The identification of a living relative of Sitting Bull with a new method opens the door to future genealogical research.
Abstract
A great-grandson of the legendary Lakota Sioux leader Sitting Bull (Tatanka Iyotake), Ernie LaPointe, wished to have their familial relationship confirmed via genetic analysis, in part, to help settle concerns over Sitting Bull’s final resting place. To address Ernie LaPointe’s claim of family relationship, we obtained minor amounts of genomic data from a small piece of hair from Sitting Bull’s scalp lock, which was repatriated in 2007. We then compared these data to genome-wide data from LaPointe and other Lakota Sioux using a new probabilistic approach and concluded that Ernie LaPointe is Sitting Bull’s great-grandson. To our knowledge, this is the first published example of a familial relationship between contemporary and a historical individual that has been confirmed using such limited amounts of ancient DNA across such distant relatives. Hence, this study opens the possibility for broadening genealogical research, even when only minor amounts of ancient genetic material are accessible.
INTRODUCTION
Tatanka Iyotake, also known as Sitting Bull, was leader of the Hunkpapa Lakota Sioux who lived from 1831 to 1890 (1). For many years, he led his people in a fight against the policies of the United States for Native Americans, and he is particularly well known for his victory over General Custer in 1876 in the Battle of the Little Big Horn, also known as the Battle of Greasy Grass. Fourteen years after this battle, Sitting Bull was shot and killed by the Indian Police while they were attempting to arrest him (2). He was buried at Fort Yates in North Dakota on what was then known as the Standing Rock Agency, now the Standing Rock Indian Reservation. However, whether his remains are still there is a matter of debate. Some part of Lakota oral history says that his supporters moved his remains to an unknown location in Canada shortly after his burial (3, 4). In 1953, relatives of Sitting Bull engaged with a mortician to open the Fort Yates gravesite, and it is presumed that they moved his skeletal remains to a new gravesite in Mobridge, South Dakota. Today, two separate official gravesites exist for Sitting Bull, one at Fort Yates and the other at Mobridge. Both sites receive visitors wishing to pay their respects. Ernie LaPointe believes his relatives moved Sitting Bull’s remains to Mobridge and has concerns about the possible commercialization and care of the gravesite. To have the right to determine the fate of the gravesite, he needs to prove that he is a relative of Sitting Bull. The familial relationship between LaPointe and Sitting Bull has until now been based on birth and death certificates, a family tree, and a review of historical records (5). Genetic evidence would constitute an additional line of evidence and thus strengthen his documentation.
Notably, without any permission or authority whatsoever, before Sitting Bull’s burial, H. Deeble, the post surgeon at Fort Yates, took both Sitting Bull’s scalp lock (the hair lock that held a feather near the crown of the head; Fig. 1) and his cloth leggings as souvenirs. Both items had been loaned by H. Deeble to the Smithsonian Institution in 1896 (6).
Fig. 1. Sitting Bull’s scalp lock.
The lock that had been taken by H. Deeble and later brought to the Smithsonian Institution. Catalog EL00226, courtesy Department of Anthropology, Smithsonian Institution.
In 2007, the museum returned the scalp lock and leggings to LaPointe and his three sisters, Sitting Bull’s great-grandchildren and closest living relatives (5). Following the repatriation, most of the lock of hair was burned in a spiritual ceremony. However, a small piece was saved for future study, and we here present the results analyzing this piece with the goal of determining whether Ernie LaPointe can be genetically identified as the great-grandchild of Sitting Bull.
Other genetic studies of historical remains have had similar goals of investigating their authenticity and/or determining their familial relationship with one or more contemporary individuals. However, the success of these previous studies has been mixed and highly debated [see e.g., (7–11)]. In addition, notably, almost all of them have relied heavily on uniparental markers with key analyses based on mitochondrial DNA and Y-chromosomal DNA (7, 9, 10, 12–16), and many of them have made use of short tandem repeats (STRs) (7, 10, 12, 13, 15, 16). These approaches have several limitations. First, uniparental markers are only useful in the context of relatedness inference if the individuals of interest are related either entirely through a maternal lineage or entirely through a paternal lineage. In addition, even if this is the case, the resolution that uniparental markers provide is usually very limited; since many human mitochondrial and Y-chromosome haplogroups are common, even unrelated individuals can have the same haplogroup just by chance. Therefore, these markers can be primarily used to rule out familial relationships. They rarely provide strong evidence supporting that two individuals are related or allow the determination of a specific familial relationship (17). Second, STRs are known to be difficult to work on for degraded DNA (18); hence, the approach requires that the quantity and quality of the DNA from the historical sample is good.
In the case of Sitting Bull, written records indicate Ernie LaPointe and Sitting Bull are related through Ernie LaPointe’s mother (table S1), ruling out the possibility of using uniparental markers for investigating whether Ernie LaPointe can be genetically identified as a descendant of Sitting Bull. Furthermore, early screens of the hair sample from Sitting Bull revealed that the amount of endogenous DNA available was extremely limited because of postmortem degradation and likely because of unrecorded treatment of the hair sample with inorganic arsenic compounds, as was common practice at the time. Thus, STR data would be technically challenging to obtain, and it would likely not provide sufficient power to resolve familial relationships as distant as the one we are interested in. We therefore took a different approach: We generated shotgun sequencing data from autosomal loci across the genome of the Sitting Bull hair sample and compared it to dense single-nucleotide polymorphism (SNP) chip data from Ernie LaPointe. A similar approach has been used successfully once before in a recent study based on genomic shallow sequencing of both a historical individual and two of his living relatives (19). However, that study benefitted from both very good preservation of the ancient molecules and from the access to close (second-degree) living relatives. In contrast, the relationship of interest here is more distant, and sample preservation is not good. Our initial intent behind the sequencing approach was to obtain a coverage of 1 to 2× for the Sitting Bull sample, allowing us to use specialized relatedness estimation methods recently developed for low-depth sequencing data to answer our question, e.g., the genotype likelihood-based method NgsRelate (20). However, the amount of endogenous data in the hair sample turned out to be so limited (mean sequencing coverage of 0.02×) that this method was not applicable. Furthermore, other methods shown to work on low-coverage data (21) only work for familial relationships of second degree or closer, such as grandparent-grandchild. Therefore, we developed and applied a new computational method tailored to low-coverage data and applicable to more distant familial relationships. In the following, we present this new method and use it to address the question regarding the familial relationship between Sitting Bull and Ernie LaPointe.
RESULTS
Sequencing the hair sample and assessing authenticity
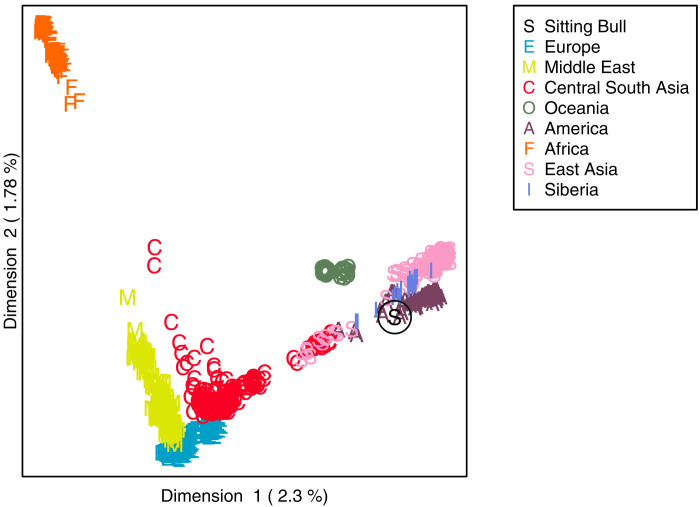
First, we extracted DNA from Sitting Bull’s hair, built indexed Illumina sequencing libraries, and sequenced them on the Illumina HiSeq 2000/2500 platforms. After mapping, we removed duplicates and filtered away bases with base quality below 20 and reads with mapping quality below 25, which left us with sequencing data covering 0.8% of the genome with a depth of coverage of 2.7, or equivalently a mean sequencing coverage of 0.02×. Next, to assess the authenticity of the sample, we estimated base specific error rates of the quality-filtered sequencing data and found markedly higher error rates at SNP loci with transitions (fig. S1), which is typical of ancient DNA due to postmortem degradation (22). The remaining error rates were all below 0.001. Thus, the results of this analysis both corroborate the authenticity of the hair sample and suggest that most of the errors in the data can be avoided by removing transitions, which we did before performing all subsequent analyses. The authenticity of the sample is also supported by results from multidimensional scaling (MDS) analyses based on the sequencing data from Sitting Bull combined with SNP chip data from worldwide populations included in the Human Genome Diversity Panel (HGDP) (23). The MDS plot obtained for joint dataset clearly shows that the sample from the hair lock clusters with Native American individuals (Fig. 2) as would be expected since the hair sample is from Sitting Bull.
Fig. 2. The Sitting Bull sample clusters with Native American individuals supporting the authenticity of the sample.
MDS plot based on sequencing data from the Sitting Bull sample combined with SNP chip data from the worldwide HGDP dataset. The S in a circle represents the Sitting Bull sample.
Assessing the familial relationship between Sitting Bull and Ernie LaPointe using a new method
To be able to assess the familial relationship between Sitting Bull and Ernie LaPointe, we genotyped 13 unrelated Lakota Sioux individuals, including Ernie LaPointe, using the extensive Illumina Omni 5M chip. Unfortunately, the sequencing data obtained from the hair sample were so limited in amount that the overlap between this and the chip data from Ernie LaPointe and the other Lakota Sioux individuals is restricted to 2259 polymorphic loci after excluding data from loci with transitions. This is not sufficient to perform reliable relatedness estimation for distantly related individuals using existing methods. Therefore, to investigate whether Ernie LaPointe can be genetically identified as the great-grandson of Sitting Bull, we developed a new probabilistic method tailored to the situation where one has limited sequencing data from a historical individual and SNP chip genotype data from a present-day individual.
Briefly, the method is a maximum likelihood method that estimates a parameter, Frel, whose value depends on how the present-day individual is related to the historical individual. More specifically, Frel is the probability that a randomly drawn allele from the historical individual, is identical by descent (IBD) with one of the present-day individuals’ two alleles in a SNP locus. Hence, if the two individuals are unrelated, then Frel is 0, and if they are related, then the expected value of Frel is 0.5m, where m is the number of meiosis between the two samples. In particular, Frel is expected to be 0.53 = 0.125 if the present-day individual is the great-grandson of the historical individual. Thus, estimating Frel for Sitting Bull and Ernie LaPointe has the potential to reveal their true relationship.
The new method for inferring Frel is based on the observation that for any given SNP locus, we can write the likelihood of Frel as
where SB is a randomly drawn allele from the historical individual of interest (in this study, Sitting Bull) with possible alleles “0” and “1,” E is the genotype of the present-day individual of interest (in this study, Ernie LaPointe) with possible values of 0, 1, and 2, counting the number of 1 alleles, f is the frequency of the 1 allele in the population, IBD means that SB is IBD with one of the alleles that make up the genotype E, and ¬IBD means the opposite. More specifically, we can write
and
Assuming independence between SNP loci (i.e., no linkage disequilibrium), this means that the log likelihood of Frel for all loci is equal to the sum of the log of the per locus log likelihoods, i.e., log of Pr(SB|E, f, Frel) from above. We obtain an estimate of Frel using numerical optimization to identify the value of Frel that maximizes this likelihood function.
Note that this method is based on two key assumptions (i) that the two individuals are from the same population and unadmixed and (ii) that allele frequencies, f, from the relevant population can be obtained. In this study, we had access to genetic data from 13 unrelated Lakota Sioux individuals, of which 5, including Ernie LaPointe, are unadmixed according to an analysis performed with the genetic ancestry clustering tool ADMIXTURE (fig. S2). The remaining eight were inferred to have some degree of European ancestry in addition to their Lakota Sioux ancestry and thus be admixed. Consequently, we could apply the method to the data from Sitting Bull and Ernie LaPointe, but we only had five individuals to base allele frequency estimates on including Ernie LaPointe. Using these allele frequencies estimates, the method provided an Frel estimate of 0.1143 (table S2 and fig. S3), which is close to what we would expect if Ernie LaPointe is Sitting Bull’s great-grandson if the population allele frequencies are known. For comparison, the Frel estimates for the other four unadmixed Lakota Sioux individuals are all below 0.002, suggesting that the Frel estimates are not in general inflated among Lakota Sioux (table S2).
Investigating what can be concluded from the obtained estimate
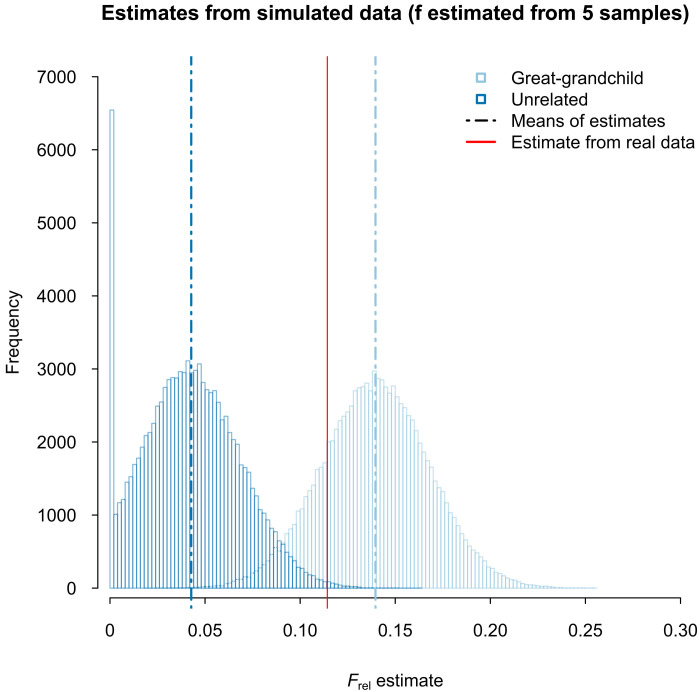
To more properly assess what can be concluded from the Frel estimate obtained for Sitting Bull and Ernie LaPointe, we performed simulations of data similar to the real data. More specifically, we simulated 100,000 datasets each consisting of low-depth sequencing data from a historical individual, SNP chip genotype data from four reference individuals, and two individuals for which we later estimated Frel: one, Eunrelated, that is unrelated to the historical individual and one, Egreat-grandchild, that is the great-grandchild of the historical individual. Each of these dataset were simulated to have the same low number of SNP loci that are polymorphic among the four reference individuals and Eunrelated or Egreat-grandchild (n = 2259) (Fig. 3). The results of applying the new method to these simulated datasets suggest that with the limited data at hand, an estimate of 0.1143 or higher can, in principle, occur even if Ernie LaPointe and Sitting Bull were not related but that this is unlikely to occur (P < 0.00389). Thus, the results from the real data combined with the simulation results show strong support for Ernie LaPointe being Sitting Bull’s great-grandson.
Fig. 3. Frel estimate for Sitting Bull and Ernie LaPointe and for simulated data.
The Frel estimate for Sitting Bull and Ernie LaPointe (red vertical line) shown in the context of the distribution of Frel estimates obtained from simulated data from individuals unrelated to a historical individual like Sitting Bull (dark blue histogram) and great-grandchildren of a historical individual like Sitting Bull (light blue histogram). Means for the two distributions are shown with dash-dotted vertical lines. The simulated data were simulated to have the same number of SNP loci on average as in real data and in the analyses of the simulated data allele frequencies were estimated from five individuals including the individual for which of Frel is estimated, just like in the analysis of the real data.
Notably, assuming the simulations are sufficiently realistic, the simulation results also show that the method has 97.0% power to detect a great-grandchild if one uses a null model of the individuals being unrelated and a 0.05 significance threshold, making it a fairly powerful method for such distant relatives given the low amount of data available. Last, the simulation results show that the estimates provided by the method are biased when applied to the simulated data; it does not, on average, provide estimates that are equal to the expected value of Frel (Fig. 3). When we reran the analyses for the same data using the true allele frequencies instead of frequencies estimated from only five individuals for the estimation of Frel, the bias was not present for the simulated great-grandchildren and markedly reduced for the unrelated individuals (fig. S4). In addition, the power to detect a great-grandchild is even larger (99.6%). This suggests that the bias is, to a large extent, caused by the fact that the sample size used to estimate allele frequencies is small and that the method would perform even better if more reference individuals were available.
DISCUSSION
This study first and foremost provides genetic support for Ernie LaPointe being the great-grandson of Sitting Bull, supporting the claim that he and his sisters were the rightful recipients of the repatriated items from the Smithsonian Institution. Second, it introduces a new approach to connecting the past to the present when very limited genetic data are available, as was the case in this study.
The new approach requires data from at least a few individuals from the same population as the individuals of interest. It also requires that none of the individuals included in the analysis are admixed or inbred, since it relies on having representative allele frequencies. Last, it relies on simulations for assessment of the certainty of the results and thus requires that data similar to that of the real data can be simulated. However, when these requirements are met the new approach is simple and easy to apply. Hence, the new approach may be valuable for future studies of remains of historical individuals to replace uniparental marker and STR-based approaches typically used so far, because these remains are often viewed as being highly sensitive by museums and the public and/or are poorly preserved, which means that future studies of these samples will have to depend on minor amounts of ancient DNA. Moreover, the new approach allows the identification of more distant relationships than other current relatedness estimation methods designed for extremely low-depth sequencing data. Hence, with the advances in ancient DNA technology and the steady interest in samples from historical figures such as the Romanovs, Richard the III, and Jesse James, the new approach may help solve interesting questions within the fields of population genetics, history, and forensics.
MATERIALS AND METHODS
Ethics statement
All Lakota Sioux participants gave oral and written consent to participate in this study. Furthermore, permission for the study was received from the closest living lineal descendants of Sitting Bull to whom the lock of hair was repatriated by the Smithsonian Institution. Three of these descendants also submitted DNA samples, and one of the three, Ernie LaPointe, gave oral and written consent to be mentioned by name in this article. We note that permission or consent for the study was not sought from the Standing Rock Sioux Tribe, which would be the modern-day tribe of Sitting Bull. However, the Tribal Chair and the Tribal Historic Preservation Officer of the Standing Rock Sioux Tribe were informed of the study and its results in August 2020 via email. They have not replied. While the authors considered seeking the tribe’s consent for the study in line with Claw et al. (24), the permission given by the lineal descendants and their desire that the study be conducted would supersede any objections raised by the tribe. In this case, the lineal descendants have a higher standing in terms of having rights to make decisions for this DNA study than the tribe. This position follows U.S. repatriation legislation where lineal descendants have priority over federally recognized tribes for making repatriation decisions. However, we also note that for this reason, we have purposely refrained from presenting analyses, such as principal components analysis, that includes the present-day Lakota Sioux individuals in a context of multiple other populations, to avoid presenting results that could be interpreted as a population genetic analysis of the tribe. The study has received approval from the research ethics committee for the Faculty of Science and the Faculty of Health and Medical Sciences at the University of Copenhagen.
The Sitting Bull hair sample
Ancient sample processing was performed in laboratories dedicated to ancient human remains (Lundbeck Foundation GeoGenetics Centre, Copenhagen, Denmark), following strict procedures to avoid contamination by modern or amplified DNA. We extracted DNA from a small hair lock following a protocol described by Rasmussen et al. (25). Three Illumina sequencing libraries were built from the DNA extract, using the procedure from Meyer and Kircher (26) with slight modifications (27). Each library was split in two halves and amplified in two steps (27) for 12 + 12 or 10 + 8 cycles, using AmpliTaq Gold (Life Technologies) and 6–base pair indexed primers. The final products were pooled and sequenced over one lane on the Illumina HiSeq 2000 platform, 93 single-read mode and one lane on the Illumina HiSeq2500 platform, 100 single-read rapid mode, at the Danish National High-Throughput DNA Sequencing Centre (Copenhagen, Denmark). The sequencing output was converted to fastq format using CASAVA, and reads with a length below 25 were discarded. The remaining reads were then trimmed and merged using AdapterRemoval (28) and mapped to hg19 (assembly hsbuilt37.1) using bwa (aln) (0.6.2) (29) with seed disabled (−l 1000). After mapping, we removed duplicates using SAMtools rmdup, and before all analyses of the sequencing data from the Sitting Bull sample, we filtered away all reads with mapping quality below 25 and bases with base quality below 20. The resulting data have a mean depth of 0.02×, and we refer to this as the quality-filtered sequencing data from Sitting Bull below.
SNP chip data from Lakota Sioux individuals
Saliva samples from 14 Lakota Sioux individuals, including Ernie LaPointe, were collected using the Oragene OG-300 DNA Self-Collection Kit (Genotek). We extracted DNA following the prepIT L2P protocol (Genotek). DNA extracts were processed for genotyping on the extensive Illumina Omni5-Quad-Exome BeadChip at AROS Applied Biotechnology (Aarhus, Denmark) to maximize the overlap with the Sitting Bull sequencing data. This led to data from 4,334,816 SNP loci. Among the 14 individuals, two were self-reported to be half-siblings, and one of these was removed from the dataset before all analyses.
Other samples
For several of the analyses, we also used genotype data from the 1000 Genomes project and HGDP and sequencing data from individual NA12778 from the 2013 release of 1000 Genomes Project (30).
Dataset used for error rate estimation
We used the quality-filtered sequencing data from the Sitting Bull hair sample and used high-quality sequencing data for the genome of individual NA12778 from the 2013 release of 1000 Genomes Project as the high-quality genome (30). To determine the ancestral alleles and, thus, the derived alleles, we used the outgroup species chimpanzee as is standard in human studies. In particular, we used the multiway alignment that includes both chimpanzee and human (pantro2 from the hg19 multiz46).
Dataset used for MDS analysis
We used the HGDP data provided with the bammds software tool (23, 31), which contain data from 644,074 SNP loci for a range of populations from across the world. From these data, we removed all transition SNP loci, since these are much more error prone (fig. S1). This left us with genotype data from 120,643 SNP loci. The quality-filtered sequencing data from the Sitting Bull hair sample overlapped 654 of these loci, and these were the reads that were included in the analysis along with the genotype data. The extraction of these reads was performed by the bammds software tool that we used for the MDS analysis.
Dataset used for ADMIXTURE analyses
We merged the Lakota Sioux SNP chip data with genomic data for 40 individuals from the CEU 1000 Genomes population keeping only the SNPs that overlap and are consistent between the two datasets. After merging, we removed SNPs with minor allele frequency below 5%, SNPs with any missingness and SNPs in high linkage disequilibrium (using the PLINK option --indep-pairwise 100 10 0.5) leaving us with data from 363,519 SNP loci.
Dataset used for estimation of Frel
We combined the quality-filtered sequencing read data from the Sitting Bull hair sample with the SNP chip data from the 13 not closely related Lakota Sioux individuals. When doing so, we first discarded all SNPs that did not overlap with any read data from Sitting Bull. This left us with data from 49,347 SNP loci. Subsequently, we removed all SNP loci with transitions, since these are much more error prone (fig. S1), which left us with 10,142 loci. Then, for each of the remaining loci, we sampled a single read from Sitting Bull, and if the read had one of the two alleles present in the SNP chip data at that locus, we included the locus in the analyses; if not, we discarded the locus, leaving us with 10,041 SNP loci. Last, we removed 8 of the 13 Lakota Sioux individuals because they were admixed (fig. S2) and subsequently discarded all SNPs that had missing data or were not polymorphic among the remaining 5 Lakota Sioux individuals. This left us with data from 2259 SNP loci to base the final analyses on.
Error estimation
We used the error estimation method from Orlando et al. (32) implemented in ANGSD (Analysis of Next Generation Sequencing Data) (33) to estimate base type–specific error rates for sequencing data from the Sitting Bull hair sample. This method relies on a comparison of the number of derived alleles in the sequencing data from the individual of interest and in a high-quality genome. The idea behind it is that any human genome should, on average, have the same number of derived alleles, and therefore, the excess of these alleles in the sequencing data from the individual of interest compared to a high-quality genome can be assumed to be due to sequencing errors and thus used as a basis for error estimation.
MDS analysis
We performed MDS analyses using the software tool bammds (version: bammds_20140602) (31). We applied the tool to the quality-filtered sequencing data from the Sitting Bull hair sample in the form of a bam file combined with the HGDP dataset that is made available along with the bammds software tool.
Admixture analyses
We used the program ADMIXTURE (34) to identify admixed individuals. When running the analyses, we assumed that the number of ancestral populations, K, is 2. We ran ADMIXTURE 50 times with different starting values and the difference in likelihood units between the highest likelihood and 10th highest likelihood was less than 0.000001, suggesting that convergence was achieved.
Simulation study
To be able to properly interpret the Frel estimate obtained for Sitting Bull and Ernie LaPointe and to estimate how powerful the new estimation method is, we simulated 100,000 datasets and applied the method to these datasets.
Data simulations
Each of the 100,000 was simulated so it mimicked the data from the analyses of the real data from of Sitting Bull and Ernie LaPointe. Specifically, each dataset consisted of low-depth sequencing data from one historical individual, SNP chip genotype data from four reference individuals, and SNP chip genotype data from two individuals for which we are interested in estimating Frel: one, Eunrelated, that is unrelated to the historical individual and one, Egreat-grandchild, that is the great-grandchild of the historical individual. Further, the number of simulated loci was chosen so the number of SNP that are polymorphic among the four reference individuals and the individual of interest (i.e., the five individuals that were used for allele frequency estimates when applying our method to the dataset) was, on average, approximately equal to the number of SNP loci used in our real data analyses, namely, 2259.
The data for each dataset were simulated by first sampling population allele frequencies from a uniform distribution between 0.1 and 0.9 (mimicking that we have data from loci on a SNP chip) for a preset number of SNP loci. Then, using these population allele frequencies, we simulated a pool of 15 haplotypes. Assuming that our loci are few and far between and thus independent, we simulated each of these haplotypes by independently for each locus sampling an allele from a Bernoulli distribution using the population allele frequency for that locus. Last, based on these haplotypes, we simulated
1) The low-depth sequencing from the historical individual by sampling two haplotypes from the pool of haplotypes without replacement and then randomly sampling an allele from one of these two haplotypes at each locus
2) The SNP chip genotype from four reference individuals by sampling four pairs of haplotypes from the pool of haplotype without replacement
3) The SNP chip data from Eunrelated by sampling two haplotypes from the haplotype pool without replacement
4) The SNP chip data from Egreat-grandchild, by simulating first a child of the historical individual based on the two haplotypes of the historical individual, then a grandchild based on the two haplotypes of the child, and lastly a great-grandchild based on the two haplotypes of the grandchild. More specifically, we simulated each of these offspring by sampling one new haplotype from the pool of haplotypes and combining this with a haplotype that was a recombination of the two haplotypes of the parent (e.g., the historical individual). The recombination was simulated by randomly sampling one allele at each locus from the two haplotypes.
When simulating the datasets, we assumed that the genotypes from the present-day individuals can be obtained reliably without any errors. In contrast, we assumed a fixed error rate of 0.001 for the historical individual and added errors by simply switching the simulated sampled allele to the other allele segregating at this locus randomly with probability equal to the error rate. We used an error rate of 0.001 to mimic the error rate estimated for nontransition loci that is below 0.001 across all categories (cf. fig. S1).
Estimating Frel from the simulated datasets
For each of the simulated datasets, our Frel estimation method was applied to (i) the simulated data from the historical individual, (ii) the simulated data from the individual, Eunrelated or Egreat-grandchild, for which we want to estimate Frel, and (iii) allele frequencies estimated from four reference individuals and Eunrelated or Egreat-grandchild (i.e., the same approach as we have used for Sitting Bull and Ernie LaPointe). The estimation method was implemented in R using the optim function with the optimization algorithm set to L-BFGS-B and lower and upper bound for Frel set to 1 × 10−12 and to 1 − (1 × 10−12), respectively.
Estimating Frel from the real data
As described above the estimation method was implemented in R using the optim function with the optimization algorithm set to L-BFGS-B and lower and upper bound for Frel set to 1 × 10−12 and to 1 − (1 × 10−12), respectively. When estimating Frel from the real data, we used genotypes from all the five unadmixed Lakota Sioux individuals for the allele frequency estimation, i.e., we included the individual for which Frel was estimated in the allele frequency estimation.
Acknowledgments
We thank E.L.’s wife, S. LaPointe, his sisters M. Little Spotted Horse-Anderson and E. Little Spotted Horse-Bates, and the anonymous participants for providing consent and endorsing this research project. Furthermore, we thank R. Nielsen for valuable input regarding the new computational method.
Funding: This work was funded by the Danish National Research Foundation, Denmark. T.S.K. was funded by a Carlsberg Foundation Young Researcher Fellowship awarded by the Carlsberg Foundation in 2019 (CF19-0712).
Author contributions: E.W. initiated and led the study. E.L. helped collect the saliva samples from the Lakota Sioux individuals and provided access to Sitting Bull’s hair repatriated by the Smithsonian. A.S.-O. performed the DNA work. J.V.M.-M. helped prepare data for analyses. I.M. and T.S.K. developed the new probabilistic approach to assessing relatedness and conducted all the analyses. W.B. provided historical context information. I.M., T.S.K., and E.W. wrote the paper with input from the remaining authors.
Competing interests: We openly acknowledge that one of the authors, E.L., has a strong personal interest in the results of this study (as described in the introduction); however, he did not take part in any part of the data analyses. The remaining authors declare that they have no competing interests.
Data and materials availability: The samples used in this study are owned by E.L., and per his request, they have been destroyed. The generated sequencing data from Sitting Bull and the generated genotype data from the Lakota Sioux individuals are also owned by E.L. and will be made available upon request in limited circumstances. Note that the genotype data from the Lakota Sioux individuals can only be used for replication purposes due to constraints in the ethical approval. Hence, these data cannot be used for general ancestry inferences, individual functional inferences, inferences of endogamy, or any other analyses beyond investigating the familial relationship between the historical individual and the modern individuals included in this study. The sequencing data from Sitting Bull can only be used for additional studies if E.L. gives permission. To obtain permission to use the data, please write E.L. (eagletalon-101@hotmail.com). Once permission is obtained, the data will be released by E.W. who is archiving it on behalf of E.L. If E.L. is not available, then his descendants should be contacted for permission.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Tables S1 and S2
REFERENCES AND NOTES
- 1.R. M. Utley, The Lance and the Shield: The Life and Times of Sitting Bull (Henry Holt & Co., 1993).
- 2.B. Yenne, Sitting Bull (Westholme Publishing, 2008). [Google Scholar]
- 3.R. Boswell, “Sitting Bull buried here?” Winnipeg Free Press, 24 February 2007, p.7.
- 4.L. Thackeray, “Bones aren’t Sitting Bull’s, some say,” Billings Gazette, 22 February, 2007.
- 5.E. LaPointe, Sitting Bull: His Life and Legacy (Gibbs Smith, 2009).
- 6.W. T. Billeck, B. Bruemmer, Assessment of a Lock of Hair and Leggings Attributed to Sitting Bull, Hunkpapa Sioux, in the National Museum of Natural History, Smithsonian Institution (National Museum of Natural History, Repatriation Office, 2007).
- 7.Charlier P., Olalde I., Solé N., Ramírez O., Babelon J.-P., Galland B., Calafell F., Lalueza-Fox C., Genetic comparison of the head of Henri IV and the presumptive blood from Louis XVI (both Kings of France). Forensic Sci. Int. 226, 38–40 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Jehaes E., Decorte R., Peneau A., Petrie J. H., Boiry P. A., Gilissen A., Moisan J. P., Van den Berghe H., Pascal O., Cassiman J. J., Mitochondrial DNA analysis on remains of a putative son of Louis XVI, King of France and Marie-Antoinette. Eur. J. Hum. Genet. 6, 383–395 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Larmuseau M. H. D., Cassiman J.-J., Decorte R., Controversial identification in a historical case is illustrative of the complexity of DNA typing in forensic research. Response to Charlier et al. Forensic Sci. Int. Genet. 9, e18–e19 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Larmuseau M. H. D., Delorme P., Germain P., Vanderheyden N., Gilissen A., Van Geystelen A., Cassiman J.-J., Decorte R., Genetic genealogy reveals true Y haplogroup of House of Bourbon contradicting recent identification of the presumed remains of two French Kings. Eur. J. Hum. Genet. 22, 681–687 (2014b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucotte G., Crépin C., Thomasset T., Paris M., Mitochondrial DNA Sequences of the Famous Karl Wilhelm Naundorff (1785 ?- 1845). Int. J. Sci. 3, 28–32 (2014). [Google Scholar]
- 12.Coble M. D., Loreille O. M., Wadhams M. J., Edson S. M., Maynard K., Meyer C. E., Niederstätter H., Berger C., Berger B., Falsetti A. B., Gill P., Parson W., Finelli L. N., Mystery solved: The identification of the two missing Romanov children using DNA analysis. PLOS ONE 4, e4838 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gill P., Ivanov P. L., Kimpton C., Piercy R., Benson N., Tully G., Evett I., Hagelberg E., Sullivan K., Identification of the remains of the Romanov family by DNA analysis. Nat. Genet. 6, 130–135 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Ivanov P. L., Wadhams M. J., Roby R. K., Holland M. M., Weedn V. W., Parsons T. J., Mitochondrial DNA sequence heteroplasmy in the Grand Duke of Russia Georgij Romanov establishes the authenticity of the remains of Tsar Nicholas II. Nat. Genet. 12, 417–420 (1996). [DOI] [PubMed] [Google Scholar]
- 15.King T. E., Fortes G. G., Balaresque P., Thomas M. G., Balding D., Maisano Delser P., Neumann R., Parson W., Knapp M., Walsh S., Tonasso L., Holt J., Kayser M., Appleby J., Forster P., Ekserdjian D., Hofreiter M., Schürer K., Identification of the remains of King Richard III. Nat. Commun. 5, 5631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogaev E. I., Grigorenko A. P., Moliaka Y. K., Faskhutdinova G., Goltsov A., Lahti A., Hildebrandt C., Kittler E. L. W., Morozova I., Genomic identification in the historical case of the Nicholas II royal family. Proc. Natl. Acad. Sci. U.S.A. 106, 5258–5263 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kayser M., Uni-parental markers in human identity testing including forensic DNA analysis. BioTechniques 43, Sxv–Sxxi (2007). [Google Scholar]
- 18.Canturk K. M., Emre R., Kınoglu K., Başpınar B., Sahin F., Ozen M., Current status of the use of single-nucleotide polymorphisms in forensic practices. Genet. Test. Mol. Biomarkers 18, 455–460 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Fernandes D., Sirak K., Novak M., Finarelli J. A., Byrne J., Connolly E., Carlsson J. E. L., Ferretti E., Pinhasi R., Carlsson J., The identification of a 1916 Irish rebel: New approach for estimating relatedness from low coverage homozygous genomes. Sci. Rep. 7, 41529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korneliussen T. S., Moltke I., NgsRelate: A software tool for estimating pairwise relatedness from next-generation sequencing data. Bioinformatics 31, 4009–4011 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn J. M. M., Jakobsson M., Günther T., Estimating genetic kin relationships in prehistoric populations. PLOS ONE 13, e0195491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briggs A. W., Stenzel U., Johnson P. L. F., Green R. E., Kelso J., Prüfer K., Meyer M., Krause J., Ronan M. T., Lachmann M., Pääbo S., Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J. Z., Absher D. M., Tang H., Southwick A. M., Casto A. M., Ramachandran S., Cann H. M., Barsh G. S., Feldman M., Cavalli-Sforza L. L., Myers R. M., Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Claw K. G., Lippert D., Bardill J., Cordova A., Fox K., Yracheta J. M., Bader A. C., Bolnick D. A., Malhi R. S., TallBear K., Garrison N. A., Chaco Canyon dig unearths ethical concerns. Hum. Biol. 89, 177–180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen M., Li Y., Lindgreen S., Pedersen J. S., Albrechtsen A., Moltke I., Metspalu M., Metspalu E., Kivisild T., Gupta R., Bertalan M., Nielsen K., Gilbert M. T., Wang Y., Raghavan M., Campos P. F., Kamp H. M., Wilson A. S., Gledhill A., Tridico S., Bunce M., Lorenzen E. D., Binladen J., Guo X., Zhao J., Zhang X., Zhang H., Li Z., Chen M., Orlando L., Kristiansen K., Bak M., Tommerup N., Bendixen C., Pierre T. L., Grønnow B., Meldgaard M., Andreasen C., Fedorova S. A., Osipova L. P., Higham T. F., Ramsey C. B., Hansen T. V., Nielsen F. C., Crawford M. H., Brunak S., Sicheritz-Pontén T., Villems R., Nielsen R., Krogh A., Wang J., Willerslev E., Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463, 757–762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer M., Kircher M., Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 6, pdb.prot5448 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Seguin-Orlando A., Schubert M., Clary J., Stagegaard J., Alberdi M. T., Prado J. L., Prieto A., Willerslev E., Orlando L., Ligation bias in illumina next-generation DNA libraries: Implications for sequencing ancient genomes. PLoS ONE 8, e78575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindgreen S., AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC. Res. Notes 5, 337 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.1000 Genomes Project Consortium , An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malaspinas A. S., Tange O., Moreno-Mayar J. V., Rasmussen M., DeGiorgio M., Wang Y., Valdiosera C. E., Politis G., Willerslev E., Nielsen R., bammds: A tool for assessing the ancestry of low-depth whole-genome data using multidimensional scaling (MDS). Bioinformatics 30, 2962–2964 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlando L., Ginolhac A., Zhang G., Froese D., Albrechtsen A., Stiller M., Schubert M., Cappellini E., Petersen B., Moltke I., Johnson P. L., Fumagalli M., Vilstrup J. T., Raghavan M., Korneliussen T. S., Malaspinas A. S., Vogt J., Szklarczyk D., Kelstrup C. D., Vinther J., Dolocan A., Stenderup J., Velazquez A. M., Cahill J., Rasmussen M., Wang X., Min J., Zazula G. D., Seguin-Orlando A., Mortensen C., Magnussen K., Thompson J. F., Weinstock J., Gregersen K., Røed K. H., Eisenmann V., Rubin C. J., Miller D. C., Antczak D. F., Bertelsen M. F., Brunak S., Al-Rasheid K. A., Ryder O., Andersson L., Mundy J., Krogh A., Gilbert M. T., Kjær K., Sicheritz-Ponten T., Jensen L. J., Olsen J. V., Hofreiter M., Nielsen R., Shapiro B., Wang J., Willerslev E., Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 499, 74–78 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 and S2