Abstract
Background
Effectively utilizing disease-relevant text information from unstructured clinical notes for medical research presents many challenges. BERT (Bidirectional Encoder Representation from Transformers) related models such as BioBERT and ClinicalBERT, pre-trained on biomedical corpora and general clinical information, have shown promising performance in various biomedical language processing tasks.
Objectives
This study aims to explore whether a BERT-based model pre-trained on disease-related clinical information can be more effective for cerebrovascular disease-relevant research.
Methods
This study proposed the StrokeBERT which was initialized from BioBERT and pre-trained on large-scale cerebrovascular disease related clinical text information. The pre-trained corpora contained 113,590 discharge notes, 105,743 radiology reports, and 38,199 neurological reports. Two real-world empirical clinical tasks were conducted to validate StrokeBERT’s performance. The first task identified extracranial and intracranial artery stenosis from two independent sets of radiology angiography reports. The second task predicted the risk of recurrent ischemic stroke based on patients’ first discharge information.
Results
In stenosis detection, StrokeBERT showed improved performance on targeted carotid arteries, with an average AUC compared to that of ClinicalBERT of 0.968 ± 0.021 and 0.956 ± 0.018, respectively. In recurrent ischemic stroke prediction, after 10-fold cross-validation on 1,700 discharge information, StrokeBERT presented better prediction ability (AUC±SD = 0.838 ± 0.017) than ClinicalBERT (AUC±SD = 0.808 ± 0.045). The attention scores of StrokeBERT showed better ability to detect and associate cerebrovascular disease related terms than current BERT based models.
Conclusions
This study shows that a disease-specific BERT model improved the performance and accuracy of various disease-specific language processing tasks and can readily be fine-tuned to advance cerebrovascular disease research and further developed for clinical applications.
Keywords: natural language processing, specific language representation model, cerebrovascular disease
1. Introduction
Cerebrovascular disease is an important cause of mortality worldwide and a major source of chronic morbidity and disability, affecting 16.9 million cases in 2010.[1] Cerebrovascular disease research relies on comprehensive clinical information, including images, laboratory data, and various clinical assessments and notes. Among these, clinical notes and radiology reports contain the most rich but under-utilized information; both are classified as unstructured electronic health records (EHR) data and usually contain jargon and abbreviations with a variety of writing styles. This presents challenges to the effective extraction and mining of meaningful clinical information to help advance medical research and improve health care. Recently, identification of various health conditions, detection of certain underlying diseases, and prediction of outcomes can be improved if unstructured EHR information showed promises using natural language processing (NLP) or similar techniques. Many research efforts have been made to unleash the power of unstructured clinical data for cerebrovascular disease research. For example, Sedghi et al applied NLP techniques and support vector machines (SVM) to predict transient ischemic attacks from medical narrative descriptions.[2] Garg et al developed an automated NLP pipeline in the EHR platform, which showed an agreement with manual TOAST subtypes classification.[3] Chen et al constructed a conditional random fields model for automatically segmenting ultrasound reports of cerebrovascular disease in Chinese patients.[4]
Word representation is an essential component in NLP models. It aims to convert words into vectors for further algorithm evaluations. In comparison with fixed word representation that are content-free with no assumption about semantics and similarity of words, a contextualized word representation considers language polysemy and relationship of words, which means it is able to capture the semantics and syntactic dependencies of words in any context. BERT (bidirectional encoder representation from transformers) is a newly developed state-of-the-art language representation model that has achieved great success in performing many NLP tasks, such as named entity recognition (NER) and question answering.[5] Compared to classic word-level vector representations (i.e., word2vec[6] and GloVe[7]), BERT used one of the deep neural network architectures called bidirectional transformers to provide contextualized word representations.[8] BERT was mainly pre-trained on general domain data like Wikipedia text and the BookCorpus dataset. Many studies have shown the effectiveness of BERT’s contextualized word representations in general domain NLP tasks.[9–12] However, NLP models designed for general purpose language understanding have shown poor performance in specific-domain text mining tasks. Therefore, Lee et al pre-trained BERT using PubMed and PubMed Central data to create BioBERT. BioBERT significantly outperformed BERT in biomedical NER, relation extraction, and question answering tasks.[13] In the clinical domain, two concurrent studies pre-trained and fine-tuned BERT-based models using the MIMIC-III database.[14] Alsentzer et al demonstrated that using clinical specific contextual embeddings (i.e. ClinicalBERT) improved upon general domain results obtained from BioBERT on clinical NER and medical natural language inference tasks.[15] In the recent work of Huang et al, their fine-tuned ClinicalBERT was superior to both the bag-of-words model and the BiLSTM (bidirectional long short-term memory network) model on 30-day hospital readmission prediction when using both discharge summaries and the first few days of clinical notes.[16] In addition, Li et al studied the effectiveness of EhrBERT, which pre-trained on 1.5 million electronic health records for biomedical or clinical entity normalization, and the results showed that EhrBERT performed better than BioBERT or BERT. Their work also found that domain-based information has an impact on the performance of BERT-based models.[17]
In this study, we explored whether a BERT-based model can be more effective in improving specific clinical tasks and advance disease research after being pre-trained on the real-world evidence (RWE) based on the disease-related clinical corpora. We developed StrokeBERT, which was initialized from the previously reported BioBERT model to inherit its previous build with diverse words and knowledge from internet and PubMed literature, and we pre-trained it with additional large-scale cerebrovascular disease-related clinical notes and reports from real world hospitals. We examined the efficacy of StrokeBERT on two challenging tasks in the cerebrovascular disease research from the real-world clinical settings, they are, the detection of high-risk patients with advanced grades of extracranial and intracranial artery stenosis and the prediction of recurrent ischemic stroke.
2. Materials and Methods
2.1. Data Source
We used clinical notes and reports contained in the Chang Gung Research Database (CGRD) from 2007 to 2018, which collected multi-institutional standardized electronic medical records from the largest private hospital system, including two medical centers, two regional hospitals, and three district hospitals from northern to southern Taiwan. This database included and represented 6.1% of outpatients and 10.2% of hospitalized patients in the Taiwanese population.[18] The diagnosis codes of ischemic stroke in CGRD have been validated.[19] In this study, we retrospectively and randomly selected two cerebrovascular disease patient datasets from CGRD for BERT-based model pre-training and the downstream fine-tuning tasks. The sample sizes of these two independent datasets were 172,051 patients for stenosis detection and 7,118 patients for the prediction of recurrent stroke. For external validation of stenosis detection in the new BERT-based model, subjects were selected from those who were admitted to Kaohsiung Veterans General Hospital (KSVGH) between 07/01/2018 and 06/30/2019 with a diagnosis of acute stroke and reports of magnetic resonance angiography (MRA). To conduct this study, ethical clearances were obtained from the Linkou Chang Gung Memorial Hospital Institutional Review Board (IRB) (201501857B0C606, 201900048B0) and from the IRB of Kaohsiung Veterans General Hospital (KSVGH20-CT3-08) in Taiwan.
2.2. Pre-training StrokeBERT
Figure 1 shows the StrokeBERT pre-training procedures. We included clinical information from the records of 77,334 patients who were admitted with cerebrovascular disease (ICD-9 code: 430–438 or ICD-10 code: I60-I69) from the CGRD pre-training dataset. The patient dataset had a total of 113,590 discharge notes, 105,743 radiology reports, and 38,199 neurological reports for ultrasonography, electroencephalograms, or psychophysiological function examinations. All clinical notes were preprocessed by removing Chinese characters/sentences (used for hospital administration), special characters (used as dividing sentences/lines), and multiple spaces. The SpaCy segmentation technique[20] was used to segment each clinical note. The final pre-training corpora contained 257,532 stroke-related clinical notes with 41,002,306 words.
Figure 1.
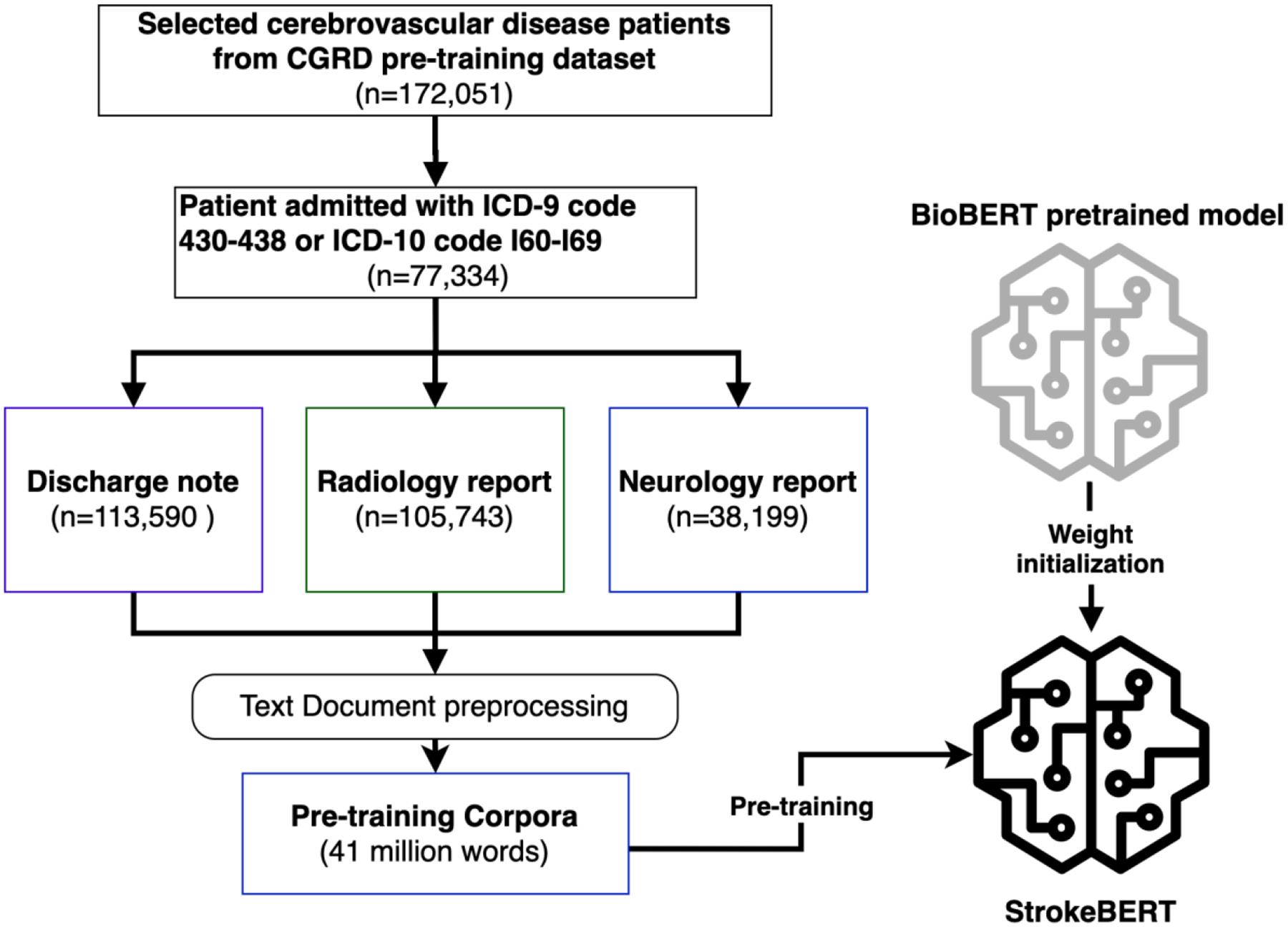
StrokeBERT pre-training process.
Taking advantage of the previously developed pre-trained models, StrokeBERT was initialized with BioBERT. For all pre-training experiments, we leverage the implementation of ClinicalBERT with Pytorch 1.0 framework [21]. The size of StrokeBERT is the same as BERTbase, which has 12 layers and each layer has 12 self-attention heads. We used the same pre-training tasks with default parameters, masked language modeling, and next sentence prediction, as in the work of Devlin et al [5], to pre-train our StrokeBERT with default parameters (e.g. masked language model probability = 0.15 and max predictions per sequence = 22). For tokenization, we used the BERT tokenizer that was based on the WordPiece algorithm[22] without lower-casing. The new model was trained for a batch size of 32, 150,000 steps with a learning rate of 5*10−5 and a maximum sequence length of 128. After pre-training, this disease-specific language representation model can be fine-tuned with one additional output layer to create task-based models such as disease identification or outcome prediction. We verified StrokeBERT’s performance in comparison with a similar pre-train model of ClinicalBERT (Bio+Clinical BERT)[15] from the medical domain corpora in the two following empirical studies. For this empirical study, the pretrain work takes about 66.8 hours on a server with 2 Intel E5-2680v4 2.4GHZ CPUs, 256GB RAM and 2 NVIDIA K80 GPUs.
2.3. Verifying the Performance of StrokeBERT in Real-world Clinical Tasks
Task I: Detection of Extracranial and Intracranial Artery Stenosis from Radiology Reports of Angiography
Carotid artery stenosis has been strongly correlated with the incidence of stroke.[23 24] In hospitals, digital subtraction angiography (DSA), computed tomographic angiography (CTA), and MRA are used to identify carotid artery stenosis and relied on the reports for final diagnosis.[25] In this empirical study, we aimed to quickly and accurately identify extracranial or intracranial artery stenosis from angiography reports using the new StrokeBERT model. There are several extracranial and intracranial artery sections mentioned in a clinical report, which can be considered as a multi-label text classification challenge. We fine-tuned StrokeBERT and ClinicalBERT[15] with one additional sigmoid output layer for comparison. For the model training, we selected TensorFlow’s sigmoid cross entropy with logits[26] as the loss function and BertAdam[27] with learning rate 5e−5 to be the optimizer; the maximum sequence length was 400; the total number of training epochs was 10 with 24 batch size. The process of fine-tuning and cross-validation is shown in Figure 2.
Figure 2.
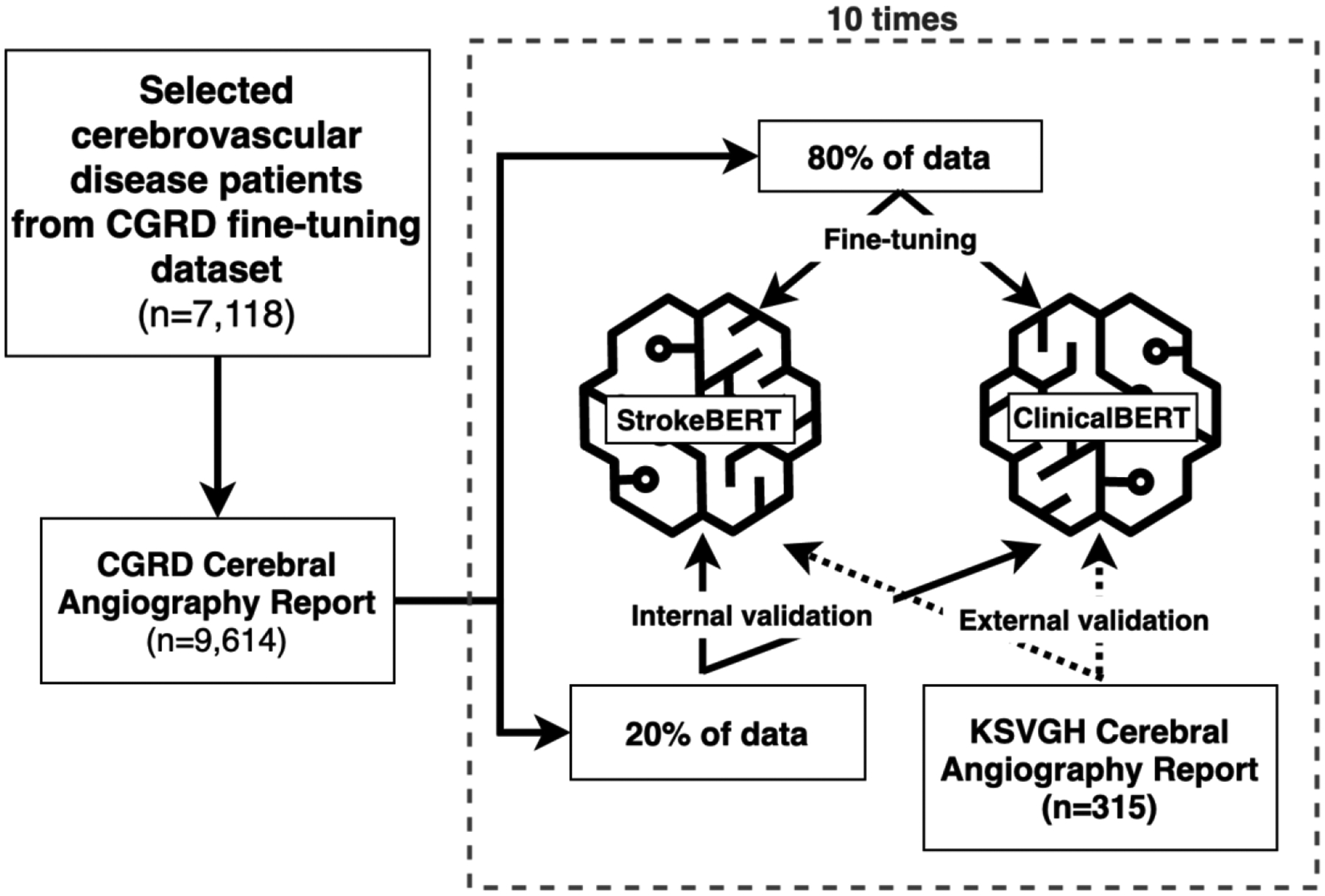
Fine-tuning process of StrokeBERT and ClinicalBERT for artery stenosis detection.
This study collected 9,614 angiography reports from 7,118 patients records with cerebrovascular diagnoses in the CGRD fine-tuning dataset. The degree of arterial stenosis was determined according to NASCET criteria.[28] To further validate the reports for our model prediction accuracy and performance, we asked two neurologists to independently label and confirm whether stenosis existed (<50% or ≥50% diameter stenosis) in 17 target arteries according to the original radiology interpreted reports. Any disagreement between the neurologists was resolved together by further discussion to build consensus. The 17 target arteries are left/right common carotid artery (LCCA, RCCA), left/right extracranial internal carotid artery (LEICA, REICA), left/right intracranial internal carotid artery (LIICA, RIICA), left/right anterior cerebral artery (LACA, RACA), left/right middle cerebral artery (LMCA, RMCA), left/right posterior cerebral artery (LPCA, RPCA), left/right extracranial vertebral artery (LEVA, REVA), left/right intracranial vertebral artery (LIVA, RIVA), and basilar artery (BA).
We used ten-times internal-external validation to evaluate the detecting capabilities of the two BERT-based models (StrokeBERT and ClinicalBERT). For each validation round, 80% of the CGRD angiography reports were used to fine-tune the BERT-based models and the other 20% were used for internal validation. To further challenge and validate the new StrokeBERT model, we included an additional 315 angiography reports from KSVGH located in southern Taiwan as an external validation dataset. The labeling process of KSVGH angiography reports was performed identically as the CGRD dataset described previously. Due to differences in the diagnosis process between the two hospital systems, the KSVGH angiography reports focused only on 11 out of 17 intracranial cerebral arteries (i.e. no LCCA, RCCA, LEICA, REICA, LEVA, and REVA). For valid comparison, we selected comparable datasets from KSVGH as an external validation dataset approved by the KSVGH IRB for this study. The model training dataset did not include the KSVGH dataset and their patients and physicians were different from those two independent hospital networks.
Task II: Investigating the Effectiveness of Recurrent Ischemic Stroke Prediction
Identification of patients at the highest risk for recurrent stroke is critical because early recurrence is associated with more severe consequences[29] and early initiation of available treatment is imperative to prevent a recurrent stroke.[30] It is known among clinicians that the discharge note is an excellent source of information that details a patient’s health conditions during the triage of the last stroke event and provides the potential to identify risks associated with recurrent stroke. This empirical study aimed to evaluate StrokeBERT’s ability in learning representations of clinical texts on the recurrent ischemic stroke prediction task.
As shown in Figure 3, we selected 3,490 patients admitted with ischemic stroke (ICD-9: 433.XX and 434.XX or ICD-10 I63.XX and I66.XX) from the CGRD fine-tuning dataset. Selected patients were classified into the recurrent ischemic stroke group and the non-recurrent ischemic stroke group. The definition of recurrent stroke in this study was that the patient was readmitted with ischemic stroke during the data collection interval described in the data sources with rehabilitation or other chronic disease admissions excluded. That is, a patient who had no admission record after discharge from the first stroke during the data collection interval was labeled as a non-recurrent ischemic stroke. A patient whose follow-up was less than the median of the recurrent stroke interval was excluded from the non-recurrent ischemic stroke dataset. After the exclusions, the recurrent stroke fine-tuning dataset contained 969 recurrent ischemic stroke patients and 731 non-recurrent ischemic stroke patients, for a total of 1,700 patients with their first discharge notes that included their chief complaint, medical history, surgery method, findings, and hospitalization. Since 2001, national accredited stroke centers have been increasingly established in medical centers of Taiwan, and CGRD data were collected among these medical centers. [31] Most of the national accredited stroke centers have a multidisciplinary team, including the outpatient department to improve acute stroke care quality. [32] One previous study showed that more than 76% of patients visited the same hospital for stroke related medical services. [33] A previous report compared the diagnosis of stroke in Taiwan’s National Health Insurance Research Database (NHIRD) with those recorded in the Taiwan Stroke Registry (TSR), a retrospective research database for stroke collected across 65 national stroke centers including CGRD, and found the positive predictive value was 88.4% with the sensitivity of 97.3%.[34]
Figure 3.
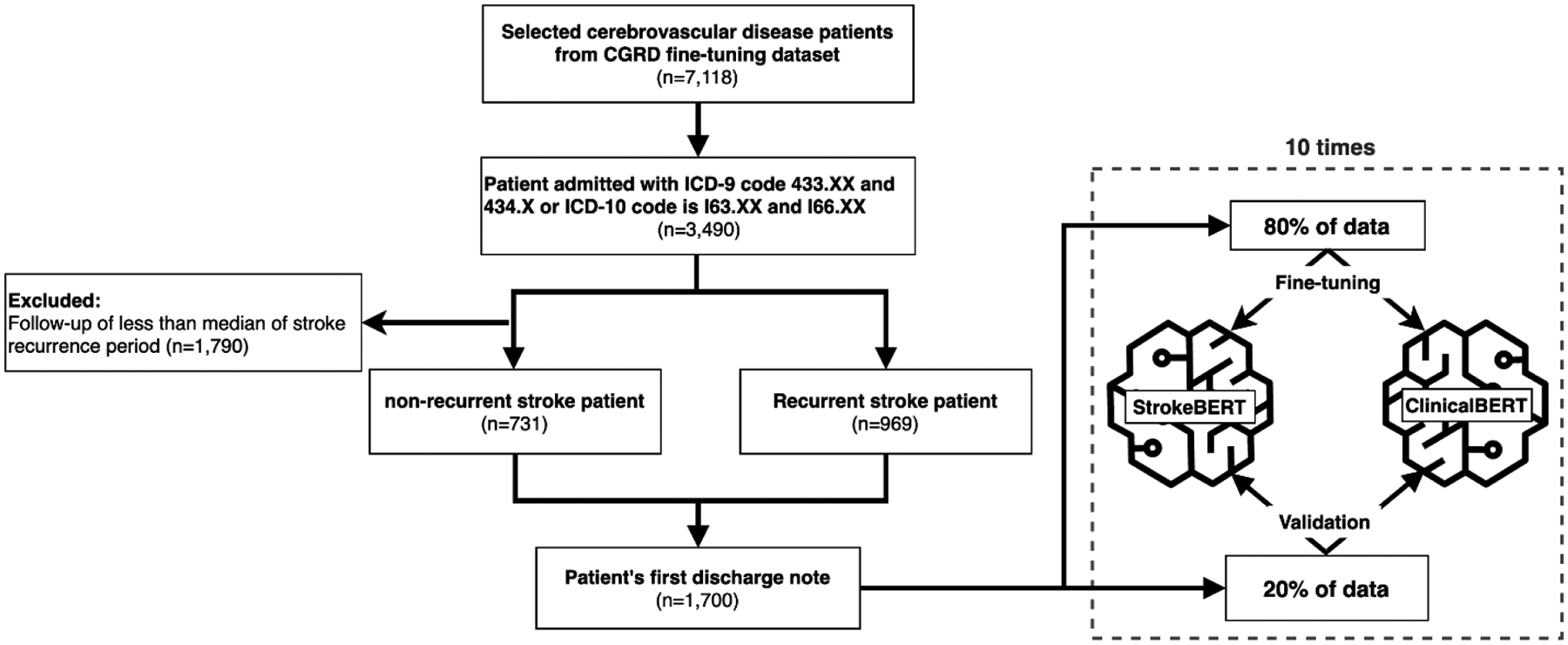
Fine-tuning process of StrokeBERT and ClinicalBERT for ischemic stroke recurrence prediction.
For valid comparison, Both StrokeBERT and ClinicalBERT were fine-tuned with one additional output layer using cross-entropy loss function.[35] The optimizer was BertAdam[27] with a learning rate of 3e−5. The maximum sequence length was 400, total number of training epochs was 10 and batch size was 24. Ten-times cross-validation was performed to assess the predictive capabilities of StrokeBERT and ClinicalBERT; 80% of data was used for BERT-based model fine-tuning, the remaining 20% was used for validation in each round (Figure 3).
3. Results
3.1. Extracting Extracranial and Intracranial Artery Stenosis Information
Table 1 depicts the performance results of StrokeBERT on identifying different targeted arteries toward stenosis. For the datasets selected from CGRD, the average AUC±SD of StrokeBERT was 0.973 ± 0.008 in intracranial arteries and 0.978 ± 0.007 in extracranial arteries. This study also performed same task using ClinicalBERT. The average AUC±SD of ClinicalBERT in intracranial arteries and in extracranial arteries were 0.971 ± 0.008 and 0.977 ± 0.007, respectively. The F1 score and area under the precision-recall curve can be found in supplementary Table S1. Overall, StrokeBERT improved ClinicalBERT in both extracranial and intracranial artery stenosis identifications, but not significantly. The different report formats and physician writing styles were observed and found to affect the results of the model being built. For example, CGRD reports use “stenosis” and “occlusion”, but KSVGH reports often used “paucity” to describe artery stenosis. The performance differences between StrokeBERT and ClinicalBERT were found slightly larger in the external validation datasets obtained from other hospitals, as expected. Despite this difference, StrokeBERT achieved better performance than other models in all intracranial arteries largely due to added clinical terminology. The average AUCs of the two BERT-based models were 0.968 ± 0.021 for StrokeBERT and 0.956 ± 0.018 for ClinicalBERT. The results also revealed that the StrokeBERT model can be trained to handle diverse words with enough variability in jargon from various data sources to allow for an accurate and precise retrieval of vital clinical information.
Table 1.
The area under the curve (AUC) of StrokeBERT in extracranial and intracranial artery stenosis detection with ten-times internal–external validation.
| Internal Validation | External Validation* | ||
|---|---|---|---|
| Intracranial | RIICA | 0.975 ± 0.006 | 0.963 ± 0.049 |
| RACA | 0.975 ± 0.009 | 0.993 ± 0.006 | |
| RMCA | 0.977 ± 0.003 | 0.989 ± 0.008 | |
| RPCA | 0.963 ± 0.016 | 0.877 ± 0.036 | |
| RIVA | 0.975 ± 0.006 | 0.995 ± 0.002 | |
| BA | 0.975 ± 0.007 | 0.901 ± 0.043 | |
| LIICA | 0.976 ± 0.007 | 0.964 ± 0.055 | |
| LACA | 0.975 ± 0.009 | 0.994 ± 0.002 | |
| LMCA | 0.974 ± 0.007 | 0.993 ± 0.004 | |
| LPCA | 0.963 ± 0.009 | 0.994 ± 0.003 | |
| LIVA | 0.973 ± 0.005 | 0.983 ± 0.019 | |
| Average | 0.973 ± 0.008 | 0.968 ± 0.021 | |
| Extracranial | RCCA | 0.969 ± 0.012 | |
| REICA | 0.987 ± 0.003 | ||
| REVA | 0.973 ± 0.006 | ||
| LCCA | 0.981 ± 0.010 | ||
| LEICA | 0.986 ± 0.003 | ||
| LEVA | 0.970 ± 0.005 | ||
| Average | 0.978 ± 0.007 |
The external validation dataset did not contain the information of extracranial carotid arteries that included left/right common carotid artery (LCCA, RCCA), left/right extracranial internal carotid artery (LEICA, REICA), left/right extracranial internal carotid artery (LEICA, REICA).
3.2. Predicting Recurrent Ischemic Stroke
The characteristics of the recurrent ischemic stroke fine-tuning dataset are shown in Table 2. Male patients account for 62.3% of the total recurrent ischemic stroke cases and 66.2% of the total non-recurrent ischemic stroke cases. The average onset age of non-recurrent ischemic stroke patients was younger than recurrent ischemic stroke patients (64.8 ± 13.0 vs. 67.0 ± 11.4). The time interval between recurrent stroke varied greatly, the average with the standard deviation is 907.0 ± 968.3 days and the median is 537 days, which represents the challenge of prediction in this diverse population.
Table 2.
Characteristics of ischemic stroke patient in recurrent ischemic stroke fine-tuning dataset.
| Recurrent ischemic stroke (n=969) | Non-recurrent ischemic stroke (n=731) | |
|---|---|---|
| Number of male (%) | 604 (62.3) | 484 (66.2) |
| First stroke onset age (mean ± SD) | 67.0 ± 11.4 | 64.8 ± 13.0 |
| Recurrence interval (days, mean ± SD) | 907.0 ± 968.3 | - |
| Median of recurrence interval (days) | 537 | - |
The receiver operating curve (ROC) analysis showed that StrokeBERT has better performance than ClinicalBERT: the average AUC±SD is 0.838 ± 0.017 for StrokeBERT and 0.808 ± 0.045 for ClinicalBERT. The smaller standard deviation of StrokeBERT’s AUC also indicates that it is more robust than ClinicalBERT on recurrent stroke prediction (figure 4). The F1 score and area under the precision-recall curve are provided in supplementary Table S2.
Figure 4.
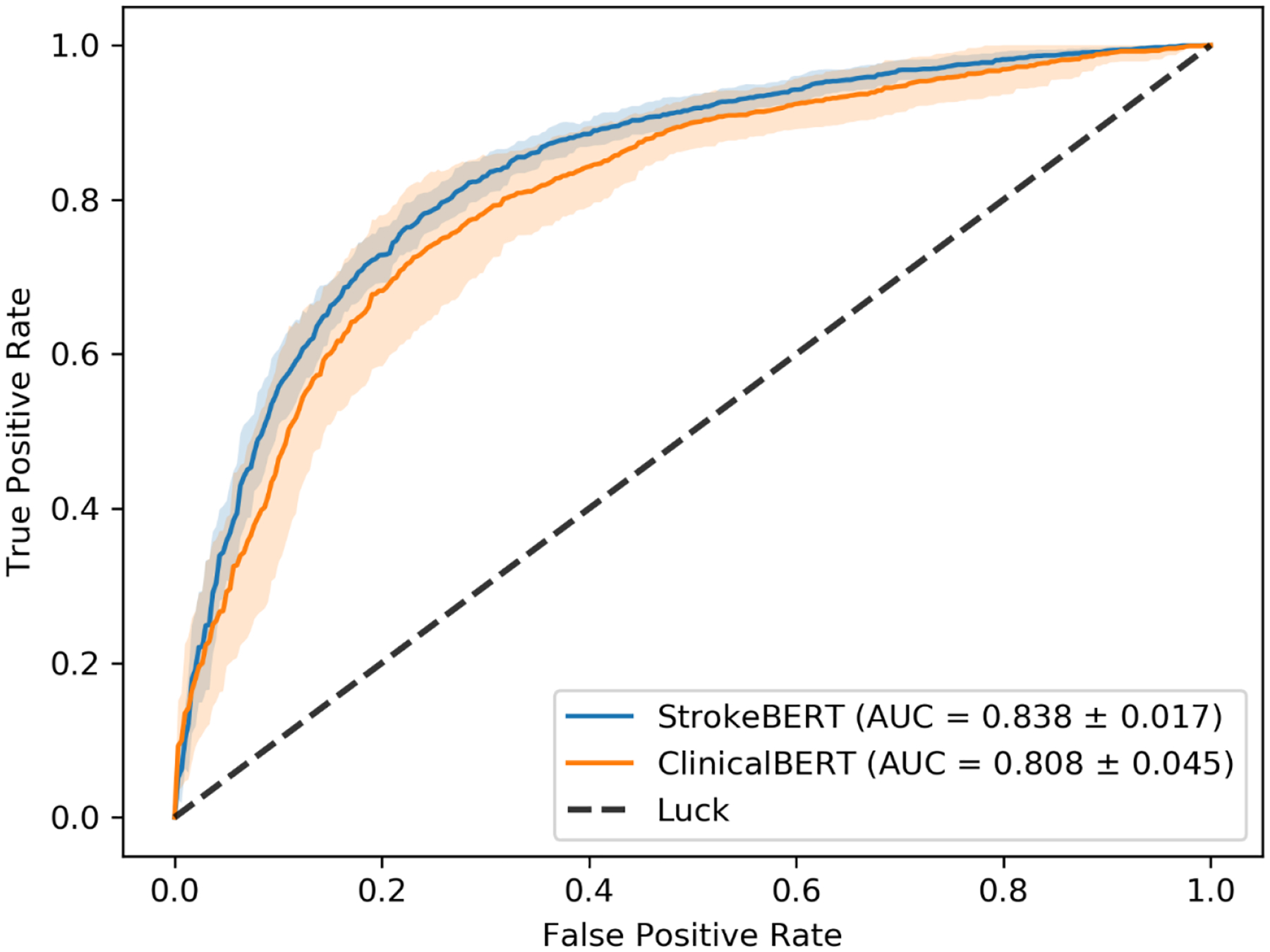
The receiver operating curve (ROC) of ischemic stroke recurrence prediction with ten-times cross-validation.
4. Discussion
We developed StrokeBERT, an improved pre-trained model using real world cerebrovascular disease-specific clinical notes, to help clinicians quickly assemble and assess patient’s health conditions and evaluate their risks for carotid artery stenosis and recurrent stroke. Taking advantage of BERT’s architecture, StrokeBERT can now be incorporated into various downstream tasks and fine-tuned as an integrated task-specific model within EHR automation (e.g. identifying carotid artery stenosis) or clinical alerts (e.g. recurrent risk of Stroke) as demonstrated in this study. We also performed analytical experiments to see how the size of the pre-training dataset and different parameter settings impact model performance. We pre-trained two additional BERT models that based only on 38,199 neurology reports or 105,743 radiology reports, respectively; and tested those models on same stenosis identification task and recurrent stroke prediction task. In summary, both neurology report-based BERT model and radiology report-based BERT model have lower AUCs than StrokeBERT in performing both tasks. The detailed measurements are displayed in supplementary Table S3 and Table S4. To further test the performance of our models affected by the parameter settings in down-stream tasks, we examined maximum sequence length and learning rate with one-time cross-validation. As shown in the supplementary Table S5 and Table S6, the learning rate and maximum sequence length significantly affected each model performance, for example, the lower learning rate used with same epoch and shorter sequence length, the lower AUC was. Regarding the parameter settings in downstream tasks, due to resource and time constraints, only maximum sequence length and learning rate with one-time cross-validation was performed in our analytical experiments.
Similar to ClinicalBERT, we found that pre-training domain-specific knowledge representation further improved the performance of the BERT-based model on specific clinical tasks. However, in this study we have found by pre-training on large-scale cerebrovascular disease clinical notes and reports, StrokeBERT was able to pay more attention to text information and events related to cerebrovascular diseases in comparison to ClinicalBERT which was pre-trained on the general medical domains. Attention scores indicating the ability to detect and identify disease relevant text information showed the potential impact and performance in downstream tasks from the model built. Figure 5 visualizes the attention scores of the word (text term) pieces in a random selected angiography report (panel A) and a discharge note (panel B). In the artery stenosis information extraction task, both ClinicalBERT and StrokeBERT showed similar attention spots on selected texts. For example, the descriptions of artery sections and degree of stenosis, such as “posterior cerebral arteries (PCA)”, “occlusion of right anterior cerebral artery”, and “azygos anterior cerebral artery”. This explains why the performance between ClinicalBERT and StrokeBERT was not significantly different. However, our study results still revealed that StrokeBERT-based models were able to identify stenosis in different extracranial and intracranial arteries from an angiography report with more disease relevant information, thus greater performance. This extracted information can be used to further train a machine-learning model to autodetect carotid artery stenosis.[36] In the real-world hospital setting, it is time-consuming to obtain relevant information on cerebral artery stenosis since clinical researchers have to label the stenosis manually from unorganized angiography reports. StrokeBERT was shown able to label the clinical reports automatically and extracted the stenosis occurred in multiple arteries simultaneously. In the recurrent ischemic stroke prediction task, both clinical-domain BERT-based models paid more attention on the descriptions of disease, symptoms, and outcomes like “ischemic cardiomyopathy with pulmonary edema”, “Hypertension“, “Diabetes mellitus”, “Chronic” and “successful percutaneous coronary intervention with drug”. However, compared to ClinicalBERT, the gender information (female) and term “rehabilitation”, which are known to be important recurrent stroke risk factors,[37] have higher attention scores in StrokeBERT. Therefore, in designing downstream tasks such as EHR automation for clinical alerts, StrokeBERT was shown to have better detection and prediction abilities. This tool could be further developed to improve public health by screening large clinical data repositories to identify patients at risk of recurrent ischemic stroke. Furthermore, it was shown that structured clinical registries can be utilized to build and ascertain various disease prediction models. [38 39] Most EHR systems contain large amount of unstructured data that can also be transformed into structured data by StrokeBERT for further clinical investigation.
Figure 5.

Visualization of attention mechanisms in StrokeBERT. Split word pieces are denoted with “##”. The warm-to-cool color spectrums indicate the attention score of the word pieces.
Many studies have explored different computational approaches for radiology report information extraction and recurrent stroke prediction, with more and more studies aimed at the annotation of diseases and conditions. However, research studies focused on identifying comprehensive intracranial and extranidal artery stenosis were found scattered. Wu et al recently published a logistic regression model, a field-aware convolution neural network (CNN), and a recurrent neural network (RNN) with an attention mechanism to identify patients with the conditions of carotid stenosis based on their ultrasound reports. These reported models achieved above 93% accuracy, with the RNN-attention model achieving near 95.4% accuracy.[38] In this study, we demonstrated that StrokeBERT achieved significantly higher performance in extracting information from radiology reports and classified patients more precisely with each extracranial and intracranial arteries stenosis. External validation for intracranial arteries in different hospital systems also well demonstrated its performance with high 0.968 AUC. As for recurrent stroke prediction, clinical variables and medical event characteristics including the ABCD2 score, brain imaging, and stroke mechanism, were previously used to predict the risk of recurrent stroke.[39] Many computational models or tools have also been developed. For instance, Leng et al evaluated the relationships between computational fluid dynamics (CFD) models and the risk of stroke recurrence;[40] the AUC of this model was reported to be 0.776. In addition, Ay et al developed a prognostic score that integrated with clinical and imaging information to quantify the early risk of recurrent stroke after ischemic stroke.[41] The AUC of their clinical-based model was 0.7 and 0.8 for their clinical and imaging combined models. As demonstrated in our study (Task II), the BERT-based NLP approach achieved superior performance with an AUC of 0.838 in recurrent stroke prediction, proving its efficacy of prediction in clinical applications. In addition, our results proved StrokeBERT’s unique ability in detecting and validating recurrent ischemic stroke in the real-world CGRD follow-up datasets for the past 10 years.
As shown by many previous studies, our results support the experience that a domain specific BERT is more efficient than a general-purpose BERT model. Several alternative approaches were also being experimented, for example, Shin et al. trained a larger BERT model, named Bio Megatron, with 345 million parameters (BERTBase has 110 million parameters) on the PubMed biomedical text corpus with about 6.1 billion words. Their study demonstrated that larger model size can further improve the performance of a domain-specific language model.[44] Another study challenged the assumption that domain-specific pretraining can benefit by starting from general-domain language models. Gu et al. showed that domain specific pretraining from scratch can significantly outperform continual pretraining from a general-domain language model. [45] These studies provided different strategy of model training that can affect the performance of domain-specific language models. In this initial study, StrokeBERT was pre-trained and fine-tuned with only limited hyperparameter optimization due to the limitations of data collections and processing power. Despite these limitations, this BERT-based platform has shown the potential to greatly reduce physician’s demands to retrospectively extract valuable information from unstructured notes during medical triage, such as patients’ past medical history, lab reports, treatments, outcomes, and complications.
Future work will focus on improving its performance of fine-tuning tasks by different machine learning algorithms and incorporating additional clinical information from more data sources to improve and solve more complex real-world medical applications for cerebrovascular diseases research and care. For example, Bacchi et al used a CNN and a RNN to predict the causes of transient ischemic attack-like conditions based on the free-text descriptions of patient’s medical history complaints. Their CNN model achieved a good predictive performance (AUC±SD; 81.9±2.0). By incorporating additional clinical information, such as magnetic resonance imaging reports, their AUC of CNN model was improved to 88.3±3.6.[42] Moreover, it has been shown that integrating clinical texts, images, and laboratory data could help deep learning models assess more complex tasks in the clinical domain.[43 44] The current research of language representation models is fast evolving in many respects. In terms of pre-training strategy, Liu et al found that the dynamic masking strategy performed slightly better than static masking during the model pre-training phase.[45] In addition, Sun et al proposed a novel language representation model enhanced by knowledge masking strategies called ERNIE (Enhanced Representation through kNowledge IntEgration), which included entity-level masking and phrase-level masking. [46] Sun et al further proposed ERNIE 2.0, a continual pre-training framework to support continual multi-task learning.[47] In addition to BERT-based models, XLNet is another advanced language representation model which was recently shown to improve over BERT on 20 NLP tasks.[48] Huang et al developed Clinical XLNet and demonstrated that it consistently outperformed other deep learning models.[49] Therefore, developing and applying other advanced language representation models and different pre-training strategies as well as building continual learning models in our disease-specific domain will be the primary focus of our future studies to advance clinical research and real world applications in the cerebrovascular diseases.
5. Conclusion
StrokeBERT is a disease-specific BERT-based model pre-trained on cerebrovascular disease-related corpora. Through validations with large multiple-center datasets and by two independent empirical tasks, we demonstrated that using disease-specific BERT (in this case, StrokeBERT) improved the performance and results of various disease-specific language processing applications. In addition, a clinical platform embedded with this StrokeBERT tool can be further developed to assist and automate physician’s notes writing tasks through auto-discovering medical terms in all aspects of EHR information to standardize and reduce potential data entry errors. As computing power continues to accelerate, and deep learning techniques become more readily accessible, disease-specific language representation models can be further improved to be a viable and powerful tool to assist in developing intelligent clinical applications and accelerating biomedical discovery in cerebrovascular diseases.
Supplementary Material
Highlights.
StrokeBERT is a disease-specific BERT-based model pre-trained on real world evidence (RWE) from cerebrovascular disease-related corpora.
The model was evaluated and validated in larger, multiple-center datasets by two independent empirical tasks (stenosis detection and stroke recurrence prediction).
Disease-specific BERT model improves results of various disease-specific language processing tasks compared to similar BERT-models pre-trained on the general domain corpora.
Acknowledgment:
This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). The authors thank the NIH Library Writing Center for manuscript editing assistance. The authors thank Google for opening BERT source codes. CKL thank the Veterans Affair Council and Kaohsiung Veterans General Hospital, Taiwan, for financial support on study abroad.
Sources of Funding:
This work is supported by funding from Intramural Research Program of National Institute of Neurological Disorders and Stroke, National Institutes of Health, USA and from the research grants of Chang Gung Memorial Hospital (CMRPG3F2211, CMRPG3J1161) and the Ministry of Science and Technology, ROC (110-2222-E-182A-001-MY2).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Code availability: Codes are available at https://github.com/whynopeoplefly/strokeBERT
Disclosures: None.
Declaration of Competing Interest: The authors have no competing interests to declare.
References
- 1.Feigin VL, Roth GA, Naghavi M, et al. Global burden of stroke and risk factors in 188 countries, during 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. The Lancet Neurology 2016;15(9):913–24 [DOI] [PubMed] [Google Scholar]
- 2.Sedghi E, Weber JH, Thomo A, Bibok M, Penn AM. Mining clinical text for stroke prediction. Network Modeling Analysis in Health Informatics and Bioinformatics 2015;4(1):16 [Google Scholar]
- 3.Garg R, Oh E, Naidech A, Kording K, Prabhakaran S. Automating ischemic stroke subtype classification using machine learning and natural language processing. Journal of Stroke and Cerebrovascular Diseases 2019;28(7):2045–51 [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Liu Q, Wei L, et al. Automatically Structuring on Chinese Ultrasound Report of Cerebrovascular Diseases via Natural Language Processing. IEEE Access 2019;7:89043–50 [Google Scholar]
- 5.Devlin J, Chang M-W, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805 2018 [Google Scholar]
- 6.Distributed representations of words and phrases and their compositionality. Advances in neural information processing systems; 2013. [Google Scholar]
- 7.Glove: Global vectors for word representation. Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP); 2014. [Google Scholar]
- 8.Attention is all you need. Advances in neural information processing systems; 2017. [Google Scholar]
- 9.Adhikari A, Ram A, Tang R, Lin J. Docbert: Bert for document classification. arXiv preprint arXiv:1904.08398 2019 [Google Scholar]
- 10.Zhang T, Kishore V, Wu F, Weinberger KQ, Artzi Y. Bertscore: Evaluating text generation with bert. arXiv preprint arXiv:1904.09675 2019 [Google Scholar]
- 11.Stance Classification, Outcome Prediction, and Impact Assessment: NLP Tasks for Studying Group Decision-Making. Proceedings of the Third Workshop on Natural Language Processing and Computational Social Science; 2019. [Google Scholar]
- 12.Tenney I, Das D, Pavlick E. Bert rediscovers the classical nlp pipeline. arXiv preprint arXiv:1905.05950 2019 [Google Scholar]
- 13.Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020;36(4):1234–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Scientific data 2016;3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alsentzer E, Murphy JR, Boag W, et al. Publicly available clinical BERT embeddings. arXiv preprint arXiv:1904.03323 2019 [Google Scholar]
- 16.Huang K, Altosaar J, Ranganath R. Clinicalbert: Modeling clinical notes and predicting hospital readmission. arXiv preprint arXiv:1904.05342 2019 [Google Scholar]
- 17.Li F, Jin Y, Liu W, Rawat BPS, Cai P, Yu H. Fine-Tuning Bidirectional Encoder Representations From Transformers (BERT)–Based Models on Large-Scale Electronic Health Record Notes: An Empirical Study. JMIR medical informatics 2019;7(3):e14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao SC, Chan YY, Kao Yang YH, et al. The Chang Gung Research Database—A multi-institutional electronic medical records database for real-world epidemiological studies in Taiwan. Pharmacoepidemiology and drug safety 2019;28(5):593–600 [DOI] [PubMed] [Google Scholar]
- 19.Lin YS, Chen TH, Chi CC, et al. Different implications of heart failure, ischemic stroke, and mortality between nonvalvular atrial fibrillation and atrial flutter—a view from a national cohort study. Journal of the American Heart Association 2017;6(7):e006406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An improved non-monotonic transition system for dependency parsing. Proceedings of the 2015 conference on empirical methods in natural language processing; 2015. [Google Scholar]
- 21.clinicalBERT, https://github.com/EmilyAlsentzer/clinicalBERT. Accessed 23 Jun. 2021.
- 22.Wu Y, Schuster M, Chen Z, et al. Google’s neural machine translation system: Bridging the gap between human and machine translation. arXiv preprint arXiv:1609.08144 2016 [Google Scholar]
- 23.Silvestrini M, Vernieri F, Pasqualetti P, et al. Impaired cerebral vasoreactivity and risk of stroke in patients with asymptomatic carotid artery stenosis. Jama 2000;283(16):2122–27 [DOI] [PubMed] [Google Scholar]
- 24.Cote R, Caron J-L. Current Concepts of Cerebrovascular Disease and Stroke Management of Carotid Artery Occlusion. Stroke 1989;20(1) [DOI] [PubMed] [Google Scholar]
- 25.Bash S, Villablanca JP, Jahan R, et al. Intracranial vascular stenosis and occlusive disease: evaluation with CT angiography, MR angiography, and digital subtraction angiography. American journal of neuroradiology 2005;26(5):1012–21 [PMC free article] [PubMed] [Google Scholar]
- 26.TensorFlow’s Sigmoid Cross Entropy with Logits, https://www.tensorflow.org/api_docs/python/tf/nn/sigmoid_cross_entropy_with_logits. Accessed 04 Mar. 2020.
- 27.BertAdam, https://github.com/huggingface/transformers/blob/694e2117f33d752ae89542e70b84533c52cb9142/README.md#optimizers. Accessed 20 Mar. 2020.
- 28.Moneta GL, Edwards JM, Chitwood RW, et al. Correlation of North American Symptomatic Carotid Endarterectomy Trial (NASCET) angiographic definition of 70% to 99% internal carotid artery stenosis with duplex scanning. Journal of vascular surgery 1993;17(1):152–59 [DOI] [PubMed] [Google Scholar]
- 29.Sacco RL, Foulkes M, Mohr J, Wolf P, Hier D, Price T. Determinants of early recurrence of cerebral infarction. The Stroke Data Bank. Stroke 1989;20(8):983–89 [DOI] [PubMed] [Google Scholar]
- 30.Kennedy J, Hill MD, Ryckborst KJ, et al. Fast assessment of stroke and transient ischaemic attack to prevent early recurrence (FASTER): a randomised controlled pilot trial. The Lancet Neurology 2007;6(11):961–69 [DOI] [PubMed] [Google Scholar]
- 31.Hsieh F-I, Chiou H-Y. Stroke: morbidity, risk factors, and care in Taiwan. Journal of stroke 2014;16(2):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeng J. Quality improvement of acute ischemic stroke patients through the Breakthrough Series (BTS) activity. J Healthcare Qual 2012;6:70–75 [Google Scholar]
- 33.Li H-W, Yang M-C, Chung K-P. Predictors for readmission of acute ischemic stroke in Taiwan. Journal of the Formosan Medical Association 2011;110(10):627–33 [DOI] [PubMed] [Google Scholar]
- 34.Hsieh C-Y, Chen C-H, Li C-Y, Lai M-L. Validating the diagnosis of acute ischemic stroke in a National Health Insurance claims database. Journal of the Formosan Medical Association 2015;114(3):254–59 [DOI] [PubMed] [Google Scholar]
- 35.CrossEntropyLoss, https://pytorch.org/docs/stable/generated/torch.nn.CrossEntropyLoss.html#torch.nn.CrossEntropyLoss. Accessed 08 Mar. 2020.
- 36.Hsu K-C, Lin C-H, Johnson KR, et al. Autodetect extracranial and intracranial artery stenosis by machine learning using ultrasound. Computers in Biology and Medicine 2020;116:103569. [DOI] [PubMed] [Google Scholar]
- 37.Andersen SD, Gorst-Rasmussen A, Lip GY, Bach FW, Larsen TB. Recurrent stroke: the value of the CHA2DS2VASc score and the essen stroke risk score in a nationwide stroke cohort. Stroke 2015;46(9):2491–97 [DOI] [PubMed] [Google Scholar]
- 38.Wu X, Zhao Y, Radev D, Malhotra A. Identification of patients with carotid stenosis using natural language processing. European Radiology 2020:1–9 [DOI] [PubMed] [Google Scholar]
- 39.Couillard P, Poppe AY, Coutts SB. Predicting recurrent stroke after minor stroke and transient ischemic attack. Expert review of cardiovascular therapy 2009;7(10):1273–81 [DOI] [PubMed] [Google Scholar]
- 40.Leng X, Scalzo F, Ip HL, et al. Computational fluid dynamics modeling of symptomatic intracranial atherosclerosis may predict risk of stroke recurrence. PLoS One 2014;9(5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ay H, Gungor L, Arsava E, et al. A score to predict early risk of recurrence after ischemic stroke. Neurology 2010;74(2):128–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bacchi S, Oakden-Rayner L, Zerner T, Kleinig T, Patel S, Jannes J. Deep learning natural language processing successfully predicts the cerebrovascular cause of transient ischemic attack-like presentations. Stroke 2019;50(3):758–60 [DOI] [PubMed] [Google Scholar]
- 43.Titano JJ, Badgeley M, Schefflein J, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nature medicine 2018;24(9):1337–41 [DOI] [PubMed] [Google Scholar]
- 44.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digital Medicine 2018;1(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Ott M, Goyal N, et al. Roberta: A robustly optimized bert pretraining approach. arXiv preprint arXiv:1907.11692 2019 [Google Scholar]
- 46.Sun Y, Wang S, Li Y, et al. Ernie: Enhanced representation through knowledge integration. arXiv preprint arXiv:1904.09223 2019 [Google Scholar]
- 47.Sun Y, Wang S, Li Y, et al. Ernie 2.0: A continual pre-training framework for language understanding. arXiv preprint arXiv:1907.12412 2019 [Google Scholar]
- 48.Xlnet: Generalized autoregressive pretraining for language understanding. Advances in neural information processing systems; 2019. [Google Scholar]
- 49.Huang K, Singh A, Chen S, et al. Clinical XLNet: Modeling Sequential Clinical Notes and Predicting Prolonged Mechanical Ventilation. arXiv preprint arXiv:1912.11975 2019 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


