To the Editor
B-acute lymphoblastic leukemia (B-ALL) comprises a wide variety of subtypes with diverse clinical and biological features and outcomes. Risk-stratified and targeted therapy according to genetic subtype has improved B-ALL outcomes [1]. Next generation sequencing (NGS) has identified several novel subtypes, including one with ZNF384 rearrangements involving ZNF384 located at position 12p13.31 [2–7]. Interestingly, patients with this subtype appear to express various leukemic phenotypes, including B-ALL (with or without aberrant expression of myeloid markers) and B/myeloid mixed phenotype acute leukemia (MPAL). In this regard, ~5% of childhood B-ALL, ~10% of adult B-ALL, and 48% of B/Myeloid MPAL cases have been observed to harbor ZNF384 rearrangements [2–4, 8]. RNA sequencing and conventional methods have identified more than 10 fusion partners of ZNF384 rearrangements [2–7], but the clinical significance of each fusion partner remains unclear due to the small number of reported cases. Here we describe the clinical characteristics and outcomes of the largest series of B-ALL cases with ZNF384 rearrangements reported to date.
We studied a total of 218 cases with ZNF384 rearrangements identified by 16 international consortia (Supplementary Table 1) belonging to the Ponte di Legno Childhood ALL Working group. Patients were diagnosed with B-ALL between 1992 and 2018 using standard morphological and immunophenotypic criteria. The focus of this study was the clinical relevance of ZNF384 fusions in B-ALL. We excluded MPAL cases because they are frequently treated off-study and can receive hybrid therapies [9]. ZNF384 rearrangements were detected by fluorescence in situ hybridization (FISH), reverse transcription polymerase chain reaction, and/or NGS as per individual study group criteria. Three groups identified 18 cases with ZNF384 rearrangements among patients with relapsed or refractory ALL, and these cases were excluded from the outcome analysis. We collated data on the clinical and biological characteristics of patients with ZNF384-rearrangements from each study group and analyzed their association with clinical outcomes.
Overall survival (OS) was calculated from the time from diagnosis to death, while event-free survival (EFS) was defined as the time from diagnosis to induction failure, relapse, a second tumor, or death; time was censored at the date of last patient contact if no event occurred. Relapse rate was defined as the time from diagnosis to relapse censoring at other events. The Kaplan–Meier method was used to estimate survival rates, and evaluation of the equality of the survivorship functions in different subgroups was performed using the two-sided log-rank test. Univariate Cox regression models were used to determine hazard ratios (HR). Other comparisons were performed using the test or Fisher’s exact test as appropriate. All probability (P) values were two-sided, and P values < 0.05 were considered statistically significant. All analyses were performed using Intercooled Stata (Statacorp 2015 Stata Statistical Software Release 14; StataCorp, College Station, TX).
Among the 218 patients with B-ALL and ZNF384 rearrangements, information on the partner gene was available for 193 cases with frequencies of 43% for EP300, located at position 22q13.2 (n = 83, 31% for TCF3 at 19p13.3 (n = 60), 9% for TAF15 at 17q12 (n = 17), 8% for CREBBP at 16p13.3 (n = 15), and 9% for others (n = 18) (Supplementary Table 2). The 18 other partner genes included six cases of EWSR1 and one case of each of ARID1B, BMP2K, CLLORF74, CCAR1, CLTC, DUX4, NIPBL, SEC24B, SMARCA2, USP25. In 2 cases the partner gene could not be determined but involvement of EP300, TCF3, TAF15, and CREBBP was excluded. Data on demographics are shown in Table 1. The female to male ratio was 1:1. The age distribution differed according to partner gene: Patients with EP300-ZNF384 were older (median age, 11 years), while patients with TCF3-ZNF384 were younger (median age, 5 years) (P < 0.001). There were no statistical differences in the distribution of data on National Cancer Institute risk group, ethnicity, leukocyte count, or CNS status according to the partner gene involved (Table 1). Immunophenotypically, the majority of the cases expressed myeloid-associated antigens CD13 (38–100%) and CD33 (78–100%), and a relatively large number of cases were negative for CD10 (14–51%) (Supplementary Table 3), as reported previously [3, 4]. Complete hematological remission was achieved in 99% of the cases. In total, 31% of patients were treated as “high risk” according to protocol, and 23% of the patients received a stem cell transplant in the first remission. Minimal residual disease (MRD) data were available for 77 patients; for 18 (23%) of these, MRD was positive (median, 2.54%; range, 0.14–25.6%) at the end of induction.
Table 1.
Demographic features of patients with B-ALL and ZNF384 rearrangements stratified by partner genes.
| Total n (%) | EP300 n (%) | TCF3 n (%) | TAF15 n (%) | CREBBP n (%) | Othera n (%) | Missingb n (%) | p value | |
|---|---|---|---|---|---|---|---|---|
| Total, n (%) | 218 (100) | 83 | 60 | 17 | 15 | 18 | 25 | |
| Sex, n(%) | ||||||||
| Male | 104 (50) | 36 (47) | 28 (47) | 11 (65) | 9 (64) | 11 (65) | 9 (36) | 0.381 |
| Female | 104 (50) | 40 (53) | 31 (53) | 6 (35) | 5 (36) | 6 (35) | 16 (64) | |
| Unknown/Missing | 10 | 7 | 1 | 0 | 1 | 1 | 0 | |
| Age (years) | ||||||||
| Median | 9.00 | 11.00 | 5.00 | 8.00 | 6.00 | 7.00 | 12.00 | |
| 1–9 | 115 (55) | 32 (42) | 44 (75) | 11 (65) | 9 (64) | 10 (59) | 9 (36) | 0.001 |
| 10–14 | 65 (31) | 28 (37) | 14 (24) | 5 (29) | 2 (14) | 4 (24) | 12 (48) | |
| 15–18 | 24 (12) | 15 (20) | 0 (0) | 1 (6) | 3 (21) | 1 (6) | 4 (16) | |
| 19–25 | 4 (2) | 1 (1) | 1 (2) | 0 (0) | 0 (0) | 2 (12) | 0 (0) | |
| Unknown/Missing | 10 | 7 | 1 | 0 | 1 | 1 | 0 | |
| Year of diagnosis | ||||||||
| 1992–2007 | 90 (43) | 26 (34) | 26 (44) | 8 (47) | 5 (36) | 2 (13) | 23 (92) | 0.171 |
| 2008–2018 | 117 (57) | 50 (66) | 33 (56) | 9 (53) | 9 (64) | 14 (88) | 2 (8) | |
| Unknown/Missing | 11 | 7 | 1 | 0 | 1 | 2 | 0 | |
| Race | ||||||||
| Asian | 62 (48) | 20 (44) | 28 (72) | 5 (45) | 4 (40) | 2 (22) | 3 (20) | 0.083 |
| White | 59 (46) | 23 (51) | 11 (28) | 6 (55) | 5 (50) | 6 (67) | 8 (53) | |
| Other | 8 (6) | 2 (4) | 0 (0) | 0 (0) | 1 (10) | 1 (11) | 4 (27) | |
| Unknown/Missing | 89 | 38 | 21 | 6 | 5 | 9 | 10 | |
| WBC count (106/L) | ||||||||
| <50,000 | 151 (74) | 59 (79) | 37 (64) | 13 (81) | 8 (57) | 15 (88) | 19 (76) | 0.088 |
| >50,000 | 54 (26) | 16 (21) | 21 (36) | 3 (19) | 6 (43) | 2 (12) | 6 (24) | |
| Unknown/Missing | 13 | 8 | 2 | 1 | 1 | 1 | 0 | |
| NCI risk group | ||||||||
| Standard Risk | 73 (35) | 24 (32) | 24 (41) | 8 (50) | 5 (36) | 8 (47) | 4 (16) | 0.524 |
| High Risk | 133 (65) | 52 (68) | 34 (59) | 8 (50) | 9 (64) | 9 (53) | 21 (84) | |
| Missing | 12 | 7 | 2 | 1 | 1 | 1 | 0 | |
| CNS3 disease at diagnosis | ||||||||
| Yes | 5 (3) | 1 (2) | 1 (2) | 0 (0) | 1 (8) | 1 (7) | 1 (4) | 0.567 |
| No | 165 (97) | 56 (98) | 45 (98) | 15 (100) | 11 (92) | 14 (93) | 24 (96) | |
| Unknown/Missing | 48 | 26 | 14 | 2 | 3 | 3 | 0 | |
| Traumatic lumbar puncture | ||||||||
| Yes, Blasts | 4 (4) | 0 (0) | 2 (6) | 0 (0) | 1 (14) | 0 (0) | 1 (20) | 0.386 |
| Yes, No Blasts | 6 (5) | 4 (9) | 2 (6) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| No | 99 (91) | 42 (91) | 29 (88) | 9 (100) | 6 (86) | 9 (100) | 4 (80) | |
| Unknown/Missing | 109 | 37 | 27 | 8 | 8 | 9 | 20 | |
| Immunophenotype | ||||||||
| B-Lineage | 205 (100) | 75 (99) | 57 (97) | 17 (100) | 14 (93) | 17 (100) | 25 (100) | N/A |
| T-Lineage | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Unknown/Missing | 13 | 8 | 3 | 0 | 1 | 1 | 0 | |
| BM Blasts at diagnosis | ||||||||
| <20 | 20 (34) | 6 (33) | 3 (21) | 2 (100) | 2 (67) | 1 (50) | 6 (32) | 0.161 |
| 20–39 | 13 (22) | 4 (22) | 6 (43) | 0 (0) | 0 (0) | 0 (0) | 3 (16) | |
| 40–59 | 11 (19) | 6 (33) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 5 (26) | |
| 60–80 | 7 (12) | 1 (6) | 3 (21) | 0 (0) | 0 (0) | 1 (50) | 2 (11) | |
| >80 | 7 (12) | 1 (6) | 2 (14) | 0 (0) | 1 (33) | 0 (0) | 3 (16) | |
| Unknown/Missing | 160 | 65 | 46 | 15 | 12 | 16 | 6 | |
| Minimal residual disease (Neg < 0.01%) | ||||||||
| Positive | 18 (23) | 5 (16) | 6 (32) | 0 (0) | 1 (17) | 4 (33) | 2 (33) | 0.491 |
| Negative | 59 (77) | 26 (84) | 13 (68) | 3 (100) | 5 (83) | 8 (67) | 4 (67) | |
| Unknown/Missing | 141 | 52 | 41 | 14 | 9 | 6 | 19 | |
| Complete remission achieved | ||||||||
| Yes | 199 (99) | 74 (100) | 56 (98) | 13 (100) | 14 (100) | 17 (100) | 25 (100) | 0.721 |
| No | 1 (1) | 0 (0) | 1 (2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Unknown/Missing | 18 | 9 | 3 | 4 | 1 | 1 | 0 | |
| Stem cell transplant received | ||||||||
| Yes | 42 (23) | 20 (26) | 13 (25) | 2 (13) | 4 (29) | 3 (20) | 0 (0) | 0.85 |
| No | 138 (77) | 58 (74) | 39 (75) | 13 (87) | 10 (71) | 12 (80) | 6 (100) | |
| Unknown/Missing | 38 | 5 | 8 | 2 | 1 | 3 | 19 | |
| Treatment risk groups | ||||||||
| Non-high risk | 144 (69) | 58 (73) | 31 (54) | 13 (76) | 9 (64) | 11 (65) | 22 (96) | 0.215 |
| High risk | 64 (31) | 22 (28) | 26 (46) | 4 (24) | 5 (36) | 6 (35) | 1 (4) | |
| Unknown/Missing | 10 | 3 | 3 | 0 | 1 | 1 | 2 | |
aThe other group includes 6 cases of EWSR1 and 1 case each of ARID1B, BMP2K, CLLORF74, CCAR1, CLTC, DUX4, NIPBL, SEC24B, SMARCA2, USP25; plus two cases where testing showed that the partner gene was not one of the four common genes.
bMissing group includes cases where information about the partner gene was not provided or where the involvement of ZNF384 was confirmed by FISH only.
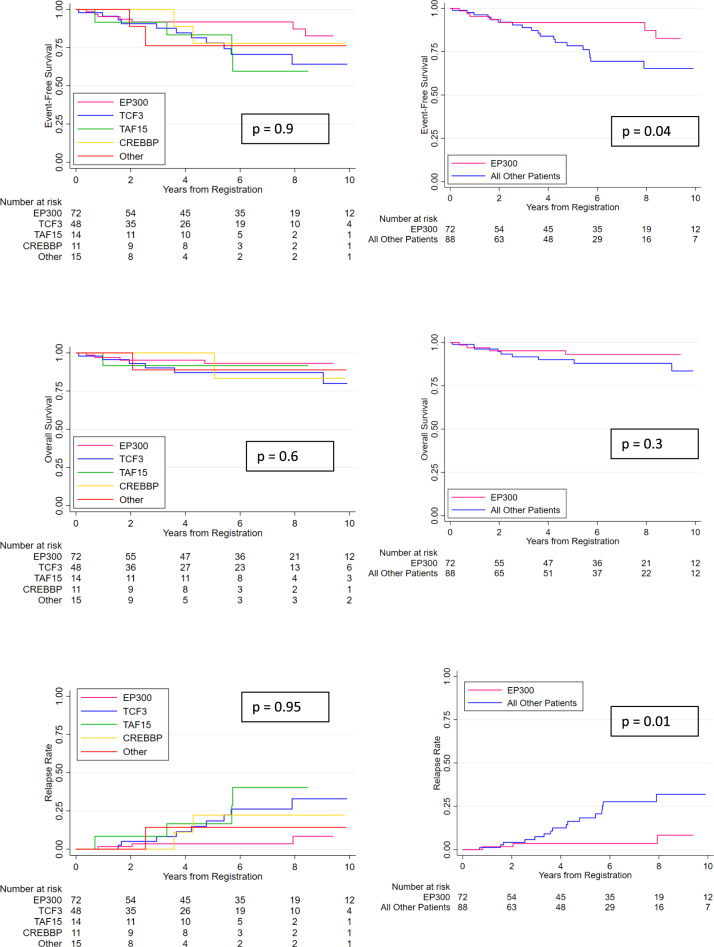
After a median follow-up of 5.8 years, the 5-year EFS rate was 85% (95% confidence interval [CI], 78–90%), and the 5-year OS rate was 91% (95% CI, 85–95%) for all patients. There was no difference in survival rate by treatment period, by country, or by region of origin (data not shown). Data on the outcome of patients with ZNF384-rearranged ALL according to partner gene are summarized in Fig. 1. Univariate and multivariate analyses revealed outcome heterogeneity by partner gene (Supplementary Table 4). Patients with an EP300-ZNF384 fusion had a significantly lower cumulative relapse rate at 5 years compared with the remaining patients, 4 % (95% CI, 1–13%) vs. 18% (11–31 %) (HR, 0.20, [95%CI, 0.06 –0.67], P = 0.01). The corresponding EFS was 92 % (95% CI, 81–97 %) vs. 78 % (95% CI, 66–87%) (P = 0.037), and OS was 93 % (95% CI, 82–97 %) vs. 90% (95% CI, 80–95%) (P = 0.289), suggesting that relapses of other ZNF384 rearrangements were salvageable. Multivariate analysis, adjusting for sex, age, WBC, and treatment period did not alter the results (supplementary Table 4).
Fig. 1. Outcomes of patients with ZNF384 rearrangement-related ALL.
Outcomes of patients with ZNF384 rearrangement-related ALL according to partner gene and EP300-ZNF384 ALL compared with all other patients. Of note, outcome data were missing for 15 patients, and further 18 and 25 patients were excluded for selection bias and missing partner gene information, respectively.
Additional genetic abnormalities were detected by multiplex ligation-dependent probe amplification (n = 91), single nucleotide polymorphism array analysis (n = 63), RNA sequencing (n = 117), whole-exome sequencing (n = 18), and whole-genome sequencing (n = 12) performed by each international consortia (Supplementary Table 5). Commonly deleted genes included those frequently recurrent in ALL [10], such as ETV6 (n = 35, 24 %), CDKN2A/2B (n = 23, 16%), and IKZF1 (n = 20, 14%); and those frequently mutated within the RAS pathway, such as FLT3 (n = 10, 14%). The distribution of deletions did not differ significantly between fusion partners (Supplementary Table 6, Supplementary Fig. 1). There were no significant associations between genomic deletions and prognosis for any of the fusion partners investigated (Supplementary Table 7). Deletions of CDKN2A/CDKN2B, which were found in 23 (16%) of the cases, tended to be associated with a higher mortality risk (HR, 3.86 [95%CI, 0.96–15.48], P = 0.057).
A recent study of monozygotic twins revealed that TCF3-ZNF384 can occur in utero, suggesting that the TCF3-ZNF384 fusion gene is crucial to initiate ALL [11]. The chromosomal translocation from which the chimeric fusion gene arises may have formed a pre-leukemic clone, which acquired additional mutations to fully transform into overt leukemia several years after birth, as has been demonstrated for ETV6–RUNX1 ALL [12]. In contrast, ALL with EP300-ZNF384 was present in patients older than those with other ZNF384-related rearrangements. The biological background for each fusion partner warrants further investigation.
In this study, 28 % of children with EP300-ZNF384 were allocated to the high-risk group because of older age, while 46% of patients with TCF3-ZNF384 were allocated to the high-risk group because of high leukocyte counts and poor initial responses. The patients with EP300-ZNF384 ALL had a lower cumulative relapse rate than the remaining patients particularly those patients with TCF3-ZNF384. However, each group of patients might benefit from MRD-directed treatment in order to avoid over- and under-treatment as well as targeted therapy. In this regard, immunotherapy, such as anti-CD19 chimeric antigen receptor T-cell therapy (CAR-T therapy), should be used carefully, if at all, in patients with ZNF384 rearrangements because of the risk of lineage switch to myeloid leukemia as a cause of relapse [13]. In fact, lineage switch from ALL to acute myeloid leukemia after CAR-T therapy has been observed in one patient with TCF3-ZNF384 [14]. In addition, relapse tends to occur late, so that the EFS curve does not plateau at 5 years (Fig. 1). Indeed, we observed two patients with TCF3-ZNF384-positive ALL relapsing more than 10 years after diagnosis [15]. Taken together, additional studies are needed to develop an optimal treatment strategy for those with poor initial responses, although allogeneic stem cell transplantation is unlikely to be indicated.
ZNF384 fusions are an enigmatic group of gene fusions which span the ALL-MPAL disease spectrum and are not readily detected and characterized by traditional genetic testing [8]. It is definitey required to screen by FISH, reverse transcription polymerase chain reaction or RNA sequencing. The major strengthen of this study is that it has collated a large well-annoatated cohort of ZNF384-fusion patients which while not uninformly treated were all diagnosed with ALL and treated as such. As with all retrospective consortia-based studies the limitations are heterogeneity in terms of recrtuiment period and treatment decisions and pathways. Nonetheless, we provide good evidence that among B-ALL patients with a ZNF384 fusion the partner gene is associated with demographic features and influences outcome, specifically with EP300-ZNF384 being associated with a lower risk of relapse. We opted to exlude MPAL cases because, historically, they have not been treated uniformly. A recent international cooperative study has defined a consensus treatment staregy for MPAL patients [9]. This initiative coupled with improved diagnostic genomic testing will enable future prospective studies to clarify the clinical relevance of the fusions in both ALL and MPAL.
Supplementary information
Acknowledgements
Research support. Grants for Clinical Cancer Research from the Ministry of Health, Labour and Welfare, Japan, Japan Agency for Medical Research and Development (AMED), Japan, Blood Cancer, the Grant of the National Center for Child Health and Development, UK, National Cancer Institute (CA021765), US, the American Lebanese Syrian Associated Charities (ALSAC), US, Czech Health Research Council (NU20-07-00322), Czech, the ERA-NET TRANSCAN EJC granted by Fondation ARC, France, the Center for Biological Resources (CRB-cancer; BB-0033-00076) of the Robert Debré Hospital, France, and the Oncode Institute and KiKa Foundation, Netherlands. Members of Study Groups. AIEOP: Valentino Conter, Andrea Biondi; Polish ALIC: Wojciech Mlynarski; BFM-A: Georg Mann, Andishe Attarbaschi, Karin Nebral, Sabine Strehl and Dagmar Schinnerl; BFM-G/CH: Martin Schrappe; COG: Stephen Hunger; DCOG: Rob Pieters; JACLS: Junichi Hara, Keizo Horibe; NOPHO: Kjeld Schmiegelow, Mats Heyman; TCCSG: Akira Ohara, Katsuyoshi Koh; TPOG: His-Che Liu; CLIP: Jan Stary; UKALL: Ajay Vora, Claire Schwab; SFCE: André Baruchel, Yves Bertrand, Marion Strullu, Catherine Paillard, Yves Reguerre, Isabelle Pellier, and Chloé Arfeuille. We thank all the members of Ponte di Legno Childhood ALL Working Group.
Author contributions
Conception and design: SH, AVM, and AM Collection of data: all authors. Data analysis and interpretation: SH, EB, AVM, and AM Statistics: EB and AVM, Manuscript writing and final approval: all authors.
Compliance with ethical standards
Conflict of interest
C.G.M.: Research support from Loxo, Abbvie, Pfizer. Consulting and speaking fees from Amgen, Illumina. C-H Pui received consulting and speaker fees from Amgen, Erytech Phamra, and Servier. H.I. received a research grant from Servier.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-021-01199-0.
References
- 1.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373:1541–52. doi: 10.1056/NEJMra1400972. [DOI] [PubMed] [Google Scholar]
- 2.Yasuda T, Tsuzuki S, Kawazu M, Hayakawa F, Kojima S, Ueno T, et al. Recurrent DUX4 fusions in B cell acute lymphoblastic leukemia of adolescents and young adults. Nat. Genet. 2016;48:569–74. doi: 10.1038/ng.3535. [DOI] [PubMed] [Google Scholar]
- 3.Liu YF, Wang BY, Zhang WN, Huang JY, Li BS, Zhang M, et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine. 2016;8:173–83. doi: 10.1016/j.ebiom.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirabayashi S, Ohki K, Nakabayashi K, Ichikawa H, Momozawa Y, Okamura K, et al. ZNF384-related fusion genes define a subgroup of childhood B-cell precursor acute lymphoblastic leukemia with a characteristic immunotype. Haematologica. 2017;102:118–29. doi: 10.3324/haematol.2016.151035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li JF, Dai YT, Lilljebjorn H, Shen SH, Cui BW, Bai L, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci USA. 2018;115:E11711–E20. doi: 10.1073/pnas.1814397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51:296–307. doi: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaliova M, Stuchly J, Winkowska L, Musilova A, Fiser K, Slamova M, et al. Genomic landscape of pediatric B-other acute lymphoblastic leukemia in a consecutive European cohort. Haematologica. 2019;104:1396–406. doi: 10.3324/haematol.2018.204974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexander TB, Gu Z, Iacobucci I, Dickerson K, Choi JK, Xu B, et al. The genetic basis and cell of origin of mixed phenotype acute leukaemia. Nature. 2018;562:373–9. doi: 10.1038/s41586-018-0436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrusak O, de Haas V, Stancikova J, Vakrmanova B, Janotova I, Mejstrikova E, et al. International cooperative study identifies treatment strategy in childhood ambiguous lineage leukemia. Blood. 2018;132:264–76. doi: 10.1182/blood-2017-12-821363. [DOI] [PubMed] [Google Scholar]
- 10.Steeghs EMP, Boer JM, Hoogkamer AQ, Boeree A, de Haas V, de Groot-Kruseman HA, et al. Copy number alterations in B-cell development genes, drug resistance, and clinical outcome in pediatric B-cell precursor acute lymphoblastic leukemia. Sci Rep. 2019;9:4634. doi: 10.1038/s41598-019-41078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bueno C, Tejedor JR, Bashford-Rogers R, Gonzalez-Silva L, Valdes-Mas R, Agraz-Doblas A, et al. Natural history and cell of origin of TC F3-ZN F384 and PTPN11 mutations in monozygotic twins with concordant BCP-ALL. Blood. 2019;134:900–5. doi: 10.1182/blood.2019000893. [DOI] [PubMed] [Google Scholar]
- 12.Greaves M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat Rev Cancer. 2018;18:471–84. doi: 10.1038/s41568-018-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novakova M, Zaliova M, Fiser K, Vakrmanova B, Slamova L, Musilova A, et al. DUX4r, ZNF384r and PAX5-P80R mutated B-cell precursor acute lymphoblastic leukemia frequently undergo monocytic switch. Haematologica. 2020. [DOI] [PMC free article] [PubMed]
- 14.Oberley MJ, Gaynon PS, Bhojwani D, Pulsipher MA, Gardner RA, Hiemenz MC, et al. Myeloid lineage switch following chimeric antigen receptor T-cell therapy in a patient with TCF3-ZNF384 fusion-positive B-lymphoblastic leukemia. Pediatr Blood Cancer. 2018;65:e27265. doi: 10.1002/pbc.27265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishimura A, Hasegawa D, Hirabayashi S, Kanabuchi S, Yamamoto K, Aiga S, et al. Very late relapse cases of TCF3-ZNF384-positive acute lymphoblastic leukemia. Pediatr Blood Cancer. 2019;66:e27891. doi: 10.1002/pbc.27891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.