Loss of SCN REV-ERBs alters oscillator period but does not render the clock arrhythmic.
Abstract
Circadian disruption, as occurs in shift work, is associated with metabolic diseases often attributed to a discordance between internal clocks and environmental timekeepers. REV-ERB nuclear receptors are key components of the molecular clock, but their specific role in the SCN master clock is unknown. We report here that mice lacking circadian REV-ERB nuclear receptors in the SCN maintain free-running locomotor and metabolic rhythms, but these rhythms are notably shortened by 3 hours. When housed under a 24-hour light:dark cycle and fed an obesogenic diet, these mice gained excess weight and accrued more liver fat than controls. These metabolic disturbances were corrected by matching environmental lighting to the shortened endogenous 21-hour clock period, which decreased food consumption. Thus, SCN REV-ERBs are not required for rhythmicity but determine the free-running period length. Moreover, these results support the concept that dissonance between environmental conditions and endogenous time periods causes metabolic disruption.
INTRODUCTION
Circadian rhythmicity is a highly conserved biological paradigm that coordinates behavior and metabolism in anticipation of a 24-hour day (1–4). In mice, endogenous molecular clocks drive an approximately 23.5-hour free-running period (Tau) in the absence of external timekeepers such as light. Light:dark (LD) cycles entrain the master clock located in the suprachiasmatic nucleus (SCN), which then coordinates behaviors, peripheral clocks, and physiological functions to maintain circadian homeostasis (1, 2). Molecular clocks are composed of transcriptional translational feedback loops (TTFLs) involving gene activation by BMAL1, which is rhythmically repressed by separate mechanisms involving PER/CRY and REV-ERB nuclear receptors. REV-ERBα and REV-ERBβ nuclear receptors are core components of the circadian TTFL in mammals (5, 6).
Several studies in vitro demonstrated that REV-ERBs are not necessary for core clock function but comprise an important secondary feedback loop that stabilizes the clock and transmits circadian output through regulation of clock-controlled genes (6–8). However, an inducible whole-body deletion of both REV-ERBs has been described to impair rhythmic behavior (9) to the same extent as deletion of PER1/2 or CRY1/2 (9–11). The difference between in vitro and in vivo function of REV-ERBs could be due to (i) different functions of REV-ERBs in peripheral clocks and the central master clock, (ii) the general impact of whole-body REV-ERB deletion on running performance confounding the capacity to evaluate free-running period (9), or (iii) the use of a Rev-erbαf/f deletion model that generates a truncated REV-ERBα protein (REV-ERBα-DBDm mice) (9, 12).
To specifically address the role of REV-ERBs in the master clock in vivo, we used a recently developed, new Rev-erbαf/fβf/f mouse model (13) to induce either whole-body or tissue-specific deletion of both REV-ERBs. Here, we show that REV-ERBs are not necessary for circadian rhythmicity of the master clock but play a major role in maintaining a free-running period close to 24 hours. Moreover, SCN double knockout (SCN-DKO) mice exhibited increased sensitivity to high-fat diet-induced obesity (DIO) and adjusting external LD cycles to match their malfunctioning pacemaker corrected these metabolic defects, providing strong evidence for the circadian desynchrony hypothesis.
RESULTS
SCN-specific deletion of REV-ERBs impairs free-running and metabolic periods
To study the specific role of REV-ERBs in the master clock, we developed a tissue-specific deletion model of REV-ERBs in the SCN. Rev-erbαf/fβf/f mice were crossed to Six3-Cre mice to induce REV-ERB deletion targeted to the SCN (SCN-DKO mice). Six3-Cre has been previously shown to be robustly expressed in the ventral anterior hypothalamus including the SCN and subparaventricular zone but not in the dorsomedial hypothalamus (14). We first confirmed at the protein level, using mice with endogenously epitope-tagged HA-REV-ERBα (15), that the peak of REV-ERBα expression in the SCN is from ZT1 to ZT6 (fig. S1, A and B), as previously published (16, 17).
To validate the model, REV-ERBα and REV-ERBβ expression was analyzed at ZT4 in punch biopsies of the SCN as well as the ventromedial hypothalamus (VMH), the dorsomedial hypothalamic nucleus (DMH), and arcuate nucleus (ARC), with the dissection method validated by the enrichment of specific markers of each hypothalamic area (fig. S1C). Analysis of the SCN punch biopsies demonstrated marked reduction of Rev-erbα and Rev-erbβ gene expression (Fig. 1A), REV-ERBα protein expression (Fig. 1B), as well as increased expression of REV-ERB target genes Bmal1 (Arntl) and Npas2 (Fig. 1A). Moreover, the loss of Rev-erbα gene expression in individual cells of the SCN was demonstrated by fluorescence in situ hybridization (FISH) (Fig. 1C). Expression of REV-ERB target genes in other hypothalamic nuclei was consistent with SCN-specific deletion of REV-ERBα and REV-ERBβ (fig. S1D). We also confirmed that REV-ERBα and REV-ERBβ expression was not affected in cerebellum, cerebral cortex, and hippocampus (fig. S1, E and F) at ZT4 and ZT10. Furthermore, circadian gene expression analysis revealed slight changes of REV-ERBα and REV-ERBβ expression in retina (fig. S1G), liver (fig. S1H), and brown adipose tissue (BAT) (fig. S1J). Circadian gene expression of Rev-erbs, Bmal1, and Npas2 was preserved in retina (fig. S1G), which, together with the modest fold changes observed in comparison to the SCN (Fig. 1A and fig. S1D), strongly suggests that REV-ERB expression in retina was maintained in SCN-DKO mice. Moreover, in accordance with quantitative polymerase chain reaction (qPCR) analysis at ZT10, REV-ERBα protein expression was slightly decreased, while BMAL1 protein expression was unchanged in liver (fig. S1I) and BAT (fig. S1K). These small changes in peripheral organs will require further investigation to delineate the direct and/or indirect role of SCN REV-ERBs on clock gene expression in peripheral organs.
Fig. 1. Generation of mice with specific deletion of REV-ERBα and REV-ERBβ in SCN.
(A) Gene expression in SCN from control (n = 10) and SCN REV-ERBα/β DKO male mice (SCN-DKO, n = 6) at ZT4. One to two independent punches were pooled together per biological replicate. Results are presented as means ± SEM and compared by Mann-Whitney test. (B) Western blot analysis of REV-ERBα protein level in SCN from control (n = 4) and SCN-SKO (Six3-Cre+, Rev-erbαflox/flox, n = 3) male mice at ZT4. Three independent punches were pooled together per biological replicate. (C) Fluorescence in situ hybridization (FISH) of SCN Rev-erbα (Nr1d1) by RNAscope in control and SCN-DKO mice at ZT4. Representative pictures are presented. Scale bars, 200 μm.
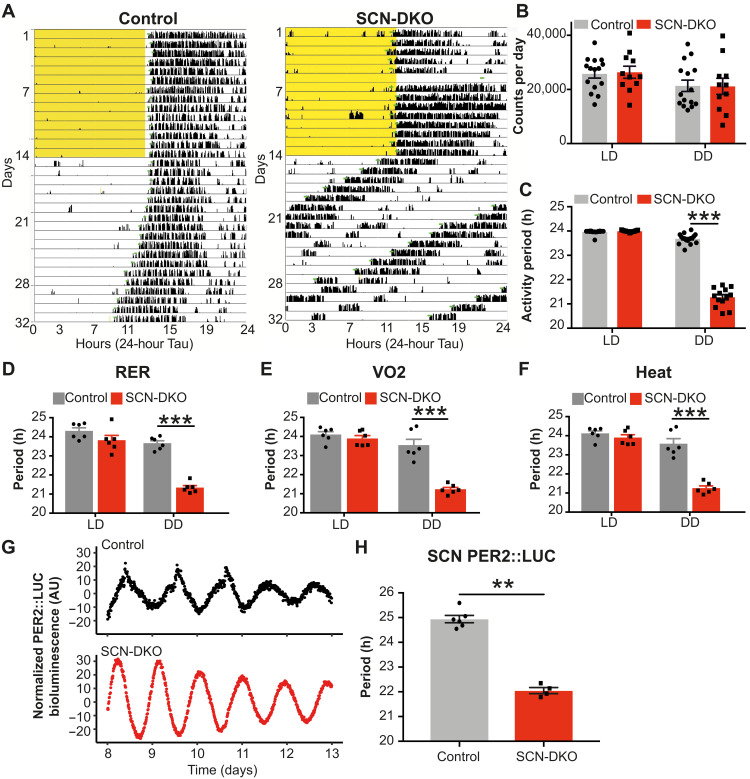
Next, we evaluated wheel-running activity of the SCN-DKO mice to delineate the specific role of REV-ERBs in the master clock rhythmicity. Both control and SCN-DKO mice are entrained by light under a regular LD cycle (12:12, T24; Fig. 2A and figs. S2A and S3, A and B, left). Moreover, locomotor activity was not impaired in SCN-DKO mice under regular LD or in constant darkness [dark:dark (DD)] (Fig. 2, A and B), enabling us to measure the free-running period in DD without a confounding reduction in activity. Contrary to deletion of other core clock genes [Cry1/2, Per1/2, Bmal1 (10, 11, 18)], SCN REV-ERBs were not necessary for the master clock rhythmicity or penetrance in DD (Fig. 2A). Nevertheless, deletion of SCN REV-ERBs strongly regulated the free-running period, which was markedly decreased to 21 hours (Fig. 2C and fig. S2A). No sex differences were observed in the function of REV-ERBs in the SCN, as female mice also displayed a 21-hour free-running period (fig. S2, B to E). Moreover, energy expenditure of SCN-DKO mice in DD also exhibited a 21-hour Tau (Fig. 2, D to F, and fig. S3, A and B, right). As expected from their shortened free-running period, SCN-DKO mice exhibited an advanced phase angle of entrainment in the 12:12 LD cycle relative to control mice (fig. S2F). To explore the SCN-intrinsic nature of the effect of REV-ERB deletion, we bred PER2::LUC mice (19) to SCN-DKO and control mice. SCN slices from the SCN-DKO-PER2::LUC mice displayed a markedly shortened period (Fig. 2, G and H), which was ~3 hours shorter than that of SCN slides from control (floxed)–PER2::LUC mice (Fig. 2, G and H), similar to the degree of shortening of the behavioral and metabolic rhythms observed in the in vivo model. Thus, expression of REV-ERBs in the SCN was not necessary for maintenance of rhythmic behavior but had a profound role in controlling the circadian period of the central pacemaker.
Fig. 2. SCN-specific deletion of REV-ERBs impairs free-running and metabolic periods.
(A) Representative actogram of control mice and SCN-DKO male mice in LD (12:12) and DD. Light cycle is highlighted in yellow. (B) Activity count per day in control (n = 15) and SCN-DKO male mice (n = 11) in LD and DD. Results are presented as means ± SEM and compared by two-way analysis of variance (ANOVA) (genotype P = 0.9479, light schedule P = 0.0002, interaction P = 0.7383). (C) Wheel-running activity period of control (n = 17) and SCN-DKO male mice (n = 13) in LD and DD. Results are presented as means ± SEM and compared by two-way ANOVA (interaction P < 0.0001) followed by Sidak’s multiple comparisons test. (D to F) Metabolic periods of respiratory-exchange ratio (RER) (D), VO2 (E), and heat (F) in control (n = 6) and SCN-DKO male and female mice (n = 6) in LD and DD. Corresponding circadian recordings are displayed in fig. S3. Results are presented as means ± SEM and compared by two-way ANOVA (interactions P < 0.05) followed by Sidak’s multiple comparisons test. (G and H) Representative examples (G) and period quantification (H) of PER2::LUC bioluminescence in the SCN from control-PER2::LUC (n = 6) and SCN-DKO-PER2::LUC (n = 4) female mice. Results are presented as means ± SEM and compared by Mann-Whitney test. AU, arbitrary units.
Whole-body deletion of REV-ERBs impairs ability to evaluate free-running period
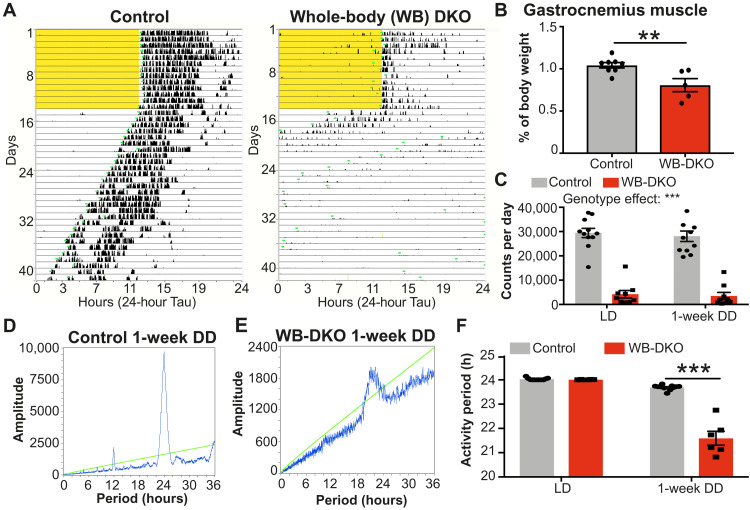
As a previous study showed that an inducible whole-body deletion of both REV-ERBs using REV-ERBα-DBDm mouse impaired rhythmic behavior (9) to the same extent as deletion of PER1/2 or CRY1/2 (9–11), we examined, in detail, the impact of whole-body deletion of both Rev-erbα and Rev-erbβ on free-running activity using the recently developed Rev-erbαf/fβf/f mouse model (13). To induce whole-body deletion of both REV-ERBs, Rev-erbαf/fβf/f mice (13) were crossed to cytomegalovirus (CMV)–Cre mice for at least three generations before exclusion of the Cre expression. As expected, Rev-erbα/β expression was decreased, while REV-ERB target genes Bmal1 (Arntl) and Npas2 were up-regulated in hypothalamus (fig. S4A), BAT (fig. S4B), and liver (fig. S4C) from the whole-body DKO mice (WB-DKO:Rev-erbαko/koβko/ko, Cre negative) compared to control mice (Rev-erbα+/+β+/+, Cre negative). WB-DKO mice weighed less (fig. S4D) and were smaller (fig. S4, E and F). Moreover, in contrast to whole-body KO mice for REV-ERBα, which exhibit increased adiposity (20), WB-DKO mice exhibited decreased epididymal white adipose tissue mass (fig. S4, G to I), suggesting a role of REV-ERBβ. Last, as previously described using REV-ERBα-DBDm mouse (9) and consistent with the function of REV-ERBs in muscle (21), WB-DKO mice exhibited a severe decrease of activity in both LD 12:12 and DD with decreased gastrocnemius muscle mass (Fig. 3, A to C). However, even with locomotor activity largely decreased, WB-DKO mice still exhibited entrainment to an LD cycle (Fig. 3A and fig. S4, J and K).
Fig. 3. Whole-body deletion of REV-ERBs impairs running capacity and ability to evaluate free-running period.
(A) Representative actogram of control mice (left) and whole-body REV-ERBα/β DKO male mice (WB-DKO, right) in LD (12:12) and DD. Light schedule is highlighted in yellow. (B) Gastrocnemius muscle weight expressed as percentage of body weight in control (n = 8) and WB-DKO male mice (n = 5). Results are presented as means ± SEM and compared by Mann-Whitney test. (C) Activity count per day in control (n = 11) and WB-DKO male mice (n = 9) in LD (12:12) and the first week of DD. Results are presented as means ± SEM and compared by two-way ANOVA (interaction P = 0.8136). (D and E) Representative chi-square periodogram of control (D) and WB-DKO (E) male mice during the first week of DD. The green chi-square oblique line indicates the significance level at P < 0.05. (F) Wheel-running activity period of control (n = 11) and WB-DKO male mice (n = 6) in LD (12:12) and during the first week of DD. Results are presented as means ± SEM and compared by two-way ANOVA (interaction P < 0.0001) followed by Sidak’s multiple comparisons test.
During the first week in DD, WB-DKO mice retained modest rhythmicity (Fig. 3, A, D, and E), which allowed us to determine a free-running period reduced to 21 hours in these animals (Fig. 3F). However, after only 1 week in DD, the pronounced locomotor defects and the low rhythm penetrance confounded the ability to accurately assess rhythmic behavior in the WB-DKO mice. The marked reduction of locomotor activity in WB-DKO mice thus represents a confounding factor in the interpretation of wheel-running activity in DD. Together with the SCN-DKO mouse model, the results obtained in WB-DKO mice demonstrate that, rather than abolishing rhythmicity, deletion of REV-ERBs in the SCN markedly shortens the circadian period of the master clock.
SCN-specific deletion of REV-ERBs alters SCN gene transcription involved in circadian entrainment
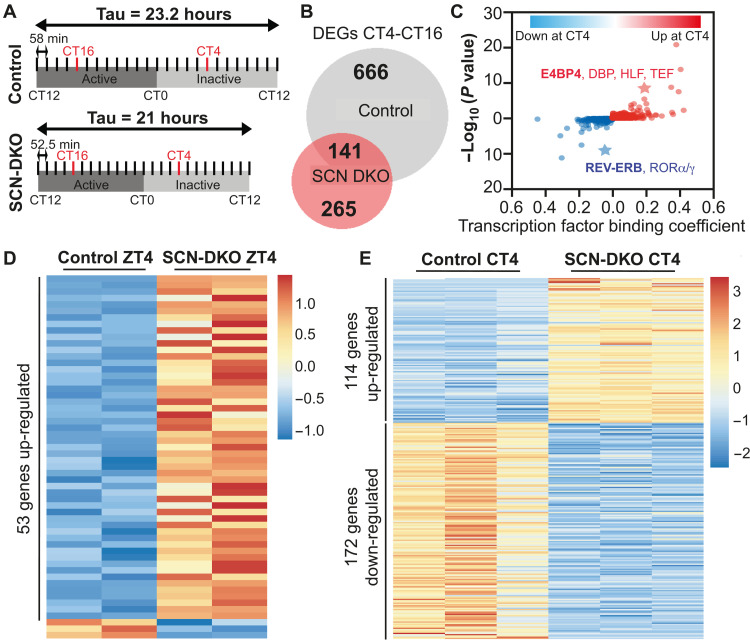
To understand how REV-ERBs regulate gene transcription in SCN, we compared gene transcription in the SCN punches from control and SCN-DKO mice housed for 3 weeks in DD and harvested at the peak [circadian time 4 (CT4)] or trough (CT16) of REV-ERB expression (Fig. 4A). In control mice, we identified 807 differentially expressed genes (DEGs) between CT4 and CT16, whereas only 406 DEGs between CT4 and CT16 were identified in SCN-DKO mice including 141 in common with control (Fig. 4B). Among the 666 CT4-CT16 DEGs lost in SCN-DKO mice, we identified two clusters corresponding to genes down-regulated or up-regulated at CT4 compared to CT16 in control mice only (fig. S5A). Consistent with the repressive function of REV-ERBs on gene expression, a machine learning algorithm for transcription factor binding analysis (TBA; https://github.com/jenhantao/tba) predicted an enrichment for the REV-ERBα (Nr1d1) motif near the genes down-regulated at CT4 in control mice only (Fig. 4C). Moreover, the E4BP4 motif was among the most enriched motifs near the genes up-regulated at CT4 in control mice only (Fig. 4C), consistent with a previous report suggesting that REV-ERBs indirectly activate the expression of genes by direct repression of the potent repressor E4BP4 (22). Among the 265 CT4-CT16 DEGs gained in SCN-DKO mice, we identified two clusters corresponding to genes down-regulated or up-regulated at CT4 compared to CT16 in SCN-DKO mice only (fig. S5B). Notably, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the CT4-CT16 DEGs lost (fig. S5C) or gained (fig. S5D) in SCN-DKO mice implicated mitogen-activated protein kinase (MAPK), calcium signaling, and circadian entrainment. Together, these analyses revealed a key role of REV-ERBs in gene transcription in SCN involved in circadian entrainment.
Fig. 4. SCN-specific deletion of REV-ERBs alters SCN gene transcription.
(A) Schematic representation of the harvest performed in DD and the method used to define CT4 and CT16 in control and SCN-DKO mice. (B) Venn diagram comparison of CT4-CT16 differentially expressed genes (DEGs) in SCN from control and SCN-DKO mice [transcripts per million (TPM) > 0.1, fold change (FC) > 1.5, false discovery rate (FDR) < 0.05]. Corresponding heatmap and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis is presented in fig. S5 (A to D). (C) Transcription factor binding analysis (machine learning algorithm applied to promoter regions) of CT4-CT16 DEGs up- or down-regulated in control mice at CT4 and lost in SCN-DKO mice. The value of each binding coefficient indicates whether the presence of a motif is correlated with the regions of interest. (D) Heatmap of the DEGs in SCN-DKO (n = 2) mice compared to control mice (n = 2) at ZT4 (TPM > 0.1, FC > 1.5, FDR < 0.05). Three to four independent SCN punches were pooled together per biological replicate. Corresponding KEGG pathway analysis is presented in fig. S5E. (E) Heatmap of the DEGs in SCN-DKO (n = 3) mice compared to control mice (n = 3) at CT4 (TPM > 0.1, FC > 1.5, FDR < 0.05). Three to four independent SCN punches were pooled together per biological replicate. Corresponding KEGG pathway analysis is presented in fig. S5F.
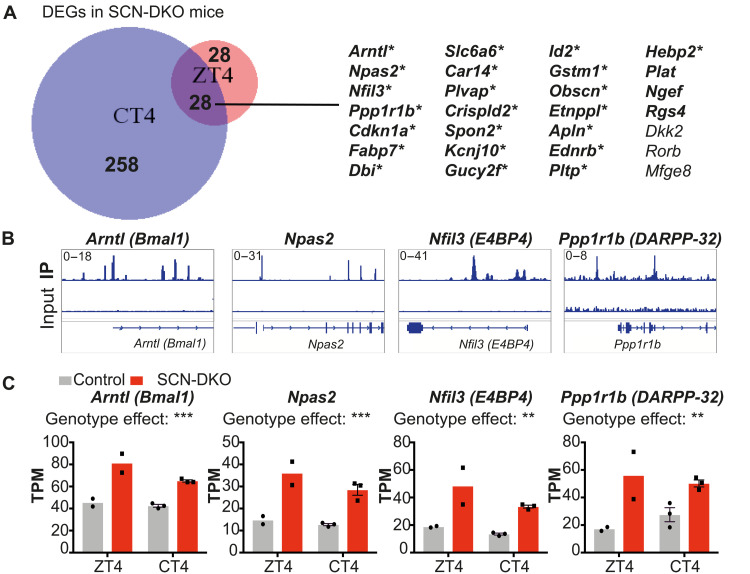
To exclude transcriptional changes potentially caused by the different free-running periods in control and SCN-DKO mice housed in DD, we also performed RNA sequencing (RNA-seq) in the SCN from control and SCN-DKO mice housed in a 24-hour LD cycle (12:12) and harvested at ZT4. We identified 56 DEGs at ZT4 (Fig. 4D) and 286 DEGs at CT4 (Fig. 4E) in SCN-DKO mice compared to control mice. KEGG analysis of the DEGs in SCN-DKO mice at ZT4 or CT4 revealed pathways involved in circadian rhythms, as well as dopaminergic signaling at ZT4 (fig. S5E), and cyclic adenosine 3′,5′-monophosphate (cAMP) and calcium signaling pathways at CT4 (fig. S5F), all of which are critical for SCN function and circadian entrainment (1, 2). Direct comparison of SCN-DKO at CT4 and ZT4 revealed a short list of 28 genes differentially expressed in both conditions (Fig. 5A). We confirmed that 22 of the 28 genes were up-regulated in SCN-DKO mice compared to control mice at both CT4 and ZT4 and contained an HA-REV-ERBα binding site (Fig. 5A) in the hypothalamus (23), strongly suggesting direct regulation by REV-ERBs [as illustrated for Arntl (BMAL1), Npas2, Nfil3 (E4BP4), and Ppp1r1b in Fig. 5, B and C]. Ppp1r1b was of particular interest because this gene encodes DARPP-32, an inhibitor of protein phosphatase 1 (PP1) (24). PP1 is the main phosphatase that functions along with casein kinase 1 (CK1) to regulate PER2 phosphorylation (25, 26). Mutation of CK1 and inhibition of PP1 have been shown to shorten circadian periods in vitro and in vivo by inducing PER2 hyperphosphorylation and degradation (26–28). These data suggest that REV-ERBs could regulate the master clock period, in part, by direct repression of Ppp1r1b expression. In this regard, we confirmed the up-regulation of Ppp1r1b in SCN-DKO mice at ZT4 by qPCR, as well as the dysregulation of Cry1 expression and minor changes of Cry2 and Per1/2 expression (fig. S5G). RNA-seq also revealed other pathways directly or indirectly regulated by REV-ERB deletion that could affect the master clock period including Cry1/2, Vipr2, dopaminergic signaling, as well as the cAMP and calcium signaling pathways (fig. S5F).
Fig. 5. Direct regulation of SCN gene transcription by REV-ERBs.
(A) Venn diagram comparison of differential gene expression in SCN-DKO mice compared to control mice at CT4 and ZT4. Genes written in bold are up-regulated in SCN-DKO mice at CT4 and ZT4; * indicates the presence of confident HA-REV-ERBα binding site in the arcuate hypothalamic nucleus. (B) Genome browser view of HA-REV-ERBα chromatin immunoprecipitation sequencing (ChIP-seq) peaks in the arcuate hypothalamic nucleus. (C) Gene expression comparison in SCN from control and SCN-DKO mice at ZT4 and CT4 (n = 2 to 3). Results are presented as means ± SEM and compared by two-way ANOVA (interaction P > 0.05).
SCN-DKO mice are entrained to 21-hour LD cycle contrary to control mice
In 1959, Pittendrigh and Bruce (29) formulated the “circadian resonance hypothesis” proposing that “LD cycles might be close enough to ensure entrainment of the endogenous clock but nevertheless sufficiently different to impair coordination of constituent intracellular processes” (29). SCN-DKO mice, harboring deletion of a core TTFL clock factor and a robust but abnormal endogenous pacemaker period, represent a unique mouse model to test the desynchrony hypothesis. In contrast with deletion of other core clock genes and their redundant isoform [Cry1/2, Per1/2, Bmal1 (10, 11, 18)], SCN REV-ERBs were not necessary for the master clock rhythmicity but had a major role in maintaining circadian pacemaker period.
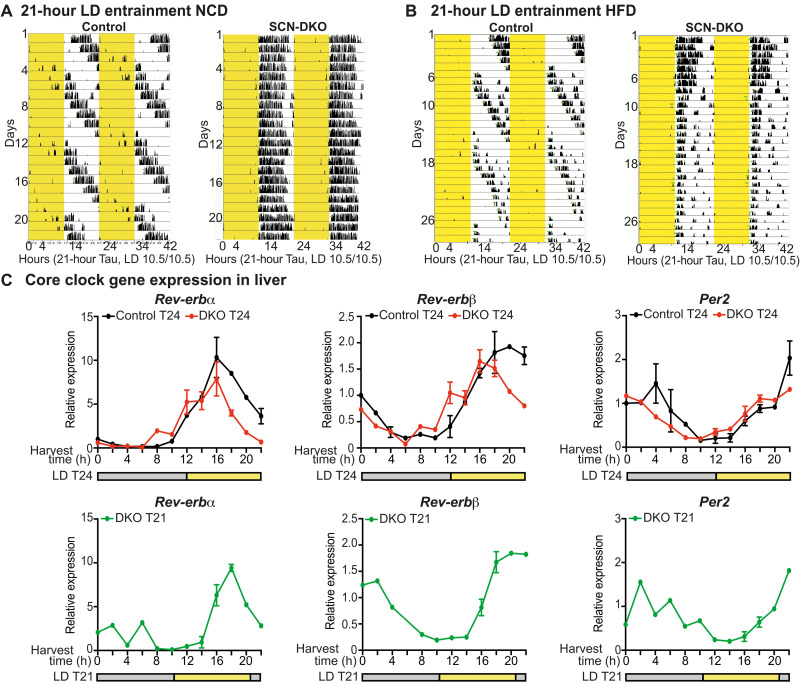
REV-ERB SCN-DKO mice were able to be entrained to LD cycle (T-cycle) T24 (12:12) (Fig. 1A) and T21 (10.5:10.5) on both a normal chow diet (NCD) (Fig. 6A) and 60% high-fat diet (HFD) (Fig. 6B), allowing a test of the circadian resonance hypothesis on metabolism, i.e., determining whether forced entrainment to a T24 rhythm impairs circadian metabolism coordination and homeostasis. While SCN-DKO mice were able to be entrained to T24 and T21 LD cycles, control mice were not entrainable to T21 LD cycles as observed in wheel-running activity on NCD and HFD (Fig. 6, A and B). Moreover, we evaluated core clock gene expression in livers from both control and SCN-DKO mice housed at T24 as well as SCN-DKO mice at T21 on HFD (Fig. 6C). Liver circadian gene expression in control mice could not be evaluated in T21 LD cycle as they are running through the LD cycle. Consistent with wheel-running activity, the liver clock maintains overall rhythmicity in SCN-DKO mice at both T24 and T21 (Fig. 6C). Together, these results confirmed that SCN-DKO mice are entrained to T21 LD cycle and the expected absence of entrainment in control mice at T21, as previously described in a 20-hour LD cycle (30). Notably, we observed that control mice housed in 21-hour LD cycle tend to show an exaggerated weight gain on HFD compared to 24-hour LD cycle (fig. S6, A and B), demonstrating that 21-hour LD cycle per se does not lead to reduced weight gain. However, as control mice are not entrained to the 21-hour LD cycle, the metabolic effect observed in control T21 mice could be triggered by mechanisms other than circadian dissonance and thus cannot be used as control for the following experiments testing the circadian resonance hypothesis on metabolism.
Fig. 6. SCN-DKO mice are entrained to 21-hour LD cycle contrary to control mice.
(A and B) Representative actogram plotted on 21-hour Tau of control and SCN-DKO male mice (n = 6) housed in LD (10.5:10.5) on NCD (A) or a 60% HFD (B). Light schedule is highlighted in yellow. (C) Circadian gene expression in liver from control (n = 1 to 3) and SCN-DKO (n = 1 to 3) male and female mice housed in T24 LD cycle (12:12) or T21 LD cycle (10.5:10.5) on HFD. Harvest time and LD cycle are presented. Results are presented as means ± SEM.
Matching LD cycle to the SCN-DKO mouse free-running period reduces their sensitivity to HFD-induced obesity
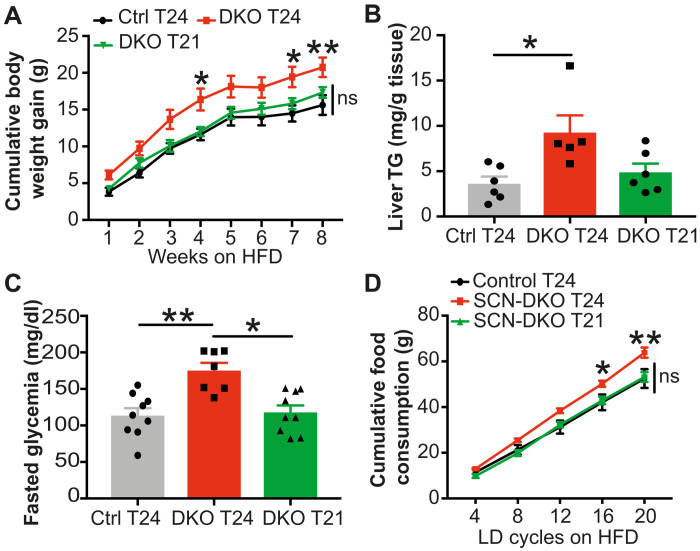
When SCN-DKO mice were housed in regular 12:12 LD cycle (T24) or Tau-like 10.5:10.5 LD cycle (T21) and fed on NCD, body weights were similar to control mice housed at T24 (fig. S6C). However, when fed with HFD, SCN-DKO mice housed at T24 rapidly gain more body weight compared to control mice housed at T24 (Fig. 7A and fig. S6D).
Fig. 7. Matching LD cycle to SCN-DKO mice free-running period reduces their sensitivity to diet-induced obesity and their food consumption.
(A) Cumulative body weight gain of control (n = 9) and SCN-DKO mice (n = 7 to 9) housed on T24 or T21 LD cycle on HFD. Results are presented as means ± SEM and compared by two-way repeated measures (RM) ANOVA (interaction P = 0.3148, genotype P = 0.0085, time P < 0.0001) followed by Sidak’s multiple comparisons test. (B) Liver triglycerides (TG) in control (n = 6) and SCN-DKO mice (n = 5 to 6) housed during 4 weeks on T24 or T21 LD cycle on HFD. Results are presented as means ± SEM and compared by Kruskal-Wallis test. (C) Blood glucose concentrations at CT2 after 14-hour fasting in control (n = 9) and SCN-DKO mice (n = 7 to 9) housed during 5 weeks on T24 or T21 LD cycle on HFD. Results are presented as means ± SEM and compared by Kruskal-Wallis test. (D) Cumulative food consumption per LD cycle in control mice housed on T24 (n = 5) or SCN-DKO mice housed on T24 or T21 (n = 4 to 6) on HFD. Results are presented as means ± SEM and compared by two-way RM ANOVA (interaction P = 0.0053) followed by Tukey’s multiple comparisons test (* indicates differences between SCN-DKO T24 and both control T24 and SCN-DKO T21). ns, not significant.
The desynchrony hypothesis predicts that the metabolic disturbances observed in SCN-DKO mice on HFD could be ameliorated by matching the external LD cycle to their dysfunctional endogenous pacemaker. SCN-DKO mice housed in 10.5:10.5 LD cycle (T21) were no longer more sensitive to DIO as observed in SCN-DKO mice housed on T24 LD (Fig. 7A and fig. S6D). Furthermore, whereas SCN-DKO mice housed on T24 LD cycle accumulated more inguinal adipose tissue than control mice, this difference was abrogated when SCN-DKO mice were housed on T21 LD cycle (fig. S6E). No changes were observed in epididymal white adipose tissue in any conditions studied (fig. S6F). SCN-DKO mice housed on a T24 LD cycle also manifest increased hepatic triglycerides (TG) compared to control mice housed at T24 LD cycle (Fig. 7B), a hallmark of nonalcoholic fatty liver disease that has become a major public health concern (31). However, the exacerbation of hepatic steatosis was corrected when the external LD cycle was adjusted to the endogenous free-running period of 21 hours in HFD-fed SCN-DKO mice (Fig. 7B). Similarly, fasted glycemia was increased in SCN-DKO mice housed on a T24 LD cycle on HFD compared to control mice housed at T24 LD cycle but rescued on a T21 LD cycle (Fig. 7C). Moreover, glucose tolerance at CT2 was decreased in SCN-DKO mice compared to control mice housed in T24 LD cycle (fig. S6G), while glucose tolerance was rescued in SCN-DKO housed in T21 LD cycle compared to SCN-DKO housed in T24 (fig. S6H). No changes were observed in insulin tolerance at CT2 (fig. S6, I and J), but SCN-DKO mice housed in T21 exhibited increased insulin tolerance compared to SCN-DKO mice housed in T24 at CT13 (fig. S6, K and L).
Previous studies of circadian desynchrony have shown that miscoordination of central and peripheral clocks leading to dysregulated feeding behavior and nonoptimal metabolic flux mediates increased sensitivity to DIO (3, 32, 33). The desynchrony between LD cycle period and internal period was associated with increased food consumption by SCN-DKO mice compared to control mice when housed in T24 LD cycle but not in T21 LD cycle (Fig. 7D and fig. S6, M and N). While it is unclear if feeding homeostasis is balanced regarding energy expenditure per LD cycle or per 24 hours, increased food consumption in SCN-DKO mice housed at T24 was observed when compared per LD cycle (Fig. 7D and fig. S6N) and per 24 hours (fig. S6M), suggesting that increased feeding in SCN-DKO mice was specific to T24 LD environment. Together, these results implicate circadian desynchrony between internal clocks and exogenous light cycles as the cause of the increased sensitivity to DIO observed in SCN-DKO mice, whose endogenous clocks run with a 21-hour period, when forced to be entrained on a 24-hour LD cycle.
DISCUSSION
We have shown that REV-ERBs are not necessary for circadian rhythmicity of the SCN master clock but play a major role in maintaining a free-running period close to 24 hours in vivo as well as ex vivo in PER2::LUC SCN slices. Moreover, the SCN-DKO mice exhibited adverse metabolic consequences when housed on 24-hour LD cycle. Adjusting external LD cycles to match their malfunctioning pacemaker period corrected the metabolic defects observed on HFD, providing strong evidence for the circadian desynchrony hypothesis (Fig. 8). These findings highlight a previously unknown function of REV-ERBs, demonstrating that SCN REV-ERBs are essential timekeepers and metabolic buffers to our 24-hour circadian world.
Fig. 8. Circadian desynchrony and metabolic dysfunction.
External LD cycles force the synchronization of the internal SCN-DKO clock, but the external/internal T-cycle dissonance increases feeding and sensitivity to DIO. This circadian dissonance can be corrected by adjusting the external LD cycle to the endogenous free-running period in mice lacking REV-ERBs in the SCN (SCN-DKO mice).
While there is no perfect model for specific Cre expression in the SCN, we confirmed that the expression of Rev-erbs was not affected in non-SCN hypothalamic nuclei as well as ventral forebrain and hindbrain structures. The mechanism underlying control of the master clock period by REV-ERBs remains to be determined, but one possibility is their direct repression of Ppp1r1b, which is known to control clock period through alteration of PER2 phosphorylation (24–26). This Ppp1r1b-dependent mechanism is consistent with the 21-hour free-running period observed in CK1 mutant mice leading to hyperphosphorylation of PER2 (26–28), but further work will be required to test this potential mechanism by which REV-ERBs could control the circadian period.
The molecular clock in mammals is organized in two TTFLs: The core loop is triggered by CLOCK/BMAL1 activity regulating Per1/2 and Cry1/2 circadian expression, while the secondary feedback loop is run by the RORα and REV-ERBα/β nuclear receptors regulating circadian expression of Bmal1. While REV-ERBα and REV-ERBβ nuclear receptors are recognized as key components of the molecular clock, several in vitro studies have demonstrated that the absence of REV-ERBs, which leads to constitutively high levels of Bmal1 expression, does not alter the circadian expression of Per1/2 and Cry1/2 (7, 8). These findings are consistent with the fact that constitutive expression of Bmal1 in Bmal1-deficient fibroblasts restores Per2 circadian expression (7). In vivo, Rev-erbα–deficient mice exhibit a modest change in circadian behavioral rhythms (6), which is likely limited by redundancy between Rev-erbα and Rev-erbβ (9). However, here, we demonstrated that whole-body deletion of both REV-ERBs results in pronounced locomotor defects and low rhythm penetrance in DD. Ultimately, this confounded the ability to accurately assess rhythmic behavior in the WB-DKO mice after 1 week in DD, as previously observed in inducible DKO mice (9).
We show here that, in stark contrast to earlier predictions (9), deletion of both REV-ERBs in the SCN master clock does not lead to an arrhythmic phenotype in DD as is seen in mice with mutations in the core clock (Per1-2−/− and Cry1-2−/−). Rather, these mice exhibit a markedly shortened free-running period, demonstrating that SCN REV-ERBs are necessary to stabilize the pacemaker period rather than maintain rhythmicity. This shortened period was retained in SCN slices studied ex vivo. Our results thus reconcile in vitro observations with mouse physiology and demonstrate the critical role of REV-ERBs in an auxiliary loop that fine-tunes the core loop. We note that while this work was under review, Ding et al. reported that mutation of REV-ERBs in GABA (γ-aminobutyric acid)–releasing (GABAergic) neurons also induced a shorter free-running period and altered glucose homeostasis (34). This work used a Rev-erbαflox mouse model that is not a KO, but produces a mutated protein that lacks the DNA-binding domain (DBD), and is massively overexpressed with potential for artifact due to squelching of the corepressors that also function with other nuclear receptors (12, 35). In addition, Ding et al. used VGAT-Cre that not only is known to target the SCN but also affects REV-ERB expression in lateral hypothalamus, which is known as a feeding and glucose sensing center (36–39). Deletion of REV-ERBs in these non-SCN hypothalamic nuclei has marked effects on diurnal food intake and leptin sensitivity (23). Thus, metabolic phenotypes of the mice in which REV-ERBs are mutated using VGAT-Cre are likely a combination of the metabolic effects caused by circadian misalignment induced by REV-ERB deletion in SCN, as demonstrated in the present study, as well as the effects of altering REV-ERBs in non-SCN hypothalamic nuclei. Last, Ding et al. also observed a shortened free-running period, which emphasizes the robust phenotype of REV-ERB deletion in the SCN and the absolute requirement of REV-ERBα’s DBD to maintain the pacemaker period.
The circadian resonance hypothesis formulated in 1959 (29) and tested in 1972 in Drosophila melanogaster (40) by Pittendrigh argues that oscillating systems perform most effectively when they are synchronized to their natural circadian frequency, i.e., when the free-running SCN period (Tau) and LD cycle (T-cycle) are aligned. A plethora of studies have demonstrated the cardiac and metabolic consequences of external circadian desynchrony induced by jetlag or shift work protocols, which model when the circadian system is contradicted and not entrained by the LD cycle (2–4, 33). The circadian resonance hypothesis, however, is more subtle, as even an LD cycle that is otherwise entrainable, but does not match an organism’s endogenous Tau, induces circadian dissonance, which leads to a reduction in circadian fitness and a shortened life span. In humans, comparative studies support the circadian resonance hypothesis paradigm, as Tau deviation from 24-hour LD during migration north correlates with susceptibility to obesity (41). Moreover, comparative studies across rodent and primate species highlight that a deviation of Tau from 24 hours is related to length of life span and obesity (41–43).
Regarding metabolism, a recent study in wild-type mice suggested that imposing an LD cycle oscillating at a Tau-like period length (23.7 hours) prevented HFD-induced circadian disruptions of locomotor activity and obesity (44), as previously suggested in wild-type mice exposed to a range of non–24-hour LD frequencies (45). Only a few of studies have corrected internal clock timing in mutant rodents by environmental change: CK1ε Tau-mutant heterozygote hamsters with a 22-hour Tau present cardiorenal pathologies when entrained to 24-hour T-cycle that was prevented by raising them on a 12:10 LD cycle from 4 to 17 months of age (46). Moreover, Zhou et al. (47) demonstrated that CK1ε (Tau/Tau) mice were protected against DIO on a 20-hour LD cycle. However, the CK1ε Tau mutations are germline and thus may have an impact on peripheral metabolic functions in addition to the brain (47, 48). The present studies using an SCN-specific model are the first to use an SCN-specific mutation of core clock components to demonstrate the consequences of disrupting circadian resonance on energy metabolism and thus more directly interrogate the metabolic consequences driven by SCN desynchrony with the LD cycle.
SCN REV-ERB DKO mice were able to be entrained to both 24- and 21-hour LD cycles. On the contrary, control mice cannot be entrained to 21-hour LD cycle, as previously described in a 20-hour LD cycle inducing a major circadian disruption and metabolic consequences (30) and thus cannot be used to test the circadian resonance hypothesis. However, we note that control mice housed in 21-hour LD cycles tended to gain more weight on HFD than control mice housed in 24-hour LD cycle, demonstrating that 21-hour LD cycles per se do not reduce body weight gain. Our results demonstrate that increased sensitivity to DIO observed in SCN-DKO at T24 LD cycle is prevented by housing them in a Tau-like T21 LD cycle.
Last, food consumption increased in SCN-DKO at T24 is prevented at T21. This key role of feeding behavior is consistent with previous reports highlighting that circadian miscoordination between central and peripheral clocks leads to dysregulated feeding behavior and increased sensitivity to HFD (3, 32, 33). Thus, this study provides further evidence that coordination between environment and SCN clock is crucial to coordinate feeding behavior and the maintenance of metabolic circadian homeostasis. Moreover, these results further support the notion that metabolic dysfunction induced by circadian dissonance is secondary to disrupted feeding behavior and, thus, could be reversed by restricted feeding, as previously demonstrated in clock-deficient mice (49). No metabolic phenotype was observed on NCD, which is consistent with studies showing that shift work exacerbates DIO (50) and HFD weakens the capacity of the master clock to be entrained by LD cycle (51), creating a vicious cycle that amplifies the consequences of circadian desynchrony between external time givers and the endogenous molecular clock period. Together, these results suggest that entrainment of SCN-DKO mice to a longer LD cycle relative to their internal clock period leads to increased food intake and weight gain. This finding is consistent with a previous study showing that housing wild-type mice under 27-hour LD cycles, which represents the same degree of external LD cycle lengthening relative to internal clock period, increased food intake under normal chow feeding conditions (45).
In sum, we have demonstrated a key role of REV-ERBs in the master clock as determinants of free-running period and highlight the key role of this auxiliary loop triggered by SCN REV-ERBs to adjust the mammalian core clock to our 24-hour circadian world. Moreover, we have provided experimental evidence that the circadian resonance hypothesis also applies to metabolic homeostasis. These results also provide a theoretical underpinning for evidence of REV-ERB dysfunction in human mental, metabolic, and cardiac diseases (52–54). More broadly, such defects could underlie human circadian desynchrony syndromes, which may be amenable to chronotherapeutic strategies to treat associated metabolic and behavioral disorders.
MATERIALS AND METHODS
Animals
As previously described, HA-REV-ERBα mice were generated and maintained on C57BL/6J genetic background (15). Reverbαflox/flox-Reverbβflox/flox mice (13) maintained on a C57BL/6J background (Jackson Labs Technologies Inc., stock 008661) were bred to Six3-Cre mice (14) (provided by S. Blackshaw) to induce SCN-specific deletion of REV-ERBs (SCN-DKO). Reverbαflox/flox mice (13) were also bred to Six3-Cre mice to induce SCN-specific deletion of REV-ERBα only (SCN-SKO). To induce whole-body deletion of REV-ERBs (WB-DKO mice), Reverbαflox/flox-Reverbβflox/flox mice were bred to CMV-Cre mice [B6.C-Tg(CMV-cre)1Cgn/J mice, Jackson Labs Technologies Inc., stock 006054]. Whole-body deletion of REV-ERBs was induced for at least three generations, and then Cre expression was excluded from the breeding scheme to avoid side effects of Cre expression. Reverbαflox/flox-Reverbβflox/flox mice (13) maintained on a C57BL/6J background (Jackson Labs Technologies Inc., stock 008661) were also bred to PER2::LUC mice (Jackson Labs Technologies Inc., stock 006852) for ex vivo recording of PER2::LUC bioluminescence. For both SCN-DKO and WB-DKO, the same primer set was designed to simultaneously detect Reverbαflox or ReverbαKO and Reverbβflox or ReverbβKO alleles in both mouse lines (9, 13). Except where otherwise noted, mice were bred and group-housed under a 12:12-hour LD cycle (lights on at 7 a.m., lights off at 7 p.m.) with free access to water and normal chow (NCD, LabDiet, 5010). All experiments were carried out on 3- to 5-month-old male littermates in SCN-DKO and HA-REV-ERBα mouse lines, or 6- to 10-week-old male littermates in WB-DKO mouse line. Animals used in the control groups are Reverbαflox/flox-Reverbβflox/flox or Reverbα+/+-Reverbβ+/+ littermates to the SCN-DKO or WB-DKO mice, respectively. Experiments performed in females are specified in the figure legends. All animal studies followed the guidelines of the Institutional Animal Care and Use Committee of the University of Pennsylvania in accordance with the guidelines of the National Institutes of Health.
Record and analysis of wheel-running activity
Mice were single-housed with a running wheel and ad libitum food and water in circadian cabinets. After 1 week of acclimation, photoentrainment was assayed in a 12:12-hour (T24) or 10.5:10.5-hour (T21) LD cycle and free-running rhythms were evaluated in constant darkness (DD) during 3 weeks. Wheel-running activity was recorded and analyzed using ClockLab software. Default parameters of ClockLab Analysis software (Actimetrics) were used to determine the onset and offset of wheel-running activity and circadian periods and to generate chi-square periodograms and actograms. To measure phase angle of entrainment, the time of onset of activity in each DD cycle was automatically detected via ClockLab software (Actimetrics) and then manually confirmed, adjusted, or excluded in a blinded fashion. Using the first seven onset times in DD, a linear regression was created in ClockLab to extrapolate back to the final day in LD. To determine the phase angle of entrainment, this extrapolated onset time was subtracted from the time of lights off.
Definition of CT in control and SCN-DKO mice
CT in control and SCN-DKO mice was determined in each mouse individually, as illustrated in Fig. 4A for gene expression analysis performed in DD at CT4 and CT16. First, Tau was individually calculated in each mouse after 3 weeks of housing in DD, and CT12 was determined from the activity traces as the onset of wheel-running activity in each mouse the day before or the day of the harvest. Then, circadian hour was calculated as circadian hour (min) = Tau/24*60, and harvest time at CT16 and CT4 was determined in each mouse as CT16 = CT12 + 4 circadian hours and CT4 = CT12 + 16 circadian hours. For experiments performed in different LD cycles (T24 versus T21), CT was determined using the same logic, i.e., one circadian hour (min) = Tau/24*60, and CT12 was defined as the onset of the active phase at lights off.
In vivo metabolic phenotyping by indirect calorimetry
Comprehensive Lab Animal Monitoring System (Oxymax Windows; Columbus Instruments, Columbus, OH, USA) was used to determine energy expenditure by indirect calorimetry [volume of O2 and CO2, respiratory-exchange ratio (RER), and heat production]. After 2 days of adaptation, experimental data were obtained for 4 days in 12:12-hour LD cycles (96 hours), directly followed by 7 days in DD (168 hours). Data acquisition and analysis were performed with the Oxymax and Prism software. Data were collected every 15 min sequentially.
Analysis of PER2::LUC luminescence in SCN slices
Control and SCN-DKO mice heterozygous for PER2::LUC were housed on 12:12 LD before brain extraction 2 to 4 hours before lights off. SCN slices were made at 200-μm thickness in cold slicing medium [1× Hanks’ balanced salt solution, penicillin-streptomycin (100 U/ml), sodium bicarbonate (375 μg/ml), and Hepes (2.38 mg/ml)] and cultured on membranes (Millipore, PICMORG50) in 35-mm dishes with 1.2 ml of culture medium [Dulbecco’s Modified Eagle’s medium (Sigma-Aldrich, D2902), penicillin-streptomycin (25 U/ml), sodium bicarbonate (352.5 μg/ml), Hepes (2.38 mg/ml), and d-glucose (3.5 mg/ml)]. For every 6 ml of culture medium, 6 μl of 0.1 M luciferin and 120 μl of 50X B-27 Plus were added. Slices were incubated in LumiCycle (Actimetrics) at 37°C. Luminescence counts were made for each sample every 10 min for 14 days, with a medium change on the seventh day. Luminescence traces were normalized by subtracting a 24-hour running mean (center-aligned) baseline. Period was measured by Lomb-Scargle periodogram on the normalized traces following the first medium change (days 8 to 14).
LD cycle modification on HFD
Mice were transferred to two satellite rooms in which LD cycle can be controlled manually and set to 12:12-hour LD or 10.5:10.5-hour LD. Mice were single-housed with ad libitum water and fed with HFD composed of 60:20:20 kcal percentage of fat/carbohydrate/protein (Research Diets, D12492i). Body weight and food consumption were manually recorded weekly and after 4, 8, 12, 16, and 20 LD cycles. For metabolic study, mice who have gained less than 1 g/week on HFD were excluded from the study and from all metabolic measurements performed (body weight gained, tissue mass, hepatic TG assay, blood glucose measurement, and food recording). For the three cohorts of mice reported in the paper, 2 control T24 mice were excluded out of 24 total, 2 control T21 mice were excluded out of 18 total, 2 SCN-DKO T24 mice were excluded out of 20 total, and 0 SCN-DKO T21 mice were excluded out of 27 mice total. For food intake measurements, mice who crumbled their food were excluded from food intake analysis: two control T24 mice were excluded out of seven total; one SCN-DKO T24 mouse was excluded out of five total; and zero SCN-DKO T21 mice were excluded out of six total. Exclusion based on technical issues related to recording food intake was not a criterion of exclusion for the entire metabolic study.
Glucose and insulin tolerance tests
For glucose tolerance test, mice were fasted for 14 hours and then injected intraperitoneally with glucose (2 g/kg) at CT2 in both mice housed at T24 and T21 LD cycles. For insulin tolerance test, mice were fasted for 4 hours and then injected intraperitoneally with insulin (0.5 U/kg) at CT2 or CT13 in both mice housed at T24 and T21 LD cycles. Blood glucose levels were monitored by tail bleed every 15 min after glucose and insulin injection.
Histology
HA-REV-ERBα staining was performed in SCN, as previously described in ARC (23). Briefly, HA-REV-ERBα mice were sacrificed at eight ZT points in 3-hour intervals (8 a.m.—ZT1, 11 a.m.—ZT4, 2 p.m.—ZT7, 5 p.m.—ZT10, 8 p.m.—ZT13, 11 p.m.—ZT16, 2 a.m.—ZT19, 5 a.m.—ZT22; n = 5 at each time point). Brains were embedded in optimum cutting temperature (OCT) compound, and 14-μm sections containing SCN were used to perform immunofluorescent staining for HA (C29F4, Cell Signaling Technology, 3724S), as previously described (23), and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired with an inverted Olympus IX81 microscope using MetaMorph software. Image-based Tool for Counting Nuclei (ITCN) plugin in ImageJ was used to quantify HA-REV-ERBα–positive cells in each area of interest and normalized to DAPI-positive nuclei.
FISH of SCN Rev-erbα (Nr1d1)
For quantification of SCN Nr1d1 mRNA expression, brains were rapidly removed and fixed in 4% paraformaldehyde in 0.1 M phosphate-buffered saline (PBS) (pH 7.4) overnight at 4°C and then transferred to a 30% sucrose solution in 0.1 M PBS before embedded in OCT compound. Subsequently, brains were sectioned on a cryostat in the coronal plane at 14-μm thickness and collected on Superfrost Plus slides (VWR) in three series of slides, each slide containing four brain sections. To identify the slices of interest, Nissl staining was performed on serial 1 sections. Sections taken through the rostral-caudal extent of the SCN were stored at −80°C until further analysis. As previously described (23), FISH was performed using RNAscope Multiplex Fluorescent Reagent kit v2 (ACDBio, 323110) per the manufacturer’s instructions. Detection was carried out using a custom probe produced by ACDBio for Rev-erbα (Nr1d1) mRNA targeting exons 4 and 5 (Mm-Nr1d1-O2-C3, 1097521-C3). Following a series of amplification steps, sections were mounted with Fluorogel DAPI-containing mounting medium (Thermo Fisher Scientific). Sections were visualized using a Keyence BZ-X800 fluorescence microscope under a 10× objective. Images presented are a representative view of a control and SCN-DKO to confirm the lack of robust Nr1d1 mRNA expression within the DKO SCN.
Hypothalamic nuclei microdissection
SCN, ARC, DMH, and VMH were dissected as previously described (23, 55). Briefly, tissue viability was maintained during the dissection using oxygenated and warm Earle’s balanced salt solution medium (EBSS; Sigma-Aldrich, D5758) complemented with sodium bicarbonate (0.44 g/100 ml EBSS) and glucose (0.884 g/100 ml EBSS). Brain sections (0.2 to 0.5 mm) of the hypothalamic area of interest were performed on TC-1 Tissue Chopper (Electron Microscopy Sciences, 93100) before micro-punches of SCN, ARC, and VMH/DMH. The specific enrichment of each hypothalamic nuclei was confirmed by reverse transcription qPCR (RT-qPCR).
Western blot
SCN, liver, and BAT samples were processed as previously described (56). For SCN, three biological replicates were pooled together. Western blot images were imaged using Bio-Rad ChemiDoc Imaging Systems. Antibodies used are as follows: REV-ERBα = Abcam 174309 (1:1000), BMAL1 = Bethyl A606-313A (1:1000), and Vinculin-HRP = Cell Signaling Technology E1E9V XP(R) (1:1000).
mRNA isolation
Two to four independent hypothalamic nuclei punches were pooled together per biological replicate and homogenized with 1 ml of QIAzol Lysis Reagent (Qiagen) using Pellet-Pestle (Kimble, 749515). For hypothalamic nuclei, total RNA was purified and collected from RNeasy Mini spin columns according to the RNeasy Lipid Mini Kit (Qiagen, 74804). For peripheral tissues, cerebellum, cerebral cortex, and hippocampus, total RNA was purified and collected from RNeasy Mini spin columns according to the RNeasy Mini Kit (Qiagen, 74106). For retina, total RNA was purified and collected from RNeasy Mini spin columns according to RNeasy Micro Kit (Qiagen, 74004).
Gene expression analysis (RT-qPCR)
Purified total RNA was used for complementary DNA (cDNA) synthesis (High Capacity cDNA Reverse Transcription Kit, Applied Biosystems, 43-688-14) and qPCR using Power SYBR Green PCR Master Mix (Applied Biosystems). The QuantStudio 6 Flex Real-Time PCR software and system were used, and the primers were validated (table S1). RT-qPCR results were analyzed by standard curve method and normalized to Tbp (all brain and retina samples) or 36B4 expression (BAT and liver).
RNA-seq library preparation and data analysis
Following procedures previously described (23), RNA integrity was examined using Agilent High Sensitivity RNA ScreenTape. One microgram of RNA samples with RNA integrity number >7 was used for RNA cleanup and library preparation with an Illumina TruSeq Stranded Total RNA Library Prep kit according to the manufacturer’s instructions. All barcoded libraries were quantified by the KAPA Library Quantification Kit (Roche) and equimolarly pooled for subsequent sequencing. High-throughput sequencing data were generated through either the Functional Genomics Core at University of Pennsylvania or Novagene. Sequencing reads were aligned to UCSC mm10 genome using STAR v 2.6 (57). Read counts were then obtained with featureCounts per standard manual’s instructions (58). We removed genes with low expression with transcripts per million (TPM) < 0.1. DEGs (cutoff defined as false discovery rate < 0.05, >0.1 TPM) were identified using DESeq2 (59). Heatmaps were generated in R with package “pheatmap” by (i) identifying DEGs between CT4 and CT16 with the aforementioned cutoffs separately for control and DKO mice, (ii) identifying CT4-CT16 DEGs that were unique to only control, and (iii) mapping their corresponding z-transformed TPM values across all biological replicates and treatment conditions. Heatmap color scheme was scaled accordingly for optimal graphical illustrations. Gene Ontology analyses and transcriptional factor enrichment were performed by Enrichr webserver (60). For TBA (61), promoter regions of 666 CT4-CT16 DEGs lost in SCN-DKO were used for prediction of factors whose motifs were enriched. TBA was performed with default parameters (https://github.com/jenhantao/tba) with multithreading and option -p to test for significance with a likelihood ratio test across five independent train-test iterations. Notably, RNA-seq experiments have been performed once at ZT4, CT4, or CT16 (n = 2 to 5). Thus, the results shown in Figs. 4 and 5 and fig. S5 do not reflect independent experiments/samples.
Statistical analysis
For graphing and statistical analysis, GraphPad software (Prism 7) was used, except for sequencing data for which RStudio software (v1.0.153) was used. For comparisons between two groups, statistical analyses were performed using Mann-Whitney test. One-way analysis of variance (ANOVA) and Holm-Sidak’s multiple comparisons test were used for the comparison between more than two groups. Last, two-way ANOVA was used for assessment of variable’s effects (time, diet, or genotype) followed by Sidak’s multiple comparisons test. Detailed statistical test(s) used and corresponding P value are described in figure legends. Data are presented as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
Acknowledgments
We gratefully acknowledge P. Dierickx, M. Kotte, and J. Bedont for valuable help and discussions and P. Jha and A. Reddy for use of the vibratome. We thank the Gene Expression and Functional Genomics Core of the Penn Diabetes Research Center (DK19525) for next-generation sequencing and L. Cheng from the Penn Molecular Pathology and Imaging core for help with histology.
Funding: M.A. was supported by American Diabetes Association Training Grants (1-18-PDF-126). L.N.W. was supported by NIH T32DK007314-40. M.R.H. was supported by NIH R01DKDK021397. M.A.L. was supported by NIH R01DK45586, the JPB Foundation, and the Cox Institute for Medical Research.
Author contributions: M.A. and M.A.L. conceived the project. M.A. and B.M.K. performed animal experiments. H.C.B.N. performed RNA-seq, ChIP-seq, and bioinformatics analysis. M.A., B.M.K., C.W.T., L.N.W., L.C.P., and B.J.C. performed research. M.C.T. performed PER2::LUC experiment. C.E.G., J.G., and M.R.H. helped perform the T-cycle and RNAscope experiments. M.A., B.M.K., H.C.B.N., and M.A.L. analyzed data. M.A. and M.A.L. wrote the manuscript. All authors edited and approved the manuscript.
Competing interests: M.A.L. is an advisory board member for Pfizer Inc., is a consultant to Madrigal and Calico, and receives support from Pfizer for research not overlapping with the work reported here. M.R.H. receives research funding from Eli Lilly & Co. and Boehringer Ingelheim that was not used in support of these studies. M.R.H. is the CEO of Cantius Therapeutics LLC that pursues biological work unrelated to the current study. The other authors declare that they have no competing interests.
Data and materials availability: Correspondence and requests for materials should be addressed to the lead contact: M.A.L. (lazar@pennmedicine.upenn.edu). There are restrictions to the availability of the mouse line generated in this study due to materials transfer agreement requirement. Mice bearing the Rev-erb alpha flox allele can be provided by M. Lazar and the University of Pennsylvania pending scientific review and a completed material transfer agreement. Requests should be submitted to lazar@pennmedicine.upenn.edu. Mice bearing the Rev-erb beta flox allele were obtained from the Institut Clinique de la Souris under a material transfer agreement that requires their permission to distribution. Requests should be submitted to ics@igbmc.fr. The codes supporting the current study are common algorithms described in Materials and Methods. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Table S2 contains all the raw data of the figures without data point values discernible in plots. The ChIP-seq data used are available at GSE148644 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148644). The RNA-seq data generated during this study are available at GSE152919 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE152919).
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
Table S1
Other Supplementary Material for this manuscript includes the following:
Table S2
REFERENCES AND NOTES
- 1.Hastings M. H., Maywood E. S., Brancaccio M., Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Maury E., Off the clock: From circadian disruption to metabolic disease. Int. J. Mol. Sci. 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arble D. M., Ramsey K. M., Bass J., Turek F. W., Circadian disruption and metabolic disease: Findings from animal models. Best Pract. Res. Clin. Endocrinol. Metab. 24, 785–800 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheer F. A., Hilton M. F., Mantzoros C. S., Shea S. A., Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc. Natl. Acad. Sci. U.S.A. 106, 4453–4458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bugge A., Feng D., Everett L. J., Briggs E. R., Mullican S. E., Wang F., Jager J., Lazar M. A., Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev. 26, 657–667 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preitner N., Damiola F., Luis-Lopez-Molina, Zakany J., Duboule D., Albrecht U., Schibler U., The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Liu A. C., Tran H. G., Zhang E. E., Priest A. A., Welsh D. K., Kay S. A., Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLOS Genet. 4, e1000023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda R., Tsuchiya Y., Koike N., Umemura Y., Inokawa H., Ono R., Inoue M., Sasawaki Y., Grieten T., Okubo N., Ikoma K., Fujiwara H., Kubo T., Yagita K., REV-ERBα and REV-ERBβ function as key factors regulating mammalian circadian output. Sci. Rep. 9, 10171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H., Zhao X., Hatori M., Yu R. T., Barish G. D., Lam M. T., Chong L. W., DiTacchio L., Atkins A. R., Glass C. K., Liddle C., Auwerx J., Downes M., Panda S., Evans R. M., Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485, 123–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng B., Albrecht U., Kaasik K., Sage M., Lu W., Vaishnav S., Li Q., Sun Z. S., Eichele G., Bradley A., Lee C. C., Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105, 683–694 (2001). [DOI] [PubMed] [Google Scholar]
- 11.van der Horst G. T., Muijtjens M., Kobayashi K., Takano R., Kanno S.-i., Takao M., de Wit J., Verkerk A., Eker A. P. M., van Leenen D., Buijs R., Bootsma D., Hoeijmakers J. H. J., Yasui A., Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Fang B., Emmett M. J., Damle M., Sun Z., Feng D., Armour S. M., Remsberg J. R., Jager J., Soccio R. E., Steger D. J., Lazar M. A., Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science 348, 1488–1492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dierickx P., Emmett M. J., Jiang C., Uehara K., Liu M., Adlanmerini M., Lazar M. A., SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc. Natl. Acad. Sci. U.S.A. 116, 12147–12152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bedont J. L., LeGates T. A., Slat E. A., Byerly M. S., Wang H., Hu J., Rupp A. C., Qian J., Wong G. W., Herzog E. D., Hattar S., Blackshaw S., Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep. 7, 609–622 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adlanmerini M., Carpenter B. J., Remsberg J. R., Aubert Y., Peed L. C., Richter H. J., Lazar M. A., Circadian lipid synthesis in brown fat maintains murine body temperature during chronic cold. Proc. Natl. Acad. Sci. U.S.A. 116, 18691–18699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. B., Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Onishi H., Yamaguchi S., Yagita K., Ishida Y., Dong X., Kimura H., Jing Z., Ohara H., Okamura H., Rev-erbalpha gene expression in the mouse brain with special emphasis on its circadian profiles in the suprachiasmatic nucleus. J. Neurosci. Res. 68, 551–557 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., Simon M. C., Takahashi J. S., Bradfield C. A., Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S., PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delezie J., Dumont S., Dardente H., Oudart H., Gréchez-Cassiau A., Klosen P., Teboul M., Delaunay F., Pévet P., Challet E., The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 26, 3321–3335 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Woldt E., Sebti Y., Solt L. A., Duhem C., Lancel S., Eeckhoute J., Hesselink M. K. C., Paquet C., Delhaye S., Shin Y., Kamenecka T. M., Schaart G., Lefebvre P., Nevière R., Burris T. P., Schrauwen P., Staels B., Duez H., Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 19, 1039–1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang B., Everett L. J., Jager J., Briggs E., Armour S. M., Feng D., Roy A., Gerhart-Hines Z., Sun Z., Lazar M. A., Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159, 1140–1152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adlanmerini M., Nguyen H. C. B., Krusen B. M., Teng C. W., Geisler C. E., Peed L. C., Carpenter B. J., Hayes M. R., Lazar M. A., Hypothalamic REV-ERB nuclear receptors control diurnal food intake and leptin sensitivity in diet-induced obese mice. J. Clin. Invest. 131, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang H. B., Horiuchi A., Watanabe T., Shih S. R., Tsay H. J., Li H. C., Greengard P., Nairn A. C., Characterization of the inhibition of protein phosphatase-1 by DARPP-32 and inhibitor-2. J. Biol. Chem. 274, 7870–7878 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Gallego M., Kang H., Virshup D. M., Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem. J. 399, 169–175 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H. M., Chen R., Kim H., Etchegaray J. P., Weaver D. R., Lee C., The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc. Natl. Acad. Sci. U.S.A. 108, 16451–16456 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng Q. J., Logunova L., Maywood E. S., Gallego M., Lebiecki J., Brown T. M., Sládek M., Semikhodskii A. S., Glossop N. R. J., Piggins H. D., Chesham J. E., Bechtold D. A., Yoo S. H., Takahashi J. S., Virshup D. M., Boot-Handford R. P., Hastings M. H., Loudon A. S. I., Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58, 78–88 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maywood E. S., Chesham J. E., Smyllie N. J., Hastings M. H., TheTau Mutation of casein kinase 1ϵ sets the period of the mammalian pacemaker via regulation of Period1 or Period2 clock proteins. J. Biol. Rhythms 29, 110–118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. S. Pittendrigh, V. G. Bruce, Daily rhythms as coupled oscillator systems and their relation to thermoperiodism and photoperiodism, in Photoperiodism and Related Phenomena in Plants and Animals, R. B. Withrow, Ed. (AAAS, 1959), p. 475–505. [Google Scholar]
- 30.Karatsoreos I. N., Bhagat S., Bloss E. B., Morrison J. H., McEwen B. S., Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. Proc. Natl. Acad. Sci. U.S.A. 108, 1657–1662 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Younossi Z. M., Non-alcoholic fatty liver disease—A global public health perspective. J. Hepatol. 70, 531–544 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Mukherji A., Kobiita A., Damara M., Misra N., Meziane H., Champy M. F., Chambon P., Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 112, E6691–E6698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West A. C., Bechtold D. A., The cost of circadian desynchrony: Evidence, insights and open questions. Bioessays 37, 777–788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding G., Li X., Hou X., Zhou W., Gong Y., Liu F., He Y., Song J., Wang J., Basil P., Li W., Qian S., Saha P., Wang J., Cui C., Yang T., Zou K., Han Y., Amos C. I., Xu Y., Chen L., Sun Z., REV-ERB in GABAergic neurons controls diurnal hepatic insulin sensitivity. Nature 592, 763–767 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding G., Li X., Hou X., Zhou W., Gong Y., Liu F., He Y., Song J., Wang J., Basil P., Li W., Qian S., Saha P., Wang J., Cui C., Yang T., Zou K., Han Y., Amos C. I., Xu Y., Chen L., Sun Z., Author correction: REV-ERB in GABAergic neurons controls diurnal hepatic insulin sensitivity. Nature 595, E2 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Myers M. G. Jr., Olson D. P., Central nervous system control of metabolism. Nature 491, 357–363 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Burdakov D., Karnani M. M., Gonzalez A., Lateral hypothalamus as a sensor-regulator in respiratory and metabolic control. Physiol. Behav. 121, 117–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karnani M., Burdakov D., Multiple hypothalamic circuits sense and regulate glucose levels. Am. J. Physiol. Regul. Integr. Comp. Physiol. 300, R47–R55 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evans M. C., Rizwan M. Z., Anderson G. M., Insulin action on GABA neurons is a critical regulator of energy balance but not fertility in mice. Endocrinology 155, 4368–4379 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Pittendrigh C. S., Minis D. H., Circadian systems: Longevity as a function of circadian resonance in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 69, 1537–1539 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wyse C. A., Does human evolution in different latitudes influence susceptibility to obesity via the circadian pacemaker?: Migration and survival of the fittest in the modern age of lifestyle-induced circadian desynchrony. Bioessays 34, 921–924 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Wyse C. A., Coogan A. N., Selman C., Hazlerigg D. G., Speakman J. R., Association between mammalian lifespan and circadian free-running period: The circadian resonance hypothesis revisited. Biol. Lett. 6, 696–698 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hozer C., Perret M., Pavard S., Pifferi F., Survival is reduced when endogenous period deviates from 24 h in a non-human primate, supporting the circadian resonance theory. Sci. Rep. 10, 18002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steckler R., Tamir S., Gutman R., Mice held at an environmental photic cycle oscillating at their tau-like period length do not show the high-fat diet-induced obesity that develops under the 24-hour photic cycle. Chronobiol. Int. 38, 598–612 (2021). [DOI] [PubMed] [Google Scholar]
- 45.West A. C., Smith L., Ray D. W., Loudon A. S. I., Brown T. M., Bechtold D. A., Misalignment with the external light environment drives metabolic and cardiac dysfunction. Nat. Commun. 8, 417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martino T. A., Oudit G. Y., Herzenberg A. M., Tata N., Koletar M. M., Kabir G. M., Belsham D. D., Backx P. H., Ralph M. R., Sole M. J., Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1675–R1683 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Zhou L., Summa K. C., Olker C., Vitaterna M. H., Turek F. W., Altered body weight regulation in CK1ε null and tau mutant mice on regular chow and high fat diets. Genet. Res. Int. 2016, 1–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loudon A. S., Meng Q. J., Maywood E. S., Bechtold D. A., Boot-Handford R. P., Hastings M. H., The biology of the circadian Ck1ε tau mutation in mice and Syrian hamsters: A tale of two species. Cold Spring Harb. Symp. Quant. Biol. 72, 261–271 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Chaix A., Lin T., Le H. D., Chang M. W., Panda S., Time-restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab. 29, 303–319.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S. M., Neuendorff N., Alaniz R. C., Sun Y., Chapkin R. S., Earnest D. J., Shift work cycle-induced alterations of circadian rhythms potentiate the effects of high-fat diet on inflammation and metabolism. FASEB J. 32, 3085–3095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maroni M. J., Capri K. M., Arruda N. L., Gelineau R. R., Deane H. V., Concepcion H. A., DeCourcey H., Monteiro De Pina I. K., Cushman A. V., Chasse M. H., Logan R. W., Seggio J. A., Substrain specific behavioral responses in male C57BL/6N and C57BL/6J mice to a shortened 21-hour day and high-fat diet. Chronobiol. Int. 37, 1–15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kang J. I., Park C. I., Namkoong K., Kim S. J., Associations between polymorphisms in the NR1D1 gene encoding for nuclear receptor REV-ERBα and circadian typologies. Chronobiol. Int. 32, 568–572 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Goumidi L., Grechez A., Dumont J., Cottel D., Kafatos A., Moreno L. A., Molnar D., Moschonis G., Gottrand F., Huybrechts I., Dallongeville J., Amouyel P., Delaunay F., Meirhaeghe A., Impact of REV-ERB alpha gene polymorphisms on obesity phenotypes in adult and adolescent samples. Int. J. Obes. 37, 666–672 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Tokat B., Kanca-Demirci D., Gul N., Satman I., Ozturk O., Ozder A., Kucukhuseyin O., Yilmaz-Aydogan H., Determination of genetic changes of Rev-erb beta and Rev-erb alpha genes in Type 2 diabetes mellitus by next-generation sequencing. Gene 763, 145058 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Atkins N., Miller C. M., Owens J. R., Turek F. W., Non-laser capture microscopy approach for the microdissection of discrete mouse brain regions for total RNA isolation and downstream next-generation sequencing and gene expression profiling. J. Vis. Exp. 3125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emmett M. J., Lim H. W., Jager J., Richter H. J., Adlanmerini M., Peed L. C., Briggs E. R., Steger D. J., Ma T., Sims C. A., Baur J. A., Pei L., Won K. J., Seale P., Gerhart-Hines Z., Lazar M. A., Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 546, 544–548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., Ma’ayan A., Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fonseca G. J., Tao J., Westin E. M., Duttke S. H., Spann N. J., Strid T., Shen Z., Stender J. D., Sakai M., Link V. M., Benner C., Glass C. K., Diverse motif ensembles specify non-redundant DNA binding activities of AP-1 family members in macrophages. Nat. Commun. 10, 414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Table S1
Table S2