Abstract
Bartonella henselae is the main causative agent of cat-scratch disease, and both B. henselae and Bartonella quintana cause angioproliferative disorders such as bacillary angiomatosis. To increase the sensitivity of Bartonella detection by PCR and to improve the species differentiation, we developed a semiquantitative, species-specific PCR-based enzyme immunoassay (EIA). The 16S rRNA gene was selected as the target sequence. Internal nucleotide sequences derived from the amplified 16S rRNA region were used to develop species-specific oligonucleotide probes for B. henselae and B. quintana. Biotin-labeled PCR products were immobilized on streptavidin-coated microtiter plates, hybridized to a digoxigenin-labeled probe, and detected with antidigoxigenin peroxidase conjugate. No cross-hybridization with other Bartonella or non-Bartonella species was observed. This EIA was as sensitive as dot blot hybridization and was 10 times more sensitive than visualization of PCR products on agarose gels. Serial dilutions of B. henselae and B. quintana suspensions demonstrated that an optical density (OD) of approximately 0.200 was equivalent to 5 CFU in the reaction mixture. By comparing the OD of the bacterial dilutions with that obtained from clinical specimens we could determine that the number of CFU in clinical samples ranged from 103 to 106 CFU/ml. The PCR-EIA developed in the present study is a rapid, sensitive, and simple method for the diagnosis of B. henselae and B. quintana infections.
The genus Bartonella presently includes 14 species, but at present, only 5 of them are known to be pathogenic for humans. B. bacilliformis is the agent of bartonellosis, a diphasic illness which is endemic in the South American Andes and which is limited to the valleys of Peru, Ecuador, and Colombia (7). Only a few reports of human infections due to B. elizabethae (8) and B. clarridgeiae (13, 15) have been published. The majority of human infections are caused by the two species B. henselae, the main agent of cat-scratch disease (CSD), and B. quintana, which causes trench fever and endocarditis, most often observed in homeless, chronic alcoholic patients. CSD occurs worldwide and is probably the most common Bartonella infection. It is a common cause of subacute, regional lymphadenopathy in mostly immunocompetent children and adults. Atypical manifestations of CSD including Parinaud’s oculoglandular syndrome, hepatic and splenic abscesses, and central nervous system and pulmonary manifestations are well characterized (2, 4).
Immunocompromised patients are more likely to have systemic infections caused by both B. henselae and B. quintana. Especially in human immunodeficiency virus-infected patients clinical manifestations like bacillary angiomatosis (BA), bacillary peliosis hepatis, osteolytic lesions, relapsing fever with bacteremia, endocarditis, and encephalitis are common (2).
Laboratory methods for the diagnosis of Bartonella infections include isolation of the organisms by culture, serological assays, histopathological examination, and molecular detection of Bartonella DNA in affected tissue (2, 21, 22). However, Bartonella species are fastidious, slowly growing bacteria, and routine bacterial culture protocols usually do not allow detection of these organisms (19). Serological testing for detection of antibodies to B. henselae in CSD patients seems to be quite reliable when titers are 1:512 or higher. The seroprevalence (usually low antibody titers) in healthy individuals is high (up to 30%), and low antibody levels (between 1:64 and 1:256) could indicate prior contact with B. henselae but could also indicate the onset or the end of illness (22). Therefore, the diagnosis of Bartonella infection in patients with low antibody titers should be confirmed histologically and/or by detection of Bartonella DNA in the affected tissue. Additionally, serological methods do not allow differentiation between B. henselae and B. quintana infections. The cross-reactivity between these two species was demonstrated to be very high (95%) in patients with CSD (22). Unfortunately, there exist no reliable data concerning serological testing in immunocompromised patients with Bartonella infections.
Recently, several PCR-based assays have been developed for detection of Bartonella DNA in clinical specimens (1, 3, 5, 10, 16, 21, 23). Successful amplification of Bartonella DNA depends not only on the primers used but also on the condition of the specimens (native and non-formalin-fixed, frozen, or formalin-fixed, paraffin-embedded specimens). Almost all primers used for diagnosis of Bartonella infections are genus specific but not species specific (1, 21). Therefore, hybridization or sequencing of the amplified DNA is required for species identification. Both methods are difficult, expensive, and time-consuming.
The aim of the present study was to develop a rapid and simple method for species-specific detection of amplified Bartonella DNA by a PCR-based enzyme immunoassay (EIA).
MATERIALS AND METHODS
Bacterial strains.
B. henselae Houston-1 (ATCC 49882) and Bartonella quintana CIP 103739 (Collection de l’Institut Pasteur, Paris, France) were used as positive controls for further testing. The strains were cultured on chocolate agar plates containing 10% defibrinated sheep blood. The plates were incubated at 37°C in a 5% carbon dioxide atmosphere for 3 to 4 days. B. bacilliformis ATCC 35685, B. elizabethae ATCC 49927, B. clarridgeiae ATCC 51784, and the following non-Bartonella species served as negative controls in the present study: Afipia felis ATCC 53690, Staphylococcus aureus, Streptococcus pyogenes, Streptococcus agalactiae, Pseudomonas aeruginosa, Escherichia coli, Enterobacter cloacae, Klebsiella pneumoniae, and Citrobacter diversus. The non-American Type Culture Collection (ATCC) strains were clinical isolates of the Institut für Medizinische Mikrobiologie und Hygiene, Freiburg, Germany.
Clinical specimens.
The PCR-EIA was evaluated with 16 clinical specimens. Lymph node biopsy specimens were obtained from nine patients with clinically, serologically, and histopathologically proven CSD. Two skin biopsy specimens were obtained from human immunodeficiency virus-infected patients with histopathologically diagnosed BA. Five lymph node specimens from patients without any evidence of CSD were used as negative controls.
Extraction of DNA.
DNA was extracted from the (mostly formalin-fixed, paraffin-embedded) lymph node and skin biopsy specimens by using a commercially available kit (Qiagen GmbH, Hilden, Germany) as proposed by the manufacturer. The extracted DNA was used as a template in the PCR assays. Purified DNAs from cultured bacterial strains of B. henselae and B. quintana were used as positive controls. Extracted DNA from non-Bartonella species and from five lymph node specimens from patients without evidence of CSD were used as negative controls.
Primers and probes.
The primer pair previously described by Relman et al. (17) was used to amplify a 296-bp fragment of the Bartonella 16S rRNA gene by PCR as described elsewhere (20). PCR products were labeled during the amplification by using the 5′-biotin-modified primer p12B-bio (Table 1). Digoxigenin end-labeled B. henselae-specific (RHp-dig) and B. quintana-specific (RQp-dig) oligonucleotide probes (Table 1) were prepared according to the sequence given by Daly et al. (8) and were used for the detection of PCR products by dot blot hybridization and EIA. The probes differed from each other by three nucleotides.
TABLE 1.
Oligonucleotides used in PCR and hybridization analyses
| Oligonucleotide | Sequence (5′ to 3′)a | Positionb |
|---|---|---|
| p12B | GAGATGGCTTTTGGAGATTA | 1215–1234 |
| p24E | CCTCCTTCAGTTAGGCTGG | 955–973 |
| p12B-bio | Biotin-GAGATGGCTTTTGGAGATTA | 1215–1234 |
| RHp-dig (B. henselae) | dig-AGCATTTGGTTGGGCACTCT | 1067–1086 |
| RQp-dig (B. quintana) | dig-ATCATTAAGTTGGGCACTCT | 1067–1086 |
Boldface letters indicate three different nucleotide pairs.
Position within the 16S rRNA gene of Bartonella species (8).
PCR amplification.
The reaction mixture consisted of bovine serum albumin (BSA; 8 ng/μl), deoxynucleoside triphosphates (200 μM each), primers (117 nM each), Taq polymerase (4 U; Pharmacia Biotech), and 5 μl of extracted DNA in 100.0 μl of TBE (Tris-borate-EDTA) buffer. The PCR was performed as described previously (20).
All oligonucleotides were synthesized and were modified at their 5′ ends with biotin or digoxigenin by Birsner & Grob Biotech (Freiburg, Germany).
EIA for detection of PCR products.
The EIA was performed as described previously by Lüneberg et al. (14) with modifications. Streptavidin-coated microtiter plates (Micro Coat GmbH, Penzberg, Germany) were washed two times with phosphate-buffered saline (PBS)–0.05% Tween. All aliquots of the PCR mixtures were analyzed in duplicate. The PCR fragments were generated with the biotinylated primer p12B-bio and the unlabeled primer p24E. A total of 12 μl of the PCR product was diluted in 10 mM sodium phosphate (pH 7.4)–100 mM NaCl to a final volume of 60 μl and was pipetted into each well for immobilization on streptavidin-coated microtiter wells. After binding of the PCR product via the incorporated biotinylated primer for 15 min, the wells were washed three times with PBS–0.05% Tween. The double-stranded PCR product was denatured with 0.1 M NaOH for 10 min. The unlabeled strands were removed by washing once with 0.1 M NaOH and three times with 0.1 M Tris-HCl (pH 7.5; Merck, Darmstadt, Germany). The immobilized single-stranded PCR product was hybridized with 0.2 pmol of 5′-digoxigenin-labeled oligonucleotide RHp-dig or RQp-dig for 2 h in a water bath at 55°C. The hybridization solution contained 0.6 M NaCl, 20 mM sodium phosphate (pH 7.4), 1 mM EDTA (Merck, Darmstadt, Germany), 0.02% Ficoll, 0.02% polyvinylpyrrolidone, 0.02% BSA (Sigma, Deisenhofen, Germany), and 0.2 pmol of the digoxigenin-labeled oligonucleotide per well. Afterward, the plates were washed three times with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; Sigma) at room temperature and twice for 5 min in 3× SSC at 55°C in a water bath. Anti-digoxigenin Fab fragments conjugated with horseradish peroxidase (Boehringer Mannheim) were diluted at a final concentration of 150 mU/ml (1:1,000 in 1% BSA-PBS), and 50 μl of this dilution was applied to the wells for 30 min at 37°C. The unbound conjugate was removed by washing with PBS–0.05% Tween three times. Finally, 50 μl of 2,2′-azino-bis-3-ethylbenz-thiazoline-6-sulfonic acid (ABTS); Boehringer Mannheim) substrate solution was added, and the mixture was incubated at 37°C with shaking for 30 min. The A405 of each well was determined with a microtitration plate reader (Ceres 900; Biotek).
Analysis of PCR products by dot blot hybridization.
Hybridization of the PCR products was performed by the protocol described by Anderson et al. (1), with modifications. The internal digoxigenin-labeled oligonucleotide probes RHp-dig and RQp-dig used as hybridization probes were the same as those used for PCR-EIA. In short, 10 μl of each PCR product was denatured for 5 min at 95°C and was then put immediately on ice. Five-microliter aliquots were spotted onto each of two nylon membranes (Qiabrane Nylon plus; Qiagen GmbH, Hilden, Germany) and air dried, and the DNA was cross-linked to the nylon membrane by UV irradiation for 3 min. The membranes were then blocked for 1 h at 61°C by using standard prehybridization solution. Hybridization was performed for 1 h at 61°C. The hybridization buffer contained 0.75 pmol/2 ml of probe RHp-dig for detection of B. henselae or 7.5 pmol/2 ml of probe RQp-dig for detection of B. quintana. The hybridized membranes were washed twice for 15 min at 61°C in 5× SSC–0.1% sodium dodecyl sulfate (SDS) to remove the surplus oligonucleotide probes. The hybridized membrane was then washed, blocked, incubated for 30 min with alkaline phosphatase-conjugated anti-digoxigenin-antibody (Boehringer Mannheim), washed again twice for 15 min, and soaked in Luminogen CSPD substrate (Boehringer Mannheim). To increase the luminescence reaction, the membrane was stored for 25 min at 37°C. Afterward the membrane was exposed to X-ray film (Kodak S-Omat AR) for 40 min, followed by development of the film.
RESULTS
PCR amplification.
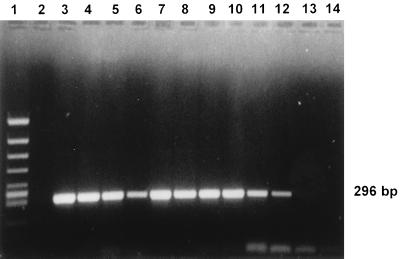
With the primers described by Relman et al. (17), the DNAs of all Bartonella species known to cause human diseases (B. henselae, B. quintana, B. bacilliformis, B. elizabethae, B. clarridgeiae) could be amplified (Fig. 1). No differences in PCR amplification were observed by using the biotinylated or the unbiotinylated oligonucleotide primer p12B. None of the other non-Bartonella bacterial strains reacted (Table 2).
FIG. 1.
Agarose gel electrophoresis of the amplified DNAs of five Bartonella species and Afipia felis, which is known to cause human infections. Lane 1, molecular size marker; lane 2, negative control; lanes 3 and 4, B. quintana; lanes 5 and 6, B. henselae; lanes 7 and 8, B. elizabethae; lanes 9 and 10, B. bacilliformis; lanes 11 and 12, B. clarridgeiae; lanes 13 and 14, A. felis.
TABLE 2.
Specificity and sensitivity of PCR, PCR-EIA, and dot blot hybridization on the basis of results with serial dilutions of B. henselae and B. quintanaa
| Strain and serial dilution (CFU/ml) | Intensityb by PCR with probes p12B-bio and p24E | OD/intensity by PCR-EIA
|
Intensityb by dot blot hybridization with probes:
|
||
|---|---|---|---|---|---|
| RHp-dig | RQp-dig | RHp-dig | RQp-dig | ||
| B. henselae Houston-1 | |||||
| 106 | ++++ | 3.187/++++ | 0.088/− | ++++ | − |
| 105 | +++ | 1.000/+++ | 0.105/− | +++ | − |
| 104 | ++ | 0.440/++ | 0.108/− | ++ | − |
| 103 | (+) | 0.197/+ | 0.082/− | + | − |
| 102 | − | 0.112/− | 0.081/− | − | − |
| 101 | − | 0.087/− | 0.080/− | − | − |
| B. quintana CIP | |||||
| 106 | ++++ | 0.099/− | 3.088/++++ | − | ++++ |
| 105 | +++ | 0.088/− | 2.572/+++ | − | +++ |
| 104 | ++ | 0.109/− | 0.655/++ | − | ++ |
| 103 | + | 0.101/− | 0.266/+ | − | + |
| 102 | − | 0.106/− | 0.107/− | − | (+) |
| 101 | − | 0.081/− | 0.078/− | − | − |
| B. bacilliformis | + | 0.088/− | 0.086/− | − | − |
| B. clarridgeiae | + | 0.089/− | 0.084/− | − | − |
| B. elizabethae | + | 0.102/− | 0.112/− | − | − |
| A. felis | − | 0.099/− | 0.081/− | − | − |
| Negative control | − | 0.099/− | 0.081/− | − | − |
| S. aureus | − | 0.084/− | 0.084/− | − | − |
| S. pyogenes | − | 0.114/− | 0.077/− | − | − |
| S. agalactiae | − | 0.082/− | 0.097/− | − | − |
| P. aeruginosa | − | 0.106/− | 0.081/− | − | − |
| E. coli | − | 0.080/− | 0.079/− | − | − |
| E. cloacae | − | 0.108/− | 0.081/− | − | − |
| K. pneumoniae | − | 0.099/− | 0.086/− | − | − |
The data presented here are the means of duplicate values.
Intensity of ethidium bromide-stained agarose gels or by dot blot hybridization: strongly positive (++++) to weakly positive (+).
EIA for detection of PCR products.
All incubation steps and concentrations of reagents were optimized with the strains of B. henselae and B. quintana mentioned above. An optical density (OD) by the PCR-EIA of ≥0.2 was a considered positive result and corresponded to twice the absorbance for the negative control. With this cutoff, B. henselae and B. quintana could be detected with the oligonucleotides RHp-dig and RQp-dig down to a concentration of 103 CFU/ml in serial dilutions, corresponding to an equivalent of 5 CFU of Bartonella species in the PCR mixture with 5 μl of DNA (Table 2). No cross-reactivity was observed between B. henselae and B. quintana or with the other Bartonella species. Thus, the oligonucleotides RHp-dig and RQp-dig appear to be specific for B. henselae and B. quintana, respectively. None of the non-Bartonella isolates hybridized with either probe.
Analysis of PCR products by dot blot hybridization.
The same oligonucleotide probes used for detection of B. henselae (RHp-dig) and B. quintana (RQp-dig) by PCR-EIA were also applied in the dot blot hybridization assay by using the same serial dilutions of both Bartonella strains. By dot blot hybridization both probes were specific for B. henselae and B. quintana, and no cross-reactivity with the Bartonella species or non-Bartonella isolates was seen (Table 2).
Determination of sensitivities of PCR-EIA and dot blot hybridization.
One aliquot of each of the 10-fold dilutions of suspensions of B. henselae and B. quintana was used for quantitative culture to determine the number of CFU, while another aliquot was analyzed in parallel by the PCR-based methods. In ethidium bromide-stained agarose gels, a band was visible from PCRs performed with 1 to 5 μl of dilutions of up to 103 CFU of B. henselae and B. quintana per ml; these corresponded to 1 to 5 CFU in the reaction mixture. The sensitivity obtained by the PCR-EIA was approximately 10-fold higher; weakly positive bands in the agarose gels gave a clear positive result in the EIA. Dot blot hybridization was also 10 times more sensitive than the test with ethidium bromide-stained gels (Table 2).
PCR-EIA and hybridization with clinical specimens.
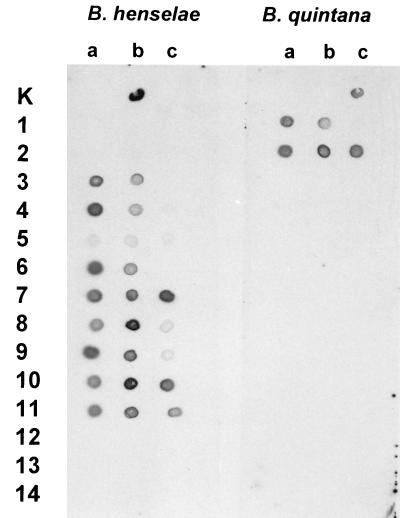
From the 16 clinical specimens which were tested by both PCR-EIA and dot blot hybridization, the lymph nodes from five control patients (without Bartonella infections) did not react by either method. The results for specimens from 11 patients with suspected Bartonella infection are shown in Fig. 2 and Table 3. The skin biopsy specimens from the two patients with BA reacted only with the B. quintana-specific probe by both methods, whereas the 9 lymph node biopsy specimens from patients with CSD gave positive results with the B. henselae-specific probe. None of the five samples without suspected Bartonella infection hybridized with either probe.
FIG. 2.
Results of dot blot hybridization with the B. henselae-specific probe RHp-dig and the B. quintana-specific probe RQp-dig with PCR products of samples from patients with BA (rows 1 and 2), patients with CSD (rows 3 to 11), and negative control patients (rows 12 to 14) in three dilution steps: a, 1:1; b, 1:5; and c, 1:50. Dot Ka, negative control; dot Kb, a B. henselae strain; dot Kc, a B. quintana strain.
TABLE 3.
ODs and calculated CFU per milliliter for the human specimens investigated
| Patient no., diagnosis | Dilution of extracted DNA | Intensity by PCR (on gels) | OD by PCR-EIAa
|
Bartonella species, calculated no. of CFU/ml | |
|---|---|---|---|---|---|
| RHp | RQp | ||||
| 1, BA | 1:1 | +++ | 0.103 | 1.677 | B. quintana, 1 × 105 |
| 1:5 | ++ | 0.088 | 0.530 | ||
| 1:50 | + | 0.093 | 0.220 | ||
| 2, BA | 1:1 | +++ | 0.091 | 1.202 | |
| 1:5 | +++ | 0.093 | 1.695 | B. quintana, 5 × 105 | |
| 1:50 | ++ | 0.093 | 0.559 | ||
| 3, CSD | 1:1 | ++ | 0.414 | 0.084 | B. henselae, 1 × 104 |
| 1:5 | ++ | 0.312 | 0.085 | ||
| 1:50 | − | 0.085 | 0.094 | ||
| 4, CSD | 1:1 | ++ | 0.558 | 0.105 | B. henselae, 4 × 104 |
| 1:5 | + | 0.260 | 0.101 | ||
| 1:50 | (+) | 0.146 | 0.098 | ||
| 5, CSD | 1:1 | + | 0.221 | 0.097 | B. henselae, 2 × 103 |
| 1:5 | + | 0.205 | 0.097 | ||
| 1:50 | (+) | 0.152 | 0.090 | ||
| 6, CSD | 1:1 | ++ | 0.515 | 0.087 | B. henselae, 2 × 104 |
| 1:5 | + | 0.307 | 0.099 | ||
| 1:50 | (+) | 0.090 | 0.103 | ||
| 7, CSD | 1:1 | ++ | 0.325 | 0.083 | |
| 1:5 | ++ | 0.358 | 0.085 | ||
| 1:50 | ++ | 0.590 | 0.086 | B. henselae, 2 × 106 | |
| 8, CSD | 1:1 | ++ | 0.393 | 0.087 | |
| 1:5 | +++ | 1.122 | 0.086 | B. henselae, 5 × 105 | |
| 1:50 | − | 0.182 | 0.089 | ||
| 9, CSD | 1:1 | +++ | 0.958 | 0.085 | B. henselae, 1 × 105 |
| 1:5 | ++ | 0.580 | 0.103 | ||
| 1:50 | − | 0.179 | 0.099 | ||
| 10, CSD | 1:1 | ++ | 0.481 | 0.090 | |
| 1:5 | +++ | 1.356 | 0.084 | B. henselae, 2 × 106 | |
| 1:50 | + | 0.347 | 0.085 | ||
| 11, CSD | 1:1 | ++ | 0.420 | 0.087 | B. henselae, 1 × 104 |
| 1:5 | ++ | 0.415 | 0.085 | ||
| 1:50 | (+) | 0.261 | 0.087 | ||
ODs are means for duplicate values.
The approximate numbers of CFU in the clinical samples were calculated as follows: with bacterial suspensions (Table 2) an OD of 0.2 corresponded roughly to 103 CFU/ml, but the increase in the OD was not a linear function of the increase in the number of CFU in bacterial dilutions. Therefore, an empiric standard curve was drawn by using the data given in Table 2 to determine the concentrations in the clinical specimens. By using the results for dilutions of 1:1, 1:5, and 1:50 of the clinical samples, the numbers of CFU were detected on the basis of the highest positive OD (Table 3). It should be emphasized that these results are obtained by comparing the ODs obtained by PCR with DNA extracted from human specimens with the ODs obtained by PCR with DNA extracted from cultured bacteria, and differences in the real and the calculated bacterial densities in human specimens cannot be excluded.
DISCUSSION
Various PCR procedures with different target sequences for detection of Bartonella species have been described (1, 3, 5, 10, 16, 21, 23). However, there are great differences in the sensitivities of these procedures. The three target genes most often used for PCR detection of Bartonella DNA are the 16S rRNA gene used in the present study, the citrate synthase gene (gltA), and the 60-kDa heat shock protein gene (htrA). Few data are available to compare these assays. In a recent study, the 16S rRNA gene PCR was slightly more sensitive than the htrA gene PCR, but false-negative results were obtained by both assays (21). However, agarose gel analysis and ethidium bromide staining do not appear to be sensitive enough for visualization of small amounts of PCR products. Dot blot hybridization or enzyme immunoassay used for the detection of the PCR-amplified DNA showed a 10-fold higher sensitivity than ethidium bromide staining in some studies (1, 9, 11, 14, 18). Fujita et al. (9) demonstrated that the OD of the EIA correlated with the number of C. albicans blastoconidia suspended in blood. Jantos et al. (12) developed a PCR-EIA for detection of Chlamydia pneumoniae which was as sensitive as Southern blot hybridization for the detection of PCR products and 100 times more sensitive than visualization of PCR products on agarose gels. Additionally, DNA extracted from human specimens may contain large amounts of cellular DNA that may inhibit amplification. Therefore, template dilution and amplification of different dilution steps may increase the sensitivity of PCR (1, 11; our results).
Detection of B. henselae- and B. quintana-amplified DNA by EIA has been reported previously by Ritzler and Altwegg (18), but no serial dilutions of Bartonella suspensions with detection of the number of CFU or clinical specimens have been investigated. When serial dilutions of the bacterial suspensions or different dilution steps of the template from clinical specimens were analyzed in our study, dot blot hybridization and EIA were 10 times more sensitive than agarose gel electrophoresis with ethidium bromide staining. We were able to detect 1 to 5 CFU of Bartonella species in the PCR mixture with bacterial dilutions and in the clinical specimens corresponding to 1 × 103 to 2 × 106 CFU/ml. Burnie et al. (6) described a semiquantitative PCR-EIA for the detection of circulating DNA in patients with disseminated candidiasis and showed that the OD also correlated with the clinical status of the patients, becoming negative if therapy was successful and progressively more positive if the patient’s condition deteriorated. However, it cannot be excluded that the OD and the calculated number of CFU per milliliter in our clinical specimens do not correspond exactly to the number of CFU in the investigated bacterial dilutions. As already mentioned, cellular DNA in human specimens may have an inhibitory effect on the amplification of the bacterial DNA, and the real number of CFU per milliliter in human specimens could be even higher.
In conclusion, our PCR-based EIA for the species-specific identification and quantification of amplified B. henselae and B. quintana DNAs was more sensitive than ethidium bromide staining of agarose gels, less time-consuming, and less labor-intensive than sequencing or dot blot hybridization for identification of amplified DNA. Species-specific results can be obtained within 24 h, and we think that this assay will improve the means of diagnosis of Bartonella infections.
ACKNOWLEDGMENTS
We thank Wolfgang Bredt for helpful discussions and critical reading of the manuscript and Karin Oberle for excellent technical assistance.
REFERENCES
- 1.Anderson B, Sims K, Regnery R, Robinson L, Schmidt M J, Goral S, Hager C, Edwards K. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J Clin Microbiol. 1994;32:942–948. doi: 10.1128/jcm.32.4.942-948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson B E, Neuman M A. Bartonella spp. as emerging human pathogens. Clin Microbiol Rev. 1997;10:203–219. doi: 10.1128/cmr.10.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avidor B, Kletter Y, Abulafia S, Golan Y, Ephros M, Giladi M. Molecular diagnosis of cat scratch disease: a two-step approach. J Clin Microbiol. 1997;35:1924–1930. doi: 10.1128/jcm.35.8.1924-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass J, Vincent J M, Person D A. The expanding spectrum of Bartonella infections. II. Cat-scratch disease. Pediatr Infect Dis J. 1997;16:163–179. doi: 10.1097/00006454-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Bergmans A M, Groothedde J W, Schellekens J F P, van Embden J D A, Ossewaarde J M, Schouls L M. Etiology of cat scratch disease: comparison of polymerase chain reaction detection of Bartonella (formerly Rochalimaea) and Afipia felis DNA with serology and skin tests. J Infect Dis. 1995;171:916–923. doi: 10.1093/infdis/171.4.916. [DOI] [PubMed] [Google Scholar]
- 6.Burnie J P, Golbang N, Matthews R C. Semiquantitative polymerase chain reaction enzyme immunoassay for diagnosis of disseminated candidiasis. Eur J Clin Microbiol Infect Dis. 1997;16:346–350. doi: 10.1007/BF01726361. [DOI] [PubMed] [Google Scholar]
- 7.Cáceres-Ríos H, Rodríguez-Tafur J, Bravo-Puccio F, Maguina-Vargas C, Sanguineti Díaz C, Ramos D C, Patarca R. Verrugua peruanan: an infectious endemic angiomatosis. Crit Rev Oncog. 1995;6:47–56. doi: 10.1615/critrevoncog.v6.i1.40. [DOI] [PubMed] [Google Scholar]
- 8.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O’Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita S, Lasker B A, Lott T J, Reiss E, Morrison C J. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33:962–967. doi: 10.1128/jcm.33.4.962-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldenberger D, Schmidheini T, Altwegg M. Detection of Bartonella henselae and Bartonella quintana by a simple and rapid procedure using broad-range PCR amplification and direct single-strand sequencing of part of the 16S rRNA gene. Clin Microbiol Infect. 1997;3:240–245. doi: 10.1111/j.1469-0691.1997.tb00604.x. [DOI] [PubMed] [Google Scholar]
- 11.Goldenberger D, Zbinden R, Perschil I, Altwegg M. Nachweis von Bartonella (Rochalimaea) henselae/B. quintana mittels Polymerase Kettenreaktion (PCR) Schweiz Med Wochenschr. 1996;126:207–213. [PubMed] [Google Scholar]
- 12.Jantos C A, Roggendorf R, Wuppermann F N, Hegemann J H. Rapid detection of Chlamydia pneumoniae by PCR-enzyme immunoassay. J Clin Microbiol. 1998;36:1890–1894. doi: 10.1128/jcm.36.7.1890-1894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kordick D L, Hilyard E J, Hadfield T L, Wilson K H, Steigerwald A G, Brenner D J, Breitschwerdt E B. Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease) J Clin Microbiol. 1997;35:1813–1818. doi: 10.1128/jcm.35.7.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lüneberg E, Jensen J S, Frosch M. Detection of Mycoplasma pneumoniae by polymerase chain reaction and nonradioactive hybridization in microtiter plates. J Clin Microbiol. 1993;31:1088–1094. doi: 10.1128/jcm.31.5.1088-1094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margileth A M, Baehren D F. Chest-wall abscess due to cat-scratch disease (CSD) in an adult with antibodies to Bartonella clarridgeiae: case report and review of the thoracopulmonary manifestation of CSD. Clin Infect Dis. 1998;27:353–357. doi: 10.1086/514671. [DOI] [PubMed] [Google Scholar]
- 16.Mouritsen C L, Litwin C M, Maiese R L, Segal S M, Segal G H. Rapid polymerase chain reaction-based detection of the causative agent of cat scratch disease (Bartonella henselae) in formalin-fixed, paraffin-embedded samples. Hum Pathol. 1997;28:820–826. doi: 10.1016/s0046-8177(97)90156-8. [DOI] [PubMed] [Google Scholar]
- 17.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 18.Ritzler M, Altwegg M. Sensitivity and specificity of a commercially available enzyme-linked immunoassay for the detection of polymerase chain reaction amplified DNA. J Microbiol Methods. 1996;27:233–238. [Google Scholar]
- 19.Sander A. Microbiological diagnosis of Bartonella species and Afipia felis. In: Schmidt A, editor. Contributions to microbiology. 1. Bartonella and Afipia species emphasizing Bartonella henselae. Basel, Switzerland: Karger; 1998. pp. 98–112. [Google Scholar]
- 20.Sander A, Bühler C, Pelz K, von Cramm E, Bredt W. Detection and isolation of two Bartonella henselae variants in domestic cats in Germany. J Clin Microbiol. 1997;35:584–587. doi: 10.1128/jcm.35.3.584-587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sander A, Posselt M, Böhm N, Ruess M, Altwegg M. Detection of Bartonella henselae DNA by two different PCR assays and determination of the genotype in histologically defined cat scratch disease. J Clin Microbiol. 1999;37:993–997. doi: 10.1128/jcm.37.4.993-997.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sander A, Posselt M, Oberle K, Bredt W. Seroprevalence of antibodies to Bartonella henselae in patients with cat scratch disease and in healthy controls: evaluation and comparison of two commercial serological tests. Clin Diagn Lab Immunol. 1998;5:486–490. doi: 10.1128/cdli.5.4.486-490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott M A, McCurley T L, Vnencak-Jones C L, Hager C, McCoy J A, Anderson B, Collins R D, Edwards K M. Cat scratch disease. Detection of Bartonella henselae DNA in archival biopsies from patients with clinically, serologically, and histologically defined disease. Am J Pathol. 1996;149:2161–2167. [PMC free article] [PubMed] [Google Scholar]