Macrophages can invade the fly brain triggered by inflammation and show phagocytotic responses reducing longevity.
Abstract
The nervous system is shielded from circulating immune cells by the blood-brain barrier (BBB). During infections and autoimmune diseases, macrophages can enter the brain where they participate in pathogen elimination but can also cause tissue damage. Here, we establish a Drosophila model to study macrophage invasion into the inflamed brain. We show that the immune deficiency (Imd) pathway, but not the Toll pathway, is responsible for attraction and invasion of hemolymph-borne macrophages across the BBB during pupal stages. Macrophage recruitment is mediated by glial, but not neuronal, induction of the Imd pathway through expression of Pvf2. Within the brain, macrophages can phagocytose synaptic material and reduce locomotor abilities and longevity. Similarly, we show that central nervous system infection by group B Streptococcus elicits macrophage recruitment in an Imd-dependent manner. This suggests that evolutionarily conserved inflammatory responses require a delicate balance between beneficial and detrimental activities.
INTRODUCTION
The central nervous system (CNS) integrates and computes external and internal stimuli and orchestrates appropriate motor responses. It comprises a large number of neurons and glial cells that interact in manifold ways. To ensure proper homeostatic function within the brain, the nervous system is separated from the circulatory system by the blood-brain barrier (BBB). The BBB guarantees metabolic and ion regulation in the brain and also represents an additional barrier preventing the invasion of pathogens.
Because of its efficient separation from circulation, the nervous system is considered an immune-privileged organ (1). Thus, additional cellular defense mechanisms have evolved to cope with infections and the removal of cellular debris within the CNS. In vertebrates, microglia, a cell type originating from the fetal yolk sac, migrate into the developing nervous system to perform immune surveillance (2) and clear superfluous cells and projections. In invertebrates, glial cells phagocytose dying neurons and cellular debris to promote tissue homeostasis (3–5). These local, (micro)glial clearance mechanisms are generally sufficient in a healthy animal but can become overloaded in case of infection or during neurodegenerative and/or autoimmune diseases. These insults can trigger the extravasation of circulating lymphocytes and macrophages across the BBB into the brain, which will clear pathogens and cellular debris but which can also result in further brain damage (6).
The immune responses that unfold following bacterial infection in vertebrates are also frequently observed in invertebrates (7, 8). Drosophila melanogaster provides a particularly well-defined model to explore the molecular underpinnings of immune reactivity (9). Flies respond to septic injury by activating a complex immune response that is highly similar to the mammalian innate immune response. The primary systemic immune cells in Drosophila are the hemocytes, most of which are migratory phagocytes or macrophages (10, 11). These cells are very efficient in clearing pathogens from the circulation through phagocytosis (12).
Another branch of the immune response consists of a large array of antimicrobial peptides (AMPs) that directly target pathogens. They are secreted into the hemolymph mainly by fat body cells, the equivalent of the vertebrate liver. Their expression is triggered by the activation of two core immune pathways, the Toll and the immune deficiency (Imd) pathways, which ultimately center on the activity of nuclear factor κB (NF-κB) transcription factors. The Toll pathway is mostly activated by Gram-positive bacteria and by fungi leading to activation of Dorsal and Dorsal-related immunity factor (Dif). The Imd pathway responds primarily to Gram-negative and bacillus-shaped Gram-positive bacteria (13) and results in the cleavage by a Death related ced-3/Nedd2-like caspase (Dredd) caspase of an inhibitory domain of Relish, also a member of the NF-κB transcription factor class (Fig. 1) (9, 13, 14). In Drosophila cell culture, this pathway can also result in the expression of the platelet-derived growth factor (PDGF)– and vascular endothelial growth factor (VEGF)–related factors 2 and 3 (Pvf2 and Pvf3), two ligands of the Drosophila PDGF/VEGF receptor homolog (Pvr) that are implicated in invasive migration of macrophages and negative regulation of Imd activity (15–17).
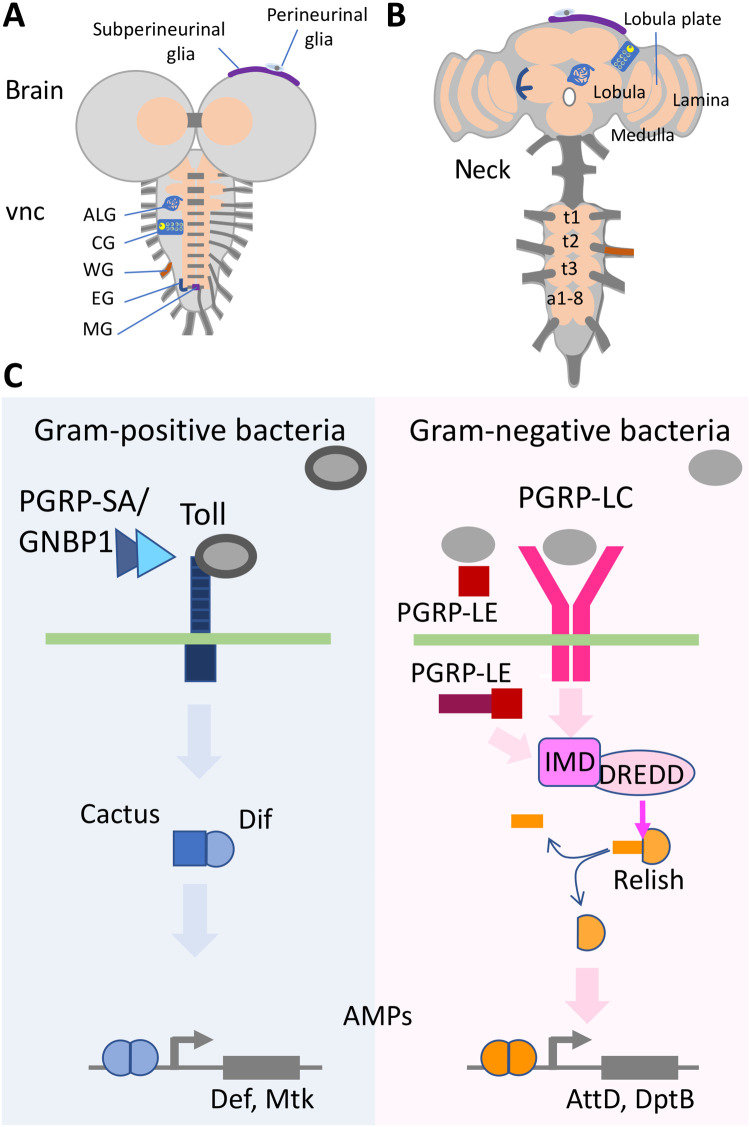
Fig. 1. Drosophila brains and immunity induction.
(A and B) Schematic view of (A) a third instar larval CNS and (B) an adult brain each composed of the ventral nerve cord (vnc) and the brain lobes. The subperineurial and the perineurial glia establish the BBB. The remaining glial cell types are astrocyte-like glia (ALG), cortex glia (CG), wrapping glia (WG), ensheathing glia (EG), and midline glia (MG). The thoracic neuromeres (t1 to t3) expand during pupal development, while the abdominal neuromers a1-8 condense. Some neuropil areas of the adult are indicated. (C) Signaling pathways directing the innate immune response. The Toll pathway is preferentially activated by Gram-positive bacteria, and the peptidoglycan recognition proteins (PGRP-LC and PGRP-LE) detect mostly Gram-negative bacteria. All relevant components are indicated. For further details, see main text.
Sensing of the pathogen by the host is the first step of this immune response and mostly relies on the activity of peptidoglycan recognition proteins (PGRPs). The Drosophila genome encodes 13 PGRPs that either show a zinc-dependent amidase activity (PGRP-LB, PGRP-LF, PGRP-SB1, PGRP-SB2, PGRP-SC1a, PGRP-SC1b, and PGRP-SC2) or are noncatalytic (PGRP-SA, PGRP-SD, PGRP-LA, PGRP-LC, PGRP-LD, and PGRP-LE). This variety allows the detection of different pathogens. In particular, Gram-positive bacteria are detected by PGRP-SA, which activates the Toll pathway in fat body cells together with the Gram-negative binding protein 1 (GNBP1) (18–21). In contrast, Gram-negative bacteria are mostly detected by transmembrane PGRP-LC, the secreted PGRP-SD, and PGRP-LE, which, depending on the isoform, can be acting as secreted or cytoplasmic receptors, which lead to activation of the Imd pathway in the fat body (22–28). In addition, PGRP-LA is involved in Imd activation, while PGRP-LB and PGRP-LF inhibit it (29). Pathogen recognition by PGRPs is thus an essential step to select an appropriate immune response.
Although it is well documented that bacterial infections occur in the fly hemolymph, it is presently unclear if they could trigger invasion of immune system cells into the fly CNS. Here, we asked whether neurons or glial cells can mount an inflammatory response upon bacterial infection and whether such a response is capable of attracting macrophages into the brain tissue. We recently developed a model of brain infection in Drosophila, using both ex vivo and in vivo models, and showed the passage of several mammalian neurotropic pathogens across the BBB (30). In particular, we have demonstrated that group B Streptococcus (GBS), bacteria known to cause meningitis in neonates, can breach the BBB and enter the larval CNS. Here, we demonstrate that upon GBS infection, PGRP-SA, PGRP-LC, and Pvf2 are up-regulated. Glial subtype–specific expression of PGRP-LC and pan-glial expression of PGRP-LE trigger invasion of the pupal brain by macrophages. Labeling of macrophages in the hemolymph by latex beads or transplantation of genetically labeled macrophages into unlabeled hosts demonstrates that hemolymph-borne macrophages invade the nervous system in reaction to the antibacterial response. The response mounted by PGRP-LE–expressing glia triggers Pvf2 expression, which is required and sufficient for macrophage invasion. Activation of the Imd pathway in neurons did not stimulate macrophage infiltration of the brain, although Pvf2 expression in either neurons or glial cells is sufficient to trigger migration of macrophages into the CNS. This indicates that only glial cells can mount an immune response. Invading macrophages are mostly found in the neuropil, where they also phagocytose synaptic material pointing to the need of balancing positive and negative consequences of invading macrophages.
RESULTS
Pan-glial expression of PGRPs triggers an immunity response
The Drosophila CNS is surrounded by hemolymph and comprises the brain lobes and the ventral nerve cord (Fig. 1A). During pupal stages, the larval nervous system is reorganized. In adults, the brain is located in the head capsule, and the ventral nerve cord is found in the thorax (Fig. 1B). These two parts of the CNS are connected by the neck region, a thick axon tract, devoid of cell bodies. The nervous system is covered by an efficient BBB that allows a well-balanced ion and metabolite homeostasis and blocks the entry of xenobiotic substances (31, 32).
So far, no macrophage infiltration has ever been reported in the Drosophila CNS. In mammals, the onset of an immune activation precedes macrophage infiltration. We thus wondered whether immune activation in the Drosophila CNS could also induce this phenomenon and provide an experimental model to dissect the cellular and molecular mechanisms supporting macrophage invasion of the CNS.
As a prerequisite, we first assayed whether immune activation could be triggered and detected in the Drosophila CNS. In other tissues, the two core immune pathways (Toll and Imd) can be activated by a number of PGRPs, with different specificities (Fig. 1C). We thus tested whether forced PGRP expression is able to induce an immune response also in the CNS tissue. We first expressed PGRP-LC, PGRP-LE, and PGRP-SA (with its cofactor GNBP1) in glial cells using repo-Gal4 and determined their efficacy in inducing an antimicrobial response, by measuring the expression of four AMPs known to act with some specificity on Gram-positive bacteria [Defensin (Def), Metchnikowin (Met)] or against Gram-negative bacteria [Attacin-D (AttD), Diptericin B (DptB)] (33), and thus more associated with the Toll or Imd pathways, respectively. The induction of PGRP-LC and PGRP-LE appeared differentially effective (see Materials and Methods, Tables 1 and 2, and fig. S1, A and B). Pan-glial expression of PGRP-LC results in an up to 15,000-fold induction of AMPs (AttD, DptB and Def, Mtk). Moreover, we noted a fivefold induction of Pvf2 in larval stages (fig. S1A). Later stages could not be analyzed because of lethality caused by PGRP-LC expression. Glial expression of PGRP-LE is less effective (fig. S1B) and causes an up to 5000-fold up-regulation of AMP expression, which becomes less prominent during pupal stages (fig. S1B). In contrast, PGRP-SA weakly induces expression of Pvf2 in larval stages and expression DptB and Mtk in pupal stages (fig. S1C). Together, our results indicate that immune activation can be triggered in the Drosophila CNS through pan-glial expression of different PGRPs.
Table 1. Fly stocks.
| Stock | Source | Identifier |
| w[*]; P{w[+mC] = UAS-PGRP-LE.FLAG}2 | Bloomington Stock Center | 33054 |
| y[1] w[*]; P{w[+mC] = UAS-PGRP-LC.x}1 | Bloomington Stock Center | 30919 |
| w[*]; P{y[+t7.7] w[+mC] = 10XUAS-mCD8-GFP}attP2 | Bloomington Stock Center | 32184 |
| w[*]; P{w[+mC] = UAS-mCD8.ChRFP}2 | Bloomington Stock Center | 27391 |
| w[1118]; P{w[+mC] = XP}Pvf2[d02444] | Bloomington Stock Center | 19631 |
| w[*]; P{w[+mC] = UAS-FLAG-Rel.68}1; TM2/TM6C, Sb[1] | Bloomington Stock Center | 55777 |
| w[*]; P{w[+mC] = UASp-Tl.PA-GFP}3/CyO | Bloomington Stock Center | 30900 |
| y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.HM04020}attP2 | Bloomington Stock Center | 31713 |
| y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.HM05154}attP2 | Bloomington Stock Center | 28943 |
| y[1] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.HMJ23540}attP40/CyO | Bloomington Stock Center | 61955 |
| y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = TRiP.HMS04479}attP40 | Bloomington Stock Center | 57035 |
| y[1] sc[*] v[1] sev[21]; P{y[+t7.7] v[+t1.8] = TRiP.HMC03539}attP2 | Bloomington Stock Center | 53310 |
| w[*]; P{w[+mC] = tubP-GAL80[ts]}10; TM2/TM6B, Tb[1] | Bloomington Stock Center | 7108 |
| w[*];P{w[+mC] = UASpalphatub84B.Dendra2} 1 M/TM3, Sb[1] | Bloomington Stock Center | 51316 |
| w[*];P{w[+mC] = UASalphatub84B.Dendra2}5 M | Bloomington Stock Center | 51315 |
| y[1] w[*]; P{w[+m*] = nSyb-GAL4.S}3 | Bloomington Stock Center | 51635 |
| w[*]; P{w[+mC] = GAL4-elav.L}3 | Bloomington Stock Center | 8760 |
| GMR14F11-Gal4 | Bloomington Stock Center | 48653 |
| P{GMR90C03-GAL4}attP2 | Bloomington Stock Center | 47122 |
| w[1118]; P{w[+m*] = GAL4}repo/TM3, Sb[1] | Bloomington Stock Center | 7415 |
| w[1118]; P{y[+t7.7] w[+mC] = GMR54C07-lexA}attP40 | Bloomington Stock Center | 61562 |
| w[*]; P{w[+mC] = nrv2-GAL4.S}3; P{nrv2-GAL4.S}8 | Bloomington Stock Center | 6797 |
| w[*]; P{y[+t7.7] w[+mC] = 13XLexAop2-mCD8-GFP}attP2 | Bloomington Stock Center | 32203 |
| UAS-PGRP-SA | This study | |
| UAS-GNBP1 | This study | |
| vasPhiC31; attP40; attP2 | Gift from S. Luschnig | |
| Sp/CyO; srpHemo-moe::3xmCherry | Gift from D. Siekhaus | (34) |
| srpHemo-moe::3xmCherry /II;PrDr/TM3 | Gift from D. Siekhaus | (34) |
| Sp/CyO; srpHemo-H2A::3xmCherry | Gift from D. Siekhaus, A | (34) |
| Rec KM1 15a 68b Hml ∆RFP /CyO | Gift from K. Brückner | |
| Tret1-1-Gal4 | Gift from S. Schirmeier | |
| Tret1-1-Gal80 | This study | |
| w[1118]; P{w[+mC] = UAS-GFP.dsRNA.R}143 | Bloomington Stock Center | 9331 |
| nrxIV454 | (85) | |
| alrm-Gal80 | (71) | |
| nrv2-Gal80 | Gift from S. Rodrigues | |
| moody-Gal80 | (86) | |
| GMR90C03-Gal80 | (86) | |
| moody-Gal4 B1 | (87) | |
| moody-Gal4 B4 | (87) | |
| Gliotactin-Gal4 [rL82-Gal4] | (88) | |
| NP2222-Gal4 | (89) | |
| NP3233-Gal4 | (89) | |
| alrm-Gal4 | (70) | |
| repo-Gal4DBD | Gift of N. Pogodalla | |
| 83E12-Gal4AD | Gift of N. Pogodalla |
Table 2. TaqMan gene expression assays for qPCR.
| Gene | TaqMan gene expression assay |
| RpL32 | Dm02151827_g1 |
| Pvf2 | Dm01814370_m1 |
| PGRP-LE | Dm01839231_g1 |
| PGRP-LC | Dm01798314_m1 |
| Relish | Dm02134843_g1 |
| Toll | Dm02151201_g1 |
| PGRP-SA | Dm01837989_g1 |
| Dif | Dm01810798_g1 |
| Dorsal | Dm01810803_g1 |
| Attacin-B | Dm02135981_g1 |
| Diptericin B | Dm01821557_g1 |
| Defensin | Dm01818074_s1 |
| Metchnikowin | Dm01821460_s1 |
Pan-glial expression of PGRP-LC and PGRP-LE but not PGRP-SA triggers macrophage invasion into the brain
Next, we asked whether the immune response triggered by pan-glial expression of PGRPs was also associated with recruitment of macrophages. To detect invading macrophages, we compared existing promoter fusions that all direct expression in macrophages. The hml-dsRed reporter shows a very weak general expression in the CNS (fig. S2, A and A′), and flies carrying this reporter have reduced viability upon pan-glial immunity induction (see below). The srpHemo-H2A::3XmCherry reporter (34) not only provides a strong nuclear labeling of macrophages but also labels a subset of CNS glia (fig. S2, B and B′). srpHemo-moe::3XmCherry (34) gives an excellent staining of macrophages and also labels only six thoracic neurons (Fig. 2A, asterisk, and fig. S2, C and C′). This construct was therefore used in subsequent experiments, if not indicated otherwise.
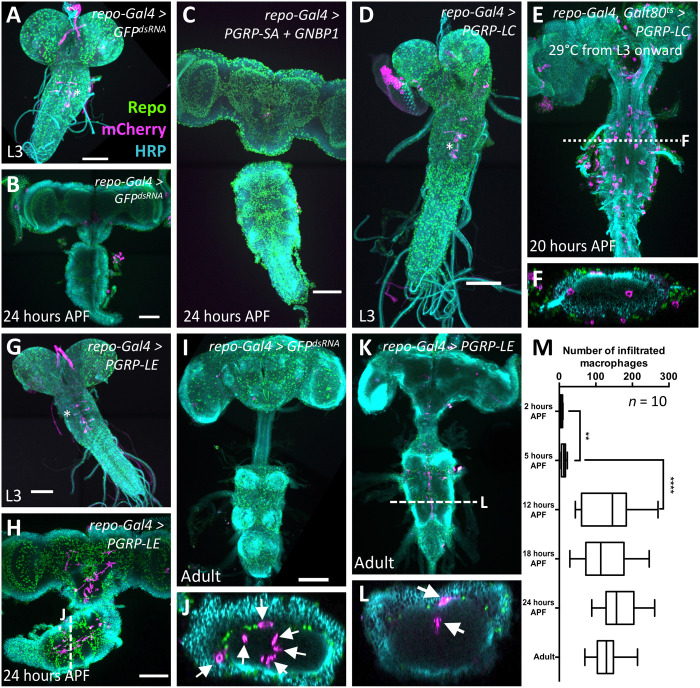
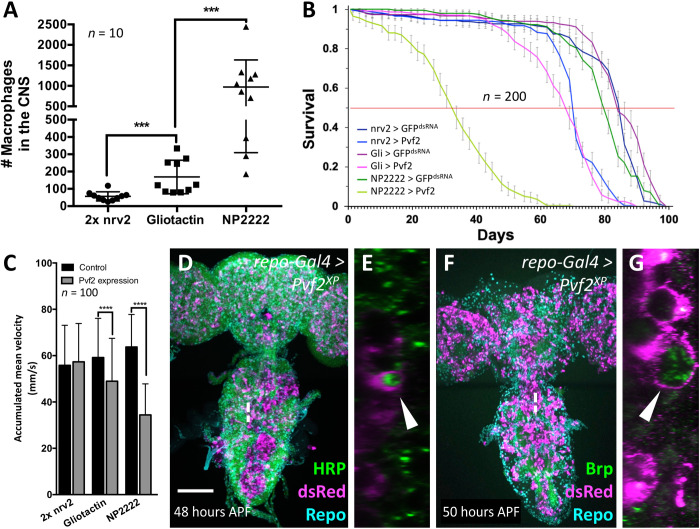
Fig. 2. Macrophages enter the brain during pupal stages upon immunity induction.
Dissected brains of indicated age stained for Repo (green) to label glial nuclei, HRP (cyan) to label neuronal membranes, and mCherry expression (magenta) directed by the macrophage marker srpHemo-moe::3xmCherry. wL3, wandering third instar larva. Adults were 7 days old. Scale bars, 100 μm. (A) Control larva expressing double-stranded RNA directed against green fluorescent protein (GFP). The asterisk indicates six neurons expressing the macrophage marker srpHemo-moe::3xmCherry. (B) Control pupa expressing double-stranded RNA directed against GFP. (C) Pupal brain expressing PGRP-SA and GNBP1 in glial cells. Note the absence of mCherry-expressing cells in the brain. (D) Larval brain expressing PGRP-LC in glial cells, no macrophages are found in the brain. The asterisk denotes neurons weakly expressing the marker srpHemo-moe::3xmCherry. (E) PGRP-LC expression is restricted to late larval and pupal stages. Macrophages invade the brain. The dashed line indicates the orthogonal section shown in (F). (G) Larva with PGRP-LE induction in glial cells. Note the elongated ventral nerve cord. The asterisk indicates neurons weakly expressing the macrophage marker. (H and J) Pupal brain expressing PGRP-LE. Note the presence of srpHemo-moe::3xmCherry–expressing cells in the brain. The white dashed line indicates the position of the orthogonal section shown in (J) (arrows indicate macrophages). (I) Adult control brain expressing GFPdsRNA. (K and L) Adult brain with immunity induction in glia. Cherry-expressing cells are found in the brain. The white dashed line indicates the orthogonal section shown in (L) (arrows indicate macrophages). (M) Quantification of the infiltration rate. To count the number of Cherry-positive cells, we used the srpHemo-H2A::3xmCherry marker. Pupae of increasing age were dissected, and the number of Cherry-expressing cells was determined using Imaris. P values are **P2–5 hours APF = 0.0034 and ****P5–12 hours APF < 0.0001 (t test); n = 10 for every time point.
First, we tested whether expression of PGRP-SA is sufficient to trigger macrophage invasion into the CNS. Since PGRP-SA requires the cofactor GNBP1 (18, 19, 21), we generated corresponding upstream activating sequence (UAS)–based transgenes and expressed them in the background of the srpHemo-moe::3XmCherry reporter in all glial cells. This expression regime does not lead to lethality and is not sufficient to trigger macrophage recruitment or invasion in larvae or pupae (Fig. 2, A to C).
Next, we determined whether PGRP-LC expression could trigger macrophage invasion of the brain. Pan-glial expression of PGRP-LC resulted in larval lethality with smaller brain lobes and no associated macrophages (Fig. 2D). To restrict pan-glial expression of PGRP-LC to the wandering third instar larval stage and early pupae, we used a temperature-sensitive Gal80 (35) (repo-Gal4, tub-Gal80ts, UAS-PGRP-LC, and srpHemo-moe::3XmCherry). This shifts the lethal phase to late pupal stages, and in these conditions, macrophages are notably found around or inside the neuropil of the pupal brain (Fig. 2, E and F). No invading macrophages are detected throughout development in the absence of the Gal4 driver.
Pan-glial expression of PGRP-LE does not cause larval lethality as expression of PGRP-LC, and female flies survive until adult stages. Upon pan-glial PGRP-LE expression, no macrophages were found in the larval brain but were found in the brain of early pupae, where they often exhibit an elongated shape (Figs. 2, G, H, and J, and 3). As observed following PGRP-LC expression, macrophages were mostly located around or inside the neuropil (Fig. 2J). No macrophages are found in the brain in the absence of a Gal4 driver.
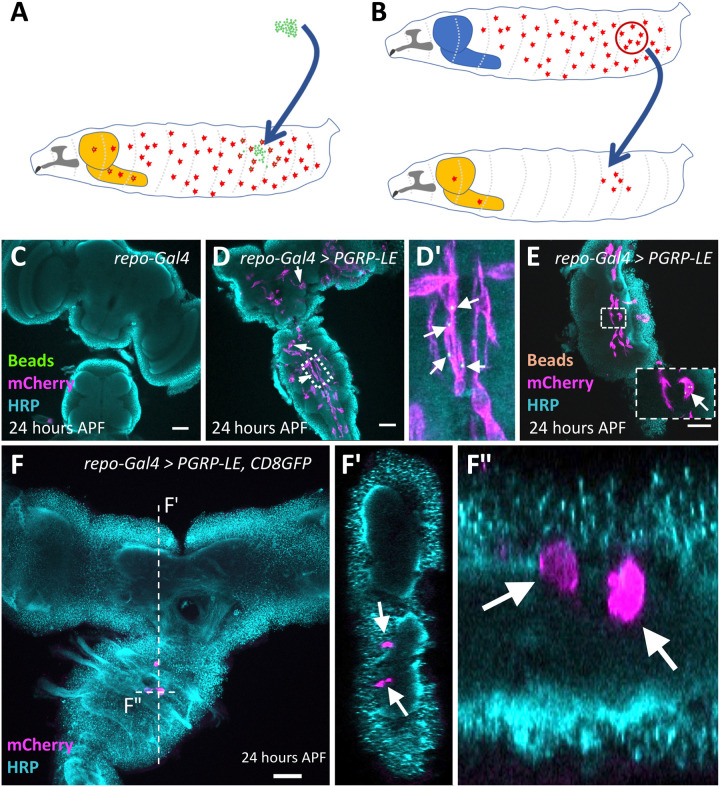
Fig. 3. Cherry-expressing cells originate from the hemolymph.
(A and B) Schematic view on the experimental strategies. The light orange–colored structure corresponds to the inflamed brain. The blue-colored brain reflects a control animal without infection. (A) Injection of fluorescent latex beads into the hemolymph of wandering third instar larvae. (B) Transplantation of macrophages expressing the srpHemo-moe::3xmCherry into the hemolymph of wandering third instar larvae. (C to F) Twenty-four hours APF pupal brains were stained for horseradish peroxidase (HRP) (cyan) and mCherry (magenta). (C) Control brain with no immunity induction. (D and E) Following pan-glial immunity induction, macrophages containing 1-μm green latex beads (D and D′) (arrows indicate macrophages containing latex beads) or 0.5-μm orange latex beads (E) (see arrows) are found in the CNS. (F and F″) Pupal brain after immunity induction and transplantation of genetically labeled macrophages (arrows). Several labeled macrophages are found in the ventral nerve cord. The position of the orthogonal sections shown in (F’) and (F″) is indicated by a white dashed line. Scale bars, 100 μm.
In conclusion, glial expression of either PGRP-LC or PGRP-LE is able to trigger invasion of macrophages, whereas expression of PGRP-SA is not able to recruit macrophages into the brain. On the basis of quantitative real-time polymerase chain reaction (qPCR) data, PGRP-LC and PGRP-LE trigger a similar immunity response, although PGRP-LC is much stronger and causes early lethality when expressed in a pan-glial manner (fig. S1). To avoid the use of an additional Gal80ts construct, we therefore continued our analysis using PGRP-LE–induced immunity.
Invading macrophages originate from the hemolymph
To determine when macrophages entered the pupal brain, we followed macrophage invasion over time. A strong increase in the number of macrophages located within the brain is noted during 5 and 12 hours after puparium formation (APF), but no further increase is seen afterward (Fig. 2, I and K to M). The number of macrophages also remains constant after eclosion, suggesting that no additional macrophages invade the CNS in adult stages (Fig. 2M).
In Drosophila, macrophages and glial cells have distinct origins but share expression of the master regulatory gene glial cells missing (36, 37). Thus, pan-glial expression of PGRP-LC or PGRP-LE might cause a transformation of the fate of resident glial cells toward a macrophage-like fate. To test whether the srpHemo-moe::3XmCherry–expressing cells found in the pupal brain are respecified glial cells or originate from the hemolymph, we performed two types of experiments. First, we injected 0.5-μm (orange) or 1-μm (green) large fluorescent latex beads into the hemocoel of late wandering larvae (Fig. 3A). These latex beads are taken up by macrophages within several hours but cannot be metabolized and, thus, remain within the cytoplasm where they can serve as a lineage marker. In a complementary set of experiments, we conducted transplantation experiments and grafted srpHemo-moe::3XmCherry–expressing late wandering third instar larvae macrophages into the hemolymph of late wandering third instar larvae expressing PGRP-LE in all glial cells (Fig. 3B).
Upon injection of fluorescently labeled latex beads into larvae expressing PGRP-LE in all glial cells, we found macrophages within the pupal brain that contained one or two of these beads (Fig. 3, C to E, arrows). This suggests that beads were endocytosed in the hemolymph before invasion of macrophages into the CNS. This was also corroborated by the results of the homochronic transplantation experiments. After the transplantation of srpHemo-moe::3XmCherry–expressing but otherwise wild-type macrophages into animals that expressed PGRP-LE in all glial cells, we found one to five Cherry-expressing macrophages in the CNS in 50% of the brains analyzed (n = 45 experiments; Fig. 3F). Together, these data suggest that hemolymph-resident macrophages are able to migrate across the BBB upon immunity induction in glial cells.
Macrophages are routed to the CNS via Pvf2
Pan-glial expression of all immunity receptors (PGRP-LC, PGRP-LE, and PGRP-SA) induces expression of Pvf2 in larval stages, with PGRP-LE being the most efficient inductor of Pvf2 expression, especially during early pupal stages (fig. S1). Likewise, Pvf2 is not significantly up-regulated in 12- to 18-hour APF pupae, which correlates to the finding that macrophage invasion terminates after 12 hours APF. Thus, we reasoned that Pvf2 expression downstream of PGRP-LE might mediate the attraction of macrophages into the brain.
To analyze whether Pvf2 expression downstream of PGRP-LE is required for invasion of macrophages, we suppressed Pvf2 expression in the background of pan-glial overexpression of PGRP-LE. Whereas PGRP-LE expression robustly attracts macrophages across the BBB, concomitant silencing of Pvf2 efficiently suppressed invasion of macrophages (Fig. 4, A to C). This indicates that the Imd pathway triggers recruitment of macrophages to the CNS via the induction of Pvf2 expression.
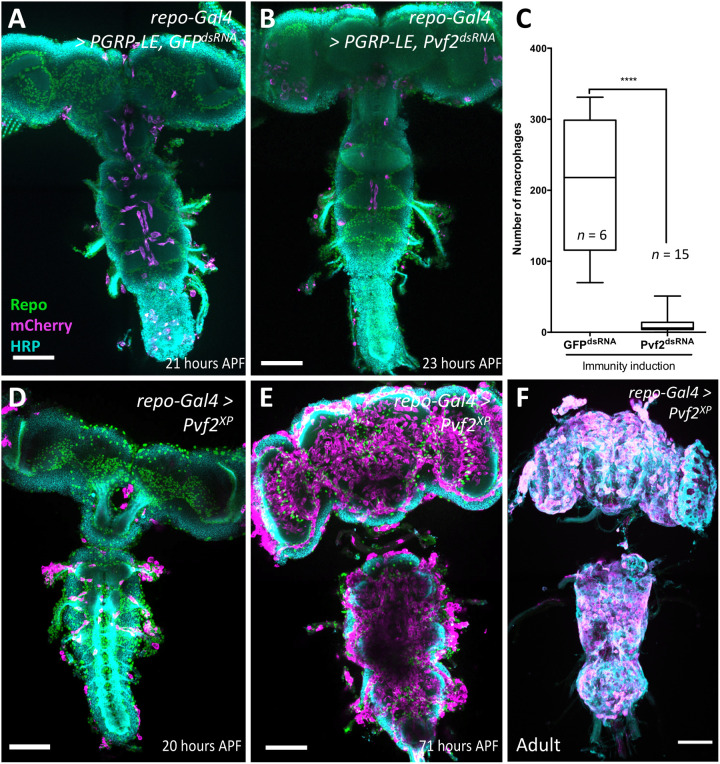
Fig. 4. Pvf2 acts downstream of PGRP-LE to direct macrophage invasion.
(A) Pupal brain with pan-glial immunity induction. Note the presence of macrophages in the CNS. (B) Pupal brain with pan-glial immunity induction and concomitant silencing of Pvf2. Note that fewer macrophages are found in the CNS. (C) Quantification of the number of invading macrophages using srpHemo-H2A::3xmCherry marker in brains with immunity induction without [see (A)] and with RNA interference–mediated Pvf2 knockdown [see (B)] (****P < 0.0001, Mann-Whitney). (D) Pan-glial expression of Pvf2 triggers the invasion of srpHemo-moe::3xmCherry–expressing macrophages into the nervous system 20 hours APF. The macrophages appear to follow axon tracts toward the neuropil. (E) Upon pan-glial Pvf2 expression, the number of macrophages invading the brain increases further during later pupal stages (71 hours APF). (F) Upon pan-glial Pvf2 expression, macrophages are still present in the adult brain (20-day-old female brain). Scale bars, 100 μm.
To test whether Pvf2 is sufficient for macrophage invasion, we next expressed Pvf2 in all glial cells. We noted a very pronounced invasion of macrophages into the pupal brain but not in the larval brain, which appeared stronger than what is observed following immunity induction by PGRP-LE expression [compare Fig. 2 (H to L) with Fig. 4 (D to F)]. Macrophages persist in the adult brain, and in addition, many macrophages are found attached to the nervous system, which is not observed following immunity induction [compare Fig. 2 (K and L) with Fig. 4F]. No invading macrophages were found in the absence of a Gal4 driver.
Expression of Pvf2 unexpectedly also induced a moderate but significant up-regulation of PGRP-LC expression during larval stages (fig. S3A). In addition, we noted increased expression of several AMPs in pupal stages, which might be a secondary effect of the invading macrophages (Fig. 4).
To further elucidate how macrophages enter the nervous system, we performed an electron microscopic analysis. In pupal brains expressing Pvf2 in all glial cells at 12 hours APF, we found macrophage-like cells that can be recognized by numerous intracellular vesicles (fig. S4, Mø), navigating between perineurial glial cells and contacting subperineurial glial cells that still have intact septate junctions (SJ) (fig. S4).
We next tested whether expression of Pvf2 is sufficient to trigger invasion of macrophages into the adult CNS. We performed Gal80ts experiments where we restricted Pvf2 expression either to pupal or to adult stages only. Macrophages were found in the CNS upon pupal expression, but no macrophages could be detected when expression of Pvf2 was confined to the adult stages. This suggests that a window of competence for invasive migration of macrophages exists only during early pupal stages (fig. S3, B to D).
Macrophages are routed to the CNS via Imd-dependent Pvf2 expression independent of c-Jun N-terminal kinase signaling
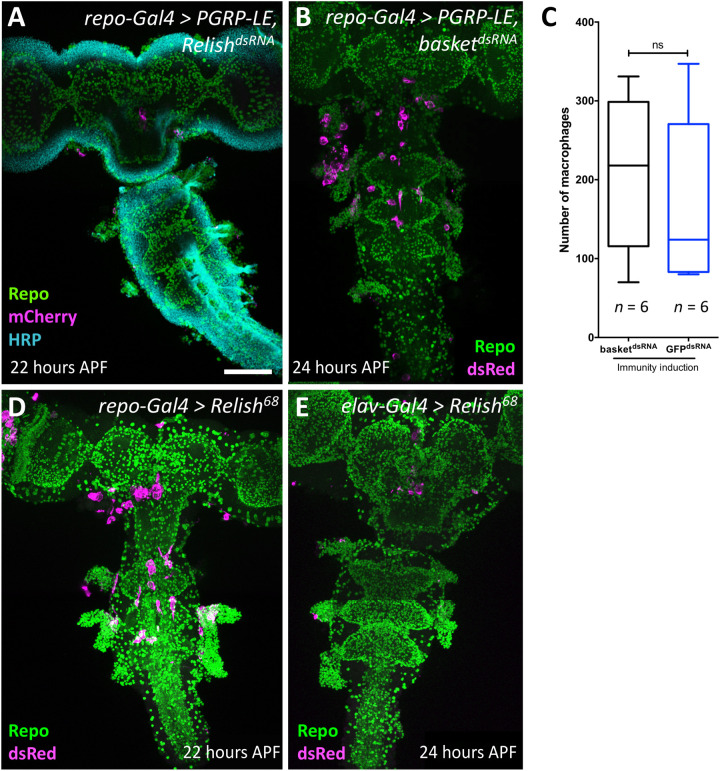
It has been recently demonstrated that activation of the Imd pathway drives c-Jun N-terminal kinase (JNK)–dependent expression of the Pvr ligand Pvf2 (15). To elucidate the signaling cascade downstream of PGRP-LE, we silenced expression of Relish, encoding a NF-κB–type transcription factor, and basket, encoding a protein kinase regulating the JNK pathway, which is also implicated in immunity induction (38), concomitantly to immunity induction. Whereas suppression of Relish blocked invasion of macrophages (n = 6; Fig. 5A), no block of macrophage invasion was observed upon suppression of the JNK pathway (n = 7; Fig. 5, B and C). Moreover, expression of activated Relish (39) in glial cells, but not in neurons, is sufficient to trigger invasion of macrophages into the brain (Fig. 5, D and E). Activation of the Toll pathway (Fig. 1C) by expression of PGRP-SA together with GNBP1 or by silencing cactus expression or by overexpression of Toll is not sufficient to recruit macrophages into the CNS (Fig. 2C and fig. S5, A and B). In conclusion, our results suggest that the Imd pathway can trigger expression of Pvf2 via Relish to guide migration of macrophages across the BBB.
Fig. 5. Relish is required for macrophage invasion.
(A, B, D, and E) Twenty-two to 24-hour-old pupal brains stained as indicated. Glial nuclei (anti-Repo, green), neuronal membranes (anti-HRP, cyan), and invading macrophages (mCherry or dsRed, magenta). Scale bar, 100 μm. (A) Upon concomitant expression of PGRP-LE and silencing of relish expression using RNA interference, no macrophages enter the brain. (B and C) No suppression of macrophage invasion induced by PGRP-LE expression is observed following concomitant suppression of basket expression. For quantification, see (C) (n = 6; P = 0.6753, Mann-Whitney). ns, not significant. (D) Pan-glial expression of activated Relish causes an invasion of macrophages into the brain. (E) Neuronal expression of activated Relish is not sufficient to trigger invasion of macrophages into the CNS.
Several glial cell types attract macrophages across the BBB
To test the relevance of the different glial cell layers, we used a set of specific Gal4 and Gal80 lines that allow targeting of specific glial cell types (see Materials and Methods, Table 3).
Table 3. Expression patterns of different driver lines.
| Driver | Expression pattern |
| repo-Gal4 | All glia (CNS and peripheral nervous system) |
| nrv2-Gal4 | All glia except perineurial and subperineurial glia |
| Tret1-1-Gal4 | Perineurial glia |
| moody-Gal4 B1 | Subperineurial glia (strong) |
| moody-Gal4 B4 | Subperineurial glia (weak) |
| Gliotactin-Gal4 | Subperineurial glia and very few neurons |
| NP2222-Gal4 | Cortex glia |
| alrm-Gal4 | Astrocyte-like glia |
| NP3233-Gal4 | Astrocyte-like glia |
| GMR83E12-Gal4 | Ensheathing glia |
| nSyb-Gal4 | Neurons (strong) |
| elav-Gal4 | Neurons |
| GMR14F11 | Mushroom bodies |
| GMR90C03-Gal4 | All glia of the CNS |
| nrv2-Gal4, GMR90C03-Gal80 | Peripheral wrapping glia |
| GMR54C07-LexA | Subperineurial glia |
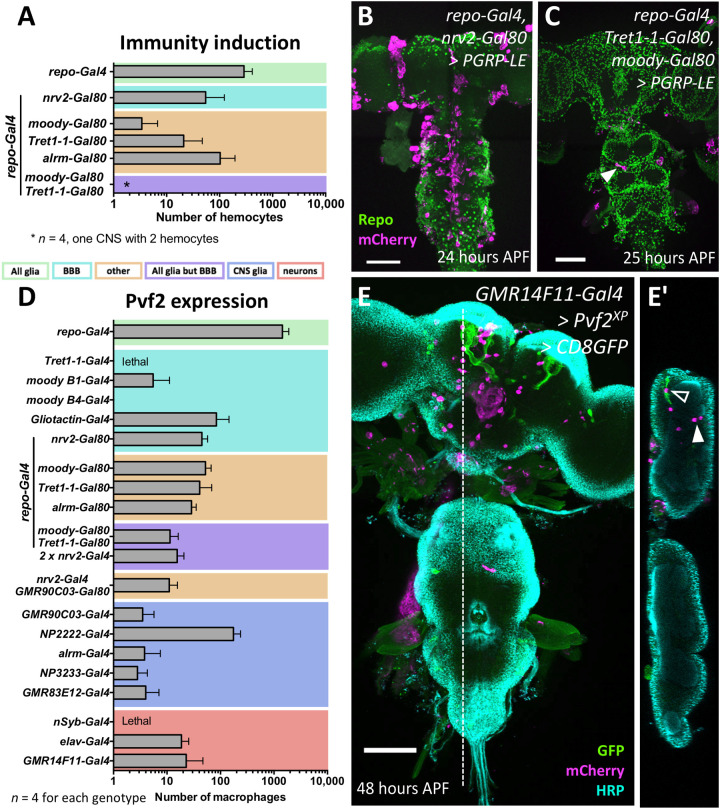
When we expressed PGRP-LE in all glial cells, about 150 macrophages were found in each pupal brain (Fig. 6A). Expression in cortex glia, ensheathing glia, or astrocyte-like glia was not required for invasion (Fig. 6, A and B). However, when we excluded PGRP-LE expression from the cells of the BBB by combining repo-GAL4 with Tret1-1-Gal80 expressed in perineurial glia or moody-Gal80 expressed in subperineurial glia, the number of invading macrophages is reduced by 90 to 95% (Fig. 6, A and C). Unexpectedly, when we induced expression of PGRP-LE only in any one of the glial cell types individually (using Tret1-1-Gal4, or Gli-Gal4, or NP2222-Gal4, or alrm-Gal4, or 83E12-Gal4, or nrv2-Gal4; see Materials and Methods, Table 3) or in all neurons (elav-Gal4), no invasion of the brain by macrophages was noted (fig. S5C). Although pan-glial expression of PGRP-LC results in larval lethality, expression of PGRP-LC in subperineurial glial cells using Gliotactin-Gal4 is able to trigger invasion of few macrophages into the nervous system (fig. S5D). In conclusion, these experiments show that immunity induction in the BBB-forming glial cells appears most relevant during this recruitment, but additional glial cell types are likely to be involved, too.
Fig. 6. Cell type–specific induction of immunity induces invasion of macrophages.
(A) Pan-glial activation of immunity response triggers invasion of many macrophages into the CNS (n = 10 for repo>>PGRP-LE; n = 4 for all other genotypes). When pan-glial PGRP-LE expression was blocked in various glial subtypes, the number of invading macrophages is reduced. The color coding of the different glial subtypes is indicated below. For further information about the different Gal4 drivers, see Materials and Methods, Table 3. (B) Concomitant silencing in cortex and ensheathing glia using nrv2-Gal80 causes a similar reduction in the number of invading macrophages as concomitant silencing in astrocyte-like glial cells using alrm-Gal80. (C) Upon PGRP-LE expression in all glial cells but the BBB, only very few macrophages entered the brain (arrowhead) (repo-Gal4, moody-Gal80, and Tret1-1-Gal80 UAS-PGRP-LE). (D) Average number of invading macrophages in different expression regimes (n = 10 for repo>>Pvf2; otherwise, n = 4). Color coding is as in (A). In all expression regimes, macrophages enter the brain, except for moodyB4-Gal4–driven Pvf2 expression. (E) Notably, even expression of Pvf2 in only few neurons (GMR14F11-Gal4 is active in the mushroom bodies only, shown by concomitant expression of UAS-CD8-GFP), is able to recruit macrophages into the brain lobes [open arrowhead indicates macrophage associated with mushroom body, and filled arrowhead indicates macrophage located in some distance (E and E′)]. Scale bars, 100 μm.
Above, we showed that PGRP-LE requires Pvf2 expression to induce macrophage infiltration into the brain. We therefore expressed Pvf2 in different glial cell types and neuronal cell types (Fig. 6D). Expression of Pvf2 using the Tret1-1-Gal4 driver resulted in lethality. Expression of Pvf2 in the subperineurial glial cells using moody-Gal4 or Gliotactin-Gal4 triggered infiltration of macrophages into the nervous system. However, when we expressed Pvf2 in all glial cells but the glial cells of the BBB (or subperineurial or perineurial glia separately), we still noted infiltration of the brain (Fig. 6D). Moreover, expression of Pvf2 in cortex or in ensheathing, or in astrocyte-like glia is able to trigger infiltration of macrophages. When macrophages are attracted to the brain by Pvf2 expression in subperineurial glial cells, they nevertheless migrated toward the neuropil, suggesting that macrophage attraction and migration within the CNS are controlled by different signaling pathways.
While high-level expression of Pvf2 in neurons using nsyb-Gal4 results in larval lethality, lower expression of Pvf2 with elav-Gal4 also triggers invasion of a moderate number of macrophages into the CNS. Likewise, we were able to recruit macrophages into the brain using the mushroom body–specific Gal4 driver GMR14F11-Gal4 (Fig. 6, D and E). In this case, macrophages are mostly found close to the mushroom body neuropil. In summary, these data suggest that infiltration of the brain by macrophages can be regulated by neuronal and glial cells.
The BBB is intact during development
Macrophages are able to invade the CNS mostly in the first 12 hours of pupal development. During this time, the animal does not move, and thus, the BBB might be open, allowing entry of macrophages upon immunity induction. To directly determine whether the integrity of the BBB is compromised during this time period, we performed dye penetration experiments using fluorescently labeled dextran of 10- and 70-kilodalton (kDa) size on differentially aged animals. In all cases, filet preparations were performed with great caution, avoiding any disruption of the CNS or the different segmental nerves. The specimens were opened at the dorsal midline, and the gut and adhering fat body were carefully removed by gentle pipetting. Labeled dextran was added, and penetration into the CNS was recorded for 40 min. Seventy kDa labeled dextran was efficiently excluded in all developmental stages tested (wandering third instar larvae, 5, 24, and 48 hours APF) (fig. S6, A and B). Penetration of 10-kDa labeled dextran increased in the first 24 hours APF but decreased again between 24 and 48 hours APF (fig. S6, C and D). The lack of penetration of 70 kDa suggests that the BBB remains intact during early pupal development. The slight penetration of 10-kDa dextran might indicate that septate junctions that normally provide the exclusion of solutes open transiently.
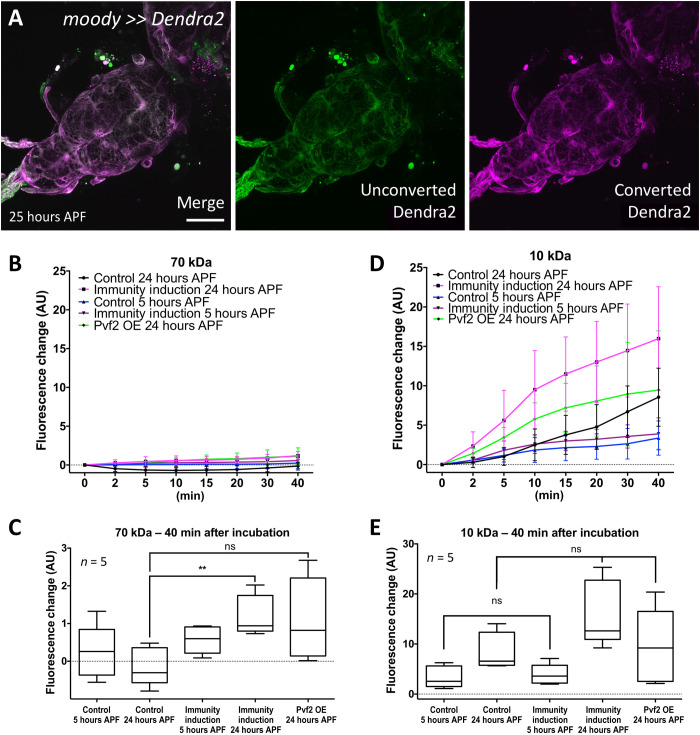
To test whether the BBB-forming subperineurial glial cells are present throughout development, we expressed CD8-GFP using the subperineurial glia–specific enhancer element GMR54C07 (40) (GMR54C07-LexA LexAOP-CD8-GFP). Throughout development, green fluorescent protein (GFP) expression is detected in a sheath around the nervous system, indicating that a BBB is present in all developmental stages (fig. S7). In addition, we expressed the photoconvertible protein Dendra2 in subperineurial glia, which we photoconverted in wandering third instar larvae and imaged at 25 hours APF. Since the photoconverted Dendra2 protein remains in the subperineurial glia, this indicates that the BBB does not disintegrate in the first 25 hours of pupal development (Fig. 7A).
Fig. 7. Immunity induction does not affect BBB integrity.
(A) The BBB-forming cells are not replaced during onset of pupal development. Confocal image of a 24- to 25-hour APF pupal brain. Larvae with the genotype (moody-Gal4 UAS-tub-Dendra2) were subjected to photoconversion of the subperineurial glia covering the ventral nerve cord. The resultant red fluorescent Dendra2 protein can be detected in pupal brain 25 hours APF. Scale bar, 100 μm. (B) Quantification of dye uptake experiments using fluorescein-labeled 70-kDa dextran in control (repo-Gal4, UAS-GFPdsRNA) and after pan-glial immunity induction (repo-Gal4, UAS-PGRP-LE). Datasets were obtained 5 and 24 hours APF. In addition, we included 24-hour APF pupal brains expressing Pvf2. AU, arbitrary units. OE, overexpression. (C) Quantification of changes in fluorescence uptake after 40 min. A slight but significant increase in fluorescein-labeled dextran can be detected following immunity induction (**P = 0.0079) but not following Pvf2 expression (P = 0.0556). (D) Quantification of dye uptake experiments using Texas Red–labeled 10-kDa dextran in control and after pan-glial immunity induction. The same genotypes and time points as in (A) were used. A large variability of data points was found resulting in large error bars. In all genotypes analyzed, an increase in Texas-red–labeled dextran in the CNS can be detected. (E) Quantification of changes in Texas-red uptake after 40 min. A significant increase in Texas-red–labeled dextran was found for control brains (P = 0.0317) and those with a pan-glial immunity induction (P = 0.0079). However, the levels of Texas-red–labeled dextran were not significantly different between control and pan-glial immunity induction (P5hAPF = 0.667 and P24hAPF = 0.095). Likewise, no differences in Texas-red–labeled dextran uptake were noted in 24-hour APF brains expressing Pvf2 (P = 0.944). n = 5.
In a next step, we assayed the tightness of the BBB following immunity induction by pan-glial PGRP-LE expression. Here, we chose 5 and 24 hours APF and detected a slight but significant increase in BBB permeability at 24 hours APF for 70-kDa dextran (P = 0.0079; Fig. 7, B and C). This increase in BBB permeability could be determined since 70-kDa dextran does not penetrate in the control, unlike the smaller 10-kDa dextran. Here, we also noted a slight increase in permeability compared with the control; however, possibly because of the general leakage noted in the control, this increase was not significant (P = 0.0952; Fig. 7, D and E). Similar results were obtained following pan-glial expression of Pvf2. The slight increase in dextran permeability upon immunity induction or Pvf2 expression might indicate that transmigration of macrophages across the BBB or other signaling consequences of the activation of these pathways disrupt the integrity of the subperineurial glial cells in a locally and temporally confined manner.
Macrophage invasion reduces longevity and causes brain damage
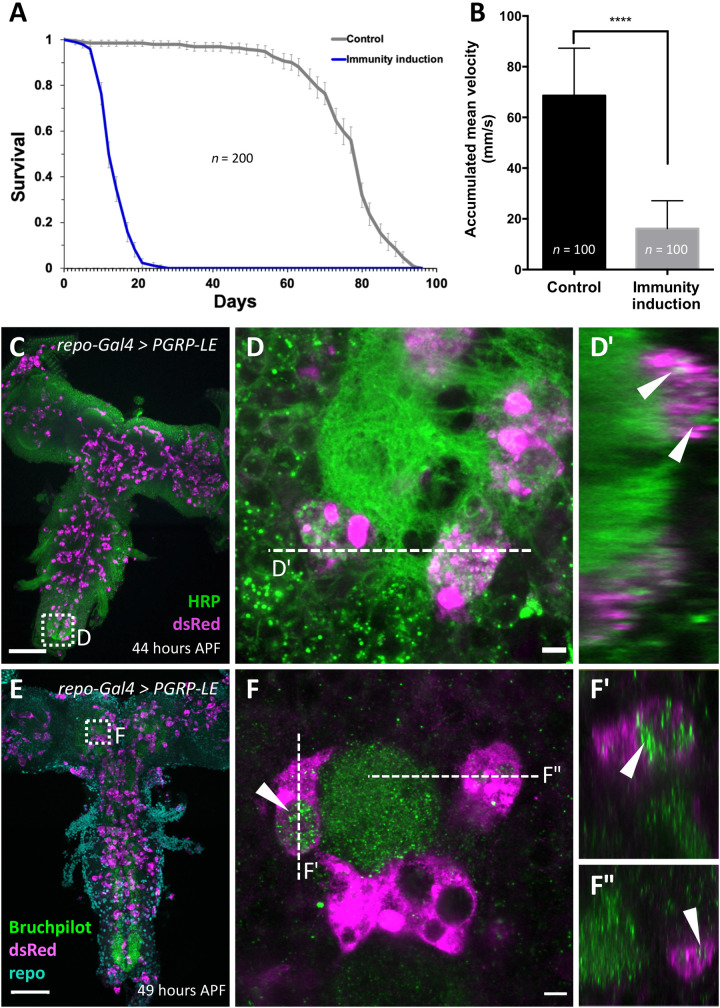
Immune cell invasion often contributes to severity of the human brain diseases. Above, we have shown that macrophage invasion can be a consequence of immunity induction. Upon pan-glial PGRP-LE expression, adult flies eclose, allowing us to assess functional consequences. These flies exhibit a markedly reduced life span and die on average after 11 days (Fig. 8A). Already after 1 week, flies with immunity induction show reduced locomotor abilities. The rapid iterative negative geotaxis (RING) assay exploits the negative geotaxis shown by adult flies (41). The climbing ability of 1-week-old control flies (repo-Gal4 UAS-CD8-Cherry) is almost four times as pronounced as the ability of 1-week-old flies with a pan-glial immunity induction (Fig. 8B). Thus, expression of PGRP-LE triggers reduced longevity and climbing abilities and macrophage invasion into the brain. To test whether macrophages might be responsible for the observed fitness deficits, we stained specimens with horseradish peroxidase (HRP), labeling all neuronal membranes (42) or Bruchpilot (Brp), a prominent synaptic marker (43, 44). Both HRP and Brp signal was frequently found inside invading macrophages in the neuropil, suggesting that these cells also phagocytose parts of neuronal cells including synaptic material (Fig. 8, C to F).
Fig. 8. Pan-glial immunity induction affects longevity and climbing ability.
(A) Longevity of flies with pan-glial immunity induction compared to control flies (repo-Gal4, UAS-CD8Cherry versus repo-Gal4 UAS-PGRP-LE). The viability is markedly reduced (n = 200 females; P = 3.75 × 10−92). (B) Same genotypes as in (A). The climbing ability of 7-day-old flies is markedly reduced upon immunity induction (****P < 0.0001). (C to F) Two-day-old pupae with immunity induction stained for macrophages using [hml∆dsRed] and subsequent anti-dsRed staining, and either anti-HRP (green) (C and D) to label neuronal membranes or anti-Brp (green) (E and F) to label synapses. Note that macrophages harbor vesicles containing neuronal membrane material [arrowheads in (D′), (F′), and (F″)]. Scale bars, 100 μm (C and E) and 5 μm (D and F).
To rule out that immunity induction causes neuronal deficits by other means, we analyzed animals with different Pvf2 expression regimes characterized by different numbers of macrophages in the CNS (Fig. 9A). Upon Pvf2 expression using nrv2-Gal4, on average, 57 macrophages are found in the CNS, whereas expression directed using Gli-Gal4 resulted in on average 168 macrophages and expression using NP2222-Gal4 resulted on average in 971 macrophages in the brain (n = 10 brains each; Fig. 9A). Depending on the expression regime, we noted differential effects on longevity. Animals expressing the highest amount of Pvf2 had the shortest life expectancy (Fig. 9B). Likewise, we noted a decrease in climbing abilities depending on the level of Pvf2 expression (Fig. 9C). Macrophages that are recruited to the brain via Pvf2 expression are still able to phagocytose neuronal material as shown for macrophages recruited to the brain via immunity induction (Fig. 9, D and E). This suggests that the loss of synapses caused by invading macrophages is responsible for the observed reduction in longevity and the loss of locomotor abilities, but further experiments are required to rule out that Pvf2 overexpression has general detrimental effects in neurons.
Fig. 9. CNS resident macrophages affect survival and phagocytose neuronal membranes.
(A) Number of macrophages invading the brain of 1-week-old adult flies expressing Pvf2 with different glial cell–specific Gal4 drivers. The P values are ***Pnrv2 vs. Gli = 0.0001 and ***Pgli vs. NP2222 = 0.0002 (Mann-Whitney). (B) Same genotypes as in (A). Longevity of female flies with glial subtype–specific Pvf2 expression compared to control flies. The corresponding Gal4 driver element is indicated in the figure. For control, we expressed GFPdsRNA. The viability is reduced upon Pvf2 expression in all glial subtypes tested and inversely correlates with the number of invading macrophages (n = 200 females in groups of 20 each; ****Pnrv2 = 4.425 × 10−32, ****PGli = 5.2984 × 10−41, and ****PNP2222 = 1.9936 × 10−89). (C) Same genotypes as in (A). The climbing ability of 7-day-old females is reduced upon Pvf2 expression and again inversely correlates with the number of invaded macrophages (Pnrv2 = 0.237, ****PGli < 0.0001, and ****PNP2222 < 0.0001). (D to G) Two-day-old pupae with pan-glial Pvf2 expression stained for macrophages using [hml∆dsRed] and subsequent anti-dsRed staining, and either anti-HRP (green) (D and E) to label neuronal membranes or anti-Brp (green) (F and G) to label synapses. Note that macrophages harbor vesicles containing neuronal membrane material (arrowheads). Scale bar, 100 μm.
CNS infection by GBS is accompanied by an immune response and triggers macrophage entry
The above data suggest that activation of the Imd pathway is sufficient to trigger macrophage invasion into the pupal brain. We wondered whether these findings were transferable to a pathological condition known to be associated with macrophage infiltration into the CNS.
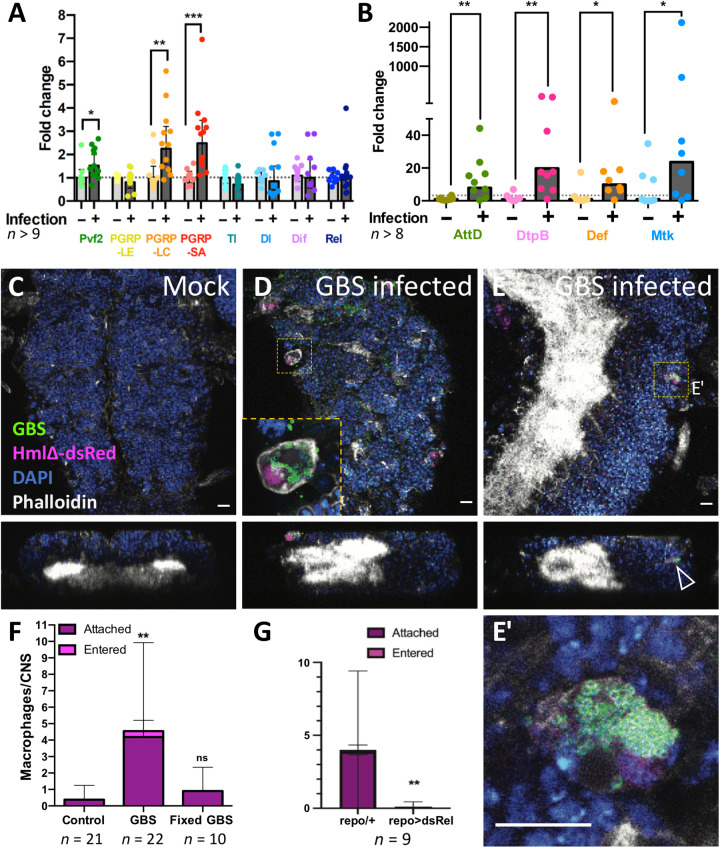
We recently developed a model of brain infection in Drosophila and showed that Gram-positive GBS bacteria were able to enter the Drosophila larval brain (30). We thus studied whether GBS infection of the CNS elicits similar antibacterial responses as determined for PGRP overexpression. We injected GBS into the hemolymph of late third instar larvae that stopped feeding and started wandering (see Materials and Methods). GBS-injected larvae survived up to 5 hours after infection (30), and we were able to perform qPCR experiments on brains dissected 4 to 5 hours after injection. We observed more than a twofold up-regulation of PGRP-LC and PGRP-SA (Fig. 10A), which activate the Imd or Toll pathway, respectively. In addition, we noted a 50% up-regulation of Pvf2. While Toll, as well as dorsal (dl), Dorsal-related immunity factor (Dif), and Relish (Rel), is not up-regulated (Fig. 10A), the expression of the four AMPs studied above (Def, Met, AttD, and DptB) is significantly up-regulated upon GBS infection (Fig. 10B). These data suggest that infection with GBS triggers an immune response within the CNS, where macrophages might be recruited via PGRP-LC activity.
Fig. 10. GBS infection leads to immunity induction and macrophage recruitment in early pupal brains.
(A and B) Wandering third instar Drosophila larvae injected with GBS were dissected 4 to 5 hours after infection, and the brains were subjected to qPCR. Expression of the genes PGRP-LC, PGRP-SA, Pvf2, and all AMPs tested (AttD, DtpB, Def, and Mtk) was up-regulated upon infection. Mann-Whitney test for Pvf2, *P = 0.0227; PGRP-LC, **P = 0.0024; PGRP-SA, ***P = 0.0002; AttD, **P = 0.0055; DtpB, **P = 0.0025; Def, *P = 0.0207; Mtk, *P = 0.0499. Unpaired t test for PGRP-LE, P = 0.5297; Tl, P = 0.0988; Dl, P = 0.6202; Dif, P = 0.6758. (C to E) Macrophage recruitment and infiltration to GBS-infected pupal brains. Confocal images (top and orthogonal views) showing Drosophila CNS of hml-dsRed pupae 4 to 5 hours after injection in the hemolymph of either (C) Mock or (D and E) GBS. Macrophages (hml-dsRed in magenta) containing GBS (anti-GBS in green) are detected attached to the CNS (D, inset) or inside the CNS (E). (E′) is a close-up of the dotted boxes from (E). The arrowhead points to a macrophage within the CNS cortex. Phalloidin is in white, and 4′,6-diamidino-2-phenylindole (DAPI) is in blue. Scale bars, 10 μm. (F) Quantification of macrophage localization in the whole CNS 4 to 5 hours after injection in the hemolymph of mock (control), GBS, or formaldehyde-fixed GBS. Kruskal-Wallis test followed by Dunn’s multiple comparisons test on brain-associated (attached + entered) macrophages was performed generating adjusted P values: **P(control vs. GBS) = 0.0011 and P(control vs. fixed-GBS) = 0.7165. Control, n = 21 CNS; GBS, n = 22 CNS; fixed-GBS, n = 10 CNS. (G) Number of srpHemo-moe::3xmCherry–expressing macrophages that attach to the brain upon GBS infection of repo-Gal4 animals and repo-Gal4>UAS-relishdsRNA animals [n = 9; **P = 0.0078, Mann-Whitney on brain-associated (attached + entered) macrophages].
To test whether GBS infection also leads to a recruitment of macrophages, we used the hml-dsRed marker to follow macrophages during infection (45), in control and GBS-injected wandering third instar larvae, and analyzed early pupal brains 5 hours after injection. In mock-injected larvae, only few macrophages are attached to the CNS, and no macrophages were ever found inside the brain (Fig. 10, C and F). Upon GBS injection, the number of macrophages attached to the CNS surface increased more than 10-fold (Fig. 10, D to F), indicating that they are attracted by the infected tissue. Notably, we also detected macrophages inside the CNS, which did not show morphological hallmarks characteristic of cell death (Fig. 10, E and E′). Macrophages were filled with bacteria in line with their primary role as phagocytic immune cells (Fig. 10, D and E′). In addition, we used formaldehyde-fixed GBS as control. In this context, macrophages are not recruited (Fig. 10F). This implies that live bacteria are required for macrophage recruitment to the brain. To test whether relish is also required for macrophage recruitment in the GBS infection model as it is in the PGRP-LE overexpression situation, we silenced relish expression in glial cells, which indeed blocked macrophage recruitment in response to GBS infection (Fig. 10G). This shows that the Imd pathway is necessary for macrophage recruitment to the brain in response to GBS infection.
To test whether other bacterial pathogens could elicit a similar immune response in the CNS, we performed ex vivo infection assays using Pseudomonas aeruginosa, a Gram-negative species. While we were able to detect bacteria attached to the brain, P. aeruginosa does not appear to enter the CNS (fig. S8, A to F). P. aeruginosa infection is also not able to trigger increased expression of PGRP-LC, PGRP-LE, and PGRP-SA. In addition, expression of AMPs is not increased by infection with P. aeruginosa but rather appeared to be decreased for AttD and Def (fig. S8G). This difference in response between the two pathogens could be due to the Gram status, the bacterial species per se, the difference in neurotropism, or the in vivo versus ex vivo systems.
To conclude, we show that both GBS and P. aeruginosa reach the CNS, while only GBS appears able to enter this tissue. Notably, in response to GBS invasion, Drosophila immune cells can overcome the BBB via glial activation of the Imd pathway, a situation reminiscent to our findings using an overexpression system.
DISCUSSION
Here, we establish a novel model of neuroinflammation in Drosophila using forced expression of PGRPs. This paradigm is capable of mounting an inflammatory reaction as seen by the induction of AMP expression and results in a recruitment of macrophages to the brain. Similarly, we show that GBS bacteria present in the hemolymph can enter the brain to elicit an antimicrobial response, which also triggers macrophage invasion from the hemolymph to the CNS, although at very reduced numbers. GBS brain infection unexpectedly activates both the Toll and the Imd pathways, the latter being associated more classically with infection of Gram-negative bacteria. Thus, immunity reactions in the brain may differ from the remaining body. In agreement with this notion, we failed to elicit a clear activation of the Imd pathway following infection with Gram-negative P. aeruginosa but rather observed the activation of Toll pathway components.
To enter the brain, GBS move across the BBB in a low-density lipoprotein receptor LpR2-dependent manner (30). Since infection also develops in an ex vivo system, GBS do not appear to use macrophages as Trojan horses to cross the BBB. This barrier is formed by glial cells and stays intact during pupal stages. These glial cells express PGRPs (46) that bind bacterial surface proteins and induce the Imd immunity pathway. Moreover, upon GBS infection, PGRP-LC and PGRP-SA expression is increased.
While PGRP-SA is involved in Toll pathway activation, its expression, together with its cofactor GNBP1, failed to mount an invasion of the brain by macrophages. Correlating with this, we noted a lack of Pvf2 induction by PGRP-SA/GNBP1 in pupal stages. In contrast, PGRP-LC and PGRP-LE are both involved in the Imd pathway, and its activation is sufficient and necessary to attract macrophages across an intact BBB during pupal stages. It has been shown that PGRP-LC is activated by a diaminopimelic acid type of peptidoglycans (47), which are only found on Gram-negative bacteria and not present on GBS, and it is thus unclear how PGRP-LC (or PGRP-LE) might affect the Imd pathway. However, it has been recently shown that PGRP-LC can also bind Lys-type peptidoglycans, which are typically found on Gram positive bacteria (48). Moreover, overexpression of PGRP-LE in the absence of any bacteria is also sufficient to trigger Imd pathway activation.
In Drosophila pupae, macrophages reside in the hemolymph. As vertebrate microglia, they are not derived from neural progenitors but originate from the mesoderm. Both Drosophila lineages, macrophages, and glial cells require the activation of the transcription factor Glial cells missing (36, 49). Moreover, macrophages and glial cells both express receptors recognizing apoptotic cells or bacteria: Draper, Simu, and PGRPs. Only the Croquemort receptor, which recognizes Gram-positive cells and apoptotic cells, is specifically expressed by macrophages (12). To exclude that glial expression of PGRP-LE induces a fate switch that forces specific CNS glial cells to adopt a hemocyte-like fate as suggested before (50), we conducted transplantation experiments using genetically marked macrophages. In addition, we labeled macrophages in the hemolymph by injecting fluorescently labeled latex beads while mounting an immunity response. These data clearly demonstrate that macrophages are recruited to the CNS from the hemolymph.
Recruitment of macrophages to the brain is mostly occurring in the first 12 hours of pupal development. During this stage, ecdysone-triggered histolysis starts, which also leads to a disintegration of larval tissues including the gut (51). Thus, during early pupal stages, bacteria have a higher chance to escape from the gut to possibly invade the remaining tissues. In addition, ecdysone triggers motility of macrophages during exactly this period (52). Moreover, the extracellular matrix that surrounds the entire nervous system (53) is remodeled during pupal development. For remodeling, macrophages adhere to the existing matrix and thus are in the position to easily enter the brain provided that the respective immunity signal is presented. Further studies are needed to understand the molecular mechanisms constraining invasive migration to the first hours of pupal life.
Invasive migration of macrophages in stage 12 embryos and invasive migration of border cells in the fly ovary require expression of Pvf2/3 (54, 55–57). In addition, Pvf2/3 acts as a survival factor, is able to stimulate macrophage proliferation, and is thought to act as a chemoattractant for macrophages in the embryonic CNS (58–63). Here, we demonstrated that PGRP-LC and PGRP-LE both activate the NF-κB protein Relish in glial cells to trigger Pvf2-dependent invasion of macrophages into the brain. A similar relish-dependent Pvf2 activation has been described for Drosophila cells of the adult midgut (17).
Notably, expression of Pvf2 in astrocyte-like or ensheathing glia that reside deep within the brain, as well as in neurons, is sufficient to induce the invasion of the brain by macrophages, which raises the questions of what the attractive signal is and how it reaches the macrophages. Pvf2 expressed by neuropil-associated glia is not expected to leave the CNS due to the BBB. The highest expression of Pvr in the adult brain is in the BBB itself (46). Thus, secretion of Pvf2 from neurons or glial cells within the CNS might activate Pvr in these glial cells to control transmigration of macrophages across the BBB.
During normal development of the fly embryo, macrophages migrate across a tissue barrier in a defined time window depending on Minerva, an atypical major facilitator superfamily member (56, 63). Here, we have shown that the BBB stays intact in early pupal stages, which is the window of invasiveness. However, we noted subtle but significant changes in the permeability of smaller dextran molecules during this phase, in a similar fashion in wild type and under immunity induction. This suggests that the BBB is transiently opened similar to what has been described for the moody mutant phenotype (64, 65). moody mutants have disrupted septate junction strands that compromise barrier function (66). To compensate this defect, BBB-forming subperineurial glial cells form additional interdigitations that increase the length of the paracellular diffusion path and thereby restore the barrier function (66). However, when we activated the immunity response in adult moody mutants, still no macrophages entered the brain. Thus, compromised septate junction strand formation does not explain alone how macrophages enter the brain during early pupal stages.
Phagocytosis in the nervous system must be a carefully controlled and well-balanced process. In the noninfected Drosophila brain, phagocytosis of neuronal debris is mediated by glial cells. The regulation of glial responses to neuronal damage is largely encoded by the engulfment receptor Draper, which promotes phagocytosis through an immunoreceptor tyrosine–based activation motif (ITAM) (5, 67–72). Glial cells might be able to cope with bacterial infection but, in addition, trigger invasive migration of macrophages that eventually will clear invading bacteria. The activity of macrophages is, however, not restricted to bacteria and is also initiated toward synapses, and this overshooting phagocytosis might contribute to decreased longevity of the fly.
The influence of macrophages in the brain might be generally relevant to pathological conditions associated with increased neuroinflammation and immune activation in this tissue. The human neurodegenerative disease ataxia-telangiectasia (A-T) is due to mutation of the Ataxia Telangiectasia Mutated (ATM) kinase that is involved in control of genomic integrity and cell cycle. Knockdown of the Drosophila ATM homolog in glial cells causes an A-T–like neurodegeneration model associated with reduced mobility and longevity. Concomitantly, an elevated expression of the Imd pathway was observed, and inhibition of relish but not dif blocks neurodegeneration (73, 74). This might suggest that reduced mobility and longevity that is observed in many neurodegenerative disease models in Drosophila are possibly in part caused by mounting an immune signal that leads to invading macrophages.
In vertebrates, it has been well documented that inflammatory conditions in the CNS trigger the extravasation of circulating lymphocytes and macrophages across the BBB (6). The infiltration of leukocytes into the CNS is an essential step in the pathogenesis of multiple sclerosis (75–77). Breach of the BBB also occurs during bacterial infection, such as meningitis, when local immune cells recruit circulating monocytes into the CNS (78). In both cases, minimizing macrophage access to the brain via the BBB is crucial for preventing brain damage. Likewise, it has been shown that activation of microglia can contribute to synapse loss (79, 80), again indicating the importance of carefully balanced phagocytotic activity within the nervous system. In the future, studies using Drosophila might contribute to understanding how macrophages can breach the BBB and how their phagocytotic activity can be regulated to minimize CNS tissue damage.
MATERIALS AND METHODS
All experiments were conducted according to the regulations of the German or French legislation. An approval by an ethics committee is not required for Drosophila experiments.
Fly work
All flies were raised according to standard procedures at 25°C unless otherwise noted. Flies were obtained as specified in Table 1. Crosses including a tub-Gal80ts were raised at 18°C and shifted at the indicated time to 30°C to allow Gal4-directed expression. Transgenes generated in this study were inserted the landing site attP40 (81) or in the landing site attP2 (82) using standard phiC31-integration protocols (83). The negative geotaxis assay (RING assay) was performed as described with 10 females per vial that was tapped five times in a custom-made setup to insure identical forces (41). To determine the life span of flies, 10 times 20 one-day-old mated females were kept in individual vials and were transferred three times a week. The number of dead flies was monitored throughout the experiment.
Dextran uptake assay
White prepupae were collected and kept in a moist chamber at 25°C until dissection. Filet preparations were performed in phosphate-buffered saline (PBS) by carefully cutting dorsally from the posterior to the anterior site avoiding to contact the brain using a Vannas scissors. Filets were fixed on a glass slide using heptane glue. Texas Red–conjugated dextran (1.6 mM; 10 kDa) and fluorescein-conjugated dextran (1.6 mM; 70 kDa) in H2O were applied simultaneously to the filet, and dextran uptake was recorded for 40 min on a Zeiss 880 LSM. The same laser settings were used throughout the experiments. Quantification and statistical analyses were performed using Fiji, Excel, and Prism.
Drosophila in vivo brain infection by GBS
An overnight GBS preculture was set from glycerol stocks in Brain Heart Infusion broth (BHI) at 37°C. The bacterial preculture was diluted 1:20 in BHI and was grown for 3 hours at 37°C. GBS concentration was calculated by OD600 (optical density at 600 nm) correlation [1 OD600 = 8.8 × 108 colony-forming units (CFU)/ml], and the bacteria were pelleted through 5-min centrifugation at 3500g (at 4°C) and washed twice in PBS and twice in Drosophila Schneider’s medium, and then suspended in 100 μl of Schneider’s medium (reaching 4.4 × 1010 GBS/ml). Twenty nanoliters of concentrated GBS was injected in wandering third instar larvae using the nanoinjector Nanoject III (Drummond Scientific) to reach 8.8 × 108 CFU/ml of hemolymph. Injected late third instar larvae were kept on standard fly food plates at 30°C, where most of them enter pupariation around 2 hours after infection.
Drosophila ex vivo brain infection by P. aeruginosa
An overnight PA14 preculture was set from glycerol stocks in LB at 37°C. The bacterial preculture was diluted 1:20 in LB and was grown for 3 hours at 37°C. Ex vivo brain infections were performed as described in (30). Briefly, ~109 CFU/ml was diluted 1:10 in the brain explant culture medium I [Drosophila Schneider’s medium (Gibco, 217200-24) supplemented with 2 mM l-glutamine (Gibco, 25030-032) and 0.5 mM sodium l-ascorbate (Sigma-Aldrich, A4034)]. Brain explants were infected for 3 hours at 30°C under agitation. After 3 hours, the culture medium I was replaced by culture medium II [culture medium I supplemented with 1% fetal bovine serum (Sigma-Aldrich, F4135)] for another 3 hours.
Immunohistochemistry
For Immunohistochemistry, whole brains for all developmental stages were dissected, fixed, and stained using standard protocols. White prepupae were collected in a moist chamber and kept at 25°C until the desired age. The following antibodies were used: anti-Brp (1:100; NC82, Developmental Studies Hybridoma Bank), anti-dsRed (1:1000; Takara), anti-GFP (1:500; Abcam), anti-HRP DyLight 647 conjugated (1:500; Dianova), anti-HRP fluorescein isothiocyanate conjugated (1:500; Thermo Fisher Scientific), anti-mCherry (1:1000; Invitrogen), anti-NrxIV (1:1000) (84), anti-Repo (1:2000; gift from B. Altenhein, Cologne), and anti-GBS (30). All secondary antibodies (Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 647 coupled; 1:1000) were obtained from Invitrogen.
For bacterial infection, pupal brains were dissected at indicated time points and fixed for 20 min in 4% methanol–free formaldehyde at room temperature, washed in PBS 3 × 10 min, and permeabilized in PBT (PBS and 0.3% Triton X-100) 3 × 10 min. Brains were then incubated with rabbit anti-GBS at 4°C in blocking solution (PBT, 5% bovine serum albumin, and 2% goat serum) for 18 to 36 hours, washed with PBT, incubated with secondary antibody for 3 hours at room temperature in blocking solution, and washed with PBT. Samples were incubated with Phalloidin–Atto 647N (Sigma-Aldrich, 65906) overnight at 4°C and mounted in Mowiol mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific, 62247). All images were acquired using a Zeiss LSM 880.
Electron microscopic analyses
Pupal filets were dissected in 4% formaldehyde and fixed in 4% formaldehyde and 0.5% glutaraldehyde at room temperature overnight. The next day, filets were fixed in 2% OsO4 for 1 hour on ice and after washing with ultrapure H2O and stained in 2% uranyl acetate at room temperature for 30 min. Following dehydration, specimens were embedded in epon. Ultrathin sections were obtained using a Leica UC7 Ultramicrotome and were imaged on a Zeiss TEM900 equipped with an SIS Morada digital camera.
Transplantation and bead injection
For bead injection and transplantation, appropriate wandering third instar larvae were collected and immobilized by incubation on ice. A Nanoject II (Drummond Scientific) was used for both experiments. For bead injection, larvae were injected into the posterior lateral side with fluorescently labeled latex beads (Sigma-Aldrich, #L4655 and #L5530) diluted 1:100 in PBS. After injection, larvae were placed in a fresh vial with standard food and kept at room temperature. Pupae were collected, dissected, and analyzed. For transplantation, hemolymph of larvae with labeled macrophages was aspirated and directly injected into larvae with a pan-glial immunity induction. Injected larvae were placed on fresh food, and pupae were dissected 40 to 46 hours after injection.
Molecular genetics
Tret1-1-Gal80 was cloned using an entry clone containing the promoter region of Tret1-1 (gift from S. Schirmeier) and a Gateway plasmid containing the Gal80 open reading frame. For cloning of UAS-PGRP-SA and UAS-GNBP1, the open reading frames of both genes were amplified from pupal complementary DNA (cDNA) using 5′-CACCATGCAGCCGGTTCGATT as forward and 5′-TTAGGGATTTGAGAGCCAGTGC as reverse primer for PGRP-SA, and 5′-CACCATGCCAGGATTGTGCA as forward and 5′-TCAGTTGGCGAAGACACGAACA as reverse primer for GNBP1. Both amplified genes were inserted into the pENTR/D-TOPO vector, followed by a subsequent gateway reaction into the pUAST-attB-rfa vector.
To determine transcriptional changes following bacterial infection and immunity induction, qPCR was performed. RNA was isolated from wandering third instar larvae or pupal brains with a pan-glial immunity induction. For bacterial infections, the brain was dissected 4 hours after inoculation. RNA was isolated using RNeasy mini (QIAGEN), and cDNA was synthesized using QuantiTect (QIAGEN) according to the manufacturer’s instructions. qPCR for all samples was performed using a TaqMan gene expression assay in a StepOne Real-Time PCR System (Thermo Fisher Scientific; see Table 2) together with a TaqMan Universal PCR Master Mix II (Life Technologies). RpL32 was used as a housekeeping gene. For all samples, a minimum of four biological replicates were analyzed. For expression patterns directed by all Gal4 lines used, see Table 3.
Quantification and statistical analysis
Infiltrated macrophages were quantified using Imaris (version 8.4.1, Oxford Instruments). Statistical analysis and calculations for qPCR analysis were acquired using Excel and Prism 6.0. Figure 10 represents pooled data of independent experiments. Normality/lognormality was tested using the D’Agostino–Pearson test. We analyzed each gene independently as pairs. For the normal values, we performed a t test, and for the values not following a normal law, we chose Mann-Whitney.
Acknowledgments
We are indebted to D. Siekhaus, K. Brückner, B. Lemaitre, S. Schirmeier, and N. Pogodalla for sharing many fly strains. We are thankful to D. Briand for help during GBS infection and to L. Krift for help during the transplantation experiments. We are thankful to T. Zobel for help with macrophage counting and J. Stewen for help with the qPCR. We thank S. Rumpf and members of the Klämbt lab for critical reading of the manuscript and help throughout the project.
Funding: This work was supported by an H. Hertz fellowship to B.W., the German research foundation (DFG SFB 1009) to C.K., and a starting package from Institut Pasteur/LabEx Revive to P.S.
Author contributions: Conceptualization: B.W., P.S., M.A.L., and C.K. Methodology: B.W., D.F., B.B., and S.R. Investigation: B.W., D.F., B.B., and S.R. Writing (original draft): B.W. and C.K. Writing (review and editing): B.W., B.B., P.S., and C.K. Supervision: P.S. and C.K. Funding acquisition: P.S. and C.K.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
REFERENCES AND NOTES
- 1.Niederkorn J. Y., See no evil, hear no evil, do no evil: The lessons of immune privilege. Nat. Immunol. 7, 354–359 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Saijo K., Glass C. K., Microglial cell origin and phenotypes in health and disease. Nat. Rev. Immunol. 11, 775–787 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Hilu-Dadia R., Kurant E., Glial phagocytosis in developing and mature Drosophila CNS: Tight regulation for a healthy brain. Curr. Opin. Immunol. 62, 62–68 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Logan M. A., Glial contributions to neuronal health and disease: New insights from Drosophila. Curr. Opin. Neurobiol. 47, 162–167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Logan M. A., Hackett R., Doherty J., Sheehan A., Speese S. D., Freeman M. R., Negative regulation of glial engulfment activity by Draper terminates glial responses to axon injury. Nat. Neurosci. 15, 722–730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engelhardt B., Ransohoff R. M., Capture, crawl, cross: The T cell code to breach the blood-brain barriers. Trends Immunol. 33, 579–589 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Segrist E., Cherry S., Using diverse model systems to define intestinal epithelial defenses to enteric viral infections. Cell Host Microbe 27, 329–344 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ermolaeva M. A., Schumacher B., Insights from the worm: The C. elegans model for innate immunity. Semin. Immunol. 26, 303–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemaitre B., Hoffmann J., The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Davidson A. J., Wood W., Phagocyte responses to cell death in flies. Cold Spring Harb. Perspect. Biol. 12, a036350 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banerjee U., Girard J. R., Goins L. M., Spratford C. M., Drosophila as a genetic model for hematopoiesis. Genetics 211, 367–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melcarne C., Lemaitre B., Kurant E., Phagocytosis in Drosophila: From molecules and cellular machinery to physiology. Insect Biochem. Mol. Biol. 109, 1–12 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Lemaitre B., Reichhart J. M., Hoffmann J. A., Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. U.S.A. 94, 14614–14619 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myllymäki H., Valanne S., Rämet M., The Drosophila imd signaling pathway. J. Immunol. 192, 3455–3462 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Bond D., Foley E., A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLOS Pathog. 5, e1000655 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragab A., Buechling T., Gesellchen V., Spirohn K., Boettcher A.-L., Boutros M., Drosophila Ras/MAPK signalling regulates innate immune responses in immune and intestinal stem cells. EMBO J. 30, 1123–1136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sansone C. L., Cohen J., Yasunaga A., Xu J., Osborn G., Subramanian H., Gold B., Buchon N., Cherry S., Microbiota-dependent priming of antiviral intestinal immunity in Drosophila. Cell Host Microbe 18, 571–581 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pili-Floury S., Leulier F., Takahashi K., Saigo K., Samain E., Ueda R., Lemaitre B., In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J. Biol. Chem. 279, 12848–12853 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Gobert V., Gottar M., Matskevich A. A., Rutschmann S., Royet J., Belvin M., Hoffmann J. A., Ferrandon D., Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 302, 2126–2130 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Michel T., Reichhart J. M., Hoffmann J. A., Royet J., Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414, 756–759 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Bischoff V., Vignal C., Boneca I. G., Michel T., Hoffmann J. A., Royet J., Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 5, 1175–1180 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Kaneko T., Yano T., Aggarwal K., Lim J.-H., Ueda K., Oshima Y., Peach C., Erturk-Hasdemir D., Goldman W. E., Oh B.-H., Kurata S., Silverman N., PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat. Immunol. 7, 715–723 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Iatsenko I., Kondo S., Mengin-Lecreulx D., Lemaitre B., PGRP-SD, an extracellular pattern-recognition receptor, enhances peptidoglycan-mediated activation of the Drosophila Imd pathway. Immunity 45, 1013–1023 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Valanne S., Kallio J., Kleino A., Rämet M., Large-scale RNAi screens add both clarity and complexity to Drosophila NF-κB signaling. Dev. Comp. Immunol. 37, 9–18 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Gottar M., Gobert V., Michel T., Belvin M., Duyk G., Hoffmann J. A., Ferrandon D., Royet J., The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416, 640–644 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Gendrin M., Zaidman-Rémy A., Broderick N. A., Paredes J., Poidevin M., Roussel A., Lemaitre B., Functional analysis of PGRP-LA in Drosophila immunity. PLOS ONE 8, e69742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takehana A., Katsuyama T., Yano T., Oshima Y., Takada H., Aigaki T., Kurata S., Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. U.S.A. 99, 13705–13710 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takehana A., Yano T., Mita S., Kotani A., Oshima Y., Kurata S., Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 23, 4690–4700 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maillet F., Bischoff V., Vignal C., Hoffmann J., Royet J., The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe 3, 293–303 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Benmimoun B., Papastefanaki F., Périchon B., Segklia K., Roby N., Miriagou V., Schmitt C., Dramsi S., Matsas R., Spéder P., An original infection model identifies host lipoprotein import as a route for blood-brain barrier crossing. Nat. Commun. 11, 6106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hindle S. J., Munji R. N., Dolghih E., Gaskins G., Orng S., Ishimoto H., Soung A., DeSalvo M., Kitamoto T., Keiser M. J., Jacobson M. P., Daneman R., Bainton R. J., Evolutionarily conserved roles for blood-brain barrier xenobiotic transporters in endogenous steroid partitioning and behavior. Cell Rep. 21, 1304–1316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limmer S., Weiler A., Volkenhoff A., Babatz F., Klämbt C., The Drosophila blood-brain barrier: Development and function of a glial endothelium. Front. Neurosci. 8, 365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson M. A., Lemaitre B., New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 62, 22–30 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Gyoergy A., Roblek M., Ratheesh A., Valoskova K., Belyaeva V., Wachner S., Matsubayashi Y., Sánchez-Sánchez B. J., Stramer B., Siekhaus D. E., Tools allowing independent visualization and genetic manipulation of Drosophila melanogaster macrophages and surrounding tissues. G3 8, 845–857 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L., Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Bernardoni R., Vivancos V., Giangrande A., Glide/gcm is expressed and required in the scavenger cell lineage. Dev. Biol. 191, 118–130 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Bazzi W., Cattenoz P. B., Delaporte C., Dasari V., Sakr R., Yuasa Y., Giangrande A., Embryonic hematopoiesis modulates the inflammatory response and larval hematopoiesis in Drosophila. eLife 7, e34890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delaney J. R., Stöven S., Uvell H., Anderson K. V., Engström Y., Mlodzik M., Cooperative control of Drosophila immune responses by the JNK and NF-κB signaling pathways. EMBO J. 25, 3068–3077 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiklund M.-L., Steinert S., Junell A., Hultmark D., Stöven S., The N-terminal half of the Drosophila Rel/NF-κB factor Relish, REL-68, constitutively activates transcription of specific Relish target genes. Dev. Comp. Immunol. 33, 690–696 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Kremer M. C., Jung C., Batelli S., Rubin G. M., Gaul U., The glia of the adult Drosophila nervous system. Glia 65, 606–638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gargano J. W., Martin I., Bhandari P., Grotewiel M. S., Rapid iterative negative geotaxis (RING): A new method for assessing age-related locomotor decline in Drosophila. Exp. Gerontol. 40, 386–395 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Snow P. M., Patel N. H., Harrelson A. L., Goodman C. S., Neural-specific carbohydrate moiety shared by many surface glycoproteins in Drosophila and grasshopper embryos. J. Neurosci. 7, 4137–4144 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagh D. A., Rasse T. M., Asan E., Hofbauer A., Schwenkert I., Dürrbeck H., Buchner S., Dabauvalle M.-C., Schmidt M., Qin G., Wichmann C., Kittel R., Sigrist S. J., Buchner E., Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49, 833–844 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Kittel R. J., Wichmann C., Rasse T. M., Fouquet W., Schmidt M., Schmid A., Wagh D. A., Pawlu C., Kellner R. R., Willig K. I., Hell S. W., Buchner E., Heckmann M., Sigrist S. J., Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science 312, 1051–1054 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Makhijani K., Alexander B., Tanaka T., Rulifson E., Brückner K., The peripheral nervous system supports blood cell homing and survival in the Drosophila larva. Development 138, 5379–5391 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., Kreft L., Aibar S., Makhzami S., Christiaens V., Bravo González-Blas C., Poovathingal S., Hulselmans G., Spanier K. I., Moerman T., Vanspauwen B., Geurs S., Voet T., Lammertyn J., Thienpont B., Liu S., Konstantinides N., Fiers M., Verstreken P., Aerts S., A single-cell transcriptome atlas of the aging Drosophila brain. Cell 174, 982–998.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang C.-I., Chelliah Y., Borek D., Mengin-Lecreulx D., Deisenhofer J., Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 311, 1761–1764 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Vaz F., Kounatidis I., Covas G., Parton R. M., Harkiolaki M., Davis I., Filipe S. R., Ligoxygakis P., Accessibility to peptidoglycan is important for the recognition of Gram-positive bacteria in Drosophila. Cell Rep. 27, 2480–2492.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lebestky T., Chang T., Hartenstein V., Banerjee U., Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science 288, 146–149 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Stratoulias V., Heino T. I., MANF silencing, immunity induction or autophagy trigger an unusual cell type in metamorphosing Drosophila brain. Cell. Mol. Life Sci. 72, 1989–2004 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baehrecke E. H., Ecdysone signaling cascade and regulation of Drosophila metamorphosis. Arch. Insect Biochem. Physiol. 33, 231–244 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Sampson C. J., Amin U., Couso J. P., Activation of Drosophila hemocyte motility by the ecdysone hormone. Biol Open. 2, 1412–1420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olofsson B., Page D. T., Condensation of the central nervous system in embryonic Drosophila is inhibited by blocking hemocyte migration or neural activity. Dev. Biol. 279, 233–243 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Duchek P., Somogyi K., Jékely G., Beccari S., Rorth P., Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107, 17–26 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Parsons B., Foley E., The Drosophila platelet-derived growth factor and vascular endothelial growth factor-receptor related (Pvr) protein ligands Pvf2 and Pvf3 control hemocyte viability and invasive migration. J. Biol. Chem. 288, 20173–20183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valoskova K., Biebl J., Roblek M., Emtenani S., Gyoergy A., Misova M., Ratheesh A., Reis-Rodrigues P., Shkarina K., Larsen I. S. B., Vakhrushev S. Y., Clausen H., Siekhaus D. E., A conserved major facilitator superfamily member orchestrates a subset of O-glycosylation to aid macrophage tissue invasion. eLife 8, e41801 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ratheesh A., Biebl J., Vesela J., Smutny M., Papusheva E., Krens S. F. G., Kaufmann W., Gyoergy A., Casano A. M., Siekhaus D. E., Drosophila TNF modulates tissue tension in the embryo to facilitate macrophage invasive migration. Dev. Cell. 45, 331–346.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Munier A. I., Doucet D., Perrodou E., Zachary D., Meister M., Hoffmann J. A., Janeway C. A., Lagueux M., PVF2, a PDGF/VEGF-like growth factor, induces hemocyte proliferation in Drosophila larvae. EMBO Rep. 3, 1195–1200 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood W., Faria C., Jacinto A., Distinct mechanisms regulate hemocyte chemotaxis during development and wound healing in Drosophila melanogaster. J. Cell Biol. 173, 405–416 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gold K. S., Brückner K., Macrophages and cellular immunity in Drosophila melanogaster. Semin. Immunol. 27, 357–368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brückner K., Kockel L., Duchek P., Luque C. M., Rørth P., Perrimon N., The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell 7, 73–84 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Sears H. C., Kennedy C. J., Garrity P. A., Macrophage-mediated corpse engulfment is required for normal Drosophila CNS morphogenesis. Development 130, 3557–3565 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Siekhaus D., Haesemeyer M., Moffitt O., Lehmann R., RhoL controls invasion and Rap1 localization during immune cell transmigration in Drosophila. Nat. Cell Biol. 12, 605–610 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bainton R. J., Tsai L. T.-Y., Schwabe T., DeSalvo M., Gaul U., Heberlein U., Moody encodes two GPCRs that regulate cocaine behaviors and blood-brain barrier permeability in Drosophila. Cell 123, 145–156 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Schwabe T., Neuert H., Clandinin T. R., A network of cadherin-mediated interactions polarizes growth cones to determine targeting specificity. Cell 154, 351–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Babatz F., Naffin E., Klämbt C., The Drosophila blood-brain barrier adapts to cell growth by unfolding of pre-existing septate junctions. Dev. Cell 47, 697–710.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Ziegenfuss J. S., Biswas R., Avery M. A., Hong K., Sheehan A. E., Yeung Y.-G., Stanley E. R., Freeman M. R., Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature 453, 935–939 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Purice M. D., Ray A., Münzel E. J., Pope B. J., Park D. J., Speese S. D., Logan M. A., A novel Drosophila injury model reveals severed axons are cleared through a Draper/MMP-1 signaling cascade. eLife 6, e23611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Awasaki T., Tatsumi R., Takahashi K., Arai K., Nakanishi Y., Ueda R., Ito K., Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50, 855–867 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Doherty J., Logan M. A., Taşdemir O. E., Freeman M. R., Ensheathing glia function as phagocytes in the adult Drosophila brain. J. Neurosci. 29, 4768–4781 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tasdemir-Yilmaz O. E., Freeman M. R., Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons. Genes Dev. 28, 20–33 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.MacDonald J. M., Beach M. G., Porpiglia E., Sheehan A. E., Watts R. J., Freeman M. R., The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron 50, 869–881 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Petersen A. J., Katzenberger R. J., Wassarman D. A., The innate immune response transcription factor relish is necessary for neurodegeneration in a Drosophila model of ataxia-telangiectasia. Genetics 194, 133–142 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petersen A. J., Rimkus S. A., Wassarman D. A., ATM kinase inhibition in glial cells activates the innate immune response and causes neurodegeneration in Drosophila. Proc. Natl. Acad. Sci. 109, E656–E664 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trapp B. D., Nave K.-A., Multiple sclerosis: An immune or neurodegenerative disorder? Annu. Rev. Neurosci. 31, 247–269 (2008). [DOI] [PubMed] [Google Scholar]
- 76.O’Loughlin E., Madore C., Lassmann H., Butovsky O., Microglial phenotypes and functions in multiple sclerosis. Cold Spring Harb. Perspect. Med. 8, a028993 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goverman J., Autoimmune T cell responses in the central nervous system. Nat. Rev. Immunol. 9, 393–407 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang S., Peng L., Gai Z., Zhang L., Jong A., Cao H., Huang S.-H., Pathogenic triad in bacterial meningitis: Pathogen invasion, NF-κB activation, and leukocyte transmigration that occur at the blood-brain barrier. Front. Microbiol. 7, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sarlus H., Heneka M. T., Microglia in Alzheimer’s disease. J. Clin. Invest. 127, 3240–3249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salter M. W., Stevens B., Microglia emerge as central players in brain disease. Nat. Med. 23, 1018–1027 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Markstein M., Pitsouli C., Villalta C., Celniker S. E., Perrimon N., Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Groth A. C., Fish M., Nusse R., Calos M. P., Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stork T., Thomas S., Rodrigues F., Silies M., Naffin E., Wenderdel S., Klämbt C., Drosophila Neurexin IV stabilizes neuron-glia interactions at the CNS midline by binding to Wrapper. Development 136, 1251–1261 (2009). [DOI] [PubMed] [Google Scholar]
- 85.Edenfeld G., Volohonsky G., Krukkert K., Naffin E., Lammel U., Grimm A., Engelen D., Reuveny A., Volk T., Klämbt C., The splicing factor crooked neck associates with the RNA-binding protein HOW to control glial cell maturation in Drosophila. Neuron 52, 969–980 (2006). [DOI] [PubMed] [Google Scholar]
- 86.Kottmeier R., Bittern J., Schoofs A., Scheiwe F., Matzat T., Pankratz M., Klämbt C., Wrapping glia regulates neuronal signaling speed and precision in the peripheral nervous system of Drosophila. Nat. Commun. 11, 4491 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stork T., Engelen D., Krudewig A., Silies M., Bainton R. J., Klämbt C., Organization and function of the blood-brain barrier in Drosophila. J. Neurosci. 28, 587–597 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sepp K. J., Auld V. J., Conversion of lacZ enhancer trap lines to GAL4 lines using targeted transposition in Drosophila melanogaster. Genetics 151, 1093–1101 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Awasaki T., Lai S.-L., Ito K., Lee T., Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J. Neurosci. 28, 13742–13753 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8