Abstract
Eighty-six strains of Shigella spp. were isolated during the dry season from stool samples of children under 5 years of age in Ifakara, Tanzania. The epidemiological relationship as well as the antimicrobial susceptibility and mechanisms of resistance to ampicillin, chloramphenicol, and co-trimoxazole were investigated. Four different epidemiological tools, pulsed-field gel electrophoresis (PFGE), repetitive extragenic palindromic (REP)-PCR, plasmid analysis, and antibiogram, were compared for typing Shigella strains. Seventy-eight (90%) strains were Shigella flexneri and were distributed into four groups, by either PFGE or REP-PCR, with 51, 17, 7, and 3 strains. The four strains of Shigella dysenteriae belonged to the same group, and the four strains of Shigella sonnei were distributed in two groups with three and one strain each. Plasmid analysis showed a high level of heterogeneity among strains belonging to the same PFGE group, while the antibiogram was less discriminative. REP-PCR provided an alternative, rapid, powerful genotyping method for Shigella spp. Overall, antimicrobial susceptibility testing showed a high level of resistance to ampicillin (81.8%), chloramphenicol (72.7%), tetracycline (96.9%), and co-trimoxazole (87.9%). Ampicillin resistance was related to an integron-borne OXA-1-type β-lactamase in 85.1% of the cases and to a TEM-1-type β-lactamase in the remaining 14.8%. Resistance to co-trimoxazole was due to the presence of a dhfr Ia gene in all groups except one of S. flexneri, where a dhfr VII gene was found within an integron. Chloramphenicol resistance was associated in every case with positive chloramphenicol acetyltransferase activity. All strains were susceptible to nalidixic acid, ciprofloxacin, ceftazidime, cefotaxime, and cefoxitin. Therefore, these antimicrobial agents may be good alternatives for the treatment of diarrhea caused by Shigella in Tanzania.
Acute infectious diarrheal disease is one of the most frequent causes of childhood deaths in the developing world. Diarrheal disease accounts for approximately 25% of all deaths in children younger than 5 years of age in these areas (21). Infections caused by Shigella species are an important cause of diarrheal disease, in both developing and developed countries. Worldwide, it is estimated that shigellosis causes around 600,000 deaths per year, two-thirds of the deceased being children under 10 years of age. Shigella dysenteriae and Shigella flexneri are the predominant species in the tropics, while Shigella sonnei is the predominant species in industrialized countries (18).
Shigellosis is one of the acute diarrheal diseases for which antimicrobial therapy is effective. However, today it also presents a pressing challenge, as Shigella spp. have progressively become resistant over the past decades to most of the widely used and inexpensive antimicrobials (21). Thus, the history of the genus suggests that resistance will emerge to any antimicrobial agent used intensively (25). Antimicrobial resistance in enteric pathogens is of the greatest importance in the developing world, where the rate of diarrheal diseases is highest and indiscriminate use of antimicrobial agents is a fact.
The comparative analysis of different epidemiological markers is important in order to know which is the best for tracing the source of infection during an outbreak. Several conventional typing methods and newly introduced molecular biology typing techniques have been described (3, 5, 11, 13). On the other hand, the study of the mechanisms of resistance of the resistant pathogenic bacteria may provide insight into the means by which multiple resistance is spreading among the bacterial population.
The aim of this article is to characterize Shigella strains isolated from children under 5 years of age in Ifakara, Tanzania. The work includes comparative epidemiological typing with various epidemiological tools, as well as a determination of antimicrobial susceptibility and the molecular characterization of the mechanisms of resistance to ampicillin, chloramphenicol, and co-trimoxazole.
MATERIALS AND METHODS
Bacterial strains.
Eighty-six strains of Shigella spp. were isolated from stool samples of children under 5 years of age during the dry period (July to September) of 1997 in Ifakara, Tanzania. The children included in the study were seen at Saint Francis Designated District Hospital. Shigella spp. were identified by conventional methods (16) and by serotyping. All the strains with different plasmid patterns or antibiograms were investigated in detail to determine their mechanisms of resistance to ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole.
Antimicrobial susceptibility testing.
Susceptibility testing was performed by an agar diffusion disk method as recommended by the National Committee for Clinical Laboratory Standards (17). Mueller-Hinton agar was obtained from Becton Dickinson (Cockeysville, Md.), and antimicrobial disks were obtained from BBL Microbiology Systems (Cockeysville, Md.). Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923, and Pseudomonas aeruginosa ATCC 27853 were used as quality control organisms and tested weekly. Each time a new batch of Mueller-Hinton agar was introduced, Enterococcus faecalis ATCC 29212 was tested to detect the presence of inhibitors of trimethoprim-sulfamethoxazole. The MICs of ampicillin, chloramphenicol, trimethoprim-sulfamethoxazole, tetracycline, cefoxitin, cefotaxime, ceftazidime, nalidixic acid, and ciprofloxacin for the selected strains were determined by E-test strips (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar plates, following the manufacturer’s instructions. E. coli ATCC 25922 was used as a reference strain for quality control.
Low-frequency restriction analysis of chromosomal DNA by PFGE.
Genomic DNA was prepared as described previously (15), digested with XbaI, and separated in 1% agarose gels with a contour-clamped homogeneous-field apparatus (CHEF-DR2; Bio-Rad). It was run under 200 V, with the pulse time increasing from 5 to 8 for 20 h. Pulsed-field gel electrophoresis (PFGE) patterns were interpreted by using the criteria established by Tenover et al. (26).
REP-PCR.
Repetitive extragenic palindromic (REP)-PCR was carried out following the method previously described by Gallardo et al. (6), with some modifications. Briefly, the primer 5′ GCG CCG ICA TGC GGC ATT 3′ was used under the following conditions: 30 cycles of 1 min at 94°C, 1 min at 40°C, and 1 min at 65°C, with a final extension of 16 min at 65°C. The reaction was prepared with 5 μl of boiled bacterial suspension, 1 μl of 5 mM primer, and PCR beads (Pharmacia A.B., Uppsala, Sweden). Fifteen microliters of the PCR products was separated in a 12.5% precast polyacrylamide gel with a Genephor apparatus (Pharmacia) and silver stained.
Plasmid analysis.
Plasmid DNA was extracted from overnight bacterial cultures with the commercial kit Wizard Plus SV Minipreps DNA purification system (Promega, Madison, Wis.) according to the manufacturer’s instructions. The plasmids obtained were visualized and analyzed by 0.8% agarose gel electrophoresis.
β-Lactamase detection.
β-Lactamase analysis was performed by the following methods.
(i) Isoelectrofocusing.
Isoelectrofocusing was performed as described elsewhere (6). Gels were run in a PhastSystem apparatus (Pharmacia A.B.) and developed with nitrocefin, and the isoelectric points were determined. Several strains carrying β-lactamases of known pI were used as controls and focused in parallel with the extracts.
(ii) PCR.
All PCR amplifications of the different β-lactamase genes were carried out in a DNA Thermal Cycler 480 (Perkin-Elmer Cetus, Emeryville, Calif.), using the primers previously described (6) and under the following conditions: 30 cycles of denaturation at 94°C, annealing at 55°C, and extension at 72°C, plus a final extension of 7 min at 72°C. The PCR product was run and visualized in 0.7% agarose gels stained with ethidium bromide.
Chloramphenicol acetyltransferase detection.
The chloramphenicol acetyltransferase activity assay was performed as described elsewhere (2), with slight modifications (6). Briefly, the strains were grown overnight on MacConkey agar. A heavy suspension of bacteria in 0.2 ml of 1 M NaCl, 0.01 M EDTA, and 0.05% sodium dodecyl sulfate (pH 8) was incubated in an Eppendorf tube at 37°C for 60 min. After a short centrifugation in a microcentrifuge, 50 μl was transferred to a microtitration plate. Duplicate wells were prepared with each strain, and 50 μl of a solution containing two parts 0.2 M Tris-HCl (pH 8), 2 mM acetyl coenzyme A, and one part 10 mM 5,5-dithio-bis-(2-nitrobenzoic acid) in 0.1 M Tris-HCl, pH 8, was added to each well. A 50-μl amount of 5 mM sterile chloramphenicol (dissolved in water) was added to one well (test reaction), and an equivalent amount of water was added to the duplicate well (control). The plate was reincubated at 37°C for 5 min. The reaction was stopped by adding 1 N H2SO4 and read spectrophotometrically.
Detection of trimethoprim resistance genes.
Both dhfr Ia and dhfr VII genes were amplified under the same conditions used for β-lactamases and with the following primers: dhfr Ia upper (5′ GTG AAA CTA TCA CTA ATG G 3′) and lower (5′ TTA ACC CTT TTG CCA GAT TT 3′) and dhfr VII upper (5′ TTG AAA ATT TCA TTG ATT G 3′) and lower (5′ TTA GCC TTT TTT CCA AAT CT 3′). The sizes of the PCR products for both genes were the same, 474 bp, and included the entire gene.
Integron amplification and cloning.
Reaction mixtures for integron amplification were prepared in the same way as those for β-lactamase PCR but with the following primers: upper (5′ GGC ATC CAA GCA GCA AG 3′) and lower (5′ AAG CAG ACT TGA CCT GA 3′) (10). The conditions for amplification were as follows: 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 8 min, plus a final extension of 72°C for 16 min. Twenty-five microliters of the amplified products was run in a 1.5% agarose gel and stained with ethidium bromide. The bands were excised from the gel, and the DNA was recovered with a GeneClean kit (Bio 101, Inc., La Jolla, Calif.) and cloned into pCRII vector (Invitrogen BV, Leek, The Netherlands).
DNA sequencing.
Plasmid extraction was performed as described above. The sequencing of the plasmids with the cloned inserts was done with a Thermosequenase dye terminator sequencing kit in an automatic DNA sequencer (model 377; Applied Biosystems, Perkin-Elmer, Emeryville, Calif.) following the manufacturer’s instructions.
RESULTS
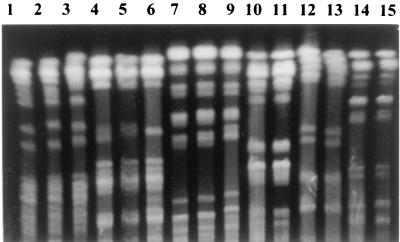
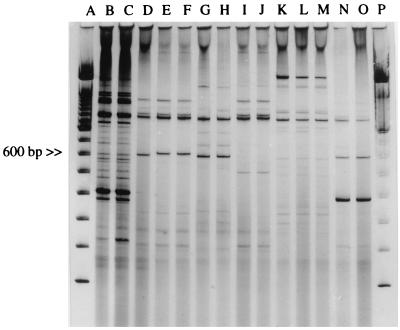
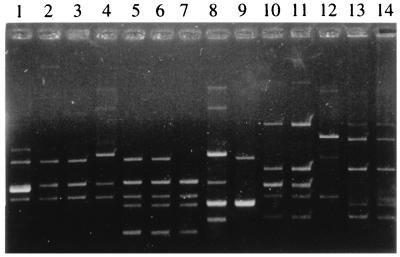
The eighty-six strains of Shigella spp. that were isolated were distributed as follows: 78 (90%) were S. flexneri, 4 (4.6%) were S. dysenteriae, and 4 (4.6%) were S. sonnei. No Shigella boydii strains were isolated. The 78 S. flexneri strains were grouped into four epidemiological groups by PFGE or REP-PCR (Fig. 1 and 2). The distribution of strains according to these epidemiological markers was as follows: 51 strains in group F-I, 17 strains in group F-II, 7 strains in group F-III, and 3 strains in group F-IV. However, the four major S. flexneri groups were subdivided into nine different subgroups based on antibiogram and plasmid analysis (Table 1). Eight different plasmid patterns were obtained among S. flexneri strains (Fig. 3). These patterns contained from three to six different plasmids each, although in some cases the difference between two patterns was due to the gain or loss of only one plasmid.
FIG. 1.
PFGE. Lanes 1, 2, and 3, S. flexneri strains belonging to group F-I; lanes 4, 5, and 6, S. flexneri strains belonging to group F-II; lanes 7, 8, and 9, S. flexneri strains belonging to group F-III; lanes 10 and 11, S. flexneri strains belonging to group F-IV; lanes 12 and 13, S. dysenteriae strains; lanes 14 and 15, S. sonnei strains.
FIG. 2.
REP-PCR. Lanes A and P, molecular size markers; lanes B and C, S. flexneri strains belonging to group F-II; lanes D, E, and F, S. flexneri strains belonging to group F-I; lanes G and H, S. flexneri strains belonging to group F-III; lanes I and J, S. flexneri strains belonging to group F-IV; lanes K, L, and M, S. dysenteriae strains; lanes N and O, S. sonnei strains.
TABLE 1.
Characterization of Shigella spp. by four different epidemiological markers
| Species | Groupa (no.) | Subgroupb (no.) | PFGE | Antibiogramc | Plasmid profile | REP-PCR group |
|---|---|---|---|---|---|---|
| S. flexneri | F-I (51) | F1 (46) | A | I | b | 1 |
| F2 (4) | A | I | a | 1 | ||
| F3 (1) | A | II | c | 1 | ||
| F-II (17) | F4 (14) | B | I | d | 2 | |
| F5 (3) | B | VI | e | 2 | ||
| F-III (7) | F6 (5) | C | I | f | 3 | |
| F7 (2) | C | III | g | 3 | ||
| F-IV (3) | F8 (1) | D | IV | h | 4 | |
| F9 (2) | D | III | h | 4 | ||
| S. sonnei | S-I (4) | S1 (3) | F | IV | j | 5 |
| S2 (1) | F | V | j | 5 | ||
| S. dysenteriae | D-I (4) | D1 (4) | E | I | i | 6 |
Distribution of strains based on PFGE and REP-PCR.
Distribution of groups according to antibiogram and plasmid analyses.
See text for phenotypes.
FIG. 3.
Plasmid patterns. Lane 1, S. flexneri strain belonging to subgroup F2; lanes 2 and 3, S. flexneri strains belonging to subgroup F1; lane 4, S. flexneri strain belonging to subgroup F3; lanes 5 and 6, S. flexneri strains belonging to subgroup F4; lane 7, S. flexneri strain belonging to subgroup F5; lane 8, S. flexneri strain belonging to subgroup F6; lane 9, S. flexneri strain belonging to subgroup F7; lanes 10 and 11, S. flexneri strains belonging to subgroups F8 and F9; lane 12, S. dysenteriae; lanes 13 and 14, S. sonnei strains belonging to subgroups S1 and S2.
On the basis of antibiotic susceptibility, six phenotypes were defined: phenotype I (Ampr Cmr Tetr Sxtr), phenotype II (Amps Cms Tets Sxts), phenotype III (Amps Cms Tetr Sxts), phenotype IV (Amps Cms Tetr Sxtr), phenotype V (Ampr Cms Tetr Sxtr), and phenotype VI (Ampr Cmr Tetr Sxts). In spite of belonging to the same clone by PFGE, REP-PCR, or plasmid analysis (Fig. 1 to 3), the four strains of S. sonnei were distributed in two groups based on the antibiogram. Three strains showed phenotype IV (group S1), and one strain showed phenotype V (group S2). The four strains of S. dysenteriae were all the same clone (Table 1).
Fourteen S. flexneri strains, three S. sonnei strains, and three S. dysenteriae strains were used for detailed investigations of the mechanisms of resistance to ampicillin, chloramphenicol, and co-trimoxazole. The MICs of ampicillin, chloramphenicol, tetracycline, co-trimoxazole, nalidixic acid, ciprofloxacin, ceftazidime, cefotaxime, and cefoxitin for these strains are shown in Table 2. For all the strains resistant to ampicillin, chloramphenicol, and tetracycline, the MICs of the drugs were >256 μg/ml, and for those resistant to co-trimoxazole, the MIC was >32 μg/ml. Overall, 92% of S. flexneri strains were resistant to ampicillin and chloramphenicol, 99% were resistant to tetracycline, and 91% were resistant to co-trimoxazole. S. dysenteriae strains were 100% resistant to ampicillin, chloramphenicol, tetracycline, and co-trimoxazole. While S. sonnei strains were all susceptible to chloramphenicol, only one of four strains was resistant to ampicillin and all showed resistance to tetracycline and co-trimoxazole. All Shigella sp. strains tested were susceptible to nalidixic acid, ciprofloxacin, cefotaxime, ceftazidime, and cefoxitin (Table 2).
TABLE 2.
Antimicrobial susceptibility of Shigella spp.
| Antimicrobial agent | MIC (μg/ml)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
S. flexneri
|
S. sonnei
|
S. dysenteriae
|
||||||||||
| F1a | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | S1b | S2 | D1c | |
| Ampicillin | >256 | >256 | 1 | >256 | >256 | >256 | 1.5 | 1.5 | 0.5 | 2 | >256 | >256 |
| Tetracycline | >256 | >256 | 1 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| Chloramphenicol | >256 | >256 | 1 | >256 | >256 | >256 | 2 | 1 | 0.75 | 1.5 | 3 | >256 |
| Co-trimoxazole | >32 | >32 | 0.032 | >32 | 0.38 | >32 | 0.125 | >32 | 0.032 | >32 | >32 | >32 |
| Nalidixic acid | 2 | 4 | 1.5 | 1.5 | 1.5 | 4 | 3 | 2 | 2 | 3 | 4 | 3 |
| Ciprofloxacin | 0.008 | 0.016 | 0.006 | 0.006 | 0.006 | 0.008 | 0.008 | 0.016 | 0.016 | 0.012 | 0.008 | 0.012 |
| Ceftazidime | 0.094 | 0.19 | 0.094 | 0.094 | 0.094 | 0.094 | 0.064 | 0.064 | 0.064 | 0.094 | 0.064 | 0.094 |
| Cefotaxime | 0.064 | 0.125 | 0.064 | 0.064 | 0.064 | <0.016 | 0.094 | 0.064 | 0.064 | 0.064 | 0.023 | 0.047 |
| Cefoxitin | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 1 | 1 | 1 |
F1 to F9, subgroups of S. flexneri based on antibiogram and plasmid analysis.
S1 and S2, subgroups of S. sonnei based on antibiogram and plasmid analysis.
D1, group of S. dysenteriae.
Isoelectric focusing was used first to detect the production of β-lactamase, and PCR with specific primers was used to corroborate the results, which are shown in Table 3. The ampicillin resistance of S. flexneri was explained in 75% of the cases by the presence of an OXA-1-type β-lactamase (pI 7.0), whereas the remaining 25% had a TEM-1-type β-lactamase (pI 5.4). The S. dysenteriae clone also carried an OXA-1-type β-lactamase, whereas the ampicillin-resistant S. sonnei strain had a TEM-1-type β-lactamase. In all cases, the OXA-1-type β-lactamase was located in an integron (data not shown).
TABLE 3.
Characteristics of Shigella sp. clinical isolatesa
| Shigella sp. | Subgroupb | β-Lactamase profile
|
CAT | DHFR | ||
|---|---|---|---|---|---|---|
| pI | TEM | OXA | ||||
| S. flexneri | F1 (4) | Ca. 7 | − | + | + | Ia |
| F2 (1) | Ca. 7 | − | + | + | Ia | |
| F3 (1) | ND | ND | ND | ND | ND | |
| F4 (2) | Ca. 7 | − | + | + | Ia | |
| F5 (1) | Ca. 7 | − | + | + | ND | |
| F6 (2) | Ca. 5.4 | + | − | + | VII | |
| F7 (1) | ND | ND | ND | ND | ND | |
| F8 (1) | ND | ND | ND | ND | Ia | |
| F9 (1) | ND | ND | ND | ND | ND | |
| S. sonnei | S1 (2) | ND | ND | ND | ND | Ia |
| S2 (1) | Ca. 5.4 | + | − | ND | Ia | |
| S. dysenteriae | D1 (3) | Ca. 7 | − | + | + | Ia |
CAT, chloramphenicol acetyltransferase; DHFR, dihydrofolate reductase; ND, not determined; −, absent; +, present.
Number of strains analyzed in each subgroup is shown in parentheses.
All strains resistant to chloramphenicol showed chloramphenicol acetyltransferase activity (Table 3), and also, all co-trimoxazole-resistant strains presented genes encoding dihydrofolate reductases (Table 3). Four of five co-trimoxazole-resistant S. flexneri epidemiological groups showed the dhfr Ia gene, and the fifth group showed a dhfr VII gene, while co-trimoxazole-resistant S. sonnei and S. dysenteriae strains also had the dhfr Ia gene.
DISCUSSION
The predominant species of Shigella during the studied period of time was S. flexneri, which is usually the predominant species in areas of endemicity, accounting for 50% of culture-positive disease (25). S. sonnei and S. dysenteriae were found in the same proportions. The most common typing procedures currently used with Shigella spp. are plasmid analysis and PFGE (7, 8, 12, 13, 24). Shigella species usually harbor a heterogenous population of plasmids, ranging in number from 2 to as many as 10 (9). Plasmid analysis has proven to be a useful typing technique (7, 8). Moreover, it is inexpensive and quick to perform, but it can be limiting if we take into account the fact that many plasmids are unstable and may be easily gained and/or lost. PFGE has a high discriminatory power, although it is cumbersome and expensive. However, it has been widely used for typing Shigella spp. (13, 24). Taking PFGE as a reference epidemiological tool, strains belonging to the same PFGE group but having different plasmid profiles and different antibiograms were observed (for instance, subgroups F4 and F5). Therefore, the mechanisms of resistance are probably carried in the missing plasmid. The contrary is also true; two strains belonging to the same PFGE group with the same plasmid profile showed different antibiograms (for instance, subgroups F8 and F9). This is probably due to an integron or transposon carrying the resistance gene integrated in the chromosome.
Recently, Liu et al. (13) compared plasmid profiles, PFGE, and enterobacterial repetitive intergenic consensus PCR for typing 20 clinical isolates of S. sonnei. PCR-based techniques have the advantages of being quick and easy to perform, and in this case they proved to be as good at discriminating epidemiologically related strains as PFGE. We found something similar with REP-PCR, another PCR-based technique, in which the amplification of the regions between REP sequences gives a good fingerprinting pattern valid for epidemiological typing. As long as the protocol is strictly followed and conditions are kept constant, this technique provides a degree of discrimination equivalent to that of PFGE with the advantages of speed, simplicity, and economy. To our knowledge, this is the first time that such a technique has been used in comparison with PFGE and plasmid profiles to type different species of Shigella.
Antimicrobial susceptibility testing showed a high degree of resistance to antibiotics most commonly used in the area (tetracycline, ampicillin, co-trimoxazole, and chloramphenicol). No resistance to quinolones and cephalosporins was observed, which can be explained by the fact that they are not used as alternative therapies in this area due to their high cost and lack of availability. However, a trend to quinolone resistance has been observed by Ries et al. (20) in S. dysenteriae strains isolated in Burundi. S. dysenteriae is considered the most resistant of the Shigella spp. (21). However, in our study S. flexneri showed the same level of resistance as S. dysenteriae. This pattern of resistance and susceptibility is commonly seen in developing countries, in contrast with strains from developed countries, which are less resistant to these antimicrobial agents (4, 27). In this study, the antimicrobial resistance pattern is not a useful epidemiological marker, due to the lack of variability in susceptibility patterns (i.e., the high level of resistance shown by most isolates). Resistance to ampicillin in S. flexneri groups F1 and F2 and S. dysenteriae (group D) is explained by the presence of an OXA-1-type β-lactamase within an integron. Group F3 S. flexneri and the one ampicillin-resistant S. sonnei (group S2) isolate had a TEM-1-type β-lactamase. Both genes have been previously described in Shigella strains isolated in Denmark and Greece (14, 22). Therefore, this is the most frequent mechanism of ampicillin resistance found in Shigella.
Besides ampicillin, the drug of choice for treating shigellosis is co-trimoxazole. Eighty-eight percent of the strains studied showed resistance to this drug, and in most cases this resistance could be explained by the presence of a dhfr Ia gene previously described in Shigella and considered the most common dihydrofolate reductase gene in the genus. In one group of S. flexneri, however, the dhfr gene found was dhfr VII, first described in E. coli (1). These genes were found inserted in an integron. Both genes were detected with specific primers to amplify the entire gene, which was further sequenced, showing in both cases 100% homology with the dhfr Ia and dhfr VII genes previously described (19, 23). Chloramphenicol resistance was explained in every case by a positive chloramphenicol acetyltransferase activity generating a high level of resistance. The use of this antibiotic has rapidly declined in many countries. However, due to the fact that it is inexpensive and presents a broad-spectrum activity it is extensively employed in developing countries, thereby ensuring strong selection pressure for the maintenance of chloramphenicol resistance.
In this study, we suggest that antibiotic resistance determinants are carried by plasmids, as well as in integrons which contain resistance genes, such as blaOXA or dhfr genes. The spread of multiresistant Shigella strains among a population in which diarrheal disease is one of the major causes of child morbidity and mortality requires greater attention to the appropriate use of antibiotics, the establishment of hygienic measures to prevent or decrease transmission, and the development of new effective drugs that can be safely used with children. Moreover, the guidelines for the treatment of shigellosis in developing countries should be updated, since in this study co-trimoxazole, one of the recommended antimicrobial agents for the treatment of shigellosis, has been shown to have little activity against Shigella spp.
ACKNOWLEDGMENTS
We thank the parents and guardians of study participants and the staff of the Ifakara Health Research and Development Centre and the St. Francis Designated District Hospital. Particular thanks go to the clinical officers whose work was of central importance.
This work was supported in part by grant SAF97/0091 and the Spanish Agency for International Co-operation (AECI-1042).
REFERENCES
- 1.Amyes S G B, Towner K J, Carter G I, Thomson C J, Young H K. The type VII dihydrofolate reductase: a novel plasmid-encoded trimethoprim-resistant enzyme from Gram-negative bacteria isolated in Britain. J Antimicrob Chemother. 1989;24:111–119. doi: 10.1093/jac/24.2.111. [DOI] [PubMed] [Google Scholar]
- 2.Azemun P, Stull T, Roberts M, Smith A L. Rapid detection of chloramphenicol resistance in Haemophilus influenzae. Antimicrob Agents Chemother. 1981;20:168–170. doi: 10.1128/aac.20.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brian M J, Van R, Townsend I, Murray B E, Cleary T G, Pickering L K. Evaluation of the molecular epidemiology of an outbreak of multiply resistant Shigella sonnei in a day-care center by using pulsed-field gel electrophoresis and plasmid DNA analysis. J Clin Microbiol. 1993;31:2152–2156. doi: 10.1128/jcm.31.8.2152-2156.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrar W E. Antibiotic resistance in developing countries. J Infect Dis. 1985;152:1103–1106. doi: 10.1093/infdis/152.6.1103. [DOI] [PubMed] [Google Scholar]
- 5.Faruque S M, Haider K, Rahman M M, Abdul Alim A R, Ahmad Q S, Albert M J, Sack R B. Differentiation of Shigella flexneri strains by rRNA gene restriction patterns. J Clin Microbiol. 1992;30:2996–2999. doi: 10.1128/jcm.30.11.2996-2999.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallardo F, Ruiz J, Marco F, Towner K J, Vila J. Increase in incidence of resistance to ampicillin, chloramphenicol and trimethoprim in clinical isolates of Salmonella serotype Typhimurium with investigation of molecular epidemiology and mechanisms of resistance. J Med Microbiol. 1999;48:367–374. doi: 10.1099/00222615-48-4-367. [DOI] [PubMed] [Google Scholar]
- 7.Gebre-Yohannes A, Drasar B S. Molecular epidemiology of plasmid patterns in Shigella flexneri types 1-6. Epidemiol Infect. 1991;107:321–334. doi: 10.1017/s0950268800048962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haider K, Huq M I, Talukder K A, Ahmad Q S. Electropherotyping of plasmid DNA of different serotypes of Shigella flexneri isolated in Bangladesh. Epidemiol Infect. 1989;102:421–428. doi: 10.1017/s0950268800030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jamieson A F, Bremner D A, Bergquist P L, Lane H E D. Characterization of plasmids from antibiotic resistant Shigella isolates by agarose gel electrophoresis. J Gen Microbiol. 1979;113:73–81. doi: 10.1099/00221287-113-1-73. [DOI] [PubMed] [Google Scholar]
- 10.Levesque C, Roy P H. PCR analysis of integrons. In: Persing D H, Smith T S, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 590–594. [Google Scholar]
- 11.Lima A A, Sidrim J J, Lima N L, Titlow W, Evans M E, Greenberg R N. Molecular epidemiology of multiple antibiotic-resistant Shigella flexneri in Fortaleza, Brasil. J Clin Microbiol. 1997;35:1061–1065. doi: 10.1128/jcm.35.5.1061-1065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litwin C M, Storm A L, Chipowsky S, Ryan K J. Molecular epidemiology of Shigella infections: plasmid profiles, serotype correlation, and restriction endonuclease analysis. J Clin Microbiol. 1991;29:104–108. doi: 10.1128/jcm.29.1.104-108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu P Y, Lau Y J, Hu B S, Shyr M J, Shi Z Y, Tsai W S, Lin Y H, Tseng C Y. Analysis of clonal relationships among isolates of Shigella sonnei by different molecular typing methods. J Clin Microbiol. 1995;33:1779–1783. doi: 10.1128/jcm.33.7.1779-1783.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maraki S, Georgiladakis A, Christidou A, Scoulica E, Tselentis Y. Antimicrobial susceptibilities and β-lactamase production of Shigella isolates in Crete, Greece, during the period 1991-1995. APMIS. 1998;106:879–883. doi: 10.1111/j.1699-0463.1998.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 15.Matushek M G, Bonten M J M, Hayden M K. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken F C, editors. Manual of clinical microbiology. 6th ed. Washington D.C: American Society for Microbiology; 1995. [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 2nd ed. Approved standard M7-A2. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 18.Preston M A, Borczyk A A. Genetic variability and molecular typing of Shigella sonnei strains isolated in Canada. J Clin Microbiol. 1994;32:1427–1430. doi: 10.1128/jcm.32.6.1427-1430.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radstrom P, Skold O, Swedberg G, Flensburg J, Roy P H, Sundstrom L. Transposon Tn5090, which carries an integron, is related to Tn7, Mu, and the retroelements. J Bacteriol. 1994;176:3257–3268. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ries A A, Wells J G, Olivola M D, Ntakibirora M, Nyandwi S, Ntibakivayo M, Ivey C B, Greene K D, Tenover F C, Wahlquist S P, Griffin P M, Tauxe R V. Epidemic Shigella dysenteriae type 1 in Burundi: panresistance and implications for prevention. J Infect Dis. 1994;169:1035–1041. doi: 10.1093/infdis/169.5.1035. [DOI] [PubMed] [Google Scholar]
- 21.Sack R B, Rahman M, Yunus M, Khan E. Antimicrobial resistance in organisms causing diarrheal disease. Clin Infect Dis. 1997;24(Suppl. 1):S102–S105. doi: 10.1093/clinids/24.supplement_1.s102. [DOI] [PubMed] [Google Scholar]
- 22.Schumacher H, Nir M, Mansa B, Grassy A. β-Lactamases in Shigella. APMIS. 1992;100:954–956. [PubMed] [Google Scholar]
- 23.Simonsen C C, Chen E Y, Levinson A D. Identification of the type I trimethoprim-resistant dihydrofolate reductase specified by the Escherichia coli R-plasmid R483: comparison with procaryotic and eucaryotic dihydrofolate reductases. J Bacteriol. 1983;155:1001–1008. doi: 10.1128/jb.155.3.1001-1008.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soldati L, Piffaretti J C. Molecular typing of Shigella strains using pulsed field gel electrophoresis and genome hybridization with insertion sequences. Res Microbiol. 1991;142:489–498. doi: 10.1016/0923-2508(91)90182-a. [DOI] [PubMed] [Google Scholar]
- 25.Tauxe R V, Puhr N D, Wells J G, Hargrett-Bean N, Blake P A. Antimicrobial resistance of Shigella isolates in the USA: the importance of international travelers. J Infect Dis. 1990;162:1107–1111. doi: 10.1093/infdis/162.5.1107. [DOI] [PubMed] [Google Scholar]
- 26.Tenover F C, Arbeit R D, Goering R V, Amickelsen P, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vila J, Gascon J, Abdalla S, Gómez J, Marco F, Moreno A, Corachan M, Jiménez de Anta T. Antimicrobial resistance of Shigella isolates causing traveler’s diarrhea. Antimicrob Agents Chemother. 1994;38:2668–2670. doi: 10.1128/aac.38.11.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]