Abstract
More than one and half years have passed, as of August 2021, since the COVID-19 caused by the novel coronavirus named SARS-CoV-2 emerged in 2019. While the recent success of vaccine developments likely reduces the severe cases, there is still a strong requirement of safety and effective therapeutic drugs for overcoming the unprecedented situation. Here we review the recent progress and the status of the drug discovery against COVID-19 with emphasizing a structure-based perspective. Structural data regarding the SARS-CoV-2 proteome has been rapidly accumulated in the Protein Data Bank, and up to 68% of the total amino acid residues encoded in the genome were covered by the structural data. Despite a global effort of in silico and in vitro screenings for drug repurposing, there is only a limited number of drugs had been successfully authorized by drug regulation organizations. Although many approved drugs and natural compounds, which exhibited antiviral activity in vitro, were considered potential drugs against COVID-19, a further multidisciplinary investigation is required for understanding the mechanisms underlying the antiviral effects of the drugs.
Keywords: coronavirus, drug repositioning, protein structure, virtual screening, biochemical screening
Significance
There is still a strong requirement of effective therapeutics for overcoming the COVID-19 pandemic. Up to 68% of the total amino acid residues encoded in the SARS-CoV-2 genome have been currently covered by the structural data. The recent activities in drug discovery against COVID-19 are reviewed in this article with emphasizing a protein structure-based approaches.
The novel coronavirus SARS-CoV-2, which emerged in 2019 in China and rapidly spread worldwide, has been causing the COVID-19 pandemic [1]. As of July 2021, there were more than 189 million confirmed cases and 4 million deaths globally. While several vaccines have been developed and the vaccination has been begun worldwide, there remains a great threat to public health and an urgent need for safety and effective therapeutic development [2]. More than one and half years since the pandemic situation, global efforts for drug discovery for combat COVID-19 are continuing. In this review, we overview the recent progress in structural analyses of the SARS-CoV-2 proteins and structure-based drug repurposing against COVID-19.
The functions of proteins encoded in the SARS-CoV-2 genome
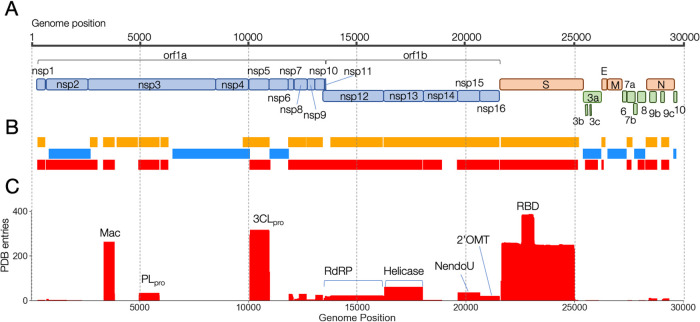
SARS-Cov-2 is an enveloped (+) single-stranded RNA coronavirus, and its genome size is ~29.9 kb in length with a 7-methyl-G 5' cap, a 3' poly-A tail, and more than 14 open reading frames (ORFs) as shown in Figure 1A. The orf1ab encodes a polyprotein of non-structural proteins, which were proteolytically processed by self-encoded two cysteine proteases, namely Papain-like protease (PLpro) in Nsp3 and main protease (Mpro or 3CLpro), into distinct 16 non-structural proteins (Nsp1 to Nsp16) essential for viral replication. The genome encodes four structural proteins, namely, spike glycoprotein (S protein), membrane (M) protein, envelope (E) protein, and nucleocapsid phosphoprotein (N protein). The genome also has at least nine ORFs for the accessory proteins (3a, 3b, 6, 7a, 7b, 8, 9b, 9c, and 10).
Figure 1 .
The genome structure of SARS-CoV-2 and map of 3D assignment. A: The genomic structure and coding proteins in the SARS-CoV-2 RNA genome. The coding proteins are colored by categories as; non-structural polyprotein (pale-blue), structural proteins (orange), and accessory proteins (green). B: Coverage of each coding region of the genome by the 3D structures deposited in the PDB. The color indicates the different types of proteins as; SARS-CoV-2 proteins (red), SARS-CoV or homologous proteins (orange), Model structures predicted by AlphaFold or CASP_commons (blue). C: The coverage depth of each genomic region by the 3D structures.
The non-structural proteins compose the viral replication and transcription complex (RTC) that are responsible for the RNA-processing, modifying, and proofreading functions maintaining the integrity of the large viral genome [3]. In addition to the two proteases, several non-structural proteins, namely, Nsp12, Nsp13, Nsp14, Nsp15, and Nsp16, have enzymatic activities for the viral replication, thus, were considered as drug targets. Nsp12 is the centerpiece of the RTC machinery and acts as RNA-dependent RNA polymerase (RdRp), which is the target protein of the approved drug Remdesivir. Nsp13, which encodes an RNA helicase, utilizes the energy derived from the hydrolysis of nucleoside triphosphates (NTPs) to unwind double-stranded (ds) RNA. Nsp14 has two catalytic domains where the N-terminus is 3'-to-5' exoribonuclease for RNA-proofreading, and the C-terminus is N7-methyltransferase (N7-MTase) for RNA modification processes, respectively. Nsp15 is a uridine-specific endoribonuclease with a C-terminal catalytic domain belonging to the EndoU nuclease family, which is highly conserved in coronaviruses. Nsp16 acts as a 2'-O-methyltransferase catalyzing the final step of the mRNA maturation where it catalyzes methylation at the ribose 2'-O position of the first nucleotide of the RNA with S-adenosyl-L-methionine (SAM) as the methyl group donor. Nsp10 plays a pivotal role for Nsp14 and Nsp16 enzymatic activities upon binding to the proteins to ensure structural stability.
The interaction of S protein, through the receptor-binding domain (RBD), to the cellular entry receptor angiotensin-converting enzyme 2 (ACE2) is the mandatory step of SARS-CoV-2 infection. Therefore, finding molecules that effectively inhibit the interaction between the virus-host proteins, including antibodies, is thought to be most promising in drug development.
Functions of the accessory proteins are largely unknown, but some of them are considered to be involved in evasion from host immune response [3–5].
Recent progress in structural determination and prediction of SARS-CoV-2 proteins
When we began to study the knowledge-based structural modeling for SARS-CoV-2 proteins in January 2020, there was no available 3D structure from the virus. But the structure data of SARS-CoV, which caused the SARS outbreak of 2003, were available in the Protein Data Bank (PDB) [6,7]. Hence, we utilized those structures as the template for homology modeling (Figure 1B). We obtained structural models for 17 proteins out of a total 26 proteins (the total number was based on the initial annotation in the reference genome of SARS-CoV-2 isolated in Wuhan city, China [8]. Note that additional 3 ORFs, namely 3b, 9b and 9c, were also considered to be expressed currently).
Since the structure of SARS-CoV-2 3CLpro, the first 3D structure of the virus, was determined by X-ray crystallography in February 2020, the 3D structure data of SARS-CoV-2 proteins have been rapidly accumulated in the PDB. According to our surveillance of the database in June 2021, more than 1300 entries had been currently released in the database, including redundant entries which cover the same protein regions, and 68% of the amino acid residues of SARS-CoV-2 were covered by at least one 3D structure of its own protein during past one and half years. Figure 1C shows the accumulation of PDB entries that cover each genome region. The largest number of structures was deposited for the RBD domain, followed by 3CLpro and Macrodomain of Nsp3, indicating the global focus on these particular domains for drug development [9,10].
Owing to the breakthrough in cryo-electron microscopy (cryo-EM) techniques, many large protein complex structures have been determined to atomic or semi-atomic resolutions. For the RTC and the trimer of the S protein of SARS-CoV-2, 3D structures were also revealed by cryo-EM method.
Computational approaches for protein structure predictions also contributed to the completion of the SARS-CoV-2 protein structures. In March 2020, the community of protein structure prediction (CASP; Critical Assessment of Techniques for Protein Structure Prediction) had launched the SARS-CoV-2 structure modeling initiative for a challenge to predict the 3D structures of the ten viral proteins and domains, namely, Nsp2, the C-terminal domain of Nsp3, Nsp4, Nsp6, ORF3a, M protein, ORF6, ORF7b, ORF8, and ORF10, for which no apparent template structure was available (https://predictioncenter.org/caspcommons/). In these challenges, about 40 participants, including groups from Japan, deposited their modeled structures of the target proteins to the CASP site by the designated deadline (all the challenges were finished by the end of May 2020). After the challenge, the structures of ORF3a and ORF8 have been experimentally determined. Particularly noteworthy was that the model structure of ORF3a deposited by AlphaFold, which utilized a deep-learning approach developed by DeepMind [11] and demonstrated startling accuracy at CASP14, showed very high similarity to that of the experimentally determined one. The other protein models by AlphaFold also assisted in the structure determinations of ORF8 and Nsp2 proteins by X-ray crystallography and cryo-EM methods, respectively, indicating that the protein structure prediction with machine learning opened the door toward the next generation of structural biology. They also provided the model structures of Nsp4, Nsp6, and M protein, of which 3D structures were not determined yet, at the website (https://deepmind.com/research/open-source/computational-predictions-of-protein-structures-associated-with-COVID-19).
SARS-CoV-2 proteins in complex with drugs in the PDB
When we built knowledge-based models of SARS-CoV-2 proteins with approved drugs, only a small set of structural data of SARS-CoV or other related homologous proteins bound to small compounds or ligands were available [12]. Here we re-examined the PDB for the SARS-CoV-2 proteins and found that some novel structures in complex with drugs/compounds were deposited as summarized in Table 1. It clearly demonstrated the consequence of the increasing interest in the mechanism of action of potential drugs against the SARS-CoV-2 proteins, as described in detail below.
Table 1 .
Drugs whose structures were determined in complex with SARS-CoV-2
| Drug | KEGG | DrugBank | Target | PDB codes | Clinical Status |
|---|---|---|---|---|---|
| Remdesivir | D11472 | DB14761 | Nsp12 (RdRp) | 7c2k, 7bv2, 7b3b, 7b3c, 7l1f | EUA for COVID-19 |
| Favipiravir | D09537 | DB12466 | Nsp12 (RdRp) | 7aap, 7ctt | Phase 3 for COVID-19 |
| Suramina) | D00808 | DB04786 | Nsp12 (RdRp) | 7d4f | Phase 2 for acute kidney injury |
| Bamlanivimab | D11936 | DB15718 | S | 7l3n | EUA for COVID-19 |
| Etesevimab | D11944 | DB15897 | S | 7c01 | EUA for COVID-19 |
| Casirivimab | D11938 | DB15941 | S | 6xdg | EUA for COVID-19 |
| Imdevimab | D11939 | DB15940 | S | 6xdg | EUA for COVID-19 |
| PF-00835231 | None | None | 3CLpro | 6xhm | Phase 1 |
| GC-376a) | None | None | 3CLpro | 6wtj, 7c8u, 7cb7, 7cbt, 7jsu, 7k0f | n.a. |
| Boceprevira) | D08876 | DB08873 | 3CLpro | 6wnp, 6xqu, 6zru, 7brp, 7c6s, 7com, 7k40 | Approved for HCV |
| Telaprevir | D09012 | DB05521 | 3CLpro | 6xqs, 6zrt, 7c7p, 7k6d, 7k6e, 7lb7 | Approved for HCV |
| Baicaleina) | C10023 | DB16101 | 3CLpro | 6m2n | Phase 2 for influenza infection |
| Carmofura) | D01784 | DB09010 | 3CLpro | 7buy | n.a. |
| Shikonina) | C17412 | None | 3CLpro | 7ca8 | n.a. |
| Ebselena) | None | DB12610 | 3CLpro | 7bak, 7bal, 7bfb | Phase 2 for COVID-19 |
| Myricetina) | C10107 | DB02375 | 3CLpro | 7b3e | n.a. |
| Ascorbic acid | D00018 | DB00126 | 3CLpro | 7mpb | Phase 3 for COVID-19 |
| Ebselena) | None | DB12610 | Nsp3 (PLpro) | 7m1y | Phase 2 for COVID-19 |
| GRL0617 | None | DB08656 | Nsp3 (PLpro) | 7jrn, 7cjm, 7cmd, 7jir | n.a. |
| GS-441524 | C22275 | DB15686 | Nsp3 (PLpro) | 7bf6 | Phase 1 for COVID-19 |
| Tipiracil | D10467 | DB09343 | Nsp15 | 6wxc | Approved for colorectal cancer |
| Sinefungin | D05846 | DB01910 | Nsp16 | 6wkq, 6yz1 | n.a. |
a) The inhibitory activity was measured and confirmed by high-throughput in vitro assays. EUA: Emergency used authorization for COVID-19 by FDA.
3CLpro—protease inhibitors
Main protease 3CLpro is one of the most-studied proteins for drug discovery since the past SARS outbreak in 2003. Therefore, many efforts have been made for structural analyses of the protein and its potential inhibitors. After the first 3D determination of 3CLpro of SARS-CoV-2, more than 300 structures were determined by X-ray crystallography. Because of the demand for understanding the inhibitory mechanism, many of the crystal structures were determined in complex with peptide mimetic compounds, natural compounds, or others.
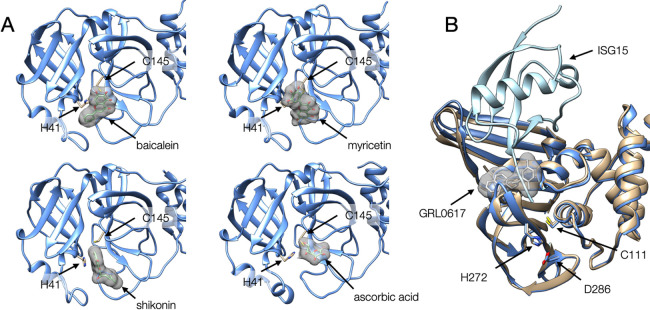
The following inhibitor compounds were co-crystalized with 3CLpro: PF-00835231 (see Table 1 for PDB ID) was originally developed more than 15 years ago against 3CLpro of SARS-CoV [13]. GC-376 was initially developed for a broad-spectrum inhibitor of 3C proteases in Picornaviruses, Noroviruses, and Coronaviruses [14]. Boceprevir and Telaprevir were originally developed as Hepatitis C virus (HCV) protease inhibitors and approved by the Food and Drug Administration (FDA) in U.S.A. These peptide-mimetic drugs, all of which were covalent inhibitors, exhibited high inhibitory activity for the 3CLpro in SARS-CoV-2 in vitro [13,15] and were utilized as lead compounds for further drug development by increasing efficacy and bioavailability [16]. Natural compounds such as baicalein, myricetin, shikonin, and ascorbic acid also bound to 3CLpro in the crystals [17] as shown in Figure 2A. These compounds, except ascorbic acid, showed inhibitor potency by in vitro high-throughput screenings [18–20] but no clinical trial conduction was announced yet for them. Despite no direct evidence for the 3CLpro inhibitory activity, ascorbic acid (vitamin C) is evaluated in phase 3 clinical trial for COVID-19 because it has anti-inflammatory properties, which implies influence of SARS-CoV-2 infection [21].
Figure 2 .
Structures of SARS-CoV-2 proteases. (A) The crystal structures of 3CLpro complexed with compounds, baicalein (upper left, [PDB ID: 6m2n], myricetin (upper right, [PDB ID: 7b3e]), shikonin (lower left, [PDB ID: 7ca8]), ascorbic acid (lower right, [PDB ID: 7mpb]). The catalytic residues (H41 and C145) are represented as stick models. (B) Superposition of the crystal structures of PLpro domains of Nsp3 complexed with its inhibitor GRL0617 (color in beige, [PDB ID: 7jrn]) and complexed with ISG15 (color in blue, [PDB ID: 6xa9]). The catalytic residues are represented by stick models.
PLpro—GRL0617
Another protease, PLpro in Nsp3, was also crucial for viral replication and evasion from host immunity. The three distinct substrates of PLpro, namely the viral polyprotein, degradative Lys48-polyubiquitin, and antiviral ISG15 signals by deubiquitination or deISGylation, make PLpro an excellent candidate for pharmacological intervention. One promising compound was GRL0617, originally discovered as a noncovalent inhibitor specifically for PLpro of SARS-CoV in 2008 [22]. The co-crystal structure of SARS-CoV-2 PLpro with GRL0617 revealed that the compound resided in a pocket in the palm region, apart from the catalytic site. When the structure was compared with another co-crystal structure of PLpro with human ISG15, GRL0617 was implied to make a steric clash to the C-terminus of ISG15, suggesting that the compound would block the C-terminus of ISG15 from binding to the catalytic site of PLpro (Figure 2B). An in vitro experiment demonstrated that GRL0617 inhibited the deubiquitination and deISG15ylation activity of PLpro, which indicated that GRL0617 was a noncovalent inhibitor of PLpro [23].
RdRp—Remdesivir/Favipiravir/Suramin
RNA-dependent RNA polymerase (RdRp/Nsp12) is one of the major drug targets of SARS-CoV-2. Remdesivir (approved drug) and Favipiravir (evaluated in phase 3 clinical trial) target RdRp. Recent advances in the cryo-EM revealed the RdRp-drug interaction modes. Remdesivir is a prodrug converted to the active drug in the triphosphate form (remdesivir-triphosphate; RTP) within cells. The complex structure of RdRp-RNA with RTP revealed that when RTP was incorporated into a new RNA product, the incorporated Remdesivir moiety made a barrier to further RNA translocation by steric hindrance, consequently stalling the RNA polymerase reaction [24,25]. The structural details of the protein-drug interaction would contribute to developing more effective drugs compared to Remdesivir. Some of the new nucleotide-analog drugs, including oral drug Molnupiravir with higher bioavailability, entered the phase 2/3 clinical trial (ClinicalTrials.gov identifier NCT04575597).
Suramin, which has been used to treat African sleeping sickness caused by trypanosomes for about 100 years, is now evaluated in phase 2 clinical trial for acute kidney injury (ClinicalTrials.gov identifier NCT04496596). Recently suramin has been shown to inhibit infection of SARS-CoV-2 in cell culture by preventing cellular entry of the virus [26]. The cryo-EM structure of SARS-CoV-2 RdRp bound to suramin revealed two binding sites. One site directly blocked the binding of the RNA template strand, and the other site made clashes with the RNA primer strand near the RdRp catalytic site [27]. The same group also demonstrated that suramin and a suramin derivative were at least 20-fold more potent than Remdesivir by a biochemical assay.
Nsp15—Tipiracil
Nsp15 is a uridine-specific endoribonuclease with the C-terminal catalytic domain belonging to the EndoU family that are highly conserved in coronaviruses. This enzyme acts in homo-hexamer. It is well known that uridylate-specific nucleolytic activity of Nsp15 on single-stranded and double-stranded (ds) RNA limits the formation of dsRNA intermediates. Thus Nsp15 compromises the ability of specific cytoplasmic viral RNA sensors to activate the IFN-I response of innate immunity to infection [28]. Although a biochemical screening identified NSC95397, which was validated as an inhibitor of the endoribonuclease, this compound did not inhibit SARS-CoV-2 growth in Vero E6 cells [29].
Recently, the structure of SARS-CoV-2 Nsp15 was determined by X-ray crystallography [30]. It formed hexamer and the overall structure resembled SARS-CoV Nsp15 because of the high sequence identity (95.7%) between the two proteins. The structure of SARS-CoV-2 Nsp15 was also determined with uracil analogs, including Tipiracil, which was a thymidine phosphorylase inhibitor approved for the treatment of refractory metastatic colorectal cancer [31]. Biochemical and whole-cell assays demonstrated that Tipiracil inhibited SARS-CoV-2 Nsp15 activity by interacting with the uridine binding pocket in the active site, suggesting that uracil and its derivatives might represent a plausible lead for nucleotide-like drug development [30].
Nsp16—Sinefungin
The viral mRNA capping is essential for efficient viral protein production, protection of viral RNA from host degradation, and evasion from the host’s innate immune responses [32]. Therefore, the enzymes in the RNA capping are now considered one of the potential targets for drug development, while it has been less focused at the earlier stage of the pandemic. Nsp16 is an enzyme that acts in the final step of mRNA maturation as described in the previous section. Nsp16 is activated upon binding to Nsp10, which is also a cofactor of Nsp14, forming a heterodimer.
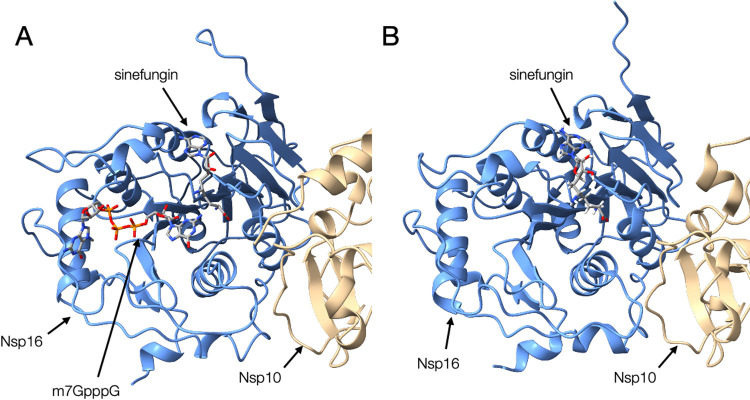
We previously built a homology model of the SARS-CoV-2 Nsp16-Nsp10 heterodimer using that of SARS-CoV as a template [33]. The template structure of SARS-CoV Nsp16 bound Sinefungin, a natural product from Streptomyces griseolus and experimentally used as antibiotics [34]. Utilizing this drug-protein complex structure, we built the model structure of SARS-CoV-2 Nsp16-Nsp10 heterodimer complexed with Sinefungin (Figure 3). Recently, the crystal structure of the Nsp16-Nsp10 heterodimer from SARS-CoV-2 was solved [35,36]. Similar to the case of SARS-CoV, the solved structure bound Sinefungin, and the complex structure is similar to our model [12]. However, because of the close structural similarity of Sinefungin to the natural metabolite SAM, it was thought to be a broad-spectrum inhibitor of methyltransferase, and inappropriate for therapeutics. Therefore attempts to design a compound that inhibits Nsp16 more specifically were reported [37].
Figure 3 .
Comparison of structures of Nsp10-Nsp16 complex. (A) The homology model of SARS-CoV-2 Nsp10 (beige) and Nsp16 (blue) complex bound to Sinefungin and m7GpppA, which were based on the structure of SARS-CoV as the template in our previous study (YP_009725311_v02_1.pdb from BSM-Arc [85] entry BSM-ID BSM00015). (B) The crystal structure of the Nsp10-Nsp16 complex from SARS-CoV-2 bound to Sinefungin [PDB ID: 6yz1].
S protein—neutralizing antibodies
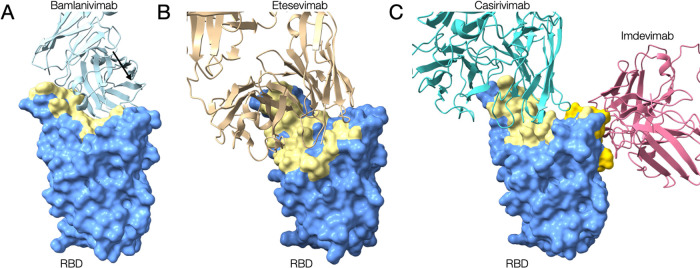
The S protein is essential for the viral entry to host cells. In the viral entry process, the receptor-binding domain (RBD) interacts with the host cell membrane receptor, angiotensin-converting enzyme 2 (ACE2). The mutations in the RBD affect the virus-host interaction and some of the mutants are assigned as variant of concern (VOC), which would have higher infectivity and/or severer clinical consequence. The interest of many researchers on this target is how to interfere with the host-viral interaction, and the most direct solution is developing neutralizing antibodies. Hence, many structural data of S protein complexed with various antibodies were deposited to the PDB. The entries contain neutralizing antibodies approved for emergency use, such as Bamlanivimab [38], Etesevimab, Casirivimab, and Imdevimab (Figure 4). Because these antibodies do not entirely share the binding interfaces, the cocktail antibody drugs are expected to be effective for viral variants, which have accumulated several mutations.
Figure 4 .
Experimentally determined structures of the RBD of S proteins from SARS-CoV-2 complexed with the approved neutralizing antibodies. (A) The cryo-EM structure of RBD complexed with Bamlanivimab [PDB ID: 7l3n]. (B) The crystal structure of RBD complexed with Etesevimab [PDB ID: 7c01]. (C) The cryo-EM structure of RBD complexed with Casirivimab and Imdevimab [PDB ID: 6xdg]. The RBD and neutralizing antibodies are represented as surface model and ribbon model, respectively. The surfaces colored yellow in RBD depict the antibodies binding regions.
Drug repurposing by in silico virtual screening
Once an atomic structure of the target protein becomes available to a high resolution, a computational molecular docking methodology enables us to virtually screen the candidate compounds that potentially inhibit or attenuate the protein functions from libraries of hundreds of thousands of compounds [39]. If an approved drug, the one with proven acceptable safety for humans, was found to be effective for COVID-19, it could make a shortcut to the early-stage of clinical trials. Therefore, many studies have employed virtual screening strategies to find potentially effective compounds from existing approved drugs for repurposing.
Many of the target viral proteins are essential enzymes for the viral replication as represented by proteases (3CLpro and PLpro) [40–47], RNA replicase (RdRp and Nsp13) [43,46,48–52], methyltransferases (Nsp14 and Nsp16) [53–56], and endoribonuclease (Nsp15) [57–59]. In the reported virtual screening studies listed in Table 2, DrugBank [60], ChEMBL [61], PubChem [62], and/or ZINC databases [63], which contained FDA-approved or clinical stage drugs, were used as compound libraries. Some studies also used databases containing natural compounds, medicinal plants, or phytochemicals such as KNApSAcK [64], MPD3 [65], IMPPAT [66], or NANPDB [67].
Table 2 .
In silico virtual screening studies and proposed drugs
| Target | Drug Libraries | Proposed potential drugs | Reference |
|---|---|---|---|
| 3CLpro | Phytochemicals with antiviral activity from 11 plants retrieving from literature and PubChem | Quercetin 3-vicianoside, Absinthin, Delphinidin 3-O-glucoside, Chrysoeriol 8-C-glucoside, Piperolactam A | Joshi et al., 2020 [40] |
| DrugBank | Proflavine, Chloroxine, Demexiptiline, Fluorouracil, Oteracil | Gao et al., 2020 [41] | |
| FDA approved drugs from DrugBank, ZINC, Selleckchem, Enamine subsets, e-Drugs and BindingDB | Metyrapone, Rufinamide, Zonisamide, Lacosamide, Apatinibe | Federico et al., 2021 [42] | |
| FDA approved drugs from ZINC | Tetracycline, Dihydroergotamine, Ergotamine, Dutasteride, Nelfinavir | Gul et al., 2020 [43] | |
| FDA approved drugs from ChEMBL, DrugBank, DrugCentral, Selleckchem | ENMD-981693, Felypressin, Brilacidin, Ritonavir, Saquinavir | Cavasotto et al., 2021 [44] | |
| FDA approved drugs from ChEMBL, DrugBank, ZINC, Selleckchem | Talampicillin, Lurasidone | Elmezayen et al., 2020 [45] | |
| KNApSAcK | Caribine, Cryptopleurine, Justicidin D, Diphyllin, Quercetin | Gani et al., 2021 [46] | |
| FDA approved drugs from DrugBank and ZINC | Ivermectin, Diosmin, Selinexor, Bromocriptine, Elbasvir, Dihydroergotamine | Yuce et al., 2021 [47] | |
| PLpro | FDA approved drugs from ChEMBL, DrugBank, DrugCentral, Selleckchem | Anatibant, Pilaralisib, Tiracizine, Zabofloxacin, Picotamide | Cavasotto et al., 2021 [44] |
| RBD of S protein |
FDA approved drugs from ChEMBL, DrugBank, DrugCentral, Selleckchem | Pralatrexate, Carumonam, Aclerasteride, Granotapide | Cavasotto et al., 2021 [44] |
| KNApSAcK | (–)-beta-sitosterol, 10-Methoxycamptothecin, Inoxanthone, Emetine, Alpha-cedrene | Gani et al., 2021 [46] | |
| Nsp12 (RdRp) |
FDA approved drugs from ZINC database | Eltrombopag, Tipranavir, Ergotamine, Conivaptan | Gul et al., 2020 [43] |
| FDA approved drugs from e-Drugs3D | Quinupristin, Cetrorelix, Dactinomycin, Rifampin, Sirolimus | Pokhrel et al., 2020 [48] | |
| ZINC15 database | Ivermectin, Rifabutin, Rifapentine, Fidaxomicin, 7-methyl-guanosine-5'-triphosphate-5'-guanosine | Parvez et al., 2020 [49] | |
| KNApSAcK | Justicidin D, 10-Methoxycamptothecin, Inoxanthone, 3-O-Caffeoylquinic acid | Gani et al., 2021 [46] | |
| Phytochemicals from Indian medicinal plants | Cordifolide A, Sitoindoside IX | Koulgi et al., 2021 [50] | |
| Nsp13 | Natural compounds from MPD3 database | ZINC257223845 | Ahmad S. et al., 2021 [51] |
| Natural compounds from IMPPAT database | (+)-Epiexcelsin, Euphorbetin, Isorhoeadine, Picrasidine M, Picrasidine N | Vivek-Ananth et al., 2021 [52] | |
| Nsp14 (N7-MTase) |
Compounds from Traditional Chinese Medicine database | TCM20111, TCM31007, TCM3495, TCM5376, TCM57025 | Selvaraj et al., 2021 [53] |
| FDA approved, worked-out-FDA or investigational-only drugs from ZINC database | Hypericin, Olysio, Sovaprevir, Celsentri, Saquinavir | Liu et al., 2021 [54] | |
| Nsp14 (ExoN) |
FDA approved, worked-out-FDA or investigational-only drugs from ZINC database | Hypericin, Bromocriptine, Tanespimycin, Idarubicin, Emend | Liu et al., 2021 [54] |
| Nsp15 | FDA approved drugs from ZINC | Citrate, Dihydroergotamine, Ergotamine, Glisoxepine, Idarubicin | Chandra et al., 2020 [57] |
| FDA approved drugs from DrugBank | Elbasvir, Paritaprevir | Sixto-Lopez Y et al., 2021 [58] | |
| Selleckchem Natural compound library | Oleuropein, Thymopentin | Vijayan R & Gourinath S 2021 [59] | |
| Nsp16 | FDA approved drugs from the PyRx 0.8 virtual screening tool | Digitoxin, Dihydroergotamine, Irinotecan, Sinefungin, Teniposide | Sharma K et al., 2020 [55] |
| Natural compounds from North African Natural Products database | Citrinamide A, 4,5-Di-p-trans-coumaroylquinic acid, Genkwanin-6-C-beta-glucopyranoside, Paraliane diterpene | Mohammad A et al., 2021 [56] |
All the target proteins except for PLpro were examined in two or more independent studies. Intriguingly, the drugs raised by each study for the same target showed limited overlap with each other. It might be due to the differences in employed compound libraries, docking tools, or different atomic coordinates of the target proteins. On the other hand, several compounds were commonly raised in the independent studies; for example, Dihydroergotamine, originally discovered as a serotonin receptor antagonist and was approved for aborting or preventing vascular headaches, was commonly found as a potential 3CLpro inhibitor in two independent studies [43,47]. Other examples were quercetin and its derivative, which were sort of flavonoids and previously reported as the candidates of 3CLpro inhibitor for SARS-CoV [68].
Some compounds were commonly raised as candidates for multiple target proteins of SARS-CoV-2. For instance, Elbasvir, which was originally discovered as an HCV NS5A inhibitor, was selected as the candidate for 3CLpro and Nsp15. Also, Ergotamine, a similar drug as Dihydroergotamine, was selected as a drug for 3CLpro, Nsp15, and RdRp in different studies, suggesting that these drugs have the potency to bind several target proteins.
In general, the drug candidates raised by virtual screening should also be verified experimentally via in vitro biochemical or cell-based assays. To our best knowledge, no drug of COVID-19 found only in the virtual-screening studies has successfully passed to clinical trials to date.
Drug repurposing by biochemical screening assays
Several independent groups performed in vitro biochemical assays for screening compounds from chemical libraries that can inhibit each viral enzyme. To identify drug-repurposing candidates, most of the studies used chemical libraries containing FDA-approved or late-stage clinical trial drugs provided by non-profit organizations such as Drug Repurposing Hub [69] and ReFRAME library [70], or purchased from commercial suppliers. These in vitro assays targeted mainly seven enzymatic activities of six SARS-CoV-2 proteins, namely, 3CLpro (protease) [15,17,19], PLpro (protease) [71], RdRp (RNA replicase) [72], Nsp13 (helicase) [73], Nsp14 (exonuclease) [74], Nsp14 (N7-MTase) [75], and Nsp15 (endonuclease) [29,76] (Table 3).
Table 3 .
In vitro screening studies and proposed repurposing drugs
| Target | Drug Libraries | Proposed candidate | Reference |
|---|---|---|---|
| 3CLpro | ~10,000 compounds of approved or clinical used drugs and natural products from commercial libraries e.g. Target Mol, Selleck, Shanghai Institute for Advanced Immunochemical Studies | Ebselen, Disulfiram, Tideglusib, Carmofur Shikonin | Jin et al., 2020 [17] |
| 18 protease inhibitors including HCV or HIV proteases | Boceprevir, GC-376 | Fu et al., 2020 [15] | |
| Various flavonoids | Baicalin, Herbacetin, Pectolinarin | Jo S. et al., 2020 [20] | |
| Dompe “Safe-In-Man” proprietary collection (containing 607 drug candidates), EU-OPENSCREEN, DrugRepurposingHub) | Thioguanosine, MG-132, Myricetin | Kuzikov et al., 2021 [19] | |
| PL pro | 5,576 compounds (3,727 approved drugs and late-stage clinical drug candidates) | No candidate found | Klemm T et al., 2020 [71] |
| RdRp | ~5000 Commercial libraries (Sigma, Selleck, Enzo, Tocris, Calbiochem, Symansis) | GSK-650394, C-646, BH3I-1, Suramin, Cefsulodin | Bertolin et al., 2021 [72] |
| Nsp13 | ~5000 Commercial libraries (Sigma, Selleck, Enzo, Tocris, Calbiochem, Symansis) | FPA124, Suramin | Zeng et al., 2021 [73] |
| Nsp14 (N7-MTase) |
~5000 Commercial libraries (Sigma, Selleck, Enzo, Tocris, Calbiochem, Symansis) | Trifluperidol, PF-03882845, Inauhzin, Lomeguatrib | Basu et al., 2021 [75] |
| Nsp14 (ExoN) | ~5000 Commercial libraries (Sigma, Selleck, Enzo, Tocris, Calbiochem, Symansis) | Patulin, Aurintricarboxylic acid | Canal et al., 2021 [74] |
| Nsp15 | ~5000 Commercial libraries (Sigma, Selleck, Enzo, Tocris, Calbiochem, Symansis) | NSC95397 | Canal et al., 2021 [29] |
| ReFRAME drug repurposing library | Exebryl-1 | Choi R et al., 2021 [76] |
Although both virtual-screening and in vitro assay approaches utilized similar drug libraries in the cases of 3CLpro, most of the compounds raised by the virtual screening studies were not re-discovered by in vitro high-throughput inhibitory assays. Exceptionally, compounds with flavonoid backbone, e.g., baicalein, quercetin, myricetin, and baicalin, were commonly observed in both lists. This result might be partly due to the limited number of drugs/compounds listed as candidates in the computational studies.
Among the drugs discovered by those in vitro assays, we confirmed that Ebselen and Disulfiram, both exhibited inhibitory activities against 3CLpro, have been entered into phase 2 clinical trials for COVID-19 (ClinicalTrials.gov identifiers NCT04484025 and NCT04485130, respectively).
Drug repurposing by cell-based screening
The cell-based assay is most widely used for the drug screening because of its high-applicability to throughput assays compared to biochemical approaches. Usually, the cytopathogenic effect (CPE), the changes in host cell morphology caused by the target infecting virus, is measured as infection level in this method. The antiviral activity can be evaluated by measuring a change of the degree of CPE with/without a compound. At the earlier stage of the COVID-19 pandemic, many approved drugs, including Chloroquine/Hydroxychloroquine [77], Ivermectin [78], Nelfinavil [79] and Cepharanthine [79,80], were identified as potent anti-COVID-19 drugs by the cell-based assays. Many of these drugs were entered into clinical trials including ongoing ones. The FDA granted an emergency use authorization (EUA) for chloroquine and hydroxychloroquine for the treatment of hospitalized patients with COVID-19 in March 2020. However, the emergency use was revoked in June based on the follow-up assessment of EUA drug and the clinical trial results, in which it was demonstrated that the drugs did not show any effectiveness in reducing either mortality or morbidity [81]. For Ivermectin, many clinical trials are ongoing in several countries including Japan, while WHO has recommended its limited use within clinical trials because of less than sufficient evidence for the drug efficacy against COVID-19 [82]. Nelfinavir, which is a viral protease inhibitor and approved for HIV infection, has been entered into a clinical trial in Japan [83].
With the cell-based assays, compounds can be evaluated under the presence of human and viral genome/proteome, which endorses promising feasibility for the drug candidates. On the other hand, due to the nature of this method, target proteins and inhibition mechanisms of the identified potential drugs in host cells tend to be uncertain. A recent report demonstrated that some antiviral drugs accumulated in an intracellular compartment, such as endosomes or lysosomes, can directly or indirectly inhibit lipid processing. Consequently, the accumulation caused toxic phospholipidosis, which is known to be associated with inhibition of coronavirus replication. It was suggested to be the inhibition mechanism of several candidates found in the cell-based assays. In this case, the potential drugs do not directly interfere with viral proteins, and likely cause cell-toxic side effects [84]. In fact, Hydroxychloroquine or similar drugs, which caused phospholipidosis in in vitro assays, eventually failed clinical trials. Also it should be noted that phospholipidosis induced by drugs could be overlooked in the period of clinical trials because it is typically an in vivo side effect that appears after chronic administration.
Although not all the drugs found in cell-based assays would have the phospholipidosis effect, mechanisms underlying the antiviral activities of the drugs remain largely elusive. In order to verify the underlying mechanisms of drugs, a multidisciplinary approach combining in silico, in vitro, and in vivo assays is promising.
Conclusion
Owing to the global efforts of experimental determination of SARS-CoV-2 protein structures, nearly 70% of the viral protein structurome has been covered within one and half years. Notably, the recent startling success of the machine learning-based structural prediction significantly contributed to the progress of the structurome. Consequently, an enormous number of in silico virtual screenings based on the viral protein structures or in vitro biochemical screening studies have been reported for repurposing approved drugs in a very short period. However, only a few of the detected drug candidates have successfully passed clinical trials and been approved at this point of time, highlighting a large gap lying between in silico/in vitro and in vivo/clinical studies for drug repurposing. Remdesivir is the sole approved COVID-19 therapeutic drug directory-targeted to the SARS-CoV-2 proteins. This fact might represent a potential limitation in repurposing the existing drugs, which were not particularly designed for targeting the SARS-CoV-2 proteins. Accordingly, the global efforts would be shifted to a specific design of new chemical entities against COVID-19. In fact, a couple of 3CLpro inhibitors, which have been newly designed by pharmaceutical companies including one in Japan, are being evaluated in phase 1 clinical trials. In such cases, the accumulation of the viral protein structures in complex with known drugs would contribute as primers to a new drug design not only in developing more effective therapeutic against COVID-19, but also preparing weapons against emerging new viruses in the near future.
Acknowledgement
This work was partly supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis of Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED (JP20am0101111 and JP20am0101069), and Grants-in-Aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology-Japan (JP21H03547 and JP19K12211).
Conflicts of Interest
The authors declare no competing financial interests.
Author Contributions
A.H., C.S., M.S., S.N., M.O., S.K., T.S. wrote the manuscript.
References
- [1].Wang, C., Horby, P. W., Hayden, F. G., Gao, G. F.. A novel coronavirus outbreak of global health concern. Lancet 395, 470–473 (2020). https://doi.org/10.1016/s0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].WHO Solidarity Trial Consortium, Pan, H., Peto, R., Henao-Restrepo, A.-M., Preziosi, M.-P., Sathiyamoorthy, V., et al. Repurposed antiviral drugs for Covid-19—interim WHO solidarity trial results. N. Engl. J. Med. 384, 497–511 (2021). https://doi.org/10.1056/nejmoa2023184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].V’kovski, P., Kratzel, A., Steiner, S., Stalder, H., Thiel, V.. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 19, 155–170 (2021). https://doi.org/10.1038/s41579-020-00468-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gordon, D. E., Jang, G. M., Bouhaddou, M., Xu, J., Obernier, K., White, K. M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468 (2020). https://doi.org/10.1038/s41586-020-2286-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gordon, D. E., Hiatt, J., Bouhaddou, M., Rezelj, V. V., Ulferts, S., Braberg, H., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370, eabe9403 (2020). https://doi.org/10.1126/science.abe9403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Burley, S. K., Berman, H. M., Bhikadiya, C., Bi, C., Chen, L., Di Costanzo, L., et al. Protein Data Bank: The single global archive for 3D macromolecular structure data. Nucleic Acids Res. 47, D520–D528 (2019). https://doi.org/10.1093/nar/gky949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kinjo, A. R., Bekker, G., Suzuki, H., Tsuchiya, Y., Kawabata, T., Ikegawa, Y., et al. Protein Data Bank Japan (PDBj): updated user interfaces, resource description framework, analysis tools for large structures. Nucleic Acids Res. 45, D282–D288 (2017). https://doi.org/10.1093/nar/gkw962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu, F., Zhao, S., Yu, B., Chen, Y. M., Wang, W., Song, Z. G., et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). https://doi.org/10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Schuller, M., Correy, G. J., Gahbauer, S., Fearon, D., Wu, T., Díaz, R. E., et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 7, eabf8711 (2021). https://doi.org/10.1126/sciadv.abf8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Douangamath, A., Fearon, D., Gehrtz, P., Krojer, T., Lukacik, P., Owen, C. D., et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 11, 5047 (2020). https://doi.org/10.1038/s41467-020-18709-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). https://doi.org/10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hijikata, A., Shionyu-Mitsuyama, C., Nakae, S., Shionyu, M., Ota, M., Kanaya, S., et al. Knowledge-based structural models of SARS-CoV-2 proteins and their complexes with potential drugs. FEBS Lett. 594, 1960–1973 (2020). https://doi.org/10.1002/1873-3468.13806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hoffman, R. L., Kania, R. S., Brothers, M. A., Davies, J. F., Ferre, R. A., Gajiwala, K. S., et al. Discovery of ketone-based covalent inhibitors of Coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 63, 12725–12747 (2020). https://doi.org/10.1021/acs.jmedchem.0c01063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim, Y., Lovell, S., Tiew, K.-C., Mandadapu, S. R., Alliston, K. R., Battaile, K. P., et al. Broad-spectrum antivirals against 3C or 3C-like proteases of Picornaviruses, Noroviruses, and Coronaviruses. J. Virol. 86, 11754–11762 (2012). https://doi.org/10.1128/jvi.01348-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fu, L., Ye, F., Feng, Y., Yu, F., Wang, Q., Wu, Y., et al. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 11, 4417 (2020). https://doi.org/10.1038/s41467-020-18233-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Vandyck, K., Deval, J.. Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr. Opin. Virol. 49, 36–40 (2021). https://doi.org/10.1016/j.coviro.2021.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582, 289–293 (2020). https://doi.org/10.1038/s41586-020-2223-y [DOI] [PubMed] [Google Scholar]
- [18].Su, H. X., Yao, S., Zhao, W. F., Li, M. J., Liu, J., Shang, W. J., et al. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 41, 1167–1177 (2020). https://doi.org/10.1038/s41401-020-0483-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jo, S., Kim, S., Kim, D. Y., Kim, M. S., Shin, D. H.. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme Inhib. Med. Chem. 35, 1539–1544 (2020). https://doi.org/10.1080/14756366.2020.1801672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kuzikov, M., Costanzi, E., Reinshagen, J., Esposito, F., Vangeel, L., Wolf, M., et al. Identification of inhibitors of SARS-CoV-2 3CL-pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol. Transl. Sci. 4, 1096–1110 (2021). https://doi.org/10.1021/acsptsci.0c00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Carr, A. C. A new clinical trial to test high-dose vitamin C in patients with COVID-19. Crit. Care 24, 113 (2020). https://doi.org/10.1186/s13054-020-02851-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ratia, K., Pegan, S., Takayama, J., Sleeman, K., Coughlin, M., Baliji, S., et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U.S.A. 105, 16119–16124 (2008). https://doi.org/10.1073/pnas.0805240105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fu, Z., Huang, B., Tang, J., Liu, S., Liu, M., Ye, Y., et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 12, 488 (2021). https://doi.org/10.1038/s41467-020-20718-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yin, W., Mao, C., Luan, X., Shen, D. D., Shen, Q., Su, H., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368, 1499–1504 (2020). https://doi.org/10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kokic, G., Hillen, H. S., Tegunov, D., Dienemann, C., Seitz, F., Schmitzova, J., et al. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 12, 279 (2021). https://doi.org/10.1038/s41467-020-20542-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Salgado-Benvindo, C., Thaler, M., Tas, A., Ogando, N. S., Bredenbeek, P. J., Ninaber, D. K., et al. Suramin inhibits SARS-CoV-2 infection in cell culture by interfering with early steps of the replication cycle. Antimicrob. Agents Chemother. 64, e00900-20 (2020). https://doi.org/10.1128/aac.00900-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yin, W., Luan, X., Li, Z., Zhou, Z., Wang, Q., Gao, M., et al. Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin. Nat. Struct. Mol. Biol. 28, 319–325 (2021). https://doi.org/10.1038/s41594-021-00570-0 [DOI] [PubMed] [Google Scholar]
- [28].Mandilara, G., Koutsi, M. A., Agelopoulos, M., Sourvinos, G., Beloukas, A., Rampias, T.. The Role of Coronavirus RNA-processing enzymes in innate immune evasion. Life 11, 571 (2021). https://doi.org/10.3390/life11060571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Canal, B., Fujisawa, R., McClure, A. W., Deegan, T. D., Wu, M., Ulferts, R., et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp15 endoribonuclease. Biochem. J. 478, 2465–2479 (2021). https://doi.org/10.1042/bcj20210199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kim, Y., Wower, J., Maltseva, N., Chang, C., Jedrzejczak, R., Wilamowski, M., et al. Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2. Commun. Biol. 4, 193 (2021). https://doi.org/10.1038/s42003-021-01735-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kish, T., Uppal, P.. Trifluridine/Tipiracil (Lonsurf) for the Treatment of Metastatic Colorectal Cancer. P T 41, 314–325 (2016). [PMC free article] [PubMed] [Google Scholar]
- [32].Decroly, E., Ferron, F., Lescar, J., Canard, B.. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 10, 51–65 (2011). https://doi.org/10.1038/nrmicro2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Decroly, E., Debarnot, C., Ferron, F., Bouvet, M., Coutard, B., Imbert, I., et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2'-o-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 7, e1002059 (2011). https://doi.org/10.1371/journal.ppat.1002059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kuroda, Y., Yamagata, H., Nemoto, M., Inagaki, K., Tamura, T., Maeda, K.. Antiviral effect of sinefungin on in vitro growth of feline herpesvirus type 1. J. Antibiot. (Tokyo) 72, 981–985 (2019). https://doi.org/10.1038/s41429-019-0234-4 [DOI] [PubMed] [Google Scholar]
- [35].Viswanathan, T., Arya, S., Chan, S. H., Qi, S., Dai, N., Misra, A., et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 11, 3718 (2020). https://doi.org/10.1038/s41467-020-17496-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rosas-Lemus, M., Minasov, G., Shuvalova, L., Inniss, N. L., Kiryukhina, O., Brunzelle, J., et al. High-resolution structures of the SARS-CoV-2 2'-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 13, eabe1202 (2020). https://doi.org/10.1126/scisignal.abe1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bobiļeva, O., Bobrovs, R., Kaņepe, I., Patetko, L., Kalniņš, G., Šišovs, M., et al. Potent SARS-CoV-2 mRNA cap methyltransferase inhibitors by bioisosteric replacement of methionine in SAM cosubstrate. ACS Med. Chem. Lett. 12, 1102–1107 (2021). https://doi.org/10.1021/acsmedchemlett.1c00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jones, B. E., Brown-Augsburger, P. L., Corbett, K. S., Westendorf, K., Davies, J., Cujec, T. P., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 13, eabf1906 (2021). https://doi.org/10.1126/scitranslmed.abf1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ma, D.-L., Chan, D. S.-H., Leung, C.-H.. Drug repositioning by structure-based virtual screening. Chem. Soc. Rev. 42, 2130–2141 (2013). https://doi.org/10.1039/c2cs35357a [DOI] [PubMed] [Google Scholar]
- [40].Joshi, T., Joshi, T., Sharma, P., Mathpal, S., Pundir, H., Bhatt, V., et al. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur. Rev. Med. Pharmacol. Sci. 24, 4529–4536 (2020). https://doi.org/10.26355/eurrev_202004_21036 [DOI] [PubMed] [Google Scholar]
- [41].Gao, K., Nguyen, D. D., Chen, J., Wang, R., Wei, G. W.. Repositioning of 8565 existing drugs for COVID-19. J. Phys. Chem. Lett. 11, 5373–5382 (2020). https://doi.org/10.1021/acs.jpclett.0c01579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Federico, L. B., Silva, G. M., da Silva Hage-Melim, L. I., Gomes, S. Q., Barcelos, M. P., Galindo Francischini, I. A., et al. Identification of known drugs as potential SARS-CoV-2 Mpro inhibitors using ligand- and structure-based virtual screening. Future Med. Chem. 13, 1353–1366 (2021). https://doi.org/10.4155/fmc-2021-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gul, S., Ozcan, O., Asar, S., Okyar, A., Barıs, I., Kavakli, I. H.. In silico identification of widely used and well-tolerated drugs as potential SARS-CoV-2 3C-like protease and viral RNA-dependent RNA polymerase inhibitors for direct use in clinical trials. J. Biomol. Struct. Dyn. 39, 6772–6791 (2021). https://doi.org/10.1080/07391102.2020.1802346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cavasotto, C. N., Di Filippo, J. I.. In silico Drug Repurposing for COVID-19: Targeting SARS-CoV-2 proteins through docking and consensus ranking. Mol. Inform. 40, e2000115 (2021). https://doi.org/10.1002/minf.202000115 [DOI] [PubMed] [Google Scholar]
- [45].Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., Yelekçi, K.. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 39, 2980–2992 (2021). https://doi.org/10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gani, M. A., Nurhan, A. D., Maulana, S., Siswodihardjo, S., Shinta, D. W., Khotib, J.. Structure-based virtual screening of bioactive compounds from Indonesian medical plants against severe acute respiratory syndrome coronavirus-2. J. Adv. Pharm. Technol. Res. 12, 120–126 (2021). https://doi.org/10.4103/japtr.japtr_88_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yuce, M., Cicek, E., Inan, T., Dag, A. B., Kurkcuoglu, O., Sungur, F. A. Repurposing of FDA-approved drugs against active site and potential allosteric drug-binding sites of COVID-19 main protease. Proteins prot.26164 (2021). https://doi.org/10.1002/prot.26164 [DOI] [PMC free article] [PubMed]
- [48].Pokhrel, R., Chapagain, P., Siltberg-Liberles, J.. Potential RNA-dependent RNA polymerase inhibitors as prospective therapeutics against SARS-CoV-2. J. Med. Microbiol. 69, 864–873 (2020). https://doi.org/10.1099/jmm.0.001203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Parvez, M. S. A., Karim, M. A., Hasan, M., Jaman, J., Karim, Z., Tahsin, T., et al. Prediction of potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2 using comprehensive drug repurposing and molecular docking approach. Int. J. Biol. Macromol. 163, 1787–1797 (2020). https://doi.org/10.1016/j.ijbiomac.2020.09.098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Koulgi, S., Jani, V., Mallikarjunachari Uppuladinne, V. N., Sonavane, U., Joshi, R.. Natural plant products as potential inhibitors of RNA dependent RNA polymerase of Severe Acute Respiratory Syndrome Coronavirus-2. PLoS One 16, e0251801 (2021). https://doi.org/10.1371/journal.pone.0251801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ahmad, S., Waheed, Y., Ismail, S., Bhatti, S., Abbasi, S. W., Muhammad, K.. Structure-based virtual screening identifies multiple stable binding sites at the RecA domains of SARS-CoV-2 helicase enzyme. Molecules 26, 1446 (2021). https://doi.org/10.3390/molecules26051446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Vivek-Ananth, R. P., Krishnaswamy, S., Samal, A. Potential phytochemical inhibitors of SARS-CoV-2 helicase Nsp13: a molecular docking and dynamic simulation study. Mol. Divers. (2021). https://doi.org/10.1007/s11030-021-10251-1 [DOI] [PMC free article] [PubMed]
- [53].Selvaraj, C., Dinesh, D. C., Panwar, U., Abhirami, R., Boura, E., Singh, S. K.. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J. Biomol. Struct. Dyn. 39, 4582–4593 (2021). https://doi.org/10.1080/07391102.2020.1778535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu, C., Zhu, X., Lu, Y., Zhang, X., Jia, X., Yang, T.. Potential treatment with Chinese and Western medicine targeting NSP14 of SARS-CoV-2. J. Pharm. Anal. 11, 272–277 (2021). https://doi.org/10.1016/j.jpha.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sharma, K., Morla, S., Goyal, A., Kumar, S.. Computational guided drug repurposing for targeting 2'-O-ribose methyltransferase of SARS-CoV-2. Life Sci. 259, 118169 (2020). https://doi.org/10.1016/j.lfs.2020.118169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Al-Mulla, F., Abubaker, J.. Molecular simulation-based investigation of highly potent natural products to abrogate formation of the nsp10–nsp16 complex of sars-cov-2. Biomolecules 11, 573 (2021). https://doi.org/10.3390/biom11040573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chandra, A., Gurjar, V., Qamar, I., Singh, N.. Identification of potential inhibitors of SARS-COV-2 endoribonuclease (EndoU) from FDA approved drugs: a drug repurposing approach to find therapeutics for COVID-19. J. Biomol. Struct. Dyn. 39, 4201–4211 (2021). https://doi.org/10.1080/07391102.2020.1775127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sixto-López, Y., Martínez-Archundia, M.. Drug repositioning to target NSP15 protein on SARS-CoV-2 as possible COVID-19 treatment. J. Comput. Chem. 42, 897–907 (2021). https://doi.org/10.1002/jcc.26512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Vijayan, R., Gourinath, S.. Structure-based inhibitor screening of natural products against NSP15 of SARS-CoV-2 revealed thymopentin and oleuropein as potent inhibitors. J. Proteins Proteom. 12, 71–80 (2021). https://doi.org/10.1007/s42485-021-00059-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wishart, D. S., Feunang, Y. D., Guo, A. C., Lo, E. J., Marcu, A., Grant, J. R., et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018). https://doi.org/10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gaulton, A., Hersey, A., Nowotka, M. L., Patricia Bento, A., Chambers, J., Mendez, D., et al. The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954 (2017). https://doi.org/10.1093/nar/gkw1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 49, D1388–D1395 (2021). https://doi.org/10.1093/nar/gkaa971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sterling, T., Irwin, J. J.. ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015). https://doi.org/10.1021/acs.jcim.5b00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Afendi, F. M., Okada, T., Yamazaki, M., Hirai-Morita, A., Nakamura, Y., Nakamura, K., et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53, e1 (2012). https://doi.org/10.1093/pcp/pcr165 [DOI] [PubMed] [Google Scholar]
- [65].Mumtaz, A., Ashfaq, U. A., Ul Qamar, M. T., Anwar, F., Gulzar, F., Ali, M. A., et al. MPD3: a useful medicinal plants database for drug designing. Nat. Prod. Res. 31, 1228–1236 (2017). https://doi.org/10.1080/14786419.2016.1233409 [DOI] [PubMed] [Google Scholar]
- [66].Mohanraj, K., Karthikeyan, B. S., Vivek-Ananth, R. P., Chand, R. P. B., Aparna, S. R., Mangalapandi, P., et al. IMPPAT: A curated database of Indian Medicinal Plants, Phytochemistry and Therapeutics. Sci. Rep. 8, 4329 (2018). https://doi.org/10.1038/s41598-018-22631-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ntie-Kang, F., Telukunta, K. K., Döring, K., Simoben, C. V., Moumbock, A. F. A., Malange, Y. I., et al. NANPDB: A resource for natural products from northern African sources. J. Nat. Prod. 80, 2067–2076 (2017). https://doi.org/10.1021/acs.jnatprod.7b00283 [DOI] [PubMed] [Google Scholar]
- [68].Mani, J. S., Johnson, J. B., Steel, J. C., Broszczak, D. A., Neilsen, P. M., Walsh, K. B., et al. Natural product-derived phytochemicals as potential agents against coronaviruses: A review. Virus Res. 284, 197989 (2020). https://doi.org/10.1016/j.virusres.2020.197989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Corsello, S. M., Bittker, J. A., Liu, Z., Gould, J., McCarren, P., Hirschman, J. E., et al. The Drug Repurposing Hub: A next-generation drug library and information resource. Nat. Med. 23, 405–408 (2017). https://doi.org/10.1038/nm.4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Janes, J., Young, M. E., Chen, E., Rogers, N. H., Burgstaller-Muehlbacher, S., Hughes, L. D., et al. The ReFRAME library as a comprehensive drug repurposing library and its application to the treatment of cryptosporidiosis. Proc. Natl. Acad. Sci. U.S.A. 115, 10750–10755 (2018). https://doi.org/10.1073/pnas.1810137115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Klemm, T., Ebert, G., Calleja, D. J., Allison, C. C., Richardson, L. W., Bernardini, J. P., et al. Mechanism and inhibition of the papain‐like protease, PLpro, of SARS‐CoV‐2. EMBO J. 39, e106275 (2020). https://doi.org/10.15252/embj.2020106275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bertolin, A. P., Weissmann, F., Zeng, J., Posse, V., Milligan, J. C., Canal, B., et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp12/7/8 RNA-dependent RNA polymerase. Biochem. J. 478, 2425–2443 (2021). https://doi.org/10.1042/bcj20210200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zeng, J., Weissmann, F., Bertolin, A. P., Posse, V., Canal, B., Ulferts, R., et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp13 helicase. Biochem. J. 478, 2405–2423 (2021). https://doi.org/10.1042/bcj20210201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Canal, B., McClure, A. W., Curran, J. F., Wu, M., Ulferts, R., Weissmann, F., et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp14/nsp10 exoribonuclease. Biochem. J. 478, 2445–2464 (2021). https://doi.org/10.1042/bcj20210198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Basu, S., Mak, T., Ulferts, R., Wu, M., Deegan, T., Fujisawa, R., et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of Nsp14 RNA cap methyltransferase. Biochem. J. 478, 2481–2497 (2021). https://doi.org/10.1042/bcj20210219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Choi, R., Zhou, M., Shek, R., Wilson, J. W., Tillery, L., Craig, J. K., et al. High-throughput screening of the ReFRAME, pandemic box, and COVID box drug repurposing libraries against SARS-CoV-2 nsp15 endoribonuclease to identify small-molecule inhibitors of viral activity. PLoS One 16, e0250019 (2021). https://doi.org/10.1371/journal.pone.0250019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Venkadapathi, J., Govindarajan, V. K., Sekaran, S., Venkatapathy, S.. A minireview of the promising drugs and vaccines in pipeline for the treatment of COVID-19 and current update on clinical trials. Front. Mol. Biosci. 8, 637378 (2021). https://doi.org/10.3389/fmolb.2021.637378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., Wagstaff, K. M.. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 178, 104787 (2020). https://doi.org/10.1016/j.antiviral.2020.104787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ohashi, H., Watashi, K., Saso, W., Shionoya, K., Iwanami, S., Hirokawa, T., et al. Potential anti-COVID-19 agents, cepharanthine and nelfinavir, and their usage for combination treatment. iScience 24, 102367 (2021). https://doi.org/10.1016/j.isci.2021.102367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jeon, S., Ko, M., Lee, J., Choi, I., Byun, S. Y., Park, S., et al. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 64, e00819-20 (2020). https://doi.org/10.1128/aac.00819-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Self, W. H., Semler, M. W., Leither, L. M., Casey, J. D., Angus, D. C., Brower, R. G., et al. Effect of hydroxychloroquine on clinical status at 14 days in hospitalized patients with COVID-19: a randomized clinical trial. JAMA 324, 2165–2176 (2020). https://doi.org/10.1001/jama.2020.22240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Therapeutic and COVID-19: living guideline. https://app.magicapp.org/#/guideline/nBkO1E/section/LAQX7L
- [83].Hosogaya, N., Miyazaki, T., Fukushige, Y., Takemori, S., Morimoto, S., Yamamoto, H., et al. Efficacy and safety of nelfinavir in asymptomatic and mild COVID-19 patients: a structured summary of a study protocol for a multicenter, randomized controlled trial. Trials 22, 309 (2021). https://doi.org/10.1186/s13063-021-05282-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Tummino, T. A., Rezelj, V. V., Fischer, B., Fischer, A., O’Meara, M. J., Monel, B., et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science 373, 541–547 (2021). https://doi.org/10.1126/science.abi4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bekker, G.-J., Kawabata, T., Kurisu, G.. The Biological Structure Model Archive (BSM-Arc): an archive for in silico models and simulations. Biophys. Rev. 12, 371–375 (2020). https://doi.org/10.1007/s12551-020-00632-5 [DOI] [PMC free article] [PubMed] [Google Scholar]