Abstract
Background:
Polygenic risk scores (PRSs) will have important utility for asthma and other chronic diseases as a tool for predicting disease incidence and sub-phenotypes.
Objective:
We utilized findings from a large multi-ancestry GWAS of asthma to compute a PRS for asthma with relevance for racially diverse populations.
Methods:
We derived two PRSs for asthma using a standard approach (based on genome-wide significant variants) and a lasso sum regression approach (allowing all genetic variants to potentially contribute). We used data from the racially diverse Kaiser Permanente GERA cohort (68,638 Non-Hispanic Whites, 5,874 Hispanics, 6,870 Asians and 2,760 Blacks). Race was self-reported by questionnaire.
Results:
For the standard PRS, Non-Hispanic Whites showed the highest odds ratio for a standard deviation increase in PRS for asthma OR=1.16 (95% CI 1.14 to 1.18). The standard PRS was also associated with asthma in Hispanic OR=1.12 (95% CI 1.05 to 1.19), and Asian subjects OR=1.10 (95% CI 1.04 to 1.17), with a trend toward increased risk in Blacks OR=1.05 (95% CI 0.97 to 1.15). We detected an interaction by sex, with men showing higher risk of asthma with an increase in PRS as compared to women. The lasso sum regression derived PRS showed stronger associations with asthma in Non-Hispanic White subjects (OR=1.20 (95% CI 1.18 to 1.23)), Hispanics (OR=1.17 (95% 1.10 to 1.26), Asians (OR=1.18 (95% CI 1.10 to 1.27) and Blacks (OR=1.10 (95% CI 0.99 to 1.22)).
Conclusions:
PRSs across multiple racial/ethnic groups were associated with increased asthma risk, suggesting that PRSs have potential as a tool for predicting disease development.
Graphical Abstract

“Polygenic risk scores (PRSs) will have important utility for asthma and other chronic diseases as a tool for predicting disease incidence and sub-phenotypes. In this work, we utilized findings from a large trans-ethnic GWAS of asthma to compute a PRS for asthma with relevance for ethnically diverse populations.”
INTRODUCTION
Asthma affects over 350 million individuals globally and is associated with both significant health care costs and reduced quality of life. 1 The genetic contribution to asthma risk is known to be quite large, with heritability estimates ranging from 70 to 90%. 2,3 While numerous genome wide association studies (GWAS) of asthma have been conducted, 4,5 few studies 6,7 attempt to transform these findings into a summary measure (i.e. a risk score) that captures an individual’s genetic risk for asthma. Polygenic risk scores (PRSs) have great potential as a clinical tool for predicting disease development8,9 and disease sub-phenotypes 10, as a prognostic indicator, 10 and as an overall index of genetic risk for use in gene by environment interaction studies. Increased visits to physicians for individuals who are more likely to develop asthma could prevent morbidity from asthma by being able to initiate treatment earlier. Furthermore, knowledge of which individuals with asthma are most likely to have severe disease could allow earlier treatment with controller medications.
Development of a PRS presents several challenges. First, the high dimensionality of genome wide association study (GWAS) findings must be dealt with, and correlation patterns for individual variants associated with the disease outcome must be accounted for. Furthermore, the underlying GWAS studies that inform PRS calculations must be sufficiently powered to identify variants that contribute to the chronic disease phenotype of interest. Another critical issue is the lack of genetic association studies in minority populations, which in turn hinders the development of PRSs that include key genetic variants specific to these populations. Although this issue is gradually improving, it still represents an important challenge for the development of accurate PRSs, particularly since the most understudied populations (Blacks and Hispanics) are at greater risk for developing many chronic diseases, including asthma. Our aim was to compute a multiancestry PRS for asthma, utilizing the findings from the largest multiancestryGWAS study conducted thus far by Demenais et al. 11, that would be relevant for Non-Hispanic Whites, Asians, Hispanics and Blacks.
METHODS
Overview
In order to manage the high dimensionality of the original GWAS used to inform our PRS calculations, we took two approaches (Figure 1). First, we calculated a PRS using only genome-wide significant variants for use in the computation. Next, we used a lasso sum regression approach, which allows the potential for all of the genetic variants to contribute to the PRS. We validated PRS parameters and tested the association of the PRS with asthma using data from the Kaiser Permanente Northern California Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort, a large racially diverse cohort.12
Figure 1.
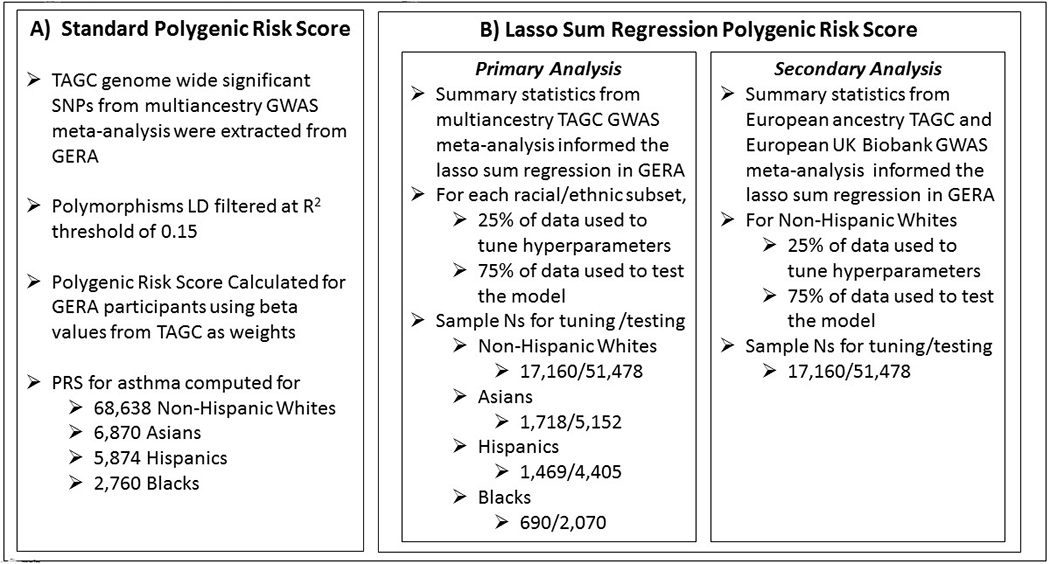
Overview of Methodology
Study populations and phenotyping
We used data from the Kaiser Permanente Northern California (KPNC) Genetic Epidemiology Research on Adult Health and Aging Cohort (GERA) cohort. 12–14 The Research Program on Genes, Environment, and Health (RPGEH) GERA cohort includes electronic medical record and genotype information for adult male and female members of the Kaiser Permanente Medical Care Plan. Sample collection and genotyping of the GERA population (dbGaP: phs000674.v1.p1) were previously described. 12–14 We searched electronic medical records and survey information from the GERA population to select adult subjects of at least 21 years of age. Data on participant’s race/ethnicity was based on self-report or derived from KP electronic medical records if self-reported race was missing in survey data. We defined asthma cases as subjects with one or more of the following electronic medical record entries: physician-diagnosed asthma, self-reported asthma, or a report of an asthma exacerbation (hospitalization, ICU/emergency department visit due to asthma); in contrast, controls (individuals without asthma) had negative reports (“0” or “No/None”) for all of the aforementioned records. This case definition has been used previously in our multiancestry GWAS of asthma in GERA. (13) We excluded subjects with the following comorbidities: chronic obstructive pulmonary disease (COPD), pulmonary embolism, primary pulmonary hypertension, cystic fibrosis, and bronchiectasis. Of all the GERA subjects with available genotyping data, asthma case status information, and relevant covariates, 14,755 asthma cases and 53,883 controls were of Non-Hispanic White race/ethnicity, 670 cases and 2,090 controls that were Black, 1,446 cases and 4,428 controls of Hispanic race/ethnicity and 1,425 cases and 5,445 controls of Asian race/ethnicity. The Kaiser Permanente Northern California Institutional Review Board (study #CN-13-1643-H) and the University of California San Francisco Human Research Protection Program Committee on Human Research (study #13–12476) approved this research project. All participants provided written informed consent. This study conforms to the standards of the Declaration of Helsinki.
Genotyping, Quality Control and Imputation
Genotyping was performed using race/ethnicity specific Affymetrix Axiom arrays as previously described.15,16 After performing quality control (QC), approximately 94% of samples and more than 98% of genetic markers passed, with an initial genotyping call rate ≥ 97%, allele frequency difference ≤ 0.15 between males and females for autosomal markers, and genotype concordance rate > 0.75 across duplicate genetic markers. PLINK v.1.94 (https://www.cog-genomics.org/plink2) was used to perform additional QC procedures. Imputation for GERA has been previously described elsewhere.12,13
Standard Polygenic risk score
We computed a PRS for asthma for subjects across all racial/ethnic groups, using genome wide significant polymorphisms identified from the Trans-National Asthma Genetic Consortium (TAGC) from Demenais et al.11 Using National Human Genome Research Institute (NHGRI), we accessed the publically available list of over 800 “multiancestry” genetic polymorphisms (at 18 loci) that were genome wide significant in the Demenais et al.11 We extracted genotyping data on these polymorphisms for GERA subjects. We LD (linkage disequilibrium) filtered the extracted polymorphisms using an R squared threshold of 0.15. Following LD filtering, 41 polymorphisms were remaining. We created a PRS for each participant by using the beta values (published in Demenais et al. 11) as weights. In our PRS calculation we accounted for the number of risk alleles an individual has. We chose to always model the risk allele (which was not necessarily the minor allele). The PRS was calculated as follows using genotype data for m SNPs, based on estimated SNP effect sizes (βj) from GWAS summary data,
where xij is the genotype for the ith individual and the jth SNP (encoded as 0,1, or 2). Using PROC LOGISTIC in SAS statistical software, we computed odds ratios for asthma with a standard deviation increase in PRS. We also performed comparisons relating odds of asthma across deciles of the PRS (vs. the 5th decile). We performed logistic regression within each strata of race/ethnicity, and adjusted our models for age, sex and body mass index (BMI). We computed the ROC AUC (area under the curve) for predicting asthma case status using the standard PRS.
Lasso Sum Regression Polygenic risk score
Primary analysis.
In addition to deriving a PRS based on a priori polymorphisms (as described above), we also computed the PRS using Lasso Sum Regression (where all SNPs (single nucleotide polymorphisms) have the potential to contribute to the PRS). Lasso Sum Regression uses summary statistics and an LD reference panel in a penalized regression framework to derive the PRS. To implement lasso sum regression, we used the R package lassosum, 17 and specified linkage disequilibrium blocks using LD regions as previously defined 18 for the European, Hispanic, East Asian and African American populations. The lassosum algorithm identifies the optimal values for parameters l and s by maximizing the correlation of the PRS with the validation phenotype data. We used summary statistics from the multiancestry GWAS of asthma in Demenais et al. to inform the lasso sum regression. For each racial/ethnic subset, we used 25% of the data to tune the hyperparameters of the lasso sum, and the remaining 75% to test the model. We computed odds ratios for asthma with a standard deviation increase in lasso-sum derived PRS and adjusted our models for age, sex and BMI using PROC LOGISTIC in SAS statistical software. We computed the ROC AUC (area under the curve) for predicting asthma case status using the lasso sum derived PRS.
Secondary analysis.
In order to enhance the potential for rare variants to contribute to the PRS, we conducted a secondary lasso sum regression analysis in Non-Hispanic White participants, that was trained on a much larger population. For this secondary analysis, we expanded the population included in the GWAS that informs the lasso sum regression PRS, by combining the largest published GWAS of asthma in Non-Hispanic Whites (Demenais et al.) with a GWAS of asthma in Non-Hispanic White participants from the UK biobank. Specifically, we conducted a GWAS of asthma on 401,837 Non-Hispanic White subjects in the UK Biobank 19 (https://www.leelabsg.org/resources) (26,332 cases and 375,505 controls), adjusted for sex, birth year, and the first four genotype principal components. Asthma was defined using a phenotypic scheme called PheCodes 20 which incorporate ICD-9 codes for asthma in the UK biobank.19 We meta-analyzed results from this GWAS with the Non-Hispanic White GWAS results from Demenais et al. (TAGC consortium). Summary statistics from this GWAS were used to inform our lasso sum regression. This GWAS meta-analysis identified 45 independent loci associated with asthma diagnosis (compared to 24 in Demenais et al. alone and 27 in UK biobank alone), and SNP based heritability (h2) as calculated by LDSC (LD score regression) was 3%. We conducted lasso sum regression using the R package lassosum,17 and specified linkage disequilibrium blocks using LD regions as previously defined 18 for the European population and the hg19 genome. As described above, GERA subjects were split into two analytical data sets for tuning (25%) and testing (75%) of the lasso sum regression model.
RESULTS
GERA Subject Characteristics
The majority of GERA subjects (82%, N=68,638) were of Non-Hispanic White race/ethnicity and the remaining 18% were minorities. Minority subjects comprised of 8% Asians (N=6,870), 7% Hispanics (N=5,874), and 3% (N= n=2,760) Blacks. Prevalence of asthma was 22% in Non-Hispanic Whites, 21% in Asians, 25% in Hispanics and 24% in Blacks. Across all racial/ethnic groups, subjects with asthma had higher BMI, were more likely to be female, and were more likely to have co-morbidities including allergic rhinitis, acute upper respiratory infections, and lower respiratory infections (Table 1).
Table 1.
Characteristics of GERA Subjects
| Variable | Non-Hispanic Whites (N=68,638) | Asians (N=6,870) | Hispanics (N=5,874) | Blacks (N=2,760) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Asthma Cases | Non-Asthmatic Controls | P Value | Asthma Cases | Non-Asthmatic Controls | P Value | Asthma Cases | Non-Asthmatic Controls | P Value | Asthma Cases | Non-Asthmatic Controls | P Value | |
|
|
||||||||||||
| Sample Size (N) | 14,755 | 53,883 | 1425 | 5445 | 1446 | 4428 | 670 | 2090 | ||||
|
| ||||||||||||
| Age in yrs. Mean (range) | 63.7 (18–90) | 64.6 (18–90) | <0.001 | 54.5 (19–89) | 54.8 (19–89) | 0.35 | 54.3 (19–90) | 54.3 (19–88) | 0.88 | 56.6 (19–90) | 58 (19–90) | 0.02 |
| % Male | 36% | 43% | <0.001 | 39% | 42% | 0.02 | 31% | 40% | <0.001 | 34% | 45% | <0.001 |
| BMI mean (range) | 28.5 (13–71) | 27 (12–77) | <0.001 | 25.5 (13–54) | 24.2 (15–50) | <0.001 | 29.2 (14–58) | 27.5 (16–75) | <0.001 | 30.6 (14–58) | 28.7 (17–57) | <0.001 |
| Comorbidities: | ||||||||||||
| Allergic rhinitis | 33% | 16% | <0.001 | 40% | 19% | <0.001 | 42% | 21% | <0.001 | 38% | 19% | <0.001 |
| Congestive Heart Failure | 3% | 2% | <0.001 | 2% | 1% | 0.001 | 2% | 1% | 0.21 | 4% | 3% | 0.36 |
| GERD | 25% | 17% | <0.001 | 16% | 12% | 0.001 | 28% | 18% | <0.001 | 26% | 17% | <0.001 |
| Acute respiratory infections | 52% | 37% | <0.001 | 49% | 34% | <0.001 | 59% | 44% | <0.001 | 54% | 39% | <0.001 |
| Chronic upper respiratory infections | 18% | 10% | <0.001 | 14% | 8% | <0.001 | 18% | 10% | <0.001 | 21% | 9% | <0.001 |
| Allergies | 71% | 51% | <0.001 | 75% | 55% | <0.001 | 74% | 48% | <0.001 | 71% | 46% | <0.001 |
Standard Polygenic Risk Score
Forty one genetic variants derived from the Demenais et al. multiancestry GWAS of asthma that remained after LD pruning (R2=0.15) are shown in Table 2. Odds ratios (taken from Demenais et al.) for individual variants ranged from 1.05 to 1.16. Many previously identified asthma loci, including IL18R1, TSLP, IL13, IL33, and GSDMA are represented in this set of gene variants. After computing a weighted PRS for each individual in the GERA population, we determined the mean and standard deviation of the PRS within each racial ethnic group. For Non-Hispanic Whites, the mean and standard deviation were (3.61 ± 0.51), for Asians (3.77 ± 0.44), for Hispanics (3.65 ± 0.47) and for Blacks (3.72 ± 0.46). Results of adjusted logistic regression models, showing associations of the PRS (population specific standard deviation increase) with odds of asthma are shown in Table 3. Non-Hispanic Whites showed the highest odds ratio for a standard deviation increase in PRS for the asthma outcome OR=1.16 (95% CI 1.14 to 1.18). Odds ratios were somewhat lower for the Asian and Hispanic populations, but were still statistically significant. In Black subjects, we observed a trend toward increased odds of asthma with a standard deviation increase in PRS, that did not reach statistical significance at p<0.05. Unadjusted models showed similar findings to those presented in Table 3, (Non-Hispanic White unadjusted OR=1.16 (95% CI 1.14 to 1.18), Asian unadjusted OR=1.08 (95% CI 1.02 to 1.15), Hispanic unadjusted OR=1.13 (95% CI 1.06 to 1.19), Black unadjusted OR=1.05 (95% CI 0.97 to 1.15). In Non-Hispanic White subjects we detected an interaction of PRS by sex (p=0.003); with male subjects showing the greatest association of the PRS with asthma case status (OR=1.20, 95% CI 1.17 to 1.25 for males; OR=1.14, 95% CI 1.11 to 1.26 for females). We did not observe any evidence of interaction between sex and PRS for asthma case status in the minority populations (p>0.05), perhaps due to lower statistical power. We computed Area under the receiver operating curve (ROC AUC) values for classifying individuals as asthma cases for each of the racial/ethnic populations. The ROC AUC was 0.54 for Non-Hispanic Whites, 0.52 for Asians, 0.53 for Hispanics and 0.51 for Blacks. We also plotted odds of asthma for deciles of the standard PRS (with 5th decile as the reference), as shown in Figure 2. A clear dose-response pattern across multiple deciles was demonstrated for Non-Hispanic Whites, which were by far the largest grouping of participants studied. We did not observe a clear dose-response across deciles for the other races. However, in Asians, associations for the highest and lowest PRS deciles suggested these extremes were linked to increased and decreased asthma risk, respectively. For Non-Hispanic Whites, the 1st decile of the PRS vs. the 5th decile showed a protective association (OR=0.81 (95% CI 0.75 to 0.89)), whereas the 7th, 8th, 9th, and 10th deciles were all significantly associated with increased risk. (The largest odds ratio was for the 10th decile vs. the 5th, OR=1.39 (95% CI 1.28 to 1.51). In Hispanics, the 10th decile vs. the 5th was associated with increased asthma risk (OR=1.62, (95% CI 1.25 to 2.11)). In Asians, we observed a protective association for the lowest decile of the PRS (OR=0.75 (95% CI 0.57 to 0.99)).
Table 2.
GWAS variants (from Demenais et al) used for computing standard polygenic risk score in GERA
| Gene/Nearest Gene | Chr | SNP | Risk Allele | Beta | Odds Ratio |
|---|---|---|---|---|---|
| IL1R1 | 2 | rs2160227 | G | 0.076 | 1.08 |
| IL18R1 | 2 | rs1921622 | A | 0.084 | 1.09 |
| IL18R1 | 2 | rs6710528 | C | 0.087 | 1.09 |
| SLC9A4 | 2 | rs17027258 | G | 0.092 | 1.10 |
| SLC25A46 | 5 | rs13018263 | T | 0.098 | 1.10 |
| SLC25A46 | 5 | rs244923 | T | 0.120 | 1.13 |
| TSLP | 5 | rs10056340 | G | 0.082 | 1.09 |
| TSLP | 5 | rs1837253 | C | 0.146 | 1.16 |
| WDR36 | 5 | rs6594499 | C | 0.099 | 1.10 |
| CAMK4 | 5 | rs10057913 | A | 0.096 | 1.10 |
| C5orf56 | 5 | rs2070729 | A | 0.071 | 1.07 |
| IL13 | 5 | rs1295686 | T | 0.111 | 1.12 |
| SEPT8, SOWAHA | 5 | rs30513 | T | 0.112 | 1.12 |
| GNPDA1, NDFIP1 | 6 | rs12655443 | A | 0.070 | 1.07 |
| ZKSCAN3 | 6 | rs6922111 | T | 0.079 | 1.08 |
| PBMUCL2, C6orf15 | 6 | rs3130955 | A | 0.073 | 1.08 |
| CDSN | 6 | rs3094216 | G | 0.078 | 1.08 |
| HLA-B | 6 | rs2442719 | T | 0.067 | 1.07 |
| MICA, HCP5 | 6 | rs2596464 | C | 0.089 | 1.09 |
| MICB | 6 | rs2855812 | T | 0.097 | 1.10 |
| AGPAT1 | 6 | rs1061808 | G | 0.072 | 1.07 |
| HLA-DRA | 6 | rs2239803 | T | 0.084 | 1.09 |
| HLA-DRA,-HLA-DRB5 | 6 | rs9269080 | A | 0.082 | 1.09 |
| HLA-DQB1,-HLA-DQA2 | 6 | rs6457614 | T | 0.116 | 1.12 |
| TAP2 | 6 | rs2239701 | C | 0.078 | 1.08 |
| BACH2 | 8 | rs12212193 | A | 0.079 | 1.08 |
| TPD52, ZBTB10 | 9 | rs11786704 | A | 0.077 | 1.08 |
| RANBP6, IL33 | 9 | rs386880 | T | 0.082 | 1.09 |
| RANBP6, IL33 | 9 | rs450108 | C | 0.091 | 1.10 |
| IL33 | 9 | rs12551256 | A | 0.083 | 1.09 |
| SFMBT2 | 10 | rs2589563 | T | 0.080 | 1.08 |
| C11orf30, LRRC32 | 11 | rs2155219 | T | 0.106 | 1.11 |
| STAT6 | 15 | rs167769 | T | 0.077 | 1.08 |
| RORA | 15 | rs2279292 | T | 0.104 | 1.11 |
| SMAD3 | 15 | rs12708492 | T | 0.085 | 1.09 |
| CLEC16A | 16 | rs17673553 | A | 0.097 | 1.10 |
| FBXL20 | 17 | rs4795355 | C | 0.082 | 1.09 |
| IKZF3 | 17 | rs3816470 | A | 0.165 | 1.18 |
| GSDMA | 17 | rs7212938 | G | 0.149 | 1.16 |
| MED24 | 17 | rs8065443 | A | 0.095 | 1.10 |
| ZNF652, PHB | 17 | rs17637472 | A | 0.078 | 1.08 |
Table 3.
Odds ratios (95% CI) for Polygenic Risk Score and Asthma in GERA*
| Non-Hispanic Whites (N=51,478) | Asians (N=5,152) | Hispanics (N=4,405) | African Americans (N=2,070) | |
|---|---|---|---|---|
| Standard PRS | 1.16 (1.14 to 1.18) | 1.10 (1.04 to 1.17) | 1.12 (1.05 to 1.19) | 1.05 (0.97 to 1.15) |
| Lasso Sum PRS (Primary Analysis) | 1.20 (1.18 to 1.23) | 1.18 (1.10 to 1.27) | 1.17 (1.10 to 1.26) | 1.10 (0.99 to 1.22) |
| Lasso Sum PRS (Secondary Analysis) | 1.28 (1.25 to 1.31) | --- | -- | -- |
Odds ratios are for a standard deviation increase in PRS within a given population. All models have been adjusted for age, sex and BMI
Figure 2.
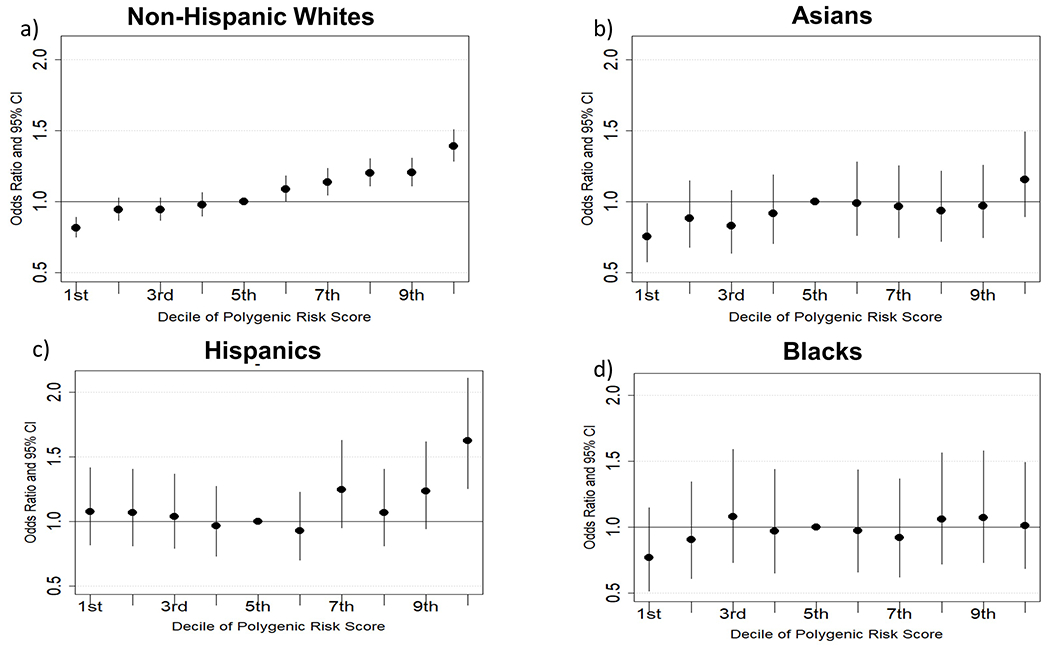
Odd ratios (95% Cis) for asthma with each decile of the polygenic risk score (5th decile as the reference) for Non-Hispanic Whites a), Asians b) Hispanics c) and Blacks d).
In a sensitivity analysis, we derived a standard PRS using only results from the Demenais et al. European meta-analysis. Odds ratios and ROC AUCs for the polygenic risk score were very similar to the results for the multiancestry standard PRS (data not shown).
Lasso Sum Polygenic Risk Score
Primary Analysis.
We report the lasso sum regression PRSs calculated for each racial/ethnic group in Table 3. We observed slightly increased odds ratio estimates for Non-Hispanic Whites, Asians, Hispanics and Blacks using lasso sum regression. ROC AUCs for prediction of asthma case status based on the lasso sum regression PRS were as follows for Non-Hispanic Whites (0.55), Asians (0.54), Hispanics (0.55) and Blacks (0.53). SNPs included in the final lasso sum regression models, along with beta values for each SNP are available in Supplementary Files. Parameters for l and s were 0.004 and 0.9, respectively.
Secondary analyses.
We used results of our meta-analysis of the European asthma GWAS from Demenais et al. (19,954 asthma cases and 107,715 controls) and an asthma GWAS performed in the UK Biobank (26,332 asthma cases and 375,505 controls) to inform an asthma PRS lasso sum regression model in Non-Hispanic White GERA subjects. This meta-analysis identified 45 significant loci associated with asthma, compared to 24 from Demenais et al. alone and 27 in the UK Biobank GWAS alone. A Manhattan plot of this meta-analysis is shown in figure 3. After testing and validating our lasso sum regression model we identified 405,088 SNPs across the genome that contributed to the best PRS. In logistic regression models, a standard deviation increase in the lasso sum derived PRS was associated with an OR=1.28 (95% CI 1.25 to 1.31) in models adjusted for age, sex and BMI (table 3). We did not detect a sex by PRS interaction in models for asthma using the lasso sum derived PRS (p=0.93 for interaction term). The ROC AUC for the lasso sum derived PRS for asthma in Non-Hispanic Whites was 0.5724, which was significantly higher than the ROC AUC for the standard PRS of 0.54 (p<0.001 for comparison). SNPs included in the final lasso sum regression model, along with beta values for each SNP are available in Supplementary Files.
Figure 3. Asthma GWAS Meta-analysis Results for TAGC and UK Biobank Populations.
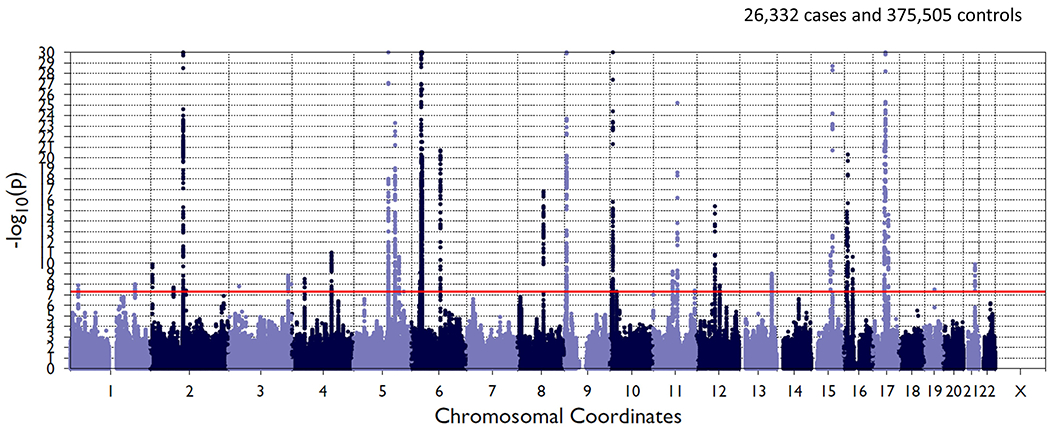
Manhattan plot of meta-analysis results is shown with chromosal position along the abscissa and −log(p-value) on the ordinate axis. Red horizontal line indicates the significance threshold for genome-wide significance.
DISCUSSION
In this study, we derived a PRS for asthma in a large racially diverse population using two different approaches, a standard PRS calculation based on 41 known GWAS variants, and a lasso sum regression technique that identified hundreds of thousands of genetic variants with the potential to contribute to asthma risk. We report three main findings. First, our standard PRS was associated with asthma risk in Non-Hispanic Whites, Hispanics and Asians. Second, in Non-Hispanic Whites our standard PRS showed stronger associations with asthma in males as compared to females. Third, the predictive ability of the lasso sum regression PRS showed slight improvements over the standard PRS in Non-Hispanic White subjects.
Our use of multiancestry GWAS data for the basis of a PRS represents an important stride forward, given that past PRSs for asthma phenotypes have been informed by GWAS in Non-Hispanic Whites alone. 21 The importance of GWAS in minority populations is now well-recognized, especially given our knowledge that critical disease-associated variants may be missed if they are absent or of low frequency in populations of European ancestry. 22 In fact, a simulation study conducted by Cavazos et al. demonstrated that inclusion of GWAS results from minority populations increases both the accuracy of the PRS and its ability to predict disease risk equitably across diverse populations. 23 Inclusion of loci from the largest multiancestry GWAS of asthma instead of its European-only subset increased the number of genetic variants considered (Demenais et al.)11 in our derivation of the PRS. We observed increased odds of asthma with higher PRSs for Hispanics and Asians in GERA, however, associations were strongest for Non-Hispanic White subjects. This finding is likely due to the fact that the populations forming the basis of our PRS weighting scheme (Demenais et al.)11 and testing data set (GERA) both had an over-representation of Non-Hispanic Whites (thereby potentially weighing the PRS more towards genetic variants in Non-Hispanic Whites, and enhancing power to test the relationship of the PRS with asthma in a Non-Hispanic White population). In fact, results for our calculation of the standard PRS based on genome wide significant variants from the European meta-analysis from Demenais et al. were very similar to the standard PRS based on the multiancestry meta-analysis. Larger genome wide association studies of asthma in minority populations will be an important factor for further improvement in accuracy of a multiancestry PRS for asthma.
Interestingly, we observed a stronger association of the standard PRS in males as compared to females. This difference may have its origins in sex-specific asthma phenotype variations. Males are at higher risk of childhood onset asthma, 24 which is known to have a larger genetic component.25 On the other hand, females are more likely to develop adult onset asthma, 24 which is believed to be more environmental in origin. Sex-specific variation in age of onset within the GERA cohort may have contributed to this interaction. However, given that these subjects were enrolled as adults, we did not have access to accurate information on age of asthma onset. It is also important to note that the original multiancestry GWAS from Demenais et al. may have an over-representation of genetic variants associated with childhood onset asthma (27 of the 66 GWAS studies in Demenais et al. were conducted in pediatric populations). (In fact, in the Demenais et al. study, it was shown that a number of loci were specifically associated with childhood but not adult asthma). When we expanded the number of genetic loci for our lasso sum regression models, to include not only the Demenais et al. GWAS summary statistics but also results from a much larger GWAS of asthma in adults, associations of the PRS with asthma were similar in men and women.
Two independent predictors of asthma in adults, sex and BMI, did not confound the association between the PRS (either the standard derived score or the lasso sum regression score) and asthma risk. In Non-Hispanic Whites, comparing odds of asthma in the 10th decile of the standard PRS vs. 5th decile revealed an odds ratio of 1.4. This increase in risk is similar in magnitude to the increase in adult asthma risk associated with being overweight (OR=1.38, 95% CI 1.17 to 1.62 vs. normal weight) 26. The fact that associations between our PRS and asthma show comparable magnitude as compared to other risk factors, even after adjustment for other risk factors, demonstrates that the PRS has and important and unique contribution to adult asthma risk.
In addition to calculating the PRS with select genome wide significant variants (the “standard” PRS), we also used an updated methodology, lasso sum regression, to compute the PRS in GERA. The lasso sum regression technique has several advantages of the standard PRS calculation; 1) it accounts for LD patterns in the genotype data (i.e. prior LD pruning is unnecessary) 2) it allows the potential of all GWAS variants to contribute to the PRS (not just the genome wide significant variants) and 3) more optimal SNP “weights” are calculated (a priori beta values from GWAS summary statistics are used as weights). We found that, as compared to the standard PRS, the lasso sum regression derived PRS showed a small increase in terms of predictive ability. The improved performance of the lasso sum derived PRS as compared to the standard score likely reflects several factors: a greater number of genetic variants were included, variants not meeting genome wide significance contributed to the score, a larger GWAS with a higher fraction of adult subjects was used to inform the PRS calculation, and the contribution of GWAS variants to the PRS was optimized by a tuning parameter in the lasso sum regression model. Given its improved performance as compared to the standard PRS, we conclude that this updated technique might be advantageous in a research setting, and, down the line in a clinical setting. With the rapid decline in the cost of genotyping millions of SNP variants, identification of an individual’s PRS using this high-dimensional technique could be feasible in a clinical setting.
While all models demonstrated that the PRS is associated with increased odds of adult asthma, the modest ROC AUCs for the PRS indicate that its ability to discriminate between asthma cases and controls is limited. Factors that may limit the predictive ability of the PRS include the following: 1) asthma phenotype heterogeneity in both the external GWAS that informed our study, as well as in the GERA cohort itself, may diminish observed genetic associations that inform the PRS 2) lack of data on age of asthma onset in GERA, which could have helped us refine the PRS 3) the absence of exposure contributions that may interact with the PRS and/or SNP by SNP interactions within the PRS calculation itself. Future studies that base PRS calculations on GWAS of molecular phenotypes of asthma may show enhanced prediction of asthma case status for these well-defined phenotypes. Ultimately, the most clinically meaningful PRSs for asthma phenotypes will be those that predict asthma medication responses.
A study on derivation of a genetic risk score for asthma, in two pediatric cohorts, did not uncover any predictive utility for a multiancestry GWAS-variant (“standard”) polygenic risk score. 27 This study also leveraged the TAGC consortium findings to derive weights for the polygenic risk score; however, it did not leverage a technique that allows the potential for all genetic variants to contribute to the score calculation. This could be why our score shows modest predictive accuracy (as compared to no predictive utility at all). Two recent studies using alterative high-dimensional PRS derivation methods (LDpred and EB-PRS) for asthma risk also allowed all genetic variants to potentially contribute to the score. 6,7 These studies, both conducted in populations of European ancestry, found very similar (modest) predictive accuracy levels when compared to our lasso sum regression results.
Our study had several strengths, including the use of a multiancestry GWAS, rather than a Non-Hispanic White GWAS alone, to inform a PRS for asthma. We were able to test our PRSs in a large racially diverse population. Lastly, we were able to test two different approaches for PRS derivation in our work. One important weakness of our study also deserve mention. Minority participants, while included in our work, were not as large as the fraction of Non-Hispanic Whites which may have reduced our power to detect associations. In Non-Hispanic White participants, we had the opportunity to conduct an additional lasso sum regression analysis with summary data from over five hundred thousand individuals. Unfortunately, we did not have access to additional minority populations to perform a similar analysis in these racial/ethnic groups. An additional potential weakness is that the slight increases in effect sizes and ROC AUCs for the lasso sum regression vs. the standard PRS approach may simply be due to inclusion of additional variants in the model and may not necessarily reflect superior methodology.
In summary, we leveraged data from a large racially diverse cohort to derive a PRS associated with increased odds of asthma in adults. Overall, the PRS had limited accuracy for predicting asthma case vs. control status. Future work will focus on PRSs for asthma-associated phenotypes (lung function, airways hyper-responsiveness, asthma/COPD overlap), and genetic scores for asthma treatment responses.
Supplementary Material
Key Messages:
We used two methods to develop polygenic risk scores for asthma in a racially diverse population.
Standard polygenic risk scores were associated with asthma risk in Non-Hispanic White, Hispanic and Asian people.
Polygenic risk scores developed using lasso sum regression showed stronger associations in all groups
Funding:
Funded by National Institutes of Health (NIH) grants: R01HD085993, K01HL125858 This research has been conducted using the UK Biobank Resource under application number 20915 (M.H.C.). Sponsors (NIH) did not have any role in development and conduct of the research.
Conflict of Interest Statement:
MHC has received consulting fees from Genentech and grant support from GSK. All other authors report no conflicts of interest.
Abbreviation List:
- PRS
Polygenic Risk Score
- GWAS
Genome wide association study
Data Availability:
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
- 1.GBD 2015 Chronic Respiratory Disease Collaborators. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med. 2017;5:691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Backer V. Estimates of asthma heritability in a large twin sample. Clin Exp Allergy. 2010;40:1054–61. [DOI] [PubMed] [Google Scholar]
- 3.McGeachie MJ, Stahl EA, Himes BE, Pendergrass SA, Lima JJ, Irvin CG, et al. Polygenic heritability estimates in pharmacogenetics: focus on asthma and related phenotypes. Pharmacogenet Genomics. 2013;23:324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ober C Asthma Genetics in the Post-GWAS Era. Ann Am Thorac Soc. 2016;13 Suppl 1:S85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim KW, Ober C. Lessons Learned From GWAS of Asthma. Allergy Asthma Immunol Res. 2019;11:170–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Z, Hasegawa K, Ma B, Fujiogi M, Camargo CA, Liang L. Association of asthma and its genetic predisposition with the risk of severe COVID-19. J Allergy Clin Immunol. 2020;146:327–329.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song S, Jiang W, Hou L, Zhao H. Leveraging effect size distributions to improve polygenic risk scores derived from summary statistics of genome-wide association studies. PLoS Comput Biol. 2020;16:e1007565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pazoki R Methods for Polygenic Traits. Methods Mol Biol. 2018;1793:145–56. [DOI] [PubMed] [Google Scholar]
- 9.Ho DSW, Schierding W, Wake M, Saffery R, O’Sullivan J. Machine Learning SNP Based Prediction for Precision Medicine. Front Genet. 2019;10:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet. 2019; [DOI] [PubMed] [Google Scholar]
- 11.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmüller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kvale MN, Hesselson S, Hoffmann TJ, Cao Y, Chan D, Connell S, et al. Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banda Y, Kvale MN, Hoffmann TJ, Hesselson SE, Ranatunga D, Tang H, et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapham K, Kvale MN, Lin J, Connell S, Croen LA, Dispensa BP, et al. Automated Assay of Telomere Length Measurement and Informatics for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann TJ, Kvale MN, Hesselson SE, Zhan Y, Aquino C, Cao Y, et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann TJ, Zhan Y, Kvale MN, Hesselson SE, Gollub J, Iribarren C, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mak TSH, Porsch RM, Choi SW, Zhou X, Sham PC. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol. 2017;41:469–80. [DOI] [PubMed] [Google Scholar]
- 18.Berisa T, Pickrell JK. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50:1335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belsky DW, Sears MR, Hancox RJ, Harrington H, Houts R, Moffitt TE, et al. Polygenic risk and the development and course of asthma: an analysis of data from a four-decade longitudinal study. Lancet Respir Med. 2013;1:453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavazos TB, Witte JS. Inclusion of variants discovered from diverse populations improves polygenic risk score transferability. HGG Adv. 2021;2:100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dharmage SC, Perret JL, Custovic A. Epidemiology of Asthma in Children and Adults. Front Pediatr. 2019;7:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira MAR, Mathur R, Vonk JM, Szwajda A, Brumpton B, Granell R, et al. Genetic Architectures of Childhood-and Adult-Onset Asthma Are Partly Distinct. Am J Hum Genet. 2019;104:665–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beuther DA, Sutherland ER. Overweight, obesity, and incident asthma: a meta-analysis of prospective epidemiologic studies. Am J Respir Crit Care Med. 2007;175:661–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dijk FN, Folkersma C, Gruzieva O, Kumar A, Wijga AH, Gehring U, et al. Genetic risk scores do not improve asthma prediction in childhood. J Allergy Clin Immunol. 2019;144:857–860.e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


