Summary
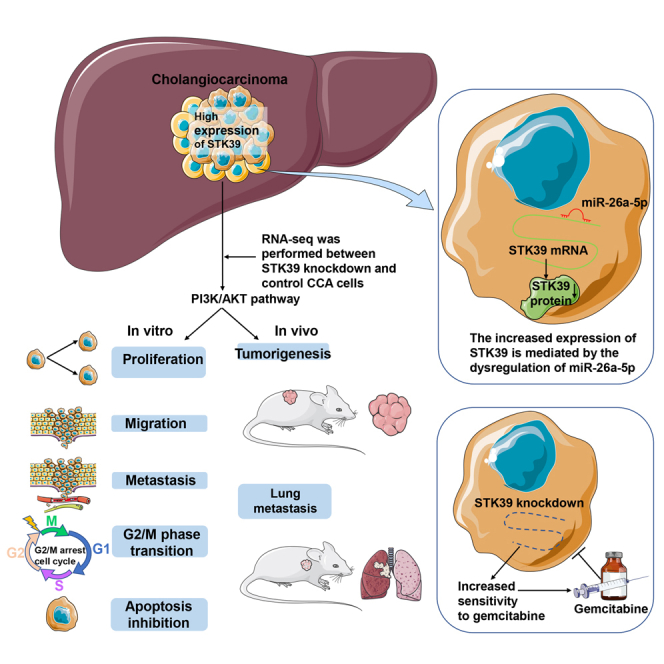
Serine/threonine kinase 39 (STK39) is overexpressed in various tumor tissues and plays an essential role in tumor progression. In this study, we investigated the clinical value, as well as the potential functions and mechanisms of STK39 in cholangiocarcinoma (CCA). The results showed that STK39 was overexpressed in CCA and negatively associated with the prognosis of patients with CCA. Functionally, STK39 knockdown suppressed cell proliferation, migration, and invasion, while STK39 overexpression facilitated tumor aggressiveness. The tumor-promoting effects of STK39 in CCA were also validated by in vivo experiments. Mechanistically, RNA-seq analysis identified that STK39 enhanced the progression of CCA by activating PI3K/AKT signaling pathway. Furthermore, overexpression of STK39 could induce gemcitabine resistance in CCA cells. Moreover, the increased expression of STK39 may be mediated by the dysregulation of miR-26a-5p. In summary, STK39 could be served as a valuable prognostic candidate and a potential therapeutic target of CCA.
Subject areas: Biological sciences, Cancer, Molecular biology
Graphical abstract
Highlights
-
•
STK39 was overexpressed in CCA, negatively associated with the prognosis of patients with CCA
-
•
STK39 knockdown suppressed cell proliferation and invasion. STK39 overexpression facilitated tumor aggressiveness
-
•
STK39 mediates oncogenic effects on CCA cells by activating the PI3K/AKT signaling pathway
-
•
STK39 reduces CCA sensitivity to gemcitabine. Increased expression of STK39 may be mediated by dysregulation of miR-26a-5p
Biological sciences; Cancer; Molecular biology
Introduction
Cholangiocarcinoma (CCA) is a common type of malignant tumor originating from the epithelium of the bile duct. The morbidity and mortality rates of CCA have increased over the past 20 years (Shin et al., 2010; Tyson and El-Serag, 2011). Currently, the morbidity rate is almost equal to the mortality rate (Khan et al., 2019; Shaib et al., 2004; Bertuccio et al., 2019). Due to lack of typical clinical symptoms, most patients are diagnosed at advanced stages or with extrahepatic metastasis (Bridgewater et al., 2014). Surgical resection is considered the gold standard treatment. However, early lymph node metastasis, low rate of surgical resection, and low sensitivity to chemotherapy are all critical causes for the poor prognosis of CCA (Valle et al., 2016; Rodrigues et al., 2021). Therefore, research on the molecular mechanism of CCA proliferation, migration, and invasion is necessary to identify potential therapeutic targets and prognostic predictors.
Serine/threonine kinase 39 (STK39, also named SPAK), is derived from the Ste20-like kinase family (Ramoz et al., 2008). It plays an important role in ion homeostasis by regulating the cation chloride cotransporters’ activities, which is indispensable in the transportation of renal salt and the stability of the circulatory system in mammalians (Richardson et al., 2011; McCormick et al., 2011). STK39 contains an N-terminal series of proline and alanine repeats (PAPA box), followed by a serine/threonine kinase catalytic domain (Gagnon et al., 2006; Tsutsumi et al., 2000). Abnormal expression of STK39 has been correlated with various human diseases, such as hypertension (Wang et al., 2009), stroke (Josiah et al., 2021), and type I diabetes (Forgetta et al., 2020). In addition, many studies have pointed out that STK39 acts as an important regulator in the occurrence and progression of malignancies (Li et al., 2016; Huang et al., 2017; Yang et al., 2018), including hepatocellular carcinoma (HCC) (Zhang et al., 2021). However, limited research has explored the molecular mechanism of STK39 in CCA.
In this study, we explored the underlying molecular mechanism of STK39 played in the progression of CCA. Our data showed that STK39 was obviously upregulated in CCA, and patients with high level of STK39 were more likely to suffer poor prognosis. Further studies revealed that STK39 knockdown decreased the tumor-promoting phenotypes. Significantly, we demonstrated that STK39 exerts its oncogenic function in CCA by activating PI3K/AKT signaling pathway. Interestingly, STK39 overexpression decreased the sensitivity of CCA cells to gemcitabine, and the increased expression of STK39 may be mediated by the dysregulation of miR-26a-5p.
Results
STK39 is upregulated in CCA tissues
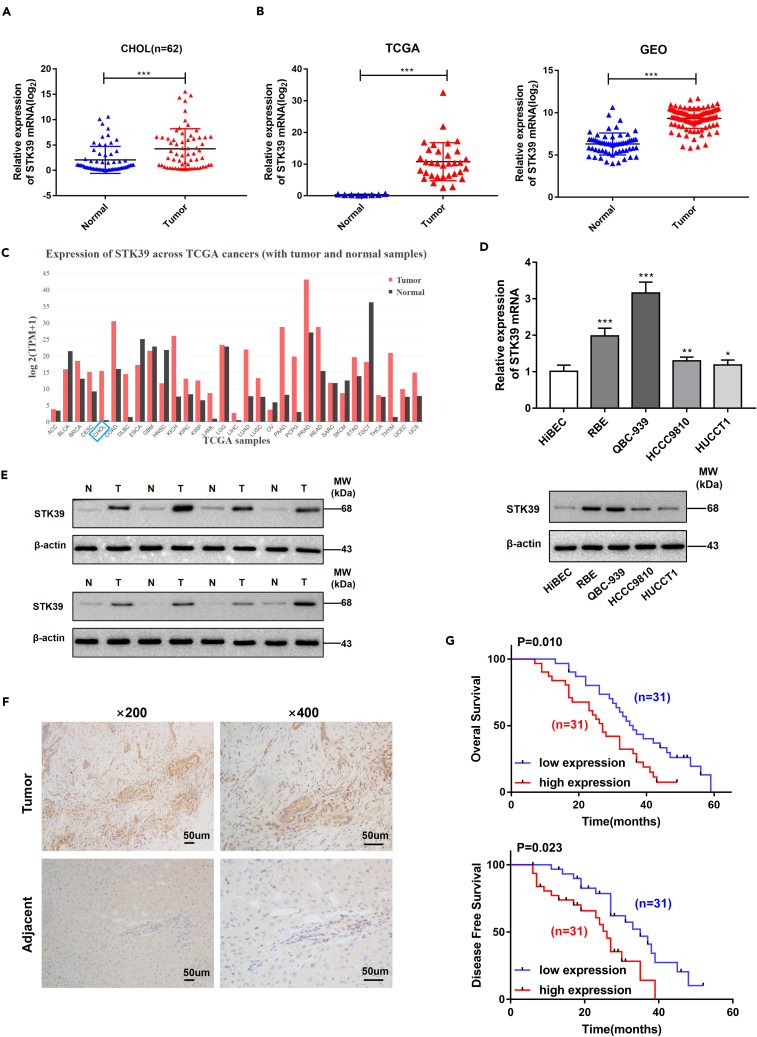
To evaluate the mRNA levels of STK39, 62 pairs of CCA and matched normal tissues were determined by qRT-PCR. The data suggested that STK39 was significantly upregulated in CCA tissues compared with normal tissues (Figure 1A). These results were consistent with data obtained from The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus dataset (GEO, GSE26566) (Figure 1B). Moreover, a pan-cancer analysis of the TCGA database demonstrated high levels of STK39 in a variety of cancer types, including liver hepatocellular carcinoma (LIHC) and cholangiocarcinoma (CHOL) (Figure 1C). In addition, CCA cell lines revealed higher levels of STK39 compared with normal bile duct cells (HiBEC) (Figure 1D). To further verify the above findings, we randomly chose eight pairs of CCA and matched normal tissues for Western blot analysis. The results revealed a significantly higher expression of STK39 in CCA tissues (Figure 1E). These results were consistent with typical immunohistochemistry (IHC, Figure 1F) analysis, which also revealed abundant protein levels of STK39 in CCA tissues. To explore the clinicopathological significance of STK39, we divided the 62 CCA cases into two groups based on the median expression of STK39. As indicated in Table 1, the high expression group appeared to develop larger tumor size, advanced T stage, and higher risks of LN metastasis (p value<0.05). There was no significant association with age, gender, differentiation, vascular invasion, and microvascular invasion (MVI). More importantly, patients in the high expression group were negatively correlated with shorter survival than those in the low expression group (Figure 1G). The above data suggest that STK39 may function as a meaningful prognostic biomarker for patients with CCA.
Figure 1.
STK39 is upregulated in CCA tissues and cell lines and is associated with poor prognosis
(A) The analysis of STK39 mRNA levels in 62 pairs of human CCA tissues and matched normal tissues.
(B) Analysis of STK39 expression in human CCA tissues and adjacent normal tissues in TCGA and GEO database.
(C) Pan-cancer analysis using TCGA data set showed that STK39 is mainly overexpressed in liver cancer (both CHOL and LIHC) and few other cancer types.
(D) STK39 mRNA and protein expression in one human intrahepatic biliary epithelial cell line (HiBEC) and four CCA cell lines were assessed by qRT-PCR and western blotting.
(E) The expression of STK39 in human HCC tissues (T) and matched normal tissues (N) were assessed by western blotting.
(F) Representative images of STK39 expression in CCA tissues by IHC staining, scale bar, 50μm.
(G) Elevated expression of STK39 was negatively correlated with the overall survival (OS) and disease-free survival (DFS) of patients with CCA. ∗p<0.05; ∗∗p<0.01; ∗∗∗p<0.001. Data were shown as mean ± SEM.
Table 1.
The clinicopathological relevance analysis of STK39 expression in patients with cholangiocarcinoma
| Characteristics | Low group |
High group |
P value |
|---|---|---|---|
| (N = 31) | (N = 31) | ||
| Gender | |||
| Male | 16 | 19 | 0.442 |
| Female | 15 | 12 | |
| Age (years) | |||
| <55 | 10 | 13 | 0.430 |
| ≥55 | 21 | 18 | |
| Tumor multiplicity | |||
| Single | 19 | 16 | 0.304 |
| Multiple | 12 | 15 | |
| Differentiation | |||
| Well to moderate | 20 | 14 | 0.126 |
| Poor | 11 | 17 | |
| Diameter (cm) | |||
| ≤5 | 17 | 9 | 0.035∗ |
| >5 | 14 | 22 | |
| TNM stage | |||
| I + II | 19 | 11 | 0.037∗ |
| III + IV | 12 | 20 | |
| Vascular invasion | |||
| Negative | 24 | 27 | 0.319 |
| Positive | 7 | 4 | |
| Microvascular invasion | |||
| Negative | 21 | 15 | 0.123 |
| Positive | 10 | 16 | |
| Lymph node metastasis | |||
| Negative | 22 | 12 | 0.011∗ |
| Positive | 9 | 19 | |
Annotation: ∗p<0.05.
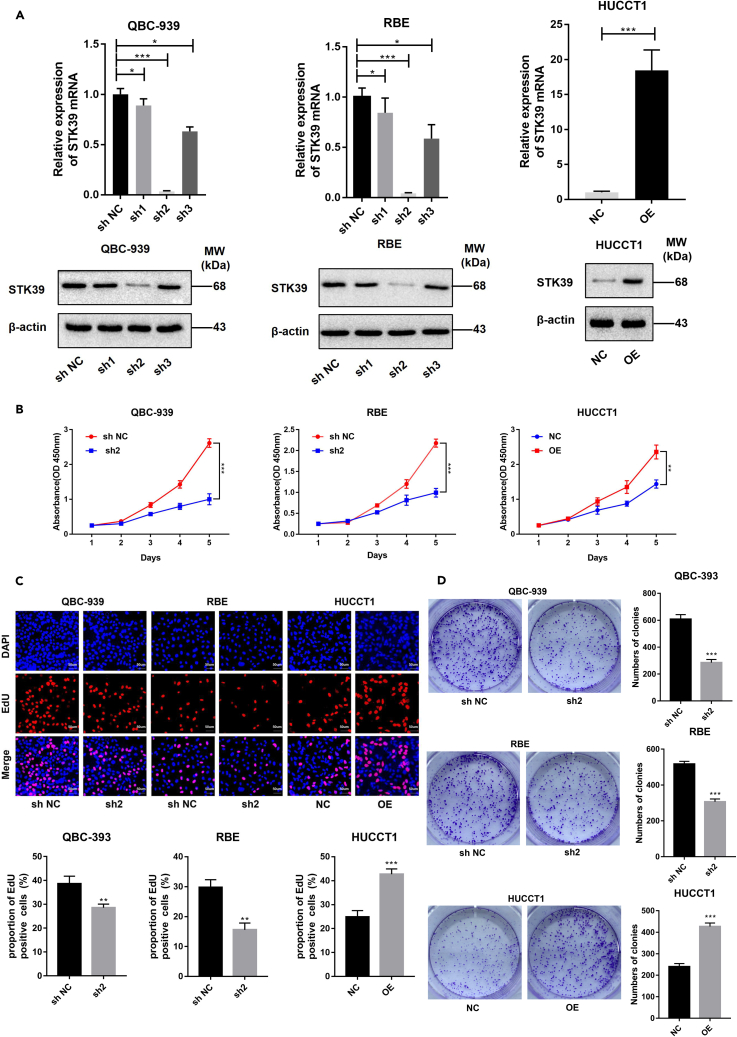
STK39 promotes the proliferation of CCA in vitro
As illustrated in Figure 1D, QBC-939 and RBE cells exhibited a higher expression level of STK39, while HUCCT1 cells showed a lower expression level. Therefore, we chose the three cell lines for further studies to explore the biological significance of STK39 in CCA. We then established stable STK39-knockdown and STK39-overexpression CCA cell lines by infection with STK39 shRNA and lentiviral vector, respectively (Figure 2A). Among the three STK39 shRNAs, only shRNA2 transfection effectively suppressed STK39 expression. Consequently, cells transfected with shRNA2 were chosen for further analysis. In order to examine the effect of STK39 on cell proliferation, CCK-8, EdU, and colony formation assays were then performed. The CCK-8 and EdU assay (Figures 2B and 2C) showed that STK39 knockdown significantly impaired the proliferation of QBC-939 and RBE cells, while overexpression of STK39 enhanced the proliferation of HUCCT1 cells. In line with our expectations, the growth inhibition effect caused by STK39 knockdown in CCA cells has also been confirmed by colony formation assay, whereas overexpression of STK39 facilitated the cell proliferation (Figure 2D). Taken together, our data demonstrated that STK39 could enhance the proliferation of CCA in vitro.
Figure 2.
STK39 overexpression promotes proliferation of CCA
QBC-939 and RBE cells were transfected with STK39 shRNA or control shRNA (sh NC); HUCCT1 cells were infected with STK39 overexpression lentivirus (OE) or control lentivirus (NC).
(A) STK39 expression in QBC-939, RBE, and HUCCT1 cells was analyzed by qPCR and Western blot.
(B) The proliferation of STK39 stable knockdown or overexpression cells was determined by CCK8 assay at 0, 24, 48, 72, and 96 h.
(C) EdU incorporation of STK39 stable knockdown or overexpression cells was detected, scale bar, 50μm.
(D) The colony formation of STK39 stable knockdown or overexpression cells was determined. ∗p<0.05; ∗∗p<0.01; ∗∗∗p<0.001. Data were shown as mean ± SEM.
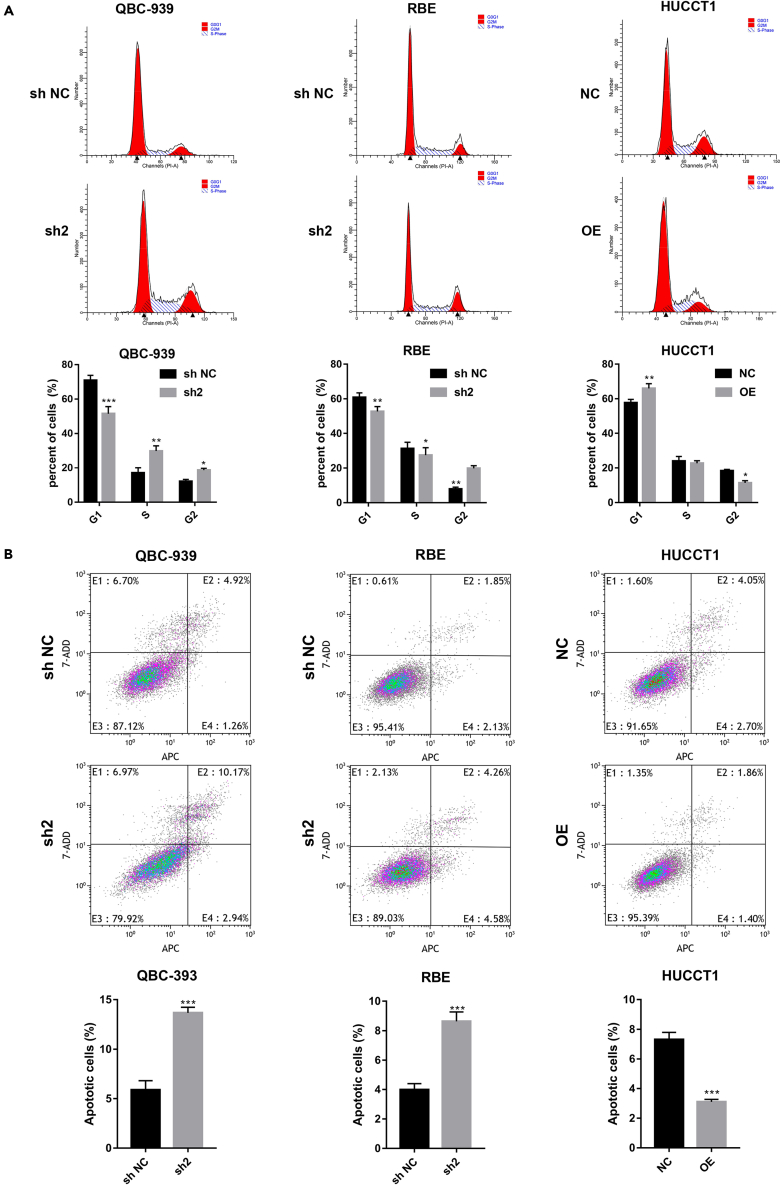
STK39 induces G2/M phase transition and inhibits CCA apoptosis
Since cell cycle and apoptosis rate can also reflect the cell proliferation activity, we then performed cell cycle analysis to explore whether STK39 has a regulatory effect on the cell cycle. The flow cytometry analysis showed that STK39 knockdown decreased the percentage of cells in the G1 phase and caused cell-cycle arrest in the G2/M phase. However, STK39 overexpression showed an opposite trend (Figure 3A). Silencing of STK39 has also been shown to induce apoptosis. As shown in Figure 3B, STK39 knockdown enhanced cell apoptosis in both QBC-939 and RBE cells. The above results suggest that STK39 may promote the growth of CCA by stimulating G2/M cell cycle transition, as well as apoptosis inhibition.
Figure 3.
Knockdown of STK39 induces CCA cells apoptosis and cell-cycle arrest
QBC-939 and RBE cells were transfected with STK39 shRNA (sh2) or control shRNA (sh NC); HUCCT1 cells were infected with STK39 overexpression lentivirus (OE) or control lentivirus (NC).
(A) Cell cycle analysis of STK39 stable knockdown or overexpression cells.
(B) The percentage of apoptotic cells of STK39 stable knockdown or overexpression cells was analyzed by Annexin V-FITC/PI staining assay. ∗p<0.05; ∗∗p<0.01; ∗∗∗p<0.001. Data were shown as mean ± SEM.
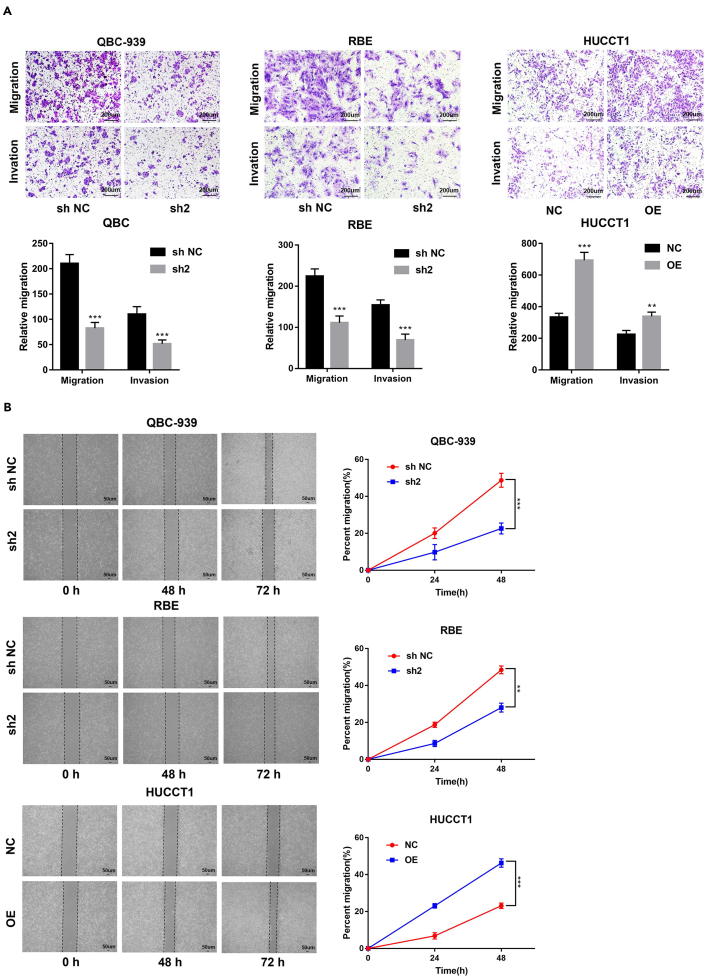
STK39 promotes CCA cell migration and invasion
To further investigate the migration and invasion ability of STK39 in CCA, transwell, and wound-healing assays were performed. As shown in Figure 4A, cell migration and invasion were suppressed by STK39 knockdown. In contrast, the opposite results were found in STK39 overexpression cells (Figure 4A). Meanwhile, the wound-healing assay showed that STK39 knockdown attenuated the migration ability of CCA cells, but this ability was markedly enhanced by STK39 overexpression (Figure 4B). Taken together, these findings indicate that STK39 enhances the motility of CCA cells.
Figure 4.
STK39 promotes the migration and invasion of CCA cells
QBC-939 and RBE cells were transfected with STK39 shRNA (sh2) or control shRNA (sh NC); HUCCT1 cells were infected with STK39 overexpression lentivirus (OE) or control lentivirus (NC).
(A) Invasive or migrated cells were measured by transwell assay with or without matrix, scale bar, 200μm.
(B) Cell migration ability was evaluated by wound healing assay, scale bar, 50μm. ∗p<0.05; ∗∗p<0.01; ∗∗∗p<0.001. Data were shown as mean ± SEM.
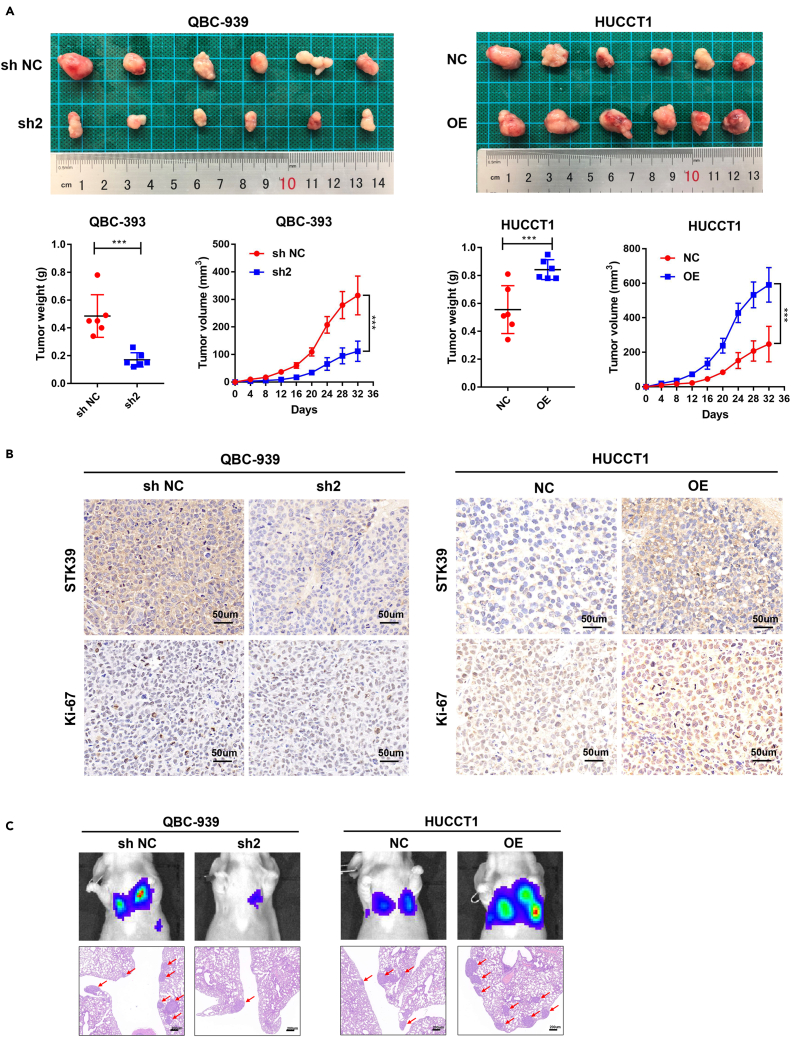
STK39 promotes tumorigenesis and metastasis of CCA in vivo
STK39 stable knockdown (QBC-939) or overexpression (HUCCT1) cell lines were injected into nude mice to verify the in vivo effect of STK39 on CCA tumorigenicity. As shown in Figure 5A, the subcutaneous tumors generated from STK39 knockdown cells were dramatically smaller and lighter than the NC group. In contrast, STK39 overexpression resulted in larger and heavier tumors than in the NC group. Meanwhile, by staining the tumors with STK39 and Ki-67, we further confirmed the positive effect of STK39 on CCA proliferation (Figure 5B). Additionally, the in vivo metastatic assay showed that downregulation of STK39 decreased detectable lung metastasis lesions compared with the NC group, while overexpression of STK39 showed the opposite trend (Figure 5C). The above in vivo experiments show that STK39 could promote tumorigenesis and metastasis in CCA.
Figure 5.
STK39 knockdown suppressed CCA cell proliferation and metastasis in vivo
(A) Nude mice were subcutaneously injected with STK39 stable knockdown or overexpression cells. The tumor volume and average weight were determined.
(B) IHC analysis of STK39 and Ki-67 in the tumors derived from mice, scale bar, 50μm.
(C) STK39 stable knockdown or overexpression cells were injected into the tail vein of nude mice to induce lung metastasis, and representative bioluminescent images were captured; liver metastatic nodules were subjected to HE staining as indicated, scale bar, 200μm. ∗p<0.05; ∗∗p<0.01; ∗∗∗p<0.001. Data were shown as mean ± SEM.
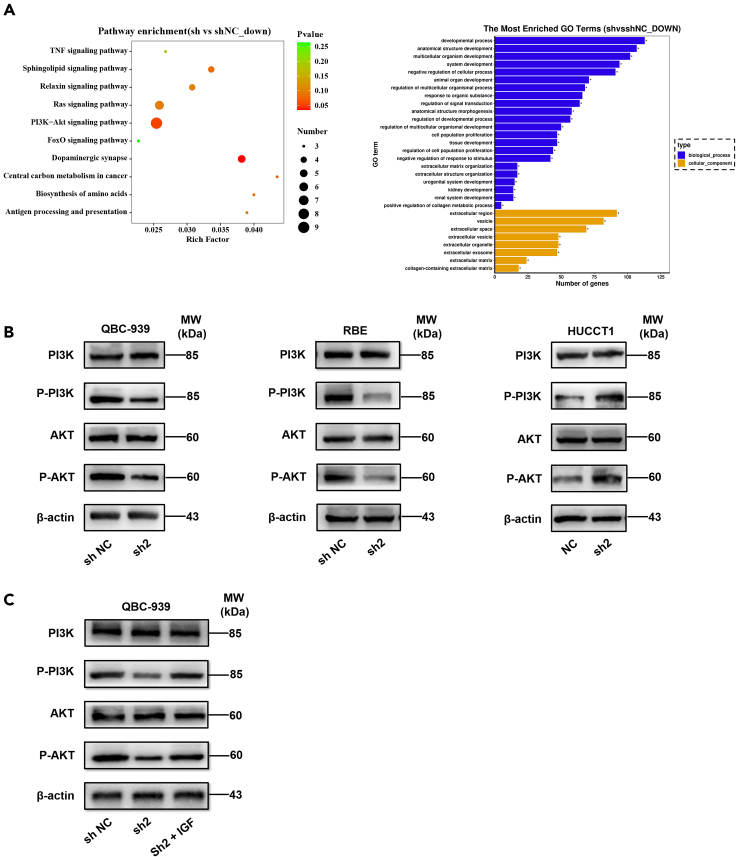
STK39 associated pathways in CCA
To inspect the potential molecular mechanisms of STK39 participating in the progression of CCA, we performed RNA-sequencing using control QBC-939 and STK39-knockdown QBC-939 cells. We listed the top 10 of pathway enrichment and found there was a strong correlation between the PI3K/AKT pathway and the downstream of STK39 (Figure 6A). Furthermore, several studies have discovered that the progression of CCA was closely correlated with the PI3K/AKT pathway (Klungsaeng et al., 2020; Zhang et al., 2018a; Jang et al., 2020). Therefore, we focused on whether STK39 could activate the PI3K/AKT pathway. We then performed western blot and the analysis showed that proteins including P-PI3K (p85) and P-AKT (Ser473), which were related to the PI3K/AKT pathway, were obviously reduced in STK39 stable knockdown cells, while significantly increased in STK39 overexpression cells (Figure 6B). However, no changes were observed in the total levels of PI3K (p85) and AKT. These findings suggest that STK39 could positively regulate the PI3K/AKT signaling pathway. These data suggest that STK39 could regulate the PI3K/AKT signaling pathway positively.
Figure 6.
STK39 mediates oncogenic effects on CCA cells through activating the PI3K/AKT signaling pathway
(A) Pathway enrichment analysis of differentially expressed genes in RNA-sequence data.
(B) Levels of PI3K, AKT, P-PI3K (p85), and P-AKT (Ser473) were examined by western blotting in STK39 stable knockdown or overexpression cells.
(C) Stable knockdown of STK39 in QBC-939 cells by shRNA was treated with or without indicated concentrations of IGF-1. The levels of PI3K, AKT, P-PI3K (p85), and P-AKT (Ser473) were determined by western blotting.
We further conducted a series of in vitro experiments to determine whether the tumor-promoting effect of STK39 on CCA is dependent on the PI3K/AKT pathway. It is worth noting that when we treated the STK39-knockdown QBC-939 cells with human recombinant IGF-1 (Biovision, CA, USA), an activator of the PI3K/AKT pathway, the reduced phosphorylation of PI3K and AKT (Ser473) was significantly restored (Figure 6C). Moreover, the positive effect of STK39 on tumor progression was rescued by the application of IGF-1 (Figures S1A–S1G). Collectively, these results indicate that STK39 may enhance the progression of CCA by activating PI3K/AKT pathway.
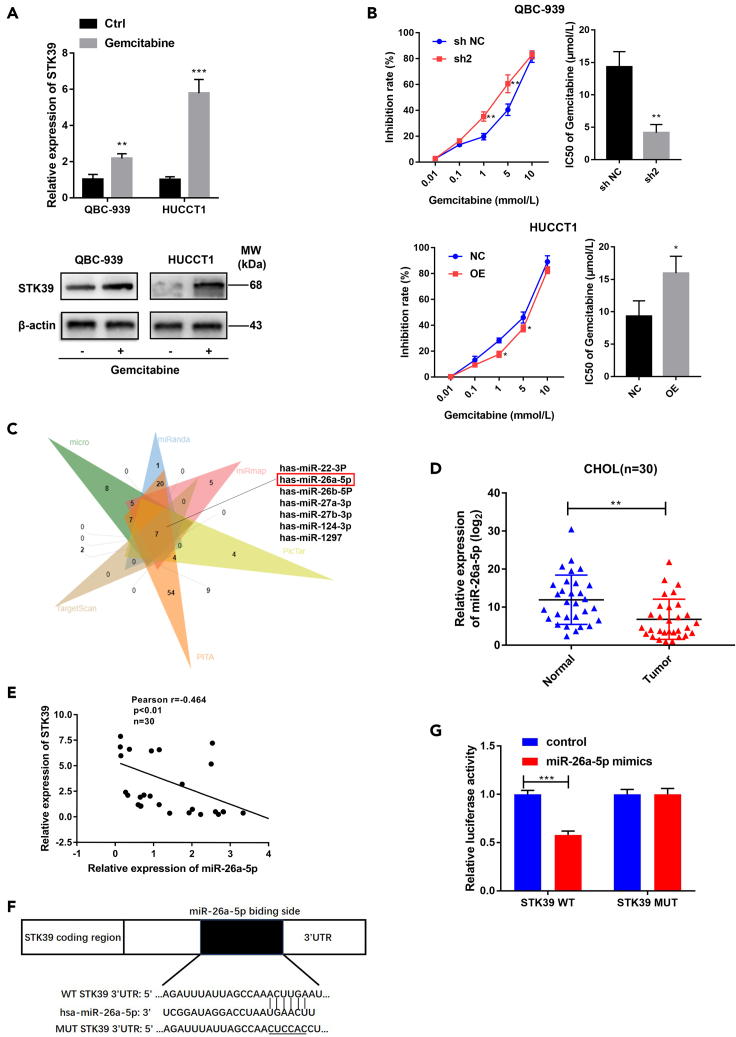
STK39 knockdown increase the CCA sensitivity to gemcitabine
It has been reported that the activation of PI3K/AKT signaling pathway is an important reason for gemcitabine-resistant of pancreatic cancer (Lan et al., 2019; Cui et al., 2021). Therefore, we focused on whether the abnormal expression of STK39 had correlations with gemcitabine resistance. Surprisingly, we found that STK39 was highly expressed in gemcitabine-resistant CCA cells (Figure 7A). Meanwhile, STK39 knockdown increased the gemcitabine inhibition rate of CCA and decreased the half-maximal inhibitory concentration (IC50) value; in contrast, opposite results were showed in STK39-overexpression cells (Figure 7B). These findings suggest that STK39 can reduce CCA sensitivity to gemcitabine, which brings us new ideas in the treatment of patients with gemcitabine resistance.
Figure 7.
The role of STK39 in gemcitabine resistance and the potential regulation of Mir-26a on STK39
(A) Human CCA cell lines were treated with gemcitabine, and the protein expression of STK39 was assessed by qRT-PCR and Western blotting.
(B) The effect of STK39 knockdown/overexpression on gemcitabine resistance.
(C) The prediction of STK39 upstream regulators was performed using TargetScan, PITA, PicTar, miRmap, miRanda, and micro-algorithms.
(D) qRT-PCR was performed to assess the expression level of miR-26a-5p in 30 paired CCA tissues (randomly selected from the previous 62 patients).
(E) Spearman correlation analysis was conducted to confirm the correlations between the STK39 mRNA and miR-26b-5p expression levels in the 30 CCA samples (r = −0.464, p< 0.01).
(F) The predicted miR-26b-5p targeting sequence in the STK39 3ʹUTR (WT STK39 3ʹUTR). The sequences of the STK39 3ʹUTR were mutated (MUT STK39 3ʹUTR).
(G) Dual-luciferase reporter assay of the cells transfected with WT STK39 3ʹUTR or MUT STK39 3ʹUTR together with 40 nM of the miR-26b-5p mimic or negative control oligoribonucleotides. Data were shown as mean ± SEM.
The increased expression of STK39 may be mediated by the dysregulation of miR-26a-5p
The binding of miRNA to the target mRNA usually leads to translational inhibition, and dysregulation of miRNAs has been associated with tumorigenicity and tumor progression as miRNAs can regulate the expression of oncogenes or tumor suppressor genes (Di Leva et al., 2014). To explore the molecular mechanism by which STK39 was regulated, an analysis of multiple bioinformatics databases was conducted to predict the potential miRNAs, which might be the upstream regulators of STK39. By intersecting the predicted results of six computational algorithms (TargetScan, PITA, PicTar, miRmap, miRanda, and micro), seven miRNAs (Figure 7C) were identified to have a putative binding site for STK39. Of these seven miRNAs, miR-26a-5p has been previously reported to be downregulated and negatively correlated with the progression of various malignances (Li et al., 2020; Wang et al., 2020; Damanti et al., 2021), including HCC (Lin et al., 2020). Therefore, we hypothesized that miR-26a-5p was also downregulated in CCA, and it may have the ability to bind to STK39. Thus, we selected miR-26a-5p for further study. To validate the above speculations, we first performed qRT-PCR and found that the expression of miR-26a-5p was downregulated (Figure 7D) and negatively associated with STK39 expression (Figure 7E) in 30 paired tissues randomly selected from the previous 62 patients with CCA. After that, a dual-luciferase reporter assay was applied to verify whether miR-26a-5p could bind to STK39. The results indicated that the transfection of miR-26a-5p mimics led to a significant decrease in luciferase activity of the WT- STK39-3’ UTR. However, almost no changes were observed in the luciferase activity of the MUT STK39-3’ UTR (Figures 7F and 7G). These results show that miR-26a-5p can regulate STK39 directly, and downregulation of miR-26b-5p may be responsible for the increased expression of STK39 in CCA tissues, which in turn promotes the progression of CCA.
Discussion
As one of the most lethal malignant tumors, CCA is relatively rare in clinic. The prognosis of CCA is apparently worse than HCC, with a 5-year survival of less than 10% (Clements et al., 2020). Therefore, there is an urgent need to identify the potential diagnostic markers or molecular pathways involved in the carcinogenesis of CCA.
In recent years, the molecular mechanism of STK39 in the tumorigenicity and tumor progression of several cancer types has been studied, including HCC (Zhang et al., 2021). However, the genetic characteristics and molecular mechanisms of STK39 in CCA are still remains to be elucidated. This study was the first study to reveal that STK39 was significantly upregulated in CCA. Meanwhile, the upregulation of STK39 was significantly correlated with poor outcomes of patient with CCA. By in vitro and in vivo experiments, we discovered that STK39 suppression significantly inhibited the growth, invasion, and metastasis of CCA. In addition, STK39 knockdown led to cell cycle progression and apoptosis, while STK39 overexpression showed the opposite trend, suggesting the oncogenic role of STK39. These observations indicate that STK39 may function as an important tumor promoter in the progression of CCA.
To investigate the mechanisms by which STK39 aggravates the progression of CCA, we performed RNA-seq and analyzed pathway enrichment. The pathway enrichment analysis results showed that the PI3K/AKT signaling pathway had a strong association with the downstream of STK39. The PI3K/AKT pathway is a classical activated signaling pathway in many cancer types (Alzahrani, 2019). Numerous studies have pointed out that the PI3K/AKT pathway activation is critical for carcinogenesis and tumor progression, including CCA (Wang et al., 2018; Duan et al., 2019; Zhu and Wei, 2020). Therefore, we speculate that STK39 might exert its tumor-promoting effect by activating the PI3K/AKT pathway. To validate this speculation, we detected the expression of proteins related to the PI3K/AKT pathway, which showed that knockdown of STK39 caused decreased phosphorylation of PI3K and AKT (Ser473). However, the total protein levels of PI3K and AKT remained unchanged. By using IGF-1, which can activate the PI3K/AKT pathway, the tumor-promoting effect of STK39 showed to be significantly rescued. The above data suggest that STK39 enhances the progression of CCA via activating PI3K/AKT signaling pathway.
Gemcitabine is one of the first-line chemotherapy drugs in the treatment of advanced CCA. However, the application of gemcitabine is limited usually by chemotherapy resistance. It has been reported that the activation of PI3K/AKT signaling pathway is an important reason for chemotherapy resistance. Lan et al. discovered that quercetin could increase GEM-induced cytotoxicity by suppressing the PI3K/AKT/mTOR axis in pancreatic cancer (Lan et al., 2019). Cui et al. suggested that the inhibition of PI3K/AKT/mTOR signaling pathway may be a promising strategy to target the growth and activity of GEM-resistant pancreatic cancer cells by regulating glucose metabolism (Cui et al., 2021). Based on the above findings that STK39 can activate the PI3K/AKT signaling pathway, we speculate that STK39 may have a certain effect on the chemosensitivity of CCA. Our data showed that silencing of STK39 increased the sensitivity of CCA to gemcitabine, while overexpression showed the opposite trend. These findings may provide new insights into the underlying mechanisms of CCA chemoresistance to gemcitabine.
Compared with searching for the downstream target regulated by STK39, it is equally important to explore the molecular mechanisms by which STK39 was upregulated in CCA. It has been found that miRNAs could inhibit tumorigenesis by downregulating the expression of oncogenes. In this study, we employed several computational algorithms to analyze the miRNAs which may bind to STK39 directly, and miR-26a-5p drew our attention. Our data indicate that miR-26a-5p can directly and negatively regulate the expression of STK39, and the increased expression of STK39 may be caused by the low expression of miR-26a-5p in CCA tissues. The design of novel miRNA-based therapeutic targets may provide new insights into CCA treatment.
Taken together, STK39 could be used as a prognostic predicator and a potential therapeutic target for CCA, and it also provides new insight into the diseases caused by the activation of PI3K/AKT pathway and the underlying mechanisms of CCA chemoresistance to gemcitabine.
Limitations of the study
Limitations still existed in the current study for the small sample size, and all the cases were obtained from a single center. Therefore, the findings of this study need to be explored in a large sample size to further support this study. In addition, further investigations are required to elucidate the underlying mechanisms of how STK39 activates the PI3K/AKT pathway and by which STK39 increase the gemcitabine sensitivity in CCA.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-SPAK antibody (rabbit monoclonal) | Abcam | Cat# ab128894, RRID:AB_11156946 |
| Anti-PI3K antibody (rabbit monoclonal) | Abcam | Cat# ab189403, RRID:AB_2651184 |
| Anti- Phospho-PI3K antibody (rabbit monoclonal) | Antibodies-Online | Cat# ABIN461415, RRID:AB_10789447 |
| Anti-AKT antibody (rabbit monoclonal) | Cell Signaling Technology | Cat# 4071, RRID:AB_1031106 |
| Anti-Phospho-AKT antibody (rabbit monoclonal) | Antibodies-Online | Cat# ABIN461276, RRID:AB_10789616 |
| Anti-β-actin antibody (Rabbit monoclonal) | Cell Signaling Technology | Cat# 58169, RRID:AB_2750839 |
| Secondary antibody (rabbit polyclonal) | Abcam | Cat# ab7090, RRID:AB_955417 |
| Anti-Ki67 antibody(Mouse monoclonal) | Abcam | Cat# ab6526, RRID:AB_305543 |
| Biological samples | ||
| Human CCA tissues and adjacent normal tissues (62 paired) | CCA cases who underwent a primary attempt of radical resection at NJMU | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| IGF-1 | Biovision | N/A |
| Gemcitabine | Eli Lilly Pharmaceuticals | N/A |
| Critical commercial assays | ||
| Cell Counting Kit-8 | DoJindo | Cat# CK0413 |
| RNA Quick Purification Kit | YiShanbio | RN001 |
| RIPA kit | Beyotime | P0013B |
| ECL Plus | Merk millipore | WBKLS0100 |
| Cell-Light EdU Apollo567 In Vitro Kit | Ribobio | C10310-1 |
| Matrigel | BD | 354277 |
| Cell Cycle Analysis Kit | Beyotime | C1052 |
| Lipofectamine 3000 | Invitrogen | L3000-015 |
| Annexin V-PE/7-AADApoptosis Detection Kit | Vazyme | A213-02 |
| Deposited data | ||
| Bioinformatic datasets | GEO | GSE26566 |
| RNA-seq on STK39-knockdown and control QBC-939 cells | SPR | PRJNA762569 |
| Experimental models: Cell lines | ||
| HUCCT1 | Chinese Academy of Sciences Cell Bank | RRID: CVCL_0324 |
| QBC-939 | Chinese Academy of Sciences Cell Bank | RRID: CVCL_6942 |
| RBE | Chinese Academy of Sciences Cell Bank | RRID: CVCL_4896 |
| HCCC9810 | Chinese Academy of Sciences Cell Bank | RRID: CVCL_6908 |
| HiBEC | Chinese Academy of Sciences Cell Bank | N/A |
| QBC-939 STK39 knockdown cell line | This paper | N/A |
| HUCCT1 STK39 overexpressing cell line | This paper | N/A |
| Gemcitabine-resistant QBC-939 cell line | This paper | N/A |
| Gemcitabine-resistant HUCCT1 cell line | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: BALB/c nude mice (6-week-old, female) | Model Animal Research Center of NJMU | N/A |
| Oligonucleotides | ||
| STK39 sequencing: Forward: 5′-TGCCAGACCAGTATGGATG AA-3′ Reverse: 5′-GGTGTAATAGGTCACTACG TTGG-3′ |
Realgene, Nanjing, China | N/A |
| β-actin sequencing: Forward: 5′-ATTGCCGACAGGATGCAGAA-3′ Reverse: 5′-GCTGATCCACATCTGCTGG AA-3′ |
Realgene, Nanjing, China | N/A |
| miR-26a-5p sequencing: Forward: 5′-TGGTTCGTGGGTTCAAGTA ATCGATGGC-3′ Reverse: 5′-GCAGGGTCCGAGTATTCG-3′ |
Realgene, Nanjing, China | N/A |
| shSTK39-1: 5′-GTCAGATTCACAGGGATTT-3′ | Shanghai Genechem Co., Ltd | N/A |
| shSTK39-2: 5′-AGGCAATAATAGCAACAAT-3′ | Shanghai Genechem Co., Ltd | N/A |
| STK39 shRNA 3: 5′-ACGGCAAGTCCTTTAG AAA-3′ |
Shanghai Genechem Co., Ltd | N/A |
| sh NC: TTCTCCGAACGTGTCACGT | Shanghai Genechem Co., Ltd | N/A |
| Recombinant DNA | ||
| Ubi-MCS-3FLAG-SV40-puromycin STK39 | Shanghai Genechem Co., Ltd | N/A |
| Software and algorithms | ||
| GraphPad Prism 5.0 | GraphPad | https://graphpad.com |
| ImageJ | NIH | http://imagej.nih.gov/ij/ |
| FlowJo | Treestar Inc. | https://www.flowjo.com |
| Other | ||
| Cytoflex flow cytometer | Beckman | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be contacted directly to and will be fulfilled by the Lead Contact, Xuehao Wang (wangxh@njmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Patients and tissue samples
A total of 62 paired CCA tissues and adjacent normal tissues were collected from CCA cases (35 males and 27 females), aged between 33 and 74 years (average, 52.8 years), who underwent a primary attempt of radical resection between July 2016 and June 2019 at the First Affiliated Hospital of Nanjing Medical University. Patients included in this study were not subject to preoperative chemotherapy or radiation therapy. The pathological characteristics of CCA were confirmed by two pathologists. This study was approved by the Ethical Committee of the First Affiliated Hospital of Nanjing Medical University (2019-SRFA-276).
Cell lines and cell culture
HUCCT1 (RRID: CVCL_0324), QBC-939(RRID: CVCL_6942), RBE (RRID: CVCL_4896), HCCC9810 (RRID: CVCL_6908), and HiBEC cell lines were obtained from purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). The cell lines were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. The cells were maintained at 37°C in an incubator supplemented with 5% CO2.
Cell treatment and generation of gemcitabine-resistant cells
The expression of STK39 was stably knocked down/overexpressed by transfection with lentiviral vectors specific to human STK39 (Table S1) at an appropriate multiplicity of infection (MOI). Stably transfected cells were selected by puromycin according to protocols. Two cell lines (QBC-939 and HUCCT1) were chosen to establish gemcitabine-resistant CCA cells. Briefly, cells were exposed to graded drug concentrations (From 10μmol/L to 15mmol/L) step by step to induce gemcitabine resistance. After 12 months of induction, two gemcitabine-resistant CCA cell lines were generated. Gemcitabine was purchased from Eli Lilly Pharmaceuticals (Indianapolis, IN, USA).
In vivo animal assay
The animal study was approved by the Nanjing Medical University (NJMU) Institutional Animal Care and Use Committee. BALB/c nude mice (6-week-old, female) were obtained from the Model Animal Research Center of NJMU. The mice were randomly divided into four groups for tumor xenograft experiments: sh-STK39 group, sh-NC-STK39 group, OE-STK39 group, and OE-NC-STK39 group (six mice per group). Mice were subcutaneously injected with 1×106 HUCCT1 cells stably expressing OE-STK39/OE-NC-STK39 or QBC-939 cells transfected with sh-STK39/sh-NC-STK39 into the left inguinal region. Tumor size was recorded every four days using calipers, and the mice were euthanized after 32 days. The tumor volume was analyzed using the following formula: volume = 1/2 × width2 × length. Transfected cells were injected into the tail vein of the mice to assess the effect of STK39 on lung metastasis. After three weeks, the bioluminescent images of mice were captured. The mice were euthanized, and the lungs were removed for hematoxylin & eosin (HE) staining.
Method details
Immunohistochemistry (IHC)
CCA tissues and paired adjacent normal tissues were made into Paraffin-embedded tissue and then cut into 5-μm-thick slices. Then the slices were placed in 3% H2O2 solution for 20 min. After that, the slices were blocked with 4% BSA for 25min, and incubated with diluted primary antibody at 4°C overnight. After incubated with the secondary antibody of the related species for 90 min, diaminobenzidine solution was added to each slice, and then hematoxylin was used to stained with slices. Finally, the slices were evaluated in a blinded manner.
RNA extraction and quantitative real-time PCR(qRT-PCR)
Total RNA was isolated using RNA Quick Purification kit (YiShanbio, Shanghai, China) according to the manufacturer's protocol. Reverse transcription was conducted using PrimeScript RT Reagent kit. β-actin was regarded as an endogenous control gene. The sequence-specific primers are shown in Table S2. The expression of STK39 mRNA was calculated using the 2−ΔΔCt method.
Protein extraction and western blotting
RIPA kit (Beyotime, Shanghai, China) was used to extract total protein. Proteins were separated by SDS-PAGE and subsequently transferred onto PVDF membranes. The membranes were incubated with the corresponding antibodies at 4°C overnight. β-actin was used as a loading control. ECL Plus (EMD Millipore, Billarica, MA, USA) was used to detect ECL signals.
Antibodies
The following antibodies were used for immunostaining: anti-SPAK antibody (Abcam, ab128894) and anti-Ki67 antibody (Abcam, ab6526). The following antibodies were used for western blotting: anti-SPAK antibody (Abcam, ab128894), anti-PI3K antibody (Abcam, ab189403), anti-P-PI3K antibody (Antibodies-Online, ABIN461415), anti-AKT antibody (Cell Signaling Technology, 4071), anti-P-AKT antibody (Antibodies-Online, ABIN461276), anti-β-actin antibody (Abcam, ab7090), and secondary antibody (Abcam, ab6526)
CCK-8 cell proliferation assay
Cell Counting Kit-8 were purchased from MCE (Shanghai, China). Cells were seeded in 96-well plates at a density of 1×103 cells/well and cultured in complete culture medium (100 ul/well) over night. At 0, 24, 48, 72, 96 and 120 h, 10 μl of CCK-8 solution was added to each well. After 2 h of incubation, the absorbance value was detected with a spectrophotometer (Thermo Scientific, Pittsburgh, PA, USA) at 450 nm.
Colony formation assay
Approximately 800 cells were plated into 6-well plates containing a complete culture medium (3 ml/well). After ten days, each well was first washed using phosphate buffer saline (PBS) and then fixed with ethyl alcohol for 1 min. After that, cells were stained with 1% crystal violet for 20 min. Colonies were counted visually.
5-ethynyl-2′-deoxyuridine (EdU) proliferation assay
The EdU assay was performed by using an EdU incorporation assay kit (Ribobio, Shanghai, China) to evaluate the cell growth ability. Cells (5×104/well) were seeded into 24-well plates and cultured for 12h, then incubated with EdU for 2h, and the rest steps are as previously described (Zhang et al., 2018b).
Transwell assay
For migration assay, approximately 5×104 cells were suspended in a 300ul FBS-free medium and then sown on the upper chamber. Further, 600ul medium with 10% FBS was placed in the lower chamber. After a complete incubation of about 36 h, noninvasive cells were removed by cotton-tipped swabs. The migrated cells were stabilized with 4% paraformaldehyde and stained with 0.1% crystal violet for 20 min. Five fields were randomly imaged under a bright-field microscope, and the number of cells was counted. Invasion assay was performed as migration assay, except that the upper chamber was coated with Matrigel (BD Bioscience Pharmingen).
Wound healing assay
Cells of suitable density were sown in 6-well plates and cultured with an FBS-free medium to grow a monolayer for 24h. A 200-μL pipette tip was then used to scratch a wound in each well. Images of the wound were captured at 0, 24, and 48 h.
Cell cycle analysis
Cells were harvested at 48 h and fixed overnight with 70% ethanol at 4°C. After being suspended in PBS, cells were stained with 500 μl PI staining solution (Beyotime, Shanghai, China) for 30 min. Then, cells were analyzed with a flow cytometer (Beckman, China).
Cell apoptosis analysis
Cell apoptosis was detected using a Cytoflex flow cytometer (Beckman, China). Briefly, Annexin V-FITC kit (Lianke, Hangzhou, China) was used to measure cell apoptosis according to the manufacture’s protocol. Data were analyzed using ModFit software.
RNA-sequence (RNA-seq)
The global gene expression profiles of STK39-knockdown and control QBC-939 cells were examined by RNA-seq in Tiangen Biotech (Beijing, China). Enrichr tool was used to analyze the biological process. Data were deposited at NCBI Squence Read Archive (SPR) database, and the accession number is PRJNA762569.
Prediction and validation of STK39 upstream regulators
The analysis of STK39 predicted upstream regulators was performed using TargetScan, PITA, PicTar, miRmap, miRanda, and micro algorithms. Luciferase Reporter Assay was performed to detect whether STK39 was a direct target gene of miR-26a-5p. Wild-type (WT) or mutant (MUT) STK39 3ʹUTR Dual-luciferase report plasmids were constructed by RiboBio. The plasmids were co-transfected with miR-26a-5p mimic or negative control using Lipofectamine 3000 (Invitrogen). After 24 h of transfection, Dual-Luciferase Reporter Assay System (Promega, USA) was used to determine the luciferase activity.
Quantification and statistical analysis
Data are shown as the mean ± standard deviation (SD). Each experiment was repeated triple times. Statistical analysis was performed using GraphPad Prism software (version 5.0; San Diego, CA, USA). The chi-square test was adopted for two groups comparison. Kaplan-Meier analysis was used for survival analysis. A Spearman correlation test was used to analyze the relationship between miR-26a-5p and STK39. P < 0.05 was considered statistically significant.
Additional resources
This study has not generated or contributed to a new website/forum and is not part of a clinical trial.
Acknowledgments
We are grateful to our colleagues for their valuable suggestions and technical assistance for this study. This work was supported by research grants from the Major Program of the National Natural Science Foundation of China (81530048 and 31930020); the National Natural Science Foundation of China (81972768 and 81870488); the Southeast University and Nanjing Medical University Cooperative Research Project, China (2019DN0003).
Author contributions
Conception and design, X.H.W., J.D.W., and W.W.T.; development of methodology: X.P.H., Y.Z., Y.W.L., D.W.R., G.Q.S., and G.S.S.; collection and acquisition of data: X.P.H.; analysis of data: X.P.H., D.W.R., G.Q.S., and G.S.S.; writing, review, and/or revision of the manuscript: X.P.H., Y.Z., X.H.W., J.D.W., and W.W.T. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103223.
Contributor Information
Weiwei Tang, Email: 1243773473twww@sina.com.
Jindao Wu, Email: wujindao@njmu.edu.cn.
Xuehao Wang, Email: wangxh@njmu.edu.cn.
Supplemental information
Data and code availability
The bioinformatic datasets used in this manuscript were obtained from Gene Expression Omnibus (GEO) and the accession number is GSE26566.
All raw reads of RNASeq data reported in this paper were stored in NCBI sequence Read Archive (SRA) database, and the accession number is PRJNA762569.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alzahrani A.S. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin. Cancer Biol. 2019;59:125–132. doi: 10.1016/j.semcancer.2019.07.009. [DOI] [PubMed] [Google Scholar]
- Bertuccio P., Malvezzi M., Carioli G., Hashim D., Boffetta P., El-Serag H.B., La Vecchia C., Negri E. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J. Hepatol. 2019;71:104–114. doi: 10.1016/j.jhep.2019.03.013. [DOI] [PubMed] [Google Scholar]
- Bridgewater J., Galle P.R., Khan S.A., Llovet J.M., Park J.W., Patel T., Pawlik T.M., Gores G.J. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J. Hepatol. 2014;60:1268–1289. doi: 10.1016/j.jhep.2014.01.021. [DOI] [PubMed] [Google Scholar]
- Clements O., Eliahoo J., Kim J.U., Taylor-Robinson S.D., Khan S.A. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: a systematic review and meta-analysis. J. Hepatol. 2020;72:95–103. doi: 10.1016/j.jhep.2019.09.007. [DOI] [PubMed] [Google Scholar]
- Cui J., Guo Y., Wu H., Xiong J., Peng T. Everolimus regulates the activity of gemcitabine-resistant pancreatic cancer cells by targeting the Warburg effect via PI3K/AKT/mTOR signaling. Mol. Med. 2021;27:38. doi: 10.1186/s10020-021-00300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damanti C.C., Gaffo E., Lovisa F., Garbin A., Di Battista P., Gallingani I., Tosato A., Pillon M., Carraro E., Mascarin M. MiR-26a-5p as a reference to normalize MicroRNA qRT-PCR levels in plasma exosomes of pediatric hematological malignancies. Cells. 2021;10:101. doi: 10.3390/cells10010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H.X., Li B.W., Zhuang X., Wang L.T., Cao Q., Tan L.H., Qu G.F., Xiao S. TCF21 inhibits tumor-associated angiogenesis and suppresses the growth of cholangiocarcinoma by targeting PI3K/Akt and ERK signaling. Am. J. Physiol. Gastrointest.Liver Physiol. 2019;316:G763–G773. doi: 10.1152/ajpgi.00264.2018. [DOI] [PubMed] [Google Scholar]
- Forgetta V., Manousaki D., Istomine R., Ross S., Tessier M.C., Marchand L., Li M., Qu H.Q., Bradfield J.P., Grant S.F.A. Rare genetic variants of large effect influence risk of type 1 diabetes. Diabetes. 2020;69:784–795. doi: 10.2337/db19-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon K.B., England R., Delpire E. Characterization of SPAK and OSR1, regulatory kinases of the Na-K-2Cl cotransporter. Mol.Cell Biol. 2006;26:689–698. doi: 10.1128/MCB.26.2.689-698.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Zhou Y., Cao Y., Tao J., Zhou Z.H., Hang D.H. STK39, overexpressed in osteosarcoma, regulates osteosarcoma cell invasion and proliferation. Oncol.Lett. 2017;14:4599–4604. doi: 10.3892/ol.2017.6728. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jang D.K., Lee Y.G., Chan Chae Y., Lee J.K., Paik W.H., Lee S.H., Kim Y.T., Ryu J.K. GDC-0980 (apitolisib) treatment with gemcitabine and/or cisplatin synergistically reduces cholangiocarcinoma cell growth by suppressing the PI3K/Akt/mTOR pathway. Biochem.Biophys. Res. Commun. 2020;529:1242–1248. doi: 10.1016/j.bbrc.2020.06.011. [DOI] [PubMed] [Google Scholar]
- Josiah S.S., Meor Azlan N.F., Zhang J. Targeting the WNK-SPAK/OSR1 pathway and cation-chloride cotransporters for the therapy of stroke. Int.J.Mol.Sci. 2021;22:1232. doi: 10.3390/ijms22031232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S.A., Tavolari S., Brandi G. Cholangiocarcinoma: epidemiology and risk factors. Liver Int. 2019;39:19–31. doi: 10.1111/liv.14095. [DOI] [PubMed] [Google Scholar]
- Klungsaeng S., Kukongviriyapan V., Prawan A., Kongpetch S., Senggunprai L. Targeted modulation of FAK/PI3K/PDK1/AKT and FAK/p53 pathways by cucurbitacin b for the antiproliferation effect against human cholangiocarcinoma cells. Am. J.Chin. Med. 2020;48:1475–1489. doi: 10.1142/S0192415X2050072X. [DOI] [PubMed] [Google Scholar]
- Lan C.Y., Chen S.Y., Kuo C.W., Lu C.C., Yen G.C. Quercetin facilitates cell death and chemosensitivity through RAGE/PI3K/AKT/mTOR axis in human pancreatic cancer cells. J. Food Drug Anal. 2019;27:887–896. doi: 10.1016/j.jfda.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G., Garofalo M., Croce C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhu W., Xiong L., Yu X., Chen X., Lin Q. Role of high expression levels of STK39 in the growth, migration and invasion of non-small cell type lung cancer cells. Oncotarget. 2016;7:61366–61377. doi: 10.18632/oncotarget.11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang P., Wu L.L., Yan J., Pang X.Y., Liu S.J. miR-26a-5pinhibit gastric cancer cell proliferation and invasion through mediated Wnt5a. Onco Targets Ther. 2020;13:2537–2550. doi: 10.2147/OTT.S241199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Jian Z., Jin H., Wei X., Zou X., Guan R., Huang J. Long non-coding RNA DLGAP1-AS1 facilitates tumorigenesis and epithelial-mesenchymal transition in hepatocellular carcinoma via the feedback loop of miR-26a/b-5p/IL-6/JAK2/STAT3 and Wnt/beta-catenin pathway. Cell Death Dis. 2020;11:34. doi: 10.1038/s41419-019-2188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick J.A., Mutig K., Nelson J.H., Saritas T., Hoorn E.J., Yang C.L., Rogers S., Curry J., Delpire E., Bachmann S. A SPAK isoform switch modulates renal salt transport and blood pressure. Cell Metab. 2011;14:352–364. doi: 10.1016/j.cmet.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramoz N., Cai G., Reichert J.G., Silverman J.M., Buxbaum J.D. An analysis of candidate autism loci on chromosome 2q24-q33: evidence for association to the STK39 gene. Am. J. Med. Genet.B Neuropsychiatr.Genet. 2008;147B:1152–1158. doi: 10.1002/ajmg.b.30739. [DOI] [PubMed] [Google Scholar]
- Richardson C., Sakamoto K., de los Heros P., Deak M., Campbell D.G., Prescott A.R., Alessi D.R. Regulation of the NKCC2 ion cotransporter by SPAK-OSR1-dependent and -independent pathways. J.Cell Sci. 2011;124:789–800. doi: 10.1242/jcs.077230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues P.M., Olaizola P., Paiva N.A., Olaizola I., Agirre-Lizaso A., Landa A., Bujanda L., Perugorria M.J., Banales J.M. Pathogenesis of cholangiocarcinoma. Annu. Rev. Pathol. 2021;16:433–463. doi: 10.1146/annurev-pathol-030220-020455. [DOI] [PubMed] [Google Scholar]
- Shaib Y.H., Davila J.A., McGlynn K., El-Serag H.B. Rising incidence of intrahepatic cholangiocarcinoma in the United States: a true increase? J. Hepatol. 2004;40:472–477. doi: 10.1016/j.jhep.2003.11.030. [DOI] [PubMed] [Google Scholar]
- Shin H.R., Oh J.K., Masuyer E., Curado M.P., Bouvard V., Fang Y.Y., Wiangnon S., Sripa B., Hong S.T. Epidemiology of cholangiocarcinoma: an update focusing on risk factors. Cancer Sci. 2010;101:579–585. doi: 10.1111/j.1349-7006.2009.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi T., Ushiro H., Kosaka T., Kayahara T., Nakano K. Proline- and alanine-rich Ste20-related kinase associates with F-actin and translocates from the cytosol to cytoskeleton upon cellular stresses. J. Biol. Chem. 2000;275:9157–9162. doi: 10.1074/jbc.275.13.9157. [DOI] [PubMed] [Google Scholar]
- Tyson G.L., El-Serag H.B. Risk factors for cholangiocarcinoma. Hepatology. 2011;54:173–184. doi: 10.1002/hep.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle J.W., Borbath I., Khan S.A., Huguet F., Gruenberger T., Arnold D., Committee E.G. Biliary cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2016;27:v28–v37. doi: 10.1093/annonc/mdw324. [DOI] [PubMed] [Google Scholar]
- Wang Y., O'Connell J.R., McArdle P.F., Wade J.B., Dorff S.E., Shah S.J., Shi X., Pan L., Rampersaud E., Shen H. From the cover: whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc. Natl. Acad. Sci. U. S. A. 2009;106:226–231. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liang Y., Yang G., Lan Y., Han J., Wang J., Yin D., Song R., Zheng T., Zhang S. Tetraspanin 1 promotes epithelial-to-mesenchymal transition and metastasis of cholangiocarcinoma via PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018;37:300. doi: 10.1186/s13046-018-0969-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Liu T., Xue W., Fang Y., Chen X., Xu L., Zhang L., Guan K., Pan J., Zheng L. ARNTL2 promotes pancreatic ductal adenocarcinoma progression through TGF/BETA pathway and is regulated by miR-26a-5p. Cell Death Dis. 2020;11:692. doi: 10.1038/s41419-020-02839-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Mao W., Rodriguez-Aguayo C., Mangala L.S., Bartholomeusz G., Iles L.R., Jennings N.B., Ahmed A.A., Sood A.K., Lopez-Berestein G. Paclitaxel sensitivity of ovarian cancer can be enhanced by knocking down pairs of kinases that regulate MAP4 phosphorylation and microtubule stability. Clin.Cancer Res. 2018;24:5072–5084. doi: 10.1158/1078-0432.CCR-18-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ji G., Han S., Shao Z., Lu Z., Huo L., Zhang J., Yang R., Feng Q., Shen H. Tip60 suppresses cholangiocarcinoma proliferation and metastasis via PI3k-AKT. Cell Physiol. Biochem. 2018;50:612–628. doi: 10.1159/000494183. [DOI] [PubMed] [Google Scholar]
- Zhang Y., You W., Zhou H., Chen Z., Han G., Zuo X., Zhang L., Wu J., Wang X. Downregulated miR-621 promotes cell proliferation via targeting CAPRIN1 in hepatocellular carcinoma. Am. J. Cancer Res. 2018;8:2116–2129. [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang X., Fang D., Xu P., Mo X., Hu C., Abdelatty A., Wang M., Xu H., Sun Q. STK39 is a novel kinase contributing to the progression of hepatocellular carcinoma by the PLK1/ERK signaling pathway. Theranostics. 2021;11:2108–2122. doi: 10.7150/thno.48112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Wei Y. Antitumor activity of celastrol by inhibition of proliferation, invasion, and migration in cholangiocarcinoma via PTEN/PI3K/Akt pathway. Cancer Med. 2020;9:783–796. doi: 10.1002/cam4.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bioinformatic datasets used in this manuscript were obtained from Gene Expression Omnibus (GEO) and the accession number is GSE26566.
All raw reads of RNASeq data reported in this paper were stored in NCBI sequence Read Archive (SRA) database, and the accession number is PRJNA762569.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.