Abstract
Four Australian hospital laboratories evaluated the performance of the Abbott LCx Mycobacterium tuberculosis assay with 2,347 specimens (2,083 respiratory and 264 nonrespiratory specimens) obtained from 1,411 patients. A total of 152 specimens (6.5%) were culture positive for Mycobacterium tuberculosis complex (MTBC); of these, 79 (52%) were smear positive. After resolution of discrepant data, the overall sensitivity, specificity, and positive and negative predictive values for the LCx assay were 69.7, 99.9, 99.1, and 97.7% respectively. For smear-positive respiratory specimens that were culture positive for MTBC, the values were 98.5, 100, 100, and 98.4%, respectively, while the values for smear-negative respiratory specimens were 41.5, 99.9, 96.4, and 98%, respectively. Relative operating characteristic curves were constructed to demonstrate the relationship between sensitivity and specificity for a range of possible cutoff values in the LCx assay. These graphs suggested that the assay sensitivity for respiratory samples could be increased from 70.2 to 78.6%, while the specificity would be reduced from 99.9 to 99.4% by inclusion of a grey zone (i.e., LCx assay values of between 0.2 and 0.99). An algorithm is presented for the handling of specimens with LCx assay values within this grey zone.
Culture is usually required for the laboratory confirmation of tuberculosis. The resulting isolate is also necessary to identify the organism to the species level, to determine drug susceptibility, and to obtain a molecular profile for epidemiological purposes. Unfortunately, substantial time delays occur (21) because conventional methods may require up to 8 weeks for cultures to become positive, although the radiometric systems (2) and the newer, nonradiometric, continuous monitoring systems (4, 29, 37, 44) have reduced the time to culture positivity. During this delay, patients with suspected tuberculosis and smears negative for acid-fast bacilli may be subjected to bronchoscopy or other invasive procedures to obtain a diagnosis or may be commenced on antituberculosis therapy. The worldwide reemergence of tuberculosis, the rise of multidrug-resistant Mycobacterium tuberculosis (12, 33), and the ongoing transmission of tuberculosis within and between high-risk groups (16, 40) have accelerated the search for more sensitive and rapid diagnostic laboratory methods.
The development of PCR and other nucleic acid amplification techniques has led to the introduction of in-house and commercial assays for the detection of M. tuberculosis complex (MTBC) directly from processed clinical samples. The advantages of commercial systems are that they are optimized and validated tests, they specifically identify the amplified product, and they use simplified protocols with greater automation.
The Abbott LCx Mycobacterium tuberculosis (LCx) assay (Abbott Diagnostics Division, Abbott Park, Ill.) uses the ligase chain reaction for the amplification of a segment of the single-copy gene that encodes protein antigen b. The gene is specific for members of the MTBC (38). The LCx assay was designed for use with processed respiratory specimens, although two studies have evaluated the assay with nonrespiratory specimens (18, 31). The aims of the present study were to evaluate the performance of the LCx assay with respiratory and nonrespiratory specimens and to review the setting of the sample rate/cutoff value, presently set at a value of 1.0, to determine whether the cutoff value may be reduced to improve test sensitivity without unduly compromising specificity.
MATERIALS AND METHODS
Clinical specimens and patients.
Four experienced mycobacteriology laboratories in Australia evaluated the LCx assay (Abbott Laboratories). Respiratory and nonrespiratory specimens were collected from patients under investigation for tuberculosis. Once the specimens had reached the laboratory, they were stored at 4°C until they were processed. After processing, an aliquot of sample was stored at −20 or −70°C until testing by the LCx assay. For specimens to be included in the study, microscopy, culture, and LCx assay results plus details about the patients including antituberculosis therapy were required. Specimens for which cultures were discontinued due to contamination were excluded from the study.
Decontamination and culture protocols.
All four laboratories used digestion and decontamination protocols for specimens likely to contain contaminating organisms. The processing and culture protocols used in each of the four participating laboratories are summarized in Table 1. BACTEC 12B vials containing 0.1 ml of PANTA-plus supplement were inoculated according to the manufacturer’s recommendations (0.5 ml for decontaminated specimens; up to 1.0 ml for specimens from usually sterile sites) onto Löwenstein-Jensen slants, which received 0.2 to 0.4 ml of specimen. MB/BacT vials containing 0.5 ml of antibiotic supplement including vancomycin) were inoculated according to the manufacturer’s recommendations (up to 0.5 ml for all specimen types). Inoculated BACTEC 12B and MB/BacT vials were incubated for a minimum of 6 weeks, although smear-positive specimens were incubated for an additional 4 weeks. Löwenstein-Jensen media were incubated for a minimum of 10 to 12 weeks, regardless of the smear status. Samples from sterile sites were cultured without decontamination. Tissues were macerated in sterile saline, and material from swabs was resuspended in sterile saline. Cultures of specimens obtained from superficial sites were incubated at 30 to 32°C and at 35 to 36°C. Laboratories that used the fluorochrome staining technique confirmed the results for all new smear-positive specimens by overstaining with the Ziehl-Neelsen stain. Smears were quantified by a recognized reporting scheme (22).
TABLE 1.
Processing and culture protocols used by the participating laboratories
| Laboratory | Decontaminationa | Stain for microscopyb | Inoculation procedurec
|
|
|---|---|---|---|---|
| Respiratory specimens | Nonrespiratory specimens | |||
| A | 2% NaOH-NALC | Auramine | BACTEC 12B ± L-J | BACTEC 12B + L-J |
| B | 2% NaOH-NALC | Auramine | BACTEC 12B + L-J | BACTEC 12B + L-J |
| C | 2% NaOH-NALC | Auramine | MB/BacT + L-J | MB/BacT + L-J |
| D | 2% NaOH | Z-N | L-J (±BACTEC 12B) | BACTEC 12B + L-J |
NALC, N-acetyl-l-cysteine. Laboratories A and D used 1 M phosphoric acid to halt digestion and decontamination, while laboratories B and C used phosphate-buffered saline (up to a 50-ml total volume). Specimens were concentrated by centrifugation (2,300 to 3,220 × g).
Smear samples were stained by the Ziehl-Neelsen (Z-N) technique, the auramine fluorochrome method (with all new smear-positive specimens confirmed by Ziehl-Neelsen overstaining).
After processing, respiratory and nonrespiratory specimens were inoculated into BACTEC 12B vials (Becton Dickinson, Sparks, Md.), MB/BacT broth (Organon Teknika Corp. Durham, N.J.), and/or onto Löwenstein-Jensen (L-J) slants. Details on the incubation temperatures and reading schedules used in each laboratory are available from the authors upon request. ±, with or without.
Identification of mycobacteria.
All isolates were checked for acid fastness. Conventional identification procedures based on physical and biochemical properties (22), multiplex PCR (13), commercial probes for culture confirmation (Accuprobe; Gen-Probe, Inc., San Diego, Calif.) (17, 25), or 16S rRNA gene sequence analysis (35) were used for the identification of mycobacterial or nocardial isolates.
LCx assay protocol.
The LCx assay was performed according to the manufacturer’s recommendations and comprised three stages: specimen preparation to remove potential amplification inhibitors, amplification by LCR technology, and detection via a microparticle enzyme immunoassay (MEIA). Duplicate negative and calibrator controls are included in each run. The MEIA result for each sample is compared with the averaged LCx assay calibrator control value and is expressed as the ratio of the sample rate/cutoff (S/CO) value multiplied by 0.3. Samples with an S/CO value of 1.0 or greater were considered positive. The four participating laboratories also included a positive control (e.g., M. bovis BCG) and a negative control (e.g., an atypical mycobacterium) in each run to confirm the assay performance.
Resolution of discrepant results.
A discrepancy occurred when the results of culture and the LCx assay differed from each other. In these situations, a review of the patient’s medical records was undertaken to determine a clinical diagnosis of tuberculosis or disease caused by MTBC on the basis of symptoms, signs, X rays, results of other laboratory investigations, and response to antituberculosis treatment. In this analysis for resolution of discrepant results, the LCx assay results were compared with the combined results of the “gold standard” of culture and the final clinical diagnosis.
Statistical analysis.
The data were tabulated and analyzed with a commercial computer program (Microsoft Excel 97; Microsoft Corporation, Redmond, Wash.). Epi Info (version 6.04b; Centers for Disease Control and Prevention, Atlanta, Ga.) was used to perform chi-square analyses to compare the performance characteristics of the LCx assay in each of the participating laboratories and to calculate the confidence intervals surrounding various estimates of sensitivity and specificity. Relative operating characteristic (ROC) curves were also constructed by plotting the sensitivity and specificity of the LCx assay for a range of cutoff values (5).
RESULTS
Specimen and patient analysis.
A total of 2,347 specimens were examined by microscopy, culture, and the LCx assay. Of these, 2,083 specimens were of respiratory origin (sputum or tracheal aspirates, 1,659; bronchoscopy specimens, 420; and gastric aspirates, 4). There were 264 nonrespiratory specimens (biopsy specimens, 98; pleural fluid, 49; lymph node, 36; other sterile site aspirates, 30; pus, 14; cerebrospinal fluid, 11; urine, 8; feces, 3), plus another 15 specimens from miscellaneous sites. The specimens were obtained from 1,411 patients; for 1,332 (94.4%) of the patients three or fewer specimens were tested in the study. The mean numbers of samples tested from each patient were 1.6 (standard deviation [SD], 1.1), 1.7 (SD, 1.1), and 1.2 (SD, 0.7) for all specimens combined, for respiratory samples, and for samples from nonrespiratory sites, respectively.
Culture and smear results.
Of the 2,347 specimens, 152 (6.5%) specimens (119 respiratory and 33 nonrespiratory specimens) from 98 patients were culture positive for MTBC (M. tuberculosis, 150; M. bovis BCG, 2). Three or fewer specimens were tested for 94% of the patients with MTBC culture-positive specimens. Of the MTBC culture-positive specimens, 79 (52%) had a positive smear result (respiratory specimens, 66; nonrespiratory specimens, 13).
Atypical mycobacteria were isolated from 112 specimens. The species isolated included M. avium complex (n = 67), M. abscessus (n = 17), M. fortuitum (n = 5), M. chelonae (n = 2), M. kansasii (n = 1), M. terrae (n = 1), and M. ulcerans (n = 1). A further 18 single isolates were not identified. Nocardia asteroides was recovered from a single respiratory specimen. None of these specimens were positive by the LCx assay.
Initial analysis of Abbott LCx assay.
The results of the LCx assay and microscopy were compared with those of culture and then with the results of the revised gold standard at four Australian hospital laboratories. No significant differences were noted in the performance of the LCx assay between the four laboratories (data not shown). For example, there was no statistical difference between the four estimates of the sensitivity of the LCx assay when culture was used as the gold standard (χ2 = 1.71, degrees of freedom = 3, P = 0.63). The pooled results from the four laboratories are therefore presented in the following analyses. For all specimens, when culture was used as the gold standard, microscopy had a sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of 52, 96.2, 48.8, and 96.7%, respectively. When stratified on the basis of specimen site, the sensitivities of microscopy for respiratory and nonrespiratory specimens were 55.5 and 39.4%, respectively.
When the LCx assay was compared to culture, the sensitivity, specificity, PPV, and NPV for all specimens were 75, 98.9, 80.9, and 98.3%, respectively. When stratified by specimen site, the values for respiratory specimens were 77.3, 98.9, 80.7, and 98.6%, respectively, and the values for nonrespiratory specimens were 66.7, 97.8, 81.5, and 95.4%, respectively (Table 2). The performance of the LCx assay for smear-positive and smear-negative respiratory specimens is presented in Table 2. Notably, the sensitivity for smear-positive respiratory specimens was 98.5%, but it was only 50.9% for those that were smear negative. A similar analysis by smear status was not performed for the nonrespiratory specimens because of the smaller sample size.
TABLE 2.
Initial comparison of LCx assay results with culture results
| Specimen (no.) | No. of specimens MTBC positive by culture
|
No. of specimens MTBC negative by culture
|
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | ||
|---|---|---|---|---|---|---|---|---|
| LCx assay positive | LCx assay negative | LCx assay positive | LCx assay negative | |||||
| All (2,347) | 114 | 38 | 27 | 2,168 | 75.0 | 98.8 | 80.9 | 98.3 |
| Respiratory (2,083) | 92 | 27 | 22 | 1,942 | 77.3 | 98.9 | 80.7 | 98.6 |
| Smear positive (139) | 65 | 1 | 10 | 63 | 98.5 | 86.3 | 86.7 | 98.4 |
| Smear negative (1,944) | 27 | 26 | 12 | 1,879 | 50.9 | 99.4 | 69.2 | 98.6 |
| Nonrespiratory (264) | 22 | 11 | 5 | 226 | 66.7 | 97.8 | 81.5 | 95.4 |
Discrepant analysis of Abbott LCx assay.
Laboratory and clinical data for specimens with discordant results were reviewed. This analysis for resolution of discrepant results involved the exclusion and reclassification of some respiratory and nonrespiratory specimens: 42 specimens from patients on treatment longer than 7 days were excluded (24 LCx assay-positive and culture-negative specimens, 17 LCx assay-negative and culture-negative specimens, and 1 LCx assay-positive and culture-positive specimen), 2 culture-negative specimens with high LCx assay values were reclassified as MTBC positive on the basis of clinical criteria, as were 12 LCx assay-negative and culture-negative samples. Of the 14 specimens reclassified as MTBC positive, 9 were from patients from whom samples were collected from the same or a similar site and whose samples were culture positive for MTBC. The remaining five specimens came from patients without laboratory confirmation of tuberculosis but whose clinical presentation, radiographic findings, and response to treatment resulted in a final diagnosis of tuberculosis. In the analysis of the data for resolution of discrepant results, the sensitivity, specificity, PPV, and NPV of the LCx assay were 69.7, 99.9, 99.1, and 97.7%, respectively. For respiratory specimens, the values were 70.2, 99.9, 98.9, and 98.0%, respectively, and for nonrespiratory specimens, the values were 67.6, 100, 100, and 95.3%, respectively (Table 3).
TABLE 3.
Assessment of LCx assay results after analysis for resolution of discrepant results (culture result plus clinical diagnosis)
| Specimen (no.) | No. of specimens MTBC positive by culture
|
No. of specimens MTBC negative by culture
|
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | ||
|---|---|---|---|---|---|---|---|---|
| LCx assay positive | LCx assay negative | LCx positive | LCx assay negative | |||||
| All (2,347) | 115 | 50 | 1 | 2,139 | 69.7 | 99.9 | 99.1 | 97.7 |
| Respiratory (2,083) | 92 | 39 | 1 | 1,915 | 70.2 | 99.9 | 98.9 | 98.0 |
| Smear positive (139) | 65 | 1 | 0 | 60 | 98.5 | 100 | 100 | 98.4 |
| Smear negative (1,944) | 27 | 38 | 1 | 1,855 | 41.5 | 99.9 | 96.4 | 98.0 |
| Nonrespiratory (264) | 23 | 11 | 0 | 224 | 67.6 | 100 | 100 | 95.3 |
The analysis for resolution of discrepant results also found that the LCx assay performed better for smear-positive respiratory specimens than for smear-negative respiratory specimens (Table 3). For smear-positive respiratory specimens, the values were 98.5, 100, 100, and 98.4%, respectively, and for smear-negative respiratory specimens, the values were 41.5, 99.9, 96.4, and 98.0%, respectively.
The estimated sensitivities of the LCx assay for smear-positive and smear-negative nonrespiratory samples were 84.6% (95% confidence interval, 53.7 to 97.2%) and 57.1% (95% confidence interval, 34.4 to 77.4%), respectively. These confidence intervals are wide because only 34 nonrespiratory specimens had a final diagnosis of tuberculosis (Table 3).
ROC curve analysis.
In the LCx assay, samples with results that fall above or below an established cutoff value are defined as positive or negative, respectively. The cutoff value is the mean rate for the LCx assay calibrator duplicates multiplied by 0.30. When the S/CO value is 1.0 or greater, the LCx assay result is interpreted as positive, while S/CO values of less than 1.0 indicate a negative result.
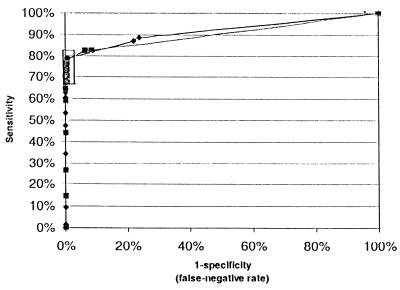
In addition to evaluating the performance of the LCx assay at this one predetermined cutoff value, ROC curves were constructed for respiratory and nonrespiratory specimens by using the resolved data set to show the correlation of sensitivity and specificity over a range of cutoff values. Inspection of Fig. 1 suggested that lowering of the cutoff to 0.2 could improve sensitivity with only a marginal reduction in specificity for both specimen types. This impression was confirmed by reanalyzing the LCx results for respiratory specimens (of which this study had a large cohort of 2,083 specimens) and comparing the sensitivity and specificity of the assay at different cutoff values (Table 4). Inclusion of a grey zone of LCx assay values of from 0.2 to 0.99 improved the sensitivity by 8.4%, while it reduced the specificity by only 0.5%. Overall, 21 respiratory samples, including 11 positive for MTBC by analysis for resolution of discrepant results, and 3 nonrespiratory samples, including 2 positive for MTBC by analysis for resolution of discrepant results, produced LCx assay values within this grey zone. Of the 13 specimens whose values fell within this grey zone but that were ultimately classified as MTBC positive, only one respiratory specimen was smear positive.
FIG. 1.
ROC curve for LCx assay for respiratory and nonrespiratory specimens. These ROC curves were constructed on the basis of the performance of the Abbott LCx assay in the analysis for resolution of discrepant results for isolates from respiratory (⧫) and nonrespiratory (■) specimens. The boxed area represents the performance of the assay for cutoff values of 0.1 to 1.0. Table 4 describes the sensitivity and specificity of the assay for various cutoff values within this range.
TABLE 4.
Performance of LCx assay at various cutoff values when testing respiratory specimens
| Cutoff value | Sensitivity (% [95% confidence interval]) | Specificity (% [95% confidence interval]) |
|---|---|---|
| 0.2 | 78.6 (70.4–85.1) | 99.4 (98.9–99.7) |
| 0.5 | 75.6 (67.1–82.5) | 99.5 (99.0–99.7) |
| 0.75 | 72.5 (63.9–79.8) | 99.8 (99.5–99.9) |
| 1.0 | 70.2 (61.5–77.7) | 99.9 (99.7–100) |
The use of this grey zone was then investigated by reviewing the S/CO values for both respiratory and nonrespiratory specimens. The mean S/CO value for specimens positive for MTBC by analysis for resolution of discrepant results was 3.05 (range, 0.03 to 7.56). Of the 50 specimens that produced false-negative LCx assay results, 13 had values within the grey zone. Adequate volumes remained for retesting of five of these specimens; the grey zone LCx assay values were reproducible for all five specimens, with the results of repeat LCx tests ranging from 0.45 to 1.16.
The mean S/CO value for 2,140 specimens negative for MTBC by analysis for resolution of discrepant results was 0.06 (range, 0.03 to 1.08). Only 11 of these MTBC-negative specimens had S/CO values between 0.2 and 0.99. Sufficient volumes remained for nine of the specimens to be retested; eight returned an S/CO value of 0.04 or less. The other specimen had an initial S/CO value of 0.73 and a value of 0.66 on repeat testing. This sputum specimen was negative by microscopy and culture, and an additional sputum sample was collected and was negative by all assays, including the LCx assay.
Finally, one sputum specimen reproducibly gave an LCx assay value of 1.08 that was defined as a false-positive result. The specimen was collected from an elderly nursing home resident who had been exposed to a smear-positive tuberculosis patient. This individual who had contact with a patient with tuberculosis has been monitored for more than 12 months, and the results of all other microbiological and radiological investigations have remained negative.
DISCUSSION
This collaborative study is the most extensive evaluation of the LCx assay published to date and was based on the testing of 152 culture-positive samples from among 2,083 respiratory and 264 nonrespiratory specimens. After resolution of discrepant results, the sensitivity, specificity, PPV, and NPV for respiratory specimens were 70.2, 99.9, 98.9, and 98.0%, respectively. These findings agree with previous evaluations of the LCx assay, which reported values of 77.1 to 90.2, 98.4 to 100, 72.8 to 100%, and 90.5 to 99.5%, respectively (3, 20, 24, 28, 31, 36, 39, 45). The assay sensitivities for smear-positive and smear-negative respiratory specimens were 98.5 and 41.5%, respectively (Table 3). Again, these values are similar to those reported previously (i.e., 92.1 to 100 and 36.8 to 73.3%, respectively).
On the basis of sensitivity and specificity, the performance of the LCx assay is comparable to those of other commercially available nucleic acid amplification tests (NAATs). For resolved data, the sensitivities for smear-positive and smear-negative pulmonary specimens were 93 to 100 and 43 to 81%, respectively, for the Gen-Probe amplified Mycobacterium tuberculosis direct test (8, 9, 14, 26, 30, 41, 42), 89 to 97.6 and 42.9 to 74%, respectively, for the Roche Mycobacterium tuberculosis Amplicor test (6, 7, 10, 14, 15, 27), and 90 to 96 and 48 to 68%, respectively, for the Cobas Amplicor test (32, 34). Although the LCx assay is not licensed for use with nonrespiratory specimens, the sensitivities were 84.6 and 57.1% for smear-positive and smear-negative specimens, respectively, in the present evaluation; these findings were comparable to those of other studies investigating the performance of the LCx assay with nonrespiratory specimens (81.8 to 100 and 35.3 to 71.1%, respectively (18, 31).
However, comparison of NAATs for the detection of MTBC nucleic acid directly from clinical specimens is problematic for several reasons. The proportion of smear-positive and culture-positive specimens in a study cohort affects the estimate of assay sensitivity because the ability of an NAAT to detect MTBC DNA is directly related to the concentration of bacilli in the specimen (3, 27, 28). For example, Moore and Curry (28) found that all specimens with >500 CFU of MTBC per ml were LCx assay positive, but only 44% were positive when specimens contained <500 CFU of MTBC per ml. Ausina et al. (3) found that all 18 specimens with <100 CFU of MTBC per ml were negative by the LCx assay. Similar findings have been noted for the Gen-Probe AMTDT and Roche Amplicor assays (27). Hence, studies with a high rate of smear-positive and culture-positive samples have found the LCx assay to be about 90% sensitive (3, 24, 39), while studies with lower rates of strongly positive results for samples have reported sensitivities of about 78% (20, 28).
NAAT evaluations may also be biased as a result of the inclusion of multiple specimens from individual patients (11). More than one specimen from each patient should be tested to provide sufficient opportunity for the detection of MTBC (24, 26, 32), especially in specimens smear negative for acid-fast bacilli but culture positive for MTBC. However, when more than three specimens are tested from a given patient, bias may be introduced (11). The present study has attempted to avoid these biases. Consecutive samples submitted to the participating laboratories were tested; the sample cohort was not enriched with positive samples; hence, the prevalence of culture-positive samples was relatively low (i.e., 6.5%) and the sensitivity of the present study is among the lower of published estimates (3, 20, 24, 28, 31, 36, 39, 45). Furthermore, an average of only 1.6 specimens were collected per patient, and the 152 culture-positive specimens were collected from 98 patients (for 94% of these patients three or fewer samples were collected, with an average of 1.5 specimens collected from each patient).
Studies have also varied in their handling of discrepant results. Several evaluations have defined culture-negative, LCx assay-positive specimens from patients on antituberculosis treatment as having false-negative culture results (3, 18, 20, 24, 36, 39). At present, there is no clinical utility in detecting MTBC DNA or RNA in specimens from currently (or previously) treated patients. Amplification tests may remain positive for extended periods of time and are not useful for the monitoring of patients undergoing treatment (3, 10, 14, 19, 43). Specimens from patients who had received antituberculosis therapy for 7 days or longer were therefore excluded from the analysis for resolution of discrepant results. Similarly, six specimens from patients who had completed a course of antituberculosis treatment and who were considered clinically cured but who remained LCx assay positive were not included in the analysis for resolution of discrepant results. The reproducible borderline-positive LCx assay result obtained for the elderly individual who had contact with a patient with tuberculosis was defined as false positive on clinical, radiological, and laboratory grounds. Alonso et al. (1) have also reported reproducible borderline-positive LCx assay results when they tested a sputum sample from a person who had been in contact with a patient with tuberculosis.
Construction of ROC curves suggested that reducing the cutoff value to 0.2 could improve the sensitivity of the LCx assay. Only a marginal loss of specificity would result because the LCx assay values for specimens from patients negative for MTBC cluster around a low mean value (0.06 in the present study). Only 11 (0.5%) of these specimens had S/CO values within the grey zone of 0.2 to 0.99. Yuen et al. (45) reported a mean LCx assay value for negative samples of 0.035 and also noted the wide separation of readings for positive and negative samples in the LCx assay. They suggested the use of a lower cutoff but did not propose a particular grey zone.
While a larger evaluation of specimens whose results fall in the grey zone is necessary, a simple algorithm for classification of these specimens as positive or negative is proposed. Repeat testing of nine samples from the present study with initial LCx assay values between 0.2 and 0.99 demonstrated that, for specimens from patients without tuberculosis, repeat testing produced a true-negative result for eight samples. In contrast, specimens culture positive for MTBC with an initial LCx assay result between 0.2 and 0.99 repeatedly produced S/CO values within the grey zone. The preponderance of smear-negative, culture-positive specimens in this group suggests that inclusion of a grey zone may facilitate detection of samples containing smaller numbers of bacilli. Our laboratories now perform repeat tests for any specimen with LCx assay values between 0.2 and 0.99; those that give a negative result on repeat testing are reported as such. Specimens that repeatedly produce S/CO values within the grey zone are reported as “equivocal” (until further experience is gained with these borderline samples), but the clinician is informed personally of the high likelihood of the presence of MTBC. The handling of specimens with results in the grey zone highlights the fact that effective communication between the clinician and the laboratory is even more important when new laboratory tests are being introduced.
The LCx assay integrates well into the routine diagnostic laboratory. Because the amplification and detection steps are automated, once specimen preparation is completed, the assay requires minimal involvement from laboratory staff, with the time to test completion being approximately 5 h. Specimen preparation involves two washing steps and appears to reduce the potential for specimen inhibitors to interfere with the assay (23). An internal amplification control is not included in the assay, so a second tube should be spiked with whole MTBC cells to check for the presence of inhibitors. Additionally, the assay may be held at almost any step of the procedure, further enhancing its integration into the laboratory work flow. The kit instruction sheet and equipment manuals are comprehensive and easy to follow, making the assay protocol straightforward.
In conclusion, the Abbott LCx assay is a well-presented, straightforward, rapid, and reliable semiautomated NAAT for the detection of MTBC directly from respiratory and nonrespiratory specimens. The LCx assay detects MTBC in almost all smear-positive, culture-positive and nearly half of smear-negative, culture-positive specimens. With modification of the cutoff value, a further improvement in test performance is possible. It is a relevant adjunct test in circumstances in which a rapid diagnosis of tuberculosis may have a substantial impact upon patient management and public health considerations. Microscopy and culture remain mandatory components of mycobacterial investigations.
ACKNOWLEDGMENTS
We thank Abbott Diagnostics for supplying the LCx assay kits. We acknowledge the technical assistance of Allan Goodwin and Shirley Ellis.
I.B. is supported by a Neil Hamilton Fairley Fellowship (fellowship 987069) awarded by the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Alonso P, Orduña A, Bratos M A, San Miguel A, Rodriguez Torres A. Clinical evaluation of a commercial ligase-based gene amplification method for detection of Mycobacterium tuberculosis. Eur J Clin Microbiol Infect Dis. 1998;17:371–376. doi: 10.1007/BF01691563. [DOI] [PubMed] [Google Scholar]
- 2.Anargyros P, Astill D S J, Lim I S L. Comparison of improved BACTEC and Lowenstein-Jensen media for culture of mycobacteria from clinical specimens. J Clin Microbiol. 1990;28:1288–1291. doi: 10.1128/jcm.28.6.1288-1291.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausina V, Gamboa F, Gazapo E, Manterola J M, Lonca J, Matas L, Manzano J R, Rodrigo C, Cardona P J, Padilla E. Evaluation of the semiautomated Abbott LCx Mycobacterium tuberculosis assay for direct detection of Mycobacterium tuberculosis in respiratory specimens. J Clin Microbiol. 1997;35:1996–2002. doi: 10.1128/jcm.35.8.1996-2002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badak Z F, Kiska D L, Setterquist S, Hartley C, O’Connell M A, Hopfer R L. Comparison of the mycobacteria growth indicator tube with BACTEC 460 for detection and recovery of mycobacteria from clinical specimens. J Clin Microbiol. 1996;34:2236–2239. doi: 10.1128/jcm.34.9.2236-2239.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck J R, Shultz E K. The use of relative operating characteristic (ROC) curves in test performance evaluation. Arch Pathol Lab Med. 1986;110:13–20. [PubMed] [Google Scholar]
- 6.Bennedsen J, Thomsen V E, Pfyffer G E, Funke G, Feldmann K, Beneke A, Jenkins P A, Hegginbothom M, Fahr A, Hengstler M, Cleator G, Klapper P, Wilkins E G L. Utility of PCR in diagnosing pulmonary tuberculosis. J Clin Microbiol. 1996;34:1407–1411. doi: 10.1128/jcm.34.6.1407-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann J S, Woods G L. Clinical evaluation of the Roche AMPLICOR PCR Mycobacterium tuberculosis test for detection of M. tuberculosis in respiratory specimens. J Clin Microbiol. 1996;34:1083–1085. doi: 10.1128/jcm.34.5.1083-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodmer T, Gurtner A, Schopfer K, Matter L. Screening of respiratory tract specimens for the presence of Mycobacterium tuberculosis by using the Gen-Probe amplified Mycobacterium Tuberculosis Direct Test. J Clin Microbiol. 1994;32:1483–1487. doi: 10.1128/jcm.32.6.1483-1487.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodmer T, Möckl E, Mühlemann K, Matter L. Improved performance of Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test when 500 instead of 50 microliters of decontaminated sediment is used. J Clin Microbiol. 1996;34:222–223. doi: 10.1128/jcm.34.1.222-223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpentier E, Drouillard B, Dailloux M, Moinard D, Vallee E, Dutilh B, Maugein J, Bergogne-Berezin E, Carbonnelle B. Diagnosis of tuberculosis by Amplicor Mycobacterium tuberculosis test: a multicenter study. J Clin Microbiol. 1995;33:3106–3110. doi: 10.1128/jcm.33.12.3106-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charache P. Comparison of the amplified Mycobacterium tuberculosis (MTB) direct test, Amplicor MTB PCR, and IS6110-PCR for detection of MTB in respiratory specimens. Clin Infect Dis. 1996;23:1107–1108. doi: 10.1093/clinids/23.5.1099. . (Editorial response.) [DOI] [PubMed] [Google Scholar]
- 12.Cohn D L, Bustreo F, Raviglione M C. Drug-resistant tuberculosis: review of the worldwide situation and the WHO/IUATLD global surveillance project. Clin Infect Dis. 1997;24:S121–S130. doi: 10.1093/clinids/24.supplement_1.s121. [DOI] [PubMed] [Google Scholar]
- 13.Cousins D V, Wilton S D, Francis B R, Gow B L. Use of polymerase chain reaction for rapid diagnosis of tuberculosis. J Clin Microbiol. 1992;30:255–258. doi: 10.1128/jcm.30.1.255-258.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalovisio J R, Montenegro-Jones S, Kemmerly S A, Genre C F, Chambers R, Freer D, Pankey G A, Failla D M, Haydel K G, Hutchinson L, Lindley M F, Nunez B M, Praba A, Eisenach K D, Cooper E S. Comparison of the amplified Mycobacterium tuberculosis (MTB) direct test, Amplicor MTB PCR, and IS6110-PCR for detection of MTB in respiratory specimens. Clin Infect Dis. 1996;23:1099–1106. doi: 10.1093/clinids/23.5.1099. [DOI] [PubMed] [Google Scholar]
- 15.D’Amato R F, Wallman A A, Hochstein L H, Colaninno P M, Scardamaglia M, Ardila E, Ghouri M, Kim K, Patel R C, Miller A. Rapid diagnosis of pulmonary tuberculosis by using Roche AMPLICOR Mycobacterium tuberculosis PCR test. J Clin Microbiol. 1995;33:1832–1834. doi: 10.1128/jcm.33.7.1832-1834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dwyer B, Jackson K, Raios K, Sievers A, Wilshire E, Ross B. DNA restriction fragment analysis to define an extended cluster of tuberculosis in homeless men and their associates. J Infect Dis. 1993;167:490–494. doi: 10.1093/infdis/167.2.490. [DOI] [PubMed] [Google Scholar]
- 17.Evans K D, Nakasone A S, Sutherland P A, de la Maza L M, Peterson E M. Identification of Mycobacterium tuberculosis and Mycobacterium avium-M. intracellulare directly from primary BACTEC cultures by using acridinium-ester-labeled DNA probes. J Clin Microbiol. 1992;30:2427–2431. doi: 10.1128/jcm.30.9.2427-2431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamboa F, Dominguez J, Padilla E, Manterola J M, Gazapo E, Lonca J, Matas L, Hernandez A, Cardona P J, Ausina V. Rapid diagnosis of extrapulmonary tuberculosis by ligase chain reaction amplification. J Clin Microbiol. 1998;36:1324–1329. doi: 10.1128/jcm.36.5.1324-1329.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamboa F, Manterola J M, Viñado B, Matas L, Gimenez M, Lonca J, Manzano J R, Rodrigo C, Cardona P J, Padilla E, Dominguez J, Ausina V. Direct detection of Mycobacterium tuberculosis complex in nonrespiratory specimens by Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test. J Clin Microbiol. 1997;35:307–310. doi: 10.1128/jcm.35.1.307-310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrino M G, Glupczynski Y, Degraux J, Ninet H, Delmée M. Evaluation of the Abbott LCx Mycobacterium tuberculosis assay for direct detection of Mycobacterium tuberculosis complex in human samples. J Clin Microbiol. 1999;37:229–232. doi: 10.1128/jcm.37.1.229-232.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huebner R E, Good R C, Tokars J I. Current practices in mycobacteriology: results of a survey of state public health laboratories. J Clin Microbiol. 1993;31:771–775. doi: 10.1128/jcm.31.4.771-775.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kent P T, Kubica G P. Public health mycobacteriology: a guide to the level III laboratory. Atlanta, Ga: Centers for Disease Control; 1985. [Google Scholar]
- 23.Leckie G W, Erickson D D, He Q, Facey I E, Lin B-C, Cao J, Halaka F G. Method for reduction of inhibition in a Mycobacterium tuberculosis-specific ligase chain reaction DNA amplification assay. J Clin Microbiol. 1998;36:764–767. doi: 10.1128/jcm.36.3.764-767.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindbråthen A, Gaustad P, Hovig B, Tønjum T. Direct detection of Mycobacterium tuberculosis complex in clinical samples from patients in Norway by ligase chain reaction. J Clin Microbiol. 1997;35:3248–3253. doi: 10.1128/jcm.35.12.3248-3253.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lumb R, Lanser J A, Lim I S. Rapid identification of mycobacteria by the Gen-Probe Accuprobe system. Pathology. 1993;25:313–315. doi: 10.3109/00313029309066597. [DOI] [PubMed] [Google Scholar]
- 26.Miller N, Hernandez S G, Cleary T J. Evaluation of Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test and PCR for direct detection of Mycobacterium tuberculosis in clinical specimens. J Clin Microbiol. 1994;32:393–397. doi: 10.1128/jcm.32.2.393-397.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore D F, Curry J I. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by Amplicor PCR. J Clin Microbiol. 1995;33:2686–2691. doi: 10.1128/jcm.33.10.2686-2691.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore D F, Curry J I. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by ligase chain reaction. J Clin Microbiol. 1998;36:1028–1031. doi: 10.1128/jcm.36.4.1028-1031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfyffer G E, Cieslak C, Welscher H M, Kissling P, Rüsch-Gerdes S. Rapid detection of mycobacteria in clinical specimens by using the automated BACTEC 9000 MB system and comparison with radiometric and solid-culture systems. J Clin Microbiol. 1997;35:834–841. doi: 10.1128/jcm.35.9.2229-2234.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfyffer G E, Kissling P, Wirth R, Weber R. Direct detection of Mycobacterium tuberculosis complex in respiratory specimens by a target-amplified test system. J Clin Microbiol. 1994;32:918–923. doi: 10.1128/jcm.32.4.918-923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piersimoni C, Callegaro A, Scarparo C, Penati V, Nista D, Bornigia S, Lacchini C, Scagnelli M, Santini G, De Sio G. Comparative evaluation of the new Gen-Probe Mycobacterium tuberculosis amplified direct test and the semiautomated Abbott LCx Mycobacterium tuberculosis assay for direct detection of Mycobacterium tuberculosis complex in respiratory and extrapulmonary specimens. J Clin Microbiol. 1998;36:3601–3604. doi: 10.1128/jcm.36.12.3601-3604.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajalahti I, Vuorinen P, Nieminen M M, Miettinen A. Detection of Mycobacterium tuberculosis complex in sputum specimens by the automated Roche Amplicor Mycobacterium Tuberculosis Test. J Clin Microbiol. 1998;36:975–978. doi: 10.1128/jcm.36.4.975-978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raviglione M C, Snider D E, Kochi A. Global epidemiology of Tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 34.Reischl U, Lehn N, Wolf H, Naumann L. Clinical evaluation of the automated COBAS AMPLICOR MTB assay for testing respiratory and nonrespiratory specimens. J Clin Microbiol. 1998;36:2853–2860. doi: 10.1128/jcm.36.10.2853-2860.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogall T, Flohr T, Böttger E C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990;40:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 36.Rohner P, Jahn E I M, Ninet B, Ionati C, Weber R, Auckenthaler R, Pfyffer G E. Rapid diagnosis of pulmonary tuberculosis with the LCx Mycobacterium tuberculosis assay and comparison with conventional techniques. J Clin Microbiol. 1998;36:3046–3047. doi: 10.1128/jcm.36.10.3046-3047.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohner P, Ninet B, Metral C, Emler S, Auckenthaler R. Evaluation of the MB/BacT system and comparison to the BACTEC 460 system and solid media for isolation of mycobacteria from clinical specimens. J Clin Microbiol. 1997;35:3127–3131. doi: 10.1128/jcm.35.12.3127-3131.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sjöbring U, Mecklenburg M, Andersen Å B, Miörner H. Polymerase chain reaction for detection of Mycobacterium tuberculosis. J Clin Microbiol. 1990;28:2200–2204. doi: 10.1128/jcm.28.10.2200-2204.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tortoli E, Lavinia F, Simonetti M T. Evaluation of a commercial ligase chain reaction kit (Abbott LCx) for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary specimens. J Clin Microbiol. 1997;35:2424–2426. doi: 10.1128/jcm.35.9.2424-2426.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Deutekom H, Gerritsen J J J, van Soolingen D, van Ameijden E J C, van Embden J D A, Coutinho R A. A molecular epidemiological approach to studying the transmission of tuberculosis in Amsterdam. Clin Infect Dis. 1997;25:1071–1077. doi: 10.1086/516072. [DOI] [PubMed] [Google Scholar]
- 41.Vlaspolder F, Singer P, Roggeveen C. Diagnostic value of an amplification method (Gen-Probe) compared with that of culture for diagnosis of tuberculosis. J Clin Microbiol. 1995;33:2699–2703. doi: 10.1128/jcm.33.10.2699-2703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuorinen P, Miettinen A, Vuento R, Hällstrom O. Direct detection of Mycobacterium tuberculosis complex in respiratory specimens by Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test and Roche Amplicor Mycobacterium Tuberculosis Test. J Clin Microbiol. 1995;33:1856–1859. doi: 10.1128/jcm.33.7.1856-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wobeser W L, Kradjen M, Conly J, Simpson H, Yim B, D’Costa M, Fuksa M, Hian-Cheong C, Patterson M, Phillips A, Bannatyne R, Haddad A, Brunton J L, Kradjen S. Evaluation of Roche Amplicor PCR assay for Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:134–139. doi: 10.1128/jcm.34.1.134-139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woods G L, Fish G, Plaunt M, Murphy T. Clinical evaluation of the Difco ESP culture system II for growth and detection of mycobacteria. J Clin Microbiol. 1997;35:121–124. doi: 10.1128/jcm.35.1.121-124.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuen K-Y, Yam W-C, Wong L-P, Seto W-H. Comparison of two automated DNA amplification systems with a manual one-tube nested PCR assay for diagnosis of pulmonary tuberculosis. J Clin Microbiol. 1997;35:1385–1389. doi: 10.1128/jcm.35.6.1385-1389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]