Old genetic variants were key to the ability of bottlenose dolphins to repeatedly adapt to coastal waters across the world.
Abstract
Studying repeated adaptation can provide insights into the mechanisms allowing species to adapt to novel environments. Here, we investigate repeated evolution driven by habitat specialization in the common bottlenose dolphin. Parapatric pelagic and coastal ecotypes of common bottlenose dolphins have repeatedly formed across the oceans. Analyzing whole genomes of 57 individuals, we find that ecotype evolution involved a complex reticulated evolutionary history. We find parallel linked selection acted upon ancient alleles in geographically distant coastal populations, which were present as standing genetic variation in the pelagic populations. Candidate loci evolving under parallel linked selection were found in ancient tracts, suggesting recurrent bouts of selection through time. Therefore, despite the constraints of small effective population size and long generation time on the efficacy of selection, repeated adaptation in long-lived social species can be driven by a combination of ecological opportunities and selection acting on ancestral standing genetic variation.
INTRODUCTION
Understanding the processes that allow species to extend their ranges and adapt to novel environments is a long-standing question in biology, in which interest now extends well beyond this disciplinary field because of the potential effect of global change on species ranges. The colonization of novel environments may result in new selective pressures on individuals and promote local adaptation (1). However, linking genetic divergence to local adaptation is particularly challenging as genetic differentiation may also arise because of demographic history (2) and other selective processes such as background selection (3). Replicate adaptation of different populations to similar environments is often considered strong evidence of the repeated action of natural selection (4). Hence, we can study repeated evolution to gain insights into the mechanisms driving genetic variation and adaptation.
Iconic examples of repeated evolution include adaptation to similar environments—i.e., parallel adaptation to freshwater environments from marine habitats in threespine sticklebacks, Gasterosteus aculeatus (5); adaptation to the same host species in stick insects, Timema cristinae (6); high-altitude adaptation in multiple human, Homo sapiens, populations (7); and different light conditions in cichlid fish (8)—or similar responses to comparable stressors [e.g., myxoma virus (9)]. Our understanding of the mechanisms involved in repeated phenotypic evolution has recently shifted from a binary view of identical versus idiosyncratic processes to a continuum ranging from parallel (i.e., selection on the same variants), to convergent (selection on different variants in the same genes or in pathways with similar functions), to nonparallel (i.e., selection specific to one population) (6, 10–14).
Adaptation to novel habitats may occur rapidly if the genetic substrate which selection acts upon was already segregating in the ancestral population [i.e., standing genetic variation (SGV) (5, 6, 9, 10, 15, 16)], as balanced polymorphisms (17), or introgressed from a locally adapted outgroup (18). In the latter scenario, gene flow may be an important driver of recurrent adaptation, implying that independent evolution is not necessarily a prerequisite (18). Alleles present as SGV may have been selected in past environments, potentially increasing their chances to be the recurrent target of natural selection (15). Recent studies have highlighted that the origin of the alleles that enable populations to recurrently adapt to similar environments may be much older than the divergence of the populations themselves (16, 19, 20). For example, the reservoir of freshwater-adaptive alleles in marine populations of threespine sticklebacks, which have been recurrently selected after freshwater colonization during the past 12,000 years and presumably during previous interglacials, has been segregating for millions of years (20).
With the rare exception of humans (7, 21), reported cases of parallel and convergent evolution (repeated adaptation) among populations almost exclusively involve relatively short-lived species (5, 6, 8, 9, 16). In long-lived species, such as large mammals, long generation time, low fecundity, and small effective population size may reduce the efficacy of selection (15, 22). In humans, cultural innovations such as animal domestication and the colonization of new habitats, such as high altitude, likely exposed extant neutral genetic variation to novel selective regimes (7, 21). In other long–life span social mammals, the colonization of new ecological niches followed by stable transmission of learned behaviors, such as foraging strategies or habitat preferences, may also create opportunities for natural selection to act upon locally adaptive genetic variation, although examples are scarce [but see killer whales, Orcinus orca (23)].
Here, we tested for repeated evolution driven by ecological niche specialization in a highly social marine mammal, the common bottlenose dolphin, Tursiops truncatus, which has a worldwide temperate and tropical distribution. Two ecotypes of common bottlenose dolphins (“pelagic” or “offshore” and “coastal”) have repeatedly formed in multiple regions of the world (24–28). Coastal populations were suggested to have been founded from pelagic source populations (24–26, 29). They are thus an excellent study system to test whether repeated evolution occurred and involved the same molecular processes during the repeated colonization of coastal habitats.
Throughout their range, coastal populations have different diets compared with pelagic populations (30, 31) and can display phenotypic traits adapted to coastal waters, in particular, for feeding (31–33). Coastal populations in distinct regions of the world can share some morphological traits such as larger teeth, rostra, and internal nares when compared to pelagic populations. They can also show some population-specific traits, such as body size in western North Atlantic (WNA) coastal bottlenose dolphins, which are smaller than their pelagic counterparts, while in other regions, the pattern is reversed or there are no discernable differences (30–33). There is no overall convergence in the morphology of the species in coastal habitats across its range. However, coastal populations tend to share more cryptic behavioral phenotypes, such as strong site fidelity and reduced dispersal (26, 28, 34), and stable foraging ecology, which can be socially transmitted from mother to calves and from conspecifics in the same social group (35, 36). The colonization of coastal habitats may have created opportunities for local adaptation to arise, and social behavior and learning abilities may have facilitated the transmission of advantageous learned behaviors, such as habitat-specific foraging techniques.
In this study, we tested the hypothesis that SGV was repeatedly targeted by selection when the common bottlenose dolphins colonized the coastal environment. We first identified population structure and demographic history. We showed that the pelagic and coastal ecotype pairs have a complex non–tree-like history. Then, we assessed the evidence of parallel selection to the coastal habitat across the genome and identified candidate genes under parallel linked selection, potentially involved in cognitive abilities and feeding.
RESULTS AND DISCUSSION
Genetic structure
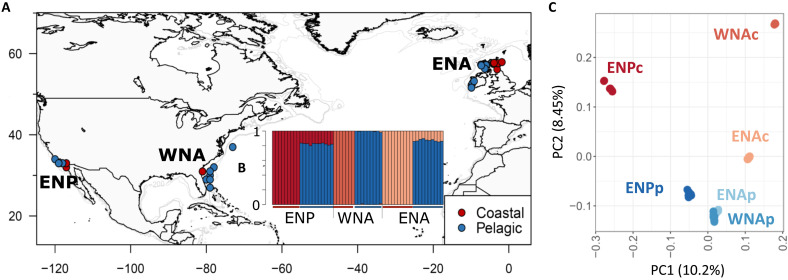
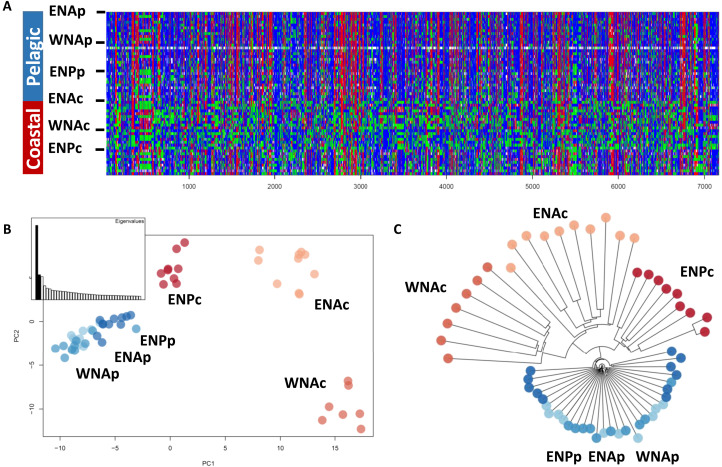
We analyzed the genomic variation at single-nucleotide polymorphisms (SNPs) by sequencing the whole genomes of 57 common bottlenose dolphins (Fig. 1A) at a mean sequencing depth (± SD) of 10.56X (± 2.18), after read quality and variant filtering (see table S1 and Materials and Methods). Our sampling includes 10 pelagic and 10 coastal individuals from the eastern North Atlantic (ENAp and ENAc), 10 pelagic and 7 coastal individuals from the western North Atlantic (WNAp and WNAc), and 11 pelagic and 9 coastal individuals from California, eastern North Pacific (ENPp and ENPc). Whenever possible, we used genotype likelihoods in the analyses to account for the uncertainty inherently present in low-depth sequencing data (37, 38) or used methods based on allele frequencies (table S2).
Fig. 1. Sampling location and population structure of coastal and pelagic common bottlenose dolphins.
(A) Map of sample locations of the common bottlenose dolphin ecotypes, in the eastern North Atlantic (ENA), western North Atlantic (WNA), and eastern North Pacific (ENP). (B) Ancestry proportions for each of the 57 individuals inferred in NGSAdmix (39) for a number of clusters, K = 4, identified as the highest level of structure using the Evanno method (75). (C) PCA showing the first and second PCs. The proportion of genetic variance captured by each component is indicated between parentheses.
The genetic structure obtained from a principal components analysis (PCA) (38) and the individual-based ancestry and clustering analysis of NGSAdmix (39) based on a set of 798,572 unlinked high-quality SNPs indicated that the samples assigned a priori to a population clustered together (Fig. 1, B and C, and figs. S2 and S3). The analyses showed two major axes of differentiation: Atlantic versus Pacific (PC1) and pelagic versus coastal (PC2). Coastal populations drove this pattern in both cases: ENPc along PC1 and WNAc along PC2 (Fig. 1C). The ENAc population was intermediate between the WNAc and Atlantic pelagic populations. Genetic differentiation among the three pelagic populations was less than between any pelagic and coastal populations, even those in parapatry (Fig. 1, B and C, figs. S2 and S3, and pairwise FST in table S3). These patterns of differentiation suggest that each coastal population has a history that includes both population-specific drift and drift shared with the other coastal populations and/or differences in gene flow with parapatric pelagic populations.
Evolutionary relationships among ecotypes
To test whether our three geographic pairs of coastal and pelagic ecotypes represent independent divergence events, we reconstructed their population histories using approaches that draw inferences from covariance in allele frequencies among populations. We first explored the evolutionary relationships among populations and potential admixture events using TreeMix (40), using the Indo-Pacific bottlenose dolphin, Tursiops aduncus, as an outgroup to root the tree. The TreeMix results may be difficult to interpret as our samples cover only a part of the geographical distribution of the species, implying that there are ghost populations in the tree inference. Thus, caution should be taken against any strong interpretation on branching orders and migration edges.
All the internal branch lengths were relatively short in the TreeMix analyses, consistent with populations rapidly radiating from a shared ancestral population. Branch lengths support marginal drift in the pelagic populations, in contrast to stronger drift in allele frequencies in the coastal populations, from a shared common ancestral gene pool (Fig. 2A, figs. S4 and S5, and Supplementary Text). Given this, the allele frequencies in present-day pelagic populations will be closer to the common shared ancestral population than to any of the present-day coastal populations. The best supported population tree had two migration edges, supporting the existence of conflicting genealogies across the genome (Fig. 2A). There was no clear single bifurcation between pelagic and coastal ecotypes or between Pacific and Atlantic populations. There were also no clear independent pelagic and coastal bifurcations associated with each geographic region.
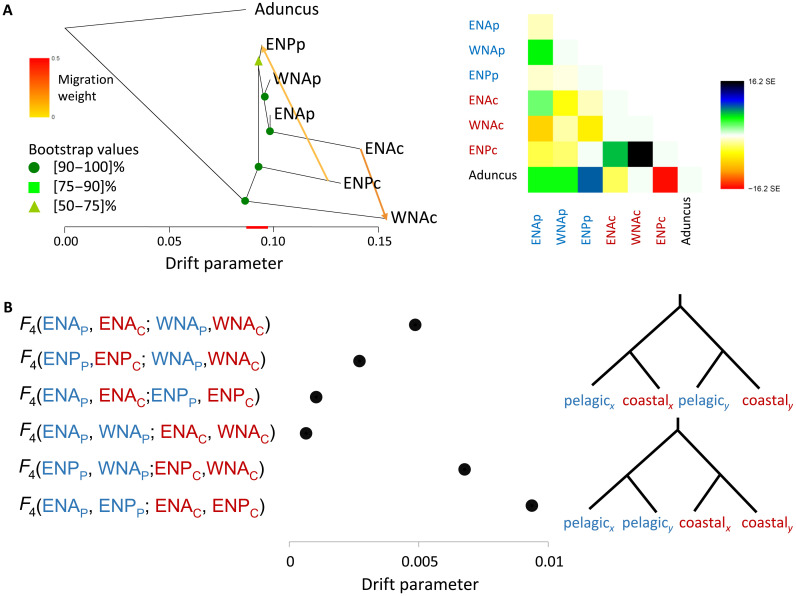
Fig. 2. Admixture among populations of common bottlenose dolphins.
(A) Left: TreeMix consensus tree and bootstrap values displaying the relationships among populations as a bifurcating maximum-likelihood tree with two migration edges (M = 2), inferred as the best topology. Branch lengths on the horizontal axis represent the amount of genetic drift that has occurred along each branch. Bootstrap supports for each of the nodes are indicated. Right: Residual fit of the observed versus the predicted squared allele frequency difference, expressed as the number of SE of the deviation. SE values are represented by colors according to the palette on the right. Residuals above zero indicate populations that are more closely related to each other in the data than in the best-fit tree and have potentially undergone admixture. Negative residuals represent populations that are less closely related in the data than represented in the best-fit tree. (B) Patterns of allele sharing expressed using the F4 statistics of the form F4(pelagicx, coastalx; pelagicy, coastaly) or F4(pelagicx, pelagicy; coastalx, coastaly). All SE estimations are less than 1 × 10−4, and all F4 statistics were significant on the basis of z scores greater than 3, which is the equivalent of a significance of P < 0.0026. The red line on the x axis of the TreeMix graph in (A) represents the extent of variation in the shared drift parameter represented on the x axis in the F4 statistics plot in (B) and illustrates that drift in coastal populations is largely population specific. The placement of the red line on the x axis is arbitrary and not indicative of where on the tree shared drift is inferred to have occurred.
We further tested whether geographic pairs of pelagic and coastal ecotypes had evolved independently by exploring patterns of correlated changes in allele frequencies between two pairs of populations, estimated using F4 statistics (41, 42) of the form (pelagicx, coastalx; pelagicy, coastaly), where x and y represent different geographic regions. We also calculated F4 statistics comparing drift between the same ecotypes from different geographic regions, F4(pelagicx, pelagicy; coastalx, coastaly), therefore testing for a single origin of each ecotype. Tests of both forms were all significantly positive (Fig. 2B and Supplementary Text), indicating that none of the topologies were perfect representations of the relationships among the four populations.
Statistics of the form F4(XNAp,ENPp;XNAc,ENPc), with X representing east or west, had the highest values, which were much higher than those of F4(XNAp,XNAc;ENPp,ENPc), which were close to 0, indicating relatively independent evolution of the ecotype pairs in the Atlantic and the Pacific. In contrast, F4(ENAp,WNAp;ENAc,WNAc) was the lowest of all and lower than F4(ENAp,ENAc;WNAp,WNAc), suggesting that the two Atlantic coastal populations may be derived from the same ancestral pelagic population and/or experienced gene flow. Thus, TreeMix and F4 statistics both support complex non–tree-like relationships. However, our TreeMix, NGSAdmix, and PCA results indicate that each coastal population has experienced independent histories to such an extent that allele frequencies are clearly differentiated among all coastal populations. Ultimately, a more comprehensive geographical sampling together with the temporal resolution offered by haplotype-based inferences may be needed to fully resolve these relationships (43).
Demographic history
We then estimated historical variation in effective population size (Ne) for each population, which reflects changes in genetic diversity because of variation in population size, population structure, gene flow (44), and linked selection (i.e., background selection or selective sweeps) (22, 45). For that purpose, we used the coalescent-based approach SMC++ (46) to carry out two analyses, one with the putatively neutral regions as identified by Flink [see details below (47)] and another with all regions. As results were very similar, we present here the results with the putatively neutral regions (Fig. 3A and fig. S6); those with all regions can be found in the Supplementary Materials (fig. S7, A and B). We found that pelagic populations experienced demographic expansions, ~150,000 to 120,000 years before present (yBP), followed by a period of more stable Ne than the coastal populations (Fig. 3A and figs. S6 and S7, A and B). Population expansion in all populations during the first part of the last glacial period, which started ~115,000 yBP, may reflect changes in connectivity, rather than an increase in Ne, as suitable habitat became scarce (29).
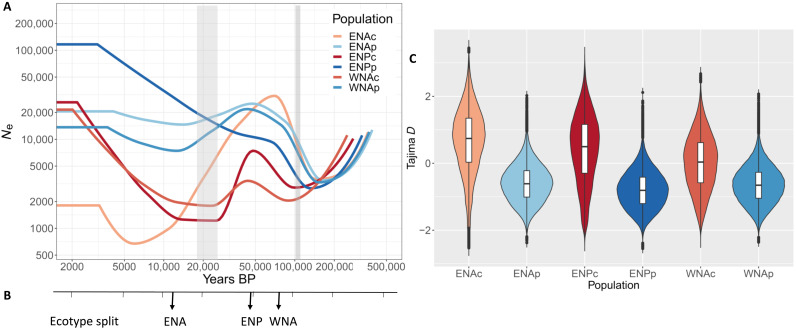
Fig. 3. Demographic history of common bottlenose dolphin populations.
(A) Changes in effective population size through time inferred for each common bottlenose dolphin population using SMC++, using a mutation rate of 2.56 × 10−8 substitution per nucleotide per generation (83) and a generation time of 21.1 years (81). The timing of the onset of the last glacial period (110,000 yBP) and the Last Glacial Maximum (26,500 to 19,000 yBP) are indicated in gray shading. (B) Split time between ecotypes within each region estimated using SMC++. Populations are eastern North Atlantic coastal (ENAc), eastern North Atlantic pelagic (ENAp), eastern North Pacific coastal (ENPc), eastern North Pacific pelagic (ENPp), western North Atlantic coastal (WNAc), and western North Atlantic pelagic (WNAp). (C) Tajima’s D estimated for each population; the violin plots indicate the kernel probability density of the data, the box indicates the interquartile range, and the horizontal marker indicates the median of the data.
Coastal populations experienced more erratic fluctuations in Ne than the pelagic populations. Ne in the ENA coastal population closely followed the same trajectory as the ENA pelagic population up to 50,000 yBP, suggesting that they were a single ancestral population up to this point. In contrast, the WNA and ENP coastal populations displayed different Ne changes than their pelagic counterparts from ~150,000 and ~115,000 yBP, respectively. We observe a long period of low Ne in the WNA and ENP coastal populations from 25,000 to 12,000 yBP. The ENA coastal population showed a steady decrease from 70,000 to 7000 yBP. This suggests that during the Last Glacial Maximum (LGM), coastal populations had low Ne or were fragmented. Thus, all coastal populations went through an inferred reduction in Ne followed by a postglacial expansion. However, other mechanisms, which are not accounted for in SMC++, could be confounding factors in these demographic inferences; similar Ne trajectories can result from changes in gene flow and population structure (44) and/or linked selection (22, 45).
Reduced nucleotide diversity, Watterson’s theta, and consequently positive Tajima’s D estimates—in particular, for the ENAc and ENPc populations (Fig. 3C and figs. S8, A and B, and S9)—and large amount of drift (Fig. 2A and fig. S5) may also indicate that the coastal populations have experienced reductions in Ne and suggest that they are derived from larger ancestral populations. Again, other processes may influence those estimates such as gene flow, which can lead to positive Tajima’s D estimates (48). Nevertheless, access to novel, previously ice-covered shallow coastal habitats during past climate change such as at the end of the LGM in the ENA (29, 30) or during warm interstadials in the WNA and ENP has likely created opportunity for ecological differentiation. Coastal habitats provide a mosaic of environments and different and potentially more stable food resources (30, 49).
We estimated divergence time between the two ecotypes within each region using SMC++ on putatively neutral regions. The oldest divergence between pelagic and coastal ecotypes occurred in the WNA (around 80,000 yBP), and the youngest was around 12,000 yBP, during a postglacial divergence in the ENA (Fig. 3B and fig. S10, A and B). We acknowledge that these estimations do not consider gene flow, which may have occurred between ecotypes since their divergences and possibly not at the same rate in all three regions.
Overall, our analyses reveal a complex, reticulated evolutionary history of common bottlenose dolphins, with the pelagic populations being genetically similar to the common ancestral population. In contrast, each coastal population has experienced strong population-specific drift. Local PCA (50), a method that describes heterogeneity in patterns of relatedness among populations (fig. S12 and Supplementary Text), indicated that the dolphin genomes were composed of regions with different evolutionary histories. This analysis supports both the demographic histories suggested by admixture plot and PCA results and those inferred by the F4 statistics and TreeMix where the ENA and WNA coastal populations were more closely related than expected under entirely independent ecotype splits on each side of the Atlantic (Fig. 2, A and B, and fig. S5). Furthermore, on PC3 and PC4, coastal populations from the Atlantic and Pacific clustered together and likewise for the pelagic populations, potentially suggesting parallel ecotype-based processes (fig. S12).
Mechanisms of repeated evolution to coastal habitat
To test whether the above results can indicate repeated selective sweeps associated with coastal habitat, we used Flink (47), an extension of BayeScan (51) that takes linkage among loci into account. It uses a hierarchical island model, and we considered ecotype (coastal versus pelagic) as the top hierarchical level. Although details about the evolutionary history of the species need to be further studied, our results support this hierarchical structure. In particular, NGSAdmix results with K = 2 to 4 group all pelagic populations in the same genetic cluster (Fig. 1 and fig. S2C). Furthermore, TreeMix and F4 statistics results group the Atlantic populations by ecotype (Fig. 2, A and B, and fig. S5). Our analyses show notable differences in patterns of inferred selection involving mainly divergent selection between coastal and pelagic ecotypes (higher hierarchy) and both divergent selection and selection homogenizing allele frequencies (less genetic differentiation than expected under neutrality) among coastal populations (Fig. 4, A and B, Supplementary Text, and figs. S13A and S14A). For the sake of brevity, in what follows, we describe the latter as “homogenizing selection” [but note that it is referred to as “balancing selection” in Flink (47) and BayeScan (51)].
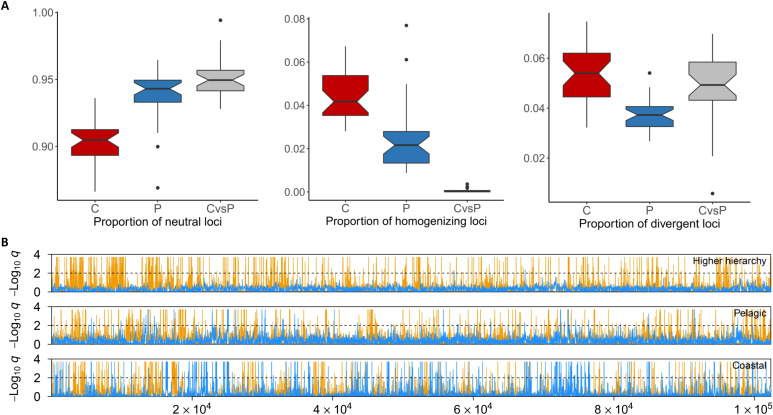
Fig. 4. Patterns of selection within and between common bottlenose dolphin ecotypes.
(A) Boxplots of the genomic patterns of selection that are the proportion of neutral, homogenizing, and divergent loci within coastal (C) and pelagic (P) ecotypes and between the two ecotypes (CvsP). (B) Patterns of selection (divergent: yellow, homogenizing: blue) inferred using Flink from one super-scaffold for the different hierarchical groupings that are between coastal and pelagic populations (top), among pelagic populations (middle), and among coastal populations (bottom). The y axis indicates the locus-specific FDR for divergent (orange) and homogenizing (blue) selection. The black dashed line shows the 1% FDR threshold, above which we consider a locus under selection.
Divergent selection patterns may be inflated by false positives associated with drift in coastal populations. We therefore conservatively consider only the 7165 SNPs inferred to be evolving under both homogenizing selection among coastal populations and divergent selection between ecotypes, as putative loci underlying parallel evolution to geographically distant coastal habitats (Fig. 5, A to C, and Supplementary Text), and focus on those variants in the rest of our study. Considering the possible origins of the variants inferred to be underlying parallel evolution, we find that most (87%) were polymorphic, i.e., present as SGV, in the pelagic populations, and 57% were polymorphic in all three pelagic populations. This suggests that each of the coastal populations had access to much of the same SGV, which was responsible for the homogenization of allele frequencies across geographically distant coastal populations. This same SGV would also be responsible for the differentiation between coastal and pelagic populations at these genomic sites. This can be visualized in the PCA and unrooted neighbor-joining tree based on these 7165 SNPs, where the populations cluster by ecotype (Fig. 5, B and C). In these analyses, the Atlantic coastal populations are more closely related to each other than to the ENPc population, and we therefore acknowledge that some of those 7165 SNPs may be under selection within the Atlantic only, possibly due to their partially shared ancestry or more similar SGV within oceans.
Fig. 5. Patterns of genetic variation of the 7165 SNPs under parallel linked selection to coastal habitat, i.e., under both homogenizing selection among coastal populations and divergent selection between ecotypes.
These SNPs included closely linked sites scattered across the genome in 362 regions separated by at least 100 kb. (A) Plot of the homozygote reference genotypes in blue, heterozygote in green, and homozygote for the alternate allele in red. (B) PCA and (C) neighbor-joining distance tree showing the genetic structure of the common bottlenose dolphin samples for this particular SNP set.
Note that Flink (and any other genome-scan method) is more likely to detect sites linked to the targets of selection rather than the targets themselves (47). These 7165 SNPs are linked among themselves [median distance of 54 base pairs (bp)] into 362 distinct clusters, which are separated by at least 100 kb and, therefore, may represent linked selection acting upon a much smaller number of haplotypes. The genotypes at those SNPs are mainly heterozygous (Fig. 5A), and the site frequency spectrums (SFSs) of the variable sites are shifted toward intermediate allele frequencies in all three coastal populations (fig. S18), in contrast to the SFS for all the SNPs (fig. S9). These two observations are consistent with selective sweeps from SGV. Although loci directly under selection are expected to quickly become fixed for the beneficial allele, linked neutral loci are expected to have alleles at intermediate frequencies (15) and therefore a high prevalence of heterozygous genotypes, such as observed in the coastal dolphin populations (Fig. 5A).
In addition, under incomplete soft selective sweeps from SGV, we would expect both loci directly under selection and neutral loci closely linked to the selected variant to have intermediate frequencies, again as we see in the coastal populations. Incomplete sweeps are expected when effective population size is reduced and under low migration, such as in human populations (52) and the coastal bottlenose dolphin populations studied here. We therefore hypothesize that these 7165 SNPs likely include hitchhiking SNPs closely linked to the selected variants. However, we cannot exclude that some of them may be under balancing selection, i.e., due to heterozygote advantage/heterosis or frequency-dependent selection. We hereafter refer to these 7165 SNPs as evolving under parallel linked selection across geographically distant coastal populations, as they may include both the targets of selection and sites in tight linkage to them.
Evidence of ancient origins of selected variants
Given our finding of parallel linked selection in coastal populations separated across spatial scales, we hypothesize that this process may have occurred recurrently during coastal habitat colonization in previous interglacial periods. Under this proposed scenario, the present-day coastal populations of bottlenose dolphins would be only the latest of a series of postglacial colonizers to make use of these alleles. While we do not have genomic data from previous interglacials with which to investigate this hypothesis, we can make a prediction that SGV that has been subject to recurrent bouts of selection across multiple interglacials would be found in tracts of older ancestry. By this, we mean that the estimates of the time to the most recent common ancestor (TMRCA) of coastal and pelagic populations would be older for tracts containing SNPs under parallel linked selection than the genome-wide average.
To identify such tracts in coastal dolphin genomes, we searched for clusters of dense private mutations (53) segregating in each coastal population relative to parapatric pelagic populations (see Materials and Methods, Supplementary Text, and fig. S11), taking variation in mutation rate along the genome into account. Regions of high densities of mutations private to each coastal population relative to the parapatric pelagic populations are indicative of an older TMRCA, and we hereafter refer to such tracts as “ancient.” We found ancient tracts in all three coastal populations (tables S4 and S5 and Supplementary Text), with the length of all those tracts being between 10 and 25 Mb (table S4). The inferred TMRCA of these ancient tracts (0.6 to 2.3 million years) was much older than those of the rest of the genome (0.1 to 0.4 million years) (table S5). The divergence dates of T. aduncus and T. truncatus estimated by Moura et al. (54) and McGowen et al. (55) are close to the TMRCA of the ancient tracts found in the WNA coastal individuals (1.0 to 2.6 million years; table S5), after correcting for the different mutation rates used between studies.
We found that a large proportion (66%) of the 7165 candidate SNPs under parallel linked selection in coastal populations were found in these ancient tracts. In contrast, only an average of 22% (range, 21.2 to 22.7; SD, 0.32) of 100 random samples of the same number of putatively neutral SNPs were found in the ancient tracts (fig. S19). The ancient tracts containing coastal-associated alleles could have been introgressed from an unsampled “ghost” population (56, 57), which diverged from the sampled populations a long time ago, so that the introgressed regions contain mutations, which accumulated in the ghost population over time, likely close to the split time between T. truncatus and T. aduncus. The spread of these ancient alleles may have also occurred by gene flow between coastal populations. However, we do not have further support for these two hypotheses, and it is difficult to explain how gene flow could have happened between coastal populations in the Pacific and the Atlantic. Alternatively, coastal populations could have independently diverged from the same or closely related ancestral pelagic populations, making repeated adaptation through shared initial ancient SGV a possible alternative explanation for our results. This hypothesis may hold particularly for the Atlantic populations, given their partially shared ancestry, and could explain the stronger patterns of parallel linked selection within the Atlantic (Fig. 5, B and C).
Another parsimonious hypothesis, considering the relatively independent demography of ecotype pairs in the Atlantic and Pacific, and given the prevalence of these SNPs as SGV in the pelagic populations, is that coastal adaptation occurred in different oceans by repeated selection through space and time on ancient SGV, which persisted at low frequencies in the large pelagic populations. There are precedents for such recurrent use of old SGV in nature; ancient polymorphisms have enabled rapid parallel ecotype formation in saltmarsh beetles (16) and in threespine sticklebacks (20, 58). In sticklebacks, freshwater-adapted alleles have persisted as SGV in the large marine populations as a result of episodic recurrent gene flow from freshwater populations (the so-called transporter hypothesis) (20, 59).
A similar mechanism could apply to our study system; most of the SNPs inferred to be evolving under parallel linked selection in coastal populations are located in ancient tracts. The age of these tracts (0.6 to 2.3 million years) predates the start of the last glacial period (115,000 to 11,700 yBP) and of many other previous Quaternary glacial periods (table S5). We therefore speculate that ancient tracts containing variants evolving under parallel linked selection may have contributed to the recurrent colonization of newly available coastal habitats by bottlenose dolphins during past interglacial periods. In addition, we propose that migration back into the pelagic populations potentially retained these ancient tracts as standing variation at low or intermediate frequency within the pelagic populations. We see this akin to the “sieving” of balanced polymorphism during the speciation process proposed by Guerrero and Hahn (17). Together, our results contribute toward the emerging hypothesis that old polymorphisms may allow rapid ecotype formation when new ecological opportunities arise and, ultimately, ecological speciation (19).
Patterns of selection and ecology and behavior
Although the exact evolutionary scenario involved in repeated evolution still requires further investigation, likely involving extensive sampling across the range of the species, our results together with previous studies on human populations represent rare examples of species with long generation time for which repeated evolution from SGV has been uncovered (7, 21). In humans, similar stable lifestyles [e.g., life in high altitudes (7)] or same cultural revolutions [e.g., cattle domestication (21)] likely created opportunities for cryptic phenotypes such as resistance to hypoxia or lactase persistence to become beneficial and for convergent phenotypic adaptation to occur. Nonhuman examples of socially driven local adaptation are scarce, but killer whale ecotypes have likely evolved as a result of demographic history, ecological opportunity, and gene-culture interactions (23). Coastal bottlenose dolphins (Tursiops sp.) also exhibit complex behaviors, such as stable habitat specializations or social learning of foraging techniques, that strongly influence their patterns of genetic variability (27, 30, 36), and we hypothesize that these also facilitated their ability to adapt to novel conditions.
Although further investigation is warranted as many complex traits may be polygenic (60) and it is difficult to prove causal relations between behavioral/ecological traits and genes under selection, the 7165 SNPs inferred to have evolved under parallel linked and recurrent selection overlapped with 45 genes. Those include genes related to behavioral and ecologically relevant functions, and thus cryptic phenotypic variations (Supplementary Text and table S7). We found genes related to cognitive abilities, learning, and memory [RELN (61, 62) and ADARB2 (63)]. RELN encodes for the reelin protein, which has a role in the modulation of synaptic transmission in response to experience (61, 62). Coastal bottlenose dolphins (Tursiops sp.) develop habitat-specific foraging techniques, which are transmitted maternally or in social groups (35, 36) and may require genetic adaptations for increased cognitive abilities. RELN has been found under positive selection in sea otters, which also exhibit maternally transmitted foraging behavior (64). Other ecologically relevant genes include those involved in lipid metabolism and storage [AGK, LPIN2, and KLB (65)], which may be involved in adaptation to the differing diets documented in coastal populations, mainly involving large fish, and pelagic populations, which primarily eat pelagic fish and squid (30, 31).
We acknowledge that there may be other processes contributing to convergent ecotype adaptation, driven by selection on SGV or new mutations in other regions of the genome, and which are not shared by different ecotype pairs but that were not detected in our analysis, focusing on testing for parallel patterns of evolution. In addition, we observed that 113,530 SNPs were under divergent selection among coastal populations, and although these may include false positives due to extensive drift in coastal populations, this suggests divergent selection linked to heterogeneous habitat. This is not unexpected given the environmental variations of coastal habitats across the bottlenose dolphins’ range. Our findings corroborate other studies highlighting that other processes such as environmental heterogeneity may contribute to genotypic and phenotypic variation within ecotypes (11–13). This holds even for the most emblematic example of parallel evolution, the threespine sticklebacks, where deviation from parallel adaptation may be the result of geographic distance, stochastic processes, and adaptation to environmental variation within habitat types (12, 13).
To conclude, we find that selection acting upon ancient SGV fueled repeated adaptation of common bottlenose dolphins to coastal environments. Recurrent bouts of selection on genetic variation may promote adaptation to coastal habitat via reusing linked variants with minimal pleiotropic effects, thereby facilitating their persistence at low frequency in source populations and enabling repeated evolution of derived populations at the range margins (66). Our study contributes to the growing body of evidence that ancient polymorphisms are a major substrate for rapid ecological adaptive divergence (19) and can have a key role in local adaptation of long-lived organisms. We propose that such variation has been the source of past adaptation during the glacial cycles and may prove to be critical for species to cope with the current rapid directional environmental changes driven by current global climatic change.
MATERIALS AND METHODS
Ethics statement
Samples were collected under permits MMPA Permit 779-1633 and MMPA Permit 779-1339 for the WNA and NMFS 14097 for the ENP. They were shipped from the Southwest Fisheries Science Center, NOAA Fisheries, USA, to the University of St. Andrews, UK, under CITES institutional permits US057 and GB035, and the U.S. Fish and Wildlife Service permit 16US690343/9 and to BGI in Hong Kong, China, under CITES export permit 547016/01.
Sample collection
Epidermal tissue samples were collected from 57 bottlenose dolphins (Fig. 1A, table S1, and Supplementary Text).
Laboratory procedures
DNA extraction protocol procedures are detailed in the Supplementary Text. Library and whole-genome resequencing was performed at the Beijing Genomics Institute (BGI). Illumina libraries were built on 300-bp DNA fragments and sequenced on an Illumina HiSeq X Ten platform (Supplementary Text).
Data processing and filtering
The read trimming and mapping and data filtering are described in detail in the Supplementary Text. Sequencing reads were processed with Trimmomatic v.0.32 (67) using default parameters, and sequence reads shorter than 75 bp were discarded. The remaining filtered reads were first mapped to a modified version of a published common bottlenose dolphin mitochondrial genome (GenBank: KF570351.1) (68). Reads that did not map to the mitochondrial genome were then mapped to the reference common bottlenose dolphin genome assembly (GenBank: GCA_001922835.1, NIST Tur_tru v1) using BWA mem (v.0.7.15) with default options (69).
Picard-tools v.2.1.0 (70) was used to add read groups, merge the bam files from each individual from the different lanes, and remove duplicate reads. Then, indel realignment was performed using GATK v.3.6.0 (71). We kept only the mapped reads with a mapping quality of at least 30 and removed repeated regions as identified using RepeatMasker (72), regions of excessive coverage, and the sex chromosomes (see details in the Supplementary Text).
SNP calling using genotype likelihoods
We called SNPs taking genotype uncertainty into account by calculating genotype likelihoods in ANGSD v.0.913 (73) using the samtools model (GL 1) and keeping SNPs with a minimum allele frequency (MAF) of 0.05 and having data in a minimum of 75% of the individuals. In ANGSD, all analyses described below were run considering only SNPs with a phred quality and a mapping quality score of 30. We further filtered the bam files by excluding SNPs that showed both significant deviation from Hardy-Weinberg equilibrium and an inbreeding coefficient (F) value <0 within populations in ANGSD, as these can be the result of paralogs or other mapping artifacts.
Linkage disequilibrium pruning and population structure analyses
We excluded one individual (sample 7Tt182 from the WNA pelagic population) from the population structure analyses, which were not based on population allele frequencies, as this individual had a coverage much lower than the others (table S1). We used NgsLD (74) to obtain a set of unlinked SNPs (Supplementary Text). Population structure analyses, admixture analysis in NGSAdmix (39), and PCAs in PCAngsd (38) were run using a set of 798,572 unlinked SNPs. Note that all SNPs were included in the other analyses. NGSAdmix was run 10 times for each K value between 2 and 8, using a tolerance for convergence of 1 × 10−10 and a minimum likelihood ratio value of 1 × 10−6. Consistency between runs was checked, and the runs with the highest likelihood were plotted. The highest level of structure was identified using the Evanno method (75).
Ancestral state reconstruction
We describe how we reconstructed the ancestral state of alleles in the Supplementary Text.
Genotype calling
We called variants (i.e., generation of a vcf file) using samtools v.1.2 mpileup and bcftools multiallelic and rare-variant calling, option –m on the filtered bam files (76, 77). Variable sites with a minimum mapping quality of 30, a phred score quality of 30, and genotype quality of 20 were retained in vcftools v.0.1.16 (78). We kept SNPs with a minimum MAF of 0.05 and having genotype data in a minimum of 75% of all the individuals and a minimum of five individuals in each of the six populations. The vcf file was also filtered for monomorphic and nonbiallelic sites, totaling 2,003,833 SNPs. Coverage was estimated using vcftools. A vcf file was used as an input for the analyses described below apart from the unfolded SFS and diversity estimates, which were estimated using genotype likelihoods in ANGSD, and the ancient ancestry analyses, which were based on pseudohaploid calls (random sampling of an allele at each site; see details below and table S2).
Admixture analyses
We reconstructed the relationships of the coastal and pelagic ecotypes from the different regions as a maximum likelihood bifurcating tree using TreeMix version 1.13 (Supplementary Text) (40). We ran TreeMix using one individual T. aduncus [SRX2653496/SRR5357656 (79)] as a root. Reads of this T. aduncus individual were mapped to the common bottlenose dolphin reference genome assembly as described above and processed as described earlier for our data. The vcf file was further filtered for sites with missing data in T. aduncus. We first ran TreeMix 10 times for each value of M (migration events) ranging from 0 to 10 (-global -k 1000). We estimated the optimal number of migration events to two using the optM R package (https://cran.r-project.org/web/packages/OptM/index.html) and a possible suboptimal number of five migration events. We then ran TreeMix 100 times for 0 (as null model) to five migration events and obtained a consensus tree and bootstrap values using the BITE R package (80). The residual covariance matrix was estimated for each M value and the consensus tree using TreeMix.
We then estimated F4 statistics to test whether geographic pairs of pelagic and coastal ecotypes had evolved independently (41, 42). The F4 statistics can be used to test whether a given tree describes accurately the relationships among four test populations and to detect admixture events (see Supplementary Text for details). F4 statistics were computed for each possible combination of population using the fourpop function in TreeMix version 1.13 (40). We accounted for linkage disequilibrium by jackknifing in windows of 1000 SNPs. This block jackknife was used to obtain an SE on the estimate of the F4 statistics and test for significance using a z score.
Demographic history
We computed demographic history, that is, changes in effective population sizes (Ne) through time and ecotype splits within a region and splits of the different pelagic ecotypes, using the program SMC++ v.1 (46). Details of the analysis procedure, run on autosome scaffolds that were more than 10 Mb and on a vcf file not filtered for any MAF, are provided in the Supplementary Text. Briefly, the repeated regions and excessive coverage regions were included as a mask file so that they were not misidentified as very long runs of homozygosity. The analysis was run both using all regions and taking out all the regions under selection, as identified with Flink (see below). Regions under selection were defined as 50 kb around each outlier SNPs under any type of selection. Regions under selection were included in the mask file when they were taken out from the dataset. Population size histories and split times between ecotypes in each region were estimated using the default settings, a generation time of 21.1 years for the species (81), and two different mutation rates. Mutation rates were (i) 9.10 × 10−10 substitutions per site per year that is 1.92 × 10−8 substitution per nucleotide per generation (82) and (ii) 1.21 × 10−9 substitution rate per site per year (83) that is 2.56 × 10−8 substitution per nucleotide per generation. Results were plotted in R v.3.6.1 (Supplementary Text) (84).
Diversity and population structure statistics
We estimated the unfolded SFS and the 2D-SFS using the ancestral state and nucleotide diversity, Watterson’s theta, and Tajima’s D for each population using ANGSD v.0.921 (see details in the Supplementary Text). We calculated nucleotide diversity and Watterson’s theta for each site, and then we estimated both the latter and Tajima’s D using a sliding-window size of 50 kb and a step size of 10 kb. We estimated mean pairwise-weighted FST using vcftools across all sites.
Ancient ancestry analyses
Ancient tracts introgressed into the coastal ecotype from a divergent lineage after splitting from the pelagic source population or differentially sorted from structure in an ancestral population will contain clusters of private alleles, and the density of which will depend on the divergence time of the introgressing and receiving lineages (fig. S11 and Supplementary Text) (56, 85). We therefore set out to screen for genomic tracts of consecutive or clustered private (i.e., relative to the allopatric pelagic individuals) alleles in each of the individuals from the coastal ecotype, taking variation in mutation rate along the genome into account. To ensure that the results are comparable despite variation in coverage between samples at some sites, we randomly sampled a single allele at each site from each diploid modern genome in all scaffolds longer than 1 Mb using ANGSD. We did not apply any MAF filter for this analysis. For the outgroup, we used all variants found in a dataset consisting of all non-allopatric pelagic samples (fig. S11).
We then used a hidden Markov model (HMM) to classify 1-kb windows into “nonancient” and ancient states based on the density of private alleles (53). The background mutation rate was estimated in windows of 100 kb, using the variant density in all individuals. We then weighted each 1-kb window by the proportion of sites not masked by our RepeatMasker and CallableLoci bed files. The HMM was trained using a set of starting parameters based on those used for humans (53). We trained the model across five independent runs, varying the starting parameters each time to ensure the consistency of the final parameter input. Posterior decoding then determines whether consecutive 1-kb windows change or retain state (ancient or nonancient) depending on the posterior probability.
Considering windows inferred as ancient with posterior probabilities of >0.8 (53, 56), we identified >1000 ancient tracts totaling >10 Mb in each coastal genome tested (table S4). The emission probabilities of the HMM are modeled as Poisson distributions with means of λAncient = μ ∙ L ∙ TAncient for the introgressed state and λIngroup = μ ∙ L ∙ TIngroup for the nonancient (or ingroup) state (54), where L is the window size (1000 bp) and μ is the mutation rate [1.92 × 10−8 and 2.56 × 10−8 substitutions per nucleotide per generation (82, 83)]. This allowed us to estimate the mean TMRCA of the ancient and ingroup windows with the corresponding segments in the outgroup dataset.
Patterns of structuration across the genome
We used Local PCA (50) to describe the three major patterns of relatedness (“corners”) among populations on four PCs for the 56 scaffolds longer than 10 Mb using the default options [two PCs and two multidimensional scaling (MDS) coordinates], the R codes available on GitHub and bins of 100 SNPs. We plotted the pairwise plots of the first four PCs for each of the three corners.
Selection analyses
We used Flink (47) to test for selection to pelagic versus coastal habitat. Flink is an extension of BayeScan (51), respectively describing selection and drift, which takes linkage among loci into account. Specifically, it applies an HMM to identify the effect of selection at linked markers using correlation in the loci-specific elements along the genome. Flink was run grouping the populations into two groups: pelagic and coastal (higher hierarchical level). Each group was composed of the three populations from each region. Scaffolds were grouped into super-scaffolds, so that each contains at least 50,000 SNPs, but each scaffold was considered independent in the analysis. In Flink, the function estimate was run, and parameter settings are described in the Supplementary Text. The number of iterations was set to 500,000, the burnin to 300,000, and the thinning to 100. We considered a locus under selection when it is within the 1% false discovery rate (FDR) threshold. Given the uncertainty about evolutionary relationships of dolphin populations, we also ran Flink using the three regions as the higher hierarchical level. This resulted in much more prevalent selection with between ~165,000 and ~195,000 SNPs under divergent selection between ecotypes within each geographical region, potentially including many false positives because of deviation from the assumed hierarchical structure model. The approach presented here, considering ecotype as the higher hierarchical level, is therefore more conservative.
To get further insights into the results obtained by Flink, we plotted the raw genotypes of all the SNPs, SNPs under homogenizing selection in the coastal populations, SNPs under divergent selection between ecotypes, and SNPs under both homogenizing selection in the coastal populations and divergent between ecotypes (defined as the SNPs under parallel linked selection) in R (see details in the Supplementary Text). We also plotted a neighbor-joining distance tree and a PCA for the SNPs under each type of selection. To determine the origin of the SNPs under selection, we defined how many were also polymorphic in the pelagic populations and compared the 2D-SFS between all pairs of populations, estimated in ANGSD (see details in Supplementary Text). Then, we defined how many SNPs under the different types of selection were found in ancient tracts. We compared the results with 100 random samples of the same number of putatively neutral SNPs found in ancient tracts.
We identified the genes directly overlapping with the SNPs under parallel linked selection using the reference genome annotation file. We describe how we determined the putative functions of the genes under selection in the Supplementary Text.
Acknowledgments
We thank everyone involved in the sample collection including C. Ryan. We thank J. Ward and P. Thorpe from the St Andrews Bioinformatics Unit for help with software installations and troubleshooting. We thank the associate editor and the reviewers for the constructive comments that greatly improved this manuscript.
Funding: Funding for sequencing was provided by the Total Foundation awarded to M.L., the University of Groningen awarded to M.C.F., and the Marine Alliance for Science and Technology for Scotland and The Russell Trust awarded to O.E.G. M.L. was supported by a Fyssen Fellowship, Total Foundation, Systematics Research Fund, Godfrey Hewitt mobility award from the European Society for Evolutionary Biology (ESEB), People’s Trust for Endangered Species, Lerner-Gray Grants for Marine Research from the American Museum of Natural History, and the University of St Andrews. M.G. was funded through a Swiss National Science Foundation grant (31003A_173062) to D.W. A.D.F. was supported by European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 663830. Bioinformatics and computational biology analyses were supported by the University of St Andrews Bioinformatics Unit, which is funded by a Wellcome Trust ISSF award (grant 105621/Z/14/Z).
Author contributions: M.L. initiated the study. M.L., M.C.F., A.D.F., and O.E.G. conceived the analyses. F.A., S.B., A.B., J.O., K.M.R., and P.E.R. collected the samples. K.M.R., P.E.R., and M.C.F. performed the DNA extractions. M.L., M.G., M.N., R.F., B.S.-B., M.C.F., A.D.F., and O.E.G. ran the analyses, and A.D.F., O.E.G., and M.C.F. provided supervision. M.L. wrote the manuscript with input from all coauthors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Biosample and SRA accession numbers are provided in table S1; data are part of the Bioproject PRJNA724031: www.ncbi.nlm.nih.gov/bioproject/724031. Input files are available on the DataSuds repository: https://doi.org/10.23708/Q2MYHE. Custom codes to generate the input files for the ancient tract analysis can be found at https://github.com/besibo/GhostAncestry. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S19
Tables S1 to S7
References
REFERENCES AND NOTES
- 1.Kawecki T. J., Ebert D., Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241 (2004). [Google Scholar]
- 2.Li J., Li H., Jakobsson M., Li S., Sjödin P., Lascoux M., Joint analysis of demography and selection in population genetics: Where do we stand and where could we go? Mol. Ecol. 21, 28–44 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Charlesworth B., Nordborg M., Charlesworth D., The effects of local selection, balanced polymorphism and background selection on equilibrium patterns of genetic diversity in subdivided populations. Genet. Res. 70, 155–174 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Conte G. L., Arnegard M. E., Peichel C. L., Schluter D., The probability of genetic parallelism and convergence in natural populations. Proc. Biol. Sci. 279, 5039–5047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E., Johnson J., Swofford R., Pirun M., Zody M. C., White S., Birney E., Searle S., Schmutz J., Grimwood J., Dickson M. C., Myers R. M., Miller C. T., Summers B. R., Knecht A. K., Brady S. D., Zhang H., Pollen A. A., Howes T., Amemiya C.; Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team, Lander E. S., Di Palma F., Lindblad-Toh K., Kingsley D. M., The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soria-Carrasco V., Gompert Z., Comeault A. A., Farkas T. E., Parchman T. L., Johnston J. S., Buerkle C. A., Feder J. L., Bast J., Schwander T., Egan S. P., Crespi B. J., Nosil P., Stick insect genomes reveal natural selection’s role in parallel speciation. Science 344, 738–742 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Foll M., Gaggiotti O. E., Daub J. T., Vatsiou A., Excoffier L., Widespread signals of convergent adaptation to high altitude in Asia and America. Am. J. Hum. Genet. 95, 394–407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seehausen O., Terai Y., Magalhaes I. S., Carleton K. L., Mrosso H. D. J., Miyagi R., van der Sluijs I., Schneider M. V., Maan M. E., Tachida H., Imai H., Okada N., Speciation through sensory drive in cichlid fish. Nature 455, 620–626 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Alves J. M., Carneiro M., Cheng J. Y., Lemos de Matos A., Rahman M. M., Loog L., Campos P. F., Wales N., Eriksson A., Manica A., Strive T., Graham S. C., Afonso S., Bell D. J., Belmont L., Day J. P., Fuller S. J., Marchandeau S., Palmer W. J., Queney G., Surridge A. K., Vieira F. G., McFadden G., Nielsen R., Gilbert M. T. P., Esteves P. J., Ferrand N., Jiggins F. M., Parallel adaptation of rabbit populations to myxoma virus. Science 363, 1319–1326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson K. A., Osmond M. M., Schluter D., Parallel genetic evolution and speciation from standing variation. Evol. Lett. 3, 129–141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bolnick D. I., Barrett R. D. H., Oke K. B., Rennison D. J., Stuart Y. E., (Non)parallel evolution. Annu. Rev. Ecol. Evol. Syst. 49, 303–330 (2018). [Google Scholar]
- 12.Stuart Y. E., Veen T., Weber J. N., Hanson D., Ravinet M., Lohman B. K., Thompson C. J., Tasneem T., Doggett A., Izen R., Ahmed N., Barrett R. D. H., Hendry A. P., Peichel C. L., Bolnick D. I., Contrasting effects of environment and genetics generate a continuum of parallel evolution. Nat. Ecol. Evol. 1, 0158 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Fang B., Kemppainen P., Momigliano P., Feng X., Merilä J., On the causes of geographically heterogeneous parallel evolution in sticklebacks. Nat. Ecol. Evol. 4, 1105–1115 (2020). [DOI] [PubMed] [Google Scholar]
- 14.De Lisle S. P., Bolnick D. I., A multivariate view of parallel evolution. Evolution 74, 1466–1481 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Barrett R. D. H., Schluter D., Adaptation from standing genetic variation. Trends Ecol. Evol. 23, 38–44 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Van Belleghem S. M., Vangestel C., De Wolf K., De Corte Z., Möst M., Rastas P., De Meester L., Hendrickx F., Evolution at two time frames: Polymorphisms from an ancient singular divergence event fuel contemporary parallel evolution. PLOS Genet. 14, e1007796 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerrero R. F., Hahn M. W., Speciation as a sieve for ancestral polymorphism. Mol. Ecol. 26, 5362–5368 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Rougemont Q., Gagnaire P.-A., Perrier C., Genthon C., Besnard A.-L., Launey S., Evanno G., Inferring the demographic history underlying parallel genomic divergence among pairs of parasitic and nonparasitic lamprey ecotypes. Mol. Ecol. 26, 142–162 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Marques D. A., Meier J. I., Seehausen O., A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34, 531–544 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Nelson T. C., Cresko W. A., Ancient genomic variation underlies repeated ecological adaptation in young stickleback populations. Evol. Lett. 2, 9–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tishkoff S. A., Reed F. A., Ranciaro A., Voight B. F., Babbitt C. C., Silverman J. S., Powell K., Mortensen H. M., Hirbo J. B., Osman M., Ibrahim M., Omar S. A., Lema G., Nyambo T. B., Ghori J., Bumpstead S., Pritchard J. K., Wray G. A., Deloukas P., Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 39, 31–40 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlesworth B., Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10, 195–205 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Foote A. D., Vijay N., Ávila-Arcos M. C., Baird R. W., Durban J. W., Fumagalli M., Gibbs R. A., Hanson M. B., Korneliussen T. S., Martin M. D., Robertson K. M., Sousa V. C., Vieira F. G., Vinař T., Wade P., Worley K. C., Excoffier L., Morin P. A., Gilbert M. T. P., Wolf J. B. W., Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat. Commun. 7, 11693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoelzel A. R., Potter C. W., Best P. B., Genetic differentiation between parapatric “nearshore” and “offshore” populations of the bottlenose dolphin. Proc. Biol. Sci. 265, 1177–1183 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natoli A., Peddemors V. M., Hoelzel A. R., Population structure and speciation in the genus Tursiops based on microsatellite and mitochondrial DNA analyses. J. Evol. Biol. 17, 363–375 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Louis M., Viricel A., Lucas T., Peltier H., Alfonsi E., Berrow S., Brownlow A., Covelo P., Dabin W., Deaville R., de Stephanis R., Gally F., Gauffier P., Penrose R., Silva M. A., Guinet C., Simon-Bouhet B., Habitat-driven population structure of bottlenose dolphins, Tursiops truncatus, in the North-East Atlantic. Mol. Ecol. 23, 857–874 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Costa A. P. B., Fruet P. F., Secchi E. R., Daura-Jorge F. G., Simões-Lopes P. C., Di Tullio J. C., Rosel P. E., Ecological divergence and speciation in common bottlenose dolphins in the western South Atlantic. J. Evol. Biol. 34, 16–32 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Lowther-Thieleking J. L., Archer F. I., Lang A. R., Weller D. W., Genetic differentiation among coastal and offshore common bottlenose dolphins, Tursiops truncatus, in the eastern North Pacific Ocean. Mar. Mamm. Sci. 31, 1–20 (2015). [Google Scholar]
- 29.Nykänen M., Kaschner K., Dabin W., Brownlow A., Davison N. J., Deaville R., Garilao C., Kesner-Reyes K., Gilbert M. T. P., Penrose R., Islas-Villanueva V., Wales N., Ingram S. N., Rogan E., Louis M., Foote A. D., Postglacial colonization of northern coastal habitat by bottlenose dolphins: A marine leading-edge expansion? J. Hered. 110, 662–674 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Louis M., Fontaine M. C., Spitz J., Schlund E., Dabin W., Deaville R., Caurant F., Cherel Y., Guinet C., Simon-Bouhet B., Ecological opportunities and specializations shaped genetic divergence in a highly mobile marine top predator. Proc. Biol. Sci. 281, 28120141558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.J. G. Mead, C. W. Potter, Recognizing two populations of the bottlenose dolphin (Tursiops truncatus) of the Atlantic coast of North America-morphologic and ecologic considerations (1995); https://repository.si.edu/bitstream/handle/10088/2652/mead_and_potter1995.pdf?sequence=1.
- 32.Costa A. P. B., Rosel P. E., Daura-Jorge F. G., Simões-Lopes P. C., Offshore and coastal common bottlenose dolphins of the western South Atlantic face-to-face: What the skull and the spine can tell us. Mar. Mamm. Sci. 32, 1433–1457 (2016). [Google Scholar]
- 33.Perrin W. F., Thieleking J. L., Walker W. A., Archer F. I., Robertson K. M., Common bottlenose dolphins (Tursiops truncatus) in California waters: Cranial differentiation of coastal and offshore ecotypes. Mar. Mamm. Sci. 27, 769–792 (2011). [Google Scholar]
- 34.Rosel P. E., Hansen L., Hohn A. A., Restricted dispersal in a continuously distributed marine species: Common bottlenose dolphins Tursiops truncatus in coastal waters of the western North Atlantic. Mol. Ecol. 18, 5030–5045 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Machado A. M. S., Cantor M., Costa A. P. B., Righetti B. P. H., Bezamat C., Valle-Pereira J. V. S., Simões-Lopes P. C., Castilho P. V., Daura-Jorge F. G., Homophily around specialized foraging underlies dolphin social preferences. Biol. Lett. 15, 20180909 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopps A. M., Ackermann C. Y., Sherwin W. B., Allen S. J., Bejder L., Krützen M., Cultural transmission of tool use combined with habitat specializations leads to fine-scale genetic structure in bottlenose dolphins. Proc. Biol. Sci. 281, 20133245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meisner J., Albrechtsen A., Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210, 719–731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skotte L., Korneliussen T. S., Albrechtsen A., Estimating individual admixture proportions from next generation sequencing data. Genetics 195, 693–702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pickrell J. K., Pritchard J. K., Inference of population splits and mixtures from genome-wide allele frequency data. PLOS Genet. 8, e1002967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich D., Thangaraj K., Patterson N., Price A. L., Singh L., Reconstructing Indian population history. Nature 461, 489–494 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D., Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leitwein M., Duranton M., Rougemont Q., Gagnaire P.-A., Bernatchez L., Using haplotype information for conservation genomics. Trends Ecol. Evol. 35, 245–258 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Mazet O., Rodríguez W., Grusea S., Boitard S., Chikhi L., On the importance of being structured: Instantaneous coalescence rates and human evolution—Lessons for ancestral population size inference? Heredity 116, 362–371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrider D. R., Shanku A. G., Kern A. D., Effects of linked selective sweeps on demographic inference and model selection. Genetics 204, 1207–1223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terhorst J., Kamm J. A., Song Y. S., Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49, 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galimberti M., Leuenberger C., Wolf B., Szilágyi S. M., Foll M., Wegmann D., Detecting selection from linked sites using an F-model. Genetics 216, 1205–1215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen R., Statistical tests of selective neutrality in the age of genomics. Heredity 86, 641–647 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Bailey H., Thompson P., Effect of oceanographic features on fine-scale foraging movements of bottlenose dolphins. Mar. Ecol. Prog. Ser. 418, 223–233 (2010). [Google Scholar]
- 50.Li H., Ralph P., Local PCA shows how the effect of population structure differs along the genome. Genetics 211, 289–304 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Foll M., Gaggiotti O., A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics 180, 977–993 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephan W., Selective sweeps. Genetics 211, 5–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skov L., Hui R., Shchur V., Hobolth A., Scally A., Schierup M. H., Durbin R., Detecting archaic introgression using an unadmixed outgroup. PLOS Genet. 14, e1007641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moura A. E., Shreves K., Pilot M., Andrews K. R., Moore D. M., Kishida T., Möller L., Natoli A., Gaspari S., McGowen M., Chen I., Gray H., Gore M., Culloch R. M., Kiani M. S., Willson M. S., Bulushi A., Collins T., Baldwin R., Willson A., Minton G., Ponnampalam L., Hoelzel A. R., Phylogenomics of the genus Tursiops and closely related Delphininae reveals extensive reticulation among lineages and provides inference about eco-evolutionary drivers. Mol. Phylogenet. Evol. 146, 106756 (2020). [DOI] [PubMed] [Google Scholar]
- 55.McGowen M. R., Tsagkogeorga G., Álvarez-Carretero S., Dos Reis M., Struebig M., Deaville R., Jepson P. D., Jarman S., Polanowski A., Morin P. A., Rossiter S. J., Phylogenomic resolution of the cetacean tree of life using target sequence capture. Syst. Biol. 69, 479–501 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duranton M., Allal F., Valière S., Bouchez O., Bonhomme F., Gagnaire P.-A., The contribution of ancient admixture to reproductive isolation between European sea bass lineages. Evol. Lett. 4, 226–242 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foote A. D., Martin M. D., Louis M., Pacheco G., Robertson K. M., Sinding M. S., Amaral A. R., Baird R. W., Baker C. S., Ballance L., Barlow J., Brownlow A., Collins T., Constantine R., Dabin W., Rosa L. D., Davison N. J., Durban J. W., Esteban R., Ferguson S. H., Gerrodette T., Guinet C., Bradley Hanson M., Hoggard W., Matthews C. J. D., Samarra F. I. P., de Stephanis R., Tavares S. B., Tixier P., Totterdell J. A., Wade P., Excoffier L., Gilbert M. T. P., Wolf J. B. W., Morin P. A., Killer whale genomes reveal a complex history of recurrent admixture and vicariance. Mol. Ecol. 28, 3427–3444 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Colosimo P. F., Hosemann K. E., Balabhadra S., Villarreal G. Jr., Dickson M., Grimwood J., Schmutz J., Myers R. M., Schluter D., Kingsley D. M., Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307, 1928–1933 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Schluter D., Conte G. L., Genetics and ecological speciation. Proc. Natl. Acad. Sci. U.S.A. 106, 9955–9962 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pritchard J. K., Pickrell J. K., Coop G., The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20, R208–R215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weeber E. J., Beffert U., Jones C., Christian J. M., Forster E., Sweatt J. D., Herz J., Reelin and ApoE receptors cooperate to enhance hippocampal synaptic plasticity and learning. J. Biol. Chem. 277, 39944–39952 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Herz J., Chen Y., Reelin, lipoprotein receptors and synaptic plasticity. Nat. Rev. Neurosci. 7, 850–859 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Mladenova D., Barry G., Konen L. M., Pineda S. S., Guennewig B., Avesson L., Zinn R., Schonrock N., Bitar M., Jonkhout N., Crumlish L., Kaczorowski D. C., Gong A., Pinese M., Franco G. R., Walkley C. R., Vissel B., Mattick J. S., Adar3 is involved in learning and memory in mice. Front. Neurosci. 12, 243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beichman A. C., Koepfli K.-P., Li G., Murphy W., Dobrynin P., Kliver S., Tinker M. T., Murray M. J., Johnson J., Lindblad-Toh K., Karlsson E. K., Lohmueller K. E., Wayne R. K., Aquatic adaptation and depleted diversity: A deep dive into the genomes of the sea otter and giant otter. Mol. Biol. Evol. 36, 2631–2655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aulchenko Y. S., Pullen J., Kloosterman W. P., Yazdanpanah M., Hofman A., Vaessen N., Snijders P. J. L. M., Zubakov D., Mackay I., Olavesen M., Sidhu B., Smith V. E., Carey A., Berezikov E., Uitterlinden A. G., Plasterk R. H. A., Oostra B. A., van Duijn C. M., LPIN2 is associated with type 2 diabetes, glucose metabolism, and body composition. Diabetes 56, 3020–3026 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Oomen R. A., Kuparinen A., Hutchings J. A., Consequences of single-locus and tightly linked genomic architectures for evolutionary responses to environmental change. J. Hered. 111, 319–332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moura A. E., Nielsen S. C. A., Vilstrup J. T., Moreno-Mayar J. V., Gilbert M. T. P., Gray H. W. I., Natoli A., Möller L., Hoelzel A. R., Recent diversification of a marine genus (Tursiops spp.) tracks habitat preference and environmental change. Syst. Biol. 62, 865–877 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Broad Institute, Picard tools (2016).
- 71.G. A. Van der Auwera, B. D. O’Connor, Genomics in the Cloud: Using Docker, GATK, and WDL in Terra (O’Reilly Media Inc., 2020).
- 72.A. F. A. Smit, R. Hubley, P. Green, RepeatMasker Open-4.0. 2013–2015 (2015).
- 73.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fox E. A., Wright A. E., Fumagalli M., Vieira F. G., ngsLD: Evaluating linkage disequilibrium using genotype likelihoods. Bioinformatics 35, 3855–3856 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Evanno G., Regnaut S., Goudet J., Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.P. Danecek, S. Schiffels, R. Durbin, Multiallelic calling model in bcftools (-m) (2014); https://samtools.github.io/bcftools/call-m.pdf.
- 78.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., McVean G., Durbin R.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vijay N., Park C., Oh J., Jin S., Kern E., Kim H. W., Zhang J., Park J.-K., Population genomic analysis reveals contrasting demographic changes of two closely related dolphin species in the last glacial. Mol. Biol. Evol. 35, 2026–2033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.M. Milanesi, S. Capomaccio, E. Vajana, L. Bomba, J. F. Garcia, P. Ajmone-Marsan, L. Colli, BITE: An R package for biodiversity analyses. bioRxiv, 181610 (2017). 10.1101/181610. [DOI]
- 81.B. L. Taylor, S. J. Chivers, J. Larese, W. F. Perrin, Generation length and percent mature estimates for IUCN assessments of cetaceans (Administrative Report LJ-07-01). National Marine Fisheries Service, Southwest Fisheries Science Center, NOAA, La Jolla, CA, USA (2007).
- 82.Dornburg A., Brandley M. C., McGowen M. R., Near T. J., Relaxed clocks and inferences of heterogeneous patterns of nucleotide substitution and divergence time estimates across whales and dolphins (Mammalia: Cetacea). Mol. Biol. Evol. 29, 721–736 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Yim H.-S., Cho Y. S., Guang X., Kang S. G., Jeong J.-Y., Cha S.-S., Oh H.-M., Lee J.-H., Yang E. C., Kwon K. K., Kim Y. J., Kim T. W., Kim W., Jeon J. H., Kim S.-J., Choi D. H., Jho S., Kim H.-M., Ko J., Kim H., Shin Y.-A., Jung H.-J., Zheng Y., Wang Z., Chen Y., Chen M., Jiang A., Li E., Zhang S., Hou H., Kim T. H., Yu L., Liu S., Ahn K., Cooper J., Park S.-G., Hong C. P., Jin W., Kim H.-S., Park C., Lee K., Chun S., Morin P. A., O’Brien S. J., Lee H., Kimura J., Moon D. Y., Manica A., Edwards J., Kim B. C., Kim S., Wang J., Bhak J., Lee H. S., Lee J.-H., Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 46, 88–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.R Core Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing (Vienna, Austria, 2019); www.R-project.org/.
- 85.Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E., Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16, 359–371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McVean G., A genealogical interpretation of principal components analysis. PLOS Genet. 5, e1000686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jombart T., Ahmed I., adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cuéllar-Rivas E., Pustovrh-Ramos M. C., The role of enamelysin (mmp-20) in tooth development: Systematic review. Rev. Fac. Odontol. Univ. Antioq. 27, 154–176 (2015). [Google Scholar]
- 89.Rosel P. E., Block B. A., Mitochondrial control region variability and global population structure in the swordfish, Xiphias gladius. Mar. Biol. 125, 11–22 (1996). [Google Scholar]
- 90.Miller S. A., Dykes D. D., Polesky H. F., A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16, 1215–1215 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.R. Sinha, G. Stanley, G. S. Gulati, C. Ezran, K. J. Travaglini, Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. bioRxiv (2017); www.biorxiv.org/content/biorxiv/early/2017/04/09/125724.full.pdf.
- 92.Morin P. A., Parsons K. M., Archer F. I., Ávila-Arcos M. C., Barrett-Lennard L. G., Dalla Rosa L., Duchêne S., Durban J. W., Ellis G. M., Ferguson S. H., Ford J. K., Ford M. J., Garilao C., Gilbert M. T. P., Kaschner K., Matkin C. O., Petersen S. D., Robertson K. M., Visser I. N., Wade P. R., Ho S. Y. W., Foote A. D., Geographic and temporal dynamics of a global radiation and diversification in the killer whale. Mol. Ecol. 24, 3964–3979 (2015). [DOI] [PubMed] [Google Scholar]
- 93.S. Andrews, Others, FastQC: A quality control tool for high throughput sequence data (2010).
- 94.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lindblad-Toh K., Garber M., Zuk O., Lin M. F., Parker B. J., Washietl S., Kheradpour P., Ernst J., Jordan G., Mauceli E., Ward L. D., Lowe C. B., Holloway A. K., Clamp M., Gnerre S., Alföldi J., Beal K., Chang J., Clawson H., Cuff J., Di Palma F., Fitzgerald S., Flicek P., Guttman M., Hubisz M. J., Jaffe D. B., Jungreis I., Kent W. J., Kostka D., Lara M., Martins A. L., Massingham T., Moltke I., Raney B. J., Rasmussen M. D., Robinson J., Stark A., Vilella A. J., Wen J., Xie X., Zody M. C.; Broad Institute Sequencing Platform and Whole Genome Assembly Team, Worley K. C., Kovar C. L., Muzny D. M., Gibbs R. A.; Baylor College of Medicine Human Genome Sequencing Center Sequencing Team, Warren W. C., Mardis E. R., Weinstock G. M., Wilson R. K.; Genome Institute at Washington University, Birney E., Margulies E. H., Herrero J., Green E. D., Haussler D., Siepel A., Goldman N., Pollard K. S., Pedersen J. S., Lander E. S., Kellis M., A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moura A. E., Kenny J. G., Chaudhuri R., Hughes M. A., Welch A. J., Reisinger R. R., de Bruyn P. J. N., Dahlheim M. E., Hall N., Rus Hoelzel A., Population genomics of the killer whale indicates ecotype evolution in sympatry involving both selection and drift. Mol. Ecol. 23, 5179–5192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Foote A. D., Liu Y., Thomas G. W. C., Vinař T., Alföldi J., Deng J., Dugan S., van Elk C. E., Hunter M. E., Joshi V., Khan Z., Kovar C., Lee S. L., Lindblad-Toh K., Mancia A., Nielsen R., Qin X., Qu J., Raney B. J., Vijay N., Wolf J. B. W., Hahn M. W., Muzny D. M., Worley K. C., Gilbert M. T. P., Gibbs R. A., Convergent evolution of the genomes of marine mammals. Nat. Genet. 47, 272–275 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li H., Durbin R., Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.H. Wickham, ggplot2: Elegant Graphics for Data Analysis (Springer, 2016). [Google Scholar]
- 100.H. Wickham, Scales: Scale functions for visualization, R package version 0. 4. 0 (2016); https://CRAN.R-project.org/package=scales.
- 101.E. Neuwirth, RColorBrewer: ColorBrewer palettes, R package version 1.1–2, The R Foundation (2014).
- 102.Nielsen R., Korneliussen T., Albrechtsen A., Li Y., Wang J., SNP calling, genotype calling, and sample allele frequency estimation from new-generation sequencing data. PLOS ONE 7, e37558 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Knaus B. J., Grünwald N. J., vcfr: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 17, 44–53 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Paradis E., Schliep K., ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 105.Maglott D., Ostell J., Pruitt K. D., Tatusova T., Entrez Gene: Gene-centered information at NCBI. Nucleic Acids Res. 39, D52–D57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.The UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.O’Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., Astashyn A., Badretdin A., Bao Y., Blinkova O., Brover V., Chetvernin V., Choi J., Cox E., Ermolaeva O., Farrell C. M., Goldfarb T., Gupta T., Haft D., Hatcher E., Hlavina W., Joardar V. S., Kodali V. K., Li W., Maglott D., Masterson P., McGarvey K. M., Murphy M. R., O’Neill K., Pujar S., Rangwala S. H., Rausch D., Riddick L. D., Schoch C., Shkeda A., Storz S. S., Sun H., Thibaud-Nissen F., Tolstoy I., Tully R. E., Vatsan A. R., Wallin C., Webb D., Wu W., Landrum M. J., Kimchi A., Tatusova T., DiCuccio M., Kitts P., Murphy T. D., Pruitt K. D., Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T. I., Nudel R., Lieder I., Mazor Y., Kaplan S., Dahary D., Warshawsky D., Guan-Golan Y., Kohn A., Rappaport N., Safran M., Lancet D., The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics 54, 1.30.1–1.30.33 (2016). [DOI] [PubMed] [Google Scholar]
- 109.Hamosh A., Scott A. F., Amberger J. S., Bocchini C. A., McKusick V. A., Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33, D514–D517 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S19
Tables S1 to S7
References