Summary
Immune activation has been shown to play a critical role in the development of schizophrenia; however its underlying mechanism remains unknown. Our report demonstrates a high-quality protein interaction network for schizophrenia (SCZ Network), constructed using our “neighborhood walk” approach in combination with “random walk with restart”. The spatiotemporal expression pattern of the genes in this disease network revealed two developmental stages sensitive to perturbation by immune activation: mid-to late gestation, and adolescence. Furthermore, we induced immune activation at these stages in mice, carried out transcriptome sequencing on the mouse brains, and illustrated clear potential molecular pathways and key regulators correlating maternal immune activation during gestation and an increased risk for schizophrenia after a second immune activation at puberty. This work provides not only valuable resources for the study on molecular mechanisms underlying schizophrenia, but also a systematic strategy for the discovery of molecular pathways of complex mental disorders.
Subject areas: Clinical neuroscience, Immunology, Bioinformatics, Biocomputational method, Systems biology
Graphical abstract

Highlights
-
•
A high-quality molecular network for schizophrenia (SCZ Network)
-
•
A landscape of molecular pathways linking immune activation and schizophrenia
-
•
The spatiotemporal network dynamics revealing stages susceptible to immune activation
-
•
Identification of the molecular pathways and regulators in the immune-activated brain
Clinical neuroscience; Immunology; Bioinformatics; Biocomputational method; Systems biology
Introduction
Schizophrenia (SCZ) has a lifetime incidence rate around 1%, and is associated with patient disability and severe socioeconomic burdens for families and our entire society (Knapp et al., 2004; Saha et al., 2007). It is a complex polygenic disorder with a wide range of psychiatric symptoms that are generally categorized as positive, negative, or cognitive (International Advisory Group for the Revision of ICD-10 Mental and Behavioural Disorders, 2011; Janca et al., 1993). Schizophrenia has a heritability of about 80% (Cannon et al., 1998; Farmer et al., 1987) and its genetic risk factors show very high heterogeneity. Studies of functional genetics have identified several schizophrenia susceptibility genes, such as DISC1 (Blackwood et al., 2001), PDE4B (Millar et al., 2005), NPAS3 (Yu et al., 2014), and NRG1 (Harrison and Law, 2006). Many SCZ-associated loci have been identified through genome-wide association studies (GWASs) (O'Donovan et al., 2008; Polednak, 1991; Ripke et al., 2013; Schizophrenia Psychiatric Genome-Wide Association Study, 2011). Recently, two large cohort GWASs have identified robust and replicable common alleles with significant associations to schizophrenia (Pardinas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics, 2014). The major histocompatibility complex (MHC) has been found to be associated with schizophrenia by several groups (International Schizophrenia Consortium et al., 2009; Pardinas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics, 2014; Shi et al., 2009; Stefansson et al., 2009; Walters et al., 2013). Some hotspots of genomic deletions or duplications have been identified at 1q21.1, 15q11.2 and 15q13.3, 22q11.2, and 16p11.2 (Rees et al., 2014; Szatkiewicz et al., 2014). In addition, whole exome sequencing discovered de novo mutations, which are notably enriched in glutamatergic postsynaptic proteins comprising of activity-regulated cytoskeleton-associated protein (ARC) and N-methyl-D-aspartate receptor (NMDAR) complexes (Fromer et al., 2014; Xu et al., 2011), suggesting that a dynamic synapse regulation may play a role in the disease development. Recent transcriptomic studies on the postmortem brain samples of the patients have identified SCZ-dysregulated genes in the brain, which are enriched for synaptic genes and immune genes (Bowen et al., 2019; Fromer et al., 2016).
A great challenge of studying the molecular mechanisms underlying schizophrenia and of identifying drug targets is that some loci are common variants, and each may only play a small role in the pathogenesis. Rare variants are found to significantly increase the disease burden (Fromer et al., 2014; Singh et al., 2017; Xu et al., 2011), and they are suggested to have bigger effects and thus higher penetrance (Singh et al., 2017). Another challenge is that “gene-environment interaction” and “environment-environment interaction” may play a critical role in schizophrenia (Khan and Powell, 2018). The “Two-Hit Hypothesis” of schizophrenia, proposed in 1999, suggests that schizophrenia needs two events (“hits”) for its development: the first event (i.e., basic requirement) represents genetic factors such as germline mutations; and the second consists of environmental factors such as viral infections and social stressors (Bayer et al., 1999; Feigenson et al., 2014). Later on, it has been suggested that in-utero or early life factors, such as maternal infection and stress, may also serve as “the first hit” (Estes and McAllister, 2016).
In any case, accumulating evidence points to the important role of abnormal immune activation in the etiology of schizophrenia. Proinflammatory cytokine levels have been consistently reported to be increased in the serum (Goldsmith et al., 2016; Miller et al., 2011) and also in the prefrontal cortex (Fillman et al., 2013; van Kesteren et al., 2017) of schizophrenia patients. Prenatal exposure to maternal immune activation increases the risk of schizophrenia later in adult life (Brown, 2012; Brown and Derkits, 2010; Estes and McAllister, 2016); adolescent stress is thought to be another driving factor (Gomes and Grace, 2017). Moreover, as previously mentioned, genome-wide association studies have identified loci covering genes involved in immune function, such as the major histocompatibility complex (MHC) (International Schizophrenia Consortium et al., 2009; Pardinas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics, 2014; Shi et al., 2009; Stefansson et al., 2009; Walters et al., 2013). Transcriptome sequencing of postmortem brain samples found transcriptional alteration associated with widespread increasing inflammation in the brain of schizophrenia patients (Lanz et al., 2019). A recent study found increased microglia-mediated synaptic pruning in SCZ patient-derived neural culture (Sellgren et al., 2019). All these lines of evidence point to the critical role of abnormal immune activation in schizophrenia. However, studying the molecular mechanism that links abnormal immune activation to schizophrenia remains a major challenge.
Despite the large number of genes involved in SCZ, patients display similar core phenotypic features, suggesting that SCZ risk genes may share a common molecular network. Therefore, a high-quality molecular network underlying this disease is urgently needed for illustrating the molecular mechanisms on how genetic and environmental factors may lead to the development of schizophrenia. Proteome-scale studies of protein interactions have shed light on the etiology of diseases and putative therapeutic targets (Cusick et al., 2005; Gonzalez and Kann, 2012). High-throughput mapping of protein interactions (Havugimana et al., 2012; Hein et al., 2015; Rolland et al., 2014; Yang et al., 2016) and literature curation of small-scale studies (Cusick et al., 2009) have accumulated huge amounts of protein-protein interaction data. Recently, the interactions from different sources have been integrated into a comprehensive scored human interactome network containing about 600,000 interactions (Li et al., 2017). This vast amount of protein interaction data provides a potential molecular network for illustrating pathways underlying complex disorders such as schizophrenia. “Random walk” was first developed to study the network topology (Lovász, 1993), and later “random walk with restart” (RWR) was introduced for Internet search engines (Page, 1998). More recently, RWR has been utilized to search for novel disease candidate genes, using known disease genes as seeds in a molecular network (Kohler et al., 2008; Valdeolivas et al., 2019).
Here, we have developed an approach to identifying disease-associated neighborhoods across the human interactome network, which we termed as “neighborhood walk” (NW). Utilizing the NW approach in combination with RWR, we have constructed a high-quality protein interaction network for schizophrenia (SCZ Network). This network is enriched in signaling pathways regulating immune response as well as synapse formation and modulation. By integrating the spatiotemporal gene expression data of human brain with the SCZ Network, we found that the immune response genes in this network are enriched at two developmental stages (mid- to late fetal stage and puberty stage). Therefore, it is likely that at these two developmental stages, the SCZ Network is most sensitive to the perturbation of immune activation. To further verify that the potential schizophrenia signaling pathways are sensitive to the perturbation of immune activation during the two critical stages, we induced immune activation at these two stages and carried out transcriptome sequencing on immune-activated mouse brains. By integrating the transcriptome data from immune-activated mouse brains with the SCZ Network, we have identified potential molecular pathways and key regulators, which may illustrate a detailed view of the mechanisms on how maternal immune activation during fetal development may increase the risk for schizophrenia with a second immune activation at puberty.
Results
Constructing a protein-protein interaction network for schizophrenia using the “neighborhood walk” and “random walk with restart” methods
To collect high-confidence schizophrenia candidate genes, we used a recently updated comprehensive database for schizophrenia genetic research (SZDB) (Wu et al., 2017, 2020). The SZDB (version 2.0) collects a total of 7,903 schizophrenia candidate genes from literature, which were identified by 7 different methods in 87 studies including newly published genome scale studies (Wu et al., 2017, 2020). We re-evaluated each source data and selected a core set of 1,836 high-confidence SCZ candidate genes, including 223 genes from 4 linkage studies, 529 genes from 2 large-cohort genome-wide association studies, 153 genes from 77 copy number variation studies, 543 genes from 1 transcriptomic study on postmortem brains, 63 genes from 1 imputation study, 307 genes from 3 studies on differential methylation and 165 genes from 14 studies on exome sequencing (Figures S1A and S1B). Of the 1,836 high-confidence SCZ candidate genes, 1,720 are protein-coding genes, which were used as seed genes to expand SCZ candidate genes. Of these 1,720 seed genes, 55.4% (953 of 1,720) are reported in two or more studies (Figure 1A), while 92.3% (1,588 of 1,720) are found by only one of the seven methods (Figure 1B), suggesting that each method may have its bias. To test the functional coherence of the selected seed genes, we mapped these 1,720 seeds to human interactome to measure their interconnectivity. The same number of randomly chosen coding genes from 7,903 candidate genes was used as control. Compared with the candidate genes in the source data, selected seed genes have a significantly larger main interconnected network (empirical p = 0.001, Figure 1C), suggesting that these seed genes are more functionally related.
Figure 1.
Construction of SCZ Network from human interactome using Neighborhood walk (NW) and Random walk with Restart (RWR) methods (See also Figure S1)
(A) The distribution of the number of studies (PubMed IDs) reporting the SCZ association of each of the core set of 1,720 high-confidence SCZ candidate genes: 767 genes are supported by only one study, 708 genes by two studies, and 245 genes by at least 3 studies.
(B) Distribution of the methods for identifying the SCZ association of each of the core set of 1,720 high-confidence SCZ candidate genes: 1,588 genes supported by one method, 119 genes supported by two methods and 13 genes supported by at least 3 methods. The methods include linkage analysis, genome-wide association studies (GWAS), copy number variations (CNV), differentially expressed genes (DEG), imputation analysis, differentially methylated genes (DMG) and whole exome sequencing (WES).
(C) The interconnectivity of selected core set of SCZ candidate genes (seeds). The core set of 1,720 high-confidence SCZ candidate genes were mapped to human interactome and an interconnected network of 1,298 seed genes was extracted. The same number of randomly chosen coding genes from 7,903 SCZ candidate genes in the source data were used as control. Compared with the 1,000 random samplings of SCZ candidate genes in the source data, selected seed genes have a significantly larger interconnected network (empirical p = 0.001).
(D)Mapping seeds onto the human interactome. The 1,720 high-confidence SCZ candidate genes (as seeds) were mapped onto the comprehensive human interactome network which contains 17,210 nodes (1,592 seeds, 15,618 nonseeds) and 597,859 edges.
(E) A schematic for RWR and NW. Top: the genes in the human interactome network were ranked based on their proximity with “seed genes” using “random walk with restart (RWR)” approach to expanding SCZ candidate genes. Bottom: The neighborhoods in the human interactome network were defined and ranked based on their “seed ratio” using “neighborhood walk (NW)” approach.
(F) Extracting SCZ Network from human interactome. Comparison between RWR-extracted network and NW-extracted network is shown as the Venn diagram. RWR-extracted subnetwork (RWR-Subnet): the genes with RWR score more than 1e-05 were included as expanded SCZ candidate genes and a network containing the 1,592 seed genes and 5,920 expanded SCZ candidate genes were extracted from the human interactome. NW-extracted subnetwork (NW-Subnet): The neighborhoods with seed ratio ≥75% were extracted from the human interactome and these neighborhoods were further combined into a network containing 1,592 seed genes and 3,858 expanded candidate genes. The SCZ Network: The intersection of RWR-extracted subnetwork and NW-extracted subnetwork was taken as the disease network for schizophrenia, containing 4,573 nodes (1,592 seeds; 2,981 candidates) and 192,392 edges.
From a network perspective, products of the SCZ disease genes should interconnect at the molecular level to determine the complex traits of this disease. To identify additional SCZ candidate genes that may function together with the known SCZ candidate genes, we mapped the 1,720 high-confidence SCZ candidate genes onto a high-quality comprehensive human interactome network (Figure 1D), which contains 17,210 protein-coding genes (nodes) and about 600,000 interactions (edges) (Table S2) (Li et al., 2017). Of the 1,720 high-confidence SCZ candidate genes, 1,592 were located in the human interactome network. These 1,592 seed genes were more connected than randomly picked nodes with the same degrees (Figures S1C and S1D), suggesting that the seed genes were functionally related. Using these seed genes, we applied two strategies and combined the results to construct a protein interaction network for schizophrenia (Figure 1E). Firstly, we carried out the “random walk with restart” (RWR) approach to rank the genes according to their similarity with these seed genes in the human interactome network. Secondly, we developed a method, “neighborhood walk” (NW), to rank the neighborhoods according to their seed ratios and p-value, which were considered as the levels of association to schizophrenia.
RWR has been adapted for ranking novel candidate genes using known disease genes as seeds in molecular networks (Kohler et al., 2008; Valdeolivas et al., 2019). Here, we used RWR to expand SCZ candidate genes in the human interactome (Figure 1E). We extracted an RWR-retrieved subnetwork containing 7,512 genes (including 1,592 seed genes and 5,920 expanded SCZ candidate genes) using RWR score = 1 × 10−5 as the cutoff for choosing the expanded candidate genes (Figure 1F and Table S3).
The neighborhood walk (NW) approach was based on the idea that functionally related genes are more likely to cluster together in the same neighborhood in a molecular network. Therefore, the neighborhoods with higher level of seed density are more likely to be associated with schizophrenia. Using NW approach to searching for neighborhoods in the human interactome network based on seed density (Figure S1E, See STAR Methods for details), we were able to identify the network neighborhoods that may be involved in schizophrenia. Briefly, we initially mapped the SCZ candidate genes as “seeds” onto the human interactome network and assigned a risk score (R-score) to each seed gene according to the number of methods used in identifying the gene as an SCZ candidate gene (Table S3). For each nonseed gene, we calculated a guilt score (G-score = the average R-score of its interactors) (Table S3). For each edge, we calculated an edge weight between the two interacting nodes (W = the sum of the R-score and/or G-score) and a weighted edge length (WEL = 1/W) (Table S3), and for any two nodes in the network, we calculated a weighted shortest distance (Figure S1E). To search for SCZ-associated neighborhoods, we started the “walking” from each seed and stopped at a potential neighborhood border at a certain walking distance (WSD) from the origin, i.e., if further “walking” to the next sphere made the seed ratio in the neighborhood drop significantly (Figure S1E). We obtained 1,592 potential SCZ-associated neighborhoods (one for each seed) and ranked them according to the seed ratio and p-value (Table S3). Three neighborhoods were chosen as examples to be visualized in the background of human interactome network using Cytoscape, and functional enrichment was performed to identify the main functional terms (Figure S1F), showing topological and functional coherence. The neighborhoods with seed ratio ≥75% were selected and combined into a subnetwork (NW-based Subnetwork), which contains 1,592 seed genes and 3,858 expanded SCZ candidate genes.
The NW approach was designed based on the hypothesis that disease genes of a complex disease were functionally converged and thus clustered together in neighborhoods of the human interactome network. In order to test this hypothesis, we randomly picked 1,592 genes with exactly the same degrees as the 1,592 seeds and, starting with the degree-controlled fake seeds, we performed the same procedure using the same cutoff of 75% seed ratio for selecting “disease”-associated neighborhoods. The procedure was repeated for 100 times, and the best result showed only three very small retrieved neighborhoods (a total of eight genes, including 7 fake seeds and 1 “expanded candidate gene”) (Figure S1G). This result demonstrates that NW exclusively retrieves disease-associated neighborhoods based on functional convergence of disease genes. We also performed RWR for 100 times starting with 1,592 fake seeds using the cutoff 10ˆ-5 (Figure S1H). Using fake seeds, RWR retrieved more “expanded candidate genes” than using real seeds, suggesting that RWR has no preference for seed genes with functional convergence.
NW-retrieved subnetwork and RWR-retrieved subnetwork have 54.5% (4,573/8,389) nodes in common (Figure 1F). We took the intersection of these two subnetworks as the final network for schizophrenia (SCZ Network) (Figure 1F, Table S4) for downstream analysis, which contains 1,592 seed genes and 2,981 expanded candidate genes. This network only has one major component and four isolated nodes.
Evaluation on the SCZ Network
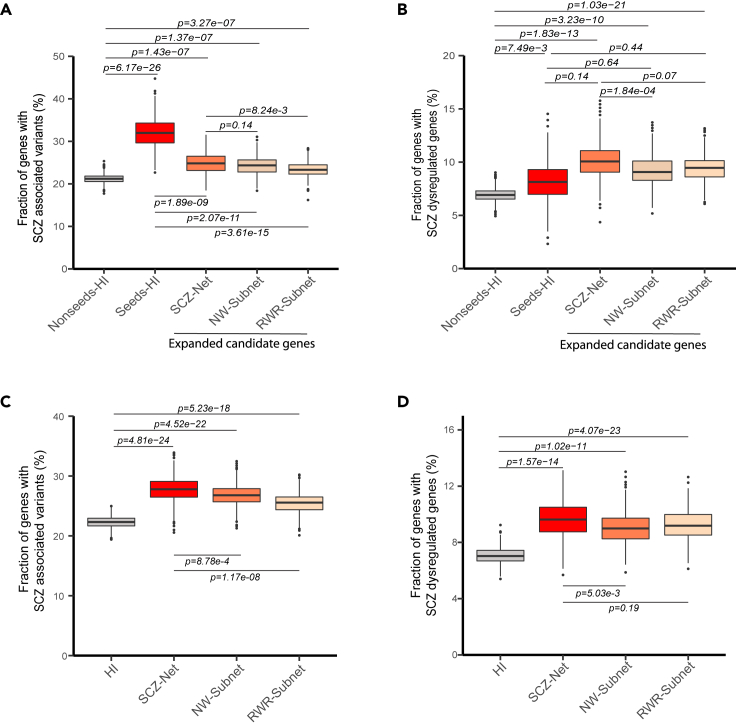
In order to evaluate the quality of the expanded candidate genes in the SCZ Network, we used independent datasets to compare SCZ relevance between the expanded candidate genes and the seed genes. We obtained the independent SCZ-associated gene sets from three recently published systematic studies (Table S5), including: (1) genomic variants associated with schizophrenia (Howrigan et al., 2020; Rees et al., 2020); (2) dysregulated genes in the postmortem brain of schizophrenia patients (SCZ-dysregulated genes) (Bowen et al., 2019). We calculated the recovery ratios of these two SCZ-associated gene sets by the seed genes, the expanded candidate genes in the RWR- or NW-extracted subnetworks, the intersection of these two subnetworks, and the nonseed genes in the background network (the human interactome network) respectively (Figures 2A and 2B). Compared to the nonseed genes in the background network, both the seed genes and the expanded candidate genes in the RWR- or NW-extracted subnetworks show significant enrichment of SCZ-associated variants (Two-tailed Fisher’s exact tests, p = 6.17e−26 for seed genes, p = 1.37e−07 for the expanded candidates by NW, p = 3.27e−07 for the expanded candidates by RWR, p = 1.43e-7 for the expanded candidate genes identified by both, Figure 2A). In addition, compared to the nonseed genes in the background network, both the seed genes and the expanded candidate genes by RWR and NW show significant enrichment of SCZ-dysregulated genes (Two-tailed Fisher’s exact tests, p = 7.49e−3 for seed genes, p = 3.23e−10 for the expanded candidates by NW, p = 1.03e−21 for the expanded candidates by RWR, p = 1.83e-13 for the expanded candidate genes identified by both, Figure 2B). Furthermore, the seed genes are more enriched with SCZ-associated variants than the expanded candidates (Two-tailed Fisher’s exact test, p = 2.07e-11 for the expanded candidate genes by NW, p = 3.61e-15 for the expanded candidate genes by RWR, p =1.89e-9 for the expanded candidate genes identified by both, Figure 2A), but show similar enrichment of SCZ-dysregulated genes compared with the expanded candidate genes (Two-tailed Fisher’s exact test, p = 0.64 for the expanded candidate genes by NW, p = 0.44 for the expanded candidate genes by RWR, p = 0.14 for the expanded candidate genes identified by both, Figure 2B). These results suggest that previous studies might be biased towards genetic variants, while the RWR or NW methods did not exhibit such bias. Using a core set of high-quality SCZ candidate genes as seed genes, RWR and NW performed equally well according to the enrichment of SCZ-variants in the expanded candidate genes (Two-tailed Fisher’s exact test, p = 0.32 for SCZ-variants, p = 0.72 for SCZ-DEGs). Furthermore, the expanded candidate genes by both RWR and NW show greater enrichment of the SCZ-associated genes than those expanded by RWR alone (Two-tailed Fisher’s exact test, p = 8.24e-3 for SCZ-associated variants, Figure 2A) or NW alone (Two-tailed Fisher’s exact test, p = 1.84e-4 for dysregulated genes) (Figure 2B), showing that the combination of these two methods has better performance than either one. These results support our decision on choosing the intersection of RWR-retrieved subnetwork and NW-retrieved subnetwork as high-confidence SCZ Network.
Figure 2.
Validation of the SCZ Network using independent data sets of variants and dysregulated genes associated with schizophrenia (See also Figure S2)
(A) Box and whisker graph for the fractions of SCZ-associated variants in the expanded candidate genes in SCZ Network (SCZ-Net), NW-extracted subnetwork (NW-Subnet) and RWR-extracted subnetwork (RWR-Subnet), compared with those in the seeds in human interactome (seeds-HI) and the nonseeds in human interactome (nonseeds-HI).
(B) Box and whisker graph for the fractions of SCZ-dysregulated genes in the expanded candidate genes in SCZ Network (SCZ-Net), NW-extracted subnetwork (NW-Subnet) and RWR-extracted subnetwork (RWR-Subnet), compared with those in the seeds in human interactome (seeds-HI) and the nonseeds in human interactome (nonseeds-HI).
(C) Box and whisker graph for the fractions of SCZ-associated variants in the SCZ Network (SCZ-Net), NW-extracted subnetwork (NW-Subnet) and RWR-extracted subnetwork (RWR-Subnet), compared with those in HI (human interactome).
(D) Box and whisker graph for the fractions of SCZ-dysregulated genes in the SCZ Network (SCZ-Net), NW-extracted subnetwork (NW-Subnet) and RWR-extracted subnetwork (RWR-Subnet), compared with those in HI (human interactome). P-values were calculated using Two-sided Fisher’s exact test. P-values on lines above or below the boxes are calculated from comparisons between the two groups. Lines extending vertically from the boxes indicate variability outside the upper and lower quartiles, estimated using bootstrapping method with 1,000 resamplings.
We further compared the recovery ratios of the SCZ-associated gene sets by the SCZ Network with those by RWR/NW-extracted subnetworks or the background network (Figures 2C and 2D). Compared to the background network, the extracted subnetworks using RWR, NW or both (SCZ Network) show significant enrichment with SCZ-associated variants (p = 5.23e-18 for RWR-extracted subnetwork; p = 4.52e-22 for NW-extracted subnetwork, p = 4.81e-24 for SCZ Network, Figure 2C) and significant enrichment with SCZ-dysregulated genes (p = 4.07e-23 for RWR-extracted subnetwork; p = 1.02e-11 for NW-extracted subnetwork, p = 1.57e-14, Figure 2D). Compared to RWR/NW-extracted subnetworks, SCZ Network showed both increased enrichment with SCZ-associated variants (Two-tailed Fisher’s exact tests, p = 8.78e-4 compared to NW-extracted network, p = 1.17e-08 compared to RWR-extracted subnetwork, Figure 2C) and increased enrichment with SCZ-dysregulated genes (Two-tailed Fisher’s exact tests, p = 5.03e-3 compared to NW-based subnetwork, Figure 2D). All these results demonstrate that both RWR and NW approaches have identified additional SCZ candidate genes, which show significant enrichment of SCZ-associated genes, and that the combination of RWR and NW had significantly better performance than RWR or NW alone, suggesting that the SCZ Network constructed using the combination of RWR and NW methods may be a highly SCZ-relevant molecular network.
We further performed enrichment analysis on MetaCore disease biomarkers of mental disorders for the expanded candidate genes and seed genes in the SCZ Network. We found that biomarkers for several mental disorders, including schizophrenia, schizophrenia spectrum disorder, major depressive disorder, neurodevelopmental disorders, bipolar disorders, autism spectrum disorder, etc., are significantly enriched with both the seed genes and the expanded candidate genes (Figure S2, Table S5). These results further confirm the high quality of the SCZ Network, and also suggest a potential common molecular mechanism for the comorbidity of these mental disorders. The expanded candidate genes and seed genes are enriched with biomarkers for ‘mental disorders’ in equal magnitude, but the seed genes show greater enrichment for ‘schizophrenia’, probably due to the bias of schizophrenia biomarkers towards well known SCZ candidate genes.
SCZ network is enriched in synaptic and immune signaling pathways
In order to explore the molecular mechanisms underlying schizophrenia, we performed pathway enrichment analysis using MetaCore and KEGG databases. MetaCore pathway database is a comprehensive pathway database, and therefore, the enriched MetaCore pathways may provide a list of potential SCZ-associated pathways in the SCZ Network (Table S6-1), which may help further illustrate the molecular mechanisms of schizophrenia. We examined literature for the 10 most significantly enriched pathways, including ROS-induced cellular signaling, GPCR signaling, MET and RON signaling, cytokine/JAK/STAT signaling, GDNF signaling, EGFR signaling, etc., (Figure S3A), and found that each of these pathways is well reported to associate with both schizophrenia and immunity by multiple studies. Interestingly, all these pathways are also reported to be involved in synaptogenesis or synapse modulation (See Table S6-2 for literature information). These top enriched MetaCore pathways, each suggested by literature to be associated with immune activation, synaptic regulation, and schizophrenia, may provide us with a direction for further exploring the SCZ Network. The enriched KEGG pathways would provide functional information of the network (Table S6-3). We compared the top enriched KEGG pathways between the seed genes, the expanded candidate genes and all genes (Figure 3A). Among these pathways, some pathways such as ‘MAPK signaling’, ‘cAMP signaling’, ‘ErbB signaling’, ‘GABAergic synapse’, ‘dopaminergic synapse’, ‘Alzheimer's disease’, and ‘circadian entrainment’ are common between the seed genes and the expanded candidate genes (Figure 3A); some pathways such as ‘calcium signaling’, ‘neuroactive ligand-receptor interaction’ and ‘nicotine addiction’ are only found for seed genes (Figure 3A), while other pathways such as ‘viral infections’, ‘bacterial infections’, ‘viral carcinogenesis’, ‘pancreatic cancer’, ‘cellular senescence’ and ‘ubiquitin-mediated proteolysis’ are found for the expanded candidate genes (Figure 3A). Interestingly, the pathways that are only enriched with the expanded candidate genes are mostly related to immune response and cancer, suggesting that the network approach may compensate the genetic approaches in identifying some SCZ candidate genes involved in immunity. All these top enriched KEGG pathways are documented to associate with schizophrenia or psychiatric disorders (See Table S6-4 for literature information). The literature information on the top enriched MetaCore and KEGG pathways suggests a close link between immune activation, synapse function, and schizophrenia. It is consistent with previous studies which discovered the involvement of immune system (Brown, 2012; Brown and Derkits, 2010; Estes and McAllister, 2016; Fillman et al., 2013; van Kesteren et al., 2017) and synaptic loss and impaired synaptic plasticity (Boksa, 2012; Forsyth and Lewis, 2017; Keshavan et al., 1994; Osimo et al., 2019; Sellgren et al., 2019; Stephan et al., 2006) in the development of schizophrenia.
Figure 3.
SCZ network is enriched for synaptic and immune function (See also Figure S3)
(A) Functional enrichment analyses on the SCZ Network for the seed genes (known candidate genes) and the expanded candidate genes. The enriched KEGG pathways are those with overrepresentation of genes in the SCZ Network. Only top enriched KEGG pathways are shown. The sizes of the dots represent gene ratios and the colors represent the adjusted p-values. The pathways in immune response are in blue font, the synaptic signaling pathways (MAPK, cAMP or ErbB signaling pathways) are in green font; the synapse pathways are in cyan, other pathways related to cancer, Alzheimer's disease or circadian entrainment are in black.
(B) Synaptic and immune genes in the SCZ Network. Left pie chart is split into 4 categories: (1) immune genes, (2) synaptic genes, (3) the genes annotated as both immune and synaptic genes, and (4) other genes. The bars on the right show the proportions of the seeds and the expanded candidate genes in categories (i), (ii) and (iii).
(C) Enrichment of synaptic genes in the seed genes and the expanded SCZ candidate genes compared to the human interactome (p = 4.88e-47 for all genes in SCZ Network, p = 2.37e-27 for seed genes, p = 2.39e-15 for the expanded candidates). The seed genes are significantly more enriched with synaptic genes than the expanded candidates (p = 1.98e-4).
(D) Enrichment of immune genes in the seed genes and the expanded candidate genes compared to the human interactome (p = 1.44e-42 for all genes in SCZ Network, p = 0.09 for seed genes, p = 4.22e-47 for the expanded candidates). The expanded candidate genes are significantly more enriched immune genes than seed genes (p = 1.37e-09).
P-values were calculated using Two-sided Fisher’s exact test. P-values on lines above the top of boxes are from comparisons between two groups. Lines extending vertically from the boxes indicate variability outside the upper and lower quartiles, estimated using bootstrapping method with 1,000 resamplings.
Consistent with the pathway enrichment results, the SCZ Network contains 10.4% (474/4,573) synaptic genes and 24.3% (1,109/4,573) immune genes (Figure 3B, Tables S6-5–S6-8), showing significant enrichment (Fisher’s exact tests, p = 4.88e-47 for synaptic genes, p =1.44e-42 for immune genes, Figures 3C and 3D), compared to the background network which contains 4.2% (532/12,637) synaptic genes and 15.0% (1,901/12,637) immune genes. Compared to other genes in the background network (the human interactome), both the seed genes and the expanded candidate genes are significantly enriched with synaptic genes (Fisher’s exact tests, p = 2.37e-27 for seed genes; p = 2.39e-15 for the expanded candidate genes, Figure 3C), but the seed genes show stronger enrichment than the expanded candidate genes (Fisher’s exact test, p = 1.98e-4, Figure 3C). However, only the expanded candidate genes are significantly enriched with immune genes (Fisher’s exact test, p = 0.09 for seed genes; p = 4.22e-47 for the expanded candidate genes, Figure 3D). These results demonstrate that apart from synaptic genes our network approach has captured many additional candidate genes involving in SCZ-related immune response that have not been identified in previous genetic studies. The interconnection among the immune genes, synaptic genes and other genes in the SCZ Network (Figure S3B) suggests a critical role of immune genes in the pathogenesis of schizophrenia, which provides the basis for systematic investigation on the molecular mechanisms linking immune activation to schizophrenia.
Spatiotemporal expression patterns of genes in SCZ network associates with two susceptible developmental stages
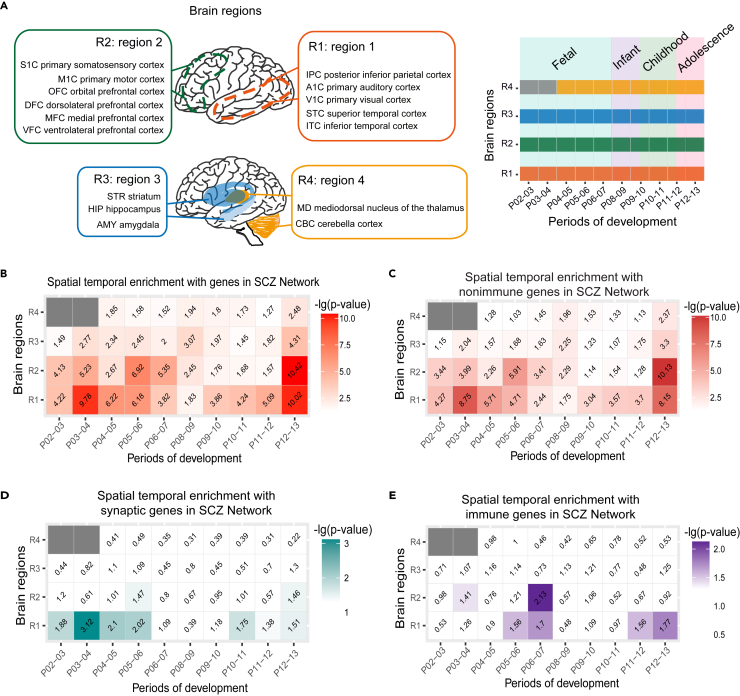
We are interested in understanding whether the spatiotemporal dynamics of the SCZ Network might reveal the stages and brain regions sensitive to immune activation and, more importantly, the underlying molecular pathways. We utilized the expression data from 4 regions of human brains at different developmental periods (periods 2-13, P2-P13) (Kang et al., 2011; Lin et al., 2015) to calculate the overrepresentation of the genes in the SCZ Network in different spatiotemporal phases (Figures 4A and S4; see STAR Methods and Table S7 for details). The p-values for the enrichments were calculated using one-side Fisher’s exact test. Region 1 includes: posterior inferior parietal cortex, primary auditory (A1) cortex, primary visual (V1) cortex, superior temporal cortex, and inferior temporal cortex; region 2 includes: primary somatosensory cortex, primary motor cortex, orbital prefrontal cortex, dorsolateral prefrontal cortex, medial prefrontal cortex, and ventrolateral prefrontal cortex; region 3 includes: striatum, hippocampus, and amygdala; region 4 includes: mediodorsal nucleus of the thalamus, and cerebella cortex (Figure 4A).
Figure 4.
Spatiotemporal dynamic expression of the genes in the SCZ Network during brain development (See also Figure S4)
(A) A schematic of human spatiotemporal expression data source: 4 brain regions and 10 developmental periods. Region 1 includes: posterior inferior parietal cortex, primary auditory (A1) cortex, primary visual (V1) cortex, superior temporal cortex, inferior temporal cortex; region 2 includes: primary somatosensory cortex, primary motor cortex, orbital prefrontal cortex, dorsolateral prefrontal cortex, medial prefrontal cortex and ventrolateral prefrontal cortex; region 3 includes: striatum, hippocampus and amygdala; region 4 includes: mediodorsal nucleus of the thalamus and cerebella cortex. Developmental periods including: early fetal development, early mid-fetal development, late mid-fetal development, late fetal development, neonatal and early infancy, late infancy, early childhood, middle and late childhood, adolescence and young adulthood (see Figure S4).
(B–E) Enrichment of different categories of genes of the SCZ Network in spatiotemporal phases. The heatmap shows the significance levels of overrepresentation of the genes of the SCZ Network in the spatiotemporal phases. One-sided Fisher’s exact test was used for the analyses of enrichments. The numbers in the squares represent the −log10(p-value). (B) Enrichment of all genes of the SCZ Network in spatiotemporal phases; (C) Enrichment of the non-immune genes of the SCZ Network in spatiotemporal phases; (D) Enrichment of the synaptic genes of the SCZ Network in spatiotemporal phases; (E) Enrichment of the immune genes of the SCZ Network in spatiotemporal phases.
The genes in the SCZ Network are significantly enriched in the regions 1 and 2 (the cortex regions) during most of development periods (log10-transformed p-values shown in Figure 4B). Stronger enrichment is found in region 1 during periods 3-6, 11-13 (P3-6: early fetal to late mid-fetal development, p = 1.66e-10 ∼ 6.60e-07; P11-13: middle childhood to young adulthood, p = 8.05e-06 ∼ 9.59e-11), in region 2 during periods 3-4, 5-7 (P3-4: early fetal to early mid-fetal development, p = 5.87e-06; P5-7: early mid- to late fetal development, p = 1.20e-07 ∼ 4.45e-06) and periods 12-13 (adolescence to young adulthood, p = 3.77e-11) and in region 3 during periods 12-13 (adolescence to young adulthood, p = 4.85e-5) (Figure 4B). The overrepresentation of the genes in the SCZ Network in the stages of early- to late fetal development and adolescence to young adult suggests that this network may be more susceptible to perturbation during fetal development and adolescence. Since 76% of the genes in the SCZ Network are nonimmune genes, the nonimmune genes show a very similar pattern as all genes in the network (Figure 4C).
Accumulating evidence suggests that synaptic plasticity impairment during development may play a role in the development of schizophrenia (Boksa, 2012; Forsyth and Lewis, 2017; Keshavan et al., 1994; Osimo et al., 2019; Sellgren et al., 2019; Stephan et al., 2006). We further explored the spatial temporal expression pattern of the synaptic genes of the SCZ Network (Figure 4D). Synaptic genes are significantly enriched in region 1 during periods 2-6 (P2-6: early fetal to late mid-fetal development, p = 7.6e-4 ∼ 0.01) and 10-13 (P10-13: early childhood to young adulthood, p = 0.04–0.02); in region 2 during periods 5-6 (P5-6: early-to-late midfetal development, p = 0.03) and periods 12-13 (P12-13: adolescence to young adulthood, p = 0.03) (Figure 4E). The overrepresentation of synaptic genes during periods from early fetal to late mid-fetal development and from early childhood to young adult suggests these stages are critical for synaptic development. In addition, these stages are coincident with the two critical stages for cortex development, in which cell differentiation and synapse remodeling may be influenced by the environment factors, respectively (Selemon and Zecevic, 2015).
To further identify the brain regions and developmental stages that are sensitive to immune activation, we further calculated the overrepresentation of the immune genes of the SCZ Network in the spatiotemporal phases. The immune genes are more significantly enriched in region 1 during periods 5-7, 11-13 (P5-7: early mid- to late fetal development p = 0.02–0.03; P11-13: middle childhood to young adulthood, p = 0.02–0.03); in region 2 during periods 6-7 (P6-7: late mid-to late fetal development, p = 0.007) (Figure 4E). The overrepresentation of immune genes in this network during both early- to mid-fetal development and peripuberty suggests that it may be more easily affected by immune activation during these stages. These results are consistent previous findings that fetal and peripubertal developmental stages are the two key stages for schizophrenia: (1) maternal infection (Brown, 2012; Brown and Derkits, 2010; Estes and McAllister, 2016), maternal stress (Gomes et al., 2019) and placental diseases (Kendell et al., 1996) during gestation increase the risk of schizophrenia in the offspring; (2) adolescent stress is also a risk factor for schizophrenia (Gomes and Grace, 2017). These results suggest that abnormal immune activation during these stages may perturb the molecular network in the brain, potentially leading to schizophrenia. On the other hand, many environmental factors, including infection and stress, induce peripheral and central immune activation (Uhlhaas, 2011).
Based on network topology, the immune genes, synaptic genes and other genes are interconnected with each other, and therefore immune activation may affect brain development and synapse remodeling. Recent studies show that immune genes in the brain may regulate neuronal synapse remodeling (Boulanger, 2009; Morimoto and Nakajima, 2019). The complement component 4 (C4) participates in the synaptic pruning regulation (Sekar et al., 2016). The immune activation increases synaptic pruning through microglia (Sellgren et al., 2019). Our results are consistent with previous findings of the critical role of immune system in brain development (Morimoto and Nakajima, 2019; Schepanski et al., 2018). Overall, our findings illustrated the connection between immune activation and synapse plasticity during peripubertal developmental stages, further confirming that fetal and peripubertal developmental stages are critical for schizophrenia development (Mottahedin et al., 2017). Since the typical onset time of schizophrenia is the transition period from adolescence to adulthood (Uhlhaas, 2011), it may be possible that maternal immune activation during fetal development may prime the brain to be susceptible to a second immune activation at adolescence.
Immune activation during fetal development and puberty dysregulates signaling pathways in SCZ network
In order to further investigate how immune activation might be involved in the molecular mechanism underlying schizophrenia, we induced immune activation twice in mice: first during mid-late fetal development stage by intraperitoneal injection (IPI) of lipopolysaccharide (LPS) into pregnant mice at G16 (16th day after gestation), and a second time during puberty by IPI of LPS into the male offspring at P30 (30th day after birth) (Figure 5A). We used saline as control, and designed the experiments in three combinations of saline and LPS treatments: (1) S-S (control group): saline at G16 + saline at P30; (2) L-S: LPS at G16 + saline at P30; (3) S-L: saline at G16 + LPS at P30; (4) L-L: LPS at G16 + LPS at P30 (Figure 5A).
Figure 5.
Immune activation during fetal development and puberty dysregulates signaling pathways in SCZ Network (See also Figures S5–S7)
(A) A schematic of experimental design. The first intraperitoneal injection (IPI) of LPS or saline to the pregnant mice was performed on G16 (Gestation Day 16) and the second IPI of LPS or saline to the offspring was performed on P30 (Postnatal Day 30). The combination of LPS and saline treatments: saline at G16 + saline at P30 (S-L); LPS at G16 + saline at P30 (L-S); saline at G16 + LPS at P30 (S-L); LPS at G16 + LPS at P30 (L-L).
(B) The Venn diagram of the DEGs in the L-S group (yellow), S-L group (cyan) and L-L group (blue) compared with S-S group (p < 0.05 and |FC|≥1.3).
(C–E) The threshold-free comparison of expression alteration between three treatment groups by rank-rank hypergeometric overlap (RRHO). The log10-transformed hypergeometric p-values are plotted in a heatmap as indicated by an accompanying color scale, mapping the degree of the statistical significance of overlaps between two differential transcriptomes (two ranked gene lists on the X-axis or Y-axis). Each pixel in a RRHO map represents the log10-transformed hypergeometric p-value for the overlap of subsections of two ranked lists from a point on the map (corresponding to a rank threshold pair: Rank X, Y) to either bottom left corner (Rank 1, 1) or top right corner (Rank N, N). The numbers on color scales indicate −log10(p-value). The genes along each axis are sorted from most significantly up-regulated to most down-regulated from the lower left corners. The RRHO maps: L-S group versus L-L group (C); L-S group versus S-L group (D); S-L group versus L-L group (E).
(F) Over-represented disease biomarkers of mental disorders in the DEGs of L-S, S-L and L-L groups identified via MetaCore “Diseases (by Biomarkers)”. The X-axis represents −log10 (FDR). Y-axis lists the enriched diseases. Vertical red dashed line shows the cutoff of FDR = 0.05.
(G) The node degree distribution of SCZ Network with top hub DEGs highlighted. Y-axis represents the gene count and X-axis represents the degree. Top 10% of genes have degrees ≥212 as indicated by the red line. The 10 DEGs among the top hub genes are marked (The degrees of all the DGEs in the SCZ Network are listed in Table S9).
(H) The MetaCore pathways enriched with DEGs. The Venn diagrams show the overlaps between the enriched pathways with DEGs and the enriched pathways with genes in SCZ Network: 18 of 20 (90%), 69 of 85 (81.2%) and 95 of 114 (83.3%). A total number of 100 enriched pathways in L-S, S-L or L-L groups are overlapped with enriched pathways of SCZ Network. These 100 enriched pathways are clustered into 18 groups by calculating the distance between two pathways based on shared genes within SCZ Network. The dot plot shows the enrichment of the pathways enriched with DEGs in L-S, S-L, L-L groups and with genes in SCZ Network. The red, yellow, cyan or blue dots show the −log10 transformed FDR value of pathways in SCZ network, L-S, S-L, and L-L groups, respectively. Vertical purple dashed line shows the cutoff of FDR = 0.05. The 18 representative pathways are listed on the bottom right corner. The information of all pathways is listed in Table S9.
We carried out poly-A selected RNA-seq sequencing on the whole brain at P32 to evaluate transcriptional changes and then performed the differential expression analysis using DeSeq2 (Figures S5A and S5B, STAR Methods). The differentially expressed genes (DEGs) identified were 170 from L-S group, 474 from S-L group and 927 from L-L group using S-S group as control (p< 0.05 and |Fold Change| ≥1.3, Figure 5B, Table S8), suggesting that L-L group have greater immune response than S-L group or L-S group. The overlap of DEGs is 22.9% (39/170) between L-S and L-L group, while 52.3% (248/474) between S-L and L-L group (Figure 5B). We then looked at the genome-wide comparison of differential expression patterns at threshold-free manner among these three groups using rank-rank hypergeometric pair-wise comparison (Plaisier et al., 2010), which showed co-regulation of genes among the three groups (Figures 5C–5E).
We further performed MetaCore disease biomarker enrichment analysis in the DEGs of L-S, S-L and L-L groups (FDR <0.05). The top enriched mental diseases include ‘schizophrenia’, ‘schizophrenia spectrum and other psychotic disorders’, ‘bipolar’, ‘bipolar related disorder’, ‘major depressive disorder’ and ‘depressive disorder’ (Figure 5F, Table S9-1), suggesting that gene dysregulation by maternal and adolescent immune activation may underly multiple psychotic disorders. As described above (Figure S2), the SCZ Network is also enriched for biomarkers of these same mental diseases. These results not only confirm the current perspective on the role of immune activation in the pathogenesis of psychiatric disorders, but also suggest potential common mechanisms for the comorbidity between these mental disorders reported in epidemiological studies (Buckley et al., 2009).
To evaluate the potential effects that the gene differential expression induced by immune activation may have on the SCZ Network, we mapped the DEGs on the SCZ Network and ranked the node degrees (Figure 5G, Table S9-2). Of 184 DEGs that are in the SCZ Network, 28 DEGs have a node degree above the average while 8 DEGs (ACTA2, ALDH1A2, PLK1, MYH11, CBL, MAP1LC3A, AURKB and RPL18A) are among the top 10% hub genes (Figure 5G). These hub genes may play a key role in linking immune activation to schizophrenia. The literature mining for these top hub DEGs identified many studies about their association with schizophrenia, psychiatric disorders or nervous system related functions (See Table S9-3 for literature information). For example, ACTA2 is significantly downregulated in the postmortem cortex of schizophrenia patients (Mladinov et al., 2016), and downregulation of ACTA2 inhibits neural stem cell migration (Zhang et al., 2020). The deletion of 15q11-13, which covers gene ALDH1A2, is suggested to be involved schizophrenia (Reay and Cairns, 2020). ALDH1A2 is found to associate with schizophrenia in a large scale genome-wide association study (Reay et al., 2020).
In order to understand the molecular mechanisms on how the immune activation during gestation (the first hit) and during puberty (the second hit) may increase the risk for schizophrenia, we searched for MetaCore pathways enriched with DEGs of each group. Using a cutoff of FDR < 0.05, we identified 20 enriched pathways for DEGs in L-S group, 85 for S-L group and 104 for L-L group. Of the enriched pathways, 90.0% (18/20) in L-S group, 81.2% (69/85) in S-L group and 83.3% (95/114) in L-L group are overlapping with the enriched pathways in SCZ Network (Figures 5H and S6, Tables S9-4, S9-5, and S9-6). A total number of 121 enriched pathways are identified in the three groups of DEGs, and 100 are also enriched in SCZ Network (Table S9-7). We also carried out pathway enrichment analysis on the seed genes and the expanded candidate genes of the SCZ Network separately, and found that the seed genes and the expanded candidate genes had similar proportion of enriched pathways overlapping with those of DEGs (Figure S7). These results suggest that the majority of the dysregulated signaling pathways in the brain induced by immune activation during fetal development and/or during puberty may be involved in schizophrenia, consistent with the accumulating evidence that maternal and peripubertal immune activation increase the risk of schizophrenia (Brown, 2012; Brown and Derkits, 2010; Estes and McAllister, 2016; Gomes and Grace, 2017; Gomes et al., 2019; Kendell et al., 1996).
The enrichment of these pathways was compared between the three groups and the SCZ Network (Figures 5H and S6). L-L group shows better enrichment in most of these pathways than L-S and S-L groups (Figures 5H and S6), suggesting that maternal immune activation during fetal development may increase the risk of schizophrenia after a second immune activation at puberty. In order to identify the main categories of pathways linking immune activation to schizophrenia, we clustered the enriched pathways based on shared genes within SCZ Network (Figures 5H and S6). Eighteen pathway clusters were identified, and one representative pathway was selected for each cluster. Some of the clusters, i.e., C2, C5, C17 and C18, are highly enriched with genes in SCZ Network (Figure S6), suggesting that the disruption of pathway in these clusters may have more impact on the molecular network. The “first hit” and the “second hit” may have different effects on the pathways. The “first hit” alone affects only a small number of clusters, including the WNT signaling pathways in clusters C14 and C15 (Figure S6), which are important for fetal brain development, while the “second hit” alone affects most of the clusters. On the other hand, the vast majority of clusters except a few, are more affected by ‘two hits’ (L-L group) than by the “second hit” alone. Examples of the exception include cluster C7 and C10, which are more affected by “second hit” (S-L group) than by ‘two hits’ (L-L group) (Figure S6), suggesting that the “first hit” may have primed the brain to response differently to the “second hit”. All these 18 representative pathways are well supported to associate with schizophrenia based on literature (See Table S9-8 for literature information). For example, pathways for antigen presentation by MHC (International Schizophrenia Consortium et al., 2009; Pardinas et al., 2018; Schizophrenia Working Group of the Psychiatric Genomics, 2014; Shi et al., 2009; Stefansson et al., 2009; Walters et al., 2013), canonical (beta-catenin dependent) and noncanonical (beta-catenin independent) WNT signaling (Hoseth et al., 2018; Hussaini et al., 2014) are well-documented to be involved in schizophrenia.
Key regulators for immune activation in mouse brain
The comparison of the brain transcriptome profiles among the L-S, S-L and L-L groups, suggest that injection of LPS to the mother during mid-late gestation stage may induce latent sensitization of the brain to a second injection of LPS to the offspring at adolescence (Figures 5 and S5). We assumed that the response patterns of the brain to immune activation is established and maintained by the gene expression programs through transcription factors, cofactors and chromatin regulators. In order to identify the major transcription factors (TFs) that may regulate the transcriptional alteration in these three groups, we searched for enriched TF-binding motifs in the promoter regions of the DEGs using HOMER (Heinz et al., 2010). We found a total number of 24 TF-binding motifs corresponding to 26 TFs after removing the motifs whose corresponding TFs were not expressed in any of the transcriptomes in L-S, S-L, L-L or control groups (Figures 6A and S8A). These 24 motifs are enriched in up-regulated genes (ISRE, ZBTB12, MAFA and KLF9), down-regulated genes (NF1-halfsite, ARNT:AHR, NR2E1, SOX7, SOX17, SMAD4, SIX1, TWIST1, HIF1A and PAX3:FOXO1) or both (IRF2, IRF8, ZBTB1, PRDM1, SIX2, SPI1, IRF3, IRF1 and NR2F2): 6 for L-S group, 6 for S-L group and 16 for L-L group (Figures 6A, S8B, Tables S10-1 and S10-2). There are 3 common motifs (ISRE, ARNT:AHR and NF1-halfsite) between L-L and S-L group, and 1 common motif (IRF2) between L-L and L-S groups, but no common motifs between L-S and S-L groups (Figures 6A and S8A).
Figure 6.
Key regulators for immune activation in mouse brain (See also Figure S8)
(A) The 24 binding motifs and corresponding transcription factors (TFs) predicted for DEGs of L-S, S-L and L-L groups using HOMER. The motif patterns are shown on the left. The colored bars on the right indicate the target DEGs in L-S, S-L and L-L groups.
(B) The mRNA expression levels (detected in the bulk RNA-Seq) of the identified TFs. Dot size shows absolute value of log2(fold change) in L-S, S-L and L-L groups. The colors represent the up-regulation (red) or down-regulation (blue). The asterisks indicate the p-values <0.5.
(C) The 24 transcription factors (TFs) were clustered into 6 groups based on the correlation between their shared targets using Pearson correlation coefficient. Red and blue show the comparison pairs with a higher and lower correlation coefficient respectively. The colored bars on the top show the groups the DEGs belong to.
(D) The Venn diagrams show the overlaps between the pathways enriched with the target DEGs of the TF groups and the pathways enriched with the genes in SCZ Network.
(E) The comparison of the top enriched pathways among the 6 TF groups. The enrichment of the top 10 enriched pathways in each TF group was compared with enrichment of these pathways in other TF groups and SCZ Network. The sizes of the dots represent gene ratios and the colors represent FDR adjusted p-values.
(F) Integrated protein-protein interactions and regulatory cascades of the identified TFs. Node size represents the degree. Red, yellow and grey node colors represent seeds, expanded candidates or other genes in SCZ Network, respectively. Cyan arrow edges and grey solid edges show the TF-to-target relationship and protein-protein interaction respectively.
(G) The node degree distribution of SCZ Network with the identified TFs highlighted. The 11 of 26 identified TFs are in the SCZ Network.
We checked the expression alteration of the 26 TFs corresponding to these 24 motifs by examining the transcriptomes of mouse brain of S-S, L-S, S-L, and LL groups. Of the 26 TF genes, 10 (Irf8, Spi1, Arnt, Nf1, Smad4, Six1, Six2, Twist1, Klf9 and Foxo1) in L-L group and 5 (Spi1, Nf1, Six2, Twist1 and Klf9) in S-L group showed significant expression alteration (Wald test, p < 0.05, Figure 6B, Table S10-3), but no TFs in L-S group showed significant expression change (Figure 6B, Table S10-3). The genes Irf8, Arnt, Smad4, Six1, and Foxo1 only showed significant expression change in L-L group, not in S-L group, suggesting that both “first hit” and “second hit” may be needed for expression alteration of these 5 TFs. We noticed that expression levels of some TFs were altered at an extent below the differential expression cutoff for TFs (p = 0.05) in L-S group. For example, Six1 showed 1.51-fold upregulation (Wald test, p = 0.13) in L-S group and a 1.14-fold change (p = 0.69, Wald test) in S-L group, but a fold change of 2.27 (Wald test, p = 0.001) in L-L group. These results demonstrated that, “first hit” or “second hit” alone was not able to significantly alter the expression level of some TFs while the combination of “two hits” was able to change their expression, suggesting that the “first hit” may prime the brain to make it more sensitive to a “second hit”. Such priming phenomenon has also been observed in early life stress in generating mouse model for depressive disorder, which makes the individuals more sensitive to social stress in adult life (Pena et al., 2017).
In order to find potential collaborative regulation among the 26 TFs during immune activation, we clustered the TF-binding motifs by calculating the correlation coefficient between two TF-binding motifs based on their shared target DEGs, and identified 6 groups including 5 clusters and 1 singleton (Figure 6C). We further performed MetaCore pathway enrichment analysis (FDR <0.05) for the TF targets in each group and compared the enriched pathways with those identified in SCZ Network (Tables S10-4–S10-9). The majority of the enriched pathways were also enriched with SCZ Network: Group I: 88.9% (40/45); Group II: 98% (49/50); Group III: 86.8% (59/68); Group IV: 88.5% (46/52); Group V: 80% (20/25); Group VI: 75% (3/4). We compared the top 10 enriched pathways from each group and found that each of these TF groups regulate both distinct and common pathways (Figure 6E). For example, ‘Antigen presentation by MHC class I: cross-presentation’ is shared by Group 1 and Group 3, and ‘Immune response_IFN-gamma in macrophages activation’ is unique to Group 1. Most of these top enriched pathways (73%) are reported to be associated with schizophrenia or psychotic disorders (See Table S10-10 for literature information). Therefore, together with the pathway clusters identified for the 100 SCZ-associated pathways (Figures 5H and S6; Table S9), these 6 TF groups and their target pathways (Figures 6C–6E; Table S10) provide a framework for studying the immune activation-induced transcriptional regulation of the pathways involved in schizophrenia.
In order to understand how the TFs may regulate each other to coordinate the immune responses after the “first hit” and/or the “second hit”, we mapped potential transcriptional regulation among these TFs using HOMER motif scan results for the promoter regions of the TF genes, and also integrated with the protein-protein interactions among these TFs using interaction data of the SCZ Network. This network contains 14 transcriptional regulatory relationships and 8 protein-protein interactions among 15 TFs (Figure 6F), where SIX1 and SIX2 are TF-TF regulatory hubs, while SMAD4 is protein interaction hub, suggesting that SIX1, SIX2 and SMAD4 may play critical role in immune activation (see Table S10-11 for literature information). SIX1 can be regulated by 5 TFs (SIX2, PRDM1, IRF8, IRF3 and SPI1) while SIX2 can regulate three genes (SIX1, TWIST1 and IRF8) and can be regulated by itself and two other TFs (ZBTB18 and NR2F2). As described above, SIX1 is only upregulated in mice with LPS treatments during both gestation and puberty (Figure 6B), suggesting that the priming by “first hit” during embryonic development may be necessary for the transcriptional upregulation of this genes after the “second hit” during puberty. It is reported in a recent study that SIX1 and SIX2 control the activity of inflammatory genes such as interferon (IFN) in response to noncanonical NF-κB pathway activation to protect cell from inflammatory damage (Liu et al., 2019). Multiple studies also reported the dysregulation of NF-κB signaling pathway in cortical immune activation in schizophrenia (Roussos et al., 2013; Volk et al., 2019). As an interaction hub (Figures 6F and 6G), SMAD4 binds 4 TFs (IRF3, NR2F2, NF1 and FOXO1). IRF3 (Xu et al., 2014), NR2F2 (Qin et al., 2013), NF1 (Kretova et al., 2014), and FOXO1 (Ellis et al., 1989), each can bind to SMAD4 to suppress SMAD transcriptional complexes, which is at the core of the TGF-β signaling pathways (Mullen et al., 2011). Genetic variants in TGF-β are found to associate with schizophrenia (Jia et al., 2010) while TGF-β signaling promotes neuronal axon formation (Yi et al., 2010) and dendritic growth (Luo et al., 2016). All these lines of evidence support our prediction on the critical roles of SIX1, SIX2, and SMAD4 in the immune activation associated with schizophrenia. Systematic empirical investigation into the roles of these transcription factors and signaling pathways in an immune response may illustrate a more detailed molecular mechanisms on how immune activation in the brain may lead to schizophrenia.
Discussion
The most significant advances in searching for risk factors of schizophrenia over the last decade include: first, a large number of schizophrenia-associated genes have been identified by genome wide studies (Bowen et al., 2019; Fromer et al., 2014; Howrigan et al., 2020; Pardinas et al., 2018; Rees et al., 2020; Schizophrenia Working Group of the Psychiatric Genomics, 2014; Xu et al., 2011); and second, immune activation has been repeatedly proven to be associated with this disease (Brown, 2012; Brown and Derkits, 2010; Fillman et al., 2013; Goldsmith et al., 2016; Miller et al., 2011; van Kesteren et al., 2017). Proinflammatory cytokine levels have been consistently reported to be increased in the serum of SCZ patients (Goldsmith et al., 2016; Miller et al., 2011) and also in their prefrontal cortex (Fillman et al., 2013). Maternal infection or stress during pregnancy greatly increases the risk of schizophrenia in the offspring (Brown, 2012; Brown and Derkits, 2010; Estes and McAllister, 2016; Gomes et al., 2019), while adolescent stress is also reportedly a risk factor (Gomes and Grace, 2017; Harrop and Trower, 2001; Trotman et al., 2013). The “environment-environment interaction” model of the “two-hit” hypothesis is proposed explanation for the etiology (Khan and Powell, 2018). What is needed is a systematic way to illustrate the underlying molecular mechanism, particularly how immune system contributes to the disease onset. Here we presented a systematic search for molecular pathways linking immune activation to schizophrenia by extracting a high-quality protein interaction network for schizophrenia (SCZ Network) using our in-house developed “neighborhood walk” (NW) approach in combination with “random walk with restart” (RWR).
Starting from a core set of 1,720 high-confidence SCZ candidate genes, we extracted the SCZ Network from a high-quality comprehensive human interactome network (Li et al., 2017) using NW and RWR (Figure 1). This is the first time that NW approach is applied in searching for disease-associate neighborhoods in a molecular network. NW and RWR show comparable performance (Figure 2). Although both RWR and NW start walking from seeds in the network, they retrieve disease candidate genes based on different principles. RWR walks from a seed to all directions with a restart probability, to examine the proximity between each individual seed and all the other nodes based on the topology of the network. Therefore, RWR does not depend on the functional convergence of the disease genes. Starting from randomly picked degree-controlled fake seeds, RWR retrieved more “disease” candidate genes than starting from real seeds (Figure S1H), probably because the fake seeds were not functionally related and thus were topologically scattered in the network. However, NW walks from a seed to explore the disease association of the neighborhood. It depends on the functional convergence of the seed genes in neighborhoods of the network. Starting with randomly picked degree-controlled fake seeds, NW was not able to retrieve sizable “disease”-associated neighborhoods (Figure S1G), because functionally unrelated genes were scattered in the network, not clustered in neighborhoods of the network. The common nodes of these two subnetworks show stronger enrichment with SCZ-associated genes than either subnetwork alone (Figure 2). Therefore, we selected the common genes identified by these two methods, which generated a more reliable molecular network for schizophrenia than each method alone. The expanded candidate genes and the seed genes in the SCZ Network have enrichment of equal magnitude with the SCZ-dysregulated genes in the brain (Figure 2B), confirming the high quality of the SCZ Network. MetaCore and KEGG pathway enrichment analyses identified underlying signaling pathways (Tables S6-1 and S6-3). We examined the literature for the top ten enriched MetaCore pathways (see Table S6-2 for literature information) and found that all these top pathways are involved in schizophrenia and immune regulation. Most interestingly, these same pathways are also reported to be involved in synaptogenesis and synapse modulation. The literature search for the top enriched KEGG pathways reached similar conclusion (see Table S6-4 for literature information). These findings together suggest that the molecular signaling pathways in the SCZ Network that are involved in immune regulation may contribute to the development of schizophrenia through regulating the formation and modulation of synapses. These results not only further confirm the high quality of the SCZ Network, but also suggest that molecular processes of immune activation and synapse remodeling might be a fundamental mechanism for schizophrenia.
The spatiotemporal expression pattern of the genes of the SCZ Network in the brain during development demonstrates that this molecular network is overrepresented in the expressed genes during both “early- to late fetal development” and “adolescence to early adulthood” (Figure 4B), suggesting that this molecular network may be more susceptible to perturbation during these developmental stages. Additionally, immune genes are more likely to be expressed in cortex regions from mid- to late fetal development and from adolescence to early adulthood (Figure 4E), further suggesting that immune activation during these two sensitive developmental stages may disturb the molecular network. The spatiotemporal expression pattern of the SCZ Network is coincident with the structural and functional abnormalities found in the cortex of the schizophrenia patients (Karlsgodt et al., 2010), and with the developmental stages of cortex in animal models (Selemon and Zecevic, 2015), suggesting that the normal function of this molecular network may be involved in the structural and functional development of cortex regions, and that immune activation at these critical stages may disturb the cortex development, leading to schizophrenia. The spatiotemporal expression pattern of the immune genes in the SCZ Network fits well with “environment-environment interaction” model of the “two-hit” hypothesis on the etiology (Khan and Powell, 2018), and it is also consistent with the clinical finding on increased immune response in patients (Fillman et al., 2013; Goldsmith et al., 2016; Miller et al., 2011), suggesting a “twice immune-activation” mechanism for this model.
To explore “twice immune-activation” mechanism, we have built a “two-hit” mouse model using LPS to mimic infection during gestation and during puberty. We induced immune activation twice in mice, first during mid-late fetal development stage by intraperitoneal injection (IPI) of lipopolysaccharide (LPS) in to the pregnant mice at G16 (16th day after gestation), and a second time during puberty by IPI of LPS into the male offspring at P30 (30th day after birth) (Figure 5A), and carried out transcriptome sequencing on the brain at P32 to explore the differentially expressed genes (DEGs) (Figure 5B). The DEGs of L-S, S-L or L-L groups are enriched in biomarkers of ‘schizophrenia’ and other mental disorders such as ‘bipolar’, ‘bipolar related disorder’, ‘major depressive disorder’ and ‘depressive disorder’ (Figure 5F), suggesting that immune activation may underlie multiple psychotic disorders. The differentially expressed genes in three treatment groups are enriched in a total of 121 pathways (Tables S9-4–S9-6). Surprisingly, 82.6% (100/121) of the enriched pathways of the differentially expressed genes induced by immune activation are also identified in SCZ Network (Table S9-7). These pathways are clustered into 18 groups according to their shared genes in SCZ Network (Figures 5H and S6). All the representative pathways from the 18 clusters are documented to associate with schizophrenia or other psychiatric disorders (Table S9-8), providing a framework for investigating the mechanisms on how immune activation in the brain may lead to schizophrenia.
The mice with “two hits” showed greater transcription alteration than the mice with the “first hit” or the “second hit” alone (Figure 5B, Table S8), suggesting that the “first hit” may have effects on the brain to make it more sensitive to the “second hit”. We searched for enriched binding motifs in the promoter regions of the differentially expressed genes and found 24 TF-binding motifs corresponding to 26 transcription factors (Figure 6A). These transcription factors can be clustered into 6 groups, each regulating a group of pathways (Figure 6C). Majority of pathways identified for each group of TFs overlap with the signaling pathways identified in the SCZ Network (Figure 6D; Tables S10-4–S10-9), further providing a framework for studying the key regulators of these immune-activated pathways linking to schizophrenia. The network of TFs with regulatory edges and protein interaction edges suggest that SIX1 and SIX2 are regulatory hubs and SMAD4 is an interaction hub (Figures 6F and 6G). SIX1 and SIX2 are reported to be involved in cortical immune activation through NF-κB pathway (Liu et al., 2019; Roussos et al., 2013; Volk et al., 2019). SMAD4 transcription factor is at the core of the TGF-β signaling pathways (Mullen et al., 2011), regulating neuronal axon formation (Yi et al., 2010) and dendritic growth (Luo et al., 2016).
In summary, in order to lay the foundation for studying molecular mechanism underlying schizophrenia, we consolidated a core set of high-confidence SCZ candidate genes and then constructed a high-quality SCZ Network from a comprehensive interactome dataset using “neighborhood walk” and “random walk with restart”. The dynamic spatiotemporal expression patterns of the genes of the SCZ Network in human brain revealed “mid- to late fetal development” and “adolescence to early adulthood” as critical stages when the molecular network is susceptible to environmental insults. The top enriched pathways suggested a tight link between immune activation, synapse remodeling and schizophrenia. The transcriptome sequencing on the brain of mice with immune activation at late fetal developmental stage or puberty revealed that over 80% of the dysregulated pathways by immune activation overlap with pathways in the SCZ Network. The dysregulated pathways further supported the molecular link between immune signaling, synaptic function, and schizophrenia. HOMER scan on the promoter regions of the dysregulated genes identified a small number of potential key regulators, such as SIX1, SIX2, and SMAD4, of the SCZ-associated immune activation in the brain. Our work provides a strategy for illustrating the molecular mechanism underlying complex psychiatric disorders and a rich resource for disease candidate genes as well as systematically evaluated signaling pathways to study the pathogenesis and therapeutics of schizophrenia.
Limitations of the study
This study only provides a landscape of genes and pathways that may be involved in the development of schizophrenia, especially the molecular pathways and regulators that may link the immune activation to this disease. A limitation of our study is the lack of deeper understanding of the identified pathways and key regulators using cell models and mouse models. Another limitation is that our in-house developed NW method has not been tested in different datasets. In theory, NW retrieves disease candidate genes based on their convergence in the neighborhoods of the network, and therefore functionally unrelated genes may be left out of the selected disease-associated neighborhoods. Although we have demonstrated the validity of NW in retrieving SCZ candidate genes, this method needs to be further tested in the study of other complex diseases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Lipopolysaccharide | Sigma-Aldrich | L2630-10MG, lot: #099M4002V |
| Critical commercial assays | ||
| RNeasy Plus Universal Mini Kit | Qiagen | Cat.NO.73404 |
| Deposited data | ||
| Raw and analyzed data | This paper | GSE156568 |
| Mouse reference genome, GRCm38 | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/grc/mouse |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6J | Southern Medical University | N/A |
| Software and algorithms | ||
| R (version 3.6.0) | The R Project for Statistical Computing | https://cran.r-project.org/bin/windows/base/old/ |
| RandomWalkRestartMH (version 1.6.0) | Valdeolivas et al., 2019 | http://bioconductor.org/packages/RandomWalkRestartMH/ |
| Neighborhood walk | This paper | Supplemental information |
| STAR (version 2.5.2b). | Dobin et al., 2013 | https://github.com/alexdobin/STAR/releases |
| HTSeq (version 0.11.2) | Anders et al., 2015 | https://htseq.readthedocs.io/en/master/ |
| HOMER (version v4.11.1) | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Other | ||
| Resource data for selecting core set of high-confidence SCZ candidate genes | This paper | STAR Methods and supplemental information |
| Construction and validation of SCZ Network, related resources | This paper | STAR Methods and supplemental information |
| SCZ Network analyses and related resources | This paper | STAR Methods and supplemental information |
| RNA-seq data analyses and related resources | This paper | STAR Methods and supplemental information |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xinping Yang (xpyang1@smu.edu.cn)
Materials availability
This study did not generate new unique reagents. All renewable reagents and detailed protocols will be available upon request.
Experimental model and subject details
Animal
The C57BL/6 mice were housed under standard conditions (12 h/12 h light/dark cycle, access to dry food and water ad libitum). All the experimental procedures involving mice were approved by the Committee on Animal Care and Use at the Southern Medical University.
Immune activation in mice during mid-fetal stage and adolescent stage
Immune activation was induced twice in mice, first during mid-late fetal development stage by intraperitoneal injection (IPI) of lipopolysaccharide (LPS) (150ug/kg) in to the pregnant mice at G16 (16th day after gestation) and a second time during puberty by IPI of LPS into the male offspring at P30 (30th day after birth). We then carried out poly-A selected RNA-seq sequencing on the brain at P32 to evaluate transcriptional changes. We used saline as control for LPS, and set the experiments in three combination of saline and LPS treatments: (1) S-S (control group): saline at G16 + saline at P30; (2) L-S: LPS at G16 + saline at P30; (3) S-L: saline at G16 + LPS at P30; (4) L-L: LPS at G16 + LPS at P30. Lipopolysaccharide (LPS) was purchased from Sigma, USA.
Method details
Collection of Schizophrenia candidate genes
A total number of 1,720 schizophrenia candidate genes (protein coding genes) were collected from 7 methods in 87 studies pooled by SZDB: copy number variations (CNVs), differential expressed genes (DEGs), differential methylated genes (DMGs), whole exome sequencing (WES), genome-wide association studies (GWASs), imputation and linkage (Table S1). For genes from CNVs, DMGs and WES, only genes supported by two or more publications are kept. For genes from DEGs, only genes with FDR ≤0.05 are kept.
Human interactome data
The comprehensive human interactome data were downloaded from InBioMap (https://www.intomics.com/inbio/map.html) (Table S2).
Random walk with restart (RWR)
RWR was performed using R package “RandomWalkRestartMH” (version 1.6.0) (Valdeolivas et al., 2019). The cutoff score ≥ 1e-05 from RWRM results was used for identifying disease candidate genes.
Neighborhood walk (NW)
NW was performed using in-house developed algorithm.
-
i.
We first mapped the 1,720 high-confidence SCZ candidate genes as “seeds” onto the human interactome network (Table S2).
-
ii.
For each seed, we assigned a risk score (R-score = 1-1.4−x, x is the number of methods used in identifying the gene as SCZ candidate gene in literature). We used “R-score = 1-1.4−x” for scoring seeds because it was shown to give seeds proper weights (Calderone et al., 2013). For each nonseed, we calculated a guilt score (G-score = the average R-score of its interactors) (Table S3).
-
iii.
For each edge, we calculated an edge weight (W = the sum of the R-score and/or G-score of the two interacting nodes) and a weighted edge length (WEL) (WEL = 1/W) (Table S3).
-
iv.
For any two nodes in the network, we calculated a weighted shortest distance (WSD): (1/Wi is the weighted edge length of an edge in a given shortest path) (Figure S1E).
-
v.
For each seed as an origin, we started the “walking” from the origin and stopped at a potential neighborhood border (a sphere with certain walking distance) if further “walking” to next sphere made the seed ratio in the neighborhood drop significantly, (walking distance between two spheres is 1.0) (Figure S1E);
-
vi.
We obtained 1,592 ranked neighborhoods (one for each seed) based on seed ratio (Table S3). The neighborhoods with seed ratio from 75% to 100% were considered as SCZ-related network neighborhoods.
-
vii.
We extracted an NW-retrieved Subnetwork by fusing the 1,043 neighborhoods with the seed ratio ≥75%, which contains 1,592 seed genes (high-confidence candidate genes) and 3,858 additional SCZ candidate genes (nonseed genes in the network).
Independent datasets for validation of SCZ network
In order to compare the SCZ relevance of the expanded candidate genes and seed genes in SCZ network, we calculated the enrichment of with dysregulated genes or variants identified in SCZ patients collected from recent literature. These data sources have not been used for collection of seed genes and therefore they contain SCZ-associated genes (dysregulated genes or genes harboring variants) unbiased towards seeds or nonseeds. A total number of 1,682 brain differentially expressed genes (DEGs) (Table S5) between schizophrenia patients and normal subjects were collected from the study by Bowen et al. (2019), and a total number of 4,217 exome variants (Table S5) identified in the postmortem brains of SCZ patients were collected from the studies by Howrigan et al. (2020) and Rees et al. (2020). The three studies are listed below:
-
1
.Bowen, E. F. W. et al. DLPFC transcriptome defines two molecular subtypes of schizophrenia. Transl Psychiatry 9, 147, https://doi.org/10.1038/s41398-019-0472-z (2019).
-
2
.Howrigan, D. P. et al. Exome sequencing in schizophrenia-affected parent-offspring trios reveals risk conferred by protein-coding de novo mutations. Nature neuroscience 23, 185-193, https://doi.org/10.1038/s41593-019-0564-3 (2020).
-
3
.Rees, E. et al. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nature neuroscience 23, 179-184, https://doi.org/10.1038/s41593-019-0565-2 (2020).
Tools and data sources for functional enrichment analysis
-
1
.KEGG pathway enrichment analysis were performed using R package “clusterProfiler” (version 3.14.3) (Yu et al., 2012).
-
2
.Disease Enrichment was performed using MetaCore Disease (by biomarker) from Clarivate Analytics (version 20.2).
-
3.Collection of immune genes: 3,904 immune genes (Table S6) were collected from:
-
i.InnateDB (Innate Immunity Genes, https://www.innatedb.com) (Breuer et al., 2013);
-
ii.GO-immune-response (http://amigo.geneontology.org/amigo/term/GO:0006955);
- iii.
- iv.
-
v.IMGT/GENE-DB (the international ImMunoGeneTics information system): (http://www.imgt.org/genedb/).
-
i.
-
4
.Collection of synaptic transmission genes: 1,068 synaptic transmission genes (Table S6) were collected from GO (http://amigo.geneontology.org/amigo/landing) using key word “synaptic transmission”.
The spatiotemporal expression of genes in human brain
The brain transcriptome data was generated across 13 dissection stages (Kang et al., 2011). Spatiotemporal co-expression was calculated according to a previous study (Lin et al., 2015), as described below. Since well-defined anatomical brain structures are limited during early embryonic development, the first period (4-8 postconceptional weeks, PCW) was removed from further analysis. The dissection stages were merged into ten partially-overlapping developmental periods ranging from 8 PCW to 40 years of age (Figures 4A and S4). Brain regions were grouped into four clusters using hierarchical clustering based on brain transcriptional similarity to reflect actual topological proximity and functional segregation (Willsey et al., 2013). As a result, 38 spatiotemporal regions were defined after eliminating one region from P2-P4 (P2-4, R4) due to lack of transcriptome data. Co-expression networks were used to identify phase-specific functional relationships between genes. A co-expression between two genes was defined as positive if the pair-wise Pearson Correlation Coefficient (PCC) value was > 0.8. Using this approach, 38 different spatiotemporal co-expression networks were generated and the co-expressed gene pairs were used for the enrichment analyses of SCZ network (Table S7).
RNA preparation
For each group, six pregnant mice were treated saline or LPS (maternal immune activation); and the male offspring were treated with saline or LPS (immune activation at puberty). For each group, four whole-brain RNA samples were prepared. Each RNA sample was extracted from the whole brain of one adult mouse according to the manufacturer's protocol (RNAeasy Mini Kit, Qiagen, Germantown, MD, USA). The quality and yield of the isolated RNAs was assessed using a NanoDrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Only RNAs with a high RNA integrity number (RIN > 9) were selected and used for the subsequent sequencing.
RNA-Seq data analysis and differential motifs
RNA sequencing was performed at Berry Genomics (Beijing, China) using Illumina NovaSeq. Sequenced reads were mapped to GRCm38 whole genome using STAR_2.5.2b. Raw counts are calculated by HTSeq-count version 0.11.2. Pair-wised analyses are performed, genes with >3 raw counts in all four samples of case or control were considered as expressed genes and used for downstream analysis (Table S8). The principal components analysis (PCA) is performed by plotPCA function using normalized count by variance stabilizing transformation function of DEseq2 (v1.20.0).
Differential expression analysis on two groups was performed using the DESeq2. Differentially expressed genes (DEGs) were determined using a cutoff of p < 0.05 and |FC|≥1.3 for DESeq2 (Table S8). Functional annotation for pathways was performed using MetaCore from Clarivate Analytics (version 20.2).
The threshold-free comparison of expression alteration between different treatment groups was carried out using R package “RRHO” (version 1.26.0). Full threshold-free gene list of differential transcriptomes (L-S group, S-L group or L-L group compared to S-S group) were ranked by the −log10(p-value) multiplied by the sign of the fold change from the DESeq2 analysis.
The log10-transformed hypergeometric p-value are plotted to map the degree of the statistically significant overlap between two differential transcriptomes (two ranked gene lists). Each pixel in a RRHO map represents the signed log10-transformed hypergeometric p-value for the overlap of subsections of two ranked lists from a point on the map (corresponding to a rank threshold pair: Rank X, Y) to either bottom left corner (Rank 1, 1) or top right corner (Rank N, N). Genes along each axis are sorted from most significantly up-regulated to most down-regulated from the lower left corners.
Pathways are clustered by calculating the distance between two pathways based on shared genes within SCZ Network using dist() function with method “euclidean” and hclust() function with method “ward.D” in R package “stats” (version 3.6.3). The dendrogram tree was plot using R packages dendextend (version 1.14.0) and ggdendro (version 0.1-20).
HOMER (version v4.11.1) motif analysis was used to predict differential motifs (enriched transcription factor-binding sites) located within −2,000 to +1,000 bases of the TSS of DEGs.
TF correlation was calculated by their shared target DEGs using cor() function with method “pearson” in R package “stats” (version 3.6.3). Then plot was performed by corrplot package (version 0.84) using hierarchical clustering order.
Quantification and statistical analysis
The p-values in the enrichment analysis were calculated using Fisher's Exact Test. And the p-values in the differentially expressed analysis were calculated using Wald significance tests.
Acknowledgments
The work was supported by the Science and Technology Planning Project of Guangdong Province (Grant No. 2017B020227010), the National Natural Science Foundation of China (Grant No. 82071503 and Grant No. 81571097), the Natural Science Foundation of Guangdong Province (Grant No. 2016A030308020), the Science and Technology Projects of Guangzhou (Grant No. 201704020116) and the Guangdong Key Project in “Development of new tools for diagnosis and treatment of Autism” (Grant No. 2018B030335001). The funding organizations had no role in the design and conduct of the study; the collection, management, analysis, and interpretation of the data; and the preparation, review, or approval of the manuscript.
Author contributions
X.Y. conceived and supervised the project; Y.H. supervised the project; Y.G. performed the computational analysis; Y.G., Y.L., X.L., S.L acquired the experimental data. Z.R. ,Xiaoxue.Y. participated in the analysis; Y.G., X.Y., and Y.H. prepared the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103209.
Supplemental information
(S1-1) All SCZ-candidate genes from original source datasets.
(S1-2) Core set of high-confidence SCZ-candidate genes.
(S2-1) Human interactome data (nodes).
(S2-2) Human interactome data (edges).
(S3-1) RWR ranking metrics (RWR score).
(S3-2) NW ranking metrics (R-scores and G-scores).
(S3-3) NW ranking metrics (nodes distance and edge weight).
(S3-4) NW ranking metrics (all neighborhoods).
(S4-1) RWR-extracted Network.
(S4-2) NW-extracted Network.
(S4-3) SCZ Network.
(S5-1) Differentially expressed genes (DEGs) in schizophrenic brains.
(S5-2) Exome variants identified in SCZ patients.
(S5-3) Enrichment results.
(S5-4) Over-represented MetaCore disease biomarkers for mental disorders in the expanded candidate genes and seed genes in the SCZ Network.
(S6-1) MetaCore pathways enriched with genes in the SCZ network.
(S6-2) Literature information of top enriched MetaCore pathways in the SCZ network.
(S6-3) KEGG pathways enriched with genes in the SCZ network.
(S6-4) Literature information of top enriched KEGG pathways in the SCZ network.
(S6-5) Immune genes.
(S6-6) Synaptic transmission genes.
(S6-7) Overrepresentation of immune genes in SCZ Network.
(S6-8) Overrepresentation of synaptic transmission genes in SCZ Network.
(S7-1) Co-expressed genes from 4 regions of the brain at different periods.
(S7-2) Overrepresentation of the SCZ Network genes in spatiotemporal phases.
(S7-3) Overrepresentation of non-immune genes of the SCZ Network in spatiotemporal phases.
(S7-4) Overrepresentation of immune genes of the SCZ Network in spatiotemporal phases.
(S7-5) Overrepresentation of synaptic transmission genes of the SCZ Network in spatiotemporal phases.
(S8-1) The expressed genes of L-S group.
(S8-2) The expressed genes of S-L group.
(S8-3) The expressed genes of L-L group.
(S8-4) The differentially expressed genes of L-S group.
(S8-5) The differentially expressed genes of S-L group.
(S8-6) The differentially expressed genes of L-L group.
(S9-1) Over-represented MetaCore disease biomarkers for mental disorders in the expanded candidate genes and the seed genes in three treatment groups.
(S9-2) Degree of DEGs in SCZ network.
(S9-3) Literature information of DEGs among the top 10% hub genes.
(S9-4) Metacore pathway maps enriched with DEGs in L-S group.
(S9-5) Metacore pathway maps enriched with DEGs in S-L group.
(S9-6) Metacore pathway maps enriched with DEGs in L-L group.
(S9-7) The 100 pathways enriched with three groups of DEGs and with genes in the SCZ Network.
(S9-8) Literature information for 18 representative pathways.
(S10-1) All motifs with enrichment pvalues by HOMER.
(S10-2) The binding motifs in the promoter regions of the DEGs.
(S10-3). Gene expression data of the corresponding TFs of the enriched motifs.
(S10-4) Metacore pathway maps enriched with targets in TF group I.
(S10-5) Metacore pathway maps enriched with targets in TF group II.
(S10-6) Metacore pathway maps enriched with targets in TF group III.
(S10-7) Metacore pathway maps enriched with targets in TF group IV.
(S10-8) Metacore pathway maps enriched with targets in TF group V.
(S10-9) Metacore pathway maps enriched with targets in TF group VI.
(S10-10) Literature information of top enriched pathways for target DEGs of 6 TF group.
(S10-11) Literature information of the predicted TFs in the SCZ Network.
Data and code availability
Data availability
Detailed analyzed data are included in supplemental excel tables. RNA-seq data is deposited at GEO (GEO: GSE156568).
Code availability
The original code for the calculation in the “neighborhood walk” approach is uploaded as ZIP file (Data S11).
Other source availability
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Anders S., Theodor Pyl P., Huber W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T.A., Falkai P., Maier W. Genetic and non-genetic vulnerability factors in schizophrenia: the basis of the “two hit hypothesis”. J. Psychiatr. Res. 1999;33:543–548. doi: 10.1016/s0022-3956(99)00039-4. [DOI] [PubMed] [Google Scholar]
- Blackwood D.H., Fordyce A., Walker M.T., St Clair D.M., Porteous D.J., Muir W.J. Schizophrenia and affective disorders--cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: clinical and P300 findings in a family. Am. J. Hum. Genet. 2001;69:428–433. doi: 10.1086/321969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boksa P. Abnormal synaptic pruning in schizophrenia: urban myth or reality? J. Psychiatry Neurosci. 2012;37:75–77. doi: 10.1503/jpn.120007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger L.M. Immune proteins in brain development and synaptic plasticity. Neuron. 2009;64:93–109. doi: 10.1016/j.neuron.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Bowen E.F.W., Burgess J.L., Granger R., Kleinman J.E., Rhodes C.H. DLPFC transcriptome defines two molecular subtypes of schizophrenia. Transl Psychiatry. 2019;9:147. doi: 10.1038/s41398-019-0472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer K., Foroushani A.K., Laird M.R., Chen C., Sribnaia A., Lo R., Winsor G.L., Hancock R.E., Brinkman F.S., Lynn D.J. InnateDB: systems biology of innate immunity and beyond--recent updates and continuing curation. Nucl. Acids Res. 2013;41:D1228–D1233. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.S. Epidemiologic studies of exposure to prenatal infection and risk of schizophrenia and autism. Developmental Neurobiol. 2012;72:1272–1276. doi: 10.1002/dneu.22024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.S., Derkits E.J. Prenatal infection and schizophrenia: a review of epidemiologic and translational studies. Am. J. Psychiatry. 2010;167:261–280. doi: 10.1176/appi.ajp.2009.09030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley P.F., Miller B.J., Lehrer D.S., Castle D.J. Psychiatric comorbidities and schizophrenia. Schizophrenia Bull. 2009;35:383–402. doi: 10.1093/schbul/sbn135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone A., Castagnoli L., Cesareni G. mentha: a resource for browsing integrated protein-interaction networks. Nat. Methods. 2013;10:690–691. doi: 10.1038/nmeth.2561. [DOI] [PubMed] [Google Scholar]
- Cannon T.D., Kaprio J., Lonnqvist J., Huttunen M., Koskenvuo M. The genetic epidemiology of schizophrenia in a Finnish twin cohort. A population-based modeling study. Arch. Gen. Psychiatry. 1998;55:67–74. doi: 10.1001/archpsyc.55.1.67. [DOI] [PubMed] [Google Scholar]
- Cusick M.E., Klitgord N., Vidal M., Hill D.E. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14:R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- Cusick M.E., Yu H., Smolyar A., Venkatesan K., Carvunis A.R., Simonis N., Rual J.F., Borick H., Braun P., Dreze M. Literature-curated protein interaction datasets. Nat. Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras1 T., R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.S., Jr., Ramamurthy S., Schoenfeld L.S., Hoffman J., Walsh N.E. Diagnostic epidural opioid technique. Clin. J. Pain. 1989;5:211–215. doi: 10.1097/00002508-198909000-00003. [DOI] [PubMed] [Google Scholar]
- Estes M.L., McAllister A.K. Maternal immune activation: implications for neuropsychiatric disorders. Science. 2016;353:772–777. doi: 10.1126/science.aag3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer A.E., McGuffin P., Gottesman Twin concordance for DSM-III schizophrenia. Scrutinizing the validity of the definition. Arch. Gen. Psychiatry. 1987;44:634–641. doi: 10.1001/archpsyc.1987.01800190054009. [DOI] [PubMed] [Google Scholar]
- Feigenson K.A., Kusnecov A.W., Silverstein S.M. Inflammation and the two-hit hypothesis of schizophrenia. Neurosci. Biobehav. Rev. 2014;38:72–93. doi: 10.1016/j.neubiorev.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillman S.G., Cloonan N., Catts V.S., Miller L.C., Wong J., McCrossin T., Cairns M., Weickert C.S. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol. Psychiatry. 2013;18:206–214. doi: 10.1038/mp.2012.110. [DOI] [PubMed] [Google Scholar]
- Forsyth J.K., Lewis D.A. Mapping the consequences of impaired synaptic plasticity in schizophrenia through development: an integrative model for diverse clinical features. Trends Cogn. Sci. 2017;21:760–778. doi: 10.1016/j.tics.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M., Pocklington A.J., Kavanagh D.H., Williams H.J., Dwyer S., Gormley P., Georgieva L., Rees E., Palta P., Ruderfer D.M. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M., Roussos P., Sieberts S.K., Johnson J.S., Kavanagh D.H., Perumal T.M., Ruderfer D.M., Oh E.C., Topol A., Shah H.R. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016;19:1442–1453. doi: 10.1038/nn.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith D.R., Rapaport M.H., Miller B.J. A meta-analysis of blood cytokine network alterations in psychiatric patients: comparisons between schizophrenia, bipolar disorder and depression. Mol. Psychiatry. 2016;21:1696–1709. doi: 10.1038/mp.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes F.V., Grace A.A. Adolescent stress as a driving factor for schizophrenia development-a basic science perspective. Schizophrenia Bull. 2017;43:486–489. doi: 10.1093/schbul/sbx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes F.V., Zhu X., Grace A.A. Stress during critical periods of development and risk for schizophrenia. Schizophr Res. 2019;213:107–113. doi: 10.1016/j.schres.2019.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M.W., Kann M.G. Chapter 4: protein interactions and disease. PLoS Comput. Biol. 2012;8:e1002819. doi: 10.1371/journal.pcbi.1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P.J., Law A.J. Neuregulin 1 and schizophrenia: genetics, gene expression, and neurobiology. Biol. Psychiatry. 2006;60:132–140. doi: 10.1016/j.biopsych.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Harrop C., Trower P. Why does schizophrenia develop at late adolescence? Clin. Psychol. Rev. 2001;21:241–265. doi: 10.1016/s0272-7358(99)00047-1. [DOI] [PubMed] [Google Scholar]
- Havugimana P.C., Hart G.T., Nepusz T., Yang H., Turinsky A.L., Li Z., Wang P.I., Boutz D.R., Fong V., Phanse S. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol.Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoseth E.Z., Krull F., Dieset I., Morch R.H., Hope S., Gardsjord E.S., Steen N.E., Melle I., Brattbakk H.R., Steen V.M. Exploring the Wnt signaling pathway in schizophrenia and bipolar disorder. Transl Psychiatry. 2018;8:55. doi: 10.1038/s41398-018-0102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howrigan D.P., Rose S.A., Samocha K.E., Fromer M., Cerrato F., Chen W.J., Churchhouse C., Chambert K., Chandler S.D., Daly M.J. Exome sequencing in schizophrenia-affected parent-offspring trios reveals risk conferred by protein-coding de novo mutations. Nat. Neurosci. 2020;23:185–193. doi: 10.1038/s41593-019-0564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussaini S.M., Choi C.I., Cho C.H., Kim H.J., Jun H., Jang M.H. Wnt signaling in neuropsychiatric disorders: ties with adult hippocampal neurogenesis and behavior. Neurosci. Biobehav Rev. 2014;47:369–383. doi: 10.1016/j.neubiorev.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Advisory Group for the Revision of ICD-10 Mental. Behavioural Disorders A conceptual framework for the revision of the ICD-10 classification of mental and behavioural disorders. World Psychiatry Off. J. World Psychiatr. Assoc. 2011;10:86–92. doi: 10.1002/j.2051-5545.2011.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium. Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janca A., Ustun T.B., Early T.S., Sartorius N. The ICD-10 symptom checklist: a companion to the ICD-10 classification of mental and behavioural disorders. Social Psychiatry Psychiatr. Epidemiol. 1993;28:239–242. doi: 10.1007/BF00788743. [DOI] [PubMed] [Google Scholar]
- Jia P., Wang L., Meltzer H.Y., Zhao Z. Common variants conferring risk of schizophrenia: a pathway analysis of GWAS data. Schizophr Res. 2010;122:38–42. doi: 10.1016/j.schres.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M., Pletikos M., Meyer K.A., Sedmak G. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt K.H., Sun D., Cannon T.D. Structural and functional brain abnormalities in schizophrenia. Curr. Dir. Psychol. Sci. 2010;19:226–231. doi: 10.1177/0963721410377601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendell R.E., Juszczak E., Cole S.K. Obstetric complications and schizophrenia: a case control study based on standardised obstetric records. Br. J. Psychiatry J. Ment. Sci. 1996;168:556–561. doi: 10.1192/bjp.168.5.556. [DOI] [PubMed] [Google Scholar]
- Keshavan M.S., Anderson S., Pettegrew J.W. Is schizophrenia due to excessive synaptic pruning in the prefrontal cortex? The Feinberg hypothesis revisited. J. Psychiatr. Res. 1994;28:239–265. doi: 10.1016/0022-3956(94)90009-4. [DOI] [PubMed] [Google Scholar]
- Khan A., Powell S.B. Sensorimotor gating deficits in “two-hit” models of schizophrenia risk factors. Schizophrenia Res. 2018;198:68–83. doi: 10.1016/j.schres.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp M., Mangalore R., Simon J. The global costs of schizophrenia. Schizophrenia Bull. 2004;30:279–293. doi: 10.1093/oxfordjournals.schbul.a007078. [DOI] [PubMed] [Google Scholar]
- Kohler S., Bauer S., Horn D., Robinson P.N. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretova M., Sabova L., Hodny Z., Bartek J., Kollarovic G., Nelson B.D., Hubackova S., Luciakova K. TGF-beta/NF1/Smad4-mediated suppression of ANT2 contributes to oxidative stress in cellular senescence. Cell Signal. 2014;26:2903–2911. doi: 10.1016/j.cellsig.2014.08.029. [DOI] [PubMed] [Google Scholar]
- Lanz T.A., Reinhart V., Sheehan M.J., Rizzo S.J.S., Bove S.E., James L.C., Volfson D., Lewis D.A., Kleiman R.J. Postmortem transcriptional profiling reveals widespread increase in inflammation in schizophrenia: a comparison of prefrontal cortex, striatum, and hippocampus among matched tetrads of controls with subjects diagnosed with schizophrenia, bipolar or major depressive disorder. Transl Psychiatry. 2019;9:151. doi: 10.1038/s41398-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wernersson R., Hansen R.B., Horn H., Mercer J., Slodkowicz G., Workman C.T., Rigina O., Rapacki K., Staerfeldt H.H. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat. Methods. 2017;14:61–64. doi: 10.1038/nmeth.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G.N., Corominas R., Lemmens I., Yang X., Tavernier J., Hill D.E., Vidal M., Sebat J., Iakoucheva L.M. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. 2015;85:742–754. doi: 10.1016/j.neuron.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Mar K.B., Hanners N.W., Perelman S.S., Kanchwala M., Xing C., Schoggins J.W., Alto N.M. A NIK-SIX signalling axis controls inflammation by targeted silencing of non-canonical NF-kappaB. Nature. 2019;568:249–253. doi: 10.1038/s41586-019-1041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovász L. Random walks on graphs: a survey, combinatorics, Paul erdos is eighty. Bolyai Soc. Math. Stud. 1993;2:4. [Google Scholar]
- Luo S.X., Timbang L., Kim J.I., Shang Y., Sandoval K., Tang A.A., Whistler J.L., Ding J.B., Huang E.J. TGF-beta signaling in dopaminergic neurons regulates dendritic growth, excitatory-inhibitory synaptic balance, and reversal learning. Cell Rep. 2016;17:3233–3245. doi: 10.1016/j.celrep.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar J.K., Pickard B.S., Mackie S., James R., Christie S., Buchanan S.R., Malloy M.P., Chubb J.E., Huston E., Baillie G.S. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Miller B.J., Buckley P., Seabolt W., Mellor A., Kirkpatrick B. Meta-analysis of cytokine alterations in schizophrenia: clinical status and antipsychotic effects. Biol. Psychiatry. 2011;70:663–671. doi: 10.1016/j.biopsych.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladinov M., Sedmak G., Fuller H.R., Babic Leko M., Mayer D., Kirincich J., Stajduhar A., Borovecki F., Hof P.R., Simic G. Gene expression profiling of the dorsolateral and medial orbitofrontal cortex in schizophrenia. Transl Neurosci. 2016;7:139–150. doi: 10.1515/tnsci-2016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K., Nakajima K. Role of the immune system in the development of the central nervous system. Front Neurosci. 2019;13:916. doi: 10.3389/fnins.2019.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottahedin A., Ardalan M., Chumak T., Riebe I., Ek J., Mallard C. Effect of neuroinflammation on synaptic organization and function in the developing brain: implications for neurodevelopmental and neurodegenerative disorders. Front. Cell. Neurosci. 2017;11:190. doi: 10.3389/fncel.2017.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen A.C., Orlando D.A., Newman J.J., Loven J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., DeKoter R.P., Young R.A. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donovan M.C., Craddock N., Norton N., Williams H., Peirce T., Moskvina V., Nikolov I., Hamshere M., Carroll L., Georgieva L. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Osimo E.F., Beck K., Reis Marques T., Howes O.D. Synaptic loss in schizophrenia: a meta-analysis and systematic review of synaptic protein and mRNA measures. Mol. Psychiatry. 2019;24:549–561. doi: 10.1038/s41380-018-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.B.a.L. The anatomy of a large-scale hypertextual web search engine. Comput. Netw. 1998;30:107–117. [Google Scholar]
- Pardinas A.F., Holmans P., Pocklington A.J., Escott-Price V., Ripke S., Carrera N., Legge S.E., Bishop S., Cameron D., Hamshere M.L. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena C.J., Kronman H.G., Walker D.M., Cates H.M., Bagot R.C., Purushothaman I., Issler O., Loh Y.E., Leong T., Kiraly D.D. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science. 2017;356:1185–1188. doi: 10.1126/science.aan4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisier S.B., Taschereau R., Wong J.A., Graeber T.G. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucl. Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polednak A.P. Black-white differences in infant mortality in 38 standard metropolitan statistical areas. Am. J. Public Health. 1991;81:1480–1482. doi: 10.2105/ajph.81.11.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Wu S.P., Creighton C.J., Dai F., Xie X., Cheng C.M., Frolov A., Ayala G., Lin X., Feng X.H. COUP-TFII inhibits TGF-beta-induced growth barrier to promote prostate tumorigenesis. Nature. 2013;493:236–240. doi: 10.1038/nature11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay W.R., Atkins J.R., Quide Y., Carr V.J., Green M.J., Cairns M.J. Polygenic disruption of retinoid signalling in schizophrenia and a severe cognitive deficit subtype. Mol. Psychiatry. 2020;25:719–731. doi: 10.1038/s41380-018-0305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reay W.R., Cairns M.J. The role of the retinoids in schizophrenia: genomic and clinical perspectives. Mol. Psychiatry. 2020;25:706–718. doi: 10.1038/s41380-019-0566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E., Han J., Morgan J., Carrera N., Escott-Price V., Pocklington A.J., Duffield M., Hall L.S., Legge S.E., Pardinas A.F. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat. Neurosci. 2020;23:179–184. doi: 10.1038/s41593-019-0565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees E., Walters J.T., Georgieva L., Isles A.R., Chambert K.D., Richards A.L., Mahoney-Davies G., Legge S.E., Moran J.L., McCarroll S.A. Analysis of copy number variations at 15 schizophrenia-associated loci. Br. J. PsychiatryJ. Ment. Sci. 2014;204:108–114. doi: 10.1192/bjp.bp.113.131052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S., O'Dushlaine C., Chambert K., Moran J.L., Kahler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T., Tasan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P., Katsel P., Davis K.L., Giakoumaki S.G., Lencz T., Malhotra A.K., Siever L.J., Bitsios P., Haroutunian V. Convergent findings for abnormalities of the NF-kappaB signaling pathway in schizophrenia. Neuropsychopharmacology. 2013;38:533–539. doi: 10.1038/npp.2012.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S., Chant D., McGrath J. A systematic review of mortality in schizophrenia: is the differential mortality gap worsening over time? Arch. Gen. Psychiatry. 2007;64:1123–1131. doi: 10.1001/archpsyc.64.10.1123. [DOI] [PubMed] [Google Scholar]
- Schepanski S., Buss C., Hanganu-Opatz I.L., Arck P.C. Prenatal immune and endocrine modulators of offspring's brain development and cognitive functions later in life. Front Immunol. 2018;9:2186. doi: 10.3389/fimmu.2018.02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Psychiatric Genome-Wide Association Study Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selemon L.D., Zecevic N. Schizophrenia: a tale of two critical periods for prefrontal cortical development. Translational psychiatry. 2015;5:e623. doi: 10.1038/tp.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellgren C.M., Gracias J., Watmuff B., Biag J.D., Thanos J.M., Whittredge P.B., Fu T., Worringer K., Brown H.E., Wang J. Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat. Neurosci. 2019;22:374–385. doi: 10.1038/s41593-018-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Levinson D.F., Duan J., Sanders A.R., Zheng Y., Pe'er I., Dudbridge F., Holmans P.A., Whittemore A.S., Mowry B.J. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T., Walters J.T.R., Johnstone M., Curtis D., Suvisaari J., Torniainen M., Rees E., Iyegbe C., Blackwood D., McIntosh A.M. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat. Genet. 2017;49:1167–1173. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H., Ophoff R.A., Steinberg S., Andreassen O.A., Cichon S., Rujescu D., Werge T., Pietilainen O.P., Mors O., Mortensen P.B. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan K.E., Baldeweg T., Friston K.J. Synaptic plasticity and dysconnection in schizophrenia. Biol. Psychiatry. 2006;59:929–939. doi: 10.1016/j.biopsych.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Szatkiewicz J.P., O'Dushlaine C., Chen G., Chambert K., Moran J.L., Neale B.M., Fromer M., Ruderfer D., Akterin S., Bergen S.E. Copy number variation in schizophrenia in Sweden. Mol. Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman H.D., Holtzman C.W., Ryan A.T., Shapiro D.I., MacDonald A.N., Goulding S.M., Brasfield J.L., Walker E.F. The development of psychotic disorders in adolescence: a potential role for hormones. Horm. Behav. 2013;64:411–419. doi: 10.1016/j.yhbeh.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas P.J. The adolescent brain: implications for the understanding, pathophysiology, and treatment of schizophrenia. Schizophrenia Bull. 2011;37:480–483. doi: 10.1093/schbul/sbr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdeolivas A., Tichit L., Navarro C., Perrin S., Odelin G., Levy N., Cau P., Remy E., Baudot A. Random walk with restart on multiplex and heterogeneous biological networks. Bioinformatics. 2019;35:497–505. doi: 10.1093/bioinformatics/bty637. [DOI] [PubMed] [Google Scholar]
- van Kesteren C.F., Gremmels H., de Witte L.D., Hol E.M., Van Gool A.R., Falkai P.G., Kahn R.S., Sommer I.E. Immune involvement in the pathogenesis of schizophrenia: a meta-analysis on postmortem brain studies. Transl. Psychiatry. 2017;7:e1075. doi: 10.1038/tp.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk D.W., Moroco A.E., Roman K.M., Edelson J.R., Lewis D.A. The role of the nuclear factor-kappaB transcriptional complex in cortical immune activation in schizophrenia. Biol. Psychiatry. 2019;85:25–34. doi: 10.1016/j.biopsych.2018.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters J.T., Rujescu D., Franke B., Giegling I., Vasquez A.A., Hargreaves A., Russo G., Morris D.W., Hoogman M., Da Costa A. The role of the major histocompatibility complex region in cognition and brain structure: a schizophrenia GWAS follow-up. Am. J. Psychiatry. 2013;170:877–885. doi: 10.1176/appi.ajp.2013.12020226. [DOI] [PubMed] [Google Scholar]
- Willsey A.J., Sanders S.J., Li M., Dong S., Tebbenkamp A.T., Muhle R.A., Reilly S.K., Lin L., Fertuzinhos S., Miller J.A. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Li X., Liu J., Luo X.J., Yao Y.G. SZDB2.0: an updated comprehensive resource for schizophrenia research. Hum. Genet. 2020;139:1285–1297. doi: 10.1007/s00439-020-02171-1. [DOI] [PubMed] [Google Scholar]
- Wu Y., Yao Y.G., Luo X.J. SZDB: a database for schizophrenia genetic research. Schizophrenia Bull. 2017;43:459–471. doi: 10.1093/schbul/sbw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Roos J.L., Dexheimer P., Boone B., Plummer B., Levy S., Gogos J.A., Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat. Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Bailey-Bucktrout S., Xi Y., Xu D., Du D., Zhang Q., Xiang W., Liu J., Melton A., Sheppard D. Innate antiviral host defense attenuates TGF-beta function through IRF3-mediated suppression of Smad signaling. Mol.Cell. 2014;56:723–737. doi: 10.1016/j.molcel.2014.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Coulombe-Huntington J., Kang S., Sheynkman G.M., Hao T., Richardson A., Sun S., Yang F., Shen Y.A., Murray R.R. Widespread expansion of protein interaction capabilities by alternative splicing. Cell. 2016;164:805–817. doi: 10.1016/j.cell.2016.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J.J., Barnes A.P., Hand R., Polleux F., Ehlers M.D. TGF-beta signaling specifies axons during brain development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Arbez N., Nucifora L.G., Sell G.L., Delisi L.E., Ross C.A., Margolis R.L., Nucifora F.C., Jr. A mutation in NPAS3 segregates with mental illness in a small family. Mol. Psychiatry. 2014;19:7–8. doi: 10.1038/mp.2012.192. [DOI] [PubMed] [Google Scholar]
- Zhang J., Jiang X., Zhang C., Zhong J., Fang X., Li H., Xie F., Huang X., Zhang X., Hu Q. Actin alpha 2 (ACTA2) downregulation inhibits neural stem cell migration through rho GTPase activation. Stem Cells Int. 2020;2020:4764012. doi: 10.1155/2020/4764012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(S1-1) All SCZ-candidate genes from original source datasets.
(S1-2) Core set of high-confidence SCZ-candidate genes.
(S2-1) Human interactome data (nodes).
(S2-2) Human interactome data (edges).
(S3-1) RWR ranking metrics (RWR score).
(S3-2) NW ranking metrics (R-scores and G-scores).
(S3-3) NW ranking metrics (nodes distance and edge weight).
(S3-4) NW ranking metrics (all neighborhoods).
(S4-1) RWR-extracted Network.
(S4-2) NW-extracted Network.
(S4-3) SCZ Network.
(S5-1) Differentially expressed genes (DEGs) in schizophrenic brains.
(S5-2) Exome variants identified in SCZ patients.
(S5-3) Enrichment results.
(S5-4) Over-represented MetaCore disease biomarkers for mental disorders in the expanded candidate genes and seed genes in the SCZ Network.
(S6-1) MetaCore pathways enriched with genes in the SCZ network.
(S6-2) Literature information of top enriched MetaCore pathways in the SCZ network.
(S6-3) KEGG pathways enriched with genes in the SCZ network.
(S6-4) Literature information of top enriched KEGG pathways in the SCZ network.
(S6-5) Immune genes.
(S6-6) Synaptic transmission genes.
(S6-7) Overrepresentation of immune genes in SCZ Network.
(S6-8) Overrepresentation of synaptic transmission genes in SCZ Network.
(S7-1) Co-expressed genes from 4 regions of the brain at different periods.
(S7-2) Overrepresentation of the SCZ Network genes in spatiotemporal phases.
(S7-3) Overrepresentation of non-immune genes of the SCZ Network in spatiotemporal phases.
(S7-4) Overrepresentation of immune genes of the SCZ Network in spatiotemporal phases.
(S7-5) Overrepresentation of synaptic transmission genes of the SCZ Network in spatiotemporal phases.
(S8-1) The expressed genes of L-S group.
(S8-2) The expressed genes of S-L group.
(S8-3) The expressed genes of L-L group.
(S8-4) The differentially expressed genes of L-S group.
(S8-5) The differentially expressed genes of S-L group.
(S8-6) The differentially expressed genes of L-L group.
(S9-1) Over-represented MetaCore disease biomarkers for mental disorders in the expanded candidate genes and the seed genes in three treatment groups.
(S9-2) Degree of DEGs in SCZ network.
(S9-3) Literature information of DEGs among the top 10% hub genes.
(S9-4) Metacore pathway maps enriched with DEGs in L-S group.
(S9-5) Metacore pathway maps enriched with DEGs in S-L group.
(S9-6) Metacore pathway maps enriched with DEGs in L-L group.
(S9-7) The 100 pathways enriched with three groups of DEGs and with genes in the SCZ Network.
(S9-8) Literature information for 18 representative pathways.
(S10-1) All motifs with enrichment pvalues by HOMER.
(S10-2) The binding motifs in the promoter regions of the DEGs.
(S10-3). Gene expression data of the corresponding TFs of the enriched motifs.
(S10-4) Metacore pathway maps enriched with targets in TF group I.
(S10-5) Metacore pathway maps enriched with targets in TF group II.
(S10-6) Metacore pathway maps enriched with targets in TF group III.
(S10-7) Metacore pathway maps enriched with targets in TF group IV.
(S10-8) Metacore pathway maps enriched with targets in TF group V.
(S10-9) Metacore pathway maps enriched with targets in TF group VI.
(S10-10) Literature information of top enriched pathways for target DEGs of 6 TF group.
(S10-11) Literature information of the predicted TFs in the SCZ Network.
Data Availability Statement
Data availability
Detailed analyzed data are included in supplemental excel tables. RNA-seq data is deposited at GEO (GEO: GSE156568).
Code availability
The original code for the calculation in the “neighborhood walk” approach is uploaded as ZIP file (Data S11).
Other source availability
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.