Summary
Apolipoprotein E4 (APOE4) is the strongest genetic risk factor for sporadic Alzheimer’s disease (AD). APOE4 is known to affect the function of microglia, but to what extent this gene drives microglial gene expression has thus far not been examined. Using a transgenic mouse model of AD that expresses human APOE, we identify a unique transcriptional profile associated with APOE4 expression. We also show a sex and APOE interaction, such that both female sex and APOE4 drive expression of this gene profile. We confirm these findings in human cells, using microglia derived from induced pluripotent stem cells (iMGL). Moreover, we find that these interactions are driven in part by genes related to metal processing, and we show that zinc treatment has APOE genotype-dependent effects on iMGL. These data identify a sex- and APOE4-associated microglial transcription profile and highlight the importance of considering interactive risk factors such as sex and environmental exposures.
Subject areas: Molecular biology, Biology of gender, Transcriptomics
Graphical abstract
Highlights
-
•
Sex and APOE genotype drive microglial gene expression profiles in APOE-FAD mice
-
•
Human iPSC-derived microglia show an APOE-dependent transcriptional profile
-
•
APOE genotype and sex are associated with heavy metal processing genes
-
•
Zinc exposure affects iPSC-derived microglia in an APOE-genotype-dependent manner
Molecular biology; Biology of gender; Transcriptomics
Introduction
Alzheimer’s disease (AD) is a progressive, neurodegenerative disorder with only very limited and often ineffective treatment options. AD is characterized by the accumulation of β-amyloid (Aβ) plaques and neurofibrillary tangles composed of hyperphosphorylated tau, which are thought to drive neuron loss (LaFerla, 2010; Serrano-Pozo et al., 2011). In addition, there is strong evidence of chronic neuroinflammation and glial activation, and this has been proposed as a potential driving force of the disease (Glass et al., 2010; Wyss-Coray and Rogers, 2012), as increases in cerebrospinal fluid inflammatory markers can be detected even before Aβ pathology (Eikelenboom et al., 2011).
Neuroinflammation is largely regulated by two major cell types in the brain: astrocytes and microglia. Increasing evidence suggests that microglia are a key player in AD pathogenesis; however, there is conflicting evidence as to whether they are protective or detrimental (Hansen et al., 2018). For example, microglia are found localized around Aβ plaques in the AD brain, which initially may be beneficial (Yeh et al., 2016). However, as microglia become less efficient at phagocytosing Aβ with age and disease (Spittau, 2017), their chronic activation and release of pro-inflammatory cytokines may become harmful and exacerbate neuron loss (Blasko et al., 2004; Mosher and Wyss-Coray, 2014). Patients with AD (Friedman et al., 2018) have microglia with a unique transcriptional signature, referred to as disease- associated microglia (DAM), and this has been recapitulated in the commonly used 5xFAD familial mouse model of AD compared with wild-type mice (Keren-Shaul et al., 2017). These cells exhibit a downregulation of genes associated with homeostatic mechanisms and increased expression of genes associated with immune responses and phagocytosis (Keren-Shaul et al., 2017). They have also been presumed to be beneficial in slowing the spread of Aβ (Deczkowska et al., 2018; Keren-Shaul et al., 2017).
The main risk factor for late-onset sporadic AD, the E4 allele of apolipoprotein E (APOE), is well recognized to directly affect both immune and microglial functions. Apolipoprotein E4 (APOE4) is associated with increased Aβ deposition and cognitive decline (Villemagne et al., 2013) as well as with earlier disease onset (van der Flier et al., 2011), compared with the neutral risk isoform, APOE3. APOE4 is associated with increased inflammation, both in healthy carriers (Colton et al., 2004; Gale et al., 2014) and in patients with AD (Olgiati et al., 2010), and APOE4 carriers demonstrate greater cognitive decline due to pro-inflammatory cytokines (Schram et al., 2007). Humans (Egensperger et al., 1998) and AD transgenic mice carrying APOE4 have increased numbers of both total and reactive microglia (Moser and Pike, 2017; Rodriguez et al., 2014). Of interest, the effects of APOE4 differ by sex, with females showing an exacerbation of APOE4-related AD outcomes (Altmann et al., 2014; Cacciottolo et al., 2016).
Although recent work demonstrates effects of APOE4 on global gene expression in the brain (Zhao et al., 2020), it is unclear to what extent APOE4 alters microglial gene transcription and, if so, how changes relate to the already characterized DAM phenotype. To address this role of APOE4, we analyzed microglia isolated from the APOE-FAD transgenic mouse model of AD (Youmans et al., 2012). These mice are a cross between the 5xFAD mouse and the human APOE mouse that has mouse APOE replaced with either human APOE3 or APOE4. We also analyzed human microglia differentiated from induced pluripotent stem cells (iMGL) from healthy older APOE3 or APOE4 adult donors. In addition, to address whether sex differentially affects APOE4-specific microglial gene expression, mouse and human microglia of both sexes were profiled. As may be expected, RNA sequence (RNA-Seq) analysis shows that the microglial transcription profile from APOE4 partially overlaps with the previous set of identified DAM genes. But, notably, APOE4 also drives its own distinct microglial transcription profile. Furthermore, we show that sex differentially affects this APOE4 transcriptional profile. This unique microglial transcription phenotype driven by an interaction of APOE4 and sex highlights the importance of considering genetic background and sex in disease modeling.
Results
FAD transgenes drive a unique disease-associated transcriptional profile in microglia isolated from mice expressing human APOE
We first sought to determine whether there is a DAM phenotype in FAD mice carrying human APOE (APOE-FAD), and to what extent this gene expression profile overlaps with the DAM profile previously described in 5xFAD mice by Keren-Shaul et al. Microglia were extracted from whole brain of human APOE and APOE-FAD mice. The sex, genotype, and age of each mouse are detailed in Table S1. Following perfusion with ice-cold phosphate buffered saline (PBS) to remove any blood immune cell types, CD11b+ cells were labeled with magnetic beads and isolated by sorting on a MACS column (Figure S1A). This protocol (Garcia et al., 2014) produced a highly purified microglial population, as evidenced by high IBA1 expression but low levels of the astrocytic and neuronal markers GFAP and MAP2 (Figure S1B). Samples were processed for RNA-Seq and analyzed using principal component analysis (PCA).
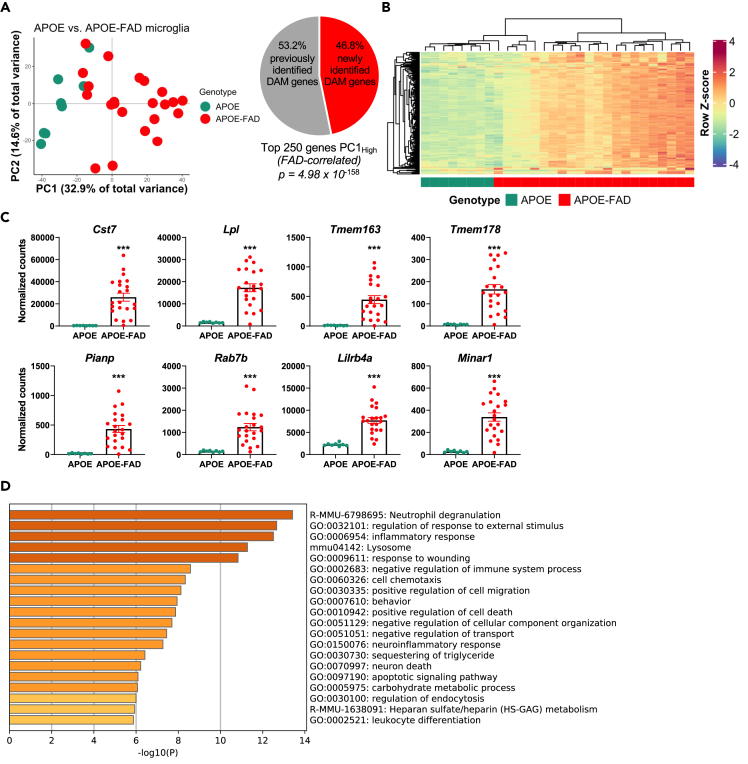
In order to understand global differences in microglial gene transcription between APOE and APOE-FAD mice, we collapsed across APOE genotype and sex in this analysis. PCA performed using all expressed genes demonstrated that PC1 separated APOE-FAD from APOE mice (Figure 1A). The top 250 ranked genes from PC1; thus, the top 250 genes associated with APOE-FAD mice along PC1, were then compared with the DAM genes previously identified by Keren-Shaul et al. Although 53.2% of 133 genes from PC1 overlap with the DAM genes that were previously reported as differentially expressed between wild-type and 5xFAD mice, the other 117 genes are unique to the humanized APOE-FAD mouse model used here, suggesting that these genes are related to human APOE expression (Figure 1A). The top 250 genes associated with PC1, along with log2 fold changes between APOE and APOE-FAD animals and adjusted p values, are detailed in Table S2.
Figure 1.
FAD transgenes drive a microglial transcription profile in mice carrying human APOE
(A) PC1 separates APOE from APOE-FAD mice. Of the top 250 genes associated with APOE-FAD mice along PC1, 53.2% are disease-associated microglia genes that have been previously identified, whereas 46.8% are unique to this mouse model with human APOE. Of these 250 genes, 242 are statistically significantly different at a Benjamini-Hochberg false discovery rate (FDR) adjusted p value of p < 0.05; these 242 genes are referred to as DAM-APOE genes and used for subsequent comparisons.
(B) Heatmap of the top 250 genes associated with APOE-FAD versus APOE mice along PC1. Gene expression levels were normalized, scaled, and centered and are displayed as Z scores.
(C) Gene expression (in normalized counts) of select disease-associated microglia markers, showing increased expression of these genes in APOE-FAD relative to APOE mice. Data are presented as mean (±SEM) values; n = 8–20/group. APOE mice are shown in green; APOE-FAD mice are shown in red. ∗∗∗ Benjamini-Hochberg adjusted p < 0.001 relative to APOE mice.
(D) Gene ontology (GO) analysis showing enrichment of GO terms in DAM-APOE genes.
To assess enrichment, hypergeometric ratio testing was performed, demonstrating that the degree of overlap between these gene sets is highly significant at an adjusted p < 4.98 × 10−158. Based on differential gene expression (DGE) analysis, we found that 242 of the top 250 ranked PC1 genes are significantly different between APOE-FAD and APOE mice (p < 0.05; false discovery rate [FDR] < 5%). As expected, these genes are upregulated in APOE-FAD compared with APOE mice (Figures 1B and 1C). Increased expression of previously identified DAM genes in APOE-FAD as compared with APOE mice was confirmed by rt-PCR (Figure S1C). Thus, we define the 242 genes significantly driving the separation of APOE and APOE-FAD mice along PC1 as DAM-APOE genes and use this set of genes for all subsequent analyses. These results demonstrate that human APOE shifts the transcriptional signature associated with FAD transgenes and highlight the importance of considering this genetic risk factor in transcriptional profiling.
In order to identify the biological processes differentially regulated in microglia from APOE and APOE-FAD mice, a gene ontology (GO) analysis (Zhou et al., 2019) was performed on the 242 DAM-APOE genes (Figure 1D). Several immune processes including the GO terms “inflammatory response” (GO 0006954), “negative regulation of immune system process” (GO 0002683), “neuroinflammatory response” (GO 0150076), “leukocyte differentiation” (GO 0002521), and the reactome gene set “neutrophil degranulation” (R-MMU-6798695) were overrepresented in the DAM-APOE genes. Moreover, a number of GO terms relating to cellular movement/responses including “regulation of response to external stimulus” (GO 0032101), “response to wounding” (GO 0009611), “cell chemotaxis” (GO 0060326), and “positive regulation of cell migration” (GO 0030335) were also overrepresented in this gene set. GO terms related to cell death and/or turnover were also upregulated; specifically, “positive regulation of cell death” (GO 0010942), “negative regulation of cellular component organization” (GO 0051129), “negative regulation of transport” (GO 0051051), “neuron death” (GO 0070997), and “apoptotic signaling pathway” (GO 0097109). We also found that genes associated with “sequestering of triglyceride” (GO 0030730) and “carbohydrate metabolic process” (GO 0005975) were upregulated in APOE-FAD versus APOE mice. Lipid metabolism pathways have previously been identified as part of the DAM signature (Keren-Shaul et al., 2017; Marschallinger et al., 2020), and microglia from 5xFAD mice were shown to have impairments in metabolic processes (Baik et al., 2019). One of the DAM-APOE genes was recently shown to be a negative regulator of microglial phagocytosis (Pluvinage et al., 2019), suggesting that DAM-APOEs may be defective in this function.
We performed ingenuity pathway analysis in order to identify potential upstream regulators of the DAM-APOE transcription profile (Figure S2). The top predicted upstream regulators were heavily enriched for proinflammatory cytokines, chemokines, and interleukins (Figure S2). Of interest, APOE was also predicted to be one of the top upstream regulators, suggesting that dysregulated APOE signaling contributes to the disease-associated DAM-APOE transcription profile.
In summary, we find a clear separation between APOE and APOE-FAD mice along PC1 and show that, although half of the genes driving this separation have previously been identified as DAMs in the 5xFAD mouse, the other half are unique to this humanized APOE mouse model.
Sex and APOE genotype interact in driving AD-related pathology and the DAM-APOE signature in APOE-FAD mice
In order to examine how human APOE4 affects AD-related pathology and microglia, we assessed APOE-FAD mice between 7 and 9 months, an age at which significant Aβ pathology has developed. Because APOE3 has neutral AD risk, APOE3-FAD transgenic mice were used as the comparative group. Mice are homozygous for APOE4 or APOE3 and heterozygous for the FAD transgenes. APOE genotype and sex have previously been shown to interact in driving AD risk (Altmann et al., 2014) and AD-related pathology (Altmann et al., 2014; Cacciottolo et al., 2016; Stephen et al., 2019), thus both male and female APOE4-FAD and APOE3-FAD mice were included.
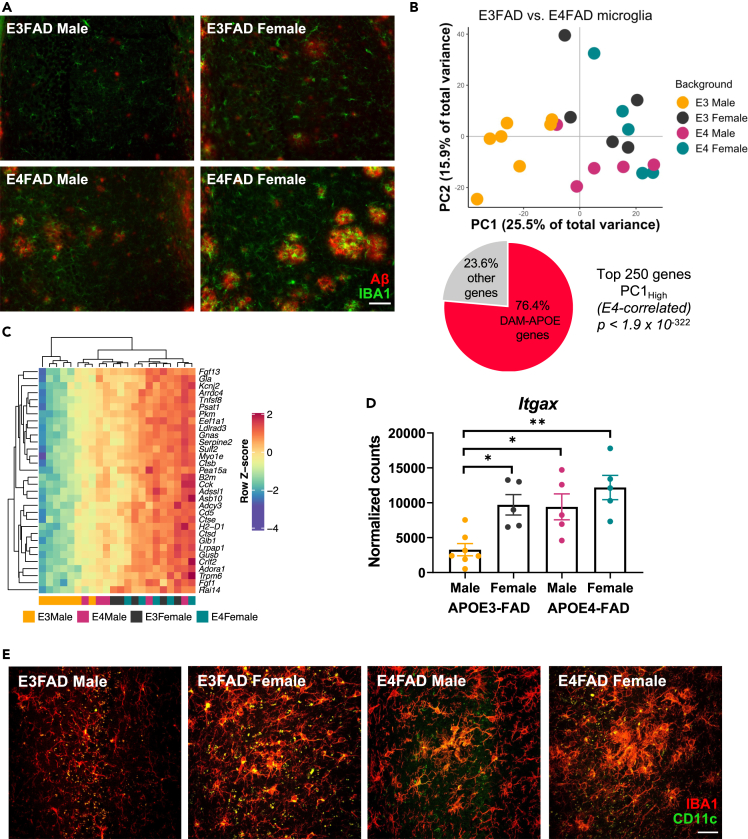
Immunohistochemical staining of the hippocampus was performed for Aβ plaques and IBA1+ microglia (Figure 2A). Consistent with previous reports (Moser and Pike, 2017; Rodriguez et al., 2014; Stephen et al., 2019; Youmans et al., 2012), quantification showed that Aβ plaque load in subiculum, CA1, and CA2/3 of the hippocampus was significantly greater in APOE4-FAD mice compared with control APOE3-FAD (Figures S3A–S3C). Moreover, there was a significant effect of sex in APOE4-FAD mice, with females having increased Aβ pathology in CA1 and CA2/3 (Figures S3B and S3C). Although there were no effects of genotype or sex on IBA1+ microglial number in the subiculum or CA1 (Figures S3D and S3E), microglia numbers in CA2/3 were significantly increased in APOE4-FAD mice compared with control APOE3-FAD (Figure S3F). In addition, female mice had a significantly greater density of microglia in CA2/3 than did male mice, across both APOE3- and APOE4-FAD genotypes. Collectively, these results confirm that APOE4 is associated with increases in Aβ pathology across all regions of the hippocampus and in microglial number in CA2/3 and that both are exacerbated in female mice.
Figure 2.
Sex and APOE genotype drive amyloid pathology and microglial gene expression profiles in APOE-FAD mice
(A) Representative images of immunohistochemical staining for amyloid-β (red) and the microglial marker IBA1 (green) in male and female APOE3- and APOE4-FAD mice. Scale bar, 50 μm.
(B) Samples separate by APOE and sex along PC1, with APOE3-FAD males clustering separately from mice of both sexes and APOE3-FAD females. A large proportion of the genes driving this separation (76.4%) are DAM-APOE genes. APOE3-FAD males are shown in yellow, APOE3-FAD females in gray, APOE4-FAD males in magenta, and APOE4-FAD females in teal.
(C) Heatmap of the DAM-APOE genes associated with sample clustering along PC1. Gene expression levels were normalized, scaled, and centered and are displayed as Z scores.
(D and E) Levels of the DAM-APOE marker, CD11c, are increased in APOE3-FAD females and in APOE4-FAD mice of both sexes, both at the (D) transcript and (E) protein levels. (D) Gene expression of Itgax in normalized counts; data are presented as mean (±SEM) values; n = 5–7/group. ∗p < 0.05, ∗∗p < 0.01; one-way ANOVA with Tukey’s multiple comparisons test correction. (E) Representative immunofluorescence images of microglia (IBA1, red) and CD11c (green) in the hippocampal subiculum of male and female APOE3- and APOE4-FAD mice. Scale bar, 25 μm.
We next sought to determine whether the APOE × sex interaction we observed in AD-related pathology (Figures 2A and S3) extends to microglial gene expression, and further, to what extent this may be driven by the DAM-APOE genes identified in Figure 1. Therefore, we compared transcriptional signatures between microglia from male and female APOE3-FAD and APOE4-FAD mice. Indeed, both sex and APOE genotype contribute to the separation observed along PC1, such that APOE4-FAD mice of both sexes and APOE3-FAD females clustered separately from APOE3-FAD males (Figure 2B). Moreover, of the top 250 positive gene loadings along PC1, 191 genes, or 76% were DAM-APOE genes. Hypergeometric ratio testing demonstrates that this degree of overlap is highly statistically significant at an adjusted p < 1.9 × 10−322.
Individual gene expression patterns mimic the separation observed along PC1 in Figure 2B. That is, the 32 DAM-APOE genes found in the top 100 positive gene loadings along PC1 show increased expression in APOE3-FAD females and in APOE4-FAD males and females (Figure 2C). RNA-Seq results were confirmed by rt-PCR in a subset of APOE3-FAD and APOE4-FAD microglial samples (Figure S4).
The separation observed along PC1 is not due to the influence of X and Y chromosome-linked genes, as only 7 of the top 250 genes associated with separating by sex along PC1 are sex chromosome-linked genes, and this does not reach statistical significance (p = 0.629). In addition, we removed 730 X and Y chromosome-linked genes from the total of 23,768 observed genes and re-ran PCA. As shown in Figure S5A, the samples cluster in a highly similar manner to the pattern observed when these genes are included in Figure 2B. Thus, the separation we observe by APOE genotype and sex is not driven by X and Y chromosome-linked genes.
To confirm that the pattern of results obtained at the gene transcript level is consistent with protein level changes, we immunohistochemically stained hippocampal sections for CD11c. Indeed, the same pattern holds true, such that APOE3-FAD males show lower expression of this marker than APOE3-FAD females and APOE4-FAD animals of both sexes, both in Itgax gene expression (Figure 2D) and protein levels (Figure 2E). Thus, sex and APOE genotype individually affect expression of this microglial phenotype, with the APOE4 allele and female sex being associated with increased DAM-APOE expression. Given known sex and APOE interactions on AD risk in the epidemiological literature, this finding is significant, not only in establishing a mouse model of this interaction but also in identifying microglia as a critical cell type in which effects of these risk factors may converge.
Although we clearly establish the role of human APOE in DAM gene expression, the temporal nature of the DAM-APOE phenotype remains to be addressed. That is, APOE4 and female sex may affect microglial function, which then drives Aβ pathology, or alternatively, these two factors could drive increases in Aβ, which could then in turn affect microglial gene expression. In order to begin addressing whether the APOE × sex interaction occurs in a non-disease background, we performed PCA on samples isolated from APOE mice that do not express FAD transgenes. Although there is no clear separation by sex in these animals in the first six principal components analyzed, which represents 95% of the cumulative variance in the data (Figure S5B), there is a separation by APOE genotype along PC3 (Figure S5C). Of the top 250 positive gene loadings along PC3, nine genes are DAM-APOE genes and show increased expression in microglia from APOE4 mice (Figure S5D). Hypergeometric ratio testing demonstrates that this degree of overlap is statistically significant at an adjusted p = 0.001. The finding that there is some separation by APOE genotype in APOE mice without FAD transgenes suggests that the effects of APOE genotype on microglial transcription profiles is not entirely dependent on Aβ pathology.
It has been reported that disease-associated gene expression signatures characterized in mouse microglia are divergent from those found in human microglia (Hasselmann et al., 2019). Thus, we do not know the extent to which the effects of APOE and sex we observed in our mouse model translate to human microglia. To address this, as well as to further examine APOE genotype × sex interactions in non-disease states, we utilized human iPSC-derived microglia (iMGL) from healthy, cognitively normal donors, who reached age 85 years without experiencing significant health problems.
Human iPSC-derived microglia show an APOE4-dependent transcriptional signature that is enriched for DAM-APOE genes
iMGL from healthy APOE3 and APOE4 carriers were used (1) to examine whether our findings in the APOE-FAD mouse model translate to human microglia and (2) to determine whether APOE4 intrinsically affects microglia, independent of neurodegeneration and pathology. The iPSC lines used are shown in the key resources table and include five APOE4 noncarriers (E3/E3), as well as four heterozygous carriers (E3/E4) and one homozygous APOE4 carrier. The following lines were differentiated and processed for RNA-Seq twice: EDi029, EDi035, EDi036, EDi023, and CS8FP5iCTR; all other lines were differentiated and analyzed once. iPSC lines were differentiated following an established protocol (Douvaras et al., 2017), as illustrated in Figure S6A. The resulting cells expressed canonical microglia markers including P2ry12, IBA1, and CD11c (Figure S6B).
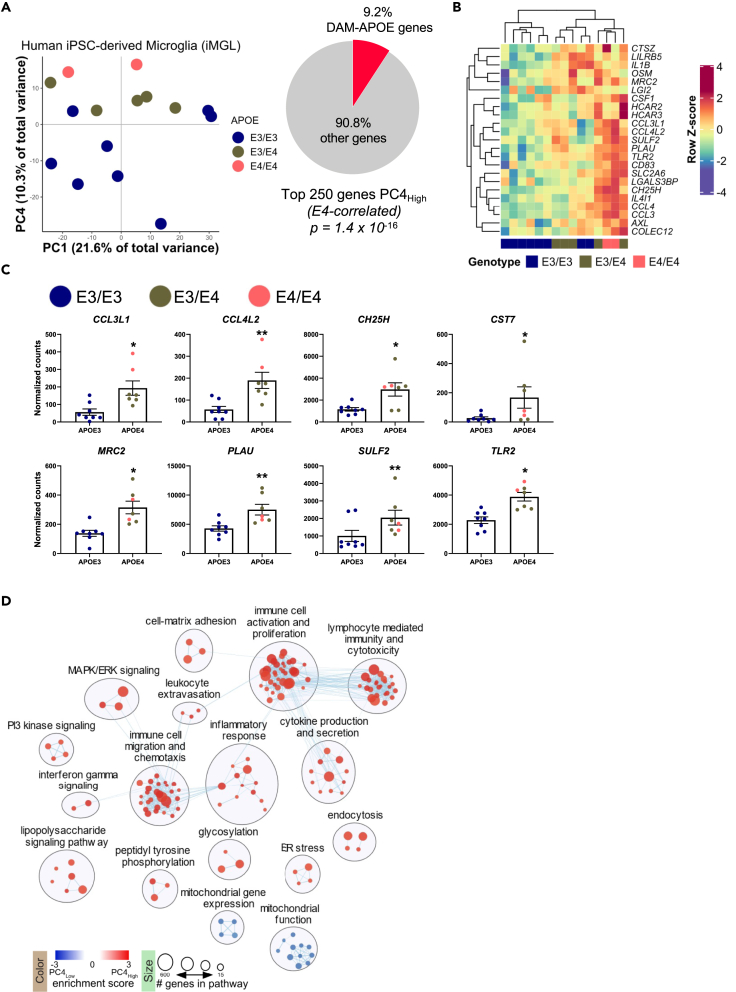
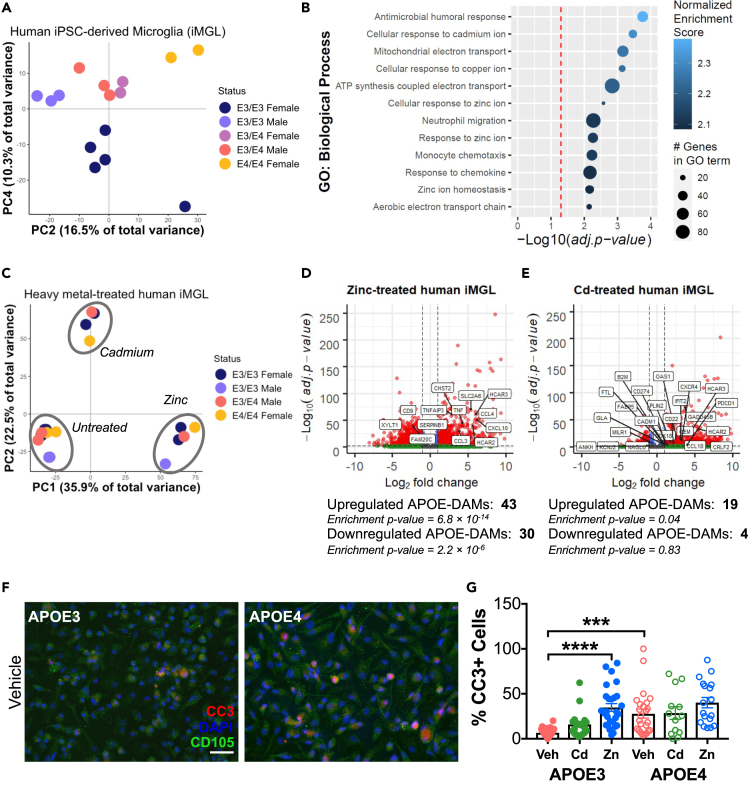
iMGL were collected and processed for RNA-Seq, and PCA was performed on individual samples using all expressed genes. Results demonstrated that samples separated by APOE genotype along PC4 (Figure 3A), with APOE3 homozygotes clustering at the bottom of PC4, APOE4 heterozygotes clustering in the middle, and APOE4 homozygous samples at the top. Although some degree of separation based on APOE genotype was observed along other PCs, PC4 showed the greatest degree of enrichment for DAM-APOE genes (Figure S7A). Of the 242 genes identified as DAM-APOE in the APOE-FAD mouse model, 223 genes have a human ortholog. Comparing these 223 DAM-APOE orthologous genes to the top 250 genes associated with APOE4 along PC4 revealed that 23 genes or 9.2% of genes overlapped. Based on hypergeometric ratio testing, this degree of overlap is highly significant at an adjusted p < 1.39 × 10−16 (Figure 3A).
Figure 3.
Human iPSC-derived microglia show an APOE-dependent transcriptional profile, enriched for DAM-APOE genes
(A) Samples separate by APOE genotype along PC4, with APOE4 iMGL clustering at the top and APOE3 iMGL at the bottom of PC4. Of the top 250 genes associated with APOE4, 23 genes (9.2%) are human orthologs of DAM-APOE genes found in mouse.
(B) Heatmap of the 23 DAM-APOE genes found in the top 250 genes driving PC4. Gene expression levels were normalized, scaled, and centered and are displayed as Z scores.
(C) Gene expression of the eight DAM-APOE genes meeting a cutoff of FDR <0.05 in DGE analysis; data are presented as mean (±SEM) values; n = 7–8/group. ∗p < 0.05, ∗∗p < 0.01; Benjamini-Hochberg false discovery rate adjusted p value. For (A)–(C), E3/E3 samples are shown in blue, E3/E4 in green, and E4/E4 in pink.
(D) Pathway enrichment analysis of ranked PC4 gene loadings showing an enrichment map summary of significantly enriched gene sets determined using GSEA software.
Comparing the 250 genes associated with APOE4 along PC4 to other previously published human microglial transcription profiles reveals little overlap. Specifically, these APOE4-associated genes were compared with gene sets found to be increased in AD patient microglia isolated from both fusiform gyrus and temporal cortex (Friedman et al., 2018). None of the 24 genes increased in fusiform gyrus-derived microglia (p = 1), and only 1 of 69 genes increased in temporal cortex microglia (p = 0.51), overlapped with APOE4-associated genes. We also compared genes associated with APOE4 in iMGL and a human microglial transcription profile associated with aging (Olah et al., 2018). Specifically, genes that were significantly different between middle-aged and aged human microglia at adjusted p < 0.05 with a logFC ≥2.0 were selected from the previously published dataset. Of the 1,034 genes that met these criteria, 24 genes, or 2.3%, overlapped with the APOE4 iMGL genes (p = 0.00018). This suggests that the effects of APOE4 observed in iMGL correspond more with an aging rather than a neurodegeneration phenotype; however, the greatest degree of overlap is with the mouse DAM-APOE genes identified in the current study (Figure S7B).
As shown in Figure 3B, APOE4 is associated with increased expression of the 23 DAM-APOE genes that are enriched in PC4. Furthermore, DGE analysis comparing APOE3 with APOE4 iMGL (including APOE4 heterozygous and homozygous lines) reveals that 8 of the 23 DAM-APOE genes are statistically significantly different between APOE3 and APOE4 (FDR adjusted p value < 0.05). As expected, expression of these genes is increased in APOE4 iMGL (Figure 3C). Three of these genes (CCL3L1, CCL4L2, and CST7) were also found to be increased in APOE4 versus APOE3 mice (Figure S5D). These results demonstrate that findings in the APOE-FAD mouse model translate to human iPSC-derived microglia. Of importance, we identify an APOE4-dependent gene set that is robust across species. Understanding the role of these genes in AD may identify novel mechanisms of degeneration and uncover new potential therapeutic targets.
We next evaluated the molecular pathways associated with PC4 using Gene Set Enrichment Analysis (GSEA) on the ranked PC gene loadings. Pathways associated with inflammation and immune cell activation, proliferation, and migration were significantly enriched in positive gene loadings along PC4 associated with the APOE4 genotype (Figure 3D). Taken together, these results suggest that microglia with an APOE4 allele have an activated DAM-APOE transcription profile and are in a heightened inflammatory state.
APOE4 iMGL show increased phagocytic protein expression and morphological alterations
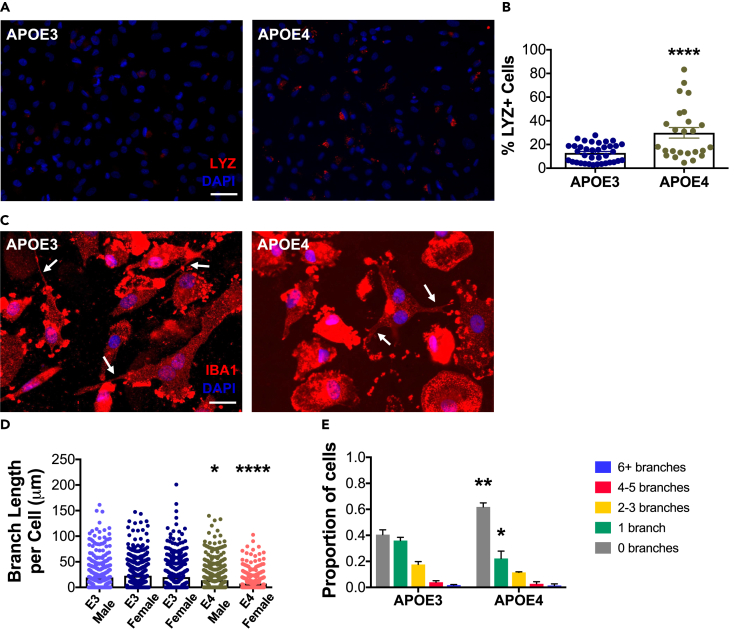
Immunocytochemistry (ICC) was performed and quantified in a subset of human cell lines. Both heterozygous and homozygous iMGL were included in the APOE4 condition. Staining and quantification of the phagocytosis marker lysozyme C (Figures 4A and 4B) revealed a significant effect of APOE4, with only 12.83% (±1.26) of APOE3 iMGL, but 29.85% (±4.47) of APOE4 iMGL staining for lysozyme C (p < 0.0001). Thus, even under baseline conditions, iMGL from APOE4 carriers show significantly increased levels of this phagocytic marker compared with those from APOE3 carriers.
Figure 4.
iPSC-derived microglia (iMGL) from APOE4 carriers show increased protein expression of lysozyme C and altered microglial morphology
(A) Representative immunofluorescence images of lysozyme C staining. Scale bar, 50 μm.
(B) The percentage of cells expressing lysozyme C is increased in APOE4 relative to APOE3 iMGL. Data are presented as mean (±SEM) values. APOE3 samples are shown in blue and APOE4 samples in green. ∗∗∗∗p < 0.0001; unpaired t test.
(C) Representative images of IBA1 staining used for branch length analysis. Scale bar, 25 μm; arrows point to representative branches.
(D) APOE4 iMGL have significantly shorter branch length per cell compared with APOE3 iMGL. APOE4 iMGL include both heterozygous and homozygous lines. Data are shown as the mean branch length per cell (±SEM); APOE3 males and females are shown in light and dark blue, respectively, whereas APOE4 males are shown in green and APOE4 females are in pink. ∗p < 0.05, ∗∗∗∗p < 0.0001; Bonferroni’s multiple comparisons adjusted p values relative to APOE3 iMGL.
(E) A greater proportion of APOE4 iMGL have zero branches, whereas a decreased proportion have one branch. Data are shown as the proportion of cells characterized as having a given number of branches (±SEM). ∗∗p < 0.01 relative to the number of APOE3 cells with zero branches; ∗p < 0.05 relative to the number of APOE3 cells with 1 branch; t test with Sidak-Bonferroni multiple comparisons test correction.
Resting-state microglia have extensive branching with long, thin processes that allow them to survey the brain parenchyma for potential damage. As APOE4 microglia have previously been shown to have reduced branching (Stephen et al., 2019), we next examined whether iMGL also demonstrate this APOE-dependent phenotype. Representative images of IBA1 staining are shown in Figure 4C. Tracing iMGL processes revealed that APOE4 cells have significantly shorter processes with an average branch length of 14.09 μm (±1.03) per cell as compared with 23.81 μm (±1.54) per cell observed in APOE3 iMGL (Figure 4D; p < 0.0001). Moreover, we found that a significantly greater proportion of APOE4 iMGL have zero branches (p < 0.01), but significantly fewer cells have one branch (p < 0.05), relative to APOE3 iMGL (Figure 4E). These results suggest that APOE4 iMGL may have a more reactive phenotype, as reductions in branching are known to occur as microglia transition from a resting to a reactive state. In addition, microglial branching allows for more efficient surveillance, since each cell can monitor a greater surface area. Thus, our data suggest that iMGL from APOE4 carriers not only have an altered transcriptional phenotype but may also be more reactive and have functional deficits in patrolling the nervous tissue, potentially allowing greater buildup of the Aβ plaques that define AD.
APOE and sex interactions are partially driven by genes involved in metal processing
Because our results in mice demonstrated that microglial transcription profiles showed significant interactions between APOE and sex, we performed an additional PCA on iMGL samples, using both APOE genotype and sex as variables. Remarkably, our data demonstrate that iMGL exhibit the same APOE by sex interaction as we observed in mouse microglia (Figure 2). That is, along PC2, iMGL from APOE3 males cluster together and separately from APOE3 females and from APOE4 iMGL of both sexes (Figure 5A). Moreover, we observe further separation by APOE and sex, such that the groups clustering together along PC2 can be divided into E3/E3 males, then E3/E4 males and females, and finally at the top of PC2, E4/E4 females. To ensure that the observed separation is not driven exclusively by the homozygous female APOE4 iMGL, an additional PCA was performed excluding these samples. As shown in Figure S8A, the remaining samples cluster quite similarly, with APOE3 male iMGL clustering separately from APOE3 female and APOE4 male and female iMGL, along PC2. Thus, we include the APOE4/E4 samples in all further analyses. Of the top 250 positive gene loadings along PC2, 25 genes are X or Y chromosome-linked genes (p = 4.76 × 10−7). However, we find that samples cluster in a highly similar manner when we remove the 772 sex chromosome-linked genes from the 24,289 total observed genes results (Figure S8B), suggesting that these genes are not the primary drivers of the observed clustering of samples.
Figure 5.
Separation by APOE genotype and sex is associated with genes related to zinc and cadmium, which differentially affect APOE3 and APOE4 iMGL
(A) Samples separate by both APOE genotype and sex along PC2, such that male APOE3 iMGL cluster together and separately from other samples.
(B) Gene ontology analysis using the top PC2 gene loadings shows enrichment for genes related to metal processing, particularly cadmium and zinc.
(C) Cadmium- and zinc-treated iMGL separate from vehicle-treated cells along PC1 and PC2.
(D and E) Differential gene expression analysis reveals significant enrichment for DAM-APOE genes in response to zinc (D) but not cadmium (E).
(F) Representative immunofluorescence images of CD105 (green) and cleaved caspase 3 (CC3; red) in vehicle-treated APOE3 and APOE4 iMGL. Scale bar, 50 μm.
(G) The percentage of CD105+ cells expressing CC3 is increased in vehicle-treated APOE4 iMGL and in response to zinc treatment in APOE3 iMGL. Vehicle-treated samples are shown in pink, cadmium-treated in green, and zinc-treated in blue; APOE3 iMGL are shown as closed circles and APOE4 iMGL as open circles. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; Bonferroni’s multiple comparisons adjusted p values relative to vehicle-treated APOE3 iMGL.
However, of the top 250 positive gene loadings along PC2 for the mouse study, only 10 genes overlap with the 223 human DAM-APOE orthologs. Although this represents a significant degree of overlap (p = 1.1 × 10−4), GO analyses revealed that the pathways enriched for in the 250 PC2 positive gene loadings are distinct from those associated with DAM-APOE genes. Of note, a number of GO terms related to metal processing were enriched for in the 250 genes driving PC2 variance (Figure 5B), including several terms related to cellular responses to cadmium and zinc ions. This finding is particularly interesting not only because it has long been suggested that high exposure to metals may be a risk factor for AD (Killin et al., 2016) but also because a number of studies have suggested interactions between APOE4 and metals (Godfrey et al., 2003; Moir et al., 1999; Zhang et al., 2020).
Because results of the GO analysis specifically implicated cadmium and zinc pathways, we treated a subset of iMGL lines (three homozygous for APOE3, one heterozygous for APOE4, and one homozygous for APOE4) with these metals and then performed RNA-Seq. PCA was performed on individual samples treated with either 30 μM zinc or 7.5 μM cadmium for 24 h. Samples separated by treatment along PC1 and PC2, such that zinc-treated samples separated from vehicle samples along PC1 and cadmium-treated samples separated from vehicle samples along PC2 (Figure 5C). DGE analysis revealed that zinc treatment was associated with 43 significantly upregulated (p = 6.8 × 10−14) and 30 significantly downregulated DAM-APOE genes (p = 2.2 × 10−6; Figure 5D). Thus, the gene set showing differential regulation in response to zinc treatment is highly enriched for DAM-APOE markers. However, this was not true to the same extent for cadmium-treated iMGL, in which only 19 of the upregulated (p = 0.04) and 4 of the downregulated (p = 0.83) genes were part of the human DAM-APOE orthologs (Figure 5E).
To further examine potential APOE genotype-dependent responses to metal treatments, ICC was performed using cleaved caspase 3 (CC3) as a marker for cell death (Figures 5F and S8C). As shown in Figure 5G, there was a significant effect of APOE genotype on levels of CC3 even at baseline (p = 0.0002), with the percentage of APOE4 iMGL expressing CC3 (27.87% ± 4.89) being over four times as much as that of APOE3 iMGL (6.74% ± 0.74). Although cadmium treatment did not increase cell death in iMGL of either genotype, zinc treatment was associated with a significant increase in CC3-positive cells, specifically in APOE3 iMGL (p < 0.0001). Thus, APOE4 is associated with an inherent increase in iMGL cell death compared with APOE3, but this is not exacerbated by zinc treatment as it is in APOE3 iMGL, pointing to a potential gene by environment interaction in these cells.
Discussion
Microglia are key players in neurodegenerative diseases and have unique transcriptional phenotypes in both animal models of AD (Keren-Shaul et al., 2017) and in human samples (Friedman et al., 2018; Patir et al., 2019). However, the extent to which various AD risk factors, including genetics, sex, and environmental factors affect microglial transcription profiles has thus far not been thoroughly examined. Because the main genetic risk factor for AD, APOE4, is known to alter glial function, we wanted to understand how APOE affects microglial gene transcription profiles. We define the microglial transcription signature in mice carrying human APOE transgenes, which only partially overlaps with reported DAM genes, and show that APOE genotype and sex interact in driving expression of these genes. We find significant overlap of this DAM-APOE signature between microglia from an AD mouse model and iMGL differentiated from human iPSCs.
Our findings add to a growing literature demonstrating shifts in microglial gene expression across various states and diseases, including transcriptional signatures associated with specific DAM subpopulations (Rangaraju et al., 2018), with different stages of neurodegeneration (Mathys et al., 2017), and with aging in both human (Olah et al., 2018) and mouse (Hammond et al., 2019) microglia. Recent reports have also demonstrated a role for APOE in the context of microglial gene expression (Liu et al., 2020; Sala Frigerio et al., 2019). For instance, TREM2-dependent APOE signaling is necessary for expression of the DAM phenotype, and blocking this pathway restores homeostatic microglia while reducing neuron loss (Krasemann et al., 2017). Moreover, a common APOE-dependent transcriptional signature was identified in mouse microglia across different models of Aβ and tau accumulation, as well as in aging (Kang et al., 2018).
One critical question is the ultimate function of cells that upregulate DAM gene expression. Although some have suggested that they could restrict further progression of pathology (Deczkowska et al., 2018), this may depend on the relative prevalence of various subpopulations (Rangaraju et al., 2018) and on disease stage, as DAMs may initially phagocytose Aβ plaques but their chronic activation could later result in further neurodegeneration. Indeed, the DAM transcription profile is remarkably similar to a gene signature found in microglia during development, when these cells are engaged in synaptic pruning (Li et al., 2019). Thus, it has been suggested that DAMs are the result of a reactivation of developmental programs in an effort to eliminate pathological protein buildup, but uncontrolled proliferation of these cells ultimately may lead to further neuronal loss. We found that APOE4 iMGL have increased levels of the apoptotic protein CC3 and decreased branching, suggesting that these cells may be more vulnerable and less effective at immune surveillance than their APOE3 counterparts. In addition, APOE4 iMGL were previously shown to be less effective at both phagocytosing fluorescent beads (Konttinen et al., 2019) and clearing Aβ in an AD organoid model (Lin et al., 2018). Indeed, the increased Aβ deposition found in APOE4 mice has been shown to be due to reduced clearance rather than increased production of Aβ (Castellano et al., 2011), and this could be due to APOE4’s effects on microglia. Taken together with our findings, this suggests that APOE4 microglia may have a reduced functional capacity. However, additional work is needed to definitively conclude whether DAM-APOE gene expression results in a helpful or harmful phenotype, and along what timeline.
Another critical question is to what extent DAMs and the DAM-APOE genes identified here are a result versus a driver of disease. Our findings in mice revealed that the DAM-APOE gene signature was increased with both sex and APOE4. However, both of these factors are also associated with higher levels of Aβ, as demonstrated here and previously (Cacciottolo et al., 2016; Stephen et al., 2019), so that the DAM-APOE signature could be a result of this increased Aβ pathology. Alternatively, APOE4 and female sex may drive transcriptional changes even in the absence of disease, resulting in a decrease in microglial function that allows for greater Aβ accumulation throughout life. Indeed, our findings in APOE mice that do not express FAD transgenes and in human iMGL generated from healthy, cognitively normal older adults suggest that this may be the case. Our findings of transcriptional differences between APOE3 and APOE4 iMGL and of significant enrichment for the DAM-APOE genes we identified in mouse suggest that there are inherent differences in APOE4 microglia, even in the absence of neurodegeneration. However, APOE4 iMGL may show even greater transcriptional changes or enrichment of DAM-APOE genes in the context of AD. We plan to address this in future work both by exposing iMGL from healthy donors to Aβ as well as by generating iMGL from APOE4-carrying patients with AD.
The data presented here also point to the importance of considering interactive risk factors. For one, we confirm APOE genotype and sex interactions in AD and demonstrate that these effects exist at the level of microglial gene signatures. Moreover, our finding that APOE4 iMGL from healthy older adults show transcriptional differences associated with a disease phenotype points to the role of modifying factors in determining disease outcomes. For example, it may be the case that microglia are inherently different in APOE4 carriers but this does not result in AD pathogenesis in the absence of a “second hit.”
Indeed, interactions between APOE4 and a number of environmental or lifestyle risk factors have been reported, including exposure to hazardous substances like heavy metals (Killin et al., 2016). For example, the adverse effects of metal exposure are exacerbated in children carrying APOE4 (Ng et al., 2013; Snoj Tratnik et al., 2017), and there is a higher frequency of APOE4 in adults showing symptoms of mercury toxicity (Godfrey et al., 2003). Experimentally, cadmium induces greater cognitive deficits in mice carrying APOE4 (Zhang et al., 2020), and zinc and copper induce greater Aβ aggregation in the presence of APOE4 (Moir et al., 1999).We found that the genes associated with separating iMGL by both APOE genotype and sex were related to metal processing pathways. Moreover, our results demonstrate an interaction between APOE genotype and metal exposure, where zinc treatment increases cell death specifically in APOE3 iMGL, whereas cells from APOE4 carriers display greater cell death at baseline but do not show a further increase with zinc exposure.
There are a number of other environmental or lifestyle risk factors that have been shown to interact with APOE4, including high-fat diet (Moser and Pike, 2017), air pollution (Cacciottolo et al., 2017), cigarette smoking (Durazzo et al., 2016), and concussions (Merritt et al., 2018). However, gene by environment interactions, and especially the potential mechanisms underlying them, have largely been understudied in diseases like AD. The iPSC lines used in the current study are an excellent model for further exploring interactions not only between APOE and metal exposure but also between APOE and other risk factors. Studying these interactions in a highly controlled experimental setting will allow for dissecting the molecular pathways that may be involved as well as for ultimately testing targeted therapeutics.
Moreover, the use of iPSCs provides the ability to address these interactions in a human background. This is particularly important in the context of studying microglia, as mice are a better model for understanding neuronal processes, rather than cells that are part of the immune system (Galatro et al., 2017; Gosselin et al., 2017; Penney et al., 2020). Specifically, previous studies have shown that rodent microglia do not fully recapitulate many biological aspects of human microglia, both transcriptionally and in terms of their responses to aging and neurodegeneration (Friedman et al., 2018; Gosselin et al., 2017; Hasselmann et al., 2019; Xu et al., 2020). Strikingly, our results indicate a remarkable degree of transcriptional overlap between mouse and human microglia in regard to APOE4, suggesting that the effects of human APOE isoforms may be conserved in human and rodent microglia.
One important distinction between the current study and previously reported data on DAM genes is that we used bulk RNA-Seq, whereas Keren-Shaul et al. used single-cell RNA-Seq. Thus, we cannot determine whether the DAM-APOE phenotype we identify represents global microglial gene expression or is specific to a subset of microglia. However, Keren-Shaul et al. found that, by 8 months of age, the majority of microglia from 5xFAD mice had assumed a DAM profile. Thus, given that we are using mice on the same 5xFAD background at 7–9 months of age, it is likely that a large proportion of these cells will also have assumed this phenotype. However, future studies should investigate these changes by single-cell RNA-Seq to determine the extent to which the DAM-APOE genes identified here are expressed in different subtypes and to what extent this differs between APOE3 and APOE4 carriers.
Although our model comes closer to the human condition, previous work has demonstrated that iMGL are rather immature and more similar to fetal than adult cells (Hasselmann et al., 2019). Using chimeric models in which iMGL are transplanted into mouse models has been shown to result in more mature and functional microglia phenotypes (Hasselmann et al., 2019; Xu et al., 2020). Future studies transplanting iMGL from APOE3 and APOE4 iPSC lines into mouse models may result in more mature, adult-like cells and could thus be valuable in the context of aging and neurodegeneration.
One limitation of the current study is the lack of a homozygous male APOE4 iPSC line. The effects of APOE4 are exacerbated in female carriers (Altmann et al., 2014); thus, although we observe a gene-dosage effect in the female APOE4 iMGL in several analyses, it is possible that the effects of two APOE4 alleles would not be as strong in a male line. It will also be important to determine the effects of sex and APOE4 in further functional assays as well as in other AD-relevant cell types derived from iPSCs, including neurons and astrocytes. In fact, astrocytes are the main source of APOE in the brain (Boyles et al., 1985), and effects of APOE4 have been shown in iPSC-derived astrocytes (Zhao et al., 2017). Moreover, reactive, neurotoxic astrocytes are induced by activated, pro-inflammatory microglia (Liddelow et al., 2017); thus, co-culture studies will be critical for understanding how the DAM-APOE phenotype we identify here affects other cell types. One especially useful tool in this regard is the organ-chip model our laboratory has previously used (Sances et al., 2018; Vatine et al., 2019), as this allows for examining the interactions between multiple cell types in a 3-dimensional environment.
In summary, our use of complimentary in vivo and in vitro approaches identified a microglial transcription profile associated with human APOE. Moreover, the demonstrated interactions between APOE and sex, as well as between APOE and metal exposure, point to the importance of considering various interactive risk factors. These findings provide the foundation for future work pursuing the functional consequences of these microglial phenotypes as well as the mechanisms underlying gene expression changes, which can ultimately be used to identify therapeutic targets.
Limitations of the study
Although our study demonstrates that APOE genotype and sex affect microglial transcription profiles and drive expression of previously identified DAM genes, we cannot determine whether all microglia are affected or if this represents a specific subset. We find significant overlap between transcriptional signatures in mice expressing human APOE3 or APOE4 and in human iMGL; however, human iMGL have been demonstrated to be rather immature. Thus, it is unclear to what extent mature, human, in vivo microglia behave in similar ways. An additional limitation of this study is the lack of a homozygous male APOE4 iPSC line. This is an exceedingly rare cell line, as less than 2% of the population are homozygous APOE4 carriers and we are generating iMGL specifically from healthy, non-demented older adults. Future studies should also determine the extent to which the changes we observe in gene expression and in microglial morphology as well as in the response to heavy metal treatments lead to functional changes in these cells. Finally, the mechanism by which APOE4 microglia show altered transcriptional phenotypes has yet to be elucidated and should be pursued in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-β amyloid | Invitrogen | Cat# 71-5800 RRID: AB_2533989 |
| Rabbit polyclonal anti-IBA1 | Wako Chemicals | Cat# 019-19741 RRID: AB_839504 |
| Armenian Hamster monoclonal anti-CD11c | Novus Biologicals | Cat# NB110-97871 RRID: AB_1290725 |
| Rabbit polyclonal anti-P2ry12 | Atlas Antibodies | Cat# HPA013796 RRID: AB_1854884 |
| Goat polyclonal anti-lysozyme C | Santa Cruz Biotechnologies | Cat# sc27958 RRID: AB_2138790 |
| Mouse monoclonal anti-CD105 | R&D Systems | Cat# MAB1097 RRID: AB_35739 |
| Rabbit monoclonal anti-cleaved caspase 3 (ASP175) (5A1E) | Cell Signaling Technology | Cat# 9664 RRID: AB_2070042 |
| Horse anti-rabbit biotinylated secondary | Vector Laboratories | Cat# BA-1100 RRID: AB_2336201 |
| Goat anti-mouse secondary Alexa Fluor 488 | abcam | Cat# ab150113 RRID: AB_2576208 |
| Goat anti-rabbit secondary Alexa fluor 594 | Invitrogen | Cat# R37117 RRID: AB_2556545 |
| Goat anti-Armenian hamster secondary Alexa fluor 488 | Jackson Immuno-Research Labs | Cat# 127-545-099 RRID: AB_2338996 |
| DAPI | Invitrogen | Cat# 3571 RRID: AB_2307445 |
| Vectastain ABC kit, standard | Vector Laboratories | Cat# PK-6100 RRID: AB_2336819 |
| DAB peroxidase substrate kit, 3,3’-diaminobenzidine | Vector Laboratories | Cat# SK-4100 RRID: AB_2336382 |
| Vectashield hardset antifade mounting medium | Vector Laboratories | Cat# H-1400 RRID: AB_2336787 |
| CD11b microbeads, human & mouse | Miltenyi Biotec | Cat# 130-049-601 RRID: N/A |
| CD14 microbeads, human | Miltenyi Biotec | Cat# 130-050-201 RRID: AB_2665482 |
| Chemicals, peptides, and recombinant proteins | ||
| Formic acid, reagent grade | Sigma Aldrich | F0507 |
| Bovine Serum Albumin | Sigma Aldrich | A7906 |
| Matrigel | BD Biosciences | 356234 |
| mTeSR1 | StemCell Technologies, Inc. | 85851 |
| Accutase | EMD Millipore | SCR005 |
| bFGF | EMD Millipore | 01-106 |
| Recombinant human BMP4 protein | R&D Systems | 314-BP-010 |
| Recombinant human FLT3 protein | R&D Systems | 308-FK-100 |
| Recombinant human GM-CSF protein | R&D Systems | 215-GM-050 |
| Recombinant human IL3 protein | R&D Systems | 203-IL-010 |
| Recombinant human IL34 protein | R&D Systems | 5265-IL-010 |
| Recombinant human M-CSF protein | R&D Systems | 216-MC-025 |
| Recombinant human SCF protein | R&D Systems | 255-SC-010 |
| Recombinant human VEGF protein | R&D Systems | 239-VE-010 |
| Recombinant human thrombopoietin protein | R&D Systems | 288-TP-005 |
| StemPro 34 Serum Free Media | Gibco | 10639011 |
| RPMI 1640 Media | Gibco | 11875093 |
| GlutaMAX Supplement | Gibco | 35050061 |
| Cadmium Chloride | Sigma Aldrich | 202908 |
| Zinc Chloride | Sigma Aldrich | 229997 |
| Critical commercial assays | ||
| Neural Tissue Dissociation Kit (P) | Miltenyi Biotec | 130-092-628 |
| Direct-zol RNA Miniprep Kit | Zymo Research | R2051 |
| SMART-Seq v4 Ultra Low Input RNA Kit | Takara | 091817 |
| Deposited data | ||
| Raw data files for RNAseq | NCBI Gene Expression Omnibus | GSE163857 |
| Experimental models: Cell lines | ||
| EDi036-A; female, 79 years, E3/E3 | iPSC Core, Cedars Sinai Medical Center | EDi036 |
| EDi034-A; female, 79 years, E3/E3 | iPSC Core, Cedars Sinai Medical Center | EDi034 |
| EDi035-A; female, 79 years, E3/E3 | iPSC Core, Cedars Sinai Medical Center | EDi035 |
| EDi028-A; male, 79 years, E3/E3 | iPSC Core, Cedars Sinai Medical Center | EDi028 |
| EDi029-A; male 80 years, E3/E3 | iPSC Core, Cedars Sinai Medical Center | EDi029 |
| EDi042-A; female, 79 years, E3/E4 | iPSC Core, Cedars Sinai Medical Center | EDi042 |
| EDi044-A; female, 80 years, E3/E4 | iPSC Core, Cedars Sinai Medical Center | EDi044 |
| EDi023-A; female, 79 years, E4/E4 | iPSC Core, Cedars Sinai Medical Center | EDi023 |
| EDi022-A; male; 79 years, E3/E4 | iPSC Core, Cedars Sinai Medical Center | EDi022 |
| CS8FP5iCTR; male, 45 years, E3/E4 | iPSC Core, Cedars Sinai Medical Center | CS8FP5iCTR |
| Experimental models: Organisms/strains | ||
| Mouse: APOE-FAD | Generated in the laboratory of Dr. Mary Jo LaDu | N/A |
| Software and algorithms | ||
| Prism 7 | GraphPad Software | |
| R statistical software package version 1.1.463 | R Foundation | |
| Other | ||
| MACS SmartStrainers (70um) | Miltenyi Biotec | 130-098-462 |
| MS Columns | Miltenyi Biotec | 130-042-201 |
| MACS Separation Buffer | Miltenyi Biotec | 130-091-221 |
| OctoMACS Separator | Miltenyi Biotec | 130-042-109 |
Resource availability
Lead contact
Requests for further information and resources or reagents should be directed to and will be addressed by the Lead Contact of the study, Dr. Clive N. Svendsen, PhD (clive.svendsen@cshs.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
A colony of APOE-FAD mice were bred and maintained at vivarium facilities at the University of Southern California, from breeder mice generously provided by Dr. Mary Jo LaDu (University of Illinois at Chicago). EFAD mice are a cross between the 5xFAD mouse model that carries five familial Alzheimer’s disease (FAD) mutations (APP K670N/M671L + I716V + V717I and PS1 M146L + L286V) and the APOE-targeted replacement mouse, in which the coding region of the mouse APOE gene is replaced by that of human APOE3 or APOE4. Mice are homozygous for human APOE and heterozygous for the FAD transgenes, such that approximately half of each litter will carry the FAD transgenes (APOE-FAD mice) and half will not (APOE mice). All animals were group-housed under a 12 h light/dark cycle with ad libitum access to food and water. All animal procedures were conducted under protocols approved by the University of Southern California Institutional Animal Use and Care Committee and in accordance with National Institute of Health standards.
For immunohistochemical studies, male and female APOE3- and APOE4-FAD mice, aged 7-9 months, were anesthetized with inhalant isoflurane and were transcardially perfused with ice-cold 0.1 M PBS. The brains were rapidly removed and post-fixed by immersion in 4% paraformaldehyde (PFA)/0.1 M PBS for 48 h at 4°C, before being stored in 0.1 M PBS/0.03% NaN3 until sectioning for immunohistochemistry. For isolation of microglia, 7–9 month-old mice were similarly anesthetized and transcardially perfused, and brains were stored in 5 mL ice-cold Hank’s balanced salt solution (HBSS) without Cl and Mg, for a maximum of 1 h before dissociation.
Cell lines
All iPSC lines were generated at the iPSC Core at Cedars-Sinai Medical Center. Parent cells were transfected with non-integrating oriP/EBNA1 plasmids that rely on episomal expression of reprogramming factors. Details regarding the disease status, sex, age at collection, and APOE genotype for each iPSC line are provided in the key resources table. All cell lines and protocols were approved for use under the institutional review board (IRB) and stem cell research oversight committee (SCRO) protocols 21505 and 32834.
Method details
Microglial isolation
Microglia were isolated from whole brain following the protocol by Garcia et al. (2014), and using the Neural Tissue Dissociation Kit P (#130-092-628, Miltenyi Biotec). Whole brains were dissociated in a 35 mm petri dish with HBSS using a scalpel to roughly chop the tissue. Homogenated tissue was then centrifuged at 300 × g for 2 min at room temperature and the supernatant was aspirated. Tissue was resuspended in Enzyme Mix 1 and incubated for 15 min at 37°C under slow, continuous rotation. After adding Enzyme Mix 2, tissue was further dissociated using a 1 mL pipette tip, and then incubated again for 10 min at 37°C under rotation. A final dissociation step was performed by pipetting slowly using 2 glass Pasteur pipettes of decreasing diameter, after which samples were incubated at 37°C for another 10 min. The cell suspension was added to a 70 μm MACS SmartStrainer (#130-098-462, Miltenyi Biotec), after which 10 mL HBSS were added and the cell suspension was centrifuged at 300 × g for 10 min at 4°C.
Myelin removal was then performed by adding 5 mL of 30% Percoll solution to the cell pellet and centrifuging at 700 × g for 15 min at 4°C. The upper myelin layer was carefully removed using a 1 mL pipette tip, before the remaining Percoll solution was removed using a serological pipette. Cells were suspended in 10 mL HBSS to wash out any remaining Percoll solution, centrifuged at 300 × g for 10 min at 4°C, and the supernatant was discarded.
Cell pellets were then resuspended in 90 μL MACS buffer (#130-091-221, Miltenyi Biotech) and 10 μL CD11b microbeads (#130-049-601, Miltenyi Biotec), and incubated for 15 min at 4°C. After adding 2 mL MACS buffer, cells were again centrifuged for 10 min at 300 × g, the supernatant was removed, and the pellet resuspended in 500 uL MACS buffer. MS columns (#130-042-201, Miltenyi Biotec) were placed in the magnetic field of an OctoMACS Separator (#130-042-109, Miltenyi Biotec) and rinsed with 500 μL MACS buffer before applying the cell suspension to the column. Columns were washed 3 times with 500 μL MACS buffer each. The column was removed from the separator and placed on a collection tube. 1 mL MACS buffer was added to the column and the magnetically labeled cell fraction was immediately flushed out using a plunger supplied with the MS columns.
iMicroglia differentiation from iPSCs
Reprogramming was conducted by non-integrating methods. All iPSC lines were fully reprogrammed as demonstrated by staining for alkaline phosphatase and other pluripotency markers and passed the ‘PluriTest’ as shown by low gene expression of ‘novelty’ markers and high expression of ‘pluripotency’ markers. Southern blotting and genomic PCR analyses confirmed the absence of plasmid gene expression after several passages, demonstrating that reprogramming plasmids were not integrated.
iPSCs were differentiated into microglia following the protocol by Douvaras et al. (2017) with slight modifications, as described here. Briefly, iPSCs were grown in 1 mg/6-well plate Matrigel (#354230, Corning) in mTeSR (#5850, StemCell Technologies Inc.) at 37°C and with 5% O2 until they reached 80-90% confluency. Cells were passaged using a StemPro EZPassage Disposable Stem Cell Passaging Tool (#23181010, Life Technologies), and plated on matrigel in mTeSR. Cells were maintained over the next 2–4 days, and cells that were differentiating, growing too close to other colonies, or abnormally shaped were removed. Once wells had roughly 10–12 colonies measuring 1.0 mm in diameter on average, differentiation was started using stage 1 media, consisting of 80 ng/ml bone morphogenetic protein 4 (BMP4; #314-BP-010, R&D Systems) in mTeSR. Media was changed daily for 4 days, at which point cells were switched to stage 2 media, consisting of 25 ng/ml basal fibroblast growth factor (bFGF; #01-106, Millipore Sigma), 100 ng/ml stem cell factor (SCF; #255-SC-010, R&D Systems) and 80 ng/ml vascular endothelial growth factor (VEGF; #293-VE-010, R&D Systems) in StemPro-34 serum free medium (#10639011, Gibco). Two days later, cells were switched to stage 3 media, consisting of StemPro-34 supplemented with 50 ng/ml SCF, 50 ng/ml interleukin-3 (IL-3; #203-IL-010, R&D Systems), 5 ng/ml thrombopoietin (TPO; #288-TP-005, R&D Systems), 50 ng/mL macrophage colony stimulating factor (M-CSF; #216-MC-010, R&D Systems), and 50 ng/mL FMS-related tyrosine kinase 3 ligand (Flt3; #308-FK-005, R&D Systems). Media was changed 4 days later and replaced with stage 3 media until day 12–14, when cells were switched to stage 4 media, consisting of 50 ng/ml M-CSF, 50 ng/ml Flt3 ligand, and 25 ng/ml granulocyte-macrophage colony stimulating factor (GM-CSF; #215-GM-010, R&D Systems) in StemPro-34 medium. Four days later, the floating cells in the supernatant were collected, pelleted, resuspended in fresh stage 4 media, and returned to their respective plate, with even distribution among the wells. These collection feeds were performed every 4 days, with floating cells being taken for sorting rather than resuspending, during every other feed. After sorting for CD14+ cells, these were plated at a density of 50 ×103 cells/cm2 in RPMI-1640 medium (#11875093, Gibco) supplemented with 2mM GlutaMAX (#35050061, Gibco), 10 ng/ml GM-CSF, and 100 ng/ml interleukin -34 (IL-34; #5265-IL-010, R&D Systems). Half-media changes were performed every other day for 2 weeks, before cells were treated with either 30 μM zinc or 7.5 μM cadmium for 24 h. Cells were then either fixed in PFA for 10 min for subsequent ICC or enzymatically harvested by Accutase (#07920, StemCell Technologies) treatment for 10 min at 37°C, collected and pelleted, and stored at −80°C until RNA extraction.
Immunohistochemistry and immunocytochemistry
Immunohistochemistry was performed as previously described (Moser and Pike, 2017; Moser et al., 2018, 2019), and as detailed for each antibody below. For amyloid-β (Aβ) and allograft inflammatory factor 1 (IBA1), a standard avidin/biotin peroxidase approach using ABC Vector Elite kits (Vector Laboratories) was performed, while double labeling for IBA1 and integrin alpha X (CD11c) was performed using fluorescent secondary antibodies. After fixation, hemibrains were flash frozen on dry ice and sectioned exhaustively in the horizontal plane at 30 μm using a microtome (Leica Biosystems). Every eighth section was stained for Aβ, IBA1, and IBA1/CD11c.
For Aβ immunohistochemistry, sections were pretreated with 95% formic acid for 5 min, rinsed in tris-buffered saline (TBS), and treated with 3% H2O2/3% methanol for 10 min to block endogenous peroxidases. After 3 washes in 0.1% Triton-X/TBS for 10 min each, sections were incubated in a blocking solution containing 2% bovine serum albumin (BSA) in TBS for 30 min. Sections were then incubated in blocking solution containing Aβ primary antibody (#71-5800, 1:300, Invitrogen) at 4°C overnight. Sections were then rinsed, incubated in biotinylated secondary antibody diluted in blocking solution, and immunoreactivity was visualized using 3,3′-diaminobenzidine (Vector Laboratories).
IBA1 immunohistochemistry was performed similarly, except for the following modifications. For antigen retrieval, sections were boiled in 10 mM ethylenediamine tetraacetic acid (EDTA) pH 6.0 for 10 min, then rinsed in water 3 times for 5 min each. After washing 3 times in 0.2% Triton-X/TBS and blocking for 1 h, sections were incubated overnight at 4°C in primary antibody directed against IBA1 (#019-19741, 1:500, Wako Chemicals). For IBA1/CD11c double labeling, antigen retrieval was performed using boiling EDTA and sections were incubated for 2 days at 4°C in primary antibodies directed against IBA1 (1:500) and CD11c (#NB110-97871, 1:500, Novus Biologicals). On the following day, for both protocols, sections were washed in 0.2% Triton-X/TBS 3 times for 5 min each and then incubated with Alexa-Fluor conjugated antibodies diluted 1:500 in blocking solution for 2 h at room temperature. Sections were then washed and stained with DAPI (#D3571, 1:10,000, Invitrogen) before being mounted and coverslipped.
For ICC, sorted CD14+ cells were plated on poly-lysine-coated coverslips for 2 weeks before being fixed in 4% PFA. Cells were then rinsed in PBS, and permeabilized using 0.1% Triton X-100 in PBS for 5 min. After blocking in 2% BSA for 1h, cells were incubated overnight at 4°C in primary antibodies directed against IBA1 (1:500, Wako), CD11c (1:500, Novus Biologicals), purinergic receptor P2Y (P2ry12; #HPA013796, 1:500, Atlas Antibodies), lysozyme C (LYZ; #sc27958, 1:100, Santa Cruz Biotechnology), endoglin (CD105; #MAB1097, 1:100, R&D Systems), or cleaved caspase 3 (CC3; #9664, 1:1000, Cell Signaling Technology). Coverslips were then rinsed with PBS and incubated with Alexa-Fluor conjugated antibodies diluted 1:500 in blocking solution for 2 h at room temperature before staining with DAPI.
Cell quantification
Quantification of the area occupied by Aβ immunoreactivity (Aβ load) was achieved by capturing images of nonoverlapping fields at 20× magnification. Images were digitally captured of the hippocampal subregions subiculum (3 fields), CA1 (3 fields), and CA2/3 (3 fields) across 4 sections per animal, using a Leica AF3500 microscope. Pictures were converted to grayscale and thresholded to yield binary images separating positive and negative staining using NIH ImageJ 2.0. Aβ load was defined as the percent of the total area occupied by positive immunolabeling.
Microglial cell number was quantified via non-biased stereological analysis using the optical fractionator method (Stereo Investigator, MBF Biosciences) paired with a Leica microscope. Hippocampal subiculum, CA1, and CA2/3 were traced across two tissue sections per animal. IBA1+ cells were counted with a 63× objective, using a counting frame size of 150 × 150 μm and a 1000 × 1000 μm distance between the counting frames. The number of IBA1 positive cells per area was recorded and the total number of microglia per area of the subiculum, CA1 region, and CA3 region was calculated.
For IBA1/CD11c immunohistochemistry, confocal images were acquired of CA1 of the hippocampus using an A1 confocal microscope (Nikon) at 63× magnification and 0.5 μm. Figure 3D was generated using maximum projection for each channel in NIH ImageJ.
Quantification of ICC staining was performed by capturing images of non-overlapping fields across two coverslips, at 20× magnification for LYZ and CC3/CD105 staining and using a 40× oil objective for IBA1 branch analysis. The percent of DAPI+ cells expressing LYZ as well as the percent of CD105+/DAPI+ cells expressing CC3 were quantified. The simple neurite tracer plugin in NIH ImageJ was used to trace IBA1+ cell branches and the average branch length per cell was calculated. Additionally, the number of branches per cell was quantified. All IHC and ICC quantification was performed by blinded observers.
Transcriptional analysis
Total RNA was extracted from microglia isolated from EFAD mice and from iMGL using a Direct-zol RNA Miniprep kit (#R2051, Zymo Research), with on-column DNAse digestion, according to manufacturer’s instructions. Library preparation and sequencing was performed by the Cedars-Sinai Genomics Core. Briefly, integrity of extracted RNA was tested using an Agilent 2100 Bioanalyzer (Agilent Technologies) and only samples with RIN values above 8 were included. RNA concentration was assessed using a Qubit fluorometer (ThermoFisher Scientific) and up to 1 ug of total RNA was used for poly-A mRNA selection. cDNA was synthesized using reverse transcriptase (Invitrogen) and random primers. cDNA was then converted into double-stranded DNA and enriched with PCR for library preparation. Agencourt AMPure XP beads (#A63880, Beckman Coulter) were used to purify the PCR-amplified library and the concentration and quality were assessed. Finally, sample libraries were prepared with SMART-Seq v4 Ultra Low Input RNA kit (Takara), and were multiplexed and sequenced on a NextSeq 500 platform (Illumina) using 75 bp single-end sequencing. On average, about 27 million reads were generated from each sample for mouse microglia and an average of 36 million reads per sample for iMGL.
Raw reads obtained from RNAseq were quantified using Salmon (v1.1.0) in selective alignment mode and auto-detect library set to true. Transcriptome indicies were generated with Salmon using default parameters and the human GRCh38.p13 or mouse GRCm38.p6 transcriptome reference downloaded from http://www.gencodegenes.org. Expression counts for each gene, in raw counts and transcripts per million (TPM), were generated from the Salmon outputs using tximport package for R.
Differential gene expression and normalization were performed using DESeq2. Gene enrichment analysis was performed using the Metascape (http://metascape.org/) online tool with all parameters set to default or with Gene Set Enrichment Analysis (GSEA) software. GSEA was conducted on the ranked gene loadings for selected principal components and run against the “C5: gene ontology v7.0” gene sets downloaded from www.gsea-msigdb.org. Results were plotted with R or imported into Cytoscape (v3.7.2) for visualization and annotation using the AutoAnnotate package.
Quantification and statistical analysis
For determining enrichment of DAM-APOE genes in principal components, hypergeometric tests were performed in R using the dhyper function to determine the sum of the probabilities of the observed result or anything greater to assess significance. For the analysis of Aβ load and IBA1+ cell numbers, two-way ANOVA was run using Prism version 7 (GraphPad Software). Significant main effects were further analyzed by planned comparisons between groups of interest using the Bonferroni correction. For all testing the significance was set at a threshold of p < 0.05. All data are represented as the mean ± standard error of the mean (SEM).
Acknowledgments
The authors thank Drs. Mary Jo LaDu, Todd Morgan, and Amy Christensen for contributing APOE-FAD mice; Dr. Soshana P. Svendsen for critical review and editing of the manuscript; and Julio Orellana for help with artwork. This work was funded by the Cedars-Sinai Board of Governors Regenerative Medicine Institute (C.N.S.), the Cedars-Sinai Center for Research in Women’s Health and Sex Differences (V.A.M.), NIH grant RF1 AG058068 (C.J.P.), and Alzheimer’s Association SAGA-17-419408 (C.J.P.).
Author contributions
V.A.M.: conceptualization, methodology, validation, formal analysis, experimental assays, original manuscript draft preparation, project administration. M.J.W.: software, formal analysis, data curation, original manuscript draft preparation. S.J.H.: experimental assays, original manuscript draft preparation. R.M.L.: experimental assays, original manuscript draft preparation. C.J.P.: conceptualization, methodology, resources, manuscript review and editing, supervision, funding. C.N.S.: conceptualization, methodology, resources, manuscript review and editing, supervision, funding.
Declaration of interests
The authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103238.
Contributor Information
V. Alexandra Moser, Email: alexandra.moser@cshs.org.
Clive N. Svendsen, Email: clive.svendsen@cshs.org.
Supplemental information
The top 250 PC1-high genes along with log2fold changes between APOE and APOE-FAD animals and adjusted p-values are shown
Data and code availability
-
•
Sequencing data reported in this paper have been deposited at GEO and are publicly available as of the date of publication. The accession number is GEO: GSE163857.
-
•
All original code is available upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Altmann A., Tian L., Henderson V.W., Greicius M.D., Alzheimer’s Disease Neuroimaging Initiative Investigators Sex modifies the APOE-related risk of developing Alzheimer disease. Ann. Neurol. 2014;75:563–573. doi: 10.1002/ana.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik S.H., Kang S., Lee W., Choi H., Chung S., Kim J.-I., Mook-Jung I. A breakdown in metabolic reprogramming causes microglia dysfunction in Alzheimer’s disease. Cell Metab. 2019;30:493–507.e6. doi: 10.1016/j.cmet.2019.06.005. [DOI] [PubMed] [Google Scholar]
- Blasko I., Stampfer-Kountchev M., Robatscher P., Veerhuis R., Eikelenboom P., Grubeck-Loebenstein B. How chronic inflammation can affect the brain and support the development of Alzheimer’s disease in old age: the role of microglia and astrocytes. Aging Cell. 2004;3:169–176. doi: 10.1111/j.1474-9728.2004.00101.x. [DOI] [PubMed] [Google Scholar]
- Boyles J.K., Pitas R.E., Wilson E., Mahley R.W., Taylor J.M. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J. Clin. Invest. 1985;76:1501–1513. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciottolo M., Christensen A., Moser A., Liu J., Pike C.J., Smith C., LaDu M.J., Sullivan P.M., Morgan T.E., Dolzhenko E. The APOE4 allele shows opposite sex bias in microbleeds and Alzheimer’s disease of humans and mice. Neurobiol. Aging. 2016;37:47–57. doi: 10.1016/j.neurobiolaging.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciottolo M., Wang X., Driscoll I., Woodward N., Saffari A., Reyes J., Serre M.L., Vizuete W., Sioutas C., Morgan T.E. Particulate air pollutants, APOE alleles and their contributions to cognitive impairment in older women and to amyloidogenesis in experimental models. Transl. Psychiatry. 2017;7:e1022. doi: 10.1038/tp.2016.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano J.M., Kim J., Stewart F.R., Jiang H., DeMattos R.B., Patterson B.W., Fagan A.M., Morris J.C., Mawuenyega K.G., Cruchaga C. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci. Transl. Med. 2011;3:89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton C.A., Needham L.K., Brown C., Cook D., Rasheed K., Burke J.R., Strittmatter W.J., Schmechel D.E., Vitek M.P. APOE genotype-specific differences in human and mouse macrophage nitric oxide production. J. Neuroimmunol. 2004;147:62–67. doi: 10.1016/j.jneuroim.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Deczkowska A., Keren-Shaul H., Weiner A., Colonna M., Schwartz M., Amit I. Disease-associated microglia: a universal immune sensor of neurodegeneration. Cell. 2018;173:1073–1081. doi: 10.1016/j.cell.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Douvaras P., Sun B., Wang M., Kruglikov I., Lallos G., Zimmer M., Terrenoire C., Zhang B., Gandy S., Schadt E. Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Rep. 2017;8:1516–1524. doi: 10.1016/j.stemcr.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazzo T.C., Mattsson N., Weiner M.W., Alzheimer’s Disease Neuroimaging Initiative Interaction of cigarette smoking history with APOE genotype and age on amyloid level, glucose metabolism, and neurocognition in cognitively normal elders. Nicotine Tob. Res. 2016;18:204–211. doi: 10.1093/ntr/ntv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egensperger R., Kösel S., von Eitzen U., Graeber M.B. Microglial activation in Alzheimer disease: association with APOE genotype. Brain Pathol. 1998;8:439–447. doi: 10.1111/j.1750-3639.1998.tb00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikelenboom P., Veerhuis R., van Exel E., Hoozemans J.J.M., Rozemuller A.J.M., van Gool W.A. The early involvement of the innate immunity in the pathogenesis of late-onset Alzheimer’s disease: neuropathological, epidemiological and genetic evidence. Curr. Alzheimer Res. 2011;8:142–150. doi: 10.2174/156720511795256080. [DOI] [PubMed] [Google Scholar]
- van der Flier W.M., Pijnenburg Y.A., Fox N.C., Scheltens P. Early-onset versus late-onset Alzheimer’s disease: the case of the missing APOE ϵ4 allele. Lancet Neurol. 2011;10:280–288. doi: 10.1016/S1474-4422(10)70306-9. [DOI] [PubMed] [Google Scholar]
- Friedman B.A., Srinivasan K., Ayalon G., Meilandt W.J., Lin H., Huntley M.A., Cao Y., Lee S.-H., Haddick P.C.G., Ngu H. Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 2018;22:832–847. doi: 10.1016/j.celrep.2017.12.066. [DOI] [PubMed] [Google Scholar]
- Galatro T.F., Holtman I.R., Lerario A.M., Vainchtein I.D., Brouwer N., Sola P.R., Veras M.M., Pereira T.F., Leite R.E.P., Möller T. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat. Neurosci. 2017;20:1162–1171. doi: 10.1038/nn.4597. [DOI] [PubMed] [Google Scholar]
- Gale S.C., Gao L., Mikacenic C., Coyle S.M., Rafaels N., Murray Dudenkov T., Madenspacher J.H., Draper D.W., Ge W., Aloor J.J. APOε4 is associated with enhanced in vivo innate immune responses in human subjects. J. Allergy Clin. Immunol. 2014;134:127–134. doi: 10.1016/j.jaci.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J.A., Cardona S.M., Cardona A.E. Isolation and analysis of mouse microglial cells. Curr. Protoc. Immunol. 2014;104:14–35. doi: 10.1002/0471142735.im1435s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C.K., Saijo K., Winner B., Marchetto M.C., Gage F.H. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey M.E., Wojcik D.P., Krone C.A. Apolipoprotein E genotyping as a potential biomarker for mercury neurotoxicity. J. Alzheimers Dis. 2003;5:189–195. doi: 10.3233/jad-2003-5303. [DOI] [PubMed] [Google Scholar]
- Gosselin D., Skola D., Coufal N.G., Holtman I.R., Schlachetzki J.C.M., Sajti E., Jaeger B.N., O’Connor C., Fitzpatrick C., Pasillas M.P. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:6344. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–271.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D.V., Hanson J.E., Sheng M. Microglia in Alzheimer’s disease. J. Cell Biol. 2018;217:459–472. doi: 10.1083/jcb.201709069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann J., Coburn M.A., England W., Figueroa Velez D.X., Kiani Shabestari S., Tu C.H., McQuade A., Kolahdouzan M., Echeverria K., Claes C. Development of a chimeric model to study and manipulate human microglia in vivo. Neuron. 2019;103:1016–1033.e10. doi: 10.1016/j.neuron.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.S., Ebbert M.T.W., Baker K.E., Cook C., Wang X., Sens J.P., Kocher J.-P., Petrucelli L., Fryer J.D. Microglial translational profiling reveals a convergent APOE pathway from aging, amyloid, and tau. J. Exp. Med. 2018;215:2235–2245. doi: 10.1084/jem.20180653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B. A unique microglia type associated with restricting development of alzheimer’s disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Killin L.O.J., Starr J.M., Shiue I.J., Russ T.C. Environmental risk factors for dementia: a systematic review. BMC Geriatr. 2016;16:175. doi: 10.1186/s12877-016-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konttinen H., Cabral-da-Silva M.E.C., Ohtonen S., Wojciechowski S., Shakirzyanova A., Caligola S., Giugno R., Ishchenko Y., Hernández D., Fazaludeen M.F. PSEN1ΔE9, APPswe, and APOE4 confer disparate phenotypes in human iPSC-derived microglia. Stem Cell Rep. 2019;13:669–683. doi: 10.1016/j.stemcr.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47:566–581.e9. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFerla F.M. Pathways linking Abeta and tau pathologies. Biochem. Soc. Trans. 2010;38:993–995. doi: 10.1042/BST0380993. [DOI] [PubMed] [Google Scholar]
- Li Q., Cheng Z., Zhou L., Darmanis S., Neff N.F., Okamoto J., Gulati G., Bennett M.L., Sun L.O., Clarke L.E. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207–223.e10. doi: 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.-S., Peterson T.C. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.-T., Seo J., Gao F., Feldman H.M., Wen H.-L., Penney J., Cam H.P., Gjoneska E., Raja W.K., Cheng J. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron. 2018;98:1141–1154. doi: 10.1016/j.neuron.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Zhu B., Liu Y., Zhang X., Yin J., Li X., Jiang L., Hodges A.P., Rosenthal S.B., Zhou L. Multi-omic comparison of Alzheimer’s variants in human ESC-derived microglia reveals convergence at APOE. J. Exp. Med. 2020;217:e20200474. doi: 10.1084/jem.20200474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschallinger J., Iram T., Zardeneta M., Lee S.E., Lehallier B., Haney M.S., Pluvinage J.V., Mathur V., Hahn O., Morgens D.W. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020;23:194–208. doi: 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H., Adaikkan C., Gao F., Young J.Z., Manet E., Hemberg M., De Jager P.L., Ransohoff R.M., Regev A., Tsai L.-H. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 2017;21:366–380. doi: 10.1016/j.celrep.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt V.C., Rabinowitz A.R., Arnett P.A. The influence of the apolipoprotein E (APOE) gene on subacute post-concussion neurocognitive performance in college athletes. Arch. Clin. Neuropsychol. 2018;33:36–46. doi: 10.1093/arclin/acx051. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Atwood C.S., Romano D.M., Laurans M.H., Huang X., Bush A.I., Smith J.D., Tanzi R.E. Differential effects of apolipoprotein E isoforms on metal-induced aggregation of A beta using physiological concentrations. Biochemistry. 1999;38:4595–4603. doi: 10.1021/bi982437d. [DOI] [PubMed] [Google Scholar]
- Moser V.A., Pike C.J. Obesity accelerates Alzheimer-related pathology in APOE4 but not APOE3 mice. Eneuro. 2017;4 doi: 10.1523/ENEURO.0077-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser V.A., Uchoa M.F., Pike C.J. TLR4 inhibitor TAK-242 attenuates the adverse neural effects of diet-induced obesity. J. Neuroinflamm. 2018;15:306. doi: 10.1186/s12974-018-1340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser V.A., Christensen A., Liu J., Zhou A., Yagi S., Beam C.R., Galea L., Pike C.J. Effects of aging, high-fat diet, and testosterone treatment on neural and metabolic outcomes in male brown Norway rats. Neurobiol. Aging. 2019;73:145–160. doi: 10.1016/j.neurobiolaging.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher K.I., Wyss-Coray T. Microglial dysfunction in brain aging and Alzheimer’s disease. Biochem. Pharmacol. 2014;88:594–604. doi: 10.1016/j.bcp.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., Lin C.-C., Hwang Y.-H., Hsieh W.-S., Liao H.-F., Chen P.-C. Mercury, APOE, and children’s neurodevelopment. Neurotoxicology. 2013;37:85–92. doi: 10.1016/j.neuro.2013.03.012. [DOI] [PubMed] [Google Scholar]
- Olah M., Patrick E., Villani A.-C., Xu J., White C.C., Ryan K.J., Piehowski P., Kapasi A., Nejad P., Cimpean M. A transcriptomic atlas of aged human microglia. Nat. Commun. 2018;9:539. doi: 10.1038/s41467-018-02926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olgiati P., Politis A., Malitas P., Albani D., Dusi S., Polito L., De Mauro S., Zisaki A., Piperi C., Stamouli E. APOE epsilon-4 allele and cytokine production in Alzheimer’s disease. Int. J. Geriatr. Psychiatry. 2010;25:338–344. doi: 10.1002/gps.2344. [DOI] [PubMed] [Google Scholar]
- Patir A., Shih B., McColl B.W., Freeman T.C. A core transcriptional signature of human microglia: derivation and utility in describing region-dependent alterations associated with Alzheimer’s disease. Glia. 2019;67:1240–1253. doi: 10.1002/glia.23572. [DOI] [PubMed] [Google Scholar]
- Penney J., Ralvenius W.T., Tsai L.-H. Modeling Alzheimer’s disease with iPSC-derived brain cells. Mol. Psychiatry. 2020;25:148–167. doi: 10.1038/s41380-019-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluvinage J.V., Haney M.S., Smith B.A.H., Sun J., Iram T., Bonanno L., Li L., Lee D.P., Morgens D.W., Yang A.C. CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature. 2019;568:187–192. doi: 10.1038/s41586-019-1088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju S., Dammer E.B., Raza S.A., Rathakrishnan P., Xiao H., Gao T., Duong D.M., Pennington M.W., Lah J.J., Seyfried N.T. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Mol. Neurodegener. 2018;13:24. doi: 10.1186/s13024-018-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez G.A., Tai L.M., LaDu M.J., Rebeck G.W. Human APOE4 increases microglia reactivity at Aβ plaques in a mouse model of Aβ deposition. J. Neuroinflamm. 2014;11:111. doi: 10.1186/1742-2094-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala Frigerio C., Wolfs L., Fattorelli N., Thrupp N., Voytyuk I., Schmidt I., Mancuso R., Chen W.-T., Woodbury M.E., Srivastava G. The major risk factors for Alzheimer’s disease: age, sex, and genes modulate the microglia response to aβ plaques. Cell Rep. 2019;27:1293–1306.e6. doi: 10.1016/j.celrep.2019.03.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sances S., Ho R., Vatine G., West D., Laperle A., Meyer A., Godoy M., Kay P.S., Mandefro B., Hatata S. Human iPSC-derived endothelial cells and microengineered organ-chip enhance neuronal development. Stem Cell Rep. 2018;10:1222–1236. doi: 10.1016/j.stemcr.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schram M.T., Euser S.M., de Craen A.J.M., Witteman J.C., Frölich M., Hofman A., Jolles J., Breteler M.M.B., Westendorp R.G.J. Systemic markers of inflammation and cognitive decline in old age. J. Am. Geriatr. Soc. 2007;55:708–716. doi: 10.1111/j.1532-5415.2007.01159.x. [DOI] [PubMed] [Google Scholar]
- Serrano-Pozo A., Frosch M.P., Masliah E., Hyman B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011;1:a006189. doi: 10.1101/cshperspect.a006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoj Tratnik J., Falnoga I., Trdin A., Mazej D., Fajon V., Miklavčič A., Kobal A.B., Osredkar J., Sešek Briški A., Krsnik M. Prenatal mercury exposure, neurodevelopment and apolipoprotein E genetic polymorphism. Environ. Res. 2017;152:375–385. doi: 10.1016/j.envres.2016.08.035. [DOI] [PubMed] [Google Scholar]
- Spittau B. Aging microglia-phenotypes, functions and implications for age-related neurodegenerative diseases. Front. Aging Neurosci. 2017;9:194. doi: 10.3389/fnagi.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen T.L., Cacciottolo M., Balu D., Morgan T.E., LaDu M.J., Finch C.E., Pike C.J. APOE genotype and sex affect microglial interactions with plaques in Alzheimer’s disease mice. Acta Neuropathol. Commun. 2019;7:82. doi: 10.1186/s40478-019-0729-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatine G.D., Barrile R., Workman M.J., Sances S., Barriga B.K., Rahnama M., Barthakur S., Kasendra M., Lucchesi C., Kerns J. Human iPSC-derived blood–brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell. 2019;24:995–1005.e6. doi: 10.1016/j.stem.2019.05.011. [DOI] [PubMed] [Google Scholar]
- Villemagne V.L., Burnham S., Bourgeat P., Brown B., Ellis K.A., Salvado O., Szoeke C., Macaulay S.L., Martins R., Maruff P. Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study. Lancet Neurol. 2013;12:357–367. doi: 10.1016/S1474-4422(13)70044-9. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T., Rogers J. Inflammation in Alzheimer disease-a brief review of the basic science and clinical literature. Cold Spring Harb. Perspect. Med. 2012;2:a006346. doi: 10.1101/cshperspect.a006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Li X., Boreland A.J., Posyton A., Kwan K., Hart R.P., Jiang P. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun. 2020;11:1577. doi: 10.1038/s41467-020-15411-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh F.L., Wang Y., Tom I., Gonzalez L.C., Sheng M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron. 2016;91:328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- Youmans K.L., Tai L.M., Nwabuisi-Heath E., Jungbauer L., Kanekiyo T., Gan M., Kim J., Eimer W.A., Estus S., Rebeck G.W. APOE4-specific changes in Aβ accumulation in a new transgenic mouse model of Alzheimer disease. J. Biol. Chem. 2012;287:41774–41786. doi: 10.1074/jbc.M112.407957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Wang H., Abel G.M., Storm D.R., Xia Z. The effects of gene-environment interactions between cadmium exposure and apolipoprotein E4 on memory in a mouse model of Alzheimer’s disease. Toxicol. Sci. 2020;173:189–201. doi: 10.1093/toxsci/kfz218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Davis M.D., Martens Y.A., Shinohara M., Graff-Radford N.R., Younkin S.G., Wszolek Z.K., Kanekiyo T., Bu G. APOE ε4/ε4 diminishes neurotrophic function of human iPSC-derived astrocytes. Hum. Mol. Genet. 2017;26:2690–2700. doi: 10.1093/hmg/ddx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Ren Y., Yamazaki Y., Qiao W., Li F., Felton L.M., Mahmoudiandehkordi S., Kueider-Paisley A., Sonoustoun B., Arnold M. Alzheimer’s risk factors age, APOE genotype, and sex drive distinct molecular pathways. Neuron. 2020;106:727–742.e6. doi: 10.1016/j.neuron.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The top 250 PC1-high genes along with log2fold changes between APOE and APOE-FAD animals and adjusted p-values are shown
Data Availability Statement
-
•
Sequencing data reported in this paper have been deposited at GEO and are publicly available as of the date of publication. The accession number is GEO: GSE163857.
-
•
All original code is available upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.