Summary
Genetic studies of autism have revealed causal roles for chromatin remodeling gene mutations. Chromodomain helicase DNA binding protein 8 (CHD8) encodes a chromatin remodeler with significant de novo mutation rates in sporadic autism. However, relationships between CHD8 genomic function and autism-relevant biology remain poorly elucidated. Published studies utilizing ChIP-seq to map CHD8 protein-DNA interactions have high variability, consistent with technical challenges and limitations associated with this method. Thus, complementary approaches are needed to establish CHD8 genomic targets and regulatory functions in developing brain. We used in utero CHD8 Targeted DamID followed by sequencing (TaDa-seq) to characterize CHD8 binding in embryonic mouse cortex. CHD8 TaDa-seq reproduced interaction patterns observed from ChIP-seq and further highlighted CHD8 distal interactions associated with neuronal loci. This study establishes TaDa-seq as a useful alternative for mapping protein-DNA interactions in vivo and provides insights into the regulatory targets of CHD8 and autism-relevant pathophysiology associated with CHD8 mutations.
Subject areas: Genomics, Molecular neuroscience, Biotechnology, Genomic analysis
Graphical abstract

Highlights
-
•
ChIP-seq maps protein-DNA interactions but can have significant technical limitations
-
•
DamID maps protein-DNA interactions using genome-wide DNA adenine methylation signals
-
•
We used Targeted DamID in vivo in embryonic mouse brain to map CHD8 interactions
-
•
TaDa-seq resolved neurodevelopmental CHD8 interactions at promoters and enhancers
Genomics; Molecular neuroscience; Biotechnology; Genomic analysis
Introduction
Neurodevelopmental disorders (NDDs) including autism spectrum disorder (ASD) and intellectual disability (ID) are complex disorders caused by genetic and environmental factors that disrupt brain development. Genetic studies have identified an overlapping set of genes that, when mutated, greatly increase risk for both ASD and ID (O’Roak et al., 2012a, 2012b; Parikshak et al., 2013; De Rubeis et al., 2014; Iossifov et al., 2014; Sanders et al., 2015; Vissers et al., 2016; Satterstrom et al., 2020). Of these shared risk gene sets, a striking and surprising finding has been the strong enrichment of case mutations in genes that encode proteins involved in chromatin remodeling (O'Roak et al., 2012a; De Rubeis et al., 2014). One of these genes, with among the highest number of identified ASD and ID case mutations, is Chromodomain Helicase DNA binding protein 8 (CHD8). Characterization of patient phenotypes associated with loss-of-function CHD8 mutations has revealed a syndrome-like pattern of pathology. These patients commonly feature symptoms meeting stringent ASD diagnosis, a spectrum of ID and cognitive impairment, macrocephaly, gastrointestinal and sleep disturbances, and other symptoms (Bernier et al., 2014; Ostrowski et al., 2019; Douzgou et al., 2019; Yasin et al., 2019; An et al., 2020). The function of CHD8 and other NDD-associated chromatin remodeling proteins in developing brain remains poorly characterized, representing a major barrier to understanding the neurodevelopmental mechanisms of NDDs.
Chromatin remodelers impact the packaging and functional readout of DNA through interactions with chromatin (Hargreaves and Crabtree, 2011). The dominant approach to understanding molecular function of DNA-associated proteins is to map their specific genomic targets, primarily by chromatin immunoprecipitation followed by sequencing (ChIP-seq). ChIP-seq has been successfully applied to identify targets of ASD/ID-associated chromatin remodelers, including when profiling fetal brain tissue. However, ChIP-seq requires specific and sensitive antibodies, sufficient sample, and processing steps, specifically cross-linking and fragmentation, that can introduce signal artifacts (Marinov et al., 2014). Furthermore, ChIP-seq performs best with strong, typically direct, interactions between the protein and target DNA (Furey, 2012). This is a significant drawback, as many chromatin remodelers interact indirectly with DNA and ChIP-seq grade antibodies are not always available. Thus, a major limitation to studies of NDD-associated chromatin remodelers has been the challenges presented in identifying genomic interactions by ChIP-seq. One common alternative strategy to ChIP-seq has been to introduce epitope-tagged versions of these proteins to improve immunoprecipitation (Attanasio et al., 2014). Although this strategy overcomes some barriers, often there are still technical obstacles. For example, epitope tags may address the lack of ChIP-seq grade antibodies, but issues still remain for weak or indirect protein-DNA interactions and artifacts introduced by cross-linking and fragmentation (Furey, 2012).
The growing list of studies that report ChIP-seq-derived genomic binding patterns of CHD8 across human and mouse brain tissues and in vitro models exemplifies the challenges of applying ChIP-seq to understand chromatin remodeler function (Ceballos-Chávez et al., 2015; Cotney et al., 2015; de Dieuleveult et al., 2016; Gompers et al., 2017; Platt et al., 2017; Katayama et al., 2016; Sugathan et al., 2014). Our meta-analysis of published CHD8 ChIP-seq datasets found strong concordance across datasets for the strongest genomic interactions (Wade et al., 2019). However, there was extensive variability in the number and genomic distribution of CHD8 ChIP-seq peaks. This was true even among studies that examined similar tissue types, e.g., adult mouse cortex (Gompers et al., 2017; Platt et al., 2017; Katayama et al., 2016), and for studies that used the same antibodies and general methods. Thus, biological inferences regarding CHD8 function have varied considerably based on which ChIP-seq dataset is used. This is reflected in CHD8 publications that highlight various patterns: at one end, widespread binding including at the majority of promoters (Cotney et al., 2015; Katayama et al., 2016; Sugathan et al., 2014); at the other end, more limited binding primarily at promoters of genes involved in basic cell functions (Gompers et al., 2017; Platt et al., 2017). These contrasting ChIP-seq findings demonstrate the need for complementary methods to map genomic interactions for CHD8 and, more generally, for chromatin remodelers and other difficult to ChIP proteins.
Motivated by the need for approaches that avoid antibody-based limitations and technical issues that can be associated specifically with ChIP-seq, we decided to use Targeted DamID (TaDa) (Southall et al., 2013; Marshall et al., 2016) to map CHD8 targets in vivo in fetal mouse cortex. In TaDa, a protein of interest (here CHD8) is fused to an E. coli DNA adenine methyltransferase domain (Dam). Wherever the Dam fusion protein interacts with the genome, the methylase catalyzes methylation of adenine within the sequence GATC. As endogenous adenine methylation is extremely rare in eukaryotes (Koziol et al., 2015; Wu et al., 2016; Zhang et al., 2015; Douvlataniotis et al., 2020), the genomic interaction targets of the protein of interest can be identified by mapping adenine methylation in the genome. This approach does not require cell sorting, fixation, cross-linking, or affinity purification, as interactions are mapped via restriction digestion at methylated GATC sites, followed by DNA sequencing (Aughey et al., 2019; van den Ameele et al., 2019). TaDa has been used successfully to map genome-wide binding of transcription factors, chromatin proteins, and RNA polymerase in Drosophila and mammalian cells (for example, Southall et al., 2013; Marshall et al., 2016; Marshall and Brand, 2015; Otsuki and Brand, 2018; Cheetham et al., 2018; Marshall and Brand, 2017; Tosti et al., 2018) as well as to map non-coding RNA interactions within the genome (Cheetham and Brand, 2018).
Here, we delivered TaDa constructs by in utero electroporation (IUE) to perform CHD8 TaDa-seq in the developing mouse brain in vivo. Our results show the feasibility and value of this approach, resolving CHD8 interactions in embryonic mouse cerebral cortex. More broadly, our study highlights a novel approach toward mapping genomic binding patterns of proteins that are challenging or intractable to ChIP-seq.
Results
Cloning and in utero electroporation of CHD8 TaDa plasmid into embryonic mouse cortex
To study CHD8 binding patterns in embryonic neurodevelopment, we used an established Targeted DamID-seq (TaDa-seq) protocol (Marshall et al., 2016), combined with in utero electroporation of embryonic day (E) 13.5 mouse brain (Figures 1A–1D). A full-length human CHD8 ORF was cloned into the TaDa construct and sequence verified. As Dam activity can be toxic to cells, the experimental TaDa constructs are designed to express extremely low levels of the CHD8-Dam fusion proteins due to a bicistronic strategy that requires ribosomal reentry for expression of the Dam fusion protein (Southall et al., 2013; Figure 1A; STAR Methods). To differentiate CHD8-driven interactions from non-specific adenine methylation, Dam-only constructs serve as a control, with the same methods used for IUE delivery, library generation, and sequencing. Recruitment of CHD8 TaDa to specific genomic loci, either directly or through interaction with other proteins, is based on increased CHD8 TaDa-seq-normalized coverage compared with Dam-only control (Figure 1E). In addition to serving as a control, the Dam-only experiments capture genome-wide signatures of accessible chromatin (Aughey et al., 2018).
Figure 1.
Overview of the targeted DamID followed by sequencing (TaDa-seq) pipeline
(A) Plasmids used for TaDa. The top plasmid is a diagram for CHD8 TaDa experiments. The middle plasmid is a diagram for Dam-only experiments. The bottom plasmid is a diagram for the in utero electroporation control injected with the CHD8 TaDa or Dam-only plasmids.
(B) Schematic and flowchart of TaDa-seq experiments. E13.5 mouse embryos were injected with CHD8 TaDa or Dam-only plasmid and the in utero electroporation control plasmid. Four CHD8 TaDa and three Dam-only brains from the same litter were dissected. Frozen brains were then processed for the pipeline indicated in the gray boxes.
(C) Immunohistochemistry showing overlap between green fluorescence (in utero electroporation control), red fluorescence (mCherry expression upstream of the CHD8 TaDa open reading frame), and DAPI (nuclei) illustrates successful transfection of experimental plasmids.
(D) TaDa-seq computational analysis pipeline used in this study.
(E) Schematic showing example signal from CHD8 TaDa or Dam-only protein binding at genomic loci.
The CHD8 TaDa or Dam-only constructs were electroporated in utero into developing mouse cerebral cortex at E13.5 (Figure 1B). A pCAG-Venus construct was co-electroporated with CHD8 TaDa and Dam-only plasmids as a delivery control to visualize the electroporated region. Following delivery, there was a 4-day period where the constructs could be expressed in cells that took up the plasmids before tissues were collected at E17.5. Representative images of E17.5 cortex show green immunofluorescence representing Venus expression confirming in utero electroporation into developing somatosensory cortex, whereas red immunofluorescence shows expression of the primary open reading frame of the TaDa construct, mCherry (Figure 1C). Translation of the CHD8 TaDa or Dam-only open reading frames is too low to detect by immunostaining. Following IUE, transfected radial glial neural progenitor cells undergo self-renewal as well as producing intermediate progenitor cells and early neurons that will migrate to form the layers of the cortex. The incubation period represented the window during which Dam methylation occurred, resulting in Dam activity observed from ventricular zone progenitors to early cortical neurons. Representative IHC of a CHD8 TaDa IUE shows widespread cortical expression of the Venus delivery as well as sparser cellular labeling with mCherry, consistent with expectations of construct expression in developing cortical neurons (Figure 1C). No further investigation of cell-type-specific IUE delivery and construct expression was performed, although our results are consistent with expected IUE performance and construct expression in cortical glutamatergic neurons (Saito, 2006).
Genomic patterns of CHD8 TaDa-seq and representative CHD8 ChIP-seq datasets
For sequence-based analysis, we collected four CHD8 TaDa and three Dam-only samples from the same litter of MF-1 outbred mice and processed them using the TaDa-seq experimental and computational pipeline (Marshall and Brand, 2015; Marshall et al., 2016; see STAR Methods for details, Figures 1B–1D). Individual replicates and merged datasets for CHD8 TaDa-seq and Dam-only experiments were analyzed. Coverage plots were generated to show signal independently in the CHD8 TaDa-seq and Dam-only datasets, and CHD8 TaDa-seq was further normalized using Dam-only to visualize enrichment representing CHD8-specific interactions. Enriched genomic regions that were identified via comparison of CHD8 TaDa-seq to Dam-only read coverage in at least three of four CHD8 TaDa-seq replicates at high statistical stringency were considered to be high confidence CHD8 interaction regions. Following peak calling and merging, there were 142,375 enriched peaks in the Dam-only experiments and 24,533 that passed stringent significance and reproducibility criteria across CHD8 TaDa-seq experiments.
To examine specificity and relevance of CHD8 targets mapped by TaDa-seq, we compared our CHD8 TaDa-seq datasets to two published CHD8 ChIP-seq experiments performed on mouse brain. The first dataset was time and tissue matched with our TaDa-seq data, profiling E17.5 mouse cortex (Cotney et al., 2015). The second was from adult mouse cortex (Platt et al., 2017). CHD8 ChIP-seq datasets vary in results, whereas these ChIP-seq datasets were highly correlated to each other and other CHD8 ChIP-seq datasets (Wade et al., 2019). Raw sequence files were downloaded and analyzed using standard approaches to generate coverage and peak intervals (Wade et al., 2019; see STAR Methods), with 44,383 and 32,335 peaks mapped in the E17.5 and adult cortex datasets, respectively. We additionally examined patterns of enrichment between CHD8 TaDa-seq, CHD8 ChIP-seq, and epigenomic datasets generated for E16.5 mouse cortex via ENCODE (ENCODE Project Consortium, 2012).
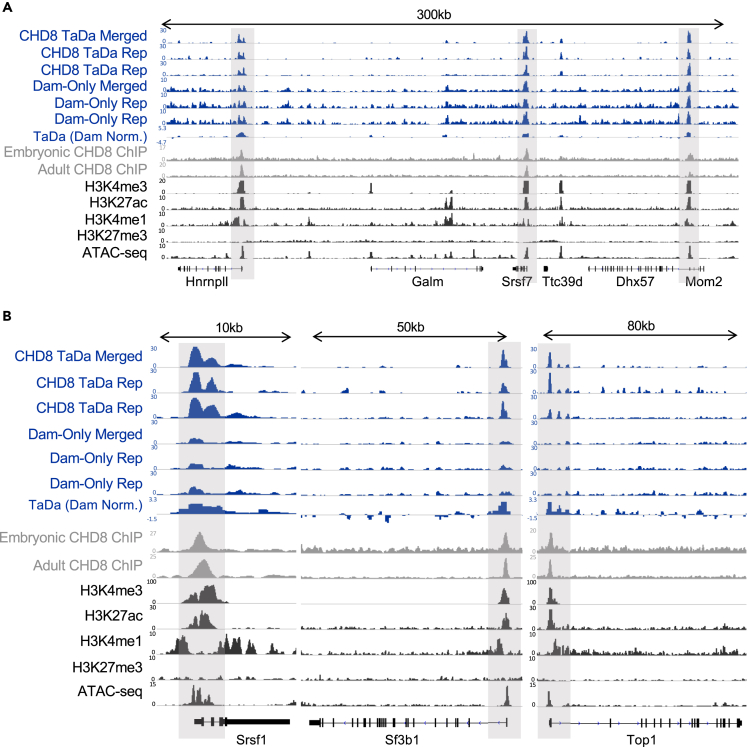
First, we examined genomic loci that were previously found to have consistent and strong CHD8 peaks across ChIP-seq datasets (Figures 2, S1), for example, promoter interactions for genes associated with RNA processing, such as Hnrnpll, Srsf7, Srsf1, and Sf3b1, or genes associated with chromatin remodeling, such as Top1 (Figures 2A and 2B). Read coverage at these loci illustrates reproducibility and specificity of CHD8 TaDa genomic interactions as compared with chromatin accessibility revealed by Dam-only controls (Figure 2A). As expected, Dam-only peaks occurred throughout these loci, indicating expected non-specific adenine methylation in regions of accessible chromatin. Comparison of CHD8 genome interactions identified by TaDa-seq with the two published CHD8 ChIP-seq datasets showed strong concordance in enrichment at these loci, indicating TaDa-seq captured reproducible interactions between CHD8 and its genomic targets.
Figure 2.
Recapitulation of CHD8 binding near promoters across the genome
(A and B) Data showing CHD8 binding at loci previously identified in CHD8 binding characterization studies, including RNA processing genes, Hnrnpll and Srsf7 in A, Srsf1 and Sf3b1 in B, and a chromatin remodeling gene, Top1, in (B). Gray boxes highlight CHD8 binding near identified promoters of interest. CHD8 TaDa, Dam-only, or Dam-only-normalized CHD8 TaDa (TaDa Dam Norm.) experiment tracks are in blue (representative biological replicates shown), CHD8 ChIP-seq experiments are in gray, and datasets of histone and chromatin accessibility signatures from the ENCODE consortium are in black. Linear representations of genes from the mouse mm10 genome are shown below coverage tracks. Height of the y axis is scaled to show the peak for each track separately. See also Figure S1. Embryonic CHD8 ChIP (Cotney et al. (2015)) embryonic CHD8 ChIP-seq, Adult CHD8 ChIP (Platt et al. (2017)) adult CHD8 ChIP-seq.
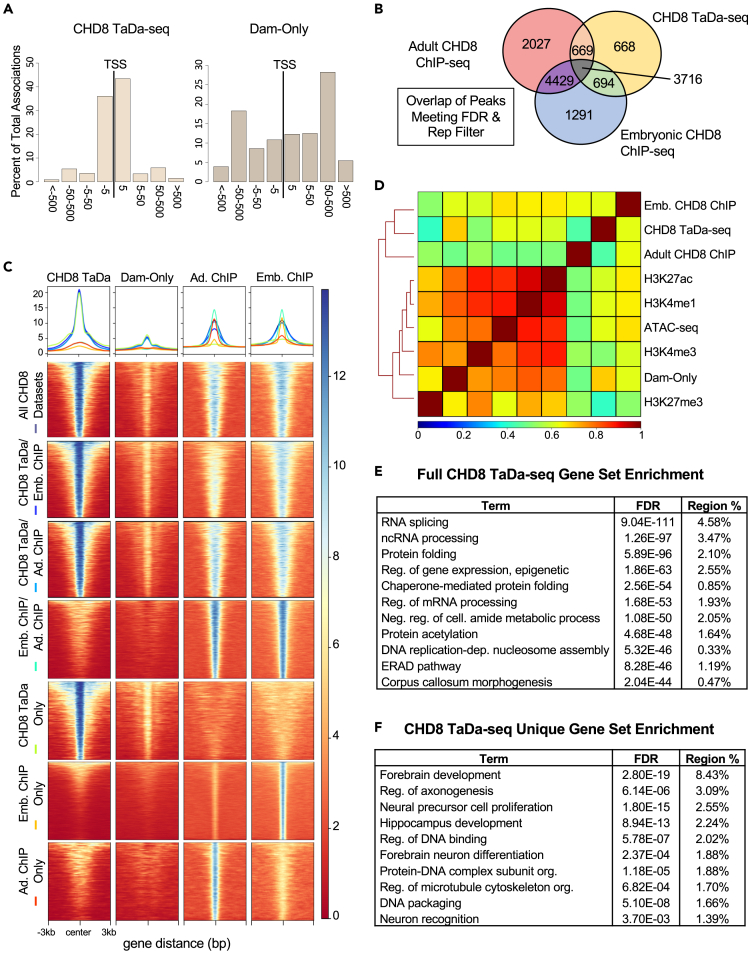
Next, we compared CHD8 TaDa-seq, Dam-only, and representative CHD8 ChIP-seq genomic binding patterns and peak overlap (Figures 3A and 3B, S2A, S2B) and plotted coverage heatmaps comparing local enrichment across overlapping and non-overlapping peak sets (Figure 3C). CHD8 TaDa-seq and CHD8 ChIP-seq both revealed CHD8 binding strongly enriched near promoters, although distal interactions were also represented to varying degrees across datasets (Figures 3A and S2A, S2B). Although many TF-binding motifs were enriched within CHD8 interaction DNA sequences, no single DNA motif was identified at CHD8 target loci defined by either TaDa-seq or ChIP-seq (Figure S3), consistent with previous work and suggesting CHD8 interactions are not generally guided by direct binding to a recognition sequence. CHD8 TaDa-seq peaks were largely present in one or both CHD8 ChIP-seq experiments (Figure 3B), with increased overlap when any CHD8 TaDa-seq peaks found in at least three replicates were included (Figure S2C). Thus, the CHD8 TaDa-seq results are consistent with previous ChIP-seq observations of CHD8 genomic binding activity. CHD8 TaDa-seq coverage was strongly correlated with CHD8 ChIP-seq coverage, even when peaks were not called in one datatype (Figure 3C). This observation, coupled with Dam-only showing a much more generalized and widespread enrichment signal at open chromatin regions, confirmed the specificity of CHD8 TaDa-seq interactions throughout the genome.
Figure 3.
Computational comparison of CHD8 binding shows correspondence in signal across TaDa-seq and ChIP-seq experiments
(A) Bar plots showing association of peaks with transcription start sites (TSS) using the GREAT online analysis tool. Bins along the x axis represent 5, 50, 500, and greater than 500 kilobases away from the nearest TSS.
(B) Venn diagram showing the number of peaks annotated to genes overlapping with CHD8 TaDa, Embryonic CHD8 ChIP-seq, and Adult CHD8 ChIP-seq using stringent CHD8 TaDa-seq peak thresholding with peaks meeting an FDR <0.00001 cutoff in at least three replicates.
(C) Genome-wide coverage heatmaps showing enrichment of signal at peaks for each dataset indicated on the left-hand side. Y axes of datasets were matched for visual comparison. Small line plots indicate the average normalized peak enrichment for each dataset with the color for each line next to each dataset name. Each peak is centered along the middle of each plot with a 3-kilobase pair window on each side. The legend indicates normalized enrichment.
(D) Genome coverage correlation heatmap showing relationship between representative CHD8 TaDa-seq, Dam-only, CHD8 ChIP-seq, and ENCODE histone mark and chromatin accessibility datasets. Data are hierarchically clustered according to genome-wide similarity as indicated by a dendrogram. Legend indicates the correlation value between datasets. H3K27me3 is a histone mark associated with repressed DNA loci. H3K4me3 is a histone mark associated with actively transcribed promoters. ATAC-seq is sequencing data of open chromatin regions. H3K4me1 and H3K27ac are histone marks associated with putative enhancers.
(E) Table showing functional annotations associated with CHD8 TaDa-seq called peaks. Region % refers to the percent of the total peak set annotated to each term.
(F) Table showing functional annotations associated with peaks only found in the CHD8 TaDa-seq dataset. Region % refers to the percent of the total peak set annotated to each term. See also Figures S2, S3, and S4. Embryonic CHD8 ChIP (Cotney et al., (2015)) embryonic CHD8 ChIP-seq, Adult CHD8 ChIP (Platt et al., (2017)) adult CHD8 ChIP-seq.
Although many of the same loci were captured, peak significance rank varied between CHD8 TaDa-seq and ChIP-seq, as previously shown with TaDa-seq in other studies (Tosti et al., 2018; Cheetham et al., 2018). There were a set of TaDa-seq peaks that were not called as peaks in either ChIP-seq dataset (Figures 3B and 3C, S2C). Despite these differences, the strongest peaks in either method were largely detected by both methods. Among peaks that were called in only the CHD8 TaDa-seq or ChIP-seq, most of these loci exhibited sub-significant signal in the other assay (Figure 3C), suggesting that these represent true interactions that are below the sensitivity threshold of these particular CHD8 ChIP-seq datasets. Of note, peaks that were called in the CHD8 TaDa-seq but not ChIP-seq were more likely to be distal compared with the full set of CHD8 TaDa-seq peaks (Figures S2A and S2B). In total, 1,719 genes were annotated by proximity to a TaDa-seq unique peak, with 241 of these genes also annotated in the adult cortex ChIP-seq and 434 in the E17.5 embryonic ChIP-seq dataset. Thus, there appear to be method-specific differences in signal and sensitivity that differently impacted detection of a set of genes with CHD8 promoters and distal regulatory interactions. Given the availability of several other CHD8 ChIP-seq datasets with varying peak set enrichment, we expanded our analysis to other CHD8 mouse studies to examine whether our TaDa-seq findings were consistent with other experiments (Katayama et al., 2016; de Dieuleveult et al., 2016; Gompers et al., 2017; Sood et al., 2020). Although CHD8 ChIP-seq experiments had variable overlap between themselves, the CHD8 TaDa-seq peak set consistently had a high level of overlap with CHD8 ChIP-seq peak sets (Figure S2D). This suggests differences in enrichment strength but strong overall overlap among interaction targets between CHD8 TaDa-seq and ChIP-seq, with high levels of detection variability across CHD8 ChIP-seq as well as between TaDa-seq and ChIP-seq. Overall, comparison with published ChIP-seq provided strong support for CHD8 TaDa-seq identifying reproducible and relevant CHD8 genomic interactions.
Genome-wide, quantitative signal between the merged CHD8 TaDa-seq and independent ChIP-seq datasets were moderately correlated (Figures 3D and S4). This is not surprising, considering that the TaDa-seq coverage profile also inherently includes some background non-specific adenine methylation. The merged CHD8 TaDa-seq and age-matched E17.5 cortex CHD8 ChIP-seq datasets were also strongly correlated with ATAC-seq and histone marks associated with open and transcriptionally active chromatin, H3K4me3/me1 and H3K27ac (Figures 3D and S4). CHD8 TaDa-seq and ChIP-seq datasets showed reduced correlation with a mark for repressive chromatin, H3K27me3. As expected, the Dam-only genome-wide signal strongly correlated with ENCODE E16.5 fetal cortex ATAC-seq datasets, confirming that Dam-only signal identifies accessible chromatin. Dam-only datasets were also strongly correlated with H3K4me3, H3K4me1, and H3K27ac marks, consistent with the relationship between accessible chromatin and transcriptionally active chromatin states at promoters and enhancers. Of note, correlation of the Dam-only coverage was much lower for the merged CHD8 TaDa-seq, indicating that the CHD8 fusion protein directed a change in the adenine methylation profile, as expected if CHD8 fusion confers interaction specificity. The four CHD8 TaDa-seq replicates sub-clustered into two pairs based on genome-wide signal (Figure S4). Two of the replicates had relatively lower CHD8-specific signal and thus clustered with the Dam-only experiments. The differences in genome-wide signal across CHD8 TaDa-seq replicates may be due to technical differences in IUE delivery, library preparation, and sequencing depth. Despite differences between replicates, all four replicates showed consistent CHD8-specific TaDa-seq enrichment patterns, as evident in example loci (Figure S1). Overall, these results support the reproducibility and biological relevance of CHD8 TaDa-seq results and suggest a primary role of CHD8 in transcriptional activation in embryonic mouse cortex.
CHD8 TaDa-seq indicates role of CHD8 in transcriptional activation of genes associated with cellular homeostasis and distal enrichment at neuronal loci
Gene ontology analysis using GREAT (McLean et al., 2010) showed CHD8 TaDa-seq peaks had the strongest enrichment for genes associated with general cellular homeostasis, and specifically with RNA splicing, protein folding, and chromatin regulation genes (Figures 3E and Table S1). This finding is consistent with previous findings of CHD8 ChIP-seq datasets (Wade et al., 2019). There was also evidence for reduced, but still significant, enrichment across all CHD8 TaDa-seq interactions at loci associated with metabolism and neuron differentiation, also in line with earlier evidence. The interactions that were called as significant peaks in TaDa-seq, but were not called as peaks in either CHD8 ChIP-seq dataset (i.e., TaDa-seq unique peaks, Figures 3B and 3C), were associated with genes annotated to terms much more strongly associated with neurodevelopment and neuronal function (Figure 3F). Peaks that were unique to either CHD8 ChIP-seq dataset did not have the same strength of functional enrichment, although some terms were enriched among associated loci for both datasets (Figure S2E).
Intersection of genes associated with CHD8 TaDa-seq peaks with transcriptomic data from a published analysis of E17.5 cortex from mice harboring heterozygous Chd8 mutations (Gompers et al., 2017) showed that genes associated with strong CHD8 TaDa-seq peaks are both highly expressed in E17.5 embryonic mouse cortex and more likely to be downregulated as a consequence of Chd8 haploinsufficiency (Figures 4A–4C). Similar general patterns were present for the two CHD8 ChIP-seq datasets (Figure S5). There was no enrichment of Dam-only interactions for downregulated genes (Figure 4B), further demonstrating that CHD8 TaDa-seq captures meaningful CHD8 genomic interactions compared with general Dam-only signal. Downregulated genes with CHD8 TaDa-seq interactions were enriched for functions associated with transcriptional regulation and RNA processing functions. CHD8 TaDa-seq interactions were less likely to be found at loci that were upregulated as a consequence of Chd8 haploinsufficiency, with some evidence for enrichment of terms associated with DNA packaging among genes with CHD8 TaDa-seq and upregulation (Figure 4D). Overall, our CHD8 TaDa-seq results provide evidence for CHD8-dependent activation of highly expressed genes associated with general cellular functions, consistent with results from individual CHD8 studies and from meta-analysis of published CHD8 ChIP-seq data (Ceballos-Chávez et al., 2015; Cotney et al., 2015; de Dieuleveult et al., 2016; Gompers et al., 2017; Platt et al., 2017; Katayama et al., 2016; Sugathan et al., 2014; Wade et al., 2019). Intersection of CHD8 TaDa-seq interactions with ASD-associated genes (Satterstrom et al., 2020) highlighted strong enrichment among ASD-associated genes that are associated with gene expression regulation functions. Although not as well represented, CHD8 TaDa-seq interactions were also present at neuronal communication and cytoskeleton genes (Figure S6A).
Figure 4.
CHD8 binding is associated with activation of highly expressed genes
(A) Box and whisker plots showing change in log fold counts per million of genes according to peak rank in CHD8 TaDa-seq (CHD8 TaDa), both CHD8 TaDa-seq and CHD8 ChIP-seq (TaDa & ChIP), or Dam-Only datasets and an E17.5 Chd8 haploinsufficiency differential gene expression dataset. Boxes were plotted according to CHD8 binding affinity bins: all genes meeting at least 0.1 count per million sequencing coverage (Expressed Genes), any genes having CHD8 binding (All Bound Genes), and the top 1,000 genes near CHD8 peaks (Top 1000 Bound). Notches indicate values within the 95% confidence interval of the median.
(B) Box and whisker plots showing log fold change of genes according to peak rank in CHD8 TaDa-seq (CHD8 TaDa), both CHD8 TaDa-seq and CHD8 ChIP-seq (TaDa & ChIP), or Dam-Only datasets and an E17.5 Chd8 haploinsufficiency differential gene expression dataset. Boxes were plotted according to CHD8 binding affinity bins: all genes meeting at least 0.1 count per million sequencing coverage (Expressed Genes), any genes having CHD8 binding (All Bound Genes), and top 1,000 genes near CHD8 peaks (Top 1000 Bound). Notches indicate values within the 95% confidence interval of the median. C-D (left) Venn diagrams indicating the number of genes overlapping between the CHD8 TaDa-seq and E17.5 Chd8 haploinsufficiency significant (p < 0.05) downregulated and upregulated datasets.
(C and D right) Tables showing functional annotations associated with genes having CHD8 binding in downregulated (C) and upregulated (D) genes from the E17.5 Chd8 haploinsufficiency dataset (p < 0.05) using goseq. Enrichment values indicate the percent of genes in the dataset that are differentially expressed and bound by CHD8 via TaDa-seq in relation to the total number of genes associated with each term. See also Figures S5 and S6.
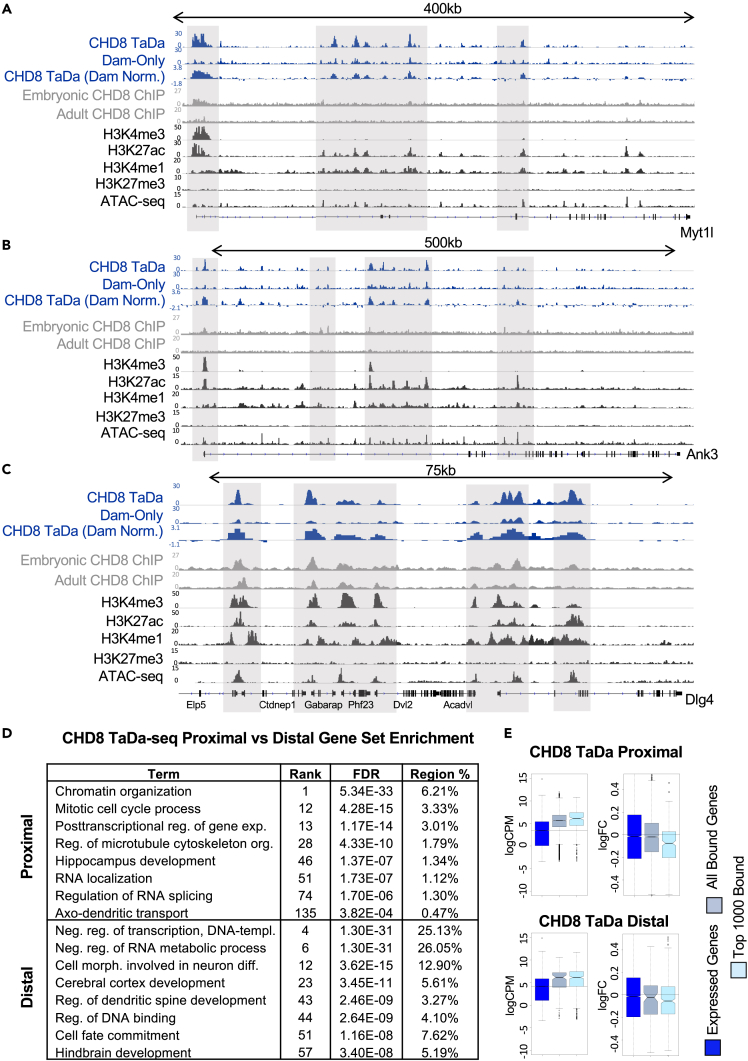
Of interest, loci with CHD8 TaDa-seq unique peaks and distal binding included many genes associated with neurodevelopmental and neuron-specific function. This suggests that CHD8 TaDa-seq captured differences in proximal versus distal targets of CHD8 in embryonic mouse cortex. Examples of neuronal genes with distal TaDa-seq CHD8 interactions include genes with dual roles in gene regulation and neurodevelopment, such as Myt1l (Figure 5A), as well as genes having more specific roles in neuronal morphology and synaptic signaling, such as Ank3 and Dlg4, which encodes PSD95 (Figures 5B and 5C). Comparison of distal CHD8 TaDa-seq peaks with marks for putative enhancers (H3K27ac, H3K4me1), open chromatin (ATAC), and transcriptional activation (H3K4me3) suggests that CHD8 TaDa-seq distal peaks are cis-regulatory elements. The distal CHD8 interactions identified in our CHD8 TaDa-seq data are somewhat captured by the representative CHD8 ChIP-seq datasets, but with reduced relative enrichment compared with the TaDa-seq signal at these sites (Figures 5A–5C).
Figure 5.
TaDa-seq identifies both promoter proximal and promoter distal CHD8 binding
(A–C) CHD8 binding near genes important for regulation of neuronal gene expression, Myt1l (A), and synaptic function, Ank3 (B) and Dlg4 (C). Gray boxes highlight CHD8 binding near select promoter and distal regions of interest overlapping with putative enhancer marks (H3K27ac and H3K4me1). CHD8 TaDa-seq experiment tracks are in blue, CHD8 ChIP-seq experiments are in gray, and datasets of histone and chromatin accessibility signatures from the ENCODE consortium are in black. Linear representations of genes from the mouse mm10 genome are shown below coverage tracks. Height of the y axis is scaled to show the peak for each track separately.
(D) Table showing functional annotations associated with promoter proximal (<1kb from TSS) (top) and promoter distal (bottom) regions. Rank refers to the rank within the dataset. A rank of 1 would mean the annotation with the smallest FDR value (aka the most significant). Region % refers to the percent of regions captured compared with the total number of peaks.
(E) Box and whisker plots showing change in log fold counts per million (left) or log fold change (right) of genes according to peak rank in CHD8 TaDa-seq proximal (top) or distal (bottom) datasets and an E17.5 Chd8 haploinsufficiency differential gene expression dataset. Boxes were plotted according to CHD8 binding affinity bins: all genes meeting at least 0.1 count per million sequencing coverage (Expressed Genes), any genes having CHD8 binding (All Bound Genes), and top 1,000 genes near CHD8 peaks (Top 1000 Bound). Notches indicate values within the 95% confidence interval of the median. See also Figures S5 and S6. Embryonic CHD8 ChIP (Cotney et al., (2015)) embryonic CHD8 ChIP-seq, Adult CHD8 ChIP (Platt et al., (2017)) adult CHD8 ChIP-seq.
To assess whether gene sets bound by CHD8 at their promoters and those targeted distally indeed are enriched for different functional categories and have evidence for CHD8 regulation, we split CHD8 TaDa-seq and ChIP-seq peaks into promoter proximal (within 1 kb of TSS) and promoter distal interactions (Figures 5D and S5). Loci with promoter binding mirrored the overall analysis (Figure 5D top). Distal CHD8 interactions were also enriched at loci associated with general regulatory function terms such as “negative regulation of transcription” and “negative regulation of RNA metabolic process.” However, loci associated with distal CHD8 interactions identified via TaDa-seq were more strongly enriched for brain development and neuronal functions, for example, “cell morphogenesis involved in neuron differentiation,” “regulation of dendritic spine development,” and “cell fate commitment” (Figure 5D bottom, Table S1). CHD8 ChIP-seq datasets similarly split into proximal and distal peaks indicate concordant patterns of increased distal binding associated with genes linked to neuronal development and function, although with reduced enrichment and lower numbers of relevant peaks compared with the TaDa-seq distal peaks (Figure S6B, Table S1). Similar to proximal interactions, loci with distal CHD8 TaDa-seq binding showed an overall significant trend toward downregulation in the age-matched published Chd8 mutant cortex RNA-seq data described above (Figure 5E), providing evidence that CHD8 TaDa-seq distal interactions have regulatory significance. The representative CHD8 ChIP-seq datasets do not exhibit an association between their distal or unique interactions and downregulation in the mutant cortex RNA-seq (Figure S5). These analyses highlight that TaDa-seq reveals distal CHD8 interactions in embryonic cortex at a subset of genes with significant relevance to neuronal development and function and that these regulatory targets appear sensitive to Chd8 haploinsufficiency.
Discussion
We successfully implemented Targeted DamID followed by sequencing (TaDa-seq) in vivo in embryonic mouse brain by in utero electroporation, characterizing genomic interactions of the NDD-relevant chromatin remodeler, CHD8. Although CHD8 ChIP-seq studies have provided valuable insights regarding CHD8 molecular function in vivo in mouse brain and across in vitro models, variable results across individual ChIP-seq experiments can confound interpretations of CHD8 activity and gene regulation. Thus, our study represents a proof-of-principle implementation of TaDa-seq in the context of in vivo mouse brain development and advances understanding of CHD8, a leading NDD risk gene. These findings open new avenues to interrogate the function of proteins that are intractable or technically challenging to study using ChIP-seq. The interactions identified here using TaDa-seq have orthogonally validated CHD8 interactions mapped using ChIP-seq and highlight the presence of CHD8 distal interactions at NDD-relevant neurodevelopmental and neuronal genes.
Implementation of TaDa-seq requires up-front steps of construct generation and delivery for expression in the cells or tissues of interest. Furthermore, the Dam methylase must be expressed and have time to methylate at genomic sites. In contrast, ChIP-seq can be performed on unmodified cells or tissues and captures interactions present at a specific time. However, ChIP-seq grade antibodies must be available, cross-linking and fragmentation is generally required, and protein-DNA interactions must be strong enough to enable sensitive capture using immunoprecipitation. Furthermore, TaDa-seq requires substantially less material than typical ChIP-seq methods (Cheetham et al., 2018; Tosti et al., 2018). We note additional general considerations and limitations with regard to implementing TaDa-seq in vivo in mouse brain. First, there is dependency on consistent and broad IUE delivery across cell types of interest in the brain, and differences in delivery to specific cell populations may impact sensitivity and reproducibility of interaction detection. Second, difference between levels of Dam-fusion protein expression and physiological expression levels of the endogenous gene (here CHD8) could result in some alteration of normal genomic interaction patterns. Third, similar to ChIP-seq, the library preparation and sequencing can introduce technical variability, so replicate experiments are critical. Finally, this method is dependent on the Dam-fusion having increased specificity compared with non-specific Dam-only control, such that reduced or low affinity of the fusion protein for the endogenous genomic targets will result in poor sensitivity and specificity. Nonetheless, while ChIP-seq remains a generally applicable method with clear temporal resolution, we here demonstrate that TaDa-seq can overcome barriers that negatively impact ChIP-seq performance and capture protein-DNA interactions that might be missed due to sensitivity thresholds of ChIP-seq. Conditional expression of the TaDa-seq constructs with cell-type specific promoters would enable identification of cell-type-specific chromatin interactions, as has been shown in Drosophila (Southall et al., 2013; Otsuki and Brand, 2018; Marshall and Brand, 2017; Cheetham and Brand, 2018). Such an approach offers the potential to address key questions regarding context-specific function and genomic interactions of chromatin remodelers and other DNA-associated proteins in the developing mouse brain.
By directly comparing CHD8 TaDa-seq and ChIP-seq, we found that TaDa-seq experiments are reproducible and perform well with regard to sensitivity and specificity, with strong overall concordance between the interactions mapped by these different methods. TaDa-seq thus joins the few published methods for resolving protein-DNA interactions genome-wide that can be deployed in vivo and do not require cross-linking or immunoprecipitation. Recently, another such method mapped transcription factor interactions at single-cell resolution by fusing transcription factors to transposase domains and locating transposition events through direct DNA sequencing (Moudgil et al., 2020). Application of TaDa-seq offers the opportunity to characterize the neurodevelopmental function of chromatin remodeler proteins implicated in NDDs, as we did here for CHD8, that might be difficult to interrogate using ChIP-seq. For example, TaDa-seq has been used to map the binding sites of kismet, the Drosophila ortholog for CHD8, which has roles in cell proliferation, synaptic transmission, axonal pruning, circadian rhythm, and memory (Gervais et al., 2019).
Overlapping sets of CHD8 interactions were largely captured by both TaDa-seq and ChIP-seq technologies, but with differences in signal strength. This could be due to general differences in performance or CHD8-specific features due to strong correlation between CHD8 binding and open chromatin at promoters. Previous comparisons of TaDa-seq and ChIP-seq in vitro and in Drosophila have found similar evidence for general concordance in target loci but divergence in quantitative strength (Southall et al., 2013; Marshall et al., 2016; Marshall and Brand, 2015; Otsuki and Brand, 2018; Cheetham et al., 2018; Tosti et al., 2018). It is possible that TaDa-seq may be less sensitive to repressive interactions where DNA is not accessible for Dam methylation, as our results suggest that peaks called in CHD8 ChIP-seq but not TaDa-seq had reduced Dam-only signal as well. On the other hand, by using adenine methylation as the readout, TaDa-seq does not seem to be as impacted by over-sampling of stronger protein-DNA interactions and may thus better capture transient or weaker interactions. Alternatively, as TaDa-seq captures adenine methylation throughout the incubation time of E13.5 to E17.5, it is possible that some of the TaDa-seq-specific CHD8 interactions are limited to stages earlier than profiled via ChIP-seq at E17.5. In summary, CHD8 interactions detected by TaDa-seq and ChIP-seq were overall highly concordant, with some evidence for assay-specific differences such that the combined interaction sets can complement each other.
Consistent with previous work, our study confirms that the majority of CHD8 interactions occur at promoters with no evidence for direct binding of CHD8 to a primary DNA motif, supporting a model of CHD8 recruitment by co-factors or transcription factors. Our results also support a direct role in transcriptional activation. CHD8 interactions were strongly correlated with open chromatin assayed by ATAC-seq and with histone marks associated with open and actively transcribed promoters. Loci with CHD8 interactions were also more likely to be downregulated owing to Chd8 haploinsufficiency. Finally, comparison between CHD8 TaDa, Dam-only, and ENCODE data clearly showed that CHD8 interactions are specific to a subset of promoter and distal loci, rather than broadly co-occurring with accessible chromatin or with the global deposition of any specific histone modifications. Our CHD8 TaDa-seq results further establish the strong enrichment of CHD8 binding near promoters of genes associated with general cellular functions involved in replication, chromatin, transcription, and translation.
The TaDa-seq data also highlight potential CHD8 involvement in distal regulation for a subset of neurodevelopmental and neuronal genes. The evidence for a brain-specific CHD8 distal interaction signature has significant potential implications for models of the role of CHD8 in brain development and function. The unexpected increased signature for CHD8 binding near distal regions in the TaDa-seq experiments indicates that using orthogonal approaches to ChIP-seq may bring novel insights due to differing detection biases. Distal CHD8-interaction regions overlap with H3K27ac, a histone mark associated with putative enhancers, suggesting a role related to distal regulatory elements. Loci with distal CHD8 interactions showed decreased E17.5 expression in a previous study, indicating these distal interactions may have regulatory relevance. It is possible that CHD8 is involved in distal chromatin remodeling or enhancer activation in the developing brain, parallel to what has been reported in the context of CHD8 in estrogen response (Ceballos-Chávez et al., 2015). Although CHD8 is an essential gene, there are opposite effects on cortex development between Chd8 null knockout mice, exhibiting microcephaly, and mice heterozygous for a Chd8 mutation, exhibiting macrocephaly (Hurley et al., 2021). It is possible that CHD8 haploinsufficiency has a specific effect on a subset of CHD8 interactions, for example, disrupting weaker interactions. Further studies are needed to explore the difference in CHD8 function in the brain at promoters versus distal sites and dosage sensitivity of these interactions in the context of CHD8 haploinsufficiency. Future studies are also necessary to determine the context-specific protein interaction partners of CHD8 to understand its role in transcriptional regulation in the brain.
In summary, this study shows the value of TaDa-seq as an alternative to ChIP-seq, to serve as a novel implementation to map protein-DNA interactions in embryonic mouse cortex. Implementation of CHD8 TaDa-seq revealed a complementary and expanded set of CHD8 target loci in the genome in developing mouse cortex, furthering understanding of the genomic function of CHD8 in neurodevelopment and the relationship between CHD8 interaction targets and ASD- and ID-relevant pathology caused by CHD8 mutations. This work serves as a model for studying other proteins, including the many chromatin remodeling factors associated with NDDs for which ChIP-seq may be technically challenging or where ChIP-seq grade antibodies are unavailable.
Limitations of the study
The TaDa-seq findings in this study are based on four biological replicates collected at a single embryonic time point and exhibit some variability in signal. We present evidence that consensus peaks observed across CHD8 TaDa-seq datasets represent reproducible CHD8 binding loci, as they have significant overlap with CHD8 ChIP-seq studies and are associated with genes that are sensitive to Chd8 haploinsufficiency. However, it is likely that many of the interactions captured in individual replicates but not across all four datasets are also true CHD8 genomic targets. Similarly, it is possible that some interactions that were specific to TaDa-seq and not present in the representative ChIP-seq datasets are not true CHD8 interaction targets or do not have regulatory relevance, as we lack an appropriate dataset for ground-truthing these findings. Furthermore, IUE delivery to embryonic mouse cortex at E13.5 preferentially results in delivery to glutamatergic neurons, and CHD8 interactions in other cell types are less likely to be recovered in our datasets. More generally, performance comparisons between CHD8 TaDa-Seq and CHD8 ChIP-seq are hampered by the lack of a gold standard set of interactions against which to test sensitivity and specificity. Finally, future studies are needed to verify regulatory relevance for the intersections identified here by CHD8 TaDa-seq that are not captured by ChIP-seq.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-GFP | Abcam | ab13970 |
| Rabbit anti-RFP | Abcam | ab62341 |
| Alexa-488 | Invitrogen | Z25002 |
| Alexa-546 | Invitrogen | Z25004 |
| Bacterial and virus strains | ||
| pCAG-mCherry-intronDam-CHD8 | This paper | N/A |
| pCAG-mCherry-intronDam | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DpnI | NEB | R0176S |
| T4 DNA ligase | NEB | M0202S |
| DpnII | NEB | R0543S |
| AlwI | NEB | R0513S |
| Critical commercial assays | ||
| Qiagen QIAamp DNA Micro Kit | Qiagen | 56304 |
| QIAquick PCR Purification Kit | Qiagen | 28104 |
| MyTaq | Bioline | BIO-21112 |
| TruSeq | Illumina | RS-122-2001 |
| Deposited data | ||
| Chd8 TaDa-seq | GEO | GSE165002 |
| Cotney et al., 2015 | GEO | GSE57369 |
| Platt et al., 2017 | GEO | PRJNA379430 |
| de Dieuleveult et al., 2016 | GEO | GSE64825 |
| Gompers et al., 2017 | GEO | GSE99331 |
| Katayama et al., 2016 | DDBJ | DRA003116 |
| Sood et al., 2020 | GEO | GSE155216 |
| Recombinant DNA | ||
| Human CHD8 ORF | Origene | RG230753 |
| Software and algorithms | ||
| TrimGalore | Martin, 2011 | https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| FastQC tool | Wingett and Andrews, 2018 | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| BWA | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net/ |
| samtools | Li et al., 2009 | http://www.htslib.org/ |
| deepTools | Ramírez et al., 2016 | https://deeptools.readthedocs.io/en/develop/index.html |
| MACS2 | Zhang et al., 2008 | https://github.com/macs3-project/MACS |
| bedtools | Quinlan and Hall, 2010 | https://bedtools.readthedocs.io/en/latest/.2 |
| GREAT | McLean et al., 2010 | http://great.stanford.edu/public/html/ |
| goseq | Young et al., 2010 | https://www.bioconductor.org/packages/release/bioc/html/goseq.html |
| HOMER | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Other | ||
| Seramag beads | Fisher Scientific | 65152105050250 |
Resource availability
Lead contact
Further information and requests for resources or reagents should be directed to the lead contact, Alex Nord, asnord@ucdavis.edu.
Materials availability
The study did not generate any unique reagents.
Method details
Targeted DamID constructs
Previously, we developed Targeted DamID (TaDa) to enable cell type-specific profiling in vivo while avoiding the potential toxicity resulting from expression of high levels of Dam methylase (Southall et al., 2013). Using TaDa, transcription of a primary open reading frame (ORF1; here mCherry) is followed by two TAA stop codons and a single nucleotide frameshift upstream of a secondary open reading frame: the coding sequence of the Dam fusion protein (ORF2; here Dam-CHD8). Translation of this bicistronic message results in expression of ORF1 as well as extremely low levels of the Dam fusion protein (ORF2) due to rare ribosomal re-entry and translational re-initiation. TaDa enables rapid, accurate and sensitive identification of genomic binding sites.
When Dam-fusion proteins are expressed in dam– bacteria, the methylase is able to methylate plasmid DNA. In transient transfection experiments, methylated plasmid DNA co-amplifies with genomic DNA and constitutes a substantial proportion of the sequencing library. For this reason, DamID was thought to be incompatible with transient transfection (Vogel et al., 2007). We introduced an intron into the coding sequence of the Dam methylase to prevent expression in bacteria but not in eukaryotes, where the intron is removed and the enzyme is expressed (J.v.d.A., S.W.C. and A.H.B., unpublished).
To generate the experimental plasmid, pCAG-mCherry-intronDam-CHD8, encoding the Dam methylase fused to the human CHD8 open reading frame (hereafter CHD8 TaDa), a full-length CHD8 isoform (Origene, RG230753) was subcloned by Gibson assembly into pCAG-mCherry-intronDam, C-terminal to the Dam methylase and a myc-tag. The control plasmid was pCAG-mCherry-intronDam (hereafter Dam-only). Plasmids were sequenced following subcloning. pCAG-Venus, encoding a variant of green fluorescent protein, served as a control for efficiency of in utero electroporation and to enable dissection of the electroporated region.
Delivery via IUE of fetal mouse cortex and generation of TaDa libraries for sequencing
MF1 mice from the same litter were in utero electroporated as previously described (Saito, 2006; Tiberi et al., 2012). CHD8 TaDa (0.5ug/ul) or Dam-only (0.5ug/ul) and electroporation-control (0.25ug/ul) plasmids were injected into the fetal brain ventricles at embryonic day (E) 13.5 before collection at E17.5. Successful electroporation was confirmed by immunohistochemistry using established methods (Tiberi et al., 2012). Primary antibodies were chicken anti-GFP 1/1000 (Abcam ab13970) and rabbit anti-RFP 1/500 (Abcam ab62341), and secondary antibodies coupled to Alexa-488 or Alexa-546 1/200 (Invitrogen). Nuclei were stained with DAPI. Images were acquired on a Leica SP8 confocal microscope and processed using ImageJ. Sample brains, 4 CHD8 TaDa and 3 Dam-only, were dissected and frozen for library processing. All mouse husbandry and experiments were carried out in a Home Office-designated facility, according to the UK Home Office guidelines upon approval by the local ethics committee (project license PPL70/8727).
Targeted DamID-seq (TaDa-seq) libraries were prepared as previously described (Marshall et al., 2016). Sample genomic DNA extraction was performed using the Qiagen QIAamp DNA Micro Kit (Qiagen, 56304). Extracted genomic DNA was digested overnight at 37°C with DpnI (NEB, R0176S) to cut adenine-methylated GATC sites. Following digestion, DNA was column purified with the QIAquick PCR Purification Kit (Qiagen, 28104) to remove un-cut genomic DNA. dsADR adaptors were blunt-end ligated to DpnI-digested fragments using T4 DNA ligase (NEB, M0202S; 2 hours at 16°C, heat inactivation at 65°C for 20 minutes) to prepare for PCR amplification. Before PCR amplification, fragments were digested with DpnII (NEB, R0543S) to cut non-methylated GATC sites and prevent amplification of unmethylated regions and purified with a 1:1.5 ratio of Seramag beads (Fisher Scientific, 65152105050250). PCR amplification of DpnII-digested fragments using MyTaq (Bioline, BIO-21112) enriched for methylated fragments before samples were sonicated and prepped for sequencing. Sonicated samples were subjected to AlwI digestion (NEB, R0513S) to remove previously ligated adaptors and initial GATC sequences from fragments. A modified TruSeq protocol was used to generate sequencing libraries involving end repair, 3′ end adenylation, sequencing adaptor ligation, and DNA fragment enrichment using a reduced number of PCR cycles. TaDa-seq libraries were sequenced on the Illumina HiSeq 1500 platform using a single-end 50bp strategy by the Gurdon Institute Next Generation Sequencing Core.
Quantification and statistical analysis
Sequenced TaDa-seq libraries were analyzed to identify genomic regions with enriched coverage for CHD8 TaDa and Dam-only libraries. Representative CHD8 ChIP-seq datasets were downloaded from the Sequence Read Archive (Cotney et al., 2015, GSE57369; Platt et al., 2017, PRJNA379430). Unaligned TaDa-seq and ChIP-seq reads were trimmed using TrimGalore (Version 0.4.2), assessed for general quality control with the FastQC tool (Version 0.11.9), and aligned to the mouse reference genome (mm10) using BWA (Version 0.7.17). Biological replicates were analyzed independently and as a single merged file generated via samtools (Version 1.10). Coverage plots were generated independently for Dam-only and CHD8 TaDa-seq replicates (deepTools, Version 3.3.1, RPKM normalization). Merged CHD8 TaDa-seq was normalized against Dam-only (Marshall and Brand, 2015) for visualization of coverage and enrichment. TaDa-seq peak calling was performed using MACS2 (Version 2.2.5) with model-based peak identification disabled, a p value cutoff set at less than 0.00001, and the merged Dam-only dataset as a control. Peak calling for individual CHD8 TaDa-seq replicates was performed against the merged Dam-only dataset to identify specific peaks that were enriched in CHD8 TaDa versus non-specific signal in Dam-only experiments. Peak calling for the Dam-only merged dataset was performed without a control dataset as Dam-only is analogous to assays of accessible chromatin. Peak calling for CHD8 ChIP-seq experiments was performed using the same MACS2 parameters, including comparison to input controls. A final set of merged CHD8 TaDa-seq peaks was obtained using bedtools intersect (Version 2.29.2) to select high confidence peaks that were present in at least 3 replicates and had a MACS2 FDR less than 0.00001.
Enriched regions from TaDa-seq and ChIP-seq datasets were annotated to genomic features using custom R scripts and combined UCSC and RefSeq transcript sets (Wade et al., 2019). CHD8 target genes were assigned to nearest transcription start site, which for distal peaks was achieved using the bedtools closest command (Version 2.29.2). Bigwig coverage files were generated using deeptools bamCoverage (Version 3.3.1). Embryonic E16.5 bigwig coverage files from the ENCODE Consortium portal (ENCODE Project Consortium, 2012; https://www.encodeproject.org/) were downloaded to compare CHD8 datasets with open chromatin and histone marks (Experiments: ENCSR428OEK, ENCSR658BBG, ENCSR587JRQ, ENCSR141ZQF, ENCSR836PUC, ENCSR129DIK). Genome-wide signal summary Spearman correlation heatmaps using the default bin size of 10 kb were generated using the multiBigwigSummary and plotCorrelation tools from deepTools (Version 3.3.1). Differences in signal intensity between CHD8 TaDa-seq replicates in the correlation heatmaps were due to differences in sequencing depth. Peak loci heatmaps were generated using the deeptools computeMatrix and plotHeatmap tools (Version 3.3.1). Intersection of called peaks was performed using bedtools intersect (Version 2.29.2) with CHD8 TaDa-seq filtered peaks and ChIP-seq datasets. Promoter-proximal versus promoter-distal and peak set concordance datasets were also obtained using the bedtools intersect tool (Version 2.29.2). Ontology analysis was performed using the GREAT online tool (McLean et al., 2010; Version 4.0.4) or goseq (Young et al., 2010; Version 1.36.0). HOMER was used to perform de novo motif discovery with default parameters (Heinz et al., 2010; Version 4.10). Comparison between CHD8 TaDa-seq and E17.5 RNA-seq data was performed using previously published RNA-seq data (Gompers et al., 2017; Wade et al., 2019).
Further analysis comparing CHD8 TaDa-seq peaks to other CHD8 mouse brain datasets was also performed. First, peak BED files were downloaded from GEO (de Dieuleveult et al., 2016: GSE64825; Gompers et al., 2017: GSE99331; Sood et al., 2020: GSE155216) or generated from raw data hosted by the DNA DataBank of Japan (Katayama et al., 2016: DRA003116; Wade et al., 2019). Then peak alignment for these datasets were converted from the mm9 to the mm10 annotation using the UCSC Genome Browser liftOver utility (Hinrichs et al., 2006). Peak overlap analysis was performed using bedtools intersect (Version 2.29.2) to identify shared peaks between datasets. Genome feature annotation and distance to TSS analyses were performed using the annotatePeaks function of HOMER (Version 4.10). Genes annotated to peaks in CHD8 ChIP-seq and TaDa-seq datasets were also identified using the HOMER annotatePeaks function and compared to autism-relevant gene sets identified in Satterstrom et al. (2020).
Acknowledgments
The authors acknowledge the National Institutes of Health (R01 MH120513 and R35 GM119831 to A.S.N., F31 MH119789 and T32 GM007377 to A.A.W.), Wellcome Trust Senior Investigator Award (103792) and Royal Society Darwin Trust Research Professorship to A.H.B., Wellcome Trust Postdoctoral Training Fellowship for Clinicians (105839) to J.v.d.A., and Herchel Smith Research Studentship to S.W.C. A.H.B. acknowledges core funding to the Gurdon Institute from the Wellcome Trust (092096) and CRUK (C6946/A14492).
Author contributions
A.A.W, A.S.N., and A.H.B. conceived of the project. J.v.d.A., S.W.C., and R.Y. performed experiments. A.A.W. performed computational analysis. A.A.W. and A.S.N. drafted the manuscript. All authors contributed to manuscript revisions.
Declaration of interests
The authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103234.
Contributor Information
Andrea H. Brand, Email: a.brand@gurdon.cam.ac.uk.
Alex S. Nord, Email: asnord@ucdavis.edu.
Supplemental information
Data and code availability
Data that support the findings of this study are available from GEO (Accession GSE165002) or upon request. Genomic coverage datasets are available as Track Hubs for visualization using the UCSC Genome Browser and analysis scripts are available at https://github.com/NordNeurogenomicsLab/.
References
- An Y., Zhang L., Liu W., Jiang Y., Chen X., Lan X., Li G., Hang Q., Wang J., Gusella J.F. De novo variants in the Helicase-C domain of CHD8 are associated with severe phenotypes including autism, language disability and overgrowth. Hum. Genet. 2020;139:499–512. doi: 10.1007/s00439-020-02115-9. [DOI] [PubMed] [Google Scholar]
- Attanasio C., Nord A.S., Zhu Y., Blow M.J., Biddie S.C., Mendenhall E.M., Dixon J., Wright C., Hosseini R., Akiyama J.A. Tissue-specific SMARCA4 binding at active and repressed regulatory elements during embryogenesis. Genome Res. 2014;24:920–929. doi: 10.1101/gr.168930.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aughey G.N., Cheetham S.W., Southall T.D. DamID as a versatile tool for understanding gene regulation. Development. 2019;146:dev173666. doi: 10.1242/dev.173666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aughey G.N., Estacio Gomez A., Thomson J., Yin H., Southall T.D. CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo. eLife. 2018;7:e32341. doi: 10.7554/eLife.32341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier R., Golzio C., Xiong B., Stessman H.A., Coe B.P., Penn O., Witherspoon K., Gerdts J., Baker C., Vulto-van Silfhout A.T. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos-Chávez M., Subtil-Rodríguez A., Giannopoulou E.G., Soronellas D., Vázquez-Chávez E., Vicent G.P., Elemento O., Beato M., Reyes J.C. The chromatin remodeler CHD8 is required for activation of progesterone receptor-dependent enhancers. Plos Genet. 2015;11:e1005174. doi: 10.1371/journal.pgen.1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham S.W., Brand A.H. RNA-DamID reveals cell-type-specific binding of roX RNAs at chromatin-entry sites. Nat. Struct. Mol. Biol. 2018;25:109–114. doi: 10.1038/s41594-017-0006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham S.W., Gruhn W.H., van den Ameele J., Krautz R., Southall T.D., Kobayashi T., Surani M.A., Brand A.H. Targeted DamID reveals differential binding of mammalian pluripotency factors. Development. 2018;145:dev170209. doi: 10.1242/dev.170209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J., Muhle R.A., Sanders S.J., Liu L., Willsey A.J., Niu W., Liu W., Klei L., Lei J., Yin J. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat. Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Dieuleveult M., Yen K., Hmitou I., Depaux A., Boussouar F., Bou Dargham D., Jounier S., Humbertclaude H., Ribierre F., Baulard C. Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature. 2016;530:113–116. doi: 10.1038/nature16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvlataniotis K., Bensberg M., Lentini A., Gylemo B., Nestor C.E. No evidence for DNA N6-methyladenine in mammals. Sci. Adv. 2020;6:eaay3335. doi: 10.1126/sciadv.aay3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzgou S., Liang H.W., Metcalfe K., Somarathi S., Tischkowitz M., Mohamed W., Kini U., McKee S., Yates L., Bertoli M. The clinical presentation caused by truncating CHD8 variants. Clin. Genet. 2019;96:72–84. doi: 10.1111/cge.13554. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey T.S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais L., van den Beek M., Josserand M., Sallé J., Stefanutti M., Perdigoto C.N., Skorski P., Mazouni K., Marshall O.J., Brand A.H. Stem cell proliferation is kept in check by the chromatin regulators kismet/CHD7/CHD8 and Trr/MLL3/4. Dev. Cell. 2019;49:556–573.e6. doi: 10.1016/j.devcel.2019.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompers A.L., Su-Feher L., Ellegood J., Copping N.A., Riyadh M.A., Stradleigh T.W., Pride M.C., Schaffler M.D., Wade A.A., Catta-Preta R. Germline Chd8 haploinsufficiency alters brain development in mouse. Nat. Neurosci. 2017;20:1062–1073. doi: 10.1038/nn.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D., Crabtree G. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F. The UCSC genome browser database: update 2006. Nucl. Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley S., Mohan C., Suetterlin P., Ellingford R., Riegman K.L.H., Ellegood J., Caruso A., Michetti C., Brock O., Evans R. Distinct, dosage-sensitive requirements for the autism-associated factor CHD8 during cortical development. Mol. Autism. 2021;12:16. doi: 10.1186/s13229-020-00409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I., O'Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Nishiyama M., Shoji H., Ohkawa Y., Kawamura A., Sato T., Suyama M., Takumi T., Miyakawa T., Nakayama K.I. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- Koziol M.J., Bradshaw C.R., Allen G.E., Costa A.S., Frezza C. Identification of methylated deoxyadenosines in genomic DNA by dA6m DNA immunoprecipitation. Bio. Protoc. 2015;6:e1990. doi: 10.21769/BioProtoc.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinov G.K., Kundaje A., Park P.J., Wold B.J. Large-scale quality analysis of published ChIP-seq data. G3 (Bethesda) 2014;4:209–223. doi: 10.1534/g3.113.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Brand A.H. damidseq_pipeline: an automated pipeline for processing DamID sequencing datasets. Bioinformatics. 2015;31:3371–3373. doi: 10.1093/bioinformatics/btv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Brand A.H. Chromatin state changes during neural development revealed by in vivo cell-type specific profiling. Nat. Commun. 2017;8:2271. doi: 10.1038/s41467-017-02385-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O.J., Southall T.D., Cheetham S.W., Brand A.H. Cell-type-specific profiling of protein-DNA interactions without cell isolation using targeted DamID with next-generation sequencing. Nat. Protoc. 2016;11:1586–1598. doi: 10.1038/nprot.2007.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudgil A., Wilkinson M.N., Chen X., He J., Cammack A.J., Vasek M.J., Lagunas T., Jr., Qi Z., Lalli M.A., Guo C. Self-reporting Transposons enable simultaneous readout of gene expression and transcription factor binding in single cells. Cell. 2020;182:992–1008.e21. doi: 10.1016/j.cell.2020.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski P.J., Zachariou A., Loveday C., Beleza-Meireles A., Bertoli M., Dean J., Douglas A.G.L., Ellis I., Foster A., Graham J.M. The CHD8 overgrowth syndrome: a detailed evaluation of an emerging overgrowth phenotype in 27 patients. Am. J. Med. Genet. C Semin. Med. Genet. 2019;181:557–564. doi: 10.1002/ajmg.c.31749. [DOI] [PubMed] [Google Scholar]
- Otsuki L., Brand A.H. Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science. 2018;360:99–102. doi: 10.1126/science.aan8795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak N.N., Luo R., Zhang A., Won H., Lowe J.K., Chandran V., Horvath S., Geschwind D.H. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R.J., Zhou Y., Slaymaker I.M., Shetty A.S., Weisbach N.R., Kim J.A., Sharma J., Desai M., Sood S., Kempton H.R. Chd8 mutation leads to autistic-like behaviors and impaired striatal circuits. Cell Rep. 2017;19:335–350. doi: 10.1016/j.celrep.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L. Large-Scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood S., Weber C.M., Hodges H.C., Krokhotin A., Shalizi A., Crabtree G.R. CHD8 dosage regulates transcription in pluripotency and early murine neural differentiation. PNAS. 2020;117:22331–22340. doi: 10.1073/pnas.1921963117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall T.D., Gold K.S., Egger B., Davidson C.M., Caygill E.E., Marshall O.J., Brand A.H. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev. Cell. 2013;26:101–112. doi: 10.1016/j.devcel.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugathan A., Biagioli M., Golzio C., Erdin S., Blumenthal I., Manavalan P., Ragavendran A., Brand H., Lucente D., Miles J. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. PNAS. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiberi L., van den Ameele J., Dimidschstein J., Piccirilli J., Gall D., Herpoel A., Bilheu A., Bonnefont J., Iacovino M., Kyba M. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nat. Neurosci. 2012;15:1627–1635. doi: 10.1038/nn.3264. [DOI] [PubMed] [Google Scholar]
- Tosti L., Ashmore J., Tan B.S.N., Carbone B., Mistri T.K., Wilson V., Tomlinson S.R., Kaji K. Mapping transcription factor occupancy using minimal numbers of cells in vitro and in vivo. Genome Res. 2018;28:592–605. doi: 10.1101/gr.227124.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ameele J., Krautz R., Brand A.H. TaDa! Analysing cell type-specific chromatin in vivo with Targeted DamID. Curr. Opin. Neurobiol. 2019;56:160–166. doi: 10.1016/j.conb.2019.01.021. [DOI] [PubMed] [Google Scholar]
- Vissers L.E., Gilissen C., Veltman J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- Vogel M.J., Peric-Hupkes D., van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Wade A.A., Lim K., Catta-Preta R., Nord A.S. Common CHD8 genomic targets contrast with model-specific transcriptional impacts of CHD8 haploinsufficiency. Front. Mol. Neurosci. 2019;11:481. doi: 10.3389/fnmol.2018.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasin H., Gibson W.T., Langlois S., Stowe R.M., Tsang E.S., Lee L., Poon J., Tran G., Tyson C., Wong C.K. A distinct neurodevelopmental syndrome with intellectual disability, autism spectrum disorder, characteristic facies, and macrocephaly is caused by defects in CHD8. J. Hum. Genet. 2019;64:271–280. doi: 10.1038/s10038-019-0561-0. [DOI] [PubMed] [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., Yin R., Zhang D., Zhang P., Liu J. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available from GEO (Accession GSE165002) or upon request. Genomic coverage datasets are available as Track Hubs for visualization using the UCSC Genome Browser and analysis scripts are available at https://github.com/NordNeurogenomicsLab/.