Summary
Cytochrome c oxidase (COX), the terminal enzyme of mitochondrial electron transport chain, couples electron transport to oxygen with generation of proton gradient indispensable for the production of vast majority of ATP molecules in mammalian cells. The review summarizes current knowledge of COX structure and function of nuclear-encoded COX subunits, which may modulate enzyme activity according to various conditions. Moreover, some nuclear-encoded subunits possess tissue-specific and development-specific isoforms, possibly enabling fine-tuning of COX function in individual tissues. The importance of nuclear-encoded subunits is emphasized by recently discovered pathogenic mutations in patients with severe mitopathies. In addition, proteins substoichiometrically associated with COX were found to contribute to COX activity regulation and stabilization of the respiratory supercomplexes. Based on the summarized data, a model of three levels of quaternary COX structure is postulated. Individual structural levels correspond to subunits of the i) catalytic center, ii) nuclear-encoded stoichiometric subunits and iii) associated proteins, which may constitute several forms of COX with varying composition and differentially regulated function.
Keywords: Mitochondria, OXPHOS, Cytochrome c oxidase, Nuclear-encoded subunits, Mitochondrial diseases
Cytochrome c oxidase
Energy demands of mammalian cells are mainly covered by ATP synthesis carried out by oxidative phosphorylation apparatus (OXPHOS) located in the central bioenergetic organelle, mitochondria. OXPHOS is composed of five multi-subunit complexes embedded in the inner mitochondrial membrane (IMM). Electron transport from reduced substrates of complexes I and II to cytochrome c oxidase (COX, complex IV, CIV) is achieved by increasing redox potential of individual active centers of complexes and carriers (ubiquinone, cytochrome c). Complex I (CI), complex III (CIII) and complex IV couple electron transport with translocation of protons across the IMM into intermembrane space (IMS). Proton gradient is then utilized by complex V (ATP synthase, CV) as a driving force for ATP production.
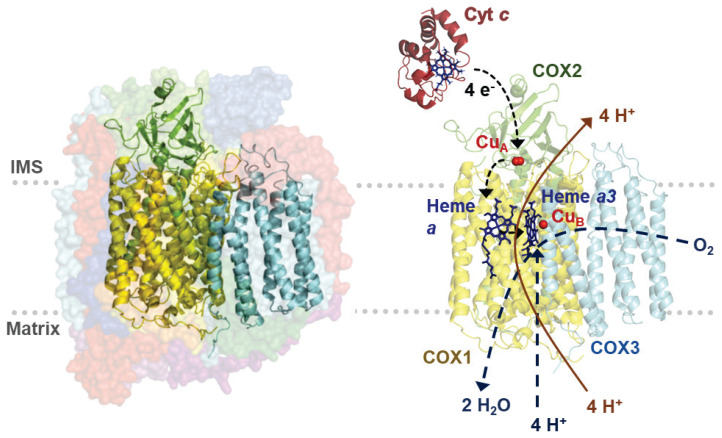
Cytochrome c oxidase (COX), the member of heme-Cu oxidases, is the terminal enzyme of mitochondrial electron transport chain (ETC) indispensable for transfer of electrons to the terminal electron acceptor, oxygen (Fig. 1). Besides the role in electron transport coupled with formation of proton gradient, COX is proposed to have important function in regulation within whole OXPHOS system.
Fig. 1.
COX structure and catalytic function scheme. Left - Model of COX crystal structure. Subunits of COX catalytic core – COX1 (yellow), COX2 (green), and COX3 (blue) are shown in cartoon mode, surrounded by nuclear-encoded subunits depicted in surface mode (semi-transparent). Right - COX catalytic core, COX1 (yellow), COX2 (green), and COX3 (blue) with substrate protein cytochrome c (red). Dashed blue lines depict oxygen conversion to water at binuclear center as a result of electron transfer from cytochrome c through metallic prosthetic cofactors and consumption of four protons. Orange line delineates proton pumping across inner mitochondrial membrane. Structural images are based on PDB files 5Z62 (COX) and 2N9J (cytochrome c), (PDB ID: 2N9J Imai et al. 2016, PDB ID: 5Z62 Zong et al. 2018).
Catalytic core of mammalian COX is composed of three mitochondria-encoded subunits (COX1, COX2, COX3), which are homologous to subunits of the prokaryotic aa3 type of terminal oxidase. COX1 subunit consists of twelve transmembrane α-helices and three catalytic centers of COX enzyme - heme a and binuclear center (BNC) composed of heme a3 and CuB atom. COX2 subunit is composed of two transmembrane α-helices and bimetallic CuA catalytic center (CuI/CuII). COX2 hydrophilic domain in IMS forms binding site for cytochrome c. COX3 subunit consists of seven transmembrane α-helices and stabilizes catalytic center of COX (Kaila et al. 2010). Without COX3 subunit, stoichiometry of pumped protons decreases in both bacterial and mitochondrial enzyme (Hosler 2004, Sharma et al. 2015).
COX catalyzes electron transfer from cytochrome c to the terminal electron acceptor, oxygen, which is reduced to water (Fig. 1). Electrons are received from cytochrome c on intermembrane side of COX enzyme following docking of cytochrome c thanks to electrostatic interactions of its conserved lysine residues with carboxyl groups of hydrophilic part of COX2 subunit (Millett et al. 1983). Further, electrons are sequentially transferred through CuA center to heme a. Heme a is called electron-queuing site due to dependency of following transfer of electrons to BNC (heme a3 and CuB) on proton translocation (Babcock and Wikström 1992). Nonpolar channel enables diffusion of oxygen into catalytic center of COX enzyme (Sharma et al. 2015, Tsukihara et al. 1996). Heme-oxygen adduct is formed by binding of oxygen into BNC and oxygen bond is immediately disrupted. Simultaneous transfer of four electrons is crucial as a precaution to reactive oxygen species (ROS) production (Ludwig et al. 2001). Then, BNC is regenerated by four reduction steps of catalytic cycle. In each step, one electron from heme a and one proton from matrix (chemical proton) are translocated into BNC, while one proton is pumped into IMS by conserved proton channels (Kaila et al. 2010). Binding kinetics of both substrates (cytochrome c and O2) is strongly influenced by the magnitude of mitochondrial membrane potential, with uncoupling increasing the enzyme turnover (Pannala et al. 2016). Enzyme catalysis may be inhibited directly at the binuclear center by non-competitive binding of cyanide (CN−) or hydrogen sulfide (H2S), as well by gases competing with oxygen binding such as NO and CO. H2S and NO may represent endogenously produced modulators of COX activity in mammals (Cooper and Brown 2008).
The presence of additional subunits in mammalian COX was initially identified by high-resolution SDS-PAGE (Kadenbach et al. 1983) and definitively approved by X-ray crystallography of bovine heart COX dimer (Tsukihara et al. 1996). These methods identified ten nuclear-encoded subunits (COX4, COX5A, COX5B, COX6A, COX6B, COX6C, COX7A, COX7B, COX7C, COX8) tightly surrounding the catalytic core. Recently, NDUFA4 has become widely accepted as fourteenth stoichiometric COX subunit (Pitceathly and Taanman 2018).
Assembly of COX enzyme
Fourteen subunits of mammalian COX monomer are encoded by both mitochondrial and nuclear genom. Therefore, expression and posttranslational modifications of individual subunits in mitochondria and cytosol need to be coordinated by crosstalk between nucleus and mitochondria (Dennerlein et al. 2017). Many auxiliary proteins are involved in assembly process, studies on S. cerevisiae uncovered dozens of nuclear-encoded assembly factors required throughout all phases of enzyme biogenesis (Fontanesi et al. 2006, Khalimonchuk and Rödel 2005, McEwen et al. 1986, Tzagoloff and Dieckmann 1990). The process seems to be conserved between yeast and mammals, where most of these factors were also found and assigned specific role in: (i) regulation of expression of catalytic core subunits (LRPPRC, TACO1, hCOA3, COX14, MITRAC7) (ii) metabolism and insertion of copper cofactors (COX17, SCO1, SCO2, COX11, COX19, COA6, COX20 (iii) heme a biosynthesis and insertion (COX10, COX15, FDX2, SURF1) and (iv) membrane insertion and processing of catalytic core subunits (OXA1l, COX18). Their importance is signified by mutations leading to inherited defects of COX enzyme in humans (Pecina et al. 2004, Rak et al. 2016).
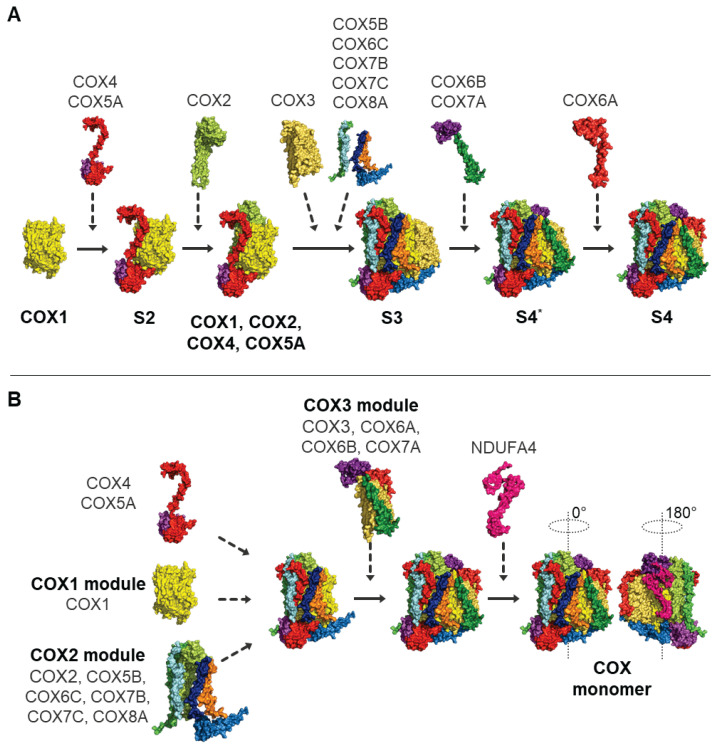
De novo COX assembly was originally viewed as a sequential process (Fig. 2A), which was described for the first time by Nijtmans et al. (Nijtmans et al. 1998) based on COX assembly kinetics, recognizing several rate-limiting steps of the process. Sequential assembly of COX was further documented by several studies based on native polyacrylamide gel electrophoresis enabling observation of individual COX assembly intermediates, accumulated due to pathogenic mutations in the genes encoding COX subunits or, more frequently, COX assembly factors (Fontanesi et al. 2006, Williams et al. 2004). Initially, COX1 subunit is synthesized and represents the first S1 intermediate. COX1 is joined by COX4 and COX5A subunits, forming second intermediate S2 (Fornůsková et al. 2010). S3 intermediate already contains subunits COX2, COX3, COX5B, COX6C, COX7B, COX7C, COX8 (Fornůsková et al. 2010, Williams et al. 2004). Then, COX7A and COX6B are added establishing last S4* intermediate, and COX6A addition finalizes COX holoenzyme (Fornůsková et al. 2010) (Fig. 2A).
Fig. 2.
Two alternative views of COX assembly. Diagrams show the original model of sequential incorporation of COX subunits (A), or assembly model assuming preassembly of individual subunit modules (B). See text for detailed description. Structural images are based on PDB file 5Z62.
Alternatively, recent theories consider COX assembly as a linear process in which different modules, not subunits, are assembled together into functional enzyme (Vidoni et al. 2017). In the modular model (Fig. 2B), each module containing COX subunits with their assembly factors and chaperons represents assembly checkpoint (Timón-Gómez et al. 2017, Vidoni et al. 2017). For instance, in early assembly stage, formation and stability of COX1 subunit associated with MITRAC complex (mitochondrial translation regulation assembly intermediate of COX) (Dennerlein and Rehling 2015), is necessary for the addition of COX4-COX5A sub-module (Mick et al. 2012, Vidoni et al. 2017). Further, COX biogenesis is regulated by assembly-controlled translational plasticity, which means that without nuclear-encoded subunits involved in early stages of COX biogenesis (COX4), translation of COX1 subunit is abolished and remains stalled as a ribosome-nascent chain complex containing MITRAC component C12ORF62 (Richter-Dennerlein et al. 2016).
The modular model significantly refined the previous sequential assembly scheme. Nevertheless, further studies using knock-out models of individual nuclear-encoded subunits are needed to precisely identify their respective role and position in enzyme assembly.
Besides de novo assembly, direct incorporation of some nuclear-encoded subunits into existing holoenzyme, similarly as found in other OXPHOS complexes, is also possible in COX. Candidate proteins for this process are subunits involved in later intermediates of the de novo assembly, namely COX6A, COX6B and COX7A, which may be incorporated into COX monomer or even COX assembled into supercomplexes (Lazarou et al. 2009). This process employs a conserved late state intermediate (LSI) complex containing COX6A and COX7A subunits with ancillary factors facilitating their incorporation. The proposed mechanism of nuclear-encoded subunits replacement in COX holoenzyme may be employed for the exchange of isoform pairs of nuclear-encoded subunits during changing conditions, or for quality control and replacement of damaged components. Perspectively, mastering this mechanism may lead to development of therapeutic approaches aiming to substitute pathogenic variants of respective subunits.
COX in supercomplexes
Individual electron transport chain (ETC) complexes were shown to form higher organization structures, termed respiratory supercomplexes. Existence of supercomplexes is consistent with the plasticity model of respiratory chain organization in IMM (Acín-Pérez et al. 2008, Enríquez 2016), which suggests coexistence of both solid-state (Chance and Williams 1955) and random collision models (Chazotte and Hackenbrock 1989). Content and ratio of various supercomplexes differ among species and tissues (Schägger and Pfeiffer 2000).
In mammals, arrangements of supercomplexes composed of Complex I, Complex III and Complex IV were studied mostly by 1-D/2-D BN-PAGE and 1-D CN-PAGE (Wittig and Schägger 2005). The most abundant supercomplex in bovine heart is I1III2IVn (n=1 – 4) supercomplex, also called respirasome (Schägger and Pfeiffer 2000). The most detailed architecture of respirasome were solved at 5.8 Å (Letts et al. 2016) and at 4.0 Å (Wu et al. 2016) in ovine and porcine heart, respectively. Moreover, the structure of human 14-subunit COX monomer containing NDUFA4 was already published (Zong et al. 2018). These structures provide strong indication that COX is incorporated into supercomplexes as a catalytically competent momomer. Recently published structure suggests the existence of a circular I2III2IV2 megacomplex (Guo et al. 2017).
Despite current studies focusing on supercomplex architecture, their precise physiological role is questioned and remains unresolved (Hirst 2018). Suggested functions include electron channeling, ROS production prevention, as well as their involvement in assembly and stabilization of individual complexes (Letts and Sazanov 2017).
COX nuclear-encoded subunits
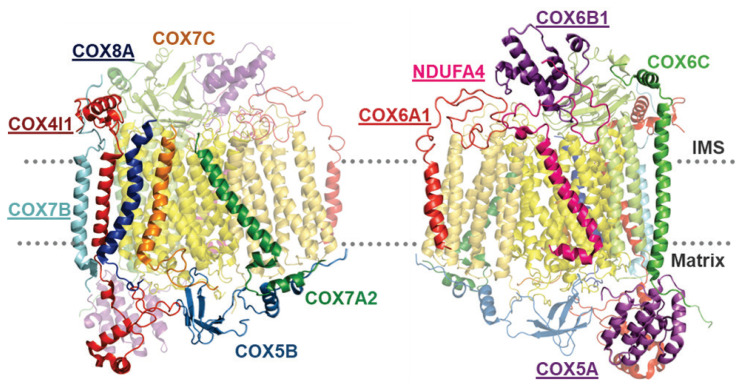
Nuclear-encoded subunits include eleven different proteins (COX5A, COX5B, COX6C, COX7C, COX4, COX6B, COX7B, COX6A, COX7A, COX8, and NDUFA4), six of which exist even in isoforms. They surround the conserved catalytic core (Fig. 3) and are not crucial for catalytic function, but may modulate COX activity under various conditions. They represent eukaryotic innovation in COX structure and function. Although numerous genes were relocated from mitochondrial genome into nucleus after endosymbiosis, COX nuclear-encoded subunits did not originate in mitochondrial genome (Szklarczyk and Huynen 2010). No homologous genes were found in either Rickettsia prowazekii (the closest taxon to the hypothetic endosymbiont), or in Reclinomonas americana genome, which retained largest number of genes in mtDNA (Andersson et al. 1998, Das et al. 2004, Lang et al. 1997). Many COX nuclear-encoded subunits may be found in genome of Drosophila melanogaster, Saccharomyces cerevisiae and Arabidopsis thaliana, which suggests appearance of these subunits before the split of eukaryotic lineages (Das et al. 2004, Pierron et al. 2012). Moreover, some nuclear-encoded subunits show two or three tissue-specific and development-specific isoforms, which arose from gene duplication. Selective pressure could have shaped specialized function and tissue-specific expression of isoform pair subunits (Little et al. 2010, Pierron et al. 2012).
Fig. 3.
Arrangement of nuclear-encoded subunits in COX monomer. Two 180° mutually-rotated images of COX monomer depict position of individually-marked nuclear-encoded subunits. Mutations in genes encoding the underlined subunits were associated with human pathologies. Images are based on PDB structure 5Z62.
Single isoform subunits COX5A, COX5B, COX6C and COX7C
Subunit COX5A, which is located on matrix side of COX enzyme, consists of five α-helices forming right-handed superhelix (Tsukihara et al. 1996). Bovine heart enzyme COX5A subunit was shown to specifically bind 3,5-diiodothyronine (T2), which abolished allosteric ATP-inhibition of COX and stimulated energetic metabolism (Cimmino et al. 1996, Lanni et al. 1996). Besides, overexpression of COX5A subunit in non-small-cell lung carcinoma (NSCLC) cells correlated with more efficient migration and invasivity of NSCLC cells (Chen et al. 2012a). Moreover, interaction of COX5A with anti-apoptotic Bcl-2 protein may increase accumulation of COX5A subunit in mitochondria and slightly increase oxidative stress, which positively affects cancer cells viability and proliferation (Chen and Pervaiz 2010).
COX5B is a matrix-facing subunit without transmembrane domain. Knockdown of COX5B in RAW 264.7 macrophage line resulted in decreased COX activity and membrane potential while ROS production was increased. COX assembly intermediates were accumulated in COX5B knockdown cells, which suggests importance of COX5B for COX assembly (Galati et al. 2009). In addition, COX5B was found to interact with regulatory subunit of cAMP-dependent protein kinase A, which inhibits COX activity (Yang et al. 1998). COX5B to COX1 ratio is prominently increased in cancer cells with higher expression of COX5A subunit (Chen et al. 2012a, Chen and Pervaiz 2010, Krieg et al. 2004). Moreover, COX5B expression was upregulated in malignant glioma (Hu and Xi 2017) and breast cancer (Gao et al. 2017b), suggesting COX5B as a novel biomarker of these pathologies.
No regulatory function of COX6C and COX7C subunits has been reported yet, despite their interaction with catalytic core subunits COX1 and COX2 (Tsukihara et al. 1996).
Subunits with tissue-specific isoforms: COX4, COX6B, COX7B and NDUFA4
COX4 is the largest nuclear-encoded subunit of COX. Interaction of its transmembrane helix with catalytic subunit COX1, as well as large membrane-extrinsic domain in mitochondrial matrix predisposes this subunit for regulatory function. Moreover, COX4 together with COX6B are crucial for docking of cytochrome c on its binding site in subunit COX2 (Sampson and Alleyne 2001). Thus, COX4 subunit represents possible tool for fine-tuning of COX enzyme and consequently the whole OXPHOS system. In mammals, COX4 subunit has two distinct isoforms (Hüttemann et al. 2001), which are expressed in tissue-dependent and oxygen-dependent manner, and thus can play a significant role in modulation of COX activity in response to changing conditions. COX4I1 isoform is expressed ubiquitously in all mammalian tissues while COX4I2 expression was found in adult and fetal lung, heart, and brain (Hüttemann et al. 2001).
COX4 subunit contains ATP/ADP binding sites, which can sense energy status of the cell and therefore regulate activity of the enzyme according to current metabolic needs (Acín-Pérez et al. 2011). ATP binding site in matrix domain mediates allosteric inhibition of enzyme activity (Acín-Pérez et al. 2011, Arnold and Kadenbach 1997, Bender and Kadenbach 2000, Lee et al. 2002, Lee et al. 2005). This regulation is abolished by phosphorylation mediated by intramitochondrial protein kinase A pathway (Acín-Pérez et al. 2011). Importantly, this regulation should not be functional in COX4I2 isoform due to presence of negatively charged residues in the region homologous to ATP binding site in COX4I1.
Increased expression of COX4I2 isoform under hypoxia was detected in numerous mammalian cell types. The switch in isoform expression is triggered by signaling pathway dependent on partial pressure of oxygen in the cell. Two possible mechanisms were proposed in literature. The first study showed that expression of COX4 isoforms in mammals is regulated by hypoxia inducible factor-1 (HIF-1α), acting at hypoxia-response elements identified in human COX4I2 gene promoter (Fukuda et al. 2007). Alternatively, novel oxygen responsive element (ORE) was discovered in proximal promoter sequence of COX4I2 gene in mouse, rat, beef and human (Hüttemann et al. 2007). Candidate ORE-binding factors were found in HEK293 cells. As activators of transcription were identified RBPJ (recombination signal sequence binding protein Jκ), whose main role is in evolutionarily conserved NOTCH signaling pathway, and CHCHD2 (coiled-coil-helix-coiled-coil-helix domain 2, alternatively MNRR1). These factors together allow hypoxic activation of COX4I2 promoter. CXXC5 (CXXC finger protein 5) was identified as a repressor of COX4I2 transcription, mainly under normoxia (Aras et al. 2013).
Recently, major progress in understanding the physiological role of COX4I2 has been achieved. Studies employing mouse COX4I2 knock-out model revealed that this isoform is essential for pulmonary hypoxic vasoconstriction (HPV) (Sommer et al. 2017). More recently, increased expression of COX4I2, along with two atypical COX subunits – COX8B and NDUFA4L2 controlled by HIF-2α was observed in glomus cells of carotid bodies in mice. Such atypical COX complex present in these specialized cells was suggested as a sensor involved in acute oxygen sensing (Gao et al. 2017a, Moreno-Dominguez et al. 2019). Mechanism of oxygen sensing both in HPV and in carotid bodies is associated with changes of mitochondrial redox status and increased ROS production during hypoxia. To facilitate its role as an oxygen sensor, oxygen affinity of the enzyme may be differentialy modulated by COX4 isoforms. Indeed, our recent results indicate that exchange of COX4I1 by COX4I2 isoform in HEK293 cells leads to two-fold reduction in COX oxygen affinity (Pajuelo Reguera et al. 2020). This represents a novel parameter that can be modified by COX subunits isoforms switch.
IMS side oriented COX6B subunit should mediate tight interaction between two monomers of COX in dimer (Tsukihara et al. 1996). COX6B1 isoform loss imposed by mild solubilization resulted in two-fold increased enzyme activity due to monomerization of COX dimer, but without changes in stoichiometry of translocated protons. Reportedly it was due to the loss of cooperative behavior of COX dimer’s monomers in cytochrome c binding kinetics (Kadenbach and Hüttemann 2015, Weishaupt and Kadenbach 1992). Second COX6B isoform (COX6B2) in mammals is specifically expressed only in testis, and thus extends a group of testes-specific mitochondrial proteins, such as ANT4 isoform of adenine nucleotide translocator (Dahout-Gonzalez 2006) or cytochrome c (Hüttemann et al. 2003). However, testis isoform of cytochrome c is not conserved in primates (Krzyzosiak et al. 2000). Contrary to COX6B1 isoform, COX6B2 contains two tyrosine residues, which hypothetically may be phosphorylated by global cAMP-dependent tyrosine phosphorylation event crucial for activation of sperm motility (Krzyzosiak et al. 2000). Moreover, two independent proteomic studies linked decreased COX6B-2 content with aberrant sperm motility (Cao et al. 2018).
COX7B1 isoform of COX7B subunit is important for COX assembly and activity (Indrieri et al. 2012). Unfortunately, function of COX7B2 isoform was not determined yet. Nevertheless, mouse COX7B2 isoform expands the group of testes-specific proteins, possibly adapting testes metabolism (Kadenbach and Hüttemann 2015). Chinese patients with nasopharyngeal cancer showed five different single nucleotide polymofphisms (SNPs) in COX7B2 gene, one of them leading to His26Gln substitution. This mutation is probably associated with higher nasopharyngeal cancer risk (Liang 2004).
NDUFA4 (COXFA4) protein, originally considered complex I subunit (Carroll et al. 2003), was found to associate with COX where it may augment enzyme activity (Balsa et al. 2012, Pitceathly et al. 2013). Moreover, NDUFA4 was recently detected in stoichiometric ratio to canonical COX subunits, and was therefore suggested as a bona fide fourteenth subunit of mammalian enzyme (Balsa et al. 2012, Pitceathly and Taanman 2018). Authors explain notorious absence of NDUFA4 subunit in COX crystal structures by loss of its association with COX during purification using high concentration of detergent dodecylmaltoside (Balsa et al. 2012, Pitceathly et al. 2013, Tsukihara et al. 1996). Indeed, structure of the 14-subunit human COX containing NDUFA4 was derived from cryo-EM structure of intact respirasome extracted by mild detergent digitonin (Zong et al. 2018). Expression of second isoform of this subunit, NDUFA4L2, was detected as COX component in glomus cells of carotid bodies. Its HIF-2α induced expression along with COX4I2 and COX8B hypothetically leads to formation of non-canonical COX complex essential for acute oxygen sensing (Moreno-Dominguez et al. 2019). Recently, a third NDUFA4 isoform, COXFA4L3 (C15orf48, Nmes1), was described in mouse testis, possibly having a role during spermatogenesis (Endou et al. 2020).
Subunits with pair of heart and liver isoforms: COX6A, COX7A and COX8
Subunits COX6A, COX7A and COX8 share tissue-specific and development-specific manner of isoforms expression. Isoform H (heart) is expressed in heart and skeletal muscle, while isoform L (liver) is ubiquitously expressed in all other tissues (Bonne et al. 1993).
COX6A subunit is in contact with COX1 subunit of opposite COX monomer in dimer and is supposed to stabilize it (Radford et al. 2002). Its two isoforms were discovered by COX6A N-terminal amino acids sequencing of subunit from mammalian liver and heart (Linder et al. 1995). Expression of heart isoform, COX6A-H, is development-specific, exchange of COX6A-L to COX6A-H occurs during the third month after the birth (Boczonadi et al. 2015, Bonne et al. 1993, Ewart et al. 1991). COX6A-L isoform in COX reconstituted from bovine kidney showed decreased proton pumping ability after addition of free palmitate, while other fatty acid or palmitoyl-CoA had no effect. This effect did not occur in heart isozyme (Lee and Kadenbach 2001). Instead, COX6A-H isoform contains ADP-binding site on its N-terminus oriented towards matrix, which mediates allosteric activation of reconstituted COX from bovine heart (Anthony et al. 1993). In contrast, high ATP content leads to decreased stoichiometry of pumped protons causing significant decrease in P/O quotient (Kadenbach et al. 1995). Proposed physiological role of this mechanism is stimulation of thermogenesis in muscle under resting conditions. COX6A-H deficient mice model showed decrease in heart muscle tissue COX activity, which correlated with decreased COX holoenzyme content. Thus, COX6A-H absence affects COX assembly or stability (Radford et al. 2002).
COX7A-H (COX7A1) isoform gene deletion in mice leads to decreased COX activity, even though the missing isoform is replaced by COX7A-L (COX7A2) in heart COX (Hüttemann et al. 2012). Moreover, COX7A subunit possesses a third isoform, SCAF1 (supercomplex assembly factor 1) (COX7A2L, COX7AR), which has similar expression pattern as COX7AL (Schmidt et al. 1999).
SCAF1 was originally described as responsible for the incorporation of COX into the supercomplexes (Lapuente-Brun et al. 2013). However, the presence of COX in the respirasomes was detected even if SCAF1 was defective (Mourier et al. 2014). Later, the preferential association of SCAF1 with CIII was identified (Perez-Perez et al. 2016). Its role in the interaction of CIII with COX was confirmed, its absence however did not necessarily lead to complete impairment of COX interaction with the respirasome (Williams et al. 2016, Lobo-Jarne et al. 2018). COX7A isoform exchange depending on the COX species involved (monomer, dimer or supercomplexes) and the existence of two types of respirasome with and without SCAF1 was proposed (Cogliati et al. 2016, Letts and Sazanov 2017). SCAF1 homology to COX7A subunits enables it to replace COX7AH or COX7AL and thus bind the complexes III and IV together to stabilize the III2IV supercomplex. This would then interact with CI forming the I1III2IV1 supercomplex (respirasome). Otherwise, COX containing COX7A2 is recruited by supercomplex I1III2 forming a variant population of respirasomes. COX7A1 containing COX was detected only in COX dimers (Cogliati et al. 2016, Letts and Sazanov 2017, Milenkovic et al. 2017).
Further, COX7A2L was described as a stress-inducible COX subunit, which is neccesary for metabolic regulation and Warburg effect induction in human breast cancer cells (Zhang et al. 2016).
COX8 subunit, which is crucial for enzyme stability (Hallmann et al. 2016), is present in three different isoforms in mammals. However, humans retained only the COX8-L isoform, while COX8-H lost its function and became a pseudogene (Goldberg et al. 2003).
COX-associated proteins
Several proteins were discovered to comigrate with COX in native electrophoreses, although they were not identified within crystal structure of COX before. They are loosely attached to COX and therefore released from the complex by solubilization preceding structural studies. Further, they may not be associated as COX subunits under all conditions, thus representing sub-stoichiometric COX components. Their role within the complex may be modulation of enzymatic function or regulation of COX incorporation into supercomplexes.
One of the associating proteins, MNRR1 (CHCHD2), was co-immunoprecipitated with COX in sub-stoichiometric ratio to COX1 and COX2 catalytic subunits. Its predicted function is regulation of dimeric COX or incorporation of COX into supercomplexes (Aras et al. 2015, Grossman et al. 2017). Moreover, MNRR1 might serve as an oxidative stress sensor, as MNRR1 absence increases ROS production. This hypothesis is supported by presence of cysteine residues, which may be differentially modified under changing redox condtions. MNRR1 role in redox signaling might also be connected with its translocation from mitochondria into nucleus and activation of COX4I2 expression (Aras et al. 2015, Hüttemann et al. 2001). Interaction with COX depends on MNRR1 phosphorylation status. Tyr99 of MNRR1 is phosphorylated by the Abl2 kinase, which enhances COX binding and regulates respiratory activity during stress (Aras et al. 2017). Stress-regulated association of CHCHD2 with CHCHD10 was implicated in neurological disorders such as Parkinson’ disease or amyotrophic lateral sclerosis, but without functional link to COX (Imai et al. 2019).
HIGD1A, which displays hypoxia-induced expression (Hayashi et al. 2015), is a homolog of yeast protein Rcf1 that promotes supercomplexes assembly (Strogolova et al. 2012). However, in mammals its function seems to be quite different. HIGD1A association with COX was supposed to cause conformational changes around heme a, which may enable proton pumping through non-canonical H-pathway (Hayashi et al. 2015). Thus, HIGD1A could contribute to sustain ATP production by OXPHOS under hypoxia (Hayashi et al. 2015). Interestingly, HIGD1A was later described as an early binding protein in COX biogenesis (Vidoni et al. 2017), indicating an alternative role in enzyme assembly. HIGD2A was firstly shown to be involved in assembly and stabilization of COX-containing supercomplexes, similar to its yeast homolog Rcf1 (Chen et al. 2012b). Recently, HIGD2A was proposed to be important even for COX3 module assembly (Hock et al. 2020, Timon-Gomez et al. 2020), while HIGD1A is involved in association of COX with complex III (Timon-Gomez et al. 2020). However, HIGD1A overexpression can alleviate HIGD2A deficiency, suggesting that the role of these two proteins is partially overlapping (Timon-Gomez et al. 2020).
Nuclear-encoded COX subunits involved in mitochondrial pathologies
Mutations in COX assembly factors genes represent frequent cause of mitochondrial diseases (Pecina et al. 2004). In these cases usually, residual COX activity is retained in spite of enzyme assembly being severely impaired (Stibůrek and Zeman 2010). The putative existence of mutations in nuclear genes encoding COX subunits had long been considered doubtful, as they could lead to total COX absence and thus more severe phenotype. Nevertheless, since 2008, rare mutations leading to hereditary human diseases, as encephalomyopathy and cardiomyopathy, were found in eight genes encoding COX nuclear-encoded subunits (Table 1).
Table 1.
Pathogenic mutations in nuclear genes encoding COX subunits
| Gene | DNA change (cDNA) | Protein/RNA change | Mutation | Disease | Cases | Publication |
|---|---|---|---|---|---|---|
| COX4I1 | c.303G>T | p.Lys101Asn | homozygous | Fanconi anemia | 1 | (Abu-Libdeh et al. 2017) |
| COX4I1 | c.454C>A | p.Pro152Thr | homozygous | Leigh syndrome with developmental regression, intellectual disability, and seizures | 2 | (Pillai et al. 2019) |
| COX4I2 | c.412G> A | p.Glu138Lys | homozygous | Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis | 4 | (Shteyer et al. 2009) |
| COX5A | c.319C>T | p.Arg107Cys | homozygous | Pulmonary arterial hypertension, lactic acidemia | 2 | (Baertling et al. 2017) |
| COX6A1 | c.247–10_247–6delCACTC | alternative splicing | homozygous | Axonal form of Charcot-Marie-Tooth syndrome | 3 | (Tamiya et al. 2014) |
| COX6B1 | c.59G>A | p.Arg20His | homozygous | Encephalomyopathy | 2 | (Massa et al. 2008) |
| COX6B1 | c.58C>T | p.Arg20Cys | homozygous | Encephalomyopathy, hydrocephalus, hypertrophic cardiomyopathy | 1 | (Abdulhag et al. 2015) |
| COX7B1 | c.196delC c.41-2A>G c.55C>T | p.Leu66Cysfs*48 p.Val14Glyfs*19 p.Gln19* |
heterozygous | X-linked microphthalmia with linear skin lesions | 3 | (Indrieri et al. 2012) |
| COX8A | c.115-1G>C | aberrant splicing | homozygous | Leigh-like syndrome with leukodystrophy and epilepsy | 1 | (Hallmann et al. 2016) |
| NDUFA4 | c.42+1G>C | aberrant splicing | homozygous | Leigh syndrome | 4 | (Pitceathly et al. 2013) |
The largest nuclear-encoded subunit COX4 has two pathogenic variants in COX4I1 isoform gene and one in COX4I2 isoform gene. Homozygous mutation in COX4I1 gene (Lys101Asn) was described in patient suffering from Fanconi anemia-like disease (Abu-Libdeh et al. 2017). Lys101Asn mutation affects conserved Lys residue localized in transmembrane helix domain contacting COX1 and COX2 catalytic core subunits. Mutation attenuated COX4I1 mRNA expression, causing decrease in COX activity and ATP production, and increased ROS production. COX4I1 protein was undetectable by Urea-SDS-PAGE/WB analysis in patient fibroblasts, which seems unlikely as loss of COX4I1 protein would completely prevent assembly of COX holoenzyme. Complementation by wild-type COX4I1 transduction into patient fibroblasts resulted in restoration of COX activity and ATP production (Abu-Libdeh et al. 2017). Recently published mutation of COX4I1 gene (Pro152Thr) was identified as a cause of pathology mimicking Leigh syndrome, typical for COX deficiencies (Pillai et al. 2019). The affected proline residue is evolutionary conserved part of C-terminal IMS domain neighbouring with COX2 and COX6C subunit near cytochrome c docking site. Its substitution by threonine leads to decreased COX activity in skeletal muscle (Pillai et al. 2019). However, the precise impact of COX4I1 Pro152Thr substitution on COX biogenesis, maintenance and function in OXPHOS context should be further characterized by biochemical and functional experiments.
COX4I2 gene revealed missense mutation (Glu138Lys) leading to exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis in 4 patients (Shteyer et al. 2009). Comparison of mRNA expression in patient fibroblasts under normoxic and hypoxic conditions showed blunted increase of COX4I2 isoform expression under hypoxia. Comparison of mRNA levels of COX4I1 and COX4I2 isoforms in whole pancreas and pancreatic islets revealed that COX4I2 isoform is mainly situated in acinar cells responsible for exocrine function of the pancreas (Shteyer et al. 2009). Detailed biochemical consequences of COX4I2 mutation remain to be characterized in detail.
The first documented case of pathology coupled with nuclear-encoded COX subunit mutation was homozygous Arg20His substitution in COX6B1, associated with infantile encephalomyopathy (Massa et al. 2008). Later, another COX6B1 substitution mutation in the same site (Arg20Cys) was identified in patient with encephalomyopathy, hydrocephalus and hypertrophic cardiomyopathy (Abdulhag et al. 2015). Described mutations lead to substitution in evolutionary conserved region that is, according to the protein modeling, crucial for COX6B1 protein conformation and its predicted contact with COX2 catalytic core subunit. Biochemical analysis of patient fibroblasts and muscle tissue harbouring Arg20His mutations revealed reduced COX6B1 protein steady-state level and decreased incorporation of mutated COX6B1 subunit into COX enzyme during assembly, recorded by presence of assembly intermediate S3 possibly preceding addition of COX6B1 subunit (Massa et al. 2008). Correspondingly, COX activity was decreased in patient cells while other OXPHOS complexes were not affected. Introduction of wild-type COX6B1 protein into mutated cells complemented the defect by increased COX holoenzyme content and COX activity as well. Even though overall outcome of both COX6B1 mutations is very similar, Arg20Cys substitution results in more severe COX deficiency than Arg20His (Abdulhag et al. 2015).
COX5A subunit mutation was found as a cause of pulmonary arterial hypertension and lactic acidemia in two patients (Baertling et al. 2017). Arg107Cys substitution is localized in conserved COX5A region contacting N-terminal matrix domain of COX4 subunit and is supposed to abolish proper COX4/COX5A interaction. Correspondingly, patient fibroblasts showed reduced content of assembled COX with accumulated S1 assembly intermediate supporting COX5A subunits importance for early stages of COX assembly process. COX defect was successfully complemented by introduction of wild-type COX5A protein increasing fully assembled COX level.
Three different mutated variants of COX7B1 gene resulting in frame-shift or stop-codon were discovered to yield an X-linked microphthalmia with linear skin lesions (Indrieri et al. 2012). To explore possible function of COX7B subunit and outcome of these mutations, authors prepared model of silenced COX7B1 subunit in HeLa cell-line. COX7B1 knock-down HeLa cells resulted in reduced amount of assembled COX, while COX assembly intermediates were not detected. Functional measurements pointed out respiratory defect following COX7B protein deficiency (Indrieri et al. 2012).
COX6A1 gene 5 bp deletion mutation was associated with Axonal form of Charcot-Marie-Tooth syndrome patients, which cause decrease in COX6A-L isoform expression resulting in impaired COX activity and ATP production (Tamiya et al. 2014). Patient phenotype of muscle atrophy was reproduced in mice model of COX6A1 knock-out. COX6A1 mutation is therefore one of dozens of genes connected to CMT pathology.
COX8A isoform mutation was described in Leigh-like syndrome patients with leukodystrophy and epilepsy (Hallmann et al. 2016). Mutation leads to aberrant splicing resulting in loss of normal COX8A transcript and protein, which affects amount of assembled COX enzyme in patient fibroblasts and skeletal muscle tissue without accumulation of COX assembly intermediates. Therefore, COX8A subunit seems to be important for COX enzyme stability and not for biogenesis. Loss of normal COX8A protein is not compensated by elevation of COX8C expression. Complementation of patient fibroblasts by wild-type COX8A refined isolated COX defect (Hallmann et al. 2016).
In the most recently described COX subunit NDUFA4 (COXFA4) gene, a homozygous mutation of donor splicing site was discovered in Leigh syndrome patients (Pitceathly et al. 2013). Mutation leads to frame-shift with premature stop-codon formation, resulting in NDUFA4 loss. Patient muscle mitochondria presented with decreased COX activity. Moreover, lower content of assembled COX was not coupled to accumulation of assembly intermediates, therefore NDUFA4 is possibly joining COX complex in the very last steps of COX assembly (Pitceathly et al. 2013), which was later confirmed by Vidoni et al. (2017).
According to the current knowledge of nuclear-encoded subunits mutations in patients we can expect that early-assembling subunits essential for COX biogenesis would not show null mutations compared to subunits involved in the final steps of assembly, as documented by substitution mutations in the case of subunits COX4, and COX5A. In contrast, even though mutations of subunits COX6A1, COX7B1, COX8A and NDUFA4 lead to aberrant splicing or frameshifts, which practically eliminate the corresponding subunits, at least residual COX activity is retained in these cases.
Conclusion
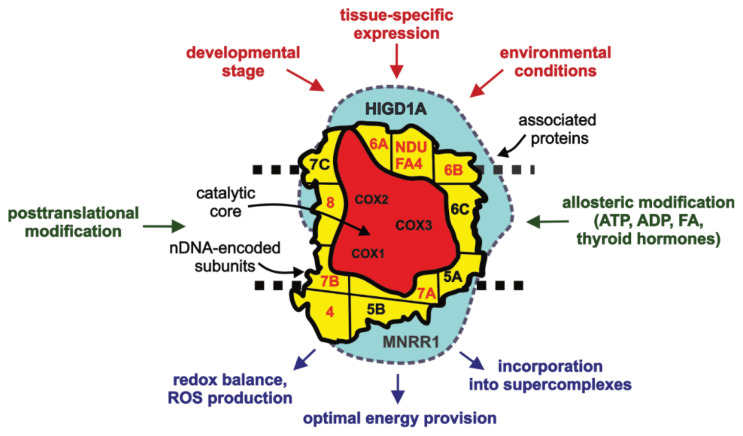
Since the canonical composition of mammalian cytochrome c oxidase was characterized by Kadenbach and coworkers (Kadenbach et al. 1983), research of COX structure and function became extremely complex with discoveries of subunits isoforms, associating proteins, posttranslational modifications or allosteric regulators. Based on the knowledge reviewed in this article, we propose three “levels” of COX structure (Fig. 4). First level is formed by irreplaceable mitochondria-encoded subunits (COX1, COX2, COX3), which form catalytic core necessary for enzymatic function. Nuclear-encoded subunits with their isoforms facilitating tissue-specific and environmental regulation of COX that are tightly bound to the catalytic core and are present in COX complex in stoichiometric ratio represent the second level. Finally, supernumerary non-stoichiometric proteins that may associate with COX only loosely or temporarily represent the final structural layer. With the pallete of different isoforms and proteins associating with COX at the second and third level of multisubunit arrangement, COX may be composed differently under diverse physiological conditions (Brzezinski and Larsson 2003). These structural modifications should optimize COX function to meet current energy demands, enable it to respond to relevant signaling pathways or regulate its interactions with other OXPHOS complexes.
Fig. 4.
Diagram of multi-level COX composition. Three-level COX composition is proposed: i) subunits of catalytic core (red), ii) stoichiometric nuclear-encoded subunits (yellow) with single (annotated in black) or two/multiple isoforms (annotated in red) iii) COX-associated proteins (blue). Factors controlling COX subunit composition on gene-expression level (upper part, in red), and in situ regulatory mechanisms (side, in green), which may act differentially on COX complexes with varying composition, modulate functional output and interactions of OXPHOS complexes (bottom part, in blue).
Disease-associated mutations in nuclear-encoded subunits of COX have started to emerge lately. Their number is expected to increase with the development of modern diagnostic techniques. Even though these pathogenic mutations were annotated in terms of sequence and impact on protein expression, their biochemical manifestation is still missing in many cases. Characterization of these mutations on functional level may lead to better understanding of the role of the respective subunits, but is complicated due to limited access to patient material. Therefore, we propose to employ cell-line based models with uniform genetic background to introduce the pathogenic gene variants. Such models may be investigated without limitations and should help uncover details on etiopathogenic mechanisms of these severe defects.
Acknowledgements
This work was supported by Grant Agency of the Czech Republic (16-13671S) and Ministry of Health of the Czech Republic (NV19-07-00149). Institutional support (Institute of Physiology) was provided by Czech Academy of Sciences (RVO:67985823).
Footnotes
Conflict of Interest
There is no conflict of interest.
References
- ABDULHAG UN, SOIFERMAN D, SCHUELER-FURMAN O, MILLER C, SHAAG A, ELPELEG O, EDVARDSON S, SAADA A. Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur J Hum Genet. 2015;23:159–164. doi: 10.1038/ejhg.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ABU-LIBDEH B, DOUIEV L, AMRO S, SHAHROUR M, TA-SHMA A, MILLER C, ELPELEG O, SAADA A. Mutation in the COX4I1 gene is associated with short stature, poor weight gain and increased chromosomal breaks, simulating Fanconi anemia. Eur J Hum Genet. 2017;25:1142–1146. doi: 10.1038/ejhg.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACÍN-PÉREZ R, FERNÁNDEZ-SILVA P, PELEATO ML, PÉREZ-MARTOS A, ENRÍQUEZ JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- ACÍN-PÉREZ R, GATTI DL, BAI Y, MANFREDI G. Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: coupled mechanisms of energy metabolism regulation. Cell Metab. 2011;13:712–719. doi: 10.1016/j.cmet.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ANDERSSON SGE, ZOMORODIPOUR A, ANDERSSON JO, SICHERITZ-PONTÉN T, ALSMARK UC, PODOWSKI RM, NASLUND AK, ERIKSSON A-SS, WINKLER HH, KURLAND CG, SICHERITZ-PONTÉN T, ALSMARK UC, PODOWSKI RM, NÄSLUND AK, ERIKSSON A-SS, WINKLER HH, KURLAND CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- ANTHONY G, REIMANN A, KADENBACH B. Tissue-specific regulation of bovine heart cytochrome-c oxidase activity by ADP via interaction with subunit VIa. P Natl Acad Sci USA. 1993;90:1652–1656. doi: 10.1073/pnas.90.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARAS S, ARRABI H, PURANDARE N, HÜTTEMANN M, KAMHOLZ J, ZÜCHNER S, GROSSMAN LI. Abl2 kinase phosphorylates Bi-organellar regulator MNRR1 in mitochondria, stimulating respiration. BBA-Mol Cell Res. 2017;1864:440–448. doi: 10.1016/j.bbamcr.2016.11.029. [DOI] [PubMed] [Google Scholar]
- ARAS S, BAI M, LEE I, SPRINGETT R, HÜTTEMANN M, GROSSMAN LI. MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion. 2015;20:43–51. doi: 10.1016/j.mito.2014.10.003. [DOI] [PubMed] [Google Scholar]
- ARAS S, PAK O, SOMMER N, FINLEY R, HÜTTEMANN M, WEISSMANN N, GROSSMAN LI. Oxygen-dependent expression of cytochrome c oxidase subunit 4-2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res. 2013;41:2255–2266. doi: 10.1093/nar/gks1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARNOLD S, KADENBACH B. Cell respiration is controlled by ATP, an allosteric inhibitor of cytochrome-c oxidase. Eur J Biochem. 1997;249:350–354. doi: 10.1111/j.1432-1033.1997.t01-1-00350.x. [DOI] [PubMed] [Google Scholar]
- BABCOCK GT, WIKSTRÖM M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- BAERTLING F, AL-MURSHEDI F, SÁNCHEZ-CABALLERO L, AL-SENAIDI K, JOSHI NP, VENSELAAR H, Van den BRAND MAM, NIJTMANS LGJ, RODENBURG RJT. Mutation in mitochondrial complex IV subunit COX5A causes pulmonary arterial hypertension, lactic acidemia, and failure to thrive. Hum Mutat. 2017;38:692–703. doi: 10.1002/humu.23210. [DOI] [PubMed] [Google Scholar]
- BALSA E, MARCO R, PERALES-CLEMENTE E, SZKLARCZYK R, CALVO E, LANDÁZURI MO, ENRÍQUEZ JA. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- BENDER E, KADENBACH B. The allosteric ATP-inhibition of cytochrome c oxidase activity is reversibly switched on by cAMP-dependent phosphorylation. FEBS Lett. 2000;466:130–134. doi: 10.1016/S0014-5793(99)01773-1. [DOI] [PubMed] [Google Scholar]
- BOCZONADI V, GIUNTA M, LANE M, TULINIUS M, SCHARA U, HORVATH R. Investigating the role of the physiological isoform switch of cytochrome c oxidase subunits in reversible mitochondrial disease. Int J Biochem Cell B. 2015;63:32–40. doi: 10.1016/j.biocel.2015.01.025. [DOI] [PubMed] [Google Scholar]
- BONNE G, SEIBEL P, POSSEKEL S, MARSAC C, KADENBACH B. Expression of human cytochrome c oxidase subunits during fetal development. Eur J Biochem. 1993;217:1099–1107. doi: 10.1111/j.1432-1033.1993.tb18342.x. [DOI] [PubMed] [Google Scholar]
- BRZEZINSKI P, LARSSON G. Redox-driven proton pumping by heme-copper oxidases. BBA-Bioenergetics. 2003;1605:1–13. doi: 10.1016/S0005-2728(03)00079-3. [DOI] [PubMed] [Google Scholar]
- CAO X, CUI Y, ZHANG X, LOU J, ZHOU J, BEI H, WEI R. Proteomic profile of human spermatozoa in healthy and asthenozoospermic individuals. Reprod Biol Endocrin. 2018;16:4–11. doi: 10.1186/s12958-018-0334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARROLL J, FEARNLEY IM, SHANNON R, HIRST J, WALKER JE. Analysis of the subunit composition of complex i from bovine heart mitochondria. Mol Cell Proteomics. 2003;2:117–126. doi: 10.1074/mcp.M300014-MCP200. [DOI] [PubMed] [Google Scholar]
- CIMMINO M, MION F, GOGLIA F, MINAIRE Y, GÉLOËN A. Demonstration of in vivo metabolic effects of 3,5-di-iodothyronine. J Endocrinol. 1996;149:319–325. doi: 10.1677/joe.0.1490319. [DOI] [PubMed] [Google Scholar]
- COGLIATI S, CALVO E, LOUREIRO M, GUARAS AM, NIETO-ARELLANO R, GARCIA-POYATOS C, EZKURDIA I, MERCADER N, VÁZQUEZ J, ENRÍQUEZ JA. Mechanism of super-assembly of respiratory complexes III and IV. Nature. 2016;539:579–582. doi: 10.1038/nature20157. [DOI] [PubMed] [Google Scholar]
- COOPER CE, BROWN GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: chemical mechanism and physiological significance. J Bioenerg Biomembr. 2008;40:533–539. doi: 10.1007/s10863-008-9166-6. [DOI] [PubMed] [Google Scholar]
- DAHOUT-GONZALEZ C. Molecular, functional, and pathological aspects of the mitochondrial ADP/ATP carrier. Physiology. 2006;21:242–249. doi: 10.1152/physiol.00005.2006. [DOI] [PubMed] [Google Scholar]
- DAS J, MILLER ST, STERN DL. Comparison of diverse protein sequences of the nuclear-encoded subunits of cytochrome c oxidase suggests conservation of structure underlies evolving functional sites. Mol Biol Evol. 2004;21:1572–1582. doi: 10.1093/molbev/msh161. [DOI] [PubMed] [Google Scholar]
- DENNERLEIN S, REHLING P. Human mitochondrial COX1 assembly into cytochrome c oxidase at a glance. J Cell Sci. 2015;128:833–837. doi: 10.1242/jcs.161729. [DOI] [PubMed] [Google Scholar]
- DENNERLEIN S, WANG C, REHLING P. Plasticity of mitochondrial translation. Trends Cell Biol. 2017;27:712–721. doi: 10.1016/j.tcb.2017.05.004. [DOI] [PubMed] [Google Scholar]
- ENDOU M, YOSHIDA K, HIROTA M, NAKAJIMA C, SAKAGUCHI A, KOMATSUBARA N, KURIHARA Y. Coxfa4l3, a novel mitochondrial electron transport chain Complex 4 subunit protein, switches from Coxfa4 during spermatogenesis. Mitochondrion. 2020;52:1–7. doi: 10.1016/j.mito.2020.02.003. [DOI] [PubMed] [Google Scholar]
- ENRÍQUEZ JA. Supramolecular Organization of Respiratory Complexes. Annu Rev Physiol. 2016;78:533–561. doi: 10.1146/annurev-physiol-021115-105031. [DOI] [PubMed] [Google Scholar]
- EWART GD, ZHANG YZ, CAPALDI RA. Switching of bovine cytochrome c oxidase subunit Via isoforms in skeletal muscle during development. Febs Lett. 1991;292:79–84. doi: 10.1016/0014-5793(91)80839-U. [DOI] [PubMed] [Google Scholar]
- FONTANESI F, SOTO IC, HORN D, BARRIENTOS A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am J Physiol-Cell Ph. 2006;291:C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- FORNŮSKOVÁ D, STIBŮREK L, WENCHICH L, VINSOVA K, HANSÍKOVÁ H, ZEMAN J. Novel insights into the assembly and function of human nuclear-encoded cytochrome c oxidase subunits 4, 5a, 6a, 7a and 7b. Biochem J. 2010;428:363–374. doi: 10.1042/BJ20091714. [DOI] [PubMed] [Google Scholar]
- FUKUDA R, ZHANG H, KIM J-W, SHIMODA L, DANG CV, SEMENZA GREGG L. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell. 2007;129:111–122. doi: 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- GALATI D, SRINIVASAN S, RAZA H, PRABU SK, KARUNAKARAN C, LOPEZ M, KALYANARAMAN B, AVADHANI NG. Role of nuclear encoded subunit Vb in the assembly and stability of cytochrome c oxidase complex: implications in mitochondrial dysfunction and ROS production. Biochem J. 2009;420:439–449. doi: 10.1042/BJ20090214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAO L, BONILLA-HENAO V, GARCIA-FLORES P, ARIAS-MAYENCO I, ORTEGA-SAENZ P, LOPEZ-BARNEO J. Gene expression analyses reveal metabolic specifications in acute O2 -sensing chemoreceptor cells. J Physiol. 2017a;595:6091–6120. doi: 10.1113/JP274684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAO S-P, SUN H-F, FU W-Y, LI L-D, ZHAO Y, CHEN M-T, JIN W. High expression of COX5B is associated with poor prognosis in breast cancer. Future Oncol. 2017b;13:1711–1719. doi: 10.2217/fon-2017-0058. [DOI] [PubMed] [Google Scholar]
- GOLDBERG A, WILDMAN DE, SCHMIDT TR, HU M, GOODMAN M, WEISS ML, GROSSMAN LI. Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates. P Natl Acad Sci USA. 2003;100:5873–5878. doi: 10.1073/pnas.0931463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROSSMAN LI, PURANDARE N, ARSHAD R, GLADYCK S, SOMAYAJULU M, HUTTEMANN M, ARAS S. MNRR1, a biorganellar regulator of mitochondria. Oxid Med Cell Longev. 2017;2017:6739236. doi: 10.1155/2017/6739236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO R, ZONG S, WU M, GU J, YANG M. Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell. 2017;170:1247–1257.e1212. doi: 10.1016/j.cell.2017.07.050. [DOI] [PubMed] [Google Scholar]
- HALLMANN K, KUDIN AP, ZSURKA G, KORNBLUM C, REIMANN J, STÜVE B, WALTZ S, HATTINGEN E, THIELE H, NÜRNBERG P, RÜB C, VOOS W, KOPATZ J, NEUMANN H, KUNZ WS. Loss of the smallest subunit of cytochrome c oxidase, COX8A, causes Leigh-like syndrome and epilepsy. Brain. 2016;139:338–345. doi: 10.1093/brain/awv357. [DOI] [PubMed] [Google Scholar]
- HAYASHI T, ASANO Y, SHINTANI Y, AOYAMA H, KIOKA H, TSUKAMOTO O, HIKITA M, SHINZAWA-ITOH K, TAKAFUJI K, HIGO S, KATO H, YAMAZAKI S, MATSUOKA K, NAKANO A, ASANUMA H, ASAKURA M, MINAMINO T, GOTO Y-I, OGURA T, KITAKAZE M, KOMURO I, SAKATA Y, TSUKIHARA T, YOSHIKAWA S, TAKASHIMA S. Higd1a is a positive regulator of cytochrome c oxidase. P Natl Acad Sci USA. 2015;112:1553–1558. doi: 10.1073/pnas.1419767112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRST J. Open questions: respiratory chain supercomplexes-why are they there and what do they do? BMC Biol. 2018;16:111. doi: 10.1186/s12915-018-0577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOCK DH, RELJIC B, ANG CS, MUELLNER-WONG L, MOUNTFORD HS, COMPTON AG, RYAN MT, THORBURN DR, STROUD DA. HIGD2A is required for assembly of the COX3 module of human mitochondrial complex IV. Mol Cell Proteomics. 2020;19:1145–1160. doi: 10.1074/mcp.RA120.002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOSLER JP. The influence of subunit III of cytochrome c oxidase on the D pathway, the proton exit pathway and mechanism-based inactivation in subunit I. BBA-Bioenergetics. 2004;1655:332–339. doi: 10.1016/j.bbabio.2003.06.009. [DOI] [PubMed] [Google Scholar]
- HU T, XI J. Identification of COX5B as a novel biomarker in high-grade glioma patients. Oncotargets Ther. 2017;10:5463–5470. doi: 10.2147/OTT.S139243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HÜTTEMANN M, JARADAT S, GROSSMAN LI. Cytochrome c oxidase of mammals contains a testes-specific isoform of subunit VIb - the counterpart to testes-specific cytochrome c? Mol Reprod Dev. 2003;66:8–16. doi: 10.1002/mrd.10327. [DOI] [PubMed] [Google Scholar]
- HÜTTEMANN M, KADENBACH B, GROSSMAN LI. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene. 2001;267:111–123. doi: 10.1016/S0378-1119(01)00385-7. [DOI] [PubMed] [Google Scholar]
- HÜTTEMANN M, KLEWER S, LEE I, PECINOVA A, PECINA P, LIU J, LEE M, DOAN JW, LARSON D, SLACK E. Mice deleted for heart-type cytochrome c oxidase subunit 7a1 develop dilated cardiomyopathy. Mitochondrion. 2012;12:294–304. doi: 10.1016/j.mito.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HÜTTEMANN M, LEE I, LIU J, GROSSMAN LI. Transcription of mammalian cytochrome c oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J. 2007;274:5737–5748. doi: 10.1111/j.1742-4658.2007.06093.x. [DOI] [PubMed] [Google Scholar]
- CHANCE B, WILLIAMS GR. A method for the localization of sites for oxidative phosphorylation. Nature. 1955;176:250–254. doi: 10.1038/176250a0. [DOI] [PubMed] [Google Scholar]
- CHAZOTTE B, HACKENBROCK CR. Lateral diffusion as a rate-limiting step in ubiquinone-mediated mitochondrial electron transport. J Biol Chem. 1989;264:4978–4985. [PubMed] [Google Scholar]
- CHEN W-L, KUO K-T, CHOU T-Y, CHEN C-L, WANG C-H, WEI Y-H, WANG L-S. The role of cytochrome c oxidase subunit Va in non-small cell lung carcinoma cells: association with migration, invasion and prediction of distant metastasis. BMC Cancer. 2012a;12:1–13. doi: 10.1186/1471-2407-12-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN YC, TAYLOR EB, DEPHOURE N, HEO JM, TONHATO A, PAPANDREOU I, NATH N, DENKO NC, GYGI SP, RUTTER J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012b;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN ZX, PERVAIZ S. Involvement of cytochrome c oxidase subunits Va and Vb in the regulation of cancer cell metabolism by Bcl-2. Cell Death Differ. 2010;17:408–420. doi: 10.1038/cdd.2009.132. [DOI] [PubMed] [Google Scholar]
- IMAI Y, MENG H, SHIBA-FUKUSHIMA K, HATTORI N. Twin CHCH Proteins, CHCHD2, and CHCHD10: key molecules of Parkinson’s disease, amyotrophic lateral sclerosis, and frontotemporal dementia. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMAI M, SAIO T, KUMETA H, UCHIDA T, INAGAKI F, ISHIMORI K. Investigation of the redox-dependent modulation of structure and dynamics in human cytochrome c. Biochem Biophys Res Commun. 2016;469:978–984. doi: 10.1016/j.bbrc.2015.12.079. [DOI] [PubMed] [Google Scholar]
- INDRIERI A, RAHDEN VAV, TIRANTI V, MORLEO M, IACONIS D, TAMMARO R, AMATO ID, CONTE I, MAYSTADT I, DEMUTH S, ZVULUNOV A, KUTSCHE K, ZEVIANI M, FRANCO B. Mutations in COX7B Cause Microphthalmia with Linear Skin Lesions, an Unconventional Mitochondrial Disease. Am J Hum Genet. 2012;91:942–949. doi: 10.1016/j.ajhg.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KADENBACH B, BARTH J, AKGÜN R, FREUND R, LINDER D, POSSEKEL S. Regulation of mitochondrial energy generation in health and disease. Biochim Biophys Acta. 1995;1271:103–109. doi: 10.1016/0925-4439(95)00016-W. [DOI] [PubMed] [Google Scholar]
- KADENBACH B, HÜTTEMANN M. The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion. 2015;24:64–76. doi: 10.1016/j.mito.2015.07.002. [DOI] [PubMed] [Google Scholar]
- KADENBACH B, JARAUSCH J, HARTMANN R, MERLE P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate-gel electrophoretic procedure. Anal Biochem. 1983;129:517–521. doi: 10.1016/0003-2697(83)90586-9. [DOI] [PubMed] [Google Scholar]
- KAILA VRI, VERKHOVSKY MI, WIKSTRÖM M. Proton-coupled electron transfer in cytochrome oxidase. Chem Rev. 2010;110:7062–7081. doi: 10.1021/cr1002003. [DOI] [PubMed] [Google Scholar]
- KHALIMONCHUK O, RÖDEL G. Biogenesis of cytochrome c oxidase. Mitochondrion. 2005;5:363–388. doi: 10.1016/j.mito.2005.08.002. [DOI] [PubMed] [Google Scholar]
- KRIEG RC, KNUECHEL R, SCHIFFMANN E, LIOTTA LA, III EFP, HERRMANN PC. Mitochondrial proteome: Cancer-altered metabolism associated with cytochrome c oxidase subunit level variation. Proteomics. 2004;4:2789–2795. doi: 10.1002/pmic.200300796. [DOI] [PubMed] [Google Scholar]
- KRZYZOSIAK J, McMILLAN G, MOLAN P, VISHWANATH R. Protein tyrosine phosphorylation during prolonged in vitro incubation of ejaculated bovine spermatozoa is regulated by the oxidative state of the medium 1. Biol Reprod. 2000;62:1615–1623. doi: 10.1095/biolreprod62.6.1615. [DOI] [PubMed] [Google Scholar]
- LANG BF, BURGER G, O’KELLY CJ. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- LANNI A, MORENO M, LOMBARDI A, GOGLIA F, GENERALE F, II F, MEZZOCANNONE V. Calorigenic effect of diiodothyronines in the rat. J Physiol. 1996;494:831–837. doi: 10.1113/jphysiol.1996.sp021536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAPUENTE-BRUN E, MORENO-LOSHUERTOS R, ACÍN-PÉREZ R, LATORRE-PELLICER A, COLAS C, BALSA E, PERALES-CLEMENTE E, QUIRÓS PM, CALVO E, RODRIGUEZ-HERNANDEZ MA, NAVAS P, CRUZ R, CARRACEDO A, LOPEZ-OTIN C, PEREZ-MARTOS A, FERNÁNDEZ-SILVA P, FERNÁNDEZ-VIZARRA E, ENRÍQUEZ JA. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- LAZAROU M, SMITH SM, THORBURN DR, RYAN MT, McKENZIE M. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. FEBS J. 2009;276:6701–6713. doi: 10.1111/j.1742-4658.2009.07384.x. [DOI] [PubMed] [Google Scholar]
- LEE I, BENDER E, KADENBACH B. Control of mitochondrial membrane potential and ROS formation by reversible phosphorylation of cytochrome c oxidase. Mol Cell Biochem. 2002;234:63–70. doi: 10.1007/978-1-4615-1087-1_7. [DOI] [PubMed] [Google Scholar]
- LEE I, KADENBACH B. Palmitate decreases proton pumping of liver-type cytochrome c oxidase. Eur J Biochem. 2001;268:6329–6334. doi: 10.1046/j.0014-2956.2001.02602.x. [DOI] [PubMed] [Google Scholar]
- LEE I, SALOMON AR, FICARRO S, MATHES I, LOTTSPEICH F, GROSSMAN LI, HU M. cAMP-dependent tyrosine phosphorylation of subunit i inhibits cytochrome c oxidase activity. J Biol Chem. 2005;280:6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- LETTS JA, FIEDORCZUK K, SAZANOV LA. The architecture of respiratory supercomplexes. Nature. 2016;537:644–648. doi: 10.1038/nature19774. [DOI] [PubMed] [Google Scholar]
- LETTS JA, SAZANOV LA. Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nature Structural and Molecular Biology. 2017;24:800–808. doi: 10.1038/nsmb.3460. [DOI] [PubMed] [Google Scholar]
- LIANG H. A rare polymorphism of the COX7B2 gene in a Cantonese family with nasopharyngeal carcinoma. Sci China Ser C. 2004;47:449. doi: 10.1360/03yc0037. [DOI] [PubMed] [Google Scholar]
- LINDER D, FREUND R, KADENBACH B. Species-specific expression of cytochrome c oxidase isozymes. Comp Biochem Phys B. 1995;112:461–469. doi: 10.1016/0305-0491(95)00093-3. [DOI] [PubMed] [Google Scholar]
- LITTLE AG, KOCHA KM, LOUGHEED SC, MOYES CD. Evolution of the nuclear-encoded cytochrome oxidase subunits in vertebrates. Physiol Genomics. 2010;42:76–84. doi: 10.1152/physiolgenomics.00015.2010. [DOI] [PubMed] [Google Scholar]
- LUDWIG B, BENDER E, ARNOLD S, HÜTTEMANN M, LEE I, KADENBACH B. Cytochrome c oxidase and the regulation of oxidative phosphorylation. ChemBioChem. 2001;2:392–403. doi: 10.1002/1439-7633(20010601)2:6<392::AID-CBIC392>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- MASSA V, FERNÁNDEZ-VIZARRA E, ALSHAHWAN S, BAKHSH E, GOFFRINI P, FERRERO I, MEREGHETTI P, ADAMO PD, GASPARINI P, ZEVIANI M. Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am J Hum Genet. 2008;82:1281–1289. doi: 10.1016/j.ajhg.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEWEN JE, KO C, KLOECKNER-GRUISSEM B, POYTON RO. Nuclear functions required for cytochrome c oxidase biogenesis in saccharomyces cerevisiae. J Biol Chem. 1986;261:11872–11879. [PubMed] [Google Scholar]
- MICK DU, DENNERLEIN S, WIESE H, REINHOLD R, PACHEU-GRAU D, LORENZI I, SASARMAN F, WERAARPACHAI W, SHOUBRIDGE EA, WARSCHEID B, REHLING P. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell. 2012;151:1528–1541. doi: 10.1016/j.cell.2012.11.053. [DOI] [PubMed] [Google Scholar]
- MILENKOVIC D, BLAZA JN, LARSSON NG, HIRST J. The enigma of the respiratory chain supercomplex. Cell Metab. 2017;25:765–776. doi: 10.1016/j.cmet.2017.03.009. [DOI] [PubMed] [Google Scholar]
- MILLETT F, De JONG C, PAULSON L, CAPALDI RA. Identification of specific carboxylate groups on cytochrome c oxidase that are involved in binding cytochrome c. Biochemistry. 1983;22:546–552. doi: 10.1021/bi00272a004. [DOI] [PubMed] [Google Scholar]
- MORENO-DOMINGUEZ A, ORTEGA-SAENZ P, GAO L, COLINAS O, GARCIA-FLORES P, BONILLA-HENAO V, ARAGONES J, HUTTEMANN M, GROSSMAN LI, WEISSMANN N, SOMMER N, LOPEZ-BARNEO J. Acute O2 sensing through HIF2alpha-dependent expression of atypical cytochrome oxidase subunits in arterial chemoreceptors. Sci Signal. 2019 doi: 10.1126/scisignal.aay9452. [DOI] [PubMed] [Google Scholar]
- MOURIER A, MATIC S, RUZZENENTE B, LARSSON NG, MILENKOVIC D. The respiratory Chain supercomplex organization is independent of COX7A2L isoforms. Cell Metab. 2014;20:1069–1075. doi: 10.1016/j.cmet.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIJTMANS LGJ, TAANMAN J-W, MUIJSERS AO, SPEIJER D, BOGERT CVANDEN. Assembly of cytochrome c oxidase in cultured human cells. Eur J Biochem. 1998;254:389–394. doi: 10.1046/j.1432-1327.1998.2540389.x. [DOI] [PubMed] [Google Scholar]
- PAJUELO REGUERA D, CUNATOVA K, VRBACKY M, PECINOVA A, HOUSTEK J, MRACEK T, PECINA P. Cytochrome c oxidase subunit 4 isoform exchange results in modulation of oxygen affinity. Cells. 2020:9. doi: 10.3390/cells9020443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PANNALA VR, CAMARA AK, DASH RK. Modeling the detailed kinetics of mitochondrial cytochrome c oxidase: Catalytic mechanism and nitric oxide inhibition. J Appl Physiol (1985) 2016;121:1196–1207. doi: 10.1152/japplphysiol.00524.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PECINA P, HOUŠŤKOVÁ H, HANSÍKOVÁ H, ZEMAN J, HOUŠTĚK J. Genetic defects of cytochrome c oxidase assembly. Physiol Res. 2004;53:S213–S223. [PubMed] [Google Scholar]
- PIERRON D, WILDMAN DE, HÜTTEMANN M, MARKONDAPATNAIKUNI GC. Cytochrome c oxidase: Evolution of control via nuclear subunit addition. BBA-Bioenergetics. 2012;1817:590–597. doi: 10.1016/j.bbabio.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PILLAI NR, ALDHAHERI NS, GHOSH R, LIM J, STREFF H, NAYAK A, GRAHAM BH, HANCHARD NA, ELSEA SH, SCAGLIA F. Biallelic variants in COX4I1 associated with a novel phenotype resembling Leigh syndrome with developmental regression, intellectual disability, and seizures. Am J Med Genet A. 2019:1–6. doi: 10.1002/ajmg.a.61288. [DOI] [PubMed] [Google Scholar]
- PITCEATHLY RDS, RAHMAN S, WEDATILAKE Y, POLKE JM, CIRAK S, FOLEY AR, SAILER A, HURLES ME, STALKER J, HARGREAVES I, WOODWARD CE, SWEENEY MG, MUNTONI F, HOULDEN H, TAANMAN J-W, HANNA M. NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Reports. 2013;3:1795–1805. doi: 10.1016/j.celrep.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PITCEATHLY RDS, TAANMAN J-W. NDUFA4 (Renamed COXFA4) Is a cytochrome -c oxidase subunit. Trends Endocrin Met. 2018;29:452–454. doi: 10.1016/j.tem.2018.03.009. [DOI] [PubMed] [Google Scholar]
- RADFORD NB, WAN B, RICHMAN A, SZCZEPANIAK LS, LI J-L, LI K, PFEIFFER K, SCHA H, GARRY DJ, MOREADITH RW, NINA B, WAN B, RICHMAN A, SZCZEPANIAK S, LI J-L, LI K, PFEIFFER K, GARRY DJ, RANDALL W. Cardiac dysfunction in mice lacking cytochrome- c oxidase subunit VIaH. Am J Physiol-Heart C. 2002;282:726–733. doi: 10.1152/ajpheart.00308.2001. [DOI] [PubMed] [Google Scholar]
- RAK M, BENIT P, CHRETIEN D, BOUCHEREAU J, SCHIFF M, EL-KHOURY R, TZAGOLOFF A, RUSTIN P. Mitochondrial cytochrome c oxidase deficiency. Clin Sci. 2016;130:393–407. doi: 10.1042/CS20150707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RICHTER-DENNERLEIN R, OELJEKLAUS S, LORENZI I, RONSÖR C, BARETH B, SCHENDZIELORZ AB, WANG C, WARSCHEID B, REHLING P, DENNERLEIN S. Mitochondrial protein synthesis adapts to influx of nuclear-encoded protein. Cell. 2016;167:471–483.e410. doi: 10.1016/j.cell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMPSON V, ALLEYNE T. Cytochrome c/cytochrome c oxidase interaction - Direct structural evidence for conformational changes during enzyme turnover. Eur J Biochem. 2001;268:6534–6544. doi: 10.1046/j.0014-2956.2001.02608.x. [DOI] [PubMed] [Google Scholar]
- SHARMA V, ALA-VANNESLUOMA P, VATTULAINEN I, WIKSTRÖM M, RÓG T. Role of subunit III and its lipids in the molecular mechanism of cytochrome c oxidase. BBA-Bioenergetics. 2015;1847:690–697. doi: 10.1016/j.bbabio.2015.04.007. [DOI] [PubMed] [Google Scholar]
- SHTEYER E, SAADA A, SHAAG A, AL-HIJAWI FA, KIDESS R, REVEL-VILK S, ELPELEG O. Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am J Hum Genet. 2009;84:412–417. doi: 10.1016/j.ajhg.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHÄGGER H, PFEIFFER K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHMIDT TR, GOODMAN M, GROSSMAN LI. Molecular evolution of the COX7A gene family in primates. Mol Biol Evol. 1999;16:619–626. doi: 10.1093/oxfordjournals.molbev.a026144. [DOI] [PubMed] [Google Scholar]
- SOMMER N, HÜTTEMANN M, PAK O, SCHEIBE S, KNOEPP F, SINKLER C, MALCZYK M, GIERHARDT M, ESFANDIARY A, KRAUT S, JONAS F, VEITH C, ARAS S, SYDYKOV A, ALEBRAHIMDEHKORDI N, GIEHL K, HECKER M, BRANDES RP, SEEGER W, GRIMMINGER F, GHOFRANI HA, SCHERMULY RT, GROSSMAN LI, WEISSMANN N. Mitochondrial complex IV subunit 4 isoform 2 is essential for acute pulmonary oxygen sensing. Circ Res. 2017;121:424–438. doi: 10.1161/CIRCRESAHA.116.310482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STIBŮREK L, ZEMAN J. Assembly factors and ATP-dependent proteases in cytochrome c oxidase biogenesis. BBA-Bioenergetics. 2010;1797:1149–1158. doi: 10.1016/j.bbabio.2010.04.006. [DOI] [PubMed] [Google Scholar]
- STROGOLOVA V, FURNESS A, ROBB-MCGRATH M, GARLICH J, STUART RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZKLARCZYK R, HUYNEN MA. Mosaic origin of the mitochondrial proteome. Proteomics. 2010;10:4012–4024. doi: 10.1002/pmic.201000329. [DOI] [PubMed] [Google Scholar]
- TAMIYA G, MAKINO S, HAYASHI M, ABE A, NUMAKURA C, UEKI M, TANAKA A, ITO C, TOSHIMORI K, OGAWA N, TERASHIMA T, MAEGAWA H, YANAGISAWA D, TOOYAMA I, TADA M, ONODERA O, HAYASAKA K. A mutation of COX6A1 causes a recessive axonal or mixed form of charcot-marie-tooth disease. Am J Hum Genet. 2014;95:294–300. doi: 10.1016/j.ajhg.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIMON-GOMEZ A, GARLICH J, STUART RA, UGALDE C, BARRIENTOS A. Distinct roles of mitochondrial higd1a and higd2a in respiratory complex and supercomplex biogenesis. Cell Reports. 2020;31:107607. doi: 10.1016/j.celrep.2020.107607. [DOI] [PubMed] [Google Scholar]
- TIMÓN-GÓMEZ A, NÝVLTOVÁ E, ABRIATA LA, VILA AJ, HOSLER J, BARRIENTOS A. Mitochondrial cytochrome c oxidase biogenesis: recent developments. Semin Cell Dev Biol. 2017;76:163–178. doi: 10.1016/j.semcdb.2017.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSUKIHARA T, AOYAMA H, TOMIZAKI T, YAMAGUCHI H, NAKASHIMA R, YAONO R, YOSHIKAWA S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- TZAGOLOFF A, DIECKMANN CL. PET Genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/MMBR.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VIDONI S, HARBOUR ME, GUERRERO-CASTILLO S, SIGNES A, DING S, FEARNLEY IM, TAYLOR RW, TIRANTI V, ARNOLD S, FERNÁNDEZ-VIZARRA E, ZEVIANI M. MR-1S Interacts with PET100 and PET117 in module-based assembly of human cytochrome c oxidase. Cell Reports. 2017;18:1727–1738. doi: 10.1016/j.celrep.2017.01.044. [DOI] [PubMed] [Google Scholar]
- WEISHAUPT A, KADENBACH B. Selective removal of subunit VIb increases the activity of cytochrome c oxidase. Biochemistry. 1992;31:11477–11481. doi: 10.1021/bi00161a028. [DOI] [PubMed] [Google Scholar]
- WILLIAMS L, VALNOT I, RUSTIN P, TAANMAN J-W. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J Biol Chem. 2004;279:7462–7469. doi: 10.1074/jbc.M309232200. [DOI] [PubMed] [Google Scholar]
- WITTIG I, SCHÄGGER H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5:4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- WU M, GU J, GUO R, HUANG Y, YANG M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell. 2016;167:1598–1609.e1510. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- YANG W-L, IACONO L, TANG W-M, CHIN K-V. Novel function of the regulatory subunit of protein kinase A: regulation of cytochrome c oxidase activity and cytochrome c release. Biochemistry. 1998;37:14175–14180. doi: 10.1021/bi981402a. [DOI] [PubMed] [Google Scholar]
- ZHANG K, WANG G, ZHANG X, HÜTTEMANN PP, QIU Y, LIU J, MITCHELL A, LEE I, ZHANG C, LEE JS, PECINA P, WU G, YANG ZQ, HÜTTEMANN M, GROSSMAN LI. COX7AR is a stress-inducible mitochondrial COX subunit that promotes breast cancer malignancy. Sci Rep-UK. 2016;6:1–12. doi: 10.1038/srep31742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZONG S, WU M, GU J, LIU T, GUO R, YANG M. Structure of the intact 14-subunit human cytochrome c oxidase. Cell Res. 2018;28:1026–1034. doi: 10.1038/s41422-018-0071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]