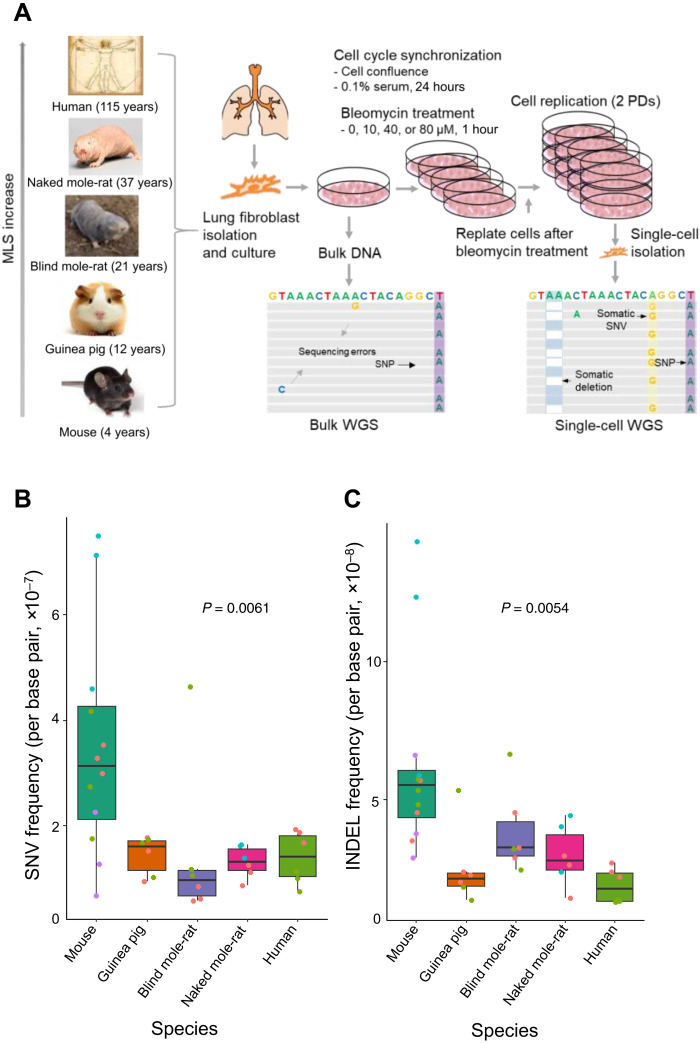
Somatic mutation rates are inversely correlated with life span in mouse, guinea pig, blind mole-rat, naked mole-rat, and humans.
Abstract
DNA mutations in somatic cells have been implicated in the causation of aging, with longer-lived species having a higher capacity to maintain genome sequence integrity than shorter-lived species. In an attempt to directly test this hypothesis, we used single-cell whole-genome sequencing to analyze spontaneous and bleomycin-induced somatic mutations in lung fibroblasts of four rodent species with distinct maximum life spans, including mouse, guinea pig, blind mole-rat, and naked mole-rat, as well as humans. As predicted, the mutagen-induced mutation frequencies inversely correlated with species-specific maximum life span, with the greatest difference observed between the mouse and all other species. These results suggest that long-lived species are capable of processing DNA damage in a more accurate way than short-lived species.
INTRODUCTION
Genome instability, the increased tendency of the genome to undergo DNA mutation, is a hallmark of aging (1). Various types of mutations, e.g., single-nucleotide variants (SNVs), small insertions and deletions (INDELs), and chromosomal aberrations, have been demonstrated to accumulate with age in different organs and tissues of humans and rodents (2). However, a definite causal relationship between mutation accumulation and the aging process has thus far not been established. If mutations are causally related to aging, then one would expect to find a correlation between the activity of genome maintenance pathways to prevent loss of genome sequence erosion and species longevity. Evidence for increased DNA repair activity in cells from long-lived, i.e., humans, cows, and elephants, as compared to short-lived, i.e., shrew, mouse, rat, and hamster, species after ultraviolet (UV) irradiation has been obtained as early as the 1970s (3). However, these results remain difficult to interpret because of major phylogenetic, lifestyle, and physiological differences other than DNA repair between these species. The repair pathway studied, i.e., nucleotide excision repair of UV lesions, is important in humans and cows that are active during the day, but much less so in nocturnal rodents (4). The problem of phylogenetic distance can, to some extent, be addressed using species with great differences in life span but are evolutionarily closely related. More recently, a panel of 18 rodent species with diverse life spans was used to study DNA double-strand break (DSB) repair, which was found to be more efficient in the longer-lived species (5).
It remains unclear how differences in DNA repair activities would affect genome sequence integrity, which is primarily determined by the accuracy, not the amount, of DNA replication and repair. The main challenge in studying mutations in somatic cells is the difficulty of measuring random mutations in a population of cells or a tissue. In vitro clonal amplification or ultrahigh depth sequencing to detect clonally amplified mutations can only be applied to stem or progenitor cells. Recent developments in single-cell sequencing have allowed to accurately assess the frequency and spectrum of SNVs and small INDELs (6, 7). Here, we used single-cell whole-genome sequencing to analyze the capacity of cells from rodent species of different life spans to maintain genome sequence integrity after treatment with a single dose of bleomycin.
The hypothesis underlying this study is that fewer mutations will be induced by a fixed dose of mutagen in cells from long-lived species than in cells from short-lived species. We chose bleomycin as the mutagen because, while primarily a clastogen, this compound is known to induce both small deletions and base substitutions (8). We studied spontaneous and bleomycin-induced somatic mutation frequency in phylogenetically close rodent species, differing as much as 10-fold in maximum life span (9). After analyzing primary lung fibroblasts from mouse, guinea pig, blind mole-rat, and naked mole-rat as compared to the same type of cells from humans as the outgroup, the results indicate that generally more mutations are induced in mouse and guinea pig cells by a fixed dose of mutagen than in the same cells from the long-lived rodents or humans. These results suggest that long-lived species may indeed be capable of processing DNA damage in a more accurate way than short-lived species.
RESULTS
Spontaneous somatic SNV and INDEL frequencies
In view of the high cost of whole-genome sequencing of multiple cells from multiple species, we selected four rodent species with the largest differences in life span: mouse (Mus musculus), guinea pig (Cavia porcellus), blind mole-rat (Nannospalax galili; i.e., Upper Galilee mountains blind mole-rat), and naked mole-rat (Heterocephalus glaber), as well as human (Homo sapiens), with maximum life spans of approximately 4, 12, 21, and 37 years for the rodents, respectively, and 115 years for human (10–13). These specific rodent species were also selected because of the availably of relatively high-quality reference genome sequences (table S1).
To identify somatic mutations and accurately estimate their frequency, single-cell whole-genome sequencing is necessary, because each single cell acquires its unique set of somatic mutations during the lifetime of the organism. We performed single-cell whole-genome sequencing using the single-cell multiple displacement amplification method that we developed earlier (see Materials and Methods) (6). This method substantially reduces potential artifactual mutations occurring during the whole-genome amplification process. We applied the method on early passage, primary lung fibroblasts from each species, isolated from young adults (Fig. 1A) (14), with single-cell amplicons that passed our locus dropout test selected for whole-genome sequencing (table S2) (6). We also sequenced DNA of bulk fibroblast populations of the same individual animals/humans to filter out germline variations. Whole-genome sequencing was performed using the Illumina HiSeq X Ten or NovaSeq platform, reaching an average sequencing depth of 27.6× across the genome (table S3). Somatic SNVs and INDELs of each cell were identified after filtering out germline variants identified from bulk DNA based on an updated version of SCcaller (see Materials and Methods) (6), capable of identification of INDELs in addition to SNVs.
Fig. 1. Spontaneous mutation frequencies in different species.
(A) Study design. Lung fibroblasts were isolated from young adult individuals of the five species, subjected to different doses of bleomycin treatment including 0 (i.e., the controls), 10, 40, and 80 μM, followed by single-cell whole-genome sequencing (scWGS). Somatic SNVs and INDELs were identified from the sequencing data by filtering out germline mutations identified from whole-genome sequencing of bulk DNA of the same individuals. MLS, maximum life span. (B and C) Spontaneous somatic SNV and INDEL frequencies of lung fibroblasts without bleomycin treatment in the five species. Data points in different colors indicate cells of different individuals analyzed for each species. Boxplot elements are defined as follows: Center line indicates median, box limits indicate upper and lower quartiles, whiskers indicate 1.5× interquartile range, and points indicate all data points.
Somatic mutation frequency was defined as the ratio of the number of somatic mutations to the number of base pairs covered in the genome with at least 20× sequencing depth, with adjustments for genome ploidy (i.e., diploid) and mutation calling sensitivity (see Materials and Methods and table S4). To determine de novo spontaneous somatic mutation frequency, we analyzed whole-genome sequences of each of three cells per individual animal/human participant from a total of 12 individuals, 4 animals for the mouse, and 2 for each other species. As shown in Fig. 1B, the median spontaneous somatic SNV frequency of mouse single cells [3.6 × 10−7 base pair (bp)−1] is about twofold higher than that of any other species, i.e., guinea pig, blind mole-rat, naked mole-rat, and human (a median SNV frequency of 1.7 × 10−7, 0.9 × 10−7, 1.4 × 10−7, and 1.7 × 10−7 bp−1 for the four species, respectively; P = 0.0061, two-tailed Student’s t test), among which no significant differences were found. Notably, these results for mouse and human cells confirm the previously observed difference between somatic mutation frequencies in dermal fibroblasts from these two species (15). The median frequencies of spontaneous somatic INDELs (Fig. 1C) were found to be about four to five times lower than those of SNVs but showed a similar pattern of a significantly higher frequency in mouse cells (5.7 × 10−8 bp−1) than in cells of the other species (4.6 × 10−8, 3.3 × 10−8, 3.4 × 10−8, and 2.6 × 10−8 bp−1 for guinea pig, blind mole-rat, naked mole-rat, and human cells, respectively). Notably, it occurred to us that inbred mice might have different somatic mutation rates than outbred animals, which is why we also analyzed cells from two four-way cross mice (UM-HET3). We did not observe a significant difference between mutation frequencies in lung fibroblasts from the two mouse strains (P = 0.535 and P = 0.155 for SNVs and INDELs, respectively). These results indicate a significantly higher spontaneous mutation frequency in lung fibroblasts from the mouse than in the cells of same type from any of the other species analyzed; among the latter, no significant differences were found.
Spontaneous somatic SNV and INDEL spectra
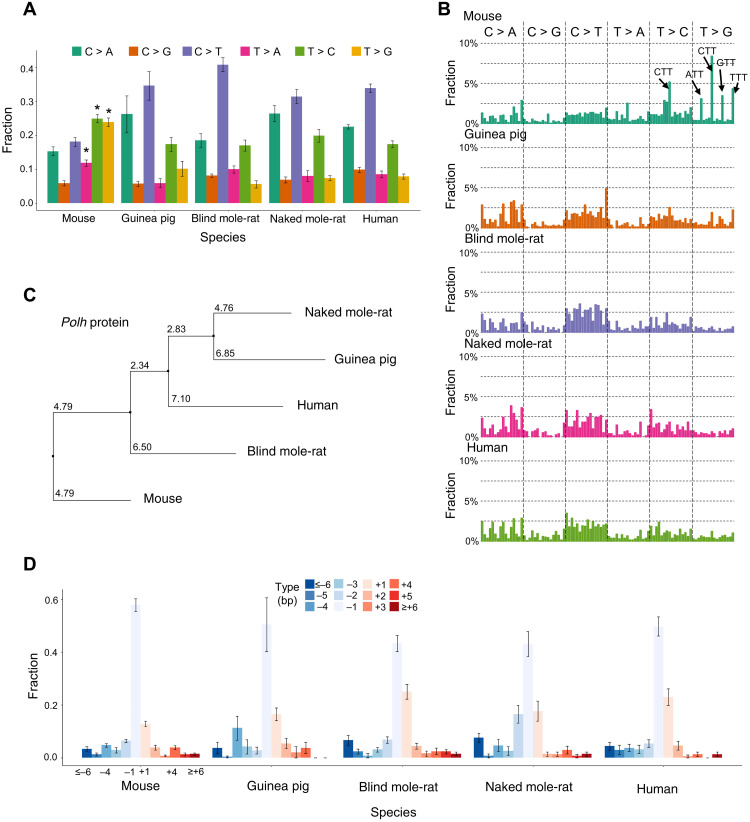
We then studied the somatic mutation spectra for possible differences between cells of these species. The most obvious species-to-species difference was found between mouse and the other species (Fig. 2A). Mouse fibroblasts had significantly more somatic SNVs at A/T bases than other species: 12, 26, and 27% of total somatic SNVs in mouse cells are T > A, T > C, and T > G, respectively, while only 9, 17, and 8% of somatic SNVs in cells of the other species are of these types (P = 0.004, P = 7.7 × 10−5, and P = 1.23 × 10−9, respectively, two-tailed Student’s t test). Notably, the most significant difference was in T > G transversions. We did not observe significant differences in mutation spectra between the inbred and outbred mouse strain. Notably, the above differences reflect the fractions of mutations of the total number of mutations in the cells. In absolute numbers, the mutations at C bases are also different between species. For example, mouse cells have the highest absolute numbers of spontaneous C > T mutations (100 per cell on average), which are likely due to deamination of 5-methylcytosine, followed by guinea pig (69 per cell; P = 0.115, two-tailed Student’s t test compared to mouse), blind mole-rat (65 per cell; P = 0.035), naked mole-rat (52 per cell; P = 0.008), and human (87 per cell; P = 0.379). These numbers inversely correlate with the maximum life span within the rodent group.
Fig. 2. Mutational spectra of spontaneous somatic mutations.
(A) Mutational spectra of spontaneous somatic SNVs. (B) Mutational spectra of spontaneous somatic SNVs in the context of their two flanking base pairs. This classified mutations into 96 categories. On the x axis, the 96 categories were sorted according to their alphabetic order. For example, the first bar from the left indicates ACA to AAA mutation, and the first from the right indicates TTT to TGT mutation. The y axis indicates the fraction of each category out of the total number of mutations. (C) A phylogeny tree of protein sequences of the Polh orthologs of the five species. Multiple sequence alignment was performed using Clustal Omega, and the phylogeny tree was calculated using the neighbor joining algorithm using Jalview based on the PID (i.e., the percentage identity between the two sequences at each aligned position) score, which indicates the number of identical residues per 100 residues. (D) Mutational spectra of spontaneous somatic INDELs. Error bars in (A) and (D) indicate SD.
By taking the two flanking bases of each mutation into consideration (Fig. 2B), we found somatic SNVs in mice, but not in the other species, to occur more frequently at TT bases. A similar mutational pattern of TT bases has been observed in human lymphoid cells [i.e., signature 9 in the Catalogue of Somatic Mutations in Cancer (COSMIC) database], which was considered the result of the error-prone DNA polymerase eta during somatic hypermutation (16). This same mutational signature was previously observed in our study of somatic mutations in normal human memory B lymphocytes, which suggests a possible shared mechanism (17). Because a mutation spectrum is a linear combination of one or more mutational signatures, we can compare our mutation spectra with known mutational signatures using correlation coefficients. On the basis of cosine similarity, the mutation spectrum found in mouse cells has the highest correlation with signature 9 (cosine correlation = 0.76), while its correlations with other COSMIC signatures are all less than 0.7. In addition, the correlations between the spectra of cells of the other four species and signature 9 are only in the range of 0.49 to 0.58.
To further test whether difference in DNA polymerase eta gene, Polh, is a cause of above unique mutation spectrum in mouse, we compared the protein sequences of the Polh orthologs from the five species, using multiple sequence alignment by Clustal Omega (18) and neighbor joining tree by Jalview (19). The mouse ortholog was found to be the outlier (Fig. 2C), which is in obvious contrast with the species phylogeny. Although we cannot exclude other underlying causes, these results suggest that DNA polymerase eta may be an important factor in the elevated mutation frequency in mouse somatic cells.
For spontaneous somatic INDELs, the mutational spectra between different species were much more similar than those of somatic SNVs. As shown in Fig. 2D, 1-bp deletion accounts for about 50% of total INDELs across cells of all species, with mouse fibroblasts having the most. The second most common INDEL is 1-bp insertion, accounting for about 20% of these events across species. This indicates that, for somatic INDELs, unlike somatic SNVs, species specificity in mutation accumulation is only observed for frequency but not spectrum.
Bleomycin-induced somatic SNVs and INDELs
To further investigate the capability of maintaining genome sequence integrity in the different rodent and human species, we analyzed mutation frequencies and spectra in the primary lung fibroblasts after treatment with three doses of bleomycin: 10, 40, and 80 μM (see Materials and Methods). Bleomycin has a cytotoxic effect and can induce DNA single-strand break and DNA DSB with a ratio of approximately 6:1, which may vary depending on the dose (20, 21). Small INDELs and SNVs are induced as a consequence of errors during DNA DSB repair (22). After bleomycin treatment, the cells were allowed to grow for two population doublings (PDs) to allow repair of the DNA damage and fixation of the mutations (Fig. 1A). A substantial difference between the species was already apparent in their proliferation abilities after the bleomycin treatment. Although bleomycin delayed cell growth in cells from all species (fig. S1, A to E), mouse cells were not even able to reach two PDs after 40 or 80 μM bleomycin treatment. Mouse cells were found to carry a substantially higher proportion (approximately 40%) of apoptotic cells compared to cells from the longer-lived species or humans under the same treatment conditions (fig. S2, A to D).
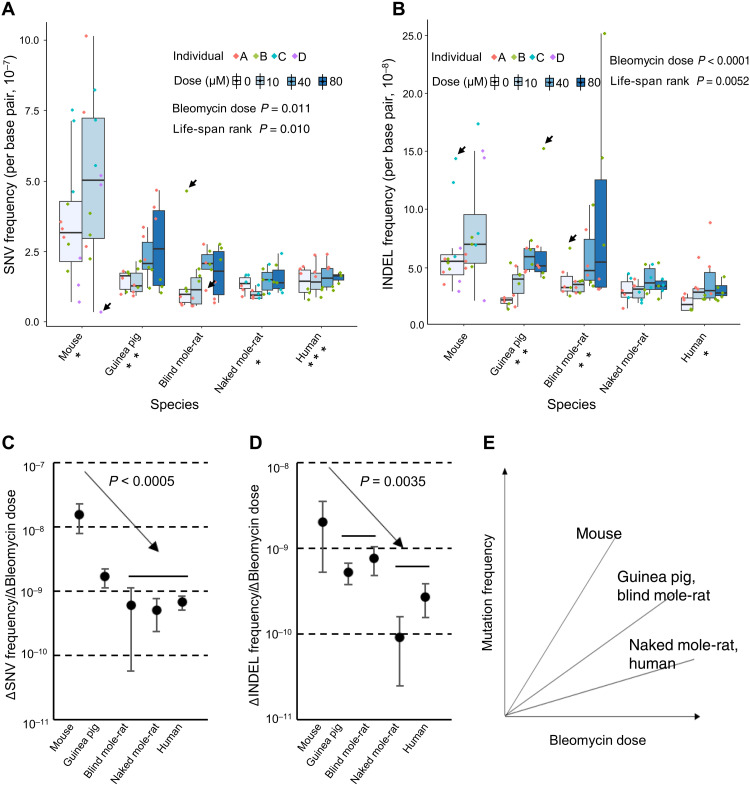
To determine somatic mutation frequency, we then isolated single fibroblasts for the three doses of bleomycin treatment for all individual animals/humans of all five species studied (for mouse only after 10 μM bleomycin) and performed whole-genome amplification, sequencing, and mutation calling as described above and in Materials and Methods. We first analyzed the resulting dataset using a linear mixed-effects regression model: Mutation frequency was modeled as a function of (i) bleomycin dose (fixed effect), (ii) the ranking of species-specific life span (mouse, 1; guinea pig, 2; blind mole-rat, 3; naked mole-rat, 4; human, 5) (fixed effect), and (iii) animal/human individual (random effect; see Materials and Methods). We found both bleomycin dose and the life-span rank to be significant factors affecting somatic SNV frequencies (P = 0.011 and P = 0.010, respectively, ANOVA; Fig. 3A) and somatic INDEL frequencies (P < 0.0001 and P = 0.0052, respectively, ANOVA; Fig. 3B). In addition, as with spontaneous mutations, we did not observe a significant difference between the two mouse strains in response to bleomycin treatment (P = 0.832 and P = 0.137 for SNVs and INDELs, respectively, sANOVA).
Fig. 3. Bleomycin-induced somatic mutation frequencies.
(A and B) Somatic SNV and INDEL frequencies of lung fibroblasts from the five species after bleomycin treatment. Data points in different colors indicate cells of different individuals analyzed of each species. *P < 0.05, **P < 0.01, and ***P < 0.001, when testing the correlation with bleomycin dose for each species individually. Boxplot elements are defined as follows: Center line indicates median, box limits indicate upper and lower quartiles, whiskers indicate 1.5× interquartile range, and points indicate all data points. Arrows indicates examples of cells with either marked higher or lower mutation frequencies than the other cells. (C and D) Increase of somatic SNV and INDEL frequencies per 1 μM bleomycin in different species. P values were estimated for the correlative pattern between mutation frequency increase per 1 μM bleomycin and rank of species-specific life spans and were based on a permutation test of 2000 repeats. (E) Schematic summary of the results in (C) and (D).
The cell-to-cell variation in mutation frequency appeared to be generally higher in mouse, guinea pig, and blind mole-rat than in the naked mole-rat or human. Examples of outliers found among cells of the first three species are indicated in Fig. 3 (A and B). Although the number of cells analyzed is not sufficient to draw firm conclusions, these results may suggest that mutation frequency among cells from longer-lived species is more stable than in those of shorter-lived species and therefore also point toward greater genome maintenance capacity.
We also analyzed mutational spectra but did not observe significant differences in spectra before and after bleomycin treatment (fig. S3, A and B). This strongly suggests that mechanisms involved in repair of bleomycin-induced DNA damage are similar to those acting upon spontaneous DNA damage.
We then tested whether the extent of the bleomycin-induced mutation frequency increase also inversely correlated with species-specific life span. Specifically, we estimated the elevated mutation frequency per 1 μM bleomycin for each species separately, using linear regression models, and compared them with the life-span rankings of the species. The results show that, for somatic SNVs, mouse cells acquired the highest number of mutations per 1 μM bleomycin, with guinea pig cells the second highest, and cells from the other species jointly at the lowest level (Fig. 3C and fig. S4A). We tested whether such a pattern of negative correlation with species-specific life span was statistically significant by a permutation test, randomizing the species that each animal/human individual belongs to. After 2000 repeats, we found that not a single repeat reflected such a pattern, corresponding to a P < 0.0005 (permutation test; 2000 repeats; one-tailed). For somatic INDELs, a similar pattern was observed with the exception that blind mole-rat cells were about the same as guinea pig cells (P = 0.0035, permutation test; 2000 repeats; one-tailed; Fig. 3D and fig. S4B). The results above indicate that genome maintenance accuracy is the highest in somatic cells of humans and naked mole-rat, followed by those of blind mole-rat and guinea pig, with that in mouse cells the lowest (Fig. 3E). Notably, the largest interspecies difference remained between mice and all other species, as was found for spontaneous mutations. This could be interpreted either in terms of the mouse as a unique species, different from all others, or as a possible plateau in maintaining genome sequence integrity readily reached in longer-lived species.
DISCUSSION
In a study conducted well before the advent of high-throughput sequencing, germline mutation rates during evolution were estimated from interspecies DNA sequence differences analyzed by thermal stability of DNA hybrids or neutral base pair substitutions in the relatively few coding regions sequenced. The results indicated different rates of DNA change among different phylogenetic groups, with the slowest rates observed for higher primates and faster rates for short-lived rodents, including mouse, rat, and hamster (23). These results are unlikely to be explained by differences in generation time and may instead reflect less-accurate DNA replication systems in shorter-lived rodents than in longer-lived primates (23, 24). Within primates, the rate of DNA sequence change in the human lineage compared to other primate lineages is the lowest. This so-called hominoid slowdown (25) coincides with the longevity increase during evolution of primates (26). Hence, the evidence suggests that genome instability as controlled by accuracy of replication and repair is a major factor in determining species-specific life span.
Evidence that germline mutation rate is a factor in the aging process has also come from studies within the human species. Recently, age-adjusted mutation rates were determined in 61 women and 61 men from the Utah CEPH (Centre d’Etude du Polymorphisme Humain) families, with higher mutation rates notably associated with higher all-cause mortality in both sexes and a shorter reproductive life span in women (27).
Germline mutation frequencies are not necessarily the same as somatic mutation frequencies. While it is reasonable to assume that the selection processes responsible for varying germline mutation rates between species have the same effect on somatic mutation rates, the latter have been shown to differ between tissues within one species (28). Moreover, somatic mutation frequencies, corrected for the difference in the number of cell divisions per generation, has been shown to be at least an order of magnitude higher than the germline mutation frequency in both mouse and human (15). Unfortunately, somatic mutation burdens are different from cell to cell and cannot be assessed by bulk sequencing. For this reason, we previously developed a single-cell mutation analysis method (6). Single-cell or clone whole-genome sequencing provides not only accurate estimation of mutational frequency per cell (17, 29–31) but also the future possibility of studying interactions among mutations within a cell, which may also have substantial functional effects relevant to aging and/or age-related diseases (32, 33).
In this present work, we directly studied spontaneous and mutagen-induced mutation frequency in primary lung fibroblasts from short- and long-lived rodent species and in human cells of the same type. The results show that cells from short-lived mice have a significantly higher frequency of somatic mutations than the same cell type from longer-lived rodents or humans. On the basis of our observation of excess mutations at TT bases in the mouse cells alone and its association with DNA polymerase eta, we found that the latter is a phylogenetic outlier among the species studied. Hence, the observed increase in SNV frequency in mouse cells can reasonably be explained, at least in part, by the more error-prone DNA polymerase eta, of which the protein sequence in mouse deviates from that in all other species. While this could still be longevity-related and DNA polymerase eta may also play a role in SNV induction in other similarly short-lived rodent species, the main conclusion remains that the life-span differences between the other species do not correlate with differences in spontaneous mutation rates.
Stress-related aging and longevity differences are often only uncovered after challenging animals or cells. Here, we treated the primary lung fibroblasts from each species with the mutagen bleomycin to provide such a challenge. The results confirm that, in mouse cells, bleomycin-induced mutation frequency is also significantly higher than in cells of other longer-lived rodent species or in human cells. This also could be explained in part by the more error-prone polymerase eta. However, for bleomycin-induced mutations, we find induced mutation loads to correlate with species-specific life span across all species tested. This result cannot be explained by polymerase eta alone and suggests that other mechanisms contribute to lower mutation frequency in long-lived animals. Multiple mechanisms could be involved from enhanced enzymatic function of DNA repair enzymes to elevated expression levels. Expression of genome maintenance genes has been found to correlate with life span in different rodent species (34, 35). Genome maintenance is complex, and it will be very difficult to tease out the species-specific pathways responsible for variation in mutagenic response. Meanwhile, this first evidence that accuracy of maintaining genome sequence integrity is correlated with species-specific life span could serve as a model for more extensive studies. These studies will become feasible once the currently still high-sequencing costs have come down and robust reference genomes are available for most rodent species.
MATERIALS AND METHODS
Animal subjects and human individuals
All rodent experiments were performed according to procedures approved by the University of Rochester Committee on Animal Resources. In this study, we used cells from four mice (M. musculus), including two inbred mice (C57BL/6; 6 months old) from the Gorbunova laboratory and two four-way cross mice (UM-HET3; 4 months old) from the Gladyshev laboratory. Two guinea pigs (C. porcellus), two blind mole-rats (N. galili; i.e., Upper Galilee mountains blind mole-rat), and three naked mole-rats (H. glaber) were used in this study, which were young adults available in the Gorbunova laboratory. On the basis of body size and weight, they are also young adults. Two frozen human (H. sapiens) primary fibroblast samples were obtained from the American Type Culture Collection (ATCC) (HS-A; 14 years old) and Lonza (HS-B; 12 years old). All species are approximately matched by specific fractions of their maximum life span (age at about 10% of the maximum life span).
Lung fibroblast isolation and cell culture
Primary lung fibroblasts were isolated following a cell isolation protocol adapted from Seluanov et al. (14). Briefly, the lung from each rodent species (young adult) was minced and incubated in Dulbecco’s modified Eagle’s medium (DMEM) F-12 medium with Liberase Blendzyme 3 (0.13 U/ml) and 1× penicillin/streptomycin (P/S) at 37°C for 40 min. Dissociated cells were washed, plated in cell culture dishes with complete DMEM F-12 medium and 15% fetal bovine serum (FBS), and cultured at 37°C, 5% CO2, and 3% O2 (32°C for naked mole rat). When reaching confluence, cells were split and replated in Eagle’s minimum essential medium (EMEM; ATCC, 30-2003) supplemented with 15% FBS and 1% P/S (100 U/ml). Lung fibroblasts were purified by further passaging in the same medium. Human lung fibroblasts were obtained from ATCC and Lonza. The human cells were cultured in the complete fibroblast growth medium (FGM; Lonza, CC-3132) as recommended by the instructions.
Bleomycin treatment
Early-passage primary lung fibroblasts were cultured in EMEM (ATCC, 30-2003) with 15% FBS and P/S (100 U/ml). When reaching confluence, cells were starved in low-serum EMEM (0.1% FBS) for cell cycle synchronization at 37°C (32°C for naked mole rat cells) for 24 hours. Then, the cells were washed twice with phosphate-buffered saline (PBS) and treated, in plain EMEM (with 1% P/S), with different doses of bleomycin (0, 10, 40, and 80 μM) at 37°C (32°C for naked mole rat cells) for 1 hour. The bleomycin (EMD Millipore Corporation, 203401-10MG) was dissolved in nuclease-free water. The cells were trypsinized, harvested, and counted after treatment. Then, the cells were replated at 0.2 million as the starting number in the complete EMEM, allowing DNA damage repair and mutation fixation over two rounds of cell replications. Notably, human cells were starved in fibroblast growth basal medium (Lonza, CC-3131) with 1% P/S without supplemented FBS. The complete FGM was used for human cell culture.
Cell replication and apoptosis
Cell growth was monitored by counting cell numbers with the Cellometer Auto T4 (Nexcelom Bioscience) twice or thrice until cells reaching two PDs after replated. The cell PD time was estimated on the basis of the cell growth curve for each species under different doses of bleomycin treatment. The results of PDs are shown in fig. S1.
The cell apoptosis was detected using the Guava Annexin Red Kit (Luminex, FCCH100108) and Guava easyCyte flow cytometers (Millipore) according to the manufacturer’s instructions. The percent of apoptotic cells was analyzed by GuavaSoft software with flow cytometer. Briefly, the cells were harvested at 0, 24, 48, 72 and 96 hours after cell treatment and replating. The cells were counted and prepared at a cell concentration of approximately between 2 × 105 and 1 × 106 cells/ml. Then, 100 μl of each cell solution was transferred into one well of 96-well plate and mixed with 100 μl of annexin reagent from the kit. The mixture was incubated at room temperature for 20 min in the dark. Then, the plate with samples was loaded in a flow cytometer, and the program “Nexin assay plus” was run. All the annexin V-positive cells were calculated as apoptotic cells. The percentage of apoptotic cells from different species at each time point was shown in fig. S2. The setting of the Nexin program and analysis software for samples from different species, conditions, or time points was consistent.
Single-cell isolation
The cells were harvested when the number reached two PDs and prepared for single-cell isolation. Single-lung fibroblasts were isolated using the CellRaft array (Cell Microsystems), as described previously (6). Briefly, we first wetted an array by adding 2 ml of cell culture medium and removing after 3 min, which was repeated three times. We then added approximately 5000 fibroblasts in 3 ml of medium to the array and incubated at 37°C, 3% O2 , and 10% CO2. After 3 hours, the fibroblasts elongated and attached to the array. We then removed the medium and washed the array with the attached fibroblasts twice with 1 ml of PBS. We then added 3 ml of fresh complete medium. We identified the rafts in the array that contained one cell by microscopy. Using a magnetic wand supplied with the CellRaft system, we then transferred each of the rafts with one cell into a 0.2-ml polymerase chain reaction (PCR) tube containing 2.5 μl of PBS. We validated with a magnifier that each tube contained a single raft, froze them on dry ice, and kept them at −80°C until usage.
Single-cell whole-genome amplification
We amplified the isolated single cells using the Single-Cell Multiple Displacement Amplification protocol reported previously (6). Briefly, for each single cell, we added 1 μl of exo-resistant random primer (Thermo Fisher Scientific) and 3 μl of lysis buffer (400 mM KOH, 100 mM dithiothreitol, and 10 mM EDTA) and incubated on ice for 10 min. We neutralized the lysis buffer by adding 3 μl of stop buffer [400 mM HCl and 600 mM tris-HCl (pH 7.5)]. We then added 32 μl of master mix containing 30 μl of multiple displacement amplification reaction buffer and 2 μl of Phi29 polymerase (REPLI-g UltraFast Mini Kit, QIAGEN), incubated for 1.5 hours at 30°C and 3 min at 65°C, and held at 4°C until purification. We purified the amplicons using AMPure XP beads (Beckman Coulter) and quantified DNA concentration with the Qubit High-Sensitivity dsDNA Kit (Thermo Fisher Scientific). Simultaneously, we amplified 1 ng of genomic DNA in 2.5 μl of PBS as positive control and 2.5 μl of PBS without any template as negative control. We performed the locus dropout test as described previously (6) with primers designed for each species separately (table S2). Three single-cell amplicons per individual per experimental condition that passed the locus dropout test were prepared for whole-genome sequencing.
DNA library preparation and whole-genome sequencing
Sequencing libraries of the bulk DNA and single-cell amplicons were constructed using the TruSeq Nano DNA HT Sample Prep Kit (Illumina) by our laboratory or by Novogene Inc. The libraries were purified using AMPure XP beads (Beckman Coulter) and subjected to quality control using Bioanalyzer 2100 (Agilent) and real-time PCR. The libraries were sequenced on the Illumina HiSeq X Ten or NovaSeq S4 sequencing platforms for 2 × 150–bp paired-end reads by Novogene.
Bulk DNA extraction
Bulk DNA, i.e., DNA of bulk fibroblast populations, of each individual was extracted from primary lung fibroblasts before the bleomycin treatment experiments, using the DNeasy Blood and Tissue Kit (QIAGEN) following the manufacturer’s instructions. We determined their concentration with the Qubit High-Sensitivity dsDNA Kit (Thermo Fisher Scientific) and their quality with 1% agarose gel electrophoresis.
Sequence alignment
For raw sequencing reads of each single-cell or bulk DNA sample, we performed adapter and quality trimming using Trim Galore (version 0.6.4). Reads before and after trimming were subjected to quality control using FastQC (version 0.11.8). We aligned the trimmed sequences to their species-specific reference genomes (see table S1 for the version of reference genomes used) using Burrows-Wheeler Aligner-Maximal Exact Match (BWA-MEM, version 0.7.17) (36) and removed PCR duplicated sequences using SAMtools (version 1.9) (37).
To perform INDEL realignment and base quality score recalibration, known INDELs and single-nucleotide polymorphisms (SNPs) are required but are not available for species except for human and mouse. To handle this, we followed the Genome Analysis Toolkit (GATK) instructions to use INDELs and SNPs called from our data: We called INDELs and SNPs using GATK (version 3.5.0) (38) HaplotypeCaller from the sequence alignment (obtained above) of each bulk DNA sample and used them as a database for INDEL realignment and base quality score recalibration for all bulk DNA and single-cell samples, which were done also using GATK (version 3.5.0). We kept chromosome contigs with a minimum length of 1 Mbp for mutation analysis to avoid potential bias in mutation calling, in short, incompletely assembled contigs in some species. The resulted sequence alignments were used for finding somatic mutations as described below.
Calling somatic SNVs and INDELs
We identified somatic SNVs and INDELs, i.e., SNVs and INDELs observed only in a single cell and not in its corresponding bulk DNA by SCcaller (version 2.0.0) that we developed for variant calling of single cells previously (6). The version 2.0.0 of SCcaller was based on the same principle of its 1.0.0 version (for SNV calling only), with the additional function to call INDELs (here, we focused on 1 to 10 bp of small INDELs) in single-cell sequencing (freely available at https://github.com/biosinodx/Sccaller). Briefly, we first called heterozygous germline SNPs using the GATK HaplotypeCaller and used them in SCcaller to correct for potential amplification bias occurred during the single-cell whole-genome amplification. We then used SCcaller to call somatic autosomal SNVs and INDELs only observed in a single cell but not in its corresponding bulk, requiring 20× depth in both the samples. For INDELs, we additionally required a genotype calling quality of ≥30 and a maximum size of 10 bp to ensure the identification of true mutations instead of false positives. Germline heterozygous SNPs and INDELs of the same single cells, i.e., those mutations also present in bulk DNA sample, were also identified using SCcaller under the same criteria. This enabled us to estimate the sensitivity of variant calling, defined as the ratio of number the germline heterozygous SNPs or INDELs observed in the single cells to the number of germline heterozygous SNPs or INDELs observed in their corresponding bulk DNA sequences. Because the number of germline heterozygous SNPs and INDELs is limited in the inbred mouse strain, C57BL/6, sensitivity in these cells is much harder to estimate than that for noninbred subjects or strains. So, we approximated the sensitivities of C57BL/6 mouse cells as the average sensitivity of all the other single cells. Somatic SNV and INDEL frequencies were determined after correcting the sensitivity, the genome coverage (≥20× and autosomal requirements), and the diploidy of a single-fibroblast genome (table S4).
Analyzing mutation frequency with linear mixed-effects regression
When analyzing mutation frequency as a function of bleomycin dose and rank of species-specific life span (mouse, 1; guinea pig, 2; blind mole-rat, 3; naked mole-rat, 4; human, 5), we used a linear mixed-effect regression model: bleomycin dose and rank of life span with fixed effects and different individuals within each species with random effects. This was performed using the “lme” function in the R package “nlme”: lme(mutation freq ~ life-span rank + bleomycin dose, random = ~1|individual) (39).
Acknowledgments
We thank Z. Ke from V. Gorbunova’s laboratory for assistance with some of the experiments.
Funding: This work is funded by National Institutes of Health grant P01 AG047200 (to V.G., J.V., A.S., and V.N.G.), National Institutes of Health grant K99/R00 AG056656 (to X.D.), National Institutes of Health grant P01 AG017242 (to J.V.), National Institutes of Health grant U01 ES029519 (to J.V.), National Institutes of Health grant U01 HL145560 (to J.V.), and National Institutes of Health grant R01 AG064223 (to V.N.G.), National Institutes of Health grant U19 AG056278 (to J.V.), and the Glenn Foundation for Medical Research (to J.V.).
Author contributions: V.G. and J.V. conceived the study. L.Z., M.L., X.T., J.A., D.F., and S.-G.L. performed the experiment. X.D. analyzed the data. L.Z., X.D., and J.V. wrote the initial draft of the manuscript. All authors participated in discussion of the results and editing the manuscript.
Competing interests: L.Z., X.D., M.L., A.Y.M., and J.V. are cofounders of SingulOmics Corp. All other authors declare that they have no competing interests.
Data and materials availability: All raw sequencing data of rodents are submitted to Sequence Read Archive (SRA) databases under ID PRJNA753541. The raw sequencing data of two human individuals have been submitted to the database of Genotypes and Phenotypes (dbGaP) under ID phs2610.v1. p1.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Legends for tables S1 to S4
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4
Supporting files 1 and 2
REFERENCES AND NOTES
- 1.Lopez-Otin C., Blasco M. A., Partridge L., Serrano M., Kroemer G., The hallmarks of aging. Cell 153, 1194–1217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., Vijg J., Somatic mutagenesis in mammals and its implications for human disease and aging. Annu. Rev. Genet. 52, 397–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hart R. W., Setlow R. B., Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc. Natl. Acad. Sci. U.S.A. 71, 2169–2173 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanawalt P. C., Revisiting the rodent repairadox. Environ. Mol. Mutagen. 38, 89–96 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Tian X., Firsanov D., Zhang Z., Cheng Y., Luo L., Tombline G., Tan R., Simon M., Henderson S., Steffan J., Goldfarb A., Tam J., Zheng K., Cornwell A., Johnson A., Yang J.-N., Mao Z., Manta B., Dang W., Zhang Z., Vijg J., Wolfe A., Moody K., Kennedy B. K., Bohmann D., Gladyshev V. N., Seluanov A., Gorbunova V., SIRT6 is responsible for more efficient DNA double-strand break repair in long-lived species. Cell 177, 622–638.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong X., Zhang L., Milholland B., Lee M., Maslov A. Y., Wang T., Vijg J., Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat. Methods 14, 491–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lodato M. A., Rodin R. E., Bohrson C. L., Coulter M. E., Barton A. R., Kwon M., Sherman M. A., Vitzthum C. M., Luquette L. J., Yandava C. N., Yang P., Chittenden T. W., Hatem N. E., Ryu S. C., Woodworth M. B., Park P. J., Walsh C. A., Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Povirk L. F., Bennett R. A., Wang P., Swerdlow P. S., Austin M. J., Single base-pair deletions induced by bleomycin at potential double-strand cleavage sites in the aprt gene of stationary phase Chinese hamster ovary D422 cells. J. Mol. Biol. 243, 216–226 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Gorbunova V., Bozzella M. J., Seluanov A., Rodents for comparative aging studies: From mice to beavers. Age (Dordr.) 30, 111–119 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbunova V., Seluanov A., Zhang Z., Gladyshev V. N., Vijg J., Comparative genetics of longevity and cancer: Insights from long-lived rodents. Nat. Rev. Genet. 15, 531–540 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong X., Milholland B., Vijg J., Evidence for a limit to human lifespan. Nature 538, 257–259 (2016). [DOI] [PubMed] [Google Scholar]
- 12.de Magalhães J. P., Costa J., A database of vertebrate longevity records and their relation to other life-history traits. J. Evol. Biol. 22, 1770–1774 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Lee B. P., Smith M., Buffenstein R., Harries L. W., Negligible senescence in naked mole rats may be a consequence of well-maintained splicing regulation. GeroScience 42, 633–651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seluanov A., Vaidya A., Gorbunova V., Establishing primary adult fibroblast cultures from rodents. J. Vis. Exp. 2010, 2033 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milholland B., Dong X., Zhang L., Hao X., Suh Y., Vijg J., Differences between germline and somatic mutation rates in humans and mice. Nat. Commun. 8, 15183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov L. B., Kim J., Haradhvala N. J., Huang M. N., Ng A. W. T., Wu Y., Boot A., Covington K. R., Gordenin D. A., Bergstrom E. N., Islam S. M. A., Lopez-Bigas N., Klimczak L. J., McPherson J. R., Morganella S., Sabarinathan R., Wheeler D. A., Mustonen V.; PCAWG Mutational Signatures Working Group, Gretz G., Rozen S. G., Stratton M. R.; PCAWG Consortium , The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L., Dong X., Lee M., Maslov A. Y., Wang T., Vijg J., Single-cell whole-genome sequencing reveals the functional landscape of somatic mutations in B lymphocytes across the human lifespan. Proc. Natl. Acad. Sci. U.S.A. 116, 9014–9019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G., Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539–539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Procter J. B., Carstairs G. M., Soares B., Mourão K., Ofoegbu T. C., Barton D., Lui L., Menard A., Sherstnev N., Roldan-Martinez D., Duce S., Martin D. M. A., Barton G. J., Alignment of biological sequences with Jalview. Methods Mol. Biol. 2231, 203–224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Povirk L. F., Wübter W., Köhnlein W., Hutchinson F., DNA double-strand breaks and alkali-labile bonds produced by bleomycin. Nucleic Acids Res. 4, 3573–3580 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Ghorai M. K., Kenney G., Stubbe J., Mechanistic studies on bleomycin-mediated DNA damage: Multiple binding modes can result in double-stranded DNA cleavage. Nucleic Acids Res. 36, 3781–3790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodgers K., McVey M., Error-prone repair of DNA double-strand breaks. J. Cell. Physiol. 231, 15–24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britten R. J., Rates of DNA sequence evolution differ between taxonomic groups. Science 231, 1393–1398 (1986). [DOI] [PubMed] [Google Scholar]
- 24.Thomas G. W., Hahn M. W., The human mutation rate is increasing, even as it slows. Mol. Biol. Evol. 31, 253–257 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Goodman M., Rates of molecular evolution: The hominoid slowdown. Bioessays 3, 9–14 (1985). [DOI] [PubMed] [Google Scholar]
- 26.Cutler R. G., Evolution of longevity in primates. J. Hum. Evol. 5, 169–202 (1976). [Google Scholar]
- 27.Cawthon R. M., Meeks H. D., Sasani T. A., Smith K. R., Kerber R. A., O’Brien E., Baird L., Dixon M. M., Peiffer A. P., Leppert M. F., Quinlan A. R., Jorde L. B., Germline mutation rates in young adults predict longevity and reproductive lifespan. Sci. Rep. 10, 10001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dollé M. E., Snyder W. K., Gossen J. A., Lohman P. H., Vijg J., Distinct spectra of somatic mutations accumulated with age in mouse heart and small intestine. Proc. Natl. Acad. Sci. U.S.A. 97, 8403–8408 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bae T., Tomasini L., Mariani J., Zhou B., Roychowdhury T., Franjic D., Pletikos M., Pattni R., Chen B.-J., Venturini E., Riley-Gillis B., Sestan N., Urban A. E., Abyzov A., Vaccarino F. M., Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science 359, 550–555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franco I., Johansson A., Olsson K., Vrtačnik P., Lundin P., Helgadottir H. T., Larsson M., Revêchon G., Bosia C., Pagnani A., Provero P., Gustafsson T., Fischer H., Eriksson M., Somatic mutagenesis in satellite cells associates with human skeletal muscle aging. Nat. Commun. 9, 800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blokzijl F., de Ligt J., Jager M., Sasselli V., Roerink S., Sasaki N., Huch M., Boymans S., Kuijk E., Prins P., Nijman I. J., Martincorena I., Mokry M., Wiegerinck C. L., Middendorp S., Sato T., Schwank G., Nieuwenhuis E. E. S., Verstegen M. M. A., van der Laan L. J. W., de Jonge J., IJzermans J. N. M., Vries R. G., van de Wetering M., Stratton M. R., Clevers H., Cuppen E., van Boxtel R., Tissue-specific mutation accumulation in human adult stem cells during life. Nature 538, 260–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vijg J., Dong X., Pathogenic mechanisms of somatic mutation and genome mosaicism in aging. Cell 182, 12–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persi E., Wolf Y. I., Horn D., Ruppin E., Demichelis F., Gatenby R. A., Gillies R. J., Koonin E. V., Mutation-selection balance and compensatory mechanisms in tumour evolution. Nat. Rev. Genet. 22, 251–262 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Ma S., Upneja A., Galecki A., Tsai Y. M., Burant C. F., Raskind S., Zhang Q., Zhang Z. D., Seluanov A., Gorbunova V., Clish C. B., Miller R. A., Gladyshev V. N., Cell culture-based profiling across mammals reveals DNA repair and metabolism as determinants of species longevity. eLife 5, e19130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacRae S. L., Croken M. M. K., Calder R. B., Aliper A., Milholland B., White R. R., Zhavoronkov A., Gladyshev V. N., Seluanov A., Gorbunova V., Zhang Z. D., Vijg J., DNA repair in species with extreme lifespan differences. Aging 7, 1171–1182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M. A., The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Pinheiro, D. Bates, S. DebRoy, D. Sarkar, R core team, nlme: Linear and nonlinear mixed effects models. R package version 3.1-152 (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Legends for tables S1 to S4
Tables S1 to S4
Supporting files 1 and 2