Summary
In as early as 1997, the World Health Organization officially recognized obesity as a chronic disease. The current epidemic of obesity and overweightness has aroused great interest in the study of adipose tissue formation. The transcription factor peroxisome proliferator-activated receptor γ (PPARγ) binds to the target gene promoter regulatory sequences, acting as a key factor in regulating the differentiation of preadipocytes in the adipose tissue, and plays an important role in regulating the adipocyte metabolism. A further understanding of the structure and expression characteristics of PPARγ, in addition to its mechanisms of action in adipocyte differentiation, may be applied to control obesity and prevent obesity-related diseases. In this article, recent studies investigating the effect of regulating PPARγ on adipocyte differentiation are reviewed. In particular, the structural characteristics, expression patterns, and molecular mechanisms of PPARγ function in adipocyte differentiation are considered.
Keywords: PPARγ, Adipocytes, White adipose tissue, Brown adipose tissue, Differentiation regulation
Introduction
The current epidemic of obesity and overweight has caused a surge of interest in the study of adipose tissue formation. Recent studies have shown that adipocytes are not only the site of lipid storage, but also the source of many adipose-derived secretory factors that can affect and regulate the metabolism of adipose tissue, other tissues, and consequently the whole organism (Divella et al. 2016). Therefore, it is of great clinical significance to study the regulatory mechanisms of adipocyte proliferation and differentiation to control the formation of adipocytes and regulate immune function.
The proliferation and differentiation of adipocytes are complex processes controlled by multiple genes, in which peroxisome proliferator-activated receptor γ (PPARγ) is the main regulator. Defects in PPARs have been linked to lipodystrophy, obesity, and insulin resistance as a result of impaired adipose tissue expansion and functionality. We believe a greater understanding of PPARs has a significant potential for improving health outcomes and interventions related to obesity, diabetes, and diet.
PPARγ
PPARγ subtypes
PPARs were first discovered in 1990, and they belong to the steroid, thyroid, and retinoic acid receptor superfamily. The name of this group of nuclear receptors was derived from their ability to stimulate the proliferation of peroxisomes in rats (Issemann and Green 1990). In humans, three nuclear isoforms were discovered (PPARα, PPARβ/δ, and PPARγ) that differ in tissue distribution and specific function. PPARα is mainly expressed in the liver, kidney, heart, brown adipose tissue (BAT), and muscle, and is closely linked to the lipid metabolism (Kersten et al. 2000). PPARβ/δ is expressed in most cell types, most widely in the brain, adipose tissue and skin, and plays roles in fatty acid oxidation and energy balance (Peters et al. 2000). PPARγ is the most widely studied subtype; it is expressed predominantly in adipose tissues, both white and brown (Vella et al. 2016). PPARγ is a key transcriptional regulatory factor for adipocyte production in humans, directly regulating adipocyte differentiation and the expression of genes related to lipid metabolism (Zhang and Li 2010). PPARγ is also expressed in monocytes, macrophages (including foam cells), cardiomyocytes, vascular smooth muscle cells, and endothelial cells, among others (Sato et al. 2005). A large number of in vitro and in vivo experiments have shown that PPARγ exerts anti-inflammatory effects by inhibiting the transcription of pro-inflammatory genes. Accordingly, activation of PPARγ signaling reduces the levels of a large number of inflammatory factors (Peng et al. 2015), thereby playing a protective role against tissue injury, including the lungs and brain (Victor et al. 2006).
The human PPARγ gene is located at position 3p25 of chromosome 3 and contains nine exons that are transcribed to generate four isomers. To date, seven diverse mRNA transcripts of PPARγ (γ1–γ7) have been identified, which are differentially generated based on initiation and alternative splicing of five exons in the terminal region and six exons in the open reading frame (Kvandova et al. 2016). PPARγ1 is expressed only in cardiac and skeletal muscle cells, vascular smooth muscle cells, and endothelial cells, while PPARγ2 and PPARγ4 isoforms are primarily expressed in adipocytes, macrophages, and the colon (Kvandova et al. 2016). PPARγ3 is expressed in adipocytes and the colon. The N-terminal of human PPARγ2 protein has 28 amino acid residues, 13 longer than the N-terminal of PPARγ1. Moreover, the ability of PPARγ2 to induce adipocyte formation is higher than that of PPARγ1 (Aprile et al. 2014).
PPARγ activation by ligands
PPARγ is a ligand-dependent transcription factor. The ligands of PPARγ are mainly a variety of free fatty acids (FFA) and their derivatives, which can be divided into natural and synthetic ligands. Natural ligands mainly come from food and metabolic products of the body, including long-chain polyunsaturated FFA such as linoleic acid, linolenic acid, and arachidonic acid, as well as some arachidonic acid derivatives, and prostaglandins (PG), including l, 5-deoxyprostaglandin J2, and pros-taglandin A. The specificity of natural ligands is low; however, specificity is higher for synthetic ligands. Synthetic ligands include the insulin-sensitizing thiazolidinediones (TZD), such as troglitazone and pioglitazone, and non-steroidal anti-inflammatory drugs, such as indomethacin and niflumic acid (Hallenborg et al. 2016).
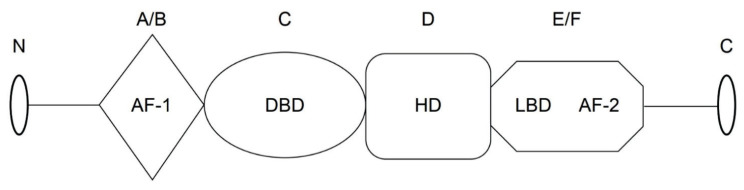
Following complex formation with a ligand, PPARs become activated and bind to the retinoid X receptor (RXR) to form a PPAR-RXR transcriptional complex (Gearing et al. 1993). Similar to other nuclear receptor proteins, the PPARs and RXRs have a standard blueprint with four structural and functional elements (Fig. 1) (Liu et al. 2016). 1) Non-ligand-dependent transcriptional activation domain (NLD); region A/B, in which the activation function-1 motif (AF-1) domain is the target for phosphorylation, and phosphorylation of AF-1 inhibits PPAR activity. 2) DNA-binding domain (DBD); region C, in which the highly conserved zinc-finger structure promotes the binding of the receptor to the DNA sequence in the promoter region of the target gene. 3) Transcriptional activity regulatory domain (HD); region D, the binding of the nuclear factor to the binding site of its cofactor affects the activity of PPAR. 4) Ligand-binding domain (LBD); region E/F, located at the c-terminal ligand-binding region forms a heterodimer, and AF-2 accelerates the binding with a cofactor, and ultimately assists in the transcription process.
Fig. 1.
Schematic representation of the domain structures of PPARγ in humans. AF-1, N-terminal ligand-independent transcriptional activation domain (A/B); DBD, DNA-binding domain (C); HD, transcriptional activity regulatory domain (D); LBD/AF-2, ligand-binding domain/activation function 2 (E/F).
The regulatory mechanism of PPARγ
Zinc fingers in the DBD bind to peroxisome proliferator response elements on the target gene promoter to regulate transcription and inhibit or activate the expression of target genes. Peroxisome proliferator response elements exist in the promoters of several genes related to glucose and lipid metabolism, such as fatty acid synthase, pyruvate carboxylase, and glucose transporter 4 (El-Jack et al. 1999). The phosphorylation state of PPARγ is regulated by its AF-1 domain (Lee and Kim 2015). PPARγ interacts with several other signaling pathways, including Janus kinase-signal transducer and activator of transcription (JAK-STAT), nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), and activator protein 1 (AP-1). PPARγ competitively binds to the synergistic activation factors CBP and P300 to inhibit AP-1 and STAT activity, blocking the production of associated pro-inflammatory cytokines (interleukin-6 (IL-6), tumor necrosis factor α (TNFα)). PPARγ can also directly bind to the P50/P56 subunit of NF-κB, forming an inhibitory transcriptional complex that inhibits the expression of NF-κB-dependent genes (Hong et al. 2003). The exact mechanisms of interactions between PPARγ and other signaling pathways remain to be fully elucidated. PPARγ is a nutrient sensor that regulates several homeostatic functions, such as glucose and lipid metabolism, by regulating the uptake, utilization, oxidation, and storage of FFA, in addition to its role in modulating inflammatory pathways.
Adipocyte differentiation
Origin of adipocytes
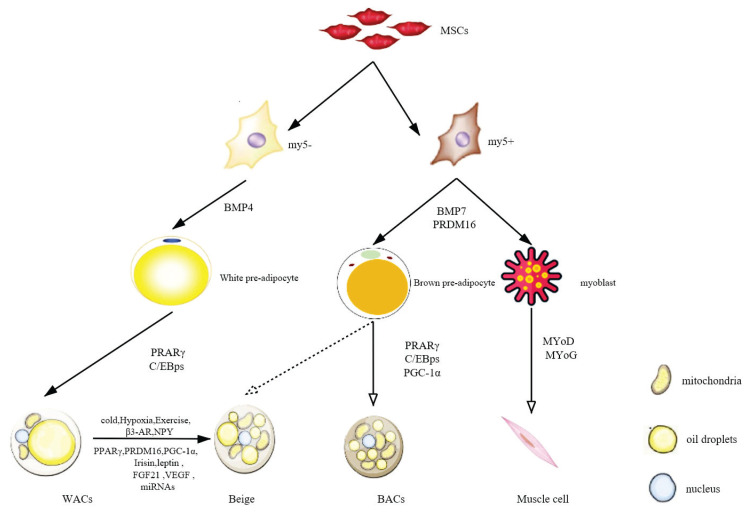
Mesenchymal stem cells (MSCs) (Fig. 2) are non-hematopoietic pluripotent stem cells with high self-renewal and multidifferentiation potential in the mesoderm. Under certain conditions, they differentiate into specific somatic cells of each hypoderm (Goudarzi et al. 2018).
Fig. 2.
Schematic of adipocyte differentiation. MSCs, mesenchymal stem cells; Myf5, myogenic regulatory factor 5; PPARγ, peroxisome proliferator-activated receptor γ; C/EBP, CCAAT enhancer-binding protein; PGC-1α, PPARγ coactivator-1α; MyoD/MyoG, myodifferentiation factor; WACs, white adipocyte cells; Beige, beige adipose tissue; BACs, brown adipocyte cells; β3-AR, β3-adrenergic receptors; NPY, neuropeptide Y; PRMD16, positive regulatory domain zinc-finger protein 16; FGF-21, fibroblast growth factor 21; VEGF, vascular endothelial growth factor.
Adipocyte differentiation can be divided into two stages. First, in the decisive stage, MSCs receive external signals and start to differentiate into precursor adipocytes. Specific molecular mechanisms are still unknown (Chu and Tao 2017). Second, terminal differentiation (the differentiation of precursor adipocytes into mature adipocytes), includes three stages: The proliferative, differentiative, and mature stages. During the proliferative stage, fusion and contact inhibition of the precursor adipocytes are followed by the entrance into the cell cycle. CCAAT enhancer-binding protein (C/EBP) family β and δ subtypes are expressed, and cell differen-tiation is initiated. In the terminal differentiative stage, resting cells move into the growth stage, and transcription factors such as PPARγ and C/EBPα are expressed at high levels, activating the expression of related genes. Cells then accumulate lipid droplets and become mature adipocytes. The mature adipocyte stage is marked by the cell being filled with a single large fat droplet, high expression of adipocyte marker genes, and secretion of cytokines that are involved in insulin sensitivity, energy balance regulation, and other processes (Tong and Hotamisligil 2001).
Differences between three types of adipocytes
White adipocyte cells (WACs)
WACs are thought to originate in the lateral mesoderm from non-myogenic factor 5 (Myf5)-expressing precursors. WACs contain large and stable lipid droplets and few mitochondria, and are found throughout the body, mainly under the skin and around internal organs. WACs store energy in the form of triglycerides and have endocrine functions. For example, WACs secrete FFA and adipocytokines, such as leptin, adiponectin, TNFα and IL-6, which act on distal tissues, including the brain, liver and muscle to regulate food intake, energy homeostasis and insulin sensitivity (Rosen et al. 2006). White adipose tissue (WAT) is therefore referred to as an active endocrine organ.
Brown adipocyte cells (BACs)
BACs and skeletal muscle cells originate from the same paraxial mesodermal cells that specifically express Myf5. Myf5 stimulates the gene transcription profile, promoting muscle differentiation, which partly explains the high metabolic activity of classic BACs (Seale et al. 2008).
BACs contain lipid droplets that are small, abundant, and dispersed throughout the cytoplasm. These cells contain a large number of mitochondria, consistent with their function of burning fat and generating heat. BAT is mainly distributed around the spine, collarbone, and adrenal glands. Extremely high mitochondrial content, dense vascular network and extensive nerve supply enable BAT to have important metabolic functions (Qian et al. 2015). Uncoupling protein 1 (UCP1) is a specific brown fat cell marker, which exists in the mitochondrial intima and regulates the thermogenesis of BAT by uncoupling the oxidative phosphorylation of ATP synthesis. Through this uncoupling, BAT protects against the cold and also burns excess fat and sugar, preventing the body from storing excess fat. Thus, both WACs and BACs play critical roles in maintaining whole-body energy homeostasis (Pellegrinelli et al. 2016).
Beige
In 1992, Cousin et al. found that if rodents are exposed to a cold environment for a prolonged time, BAC-like cells are found in their subcutaneous WAT. Like typical BACs, these adipocytes display many small lipid droplets and mitochondria scattered inside the cell and can generate heat. However, without external stimulation, BAC-like cells are not found in the normal subcutaneous WAT. This special type of adipose tissue is called the “beige adipose tissue” (Cousin et al. 1992).
The three types of adipocytes differ in gene expression patterns. Specifically, WACs express transcription factor 21 (TCF21), BACs express zinc-finger protein of the cerebellum 1 (ZIC1), and beige adipocytes express homeobox C9 (HOXC9), transmembrane protein 26 (TMEM26), cluster of diffe-rentiation 137 (CD137), and T-box transcription factor l (TBX1) (Walden et al. 2012).
There are various hypotheses on the origin of beige adipose tissue. One widely accepted theory proposes that classical BACs and skeletal muscle cells are derived from the Myf5-positive precursor lineage, and WACs and beige adipocytes are derived from Myf5-negative lineages. However, Myf5- and non-Myf5-expressing lineages are all found in WAT (Sanchez-Gurmaches et al. 2012). Expression of beige adipocyte-specific genes is also closely related to the location of the adipocytes within the tissue. Analysis of samples from human adult neck subcutaneous adipose tissue biopsies showed that, from the shallowest to the deepest tissue, the shallowest fat contains only WACs, while cells in the middle may be beige, and BACs are located in the muscle sheath. Furthermore, the deeper they observed into the biopsy sample, the higher the expression of the BAC-specific genes they found (Sanchez-Gurmaches et al. 2012). This diversity in the origin of beige adipocytes suggests that beige, white, and brown adipocytes cannot be distinguished solely by Myf5 expression (Table 1).
Table 1.
Comparison of the characteristics of the three types of adipocytes in humans.
| Characteristic | WACs | BACs | Beige |
|---|---|---|---|
| Distribution | Subcutaneous, perivisceral | Baby shoulder blade, around the kidneys | Adult neck, clavicle, armpit, around the spine, within WACs |
| Content | Majority of the body | Few and decreases with age | Few |
| Origin | Myf5 | Myf5+ | Myf5+/Myf5− |
| Molecular marker | TCF2l | ZIC1 | HOXC9, TMEM26 |
| Function | Store energy | Generate heat | Generate heat |
| Gene transcription | PPARγ2, C/EBPβ | PPARγ2, C/EBPβ, PRDM16 | PPARγ, PRDM16, PGC-1α |
BACs, brown adipose cells; WACs, white adipocyte cells; Beige, beige adipose tissue; C/EBPβ, CCAAT enhancer-binding protein β; HOXC9, homologous box C9; Myf5, myogenic factor 5; PPAR, peroxisome proliferator-activated receptor; PRDM16, positive regulatory domain zinc-finger protein 16; TCF21, transcription factor 21; TMEM26, tumor microenvironment metastasis 26; ZIC1, zinc-finger structure of cerebellum 1.
Key factors in adipocytes differentiation
PGC-1α
PPARγ coactivator-1α (PGC-1α) belongs to the PGC1 family and is a major transcriptional coactivator of mitochondrial synthesis and oxidative metabolism genes in most cells, including BAC and skeletal muscle cells. It promotes the expression of UCP1 and stimulates mitochondrial biosynthesis (Puigserver et al. 1998). In vitro experiments have demonstrated that the overexpression of PGC-1α in MSC blocks the differentiation of MSC into adipocytes. When PGC-1α is overexpressed in MSC, the expression of mitochondrial biosynthesis, respiration, and thermogenesis genes in MSC increase, suggesting that PGC-1α mediates the differentiation of MSC into BACs (Liu et al. 2004).
C/EBPs
CCAAT/enhancer-binding protein (C/EBPs) has three subtypes: C/EBPα, C/EBPβ, and C/EBPδ. C/EBPβ coexpression with positive regulatory domain zinc-finger protein 16 (PRDM16) induces differentiation of fibro-blasts into BACs. In the absence of C/EBPβ, the differentiation of fibroblasts into BACs, induced by PRDM16, is blocked, UCP1 expression is decreased, and skeletal muscle-specific gene expression is enhanced (Kajimura et al. 2009). Animal experiments have shown that high expression of C/EBPβ improves the sensitivity of cAMP, leading to an upregulation of PGC-1α and UCP1, which in turn leads to an increase in the number of beige adipocytes (Karamitri et al. 2009). Follow-up studies showed that C/EBPβ and C/EBPδ adjust the promoter element of their adjacent gene, and thereby transcriptionally activate C/EBPα. Upon activation, C/EBPα leads to PPARγ expression. Furthermore, PPARγ activity is dependent on histone acetylation 1 (HDAC1), as protease enzymatic degradation dissociates HDAC1 from a complex of C/EBPβ associated with the C/EBPα, thus promoting the expression of C/EBPβ and C/EBPα (Birsoy et al. 2008). Thus, C/EBPs and PPARγ mutually promote, and activate the PGC-1α signaling pathway through cAMP, promoting browning/beiging of adipose tissue.
PRDM16
PRDM16 is highly expressed in BAT and is not only the determinant gene of BAC directional differentiation but also the “on/off” gene for thermogenesis initiation and regulation of skeletal muscle and BAT differentiation. Therefore, PRDM16 determines the direction of differentiation for Myf5-expressing precursor cells (Harms et al. 2014). High expression of PRDM16 induces the differentiation of precursor cells to BACs and the expression of BAC-specific genes (including UCP1 and PGC-1α) by inhibiting the expression of MyoD and MyoG in myoblasts (Iida et al. 2015).
The PLDLS motif exists in the suppression domain of PRDM16, which is an evolutionarily conserved sequence consisting of five consecutive amino acids (AA 804–808). The C-terminal binding protein (CtBP) binds to the PLDLS motif to form a PRDM16/CtBP complex, and binds to specific genes in WACs to inhibit their expression. PGC-1α transposable CtBP binds to PRDM16 and forms a PRDM16/PGC-1α transcriptional complex that activates BAC-specific genes, including PGC-1α (Shingo Kajimura et al. 2008). PRDM16 promotes the expression of BAC-specific genes by recruiting related proteins (PPARα/γ, C/EBPβ and MED1) to the promoter or enhancer region of these genes. PRDM16 binding sites are found in the promoter regions of BAC-specific genes (Harms et al. 2015). The transcription complex formed by the combination of PRDM16 with C/EBPβ induces the expression of PPARγ and PGC-1α, and PRDM16 subsequently binds to PPARγ and PGC-1α, promoting the expression of UCP1 (Kajimura et al. 2009). These studies confirm that PRDM16 is the main regulator of BAC formation, by activating BAC-related genes and inhibiting the expression of WAC-related genes (Kajimura et al. 2008).
PPARγ
PPARγ plays an irreplaceable role in regulating the differentiation of WAT and BAT. MSCs expressing PPARγ2 differentiate into WACs, and MSCs expressing PPARγ2 and C/EBPβ, or PPARγ2-C/EBPβ-PRDM16 differentiate into BACs. No single cytokine has been found to promote adipocyte differentiation when PPARγ is knocked out (Lee and Ge 2014). PPARγ knockout mice result in the loss of adipogenic differentiation, which manifests as adipose atrophy and insulin resistance (Barak et al. 1999). Many of the key adipocyte-producing signaling pathways eventually also function through PPARγ and CEBPs. For example, the dephosphorylated Rb protein and Cyclin D3-Cdk6 complex enhance the transcription activity of PPARγ through phosphorylation, while Cyclin D1 inhibits its transcriptional activity (Rosen and MacDougald 2006). Cytokines, such as TNFα and IL-1, phosphorylate the AF-1 domain of PPARγ and inhibit adipocyte differentiation (Dapeng and Lixing 2010).
In their study, Ohno et al. (2012) found that complete PPARγ receptor agonists, such as rosiglitazone, binds and stabilizes the PPARγ LBD, promoting the accumulation of PRDM16. The process of adipocyte differentiation involves a very complex transcriptional regulatory network, and many mechanisms are still unknown and need further exploration.
Bone morphogenesis proteins (BMPs)
BMPs are a class of multifunctional growth factors, all of which belong to the transforming growth factor β (TGFβ) superfamily except for BMP1. Kang et al. (2009) found that five BMPs (BMP2, 4, 6, 7, and 9) effectively induce MSCs to form adipocytes in vivo and in vitro. In addition, BMP4 and BMP7 also promote the transformation of WACs into BACs.
BMP7 increases the expression of transcriptional regulators PRDM16, PGC-1α, PPARγ, and C/EBPs. In animal experiments, embryos with the depletion of BMP7 show a BAC deficiency, and adipocytes stimulated by BMP7 show a strong browning ability in vitro (Schulz et al. 2011). Continuous secretion of BMP7 promotes the browning of subcutaneous WACs and reduces the degree of obesity in mice (Townsend et al. 2014). Also, BMP7 acts on human preadipocytes to promote the transcription of UCP1, and the expression of the beige adipocyte-specific genes CD137 and TMEM26 (Okla et al. 2015).
Browning of WAT
Because browning of WAT changes the energy balance from energy storage to energy expenditure, it has become a new target for the treatment of increasingly severe and prevalent obesity, as well as metabolic syndromes (Frontini et al. 2013). The transdifferentiation of white to brown adipocyte involves a transcriptional cascade. In this transcriptional network, PPARγ, PRDM16, and PGC-1α are the core regulatory transcription factors (Kinyui and Lei 2013). Most adipocyte differentiation transforming factors act directly or indirectly through these transcription factors. Furthermore, most key adipocyte-converting signaling pathways eventually converge on them as well. The roles of each of these factors are described below.
Environmental factors
Cold
Cold temperatures stimulate not only BAC proliferation but also browning of WAT (Zhang et al. 2011). This is because the cold stimulates the transient receptor potential of peripheral sensory neurons, activates the sympathetic nervous system to release norepinephrine, and activates BAT through the PKA/p38 MAPK signaling pathway. These signaling pathways also upregulate UCP-1 through transcription factors PGC-1α and C/EBPβ. Studies have shown that cold stimulation or β3 adrenaline receptor agonists promote subcutaneous adipose tissue cell reconstruction, especially the differentiation of beige adipose cells, and increase the number and proportion of beige adipocytes in the subcutaneous tissue, thereby increasing the ability of subcutaneous fat cells to produce heat (Boss et al. 2012).
Hypoxia
Under low oxygen concentrations, the expression of PGC-1α increases, and the combination of PGC-1α and PPARγ effectively promotes the transformation of WAT into BAT, consistent with the browning of white adipocytes under normal oxygen concentrations (Bostrom et al. 2012). In a previous study, Lu et al. exposed 3T3-L1 adipocytes to transient hypoxia for 4 h/d, and found that transient hypoxia regulates adipocyte heat production, mitochondrial generation, and glucose and lipid metabolism through AMPK and its related genes, such as PGC-1α, PPARγ, and GLUT1 (Lu et al. 2015).
Exercise
During exercise, the activation of the central nervous system promotes the release of catecholamines. β-adrenergic receptor agonists promote UCP1 activation in acute (lipid breakdown) and chronic (mitochondrial synthesis, WAT browning) exercise states, increasing the expression and activity of mitochondrial synthesis genes, promoting the browning of subcutaneous white adipocytes and augmenting the WAT adipokine profile (Bostrom et al. 2012). Exercise also activates browning of white adipocytes by activating PGC-1α (Stanford et al. 2013); however, this mechanism of action remains poorly understood (Fitzgibbons et al. 2014).
Gene transcriptional regulation
PPARγ agonists
After treatment with the PPARγ agonist rosiglitazone, UCP1 and PGC-1α significantly increase in WAT in vivo, which indicates that PPARγ indirectly acts as a medium to promote browning of white adipose cells (Petrovic et al. 2010). However, PPARγ agonist-induced browning of white adipocytes is dependent on PRDM16.
PPARγ not only activates BAC gene expression but inhibits WAC gene expression during the browning of adipocytes (Vernochet et al. 2009). Moreover, PPARγ stimulation stabilizes the expression of PRDM16 mRNA, and the induced PGC-1α interacts with PRDM16 to promote BAT specific gene expression (Ohno et al. 2012). However, other studies have shown that PPARγ agonists induce brown-like cell phenotypes in undifferentiated preadipocytes, such as MSCs or poorly differentiated preadipocytes, but do not elicit brown-like changes in mature WACs (Corrales et al. 2018).
PRDM16
BAT from PRDM16 knockout mice completely loses its thermogenic function, while in vitro culture of BACs from PRDM16 knockout mice shows an almost complete loss of BAC characteristics (Seale et al. 2007). PRDM16 expression in white preadipocytes promotes differentiation into BACs, including the inhibition of white adipocyte-specific gene expression and increased expression of brown adipocyte-specific and mito-chondrial genes (Kajimura et al. 2015). PPARγ might act on PRDM16 to induce adipocyte precursors to differentiate toward BACs and regulate the expression of BAC-related thermogenic genes (Harms and Seale 2013). Furthermore, PRDM16-knockout mice are prone to obesity and insulin resistance, induced by a high-fat diet (Cohen et al. 2014).
PGC-1α
Selective PGC-1α knockout from mice results in decreased heat production by subcutaneous WACs and decreased expression of mitochondria-related genes. This suggests that PGC-1α might easily induce the browning of subcutaneous WAT. In human subcutaneous WACs, overexpression of PGC-1α leads to the presence of BACs (Mazzucotelli et al. 2007). The expression of PGC-1α in BAT is dependent on cAMP. C/EBPβ combines with the cAMP response element at the proximal end of the promoter region of PGC-1α to promote the expression of PGC-1α and mediate the browning of white adipocytes (Karamanlidis et al. 2007).
Cytokines
Fibroblast growth factor 21 (FGF21)
FGF21 is an endocrine factor that plays a key role in energy balance, glucose metabolism and lipid metabolism. Numerous studies have shown that exercise can promote the secretion of FGF21 in the muscle tissue. FGF21 is primarily derived from the liver, but is also synthesized and released from WAT, and is, therefore, considered an adipokine (Kim et al. 2013). FGF21 is a key regulator of brown differentiation of white adipocytes. A study by Veniant et al. (2015) demonstrated that FGF21 induces the browning of WACs by inducing PGC-1α expression in mouse adipocytes. Human experiments have shown that BAT also secretes FGF21 and acts on itself to promote the browning of WACs (Hondares et al. 2014)
Vascular endothelial growth factor (VEGF)
VEGF is highly expressed in the adipose tissue and plays a role in angiogenesis and energy metabolism. Hypoxia is an inducer of VEGF expression. In addition, VEGF signaling pathway is essential for the induction of beige adipocyte genes, β3 adrenergic agonists, and PPARγ after exercise (Veikkola et al. 2000).
VEGF-A-induced angiogenesis can achieve continuous and adequate oxygen and nutrient exchange during cellulite expansion, thereby improving adipose tissue function. Inhibition of VEGF-A-induced VEGFR2 activation in the early stages of weight gain, induced by a high-fat diet leads to increased systemic insulin resistance. VEGF-B is co-expressed with the nuclear-encoded mitochondrial gene, OXPHOS. PGC-1 regulates VEGF-B expression to coordinate FFA uptake and β-oxidation (Mehlem et al. 2016). VEGF-B knockout mice showed reduced lipid accumulation in the heart, muscle, and BAT, and increased lipid accumulation in WAT (Hagberg et al. 2010). This confirms the role of VEGF-B in the regulation of energy metabolism and differentiation of beige adipocytes.
Mice lacking VEGF-A tend to utilize fat, while those lacking VEGF-B tend to accumulate fat (Lee et al. 2014). Experiments in VEGF-B knockout mouse models have shown that the inactivation of VEGF-B leads to the expansion of WACs and increase in WAC-related gene expression, along with the whitening of BACs and a decrease in BAC-related gene expression. In contrast, inhibition of VEGF-A induces BAC development and expansion, increased energy expenditure, upregulation of BAC-related genes and downregulation of WAC-related genes. When VEGF-B knockout and VEGF-A inhibited are were crossed, VEGF-A and VEGF-B counteract their respective regulation of a large number of genes and effectively reverse each other’s effects (Jin et al. 2018).
Post-transcriptional regulation of genes
Non-coding RNA (ncRNA) is a general term for all functional RNAs that are not translated into proteins. In recent years, many new types of ncRNA have been discovered, and the role of some ncRNAs in gene regulatory networks has been gradually recognized. Among them, microRNAs (miRNAs) are a class of ncRNAs commonly seen in eukaryotic cells, with a length of 18~25 nt, which play key roles in the regulation of cell proliferation, differentiation, and apoptosis (Guo et al. 2010).
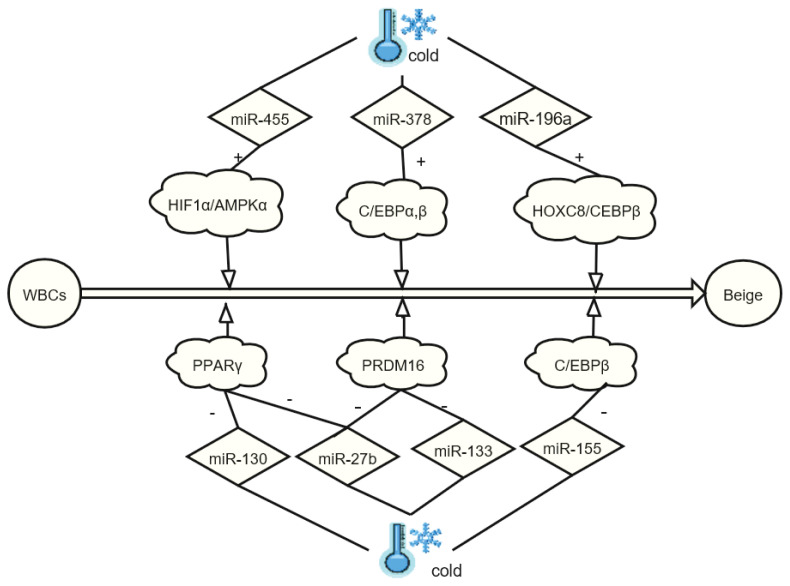
During the cold-induced browning of WACs in mice, the expression of miR-196a is significantly increased, and the expression of HOXC8 is inhibited. As a WAC gene, HOXC8 inhibits the function of C/EBP binding and thus restricts the generation of BACs. Therefore, miR-196a/HOXC8/C/EBP can be targeted as a pathway for weight loss (Mori et al. 2012). Cold also increases the expression of miR-455 in mouse brown adipose cells. In vitro experiments have shown that miR-455 regulates the differentiation and thermogenesis of brown adipose cells. Mechanistically, miR-455 mainly activates AMPK (AMP-activated protein kinase) by acting on HIF1 (hypoxia-inducible factors-1), triggering the formation of mitochondria and BACs (Zhang et al. 2015). miRNAs also bind to C/EBPs to regulate the production of BACs. Among them, miR-155 and C/EBP constitute a double closed-loop negative feedback mechanism and regulate the generation of BAT or beige adipose tissue (Chen et al. 2013). miR-378, on the other hand, regulates the browning of WACs by enhancing the binding of C/EBP to the GLUT4 promoter (Gerin et al. 2010). Moreover, miR-133 restricts browning of adipocytes by inhibiting the transcription of PRDM16 (Liu et al. 2013) and limits adipocyte differentiation by inhibiting the transcription of PPARγ (Lee et al. 2011). During exposure to cold, miR-27b expression in the adipose tissue is suppressed. Cell culture experiments have revealed that in preadipocytes undergoing browning, the suppression of miR-27b occurs at the same time as PRDM16, PPARγ and UCP1 increase, showing that miR-27b plays a negative regulatory role in the browning of WAT (Sun et al. 2014).
Because microRNAs are a large family, only a small number of microRNAs have been confirmed to play a role in the browning of WACs. For example, miR-196a, miR-378 and miR-455 play positive regulatory roles in complex diseases such as obesity and type 2 diabetes, while miR-155, miR-133, miR-133 and miR-27b play negative regulatory roles (Arias et al. 2016) (Fig. 3). With the development of RNA detection technology, more non-coding RNAs will be identified, and their regulatory mechanisms in the energy metabolism and regulatory networks formed with the transcription factors will become more evident, providing new targets for the treatment of obesity and related metabolic diseases.
Fig. 3.
Role of miRNAs in the browning of white adipose tissue. WACs, white adipocyte cell; Beige, beige adipose tissue; PPARγ, peroxisome proliferator-activated receptor γ; PRDM16, positive regulatory domain zinc-finger protein 16; C/EBPβ, CCAAT/enhancer-binding protein β.
Problems during adipocyte differentiation
At present, we have a comprehensive understanding of adipocyte differentiation; however, several important problems remain unsolved. The first problem is the origin of adipocytes, including the formation of adipocytes in the embryo, the source of the first cells of the adipose tissue, and distribution of these cells throughout the body. Answers to these questions could enable an effective reduction in the number of adipocytes or reductions in specific adipose tissue deposits, which could prevent excessive adipose tissue deposition.
The second problem is that much work remains to be done to reveal how transcription factors promote adipocyte differentiation. Although many of the transcription factors necessary for adipocyte differentiation have been identified, such as PPARγ, the mechanisms of action remain unclear. Elaboration of the mechanism of action of known transcription factors, functional verification of known key transcription factor target genes, and the identification of new transcription factors are the focus of current research.
Third, further research on the browning of WACs is necessary. BAT does not cause or promote metabolic disease. Instead, it consumes energy and reduces fat accumulation, making the browning of white adipocytes a new target in the treatment of metabolic diseases. However, browning of white adipocytes is very complex, involving the regulation of multiple genes and pathways, and is affected by hypoxia, exercise, cold, and other conditions. Therefore, further studies into are required and the browning of white adipocytes is expected to be an effective means to alleviate the development of metabolic diseases.
Conclusions
WAT, BAT and beige adipose tissue regulate the energy storage and body metabolism under the control of PPARγ, UCP1 and PGC-1α, which are closely implicated in the occurrence and development of obesity and related metabolic diseases. This paper reviewed the PPARγ in white and brown adipocyte regulation and differentiation. Browning of white adipocytes is the result of the synergistic effect of various regulatory mechanisms. Among them, PPARs, PGC-1α, PRDM16 and C/EBPs play key roles in relevant gene transcription. Moreover, the regulation of nerve and endocrine functions are accomplished through the above-mentioned transcription factors. Consequently, the browning of white fat can significantly promote systemic energy consumption and improve carbohydrate and lipid metabolism. At present, there is still a need for in-depth and detailed basic research, so that the interactions of various regulatory pathways on the browning of WAT in humans can be more clearly defined, allowing browning of white adipocytes to become an effective treatment for obesity and metabolism-related diseases.
Acknowledgements
We would like to thank Editage for English language editing.
Footnotes
Conflict of Interest
There is no conflict of interest.
References
- APRILE M, AMBROSIO MR, D’ESPOSITO V, BEGUINOT F, FORMISANO P, COSTA V, CICCODICOLA A. PPARG in human adipogenesis: differential contribution of canonical transcripts and dominant negative isoforms. PPAR Res. 2014;2014:537865. doi: 10.1155/2014/537865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ARIAS N, AGUIRRE L, FERNANDEZ-QUINTELA A, GONZÁLEZ M, LASA A, MIRANDA J, MACARULLA MT, PORTILLO MP. MicroRNAs involved in the browning process of adipocytes. J Physiol Biochem. 2016;72:509–521. doi: 10.1007/s13105-015-0459-z. [DOI] [PubMed] [Google Scholar]
- BARAK Y, NELSON MC, ONG ES, JONES YZ, RUIZ-LOZANO P, CHIEN KR, KODER A, EVANS RM. PPARγ is required for placental, cardiac, and adipose tissue development. Mol Cell. 1999;4:585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- BIRSOY K, CHEN Z, FRIEDMAN J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 2008;7:339–347. doi: 10.1016/j.cmet.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOSS O, FARMER SR. Recruitment of brown adipose tissue as a therapy for obesity-associated diseases. Front Endocrinol (Lausanne) 2012;3:14. doi: 10.3389/fendo.2012.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOSTROM P, WU J, JEDRYCHOWSKI MP, KORDE A, YE L, LO JC, RASBACH KA, BOSTRÖM EA, CHOI JH, LONG JZ, KAJIMURA S, ZINGARETTI MR, VIND BF, TU H, CINTI S, HØJLUND K, GYGI SP, SPIEGELMAN BM. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALL KM, GLASER T, ITO CY, BUCKLER AJ, PELLETIER J, HABER DA, ROSE EA, KRAL A, YEGER H, LEWIS WH, JONES C, HOUSMAN DE. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell. 1990;60:509–520. doi: 10.1016/0092-8674(90)90601-a. [DOI] [PubMed] [Google Scholar]
- CHEN Y, SIEGEL F, KIPSCHULL S, HAAS B, FRÖHLICH H, MEISTER G, PFEIFER A. MiR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun. 2013;4:1769. doi: 10.1038/ncomms2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHU DT, TAO Y. Human thermogenic adipocytes: a reflection on types of adipocyte, developmental origin, and potential application. J Physiol Biochem. 2017;73:1–4. doi: 10.1007/s13105-016-0536-y. [DOI] [PubMed] [Google Scholar]
- COHEN P, LEVY JD, ZHANG Y, FRONTINI A, KOLODIN DP, SVENSSON KJ, LO JC, ZENG X, YE L, KHANDEKAR MJ, WU J, GUNAWARDANA SC, BANKS AS, CAMPOREZ JPG, JURCZAK MJ, KAJIMURA S, PISTON DW, MATHIS D, CINTI S, SHULMAN GI, SEALE P, SPIEGELMAN BM. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COUSIN B, CINTI S, MORRONI M, RAIMBATLT S, RICQUIER D, PENICAUD L, CASTEILLA L. Occurrence of brown adipocytes in rat white adipose tissue: Molecular and morphological characterization. J Cell Sci. 1992;103:931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- DAPENG J, LIXING Z. Research progress on adipocyte differentiation and its regulation. Chin J Cell Biol. 2010;32:690–695. [Google Scholar]
- DIVELLA R, De LUCA R, ABBATE I, NAGLIERI E, DANIELE A. Obesity and cancer: the role of adipose tissue and adipo-cytokines-induced chronic inflammation. J Cancer. 2016;7:2346–2359. doi: 10.7150/jca.16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EL-JACK AK, HAMM JK, PILCH PF, FARMER SR. Reconstitution of insulin-sensitive glucose transport in fibroblasts requires expression of both PPARγ and C/EBPα. J Biol Chem. 1999;274:7946–7951. doi: 10.1074/jbc.274.12.7946. [DOI] [PubMed] [Google Scholar]
- FITZGIBBONS TP, CZECH MP. Epicardial and perivascular adipose tissues and their influence on cardiovascular disease: Basic mechanisms and clinical associations. Am Heart Assoc. 3:e000582. doi: 10.1161/jaha.113.000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRONTINI A, VITALI A, PERUGINI J, MURANO I, ROMITI C, RICQUIER D, CINTI S. White-to-brown transdifferentiation of omental adipocytes in patients affected by pheochromocytoma. Biochim Biophys Acta. 2013;1831:950–959. doi: 10.1016/j.bbalip.2013.02.005. [DOI] [PubMed] [Google Scholar]
- GEARING M, GOTTLICHER M, TEBOUL M, WIDMARK E, GUSTAFSSON JA. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc Natl Acad Sci U S A. 1993;90:1440–1444. doi: 10.1073/pnas.90.4.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERIN I, BOMMER GT, McCOIN CS, SOUSA KM, KRISHNAN V, MacDOUGALD OA. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299:E198–E206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOUDARZI F, MOHAMMADALIPOUR A, KHODADADI I, KARIMI S, MOSTOLI R, BAHABADI M, GOODARZI MT. The role of calcium in differentiation of human adipose derived stem cells to adipocytes. Mol Biotechnol. 2018;60:279–289. doi: 10.1007/s12033-018-0071-x. [DOI] [PubMed] [Google Scholar]
- GUO H, INGOLIA NT, WEISSMAN JS, BARTEL DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAGBERG CE, FALKEVALL A, WANG X, LARSSON E, HUUSKO J, NILSSON I, Van MEETEREN LA, SAMEN E, LU L, VANWILDEMEERSCH M, KLAR J, GENOVE G, PIETRAS K, STONE-ELANDER S, CLAESSON-WELSH L, YLÄ-HERTTUALA S, LINDAHL P, ERIKSSON U. Vascular endothelial growth factor b controls endothelial fatty acid uptake. Nature. 2010;464:917–921. doi: 10.1038/nature08945. [DOI] [PubMed] [Google Scholar]
- HALLENBORG P, PETERSEN RK, KOUSKOUMVEKAKI I, NEWMAN JW, MADSEN L, KRISTIANSEN K. The elusive endogenous adipogenic PPARγ agonists: lining up the suspects. Prog Lipid Res. 2016;61:149–162. doi: 10.1016/j.plipres.2015.11.002. [DOI] [PubMed] [Google Scholar]
- HARMS M, SEALE P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- HARMS MJ, ISHIBASHI J, WANG W, LIM HW, GOYAMA S, SATO T, KUROKAWA M, WON KJ, SEALE P. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. 2016;19:593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARMS MJ, LIM HW, HO Y, SHAPIRA SN1, ISHIBASHI J, RAJAKUMARI S, STEGER DJ, LAZAR MA, WON KJ, SEALE P. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 2015;29:298–307. doi: 10.1101/gad.252734.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONDARES E, GALLEGO-ESCUREDO JM, FLACHS P, FRONTINI A, CEREIJO R, GODAY A, PERUGINI J, KOPECKY P, GIRALT M, CINTI S, KOPECKY J, VILLARROYA F. Fibroblast growth factor-21 is expressed in neonatal and pheochromocytoma-induced adult human brown adipose tissue. Metabolism. 2014;63:312–317. doi: 10.1016/j.metabol.2013.11.014. [DOI] [PubMed] [Google Scholar]
- HONG G, DAVIS B, KHATOON N, BAKER SF, BROWN J. PPAR gamma-dependent anti-inflammatory action of rosiglitazone in human monocytes: suppression of TNF alpha secretion is not mediated by PTEN regulation. Biochem Biophys Res Commun. 2003;303:782–787. doi: 10.1016/s0006-291x(03)00418-2. [DOI] [PubMed] [Google Scholar]
- IIDA S, CHEN W, NAKADAI T, OHKUMA Y, ROEDER RG. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with mediator subunit MED1. Genes Devel. 2015;29:308–321. doi: 10.1101/gad.252809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ISSEMANN I, GREEN S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- JIN H, LI D, WANG X, CHEN Y, YAO Y, ZHAO C, LU X, ZHANG S, TOGO J, JI Y, ZHANG L, FENG X, ZHENG Y. VEGFa and VEGFb play balancing roles in adipose differentiation, gene expression, and function. Endocrinol. 2018;159:2036–2049. doi: 10.1210/en.2017-03246. [DOI] [PubMed] [Google Scholar]
- KAJIMURA S, SEALE P, KUBOTA K, LUNSFORD E, FRANGIONI JV, GYGI SP, SPIEGELMAN BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAJIMURA S, SEALE P, TOMARU T, ERDJUMENT-BROMAGE H, COOPER MP, RUAS JL, CHIN S, TEMPST P, LAZAR MA, SPIEGELMAN BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAJIMURA S, SPIEGELMAN BM, SEALE P. Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 2015;22:546–559. doi: 10.1016/j.cmet.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KANG Q, SONG WX, LUO Q, TANG N, LUO J, LUO X, CHEN J, BI Y, HE BC, PARK JK, JIANG W, TANG Y, HUANG J, SU Y, ZHU GH, HE Y, YIN H, HU Z, WANG Y, CHEN L, ZUO GW, PAN X, SHEN J, VOKES T, REID RR, HAYDON RC, LUU HH, HE TC. A comprehensive analysis of the dual roles of BMPs in regulating adipogenic and osteogenic differentiation of mesenchymal progenitor cells. Stem Cells Dev. 2009;18:545–559. doi: 10.1089/scd.2008.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARAMANLIDIS G, KARAMITRI A, DOCHERTY K, HAZLERIGG DG, LOMAX MA. C/EBPb reprograms white 3T3-L1 preadipocytes to a brown adipocyte pattern of gene expression. J Biol Chem. 2007;282:24660–24669. doi: 10.1074/jbc.m703101200. [DOI] [PubMed] [Google Scholar]
- KERSTEN S, DESVERGNE B, WAHLI W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
- KIM KH, KIM SH, MIN YK, YANG HM, LEE JB, LEE MS. Acute exercise induces FGF21 expression in mice and in healthy humans. PLoS One. 2013;8:e63517. doi: 10.1371/journal.pone.0063517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KINYUI AL, LEI S. Turning WAT into BAT. a review on regulators controlling the browning of white adipocytes. Biosci Rep. 2013;33:711–719. doi: 10.1042/bsr20130046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KVANDOVÁ M, MAJZÚNOVÁ M, DOVINOVÁ I. The role of PPARgamma in cardiovascular diseases. Physiol Res. 2016;65(Suppl 3):S343–S363. doi: 10.33549/physiolres.933439. [DOI] [PubMed] [Google Scholar]
- LEE EK, LEE MJ, ABDELMOHSEN K, KIM W, KIM MM, SRIKANTAN S, MARTINDALE JL, HUTCHISON ER, KIM HH, MARASA BS, SELIMYAN R, EGAN JM, SMITH SR, FRIED SK, GOROSPE M. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor γ expression. Mol Cell Biol. 2011;31:626–638. doi: 10.1128/mcb.00894-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE HS, KIM J. Constitutive expression of vascular endothelial cell growth factor (VEGF) gene family ligand and receptors on human upper and lower airway epithelial cells. Int Forum Allergy Rhinol. 2014;4:8–14. doi: 10.1002/alr.21244. [DOI] [PubMed] [Google Scholar]
- LEE JE, GE K. Transcriptional and epigenetic regulation of PPARγ expression during adipogenesis. Cell Biosci. 2014;4:29. doi: 10.1186/2045-3701-4-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE WS, KIM J. Peroxisome proliferator-activated receptors and the heart: lessons from the past and future directions. PPAR Res. 2015;2015:271983. doi: 10.1155/2015/271983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU HJ, LIAO HH, YANG Z, TANG QZ. Peroxisome proliferator-activated receptor-γ is critical to cardiac fibrosis. PPAR Res. 2016;2016:2198645. doi: 10.1155/2016/2198645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU J, WU PH, TARR PT, LINDENBERG KS, ST-PIERRE J, ZHANG CY, MOOTHA VK, JÄGER S, VIANNA CR, REZNICK RM, CUI L, MANIERI M, DONOVAN MX, WU Z, COOPER MP, FAN MC, ROHAS LM, ZAVACKI AM, CINTI S, SHULMAN GI, LOWELL BB, KRAINC D, SPIEGELMAN BM. Defects in adaptive energy metabolism with CNS-Linked hyperactivity in PGC-1α null mice. Cell. 2014;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- LIU W, BI P, SHAN T, YANG X, YIN H, WANG YX, LIU N, RUDNICKI MA, KUANG S. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9:e1003626. doi: 10.1371/journal.pgen.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU H, GAO Z, ZHAO Z, WENG J, YE J. Transient hypoxia repro-grams differentiating adipocytes for enhanced insulin sensitivity and triglyceride accumulation. Obes (Lond) 2015;40:121–128. doi: 10.1038/ijo.2015.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAZZUCOTELLI A, VIGUERIE N, TIRABY C, ANNICOTTE JS, MAIRAL A, KLIMCAKOVA E, LEPIN E, DELMAR P, DEJEAN S, TAVERNIER G, LEFORT C, HIDALGO J, PINEAU T, FAJAS L, CLÉMENT K, LANGIN D. The transcriptional coactivator peroxisome proliferator activated receptor (PPAR) gamma coactivator-1 alpha and the nuclear receptor PPAR alpha control the expression of glycerol kinase and metabolism genes independently of PPAR gamma activation in human white adipocytes. Diabetes. 2007;56:2467–2475. doi: 10.2337/db06-1465. [DOI] [PubMed] [Google Scholar]
- MEHLEM A, PALOMBO I, WANG X, HAGBERG CE, ERIKSSON U, FALKEVALL A. Pgc-1alpha coordinates mitochondrial respiratory capacity and muscular fatty acid uptake via regulation of VEGF-b. Diabetes. 2016;65:861–873. doi: 10.2337/db15-1231. [DOI] [PubMed] [Google Scholar]
- MORI M, NAKAGAMI H, RODRIGUEZ-ARAUJO G, NIMURA K, KANEDA Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012;10:e1001314. doi: 10.1371/journal.pbio.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OHNO H, SHINODA K, SPIEGELMAN BM, KAJIMURA S. PPAR gamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKLA M, HA JH, TEMEL RE, CHUNG S. BMP7 drives human adipogenic stem cells into metabolically active beige adipocytes. Lipids. 2015;50:111–120. doi: 10.1007/s11745-014-3981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PELLEGRINELLI V, CAROBBIO S, VIDAL-PUIG A. Adipose tissue plasticity: how fat depots respond differently to pathophysiological cues. Diabetologia. 2016;59:1075–1088. doi: 10.1007/s00125-016-3933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PENG Z, BAN K, WAWROSE RA, GOVER AG, KOZAR RA. Protection by enteral glutamine is mediated by intestinal epithelial cell peroxisome proliferator-activated receptor-γ during intestinal ischemia/reperfusion. Shock. 2015;43:327–333. doi: 10.1097/shk.0000000000000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETERS JM, LEE SS, LI W, WARD JM, GAVRILOVA O, EVERETT C, REITMAN ML, HUDSON LD, GONZALEZ FJ. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol Cell Biol. 2000;20:5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PETROVIC N, WALDEN TB, SHABALINA IG, TIMMONS JA, CANNON B, NEDERGAARD J. Chronic peroxisome proliferator-activated receptor gamma (PPAR gamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285:7153–7164. doi: 10.1074/jbc.m109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PUIGSERVER P, WU Z, PARK CW, GRAVES R, WRIGHT M, SPIEGELMAN BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- QIAN S, HUANG H, TANG Q. Brown and beige fat: the metabolic function, induction, and therapeutic potential. Front Med. 2015;9:162–172. doi: 10.1007/s11684-015-0382-2. [DOI] [PubMed] [Google Scholar]
- ROSEN ED, MacDOUGALD OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- ROSEN ED, SPIEGELMAN BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SANCHEZ-GURMACHES J, HUNG CM, SPARKS CA, TANG Y, LI H, GUERTIN DA. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATO N, KOZAR RA, ZOU L, WEATHERALL JM, ATTUWAYBI B, MOORE-OLUFEMI SD, WEISBRODT NW, MOORE FA. Peroxisome proliferator-activated receptor gamma mediates protection against cyelooxygenase2-induced gut dysfunctionin in a rodent model of mesenteric ischemia/reperfusion. Shock. 2005;24:462–469. doi: 10.1097/01.shk.0000183483.76972.ae. [DOI] [PubMed] [Google Scholar]
- SCHULZ TJ, HUANG TL, TRAN TT, ZHANG H, TOWNSEND KL, SHADRACH JL, CERLETTI M, McDOUGALL LE, GIORGADZE N, TCHKONIA T, SCHRIER D. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc Natl Acad Sci U S A. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEALE P, BJORK B, YANG W, KAJIMURA S, CHIN S, KUANG S, SCIME A, DEVARAKONDA S, CONROE HM, ERDJUMENT-BROMAGE H, TEMPST P. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEALE P, KAJIMURA S, YANG W, CHIN S, ROHAS LM, ULDRY M, TAVERNIER G, LANGIN D, SPIEGELMAN BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHINGO K, PATRICK S, TAKUYA T, HEDIYE EB, MARCUS PC, JORGE LR, SHERRY C, PAUL T, MITCHELL AL, BRUCE MS. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANFORD KI, MIDDELBEEK RJ, TOWNSEND KL, AN D, NYGAARD EB, HITCHCOX KM, MARKAN KR, NAKANO K, HIRSHMAN MF, TSENG YH, GOODYEAR LJ. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest. 2013;123:215–223. doi: 10.1172/jci62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN L, TRAJKOVSKI M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism. 2014;63:272–282. doi: 10.1016/j.metabol.2013.10.004. [DOI] [PubMed] [Google Scholar]
- TONG Q, HOTAMISLIGIL GS. Molecular mechanisms of adipocyte differentiation. Rev Endocr Metab Disord. 2001;2:349–355. doi: 10.1023/a:1011863414321. [DOI] [PubMed] [Google Scholar]
- TOWNSEND KL, LYNES M, COBURN J, PRITCHARD E, KWON YM, HUANG T, KAPLAN DL, TSENG YH. Silk-mediated sustained delivery of bone morphogenetic protein 7 (BMP7) to subcutaneous white adipose depot leads to browning and reversal of obesity. Diabetes. 2014;63:A64–A64. [Google Scholar]
- VEIKKOLA T, KARKKAINEN M, CLAESSON-WELSH L, ALITALO K. Regulation of angiogenesis via vascular endothelial growth factor receptors. Cancer Res. 2000;60:203–212. [PubMed] [Google Scholar]
- VELLA S, CONALDI PG, FLORIO T, PAGANO A. PPAR gamma in neuroblastoma: the translational perspectives of hypoglycemic drugs. PPAR Res. 2016;2016:1–10. doi: 10.1155/2016/3038164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VENIANT MM, SIVITS G, HELMERING J, KOMOROWSKI R, LEE J, FAN W, MOYER C, LLOYD DJ. Pharmacologic effects of FGF21 are independent of the “Browning” of white adipose tissue. Cell Metab. 2015;21:731–738. doi: 10.1016/j.cmet.2015.04.019. [DOI] [PubMed] [Google Scholar]
- VERNOCHET C, PERES SB, DAVIS KE, McDONALD ME, QIANG L, WANG H, FARMER SR. C/EBPα and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor γ agonists. Mol Cell Biol. 2009;29:4714–4728. doi: 10.1128/mcb.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VICTOR NA, WANDERI EW, GAMBOA J, ZHAO X, ARONOWSKI J, DEININGER K, LUST WD, LANDRETH GE, SUNDARARAJAN S. Altered PPARγ expression and activation after Transient focal ischemia in rats. Eur J Neurosci. 2006;24:1653–1663. doi: 10.1111/j.1460-9568.2006.05037.x. [DOI] [PubMed] [Google Scholar]
- WALDEN TB, HANSEN IR, TIMMONS JA, CANNON B, NEDERGAARD J. Recruited vs. nonrecruited molecular signatures of brown, “brite,” and white adipose tissues. Am J Physiol Endocrinol Metab. 2012;302:E19–E31. doi: 10.1152/ajpendo.00249.2011. [DOI] [PubMed] [Google Scholar]
- WANG R, LIAN X, ZHANG L, WANG ZK, SIVASAKTHIVEL S, LIU PF, CAI JH, ZHU WL. Adaptive thermogenesis of the liver in tree shrew (Tupaia belangeri) during cold acclimation. Anim Biol. 2011;61:385–401. doi: 10.1163/157075511x596873. [DOI] [Google Scholar]
- ZHANG H, GUAN M, TOWNSEND KL, HUANG TL, AN D, YAN X, XUE R, SCHULZ TJ, WINNAY J, MORI M, HIRSHMAN MF. MicroRNA-455 regulates brown adipogenesis via a novel HIF1anAMPK-PGC1 signaling network. EMBO Rep. 2015;16:1378–1393. doi: 10.15252/embr.201540837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG Y, LI H. Three important transcription factors related to lipogenesis and adipogenesis in mammal. J Northeast Agric Univ. 2010;17:62–75. [Google Scholar]