SUMMARY
Cryptosporidiosis is one of the most important causes of moderate to severe diarrhea and diarrhea-related mortality in children under 2 years of age in low- and middle-income countries. In recent decades, genotyping and subtyping tools have been used in epidemiological studies of human cryptosporidiosis. Results of these studies suggest that higher genetic diversity of Cryptosporidium spp. is present in humans in these countries at both species and subtype levels and that anthroponotic transmission plays a major role in human cryptosporidiosis. Cryptosporidium hominis is the most common Cryptosporidium species in humans in almost all the low- and middle-income countries examined, with five subtype families (namely, Ia, Ib, Id, Ie, and If) being commonly found in most regions. In addition, most Cryptosporidium parvum infections in these areas are caused by the anthroponotic IIc subtype family rather than the zoonotic IIa subtype family. There is geographic segregation in Cryptosporidium hominis subtypes, as revealed by multilocus subtyping. Concurrent and sequential infections with different Cryptosporidium species and subtypes are common, as immunity against reinfection and cross protection against different Cryptosporidium species are partial. Differences in clinical presentations have been observed among Cryptosporidium species and C. hominis subtypes. These observations suggest that WASH (water, sanitation, and hygiene)-based interventions should be implemented to prevent and control human cryptosporidiosis in low- and middle-income countries.
KEYWORDS: Cryptosporidium, molecular epidemiology, anthroponotic transmission, WASH, low- and middle-income countries
INTRODUCTION
Cryptosporidiosis is a major cause for diarrhea in young children in low- and middle-income countries. It has been recognized as one of the most important causes for moderate to severe diarrhea as well as diarrhea-related mortality in children less than 2 years in multiple recent studies (1–4). In South Asia and sub-Saharan Africa, Cryptosporidium infections contribute annually to nearly 2.9 to 4.7 million diarrheal cases in children under 2 years (5, 6). It is estimated that cryptosporidiosis-associated diarrhea caused over 48,000 deaths as well as 4.2 million disability adjusted life years (DALYs) in children less than 5 years in 2016 (7). Globally, although the mortality related to diarrhea in children under 5 years had declined by 60% from 2000 to 2016, diarrhea-associated morbidity showed a lower reduction of only 13% (8). The prevalence of pathogens unresponsive to conventional antibiotic treatments, such as Cryptosporidium spp. and rotavirus, might be responsible for the slow reduction in global diarrhea-associated morbidity.
Cryptosporidiosis is also an important cause of childhood malnutrition. Even when cryptosporidiosis is not associated with diarrhea, it can still cause severe malnutrition (9, 10). Lower weight, weight-for-age Z scores, height-for-age Z scores, and/or body mass index-for-age Z scores are found in children infected with Cryptosporidium spp. than in uninfected children (7, 10–15). Children having multiple episodes of cryptosporidiosis experience more severe stunting (10, 16). Therefore, cryptosporidiosis is an important cause of growth retardation. It has been suggested that Cryptosporidium spp. impair intestinal-epithelial barrier integrity through the induction of inflammatory responses in the small intestine and affect nutrient absorption through the destruction of intestinal epithelial cells (17–20). On the other hand, stunting at birth is associated with the occurrence of cryptosporidiosis (19, 21, 22).
Molecular epidemiological tools have been used extensively in characterizing Cryptosporidium spp. at species/genotype and subtype levels (23). While more molecular epidemiological studies of cryptosporidiosis have been conducted in developed countries, increasing numbers of studies are from low- and middle-income countries, leading to improved understanding of the epidemiology of cryptosporidiosis (6, 10, 24–38). In particular, these studies have led to the identification of anthropogenic factors involved in the acquisition of Cryptosporidium spp. in children and HIV-positive patients.
EPIDEMIOLOGICAL FEATURES OF CRYPTOSPORIDIOSIS IN LOW- AND MIDDLE-INCOME COUNTRIES
Due to higher endemicity, lower hygiene levels, and less-intensive animal farming, the epidemiological features of human cryptosporidiosis in low- and middle-income countries differ greatly from those in industrialized nations (Table 1). They include less-frequent occurrence of outbreaks, occurrence of infections at early age, more common association with HIV/AIDS, occurrence of multiple episodes of infections, and concurrence of other pathogens (39–41).
TABLE 1.
Differences in epidemiological features of human cryptosporidiosis between low- and middle-income countries and industrialized nations (23, 39, 103, 227)
| Feature | Characteristic in: |
|
|---|---|---|
| Industrialized nations | Low- and middle-income countries | |
| Endemicity | Low | High |
| Occurrence of outbreaks | High | Low |
| Susceptible population | All ages and immune statuses | Children and HIV-positive people |
| Infection in children | Late (>2 yrs) | Early (<2 yrs) |
| Major clinical symptoms | Diarrhea | Diarrhea and retarded growth |
| Asymptomatic infection | Low occurrence | High occurrence |
| Peak prevalence | Late summer and early autumn | Rainy season or cool months in the tropics |
| Major risk factors | International traveling, contact with animals or humans, swimming | Poor hygiene, overcrowding, diarrhea case in household |
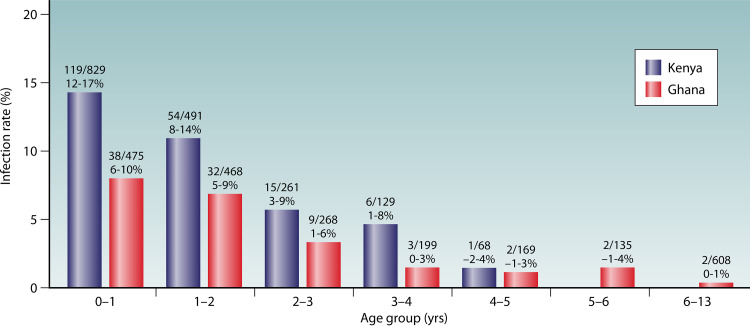
In low- and middle-income countries, cryptosporidiosis is mostly an infectious disease of young children. Pediatric cryptosporidiosis is mostly reported in children less than 2 years old (Fig. 1); recent birth cohort studies indicated as many as 77% of Bangladeshi children and 92.4% of Indian children experienced Cryptosporidium infection before the age of 2 years (9, 42). Data from the Global Enteric Multicenter Study (GEMS) conducted in several low-income countries also indicated that Cryptosporidium spp. are among the most important diarrhea-related pathogens in children under 2 years (2, 30, 31, 43). In one GEMS in children with moderate to severe diarrhea under 5 years in rural western Kenya, 88.7% of cryptosporidiosis cases occurred in children under 2 years (30). Another GEMS in Gambian children with moderate to severe diarrhea under 5 years found that 91.8% of diarrhea cases caused by Cryptosporidium spp. occurred in young children of 6 to 24 months (31). Similar results have also been found in MAL-ED (Etiology, Risk Factors and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development Project) studies (4, 13). In one MAL-ED study conducted in eight countries of Africa, Asia, and South America, nearly 65% of children under 2 years experienced Cryptosporidium infection and 54% had Cryptosporidium-associated diarrhea (13). In contrast, pediatric cryptosporidiosis in children from industrialized nations occurs later (age over 2 years) than in low- and middle-income countries (under 2 years), which is likely the result of delayed exposures to contamination under better hygiene (44).
FIG 1.
Age pattern of pediatric cryptosporidiosis in Kenya and Ghana (30, 128). Numbers above the bars denote the number of Cryptosporidium-positive cases/total number of children studied in each age group (the first line) and the 95% confidence intervals of the infection rates (the second line).
Cryptosporidiosis is also common in immunocompromised persons in low- and middle-income countries, especially HIV-positive patients (45–49). The prevalence of cryptosporidiosis in HIV-infected persons ranges from 5.6% to 25.7% in Africa, 3.7% to 45.0% in Asia, 5.6% to 41.6% in South America, and 2.6% to 15.1% in Europe (47). Higher infection rates and more severe clinical outcomes are seen in HIV-positive persons with CD4+ cell counts lower than 200 cells/μl (48). Many recent studies have reported the occurrence of cryptosporidiosis in hemodialysis patients as well as renal transplant patients in low- and middle-income countries (50–59). In contrast, human cryptosporidiosis occurs in persons of various ages and immune statuses in industrialized nations, probably as a reflection of reduced immunity. In industrialized nations, improved hygiene and better drinking source water and wastewater treatment have probably led to reduced exposure to Cryptosporidium oocysts, resulting in reduced immunity (60, 61).
Cryptosporidiosis in children is often associated with diarrhea, nausea, vomiting, abdominal cramps, low-grade fever, headache, and fatigue (60, 62). The diarrhea can be watery and voluminous but usually resolves within 1 to 2 weeks without treatment. However, the median duration of postdiarrheal shedding was about 39.5 days (95% confidence interval [CI], 30.6 to 49 days) in one study (63). The occurrence of diarrhea or other symptoms, however, was detected in only approximately one-third of Cryptosporidium-infected children in community-based studies conducted in low- and middle-income countries (42). While multiple reasons, such as prior exposure to Cryptosporidium infection and receiving colostrum, might be involved, results of a recent study in Bangladesh suggest that host genetics could play a potential role. A genetic variant within protein kinase C alpha (PRKCA) was associated with a higher risk of symptomatic cryptosporidiosis during the first year of life (64). Even subclinical cryptosporidiosis has significant adverse effects on children, as they may experience retarded growth (7, 10, 15, 65). Unlike in low- and middle-income countries, both children and adults with cryptosporidiosis in developed countries often have diarrhea (39, 60).
Cryptosporidium infections in low- and middle-income countries result mainly from poor hygiene and sanitation (66). Cohort, case-control, and cross-sectional studies have identified multiple risk factors in human cryptosporidiosis in low- and middle-income countries (Table 2). Among them, poor hygiene is the most common risk factor, followed by contact with animals, overcrowding, poor drinking water, young age, and household diarrhea. However, different studies have identified different risk factors. For example, a recent molecular epidemiological study of cryptosporidiosis in four sub-Saharan African nations has identified contact with Cryptosporidium-positive household members (risk ratio [RR] = 3.6; 95% CI, 1.7 to 7.5) or neighboring children (RR = 2.9; 95% CI, 1.6 to 5.1) rather than having positive animals (RR = 1.2; 95% CI, 0.8 to 1.9) as the risk factors (33). Contact with animals, especially calves, was a common risk factor for human cryptosporidiosis in many but not all studies (33, 66–69). It is interesting to find that animal contact was protective for Cryptosporidium infection in children in Mozambique (70). One study in Cameroon found that breastfeeding (odds ratio [OR] = 0.18; 95% CI, 0.04 to 0.90) was protective for Cryptosporidium infection in children within 6 months (71). In an investigation of a cryptosporidiosis outbreak in Botswana, hospitalization and mortality in children were associated primarily with nonbreastfeeding (72). However, prolonged breast feeding (>2 years) (OR = 2.18; 95% CI, 1.02 to 7.32) was a risk factor for pediatric cryptosporidiosis in Malaysia (73). In Zambia, male gender (OR = 2.5; 95% CI, 1.13 to 5.70), divorce (OR = 14.8; 95% CI, 1.58 to 138.4), and sharing water sources among neighbors (OR = 5.7; 95% CI, 1.15 to 27.9) were major risk factors for Cryptosporidium infection in HIV-positive persons (38). Although divergent risk factors for human cryptosporidiosis have been identified in different studies, poor hygiene was the major one in low- and middle-income countries, while swimming, contact with diarrheal persons/animals, and international travelling were the major ones in industrialized nations (39, 66, 74, 75).
TABLE 2.
Recent studies on the risk factors for human cryptosporidiosis in low- and middle-income countries
| Location | Type of study | Sample size | Study population | Major risk factor | Reference |
|---|---|---|---|---|---|
| Asia | |||||
| China | Case-control | 1,366 | Patients with and without HIV | Contact with animals | 115 |
| Cross-sectional | 1,635 | Children and adults | Overcrowding, contact with animals, infection with hepatitis B virus | 228 | |
| Cross-sectional | 321 | Children | Poor hygiene, poor drinking water | 229 | |
| Cross-sectional | 1,637 | Children with and without diarrhea | Poor hygiene | 230 | |
| Indonesia | Case-control | 4,368 | Patients with and without diarrhea | Contact with animals, overcrowding, rainfall | 231 |
| Malaysia | Cross-sectional | 276 | Children | Low birth wt, overcrowding, breastfeeding | 73 |
| Cross-sectional | 135 | Children | Old age, poor hygiene | 232 | |
| Philippines | Cross-sectional | 137 | Children and adults | Location, poor drinking water, open defecation | 233 |
| Cambodia | Cross-sectional | 498 | Children | Malnutrition, chronic medical diagnoses, contact with animals | 130 |
| Bangladesh | Case-control | 272 | Children with and without diarrhea | Young age, nonbreastfeeding, stunting | 234 |
| Cohort | 392 | Children | Malnutrition | 9 | |
| Cohort | 203 | Children | Overcrowding | 13 | |
| India | Case-control | 580 | Children with and without diarrhea | Overcrowding, stunting | 235 |
| Pakistan | Cross-sectional | 425 | Children with diarrhea | Poor hygiene, diarrhea, environmental factors | 95 |
| Iran | Cross-sectional | 171 | Children with and without diarrhea | Low birth wt, less breastfeeding, male gender | 236 |
| Case-control | 480 | Healthy persons and hemodialysis patients | Poor hygiene, diarrhea, education level, young age | 51 | |
| Lebanon | Cross-sectional | 249 | Children | Young age, digestive symptoms, diarrhea, fever | 237 |
| Cross-sectional | 412 | Patients and children | Having meals outside home, diarrhea | 238 | |
| Africa | |||||
| Ethiopia | Cross-sectional | 520 | HIV/AIDS patients | Contact with animals | 67 |
| Cross-sectional | 393 | Children with and without diarrhea | Young age | 157 | |
| Libya | Case-control | 505 | Children with and without diarrhea | Contact with animals, foreign workers from Africa, poor hygiene, poor drinking water | 239 |
| Egypt | Case-control | 100 | Children with and without diarrhea | Poor hygiene, contact with animals, diarrhea | 139 |
| Kenya | Case-control | 1,778 | Children with and without diarrhea | Young age | 30 |
| Uganda | Cross-sectional | 108 | Children and adults | Poor drinking water | 240 |
| Malawi | Case-control | 96 | Children with and without diarrhea | Contact with animals, poor hygiene, diarrhea | 241 |
| Nigeria | Cross-sectional | 692 | Children with and without diarrhea | Young age, stunting | 242 |
| Cross-sectional | 180 | Children | Young age, diarrhea | 243 | |
| South Africa | Case-control | 180 | Adults with or without HIV or diarrhea | Poor hygiene, contact with animals, poor socioeconomic state | 140 |
| Mozambique | Cross-sectional | 985 | Children with diarrhea | Nampula Province, underweight | 70 |
| Zambia | Cross-sectional | 222 | Children with diarrhea | Rainfall, breastfeeding | 244 |
| Cross-sectional | 326 | HIV/AIDS patients | Sex, marital status, sharing water sources among neighbors | 38 | |
| Angola | Cross-sectional | 351 | Children | Young age | 150 |
| Gambia | Case-control | 4,907 | Children with and without diarrhea | Poor drinking water, contact with animals, overcrowding | 31 |
| Ghana | Cross-sectional | 50 | Children and adults with HIV | Poor drinking water | 245 |
| Guinea-Bissau | Case-control | 250 | Children with and without diarrhea | Contact with animals, poor hygiene, male gender | 246 |
| Cameroon | Cross-sectional | 112 | Children | Nonbreastfeeding, poor drinking water | 71 |
| Sub-Saharan Africa | Cross-sectional | 1363 | Children | Contact with humans | 33 |
| Americas | |||||
| Cuba | Case-control | 215 | Children with and without diarrhea | Poor hygiene, contact with animals | 247 |
| Guatemala | Cohort | 130 | Children with and without diarrhea | Lack of toilet, contact with animals | 248 |
| Cross-sectional | 100 | Children with gastroenteritis | Poor hygiene, female gender | 249 | |
| Mexico | Cross-sectional | 403 | Children with diarrhea | Malnutrition, nonbreastfeeding | 250 |
| Cross-sectional | 173 | Children without diarrhea | Poor drinking water, contact with animals | 11 | |
| Cross-sectional | 132 | Children without diarrhea | Poor drinking water, overcrowding, poor hygiene, diarrhea | 251 | |
| Venezuela | Cross-sectional | 515 | Children and adults | Poor hygiene, overcrowding, young age | 252 |
| Brazil | Cohort | 189 | Children with and without diarrhea | Low birth wt, overcrowding | 253 |
| Cross-sectional | 445 | Children with diarrhea | Young age, poor hygiene, diarrhea, rainfall, male gender | 254 | |
| Peru | Cohort | 368 | Children with and without diarrhea | Lack of toilet, warm season | 255 |
Although cryptosporidiosis is highly endemic in low- and middle-income countries, it rarely causes outbreaks there, probably due to the high level of population immunity (66). A few outbreaks of human cryptosporidiosis, however, have been reported in Mexico, Brazil, Botswana, Jordan, and China, imposing additional burdens on the stretched public health system (72, 76–78). In contrast, human cryptosporidiosis in industrialized nations is best known for foodborne, waterborne, and animal contact-associated outbreaks (79, 80).
UNIQUE DISTRIBUTION OF CRYPTOSPORIDIUM SPECIES IN HUMANS IN LOW- AND MIDDLE-INCOME COUNTRIES
Although humans and other vertebrates are common hosts of Cryptosporidium spp., most species have host specificity (81). Currently, over 40 Cryptosporidium species have been recognized in mammals, birds, reptiles, amphibians, and fish (82). There are also many Cryptosporidium genotypes of unknown species status. Due to the existence of host specificity, only one to four Cryptosporidium species/genotypes are frequently found in one host species. For example, dogs are mostly infected with C. canis, cats with C. felis, rabbits with C. cuniculus, humans with C. hominis and C. parvum, sheep and goats with C. parvum, C. ubiquitum, and C. xiaoi, and cattle with C. parvum, C. bovis, C. ryanae, and C. andersoni (82). Among the known Cryptosporidium species, C. parvum is one of the few species with a wide host range and also the most important zoonotic species in humans (82).
Over 20 Cryptosporidium species and genotypes have been reported in humans, many with fewer than a handful of cases (81). Among them, C. parvum and C. hominis are two major species, being responsible for over 90% of human cryptosporidiosis cases in most areas. Other less commonly detected species include C. meleagridis, C. canis, C. felis, C. ubiquitum, C. cuniculus, C. viatorum, Cryptosporidium chipmunk genotype I, and C. muris in the order of numbers of reported cases. The remaining ones have each been occasionally detected in several cases (82).
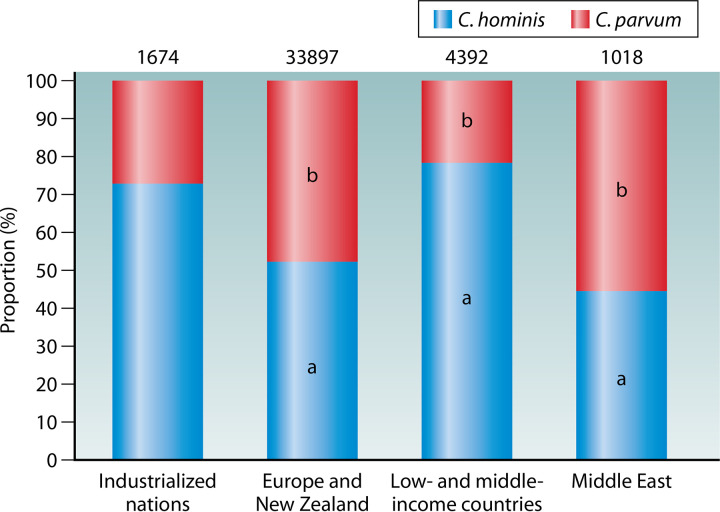
The distribution of Cryptosporidium species in humans is different between industrialized and low- and middle-income countries (Fig. 2). Molecular epidemiological studies of human cryptosporidiosis have recognized C. hominis as the dominant species in both children and HIV-positive patients in low- and middle-income countries (39). In contrast, C. hominis and C. parvum infections appear to be equally common in both immunocompromised and immunocompetent persons in European and Middle East countries as well as New Zealand (39, 75, 80, 83, 84). In some industrialized nations, such as the United States, Canada, Australia, and Japan, although most human cryptosporidiosis cases are caused by C. hominis, there is a high occurrence of C. parvum in rural areas (85–88). These results suggest that zoonotic infection is less common in low- and middle-income countries than in industrialized nations.
FIG 2.
Proportion (%) of Cryptosporidium parvum and Cryptosporidium hominis in different areas and socioeconomic conditions. Numbers above bars denote sample size (n). In the bars, “a” indicates a significant difference in proportion of C. hominis between industrialized nations and the group under comparison (P < 0.05 by chi-square test), while “b” indicates a significant difference in proportion of C. parvum between industrialized nations and the group under comparison (P < 0.05).
The distribution of other human-pathogenic Cryptosporidium species is also different between low- and middle-income countries and industrialized nations. Most human infections with C. meleagridis, C. felis, C. canis, C. viatorum, and C. muris have been reported in studies conducted in low- and middle-income countries or in persons who have traveled to these areas (10, 39, 67, 89, 90). In contrast, most human infections with C. ubiquitum, C. cuniculus, and chipmunk genotype I are from industrialized nations. In particular, C. ubiquitum and chipmunk genotype I contribute to substantial numbers of human Cryptosporidium infections in rural states in the United States, while C. cuniculus infections are reported mainly in the United Kingdom and New Zealand (83, 86, 91–94).
CHARACTERISTICS OF C. HOMINIS INFECTION IN HUMANS IN LOW- AND MIDDLE-INCOME COUNTRIES
Molecular epidemiological studies of Cryptosporidium infections in children have shown a dominance of C. hominis in low- and middle-income countries, accounting for an average of over 65% of Cryptosporidium cases. In some studies, the lack of C. hominis was possibly caused by the use of genotyping tools targeting individual species (95) or a small number of Cryptosporidium-positive specimens (96, 97). The dominance of C. hominis could be due to the importance of environmental contamination and direct person-to-person transmission in cryptosporidiosis epidemiology. This was supported by the observation of a high rate of secondary infection and infection with the same subtype within families in a case-control study of Cryptosporidium transmission in Bangladeshi households (25). Cryptosporidium hominis is also a major species in HIV-positive patients in low- and middle-income countries. There is frequently a good agreement in the distribution of Cryptosporidium species between children and HIV-positive patients in the same country.
In low- and middle-income countries, the results of molecular characterizations of C. hominis isolates have highlighted the complexity of cryptosporidiosis epidemiology. Molecular analyses of C. hominis have revealed much higher numbers of subtype families in humans in low- and middle-income countries than in industrialized nations (98). Based on sequence analysis of the 60-kDa glycoprotein (gp60) gene, C. hominis is divided into five major subtype families with very divergent sequences, including Ia, Ib, Id, Ie, and If. Each C. hominis subtype family has many subtypes that differ from each other mostly in the number of trinucleotide repeats at the 5′ end of the gene sequence. All five subtype families are common in children and HIV-positive persons in most low- and middle-income countries examined. The complexity of transmission is further supported by the occurrence of multiple subtypes within the Ia, Ib, and Id subtype families in most areas of endemicity (25, 26, 29, 33, 62, 99–102). In comparison, much lower genetic diversity of C. hominis is seen in humans in industrialized nations. In European countries, subtype family Ib contributes to over 90% of C. hominis infections (80). The high heterogeneity of C. hominis in low- and middle-income countries is considered an indication of the high intensity of cryptosporidiosis transmission in areas of endemicity (103).
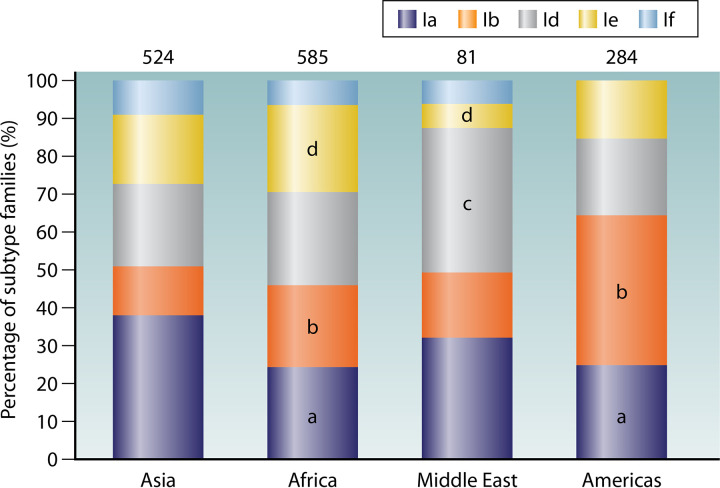
The distribution of common C. hominis subtype families varies among geographic areas (Fig. 3). In Asia, Ia is a major subtype family for C. hominis in humans, followed by Id, Ie, Ib, and If, in that order (25, 29); in Africa, the frequencies of Ia, Ib, Id, and Ie are about the same and significantly higher than that of If (33, 102, 104); in the Middle East, Id is most common, followed by Ia and Ib, with only limited occurrence of Ie and If (99, 100); in the Americas, Ib is the most common subtype family of C. hominis, followed by Ia, Id, and Ie, with the absence of If (24, 105). These differences in distribution of C. hominis subtype families possibly reflect variations in the transmission of C. hominis in humans among areas.
FIG 3.
Distribution of common Cryptosporidium hominis subtype families among low- and middle-income countries in Asia, Africa, the Middle East, and the Americas. Numbers above bars denote sample size (n). In the bars, “a,” “b,” “c,” and “d” indicate a significant difference in distribution of Ia, Ib, Id, and Ie between Asia and the group under comparison (P < 0.05 by chi-square test), respectively.
Geographical segregation is seen in the distribution of subtypes within some of the common subtype families. For instance, two major subtypes are seen within the subtype family Ib: IbA9G3 and IbA10G2. The former is common in Jordan, Tanzania, Uganda, Kenya, Bangladesh, and India, while the latter is common in Peru, Jamaica, Colombia, Argentina, and Brazil as well as South Africa. Other subtypes, such as IbA13G3, IbA10G1, IbA11G2, and IbA12G3, were reported only in limited regions. This geographic segregation in C. hominis subtypes has been confirmed by multilocus sequence type (MLST) analysis of specimens from several countries (106).
CHARACTERISTICS OF C. PARVUM INFECTION IN HUMANS IN LOW- AND MIDDLE-INCOME COUNTRIES
Cryptosporidium parvum contributes to ∼20% of cases of human cryptosporidiosis in low- and middle-income countries. As with C. hominis, there are multiple subtype families within C. parvum at the gp60 locus. Some of the C. parvum subtype families are host adapted, which is useful in tracking the sources of C. parvum infections in humans. For example, the IIa subtype family is commonly found in dairy calves, the IId subtype family is found mostly in lambs and goat kids, while the IIc subtype family is almost exclusively a human pathogen (82). Although there are more subtype families in C. parvum than in C. hominis, only 1 or 2 subtype families are responsible for most human C. parvum infections in one particular area.
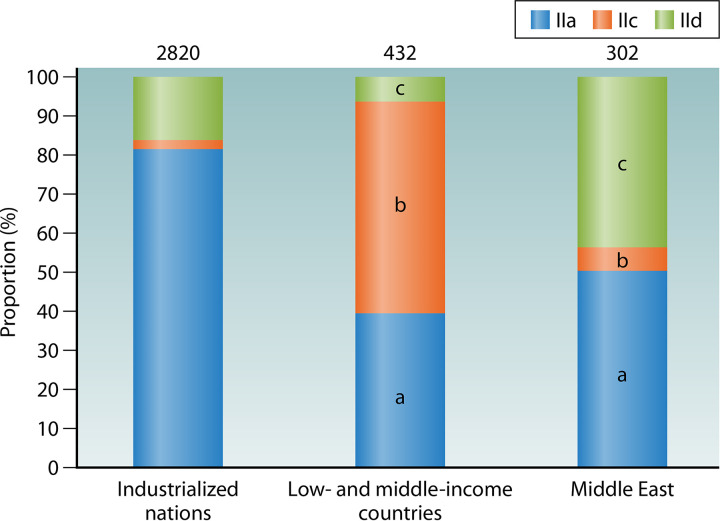
The distribution of common C. parvum subtype families varies greatly among different geographic regions and socioeconomic conditions (Fig. 4). In low- and middle-income countries, IIc contributes to over half the disease burden due to C. parvum, followed by IIa, while the contribution of IId is limited. The IIa subtypes identified in humans in low- and middle-income countries, however, were mostly from the few studies conducted in Malaysia and Ethiopia (67, 107, 108), except for a recent one in China (28), where IIa subtypes are rare in animals (109–113). In Middle East countries, some of which are highly industrialized, the disease burdens of IId and IIa are significantly higher than that of IIc. In contrast, IIa is responsible for over 80% of C. parvum infections in industrialized countries, whereas IId subtypes are seen mostly in New Zealand and Europe, and IIc infections are associated with travel to low- and middle-income countries (75, 80, 83, 98). The difference in distribution of C. parvum subtype families among geographic regions and socioeconomic conditions is probably a reflection of differences in infection sources and transmission routes.
FIG 4.
Proportion (%) of common Cryptosporidium parvum subtype families in different areas and socioeconomic conditions. Numbers above bars denote sample size (n). In the bars, “a,” “b,” and “c” indicate a significant difference in proportion of IIa, IIc, and IId between industrialized nations and the group under comparison (P < 0.05 by chi-square test), respectively.
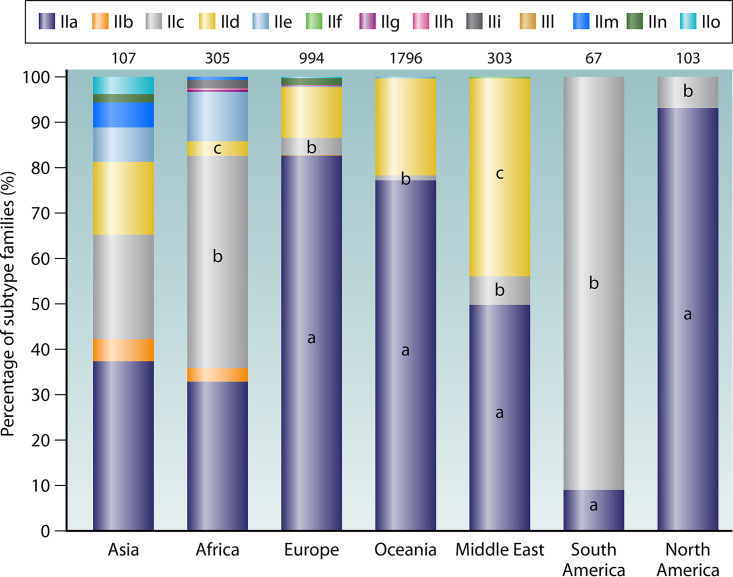
The subtype diversity of C. parvum in humans is much higher in low- and middle-income countries than in industrialized nations (Table 3; Fig. 5). As presented in Table 3, analysis of the subtype diversity using the Simpson and Shannon-Wiener indexes showed the highest subtype diversity of C. parvum in Asia, followed by Africa, the Middle East, Europe, Oceania, South America, and North America. In Africa, as many as nine subtype families are recognized, including IIa to IIe, IIg, IIh, IIi, and IIm. Among them, IIc is the most common subtype family. This is followed by the IIe subtype family, which appears to be another anthroponotic subtype family of C. parvum. The occurrence of the remaining subtype families is sporadic. The IIa subtype family, however, appears to be common in AIDS patients in Ethiopia, together with some occurrence of the IId subtype family (67).
TABLE 3.
Subtype diversity of Cryptosporidium parvum in humans in divergent geographic regions
| Location | No. of cases of indicated subtype of C. parvuma |
Simpson index | Shannon-Wiener index | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IIa | IIb | IIc | IId | IIe | IIf | IIg | IIh | IIi | IIl | IIm | IIn | IIo | |||
| Asia | 40 | 5 | 25 | 17 | 8 | 0 | 0 | 0 | 0 | 0 | 6 | 2 | 4 | 0.7678 | 1.6957 |
| Africa | 93 | 8 | 132 | 9 | 31 | 0 | 1 | 1 | 5 | 0 | 2 | 0 | 0 | 0.6578 | 1.3215 |
| Europe | 822 | 1 | 38 | 111 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 16 | 2 | 0.3019 | 0.6390 |
| Oceania | 1,387 | 0 | 20 | 384 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0.3578 | 0.5973 |
| Middle East | 151 | 0 | 19 | 132 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.5579 | 0.9016 |
| South America | 6 | 0 | 61 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1631 | 0.3015 |
| North America | 96 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1267 | 0.2483 |
Numbers represent cases of each subtype family in corresponding regions.
FIG 5.
Distribution of Cryptosporidium parvum subtype families in humans in Asia, Africa, Europe, Oceania, the Middle East, South America, and North America. Numbers above bars denote sample size (n). In the bars, “a,” “b,” and “c” indicate a significant difference in distribution of IIa, IIc, and IId between Asia and the group under comparison (P < 0.05 by chi-square test), respectively.
In Asia, up to eight subtype families have been reported, namely, IIa to IIe, IIm, IIn, and IIo. Among them, the IIa and IIc subtype families are the most common. They are followed by the IId and IIe subtype families. The common occurrence of the IIa subtype family, however, is mostly attributable to two reports in Malaysia and China (28, 108). The Chinese report was contradictory to other reports, which mostly reported the IId subtype family in the country (27, 114, 115). In fact, IIa subtypes are absent from dairy calves in China, which are exclusively infected with IId subtypes (109). Other C. parvum subtype families such as IIe, IIm, and IIo have been only occasionally reported in humans in Asia (6, 26, 32, 116, 117).
In the Middle East, the genetic diversity of C. parvum in humans is much lower. Although four subtype families are recognized, two of them have very low frequency. Among them, IId contributes to over half of the C. parvum infections. The importance of IId in human infections in Middle East countries may be related to the importance of small ruminants, which are commonly infected with C. parvum IId subtypes (118). This is followed by IIa, which accounts for almost all the remaining C. parvum infections there. In contrast, IIc and IIf subtype families were reported in only a few cases in the area (99, 100, 119–121).
In South America, only IIc and IIa have been reported in humans. Although IIc is more common than IIa there, there is an increasing occurrence of IIa in humans in recent years, especially in Colombia and Mexico, which geographically is located in North America (24, 122). The occurrence of IIa subtypes there reflects their common occurrence in dairy calves (123). The IIc subtype family, however, remains the dominant C. parvum in humans in urban areas in South America (105, 122, 124, 125).
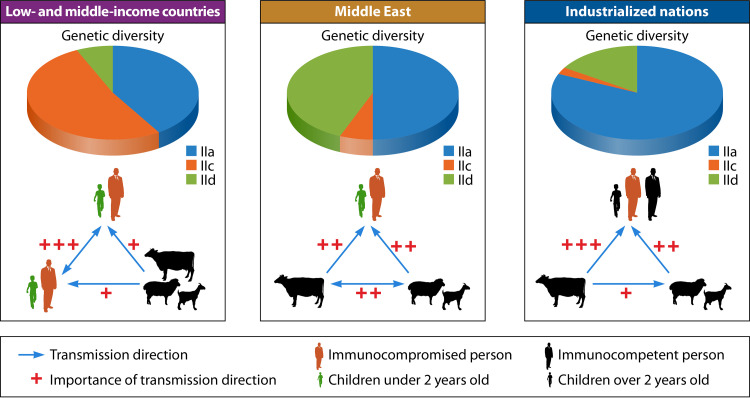
The dominance of the IIc subtype family in humans in low- and middle-income countries suggests that anthroponotic transmission plays a major role in cryptosporidiosis in this area (Fig. 6). This is especially the case in African and Asian countries and urban areas in South America, where the anthroponotic IIc subtype family is especially common in low-income countries with poor sanitation and in HIV-positive persons (126). In many Asian and African countries, the occurrence of IIc subtypes in humans is in concurrence with IIe, another anthroponotic C. parvum subtype family (26, 32, 33, 67, 104, 127–130). In addition to them, several other subtype families were also found (127, 128, 131). In contrast, the common occurrence of IIa and IId subtypes in Middle East countries suggests that zoonotic transmission of C. parvum might play a significant role in cryptosporidiosis there. This is in agreement with the common occurrence of IIa and IId subtypes in calves, lambs, and goat kids in these countries (132–138). Indeed, animal contact has been identified as a risk factor for pediatric cryptosporidiosis in Egypt (139). Zoonotic transmission appears to be important in some countries elsewhere, such as Ethiopia in Africa and Malaysia in Asia, where in addition to IIa and IId, anthroponotic transmission of IIc occurs simultaneously (67, 108). In the Ethiopian study, calf contact was identified as a risk factor for infection with the IIa subtype family (67). This is one of the rare occasions in low- and middle-income countries where zoonotic transmission of C. parvum has been explicitly implicated in playing a significant role in cryptosporidiosis epidemiology. These findings indicate that although anthroponotic transmission contributes to the majority of C. parvum infections in humans in low- and middle-income countries, zoonotic transmission is increasingly recognized in some countries where the disease is endemic. This is probably related to the increased direct or indirect contact with farm animals as part of the property relief efforts that have been under way for some years (68, 140, 141).
FIG 6.
Differences in transmission of major subtype families of Cryptosporidium parvum in humans between low- and middle-income countries and industrialized nations. The blue arrows indicate major directions of transmission, and the red plus symbols indicate their importance in cryptosporidiosis epidemiology. The relative distribution of the major subtype families is indicated in the pie chart.
In the desert country Botswana, one major outbreak of diarrhea in 2006 after heavy rains and floods had led to hundreds of deaths and thousands of infections in children less than 5 years old. It was mainly caused by cryptosporidiosis. Based on the results of molecular epidemiological investigations, C. parvum and C. hominis were found in 30 cases examined, with almost equal distribution (72). A total of five subtype families of C. hominis were found. In contrast, only two IIc subtypes (IIcA5G3a and IIcA5G3b) were found among the C. parvum-positive samples (L. Xiao, unpublished data). These data indicate that the source of contamination during the outbreak was probably of human origin. Poor sewage treatment or lack of sewage treatment might have contributed to the occurrence of the outbreak (72).
CHARACTERISTICS OF OTHER CRYPTOSPORIDIUM SPECIES IN LOW- AND MIDDLE-INCOME COUNTRIES
In addition to C. hominis and C. parvum, other species, including C. meleagridis, C. canis, C. felis, C. viatorum, and C. muris, are significant causes of human cryptosporidiosis in low- and middle-income countries (10, 24, 26, 32, 67, 103). In industrialized nations, infections with these species are frequently associated with foreign travel, as autochthonous infections with them are rare (83, 89, 142, 143). Several other species associated with zoonotic cryptosporidiosis in industrialized nations, such as C. ubiquitum, C. cuniculus, and Cryptosporidium chipmunk genotype I, are rarely detected in humans in low- and middle-income countries.
Cryptosporidium meleagridis is traditionally considered an avian species but has been found in children and HIV-positive patients in many low- and middle-income countries. In some recent studies conducted in Nigeria, Mozambique, Tunisia, Madagascar, Ghana, Bangladesh, Thailand, and Colombia, C. meleagridis was a prevalent species and contributed to 9% to 92% of the 22 to 94 cryptosporidiosis cases examined in each study (10, 25, 26, 33, 99, 129, 144). In one recent study of molecular epidemiology of cryptosporidiosis in two areas in Bangladesh, over 100 C. meleagridis infections were identified. Cryptosporidium meleagridis was found in 13% of Cryptosporidium infections in the urban area and 90% of Cryptosporidium infections in the rural area (10). Cryptosporidium meleagridis was also found at a high frequency (20 of 92 Cryptosporidium-positive cases) in HIV-positive patients in one study in Bangkok, Thailand (26). In one small-scale study in China, only C. meleagridis was found in children with diarrhea (96).
A gp60-based subtyping tool is available for the genetic characterization of C. meleagridis (145). Eight subtype families (IIIa to IIIh) and over 30 subtypes have been reported in humans (23, 145). In one study of C. meleagridis infections in HIV-positive patients in Bangkok, Thailand, nine subtypes were identified (26). Some human infections with C. meleagridis are possibly caused by zoonotic transmission, judged by the occurrence of IIIbA26G1R1b and IIIbA22G1R1c in both diarrheic children and farmed chickens in Hubei, China (96, 146). The results of one MLST analysis of C. meleagridis from children, AIDS patients, and birds in Peru did not find obvious host segregation in subtypes, suggesting that zoonotic transmission of C. meleagridis between humans and birds is possible (147). Seven of the 55 human C. meleagridis cases in the study, however, had coinfection with C. hominis, indicating that at least some of the C. meleagridis infections in humans were of anthroponotic origin.
The MLST characterization of C. meleagridis identified two major groups (group 1 and group 2) of C. meleagridis subtypes (147). They correspond to types 1 and 2 at the small-subunit (SSU) rRNA locus and IIIb and IIIc subtype families at the gp60 locus. As they also have different nucleotide sequences at the MSC6-5 and RPGR loci, they probably represent two segregated C. meleagridis populations (147). Thus far, the biological significance of the two C. meleagridis populations is not clear. In the Peruvian study, group 1 was found in both chickens and humans, and 2 of the 14 multilocus subtypes of C. meleagridis in the group were found in both AIDS patients and birds, suggesting that indeed zoonotic transmission might be involved. In contrast, group 2 was found only in humans. Nevertheless, the number of avian isolates characterized was small. Population genetic analysis of the MLST data suggests a clonal population of C. meleagridis in the study community (147).
Cryptosporidium canis is the only Cryptosporidium species found in dogs in most molecular epidemiological studies of cryptosporidiosis in companion animals (148). It has been commonly reported in humans in low- and middle-income countries. For most studies, only a few cases were positive for C. canis, but unusually high infection rates (>10% of cryptosporidiosis cases) were reported in children in Cambodia and Angola as well as HIV-positive patients in Thailand and Venezuela (26, 130, 149, 150). In Venezuela, all the C. canis-positive patients kept dogs during the survey, indicating a possible occurrence of zoonotic transmission, although no survey of dogs was done (149). Although zoonotic transmission between humans and dogs has been identified using a genotyping tool (151), the transmission from pet dogs to humans is considered a low risk (20). Currently, no subtyping tools are available for C. canis, which has seriously impeded our understanding of the transmission of C. canis between humans and dogs. In a multilocus characterization of C. canis specimens from 12 HIV-infected persons from Lima, Peru, three were coinfected with C. hominis, indicating that some of the C. canis infections in humans were probably of anthroponotic origins (152).
Similar to C. canis in canine animals, C. felis is the dominant Cryptosporidium species in cats and other felines (148). It is commonly reported in humans in low- and middle-income countries. Possible transmission of C. felis between cats and humans has been reported (153). In Peru, some of the C. felis-infected AIDS patients were coinfected with C. hominis and C. meleagridis, suggesting that not all C. felis infections in humans are the result of zoonotic transmission (152). Recently, a subtyping tool based on sequence analysis of the gp60 gene has been developed for C. felis. It was used in the confirmation of two cases of zoonotic transmission of C. felis in Sweden (154). Thus far, it has been used in the characterization of human specimens from China, India, Ethiopia, Kenya, Nigeria, Jamaica, and Peru, suggesting potential host adaption as well as geographic isolation in C. felis (155).
Cryptosporidium viatorum was first reported in travelers back to Britain from India (89) and has been reported in humans in India, China, Ethiopia, Nigeria, and Colombia (32, 34, 67, 104, 125, 156–158). In particular, a high prevalence of C. viatorum (>10% of cryptosporidiosis cases) was recognized in children and HIV-positive persons in Ethiopia and Colombia (67, 125, 157). A subtyping tool based on sequence analysis of the gp60 gene is available for C. viatorum (159). Thus, far, four subtype families have been identified, including XVa, XVb, XVc, and XVd, but only XVa subtypes have been identified in humans (159). Most XVa subtypes have only three copies of the TCA repeat in the trinucleotide repeat region of the gp60 gene. As there are minor sequence differences downstream from it, eight subtypes (namely, XVaA3a to XVaA3h) are recognized among them (23, 34, 159). Recently, these four C. viatorum subtype families, namely, XVa (XVaA6, XVaA3g, and XVaA3h), XVb (XVbA2G1), XVc (XVcA2G1a and XVcA2G1b), and XVd (XVdA3), have been reported in wild rats in Australia and China (160–162). Previously, C. viatorum was thought to be an anthroponotic species (163). As subtypes XVaA3g and XVaA3h have been found in both humans and wild rats, C. viatorum infections are probably of the rat origin (34, 143, 161). This is also supported by the occurrence of another rat intestinal Cryptosporidium species, C. occultus, in humans in low- and middle-income countries (34, 164). These two species are rarely detected in humans in industrialized nations, probably a reflection of better hygiene there.
Cryptosporidium muris is another Cryptosporidium species in rats and some other rodents but has been found in humans in a few studies in Kenya, Nigeria, Thailand, India, Saudi Arabia, Colombia, and Peru (24, 42, 144, 165–171). In agreement with this, macaque monkeys in China are commonly infected with C. muris (172). Unlike other human-pathogenic Cryptosporidium spp., C. muris is a gastric pathogen with a much longer patent period (173). A Cryptosporidium species related to it, C. andersoni, has also been found in some human cases (24). Two studies reported a high prevalence of C. andersoni in immunocompetent children and adults in China (174, 175). A multilocus subtyping tool is available for characterizing the transmission of C. muris and C. andersoni (176).
Except for the five species mentioned above, other human-pathogenic species such as C. ubiquitum, C. cuniculus, and Cryptosporidium chipmunk genotype I are seldom reported in low- and middle-income countries (39). Cryptosporidium ubiquitum is commonly found in the United States. Subtyping analysis of C. ubiquitum isolates from humans, animals, and water based on sequence analysis of the gp60 gene indicated that C. ubiquitum-infected lambs and contaminated drinking water were likely the sources for human infections (93). Cryptosporidium cuniculus was first reported in rabbits in the United States but is a common human pathogen in the United Kingdom and New Zealand, leading to one large outbreak of human cryptosporidiosis in the United Kingdom (177, 178). Cryptosporidium chipmunk genotype I was first found in rodents in the United States (179). Subtype analysis revealed that isolates from humans and wild animals shared high genetic identity at the gp60 locus, supporting the occurrence of zoonotic transmission in humans (92).
CONCURRENT INFECTIONS WITH MIXED CRYPTOSPORIDIUM SPECIES, MULTIPLE EPISODES OF INFECTIONS, AND SECONDARY TRANSMISSION
One consequence of the high prevalence and diversity of Cryptosporidium spp. in humans in low- and middle-income countries is the concurrence of mixed Cryptosporidium species. Most of the mixed infections are caused by C. hominis and C. parvum (Table 4). The clinical significance of coinfections with multiple Cryptosporidium species is not clear. In one study in India, while the distribution of Cryptosporidium species was similar between symptomatic and asymptomatic children, a much higher occurrence (8.7% of the cryptosporidiosis cases compared with 0.6%) of mixed infections with C. hominis and C. parvum was seen in symptomatic children (42). This indicates that coinfection with two or more Cryptosporidium spp. might have deleterious effects on children.
TABLE 4.
Concurrent infections with mixed Cryptosporidium species in low- and middle-income countries in recent studiesa
| Location | Study population | No. of isolates | Mixed infections (n) | Genotyping technique(s) | Reference |
|---|---|---|---|---|---|
| Asia | |||||
| India | Children | 50 | C. hominis and C. meleagridis (2) | SSU rRNA-based PCR-RFLP | 256 |
| Immunocompromised children | 53 | C. hominis and C. meleagridis (5), C. hominis and C. parvum (2) | SSU rRNA-based PCR-RFLP, gp60-based PCR and sequencing | 117 | |
| Immunocompromised patients | 71 | C. hominis and C. parvum (2) | Species-specific DHFR-based PCR and qPCR | 50 | |
| Children | 473 | C. hominis and C. parvum (14), C. andersoni and C. muris (1) | SSU rRNA-based PCR-RFLP | 42 | |
| Bangladesh | Children | 268 | C. hominis and C. parvum (1), C. hominis and C. meleagridis (13), C. parvum and C. meleagridis (1) | SSU rRNA-based pan-Cryptosporidium qPCR | 10 |
| Cambodia | Children patients | 38 | C. hominis and C. parvum (1) | SSU rRNA-based qPCR and multiplex qPCR | 130 |
| Qatar | Adults | 38 | C. hominis and C. parvum (4), C. parvum and C. meleagridis (3) | SSU rRNA-based PCR-RFLP | 180 |
| Saudi Arabia | Children | 35 | C. hominis and C. parvum (1) | SSU rRNA-based and COWP-based PCR-RFLP | 166 |
| Africa | |||||
| Tunisia | HIV+ patients | 42 | C. hominis and C. meleagridis (1) | SSU rRNA-based PCR-RFLP | 99 |
| Egypt | Diarrheal patients | 18 | C. hominis and C. parvum (3) | COWP-based PCR-RFLP | 257 |
| Children | 14 | C. hominis and C. parvum (3) | SSU rRNA-based PCR-RFLP | 139 | |
| Ethiopia | HIV+ patients | 140 | C. hominis and C. parvum (1) | SSU rRNA-based PCR-RFLP | 67 |
| Mozambique | HIV+ patients | 9 | C. hominis and C. parvum (1) | SSU rRNA-based PCR-RFLP | 129 |
| Gambia | Children | 280 | C. hominis and C. parvum (5) | TaqMan array card-based qPCR | 31 |
| Nigeria | Children | 77 | C. hominis and C. parvum (4) | SSU rRNA-based PCR-RFLP | 258 |
| Children | 44 | C. hominis and C. parvum (4) | SSU rRNA-based PCR-RFLP | 242 | |
| HIV+ patients | 4 | C. hominis and C. meleagridis (1) | SSU rRNA-based PCR-RFLP | 259 | |
| Kenya | Diarrheal children | 151 | C. hominis and C. parvum (1) | SSU rRNA-based PCR-RFLP | 260 |
| Malawi | Diarrheal children | 43 | C. hominis and C. parvum (5) | SSU rRNA-based and COWP-based PCR-RFLP | 261 |
| Uganda | Diarrheal children | 444 | C. hominis and C. parvum (18) | COWP-based PCR-RFLP | 262 |
| Americas | |||||
| Brazil | HIV+ patients | 26 | C. hominis and C. parvum (1) | SSU rRNA-based PCR-RFLP | 105 |
| Argentina | HIV+ patients | 15 | C. hominis and C. parvum (2) | SSU rRNA-based PCR-RFLP | 105 |
| Peru | Children | 156 | C. hominis and C. parvum (2), C. canis and C. meleagridis (1) | SSU rRNA-based PCR-RFLP | 124 |
| Children and HIV+ patients | 55 | C. hominis and C. meleagridis (7) | SSU rRNA-based PCR and MLST | 147 |
HIV+, HIV positive; DHFR, dihydrofolate reductase; COWP, Cryptosporidium oocyst wall protein gene.
Restriction fragment length polymorphism (RFLP) analysis of PCR products is widely used in the detection of mixed Cryptosporidium species. Using this approach, the concurrence of C. hominis in 14 of 95 C. parvum-infected children and of C. andersoni in one C. muris-infected child was identified in a recent study in India (42). Another study in Egypt identified coinfection of C. parvum in 3 of 10 C. hominis-infected children (139). Coinfections of C. parvum and C. hominis were also identified in HIV-positive patients in Argentina, Brazil, and Mozambique (105, 129). Concurrence of C. hominis and C. meleagridis was identified in children and migrant workers in Tunisia and Qatar (99, 180). Coinfections of C. hominis and C. canis/C. felis were also identified in HIV-infected patients in Peru (151).
The occurrence of mixed Cryptosporidium species may be more common than believed, as shown in some studies in which multiple molecular tools were used in the identification and characterization of Cryptosporidium spp. With the application of species-specific quantitative PCR (qPCR) in a study conducted in Bangladesh, coinfection with C. parvum or C. hominis was identified in 1 and 13 of 100 C. meleagridis-infected children, respectively. Coinfection with C. hominis was further identified in one of five C. parvum-infected children (10). The use of a similar approach identified the concurrence of C. parvum in 5 of 124 C. hominis-infected children in Gambia (31). Coinfection with C. hominis was identified in 7 of 55 C. meleagridis-positive children and AIDS patents in Peru (147). Thus, accurate identification of coinfections with multiple Cryptosporidium species can improve our understanding of the transmission of zoonotic Cryptosporidium species in humans.
Children in low- and middle-income countries can experience multiple episodes of cryptosporidiosis, possibly reflecting short-lived or incomplete protection against Cryptosporidium infection after the primary infection (39, 181). In a longitudinal birth cohort study conducted in Bangladesh, 118 of 302 Cryptosporidium-positive children had two to four episodes of infections before reaching the age of 2 years, with no significant decreases in parasite burden during repeated infections (9). In another 3-year birth cohort study in India, 81% (322/397) of children with cryptosporidiosis experienced multiple episodes of infections, with no reduction in the severity of diarrhea during repeated infections (42). Unexpectedly, there was no species-specific protection against reinfections (16, 42, 124). Protective immunity, however, could exert effects at the subtype level, as multiple episodes of C. hominis infection in children were more likely to be caused by different subtype families in a longitudinal birth cohort study conducted in Peru (124). Similar findings were obtained in another birth cohort study in Bangladesh; although four children experienced repeated infections with C. hominis, subtyping analysis revealed that they were caused by heterogeneous subtypes (29).
As one of the risk factors involved in the acquisition of cryptosporidiosis is contact with Cryptosporidium-positive patients, secondary transmission of Cryptosporidium spp. in households is expected (33, 182). One case-control study of Cryptosporidium-infected children and their family members in Bangladesh demonstrated that the secondary infection rates were 35.8% (19/53) in urban case families and 7.8% (5/64) in rural case families. This was confirmed by results of subtype analysis of the C. hominis and C. parvum involved (25). The differences in rates of secondary transmission between the urban and rural study sites were attributed to differences in the dominant Cryptosporidium species (C. hominis versus C. meleagridis) and transmission routes (anthroponotic versus zoonotic) in the two communities. Cryptosporidium meleagridis appears to be less infectious than C. hominis (25). In a multicountry study conducted in sub-Saharan Africa, identical gp60 subtypes of C. parvum or C. hominis were detected among two or more contacts in 36% of the 108 initial Cryptosporidium-positive cases followed, indicating a common occurrence of secondary transmission of Cryptosporidium spp. (33). Among them, the C. hominis subtype IeA11G3T3 was involved in a cluster that lasted over 32 days with 13 infected subjects in two neighboring households, while the C. hominis subtype IfA14G1 was detected in a cluster that lasted over 18 days with 11 infected subjects in two neighboring households.
CRYPTOSPORIDIUM GENETICS AND VIRULENCE
The clinical implications of different Cryptosporidium species and subtypes in humans are not yet clear. Studies of genotyping analyses indicated that C. hominis and C. parvum likely behaved differently in humans, with the former causing more severe clinical manifestations (183). Earlier studies in urban slums in Peru and Brazil found that children infected with C. hominis had higher oocyst shedding intensity and longer duration than those infected with C. parvum and other species (124, 184, 185). Similar results were also obtained in immunocompromised patients in India (50). Children infected with C. hominis showed significantly more severe diarrhea than those infected with other species in South India (186). In one pediatric study in Kuwait, C. hominis infections showed more severe fever and diarrhea than C. parvum infections (121). In a study conducted in Ethiopia, AIDS patients with C. hominis infections had both diarrhea and vomiting, while those with C. parvum infections had only diarrhea (67). Similarly, in immunocompromised patients in India, nausea and vomiting were more frequently found in C. hominis infections than in C. parvum infections (50). Cryptosporidium hominis also appears to be more virulent than C. meleagridis. In a study conducted in two communities in Bangladesh, diarrhea was present in approximately 30% of C. hominis infections but nearly absent in C. meleagridis infections (10).
Due to the differences in pathogenicity, C. hominis infection is expected to have more deleterious nutritional effects on infected children than C. parvum infection. In Brazil, in children under 3 months, the height-for-age Z scores (HAZ) showed significant declines with infection of C. hominis or C. parvum, but in children of 3 to 6 months following infections, only C. hominis-infected children showed continuous decline in HAZ scores, especially for asymptomatic infections (185). Similar results have also been found in children in India, where most infections were caused by C. hominis, and children with sequential infections had significantly lower HAZ scores and long-term effects on growth (16). In Sonora, Mexico, malnutrition was significantly associated with Cryptosporidium infection, especially for C. hominis (122). In one recent birth cohort study conducted in two areas in Bangladesh, however, although the dominant Cryptosporidium species were different between urban (C. hominis) and rural (C. meleagridis) areas, they had similar effects on HAZ scores and child growth (10).
Even within the same Cryptosporidium species, the clinical manifestations of cryptosporidiosis can differ among subtypes. In Peruvian children infected with C. hominis, IbA10G2 was associated with diarrhea, nausea, vomiting, and general malaise, while other subtypes were associated only with diarrhea (124). This indicates that IbA10G2 is likely more virulent than other subtypes in C. hominis. This is partially support by the fact that almost all autochthonous C. hominis infections in Europe are caused by this subtype (187). In India, most cases with subtype Ib had vomiting and/or appetite loss, while all cases with Ia and Id showed chronic diarrhea (32). Differences in the severity of diarrhea have also been found between Ia and Id subtype families of C. hominis (77, 121).
To understand the differences in clinical symptoms and pathogenicity among C. hominis subtype families, population genetics and comparative genomics analyses have been used in the characterization of isolates. In Peru, genetic recombination was identified in the virulent subtype IbA10G2 by using comparative sequence analysis of 53 isolates at 32 genetic loci across chromosome 6. Using linkage disequilibrium and recombination analyses, limited genetic recombination was identified exclusively in the gp60 gene of IbA10G2, a major subtype responsible for the outbreaks of human cryptosporidiosis in industrialized nations. Intensive transmission of the virulent subtype IbA10G2 possibly had led to genetic recombination with other subtypes. In addition, selection for the IbA10G2 type sequence was detected in a 129-kb region around the gp60 gene, which had led to reduced sequence variation within the 129-kb region in the IbA10G2 subtype, reflecting the possible involvement of the gene or other linked genes in pathogenicity (188). This was supported by comparative genomics analysis of IbA10G2 isolates and other C. hominis subtypes in the United States, which revealed the occurrence of genetic recombination in IbA10G2 at the 5′ and 3′ ends of chromosome 6 and in the gp60 region, indicating that genetic recombination likely contributed to the emergence of these hypertransmissible subtypes (189). Comparative genomics analysis of the virulent IbA10G2 and other C. hominis subtypes in Europe has identified a loss-of-function mutation in the gene (cgd6_210) encoding COWP9 in chromosome 6 in all subtypes except IbA10G2. As expected, phylogenetic analysis of IbA10G2 and other C. hominis subtypes based on 743 coding sequences indicated that all the IbA10G2 genomes formed a unique clade in spite of the existence of some heterogeneity among the IbA10G2 isolates (190). Genetic recombination in C. hominis appears to be more common in low- and middle-income countries. A comparative genomics analysis of 32 C. hominis isolates from a longitudinal cohort study of children in a Bangladeshi community identified high rates of genetic recombination in the genomes, with the area around the gp60 gene being one of the seven highly polymorphic regions. Genetic recombination was confirmed by the decay of linkage disequilibrium in the C. hominis genome over <300 bp. Because of the common occurrence of genetic recombination, the relatedness of C. hominis genomes was not segregated by gp60 subtype (29). More Cryptosporidium species and subtypes should be sequenced to better understand the population structure and genetic determinants of virulence and high transmissibility in some Cryptosporidium species and subtypes (85, 191).
IMPLICATIONS FOR WASH (WATER, SANITATION, AND HYGIENE)-BASED INTERVENTION OF CRYPTOSPORIDIOSIS IN LOW- AND MIDDLE-INCOME COUNTRIES
The results of molecular epidemiological studies suggest that anthroponotic transmission plays a major role in the transmission of Cryptosporidium spp. in humans in low- and middle-income countries (103). Since cryptosporidiosis in these countries occurs mostly in children under 2 years (2–4), targeted intervention should be implemented to control the occurrence of cryptosporidiosis in this population. This intervention should be implemented regardless of the occurrence of diarrhea and other clinical symptoms, as subclinical cryptosporidiosis can induce malnutrition and growth retardation (10).
Water, sanitation, and hygiene (WASH)-based interventions have been used effectively in the prevention and control of diarrheal diseases (192). WASH is a collection of integrated prevention and control strategies for infectious diseases which aim to improve the provision of water (e.g., safe water source), sanitation (e.g., clean toilets), and hygiene (e.g., frequent handwashing) (193–195). Application of the WASH-based interventions, such as clean drinking water, toilets for sanitation, and handwashing with soaps for hygiene, can reduce the occurrence of diarrhea and thus has been recommended as a measure to interrupt the environmental transmission of enteric pathogens such as Cryptosporidium spp. (48, 196–199). It is estimated that WASH intervention can reduce infections with diarrhea by 15 to 50%, and up to 40% nonemergency cases can be eliminated by handwashing with soaps (200–202). In children less than 5 years in low-income countries, application of WASH interventions has led to a 27 to 56% reduction in diarrhea occurrence (203, 204). Among them, handwashing appears to be the most effective long-term WASH intervention (205). It has been shown to reduce the occurrence of giardiasis in young children in rural Bangladesh (206).
WASH-based interventions may be effective against cryptosporidiosis in low- and middle-income countries (81). The risk factors associated with cryptosporidiosis occurrence, including poor hygiene, unclean drinking water, open defecation, overcrowding, and diarrhea in household (66), are key targets of WASH interventions. Therefore, good personal hygiene practices are known to reduce the transmission of Cryptosporidium spp. and other intestinal parasites in children (207). This is further supported by the negative correlation between the occurrence of the C. parvum IIc subtype family in HIV-positive individuals and the percentage of the population with access to improved sanitation (126). The use of a point-of-use drinking water filter was shown recently to reduce the occurrence of Cryptosporidium infection in children in Rwanda (208).
FUTURE PERSPECTIVES
Data from molecular epidemiological studies of human cryptosporidiosis have significantly improved our understanding of the transmission of Cryptosporidium spp. in low- and middle-income countries. We now have a better understanding of the infection sources of Cryptosporidium spp. in children and immunocompromised persons. We also have a better appreciation of the role of endemicity of cryptosporidiosis in the genetic diversity of Cryptosporidium spp., concurrence of multiple Cryptosporidium species, and multiple episodes of infections. The eminent association between cryptosporidiosis occurrence and poor hygiene has made WASH (water, sanitation, and hygiene)-based interventions an economical intervention measure against cryptosporidiosis in low- and middle-income countries.
The number of molecular epidemiological studies of cryptosporidiosis conducted in low- and middle-income countries is small, considering the fact that the disease exerts its highest toll there. Most of the data on the distribution of Cryptosporidium species and C. parvum and C. hominis subtypes in humans in low- and middle-income settings have been generated from only a handful countries in Asia and South America by established research groups. As a result, our understanding of cryptosporidiosis epidemiology may be skewed by the underrepresentation of low-income countries where Cryptosporidium transmission is most intensive and risk factors for infections might be different (66). We have yet to take advantage of the recent large scale of global disease burden and mortality studies through genetic characterizations of Cryptosporidium spp. from these well-designed epidemiological investigations (6, 31). As molecular characterizations of specimens from longitudinal birth cohort studies have provided a wealth of data on the development of species- and subtype-specific immunity, differences in the virulence of Cryptosporidium species and subtypes, and intrafamilial transmission of Cryptosporidium spp. (9, 10, 29, 42, 124), these studies should be expanded to African nations. We also need to assess vigorously the effectiveness of existing WASH-based interventions for the prevention and control of cryptosporidiosis in humans in low- and middle-income countries (192, 209, 210). Improved hygiene education is urgently needed to enable longer lasting and improved WASH behaviors (211–214). As cryptosporidiosis is a major cause of malnutrition, the development and assessment of nutritional interventions are also urgently needed (72, 206).
The application of advanced molecular tools such as comparative genomics analyses could increase significantly the depth of research in molecular epidemiology of human cryptosporidiosis. The development of procedures for whole-genome sequencing of Cryptosporidium spp. in clinical specimens without pathogen isolation and passage in laboratory animals has increased the availability of whole-genome sequence data from major human-pathogenic Cryptosporidium species. Comparative analyses of these data have begun to shed light on the genetic determinants for host adaptation in some common C. parvum subtype families and the high virulence in some C. hominis subtype families. These studies have identified genetic recombination as the driving force for the emergence of host-adapted and virulent subtypes (85, 191, 215). Thus far, the number of comparative genomics studies has only been small in low- and middle-income countries (29).
The recent development of genetic manipulation tools for Cryptosporidium spp. promotes the identification and validation of new drug targets and development of new interventions against cryptosporidiosis (216, 217). Thus far, nitazoxanide remains the only drug approved by the U.S. Food and Drug Administration for treating cryptosporidiosis (218). With the application of CRISPR/Cas9 techniques, one Cryptosporidium PI(4)K inhibitor was identified as a candidate drug against cryptosporidiosis (219). Another study applying the CRISPR/Cas9 tool showed that C. parvum salvages purine nucleotides through a single pathway, providing another target for the development of treatments for this pathogen (220). Comprehensive studies combining genetic, biochemical, and chemical techniques indicated that bicyclic azetidines can kill C. parvum in mice by inhibiting the phenylalanyl-tRNA synthetase of parasites (221). The identification of these new targets should greatly facilitate the development of new drugs against cryptosporidiosis. The recent development of genetic tagging and conditional protein degradation systems might also facilitate studies on the genetic determinants of virulence in C. hominis and the identification of vaccine candidates in the Cryptosporidium proteome (222, 223).
Understanding the reasons for the dominance of C. parvum in humans and the common occurrence of its zoonotic subtype families IIa and IId in some areas would require the use of the “one health” approach. Thus far, the transmission of C. parvum IIa and IId subtypes in the Middle East and some other Muslim countries is poorly understood. This is largely due to the lack of genetic characterization of Cryptosporidium spp. in farm animals, especially ruminants, in these areas. Systematic sampling of both humans and ruminants in the same area, comparative analysis of C. parvum subtypes from humans and animals residing in the same households, and genotyping and subtyping of Cryptosporidium spp. in drinking source water would shed light on the infection sources and transmission routes of C. parvum in these areas. This would require collaboration among public health researchers, veterinarians, and environmental scientists, which has been advocated as a new measure for the prevention and control of zoonotic cryptosporidiosis (224–226).
ACKNOWLEDGMENTS
We thank our collaborators for molecular epidemiological studies of cryptosporidiosis.
Recent studies in this area were supported in part by the Guangdong Major Project of Basic and Applied Basic Research (grant no. 2020B0301030007), the National Natural Science Foundation of China (grant no. 31820103014 and U1901208), the 111 Project (grant no. D20008), and the Innovation Team Project of Guangdong University (grant no. 2019KCXTD001).
We declare no conflicts of interest.
Biographies

Xin Yang obtained his Ph.D. degree at the College of Veterinary Medicine, Huazhong Agricultural University, in 2018 and received postdoctoral training at the South China Agricultural University between 2018 and 2020. He is currently a lecturer at Northwest A & F University. Dr. Yang’s earlier research interest was mainly in the prevention and control of gastrointestinal nematodes in small ruminants. Since 2018, he has focused primarily on the molecular epidemiology and comparative genomics of Cryptosporidium in humans and animals. Dr. Yang has published over 20 original papers in international journals.

Yaqiong Guo obtained her Ph.D. degree and postdoctoral training at East China University of Science and Technology. She is currently an associate professor at South China Agricultural University. Her research interests are primarily molecular epidemiology and comparative genomics of foodborne and waterborne parasites, such as Cryptosporidium and Cyclospora. Dr. Guo has published over 20 original papers.

Lihua Xiao obtained his M.S. from Northeast Agricultural University in China, his Ph.D. from the University of Maine, and his postdoctoral training from The Ohio State University College of Veterinary Medicine. He worked at the Centers for Disease Control and Prevention, first as a guest researcher and then as a senior staff scientist, for 25 years. He is currently a professor at South China Agricultural University. For the last 30 years, Dr. Xiao has focused largely on the taxonomy, molecular epidemiology, diagnosis, genomics, and environmental biology of Cryptosporidium, Giardia, and microsporidia in humans and animals. Dr. Xiao has published over 400 original papers, invited reviews, and book chapters.

Yaoyu Feng obtained her B.S. and M.S. from Nankai University and her Ph.D. from Tianjin University, China. She received postdoctoral training and worked as a research fellow at the National University of Singapore. From 2004 to 2016, she was a professor at Tongji University and East China University of Science and Technology. Dr. Feng is currently a professor at the South China Agricultural University, China. She has been working on the molecular epidemiology, pathogenesis, transmission, and environmental ecology of waterborne and foodborne pathogens, including Cryptosporidium, Giardia, and microsporidia. Dr. Feng has published over 170 original papers, invited reviews, and book chapters.
Contributor Information
Lihua Xiao, Email: lxiao1961@gmail.com.
Yaoyu Feng, Email: yyfeng@scau.edu.cn.
REFERENCES
- 1.Liu J, Platts-Mills JA, Juma J, Kabir F, Nkeze J, Okoi C, Operario DJ, Uddin J, Ahmed S, Alonso PL, Antonio M, Becker SM, Blackwelder WC, Breiman RF, Faruque ASG, Fields B, Gratz J, Haque R, Hossain A, Hossain MJ, Jarju S, Qamar F, Iqbal NT, Kwambana B, Mandomando I, McMurry TL, Ochieng C, Ochieng JB, Ochieng M, Onyango C, Panchalingam S, Kalam A, Aziz F, Qureshi S, Ramamurthy T, Roberts JH, Saha D, Sow SO, Stroup SE, Sur D, Tamboura B, Taniuchi M, Tennant SM, Toema D, Wu Y, Zaidi A, Nataro JP, Kotloff KL, Levine MM, Houpt ER. 2016. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, Nasrin D, Blackwelder WC, Wu Y, Farag T, Panchalingham S, Sow SO, Sur D, Zaidi AKM, Faruque ASG, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ahmed S, Qureshi S, Quadri F, Hossain A, Das SK, Antonio M, Hossain MJ, Mandomando I, Acacio S, Biswas K, Tennant SM, Verweij JJ, Sommerfelt H, Nataro JP, Robins-Browne RM, Levine MM. 2019. The incidence, aetiology, and adverse clinical consequences of less severe diarrhoeal episodes among infants and children residing in low-income and middle-income countries: a 12-month case-control study as a follow-on to the Global Enteric Multicenter Study (GEMS). Lancet Glob Health 7:e568–e584. doi: 10.1016/S2214-109X(19)30076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roth GA, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, Abdollahpour I, Abdulkader RS, Abebe HT, Abebe M, Abebe Z, Abejie AN, Abera SF, Abil OZ, Abraha HN, Abrham AR, Abu-Raddad LJ, Accrombessi MMK, Acharya D, Adamu AA, Adebayo OM, Adedoyin RA, Adekanmbi V, Adetokunboh OO, Adhena BM, Adib MG, Admasie A, Afshin A, Agarwal G, Agesa KM, Agrawal A, Agrawal S, Ahmadi A, Ahmadi M, Ahmed MB, Ahmed S, Aichour AN, Aichour I, Aichour MTE, Akbari ME, Akinyemi RO, Akseer N, Al-Aly Z, Al-Eyadhy A, Al-Raddadi RM, Alahdab F, et al. 2018. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392:1736–1788. doi: 10.1016/S0140-6736(18)32203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platts-Mills JA, Liu J, Rogawski ET, Kabir F, Lertsethtakarn P, Siguas M, Khan SS, Praharaj I, Murei A, Nshama R, Mujaga B, Havt A, Maciel IA, McMurry TL, Operario DJ, Taniuchi M, Gratz J, Stroup SE, Roberts JH, Kalam A, Aziz F, Qureshi S, Islam MO, Sakpaisal P, Silapong S, Yori PP, Rajendiran R, Benny B, McGrath M, McCormick BJJ, Seidman JC, Lang D, Gottlieb M, Guerrant RL, Lima AAM, Leite JP, Samie A, Bessong PO, Page N, Bodhidatta L, Mason C, Shrestha S, Kiwelu I, Mduma ER, Iqbal NT, Bhutta ZA, Ahmed T, Haque R, Kang G, Kosek MN, MAL-ED Network Investigators. 2018. Use of quantitative molecular diagnostic methods to assess the aetiology, burden, and clinical characteristics of diarrhoea in children in low-resource settings: a reanalysis of the MAL-ED cohort study. Lancet Glob Health 6:e1309–e1318. doi: 10.1016/S2214-109X(18)30349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 6.Sow SO, Muhsen K, Nasrin D, Blackwelder WC, Wu Y, Farag TH, Panchalingam S, Sur D, Zaidi AK, Faruque AS, Saha D, Adegbola R, Alonso PL, Breiman RF, Bassat Q, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ahmed S, Qureshi S, Quadri F, Hossain A, Das SK, Antonio M, Hossain MJ, Mandomando I, Nhampossa T, Acacio S, Omore R, Oundo JO, Ochieng JB, Mintz ED, O'Reilly CE, Berkeley LY, Livio S, Tennant SM, Sommerfelt H, Nataro JP, Ziv-Baran T, Robins-Browne RM, Mishcherkin V, Zhang J, Liu J, Houpt ER, Kotloff KL, Levine MM. 2016. The burden of Cryptosporidium diarrheal disease among children < 24 months of age in moderate/high mortality regions of Sub-Saharan Africa and South Asia, utilizing data from the Global Enteric Multicenter Study (GEMS). PLoS Negl Trop Dis 10:e0004729. doi: 10.1371/journal.pntd.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG, Colombara DV, De Hostos EL, Engmann C, Guerrant RL, Haque R, Houpt ER, Kang G, Korpe PS, Kotloff KL, Lima AAM, Petri WA, Jr, Platts-Mills JA, Shoultz DA, Forouzanfar MH, Hay SI, Reiner RC, Jr, Mokdad AH. 2018. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob Health 6:e758–e768. doi: 10.1016/S2214-109X(18)30283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.GBD 2016 Diarrhoeal Disease Collaborators. 2018. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis 18:1211–1228. doi: 10.1016/S1473-3099(18)30362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korpe PS, Haque R, Gilchrist C, Valencia C, Niu F, Lu M, Ma JZ, Petri SE, Reichman D, Kabir M, Duggal P, Petri WA, Jr. 2016. Natural history of cryptosporidiosis in a longitudinal study of slum-dwelling Bangladeshi children: association with severe malnutrition. PLoS Negl Trop Dis 10:e0004564. doi: 10.1371/journal.pntd.0004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steiner KL, Ahmed S, Gilchrist CA, Burkey C, Cook H, Ma JZ, Korpe PS, Ahmed E, Alam M, Kabir M, Tofail F, Ahmed T, Haque R, Petri WA, Jr, Faruque ASG. 2018. Species of cryptosporidia causing subclinical infection associated with growth faltering in rural and urban Bangladesh: a birth cohort study. Clin Infect Dis 67:1347–1355. doi: 10.1093/cid/ciy310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quihui-Cota L, Morales-Figueroa GG, Javalera-Duarte A, Ponce-Martinez JA, Valbuena-Gregorio E, Lopez-Mata MA. 2017. Prevalence and associated risk factors for Giardia and Cryptosporidium infections among children of northwest Mexico: a cross-sectional study. BMC Public Health 17:852. doi: 10.1186/s12889-017-4822-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quihui-Cota L, Lugo-Flores CM, Ponce-Martínez JA, Morales-Figueroa GG. 2015. Cryptosporidiosis: a neglected infection and its association with nutritional status in schoolchildren in northwestern Mexico. J Infect Dev Ctries 9:878–883. doi: 10.3855/jidc.6751. [DOI] [PubMed] [Google Scholar]
- 13.Korpe PS, Valencia C, Haque R, Mahfuz M, McGrath M, Houpt E, Kosek M, McCormick BJJ, Penataro Yori P, Babji S, Kang G, Lang D, Gottlieb M, Samie A, Bessong P, Faruque ASG, Mduma E, Nshama R, Havt A, Lima IFN, Lima AAM, Bodhidatta L, Shreshtha A, Petri WA, Jr, Ahmed T, Duggal P. 2018. Epidemiology and risk factors for cryptosporidiosis in children from 8 low-income sites: results from the MAL-ED study. Clin Infect Dis 67:1660–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garzon M, Pereira-da-Silva L, Seixas J, Papoila AL, Alves M. 2018. Subclinical enteric parasitic infections and growth faltering in infants in Sao Tome, Africa: a birth cohort study. Int J Environ Res Public Health 15:688. doi: 10.3390/ijerph15040688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogawski ET, Liu J, Platts-Mills JA, Kabir F, Lertsethtakarn P, Siguas M, Khan SS, Praharaj I, Murei A, Nshama R, Mujaga B, Havt A, Maciel IA, Operario DJ, Taniuchi M, Gratz J, Stroup SE, Roberts JH, Kalam A, Aziz F, Qureshi S, Islam MO, Sakpaisal P, Silapong S, Yori PP, Rajendiran R, Benny B, McGrath M, Seidman JC, Lang D, Gottlieb M, Guerrant RL, Lima AAM, Leite JP, Samie A, Bessong PO, Page N, Bodhidatta L, Mason C, Shrestha S, Kiwelu I, Mduma ER, Iqbal NT, Bhutta ZA, Ahmed T, Haque R, Kang G, Kosek MN, Houpt ER, MAL-ED Network Investigators. 2018. Use of quantitative molecular diagnostic methods to investigate the effect of enteropathogen infections on linear growth in children in low-resource settings: longitudinal analysis of results from the MAL-ED cohort study. Lancet Glob Health 6:e1319–e1328. doi: 10.1016/S2214-109X(18)30351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ajjampur SS, Sarkar R, Sankaran P, Kannan A, Menon VK, Muliyil J, Ward H, Kang G. 2010. Symptomatic and asymptomatic Cryptosporidium infections in children in a semi-urban slum community in southern India. Am J Trop Med Hyg 83:1110–1115. doi: 10.4269/ajtmh.2010.09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhry N, Scott F, Edgar M, Sanger GJ, Kelly P. 2021. Reversal of pathogen-induced barrier defects in intestinal epithelial cells by contra-pathogenicity agents. Dig Dis Sci 66:88–104. doi: 10.1007/s10620-020-06121-9. [DOI] [PubMed] [Google Scholar]
- 18.Coutinho BP, Oriá RB, Vieira CM, Sevilleja JE, Warren CA, Maciel JG, Thompson MR, Pinkerton RC, Lima AA, Guerrant RL. 2008. Cryptosporidium infection causes undernutrition and, conversely, weanling undernutrition intensifies infection. J Parasitol 94:1225–1232. doi: 10.1645/GE-1411.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mondal D, Minak J, Alam M, Liu Y, Dai J, Korpe P, Liu L, Haque R, Petri WA, Jr. 2012. Contribution of enteric infection, altered intestinal barrier function, and maternal malnutrition to infant malnutrition in Bangladesh. Clin Infect Dis 54:185–192. doi: 10.1093/cid/cir807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Certad G, Viscogliosi E, Chabe M, Caccio SM. 2017. Pathogenic mechanisms of Cryptosporidium and Giardia. Trends Parasitol 33:561–576. doi: 10.1016/j.pt.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Checkley W, White AC, Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA, Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar R, Ajjampur SS, Prabakaran AD, Geetha JC, Sowmyanarayanan TV, Kane A, Duara J, Muliyil J, Balraj V, Naumova EN, Ward H, Kang G. 2013. Cryptosporidiosis among children in an endemic semiurban community in southern India: does a protected drinking water source decrease infection? Clin Infect Dis 57:398–406. doi: 10.1093/cid/cit288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, Feng Y. 2017. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol 8–9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuera A, Villamizar X, Herrera G, Giraldo JC, Vasquez-A LR, Urbano P, Villalobos O, Tovar C, Ramírez JD. 2020. Molecular detection and genotyping of intestinal protozoa from different biogeographical regions of Colombia. PeerJ 8:e8554. doi: 10.7717/peerj.8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korpe PS, Gilchrist C, Burkey C, Taniuchi M, Ahmed E, Madan V, Castillo R, Ahmed S, Arju T, Alam M, Kabir M, Ahmed T, Petri WA, Haque R, Faruque ASG, Duggal P. 2019. Case-control study of Cryptosporidium transmission in Bangladeshi households. Clin Infect Dis 68:1073–1079. doi: 10.1093/cid/ciy593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sannella AR, Suputtamongkol Y, Wongsawat E, Caccio SM. 2019. A retrospective molecular study of Cryptosporidium species and genotypes in HIV-infected patients from Thailand. Parasit Vectors 12:91. doi: 10.1186/s13071-019-3348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu F, Li D, Chang Y, Wu Y, Guo Z, Jia L, Xu J, Li J, Qi M, Wang R, Zhang L. 2019. Molecular characterization of three intestinal protozoans in hospitalized children with different disease backgrounds in Zhengzhou, central China. Parasit Vectors 12:543. doi: 10.1186/s13071-019-3800-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang N, Yu X, Zhang H, Cui L, Li X, Zhang X, Gong P, Li J, Li Z, Wang X, Li X, Li T, Liu X, Yu Y, Zhang X. 2020. Prevalence and genotyping of Cryptosporidium parvum in gastrointestinal cancer patients. J Cancer 11:3334–3339. doi: 10.7150/jca.42393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilchrist CA, Cotton JA, Burkey C, Arju T, Gilmartin A, Lin Y, Ahmed E, Steiner K, Alam M, Ahmed S, Robinson G, Zaman SU, Kabir M, Sanders M, Chalmers RM, Ahmed T, Ma JZ, Haque R, Faruque ASG, Berriman M, Petri WA. 2018. Genetic diversity of Cryptosporidium hominis in a Bangladeshi community as revealed by whole-genome sequencing. J Infect Dis 218:259–264. doi: 10.1093/infdis/jiy121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delahoy MJ, Omore R, Ayers TL, Schilling KA, Blackstock AJ, Ochieng JB, Moke F, Jaron P, Awuor A, Okonji C, Juma J, Farag TH, Nasrin D, Panchalingam S, Nataro JP, Kotloff KL, Levine MM, Oundo J, Roellig DM, Xiao L, Parsons MB, Laserson K, Mintz ED, Breiman RF, O'Reilly CE. 2018. Clinical, environmental, and behavioral characteristics associated with Cryptosporidium infection among children with moderate-to-severe diarrhea in rural western Kenya, 2008–2012: the Global Enteric Multicenter Study (GEMS). PLoS Negl Trop Dis 12:e0006640. doi: 10.1371/journal.pntd.0006640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hossain MJ, Saha D, Antonio M, Nasrin D, Blackwelder WC, Ikumapayi UN, Mackenzie GA, Adeyemi M, Jasseh M, Adegbola RA, Roose AW, Kotloff KL, Levine MM. 2019. Cryptosporidium infection in rural Gambian children: epidemiology and risk factors. PLoS Negl Trop Dis 13:e0007607. doi: 10.1371/journal.pntd.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khalil S, Mirdha BR, Panda A, Singh Y, Makharia G, Paul J. 2017. Cryptosporidium species subtypes and associated clinical manifestations in Indian patients. Gastroenterol Hepatol Bed Bench 10:311–318. [PMC free article] [PubMed] [Google Scholar]
- 33.Krumkamp R, Aldrich C, Maiga-Ascofare O, Mbwana J, Rakotozandrindrainy N, Borrmann S, Caccio SM, Rakotozandrindrainy R, Adegnika AA, Lusingu JPA, Amuasi J, May J, Eibach D, CRYPTO Study Group. 9 March 2020. Transmission of Cryptosporidium spp. among human and animal local contact networks in sub-Saharan Africa: a multi-country study. Clin Infect Dis doi: 10.1093/cid/ciaa223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu N, Liu H, Jiang Y, Yin J, Yuan Z, Shen Y, Cao J. 2020. First report of Cryptosporidium viatorum and Cryptosporidium occultus in humans in China, and of the unique novel C. viatorum subtype XVaA3h. BMC Infect Dis 20:16. doi: 10.1186/s12879-019-4693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manouana GP, Lorenz E, Mbong Ngwese M, Nguema Moure PA, Maiga Ascofaré O, Akenten CW, Amuasi J, Rakotozandrindrainy N, Rakotozandrindrainy R, Mbwana J, Lusingu J, Byrne N, Melhem S, Zinsou JF, Adegbite RB, Hogan B, Winter D, May J, Kremsner PG, Borrmann S, Eibach D, Adegnika AA. 2020. Performance of a rapid diagnostic test for the detection of Cryptosporidium spp. in African children admitted to hospital with diarrhea. PLoS Negl Trop Dis 14:e0008448. doi: 10.1371/journal.pntd.0008448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galvan-Diaz AL, Bedoya-Urrego K, Medina-Lozano A, Uran-Velasquez J, Alzate JF, Garcia-Montoya G. 2020. Common occurrence of Cryptosporidium hominis in children attending day-care centers in Medellin, Colombia. Parasitol Res 119:2935–2942. doi: 10.1007/s00436-020-06782-5. [DOI] [PubMed] [Google Scholar]
- 37.Madadi S, Mahami-Oskouei M, Rafeey M, Spotin A, Aminisani N, Mahami-Oskouei L, Ghoyounchi R, Berahmat R. 2020. Comparative evaluation of Cryptosporidium infection in malnourished and well-nourished children: parasitic infections are affected by the interaction of nutritional status and socio-demographic characteristics. Comp Immunol Microbiol Infect Dis 68:101406. doi: 10.1016/j.cimid.2019.101406. [DOI] [PubMed] [Google Scholar]
- 38.Sinyangwe N, Siwila J, Muma J, Chola M, Michelo C. 2020. Factors associated with Cryptosporidium infection among adult HIV positive population in contact with livestock in Namwala District, Zambia. Front Public Health 8:74. doi: 10.3389/fpubh.2020.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao L, Cama V. 2018. Cryptosporidium and cryptosporidiosis, p 73–117. In Ortega Y, Sterling C (ed), Food microbiology and food safety. Springer, Berlin, Germany. [Google Scholar]
- 40.Gerace E, Lo Presti VDM, Biondo C. 2019. Cryptosporidium infection: epidemiology, pathogenesis, and differential diagnosis. Eur J Microbiol Immunol (Bp) 9:119–123. doi: 10.1556/1886.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan A, Shaik JS, Grigg ME. 2018. Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop 184:1–14. doi: 10.1016/j.actatropica.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 42.Kattula D, Jeyavelu N, Prabhakaran AD, Premkumar PS, Velusamy V, Venugopal S, Geetha JC, Lazarus RP, Das P, Nithyanandhan K, Gunasekaran C, Muliyil J, Sarkar R, Wanke C, Ajjampur SSR, Babji S, Naumova EN, Ward HD, Kang G. 2017. Natural history of cryptosporidiosis in a birth cohort in Southern India. Clin Infect Dis 64:347–354. doi: 10.1093/cid/ciw730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levine MM, Nasrin D, Acacio S, Bassat Q, Powell H, Tennant SM, Sow SO, Sur D, Zaidi AKM, Faruque ASG, Hossain MJ, Alonso PL, Breiman RF, O'Reilly CE, Mintz ED, Omore R, Ochieng JB, Oundo JO, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ahmed S, Qureshi S, Quadri F, Hossain A, Das SK, Antonio M, Saha D, Mandomando I, Blackwelder WC, Farag T, Wu Y, Houpt ER, Verweiij JJ, Sommerfelt H, Nataro JP, Robins-Browne RM, Kotloff KL. 2020. Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Glob Health 8:e204–e214. doi: 10.1016/S2214-109X(19)30541-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Painter JE, Hlavsa MC, Collier SA, Xiao L, Yoder JS, Centers for Disease Control and Prevention. 2015. Cryptosporidiosis surveillance—United States, 2011–2012. MMWR Suppl 64:1–14. [PubMed] [Google Scholar]
- 45.Mohebali M, Yimam Y, Woreta A. 2020. Cryptosporidium infection among people living with HIV/AIDS in Ethiopia: a systematic review and meta-analysis. Pathog Glob Health 114:183–193. doi: 10.1080/20477724.2020.1746888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utami WS, Murhandarwati EH, Artama WT, Kusnanto H. 2020. Cryptosporidium infection increases the risk for chronic diarrhea among people living with HIV in Southeast Asia: a systematic review and meta-analysis. Asia Pac J Public Health 32:8–18. doi: 10.1177/1010539519895422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang ZD, Liu Q, Liu HH, Li S, Zhang L, Zhao YK, Zhu XQ. 2018. Prevalence of Cryptosporidium, microsporidia and Isospora infection in HIV-infected people: a global systematic review and meta-analysis. Parasit Vectors 11:28. doi: 10.1186/s13071-017-2558-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmadpour E, Safarpour H, Xiao L, Zarean M, Hatam-Nahavandi K, Barac A, Picot S, Rahimi MT, Rubino S, Mahami-Oskouei M, Spotin A, Nami S, Baghi HB. 2020. Cryptosporidiosis in HIV-positive patients and related risk factors: a systematic review and meta-analysis. Parasite 27:27. doi: 10.1051/parasite/2020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu A, Gong B, Liu X, Shen Y, Wu Y, Zhang W, Cao J. 2020. A retrospective epidemiological analysis of human Cryptosporidium infection in China during the past three decades (1987–2018). PLoS Negl Trop Dis 14:e0008146. doi: 10.1371/journal.pntd.0008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dey A, Ghoshal U, Agarwal V, Ghoshal UC. 2016. Genotyping of Cryptosporidium species and their clinical manifestations in patients with renal transplantation and human immunodeficiency virus infection. J Pathog 2016:2623602. doi: 10.1155/2016/2623602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mohaghegh MA, Hejazi SH, Ghomashlooyan M, Kalani H, Mirzaei F, Azami M. 2017. Prevalence and clinical features of Cryptosporidium infection in hemodialysis patients. Gastroenterol Hepatol Bed Bench 10:137–142. [PMC free article] [PubMed] [Google Scholar]
- 52.Ghoshal U, Ranjan P, Dey A, Ghoshal UC. 2018. Intestinal cryptosporidiosis in renal transplant recipients: prevalence, species detection and comparative evaluation of SSU rRNA and Cryptosporidium oocyst wall protein genes. Indian J Med Microbiol 36:247–250. doi: 10.4103/ijmm.IJMM_18_179. [DOI] [PubMed] [Google Scholar]
- 53.Esteghamati A, Khanaliha K, Bokharaei-Salim F, Sayyahfar S, Ghaderipour M. 2019. Prevalence of intestinal parasitic infection in cancer, organ transplant and primary immunodeficiency patients in Tehran, Iran. Asian Pac J Cancer Prev 20:495–501. doi: 10.31557/APJCP.2019.20.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caner A, Zorbozan O, Tunalı V, Kantar M, Aydoğdu S, Aksoylar S, Gürüz Y, Turgay N. 2020. Intestinal protozoan parasitic infections in immunocompromised child patients with diarrhea. Jpn J Infect Dis 73:187–192. doi: 10.7883/yoken.JJID.2019.054. [DOI] [PubMed] [Google Scholar]
- 55.Taghipour A, Olfatifar M, Rostami A, Foroutan M, Vasigala V, Norouzi M. 2020. Intestinal parasites in hemodialysis patients from developing countries: a systematic review and meta-analysis. Hemodial Int 24:12–21. doi: 10.1111/hdi.12796. [DOI] [PubMed] [Google Scholar]
- 56.El-Kady AM, Fahmi Y, Tolba M, Hashim AA, Hassan AA. 2018. Cryptosporidium infection in chronic kidney disease patients undergoing hemodialysis in Egypt. J Parasit Dis 42:630–635. doi: 10.1007/s12639-018-1046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shehata AI, Hassanein F, Abdul-Ghani R. 2019. Opportunistic parasitoses among Egyptian hemodialysis patients in relation to CD4+ T-cell counts: a comparative study. BMC Infect Dis 19:480. doi: 10.1186/s12879-019-4110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zueter AM, Hijjawi NS, Hamadeneh KN, Al-Sheyab MM, Hatamleh AM. 2019. Cryptosporidiosis among hemodialysis patients in Jordan: first preliminary screening surveillance. Trop Med Infect Dis 4:131. doi: 10.3390/tropicalmed4040131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shad S, Hanif F, Ul Haq M, Luck NH, Aziz T, Mubarak M. 2019. Frequencies of common infectious organisms causing chronic diarrhea in renal transplant patients. Exp Clin Transplant 17:212–215. doi: 10.6002/ect.MESOT2018.P69. [DOI] [PubMed] [Google Scholar]
- 60.Chalmers RM, Davies AP, Tyler K. 2019. Cryptosporidium. Microbiology (Reading) 165:500–502. doi: 10.1099/mic.0.000764. [DOI] [PubMed] [Google Scholar]
- 61.Painter JE, Gargano JW, Yoder JS, Collier SA, Hlavsa MC. 2016. Evolving epidemiology of reported cryptosporidiosis cases in the United States, 1995–2012. Epidemiol Infect 144:1792–1802. doi: 10.1017/S0950268815003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Squire SA, Ryan U. 2017. Cryptosporidium and Giardia in Africa: current and future challenges. Parasit Vectors 10:195. doi: 10.1186/s13071-017-2111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMurry TL, McQuade ETR, Liu J, Kang G, Kosek MN, Lima AAM, Bessong PO, Samie A, Haque R, Mduma ER, Leite JP, Bodhidatta L, Iqbal N, Page N, Kiwelu I, Bhutta ZA, Ahmed T, Houpt ER, Platts-Mills JA. 9 October 2020. Duration of post-diarrheal enteric pathogen carriage in young children in low-resource settings. Clin Infect Dis doi: 10.1093/cid/ciaa1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wojcik GL, Korpe P, Marie C, Mentzer AJ, Carstensen T, Mychaleckyj J, Kirkpatrick BD, Rich SS, Concannon P, Faruque ASG, Haque R, Petri WA, Jr, Duggal P. 2020. Genome-wide association study of cryptosporidiosis in infants implicates PRKCA. mBio 11:e03343-19. doi: 10.1128/mBio.03343-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schnee AE, Haque R, Taniuchi M, Uddin MJ, Alam MM, Liu J, Rogawski ET, Kirkpatrick B, Houpt ER, Petri WA, Jr, Platts-Mills JA. 2018. Identification of etiology-specific diarrhea associated with linear growth faltering in Bangladeshi infants. Am J Epidemiol 187:2210–2218. doi: 10.1093/aje/kwy106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bouzid M, Kintz E, Hunter PR. 2018. Risk factors for Cryptosporidium infection in low and middle income countries: a systematic review and meta-analysis. PLoS Negl Trop Dis 12:e0006553. doi: 10.1371/journal.pntd.0006553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, Feng Y, Xiao L. 2014. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis 8:e2831. doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shalaby NM, Shalaby NM. 2015. Cryptosporidium parvum infection among Egyptian school children. J Egypt Soc Parasitol 45:125–131. doi: 10.12816/0010858. [DOI] [PubMed] [Google Scholar]
- 69.Conan A, O'Reilly CE, Ogola E, Ochieng JB, Blackstock AJ, Omore R, Ochieng L, Moke F, Parsons MB, Xiao L, Roellig D, Farag TH, Nataro JP, Kotloff KL, Levine MM, Mintz ED, Breiman RF, Cleaveland S, Knobel DL. 2017. Animal-related factors associated with moderate-to-severe diarrhea in children younger than five years in western Kenya: a matched case-control study. PLoS Negl Trop Dis 11:e0005795. doi: 10.1371/journal.pntd.0005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bauhofer AFL, Cossa-Moiane I, Marques S, Guimarães EL, Munlela B, Anapakala E, Chilaúle JJ, Cassocera M, Langa JS, Chissaque A, Sambo J, Manhique-Coutinho L, Bero DM, Kellogg TA, de Deus N. 2020. Intestinal protozoan infections among children 0–168 months with diarrhea in Mozambique: June 2014 - January 2018. PLoS Negl Trop Dis 14:e0008195. doi: 10.1371/journal.pntd.0008195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tombang AN, Ambe NF, Bobga TP, Nkfusai CN, Collins NM, Ngwa SB, Diengou NH, Cumber SN. 2019. Prevalence and risk factors associated with cryptosporidiosis among children within the ages 0–5 years attending the Limbe regional hospital, southwest region, Cameroon. BMC Public Health 19:1144. doi: 10.1186/s12889-019-7484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Creek TL, Kim A, Lu L, Bowen A, Masunge J, Arvelo W, Smit M, Mach O, Legwaila K, Motswere C, Zaks L, Finkbeiner T, Povinelli L, Maruping M, Ngwaru G, Tebele G, Bopp C, Puhr N, Johnston SP, Dasilva AJ, Bern C, Beard RS, Davis MK. 2010. Hospitalization and mortality among primarily nonbreastfed children during a large outbreak of diarrhea and malnutrition in Botswana, 2006. J Acquir Immune Defic Syndr 53:14–19. doi: 10.1097/QAI.0b013e3181bdf676. [DOI] [PubMed] [Google Scholar]
- 73.Al-Mekhlafi HM, Mahdy MA, Azlin MY, Fatmah MS, Norhayati M. 2011. Childhood Cryptosporidium infection among aboriginal communities in Peninsular Malaysia. Ann Trop Med Parasitol 105:135–143. doi: 10.1179/136485911X12899838683368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benedict KM, Collier SA, Marder EP, Hlavsa MC, Fullerton KE, Yoder JS. 2019. Case-case analyses of cryptosporidiosis and giardiasis using routine national surveillance data in the United States—2005–2015. Epidemiol Infect 147:e178. doi: 10.1017/S0950268819000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Costa D, Razakandrainibe R, Valot S, Vannier M, Sautour M, Basmaciyan L, Gargala G, Viller V, Lemeteil D, Ballet JJ, Dalle F, Favennec L, French National Network on Surveillance of Human Cryptosporidiosis. 2020. Epidemiology of cryptosporidiosis in France from 2017 to 2019. Microorganisms 8:1358. doi: 10.3390/microorganisms8091358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goncalves EM, da Silva AJ, Eduardo MB, Uemura IH, Moura IN, Castilho VL, Corbett CE. 2006. Multilocus genotyping of Cryptosporidium hominis associated with diarrhea outbreak in a day care unit in Sao Paulo. Clinics (Sao Paulo) 61:119–126. doi: 10.1590/s1807-59322006000200006. [DOI] [PubMed] [Google Scholar]
- 77.Feng Y, Wang L, Duan L, Gomez-Puerta LA, Zhang L, Zhao X, Hu J, Zhang N, Xiao L. 2012. Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg Infect Dis 18:312–314. doi: 10.3201/eid1802.110666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hijjawi N, Zahedi A, Kazaleh M, Ryan U. 2017. Prevalence of Cryptosporidium species and subtypes in paediatric oncology and non-oncology patients with diarrhoea in Jordan. Infect Genet Evol 55:127–130. doi: 10.1016/j.meegid.2017.08.033. [DOI] [PubMed] [Google Scholar]
- 79.Gharpure R, Perez A, Miller AD, Wikswo ME, Silver R, Hlavsa MC. 2019. Cryptosporidiosis outbreaks—United States, 2009–2017. MMWR Morb Mortal Wkly Rep 68:568–572. doi: 10.15585/mmwr.mm6825a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caccio SM, Chalmers RM. 2016. Human cryptosporidiosis in Europe. Clin Microbiol Infect 22:471–480. doi: 10.1016/j.cmi.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 81.Ryan U, Hijjawi N, Xiao L. 2018. Foodborne cryptosporidiosis. Int J Parasitol 48:1–12. doi: 10.1016/j.ijpara.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 82.Feng Y, Ryan UM, Xiao L. 2018. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol 34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 83.Garcia RJ, Pita AB, Velathanthiri N, French NP, Hayman DTS. 2020. Species and genotypes causing human cryptosporidiosis in New Zealand. Parasitol Res 119:2317–2326. doi: 10.1007/s00436-020-06729-w. [DOI] [PubMed] [Google Scholar]
- 84.Nazemalhosseini-Mojarad E, Feng Y, Xiao L. 2012. The importance of subtype analysis of Cryptosporidium spp. in epidemiological investigations of human cryptosporidiosis in Iran and other Mideast countries. Gastroenterol Hepatol Bed Bench 5:67–70. [PMC free article] [PubMed] [Google Scholar]
- 85.Morris A, Robinson G, Swain MT, Chalmers RM. 2019. Direct sequencing of Cryptosporidium in stool samples for public health. Front Public Health 7:360. doi: 10.3389/fpubh.2019.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loeck BK, Pedati C, Iwen PC, McCutchen E, Roellig DM, Hlavsa MC, Fullerton K, Safranek T, Carlson AV. 2020. Genotyping and subtyping Cryptosporidium to identify risk factors and transmission patterns—Nebraska, 2015–2017. MMWR Morb Mortal Wkly Rep 69:335–338. doi: 10.15585/mmwr.mm6912a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deshpande AP, Jones BL, Connelly L, Pollock KG, Brownlie S, Alexander CL. 2015. Molecular characterization of Cryptosporidium parvum isolates from human cryptosporidiosis cases in Scotland. Parasitology 142:318–325. doi: 10.1017/S0031182014001346. [DOI] [PubMed] [Google Scholar]
- 88.Abal-Fabeiro JL, Maside X, Llovo J, Bartolome C. 2015. Aetiology and epidemiology of human cryptosporidiosis cases in Galicia (NW Spain), 2000–2008. Epidemiol Infect 143:3022–3035. doi: 10.1017/S0950268815000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elwin K, Hadfield SJ, Robinson G, Chalmers RM. 2012. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000–2008. Epidemiol Infect 140:673–683. doi: 10.1017/S0950268811000860. [DOI] [PubMed] [Google Scholar]
- 90.Insulander M, Silverlas C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. 2013. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect 141:1009–1020. doi: 10.1017/S0950268812001665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, McEvoy JM. 2006. Evidence supporting zoonotic transmission of Cryptosporidium in Wisconsin. J Clin Microbiol 44:4303–4308. doi: 10.1128/JCM.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo Y, Cebelinski E, Matusevich C, Alderisio KA, Lebbad M, McEvoy J, Roellig DM, Yang C, Feng Y, Xiao L. 2015. Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. J Clin Microbiol 53:1648–1654. doi: 10.1128/JCM.03436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li N, Xiao L, Alderisio K, Elwin K, Cebelinski E, Chalmers R, Santin M, Fayer R, Kvac M, Ryan U, Sak B, Stanko M, Guo Y, Wang L, Zhang L, Cai J, Roellig D, Feng Y. 2014. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis 20:217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chalmers RM, Elwin K, Hadfield SJ, Robinson G. 2011. Sporadic human cryptosporidiosis caused by Cryptosporidium cuniculus, United Kingdom, 2007–2008. Emerg Infect Dis 17:536–538. doi: 10.3201/eid1703.100410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khan A, Shams S, Khan S, Khan MI, Khan S, Ali A. 2019. Evaluation of prevalence and risk factors associated with Cryptosporidium infection in rural population of district Buner, Pakistan. PLoS One 14:e0209188. doi: 10.1371/journal.pone.0209188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang T, Fan Y, Koehler AV, Ma G, Li T, Hu M, Gasser RB. 2017. First survey of Cryptosporidium, Giardia and Enterocytozoon in diarrhoeic children from Wuhan, China. Infect Genet Evol 51:127–131. doi: 10.1016/j.meegid.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 97.Kassouha M, Soukkarieh C, Alkhaled A. 2016. First genotyping of Cryptosporidium spp. in pre-weaned calves, broiler chickens and children in Syria by PCR-RFLP analysis. Vet Parasitol 225:86–90. doi: 10.1016/j.vetpar.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 98.Garcia RJ, French N, Pita A, Velathanthiri N, Shrestha R, Hayman D. 2017. Local and global genetic diversity of protozoan parasites: spatial distribution of Cryptosporidium and Giardia genotypes. PLoS Negl Trop Dis 11:e0005736. doi: 10.1371/journal.pntd.0005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Essid R, Menotti J, Hanen C, Aoun K, Bouratbine A. 2018. Genetic diversity of Cryptosporidium isolates from human populations in an urban area of Northern Tunisia. Infect Genet Evol 58:237–242. doi: 10.1016/j.meegid.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 100.Naguib D, El-Gohary AH, Roellig D, Mohamed AA, Arafat N, Wang Y, Feng Y, Xiao L. 2018. Molecular characterization of Cryptosporidium spp. and Giardia duodenalis in children in Egypt. Parasit Vectors 11:403. doi: 10.1186/s13071-018-2981-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Urrea-Quezada A, Sotelo-Cruz N, Gonzalez-Diaz M, Valenzuela O. 2019. Follow-up of a case of cryptosporidiosis in a toddler from Mexico: response to the treatment. J Glob Infect Dis 11:129–130. doi: 10.4103/jgid.jgid_119_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Efunshile AM, Ezeanosike O, Onyekachi ONI, Ugwu MI, Konig B, Robertson LJ. 2018. Apparent absence of Giardia infections among children under 5-years of age with acute watery diarrhoea in Abakaliki, Nigeria. Epidemiol Infect 147:e58. doi: 10.1017/S0950268818003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol 124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 104.Ukwah BN, Ezeonu IM, Ezeonu CT, Roellig D, Xiao L. 2017. Cryptosporidium species and subtypes in diarrheal children and HIV-infected persons in Ebonyi and Nsukka, Nigeria. J Infect Dev Ctries 11:173–179. doi: 10.3855/jidc.8034. [DOI] [PubMed] [Google Scholar]
- 105.Peralta RHS, Velasquez JN, Cunha FD, Pantano ML, Sodre FC, da Silva S, Astudillo OG, Peralta JM, Carnevale S. 2016. Genetic diversity of Cryptosporidium identified in clinical samples from cities in Brazil and Argentina. Mem Inst Oswaldo Cruz 111:30–36. doi: 10.1590/0074-02760150303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gatei W, Hart CA, Gilman RH, Das P, Cama V, Xiao L. 2006. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J Eukaryot Microbiol 53:S43–S48. doi: 10.1111/j.1550-7408.2006.00169.x. [DOI] [PubMed] [Google Scholar]
- 107.Adamu H, Petros B, Hailu A, Petry F. 2010. Molecular characterization of Cryptosporidium isolates from humans in Ethiopia. Acta Trop 115:77–83. doi: 10.1016/j.actatropica.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 108.Iqbal A, Lim YA, Surin J, Sim BL. 2012. High diversity of Cryptosporidium subgenotypes identified in Malaysian HIV/AIDS individuals targeting gp60 gene. PLoS One 7:e31139. doi: 10.1371/journal.pone.0031139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feng Y, Xiao L. 2017. Molecular epidemiology of cryptosporidiosis in China. Front Microbiol 8:1701. doi: 10.3389/fmicb.2017.01701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li N, Wang R, Cai M, Jiang W, Feng Y, Xiao L. 2019. Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int J Parasitol 49:569–577. doi: 10.1016/j.ijpara.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qi M, Zhang K, Huang M, Wang S, Xu C, Wang T, Jing B, Li J. 2020. Longitudinal detection of Cryptosporidium spp. in 1–10-week-old dairy calves on a farm in Xinjiang, China. Parasitol Res 119:3839–3844. doi: 10.1007/s00436-020-06904-z. [DOI] [PubMed] [Google Scholar]
- 112.Wu Y, Zhang K, Zhang Y, Jing B, Chen Y, Xu C, Wang T, Qi M, Zhang L. 2020. Genetic diversity of Cryptosporidium parvum in neonatal dairy calves in Xinjiang, China. Pathogens 9:692. doi: 10.3390/pathogens9090692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Z, Hu S, Zhao W, Guo Y, Li N, Zheng Z, Zhang L, Kváč M, Xiao L, Feng Y. 2020. Population structure and geographical segregation of Cryptosporidium parvum IId subtypes in cattle in China. Parasit Vectors 13:425. doi: 10.1186/s13071-020-04303-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lim YA, Iqbal A, Surin J, Sim BL, Jex AR, Nolan MJ, Smith HV, Gasser RB. 2011. First genetic classification of Cryptosporidium and Giardia from HIV/AIDS patients in Malaysia. Infect Genet Evol 11:968–974. doi: 10.1016/j.meegid.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 115.Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, Liu L, Feng Y, Xiao L. 2013. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol 51:557–563. doi: 10.1128/JCM.02758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hira KG, Mackay MR, Hempstead AD, Ahmed S, Karim MM, O'Connor RM, Hibberd PL, Calderwood SB, Ryan ET, Khan WA, Ward HD. 2011. Genetic diversity of Cryptosporidium spp. from Bangladeshi children. J Clin Microbiol 49:2307–2310. doi: 10.1128/JCM.00164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sharma P, Sharma A, Sehgal R, Malla N, Khurana S. 2013. Genetic diversity of Cryptosporidium isolates from patients in North India. Int J Infect Dis 17:e601–e605. doi: 10.1016/j.ijid.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 118.Santin M. 2020. Cryptosporidium and Giardia in ruminants. Vet Clin North Am Food Anim Pract 36:223–238. doi: 10.1016/j.cvfa.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 119.Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao LH. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol 43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hijjawi N, Ng J, Yang R, Atoum MF, Ryan U. 2010. Identification of rare and novel Cryptosporidium GP60 subtypes in human isolates from Jordan. Exp Parasitol 125:161–164. doi: 10.1016/j.exppara.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 121.Iqbal J, Khalid N, Hira PR. 2011. Cryptosporidiosis in Kuwaiti children: association of clinical characteristics with Cryptosporidium species and subtypes. J Med Microbiol 60:647–652. doi: 10.1099/jmm.0.028001-0. [DOI] [PubMed] [Google Scholar]
- 122.Urrea-Quezada A, Gonzalez-Diaz M, Villegas-Gomez I, Durazo M, Hernandez J, Xiao L, Valenzuela O. 2018. Clinical manifestations of cryptosporidiosis and identification of a new Cryptosporidium subtype in patients from Sonora, Mexico. Pediatr Infect Dis J 37:e136–e138. doi: 10.1097/INF.0000000000001762. [DOI] [PubMed] [Google Scholar]
- 123.Avendano C, Ramo A, Vergara-Castiblanco C, Sanchez-Acedo C, Quilez J. 2018. Genetic uniqueness of Cryptosporidium parvum from dairy calves in Colombia. Parasitol Res 117:1317–1323. doi: 10.1007/s00436-018-5818-6. [DOI] [PubMed] [Google Scholar]
- 124.Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, Gilman RH, Xiao L. 2008. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis 14:1567–1574. doi: 10.3201/eid1410.071273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sanchez A, Munoz M, Gomez N, Tabares J, Segura L, Salazar A, Restrepo C, Ruiz M, Reyes P, Qian Y, Xiao L, Lopez MC, Ramirez JD. 2017. Molecular epidemiology of Giardia, Blastocystis and Cryptosporidium among indigenous children from the Colombian Amazon Basin. Front Microbiol 8:248. doi: 10.3389/fmicb.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.King P, Tyler KM, Hunter PR. 2019. Anthroponotic transmission of Cryptosporidium parvum predominates in countries with poorer sanitation: a systematic review and meta-analysis. Parasit Vectors 12:16. doi: 10.1186/s13071-018-3263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blanco MA, Montoya A, Iborra A, Fuentes I. 2014. Identification of Cryptosporidium subtype isolates from HIV-seropositive patients in Equatorial Guinea. Trans R Soc Trop Med Hyg 108:594–596. doi: 10.1093/trstmh/tru108. [DOI] [PubMed] [Google Scholar]
- 128.Eibach D, Krumkamp R, Al-Emran HM, Sarpong N, Hagen RM, Adu-Sarkodie Y, Tannich E, May J. 2015. Molecular characterization of Cryptosporidium spp. among children in rural Ghana. PLoS Negl Trop Dis 9:e0003551. doi: 10.1371/journal.pntd.0003551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Casmo V, Lebbad M, Maungate S, Lindh J. 2018. Occurrence of Cryptosporidium spp. and Cystoisospora belli among adult patients with diarrhoea in Maputo, Mozambique. J Eukaryot Microbiol 4:e00769. doi: 10.1016/j.heliyon.2018.e00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Moore CE, Elwin K, Phot N, Seng C, Mao S, Suy K, Kumar V, Nader J, Bousfield R, Perera S, Bailey JW, Beeching NJ, Day NPJ, Parry CM, Chalmers RM. 2016. Molecular characterization of Cryptosporidium species and Giardia duodenalis from symptomatic Cambodian children. PLoS Negl Trop Dis 10:e0004822. doi: 10.1371/journal.pntd.0004822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Akiyoshi DE, Tumwine JK, Bakeera-Kitaka S, Tzipori S. 2006. Subtype analysis of Cryptosporidium isolates from children in Uganda. J Parasitol 92:1097–1100. doi: 10.1645/GE-843R.1. [DOI] [PubMed] [Google Scholar]
- 132.Nazemalhosseini-Mojarad E, Haghighi A, Taghipour N, Keshavarz A, Mohebi SR, Zali MR, Xiao L. 2011. Subtype analysis of Cryptosporidium parvum and Cryptosporidium hominis isolates from humans and cattle in Iran. Vet Parasitol 179:250–252. doi: 10.1016/j.vetpar.2011.01.051. [DOI] [PubMed] [Google Scholar]
- 133.Hijjawi N, Mukbel R, Yang R, Ryan U. 2016. Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Vet Parasitol 228:116–120. doi: 10.1016/j.vetpar.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 134.Ibrahim MA, Abdel-Ghany AE, Abdel-Latef GK, Abdel-Aziz SA, Aboelhadid SM. 2016. Epidemiology and public health significance of Cryptosporidium isolated from cattle, buffaloes, and humans in Egypt. Parasitol Res 115:2439–2448. doi: 10.1007/s00436-016-4996-3. [DOI] [PubMed] [Google Scholar]
- 135.Rahmouni I, Essid R, Aoun K, Bouratbine A. 2014. Glycoprotein 60 diversity in Cryptosporidium parvum causing human and cattle cryptosporidiosis in the rural region of Northern Tunisia. Am J Trop Med Hyg 90:346–350. doi: 10.4269/ajtmh.13-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Majeed QAH, El-Azazy OME, Abdou NMI, Al-Aal ZA, El-Kabbany AI, Tahrani LMA, AlAzemi MS, Wang Y, Feng Y, Xiao L. 2018. Epidemiological observations on cryptosporidiosis and molecular characterization of Cryptosporidium spp. in sheep and goats in Kuwait. Parasitol Res 117:1631–1636. doi: 10.1007/s00436-018-5847-1. [DOI] [PubMed] [Google Scholar]
- 137.Amer S, Zidan S, Feng Y, Adamu H, Li N, Xiao L. 2013. Identity and public health potential of Cryptosporidium spp. in water buffalo calves in Egypt. Vet Parasitol 191:123–127. doi: 10.1016/j.vetpar.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 138.Mahfouz ME, Mira N, Amer S. 2014. Prevalence and genotyping of Cryptosporidium spp. in farm animals in Egypt. J Vet Med Sci 76:1569–1575. doi: 10.1292/jvms.14-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gharieb RMA, Merwad AMA, Saleh AA, Abd El-Ghany AM. 2018. Molecular screening and genotyping of Cryptosporidium species in household dogs and in-contact children in Egypt: risk factor analysis and zoonotic importance. Vector Borne Zoonotic Dis 18:424–432. doi: 10.1089/vbz.2017.2254. [DOI] [PubMed] [Google Scholar]
- 140.Omoruyi B, Matongo F, Nkwetshana NT, Green E, Clarke AM, Ndip RN. 2011. Environmental and demographic risk factors associated with the prevalence of Cryptosporidium infection in the Alice rural settlements of the Eastern Cape Province of South Africa: a pilot study. Rev Environ Health 26:127–133. doi: 10.1515/reveh.2011.017. [DOI] [PubMed] [Google Scholar]
- 141.Sirisena UM, Iddawela WM, Noordeen F, Wickramasinghe S. 2014. Prevalence and identification of Cryptosporidium species in paediatric patients with diarrhoea. Ceylon Med J 59:75–78. doi: 10.4038/cmj.v59i3.7467. [DOI] [PubMed] [Google Scholar]
- 142.Lebbad M, Beser J, Insulander M, Karlsson L, Mattsson JG, Svenungsson B, Axen C. 2013. Unusual cryptosporidiosis cases in Swedish patients: extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. Parasitology 140:1735–1740. doi: 10.1017/S003118201300084X. [DOI] [PubMed] [Google Scholar]
- 143.Braima K, Zahedi A, Oskam C, Reid S, Pingault N, Xiao L, Ryan U. 2019. Retrospective analysis of Cryptosporidium species in western Australian human populations (2015–2018), and emergence of the C. hominis IfA12G1R5 subtype. Infect Genet Evol 73:306–313. doi: 10.1016/j.meegid.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 144.Ayinmode AB, Oliveira BCM, Obebe OO, Dada-Adgebola HO, Ayede AI, Widmer G. 2018. Genotypic characterization of Cryptosporidium species in humans and peri-domestic animals in Ekiti and Oyo States, Nigeria. J Parasitol 104:639–644. doi: 10.1645/17-74. [DOI] [PubMed] [Google Scholar]
- 145.Stensvold CR, Beser J, Axen C, Lebbad M. 2014. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. J Clin Microbiol 52:2311–2319. doi: 10.1128/JCM.00598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liao C, Wang T, Koehler AV, Fan Y, Hu M, Gasser RB. 2018. Molecular investigation of Cryptosporidium in farmed chickens in Hubei Province, China, identifies 'zoonotic' subtypes of C. meleagridis. Parasit Vectors 11:484. doi: 10.1186/s13071-018-3056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang Y, Yang W, Cama V, Wang L, Cabrera L, Ortega Y, Bern C, Feng Y, Gilman R, Xiao L. 2014. Population genetics of Cryptosporidium meleagridis in humans and birds: evidence for cross-species transmission. Int J Parasitol 44:515–521. doi: 10.1016/j.ijpara.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 148.Lucio-Forster A, Griffiths JK, Cama VA, Xiao L, Bowman DD. 2010. Minimal zoonotic risk of cryptosporidiosis from pet dogs and cats. Trends Parasitol 26:174–179. doi: 10.1016/j.pt.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 149.Certad G, Ngouanesavanh T, Hernan A, Rojas E, Contreras R, Pocaterra L, Nunez L, Dei-Cas E, Guyot K. 2006. First molecular data on cryptosporidiosis in Venezuela. J Eukaryot Microbiol 53:S30–S32. doi: 10.1111/j.1550-7408.2006.00165.x. [DOI] [PubMed] [Google Scholar]
- 150.Dacal E, Saugar JM, de Lucio A, Hernandez-de-Mingo M, Robinson E, Koster PC, Aznar-Ruiz-de-Alegria ML, Espasa M, Ninda A, Gandasegui J, Sulleiro E, Moreno M, Salvador F, Molina I, Rodriguez E, Carmena D. 2018. Prevalence and molecular characterization of Strongyloides stercoralis, Giardia duodenalis, Cryptosporidium spp., and Blastocystis spp. isolates in school children in Cubal, Western Angola. Parasit Vectors 11:67. doi: 10.1186/s13071-018-2640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xiao L, Cama VA, Cabrera L, Ortega Y, Pearson J, Gilman RH. 2007. Possible transmission of Cryptosporidium canis among children and a dog in a household. J Clin Microbiol 45:2014–2016. doi: 10.1128/JCM.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cama V, Gilman RH, Vivar A, Ticona E, Ortega Y, Bern C, Xiao L. 2006. Mixed Cryptosporidium infections and HIV. Emerg Infect Dis 12:1025–1028. doi: 10.3201/eid1206.060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Beser J, Toresson L, Eitrem R, Troell K, Winiecka-Krusnell J, Lebbad M. 2015. Possible zoonotic transmission of Cryptosporidium felis in a household. Infect Ecol Epidemiol 5:28463. doi: 10.3402/iee.v5.28463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rojas-Lopez L, Elwin K, Chalmers RM, Enemark HL, Beser J, Troell K. 2020. Development of a gp60-subtyping method for Cryptosporidium felis. Parasit Vectors 13:39. doi: 10.1186/s13071-020-3906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jiang W, Roellig DM, Lebbad M, Beser J, Troell K, Guo Y, Li N, Xiao L, Feng Y. 2020. Subtype distribution of zoonotic pathogen Cryptosporidium felis in humans and animals in several countries. Emerg Microbes Infect 9:2446–2454. doi: 10.1080/22221751.2020.1840312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ayinmode AB, Zhang H, Dada-Adegbola HO, Xiao L. 2014. Cryptosporidium hominis subtypes and Enterocytozoon bieneusi genotypes in HIV-infected persons in Ibadan, Nigeria. Zoonoses Public Health 61:297–303. doi: 10.1111/zph.12072. [DOI] [PubMed] [Google Scholar]
- 157.de Lucio A, Amor-Aramendia A, Bailo B, Saugar JM, Anegagrie M, Arroyo A, Lopez-Quintana B, Zewdie D, Ayehubizu Z, Yizengaw E, Abera B, Yimer M, Mulu W, Hailu T, Herrador Z, Fuentes I, Carmena D. 2016. Prevalence and genetic diversity of Giardia duodenalis and Cryptosporidium spp. among school children in a rural area of the Amhara Region, North-West Ethiopia. PLoS One 11:e0159992. doi: 10.1371/journal.pone.0159992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Khalil S, Mirdha BR, Paul J, Panda A, Singh Y. 2018. Molecular detection and identification of Cryptosporidium viatorum in a human immunodeficiency virus-seropositive patient. J Glob Infect Dis 10:28–29. doi: 10.4103/jgid.jgid_26_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stensvold CR, Elwin K, Winiecka-Krusnell J, Chalmers RM, Xiao L, Lebbad M. 2015. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J Clin Microbiol 53:1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhao W, Zhou H, Huang Y, Xu L, Rao L, Wang S, Wang W, Yi Y, Zhou X, Wu Y, Ma T, Wang G, Hu X, Peng R, Yin F, Lu G. 2019. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: molecular detection, species/genotype identification and implications for public health. Int J Parasitol Parasites Wildl 9:317–321. doi: 10.1016/j.ijppaw.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen YW, Zheng WB, Zhang NZ, Gui BZ, Lv QY, Yan JQ, Zhao Q, Liu GH. 2019. Identification of Cryptosporidium viatorum XVa subtype family in two wild rat species in China. Parasit Vectors 12:502. doi: 10.1186/s13071-019-3763-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Koehler AV, Wang T, Haydon SR, Gasser RB. 2018. Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus—an emerging zoonotic pathogen? Int J Parasitol Parasites Wildl 7:18–26. doi: 10.1016/j.ijppaw.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM. 2012. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int J Parasitol 42:675–682. doi: 10.1016/j.ijpara.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 164.Kvac M, Vlnata G, Jezkova J, Horcickova M, Konecny R, Hlaskova L, McEvoy J, Sak B. 2018. Cryptosporidium occultus sp. n. (Apicomplexa: Cryptosporidiidae) in rats. Eur J Protistol 63:96–104. doi: 10.1016/j.ejop.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 165.Gatei W, Wamae CN, Mbae C, Waruru A, Mulinge E, Waithera T, Gatika SM, Kamwati SK, Revathi G, Hart CA. 2006. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am J Trop Med Hyg 75:78–82. doi: 10.4269/ajtmh.2006.75.78. [DOI] [PubMed] [Google Scholar]
- 166.Al-Brikan FA, Salem HS, Beeching N, Hilal N. 2008. Multilocus genetic analysis of Cryptosporidium isolates from Saudi Arabia. J Egypt Soc Parasitol 38:645–658. [PubMed] [Google Scholar]
- 167.Muthusamy D, Rao SS, Ramani S, Monica B, Banerjee I, Abraham OC, Mathai DC, Primrose B, Muliyil J, Wanke CA, Ward HD, Kang G. 2006. Multilocus genotyping of Cryptosporidium sp. isolates from human immunodeficiency virus-infected individuals in South India. J Clin Microbiol 44:632–634. doi: 10.1128/JCM.44.2.632-634.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tiangtip R, Jongwutiwes S. 2002. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop Med Int Health 7:357–364. doi: 10.1046/j.1365-3156.2002.00855.x. [DOI] [PubMed] [Google Scholar]
- 169.Katsumata T, Hosea D, Ranuh IG, Uga S, Yanagi T, Kohno S. 2000. Short report: possible Cryptosporidium muris infection in humans. Am J Trop Med Hyg 62:70–72. doi: 10.4269/ajtmh.2000.62.70. [DOI] [PubMed] [Google Scholar]
- 170.Palmer CJ, Xiao L, Terashima A, Guerra H, Gotuzzo E, Saldias G, Bonilla JA, Zhou L, Lindquist A, Upton SJ. 2003. Cryptosporidium muris, a rodent pathogen, recovered from a human in Peru. Emerg Infect Dis 9:1174–1176. doi: 10.3201/eid0909.030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gatei W, Ashford RW, Beeching NJ, Kamwati SK, Greensill J, Hart CA. 2002. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg Infect Dis 8:204–206. doi: 10.3201/eid0802.010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen L, Hu S, Jiang W, Zhao J, Li N, Guo Y, Liao C, Han Q, Feng Y, Xiao L. 2019. Cryptosporidium parvum and Cryptosporidium hominis subtypes in crab-eating macaques. Parasit Vectors 12:350. doi: 10.1186/s13071-019-3604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chappell CL, Okhuysen PC, Langer-Curry RC, Lupo PJ, Widmer G, Tzipori S. 2015. Cryptosporidium muris: infectivity and illness in healthy adult volunteers. Am J Trop Med Hyg 92:50–55. doi: 10.4269/ajtmh.14-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jiang Y, Ren J, Yuan Z, Liu A, Zhao H, Liu H, Chu L, Pan W, Cao J, Lin Y, Shen Y. 2014. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect Dis 14:555. doi: 10.1186/s12879-014-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu H, Shen Y, Yin J, Yuan Z, Jiang Y, Xu Y, Pan W, Hu Y, Cao J. 2014. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect Dis 14:25. doi: 10.1186/1471-2334-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Feng Y, Yang W, Ryan U, Zhang L, Kvac M, Koudela B, Modry D, Li N, Fayer R, Xiao L. 2011. Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni. J Clin Microbiol 49:34–41. doi: 10.1128/JCM.01329-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Xiao L, Ryan U, Modha D, Mallaghan C. 2009. Cryptosporidium sp. rabbit genotype, a newly identified human pathogen. Emerg Infect Dis 15:829–830. doi: 10.3201/eid1505.081419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Puleston RL, Mallaghan CM, Modha DE, Hunter PR, Nguyen-Van-Tam JS, Regan CM, Nichols GL, Chalmers RM. 2014. The first recorded outbreak of cryptosporidiosis due to Cryptosporidium cuniculus (formerly rabbit genotype), following a water quality incident. J Water Health 12:41–50. doi: 10.2166/wh.2013.097. [DOI] [PubMed] [Google Scholar]
- 179.Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, Reid M, Xiao L. 2007. Cryptosporidium genotypes in wildlife from a New York watershed. Appl Environ Microbiol 73:6475–6483. doi: 10.1128/AEM.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Boughattas S, Behnke JM, Al-Sadeq D, Ismail A, Abu-Madi M. 2019. Cryptosporidium spp., prevalence, molecular characterisation and socio-demographic risk factors among immigrants in Qatar. PLoS Negl Trop Dis 13:e0007750. doi: 10.1371/journal.pntd.0007750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rogawski McQuade ET, Liu J, Kang G, Kosek MN, Lima AAM, Bessong PO, Samie A, Haque R, Mduma ER, Shrestha S, Leite JP, Bodhidatta L, Iqbal N, Page N, Kiwelu I, Bhutta Z, Ahmed T, Houpt ER, Platts-Mills JA. 2020. Protection from natural immunity against enteric infections and etiology-specific diarrhea in a longitudinal birth cohort. J Infect Dis 222:1858–1868. doi: 10.1093/infdis/jiaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Johansen ØH, Hanevik K, Thrana F, Carlson A, Stachurska-Hagen T, Skaare D, Robertson LJ. 2015. Symptomatic and asymptomatic secondary transmission of Cryptosporidium parvum following two related outbreaks in schoolchildren. Epidemiol Infect 143:1702–1709. doi: 10.1017/S095026881400243X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wang RJ, Li JQ, Chen YC, Zhang LX, Xiao LH. 2018. Widespread occurrence of Cryptosporidium infections in patients with HIV/AIDS: epidemiology, clinical feature, diagnosis, and therapy. Acta Trop 187:257–263. doi: 10.1016/j.actatropica.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 184.Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Cabrera L, Gilman RH, Lal AA. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J Infect Dis 183:492–497. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- 185.Bushen OY, Kohli A, Pinkerton RC, Dupnik K, Newman RD, Sears CL, Fayer R, Lima AA, Guerrant RL. 2007. Heavy cryptosporidial infections in children in northeast Brazil: comparison of Cryptosporidium hominis and Cryptosporidium parvum. Trans R Soc Trop Med Hyg 101:378–384. doi: 10.1016/j.trstmh.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 186.Ajjampur SS, Gladstone BP, Selvapandian D, Muliyil JP, Ward H, Kang G. 2007. Molecular and spatial epidemiology of cryptosporidiosis in children in a semiurban community in South India. J Clin Microbiol 45:915–920. doi: 10.1128/JCM.01590-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Chalmers RM, Robinson G, Elwin K, Elson R. 2019. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors 12:95. doi: 10.1186/s13071-019-3354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Li N, Xiao L, Cama VA, Ortega Y, Gilman RH, Guo M, Feng Y. 2013. Genetic recombination and Cryptosporidium hominis virulent subtype IbA10G2. Emerg Infect Dis 19:1573–1582. doi: 10.3201/eid1910.121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Guo Y, Tang K, Rowe LA, Li N, Roellig DM, Knipe K, Frace M, Yang C, Feng Y, Xiao L. 2015. Comparative genomic analysis reveals occurrence of genetic recombination in virulent Cryptosporidium hominis subtypes and telomeric gene duplications in Cryptosporidium parvum. BMC Genomics 16:320. doi: 10.1186/s12864-015-1517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sikora P, Andersson S, Winiecka-Krusnell J, Hallstrom B, Alsmark C, Troell K, Beser J, Arrighi RB. 2017. Genomic variation in IbA10G2 and other patient-derived Cryptosporidium hominis subtypes. J Clin Microbiol 55:844–858. doi: 10.1128/JCM.01798-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fan Y, Feng Y, Xiao L. 2019. Comparative genomics: how has it advanced our knowledge of cryptosporidiosis epidemiology? Parasitol Res 118:3195–3204. doi: 10.1007/s00436-019-06537-x. [DOI] [PubMed] [Google Scholar]
- 192.Ross AG, Rahman M, Alam M, Zaman K, Qadri F. 2020. Can we ‘WaSH’ infectious diseases out of slums? Int J Infect Dis 92:130–132. doi: 10.1016/j.ijid.2020.01.014. [DOI] [PubMed] [Google Scholar]
- 193.Prasad A, Gray CB, Ross A, Kano M. 2016. Metrics in urban health: current developments and future prospects. Annu Rev Public Health 37:113–133. doi: 10.1146/annurev-publhealth-032315-021749. [DOI] [PubMed] [Google Scholar]
- 194.Ross AGP, Zaman K, Clemens JD. 2019. Health concerns in urban slums: a glimpse of things to come? JAMA 321:1973–1974. doi: 10.1001/jama.2019.3774. [DOI] [PubMed] [Google Scholar]
- 195.Turley R, Saith R, Bhan N, Rehfuess E, Carter B. 2013. Slum upgrading strategies involving physical environment and infrastructure interventions and their effects on health and socio-economic outcomes. Cochrane Database Syst Rev 31:CD010067. doi: 10.1002/14651858.CD010067.pub2. [DOI] [PubMed] [Google Scholar]
- 196.Pickering AJ, Null C, Winch PJ, Mangwadu G, Arnold BF, Prendergast AJ, Njenga SM, Rahman M, Ntozini R, Benjamin-Chung J, Stewart CP, Huda TMN, Moulton LH, Colford JM, Jr, Luby SP, Humphrey JH. 2019. The WASH benefits and SHINE trials: interpretation of WASH intervention effects on linear growth and diarrhoea. Lancet Glob Health 7:e1139–e1146. doi: 10.1016/S2214-109X(19)30268-2. [DOI] [PubMed] [Google Scholar]
- 197.Chard AN, Garn JV, Chang HH, Clasen T, Freeman MC. 2019. Impact of a school-based water, sanitation, and hygiene intervention on school absence, diarrhea, respiratory infection, and soil-transmitted helminths: results from the WASH HELPS cluster-randomized trial. J Glob Health 9:020402. doi: 10.7189/jogh.09.020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.GBD Diarrhoeal Diseases Collaborators. 2017. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis 17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Julian TR. 2016. Environmental transmission of diarrheal pathogens in low and middle income countries. Environ Sci Process Impacts 18:944–955. doi: 10.1039/c6em00222f. [DOI] [PubMed] [Google Scholar]
- 200.Schmidt WP, Cairncross S. 2009. Household water treatment in poor populations: is there enough evidence for scaling up now? Environ Sci Technol 43:986–992. doi: 10.1021/es802232w. [DOI] [PubMed] [Google Scholar]
- 201.Clasen T, Alexander K, Sinclair D, Boisson S, Peletz R, Chang H, Majorin F, Cairncross S. 2015. Interventions to improve water quality for preventing diarrhoea. Cochrane Database Syst Rev 2015:CD004794. doi: 10.1002/14651858.CD004794.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fewtrell L, Kaufmann RB, Kay D, Enanoria W, Haller L, Colford JM, Jr. 2005. Water, sanitation, and hygiene interventions to reduce diarrhoea in less developed countries: a systematic review and meta-analysis. Lancet Infect Dis 5:42–52. doi: 10.1016/S1473-3099(04)01253-8. [DOI] [PubMed] [Google Scholar]
- 203.Darvesh N, Das JK, Vaivada T, Gaffey MF, Rasanathan K, Bhutta ZA, Social Determinants of Health Study Team. 2017. Water, sanitation and hygiene interventions for acute childhood diarrhea: a systematic review to provide estimates for the Lives Saved Tool. BMC Public Health 17:776. doi: 10.1186/s12889-017-4746-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dreibelbis R, Freeman MC, Greene LE, Saboori S, Rheingans R. 2014. The impact of school water, sanitation, and hygiene interventions on the health of younger siblings of pupils: a cluster-randomized trial in Kenya. Am J Public Health 104:e91–e97. doi: 10.2105/AJPH.2013.301412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kang JY, Aldstadt J. 2019. Examining time-dependent effects of water, sanitation, and hygiene (WASH) interventions using an agent-based model. Trop Med Int Health 24:962–971. doi: 10.1111/tmi.13280. [DOI] [PubMed] [Google Scholar]
- 206.Lin A, Ercumen A, Benjamin-Chung J, Arnold BF, Das S, Haque R, Ashraf S, Parvez SM, Unicomb L, Rahman M, Hubbard AE, Stewart CP, Colford JM, Jr, Luby SP. 2018. Effects of water, sanitation, handwashing, and nutritional interventions on child enteric protozoan infections in rural Bangladesh: a cluster-randomized controlled trial. Clin Infect Dis 67:1515–1522. doi: 10.1093/cid/ciy320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Omarova A, Tussupova K, Berndtsson R, Kalishev M, Sharapatova K. 2018. Protozoan parasites in drinking water: a system approach for improved water, sanitation and hygiene in developing countries. Int J Environ Res Public Health 15:495. doi: 10.3390/ijerph15030495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zambrano LD, Priest JW, Ivan E, Rusine J, Nagel C, Kirby M, Rosa G, Clasen TF. 2017. Use of serologic responses against enteropathogens to assess the impact of a point-of-use water filter: a randomized controlled trial in western province, Rwanda. Am J Trop Med Hyg 97:876–887. doi: 10.4269/ajtmh.16-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.McGinnis SM, McKeon T, Desai R, Ejelonu A, Laskowski S, Murphy HM. 2017. A systematic review: costing and financing of water, sanitation, and hygiene (WASH) in schools. Int J Environ Res Public Health 14:442. doi: 10.3390/ijerph14040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.McMichael C. 2019. Water, sanitation and hygiene (WASH) in schools in low-income countries: a review of evidence of impact. Int J Environ Res Public Health 16:359. doi: 10.3390/ijerph16030359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Gera T, Shah D, Sachdev HS. 2018. Impact of water, sanitation and hygiene interventions on growth, non-diarrheal morbidity and mortality in children residing in low- and middle-income countries: a systematic review. Indian Pediatr 55:381–393. doi: 10.1007/s13312-018-1279-3. [DOI] [PubMed] [Google Scholar]
- 212.Ejemot-Nwadiaro RI, Ehiri JE, Arikpo D, Meremikwu MM, Critchley JA. 2015. Hand washing promotion for preventing diarrhoea. Cochrane Database Syst Rev 2015:CD004265. doi: 10.1002/14651858.CD004265.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Phan HT, Tran HTT, Tran HTM, Dinh APP, Ngo HT, Theorell-Haglow J, Gordon CJ. 2018. An educational intervention to improve hand hygiene compliance in Vietnam. BMC Infect Dis 18:116. doi: 10.1186/s12879-018-3029-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.McKittrick TR, Jacobsen KH. 2014. Oral hygiene practices among middle-school students in 44 low- and middle-income countries. Int Dent J 64:164–170. doi: 10.1111/idj.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Thompson RCA, Ash A. 2019. Molecular epidemiology of Giardia and Cryptosporidium infections—what's new? Infect Genet Evol 75:103951. doi: 10.1016/j.meegid.2019.103951. [DOI] [PubMed] [Google Scholar]
- 216.Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, Cipriano MJ, Striepen B. 2015. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523:477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Vinayak S. 2020. Recent advances in genetic manipulation of Cryptosporidium. Curr Opin Microbiol 58:146–152. doi: 10.1016/j.mib.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wang B, Castellanos-Gonzalez A, White AC, Jr. 2020. Novel drug targets for treatment of cryptosporidiosis. Expert Opin Ther Targets 24:915–922. doi: 10.1080/14728222.2020.1785432. [DOI] [PubMed] [Google Scholar]
- 219.Manjunatha UH, Vinayak S, Zambriski JA, Chao AT, Sy T, Noble CG, Bonamy GMC, Kondreddi RR, Zou B, Gedeck P, Brooks CF, Herbert GT, Sateriale A, Tandel J, Noh S, Lakshminarayana SB, Lim SH, Goodman LB, Bodenreider C, Feng G, Zhang L, Blasco F, Wagner J, Leong FJ, Striepen B, Diagana TT. 2017. A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature 546:376–380. doi: 10.1038/nature22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pawlowic MC, Somepalli M, Sateriale A, Herbert GT, Gibson AR, Cuny GD, Hedstrom L, Striepen B. 2019. Genetic ablation of purine salvage in Cryptosporidium parvum reveals nucleotide uptake from the host cell. Proc Natl Acad Sci U S A 116:21160–21165. doi: 10.1073/pnas.1908239116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Vinayak S, Jumani RS, Miller P, Hasan MM, McLeod BI, Tandel J, Stebbins EE, Teixeira JE, Borrel J, Gonse A, Zhang M, Yu X, Wernimont A, Walpole C, Eckley S, Love MS, McNamara CW, Sharma M, Sharma A, Scherer CA, Kato N, Schreiber SL, Melillo B, Striepen B, Huston CD, Comer E. 2020. Bicyclic azetidines kill the diarrheal pathogen Cryptosporidium in mice by inhibiting parasite phenylalanyl-tRNA synthetase. Sci Transl Med 12:eaba8412. doi: 10.1126/scitranslmed.aba8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Choudhary HH, Nava MG, Gartlan BE, Rose S, Vinayak S. 2020. A conditional protein degradation system to study essential gene function in Cryptosporidium parvum. mBio 11:e01231-20. doi: 10.1128/mBio.01231-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wilke G, Funkhouser-Jones LJ, Wang Y, Ravindran S, Wang Q, Beatty WL, Baldridge MT, VanDussen KL, Shen B, Kuhlenschmidt MS, Kuhlenschmidt TB, Witola WH, Stappenbeck TS, Sibley LD. 2019. A stem-cell-derived platform enables complete Cryptosporidium development in vitro and genetic tractability. Cell Host Microbe 26:123–134.e8. doi: 10.1016/j.chom.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Innes EA, Chalmers RM, Wells B, Pawlowic MC. 2020. A One Health approach to tackle cryptosporidiosis. Trends Parasitol 36:290–303. doi: 10.1016/j.pt.2019.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hernandez-Gallo N, Hernandez-Florez LJ, Cortes-Vecino JA. 2018. Cryptosporidiosis and “One Health.” Rev Salud Publica (Bogota) 20:138–143. (In Spanish.) doi: 10.15446/rsap.v20n1.69959. [DOI] [PubMed] [Google Scholar]
- 226.Ryan U, Zahedi A, Paparini A. 2016. Cryptosporidium in humans and animals—a one health approach to prophylaxis. Parasite Immunol 38:535–547. doi: 10.1111/pim.12350. [DOI] [PubMed] [Google Scholar]
- 227.Ryan U, Fayer R, Xiao L. 2014. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology 141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- 228.Yang Y, Zhou Y, Cheng W, Pan X, Xiao P, Shi Y, Gao J, Song X, Chen Y, Jiang Q. 2017. Prevalence and determinants of Cryptosporidium infection in an underdeveloped rural region of Southwestern China. Am J Trop Med Hyg 96:595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yang D, Yang Y, Wang Y, Yang Y, Dong S, Chen Y, Jiang Q, Zhou Y. 2018. Prevalence and risk factors of Ascaris lumbricoides, Trichuris trichiura and Cryptosporidium infections in elementary school children in Southwestern China: a school-based cross-sectional study. Int J Environ Res Public Health 15:1809. doi: 10.3390/ijerph15091809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zheng H, He J, Wang L, Zhang R, Ding Z, Hu W. 2018. Risk factors and spatial clusters of Cryptosporidium infection among school-age children in a rural region of Eastern China. Int J Environ Res Public Health 15:924. doi: 10.3390/ijerph15050924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Katsumata T, Hosea D, Wasito EB, Kohno S, Hara K, Soeparto P, Ranuh IG. 1998. Cryptosporidiosis in Indonesia: a hospital-based study and a community-based survey. Am J Trop Med Hyg 59:628–632. doi: 10.4269/ajtmh.1998.59.628. [DOI] [PubMed] [Google Scholar]
- 232.Asady A, Ismail S, Marsitah AJ, Pakeer O. 2019. Prevalence of Cryptosporidium spp. infection among children admitted to Hospital Tengku Ampuan Afzan. Med J Malaysia 74:468–471. [PubMed] [Google Scholar]
- 233.Labana RV, Dungca JZ, Nissapatorn V. 2018. Community-based surveillance of Cryptosporidium in the indigenous community of Boliwong, Philippines: from April to December 2017. Epidemiol Health 40:e2018047. doi: 10.4178/epih.e2018047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Bhattacharya MK, Teka T, Faruque AS, Fuchs GJ. 1997. Cryptosporidium infection in children in urban Bangladesh. J Trop Pediatr 43:282–286. doi: 10.1093/tropej/43.5.282. [DOI] [PubMed] [Google Scholar]
- 235.Sarkar R, Kattula D, Francis MR, Ajjampur SS, Prabakaran AD, Jayavelu N, Muliyil J, Balraj V, Naumova EN, Ward HD, Kang G. 2014. Risk factors for cryptosporidiosis among children in a semi urban slum in southern India: a nested case-control study. Am J Trop Med Hyg 91:1128–1137. doi: 10.4269/ajtmh.14-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Khalili B, Mardani M. 2009. Frequency of Cryptosporidium and risk factors related cryptosporidiosis in under 5 year old hospitalized children due to diarrhea. Arch Clin Infect Dis 4:151–155. [Google Scholar]
- 237.Osman M, El Safadi D, Cian A, Benamrouz S, Nourrisson C, Poirier P, Pereira B, Razakandrainibe R, Pinon A, Lambert C, Wawrzyniak I, Dabboussi F, Delbac F, Favennec L, Hamze M, Viscogliosi E, Certad G. 2016. Prevalence and risk factors for intestinal protozoan infections with Cryptosporidium, Giardia, Blastocystis and Dientamoeba among schoolchildren in Tripoli, Lebanon. PLoS Negl Trop Dis 10:e0004496. doi: 10.1371/journal.pntd.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Osman M, Benamrouz S, Guyot K, El Safadi D, Mallat H, Dabboussi F, Hamze M, Viscogliosi E, Certad G. 2018. Molecular epidemiology of Cryptosporidium spp. in North Lebanon. J Infect Dev Ctries 12:34s. doi: 10.3855/jidc.10014. [DOI] [PubMed] [Google Scholar]
- 239.Saaed FMA, Ongerth JE. 2019. Giardia and Cryptosporidium in children with diarrhea, Kufra, Libya, a North African migration route city. Int J Hyg Environ Health 222:840–846. doi: 10.1016/j.ijheh.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 240.Salyer SJ, Gillespie TR, Rwego IB, Chapman CA, Goldberg TL. 2012. Epidemiology and molecular relationships of Cryptosporidium spp. in people, primates, and livestock from Western Uganda. PLoS Negl Trop Dis 6:e1597. doi: 10.1371/journal.pntd.0001597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Morse T, Grimason A, Smith H. 2011. Epidemiology of diarrhoeal disease in Malawi—a case study of cryptosporidiosis, p 474–482. The 33rd WEDC International Conference. University of Strathclyde, Glasgow, Scotland.
- 242.Molloy SF, Tanner CJ, Kirwan P, Asaolu SO, Smith HV, Nichols RA, Connelly L, Holland CV. 2011. Sporadic Cryptosporidium infection in Nigerian children: risk factors with species identification. Epidemiol Infect 139:946–954. doi: 10.1017/S0950268810001998. [DOI] [PubMed] [Google Scholar]
- 243.Nassar SA, Oyekale TO, Oluremi AS. 2017. Prevalence of Cryptosporidium infection and related risk factors in children in Awo and Iragberi, Nigeria. J Immunoassay Immunochem 38:2–9. doi: 10.1080/15321819.2016.1178652. [DOI] [PubMed] [Google Scholar]
- 244.Nchito M, Kelly P, Sianongo S, Luo NP, Feldman R, Farthing M, Baboo KS. 1998. Cryptosporidiosis in urban Zambian children: an analysis of risk factors. Am J Trop Med Hyg 59:435–437. doi: 10.4269/ajtmh.1998.59.435. [DOI] [PubMed] [Google Scholar]
- 245.Opoku YK, Boampong JN, Ayi I, Kwakye-Nuako G, Obiri-Yeboah D, Koranteng H, Ghartey-Kwansah G, Asare KK. 2018. Socio-behavioral risk factors associated with cryptosporidiosis in HIV/AIDS patients visiting the HIV referral clinic at Cape Coast Teaching Hospital. Open AIDS J 12:106–116. doi: 10.2174/1874613601812010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Molbak K, Aaby P, Hojlyng N, da Silva AP. 1994. Risk factors for Cryptosporidium diarrhea in early childhood: a case-control study from Guinea-Bissau, West Africa. Am J Epidemiol 139:734–740. doi: 10.1093/oxfordjournals.aje.a117064. [DOI] [PubMed] [Google Scholar]
- 247.Suarez Hernandez M, Gomez Ferrer R, Rodriguez Menendez G. 1999. Risks factors of Cryptosporidium in infants under one year old with acute diarrhea disease in the Province of Ciego de Avila, Cuba. Rev Esp Pediatr 55:519–522. [Google Scholar]
- 248.Cruz JR, Cano F, Caceres P, Chew F, Pareja G. 1988. Infection and diarrhea caused by Cryptosporidium sp. among Guatemalan infants. J Clin Microbiol 26:88–91. doi: 10.1128/JCM.26.1.88-91.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Laubach HE, Bentley CZ, Ginter EL, Spalter JS, Jensen LA. 2004. A study of risk factors associated with the prevalence of Cryptosporidium in villages around Lake Atitlan. Braz J Infect Dis 8:319–323. doi: 10.1590/S1413-86702004000400008. [DOI] [PubMed] [Google Scholar]
- 250.Javier Enriquez F, Avila CR, Ignacio Santos J, Tanaka-Kido J, Vallejo O, Sterling CR. 1997. Cryptosporidium infections in Mexican children: clinical, nutritional, enteropathogenic, and diagnostic evaluations. Am J Trop Med Hyg 56:254–257. doi: 10.4269/ajtmh.1997.56.254. [DOI] [PubMed] [Google Scholar]
- 251.Solorzano-Santos F, Penagos-Paniagua M, Meneses-Esquivel R, Miranda-Novales MG, Leanos-Miranda B, Angulo-Gonzalez D, Fajardo-Gutierrez A. 2000. Cryptosporidium parvum infection in malnourished and non malnourished children without diarrhea in a Mexican rural population. Rev Invest Clin 52:625–631. (In Spanish.) [PubMed] [Google Scholar]
- 252.Chacin-Bonilla L, Barrios F, Sanchez Y. 2008. Environmental risk factors for Cryptosporidium infection in an island from Western Venezuela. Mem Inst Oswaldo Cruz 103:45–49. doi: 10.1590/S0074-02762008005000007. [DOI] [PubMed] [Google Scholar]
- 253.Newman RD, Sears CL, Moore SR, Nataro JP, Wuhib T, Agnew DA, Guerrant RL, Lima AA. 1999. Longitudinal study of Cryptosporidium infection in children in northeastern Brazil. J Infect Dis 180:167–175. doi: 10.1086/314820. [DOI] [PubMed] [Google Scholar]
- 254.Pereira MD, Atwill ER, Barbosa AP, Silva SA, Garcia-Zapata MT. 2002. Intra-familial and extra-familial risk factors associated with Cryptosporidium parvum infection among children hospitalized for diarrhea in Goiania, Goias, Brazil. Am J Trop Med Hyg 66:787–793. doi: 10.4269/ajtmh.2002.66.787. [DOI] [PubMed] [Google Scholar]
- 255.Bern C, Ortega Y, Checkley W, Roberts JM, Lescano AG, Cabrera L, Verastegui M, Black RE, Sterling C, Gilman RH. 2002. Epidemiologic differences between cyclosporiasis and cryptosporidiosis in Peruvian children. Emerg Infect Dis 8:581–585. doi: 10.3201/eid0806.01-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Gatei W, Das P, Dutta P, Sen A, Cama V, Lal AA, Xiao L. 2007. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect Genet Evol 7:197–205. doi: 10.1016/j.meegid.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 257.Abd El Kader NM, Blanco M-A, Ali-Tammam M, Abd El Ghaffar AERB, Osman A, El Sheikh N, Rubio JM, de Fuentes I. 2012. Detection of Cryptosporidium parvum and Cryptosporidium hominis in human patients in Cairo, Egypt. Parasitol Res 110:161–166. doi: 10.1007/s00436-011-2465-6. [DOI] [PubMed] [Google Scholar]
- 258.Molloy SF, Smith HV, Kirwan P, Nichols RA, Asaolu SO, Connelly L, Holland CV. 2010. Identification of a high diversity of Cryptosporidium species genotypes and subtypes in a pediatric population in Nigeria. Am J Trop Med Hyg 82:608–613. doi: 10.4269/ajtmh.2010.09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ojuromi OT, Duan L, Izquierdo F, Fenoy SM, Oyibo WA, Del Aguila C, Ashafa AO, Feng Y, Xiao L. 2016. Genotypes of Cryptosporidium spp. and Enterocytozoon bieneusi in human immunodeficiency virus-infected patients in Lagos, Nigeria. J Eukaryot Microbiol 63:414–418. doi: 10.1111/jeu.12285. [DOI] [PubMed] [Google Scholar]
- 260.Mbae C, Mulinge E, Waruru A, Ngugi B, Wainaina J, Kariuki S. 2015. Genetic diversity of Cryptosporidium in children in an urban informal settlement of Nairobi, Kenya. PLoS One 10:e0142055. doi: 10.1371/journal.pone.0142055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Morse TD, Nichols RA, Grimason AM, Campbell BM, Tembo KC, Smith HV. 2007. Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol Infect 135:1307–1315. doi: 10.1017/S0950268806007758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Tumwine JK, Kekitiinwa A, Nabukeera N, Akiyoshi DE, Rich SM, Widmer G, Feng X, Tzipori S. 2003. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am J Trop Med Hyg 68:710–715. doi: 10.4269/ajtmh.2003.68.710. [DOI] [PubMed] [Google Scholar]