Abstract
Mesenchymal stem cells (MSCs) can show trisomy 7; however, the safety of these cells has not been fully investigated. The purposes of this study were to determine the ratio of patients whose synovial MSCs were transplanted clinically, to intensively investigate MSCs with trisomy 7 from a safety perspective, and to follow up the patients for 5 years after transplantation. Synovial MSCs at passage 0 were transplanted into a knee for degenerative meniscus tears in 10 patients, and the patients were checked at 5 years. The synovial MSCs were evaluated at passages 0 to 15 by G‐bands and digital karyotyping, and trisomy 7 was found in 3 of 10 patients. In those three patients, 5% to 10% of the synovial MSCs showed trisomy 7. The mRNA expressions of representative oncogenes and genes on chromosome 7 did not differ between MSCs with and without trisomy 7. Whole‐genome sequencing and DNA methylation analysis showed similar results for MSCs with and without trisomy 7. Transplantation of human synovial MSCs with trisomy 7 into eight mouse knees did not result in tumor formation under the skin or in the knees after 8 weeks in any mouse, whereas transplanted HT1080 cells formed tumors. In vitro chondrogenic potentials were similar between MSCs with and without trisomy 7. Five‐year follow‐ups revealed no serious adverse events in all 10 human patients, including 3 who had received MSCs with trisomy 7. Overall, our findings indicated that synovial MSCs with trisomy 7 were comparable with MSCs without trisomy 7 from a safety perspective.
Keywords: adult stem cells, cell culture, clinical trials, clinical translation, mesenchymal stem cells (MSCs), stem cell culture, tissue‐specific stem cells
Trisomy 7 is often found in synovial mesenchymal stem cells (MSCs). However, the safety of these cells after transplantation has not been investigated. The authors found no serious adverse events, including tumor formation, in any of our 10 patients at 5 years after transplantation of MSCs with or without trisomy 7.

Significance statement.
Trisomy 7 was found in 3 of 10 patients who underwent MSC transplantation for meniscus tears. In the three patients with trisomy 7, 5% to 10% of their synovial MSCs had trisomy 7. No abnormalities were detected in any of the safety tests conducted. No serious adverse events, including tumor formation, were observed after 5 years of follow‐up in any of the 10 patients who underwent transplantation with MSCs with or without trisomy 7. The results of this study showed that MSCs with trisomy 7 were comparable to MSCs without trisomy 7 from a safety perspective.
1. INTRODUCTION
Mesenchymal stem cells (MSCs) are widely used in clinical practice as a cell source for regenerative medicine. 1 , 2 , 3 MSCs are usually expanded in vitro to obtain sufficient numbers of cells for transplantation, but chromosomal mutations may occur during the expansion process. 4 , 5 Chromosomal abnormalities are often found in cell dysfunctions represented by Down's syndrome 6 , 7 and in epithelial tumors such as colorectal cancer 8 ; therefore, cell safety is a great concern when transplanting cells with chromosomal anomalies.
Various tests, such as soft agar colony formation assays 9 and whole‐genome sequencing tests, 10 have been conducted to predict the safety of transplanting embryonic stem (ES) cells and induced pluripotent stem (iPS) cells that have been expanded using conventional methods. The inclusion of additional tests may increase the safety of cell therapy, but more tests also increase the time and cost of the transplantation procedure. Nevertheless, appropriate safety tests should be established for various cell types.
We have developed a cell therapy for treating difficult‐to‐heal meniscus injury using MSCs derived from the synovium of the knee. 1 However, trisomy 7 is often found in synovial cells obtained from patients with osteoarthritis, a disease that occurs with aging. 11 , 12 , 13 , 14 , 15 , 16 Genes such as EGFR and HGF are coded on chromosome 7, and the detection of trisomy 7 in epithelial cells has been associated with tumor formation. 17 At present, no detailed analysis has been made of the relationship between trisomy 7 and tumor formation for synovial MSCs. The effects of transplantation of synovial MSCs with trisomy 7 are also unknown, and no related safety tests have been established.
The purpose of this study was to determine the proportion of synovial MSCs with trisomy 7 in patients who underwent clinical transplantation, the safety of transplantation of MSCs with trisomy 7, and the occurrence of tumors in patients 5 years after transplantation of these MSCs.
2. MATERIALS AND METHODS
2.1. Isolation of MSCs
This study was approved by the Medical Research Ethics Committee of Tokyo Medical and Dental University, and all human study subjects provided informed consent. For cell transplantation, human synovium was harvested from the knees of 10 donors (45 ± 8 years) during suturing of degenerative meniscus tears. Synovial MSC cultures with a high proportion of trisomy 7 were prepared by harvesting synovium during total knee arthroplasty of the knees of two donors (65‐76 years of age) with osteoarthritis. Synovium for clinical use was digested in 1 mg/mL Liberase MNP‐S GMP Grade (Roche Custom Biotech, Indianapolis, Indiana) for 3 hours, and synovia for obtaining a high proportion of MSCs with trisomy 7 was digested in 3 mg/mL collagenase (Sigma‐Aldrich by Merck KGaA, Darmstadt, Germany). After filtration through a 70‐μm cell strainer (Greiner Bio‐One GmbH, Frickenhausen, Germany), the cells were cultured at 500 to 2000 cells/cm2 in a cell culture incubator at 37° C and 5% CO2 in α‐minimum essential medium (α‐MEM; Thermo Fisher Scientific, Waltham, Massachusetts) supplemented with 1% antibiotic‐antimycotic (Thermo Fisher Scientific). For serum, we used 10% autologous serum for clinical passage zero (P0) MSCs and 10% fetal bovine serum (Thermo Fisher Scientific). The cells were counted on an automated cell counter (Luna‐FL, Logos Biosystems, Annandale, Virginia) in a disposable cell counting plate to determine the numbers of nucleated cells.
2.2. Ethical approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki, with the “Guidelines on clinical research using human stem cells” in Japan, and in conformity with the “Ministerial Ordinance on Good Clinical Practice for Medical Devices” in Japan. The study was approved by the Certified Special Committees for Regenerative Medicine at Tokyo Medical and Dental University. The protocol was enrolled in a database at the National University Hospital Council of Japan (UMIN Clinical Trials Registry) and disclosed (UMIN No. 000011881 and 000017890). The purpose of these two clinical studies was to evaluate the 24‐week safety, efficacy, and practicality of transplantation of autologous synovial MSCs to an injured medial meniscus (UMIN No. 000011881) or to an injured lateral meniscus (UMIN No. 000017890).
We were unable to recognize whether a patient had trisomy 7 at the time of transplantation. However, the presence or absence of trisomy 7, as defined by the G‐band test, was a monitoring item that did not appear to affect the performance of the transplantation since the primary MSCs were transplanted 2 weeks after harvesting the synovial MSCs and the G‐band test results for trisomy 7 were not available until several days later. Therefore, we initially obtained informed consent without mentioning trisomy 7. However, upon detecting trisomy 7, we updated the consent form as follows:
The cell preparation used for transplantation may contain cells that are positive for trisomy 7.
At the time of transplantation, the presence or absence of cells with trisomy 7 in the transplanted cell preparation is not known.
Based on previous reports, the risk of tumor formation after transplantation is low, even if the transplanted cell preparation contains some cells that are positive for trisomy 7.
The presence of cells with trisomy 7 in the transplanted cell population will be divulged only to subjects who wish to know this information.
This updated consent form was approved, informed consent was then obtained from all subjects, and the clinical studies were completed.
The purpose of the current study was to investigate the safety of using MSCs with trisomy 7 by examining patients 5 years after the transplantation. This study was positioned as a follow‐up study to UMIN000011881 and UMIN000017890. The study was approved by the Medical Research Ethics Committee of Tokyo Medical and Dental University (M2000‐2121), and genome analysis was approved by the institutional ethics committees (G2000‐211) in accordance with the ethical guidelines for human genome/gene analysis research in Japan.
2.3. Cell transplantation into human knee joints
A suspension of P0 synovial MSCs was placed on the repaired meniscus 14 days after suturing the degenerative meniscus tear. The cells were allowed to adhere to the meniscus and surrounding synovium by maintaining the knee position for 10 minutes. 1 On the next day, the patient was allowed to bend the knee and walk with a partial load.
2.4. G‐bands
MSC culture for the G‐band test was performed simultaneously with the MSC culture for transplantation. Before MSCs became confluent, they were incubated with 1 μg/mL colcemid for 2 hours at 37°C, trypsinized, and processed by standard procedures (BML Inc., Tokyo, Japan) according to the International System of Human Cytogenetic Nomenclature (ISCN, 2009). Up to 100 cells were then evaluated by experts in the G‐band test. MSCs with trisomy 7 detected in the G‐band test for P0 MSCs were considered positive for trisomy 7, unless noted otherwise.
2.5. Digital karyotyping
DNA was purified from synovial tissue and synovial MSCs at P0 to P15 using the QIAamp mini kit (Qiagen N.V., Venlo, The Netherlands). The DNA was sheared to various sizes, generally to an average of 200 bp, using the Covaris Acoustic Shearing method (Covaris, Woburn, Massachusetts). DNA yield was measured with a Qubit fluorometer (Thermo Fisher Scientific) and the Qubit ds DNA HS assay (Thermo Fisher Scientific). Sequencing libraries were prepared with the KAPA Hyper Prep Kit (KAPA Biosystems by Roche Diagnostics K.K., Basel, Switzerland). Adapter ligation reaction products were purified with Agencourt AmPure XP reagent (Beckman Coulter, Brea, California). The DNA samples were then analyzed on a bioanalyzer (Agilent Technologies, Santa Clara, California). Library amplification was monitored by real‐time polymerase chain reaction (PCR) to avoid over‐amplification. All libraries were sequenced on a HiSeq 2000 instrument (Illumina, Inc., Foster City, California) using single‐read 50‐bp reads with an index read of 8 bp. Reads were mapped to the human reference genome hg19 with Bowtie2 v.2.2.1. PCR duplicate read pairs were removed using the Picard toolkit.
2.6. Growth curve
P0 MSCs were plated at 50 cells/cm2 and cultured for 14 days. Cells at 70% to 80% confluence were then repeatedly passaged until the plateau phase. Growth curves were calculated by integrating the theoretical number of cells at the time of passage (Figure 1).
FIGURE 1.
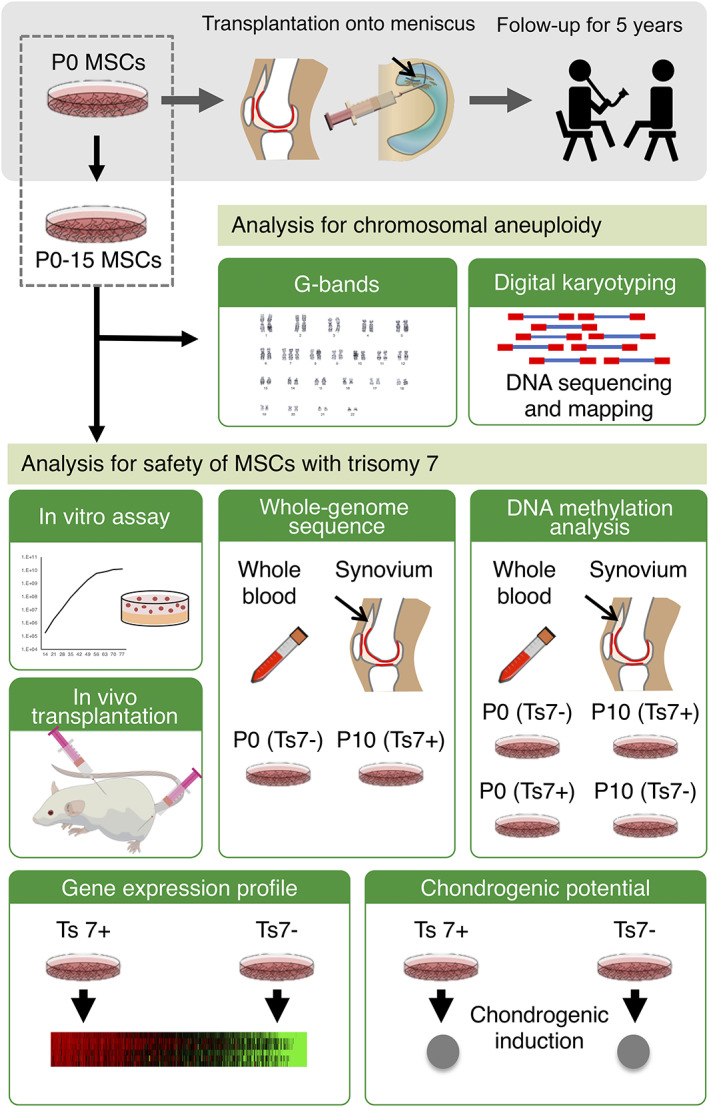
Schematic diagrams of the methods in this study. Passage 0 (P0) synovial mesenchymal stem cells (MSCs) were transplanted into the knees of 10 patients with degenerative meniscus injury. Synovial MSCs were additionally passaged up to P15. The chromosomal aneuploidy of MSCs at P0 to P15 was investigated by G‐banding and digital karyotyping using the DNA sequence. The safety of MSCs with trisomy 7 was tested by in vitro assays, in vivo transplantation, whole‐genome sequencing, and DNA methylation of P0 and P10 MSCs with/without trisomy 7. Blood and synovium were used as controls. MSCs with/without trisomy 7 were also analyzed for their gene expression profiles and chondrogenic potentials. Finally, the safety of the transplantation was evaluated after 5 years in 10 patients transplanted with MSCs with/without trisomy 7
2.7. Soft agar colony assay
MSCs at P3 that showed more than 2% trisomy 7 defined by the G‐banding tests and similar P3 MSCs without trisomy 7 were used for soft agar colony assays. HeLa cells (JCRB9004) obtained from JCRB Cell Bank (Osaka, Japan) were used as a positive control and were maintained in Dulbecco's modified Eagle medium (Sigma‐Aldrich) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic‐antimycotic. Prewarmed 2× α‐MEM containing 20% FBS, 2% antibiotic‐antimycotic, and melted 1.1% agarose (Star by Rikaken HD, Tokyo, Japan) were mixed and transferred into a well in a 6‐cm dish (Nunc by Thermo Fisher Scientific). The plate was then incubated at room temperature for 30 minutes to allow the bottom agar layer to solidify. A sample containing 3.0 × 105 cells was dissociated into a single‐cell suspension by treatment with 0.25% trypsin‐ethylenediaminetetraacetic acid (EDTA) solution (Thermo Fisher Scientific). The cell suspension in 2 × α‐MEM containing 20% FBS was then mixed with 0.66% agar and placed onto the bottom agar layer in the plate. The top agar layers were solidified to avoid gravity‐induced anchorage‐dependent cell proliferation at the bottom of the wells. The plates were incubated for 28 days at 37°C and 5% CO2. Cells in soft agar were stained with 0.1% p‐iodonitro tetrazolium violet (Dojindo Laboratories, Kumamoto, Japan) after 28 days of culture, and the colony numbers were counted.
2.8. RNA sequencing
Total RNA was extracted at P2 to P10 from MSCs showing more than 2% trisomy 7 defined by G‐banding tests and from MSCs without trisomy 7 using the RNeasy Mini Kit (Qiagen N.V.). The concentration and quality of the RNA were determined on a Quantus Fluorometer (Promega Co., Madison, Wisconsin) and an Agilent 2100 Bioanalyzer, respectively. All samples had RNA integrity number values over 7. Sequencing libraries were prepared using the Agilent SureSelect Strand‐Specific RNA Library Prep for Illumina. Briefly, poly‐A RNA was purified from 300 ng total RNA per sample using oligo dT magnetic beads. The libraries were amplified by PCR for 13 cycles and then purified with AMPure XP beads. The libraries were sequenced on an Illumina HiSeq1500 system by single‐end 50‐bp reads. Representative oncogenes were selected and analyzed by RNA sequencing (RNA‐seq). Genes on chromosome 7 were also selected among the 30 134 genes and analyzed by RNA‐seq.
2.9. Clonogenic assay
P5 synovial MSCs with and without trisomy 7 were used for clonogenic assays. Single propidium iodide‐negative cells were directly sorted by a FACS Aria III system (Becton, Dickinson and Company [BD], Franklin Lakes, New Jersey) into the wells of 96‐well plates containing 200 μL of culture medium. After 21 days, the cells were stained with 1% crystal violet stain (FUJIFILM Wako Pure Chemical Industries, Osaka, Japan), and colonies containing more than 10 cells were considered positive.
2.10. Senescence‐associated β‐galactosidase expression
P5 synovial MSCs with and without trisomy 7 were stained by SPiDER‐βGal (Dojindo Laboratories) and analyzed using a FACS Verse system (Becton Dickinson). MSCs from four donors were used. Positive ratios of SA‐β Gal cells were quantitatively evaluated.
2.11. MSC transplantation into NOD/SCID mice
In vivo tumorigenicity experiments were performed in compliance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan, June 1, 2006). Sixteen male 8‐week‐old NOD/ShiJic‐scidJcl mouse (CLEA Japan, Inc., Tokyo, Japan) were used for the experiments, which were approved by the Animal Care and Use Committee at Tokyo Medical and Dental University (approval no. A2021‐180A). P2 synovial MSCs with more than 20% trisomy 7, defined by digital karyotyping (Figure S2), were transplanted into the subcutaneous region and knee joint. HT‐1080 cells (a human sarcoma cell line obtained from JCRB Cell Bank; JCRB9113) were used as a positive control. Macroscopic and histological images were taken 8 weeks after transplantation with MSCs and when the tumor had grown to a maximum diameter of 10 mm for HT1080 cells. For histological observation of the knee, the whole knee joint was fixed in 4% paraformaldehyde, decalcified in 20% EDTA, and then embedded in paraffin wax. The specimens were stained with hematoxylin and eosin.
2.12. Whole‐genome sequence
Libraries for whole‐genome sequencing were prepared from 200 ng genomic DNA extracted from synovial tissue, P0 and P10 synovial MSCs, and whole blood samples as starting materials, following the kit manufacturer's instructions. After shearing genomic DNAs using Covaris, we made libraries with the KAPA Hyper Prep Kit (Kapa Biosystems) without PCR. Cluster generation was done with the HiSeq PE Cluster Kit v4 cBot (Illumina). Whole‐genome sequencing was performed with the HiSeq SBS Kit v4 using HiSeq2500 in the 2 × 126 PE mode. The primary base call files were converted into the FASTQ format using bcl2fastq 1.8.4 (Illumina). Adaptor sequences were removed using TrimGalore 0.4.1. Low‐quality reads were trimmed using qcleaner 3.1 (Amelieff), which has functions equivalent to those of the FASTX‐Toolkit FASTQ/A Trimmer. The sequenced reads were mapped to the reference human genome (hg19) using BWA‐MEM 0.7.10_r876. The optional parameters of BWA‐MEM were the default for Genomon and −T0 for Genomon2. The final BAM files were filtered for PCR‐duplicated reads using Novosort 1.03.01 (Novocraft Technologies, Selangor, Malaysia). We have used Genomon and Genomon2 (v2.0.5) to detect single‐nucleotide variants (SNVs) and insertions/deletions (SNVs/indels) in samples vs controls (whole blood and synovium) (Figure 1). The results of the SNV/indel results from Genomon were filtered to yield a Fisher's P value <.001, strand ratio ≠0 or 1, and variant allele frequency (VAF) of control <0.1. The results of SNVs/indels from Genomon2 were filtered to yield a P value (Fisher) ≥1.0, P value (fisher_realignment) ≥1.0, P value (EBCall) ≥3.0, strand ratio ≠0,1, and variantPairNum_tumor ≥4.
We also filtered SNVs/indels according to the following criteria: (a) the SNVs/indels in coding sequence (CDS) and splicing regions were selected; (b) synonymous SNVs were excluded; (c) the SNVs/indels for samples with a VAF more than fivefold greater than the VAF of controls and a VAF ≥0.05 were selected.
We performed copy number variant (CNV) analysis using VarScan2 (ver. 2.4.2) and Delly (ver. 0.5.6) with the final BAM files used in performing Genomon. The candidate CNVs were manually curated using GenomeJack, a genome viewer developed by Mitsubishi Space Software.
2.13. DNA methylation analyses
A 500 ng sample of genomic DNA from synovial tissue, P0 and P10 synovial MSCs, and whole blood samples was subjected to bisulfite conversion using the EZ DNA Methylation kit (Zymo Research, Irvine, California). DNA methylation profiling of the bisulfite‐converted DNA was performed using the HumanMethylation450 DNA Analysis kit (Illumina), according to the manufacturer's instructions. All DNA methylation analyses were done using RnBeads version 1.2.1 18 in R3.2.3 19 and the default parameters, unless otherwise stated. The idat files were loaded into RnBeads and quality controls were performed. The background was then subtracted using the noob method in the methylumi package, 20 and the beta values were normalized using the beta mIxture quantile dilation (BMIQ) normalization method. 21 The processed beta values of the 1000 most variable loci were visualized as a heatmap, and hierarchical clustering was performed using a correlation‐based dissimilarity metric and average linkage. Principal component analyses were performed to illustrate the global methylation patterns of the samples. The locations of promoters were defined as the regions spanning 1.5 kb upstream and 0.5 kb downstream of the transcription start site (TSS) of genes (Ensembl Genes 75). CpG islands were annotated using the CpG island track of the UCSC Genome browser. We used the region sets provided with RnBeads.
2.14. Chondrogenic differentiation
MSCs from six donors with and without trisomy 7 were harvested at P2 in a cell‐dissociation buffer. A total of 2.5 × 105 cells were transferred to a 15‐mL tube (BD Falcon, New Jersey) and cultured in chondrogenic induction medium containing 10 ng/mL transforming growth factor‐β3 (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) and 1 μg/mL bone morphogenetic protein 2 (Medtronic, Memphis, Tennessee); the medium was changed every 3 to 4 days. After 21 days, chondrogenic differentiated cells were analyzed by staining with safranin O (Fujifilm Wako), and DNA was extracted. 22 Chondrogenic ability was evaluated by wet weight, as described previously. 23 , 24
2.15. Surface markers
P0 MSCs with trisomy 7 from three donors and without trisomy 7 from three donors were analyzed by FACS Verse (BD). The cells were suspended in Hank's balanced salt solution at a density of 5 × 105 cells/mL and stained for 30 minutes on ice with antibodies, CD44, CD73, CD90, CD105, CD11b, CD14, CD31, CD45, CD146, CD271, and SSEA3 (all from BD). The data were analyzed using FACSuite software (BD).
2.16. Follow‐up
All 10 patients underwent an magnetic resonance imaging (MRI) at 2 years and again at 3 years if they requested it. They were checked by orthopedic surgeons every year until 5 years after transplantation.
2.17. Statistical analysis
Data were statistically evaluated by analysis of variance using GraphPad Prism 8 (GraphPad Software, La Jolla, California). All statistical analysis methods are described in the figure legends. Two‐tailed P values of <.05 were considered statistically significant.
3. RESULTS
3.1. Overview of the methods
Autologous P0 synovial MSCs were transplanted into the knees of 10 patients with degenerative meniscus injury. Synovial MSCs were additionally passaged up to P15 and examined for chromosomal aneuploidy, especially trisomy 7. MSCs with trisomy 7 were analyzed from the viewpoints of safety, DNA methylation, gene expression profiles, and chondrogenic potential. Finally, the safety of the 10 patients transplanted with MSCs with/without trisomy 7 was evaluated after 5 years (Figure 1).
3.2. Proportion of number of MSCs with trisomy 7 to total number of MSCs
The influence of MSC passaging on the proportion of trisomy 7 analyzed by G‐band and digital karyotyping showed two patterns: “invariant” and “increase and decrease” (Figure 2A,B). The proportion of MSCs with trisomy 7 determined by digital karyotyping was significantly correlated with the proportion determined by the G‐band method (Figure S1). Detailed chromosomal aneuploidies analyzed by G‐band and digital karyotyping are shown in Tables S1 and S2. Digital karyotyping defines the copy number of a normal disomic chromosome as 1.00. Using 5% or more as a definition of positive trisomy, trisomy 7 was found in 3 of the 10 patients (Figure 2C). The proportion of P0 MSCs with trisomy 7 was 8%, on average, based on digital karyotyping and 2%, on average, based on the G‐band method (Figure 2D). The age distributions did not differ between patients who had and did not have trisomy 7 in P0 MSCs, as determined by digital karyotyping (Figure 2E). To determine whether trisomy 7 was originally present in the original tissue or had been induced by culture manipulation, we performed digital karyotyping using DNA extracted from synovial tissue and found that trisomy 7 was also present in the original tissue (Figure S2).
FIGURE 2.
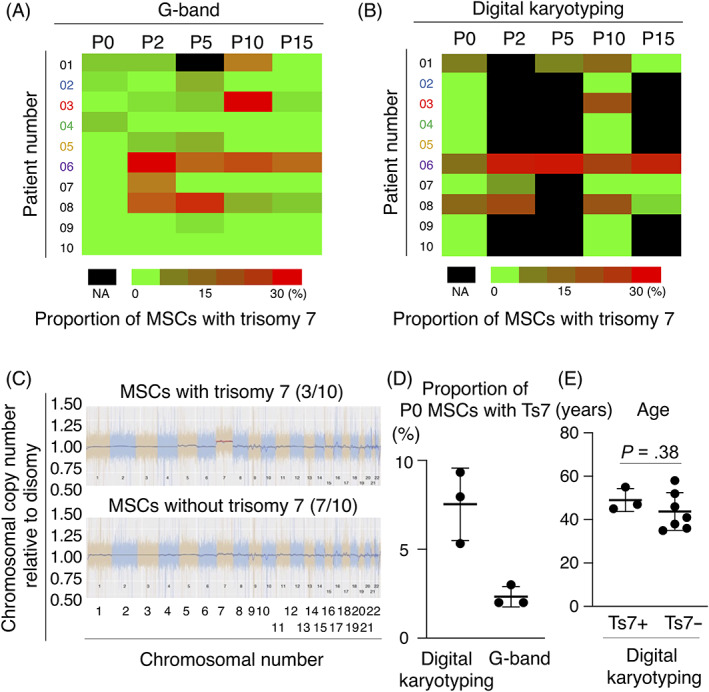
Proportion of number of MSCs with trisomy 7 to total number of MSCs. A, The proportion of MSCs with trisomy 7 in 100 cells at P0 to P15 by the G‐band method in 10 patients. B, The proportion of chromosome 7 copy number to disomic chromosome 7 copy number by digital karyotyping in P0 to P15 MSCs in 10 patients. C, Representative results of the proportion of chromosomal copy number to disomic chromosomal copy number obtained by digital karyotyping of P0 MSCs with/without trisomy 7. D, The proportion of chromosome 7 copy number to disomic chromosome 7 copy number obtained by digital karyotyping and the G‐band method in P0 MSCs with trisomy 7 in three patients. Average ± SD is shown (n = 3). E, The age distribution of patients who had and did not have trisomy 7 in P0 MSCs defined by the G‐band method. Average ± SD is shown (Ts7+; n = 3, Ts7−; n = 7). The P value was determined by the Mann‐Whitney U test. MSCs, mesenchymal stem cells; NA, not assigned; Ts7: trisomy 7; Ts7+, patients having MSCs with trisomy 7; Ts7−, patients having MSCs without trisomy 7
3.3. Conventional safety tests for MSCs with trisomy 7
The growth curves from P0 to P15 for MSCs with and without trisomy 7 did not show any abnormalities (Figure 3A). Soft‐agar colony formation assay showed colony formation in HeLa cells as a positive control but not in MSCs with trisomy 7 (Figure 3B). The mRNA expressions of the oncogenes CDKN1A, CDKN2A, MYC, and KIT did not differ between MSCs with and without trisomy 7 (Figure 3C). Clonogenic assays showed no differences in colony formation rates between MSCs with and without trisomy 7 (Figure 3D). No differences were observed in senescence‐associated beta‐galactosidase (SA‐β‐gal)‐positive cells between MSCs with and without trisomy 7 (Figure 3E). The mice transplanted with HT1080 cells under the skin and in the knees formed tumors, while those transplanted with MSCs with trisomy 7 did not form tumors in any of the eight mice after 8 weeks (Figure 3F).
FIGURE 3.
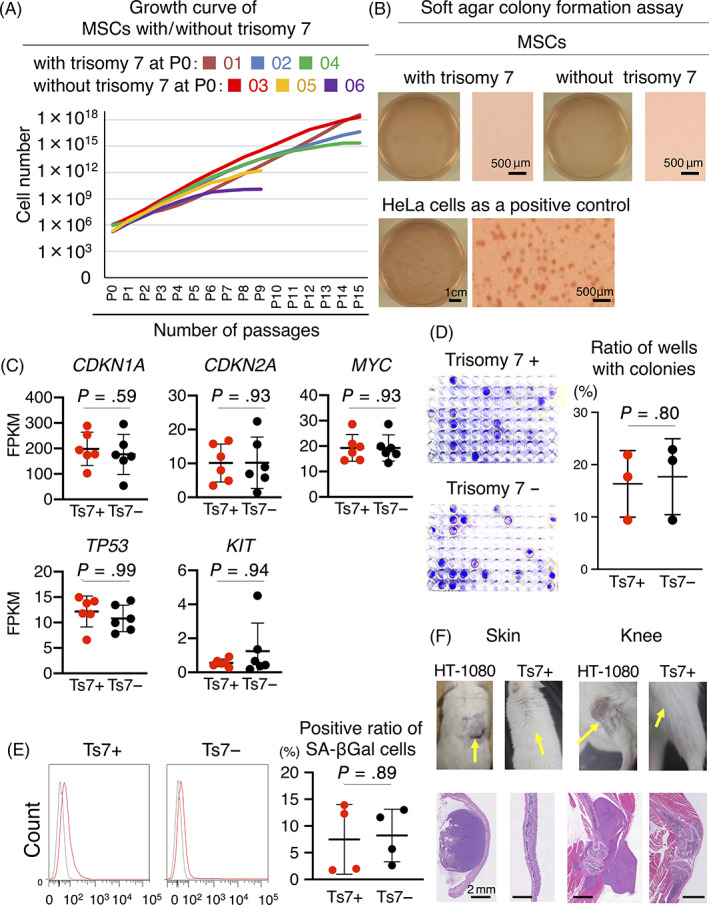
Safety tests for MSCs with trisomy 7. A, Growth curve of MSCs with/without trisomy 7. B, Soft agar colony formation assays for MSCs with/without trisomy 7 and for HeLa cells. C, mRNA expression of oncogenes. Average ± SD is shown (n = 6). P values were determined by the Mann‐Whitney U test. D, Clonogenic assay of MSCs with/without trisomy 7. One cell of patients 02 and 03 at P5 was plated in a well of a 96‐well plate. The colonies containing more than 10 cells were considered positive. Representative images of 96‐well plates at 21 days of culture and the rate of wells with colonies are shown. Average ± SD is shown (n = 3). P values were determined by the Mann‐Whitney U test. E, Senescence‐associated β‐gal assay of MSCs with/without trisomy 7 at P5. Representative histograms from flow cytometric analysis for SA‐β‐gal positive cells are shown in red and for isotype control in gray. The positive ratio of SA‐β Gal cells is also shown as average ± SD (n = 4). P values were determined by the Mann‐Whitney U test. F, Transplantation of MSCs with trisomy 7 into subcutaneous tissues and knee joints of mice. HT‐1080 cells were used as a positive control. Images were taken 8 weeks after transplantation for MSCs and when the tumor had grown to a maximum diameter of 10 mm for HT1080 cells. Injection sites are indicated by arrows. CDKN1A, cyclin‐dependent kinase inhibitor 1A; CDKN2A, cyclin‐dependent kinase inhibitor 2A; FPKM, fragments per kilobase of transcript per million fragments sequenced; MSCs, mesenchymal stem cells; MYC, MYC proto‐oncogene, bHLH transcription factor; SA‐β‐gal, senescence‐associated beta‐galactosidase; TP53, tumor protein p53; KIT, KIT proto‐oncogene, receptor tyrosine kinase
3.4. Whole‐genome sequence of MSCs with and without trisomy 7
No de novo SNVs or indels were observed for the CDS and splicing regions in the P0 MSCs without trisomy 7 or in the P10 MSCs with trisomy 7 when compared with whole blood and synovium controls (Table 1). Large chromosome abnormalities due to trisomy 7 were detected by digital karyotyping, and smaller CNVs were investigated with whole‐genome sequencing data using VarScan2 and Delly. No de novo CNVs, except for trisomy 7, were found in P0 MSCs without trisomy 7 or in P10 MSCs with trisomy 7 (Table 1).
TABLE 1.
Summary of the genome analysis
| Patient 03 | |||
|---|---|---|---|
| Control | Test sample | # of detected SNVs/indels on CDS and splicing region | # of detected CNVs except for Ts 7+ |
| Blood | Synovium | 0 | 0 |
| P0 MSCs (Ts 7−) | 0 | 0 | |
| P10 MSCs (Ts 7+) | 0 | 0 | |
| Synovium | Blood | 0 | 0 |
| P0 MSCs (Ts 7−) | 0 | 0 | |
| P10 MSCs (Ts 7+) | 0 | 0 | |
Abbreviations: CDS, coding sequence; CNV, copy number variant; indels: insertion/deletion; MSC, mesenchymal stem cell; P0, passage 0; P10, passage 10; SNV, single‐nucleotide variants; Ts7, trisomy 7.
3.5. DNA methylation in MSCs with trisomy 7
Methylation analysis was conducted on whole blood, synovium, P0 MSCs, and P10 MSCs from patients 02 and 03. In patient 02, trisomy 7 was detected in 1 of 50 in P0 MSCs by the G‐band method, but no trisomy 7 was detected at P10. In patient 03, cells with trisomy 7 were not detected at P0, but their proportion increased to 32% at P10. Each of the promoters and CpG islands was subjected to hierarchical clustering analyses using the beta values of the 1000 most variable loci (Figure 4A). Principal component analyses revealed that each cell type exhibited similar methylation patterns across patients, regardless of the presence or absence of chromosomal aneuploidy (Figure 4B).
FIGURE 4.
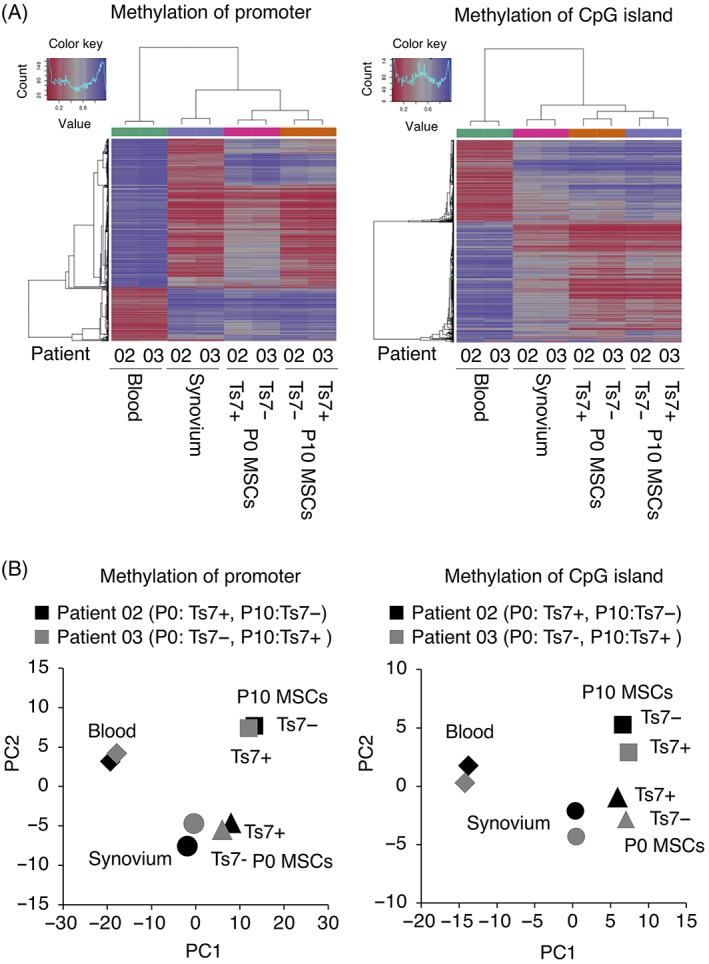
Comparison of DNA methylation between mesenchymal stem cells (MSCs) with trisomy 7 and without trisomy 7. A, Hierarchical clustering analyses using the beta values of the 1000 most variable loci for methylation of promoters and methylation of CpG islands. B, Principal component analyses using the beta values of promoters for methylation of promoters and methylation of CpG islands
3.6. Expression profiles of MSCs with trisomy 7
MSCs with more than 2% trisomy 7 in the G‐banding test were defined as positive. The expression profiles of 372 genes on chromosome 7 (Table S3) were similar between trisomy 7 positive and negative MSCs (Figure 5A). A more detailed analysis of genes on chromosome 7 also showed no differences in the expression of EGFR, HGF, IL6, PPIA, and CAV2 between the two groups (Figure 5B).
FIGURE 5.
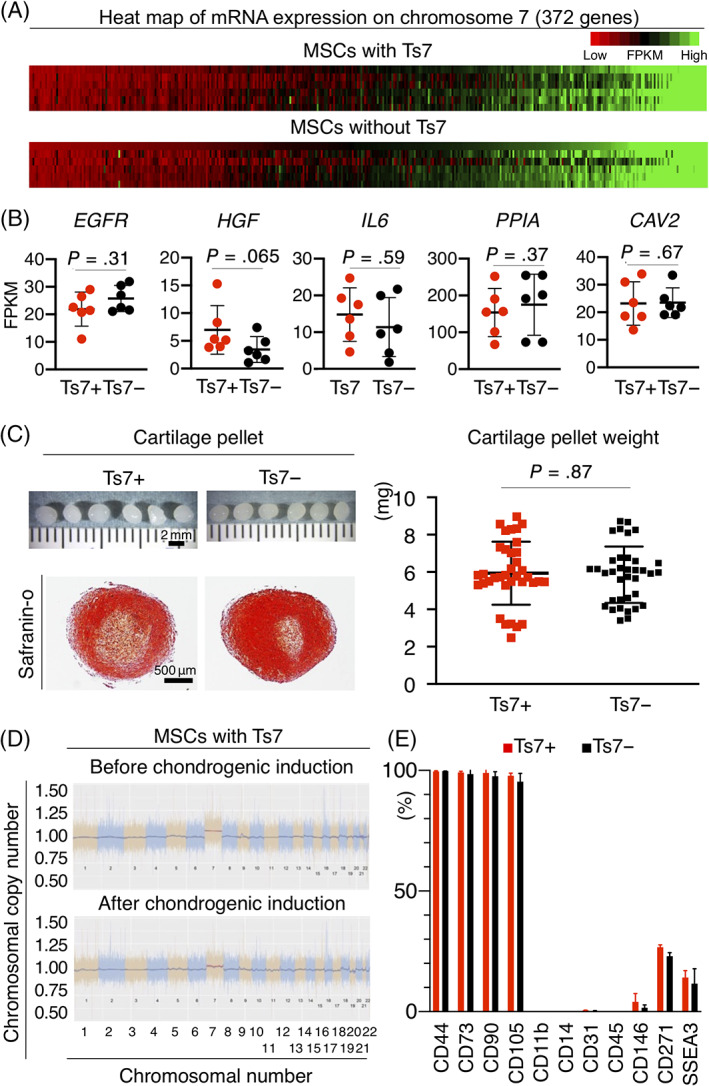
Gene expression profiles and chondrogenic potentials of MSCs with and without trisomy 7. A, Heatmap of mRNA expression on chromosome 7 (372 genes). B, The mRNA expression of representative genes on chromosome 7. Average ± SD is shown (n = 6). P values were determined by the Mann‐Whitney U test. C, Chondrogenesis potential of MSCs with trisomy 7. Representative macroscopic and safranin O‐stained histological images of cartilage pellets are shown. Quantitative analysis of the wet weight of cartilage pellets is also demonstrated. Average ± SD is shown for 36 samples from six donors. P values were determined by the Mann‐Whitney U test. D, Chromosomal copy number of MSCs with trisomy 7 before and after chondrogenic induction. Representative images for the proportion of the chromosomal copy number to the disomic chromosomal copy number obtained by digital karyotyping of P0 MSCs are shown. E, Surface markers of P0 MSCs with/without trisomy 7. Bar indicates the mean ± SD (three donors). CAV2, caveolin 2; EGFR, epidermal growth factor receptor; FPKM, fragments per kilobase of transcript per million fragments sequenced; HGF, hepatocyte growth factor; IL6, interleukin 6; MSCs, mesenchymal stem cells; PPIA, peptidylprolyl isomerase A
3.7. Chondrogenic potential of MSCs with trisomy 7
MSCs with and without trisomy 7 were differentiated into cartilage pellets and confirmed histologically (Figure 5C). Quantitative analysis showed that the wet weights of the cartilage pellets were comparable (Figure 5C). The copy number of trisomy 7 did not differ in MSCs with trisomy 7 after chondrogenic differentiation (Figure 5D).
3.8. Surface markers of MSCs with trisomy 7
The positive rates of surface markers in MSCs with trisomy 7 were more than 90% for CD44, CD73, CD90, and CD105, 10% to 30% for CD271 and SSEA3, and less than 5% for CD11b, CD14, CD31, CD45, and CD146 (Figure 5E). These rates were comparable with those observed in MSCs without trisomy 7.
3.9. Follow‐up of 10 patients for 5 years after MSC transplantation
Autologous P0 synovial MSCs, which were intensively investigated in this study, had been transplanted into the knees of 10 patients with degenerative meniscus injury. Representative MRI images showed that the meniscus tear was obscured 3 years after transplantation (Figure 6A). The 5‐year follow‐up revealed no serious adverse events, including tumor formation, in all 10 patients, including the 3 patients who had undergone transplantation with MSCs with trisomy 7 (Figure 6B).
FIGURE 6.
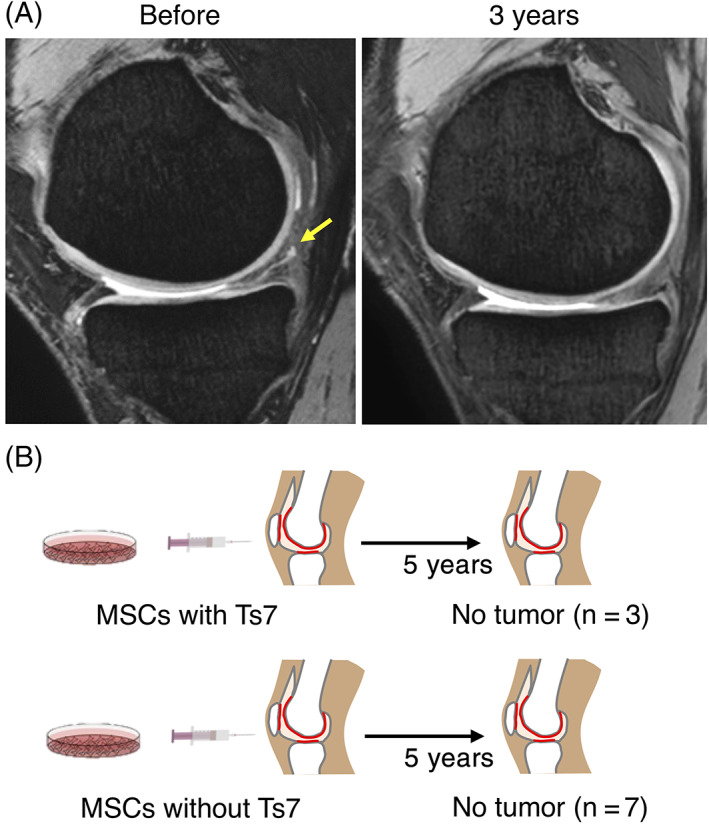
Follow‐up of 10 patients for 5 years after mesenchymal stem cell (MSC) transplantation. A, Representative magnetic resonance imaging (MRI) of the meniscus before and 3 years after cell transplantation. Yellow arrow shows a meniscus degenerative tear. (B) Follow‐up results on tumorigenesis
4. DISCUSSION
The original purpose of the two clinical studies had been to evaluate the safety, efficacy, and practicality of autologous synovial MSCs 24 weeks after transplantation to an injured meniscus. We had been unable to recognize whether the patient had trisomy 7 at the time of transplantation. After detecting trisomy 7, we updated the consent form and continued with the clinical studies. We rationalized the safety of transplantation of synovial MSCs with trisomy 7 in four ways. First, trisomy 7 is often found in synovial cells and chondrocytes in patients with osteoarthritis, 8 , 13 , 15 , 25 a condition often associated with meniscus injuries. 26 , 27 Second, previous reports suggested that trisomy 7 did not occur during cell culture but was originally present in the patient's own cells. 12 , 25 Third, the risk of tumor formation was minimal because the transplanted cells were primary synovial MSCs. Fourth, the most of the synovial MSCs attached to the synovium and did not migrate to distant organs. 28 An updated consent form was approved, informed consent was obtained from all subjects, and the clinical studies were completed.
Chromosomal aneuploidy was mainly observed as trisomy 7 in the primary MSCs and the passaged MSCs. Karyotype abnormalities, and especially trisomy 7, have been detected by G‐banding, fluorescence in situ hybridization (FISH), or array‐based comparative genomic hybridization in synovial cells and chondrocytes in patients with osteoarthritis. 11 , 12 , 13 , 14 , 15 , 16 The proportion of trisomy 7 has also been reported to increase in response to the pathological conditions from joint trauma in osteoarthritis. 25 Noncongenital trisomy 7 has been found in macrophages derived from broncho‐alveolar lavage fluid in chronic obstructive pulmonary disease, and this may reflect an inflammatory response. 25 Degenerative meniscus tears are closely associated with the onset of osteoarthritis of the knee, 29 and this could explain why our patients had a high proportion of trisomy 7. Trisomy 7 is also prevalent in other tissues, such as the colon, kidney, and skin, and the proportions of trisomy 7 can increase with age. 8 In our study, we found no significant difference in age between MSCs with and without trisomy 7, which might reflect the small age distribution in our patients.
We evaluated trisomy 7 on sequentially passaged MSCs using both digital karyotyping and the G‐band method. These evaluations, plus FISH, are the major karyotyping methods, and each has its advantages and disadvantages. The G‐band assay is a conventional method that can determine chromosome structural abnormalities, such as ploidy, aneuploidy, and translocations, but it can only visualize metaphase chromosomes and analyze, at most, 100 cells per sample. FISH can provide more direct information on the chromosomal location of genes, but the number of cells that can be observed is still limited. Digital karyotyping can reveal the proportion of chromosomal aneuploidy, regardless of mitotic phase, and can analyze data from a large number of cells. In this study, we used approximately 200 ng of DNA for digital karyotyping. Assuming that each human cell contains 6.6 pg of DNA, 30 this meant that we analyzed at least 1 × 104 cells. However, digital karyotyping analysis has two disadvantages: it cannot detect translocations and it cannot detect trisomy 7 with a positive rate of less than 5%. Therefore, we used G‐banding as the main method and digital karyotype analysis as a supportive measure.
The proportion of MSCs with trisomy 7 increased or decreased with passages in many cases. MSCs are heterogeneous cell populations and form a large number of colonies with various proliferative properties. 31 , 32 In general, highly proliferative cells become the dominant populations after multiple passages. 10 The proportion of MSCs with trisomy 7 did not continue to increase, suggesting that these cells do not have a specifically high proliferative capacity.
We performed at least six different experiments to evaluate the tumorigenicity of synovial MSCs with trisomy 7. Synovial MSCs with trisomy 7 did not acquire abnormal proliferative potential according to the growth curve analysis. In addition, synovial MSCs with trisomy 7 did not induce scaffold‐independence or clonal proliferative potential, nor did they express oncogenes or show an increase in specific senescent cells. Furthermore, similar to the safety test for iPS cells, 33 synovial MSCs with trisomy 7 did not form tumors after transplantation into the subcutaneous region and knees of NOD/SCID mice. Our intensive analysis of synovial MSCs with trisomy 7 for tumorigenesis did not reveal any specific features.
The SNV/indel and CNV analysis of whole‐genome sequence data suggested that chromosome 7 maintained a normal genetic sequence, in contrast to the dynamic alteration of the chromosomes. Analysis of DNA methylation, which was used for cell quality control, 34 showed passage‐dependent changes in cell characteristics, but no differences associated with the presence or absence of trisomy 7. However, many studies have reported an increase in chromosomes that leads to changes in cellular phenotypes. 35 , 36
Chromosome 7 contains numerous important genes, including EGFR and HGF, and several reports indicate that abnormalities in chromosomes are involved in disease. For example, cells isolated from Down syndrome patients with trisomy 21 showed significantly slower growth and altered mRNA expression profiles. 6 , 7 Similarly, ES cells with spontaneous trisomy also showed altered gene expression profiles. 37 Aneuploid cells induced with additional chromosomes could also present a different transcriptome expression profile compared with normal cells. 38 , 39 Trisomy 7 is frequently observed in colorectal cancer as an initiation event with abnormal expression of EGFR encoded by chromosome 7. 17 The association between chromosome 7 and osteoarthritis has been reported by a genome‐wide association study of 1341 Dutch Caucasian cases, in which 7q22 was detected as a novel common mutation affecting the prevalence and progression of osteoarthritis. 40 However, whether trisomy 7 is a cause or a consequence of osteoarthritis is not clear.
The expression of representative genes on chromosome 7 was similar between MSCs with and without trisomy 7 in this study. This is probably because MSCs with trisomy 7 accounted, at most, for 10% of the cell population. In addition, chromosome 7 spans about 159 million DNA base pairs and represents only 5 of the total DNA in cells, 41 so the influence of the cells with trisomy 7 may have been masked when the mRNA expression of the entire cell population was determined. If we had been able to obtain a larger proportion of MSCs with trisomy 7, we might have been able to identify the function of trisomy 7.
The chondrogenesis potential of MSCs was evaluated based on the pellet weight. During in vitro chondrogenesis of MSCs, the pellet increased in weight, the DNA yield per pellet decreased, and the amount of DNA per cell was maintained. 42 These observations mean that the increase in pellet weight can be caused by the production of extracellular matrix, rather than by cell proliferation. The pellet weight was also correlated with the expression of chondrogenic‐specific genes and the amounts of glycosaminoglycans. 32 , 43 , 44 The pellet weight is therefore a convincing indicator of in vitro chondrogenesis in a population of MSCs. 45 , 46 , 47
Trisomy 7 was still detected in the cartilage pellets differentiated from synovial MSCs with trisomy 7. The differentiation potential of cells with chromosomal aneuploidy has been reported for ES cells or tissue stem cells. Cells with trisomy artificially generated from mouse ES cells had a normal proliferative and colony‐forming potential, but no differentiation potential. 48 Conversely, cells with trisomy 7 observed in a human neural progenitor cell population showed increased proliferative capacity, improved survival, and increased expression of EGFR on chromosome 7 after transplantation into rats. 49 In other words, although a loss of differentiation ability has been verified using undifferentiated cells, such as ES cells, the differentiation ability may be maintained in tissue‐specific stem cells. Our results showing that trisomy 7 did not affect the in vitro chondrogenic differentiation suggest that trisomy 7 also has no effect on chondrogenic differentiation in vivo.
The present results suggest that transplantation of autologous synovial MSCs into the knee joint does not cause serious problems, since in vivo chromosomal aneuploidy is preserved if the number of passages is small. However, the type of problems that may arise in the case of allogeneic transplantation is not known. Even when MSCs are derived from the same cell source, different types of risks may exist depending on the route of transplantation. Therefore, each therapeutic target requires its own risk management strategy. Synovial MSCs transplanted into a joint, as in this case, remain in the joint 28 , 50 and have little effect on the whole body. However, MSCs administered intravenously can adhere to various tissues, such as the lungs, where they are maintained for a long time, 51 so they may present as yet unknown risks in the future. The results of this study do not indicate that a uniform management approach can be applied without consideration of the type or method of administration of MSCs.
This study had some limitations. The most important limitation was that the maximum proportion of trisomy 7 in synovial MSCs was only 10%. This proportion might have been too small to detect phenotypes of synovial MSCs with trisomy 7, or the phenotypes might have been silenced on the chromosome. Further clarification will require simultaneous analysis of single‐cell genome sequencing and single‐cell mRNA analysis. 52
5. CONCLUSION
Trisomy 7 was found in 3 of 10 patients who underwent MSC transplantation for meniscus tears. In the 3 patients with trisomy 7, 5% to 10% of their synovial MSCs had trisomy 7. No abnormalities were detected in any of the safety tests conducted. No serious adverse events, including tumor formation, were observed after 5 years of follow‐up in any of the 10 patients who underwent transplantation with MSCs with or without trisomy 7. Synovial MSCs with trisomy 7 were comparable with MSCs without trisomy 7 from a safety perspective, as far as we could investigate in this study.
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
M.M.: conceived the study, designed the experiments, collected and analyzed the data, drafted and wrote the paper, involved in drafting the article or revising it critically for important intellectual content, and approved the final version to be published. K.E.: drafted and wrote the paper, involved in drafting the article or revising it critically for important intellectual content, and approved the final version to be published. I.S.: drafted and wrote the paper, recruited the patients and acquired the clinical samples, provided advice regarding the research strategy, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis, involved in drafting the article or revising it critically for important intellectual content, and approved the final version to be published. H. Katano, N.O., and H. Koga: recruited the patients and acquired the clinical samples, involved in drafting the article or revising it critically for important intellectual content, and approved the final version to be published. N.A., M.N., and N.T.: provided whole‐genome sequence data and methylation analysis, wrote the manuscript, involved in drafting the article or revising it critically for important intellectual content, and approved the final version to be published. Y.H., O.O., and T.M.: conducted digital karyotyping analysis and wrote the manuscript, involved in drafting the article or revising it critically for important intellectual content, and approved the final version to be published.
Supporting information
Figure S1 Correlation of the proportion of trisomy 7 between G‐banding and digital karyotyping.
Figure S2. Representative results of chromosomal copy number to disomic chromosomal copy number obtained by digital karyotyping from synovial tissue containing blood and synovial MSCs.
Table S1. Karyotype abnormalities by G‐banding.
Table S2. Karyotype abnormalities by digital karyotyping.
Table S3‐1. List of 372 genes on chromosome 7.
Table S3‐2. List of 372 genes on chromosome 7.
Table S3‐3. List of 372 genes on chromosome 7.
Table S3‐4. List of 372 genes on chromosome 7.
ACKNOWLEDGMENTS
We thank Mika Watanabe and Kimiko Takanashi for the management of our laboratory, Ellen Roider for English editing, Reiko Kikuno for the digital karyotyping analysis, and Yuji Kohno, Yusuke Nakagawa, Koji Otabe, and Masafumi Horie for the recruitment of the patients and the acquisition of the written informed consent from the patients. This research was supported by the Japan Agency for Medical Research and Development (AMED) under Grant Numbers JP16bm0504001, JP15bk0104007, JP17bk0104038, JP16be0104003 to I.S., and by grants from the Core Center for iPS Cell Research, Research Center Network for Realization of Regenerative Medicine from AMED to N.T. N.A. is currently affiliated with BITS Co., Ltd.; N.T. and M.N. are currently affiliated with CiRA Foundation.
Mizuno M, Endo K, Katano H, et al. Transplantation of human autologous synovial mesenchymal stem cells with trisomy 7 into the knee joint and 5 years of follow‐up. STEM CELLS Transl Med. 2021;10(11):1530-1543. 10.1002/sctm.20-0491
Funding information Core Center for iPS Cell Research, Research Center Network for Realization of Regenerative Medicine; Japan Agency for Medical Research and Development (AMED)
DATA AVAILABILITY STATEMENT
Most of the data supporting the results of this study are available within the article and its supplementary materials. Requests for additional data can be sent to the corresponding author. Some of the data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Sekiya I, Koga H, Otabe K, et al. Additional use of synovial mesenchymal stem cell transplantation following surgical repair of a complex degenerative tear of the medial meniscus of the knee: a case report. Cell Transplant. 2019;28(11):1445‐1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sekiya I, Muneta T, Horie M, Koga H. Arthroscopic transplantation of synovial stem cells improves clinical outcomes in knees with cartilage defects. Clin Orthop Relat Res. 2015;473(7):2316‐2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bianco P, Cao X, Frenette PS, et al. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat Med. 2013;19(1):35‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Binato R, de Souza Fernandez T, Lazzarotto‐Silva C, et al. Stability of human mesenchymal stem cells duringin vitroculture: considerations for cell therapy. Cell Prolif. 2013;46(1):10‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim M, Rhee J‐K, Choi H, et al. Passage‐dependent accumulation of somatic mutations in mesenchymal stromal cells during in vitro culture revealed by whole genome sequencing. Sci Rep. 2017;7(1):14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brodsky JL, Aivazidis S, Coughlan CM, et al. The burden of trisomy 21 disrupts the proteostasis network in Down syndrome. Plos one. 2017;12(4):e0176307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y, Borel C, Li L, et al. Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. Nat Commun. 2017;8(1):1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Broberg K, Toksvig‐Larsen S, Lindstrand A, et al. Trisomy 7 accumulates with age in solid tumors and non‐neoplastic synovia. Genes Chromosomes Cancer. 2001;30(3):310‐315. [PubMed] [Google Scholar]
- 9. Kusakawa S, Yasuda S, Kuroda T, et al. Ultra‐sensitive detection of tumorigenic cellular impurities in human cell‐processed therapeutic products by digital analysis of soft agar colony formation. Sci Rep. 2015;5(1):17892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cai J, Miao X, Li Y, et al. Whole‐genome sequencing identifies genetic variances in culture‐expanded human mesenchymal stem cells. Stem Cell Rep. 2014;3(2):227‐233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kinne RW, Liehr T, Beensen V, et al. Mosaic chromosomal aberrations in synovial fibroblasts of patients with rheumatoid arthritis, osteoarthritis, and other inflammatory joint diseases. Arthritis Res. 2001;3(5):319‐330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dahlén A, Broberg K, Domanski HA, et al. Analysis of the distribution and frequency of trisomy 7 in vivo in synovia from patients with osteoarthritis and pigmented villonodular synovitis. Cancer Genet Cytogenet. 2001;131(1):19‐24. [DOI] [PubMed] [Google Scholar]
- 13. Broberg K, Limon J, Pålsson E, et al. Clonal chromosome aberrations are present in vivo in synovia and osteophytes from patients with osteoarthritis. Hum Genet. 1997;101(3):295‐298. [DOI] [PubMed] [Google Scholar]
- 14. Weiss KR, Georgescu HI, Gollin SM, et al. Trisomy 7 in synovial fibroblasts obtained from arthritic joints. Inflamm Res. 1999;48(suppl 2):S132‐S133. [DOI] [PubMed] [Google Scholar]
- 15. Castellanos MV, Hernández JM, Ramos L, et al. Chromosomal abnormalities are related to location and grade of osteoarthritis. Osteoarthr Cartil. 2004;12(12):982‐985. [DOI] [PubMed] [Google Scholar]
- 16. Stumm M, Boger E, Gaissmaier CG, et al. Genomic chondrocyte culture profiling by array‐CGH, interphase‐FISH and RT‐PCR. Osteoarthr Cartil. 2012;20(9):1039‐1045. [DOI] [PubMed] [Google Scholar]
- 17. Ly P, Kim SB, Kaisani AA, Marian G, Wright WE, Shay JW. Aneuploid human colonic epithelial cells are sensitive to AICAR‐induced growth inhibition through EGFR degradation. Oncogene. 2012;32(26):3139‐3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11(11):1138‐1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. R Core Team , R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. 2016. [Google Scholar]
- 20. Triche TJ, Weisenberger DJ, Van Den Berg D, et al. Low‐level processing of Illumina Infinium DNA methylation BeadArrays. Nucleic Acids Res. 2013;41(7):e90‐e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Teschendorff AE, Marabita F, Lechner M, et al. A beta‐mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakagawa Y, Muneta T, Kondo S, et al. Synovial mesenchymal stem cells promote healing after meniscal repair in microminipigs. Osteoarthr Cartil. 2015;23(6):1007‐1017. [DOI] [PubMed] [Google Scholar]
- 23. Fujisawa R, Mizuno M, Katano H, et al. Cryopreservation in 95% serum with 5% DMSO maintains colony formation and chondrogenic abilities in human synovial mesenchymal stem cells. BMC Musculoskelet Disord. 2019;20(1):316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizuno M, Katano H, Otabe K, et al. Complete human serum maintains viability and chondrogenic potential of human synovial stem cells: suitable conditions for transplantation. Stem Cell Res Ther. 2017;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kinne RW, Kunisch E, Beensen V, et al. Synovial fibroblasts and synovial macrophages from patients with rheumatoid arthritis and other inflammatory joint diseases show chromosomal aberrations. Genes Chromosomes Cancer. 2003;38(1):53‐67. [DOI] [PubMed] [Google Scholar]
- 26. Englund M. Meniscal tear—a feature of osteoarthritis. Acta Orthop Scand. 2009;75(suppl 312):1–45. [PubMed] [Google Scholar]
- 27. Ozeki N, Seil R, Krych AJ, et al. Surgical treatment of complex meniscus tear and disease: state of the art. J ISAKOS Joint Disorders Orthop Sports Med. 2021;6(1):35‐45. [DOI] [PubMed] [Google Scholar]
- 28. Ozeki N, Muneta T, Koga H, et al. Not single but periodic injections of synovial mesenchymal stem cells maintain viable cells in knees and inhibit osteoarthritis progression in rats. Osteoarthr Cartil. 2016;24(6):1061‐1070. [DOI] [PubMed] [Google Scholar]
- 29. Madry H, Kon E, Condello V, et al. Early osteoarthritis of the knee. Knee Surg Sports Traumatol Arthrosc. 2016;24(6):1753‐1762. [DOI] [PubMed] [Google Scholar]
- 30. Lo YMD, Tein MSC, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62(4):768‐775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mizuno M, Katano H, Shimozaki Y, et al. Time‐lapse image analysis for whole colony growth curves and daily distribution of the cell number per colony during the expansion of mesenchymal stem cells. Sci Rep. 2019;9(1):16835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mizuno M, Katano H, Mabuchi Y, et al. Specific markers and properties of synovial mesenchymal stem cells in the surface, stromal, and perivascular regions. Stem Cell Res Ther. 2018;9(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takei Y, Morioka M, Yamashita A, Kobayashi T, Shima N, Tsumaki N. Quality assessment tests for tumorigenicity of human iPS cell‐derived cartilage. Sci Rep. 2020;10(1):12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Almeida DC, Ferreira Marcelo RP, Franzen J, et al. Epigenetic classification of human mesenchymal stromal cells. Stem Cell Rep. 2016;6(2):168‐175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Passerini V, Ozeri‐Galai E, de Pagter MS, et al. The presence of extra chromosomes leads to genomic instability. Nat Commun. 2016;7:10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams BR, Prabhu VR, Hunter KE, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322(5902):703‐709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biancotti J‐C, Narwani K, Buehler N, et al. Human embryonic stem cells as models for aneuploid chromosomal syndromes. Stem Cells. 2010;28(9):1530‐1540. [DOI] [PubMed] [Google Scholar]
- 38. Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pavelka N, Rancati G, Zhu J, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468(7321):321‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kerkhof HJM, Lories RJ, Meulenbelt I, et al. A genome‐wide association study identifies a locus on chromosome 7q22 to influence susceptibility for osteoarthritis. Arthritis Rheum. 2010;62(2):499‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scherer SW. Human chromosome 7 circa 2004: a model for structural and functional studies of the human genome. Hum Mol Genet. 2004;13(suppl 2):R303‐R313. [DOI] [PubMed] [Google Scholar]
- 42. Sekiya I, Vuoristo JT, Larson BL, Prockop DJ. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci USA. 2002;99(7):4397‐4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kohno Y, Mizuno M, Ozeki N, et al. Yields and chondrogenic potential of primary synovial mesenchymal stem cells are comparable between rheumatoid arthritis and osteoarthritis patients. Stem Cell Res Ther. 2017;8(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagase T, Muneta T, Ju YJ, et al. Analysis of the chondrogenic potential of human synovial stem cells according to harvest site and culture parameters in knees with medial compartment osteoarthritis. Arthritis Rheum. 2008;58(5):1389‐1398. [DOI] [PubMed] [Google Scholar]
- 45. Sakaguchi Y, Sekiya I, Yagishita K, Muneta T. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum. 2005;52(8):2521‐2529. [DOI] [PubMed] [Google Scholar]
- 46. Mizuno M, Katano H, Otabe K, et al. Complete human serum maintains viability and chondrogenic potential of human synovial stem cells: suitable conditions for transplantation. Stem Cell Res Ther. 2017;8(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mochizuki T, Muneta T, Sakaguchi Y, et al. Higher chondrogenic potential of fibrous synovium‐ and adipose synovium‐derived cells compared with subcutaneous fat‐derived cells: distinguishing properties of mesenchymal stem cells in humans. Arthritis Rheum. 2006;54(3):843‐853. [DOI] [PubMed] [Google Scholar]
- 48. Zhang M, Cheng L, Jia Y, et al. Aneuploid embryonic stem cells exhibit impaired differentiation and increased neoplastic potential. EMBO J. 2016;35(21):2285‐2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Linden R, Sareen D, McMillan E, et al. Chromosome 7 and 19 trisomy in cultured human neural progenitor cells. PLoS One. 2009;4(10):e7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koga H, Muneta T, Nagase T, et al. Comparison of mesenchymal tissues‐derived stem cells for in vivo chondrogenesis: suitable conditions for cell therapy of cartilage defects in rabbit. Cell Tissue Res. 2008;333(2):207‐215. [DOI] [PubMed] [Google Scholar]
- 51. Schrepfer S, Deuse T, Reichenspurner H, Fischbein MP, Robbins RC, Pelletier MP. Stem cell transplantation: the lung barrier. Transplant Proc. 2007;39(2):573‐576. [DOI] [PubMed] [Google Scholar]
- 52. Macaulay IC, Haerty W, Kumar P, et al. G&T‐seq: parallel sequencing of single‐cell genomes and transcriptomes. Nat Methods. 2015;12(6):519‐522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Correlation of the proportion of trisomy 7 between G‐banding and digital karyotyping.
Figure S2. Representative results of chromosomal copy number to disomic chromosomal copy number obtained by digital karyotyping from synovial tissue containing blood and synovial MSCs.
Table S1. Karyotype abnormalities by G‐banding.
Table S2. Karyotype abnormalities by digital karyotyping.
Table S3‐1. List of 372 genes on chromosome 7.
Table S3‐2. List of 372 genes on chromosome 7.
Table S3‐3. List of 372 genes on chromosome 7.
Table S3‐4. List of 372 genes on chromosome 7.
Data Availability Statement
Most of the data supporting the results of this study are available within the article and its supplementary materials. Requests for additional data can be sent to the corresponding author. Some of the data are not publicly available due to privacy or ethical restrictions.


