseudovector titers, they can also have unintended functional consequences. Specifically, we found that the impact of the D614G mutation on Spike protein incorporation and vector titer was obscured by the cytoplasmic tail truncation. A similar lack of effect of the D614G mutation on titer was also reported in another study using a 21-amino-acid deletion of the Spike protein cytoplasmic tail in VSV pseudovectors (62). These findings suggest that tail-truncated Spike proteins should be used with caution for studies analyzing the impact of Spike mutations or to test potential therapeutics targeting SARS-CoV-2 entry.
The mechanism for enhanced incorporation and/or titer by D614G is not entirely understood, but structural analyses have suggested that it could impact Spike protein structure and stability, both within and between Spike monomers. For example, it has been suggested that a glycine at this location could strengthen the association between the S1/S2 subunits through an impact on the epistructure that decreases the intramolecular wrapping in the S1 subunit but promotes intermolecular wrapping between S1 and S2 (72). In an alternative model, the D614G mutation could alter the structure and/or stability of the Spike trimer by abrogating the hydrogen bond connecting D614 in the S1 subunit of one monomer with T859 in the S2 subunit of a neighboring monomer (35). These alterations were hypothesized to result in a greater tendency of the G614 monomers to form stable trimers, which, in turn, could facilitate their incorporation into virions. As evidence, a mixture of equal amounts of D614 and G614 Spike variants expressed in vector-producing cells resulted in a higher level of G614 proteins in the incorporated Spike trimers (70).
Finally, we also used the VSV pseudovectors to evaluate the impact of the D614G mutation on infectivity of different cell types and sensitivity to antibody neutralization. Consistent with previous findings using pseudoviruses (33–35, 38, 44, 45) or SARS-CoV-2 virions (42, 68), we found that the G614 variant exhibited enhanced transduction of various cell lines compared to the D614 variant, and that this correlated with increased Spike incorporation into the VSV particles (44, 70). Interestingly, the enhanced transduction effect was more profound in cells with higher ACE2 levels, which may reflect increased avidity between the receptor (ACE2) and the ligand (Spike). Finally, as previously noted, these effects were significantly abrogated when tail truncated variants were used, consistent with an upper limit for the enhancement of Spike incorporation and transduction.
In contrast, the serum neutralization studies revealed no differences in sensitivity for either residue at position 614 and in either of the cytoplasmic tail configurations. This is in agreement with the majority of reports testing the impact of the D614G mutation with full-length Spike pseudovectors or SARS-CoV-2 virus against human convalescent serum (31, 35, 44, 68), serum from convalescent animals (42), or vaccinated humans or animals (45, 68, 74).
In summary, we found that although cytoplasmic tail truncations enhance SARS-CoV-2 Spike protein incorporation into both LV and VSV vectors and enhance the titers of unconcentrated vectors, they can also mask the phenotype of the D614G mutation. Pseudotyped vectors are increasingly being used to study newly emerging SARS-CoV-2 variants, where both full-length (43, 75) and truncated Spike proteins (26, 36, 40) have been used in studies investigating the impact of mutations on Spike protein properties such as ACE2 binding, transduction efficiency, or sensitivity to neutralization. To better ensure the authenticity of the Spike protein functions being investigated in such vectors, we recommend using a full-length Spike protein and combining vector production with TFF if higher titer vectors are required.
MATERIALS AND METHODS
Plasmids.
Full-length (S) and 18-amino-acid cytoplasmic tail truncated (SΔ18) Spike proteins for the Wuhan-Hu-1 isolate of SARS-CoV-2 (GenBank accession no. MN908947.3) were provided by James Voss (The Scripps Research Institute) in a plasmid pcDNA3.3 backbone. D614G mutants were generated by site-directed mutagenesis. A VSV-G protein expression plasmid was obtained from Addgene (8454; Watertown, MA).
Cell lines.
293T, HeLa, HeLa-ACE2, Vero, VeroE6, and Huh7.5 cells were maintained in Dulbecco’s modified Eagle medium (DMEM), and Calu-3 cells were maintained in Eagle's minimum essential medium (EMEM). All media were supplemented with 4 mM glutamine and 10% fetal bovine serum (FBS). HeLa-ACE2 cells were provided by James Voss and were generated by transduction of HeLa cells with a lentiviral vector packaging a CMV-ACE2 expression cassette. The Huh7.5 cell line was provided by Jae Jung (Cleveland Clinic). All other cell lines were obtained from the ATCC.
VSV vector production, concentration, and transduction.
Replication-deficient VSVΔG vectors (76), containing expression cassettes for firefly luciferase or GFP in place of the VSV-G protein, were provided by Jae Jung and Oscar Negrete (Sandia National Laboratories), respectively. To generate Spike-pseudotyped VSV vectors, 4 × 106 293T cells were seeded in DMEM plus 10% FBS in a 10-cm plate and transfected with 15 μg of Spike expression plasmid 24 h later, using the calcium-phosphate transfection method (76). Medium was replaced 16 h later with 10 ml fresh medium, and after a further 8 h, 5 ml was removed and 2 × 108 vector genomes of VSVΔG particles were added for 1 h at 37°C. Following this incubation, cells were washed three times with phosphate-buffered saline (PBS) and incubated for a further 24 h before harvesting supernatants.
For larger-scale production, quantities were adjusted to seed 3 × 107 cells in 500-cm2 plates, transfection with 124.5 μg of Spike expression plasmid, and infection by 1.7 × 109 vector genomes of VSVΔG particles per 500-cm2 plate. To propagate VSVΔG particles, the same protocol was followed but replacing the Spike expression plasmid with the same quantity of a VSV-G expression plasmid, and no PBS washes were performed after infection by VSVΔG.
Vector supernatants were harvested and filtered through 0.45-μm syringe filters and either aliquoted or concentrated by ultracentrifugation using 20% (wt/vol) sucrose cushions for 2 h at 25,000 rpm in an SW41 or SW28 rotor (Beckman, Indianapolis, IN). Alternatively, large-scale supernatant preparations were concentrated by tangential flow filtration (TFF) using a polyethersulfone membrane hollow fiber unit with 100-kDa-molecular-weight cutoff and 115-cm2 filtration surface (Spectrum Laboratories, Rancho Dominguez, CA) and a KR2i peristaltic pump (Spectrum Laboratories). To perform buffer exchange and prevent filter blockage, every 100 ml of vector supernatant was followed by 100 ml PBS. A 10- to 12-fold concentration from the original volume to an approximately 8-ml final volume was achieved. All vectors were stored at −80°C in aliquots.
VSV-luciferase vector transductions were performed on tissue culture-treated, 96-well half-area white plates (Corning, Corning, NY), seeded with various cells lines to achieve 50% to 75% confluence at the time of transduction. Vectors were serially diluted and added to the culture to achieve final dilutions of 1:5, 1:15, 1:45, 1:135, and 1:405 and incubated at 37°C for 16 to 24 h. Transduction efficiency was quantified by measuring luciferase activity in cell lysates using Britelite plus (Perkin Elmer, Richmond, California) and following the manufacturer’s protocol. To calculate the fold change in transduction efficiency between D614 and G614 mutants, data from the 1:45 dilution points were used.
To titer VSV-GFP vectors, HeLa-ACE2 cells were seeded at 1 × 104 cells per well in 96-well plates, and the following day, 50 μl of serially diluted unconcentrated vector stocks was added. The final dilutions in the cultures were 1:2, 1:6, 1:18, and 1:27. Transduction efficiency was determined by GFP expression 16 to 24 h after transduction using flow cytometry (Guava easyCyte; MilliporeSigma, Burlington, MA) and transducing units (TU) per milliliter calculated from the dilutions showing a linear relationship between the dilution factor and the number of GFP-positive cells.
Lentiviral vector production, concentration, and transduction.
Lentiviral vectors were generated by transfection of 10-cm plates of 293T cells at 75% confluence with 2 μg of Spike expression plasmid, 10 μg of packaging plasmid pCMVdeltaR8.2 (12263; Addgene), and 10 μg of a GFP-expressing vector genome plasmid FUGW (14883; Addgene). Medium was removed 16 h later and replaced with 10 ml fresh DMEM plus 10% FBS. Supernatants were harvested 48 h after transfection, filtered through 0.45-μm syringe filters, and either aliquoted or concentrated by ultracentrifugation using 20% (wt/vol) sucrose cushions for 2 h at 25,000 rpm in an SW41 rotor (Beckman).
HeLa-ACE2 cells were transduced with Spike pseudotyped LV by seeding 1 × 104 cells per well in 96-well plates and adding 50 μl of unconcentrated vector stocks the next day. Transduction efficiency was determined by GFP expression 48 h after transduction using flow cytometry, as described above, and reported as TU per milliliter.
LV and VSV vector genome titration.
RNA from 160 μl of LV or VSV vector stocks was extracted using a viral RNA minikit (Qiagen, Hilden, Germany) and reverse transcribed into cDNA using SuperScript (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Genome copy number was determined by ddPCR for the WPRE sequences in the LV genome or for phosphoprotein (P) sequences in the VSV genome using the QX200 droplet digital PCR system (Bio-Rad, Hercules, CA) and primer/probe set WPRE-forward (CCTTTCCGGGACTTTCGCTTT), WPRE-reverse (GGCGGCGGTCACGAA), WPRE-probe (6-carboxyfluorescein [FAM]-ACTCATCGCCGCCTGCCTTGCC-6-carboxytetramethylrhodamine [TAMRA]), P-forward (GTCTTCAGCCTCTCACCATATC), P-reverse (AGCAGGATGGCCTCTTTATG), and P-probe (FAM-TCGGAGGTGACGGACGAATGTCT-IOWA BLACK). Briefly, 6.25 μl of 1:10- and 1:100-diluted cDNA was mixed with forward and reverse primers (final concentration, 900 nM), probe (final concentration, 250 nM), and 2× ddPCR supermix (Bio-Rad) and made up to 25 μl with water. Twenty microliters of each reaction mix was converted to droplets by the QX200 droplet generator, and droplet-partitioned samples were transferred to a 96-well plate and sealed. Thermal cycling was performed with the following conditions: 95°C for 10 min, 40 cycles of 94°C for 30 s, 60°C for 1 min, and 98°C for 10 min. Plates were read on a QX200 reader (Bio-Rad) and DNA copies quantified by detection of FAM-positive droplets.
Lung bud organoid differentiation and transduction.
Lung bud organoids were generated from human pluripotent stem cells (hPSCs) and validated as previously described (77). hPSC differentiation into endoderm was performed in serum-free differentiation (SFD) medium of Iscove’s modified Dulbecco’s medium–Ham’s F-12 (3:1) (Life Technologies, Carlsbad, CA) supplemented with the following: 1× N2 (Life Technologies), 0.5× B27 (Life Technologies), 50 μg/ml ascorbic acid, 1× GlutaMAX (Gibco), 0.4 μM monothioglycerol, 0.05% bovine serum albumin [BSA], 10 μM Y27632, 0.5 ng/ml human BMP4 [R&D Systems], 2.5 ng/ml human FGF2 [R&D Systems, Minneapolis, MN], and 100 ng/ml human Activin [R&D Systems]) in a 5% CO2, 5% O2 atmosphere at 37°C for 72 to 76 h. On day 4, endoderm yield was determined by the expressions of CXCR4 and c-KIT by flow cytometry. Cells used in all experiments had > 90% endoderm yield. For induction of anterior foregut endoderm, embryonic bodies were dissociated into single cells using 0.05% trypsin–0.02% EDTA and plated onto fibronectin-coated, 6-well tissue culture plates (80,000 to 105,000 cells/cm2). Cells were incubated in SFD medium supplemented with 100 ng/ml human Noggin (R&D Systems) and 10 μM SB431542 for 24 h, followed by switching to SFD medium supplemented with 10 μM SB431542 and 1 μM IWP2 (R&D Systems) for another 24 h. At the end of anterior foregut endoderm induction, cells were maintained in SFD medium supplemented with 3 μM CHIR 99021 (CHIR, R&D Systems), 10 ng/ml human FGF10, 10 ng/ml human KGF, 10 ng/ml human BMP4, and 50 nM all-trans retinoic acid for 48 h when three-dimensional cell clumps formed. Clumps were suspended by gently pipetting around the wells to form lung bud organoids, which were maintained in ultra-low-attachment multiple-well plates (Corning), fed every other day, and used for vector transduction after day 35.
To transduce lung bud organoids, 10 to 20 organoids were picked manually and transferred to 96-well U-bottom plates and transduced with 50 μl of GFP-expressing VSV vectors (1.7 × 104 TU/ml). Transduction efficiency was examined by GFP expression 24 h later by fluorescence microscopy.
Western blot analysis of Spike protein incorporation.
Vector supernatants were concentrated by ultracentrifugation (100-fold), electrophoresed on 4 to 12% Bis-Tris protein gels (Bio-Rad), and transferred to polyvinylidene difluoride membranes using a Trans-Blot turbo transfer system (Bio-Rad). Membranes were blocked with 5% milk in PBST buffer (PBS plus 0.1% Tween 20). The S1 subunit of Spike was detected using SARS-CoV-2 (COVID-19) Spike S1 antibody at 1:1,000 (9083; Prosci); the S2 subunit was detected using anti-SARS-CoV/SARS-CoV-2 (COVID-19) spike antibody clone [1A9] at 1:1,000 (GTX632604; GeneTex); and HIV-1 p24 was detected using a polyclonal anti-HIV-1 SF2 p24 rabbit antiserum at 1:6,000 (obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH; ARP-4250, contributed by DAIDS/NIAID; produced by BioMolecular Technologies). VSV M protein was detected using anti-VSV M antibody clone (23H12) (EB0011; KeraFast, Boston, MA) at 1:1,000. Horseradish peroxidase (HRP)-conjugated goat anti-mouse and goat anti-rabbit antibodies were used as secondary antibodies (Santa Cruz Biotechnology, Dallas, TX). Blots were imaged by Amersham ECL Prime Western blotting detection reagent (GE Healthcare, Chicago, IL) and Chemidoc (Bio-Rad). Densitometry was measured using ImageJ software (http://rsb.info.nih.gov/ij/).
ACE2 and TMPRSS2 cell surface expression by flow cytometry.
Cells were detached from culture flasks by 0.05% trypsin (Corning) and washed once with PBS. One million cells were resuspend in 100 μl PBS and either immediately incubated with anti-ACE2 antibody (AF933; 1:100 dilution; R&D Systems) and anti-TMPRSS2 (H4) antibody (sc-515727 PE; 1:50 dilution; Santa Cruz) or first incubated at 37°C for 6 h with shaking to allow recovery of cell surface proteins after trypsinization. Alexa Fluor 647-conjugated donkey anti-goat antibody (A21447; 1:200 dilution; Thermo Fisher Scientific, Waltham, MA) was used as a secondary antibody, and ACE2 expression was determined by flow cytometry (Guava easyCyte).
Spike protein cell surface expression.
VSV pseudovector-producing 293T cells were harvested to examine cell surface expression of the Spike protein. The S1 subunit was detected using SARS-CoV-2 (COVID-19) Spike S1 antibody at 1:100 (9083; Prosci, Fort Collins, Colorado), and the S2 subunit was detected using anti-SARS-CoV/SARS-CoV-2 (COVID-19) spike antibody clone (1A9) at 1:100 (GTX632604; GeneTex, Irvine, CA). Allophycocyanin-conjugated goat anti-mouse and donkey anti-rabbit antibodies were used as secondary antibodies (1:100 dilution; Invitrogen), and expression was detected by flow cytometry (Guava easyCyte). The expression levels of different Spike proteins were reported as mean fluorescence intensity (MFI).
Convalescent serum neutralization.
Convalescent serum from COVID-19 patients or healthy donors (collected before 30 April 2020) was obtained from Children’s Hospital Los Angeles. Convalescent sera were confirmed to be positive for IgG class antibodies against SARS-CoV-2 Spike using anti-SARS-CoV-2 ELISA (IgG) (Euroimmun, Lübeck, Germany) (78).
A suitable dose of Spike-pseudotyped VSV-Luc vectors was used in the neutralization assays to produce approximately 105 relative light units (RLU) of luciferase activity on HeLa-ACE2 cells in the absence of serum. A total of 5 × 103 HeLa-ACE2 cells were seeded in tissue culture-treated, 96-well half-area white plates (Corning) to achieve 50% to 75% confluence the following day. Convalescent or control sera were 3-fold serially diluted from 1:10 to 1:7,290, and 50 μl was incubated with the predetermined dose of the VSV-Luc vectors for 30 min at 37°C before addition of the mixture to HeLa-ACE2 cells. Cells were incubated at 37°C overnight for 16 to 24 h. Vector transduction efficiency was quantified by measuring luciferase activity as described above and neutralization (%) calculated by normalization to the values obtained on cells transduced without serum.
ACKNOWLEDGMENTS
This work was supported by a grant to P.M.C. from the W. M. Keck Foundation COVID-19 Research Fund to the Keck School of Medicine of USC.
We acknowledge the generosity of James Voss, Jae Jung, and Oscar Negrete, who provided COVID-19 reagents, cell lines, and VSVΔG particles.
Contributor Information
Paula M. Cannon, Email: pcannon@usc.edu.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Zhou P, Yang X, Lou Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang R, Di Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johns Hopkins University. 2021. Coronavirus Resource Center. Johns Hopkins Coronavirus Resource Center, Baltimore, MD. [Google Scholar]
- 3.Umakanthan S, Chattu VK, Ranade AV, Das D, Basavarajegowda A, Bukelo M. 2021. A rapid review of recent advances in diagnosis, treatment and vaccination for COVID-19. AIMS Public Health 8:137–153. 10.3934/publichealth.2021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, Agudelo M, Barnes CO, Gazumyan A, Finkin S, Hägglöf T, Oliveira TY, Viant C, Hurley A, Hoffmann HH, Millard KG, Kost RG, Cipolla M, Gordon K, Bianchini F, Chen ST, Ramos V, Patel R, Dizon J, Shimeliovich I, Mendoza P, Hartweger H, Nogueira L, Pack M, Horowitz J, Schmidt F, Weisblum Y, Michailidis E, Ashbrook AW, Waltari E, Pak JE, Huey-Tubman KE, Koranda N, Hoffman PR, West AP, Rice CM, Hatziioannou T, Bjorkman PJ, Bieniasz PD, Caskey M, Nussenzweig MC. 2020. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584:437–442. 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L. 2020. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584:115–119. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 6.Mohamed Khosroshahi L, Rokni M, Mokhtari T, Noorbakhsh F. 2021. Immunology, immunopathogenesis and immunotherapeutics of COVID-19; an overview. Int Immunopharmacol 93:107364. 10.1016/j.intimp.2020.107364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94:e00127-20. 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 5:562–569. 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masre SF, Jufri NF, Ibrahim FW, Abdul Raub SH. 2020. Classical and alternative receptors for SARS-CoV-2 therapeutic strategy. Rev Med Virol 2020:e2207–e2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, Wei D, Zhang Y, Sun XX, Gong L, Yang X, He L, Zhang L, Yang Z, Geng JJ, Chen R, Zhang H, Wang B, Zhu YM, Nan G, Jiang JL, Li L, Wu J, Lin P, Huang W, Xie L, Zheng ZH, Zhang K, Miao JL, Cui HY, Huang M, Zhang J, Fu L, Yang XM, Zhao Z, Sun S, Gu H, Wang Z, Wang CF, Lu Y, Liu YY, Wang QY, Bian H, Zhu P, Chen ZN. 2020. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther 5:283. 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–210. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bestle D, Heindl MR, Limburg H, Van TVL, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O, Rohde C, Becker S, Klenk H-D, Garten W, Steinmetzer T, Böttcher-Friebertshäuser E. 2020. TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS-CoV-2 in human airway epithelial cells and provide promising drug targets. Life Sci Alliance 3:e202000786. 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M-YM, Li L, Zhang Y, Wang XX-S. 2020. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty 9:45. 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi LE, Barbry P, Leslie A, Kiem HP, HCA Lung Biological Network, et al. 2020. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 181:1016–1035. 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Liu Q, Huang W, Li X, Wang Y. 2018. Current status on the development of pseudoviruses for enveloped viruses. Rev Med Virol 28:e1963. 10.1002/rmv.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, Zhang X. 2021. Construction and applications of SARS-CoV-2 pseudoviruses: a mini review. Int J Biol Sci 17:1574–1580. 10.7150/ijbs.59184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieterle ME, Haslwanter D, Bortz RH, Wirchnianski AS, Lasso G, Vergnolle O, Abbasi SA, Fels JM, Laudermilch E, Florez C, Mengotto A, Kimmel D, Malonis RJ, Georgiev G, Quiroz J, Barnhill J, Pirofski L, Daily JP, Dye JM, Lai JR, Herbert AS, Chandran K, Jangra RK. 2020. A replication-competent vesicular stomatitis virus for studies of SARS-CoV-2 Spike-mediated cell entry and its inhibition. Cell Host Microbe 28:486–496. 10.1016/j.chom.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11:1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell 181:281–212. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, Zhang L, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Fan C, Huang W, Xu M, Wang Y. 2020. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect 9:680–686. 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu F, Xun J, Lu L, Jiang S, Lu H, Wen Y, Huang J. 2020. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv 10.1101/2020.03.30.20047365. [DOI] [Google Scholar]
- 22.Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus ADME, van Kuppeveld FJM, Haagmans BL, Grosveld F, Bosch B-J. 2020. A human monoclonal 1 antibody blocking SARS-CoV-2 infection. Nat Commun 11:2251. 10.1038/s41467-020-16452-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt F, Weisblum Y, Muecksch F, Hoffmann HH, Michailidis E, Lorenzi JCC, Mendoza P, Rutkowska M, Bednarski E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Caskey M, Robbiani DF, Nussenzweig MC, Rice CM, Hatziioannou T, Bieniasz PD. 2020. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med 217:e20201181. 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Case JB, Rothlauf PW, Chen RE, Liu Z, Zhao H, Kim AS, Bloyet LM, Zeng Q, Tahan S, Droit L, Ilagan MXG, Tartell MA, Amarasinghe G, Henderson JP, Miersch S, Ustav M, Sidhu S, Virgin HW, Wang D, Ding S, Corti D, Theel ES, Fremont DH, Diamond MS, Whelan SPJ. 2020. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 28:475–485. 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong HL, Wu YT, Cao JL, Yang R, Liu YX, Ma J, Qiao XY, Yao XY, Zhang BH, Zhang YL, Hou WH, Shi Y, Xu JJ, Zhang L, Wang SJ, Fu BR, Yang T, Ge SX, Zhang J, Yuan Q, Huang BY, Li ZY, Zhang TY, Xia NS. 2020. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg Microbes Infect 9:2105–2113. 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu Z, Whelan SPJ, Carnahan RH, Crowe JE, Bloom JD. 2021. Complete mapping of mutations to the SARS-CoV-2 Spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29:44–57. 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klingler J, Weiss S, Itri V, Liu X, Oguntuyo KY, Stevens C, Ikegame S, Hung C, Enyindah-Asonye G, Amanat F, Bermudez-Gonzalez M, Simon V, Liu S, Lee B. 2020. Role of IgM and IgA antibodies to the neutralization of SARS-CoV-2. medRxiv 10.1101/2020.08.18.20177303. [DOI] [Google Scholar]
- 28.Luchsinger LL, Ransegnola B, Jin D, Muecksch F, Weisblum Y, Bao W, George PJ, Rodriguez M, Tricoche N, Schmidt F, Gao C, Jawahar S, Pal M, Schnall E, Zhang H, Strauss D, Yazdanbakhsh K, Hillyer CD, Bieniasz PD, Hatziioannou T. 2020. Serological assays estimate highly variable SARS-CoV-2 neutralizing antibody activity in recovered COVID19 patients. J Clin Microbiol 58:e02005-20. 10.1128/JCM.02005-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi Y, Suzuki T, Arimori T, Ikemura N, Kirita Y, Ohgitani E, Mazda O, Motooka D, Nakamura S, Matsuura Y, Matoba S, Okamoto T, Takagi J, Hoshino A. 2020. High affinity modified ACE2 receptors prevent SARS-CoV-2 infection. bioRxiv 10.1101/2020.09.16.299891. [DOI]
- 30.Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM, Zhang T, Uhl S, Bram Y, Richardson C, Zhu J, Zhao Z, Redmond D, Houghton S, Nguyen DHT, Xu D, Wang X, Jessurun J, Borczuk A, Huang Y, Johnson JL, Liu Y, Xiang J, Wang H, Cantley LC, TenOever BR, Ho DD, Pan FC, Evans T, Chen HJ, Schwartz RE, Chen S. 2021. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589:270–275. 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozono S, Zhang Y, Ode H, Sano K, Tan TS, Imai K, Miyoshi K, Kishigami S, Ueno T, Iwatani Y, Suzuki T, Tokunaga K. 2021. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun 12:848. 10.1038/s41467-021-21118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muik A, Wallisch A-K, Sänger B, Swanson KA, Mühl J, Chen W, Cai H, Maurus D, Sarkar R, Türeci Ö, Dormitzer PR, Şahin U. 2021. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science 371:1152–1153. 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daniloski Z, Jordan TX, Ilmain JK, Guo X, Bhabha G, Tenoever BR, Sanjana NE. 2021. The spike d614g mutation increases sars-cov-2 infection of multiple human cell types. Elife 10:e65365. 10.7554/eLife.65365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou T, Tsybovsky Y, Olia AS, Gorman J, Rapp M, Cerutti G 8, Chuang G-Y, Katsamba PS, Nazzari A, Sampson JM, Schön A, Wang P, Bimela J, Shi W, Teng I-T, Zhang B, Boyington JC, Sastry M, Stephens T, Stuckey J, Wang S, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD. 2020. Cryo-EM structures delineate a pH-dependent switch 4 that mediates endosomal positioning of SARS-CoV-2 spike receptor-binding domains. bioRxiv 10.1101/2020.07.04.187989. [DOI] [PMC free article] [PubMed]
- 35.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group. 2020. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JCC, Muecksch F, Rutkowska M, Michailidis H-H, Gaeble C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2020. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9:e61312. 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD. 2021. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593:130–135. 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 38.Hu J, He CL, Gao Q, Zhang GJ, Cao XX, Long QX, Deng HJ, Huang LY, Chen J, Wang K, Tang N, Huang AL. 2020. D614G mutation of SARS-CoV-2 spike protein enhances viral infectivity. bioRxiv 10.1101/2020.06.20.161323. [DOI]
- 39.Ku Z, Xie X, Davidson E, Ye X, Su H, Menachery VD, Li Y, Yuan Z, Zhang X, Muruato AE, I Escuer AG, Tyrell B, Doolan K, Doranz BJ, Wrapp D, Bates PF, McLellan JS, Weiss SR, Zhang N, Shi P-Y, An Z. 2021. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat Commun 12:469. 10.1038/s41467-020-20789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C, Lieberman JA, Oliveira TY, Yang Z, Abernathy ME, Huey-Tubman KE, Hurley A, Turroja M, West KA, Gordon K, Millard KG, Ramos V, Da Silva J, Xu J, Colbert RA, Patel R, Dizon J, Unson-O'Brien C, Shimeliovich I, Gazumyan A, Caskey M, Bjorkman PJ, Casellas R, Hatziioannou T, Bieniasz PD, Nussenzweig MC. 2021. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592:616–622. 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, de Oliveira T, Vermeulen M, van der Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN, Moore PL. 2021. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 27:622–625. 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 42.Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, Mirchandani D, Scharton D, Bilello JP, Ku Z, An Z, Kalveram B, Freiberg AN, Menachery VD, Xie X, Plante KS, Weaver SC, Shi PY. 2021. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592:116–121. 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, King NP, Veesler D, Bloom JD. 2020. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182:1295–1310.e20. 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, Rangarajan ES, Pan A, Vanderheiden A, Suthar MS, Li W, Izard T, Rader C, Farzan M, Choe H. 2020. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun 11:6013. 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weissman D, Alameh MG, de Silva T, Collini P, Hornsby H, Brown R, LaBranche CC, Edwards RJ, Sutherland L, Santra S, Mansouri K, Gobeil S, McDanal C, Pardi N, Hengartner N, Lin PJC, Tam Y, Shaw PA, Lewis MG, Boesler C, Şahin U, Acharya P, Haynes BF, Korber B, Montefiori DC. 2021. D614G Spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe 29:23–31. 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mammano F, Salvatori F, Indraccolo S, De Rossi A, Chieco-Bianchi L, Göttlinger HG. 1997. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J Virol 71:3341–3345. 10.1128/JVI.71.4.3341-3345.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christodoulopoulos I, Cannon PM. 2001. Sequences in the cytoplasmic tail of the gibbon ape leukemia virus envelope protein that prevent its incorporation into lentivirus vectors. J Virol 75:4129–4138. 10.1128/JVI.75.9.4129-4138.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Funke S, Maisner A, Mühlebach MD, Koehl U, Grez M, Cattaneo R, Cichutek K, Buchholz CJ. 2008. Targeted cell entry of lentiviral vectors. Mol Ther 16:1427–1436. 10.1038/mt.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bender RR, Muth A, Schneider IC, Friedel T, Hartmann J, Plückthun A, Maisner A, Buchholz CJ. 2016. Receptor-targeted Nipah virus glycoproteins improve cell-type selective gene delivery and reveal a preference for membrane-proximal cell attachment. PLoS Pathog 12:e1005641. 10.1371/journal.ppat.1005641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witting SR, Vallanda P, Gamble AL. 2013. Characterization of a third generation lentiviral vector pseudotyped with Nipah virus envelope proteins for endothelial cell transduction. Gene Ther 20:997–1005. 10.1038/gt.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Höhne M, Thaler S, Dudda JC, Groner B, Schnierle BS. 1999. Truncation of the human immunodeficiency virus-type-2 envelope glycoprotein allows efficient pseudotyping of murine leukemia virus retrovital vector particles. Virology 261:70–78. 10.1006/viro.1999.9847. [DOI] [PubMed] [Google Scholar]
- 52.Moore MJ, Dorfman T, Li W, Wong SK, Li Y, Kuhn JH, Coderre J, Vasilieva N, Han Z, Greenough TC, Farzan M, Choe H. 2004. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J Virol 78:10628–10635. 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aguilar HC, Anderson WF, Cannon PM. 2003. Cytoplasmic tail of moloney murine leukemia virus envelope protein influences the conformation of the extracellular domain: implications for mechanism of action of the R peptide. J Virol 77:1281–1291. 10.1128/jvi.77.2.1281-1291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khetawat D, Broder CC. 2010. A functional henipavirus envelope glycoprotein pseudotyped lentivirus assay system. Virol J 7:312–314. 10.1186/1743-422X-7-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aguilar HC, Matreyek KA, Choi DY, Filone CM, Young S, Lee B. 2007. Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J Virol 81:4520–4532. 10.1128/JVI.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zingler K, Littman DR. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases env incorporation into particles and fusogenicity and infectivity. J Virol 67:2824–2831. 10.1128/JVI.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cathomen T, Naim HY, Cattaneo R. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J Virol 72:1224–1234. 10.1128/JVI.72.2.1224-1234.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B. 2015. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science 349:191–195. 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White E, Wu F, Chertova E, Bess J, Roser JD, Lifson JD, Hirsch VM. 2017. Truncating the gp41 cytoplasmic tail of simian immunodeficiency virus decreases sensitivity to neutralizing antibodies without increasing the envelope content of virions. J Virol 92:e01688-17. 10.1128/JVI.01688-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu J, Li Z, He X, Gebre MS, Bondzie EA, Wan H, Jacob-Dolan C, Martinez DR, Nkolola JP, Baric RS, Barouch DH. 2021. Deletion of the SARS-CoV-2 Spike cytoplasmic tail increases infectivity in pseudovirus neutralization assays. J Virol 95:e00044-21. 10.1128/JVI.00044-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson MC, Lyddon TD, Suarez R, Salcedo B, LePique M, Graham M, Ricana C, Robinson C, Ritter DG. 2020. Optimized pseudotyping conditions for the SARS-COV-2 Spike glycoprotein. J Virol 94:e01062-20. 10.1128/JVI.01062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Havranek KE, Jimenez AR, Acciani MD, Fernanda M, Mendoza L, Mary J, Ballista R, Diaz DA, Brindley MA. 2020. SARS-CoV-2 Spike alterations enhance. Viruses 12:1465. 10.3390/v12121465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu X, Tao L, Zhang X. 2021. Comprehensive and systemic optimization for improving the yield of SARS-COV-2 spike pseudotyped virus. Mol Ther Methods Clin Dev 20:350–356. 10.1016/j.omtm.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin C, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC. 2014. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods 409:131–146. 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cooper AR, Patel S, Senadheera S, Plath K, Kohn DB, Hollis RP. 2011. Highly efficient large-scale lentiviral vector concentration by tandem tangential flow filtration. J Virol Methods 177:1–9. 10.1016/j.jviromet.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geraerts M, Michiels M, Baekelandt V, Debyser Z, Gijsbers R. 2005. Upscaling of lentiviral vector production by tangential flow filtration. J Gene Med 7:1299–1310. 10.1002/jgm.778. [DOI] [PubMed] [Google Scholar]
- 67.Lorenzo-Redondo R, Nam HH, Roberts SC, Simons LM, Jennings LJ, Qi C, Achenbach CJ, Hauser AR, Ison MG, Hultquist JF, Ozer EA. 2020. A clade of SARS-CoV-2 viruses associated with lower viral loads in patient upper airways. EBioMedicine 62:103112. 10.1016/j.ebiom.2020.103112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hou YJ, Chiba S, Halfmann P, Ehre C, Kuroda M, Dinnon KH, Leist SR, Schäfer A, Nakajima N, Takahashi K, Lee RE, Mascenik TM, Graham R, Edwards CE, Tse LV, Okuda K, Markmann AJ, Bartelt L, de Silva A, Margolis DM, Boucher RC, Randell SH, Suzuki T, Gralinski LE, Kawaoka Y, Baric RS. 2020. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370:1464–1468. 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou B, Thi Nhu Thao T, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, Pohlmann A, King J, Steiner S, Kelly JN, Portmann J, Halwe NJ, Ulrich L, Trüeb BS, Fan X, Hoffmann B, Wang L, Thomann L, Lin X, Stalder H, Pozzi B, de Brot S, Jiang N, Cui D, Hossain J, Wilson M, Keller M, Stark TJ, Barnes JR, Dijkman R, Jores J, Benarafa C, Wentworth DE, Thiel V, Beer M. 2021. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592:122–127. 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- 70.Michaud WA, Boland GM, Rabi SA. 2020. The SARS-CoV-2 Spike mutation D614G increases entry fitness across a range of ACE2 levels, directly outcompetes the wild type, and is preferentially incorporated into trimers. bioRxiv 10.1101/2020.08.25.267500. [DOI]
- 71.Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, Baum A, Diehl WE, Dauphin A, Carbone C, Veinotte K, Egri SB, Schaffner SF, Lemieux JE, Munro JB, Rafique A, Barve A, Sabeti PC, Kyratsous CA, Dudkina NV, Shen K, Luban J. 2020. Structural and functional analysis of the D614G SARS-CoV-2 Spike protein variant. Cell 183:739–751. 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernández A. 2020. Structural impact of mutation D614G in SARS-CoV-2 Spike protein: enhanced infectivity and therapeutic opportunity. ACS Med Chem Lett 11:1667–1670. 10.1021/acsmedchemlett.0c00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cavazzana-Calvo M, Payen E, Negre O, Wang G, Hehir K, Fusil F, Down J, Denaro M, Brady T, Westerman K, Cavallesco R, Gillet-Legrand B, Caccavelli L, Sgarra R, Maouche-Chrétien L, Bernaudin F, Girot R, Dorazio R, Mulder GJ, Polack A, Bank A, Soulier J, Larghero J, Kabbara N, Dalle B, Gourmel B, Socie G, Chrétien S, Cartier N, Aubourg P, Fischer A, Cornetta K, Galacteros F, Beuzard Y, Gluckman E, Bushman F, Hacein-Bey-Abina S, Leboulch P. 2010. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature 467:318–322. 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McAuley AJ, Kuiper MJ, Durr PA, Bruce MP, Barr J, Todd S, Au GG, Blasdell K, Tachedjian M, Lowther S, Marsh GA, Edwards S, Poole T, Layton R, Riddell S-JJ, Drew TW, Druce JD, Smith TRFF, Broderick KE, Vasan SS. 2020. Experimental and in silico evidence suggests vaccines are unlikely to be affected by D614G mutation in SARS-CoV-2 spike protein. NPJ Vaccines 5:96. 10.1038/s41541-020-00246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, Sotomayor-González A, Glasner DR, Reyes KR, Gliwa AS, Reddy NP, Sanchez C, Martin S, Federman S, Cheng J, Balcerek J, Taylor J, Streithorst JA, Miller S, Kumar GR, Sreekumar B, Chen P-Y, Schulze-Gahmen U, Taha TY, Hayashi J, Simoneau CR, McMahon S, Lidsky PV, Xiao Y, Hemarajata P, Green NM, Espinosa A, Kath C, Haw M, Bell J, Hacker JK, Hanson C, Wadford DA, Anaya C, Ferguson D, Lareau LF, Frankino PA, Shivram H, Wyman SK, Ott M, Andino R, Chiu CY. 2021. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv 10.1101/2021.03.07.21252647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitt MA. 2010. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods 169:365–374. 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen YW, Huang SX, De Carvalho ALRT, Ho SH, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova JL, Bhattacharya J, Liang AF, Palermo LM, Porotto M, Moscona A, Snoeck HW. 2017. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat Cell Biol 19:542–549. 10.1038/ncb3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chandrasekaran B, Fernandes S. 2020. A cross-sectional study examining the seroprevalence of severe acute respiratory syndrome coronavirus 2 antibodies in a university student population. Diabetes Metab Syndr 67:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]