Abstract
ATP-dependent chromatin-remodeling complexes are conserved among all eukaryotes and function by altering nucleosome structure to allow cellular regulatory factors access to the DNA. Mammalian SWI-SNF complexes contain either of two highly conserved ATPase subunits: BRG1 or BRM. To identify cellular genes that require mammalian SWI-SNF complexes for the activation of gene expression, we have generated cell lines that inducibly express mutant forms of the BRG1 or BRM ATPases that are unable to bind and hydrolyze ATP. The mutant subunits physically associate with at least two endogenous members of mammalian SWI-SNF complexes, suggesting that nonfunctional, dominant negative complexes may be formed. We determined that expression of the mutant BRG1 or BRM proteins impaired the ability of cells to activate the endogenous stress response gene hsp70 in response to arsenite, a metabolic inhibitor, or cadmium, a heavy metal. Activation of hsp70 by heat stress, however, was unaffected. Activation of the heme oxygenase 1 promoter by arsenite or cadmium and activation of the cadmium-inducible metallothionein promoter also were unaffected by the expression of mutant SWI-SNF components. Analysis of a subset of constitutively expressed genes revealed no or minimal effects on transcript levels. We propose that the requirement for mammalian SWI-SNF complexes in gene activation events will be specific to individual genes and signaling pathways.
The packaging of eukaryotic DNA into nucleosomes and higher order chromatin structure presents cells with a significant barrier to DNA utilization and necessitates mechanisms by which chromatin structure can be modified so that transcription can occur. Many multiprotein complexes with the ability to modify chromatin structure have been identified. These include histone acetyltransferases and deacetylases, which directly modify histone tail domains, and a class of energy-dependent enzymes that utilize ATP hydrolysis to alter nucleosome structure (reviewed in references 23, 30, 32, 34, 70, 83, and 84). The ATP-dependent chromatin remodeling complexes are conserved among eukaryotes, they share a related subunit that possesses DNA-stimulated ATPase activity, and each has been demonstrated to alter nucleosome structure in vitro in an ATP-dependent manner. Most of these complexes can be classified into two groups, those containing homologues of the yeast SWI2-SNF2 ATPase subunit, including yeast SWI-SNF (7, 12, 55), human SWI-SNF (hSWI-SNF) (24, 35, 82), yeast RSC (8), and Drosophila BRM complexes (54, 71), and those containing homologues of the Drosophila imitation-switch (ISWI) ATPase gene (16), including yeast ISW1 and ISW2 (76), human RSF (39), and the Drosophila NURF, CHRAC, and ACF complexes (25, 75, 78). A third group can be defined by Xenopus and human complexes containing the Mi2 protein, a related ATPase found in association with histone deacetylase activity (72, 81, 87, 90).
Although members of the ATP-dependent class of chromatin remodelers facilitate alterations in nucleosome structure in vitro, the cellular role of most of the complexes is not well defined. The yeast SWI-SNF complex is the prototype for the ATP-dependent remodeling complexes. Five of the subunits are encoded by the SWI and SNF genes that were originally isolated in screens for genes required for mating type switching or for sucrose fermentation (3, 53, 68). Subsequent work established that these genes were required for the optimal expression of a subset of inducible yeast genes (31, 41, 56, 88) and for transcription of Ty elements (11, 21, 41). The Drosophila brm protein, the ATPase subunit of the brm complex, has been shown to be a regulator of Drosophila homeotic genes (71), underscoring a role for this complex in developmentally regulated gene expression.
Human SWI-SNF complexes contain either the human BRM (hBRM) (hSNF2α) or the BRG1 (hSNF2β) homologues of the yeast SWI2-SNF2 ATPase (10, 29, 51). Components of hSWI-SNF complexes have been implicated in a range of cellular events, including gene activation, regulation of cell growth, and development and differentiation (reviewed in reference 23). Regulation of cell cycle progression may occur via interaction of BRG1-hBRM with the retinoblastoma oncoprotein (Rb) and/or cyclin E (14, 62, 65, 69). In addition, the complex or individual subunits may be targeted by viral regulatory proteins upon infection of cells by adenovirus, Epstein-Barr virus, human papillomavirus, and human immunodeficiency virus (13, 28, 37, 43, 86). The ini1 subunit has been shown to interact with the ALL-1 protein, the translocation of which is a hallmark of several types of human acute leukemias (58), and ini1 also was found to be altered in human malignant rhabdoid tumors (79), suggesting a role for ini1 as a tumor suppressor. Thus, the human SWI-SNF complex not only has a subunit that may act as a tumor suppressor (ini1) but also contains other subunits that directly interact with Rb, a known tumor suppressor. These results strongly implicate human SWI-SNF complex components in the regulation of cell growth, possibly via transcriptional control.
Several lines of evidence suggest that human SWI-SNF complexes may regulate a subset of transcriptional activation events in cells. Transient transfection of hBRM or BRG1 can increase gene induction of transfected reporter genes by some activators, while transfection of these genes mutated in the ATP binding site abrogates the increase in activation. The activators affected were limited to c-myc (9) and nuclear hormone receptors, including glucocorticoid receptor (GR), estrogen receptor, and retinoic acid receptor (10, 29, 51, 65). These findings were in agreement with earlier data showing that expression of rat GR in yeast cells required yeast SWI-SNF function (89). Additionally, physical association of BRG1 and the hSWI-SNF subunit BAF155 (hSWI3) with GR was correlated with GR-mediated activation of integrated mouse mammary tumor virus long terminal repeat (LTR) reporter genes (17), strongly implicating BRG1 in the activation of this promoter by GR. Recently, hSWI-SNF complexes were shown to bind to the C/EBPβ activator and facilitate myeloid gene activation (33). However, in other experiments, repression of reporter genes by E2F was shown to require BRG1-hBRM proteins (73), and it was recently reported that BRG1 represses transcription of the endogenous c-fos gene when introduced into cell lines that lack BRG1 and hBRM (52). Thus, SWI-SNF complexes may positively and negatively affect gene expression in mammalian cells.
Since there has been little analysis of the effects of hSWI-SNF and its components on the activation of endogenous cellular genes, we have sought to identify endogenous genes in mammalian cells that require the activity of mammalian SWI-SNF complexes. We have created cell lines that inducibly express either BRG1 or hBRM proteins that are mutated in the ATP binding site, with the objective of creating conditions in mammalian cells in which nonfunctional SWI-SNF complexes might interfere with gene activation events. Having created such lines, we then asked whether the activation of the hsp70 stress response gene was affected. We chose hsp70 for several reasons. First, the hsp70 gene is highly inducible and can be activated by a number of distinct cellular stresses, allowing us to compare activation of the same gene by different inducers. Second, there is a wealth of in vitro evidence that indicates that reconstituted stress response genes can be remodeled in vitro by Drosophila and mammalian ATP-dependent chromatin-remodeling complexes (4, 44, 74, 77, 78).
Here we report that activation of the endogenous hsp70 locus in mammalian cells is partially dependent on the SWI-SNF components BRG1 and hBRM. This is the first demonstration that any ATP-dependent chromatin-remodeling complex is involved in the activation of the stress response genes in vivo. Interestingly, activation of hsp70 by a metabolic inhibitor or by a heavy metal showed dependence on the SWI-SNF components, while activation by heat stress did not. The data suggest there are multiple pathways for activating hsp70 and that mammalian SWI-SNF complexes are selectively required during specific activation events.
MATERIALS AND METHODS
Plasmids.
pBS(KS+)CeBRG1 and pBS(KS+)CehBRM (Ce, C-terminal epitope tagged) carry BRG1 or hBRM coding sequences that contain the Flag epitope sequence at the 3′ end (S. Sif and R. E. Kingston, unpublished data). To create Flag-tagged BRG1 containing a mutation in the ATP binding site, an NsiI-BglII restriction fragment was isolated from pBJ5 BRG1 K-R (29) and cloned into NsiI-BglII-digested pBS(KS+)CeBRG1 to create pBS(KS+)CeBRG1 K-R. Clones were sequenced to verify the presence of the mutation. pBS(KS+)CeBRG1 K-R was digested with ClaI and SpeI, as well as with PvuI (which cuts only the vector) and the 5-kb insert fragment was gel purified and cloned into ClaI-SpeI cut pTet-Splice (63) to create pTS CeBRG1 K-R. Clones were sequenced to confirm the presence of the K-to-R mutation.
To create Flag-tagged hBRM containing a mutation in the ATP binding site, a 2.4-kb BglII restriction fragment was isolated from pCG hBRM-NTP (51) and cloned into BglII-digested pBS(KS+)CehBRM to create pBS(KS+)CehBRM-NTP. The mutation at the ATP binding site encodes a HindIII restriction site (51); the presence of this site in selected clones indicated that the mutation had been incorporated. pBS(KS+)CehBRM-NTP was digested with ClaI and SpeI (and PvuI to digest vector sequences) and the resulting 5-kb insert fragment cloned into ClaI-SpeI-digested pTet-Splice to create pTS CehBRM-NTP. Positive clones were then sequenced to verify the mutation.
Cell lines.
All cells were grown in Dulbecco's modified Eagle's medium-hi (DMEM-hi) (Gibco-BRL) supplemented with 10% heat-inactivated calf serum (Sigma) and 2 mM l-glutamine (Sigma). Mouse NIH 3T3 cells were purchased from the American Type Culture Collection. One-hundred-millimeter plates of NIH 3T3 cells that were six passages from receipt were transfected with 16 μg of pTet-tA, which encodes the tet-VP16 regulator (63), and 4 μg of pRSV-neo by using the calcium phosphate method (2). After 48 h, 0.12 mg of Geneticin (Gibco-BRL) per ml was added to the media. Media were changed every 36 to 48 h for 12 days, and drug-resistant colonies were picked and expanded. To test for expression of the tet-VP16 regulator, clones were washed twice with phosphate-buffered saline (PBS), and diluted into 60-mm-diameter plates in the absence or presence of 2 μg of tetracycline (Sigma) per ml. Forty-eight hours later, the plates were transiently transfected with 10 ng of pUHC13-3, a luciferase reporter gene under the control of tet operator sites (20), and 9.74 μg of pBS(SK+) as nonspecific DNA. Cells were harvested 48 h later, and cell extracts were assayed for luciferase activity by using a Luciferase Assay kit (Promega) in accordance with the manufacturer's instructions. One of the clones expressing high levels of luciferase activity in the absence but not the presence of tetracycline was selected for further use and was renamed tet-VP16.
tet-VP16 cells were maintained in 2 μg of tetracycline per ml and 75 μg of Geneticin per ml. Cells were plated in 100-mm dishes and transfected via the calcium phosphate method with 16 μg of pTS CehBRM-NTP or pTS CeBRG1 K-R and 4 μg of pRSV2-hygro, which encodes resistance to hygromycin B. After 48 h, cells were trypsinized and plated at 1:10, 1:20, 1:50, and 1:100 in the presence of 400 U of hygromycin B (Calbiochem) per ml. The media were changed every 48 h, and drug-resistant colonies were picked after 13 days. Following expansion, clones were washed twice with PBS and plated in the presence or absence of 2 μg of tetracycline per ml such that they were nearly confluent after 3 days. Western analyses of cell extracts were used to identify clones expressing 200-kDa Flag-tagged proteins in the absence but not the presence of tetracycline.
Once generated, all cell lines (except the NIH 3T3 parental line) were maintained in 2 μg of tetracycline per ml. B22, B24, H16, and H17 lines were maintained in tetracycline, Geneticin, and 350 to 400 U of hygromycin B per ml without exception. Once established, the cell lines, including NIH 3T3 cells, were passaged twice a week at dilutions generating nearly confluent plates on the next passage date. For experiments requiring growth in media lacking tetracycline, the cell lines were always washed twice with PBS before passage.
Protein extracts and analysis of protein expression.
For protein isolation, 100-mm plates of cells were scraped into 1 ml of PBS, transferred to a 1.5-ml Eppendorf tube, and centrifuged at 200 × g for 1 min. The cell pellet was frozen in liquid N2 and stored at −80°C or was immediately resuspended in 200 μl of lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40, 20% glycerol, 1 mM dithiothreitol, 1 μg of pepstatin A per ml, 4 μg of leupeptin per ml, and 1 mM phenylmethylsulfonyl fluoride. Samples were sonicated twice for 1 s and were centrifuged for 10 min at 10,000 × g at 4°C. The supernatant was removed, and protein levels were quantified by a Bradford assay using bovine serum albumin as a standard. Extracts were frozen in liquid nitrogen and stored at −80°C. For the immunoprecipitation experiment, cells were lysed by resuspension in lysis buffer, repeatedly passed through a 26-gauge needle, and then centrifuged at 10,000 × g at 4°C. The extract (1.0 mg of total protein) was incubated with 30 μl of M2 beads (Sigma) prewashed in lysis buffer. Samples were incubated overnight at 4°C on a nutator (Clay-Adams). Samples were then centrifuged at 4°C for 40 s at 1,500 × g, and the pellets were washed twice with a buffer containing 10 mM HEPES (pH 8.0), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 2 mM dithiothreitol, and 0.1% Triton X-100. The pellets were washed again in the same buffer containing 50 mM NaCl (instead of 150 mM) and lacking Triton X-100 and then were subsequently resuspended in sodium dodecyl sulfate (SDS)-sample loading buffer.
For Western analysis, 30 to 60 μg of total cellular protein was subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and then transferred to nitrocellulose. Immunoblotting was performed with secondary antibody conjugated to horseradish peroxide and ECL solutions from Amersham. Primary antibodies used for Flag epitope detection included monoclonal M2 antibody (Kodak and Sigma) for initial identification of clones expressing Flag-tagged proteins and rabbit polyclonal anti-Flag antibodies (Santa Cruz or Zymed) for all subsequent analyses. Rat monoclonal antibody against HSF1 was purchased from LabVision. Polyclonal rabbit anti-BRG1, anti-mouse BRM (mBRM), and anti-ini1 antibodies were generated against glutathione S-transferase fusion proteins to BRG1 amino acids 214 to 279 (sequence corresponding to that by Chiba et al. [10]), to mouse BRM sequences corresponding to amino acids 48 to 214 of the human BRM sequence (49) or to the entire ini1 coding sequence (28). The antiserum against hSWI3 has been described (64).
RNA isolation and expression assays.
Total cellular RNA was isolated by using TRIzol (Gibco-BRL) as described by the manufacturer. For Northern analysis, 15 μg of total cellular RNA was subjected to electrophoresis on a 1.2% agarose gel and transferred to nitrocellulose (see Fig. 3) or Nytran Plus (see Fig. 9; Schleicher & Schuell) by using 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Following UV cross-linking, the filter was prehybridized for at least 6 h at 42°C in 50% formamide, 5× SSC or 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 10% dextran sulfate, 20 mM Tris-HCl (pH 7.5), 1× Denhardt's solution, and 100 μg of sheared salmon sperm DNA per ml.
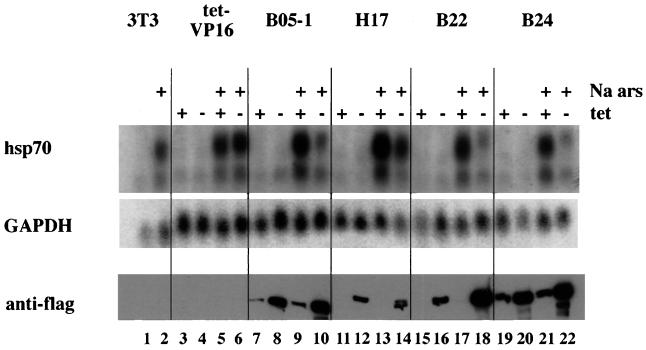
FIG. 3.
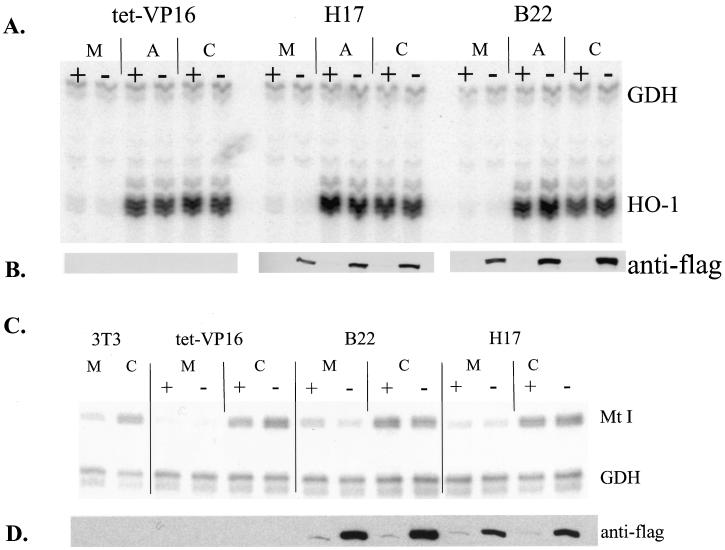
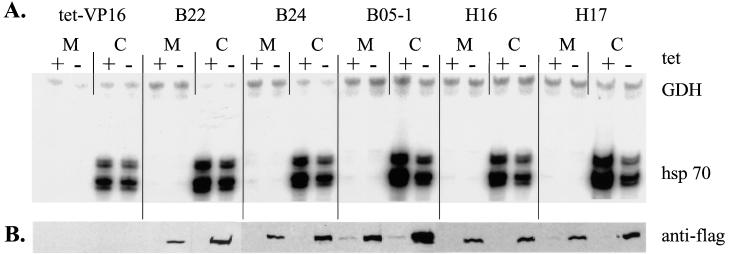
Expression of Flag-tagged mutant BRG1 or hBRM partially inhibits sodium arsenite-mediated activation of hsp70. Cell lines grown in the presence or absence of tetracycline for 4 days were treated with 100 μM sodium arsenite for 8 h. Total cellular RNA was prepared and subjected to Northern blot analysis. Blots were probed for mouse hsp70 by using a 1.8-kb restriction fragment from pM1.8 (22), stripped, and reprobed with a 700-bp EcoRI-HindIII cDNA fragment encoding human GAPDH. Quantification was performed on a Molecular Dynamics PhosphorImager using ImageQuant software. Duplicate plates were used to make protein extracts, which were used for Western analysis to confirm the expression of Flag-tagged mutant BRG1 or hBRM in each cell line.
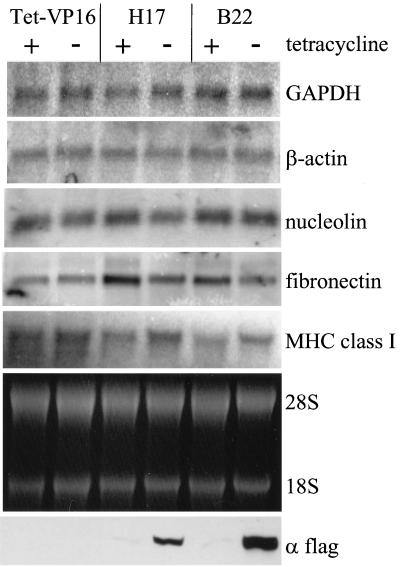
FIG. 9.
Northern analysis of constitutively expressed genes. RNA was isolated from cells maintained in the presence or absence of tetracycline for 4 days. Ethidium bromide staining of rRNA is shown. cDNA probes for rat β-actin, fibronectin and nucleolin, and for mouse GAPDH and major histocompatibility complex (MHC) class I were used to probe the blot for the corresponding mRNAs. Duplicate plates were extracted for Western analysis of Flag-tagged proteins as described above.
Gel-isolated restriction fragments were random primed with [α-32P]dATP (NEN) and DNA polymerase ± Klenow fragment (NEB), purified over a Sephadex G50 spin column, denatured, and used as a probe for Northern analyses. Following a 16- to 24-h hybridization at 42°C, the filters were rinsed in 2× SSC–0.1% SDS (see Fig. 3) or 6× SSPE–0.1% SDS (see Fig. 9) and were washed twice at room temperature for 15 min and once at 42°C for 10 min in the same buffer. Hybridization was analyzed by a PhosphorImager, and quantification was performed by using ImageQuant software (Molecular Dynamics). The filters were stripped and reprobed as indicated in the figure legends. As shown in Fig. 3, hsp70 levels were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) levels for quantitation. Primer extension analysis was performed with 15 to 20 μg of total cellular RNA as previously described (2), except that one-tenth of the suggested amount of salmon sperm DNA was used in the RNase buffer. Oligonucleotides used for hybridizations corresponded to nucleotides 660 to 635 of mouse hsp70 (22), to nucleotides 33 to 14 of the published mouse GAPDH sequence (59), to nucleotides 61 to 80 relative to the mouse heme oxygenase 1 (HO-1) start site (1), and to nucleotides 604 to 585 of mouse metallothionein I (Mt I) (19). Quantification was performed as described above.
RESULTS
To identify mammalian genes regulated by BRG1 or hBRM containing chromatin-remodeling complexes, we designed a strategy to create cell lines that inducibly express dominant negative versions of BRG1 or hBRM. We chose to utilize BRG1 and hBRM genes mutated in the ATP binding site (29, 51), since previous studies indicated that whereas transient transfection of wild-type BRG1 or hBRM could augment activation of some reporter genes, transfection of these ATP binding site mutants did not. We hypothesized that when expressed, the mutant forms would be competent for SWI-SNF complex formation, resulting in the assembly of nonfunctional complexes, and that gene activation events that required BRG1 or hBRM chromatin-remodeling complexes would therefore be impaired or inhibited.
An inducible expression system was chosen because prior studies had shown that in some cell types, BRG1 and hBRM could interact with the Rb oncoprotein and induce cell cycle arrest (14, 69) which, if universally true for all cell types, might preclude isolation of stably expressing cell lines. We utilized the tetracycline-inducible expression system described by Gossen and Bujard (20) with the modifications of Shockett et al. (63). NIH 3T3 cells were stably transformed with the tet-VP16 regulator and a gene encoding neomycin resistance. Drug-resistant colonies were screened for the ability to activate a transiently transfected luciferase reporter under the control of tet operator sites (20). One of the clones that expressed high levels of luciferase in the absence but not the presence of tetracycline was chosen for further manipulation and was named tet-VP16. tet-VP16 cells were stably transformed with a gene encoding hygromycin B resistance and a tet operator-controlled vector containing either Flag-tagged hBRM (f-hBRM) mutated at the ATP binding site or Flag-tagged BRG1 (f-BRG1) mutated at the ATP binding site. Drug-resistant clones were analyzed for expression of Flag-tagged proteins when grown in the absence but not the presence of tetracycline by Western blotting with an anti-Flag antibody. Ten of 33 drug resistant clones transformed with the f-hBRM mutant yielded tet-specific inducible expression; 5 of 27 drug-resistant clones transformed with the f-BRG1 mutant yielded tet-specific inducible expression.
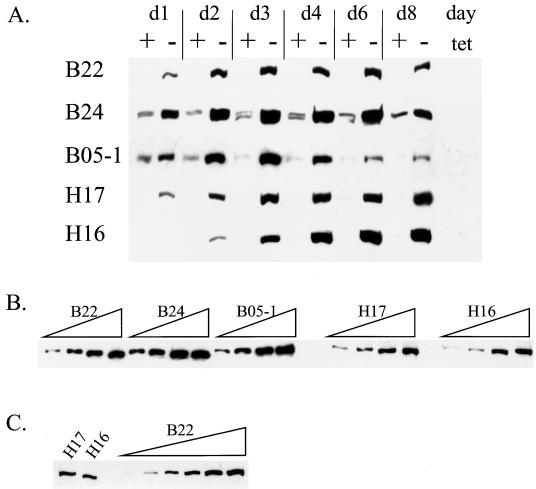
Three of the lines expressing epitope-tagged mutant BRG1 (B22, B24, and B05-1) and two lines expressing epitope-tagged mutant hBRM (H16 and H17) were chosen for further study. A time course of Flag-tagged protein expression was performed for each of the cell lines. Cells were plated at different dilutions in the presence or absence of tetracycline such that on the days indicated the plates were nearly confluent. Cell extracts were analyzed for Flag immunoreactivity by Western blotting, and the results for each of the cell lines are shown in Fig. 1A. Each of the lines yielded inducible expression; however, clear differences between the lines were observed. B24 and B05-1 cells both showed some leakiness, as low levels of f-BRG1 were present in the presence of tetracycline; however, induced f-BRG1 levels remained high at day 8 in B24 cells, whereas in B05-1 cells a peak of expression occurred at days 3 and 4, followed by a reduction in the f-BRG1 levels on days 6 and 8. The f-hBRM mutant lines both maintained Flag-tagged protein levels through day 8; however, the H16 line repeatedly took longer to induce expression than did the H17 line. It is important to note that there was some inherent variability in the amount of Flag-tagged protein expressed in the presence of tetracycline from experiment to experiment. Thus, for all experiments in which transcript levels were analyzed (see Fig. 3 to 6, 8, and 9), duplicate plates were used to make protein extracts to correlate mRNA levels with Flag-tagged protein levels.
FIG. 1.
(A) Time course of Flag-tagged protein expression. Cell lines were washed twice with PBS and diluted into media containing or lacking tetracycline such that plates were nearly confluent on the days indicated. Cells were harvested, cell pellets were extracted, and Western analysis was performed by using an anti-Flag antibody to detect levels of Flag-tagged protein. (B and C) Comparative analysis of Flag-tagged protein levels in each cell line. (B) Total cell extract (10, 20, 40, or 60 μg) from each of the B lines or each of the H lines was used for Western analysis with anti-Flag antibody. (C) Anti-Flag immunoreactivity from 60 μg of H17 and H16 extracts was compared to anti-Flag immunoreactivity from 2.5, 5, 10, 20, 40, or 60 μg of B22 extract.
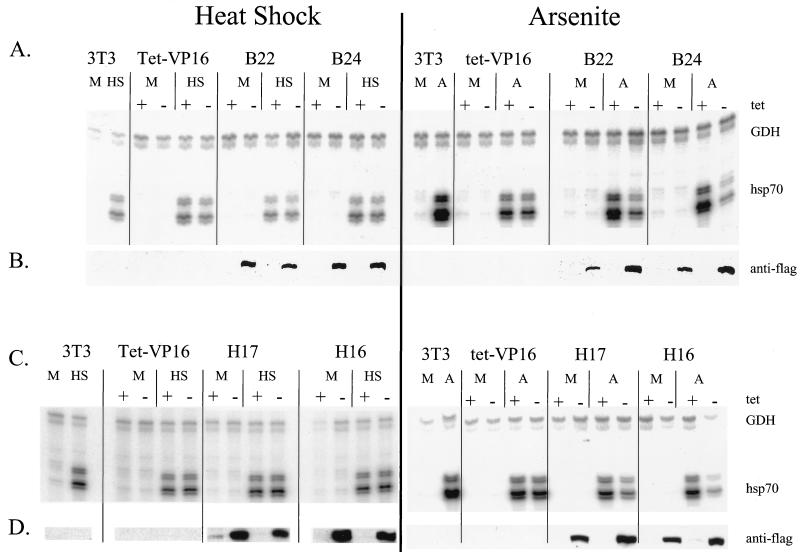
FIG. 6.
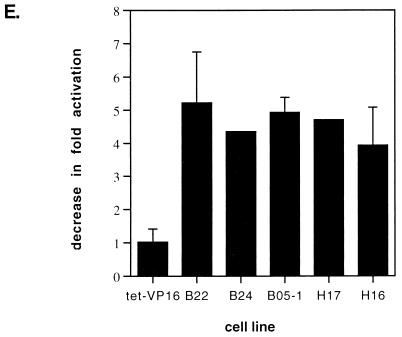
Expression of Flag-tagged mutant BRG1 or hBRM does not affect heat-shock-mediated activation of hsp70. Cell lines were placed into media either containing or lacking tetracycline. Sixty-eight hours later, half of the samples were placed at 32°C. At 84 h postpassage, the media from the cells placed at 32°C were replaced with media preheated to 42°C. The cells were then placed at 42°C for 2 h. Cell lines that had remained at 37°C were treated with 100 μM sodium arsenite for 8 h. Total cellular RNA was prepared, protein extracts were made from duplicate plates, and RNA and Western analyses were performed as described for Fig. 5. M, mock-treated cells; HS, heat-shocked cells; A, arsenite-treated cells. (E) Quantification of the decrease in activation of hsp70 by heat shock in the presence of the dominant negative proteins. Data points with error bars represent the average of three to five experiments; those without error bars are the average of two experiments.
FIG. 8.
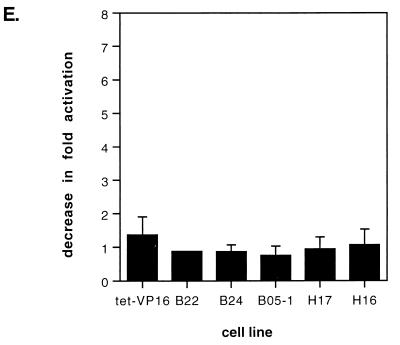
Expression of Flag-tagged mutant BRG1 or hBRM does not affect activation of HO-1 by arsenite or cadmium or cadmium-mediated activation of Mt I. Growth of cell lines and RNA and Western analyses were performed as described for Fig. 4. (A) Primer extension analysis of HO-1 expression. Cells were treated on day 4 with 20 μM CdCl2 or 100 μM sodium arsenite in serum-free media for 3 h as previously described (1); primer extension analysis used primers complementary to mouse GAPDH and mouse HO-1. (C) Primer extension analysis of Mt I expression. Cells were treated on day 4 with 20 μM CdCl2 for 8 h prior to harvesting, and primer extension analysis used primers complementary to mouse GAPDH and mouse Mt I (19). Cells were treated with cadmium chloride for 8 h instead of 24 h as was done in Fig. 5 because a time course of Cd2+-mediated induction of Mt I indicated that Mt I levels were slightly higher at 8 h than they were at 24 h (data not shown). (B and D) Western analysis to confirm the expression of Flag-tagged mutant BRG1 or hBRM in each sample. M, mock-treated cells; A, arsenite-treated cells; C, cadmium-treated cells.
To allow us to analyze all of the cell lines in parallel, we chose day 4 after removal of tetracycline as the time point at which to do experiments, as we hypothesized that this would allow the mutant proteins sufficient time to become incorporated into SWI-SNF complexes. In addition, B05-1 cells at day 4 still contained significant levels of f-BRG1.
To determine the relative amounts of Flag-tagged protein expression produced by the different lines, we analyzed increasing amounts of day 4 protein extract for Flag immunoreactivity (Fig. 1B and C). The results indicate that the f-BRG1-expressing lines produce similar amounts of Flag-tagged protein. The f-hBRM-expressing lines also expressed somewhat similar levels of Flag-tagged protein. It appeared that the H17 and H16 lines produced less Flag-tagged protein than did the f-BRG1-expressing lines, so to compare relative expression levels, 60 μg of H16 and H17 day 4 protein extract was compared to a range of B22 day 4 protein levels (Fig. 1C). The results indicate that B22 cells express approximately threefold more Flag-tagged protein than H17 cells do and approximately sixfold more protein than do H16 cells.
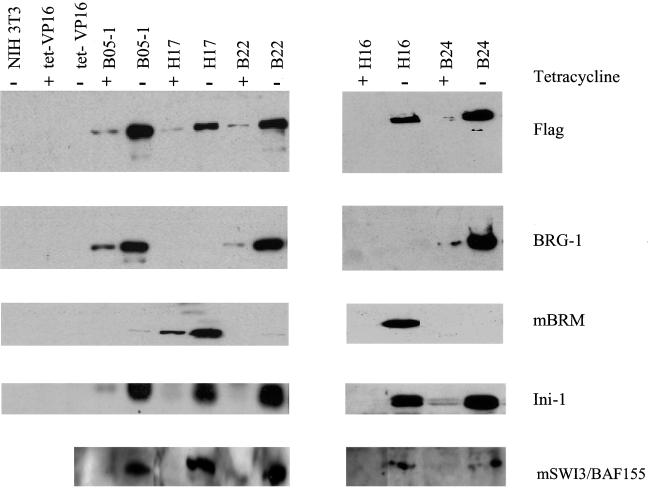
We immunoprecipitated Flag-tagged proteins from day 4 extracts to analyze whether other SWI-SNF subunits were associated with the Flag-tagged mutant proteins (Fig. 2). Bead-bound M2 anti-Flag monoclonal antibody was used for the immunoprecipitation, and the immunoprecipitated material was subjected to SDS-PAGE and Western blotting. Probing the blots with anti-Flag antibody showed the presence of inducible Flag-tagged protein in the mutant lines but not in the parental NIH 3T3 or tet-VP16 lines. The membranes were then stripped and reprobed sequentially with antibodies to BRG1, mBRM, and a human SWI3 homologue (BAF 155). Duplicate samples run on a higher percentage SDS-PAGE gel were probed for ini1. ini1 and the SWI3 homologue are components of both BRG1 and hBRM complexes (49, 50, 82). When probed with BRG1 antibody, only the B lines showed immunoreactivity dependent on the removal of tetracycline, whereas only the H lines showed immunoreactivity when probed with mBRM antibody. These results confirm the identity of the Flag-tagged protein present in each of the cell lines and demonstrate the specificity of the two antibodies. In addition, we have confirmed that hBRM and BRG1 proteins do not coimmunoprecipitate, indicating that BRG1 and hBRM likely do not associate with each other. This result is in agreement with previously reported data (82). When the membranes were probed for ini1 or the SWI3 homologue, we observed that both proteins coimmunoprecipitated with the f-BRG1 as well as the f-hBRM mutant proteins in a tet-inducible manner, indicating that both f-BRG1 and f-hBRM mutant proteins were associated with the endogenous ini1 and SWI3 proteins and were likely forming ATPase-deficient SWI-SNF complexes.
FIG. 2.
Flag-tagged mutant BRG1 and hBRM coimmunoprecipitate with endogenous ini1 and SWI3 subunits. Cell lines were grown in the presence or absence of tetracycline for 4 days. Cell pellets were extracted and immunoprecipitated with anti-Flag bound beads. The immunoprecipitated material was subjected to Western analysis with anti-Flag antibody. The filters were stripped and reprobed sequentially with antisera to BRG1, mouse BRM, and a human SWI3 homologue. Duplicate samples were run on an SDS–10% PAGE gel and probed with the ini1 antisera.
We then set out to determine whether expression of the mutant Flag-tagged proteins affected specific gene activation events. We chose to analyze the expression of the hsp70 stress response gene, since chromatin remodeling of Drosophila and mammalian hsp70 loci has been used as an in vitro assay in the identification and characterization of ATP-dependent nucleosome-remodeling factors, including NURF, and hSWI-SNF (4, 74, 77). Additionally, hsp70 expression can be activated by a number of different kinds of environmental stress, thereby affording us the opportunity to assess the involvement of BRG1- and BRM-based remodeling complexes in hsp70 induction by different stimuli.
We first analyzed the induction of hsp70 in the presence of the metabolic inhibitor, sodium arsenite. Parental and mutant cell lines were plated in duplicate in the presence or absence of tetracycline. On day 4, cells were mock treated or treated with 100 μM sodium arsenite for 8 h, at which time plates were harvested for protein or RNA isolation. Northern blot analysis of mRNA levels shows induction of hsp70 message in arsenite-treated parental NIH 3T3 and tet-VP16 cell lines (Fig. 3, lanes 1, 2, and 3 to 6). Since tet-VP16 cells contain the tet regulator but no target gene, the presence or absence of tetracycline had no effect on the level of hsp70 mRNA produced (Fig. 3, compare lanes 5 and 6). In contrast, hsp70 levels in cells expressing mutant f-BRG1 or mutant f-hBRM were reduced when tetracycline was removed, compared to hsp70 levels from cells grown in the presence of tetracycline (compare lanes 9 and 10, 13 and 14, 17 and 18, and 21 and 22). Western analysis of protein samples indicates that the decrease in hsp70 levels in the mutant cell lines correlated with expression of the mutant Flag-tagged protein. Quantification of hsp70 mRNA levels in this experiment indicated a 2.5- to 5.0-fold reduction in hsp70 induction by sodium arsenite when the mutant f-BRG1 or f-hBRM was expressed. We conclude that expression of the mutant proteins partially inhibits activation of hsp70 by sodium arsenite, likely by acting as a dominant negative. These results strongly suggest that hBRM- and/or BRG1-based chromatin-remodeling complexes contribute to the induction of the endogenous hsp70 loci upon exposure to sodium arsenite.
Since inducible hsp70 is part of a family of related genes, we were concerned that Northern analysis might reflect the expression of several different genes. We therefore repeated the experiment with a primer extension assay with oligonucleotide probes against a specific mouse hsp70 gene (hsp70.1, referred to as hsp70 hereafter) (22) and against mouse GAPDH (59) as a control. Extension products were resolved on a sequencing gel and are shown in Fig. 4. Analysis of the f-BRG1 mutant lines indicates that removal of tetracycline resulted in a decreased level of hsp70 mRNA upon sodium arsenite stimulation (Fig. 4A) and a corresponding induction in the levels of f-BRG1 mutant protein (Fig. 4B). Similarly, analysis of the f-hBRM mutant lines demonstrates that tetracycline removal resulted in a decreased level of hsp70 mRNA in response to sodium arsenite stimulation (Fig. 4C) as well as a corresponding induction in the levels of f-hBRM mutant protein (Fig. 4D). Thus, the primer extension assays confirm the results of the Northern analysis and further support a role for BRG1 and/or BRM chromatin-remodeling complexes in sodium arsenite-mediated activation of the hsp70 gene. Quantification of results from multiple experiments is presented in Fig. 4E as the decrease in fold activation due to expression of the dominant negative BRG1 or hBRM. A value of 1.0 indicates that no decrease in the level of activation occurred. In most of the cell lines, approximately four- to fivefold less hsp70 mRNA was present in the absence of tetracycline as compared to that in the presence of tetracycline.
FIG. 4.
Primer extension analysis demonstrating that expression of Flag-tagged mutant BRG1 or hBRM partially inhibits sodium arsenite-mediated activation of hsp70. Cell lines were grown as described for Fig. 4. (A and C) Total cellular RNA was used for primer extension analysis with primers complementary to mouse GAPDH (59) and mouse hsp70.1 (22). (B and D) Duplicate plates were used to make protein extracts, which were used for Western analysis to confirm the expression of Flag-tagged mutant BRG1 or hBRM in each cell line. M, mock-treated cells; A, arsenite-treated cells. (E) Quantification of the decrease in activation of hsp70 by arsenite in the presence of the dominant negative proteins. Data points with error bars represent the average of three or four experiments; those without error bars are the average of two experiments.
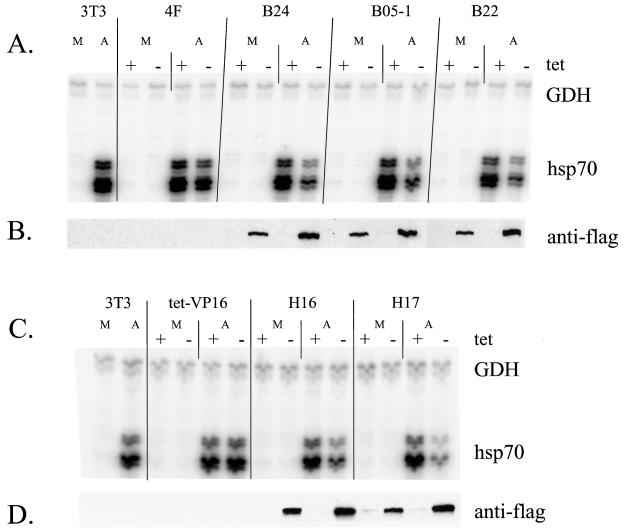
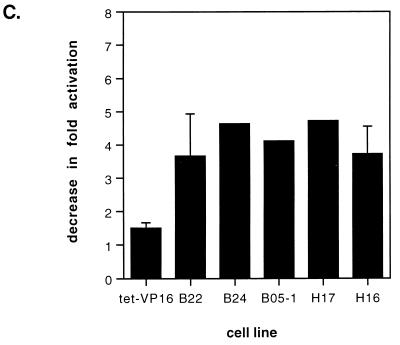
Because multiple stresses can activate the hsp70 genes, we analyzed the activation of hsp70 in response to Cd2+, a heavy metal. Cell lines were maintained in the presence or absence of tetracycline for 4 days and subsequently were exposed to 20 μM CdCl2 for 24 h. Analysis of mRNA and protein levels is presented in Fig. 5A and B. As in the case of sodium arsenite stimulation, activation of hsp70 by CdCl2 in the presence or absence of tetracycline was not significantly different in the tet-VP16 cells (lanes 3 and 4). In contrast, activation of hsp70 by CdCl2 in the mutant lines was partially inhibited when the mutant Flag-tagged protein was expressed (lanes 7 and 8, 11 and 12, 15 and 16, and 19 and 20). Quantification revealed that expression of the mutant proteins resulted in a three- to fourfold decrease in activation (Fig. 5C).
FIG. 5.
Expression of Flag-tagged mutant BRG1 or hBRM partially inhibits cadmium chloride-mediated activation of hsp70. Growth of cell lines and RNA and Western analysis were performed as described for Fig. 5 except that cells were treated on day 4 with 20 μM CdCl2 for 24 h prior to harvesting. M, mock-treated cells; C, cadmium-treated cells. (C) Quantification of the decrease in activation of hsp70 by CdCl2 in the presence of the dominant negative proteins. Data points with error bars represent the average of three or four experiments; those without error bars are the average of two experiments.
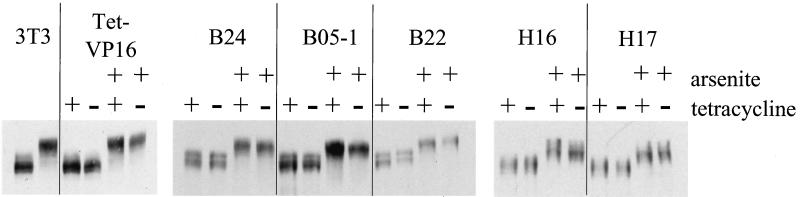
Previous studies have suggested that activation of hsp70 genes by heat shock may occur via a different pathway than does activation by other stress inducers like sodium arsenite or cadmium chloride (38, 48, 60, 91). In particular, a recent study examining purified Drosophila heat shock factor (HSF) reported that trimerization and DNA binding activities could be induced directly in vitro by heat and by oxidation, but not by a myriad of other environmental stresses, including arsenite (91). We therefore sought to test whether BRG1 or BRM complexes contribute to hsp70 activation by heat shock. Previous work in both yeasts and mammals has demonstrated that different heat shock protocols result in differences in activation of the stress response (18, 47, 66, 67, 92). Initial experiments designed to optimize heat shock in our cells revealed that greater activation of hsp70 could be achieved by transferring the cells from 37 to 32°C for 16 h prior to heat shock at 42°C than was observed when the cells were moved directly from 37 to 42°C (data not shown). Thus, we chose to focus on this particular heat shock regimen. Repeated analysis of heat shock activation of hsp70 in the presence or absence of tetracycline indicated that expression of dominant negative BRG1 or hBRM had no effect on the hsp70 levels present in either the tet-VP16 cells or in any of the mutant cell lines (data not shown). To confirm that there was no effect on hsp70 activation by this heat shock protocol, experiments in which side-by-side comparisons of heat shock and sodium arsenite stimulation were performed. Cell lines were placed in media either with or without tetracycline, and half of the samples were transferred to 32°C at day 3.3. On day 4, cells left at 37°C were treated with sodium arsenite as described above, while the cells at 32°C were subjected to heat shock by quickly replacing the media with media prewarmed to 42°C and placing the cells in a 42°C incubator for 2 h. Plates were harvested for RNA or protein extraction, and primer extension and Western analysis of the f-BRG1 mutant lines are presented in Fig. 6A and B, while analysis of the f-hBRM mutant lines are presented in Fig. 6C and D. hsp70 mRNA levels in heat-shocked B22 and B24 cells were equivalent in the presence or absence of tetracycline, whereas arsenite-treated B22 and B24 cells clearly showed a reduction in hsp70 mRNA levels in the absence of tetracycline. Similarly, no significant difference in the mRNA levels induced in heat-shocked H17 and H16 cells was observed while arsenite-treated H17 and H16 cells expressing mutant hBRM protein produced lower levels of hsp70 mRNA. Quantification of the heat shock experiments (Fig. 6E) indicated that expression of the mutant Flag-tagged proteins clearly did not cause a decrease in hsp70 levels, indicating that induction of hsp70 by heat was not affected by the presence of ATPase-deficient BRG1 or hBRM molecules in the cells. This strongly suggests that at least under the heat shock conditions employed, activation of hsp70 by heat shock is not dependent on BRG1 or BRM complexes. These results also add to the evidence that there are multiple activation pathways that can result in the induction of hsp70 mRNA.
The transcriptional activation of hsp genes in eukaryotes is mediated through the activity of heat shock factor 1 (HSF1). In unstimulated mammalian cells, HSF1 is found in a monomeric state in the cytoplasm. This form of HSF1 is incapable of binding to its cognate binding site in hsp promoters. Activation of the heat shock, or stress response, results in multiple posttranslational events that affect HSF1, including homotrimerization, translocation to the nucleus, phosphorylation, and acquisition of DNA binding and transcriptional competence (reviewed in references 40, 45, 46, 80, and 85). To rule out the possibility that the effects of dominant negative hBRM and BRG1 on activation of hsp70 by arsenite or cadmium were indirectly caused by effects on HSF1 levels, Western analyse were performed to analyze HSF1 in arsenite-treated cells (Fig. 7). The results demonstrate that neither arsenite nor induction of the dominant negative proteins affects HSF1 levels or the extent of posttranslational modifications that result in altered HSF1 mobility. Identical results were obtained when cadmium-treated cells were analyzed (data not shown). Thus, the observed decreases in hsp70 activation are not due to indirect effects of the dominant negative hBRM and BRG1 on the HSF1 transcriptional activator.
FIG. 7.
HSF1 levels are unaffected by arsenite stimulation or by expression of mutant hBRM or BRG1. Cell extracts described for Fig. 4 were utilized for Western analysis with anti-HSF1 antibody.
To further address the generality of a role for BRG1 and BRM complexes in gene activation, we analyzed the expression of the HO-1 promoter. Like hsp70, HO-1 can be activated by both arsenite and cadmium. Treatment of cells with either arsenite or cadmium revealed a significant induction of HO-1 gene expression; however, induction was unaffected by the expression of the mutant hBRM or BRG1 (Fig. 8A and B). We also analyzed expression of Mt I in CdCl2-treated cells. Mt I is induced by the presence of Cd2+ (15, 42), though the absolute activation of Mt I is lower in 3T3-based cells than in other cell types (42) and is considerably lower than the activation of hsp70 or HO-1. Primer extension analysis of Mt I revealed that the expression of f-BRG1 or f-hBRM mutant proteins had no effect on the induction of Mt I by Cd2+ (Fig. 8C and D). We conclude that not all activation events induced by arsenite or Cd2+ require BRG1 or BRM complexes, and we suggest that the involvement of BRG1 and BRM is restricted to a subset of gene activation events.
The effects of mutant hBRM and BRG1 expression on expression of constitutively active genes were also examined. Figures 3 to 6 and 8 show that levels of GAPDH were unaffected by the expression of the mutant complexes. This was confirmed by Northern analysis of RNA from cells maintained 4 days in the absence or presence of tetracycline (Fig. 9). We also determined that expression of β-actin, nucleolin, and the 28S and 18S rRNAs were largely unaffected by expression of the mutant complexes. Analysis of fibronectin mRNA levels showed a slight (less than twofold) decrease in the presence of either dominant negative protein. In contrast, analysis of major histocompatibility complex class I mRNA levels showed a slight increase in the presence of the dominant negative proteins (Fig. 9). These results suggest that mammalian SWI-SNF complexes can slightly affect the levels of constitutively expressed genes in some cases but are unlikely to play a significant role in mediating constitutive gene expression.
DISCUSSION
Prior work on mammalian SWI-SNF complexes has identified subunits, roles in cell growth and in cell cycle progression, and possible mechanisms by which the complexes cause ATP-dependent alterations of chromatin structure (reviewed in references 23, 30, 32, 83, and 84). A more limited number of studies have identified transcription factors affected by SWI-SNF complexes; these studies have largely utilized transient expression of reporter constructs to implicate SWI-SNF components in activation by nuclear hormone receptors and by the c-myc protein and in repression by the E2F regulator (9, 10, 29, 51, 73). Additionally, work by Fryer and Archer has shown that chromatin remodeling and transcriptional activation of integrated mouse mammary tumor virus LTR sequences by GR correlates with hormone-dependent association of GR and mammalian SWI-SNF components, strongly suggesting a role for mammalian BRG1 complex in activation of the viral LTR (17). However, to date only two studies have examined the regulation of an endogenous cellular gene by mammalian SWI-SNF components. Introduction of BRG1 to BRG1-deficient cell lines impaired the induction of the c-fos gene by forskolin or interleukin-6 by three- to fourfold (52), while the activation of some myeloid-specific genes was facilitated by mammalian SWI-SNF proteins (33).
Here, we provide evidence that mammalian SWI-SNF complexes contribute to the transcriptional activation of an endogenous hsp70 locus. We report that activation of hsp70 by a metabolic inhibitor or by a heavy metal, but not by heat stress, is impaired when mutant BRG1 or hBRM proteins are exogenously expressed. Additionally, activation of the HO-1 and Mt I genes by arsenite and/or cadmium was unaffected by the expression of the mutant BRG1 and hBRM proteins.
Our demonstration that mammalian SWI-SNF complexes play a role in hsp70 activation by arsenite or Cd2+ is based on observations that exogenous expression of hBRM or BRG1 proteins that are mutated in the ATP binding site can inhibit hsp70 activation. Our data strongly suggest that the mutant form of the protein is interfering with a normal function that contributes to transcriptional activation. Demonstration that the exogenously expressed BRG1 and hBRM mutant proteins could be coimmunoprecipitated with endogenous ini1 and SWI3 subunits (Fig. 2), which are common to both BRG1 and BRM complexes, suggests that the mutant proteins are being assembled into SWI-SNF complexes in the cells. We have not determined whether all of the endogenous SWI-SNF subunits are associated with the mutant proteins; therefore, we cannot state that complete complexes are forming around the nonfunctional ATPase subunits. However, stable, inducible expression of the mutant subunits coupled with their association with the endogenous ini1 and SWI3 proteins suggests that the interference in hsp70 activation that we observed is accomplished via a dominant negative mechanism. We can envision two possible mechanisms by which interference of gene expression by the putative dominant negatives may occur. In one scenario, a complete SWI-SNF complex forms around the mutant ATPase subunits, forming a SWI-SNF complex incapable of ATP binding and therefore incapable of ATP hydrolysis and ATP-dependent chromatin remodeling. This complex is likely to be targeted to appropriate places in the genome, where it would fail to function, presumably in some event involving the remodeling of nucleosome structure. Another possibility is that incomplete SWI-SNF complexes form around the mutant ATPase subunits, thereby depleting the wild-type endogenous BRG1 and BRM proteins of one or more of their associated subunits and rendering the endogenous complex nonfunctional. These possibilities are not mutually exclusive; a combination of these events may also occur.
Since the BRG1 and BRM proteins form separate complexes that appear to share the same subunits, it is not possible for us to determine whether the BRG1 complex, the BRM complex, or both complexes contribute to activation of hsp70. One could imagine, for example, that only the BRG1 complex was specifically responsible for contributing to hsp70 activation and that expression of the mutant BRG1 inhibited hsp70 activation by either of the mechanisms outlined above. However, even if the BRM complex played no role whatsoever in gene activation, expression of the BRM mutant could still sequester subunits from the endogenous BRG1 complex, thereby reducing its ability to activate hsp70. Other methods that specifically affect only one of the complexes (e.g., cell lines deficient in BRG1 or BRM) will have to be employed to distinguish which complex(es) contributes to hsp70 activation.
We have examined the involvement of mammalian SWI-SNF complexes in the activation of hsp70 by three distinct classes of stress inducers: arsenite, a metabolic inhibitor; Cd2+, a heavy metal; and heat stress. Expression of mutant SWI-SNF subunits resulted in impaired activation of hsp70 when induced by arsenite and Cd3+, but there was no effect when heat was the stress applied. The simplest interpretation of these results is that activation by arsenite and Cd2+ involves the activity of mammalian SWI-SNF complexes, but the complexes are not involved in activation by heat. An alternative explanation is that SWI-SNF complexes are involved in the activation of hsp70 in response to elevated temperature, but that there exist other activities in the cell, perhaps other complexes that affect chromatin structure, that can fully substitute when the SWI-SNF complexes are impaired. A third possibility is that the method used to accomplish heat shock (transfer from 37 to 32°C for 16 h prior to heat shock at 42°C) somehow alleviates the requirement for SWI-SNF complexes and that expression of the mutant BRG1 or hBRM proteins in the presence of other heat shock regimens impairs hsp70 activation. This possibility has not been examined experimentally. Either way, our results suggest that there are multiple mechanisms by which hsp70 activation can be accomplished and that the requirement for SWI-SNF complexes is specific to distinct activation pathways.
Multiple lines of evidence from the literature support the idea that different inducers of the stress response utilize different activation pathways. First is the obvious difference in response time to heat stress as opposed to stress by arsenite or Cd2+. The response time to each of these stimuli differs in different cell types; however, the response to heat is generally considerably quicker than is the response to most other inducers, including arsenite and Cd2+. Second, Mosser et al. (48), in kinetic studies of the response to heat and Cd2+ in HeLa cells, demonstrated that activation of the stress response by both of these inducers could occur in the presence of cycloheximide. Time courses analyzing the activation of HSF binding ability showed that in cycloheximide-treated, heat-stressed HeLa cells, activation of HSF binding occurred rapidly, then declined by 2 h post-heat shock. Addition of cadmium to these cultures caused reappearance of activated HSF, suggesting that the activation of HSF binding activity by heat and Cd2+ occurs though independent pathways (48). Other workers have reported that different stresses result in differences in HSF phosphorylation (61). In addition, specific mutations in the Schizosaccharomyces pombe HSF differentially affect the response to heat and cadmium stress, again suggesting that activation by these two inducers can occur via separate pathways (60). Finally, while it has long been recognized that HSF can be activated in vitro by heat (36, 47, 93), recent work by Zhong and colleagues has shown that direct exposure of purified, inactive Drosophila HSF to a range of other stress inducers, including arsenite, had no effect on the ability of HSF to trimerize, whereas heat directly induced HSF trimerization (91). These results suggest that HSF can directly sense heat but indirectly senses other activating agents, such as arsenite. Our results, combined with these prior data, may indicate that inducers that directly activate HSF may not require the activity of SWI-SNF complexes while inducers that indirectly activate HSF and the stress response do show a dependence on SWI-SNF complexes.
How do SWI-SNF complexes contribute to activation of hsp70 in response to arsenite or Cd2+? It has previously been shown that cycloheximide-treated cells still activate HSF and hsp transcription when stimulated by Cd2+; thus, protein synthesis is not required for the activation process (48). Demonstrating that HSF1 levels and modifications to HSF1 caused by arsenite and Cd2+ are unaffected by the dominant negative hBRM and BRG1 (Fig. 7) strongly suggests that the mutant complexes are not indirectly affecting hsp70 activation by affecting HSF1. Although we cannot rule out the possibility of an indirect mechanism whereby expression of the dominant negatives inhibits the expression of an uncharacterized, constitutively produced factor(s) that is required for hsp70 activation, we favor a more direct mechanism in which mammalian SWI-SNF complexes function during hsp70 activation by contributing to chromatin structural changes at the hsp70 locus, perhaps in combination with other ATP-dependent chromatin remodelers or other chromatin-modifying enzymes. Our previous in vitro work showing that addition of hSWI-SNF complex facilitates transcriptional elongation on reconstituted hsp70 chromatin templates (4) further supports the idea that mammalian SWI-SNF complexes may act directly at the hsp70 gene.
In the inactive state, the hsp70 locus in Drosophila and human cells contains a transcriptionally engaged RNA polymerase II (pol II) and a nascent transcript of about 25 to 50 bases (4, 57). The polymerase at these loci is stalled but is competent to resume transcription upon activation of the stress response. Prior work has shown that the presence of nucleosomes downstream of the transcription initiation site contributes to pausing by eukaryotic polymerases (26, 27), suggesting that nucleosome structure on the hsp70 gene may contribute to the formation of the paused polymerase. Activation of HSF and the stress response results in an increase in transcription initiation as well as a release of the paused polymerase, generating an increase in the level of full-length hsp70 transcripts (4, 57). In vitro studies using a reconstituted nucleosomal hsp70 template have provided evidence that hSWI-SNF complexes stimulate the ability of the HSF activation domain to promote elongation (4, 6). In vivo studies have shown that activation of the stress response also results in an alteration in chromatin structure for hundreds of base pairs downstream of the hsp70 transcription initiation site, as reflected by increased nuclease sensitivity in response to heat shock directed transcription (5). Thus, HSF facilitates hsp70 transcription by increasing both transcription initiation as well as facilitating transcription elongation. SWI-SNF complex contributions to the activation of hsp70 could occur by altering chromatin structure to facilitate transcription initiation, transcription elongation, or both.
We have demonstrated that mammalian SWI-SNF complexes contribute to the activation of the endogenous hsp70 gene and that the requirement for the complexes is specific for particular inducers of the hsp70 expression. This is the first demonstration that an ATP-dependent chromatin-remodeling complex is involved in the activation of the stress response genes in vivo. We have also demonstrated that the involvement of SWI-SNF complexes can be gene specific, as arsenite- or Cd2+-treated cells require SWI-SNF complexes for full activation of hsp70 but not for activation of the HO-1 or Mt I promoters. Additionally, expression of the mutant BRG1 or hBRM had no or minimal effects on constitutive pol I or pol II gene expression. Most constitutively expressed genes are thought to be fixed in an open chromatin structure; our results, perhaps not surprisingly, suggest that ATP-dependent chromatin-remodeling complexes are not required for the constitutive expression of these genes. We propose that the involvement of SWI-SNF complexes in gene activation in mammalian cells will be determined both by the particular chromatin structure of the induced gene as well as by the signaling pathway utilized by the cell to promote activation. We are currently examining this hypothesis by examining the role of hBRM- and BRG1-based chromatin-remodeling complexes in other gene induction events.
ACKNOWLEDGMENTS
We thank H. Su for help with tissue culture during generation of the mutant cell lines. We thank C. Muchardt, M. Yaniv, G. Kalpana, R. Morimoto, S. Jones, P. Odgren, L. Schmidt, R. Ignotz, C. Bunker, and Z. Shao for plasmids. We are grateful to P. Odgren, S. Marks, R. Ignotz, G. Stein, J. Stein, and S. Jones for sharing equipment and to C. Bunker, L. Weber, S. Jones, C. Peterson, and members of the Peterson lab for helpful discussions.
S. Sif was supported by an NIH postdoctoral fellowship. This work was supported by NIH grant RO1 GM48405 to R.E.K. and by NIH grant RO1 GM56244 to A.N.I.
REFERENCES
- 1.Alam J, Cai J, Smith A. Isolation and characterization of the mouse heme oxygenase-1 gene. J Biol Chem. 1994;269:1001–1009. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 3.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 4.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 5.Brown S A, Kingston R E. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 1997;11:3116–3121. doi: 10.1101/gad.11.23.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown S A, Weirich C S, Newton E M, Kingston R E. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998;17:3146–3154. doi: 10.1093/emboj/17.11.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns B R, Kim Y-J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant, chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 9.Cheng S W, Davies K P, Yung E, Beltran R J, Yu J, Kalpana G V. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999;22:102–105. doi: 10.1038/8811. [DOI] [PubMed] [Google Scholar]
- 10.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila Brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciriacy M, Freidel K, Löhning C. Characterization of trans acting mutations affecting Ty and Ty-mediated transcription in Saccharomyces cerevisiae. Curr Genet. 1991;20:441–448. doi: 10.1007/BF00334769. [DOI] [PubMed] [Google Scholar]
- 12.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 13.Dallas P B, Cheney I W, Liao D-W, Bowrin V, Byam W, Pacchione S, Kobayashi R, Yaciuk P, Moran E. p300/CREB binding protein related protein p270 is a component of mammalian SWI/SNF complexes. Mol Cell Biol. 1998;18:3596–3603. doi: 10.1128/mcb.18.6.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunaief J L, Strober B E, Guha S, Khavari P A, Ålin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 15.Durnam D M, Palmiter R D. Transcriptional regulation of the mouse metallothionein-I gene by heavy metals. J Biol Chem. 1981;256:5712–5716. [PubMed] [Google Scholar]
- 16.Elfring L K, Deuring R, McCallum C M, Peterson C L, Tamkun J W. Identification and characterization of Drosophila relatives of the yeast transcriptional activator SNF2/SWI2. Mol Cell Biol. 1994;14:2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 18.Gallo G, Schuetz T, Kingston R. Regulation of heat shock factor in Schizosaccharomyces pombe more closely resembles regulation in mammals than in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:281–288. doi: 10.1128/mcb.11.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glanville N, Durnam D, Palmiter R. Structure of mouse metallothionein-I gene and its mRNA. Nature. 1981;292:267–269. doi: 10.1038/292267a0. [DOI] [PubMed] [Google Scholar]
- 20.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Happel A M, Swanson M S, Winston F. The SNF2, SNF5 and SNF6 genes are required for Ty transcription in Saccharomyces cerevisiae. Genetics. 1991;128:69–77. doi: 10.1093/genetics/128.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt C, Calderwood S. Characterization and sequence of a mouse hsp70 gene and its expression in mouse cell lines. Gene. 1990;87:199–204. doi: 10.1016/0378-1119(90)90302-8. [DOI] [PubMed] [Google Scholar]
- 23.Imbalzano A. ATP dependent chromatin remodelers: complex complexes and their components. Crit Rev Eukaryot Gene Expr. 1998;8:225–255. doi: 10.1615/critreveukargeneexpr.v8.i3-4.10. [DOI] [PubMed] [Google Scholar]
- 24.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 25.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an I-SWI containing and ATP utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 26.Izban M G, Luse D S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 27.Izban M G, Luse D S. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;4:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 28.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 29.Khavari P A, Peterson C L, Tamkun J W, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 30.Kingston R, Narlikar G. ATP dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 31.Kodaki T, Hosaka K, Nikawa J, Yamashita S. The SNF2/SWI2/GAM1/TYE3/RIC1 gene is involved in the coordinate regulation of phospholipid synthesis in Saccharomyces cerevisiae. J Biochem (Tokyo) 1995;117:362–368. doi: 10.1093/jb/117.2.362. [DOI] [PubMed] [Google Scholar]
- 32.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 33.Kowenz-Leutz E, Leutz A. A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- 34.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 35.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 36.Larson J S, Schuetz T J, Kingston R E. Activation in vitro of sequence-specific DNA binding by a human regulatory factor. Nature. 1988;335:372–375. doi: 10.1038/335372a0. [DOI] [PubMed] [Google Scholar]
- 37.Lee D, Sohn H, Kalpana G V, Choe J. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature. 1999;399:487–491. doi: 10.1038/20966. [DOI] [PubMed] [Google Scholar]
- 38.Lee Y J, Curetty L, Corry P M. Differences in preferential synthesis and redistribution of HSP70 and HSP28 families by heat or sodium arsenite in Chinese hamster ovary cells. J Cell Physiol. 1991;149:77–87. doi: 10.1002/jcp.1041490111. [DOI] [PubMed] [Google Scholar]
- 39.LeRoy G, Orphanides G, Lane W S, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 40.Lis J, Wu C. Protein traffic on the heat shock promoter: parking, stalling and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- 41.Löhning C, Rosenbaum C, Ciriacy M. Isolation of the TYE2 gene reveals its identity to SWI3 encoding a general transcription factor in Saccharomyces cerevisiae. Curr Genet. 1993;24:193–199. doi: 10.1007/BF00351791. [DOI] [PubMed] [Google Scholar]
- 42.Mayo K E, Palmiter R D. Glucocorticoid regulation of metallothionein-I mRNA synthesis in cultured mouse cells. J Biol Chem. 1981;256:2621–2624. [PubMed] [Google Scholar]
- 43.Miller M E, Cairns B R, Levinson R S, Yamamoto K R, Engel D A, Smith M M. Adenovirus E1A specifically blocks SWI/SNF-dependent transcriptional activation. Mol Cell Biol. 1996;16:5737–5743. doi: 10.1128/mcb.16.10.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizuguchi G, Tsukiyama T, Wisniewski J, Wu C. Role of nucleosome remodeling factor NURF in transcriptional activation of chromatin. Mol Cell. 1997;1:141–150. doi: 10.1016/s1097-2765(00)80015-5. [DOI] [PubMed] [Google Scholar]
- 45.Morimoto R I. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- 46.Morimoto R I, Sarge K D, Abravaya K. Transcriptional regulation of heat shock genes. A paradigm for inducible genomic responses. J Biol Chem. 1992;267:21987–21990. [PubMed] [Google Scholar]
- 47.Mosser D, Kotzbauer P, Sarge K, Morimoto R. In vitro activation of heat shock transcription factor DNA binding by calcium and biochemical conditions that affect protein conformation. Proc Natl Acad Sci USA. 1990;87:3748–3752. doi: 10.1073/pnas.87.10.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mosser D D, Theodorakis N G, Morimoto R I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol Cell Biol. 1988;8:4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muchardt C, Reyes J-C, Bourachot B, Legouy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF-SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 50.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy D J, Hardy S, Engel D A. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol Cell Biol. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papoulas O, Beek S J, Moseley S L, McCallum C M, Sarte M, Shearn A, Tamkun J W. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- 55.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multiprotein complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peterson L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 57.Rougvie A E, Lis J T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 58.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce C M, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sabath D E, Broome H E, Prystowsky M B. Glyceraldehyde-3-phosphate dehydrogenase mRNA is a major interleukin 2-induced transcript in a cloned T-helper lymphocyte. Gene. 1990;91:185–191. doi: 10.1016/0378-1119(90)90087-8. [DOI] [PubMed] [Google Scholar]
- 60.Saltsman K A, Prentice H L, Kingston R E. Mutations in the Schizosaccharomyces pombe heat shock factor that differentially affect responses to heat and cadmium stress. Mol Gen Genet. 1999;261:161–169. doi: 10.1007/s004380050953. [DOI] [PubMed] [Google Scholar]
- 61.Sarge K D, Murphy S P, Morimoto R I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. . (Errata, 13:3122–3123 and 13:3838–3839). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19:1460–1469. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shockett P, Difilippantonio M, Hellman N, Schatz D G. A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc Natl Acad Sci USA. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sif S, Stukenberg P T, Kirschner M W, Kingston R E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 66.Sorger P K, Lewis M J, Pelham H R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987;329:81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- 67.Sorger P K, Pelham H R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- 68.Stern M J, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 69.Strober B E, Dunaief J L, Guha S, Goff S P. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 71.Tamkun J W, Deuring R, Scott M P, Kissinger M, Pattatucci A M, Kaufman T C, Kennison J A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcription activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 72.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 73.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. Rb and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsukiyama T, Becker P B, Wu C. ATP-dependent nuclesome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–531. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 75.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 76.Tsukiyama T, Palmer J, Landel C C, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 78.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 79.Versteege I, Sevenet N, Lange J, Rousseau-Merck M F, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 80.Voellmy R. Transduction of the stress signal and mechanisms of transcriptional regulation of heat shock/stress protein gene expression in higher eukaryotes. Crit Rev Eukaryot Gene Expr. 1994;4:357–401. [PubMed] [Google Scholar]
- 81.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 82.Wang W, Côte J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 83.Wolffe A P, Hayes J J. Chromatin disruption and modification. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 85.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 86.Wu D Y, Kalpana G V, Goff S P, Schubach W H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 88.Yoshimoto H, Yamashita I. The GAM1/SNF2 gene of Saccharomyces cerevisiae encodes a highly charged nuclear protein required for transcription of the STA1 gene. Mol Gen Genet. 1991;228:270–280. doi: 10.1007/BF00282476. [DOI] [PubMed] [Google Scholar]
- 89.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 90.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 91.Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by Drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]
- 92.Zimarino V, Tsai C, Wu C. Complex modes of heat shock factor activation. Mol Cell Biol. 1990;10:752–759. doi: 10.1128/mcb.10.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zimarino V, Wilson S, Wu C. Antibody-mediated activation of Drosophila heat shock factor in vitro. Science. 1990;249:546–549. doi: 10.1126/science.2200124. [DOI] [PubMed] [Google Scholar]