SUMMARY
To make adaptive feeding and foraging decisions, animals must integrate diverse sensory streams with multiple dimensions of internal state. In C. elegans, foraging and dispersal behaviors are influenced by food abundance, population density, and biological sex, but the neural and genetic mechanisms that integrate these signals are poorly understood. Here, by systematically varying food abundance, we find that chronic avoidance of the population-density pheromone ascr#3 is modulated by food thickness, such that hermaphrodites avoid ascr#3 only when food is scarce. The integration of food and pheromone signals requires the conserved neuropeptide receptor PDFR-1, as pdfr-1 mutant hermaphrodites display strong ascr#3 avoidance even when food is abundant. Conversely, increasing PDFR-1 signaling inhibits ascr#3 aversion when food is sparse, indicating that this signal encodes information about food abundance. In both wild-type and pdfr-1 hermaphrodites, chronic ascr#3 avoidance requires the ASI sensory neurons. In contrast, PDFR-1 acts in interneurons, suggesting that it modulates processing of the ascr#3 signal. Although a sex-shared mechanism mediates ascr#3 avoidance, food thickness modulates this behavior only in hermaphrodites, indicating that PDFR-1 signaling has distinct functions in the two sexes. Supporting the idea that this mechanism modulates foraging behavior, ascr#3 promotes ASI-dependent dispersal of hermaphrodites from food, an effect that is markedly enhanced when food is scarce. Together, these findings identify a neurogenetic mechanism that sex-specifically integrates population and food abundance, two important dimensions of environmental quality, to optimize foraging decisions. Further, they suggest that modulation of attention to sensory signals could be an ancient, conserved function of pdfr-1.
Graphical Abstract
eTOC Blurb
To forage optimally, animals must predict the future value of an existing food resource. Here, Luo and Portman show that C. elegans can assess this by weighing food abundance against population density. The authors find that food abundance engages a conserved neuromodulatory pathway to modulate avoidance of a key population density pheromone.
INTRODUCTION
Feeding and foraging provide ideal opportunities to understand the neural and genetic mechanisms underlying flexibility in innate behavior. Despite wide variation in the mechanics of foraging, the logic that guides decisions about exploiting or abandoning food resources is remarkably conserved. In animals as diverse as bees, birds, and fish, foraging decisions weigh the current benefit of feeding against the costs of exploration and the potential benefits of new food sources 1,2. When assessing the value of an existing food resource, optimal foraging requires that animals assess local food abundance and quality as well as population density, nutritional needs, and the risk of predation. Experimental studies have provided important insights into the neural mechanisms underlying these calculations and the genetic mechanisms that specify and optimize them 3.
The nematode C. elegans feeds on microbes that colonize decaying vegetation 4 and modulates its feeding and exploratory behaviors in ways that are consistent with optimal foraging theory. Foraging and dispersal in adults varies with food abundance 5,6, nutritional value 7,8, pathogenicity and toxicity 9,10, and experience 11,12. Foraging behavior exhibits substantial natural variation, owing in part to altered sensory responses 6,13–17. Furthermore, foraging and dispersal are sexually dimorphic and sensitive to nutritional state: solitary males will abandon a food source in search of mates, but only if well-fed 18–22.
In many species, foraging is sensitive to population density. Conspecifics can be competitors that reduce the future value of a crowded food source, but in other contexts, collective foraging can be advantageous 23. In C. elegans, worms will leave a food source more often if it is crowded 17,24, but some pheromones inhibit dispersal and exploration 6,15,16. Further, many natural isolates feed in groups 13 and collective foraging may be favored in patchy food environments 25,26. Thus, the role of population density in C. elegans foraging and dispersal may not be fixed. Indeed, in the wild, balancing selection can simultaneously maintain alleles that promote both strategies 16,17.
Because of its “boom-and-bust” life cycle, in which food availability and population density can vary dramatically 27, C. elegans actively monitor both of these variables when feeding. Population density in C. elegans is mainly signaled by ascarosides, derivatives of the dideoxy sugar ascarylose that serve as a modular chemical language 28–30. A prominent member of this group is ascr#3, which, together with other ascarosides, signals population density to young larvae, guiding the decision to enter the stress-induced dauer stage 31,32. ascr#3 is also a sex pheromone: it is produced predominantly by adult hermaphrodites and elicits strong male-specific attraction 33–35. Adult hermaphrodite behavioral responses to ascr#3 are typically aversive, though this depends on the context of ascr#3 presentation as well as animals’ internal state and previous experience 26,33,35–39.
In earlier studies on sexual dimorphism 34, we encountered variability in hermaphrodite ascr#3 avoidance responses, leading us to wonder whether flexible responses to this population density signal might have adaptive value. Here, we find that this is the case, identifying a neuronal and genetic mechanism whereby food abundance modulates ascr#3 aversion specifically in hermaphrodites to adaptively modulate feeding and foraging decisions.
RESULTS
Hermaphrodite pheromone avoidance is modulated by food abundance
To explore the relationship between food and pheromones, we used a quadrant-format assay 34 (Figure 1A). This assay allows robust quantitation of medium-term (1–2 hr) behavioral responses to non-volatile chemical stimuli. To control the abundance of food, we prepared standardized lawns of E. coli OP50 of three densities (Figure 1B). “T3” is a smooth, uniform lawn providing ample but not excessive food, similar to standard culture conditions. “T2” is a thin, slightly patchy lawn. “T1” is a very thin lawn with many food-free patches, providing a scarce-food environment. Notably, other factors (e.g., bacterial metabolites and environmental gases) also likely differ between these conditions.
Figure 1. Hermaphrodite ascr#3 avoidance depends on food abundance.
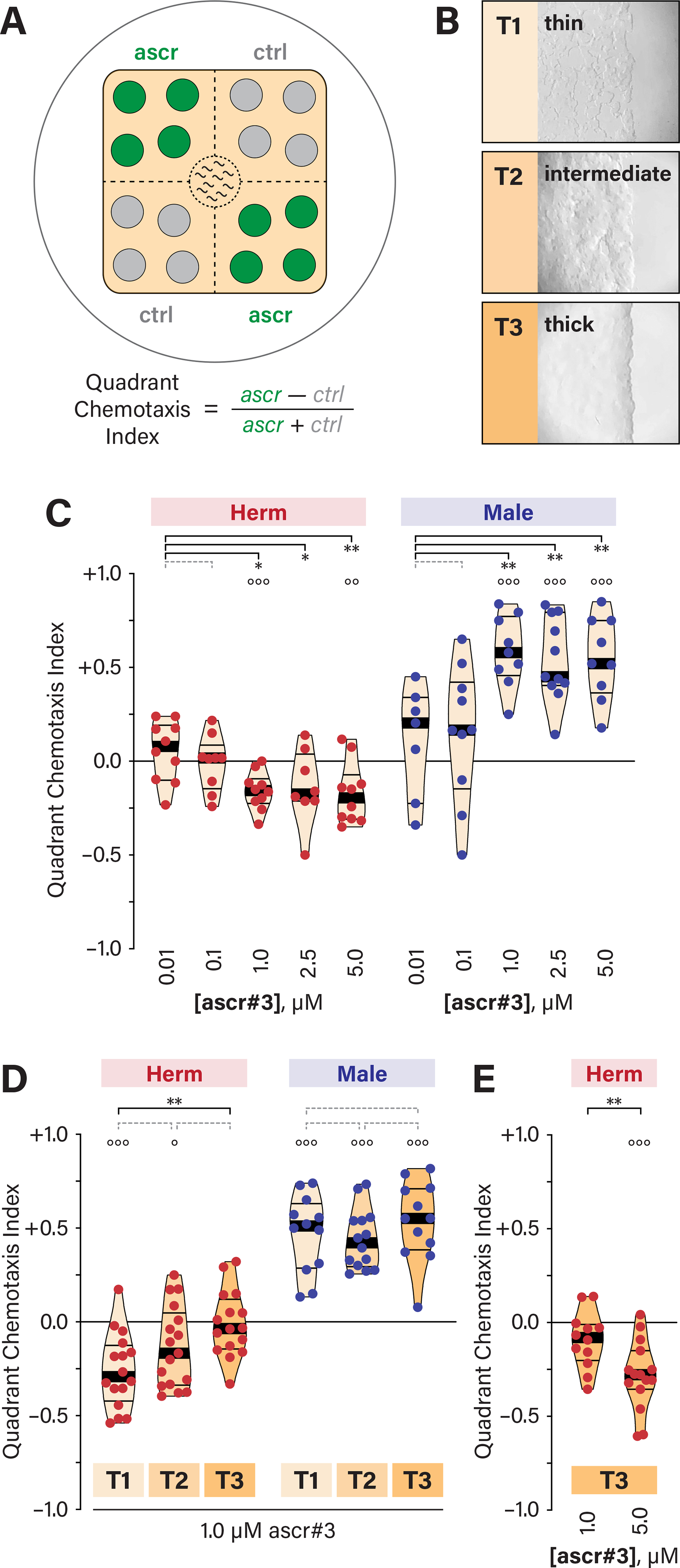
(A) The quadrant-format behavioral assay 34. (B) Images of representative bacterial lawns. T3, a thick lawn; T2, intermediate thickness; and T1, a sparse, patchy lawn. (C) Quadrant Chemotaxis Index (QCI) values for both sexes in response to a range of ascr#3 concentrations in the presence of thin food (T1). (D) QCIs for both sexes to 1.0 μM ascr#3 in three food conditions (T1, T2, and T3). (E) QCIs for adult hermaphrodites to 1.0 and 5.0 μM ascr#3 on thick food (T3). For all quadrant assay data shown in this and subsequent figures, data points are colored by sex (hermaphrodites, red; males, blue). Each point represents a single assay containing 10 worms. Violin plots are shaded according to food thickness (T1, lighter; T3, darker). The median and interquartile intervals are indicated by thick and thin lines, respectively. Statistical comparisons are indicated with black brackets and asterisks (*p ≤ 0.05; **p ≤ 0.005; ***p ≤ 0.001) or dotted gray brackets (p > 0.05). Open circles above each bar show the results of one-sample t-tests, indicating whether the observed QCI differs significantly from zero (°p ≤ 0.05; °°p ≤ 0.005; °°°p ≤ 0.001). See also Figure S1.
In hermaphrodites assayed with thin-food conditions (Figure 1C and S1), we observed modest but consistent avoidance of 1–5 μM ascr#3, but no response to lower concentrations (0.1 and 0.01 μM). In males, 1–5 μM ascr#3 elicited robust attraction, but lower concentrations (0.01 μM and 0.1 μM) had little or no effect on behavior (Figure 1C). Thus, as expected, hermaphrodites and males on sparse food exhibit concentration-dependent, qualitatively distinct responses to ascr#3.
Surprisingly, when we assayed hermaphrodite behavior in the presence of thicker T3 food, we found that ascr#3 avoidance was absent (Figure 1D). Adult males, in contrast, displayed robust ascr#3 attraction regardless of food thickness (Figure 1D). This modulation of ascr#3 aversion by food abundance could reflect active integration of food and pheromone signals by the hermaphrodite nervous system; alternatively, thick food might simply block ascr#3 detection or response. We found that more concentrated ascr#3 (5 μM) was sufficient to elicit avoidance on thick food (Figure 1E), suggesting that the nervous system actively integrates these signals.
PDF neuropeptide signaling modulates pheromone avoidance
Because the conserved neuropeptide receptor PDFR-1 40 is associated with both food-dependent and sex-specific behavioral states 19,41, we examined ascr#3 responses in pdfr-1 null mutants. Interestingly, on thin food, pdfr-1 hermaphrodites displayed markedly enhanced ascr#3 avoidance (Figure 2A, S2A). Single null mutations in pdf-1 and pdf-2, which encode ligands for PDFR-1 40, caused little change in ascr#3 avoidance. However, pdf-1; pdf-2 double mutants showed strong avoidance (Figure 2A), suggesting functional redundancy. We detected no significant difference in ascr#3 avoidance between pdfr-1 and pdf-1; pdf-2 mutants, but cannot rule out possible signaling by additional PDFR-1 ligands or ligand-independent basal PDFR-1 signaling.
Figure 2. PDFR-1 signaling couples food abundance to ascr#3 avoidance.
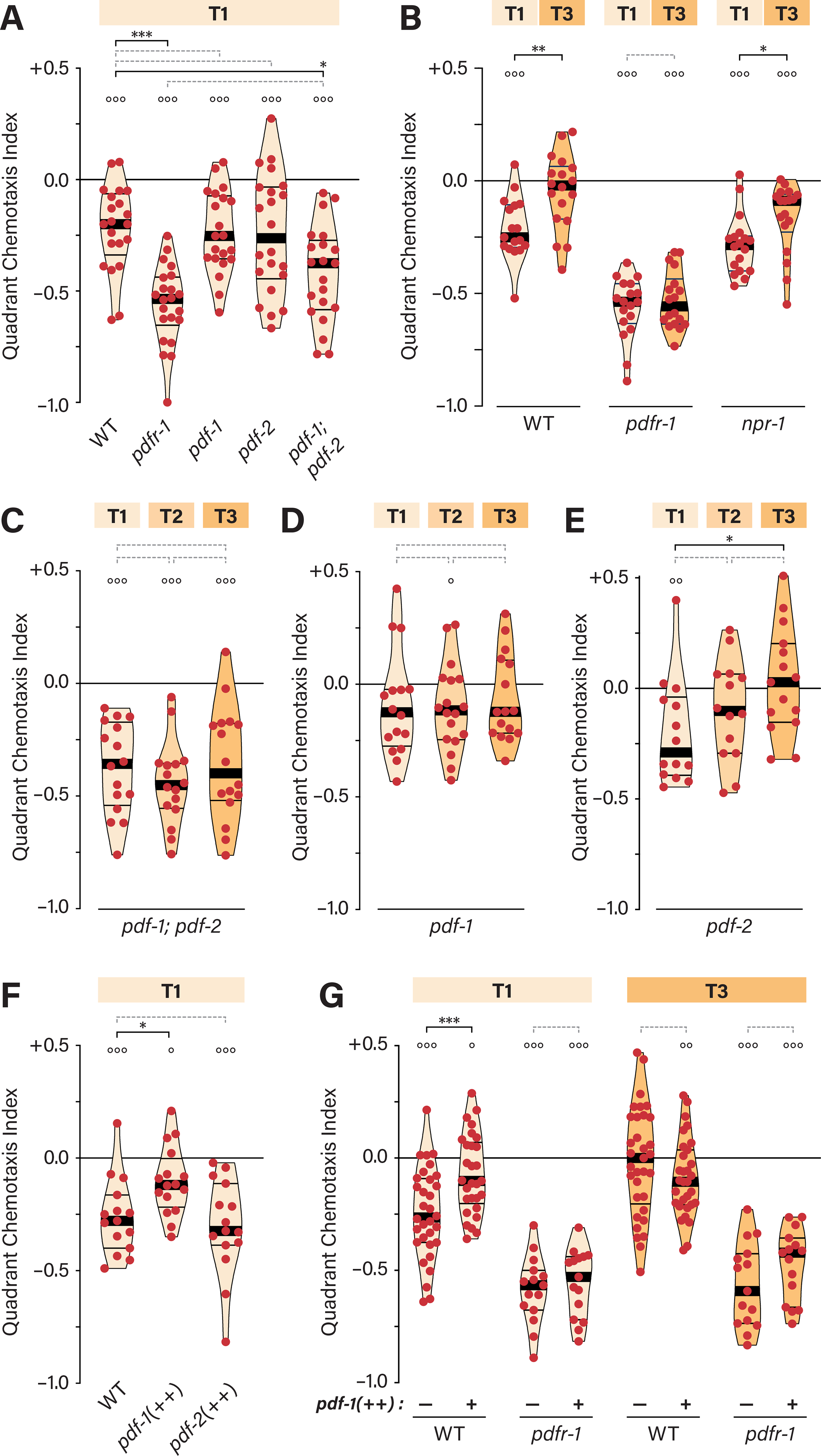
(A) QCIs for WT, pdfr-1, pdf-1, pdf-2, and pdf-1; pdf-2 hermaphrodites on T1. Unless otherwise noted (see Figure S2), all pdfr-1 strains contain pdfr-1(ok3425). (B) QCIs for WT, pdfr-1, and npr-1 hermaphrodites on T1 and T3. (C-E) QCIs for pdf-1; pdf-2 (C), pdf-1 (D), and pdf-2 (E) mutant hermaphrodites with the indicated food conditions. (F) QCIs for WT hermaphrodites and hermaphrodites overexpressing pdf-1 (pdf-1(++)) and pdf-2 (pdf-2(++)) on T1. (G) QCIs for WT, pdf-1(++), pdfr-1, and pdfr-1; pdf-1(++) hermaphrodites on T1 and T3. See also Figure S2.
Because larval exposure to ascr#3 can alter adult ascr#3 responses 38, our results might point to a role for pdfr-1 in experience-dependent plasticity. However, ascr#3 avoidance measured by the quadrant assay was independent of larval exposure to endogenously produced ascr#3, as the loss of daf-22, an enzyme necessary for synthesis of ascr#3 and other short-chain ascarosides 42, had no effect on aversion to exogenous ascr#3 in this context (Figure S2B).
PDFR-1 signaling intensity likely encodes food thickness information
Interestingly, we found that the ascr#3 responses of pdfr-1 mutants were insensitive to food thickness. Regardless of food abundance, pdfr-1 hermaphrodites showed strong aversion to ascr#3 (Figure 2B). This indicates that pdfr-1 function is necessary for the integration of food-thickness information into hermaphrodite behavioral responses to ascr#3.
Because the neuropeptide receptor npr-1 also modulates ascr#3 responses 35,36, we asked whether it might also have a role in this integration. On thin food, loss of npr-1 had no apparent effect on ascr#3 avoidance. Moreover, npr-1 mutants remained sensitive to food abundance, as ascr#3 avoidance was significantly blunted in the presence of thick food (Figure 2B). However, ascr#3 avoidance was not completely absent in npr-1 mutants on thick food, suggesting that npr-1 might have a role in the suppression of ascr#3 avoidance. Compared to pdfr-1, however, this role is subtle.
Consistent with the requirement for pdfr-1, ascr#3 avoidance was insensitive to food abundance in pdf-1; pdf-2 double mutants (Figure 2C). Surprisingly, food-dependence was also absent in pdf-1 single mutants but remained intact in pdf-2 mutants (Figures 2D, E). Thus, although ligands encoded by both genes appear to have roles in the modulation of ascr#3 aversion, pdf-1 has the primary role in linking food thickness to the strength of the aversive response.
We noted that the inhibitory function of pdfr-1 appeared to be stronger on thick food than on thin food (Figure 2B), suggesting that the intensity of PDFR-1 signaling might encode information about food thickness. To test this, we asked whether artificially increasing pdfr-1 signaling would block ascr#3 avoidance on thin food. To bring this about, we used transgenes carrying extra copies of wild-type pdf-1 or pdf-2 40. Consistent with our hypothesis, pdf-1 overexpression reduced ascr#3 avoidance on thin food (Figures 2F, G). However, overexpression of pdf-2 had no apparent effect (Figure 2F), again suggesting that pdf-1, but likely not pdf-2, conveys food thickness information. In agreement with this, overexpression of pdf-1 had no apparent effect in the presence of thick food (Figure 2G). As expected, pdfr-1 was required for the effect of pdf-1 overexpression (Figure 2G). These results indicate that the intensity of PDFR-1 signaling, driven primarily by pdf-1 activity, provides an instructive internal representation of food thickness that modulates ascr#3 avoidance.
The ASI sensory neurons are an essential component of the ascr#3 avoidance circuit
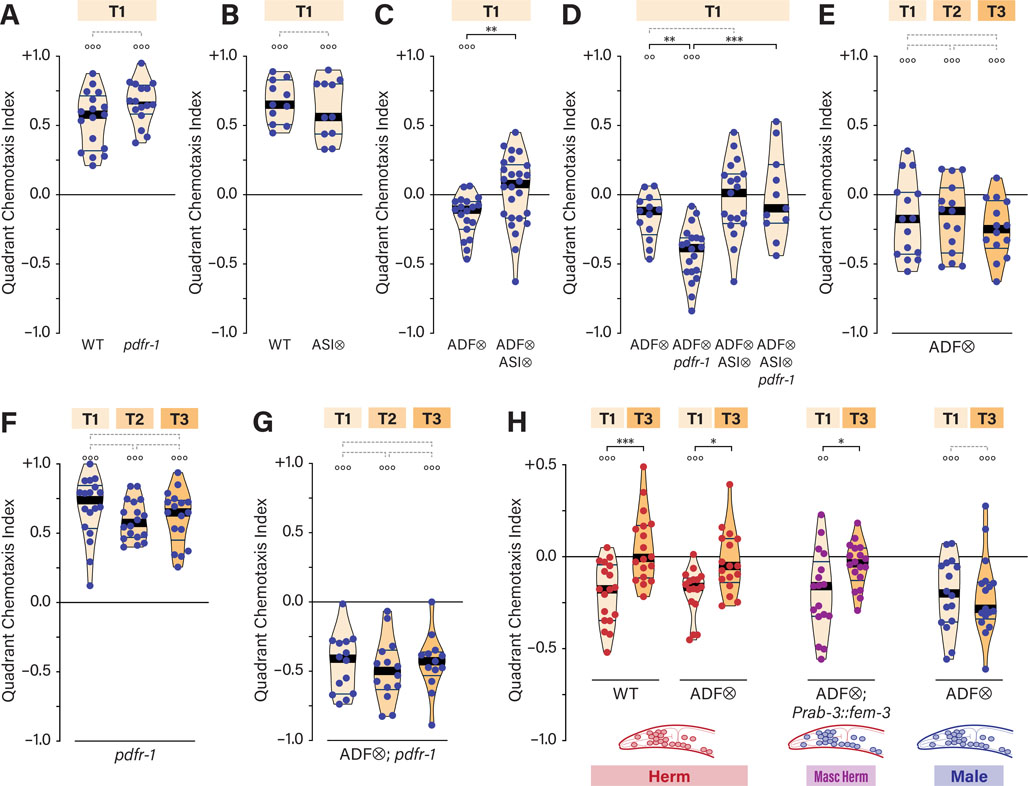
Next, we sought to characterize the neuronal basis for ascr#3 avoidance. In the absence of food, acute aversive and attractive responses to ascr#3 are mediated by the sensory neurons ADL and ASK, respectively 33,35,36,43. However, genetic ablation of these neurons had no apparent effect on ascr#3 avoidance in the quadrant assay, either in wild-type or pdfr-1 hermaphrodites (Figures 3A, B). Thus, the longer-term, on-food ascr#3 responses examined here are likely mediated by mechanisms distinct from those previously described.
Figure 3. The ASI neurons mediate pdfr-1-dependent ascr#3 avoidance.
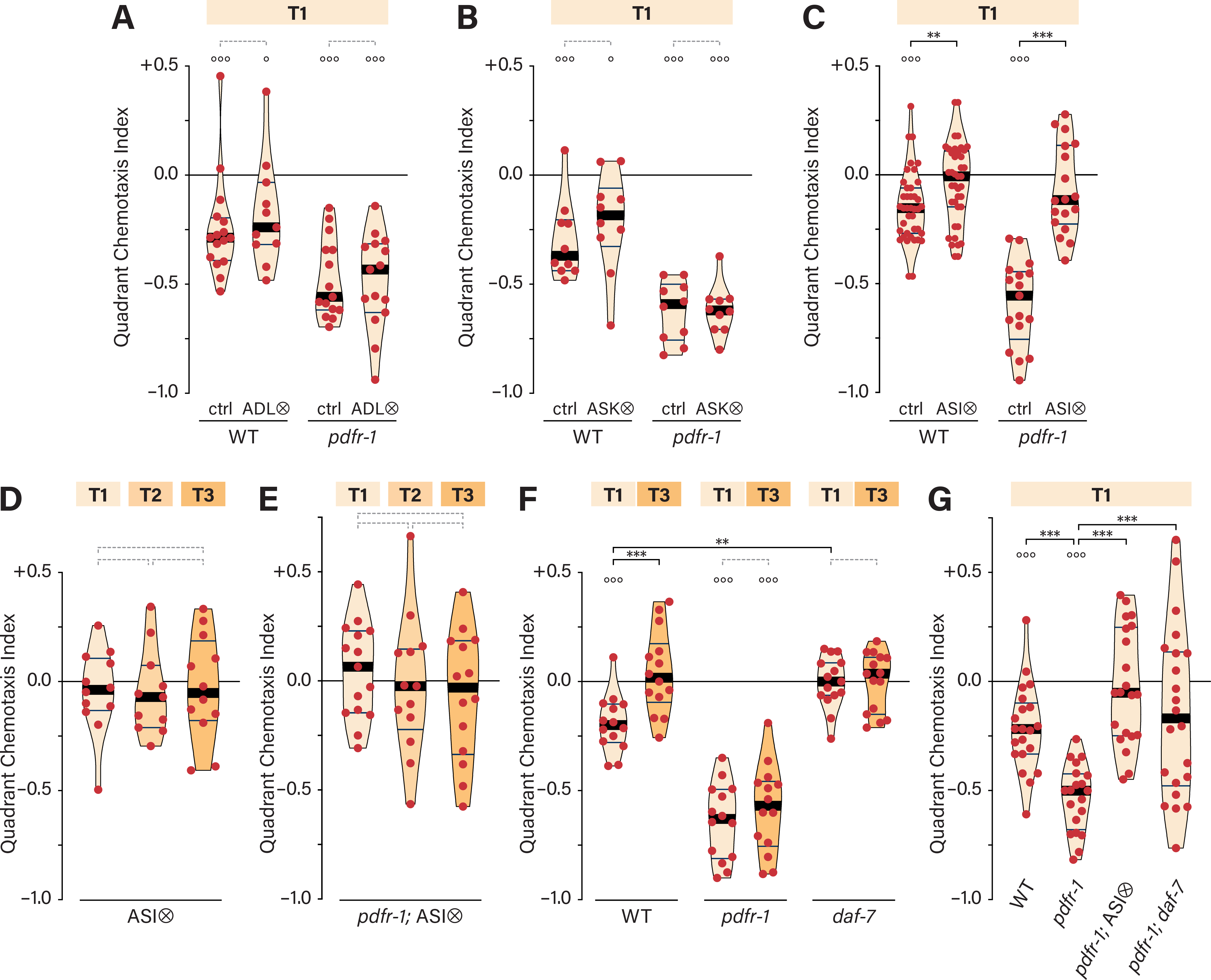
(A–C) QCIs for control and ADL-, ASK-, and ASI-ablated (ADL⨂, A; ASK⨂, B; ASI⨂, C) hermaphrodites in a wild-type or pdfr-1 background on T1. (D, E) QCIs for ASI⨂ (D) or pdfr-1; ASI⨂ (E) hermaphrodites with three food conditions. (F) QCIs for WT, pdfr-1, and daf-7 hermaphrodites with T1 and T3 food. (G) QCIs for WT, pdfr-1, pdfr-1; ASI⨂ and pdfr-1; daf-7 hermaphrodites with T1 food. See also Figure S3.
We also considered the ASI neurons, which detect ascarosides in other contexts, including the dauer-entry decision 44 and the response to the exploration-suppressing pheromone icas#9 16,45. We found that ascr#3 avoidance was abolished upon ASI ablation, both in wild-type and pdfr-1 backgrounds (Figures 3C), regardless of food thickness (Figures 3D, E). The requirement for ASI in both wild-type and pdfr-1 mutants indicates that the strong ascr#3 avoidance of pdfr-1 mutants likely results from increased activity of the same mechanism that generates avoidance in wild-type hermaphrodites, rather than the activation of a parallel mechanism.
These results suggested that ASI might directly detect ascr#3. However, using validated GCaMP transgenes and a range of pheromone concentrations, others have detected no evidence of ascr#3-evoked calcium responses in ASI (E. DiLoreto and J. Srinivasan, pers. comm.; K. Kim and P. Sengupta, pers. comm.). This is consistent with previous findings that ascarosides can engage signaling mechanisms over longer timescales without triggering calcium transients 15,32,45. The simplest interpretation of our results is that ASI directly detects ascr#3; however, it is also possible that ASI is required downstream of ascr#3 detection or in parallel with it.
We also considered the possibility that ASI detects or implements the thin-food state, since ASI-ablated hermaphrodites mimic thick-food behavior (i.e., indifference to ascr#3) on thin food (Figures 3C, D). However, this appeared not to be the case, as ASI ablation in pdfr-1 mutants did not recapitulate the moderate ascr#3 repulsion typical of pdfr-1 hermaphrodites on thick food (Figures 3C, E).
Because it is produced primarily by ASI, we also examined the TGFβ-superfamily ligand DAF-7 46,47. Like ASI-ablated hermaphrodites, daf-7 hermaphrodites exhibited no net response to ascr#3 under any conditions (Figure 3F). We also tested pdfr-1; daf-7 double mutants but observed exceptionally high variability in ascr#3 responses, suggesting a dysregulation of behavior in these animals. These results, which should be interpreted cautiously, could suggest a general requirement for daf-7 in hermaphrodite ascr#3 avoidance. If so, daf-7 might have a role in ascr#3 detection or might signal the presence or abundance of food (see Discussion).
PDFR-1 functions in multiple interneurons to modulate ascr#3 avoidance
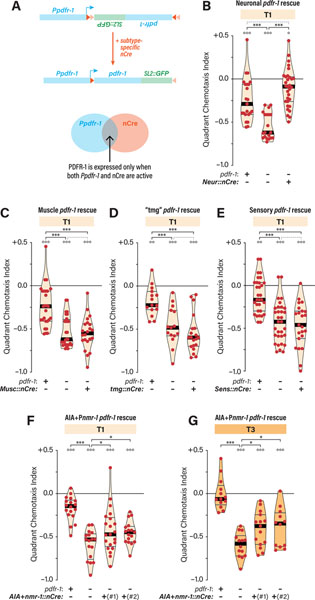
pdfr-1 is expressed in multiple head and tail neurons, as well as peripheral tissues including body-wall muscle 19,40,48. Using an established intersectional strategy 41, we tested the ability of a conditionally activatable Ppdfr-1::inv[pdfr-1::SL2::gfp] transgene, together with neuron-specific nCre constructs, to rescue the enhanced ascr#3 avoidance of pdfr-1 hermaphrodites. This transgene produces functional pdfr-1 and GFP only if the Ppdfr-1 promoter is active and nCre-mediated recombination has taken place (Figure 4A).
Figure 4. pdfr-1 acts in a distributed set of interneurons to repress ascr#3 avoidance.
(A) The strategy for Cre-based conditional pdfr-1 rescue 41. (B-G) QCIs for hermaphrodites carrying a conditional pdfr-1 transgene with the indicated pdfr-1 genotypes (+, wild-type; −, mutant) without or with nCre transgenes expressed pan-neuronally (“Neur::nCre”) (B), in muscle (“Musc::nCre”) (C), in a set of interneurons where pdfr-1 regulates motor state 41 (“tmg::nCre”) (D), in sensory neurons (“Sens::nCre”) (E), and in AIA and neurons expressing nmr-1 (“AIA+nmr-1::nCre”) (F, G), on T1 (B-F) or T3 (G) food.
Pan-neural nCre expression completely rescued the pdfr-1 defect, while muscle expression had no effect (Figures 4B, C). Next, we tested a three-transgene combination (“tmg::nCre,” driven by Ptdc-1, Pmod-1, and Pglr-3) previously shown to partially rescue the persistent dwelling of pdfr-1 mutants 41. However, we observed no rescue of ascr#3 avoidance with this transgene (Figure 4D). To ask whether pdfr-1 functions in sensory neurons, we used Posm-6::nCre, but again saw no rescuing activity (Figure 4E). This suggests that pdfr-1 is unlikely to function in ASI. Consistent with this, no GFP was detectable in animals carrying the ASI/AWA-specific Pgpa-4::nCre and the conditional pdfr-1 transgene (not shown).
We generated and tested several other nCre constructs in combination with the conditional pdfr-1 transgene. These strains exhibited no detectable GFP (Pmec-10, touch neurons) or failed to rescue the ascr#3 avoidance phenotype (Pmod-1 + Posm-6, GFP in several sensory neurons and interneurons; Ptdc-1 + Pglr-3 + Pnmr-1, GFP in many interneurons; and Pgcy-28.d, GFP in AIA interneurons) (not shown). (Note that we cannot rule out the possibility of weak, undetected rescue in these experiments.) However, with another combination, Pgcy-28.d::nCre + Pnmr-1::nCre, we observed a small but statistically significant reduction of ascr#3 avoidance in two independent transgenic lines (Figure 4F). To determine whether these effects were biologically meaningful, we assayed these lines with thick food, again observing significant rescuing activity (Figure 4G). In these animals, GFP was detected in several interneurons, including AIA, PVC, and one or more nmr-1-expressing head interneurons (AVA, AVD, AVE, AVG, and RIM). This partial rescue indicates that pdfr-1 likely has additional important sites of action outside this subset of neurons. From these results, we conclude that PDFR-1 has a distributed role in which it modulates multiple interneuronal components of the ascr#3 avoidance circuit.
The modulatory effects of food thickness are sex-specific
In males, sex-specific detection of ascr#3 by the ADF neurons promotes attraction in the quadrant assay 34. Loss of pdfr-1 or ASI had no effect on this behavior (Figures 5A, B), indicating that they are dispensable for male ascr#3 attraction.
Figure 5. Latent ASI-dependent ascr#3 avoidance in males is independent of food abundance.
(A, B) QCIs for WT and pdfr-1 (A) or ASI⨂ (B) males on T1 food. (C) QCIs for ADF⨂ and ADF⨂; ASI⨂ males on T1 food. (D) QCIs for ADF⨂, ADF⨂; pdfr-1, ADF⨂; ASI⨂, and pdfr-1; ADF⨂; ASI⨂ males on T1 food. (E-G) QCIs for ADF⨂ (E), pdfr-1 (F), and ADF⨂; pdfr-1 (G) with indicated food conditions. (H) QCIs for WT, ADF⨂, and ADF⨂; Prab-3::fem-3 hermaphrodites and ADF⨂ males on T1 and T3 food.
When ADF is genetically feminized or ablated, males display hermaphrodite-like aversive responses to ascr#3, indicating that an avoidance mechanism is unmasked 34 (Figure 5C). Simultaneous ablation of ADF and ASI ablation eliminated all response to ascr#3 in males (Figure 5C), suggesting that avoidance is mediated by a mechanism common to both sexes. Supporting this, loss of pdfr-1 enhanced the ascr#3 avoidance of ADF-ablated males, and this enhanced avoidance also required ASI (Figure 5D).
However, the behavior of ADF-ablated males was not completely equivalent to that of hermaphrodites. In particular, even in the presence of thick food, ADF-ablated males continued to manifest clear ascr#3 aversion (Figure 5E). We observed no effect of food thickness on ascr#3 attraction in pdfr-1 males or on ascr#3 aversion in ADF-ablated pdfr-1 males (Figures 5F, G). Thus, under these conditions, the modulation of ascr#3 avoidance by food thickness is a hermaphrodite-specific feature of the nervous system.
To investigate this sex difference, we genetically masculinized the hermaphrodite nervous system by pan-neuronal expression of the male sexual regulator fem-3 49. This has been shown to functionally sex-reverse many behaviors mediated by shared circuits, including chemosensation and locomotion 22,34,50–52. Simultaneously, we genetically ablated ADF to prevent it from driving ascr#3 attraction 34. Interestingly, the modulatory effects of food thickness remained intact: thick food eliminated ascr#3 aversion in masculinized hermaphrodites, just as in WT (Figure 5H). Thus, sex-specific neurons and/or non-neuronal tissues, rather than sex-shared neurons, appear to be responsible for the hermaphrodite-specificity of food-pheromone integration.
Foraging decisions that integrate food abundance and population density cues require ASI
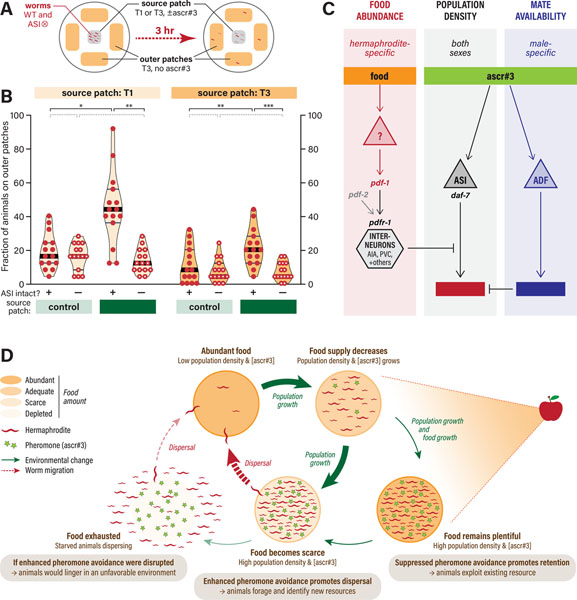
Finally, we wondered whether plasticity in ascr#3 avoidance might allow hermaphrodites to incorporate population density into their assessment of the value of an existing food resource. To explore this, we examined the dispersal of wild-type and ASI-ablated hermaphrodites from food sources of variable quality. In this assay, a thin (T1) or thick (T3) food source, supplemented with vehicle control or 1 μM ascr#3, lies at the center of a 10-cm agar plate (Figure 6A). Four peripheral patches of thick food without ascr#3, providing favorable alternative environments, lie 1.8 cm from the source patch. At the beginning of the assay, 50 young adult hermaphrodites (25 wild-type and 25 ASI-ablated, distinguishable by a fluorescent marker) were deposited on the source patch. After 3 hr, we scored the number of animals of each genotype that had migrated to the peripheral lawns.
Figure 6. Modulation of ascr#3 aversion by food thickness enables flexibility in C. elegans foraging decisions.
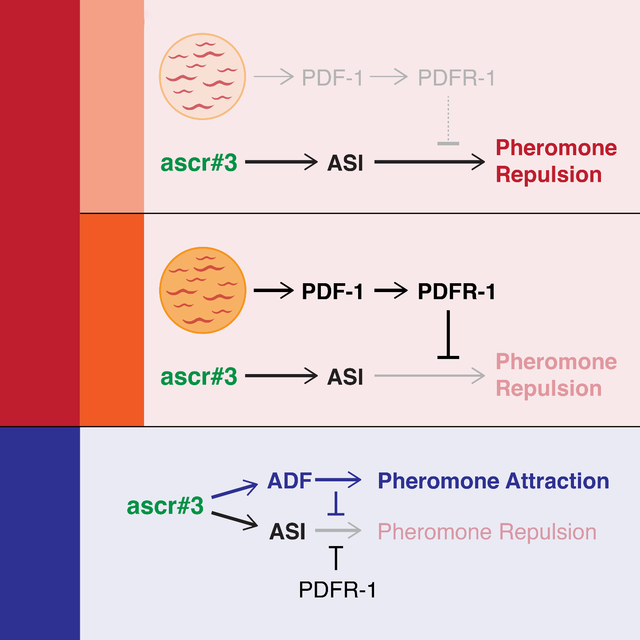
(A) Experimental design. (B) Foraging rates, shown as the frequency of animals dispersing to the outer food sources, of WT and ASI⨂ hermaphrodites placed on a source patch of thick (T3) or thin (T1) food, without (−) or with (+) ascr#3. Each data point represents a single assay containing 25 worms per genotype. Filled and open circles indicate control and ASI-ablated animals, respectively. Violin plots are shaded according to food thickness at the source patch (T1, lighter; T3, darker). The median and interquartile intervals are indicated by thick and thin black lines, respectively. Statistical comparisons between groups are indicated with black brackets and asterisks (*p ≤ 0.05; **p ≤ 0.005; ***p ≤ 0.001) or dotted gray brackets (p > 0.05). (C) A neural circuit model showing the parallel inputs of food abundance, ascr#3 via ASI, and ascr#3 via ADF. Hermaphrodite- and male-specific aspects are shown in red and blue, respectively. (D) A proposed model illustrating the adaptive value of context-dependent flexibility in C. elegans foraging behavior driven by modulation of ascr#3 avoidance.
When the source was a patch of thin food, relatively few animals migrated away, regardless of ASI ablation (Figure 6B). Supplementing the source patch with ascr#3 markedly increased dispersal rate, with nearly 50% of animals migrating to the outer patches (Figure 6B). When ASI was ablated, however, addition of ascr#3 did not increase dispersal rates above baseline (Figure 6B). Thus, consistent with our findings in the quadrant assay, ascr#3 serves as a potent driver of dispersal when food is scarce, and this effect requires ASI.
With a thick-food source, we again observed relatively few animals dispersing (Figure 6B). Adding ascr#3 caused an increase in dispersal, but the magnitude of this effect was far smaller than with thin food (Figure 6B). Again, ascr#3-mediated dispersal was dependent on ASI. Thus, even on thick food, ascr#3 can have aversive effects; however, its ability to promote dispersal is markedly blunted.
Together, these results show that flexibility in ascr#3 aversion allows hermaphrodites to modulate foraging decisions according to population density and food abundance. The ASI-dependence of this plasticity strongly suggests that it arises through the same mechanism that modulates ascr#3 aversion in the quadrant assay. Because it provides a means for animals to optimize foraging decisions, this mechanism likely has adaptive value in the wild.
DISCUSSION
In making foraging decisions, animals weigh the future value of an existing food resource against the potential risks and benefits of abandoning it. A food resource’s value depends not only on its quality and size, but also its projected rate of depletion, a primary determinant of which is local population density. How these variables are internally represented and used by neural circuits to calculate behavioral outcomes is poorly understood. Here, we show that C. elegans hermaphrodites dynamically assign a weight to the aversive population density signal ascr#3 based on food abundance, such that the salience of population density increases as food abundance diminishes (Figure 6C). This flexibility could allow hermaphrodites to disperse from a crowded, rapidly depleting food source before it is completely exhausted, avoiding potential starvation. When food is abundant, however, suppression of the population density signal could allow animals to continue to exploit an existing resource (Figure 6D). By identifying neural and genetic mediators of this process, our results provide insight into the neural mechanisms that implement behavioral plasticity and the genetically specified rules by which these mechanisms operate.
Our work builds on several studies showing that C. elegans hermaphrodites actively modulate their dispersal from an existing food resource 5,6,14,16,17,24,26. Interestingly, recent work found that adult C. elegans hermaphrodites tend to increase food-leaving behavior as their self-progeny accumulate, and that this is likely a response to daf-22-dependent ascaroside pheromones produced by L1 larvae 24. Our results indicate that the depletion of food might enhance this pheromone aversion, such that the larval-adult social interaction identified by these investigators could reflect a synergistic effect of abundant pheromone and scarce food. Another recent study provided evidence that dynamic modulation of the valence of ascaroside cues (i.e., repulsion vs. attraction) can optimize the foraging strategies of a population in a patchy food environment 26. The relationship of these processes to the ascr#3 avoidance plasticity we report here will be an interesting area for future work.
Our findings indicate hermaphrodites use two distinct sensory streams to carry pheromone and food information (Figure 6C). For the former, the ASI sensory neuron pair plays a key role. While there is no evidence that ASI exhibits a calcium response to ascr#3 stimulation, we favor the possibility that ASI directly detects ascr#3 in the behavioral paradigm employed here. Indeed, previous work indicates that several classes of C. elegans sensory neurons including ASI can detect ascaroside pheromones directly without displaying a calcium transient 15,32,45. Interestingly, these responses seem to act over a longer timescale than typical calcium-based neuronal signals, suggesting that they mediate “primer” effects of pheromones 16. However, it is also possible that ASI function is required for the effects of an ascr#3 signal from other sensory neuron(s); these possibilities are not mutually exclusive. Our results also suggest that ASI’s function depends on the TGFβ-superfamily signal DAF-7 (see below) 46,47. Interestingly, ascr#3 detection mechanisms appear to be highly context-dependent, as the behaviors elicited by ascr#3 in other settings (e.g., the acute drop test) have distinct neuronal substrates 33,35,36.
In our model, food abundance is signaled via a separate, parallel sensory stream (Figure 6C). Though the means by which food abundance is detected remain unclear, our findings strongly suggest that this variable is internally represented by the activity level of the neuropeptide receptor PDFR-1. Multiple lines of evidence support this. Animals lacking pdfr-1 or the two ligand genes pdf-1 and pdf-2 display constitutively strong ascr#3 aversion that requires ASI but is completely insensitive to food thickness. Because this aversion is stronger than that seen in WT animals under thin-food (T1) conditions, we infer that low levels of PDFR-1 signaling are active when animals are on thin food, slightly blunting ascr#3 aversion. On thick food (T3), pdfr-1 and pdf-1; pdf-2 hermaphrodites still strongly avoid ascr#3, even though wild-type hermaphrodites are essentially indifferent to it. Here, high levels of PDFR-1 signaling completely inhibit ascr#3 aversion. Our results are most consistent with the idea that pdf-1 and pdf-2 have partially redundant functions but that pdf-1-derived ligand(s) have a more important role in signaling food thickness. Such partial redundancy also exists in other contexts 53, but several studies have shown that pdf-2 can also antagonize pdf-1 40,54, indicating that the dynamics of this system are complex. Importantly, overexpressing pdf-1 in animals feeding on thin food is sufficient to cause them to behave as though they were on thick food—that is, they are now indifferent to ascr#3—while overexpressing of pdf-1 in animals feeding on thick food has no effect. Recently, pdf-2 has been implicated in signaling nutritional status information from the intestine 55, raising the intriguing possibility that pdf-1 and pdf-2 communicate external and internal information about nutrition, respectively, to pdfr-1.
How might food abundance regulate pdfr-1 signaling? We propose that the secretion of pdf-1-encoded neuropeptide(s) is regulated by a food thickness signal. Since sensory neurons are not a major site of pdf-1 expression 48,56, sensory signals might be linked to pdf-1 indirectly. One possibility is that daf-7 conveys food abundance information to pdf-1. daf-7 expression is responsive to food 46,47 and it can link food abundance to behavior, aging, and germline progenitor proliferation 57–59. However, daf-7 activity is expected to be low under thin-food conditions, where we see its strongest requirement 46,47 (Figure 3F). On thick food, where daf-7 activity should be stronger, daf-7 is not required for ascr#3 inhibition (Figure 3F). Thus, it is unlikely that daf-7 conveys food thickness information to the ascr#3 avoidance circuit. Instead, daf-7 could have a permissive role, simply signaling the presence of food to enable pheromone avoidance. Alternatively, because daf-7 is regulated by ascarosides in the dauer entry decision 46,47, daf-7 might signal ascr#3 detection by ASI. However, daf-7 is downregulated by ascaroside exposure 46,47, which would be problematic for such a model. Understanding the role of daf-7 in ascr#3 avoidance will be an important area for future work.
At least four other previously identified mechanisms might couple food abundance to pdf-1 activity. The physical presence of bacteria can be detected by the deirid sensory neuron ADE 60; dopamine released by this neuron might in turn regulate PDF-1 secretion. Bacterial food is also a rich source of chemical cues 61; the detection of these by amphid chemosensory neurons could indirectly regulate pdf-1. Further, in bacteria-rich environments, local concentrations of O2 and CO2 are decreased and increased, respectively, allowing worms to use the activity of gas-sensing neurons as proxies for food abundance 62–66. pdf-1 signaling could be downstream of such a signal. A final possibility is related to the physical structure of the sparse-food T1 environment, where bacterial patches are interspersed with many small gaps. As animals navigate this environment, the frequency with which they cross food boundaries is markedly higher than in an abundant-food environment. Recent work has shown that worms can use this information to assess the physical structure of food patches 12; an intriguing possibility is that this might occur at least in part via modulation of PDF-1 release and/or PDFR-1 signaling.
Downstream of its regulation by food abundance, we find that PDFR-1 signaling represses ascr#3 aversion by modulating a distributed set of interneurons. pdfr-1 is known to modulate the balance between roaming and dwelling states, another behavior affected by food quantity 8,41. However, restoring pdfr-1 function to neurons that mediate these effects was unable to rescue the increased ascr#3 aversion of pdfr-1 mutants, suggesting that these are distinct functions. Future work will be required to understand how pdfr-1 modulates the ascr#3 aversion circuit downstream of ASI.
Interestingly, biological sex regulates the processes we describe here in at least two ways. First, as shown previously, males-specific ascr#3 detection by ADF overrides a latent aversion drive 34. Here, we show that this aversion appears to be mediated by a sex-shared mechanism, as it requires ASI and is repressed by pdfr-1 in both sexes. However, the aversion driven by this mechanism is insensitive to food thickness: even in the presence of thick food, ADF-ablated males displayed ASI-dependent avoidance of ascr#3. Thus, some aspect of the food thickness signal—either its existence or its ability to modulate the aversive signal—is hermaphrodite-specific. Interestingly, this hermaphrodite-specific feature was not eliminated by genetic masculinization of the hermaphrodite nervous system. This suggests that the sex-specificity of food-pheromone integration depends on sex-specific neurons or sex-specific signals from peripheral tissues. Further, why males lack this sensory integration is unclear. One possibility is that, rather than encoding food thickness, pdfr-1 signaling conveys some other dimension of information in males. The importance of pdfr-1 in promoting male mate-searching behavior 19,67 strongly supports this idea.
Because of these differences, the scenarios of Figure 6D apply only to hermaphrodites. In males, ascr#3 would likely retain males under most of the conditions shown. This helps tune behavior to the distinct goals of adult males, in which mate-searching is typically favored over feeding 18,68. Nevertheless, it seems unlikely that pheromone attraction in males is completely independent of food signals. In the absence of food, males copulate infrequently and poorly; both detection and ingestion of food are important for promoting efficient mating 69,70. How males might integrate food and pheromone cues will be an interesting area for future research.
Neuromodulation is an ancient, widespread feature allowing the dynamic reconfiguration of fixed neural circuits 71. Even in simple nervous systems, myriad neuromodulatory signals exist: the C. elegans genome, for example, contains over 100 neuropeptide genes, each of which can produce multiple ligands 72 (worm.peptidegpcr.org). While important progress has been made in identifying the effects of these signals on physiology and behavior, understanding the functional significance of the modulation they bring about can be challenging. Here, we provide evidence that PDF-1/PDFR-1 signaling is an internal representation of food abundance that regulates the behavioral response to stimulation by ascr#3. Interestingly, in several systems, PDF-family neuropeptides and/or the neurons that release them can modulate arousal and sensitivity to external stimuli 73–76. Our results suggest that modulation of attention to sensory stimuli like ascr#3 could be an ancient function of this family of neuropeptides.
STAR METHODS
RESOURCE AVAILABLITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Douglas Portman (douglas.portman@rochester.edu).
Materials Availability
Plasmids and nematode strains generated in the course of this work are freely available to interested academic researchers through the Lead Contact.
Data and Code Availability
Source data obtained in the current study have not been deposited in a public repository but are available from the Lead Contact on request.
This study did not generate code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All C. elegans strains were cultured using E. coli OP50 and NGM agar as described77,78. All strains were grown at 20°C except for those containing daf-7 mutations; these were cultured at 15°C from egg to L4 stage and then assayed at 20°C. him-5(e1490) is considered the wild-type for these studies. All strains used here contained this mutation unless otherwise noted.
METHOD DETAILS
Behavioral Assays
Quadrant assay with controlled bacterial thickness
Quadrant assays were performed as described previously 34 with the following modifications. To prepare lawns of controlled thicknesses, plates were seeded with bacterial cultures of defined concentration. To prepare these, 2 mL of E. coli OP50, freshly cultured in LB media, was collected and centrifuged (12,000 g for 30 sec). After determining its mass, the bacterial pellet was vigorously resuspended in sterile water (143 μL water for each 1 mg of E. coli) to create the T3 stock. T3 stock was diluted 0.1x to create T2 stock, which was then diluted 0.1x to create T1 stock. Each assay plate was seeded with 50 μL of stock suspension and incubated at 20°C for exactly 16 hr before t = 0 min. For three random T3 stocks, bacterial density was calculated by serial dilution to be 3.5×108, 8.8×108, and 2.8×109 cfu/mL. The variation among these samples is likely amplified by extensive serial dilution, as the lawns created with these stocks appeared identical under a stereomicroscope after 16 hours of incubation. We visually inspected lawn quality for each batch of plates before use and found this procedure to reliably produce lawns appearing like those in Figure 1B.
For each assay, 30 mins before t = 0 min, ten animals were picked to the center of each plate, with the experimenter blind to genotype. At t = 0, four 1 μL drops of ascr#3 or vehicle (an equivalent volume of ethanol diluted in sterile water) were dropped into each of the four quadrants. At t = 30, 60, 90, and 120 mins, the number of animals in each quadrant was scored. For each assay, the overall Quadrant Chemotaxis Index (QCI) was calculated as the mean of the four QCI values.
Foraging assay
On the day preceding the assay, L4 hermaphrodites were picked to OP50-seeded plates (30 animals per plate) and assay plates were prepared. Guided by a custom-made transparent template, the boundaries of a center square (“source patch”) and four peripheral rectangles (“outer patches”) were drawn on the bottom of each unseeded 10-cm NGM agar plate. The shape and size of the source patch is equivalent to a single quadrant in the quadrant assay. The shape and size of each outer patch is equivalent to two adjacent quadrants. The distance from the boundary of the source patch to the nearest boundary of an outer patch is 1.8 cm. T1 or T3 bacterial stocks were made as described above. For the source patch, 12.5 μL of T1 or T3 stock suspension was dropped and carefully spread. For each outer patch, 25 μL of T3 suspension was spread. All assay plates were then incubated at 20 °C for exactly 16 hr before t = 0.
At t = 0, 10 μL ascr#3 (1 μM solution) or vehicle control (ethanol diluted in sterile water) was dropped onto the source patch under the stereomicroscope. Air bubbles were created and quickly moved around on the square food lawn to cover it completely with the solution. Once the lawn was dry (3 to 5 min), 25 WT hermaphrodites (strain DR466) and 25 ASI-ablated hermaphrodites (strain UR1110) were picked into the source patch. Each plate was incubated at 20°C for 3 hr. At t = 180 min, animals in the outer patches were counted and genotyped using GFP fluorescence.
Molecular biology and generation of transgenic strains
To express nCre in different cells, we created a series of nCre expression plasmids. Promoters were designed from sequence information from Wormbase and were amplified from purified C. elegans genomic DNA using Phusion DNA polymerase (New England Biolabs). nCre expression constructs containing these promoters and the unc-54 3’UTR were created using Gateway cloning (Thermo Fisher Scientific). All constructs were confirmed by Sanger sequencing. See Table S1 for primer sequences.
To create conditional pdfr-1 rescue strains, the nCre expression plasmid was first injected into the strain UR930 with the co-injection marker Pvha-6::mCherry(mini), a gift from K. Nehrke. The resulting transgene was then crossed into the kyEx4648 background (CX14488, generously provided by S. Flavell and C. Bargmann 41) and maintained by picking hermaphrodites with both intestinal and pharyngeal mCherry signals.
All new extrachromosomal array transgenes were created by microinjection of DNA of interest at 20 ng/μL together with co-injection marker at 50 ng/μL. At least two lines were assayed for each new array. Genetic ablation strains were confirmed by DiI staining (for ablation of ADL, ASK, and ASI) or using a fluorescent marker (for ablation of ADF). Strains were genotyped by PCR and/or Sanger sequencing (see Table S2 for details).
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise indicated, statistical significance was assessed using a two-tailed Mann-Whitney t-test with unequal variances (to compare two genotypes) or Tukey’s multiple comparison test (to compare more than two genotypes) post one-way or two-way ANOVA, corresponding to the number of factors. Asterisks indicate p values associated with these tests: *p ≤ 0.05; **p ≤ 0.005; ***p ≤ 0.001. For clarity, the brackets in each graph indicate all comparisons made; those with a statistically non-significant result (p > 0.05) are shown with dashed gray lines. For each group tested in the quadrant assay, we also carried out a one-sample t-test to ask whether there was a significant aversive or attractive response to ascr#3 (i.e., to ask whether the QCI was statistically different from zero). The resulting p values are indicated with circles above each violin plot: °p ≤ 0.05; °°p ≤ 0.005; °°°p ≤ 0.001.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli OP50 | CGC | OP50 |
| Chemicals, peptides, and recombinant proteins | ||
| ascaroside #3 (ascr#3, also known as C9 or asc-ΔC9) | Synthesized by F. Schroeder laboratory, Boyce Thomson Institute, Ithaca, NY | N/A |
| Dil (1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) | Sigma-Aldrich | Cat # 42364 |
| Phusion HF DNA Polymerase | New England Biolabs | Cat # M0530L |
| Critical commercial assays | ||
| MultiSite Gateway Three-Fragment Vector Construction Kit | Thermo Fisher Scientific | Cat # 12537-023 |
| Experimental Models: Organisms/Strains | ||
| him-5(e1490) V | CGC | DR466 |
| C. elegans wild isolate | CGC | N2 |
| npr-1(ky13) X | CGC 13 | CX4148 |
| pdfr-1(ok3425) III | CGC | VC2609 |
| pdfr-1(ok3425) III; him-5(e1490) V | This work (from DR466 and VC2609) | UR930 |
| pdf-1(tm1996) III; him-5(e1490) V | This work (from DR466 and LSC27 40) | UR954 |
| him-5(e1490) V; pdf-2(tm4780) X | This work and 54 | UR955 |
| lstEx2[pdf-1p::pdf-1::3′UTR + elt-2p::GFP] | CGC and 40 | LSC84 |
| lstEx3[pdf-2p::pdf-2::3′UTR + elt-2p::GFP] | CGC and 40 | LSC54 |
| daf-22(m130) II; him-5(e1490) V | This work (from DR466 and DR476, both from CGC) | UR978 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx595[rab-3p::Cre + vha-6p::mCherry #1] | This work and 41 | UR1363 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx596[rab-3p::Cre + vha-6p::mCherry #2] | This work and 41 | UR1364 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx597[myo-3p::Cre + vha-6p::mCherry #1] | This work and 41 | UR1365 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx598[myo-3p::Cre + vha-6p::mCherry #2] | This work and 41 | UR1366 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx599[nmr-1p::Cre + gcy-28d p::Cre + vha-6p::mCherry #1] | This work and 41 | UR1367 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx600[nmr-1p::Cre + gcy-28d p::Cre + vha-6p::mCherry #2] | This work and 41 | UR1368 |
| pdfr-1(bx142) III; him-5(e1490) V | 19 | EM938 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx601[mod-1p::Cre +tdc-1p::Cre + glr-3::Cre + vha-6p::mCherry #1] | This work and 41 | UR1369 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx602[mod-1p::Cre +tdc-1p::Cre + glr-3::Cre + vha-6p::mCherry #2] | This work and 41 | UR1370 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx603[osm-6p::Cre + vha-6p::mCherry #1] | This work and 41 | UR1371 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx604[osm-6p::Cre + vha-6p::mCherry #2] | This work and 41 | UR1372 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx605[osm-6p::Cre+ mod-1::Cre + vha-6p::mCherry #1] | This work and 41 | UR1373 |
| pdfr-1(ok3425) III; him-5(e1490) V; kyEx4648[pdfr-1::inv[pdfr-1.d-sl2-GFP]]; fsEx606[osm-6p::Cre+ mod-1::Cre + vha-6p::mCherry #2] | This work and 41 | UR1374 |
| him-5(e1490) V; oyIs84[Pgpa-4::TU#813 + Pgcy-27::TU#814 + Pgcy-27::GFP + Punc-122::dsRed] | This work and 79 (from DR466 and PY7505) | UR1110 |
| pdfr-1(ok3425) III; him-5(e1490) V; oyIs84[Pgpa-4::TU#813 + Pgcy-27::TU#814 + Pgcy-27::GFP + Punc-122::dsRed] | This work (from UR1110 and UR930) | UR1375 |
| daf-7(e1372) III; him-5(e1490) V | This work | UR913 |
| him-5(e1490) V; Ex[srh-281p::mCasp1 + myo-3p::GFP] | This work and T. Hirotsu 80 | UR1106 |
| pdfr-1(ok3425) III; him-5(e1490) V; Ex[srh-281p::mCasp1 + myo-3p::GFP] | This work (from UR1106 and UR930) | UR1376 |
| him-8(e1489) IV | CGC | CB1489 |
| pdfr-1(ok3425) III; him-8(e1489) IV | This work (from CB1489 and VC2609) | UR1223 |
| him-8(e1489) IV; qrIs2[sra-9::mCasp1] V | This work (from CB1489 and PS6025 (CGC)) | UR1216 |
| pdfr-1(ok3425) III; him-8(e1489) IV; qrIs2[sra-9::mCasp1] V | This work (from UR1223 and UR1216) | UR1231 |
| daf-7(e1372) pdfr-1(ok3425) III; him-5(e1490) V | This work (from UR930 and UR913) | UR1378 |
| him-5(e1490) V; udEx428[elt-2p::GFP + srh-142p::GFP + srh-142p::CED-3(p15) + srh-142p::CED-3(p17)] | This work and D. Ferkey 81 | UR987 |
| him-5(e1490) fsIs15[rab-3p::FEM-3(+)::outron::mCherry + unc-122p::GFP] V; udEx428[elt-2p::GFP + srh-142p::GFP + srh-142p::CED-3(p15) + srh-142p::CED-3(p17)] | This work and D. Ferkey 22,81 | UR1379 |
| pdfr-1(ok3425) III; him-5(e1490) V; udEx428[elt-2p::GFP + srh-142p::GFP + srh-142p::CED-3(p15) + srh-142p::CED-3(p17)] | This work (from UR930 and UR987) | UR1025 |
| him-5(e1490) V; oyIs84[Pgpa-4::TU#813 + Pgcy-27::TU#814 + Pgcy-27::GFP + Punc-122::dsRed]; udEx428[elt-2p::GFP + srh-142p::GFP + srh-142p::CED-3(p15) + srh-142p::CED-3(p17)] | This work (from UR987 and UR1110) | UR1380 |
| pdfr-1(ok3425) III; him-5(e1490) V; oyIs84[Pgpa-4::TU#813 + Pgcy-27::TU#814 + Pgcy-27::GFP + Punc-122::dsRed]; udEx428[elt-2p::GFP + srh-142p::GFP + srh-142p::CED-3(p15) + srh-142p::CED-3(p17)] | This work (from UR1025 and UR1375) | UR1381 |
| pdf-1(tm1996) III; him-5(e1490) V; pdf-2(tm4780) X | This work (from UR954 and UR955) | UR1388 |
| daf-22(m130) II; pdfr-1(ok3425) III; him-5(e1490) V | This work (from UR930 and UR978) | UR1389 |
| him-5(e1490) V; lstEx2[pdf-1p::pdf-1::3′UTR + elt-2::GFP] | This work and 40 (from DR466 and LSC84) | UR1390 |
| pdfr-1(ok3425) III; him-5(e1490) V; lstEx2[pdf-1p::pdf-1::3′UTR + elt-2::GFP] | This work and 40 (from UR1390 and UR930) | UR1391 |
| Oligonucleotides | ||
| See Table S1 for oligonucleotide information | ||
| Recombinant DNA | ||
| Prab-3::nCre | This work | N/A |
| Pmyo-3::nCre | This work | N/A |
| Pmod-1::nCre | This work | plasID_438 |
| Ptdc-1::nCre | This work | N/A |
| Pglr-3::nCre | This work | N/A |
| Posm-6::nCre | This work | plasID_436 |
| Pnmr-1::nCre | This work | N/A |
| Pgcy-28.d::nCre | C. Bargmann 82 | pALC07 |
| Pvha-6::mCherry | K. Nehrke 83 | pELA1 |
| Software and Algorithms | ||
| ApE, A Plasmid Editor | M. Wayne Davis | N/A |
| Prism 8 | GraphPad Software | N/A |
Highlights.
Abundant food blocks hermaphrodite avoidance of a key population density pheromone
Food abundance is encoded by the conserved PDF/PDFR-1 neuropeptide system
PDFR-1 acts in interneurons to modulate the salience of the aversive pheromone cue
Food-pheromone integration allows value assessment and promotes adaptive foraging
ACKNOWLEDGEMENTS
We thank current and past members of the Portman lab, the University of Rochester Invertebrate Biology Group, and the Western New York Worm Group for discussion and critical feedback. We are particularly grateful to E. DiLoreto and J. Srinivasan, as well as K. Kim and P. Sengupta, for communicating unpublished results, and to F. Schroeder for providing synthetic ascr#3. We thank S. Flavell, C. Bargmann, T. Hirotsu, D. Ferkey, and K. Nehrke for generously providing strains and reagents. Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). These studies were funded by NIH R01 GM130136 and R01 GM140415 to D.P.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Charnov EL (1976). Optimal foraging, the marginal value theorem. Theor Popul Biol 9, 129–136. 10.1016/0040-5809(76)90040-x. [DOI] [PubMed] [Google Scholar]
- 2.Pyke GH (1984). Optimal Foraging Theory: A Critical Review. Annual Review of Ecology and Systematics 15, 523–575. 10.1146/annurev.es.15.110184.002515. [DOI] [Google Scholar]
- 3.Calhoun AJ, and Hayden BY (2015). The foraging brain. Current Opinion in Behavioral Sciences 5, 24–31. 10.1016/j.cobeha.2015.07.003. [DOI] [Google Scholar]
- 4.Frezal L, and Felix MA (2015). C. elegans outside the Petri dish. Elife 4. 10.7554/eLife.05849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milward K, Busch KE, Murphy RJ, de Bono M, and Olofsson B (2011). Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 108, 20672–20677. 10.1073/pnas.1106134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey SC (2009). Non-dauer larval dispersal in Caenorhabditis elegans. J Exp Zool B Mol Dev Evol 312B, 224–230. 10.1002/jez.b.21287. [DOI] [PubMed] [Google Scholar]
- 7.Shtonda BB, and Avery L (2006). Dietary choice behavior in Caenorhabditis elegans. The Journal of experimental biology 209, 89–102. 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben Arous J, Laffont S, and Chatenay D (2009). Molecular and sensory basis of a food related two-state behavior in C. elegans. PloS one 4, e7584. 10.1371/journal.pone.0007584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Lu H, and Bargmann C (2005). Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184. [DOI] [PubMed] [Google Scholar]
- 10.Melo JA, and Ruvkun G (2012). Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466. 10.1016/j.cell.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pradhan S, Quilez S, Homer K, and Hendricks M (2019). Environmental Programming of Adult Foraging Behavior in C. elegans. Current biology : CB 29, 2867–2879 e2864. 10.1016/j.cub.2019.07.045. [DOI] [PubMed] [Google Scholar]
- 12.Calhoun AJ, Tong A, Pokala N, Fitzpatrick JA, Sharpee TO, and Chalasani SH (2015). Neural Mechanisms for Evaluating Environmental Variability in Caenorhabditis elegans. Neuron 86, 428–441. 10.1016/j.neuron.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bono M, and Bargmann CI (1998). Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94, 679–689. [DOI] [PubMed] [Google Scholar]
- 14.Bendesky A, Tsunozaki M, Rockman MV, Kruglyak L, and Bargmann CI (2011). Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature 472, 313–318. 10.1038/nature09821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene JS, Dobosiewicz M, Butcher RA, McGrath PT, and Bargmann CI (2016). Regulatory changes in two chemoreceptor genes contribute to a Caenorhabditis elegans QTL for foraging behavior. Elife 5. 10.7554/eLife.21454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene JS, Brown M, Dobosiewicz M, Ishida IG, Macosko EZ, Zhang X, Butcher RA, Cline DJ, McGrath PT, and Bargmann CI (2016). Balancing selection shapes density-dependent foraging behaviour. Nature 539, 254–258. 10.1038/nature19848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gloria-Soria A, and Azevedo RB (2008). npr-1 Regulates foraging and dispersal strategies in Caenorhabditis elegans. Current biology : CB 18, 1694–1699. 10.1016/j.cub.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 18.Lipton J, Kleemann G, Ghosh R, Lints R, and Emmons SW (2004). Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. The Journal of neuroscience : the official journal of the Society for Neuroscience 24, 7427–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrios A, Ghosh R, Fang C, Emmons SW, and Barr MM (2012). PDF-1 neuropeptide signaling modulates a neural circuit for mate-searching behavior in C. elegans. Nature neuroscience 15, 1675–1682. 10.1038/nn.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrios A, Nurrish S, and Emmons SW (2008). Sensory regulation of C. elegans male mate-searching behavior. Current biology : CB 18, 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wexler LR, Miller RM, and Portman DS (2020). C. elegans Males Integrate Food Signals and Biological Sex to Modulate State-Dependent Chemosensation and Behavioral Prioritization. Current biology : CB. 10.1016/j.cub.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan DA, Miller RM, Lee K, Neal SJ, Fagan KA, Sengupta P, and Portman DS (2014). Sex, age, and hunger regulate behavioral prioritization through dynamic modulation of chemoreceptor expression. Current biology : CB 24, 2509–2517. 10.1016/j.cub.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraldeau L-A, and Caraco T (2000). Social foraging theory (Princeton University Press; ). [Google Scholar]
- 24.Scott E, Hudson A, Feist E, Calahorro F, Dillon J, de Freitas R, Wand M, Schoofs L, O’Connor V, and Holden-Dye L (2017). An oxytocin-dependent social interaction between larvae and adult C. elegans. Sci Rep 7, 10122. 10.1038/s41598-017-09350-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding SS, Muhle LS, Brown AEX, Schumacher LJ, and Endres RG (2020). Comparison of solitary and collective foraging strategies of Caenorhabditis elegans in patchy food distributions. Philos Trans R Soc Lond B Biol Sci 375, 20190382. 10.1098/rstb.2019.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dal Bello M, Perez-Escudero A, Schroeder FC, and Gore J (2021). Inversion of pheromone preference optimizes foraging in C. elegans. Elife 10. 10.7554/eLife.58144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felix MA, and Duveau F (2012). Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC biology 10, 59. 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludewig AH, and Schroeder FC (2013). Ascaroside signaling in C. elegans. WormBook : the online review of C. elegans biology, 1–22. 10.1895/wormbook.1.155.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JY, Joo HJ, Park S, and Paik YK (2019). Ascaroside Pheromones: Chemical Biology and Pleiotropic Neuronal Functions. Int J Mol Sci 20. 10.3390/ijms20163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGrath PT, and Ruvinsky I (2019). A primer on pheromone signaling in Caenorhabditis elegans for systems biologists. Curr Opin Syst Biol 13, 23–30. 10.1016/j.coisb.2018.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butcher RA, Fujita M, Schroeder FC, and Clardy J (2007). Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nature chemical biology 3, 420–422. 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- 32.Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, Ragains JR, Clardy J, Touhara K, and Sengupta P (2009). Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science 326, 994–998. 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn H, Teal P, Malik R, Edison A, Sternberg P, and Schroeder F (2008). A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 454, 1115–1118. 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fagan KA, Luo J, Lagoy RC, Schroeder FC, Albrecht DR, and Portman DS (2018). A Single-Neuron Chemosensory Switch Determines the Valence of a Sexually Dimorphic Sensory Behavior. Current biology : CB 28, 902–914 e905. 10.1016/j.cub.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang H, Kim K, Neal SJ, Macosko E, Kim D, Butcher RA, Zeiger DM, Bargmann CI, and Sengupta P (2012). Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron 75, 585–592. 10.1016/j.neuron.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, and Bargmann CI (2009). A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 458, 1171–1175. 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fenk LA, and de Bono M (2017). Memory of recent oxygen experience switches pheromone valence in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 114, 4195–4200. 10.1073/pnas.1618934114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong M, Ryu L, Ow MC, Kim J, Je AR, Chinta S, Huh YH, Lee KJ, Butcher RA, Choi H, et al. (2017). Early Pheromone Experience Modifies a Synaptic Activity to Influence Adult Pheromone Responses of C. elegans. Current biology : CB 27, 3168–3177 e3163. 10.1016/j.cub.2017.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryu L, Cheon Y, Huh YH, Pyo S, Chinta S, Choi H, Butcher RA, and Kim K (2018). Feeding state regulates pheromone-mediated avoidance behavior via the insulin signaling pathway in Caenorhabditis elegans. The EMBO journal 37, e98402. 10.15252/embj.201798402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janssen T, Husson SJ, Lindemans M, Mertens I, Rademakers S, Ver Donck K, Geysen J, Jansen G, and Schoofs L (2008). Functional characterization of three G protein-coupled receptors for pigment dispersing factors in Caenorhabditis elegans. J Biol Chem 283, 15241–15249. 10.1074/jbc.M709060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, and Bargmann CI (2013). Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell 154, 1023–1035. 10.1016/j.cell.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, and Schroeder FC (2009). A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 106, 7708–7713. 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Srinivasan J, von Reuss SH, Bose N, Zaslaver A, Mahanti P, Ho MC, O’Doherty OG, Edison AS, Sternberg PW, and Schroeder FC (2012). A modular library of small molecule signals regulates social behaviors in Caenorhabditis elegans. PLoS biology 10, e1001237. 10.1371/journal.pbio.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bargmann CI, and Horvitz HR (1991). Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251, 1243–1246. [DOI] [PubMed] [Google Scholar]
- 45.McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, and Bargmann CI (2011). Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477, 321–325. 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, and Riddle DL (1996). Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274, 1389–1391. [DOI] [PubMed] [Google Scholar]
- 47.Schackwitz WS, Inoue T, and Thomas JH (1996). Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17, 719–728. [DOI] [PubMed] [Google Scholar]
- 48.Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, Varol E, Oikonomou P, Glenwinkel L, McWhirter R, et al. (2021). Molecular topography of an entire nervous system. Cell. 10.1016/j.cell.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mehra A, Gaudet J, Heck L, Kuwabara PE, and Spence AM (1999). Negative regulation of male development in Caenorhabditis elegans by a protein-protein interaction between TRA-2A and FEM-3. Genes Dev 13, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mowrey WR, Bennett JR, and Portman DS (2014). Distributed Effects of Biological Sex Define Sex-Typical Motor Behavior in Caenorhabditis elegans. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 1579–1591. 10.1523/JNEUROSCI.4352-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee K, and Portman D (2007). Neural sex modifies the function of a C. elegans sensory circuit. Current biology : CB 17, 1858–1863. 10.1016/j.cub.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 52.White J, Nicholas T, Gritton J, Truong L, Davidson E, and Jorgensen E (2007). The sensory circuitry for sexual attraction in C. elegans males. Current biology : CB 17, 1847–1857. 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 53.Herrero A, Romanowski A, Meelkop E, Caldart CS, Schoofs L, and Golombek DA (2015). Pigment-dispersing factor signaling in the circadian system of Caenorhabditis elegans. Genes Brain Behav 14, 493–501. 10.1111/gbb.12231. [DOI] [PubMed] [Google Scholar]
- 54.Meelkop E, Temmerman L, Janssen T, Suetens N, Beets I, Van Rompay L, Shanmugam N, Husson SJ, and Schoofs L (2012). PDF receptor signaling in Caenorhabditis elegans modulates locomotion and egg-laying. Molecular and cellular endocrinology 361, 232–240. 10.1016/j.mce.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 55.O’Donnell MP, Chao PH, Kammenga JE, and Sengupta P (2018). Rictor/TORC2 mediates gut-to-brain signaling in the regulation of phenotypic plasticity in C. elegans. PLoS genetics 14, e1007213. 10.1371/journal.pgen.1007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Janssen T, Husson SJ, Meelkop E, Temmerman L, Lindemans M, Verstraelen K, Rademakers S, Mertens I, Nitabach M, Jansen G, and Schoofs L (2009). Discovery and characterization of a conserved pigment dispersing factor-like neuropeptide pathway in Caenorhabditis elegans. J Neurochem 111, 228–241. 10.1111/j.1471-4159.2009.06323.x. [DOI] [PubMed] [Google Scholar]
- 57.Entchev EV, Patel DS, Zhan M, Steele AJ, Lu H, and Ch’ng Q (2015). A gene-expression-based neural code for food abundance that modulates lifespan. Elife 4, e06259. 10.7554/eLife.06259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dalfo D, Michaelson D, and Hubbard EJ (2012). Sensory regulation of the C. elegans germline through TGF-beta-dependent signaling in the niche. Current biology : CB 22, 712–719. 10.1016/j.cub.2012.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallagher T, Kim J, Oldenbroek M, Kerr R, and You YJ (2013). ASI Regulates Satiety Quiescence in C. elegans. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 9716–9724. 10.1523/JNEUROSCI.4493-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sawin ER, Ranganathan R, and Horvitz HR (2000). C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26, 619–631. [DOI] [PubMed] [Google Scholar]
- 61.Worthy SE, Haynes L, Chambers M, Bethune D, Kan E, Chung K, Ota R, Taylor CJ, and Glater EE (2018). Identification of attractive odorants released by preferred bacterial food found in the natural habitats of C. elegans. PloS one 13, e0201158. 10.1371/journal.pone.0201158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Witham E, Comunian C, Ratanpal H, Skora S, Zimmer M, and Srinivasan S (2016). C. elegans Body Cavity Neurons Are Homeostatic Sensors that Integrate Fluctuations in Oxygen Availability and Internal Nutrient Reserves. Cell Rep 14, 1641–1654. 10.1016/j.celrep.2016.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, and Bargmann CI (2004). Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430, 317–322. [DOI] [PubMed] [Google Scholar]
- 64.Cheung BH, Cohen M, Rogers C, Albayram O, and de Bono M (2005). Experience-dependent modulation of C. elegans behavior by ambient oxygen. Current biology : CB 15, 905–917. [DOI] [PubMed] [Google Scholar]
- 65.Hallem EA, Spencer WC, McWhirter RD, Zeller G, Henz SR, Ratsch G, Miller DM 3rd, Horvitz HR, Sternberg PW, and Ringstad N (2011). Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 108, 254–259. 10.1073/pnas.1017354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hallem EA, and Sternberg PW (2008). Acute carbon dioxide avoidance in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 105, 8038–8043. 10.1073/pnas.0707469105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hilbert ZA, and Kim DH (2018). PDF-1 neuropeptide signaling regulates sexually dimorphic gene expression in shared sensory neurons of C. elegans. Elife 7, e36547. 10.7554/eLife.36547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrios A (2014). Exploratory decisions of the Caenorhabditis elegans male: a conflict of two drives. Seminars in cell & developmental biology 33, 10–17. 10.1016/j.semcdb.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 69.Gruninger TR, Gualberto DG, and Garcia LR (2008). Sensory perception of food and insulin-like signals influence seizure susceptibility. PLoS genetics 4, e1000117. 10.1371/journal.pgen.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gruninger T, Gualberto D, LeBoeuf B, and Garcia L (2006). Integration of male mating and feeding behaviors in Caenorhabditis elegans. J Neurosci S 26, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marder E, O’Leary T, and Shruti S (2014). Neuromodulation of circuits with variable parameters: single neurons and small circuits reveal principles of state-dependent and robust neuromodulation. Annu Rev Neurosci 37, 329–346. 10.1146/annurev-neuro-071013-013958. [DOI] [PubMed] [Google Scholar]
- 72.Hobert O (2013). The neuronal genome of Caenorhabditis elegans. WormBook : the online review of C. elegans biology, 1–106. 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ardiel EL, Yu AJ, Giles AC, and Rankin CH (2017). Habituation as an adaptive shift in response strategy mediated by neuropeptides. NPJ Sci Learn 2, 9. 10.1038/s41539-017-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, and Kaplan JM (2013). Analysis of NPR-1 Reveals a Circuit Mechanism for Behavioral Quiescence in C. elegans. Neuron 78, 869–880. 10.1016/j.neuron.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fu Y, Tucciarone JM, Espinosa JS, Sheng N, Darcy DP, Nicoll RA, Huang ZJ, and Stryker MP (2014). A cortical circuit for gain control by behavioral state. Cell 156, 1139–1152. 10.1016/j.cell.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dubowy C, and Sehgal A (2017). Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics 205, 1373–1397. 10.1534/genetics.115.185157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stiernagle T (2006). Maintenance of C. elegans. WormBook : the online review of C. elegans biology, 1–11. 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beverly M, Anbil S, and Sengupta P (2011). Degeneracy and neuromodulation among thermosensory neurons contribute to robust thermosensory behaviors in Caenorhabditis elegans. The Journal of neuroscience : the official journal of the Society for Neuroscience 31, 11718–11727. 10.1523/JNEUROSCI.1098-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hamakawa M, Uozumi T, Ueda N, Iino Y, and Hirotsu T (2015). A role for Ras in inhibiting circular foraging behavior as revealed by a new method for time and cell-specific RNAi. BMC biology 13, 6. 10.1186/s12915-015-0114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krzyzanowski MC, Woldemariam S, Wood JF, Chaubey AH, Brueggemann C, Bowitch A, Bethke M, L’Etoile ND, and Ferkey DM (2016). Aversive Behavior in the Nematode C. elegans Is Modulated by cGMP and a Neuronal Gap Junction Network. PLoS genetics 12, e1006153. 10.1371/journal.pgen.1006153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lopez-Cruz A, Sordillo A, Pokala N, Liu Q, McGrath PT, and Bargmann CI (2019). Parallel Multimodal Circuits Control an Innate Foraging Behavior. Neuron 102, 407–419 e408. 10.1016/j.neuron.2019.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allman E, Johnson D, and Nehrke K (2009). Loss of the apical V-ATPase a-subunit VHA-6 prevents acidification of the intestinal lumen during a rhythmic behavior in C. elegans. Am J Physiol Cell Physiol 297, C1071–1081. 10.1152/ajpcell.00284.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data obtained in the current study have not been deposited in a public repository but are available from the Lead Contact on request.
This study did not generate code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.