ABSTRACT
The fungus Nosema bombycis causes significant economic losses via parasitism of an economically important insect. MicroRNAs (miRNAs) play important roles in regulating host and parasite gene expression via mRNA degradation or by inhibiting protein translation. To investigate whether microRNA-like RNAs (milRNAs) regulate N. bombycis pathogenesis and to better understand the regulatory mechanisms underlying infection, we constructed small RNA libraries from N. bombycis hyphae during the schizont proliferation period. Eleven novel milRNAs were determined by RNA sequencing and stem-loop reverse transcriptase PCR (RT-PCR) assays. Moreover, a virulence-associated milRNA, Nb-milR8, was identified as critical for N. bombycis proliferation by binding and downregulating expression of its target gene, BmPEX16, in the host during infection. Silencing of Nb-milR8 or overexpression of the target BmPEX16 gene resulted in increased susceptibility of Bombyx mori to N. bombycis infection. Taken together, these results suggest that Nb-milR8 is an important virulence factor that acts as an effector to suppress host peroxidase metabolism, thereby facilitating N. bombycis proliferation. These results provide important novel insights into interactions between pathogenic fungi and their hosts.
IMPORTANCE A thorough understanding of fungal pathogen adaptations is essential for treating fungal infections. Recent studies have suggested that the role of small RNAs expressed in fungal microsporidia genomes are important for elucidating the mechanisms of fungal infections. Here, we report 11 novel microRNA-like RNAs (milRNAs) from the fungal microsporidium Nosema bombycis and identified NB-milRNAs that adaptively regulate N. bombycis proliferation. In addition, we demonstrate that N. bombycis modulates small RNA (sRNA)-mediated infection by encoding an Nb-miR8 that downregulates the expression of the host peroxidase metabolism protein BmPEX16, which is essential for peroxisome membrane biogenesis and peroxisome assembly. These results significantly contribute to our understanding of the pathogenic mechanisms of fungi, and especially microsporidia, while providing important targets for genetical engineering-based treatment of microsporidia.
KEYWORDS: BmPEX16, Bombyx mori, Nb-milR8, Nosema bombycis, milRNAs
INTRODUCTION
Microsporidia are obligate intracellular parasitic unicellular eukaryotes (1–3). Among 200 microsporidia genera, 93 have insect hosts and are important biological control agents of agricultural pests and important pathogens of economically relevant insects (2, 4). Most parasitic pathogens have sublethal effects on their insect hosts and contribute to reduced fertility, shortened life spans, and loss of vitality (4, 5). Therefore, understanding the physiological characteristics of microsporidia and the relationship between microsporidia and their hosts are important for understanding the roles of microsporidia in agricultural production. Sequencing of microsporidia genomes has led to several investigations that have primarily focused on evaluating genomic sequence, gene evolution, and molecular function, thereby providing useful baseline data and knowledge for preventing microsporidia infections and understanding the mechanistic basis of their investigations (6, 7). Nevertheless, manipulation of microsporidia genomes by genetic modification has been minimally studied, and comprehensive treatment of microsporidia has remained elusive.
Increasing attention has been paid in recent years to understanding microRNAs (miRNAs) that are transcribed, but do not encode proteins, and can perform biological functions at the RNA level within pathogen-host interactions (8–10). miRNAs are short RNA sequences with lengths typically of 18 to 25 nucleotides (nt) and are endogenous noncoding RNAs with regulatory functions that primarily exist in eukaryotes (11). Lee et al. identified an miRNA-like RNA (milRNA) in Neurospora crassa in 2010 with similar characteristics to animal and plant milRNAs and observed that milRNA production was dependent on Dicer enzyme cleavage in addition to the activities of Dicer-like 1 (DCL-1) and other key enzymes (12). milRNAs are primarily involved in fungal stress responses, morphological regulation, and a variety of physiological processes (13–15). Most milRNAs can induce gene silencing but also inhibit host immune responses (16). For example, Fusarium oxysporum f. sp. lycopersici (Fol) Fol-milR1 is a fungal effector that suppresses host immunity by silencing a disease resistance gene in tomato plants (17). Further, some miRNAs act as virulence factors to regulate host or pathogen factors, including the fungus Valsa mali Vm-milRNA-16, which contributes to V. mali infection of apple trees by adaptively regulating virulence genes, and VA-milRNAs are virulence factors associated with infection of Rhizoctonia solani direct interference with host target genes (18, 19).
Nosema bombycis is the first described microsporidium of insect pathogens (20). The pathogen has caused huge economic losses in the Chinese and European sericulture industry, leading to the near-disappearance of European sericulture (20). Indeed, it is the primary pathogen within the sericulture industry of China, India, and Southeast Asia. N. bombycis exhibits unique biological characteristics for a eukaryote, including ribosomal similarities to those of prokaryotes (21). Given that miRNA widely exists in eukaryotic cells, it is important to explore whether N. bombycis milRNA is important for modulating its biological characteristics. Importantly, the functional and mechanistic characteristics of N. bombycis pathogenic action have not been elucidated, and it is thus a considerable challenge to identify the molecular mechanisms of interactions between these fungi and their hosts.
In this study, we identified milRNA in N. bombycis to help identify the pathogenic mechanism of N. bombycis, while further analyzing the mechanism of milRNAs and their target genes that regulate the proliferation of N. bombycis. We first analyzed the infective characteristics of Bombyx mori embryonic cell line BmE-SWU1 infected with N. bombycis by transmission electron microscopy (TEM), immunofluorescence, and reverse transcriptase PCR (RT-PCR). High-throughput whole-transcriptome sequencing was then used to identify RNA profiles of infection during schizont proliferation and abundant spore replication phases at 48 h postinfection (h p.i.). A total of 11 novel milRNAs in N. bombycis were identified, including Nb-milR1 to Nb-milR12 (excluding Nb-milR7) using high-throughput sequencing and stem-loop RT-PCR analysis. Moreover, functional characterization of Nb-milR8 and its corresponding target gene, Bombyx mori peroxisomal membrane protein (BmPEX16), indicates that milRNAs can adaptively regulate host genes to promote N. bombycis proliferation. These results provide evidence for the existence of milRNAs in N. bombycis that may provide novel insights into milRNAs, in addition to the molecular mechanisms of fungal pathogen-host interactions.
RESULTS
Proliferation of N. bombycis.
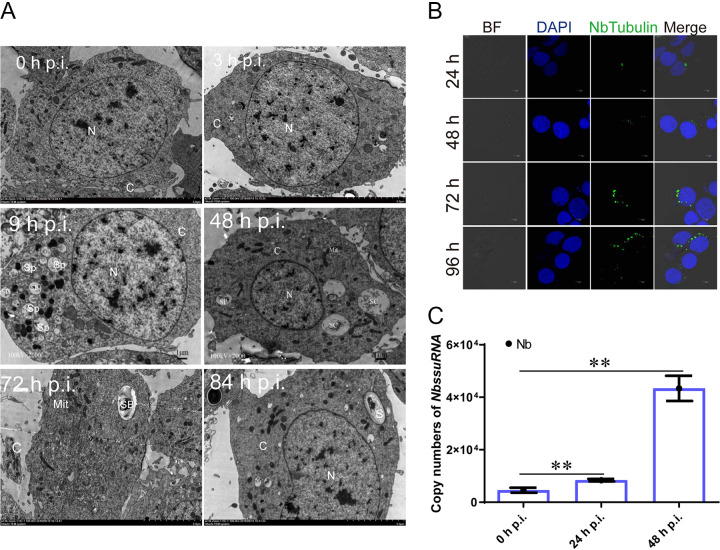
To better predict and analyze milRNA characteristics of N. bombycis, the life cycle of N. bombycis was first explored with immunofluorescence, gene expressional analysis, and TEM analysis. TEM visualization indicated that spore plasms could be observed at 3 h p.i., while spore plasm volumes increased at 9 h p.i., and bi-nucleated schizonts appeared at 48 h p.i. (Fig. 1A). The changes were followed by sporocyst appearances, cell membrane thickening, and chitinous wall formation at 72 h p.i. (Fig. 1A). Finally, mature microspores with complete walls and polar tube structures appeared at 84 h p.i. (Fig. 1A). Immunofluorescence was performed by labeling Nb-tubulin proteins, revealing that a small number of spores were injected into the cytoplasm at 24 h p.i. by ejecting polar filaments and a small amount of spore protoplasm (Fig. 1B). Subsequently, the spore plasms became larger and developed into schizonts through two divisions at 48 h p.i. RT-PCR was used to analyze genome copy numbers after N. bombycis infection. Genomic DNA copies significantly increased in number at 48 h p.i. (Fig. 1C). Thus, N. bombycis was in the schizont stage at 48 h p.i., based on the presence of a larger number of N. bombycis genome copies due to replication. This period was thus identified as a key time for screening miRNA.
FIG 1.
Infection proliferation characteristics of N. bombycis. (A) TEM micrographs illustrating the N. bombycis life cycle in BmE-SWU1 cells. N, nucleus; C, cytoplasm; Sp, spore plasm; Sc, schizont; Sb, sporoblast; S, spore. (B) Immunofluorescence assays showing N. bombycis proliferation characteristics. BF, bright fluorescence; DAPI shows B. mori and N. bombycis nuclei; green fluorescence shows the NbTubulin proteins. (C) RT-PCR analysis of N. bombycis genomic DNA copies in BmE-SWU1 cells, based on Nb SSURNA copy numbers. All data represent means of three replicates ± SD. **, P < 0.01.
Identification of milRNAs in N. bombycis.
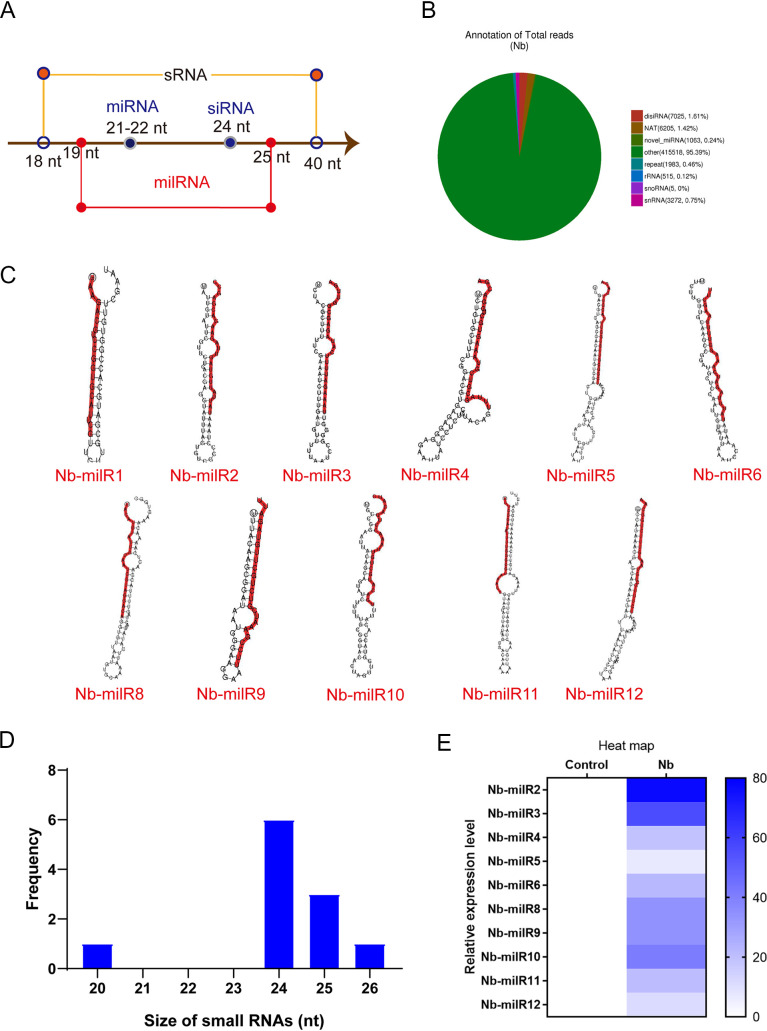
To predict N. bombycis milRNAs in BmN-SWU1 cells, RNA from the N. bombycis infected at 48 h p.i. (Nb) and uninfected (control) groups were subjected to Illumina Solexa high-throughput sequencing. A total of 12,657,370 and 19,108,751 reads were generated in the Nb and control groups, respectively, with more than 95% of reads exhibiting greater than Q20 or Q30 qualities. Thus, the sequencing data were of high quality and could be used for further analyses. Low-quality sequences, 5′ terminal junction contaminated sequences, and sequences without 3′ terminal junctions and insertion fragments were all quality filtered and removed to generate the final sequence data sets. After quality filtering, a total of 12,081,520 and 18,766,136 sRNA clean reads were obtained for the Nb and control groups, respectively (see Table S2 in the supplemental material). Clean reads for each sample were used to identify sRNAs within certain length ranges for subsequent analyses. sRNAs primarily exhibit 18- to 40-nucleotide (nt) lengths, while miRNA is 21 to 22 nt long, and milRNAs are longer than miRNAs (Fig. 2A and Fig. S1). Using this framework, milRNAs were identified based on lengths of clean reads that were obtained from the Nb group. milRNAs accounted for 0.24% of the samples when considering all sRNAs and all RNA data sets (Fig. 2B).
FIG 2.
Identification of MicroRNA-like RNAs (milRNA) in N. bombycis. (A) The length distributions of N. bombycis sRNAs. (B) The relative abundances of different sRNA classes in N. bombycis. (C) Hairpin structure characteristics for the Nb-miRNA precursor. Red lines indicate the sequences of mature Nb-milRNAs. (D) The length distribution for Nb-milRNAs in N. bombycis. (E) Expression levels of different Nb-milRNAs in N. Bombycis. All data represent the means of three replicates ± SD. **, P < 0.01.
The signature hairpin structures of miRNA precursors can be used to predict novel miRNAs. Thus, the miREvo and mirdeep2 miRNA prediction software programs were used to identify novel miRNAs (22, 23). The basic principle of miRNA identification is to probe certain lengths of sRNA alignment reference sequences, followed by analysis of secondary structures, dicer site information, free energy, and other characteristics. Subsequently, the sequence, compositions, lengths, and frequencies of sRNAs in each sample were analyzed. Among the potential Nb-milRNA genes of N. bombycis that were predicted, 11 novel miRNAs were identified, and their secondary structures were analyzed (Fig. 2C). The size distributions of the different Nb-milRNAs were also analyzed. Nb-milRNAs primarily exhibited lengths of 20 to 26 nt in N. bombycis. Over 80% of the milRNAs exhibited lengths of 24 to 25 nt (Fig. 2D). The 10 novel Nb-milRNAs were only mapped to N. bombycis (Fig. 2E and Fig. S1B), while Nb-milR1 was mapped to both groups.
Transcriptional expression of N. bombycis milRNAs.
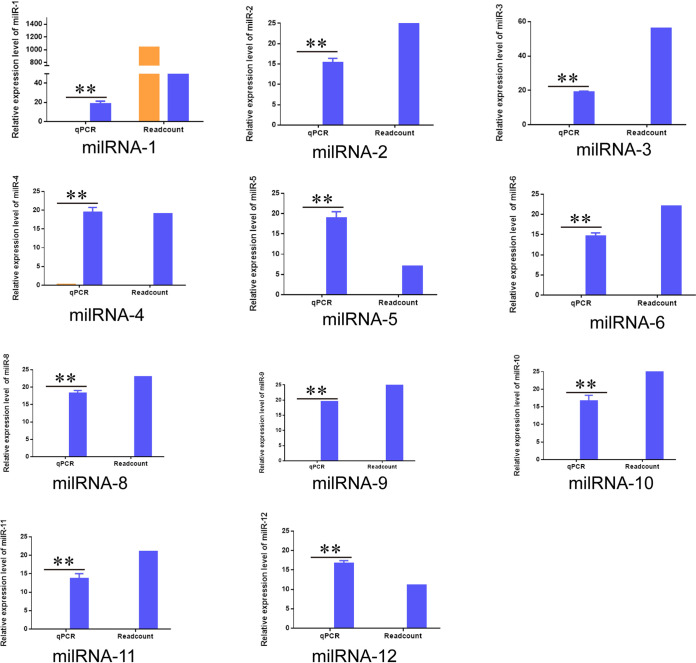
To identify milRNAs via high-throughput sequencing, total miRNAs at 48 h p.i. were extracted, and the expressions of predicted miRNAs were evaluated with stem-loop RT-PCR. The expression levels of the 11 milRNAs verified by stem-loop RT-PCR were consistent with their expression trends based on high-throughput sequencing and were only highly expressed in host cells infected with N. bombycis. Although Nb-mil1 was predicted to be expressed in infected and uninfected samples, it was also only expressed in host cells after infection with N. bombycis (Fig. 3).
FIG 3.
Transcriptional expression analysis of Nb-milRNAs. Normalized read numbers of Nb-milRNAs as transcripts per million reads (TPM) of ≥10 are shown for Nb (blue) or the control (orange) groups (i.e., “readcount” on right) in addition to expression of milRNAs that was evaluated with stem-loop RT-PCR (i.e., “qPCR” on right). All data represent the means of three replicates ± SD. **, P < 0.01.
Functional assessment of N. bombycis Nb-milR8.
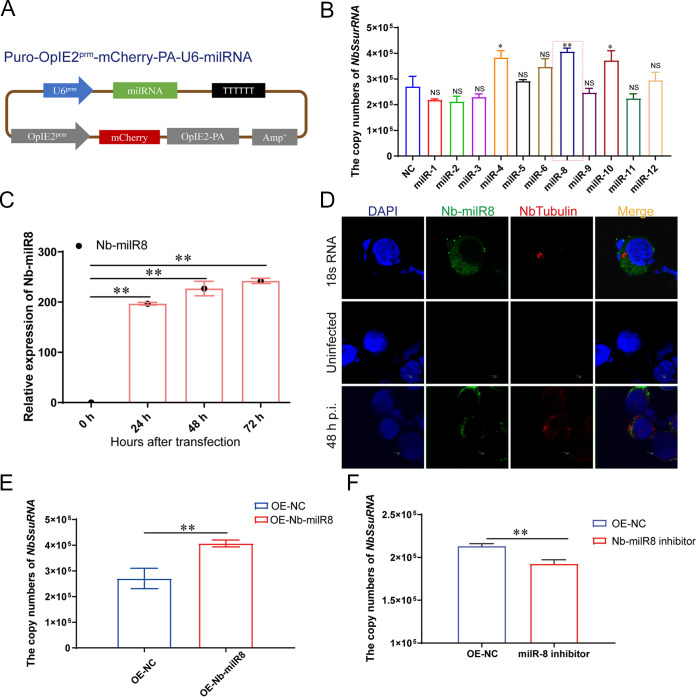
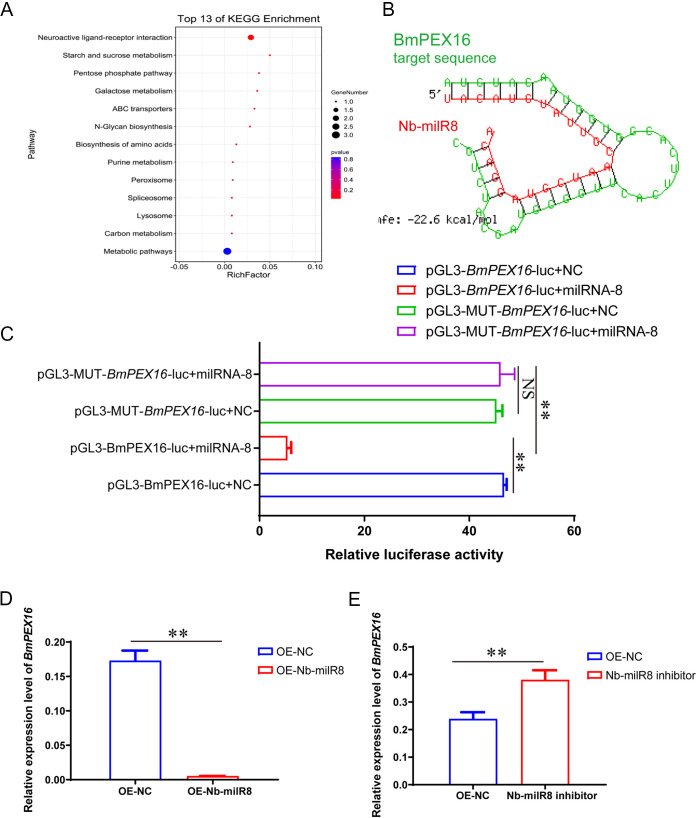
To explore the function of milRNAs on N. bombycis proliferation, target bands containing 11 Nb-milRNAs were inserted into Puro-OpIE2prm-mCherry-PA vectors to obtain recombinant Puro-OpIE2prm-mCherry-PA-U6-milRNA plasmids that would enable Nb-milRNA expression detection with fluorescence microscopy (Fig. 4A). After transfection with miRNAs into the BmE-SWU1 cells, Nb-milR4, Nb-milR8, and Nb-milR10 promoted the proliferation of N. bombycis, while Nb-milR1, Nb-milR2, Nb-milR3, Nb-milR5, Nb-milR6, Nb-milR9, Nb-milR11, and Nb-milR12 did not exhibit significant differences upon N. bombycis proliferation. Among these, Nb-milR8 was most effective toward changes in pathogen infection (Fig. 4B).
FIG 4.
Functional assessment of Nb-milR8. (A) Schematic showing the construction of the Nb-milRNA overexpression vector. (B) Effects of milRNAs on N. bombycis proliferation, as determined by genome copy numbers. Statistically significant differences were determined using Student’s t test; **, P < 0.01; *, P < 0.05; NS, ≥0.05. (C) Transcriptional levels of Nb-milR8 in BmN-SWU1 cells that were challenged with N. bombycis. (D) Localization of Nb-milR8 based on fluorescence in situ hybridization (FISH). DAPI shows the B. mori and N. bombycis nuclei; red fluorescence represents the Nb-tubulin protein; green represents the fluorescence probe for Nb-milR8. (E) Genomic DNA copy enumeration during Nb-milR8 overexpression compared to the control transformant. OE-NC indicates overexpression of the native control plasmid Puro-OpIE2prm-mCherry-PA. (F) Genomic DNA copy enumeration with the Nb-milR8 inhibitor compared to that with the control transformant. OE-NC indicates overexpression of the native control plasmid Puro-OpIE2prm-mCherry-PA. Statistically significant differences were determined using Student’s t test; **, P < 0.01. All data represent the means of three replicates.
To analyze Nb-milR8 expressional characteristics, stem-loop RT-PCR was used to analyze its expression profiles. Nb-milR8 expression levels increased after N. bombycis infection (Fig. 4C). A specific fluorescent in situ hybridization (FISH) probe was designed according to the nucleic acid sequence of Nb-milR8, allowing us to detect the Nb-milR8 expression characteristics after infection with N. bombycis within BmE-SWU1 cells. FISH analysis indicated that Nb-milR8 was expressed in the cytoplasm of N. bombycis and in host cells (Fig. 4D).
To further explore the effects of Nb-milR8 on N. bombycis proliferation, its effects on N. bombycis were evaluated after overexpression. In addition, we artificially designed an Nb-milR8 inhibitor to inhibit Nb-milR8 expression. After infection with N. bombycis, RT-PCR was used to analyze the genomic DNA copy numbers of N. bombycis. Overexpression of Nb-milR8 promoted the proliferation of N. bombycis, and Nb-milR8 inhibitor was effective at inhibiting N. bombycis proliferation (Fig. 4E and F). These results thus indicate that Nb-milR8 plays an important role in N. bombycis proliferation.
Regulation of target gene expression by Nb-milR8.
To identify the target gene of Nb-milR8 in Bombyx mori, the target gene prediction software program Miranda was used for preliminary predictions, with a threshold score set at ≥140. A total of 569 target genes were predicted for Nb-milR8 in Bombyx mori. Among these genes, 64 exhibited expression differences of over 2-fold when comparing the control and Nb groups (Fig. 2S). KEGG functional enrichment analysis indicated that eight target genes were enriched, and these were mainly associated with metabolism, neuroactive receptor ligand pathways, and peroxisome pathways (Fig. 5A). The expression levels of five of these eight genes were significantly upregulated, one gene did not exhibit significant expression differences, and two exhibited significantly downregulated expression after overexpression of Nb-milR8 (Fig. 3S). The most significant downregulated gene, BmPEX16 (NCBI reference sequence: XM_012696074.3), was chosen as a primary candidate target gene.
FIG 5.
Regulation of Nb-milR8 target gene expression. (A) KEGG enrichment analysis for the Nb-milR8 target gene. (B) Target gene prediction for Nb-milR8 was performed using the RNA hybrid program and is shown in the schematic. (C) Dual-luciferase reporter assay for the Nb-milR8 binding site. OE-NC indicates overexpression of the native control plasmid Puro-OpIE2prm-mCherry-PA. pGL3-MUT-BmPEX16-luc indicates mutation of the BmPEX16 gene bind site. (D) Relative expression of BmPEX16 after overexpression with Nb-milR8. (E) Relative expression of BmPEX16 after transfection with the Nb-milR8 inhibitor. OE-NC indicates overexpression of the native control plasmid Puro-OpIE2prm-mCherry-PA. All data represent the means of three replicates ± SD. **, P < 0.01.
The binding of Nb-milR8 and BmPEX16 was explored using the Miranda program. The seed sequence of Nb-milR8 complemented and paired with the 3′ untranscribed region (UTR) region of BmPEX16 (Fig. 5B). To further verify the binding effects of Nb-milR8 and BmPEX16, Nb-milR8 and pGL3.0-IE1-Luc-PEX16 (or a native control and pGL3.0-IE1-Luc-PEX16) were transfected into BmE-SWU1 cells, respectively. Then, 48 h posttransfection (p.t.), BmPEX16 expression was clearly regulated by Nb-milR8, and firefly luciferase activity decreased. In addition, the activity of dual luciferase decreased, indicating that BmPEX16 is indeed the target gene of Nb-milR8 and is regulated by Nb-milR8 (Fig. 5C). In contrast, no significant changes in luciferase activity were observed compared to the control after mutation of the BmPEX16 binding site. To further explore the regulatory effects of Nb-milR8 on BmPEX16, the synthetic Nb-milR8 inhibitor and the Nb-milR8 vectors were overexpressed in BmN-SWU1 cells, respectively. In addition, the expression of BmPEX16 was measured with RT-PCR. The Nb-milR8 inhibitor upregulated the expression level of the target gene after infection with N. bombycis, and overexpression of Nb-milR8 inhibited the expression level of the target gene (Fig. 5D and E). Thus, these results demonstrate that BmPEX16 is the target gene of Nb-milR8 and is regulated by it.
The target gene BmPEX16 of Nb-milR8 is required for N. bombycis proliferation.
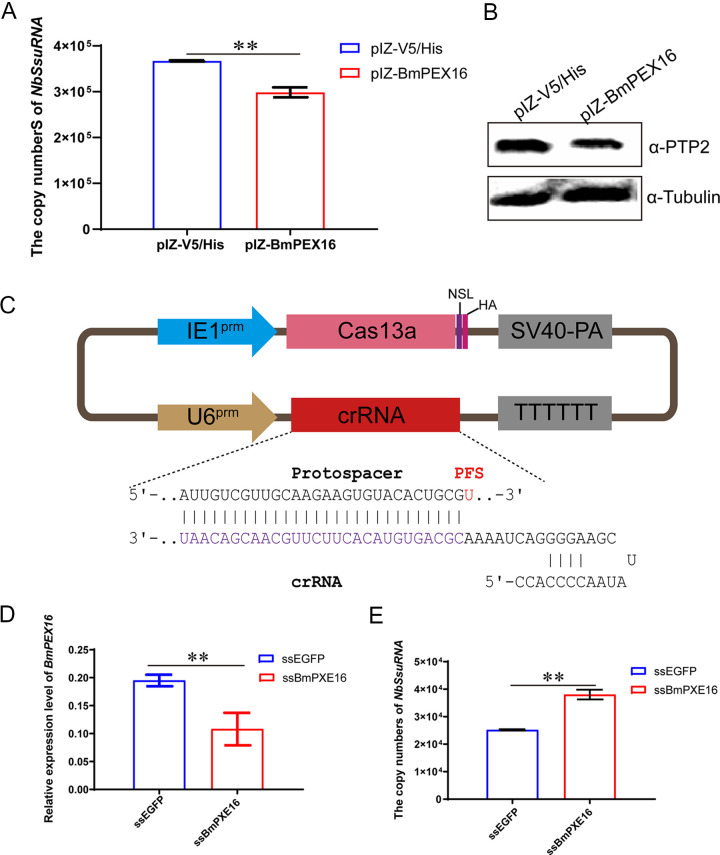
PEX16 is essential for peroxisome membrane biogenesis and may play a role in the early stages of peroxisome assembly (24, 25). To analyze peroxisome function during N. bombycis proliferation, a BmPEX16 overexpression vector was constructed and transfected into BmE-SWU1 cells. DNA and proteins were extracted from the samples after infection of N. bombycis at 48 h p.i. N. bombycis DNA copy numbers and NbPTP2 protein levels of N. bombycis were detected by quantitative PCR (qPCR) and Western blotting, respectively. Genome copy numbers for N. bombycis were downregulated, while protein expression of NbPTP2 was also downregulated after overexpressing the target gene BmPEX16. These results indicated that the proliferation of N. bombycis was inhibited by BmPEX16 expression (Fig. 6A and B).
FIG 6.
The target gene of Nb-milR8, BmPEX16, is required for N. Bombycis proliferation. (A) Analysis of N. bombycis genome copies after overexpression with BmPEX16. (B) NbPTP2 protein expression analysis after overexpression with BmPEX16 based on Western blot analysis. (C) Schematic showing the construction of a Cas13a gene editing system in B. mori. (D) Knockout efficiency of BmPEX16. (E) Genomic DNA copies of N. bombycis after knockout of BmPEX16. All data represent the means of three replicates ± SD. **, P < 0.01.
To analyze the inhibitory effect of Nb-milR8 on BmPEX16 expression, a BmPEX16 knockout vector was constructed using CRISPR/Cas13 technology (Fig. 6C). After transfecting with PSL1180-IE1-Cas13a-SV40-U6-ssPEX16, the expression of BmPEX16 was significantly reduced, as indicated by RT-PCR (Fig. 6D). To confirm the effects of the BmPEX16 knockout on N. bombycis proliferation, PSL1180-IE1-Cas13a-SV40-U6-ssPEX16 was transfected into cells for 48 h. Then, 48 h p.i., copy numbers were detected by RT-PCR, revealing that N. bombycis genome copy numbers were upregulated after knockout of the target gene BmPEX16 using the CRISPR/Cas13a system. Together, these results indicate that Nb-milR8 inhibited BmPEX16 expression by targeting its binding site and promoting the proliferation of N. bombycis.
DISCUSSION
Following the discovery of the first miRNA, lin4, in Caenorhabditis elegans in 1993, the development of biomolecular methods and bioinformatics has led to increasing numbers of miRNAs being identified in animals, plants, viruses, and even some single-celled organisms (12, 19, 26, 27). However, few milRNAs (microRNA-like RNAs) have been reported in fungi, and the mechanisms of their actions within cells are largely unknown. Microsporidia are evolutionarily unique pathogenic fungi (20). A functional RNA interference (RNAi) pathway was recently identified in the microsporidial fungus Nosema ceranae, indicating that miRNAs also exist in microsporidia (2, 28). An understanding of milRNA functions in the context of these organisms’ position within eukaryotes, in addition to the treatment of microsporidia infections, are areas of considerable interest. In this study, numerous sRNAs were identified in the schizonts of N. bombycis using high-throughput whole-genome sequencing, with additional characterization of 11 Nb-milRNA’s expression profiles (Fig. 3). Moreover, the mechanism underlying the promotion of N. bombycis proliferation by Nb-milR8 was identified as involving the regulation of the target host peroxisomal membrane protein BmPXE16 gene.
The development of high-throughput sequencing technology has led to the identification of milRNAs in Fusarium oxysporum, Rhizoctonia solani, Cordyceps guangdongensis, and Volvariella volvacea, while also considerably promoting the discovery of milRNAs with low expression and those that exhibit tissue-specific expression (29–31). In this study, the schizont phase of microsporidian infection was chosen for characterization, and a total of 11 mature milRNAs were identified and mapped, in addition to 1,063 sRNA types. Thus, milRNAs only composed 0.24% of the total sRNA pool (Fig. 2B). Likewise, only six milRNAs were identified in the microsporidian parasite Nosema ceranae, with only three being further validated, which is relatively minimal compared to other fungi (27, 32). BLAST searches of the milRNAs identified here also did not yield highly homologous milRNAs outside N. bombycis, indicating that the milRNAs arose independently from the evolution of plant, animal, and fungal miRNAs. Taken together, 11 milRNAs were validated in this study, thereby increasing the number of Nb-milRNAs known in microsporidian parasites and providing baseline data to support future research into microsporidian Nb-milRNA.
Functional RNAi pathways, Dicer, and Argonaute-gene based pathways have been documented as infection strategies promoting the selectivity of microsporidian parasitism (2). One of the most important discoveries of this study is that of diverse pathways for small RNA biogenesis within N. bombycis, including those for rRNA, tRNA, snRNA, snoRNA, milRNA, Dicer-independent siRNA (disiRNA), and small interfering RNA (siRNA) pathways (Fig. 2B). Our investigation of 11 Nb-milRNAs indicates that they are involved in regulating fungal virulence, suggesting that the biosynthesis pathway of milRNAs in N. bombycis may be similar to that of other fungi (Fig. 4). Pathogenic milRNAs are generally considered necessary components for host-pathogen interactions, with milRNAs generally thought to be helpful for promoting pathogen proliferation and replication (8, 17). For example, Vm-milRNAs adaptively regulate the virulence genes encoding sucrose nonfermenting 1 (VmSNF1), 4,5-DOPA dioxygenase extradiol (VmDODA), and a hypothetical protein (VmHy1) that enhances the virulence of V. mali in apple trees. Further, Pst-milRNA1 plays a role in virulence by inhibiting wheat pathogenicity-related genes that impair wheat resistance to Puccinia striiformis f. sp. tritici. Likewise, Vd-milR1 regulates the virulence of Verticillium dahlia through epigenetic inhibition of target gene expression (16, 19, 33). Here, we identified 11 Nb-milRNAs associated with pathogenicity of N. bombycis and confirmed the mechanism of Nb-milR8 promotion of N. bombycis proliferation via negative regulation of the peroxisome metabolic pathway through BmPXE16 gene expression (Fig. 4 and 5). The results provide insight into a unique strategy to successfully develop microsporidian parasitism of hosts wherein pathogenic fungi must disable host defenses and hijack host metabolisms.
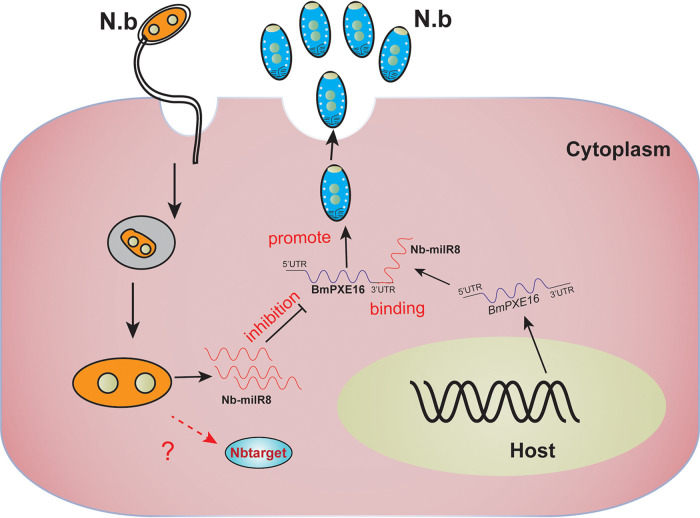
milRNAs play important roles during fungal infection by regulating the translation or mRNA stability of target genes (14, 30). Nevertheless, the lack of effective methods to identify the target genes of milRNA has led to limited functional exploration of milRNAs in fungi. In this study, a total of 569 target genes of B. mori and 110 target genes of N. bombycis were predicted for Nb-milR8, and their functional annotation revealed diverse activities in energy metabolism and peroxisome pathways (Fig. 5). To identify the interaction mechanism between the microsporidian parasite and its host, eight target genes of B. mori were identified by differential expression characteristics and functional enrichment (Fig. S2). PEX16 genes are important regulators of peroxisome biosynthesis and are involved in peroxisome formation and the energy metabolism of the yeasts Yarrowia and Saccharomyces cerevisiae (24, 34–36). Here, we also demonstrated that Nb-milR8 negatively regulated the BmPEX16 gene promoter to encourage N. bombycis proliferation during infection. These results led to the hypothesis that upregulated expression of Nb-milR8 after infection with N. bombycis leads to downregulated expression of BmPEX16 by targeting the BmPEX16 3′ UTR, thereby ultimately inhibiting host BmPEX16 expression and promoting the proliferation of N. bombycis (Fig. 7). These studies provide important target milRNAs for genetically engineering microsporidia. Infection-resistance materials could be further developed for transgenetic overexpression of milRNAs to inhibit N. bombycis proliferation or via the interference of milRNA that could then promote N. bombycis proliferation. In addition, resistance mechanisms could be targeted by focusing on the Nb-milRNA encoding gene to improve silkworm resistance to N. bombycis infection.
FIG 7.
Model for N. bombycis deployment of Nb-milR8 to promote infection by modulating BmPEX16 gene expression.
Conclusions.
This study demonstrated that N. bombycis regulated its host peroxisome pathway BmPEX16 gene via the Nb-milR8 RNA interacting with its promoter, thus enabling N. bombycis proliferation during infection. These results illustrate a regulatory mechanism of Nb-milR8 toward host metabolism that occurs during host-pathogen interactions. Consequently, these results deepen our understanding of mechanistic interactions between hosts and pathogens while providing new insights into the possible prevention of N. bombycis fungal infections.
MATERIALS AND METHODS
B. mori cell and N. bombycis.
BmE-SWU1 cell lines were cultured at 27°C in Grace medium (United States Biological, USA) (37). Cell culture medium comprised 90% Grace free medium and 10% fetal bovine serum (FBS) (Gibco, USA). The N. bombycis strain CQ1 (CVCC 102059) was obtained from the China Veterinary Culture Collection Center.
RNA extraction and sequence data analysis.
Total RNAs were extracted from the control (0 h p.i.) and the N. bombycis infection group (48 h p.i.) using an RNA isolation kit according to the manufacturer’s instructions (Omega, USA). After sample quality verification, a small RNA sample kit was used to construct a sequencing library (Yeasen, China). The unique structures of the 3′ and 5′ ends of small RNAs (i.e., 5′ ends with complete phosphate groups and 3′ ends with hydroxyl groups) allowed the use of total RNA as the starting samples, followed by direct splicing of small RNA ends and production of cDNA via reverse transcription. After PCR amplification and PAGE gel electrophoresis, a cDNA library was obtained. The different libraries were pooled and sequenced on the Illumina HiSeq TM2000/MiSeq sequencing platforms based on the effective concentrations and the requirements for target data quantities. Raw reads were first processed to obtain high-quality clean reads. The raw sequence reads will be available through the NCBI database under the accession number PRJNA760284. sRNAs of a certain length range were screened for subsequent analyses from the clean read data sets for each sample. The small RNA tags were then mapped to reference sequences using Bowtie after specifying a threshold for sequence mismatches in order to analyze their expression profiles against the reference (38).
Identification of milRNAs.
Few fungal miRNA gene data sets are present in the miRbase database, and thus characteristics of miRNA precursor hairpin structures were used to predict novel miRNAs. Specifically, the miREvo and mirdeep2 software programs were both used to predict novel miRNAs by exploring secondary structures, Dicer cleavage sites, and minimum free energy values of small RNA tags that were not annotated as described previously (22, 23). Further, custom scripts were used to obtain miRNA counts for different sequence length RNAs, in addition to base biases at each position for all identified miRNAs. The target gene prediction software program Miranda was used for preliminary predictions of Nb-milR8 target genes in N. bombycis, specifying a Miranda threshold score of ≥140.
miRNA extraction and expression analysis.
Total miRNAs at 48 h p.i. were extracted using a Molpure cell/tissue miRNA kit (Yeasen, China) according to the manufacturer’s protocols. The RNA was reverse transcribed using a specific stem-loop primer, and miRNA cDNA was used for hydrolysis probe-based RT-PCR. Primer sequences are provided in Table S1. The relative changes in expression values among groups were calculated using the 2–△CT method.
Scanning electronic microscopy (SEM).
BmE-SWU1 cells were infected with N. bombycis for 0, 3, 9, 48, 72, and 84 h, and samples were fixed for SEM analysis. Specifically, the samples were sectioned to about 10 nm with a slicer microtome instrument and then stained with acetic and citric acid before imaging with a Hitachi-SU3500 SEM instrument (SU3500, Hitachi, Japan). Images were viewed at an accelerating voltage of 100 kV using a low beam current.
Construction of milRNA expression plasmids.
To construct milRNAs, specific primers for downstream regions of milRNA were designed according to milRNA sequences. milRNAs were PCR amplified using U6 as the template. The amplified target bands of U6-milRNA and Puro-OpIE2prm-mCherry-PA were then digested using the Asc I restriction endonuclease and then ligated with linkers for 24 h. The Nb-milR8 inhibitor exhibits a complementary pairing sequence for Nb-milR8. After synthesis, the construct was similarly ligated to the Puro-OpIE2prm-mCherry-PA vector to enable competitive binding of Nb-milR8 with the target gene. The Puro-OpIE2prm-mCherry-PA-U6-milRNA plasmids were then subjected to sequencing.
Construction of Cas13a gene editing vectors.
To knockout the BmPEX16 gene of B. mori, wild-type LwCas13a and pC014 (pC014-LwCas13a-msfGFP, Addgene plasmid no. 91902) plasmids were obtained from Addgene (39). The LwCas13a fragments were cloned into pSL1180-IE1prm-SV40 vectors by digesting at the Xho I and Not I restriction sites, ultimately yielding the pSL1180-IE1prm-LwCas13a-SV40 product. Candidate ssRNA target sequences were designed using the CRISPR design software program (http://bioinfolab.miamioh.edu/CRISPR-RT/interface/C2c2.php), and U6-ssRNA expression cassettes were linked into the pSL1180-IE1prm-LwCas13a-SV40.
Dual luciferase reporter assays.
The dual luciferase expression plasmids pGL3-IE1-Fluc-PEX16 (i.e., the 3′ UTR of the target gene fused at the 3′ end of the firefly luciferase gene), pGL3-IE1-Rluc (with Rluc as an internal reference gene), and milRNA relative vectors were cotransfected into BmN-SWU1 cells. Cells were then collected at 48 h posttransfection (h p.t.), and luciferase activities were measured using a Dual-Glo luciferase assay kit (Promega, USA). Relative luciferase activity (Fluc/Rluc) was normalized to values obtained with pGL3-Basic as the control plasmid.
Immunofluorescence.
After N. bombycis infection of BmN-SWU1 cells at 24, 48, 72, and 96 h, samples were seeded on cover glasses for immunofluorescence assays (Thermo Scientific, USA) in 24-well plates (Corning, NY, USA) (40). In addition, N. bombycis at 24, 48, 72, and 96 h p.i., was stained with anti-Nb tubulin antibody and an Alexa 488 conjugated goat anti-rabbit secondary antibody. Similarly, localization of Nb-milR8 was evaluated after incubation with Nb-milR8 probes followed by fluorescence in situ hybridization (FISH) analysis. N. bombycis nuclei were then stained with DAPI for 1 h. All images were visualized with an Olympus inverted fluorescence microscope at the corresponding excitation wavelengths.
Western blotting.
Overexpression of pIZ-BmPEX16 and pIZ-V5/His vectors was induced in BmN-SWU1 cells. After 48 h p.t., samples were lysed using immunoprecipitation (IP) buffer to enable Western blot analysis (Beyotime, China). Total sample protein extracts were separated by SDS-PAGE and then blot-transferred to polyvinylidene fluoride membranes. The membranes were then incubated with rabbit α-tubulin (1:2,000) and rabbit α-PTP2 (1:2,000), followed by further incubation with horseradish peroxidase (HRP)-labeled goat anti-mouse or anti-rabbit IgG (1:5,000; Beyotime, China). Bands were visualized using the Clarity Western ECL substrate (Bio-Rad, USA).
Quantitative PCR (qPCR).
The qPCRs were conducted in a CFX96 real-time system using the SYBR select master mix reagent (Bio-Rad, Hercules, CA, USA) to determine N. bombycis genome copy numbers. Specifically, N. bombycis small subunit rRNA (SSUrRNA) gene-specific primers were used to amplify rRNA genes and estimate genome replication to ultimately evaluate genome copy numbers after 48 h of infection (41). A standard curve described by y = −3.4305× + 36.882 (R2 = 0.9989) was used to evaluate the expression levels. The sw25113 gene was used as an internal reference to determine the relative gene expression from qPCRs data. qPCRs were conducted at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s.
Statistical analyses.
Results are expressed as the means ± standard deviations (SD). A value of P < 0.01 (**) based on two-tailed tests was considered statistically significant for all analyses. All statistical analyses were conducted using the GraphPad Prism 8 software program.
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation of China (grant no. 31902214, 31872427, and 32060777), the Fundamental Research Funds for the Central Universities (SWU120008 and XDJK2020C010), the Natural Science Foundation of Chongqing (cstc2019jcyj-msxmX0096 and cstc2020jscx-msxmX0045), and the China Agriculture Research System (CARS-18). We thank Accdon for its linguistic assistance and presubmission expert review.
Z.D., N.Z., and Q.W. constructed the expression plasmid. Z.D., C.H., and N.Z. performed SEM analysis and RT-PCR. N.Z., C.H., and B.D. participated in the dual luciferase reporter, immunofluorescence, and Western blotting assays. Z.D., M.P., and C.L. conceived the experimental design and participated in data analysis. Z.D., M.P., P.C., and C.L. were involved in the preparation of the manuscript. The final manuscript was reviewed and approved by all authors.
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Cheng Lu, Email: lucheng@swu.edu.cn.
Minhui Pan, Email: pmh047@126.com.
Christina A. Cuomo, Broad Institute
REFERENCES
- 1.Weber R, Bryan RT, Schwartz DA, Owen RL. 1994. Human microsporidial infections. Clin Microbiol Rev 7:426–461. doi: 10.1128/CMR.7.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Q. 2018. Evolution of Dicer and Argonaute orthologs in microsporidian parasites. Infect Genet Evol 65:329–332. doi: 10.1016/j.meegid.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Wang M, Xu JS, Wang LL, Zhang XY, Zhou ZY. 2009. Pathogenicity and genetic divergence of two isolates of microsporidia Nosema bombycis. Yi Chuan 31:1121–1126. (In Chinese.) doi: 10.3724/sp.j.1005.2009.01121. [DOI] [PubMed] [Google Scholar]
- 4.Ptaszyńska AA, Trytek M, Borsuk G, Buczek K, Rybicka-Jasińska K, Gryko D. 2018. Porphyrins inactivate Nosema spp. microsporidia. Sci Rep 8:5523. doi: 10.1038/s41598-018-23678-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Ding S, Qin Q, Tang J, Liu L, Peng H. 2015. Morphological and phylogenetic analysis of Nosema sp. HR (Microsporidia, Nosematidae): a new microsporidian pathogen of Histia rhodope Cramer (Lepidoptera, Zygaenidae). Parasitol Res 114:983–988. doi: 10.1007/s00436-014-4264-3. [DOI] [PubMed] [Google Scholar]
- 6.Timofeev S, Tokarev Y, Dolgikh V. 2020. Energy metabolism and its evolution in Microsporidia and allied taxa. Parasitol Res 119:1433–1441. doi: 10.1007/s00436-020-06657-9. [DOI] [PubMed] [Google Scholar]
- 7.Kononchuk AG, Martemyanov VV, Ignatieva AN, Belousova IA, Inoue MN, Tokarev YS. 2021. Susceptibility of the gypsy moth Lymantria dispar (Lepidoptera: Erebidae) to Nosema pyrausta (Microsporidia: Nosematidae). Insects 12:447. doi: 10.3390/insects12050447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Awais MM, Shakeel M, Sun JC. 2021. MicroRNA-mediated host-pathogen interactions between Bombyx mori and viruses. Front Physiol 12:672205. doi: 10.3389/fphys.2021.672205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang XL, Zhang L, Li JP, Wang YJ, Duan Y, Wang J. 2015. MicroRNA-150: a potential regulator in pathogens infection and autoimmune diseases. Autoimmunity 48:503–510. doi: 10.3109/08916934.2015.1072518. [DOI] [PubMed] [Google Scholar]
- 10.Franceschini A, Meier R, Casanova A, Kreibich S, Daga N, Andritschke D, Dilling S, Ramo P, Emmenlauer M, Kaufmann A, Conde-Alvarez R, Low SH, Pelkmans L, Helenius A, Hardt WD, Dehio C, von Mering C. 2014. Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens. Proc Natl Acad Sci USA 111:4548–4553. doi: 10.1073/pnas.1402353111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu ZQ, Gao XL, Liu CN, Lv XP, Zheng SM. 2017. Analysis of microRNA expression profile in specific pathogen-free chickens in response to reticuloendotheliosis virus infection. Appl Microbiol Biotechnol 101:2767–2777. doi: 10.1007/s00253-016-8060-0. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 13.Guo JY, Wang YS, Chen T, Jiang XX, Wu P, Geng T, Pan ZH, Shang MK, Hou CX, Gao K, Guo XJ. 2020. Functional analysis of a miRNA-like small RNA derived from Bombyx mori cytoplasmic polyhedrosis virus. Insect Sci 27:449–462. doi: 10.1111/1744-7917.12671. [DOI] [PubMed] [Google Scholar]
- 14.Li XH, Fu Z, Liang HW, Wang YB, Qi X, Ding M, Sun XL, Zhou Z, Huang Y, Gu HW, Li LM, Chen X, Li DH, Zhao Q, Liu FY, Wang H, Wang J, Zen K, Zhang CY. 2018. H5N1 influenza virus-specific miRNA-like small RNA increases cytokine production and mouse mortality via targeting poly(rC)-binding protein 2. Cell Res 28:157–171. doi: 10.1038/cr.2018.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu GH, Weng ZH, Hu PP, Duan WJ, Xie BP, Sun B, Tang XY, Chen JX. 2018. Synchronous detection of ebolavirus conserved RNA sequences and ebolavirus-encoded miRNA-like fragment based on a zwitterionic copper (II) metal-organic framework. Talanta 180:396–402. doi: 10.1016/j.talanta.2017.12.045. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Sun Y, Song N, Zhao M, Liu R, Feng H, Wang X, Kang Z. 2017. Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol 215:338–350. doi: 10.1111/nph.14577. [DOI] [PubMed] [Google Scholar]
- 17.Jiang X, Qiao F, Long Y, Cong H, Sun H. 2017. MicroRNA-like RNAs in plant pathogenic fungus Fusarium oxysporum f. sp. niveum are involved in toxin gene expression fine tuning. 3 Biotech 7:354. doi: 10.1007/s13205-017-0951-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng H, Wang S, Yang W, Ding X, Li N, Chu Z, Li X. 2021. Identification of virulence associated milRNAs and their bidirectional targets in Rhizoctonia solani and maize during infection. BMC Plant Biol 21:155. doi: 10.1186/s12870-021-02930-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M, Guo Y, Tian R, Gao C, Guo F, Voegele RT, Bao J, Li C, Jia C, Feng H, Huang L. 2020. Adaptive regulation of virulence genes by microRNA-like RNAs in Valsa mali. New Phytol 227:899–913. doi: 10.1111/nph.16561. [DOI] [PubMed] [Google Scholar]
- 20.Smyk B, Cienciała M, Kosiek T. 1952. Diagnosis of Nosema bombycis infection. Med Dosw Mikrobiol 4:363–366. [PubMed] [Google Scholar]
- 21.Tang X, Zhang Y, Zhou Y, Liu R, Shen Z. 2020. Quantitative proteomic analysis of ovaries from Nosema bombycis-infected silkworm (Bombyx mori). J Invertebr Pathol 172:107355. doi: 10.1016/j.jip.2020.107355. [DOI] [PubMed] [Google Scholar]
- 22.Wen M, Shen Y, Shi S, Tang T. 2012. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics 13:140. doi: 10.1186/1471-2105-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiki Y, Yagita Y, Matsuzaki T. 2012. Peroxisome biogenesis disorders: molecular basis for impaired peroxisomal membrane assembly: in metabolic functions and biogenesis of peroxisomes in health and disease. Biochim Biophys Acta 1822:1337–1342. doi: 10.1016/j.bbadis.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 25.South ST, Gould SJ. 1999. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol 144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin YL, Ma LT, Lee YR, Lin SS, Wang SY, Chang TT, Shaw JF, Li WH, Chu FH. 2015. MicroRNA-like small RNAs prediction in the development of Antrodia cinnamomea. PLoS One 10:e0123245. doi: 10.1371/journal.pone.0123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Li M, Zhang C, Zhan N, Cheng H, Gao Y, Sun C, Deng W, Li T. 2021. Identification of microRNA-like RNAs in Cordyceps guangdongensis and their expression profile under differential developmental stages. Fungal Genet Biol 147:103505. doi: 10.1016/j.fgb.2020.103505. [DOI] [PubMed] [Google Scholar]
- 28.Heinz E, Williams TA, Nakjang S, Noel CJ, Swan DC, Goldberg AV, Harris SR, Weinmaier T, Markert S, Becher D, Bernhardt J, Dagan T, Hacker C, Lucocq JM, Schweder T, Rattei T, Hall N, Hirt RP, Embley TM. 2012. The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS Pathog 8:e1002979. doi: 10.1371/journal.ppat.1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S, Li P, Zhang J, Qiu D, Guo L. 2016. Generation of a high resolution map of sRNAs from Fusarium graminearum and analysis of responses to viral infection. Sci Rep 6:26151. doi: 10.1038/srep26151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng H, Xu M, Gao Y, Liang J, Guo F, Guo Y, Huang L. 2021. Vm-milR37 contributes to pathogenicity by regulating glutathione peroxidase gene VmGP in Valsa mali. Mol Plant Pathol 22:243–254. doi: 10.1111/mpp.13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau AYT, Xie Y, Cheung MK, Cheung PCK, Kwan HS. 2020. Genome-wide mRNA and miRNA analysis in the early stages of germ tube outgrowth in Coprinopsis cinerea. Fungal Genet Biol 142:103416. doi: 10.1016/j.fgb.2020.103416. [DOI] [PubMed] [Google Scholar]
- 32.Huang Q, Evans JD. 2016. Identification of microRNA-like small RNAs from fungal parasite Nosema ceranae. J Invertebr Pathol 133:107–109. doi: 10.1016/j.jip.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Jin Y, Zhao JH, Zhao P, Zhang T, Wang S, Guo HS. 2019. A fungal milRNA mediates epigenetic repression of a virulence gene in Verticillium dahliae. Philos Trans R Soc Lond B Biol Sci 374:20180309. doi: 10.1098/rstb.2018.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Titorenko VI, Ogrydziak DM, Rachubinski RA. 1997. Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol Cell Biol 17:5210–5226. doi: 10.1128/MCB.17.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim PK, Mullen RT. 2013. PEX16: a multifaceted regulator of peroxisome biogenesis. Front Physiol 4:241. doi: 10.3389/fphys.2013.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar KR, Wali G, Davis RL, Mallawaarachchi AC, Palmer EE, Gayevskiy V, Minoche AE, Veivers D, Dinger ME, Mackay-Sim A, Cowley MJ, Sue CM. 2018. Expanding the spectrum of PEX16 mutations and novel insights into disease mechanisms. Mol Genet Metab Rep 16:46–51. doi: 10.1016/j.ymgmr.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan MH, Xiao SQ, Chen M, Hong XJ, Lu C. 2007. Establishment and characterization of two embryonic cell lines of Bombyx mori. In Vitro Cell Dev Biol Anim 43:101–104. doi: 10.1007/s11626-006-9009-x. [DOI] [PubMed] [Google Scholar]
- 38.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F. 2017. RNA targeting with CRISPR-Cas13. Nature 550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Z, Huang L, Dong F, Hu Z, Qin Q, Long J, Cao M, Chen P, Lu C, Pan MH. 2018. Establishment of a baculovirus-inducible CRISPR/Cas9 system for antiviral research in transgenic silkworms. Appl Microbiol Biotechnol 102:9255–9265. doi: 10.1007/s00253-018-9295-8. [DOI] [PubMed] [Google Scholar]
- 41.Dong Z, Long J, Huang L, Hu Z, Chen P, Hu N, Zheng N, Huang X, Lu C, Pan M. 2019. Construction and application of an HSP70 promoter-inducible genome editing system in transgenic silkworm to induce resistance to Nosema bombycis. Appl Microbiol Biotechnol 103:9583–9592. doi: 10.1007/s00253-019-10135-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01048-21_Supp_1_seq11.pdf, PDF file, 0.4 MB (395.4KB, pdf)
Supplemental material. Download SPECTRUM01048-21_Supp_2_seq12.xlsx, XLSX file, 0.05 MB (50KB, xlsx)