Summary
Acetylation coordinates many biological processes to ensure cells respond appropriately to nutrients. However, how acetylation regulates lipid surplus-induced inflammation remains poorly understood. Here, we found that a high-fat diet (HFD) enhanced mitochondrial fatty acid β-oxidation, which enhanced acetyl-CoA levels in the liver of the large yellow croaker. The HFD activated ACLY to govern the “citrate transport” to transfer acetyl-CoA from the mitochondria to the nucleus. Elevated acetyl-CoA activated CBP to increase p65 acetylation and then aggravated inflammation. SIRT1 was deactivated with a decline in NAD+/NADH, which further aggravated inflammation. Therefore, acetylation-dependent regulation of transcription factor activity is an adaptation to proinflammatory stimuli under nutrient stress, which was also confirmed in AML12 hepatocytes. In vitro octanoate stimulation further verified that acetyl-CoA derived from fatty acid β-oxidation mediated acetylation homeostasis in the nucleus. The broad therapeutic prospects of intermediate metabolites and acetyltransferases/deacetylases might provide critical insights for the treatment of metabolic diseases in vertebrates.
Subject areas: Pathophysiology, Cellular physiology, Immunology
Graphical Abstract

Highlights
-
•
Lipid surplus improved acetyl-CoA levels and enhanced ac-p65 in the liver
-
•
Mitochondrial fatty acid β-oxidation provided acetyl-CoA for p65 acetylation
-
•
CBP and SIRT1 regulated p65 acetylation under lipid surplus
Pathophysiology; Cellular physiology; Immunology
Introduction
Lipid uptake and utilization support cell viability and functions by maintaining membrane structure, cell recognition, and energy homeostasis (Alessandrini and Facci, 2011). However, excessive lipid uptake generally induces metabolic syndrome and insulin resistance associated with chronic inflammation (Weisberg et al., 2006). During the past decades, problems involving lipid surplus-related disturbance of metabolic and immune processes have become more common due to high-fat diet (HFD) consumption (Yu et al., 2002). Hence, approaches aimed at clarifying the mechanism and finding suitable therapeutic targets are very important for human health.
Nutritional disease can be attributed to metabolic disorders caused by nutrient stress (Hotamisligil, 2006). Cells response to nutrient stress not only by activating pathways such as the AMP-activated protein kinase (AMPK) (Garcia and Shaw, 2017) and the mechanistic target of rapamycin (mTOR) pathways (Ben-Sahra and Manning, 2017), but also by altering enzymatic activity related to posttranslational modifications (PTMs). Metabolic intermediates can provide substrates for many PTM enzymes, such as acetyl-coenzyme A (acetyl-CoA) for acetylation, uridine 5′-diphospho-N-acetylglucosamine (UDP-GlcNAc) for O-GlcNAcylation, and S-adenosylmethionine for methylation (Campbell and Wellen, 2018). Correct integration of these metabolites and signals is critical to health, as protein disorders and dysregulated histone modifications are associated with multiple diseases (Kaelin and McKnight, 2013).
With the development of mass spectrometry (MS), the roles of acetylation modifications in integration of metabolism and immunity have been gradually elucidated by regulating enzymatic activity, protein subcellular localization, protein-protein interactions, and gene expression (Zaini et al., 2018). Importantly, acetylation homeostasis changes dynamically with the content of acetyl-CoA. Many lysine acetyltransferases (KATs) have a relatively high dissociation constant (KD) for acetyl-CoA (i.e., a low affinity), such that the abundance of acetyl-CoA affects the catalytic activity of KATs (Pietrocola et al., 2015). Acetyl-CoA is compartmentalized into mitochondrial and cytoplasmic/nuclear pools (Takahashi et al., 2006). In general, glucose, glutamine, and fatty acids contribute to the production of acetyl-CoA in mitochondria (Donohoe et al., 2012; Wellen et al., 2009; Yang et al., 2014a). The generation of cytoplasmic/nuclear acetyl-CoA depends mainly on the export of citrate from mitochondria, followed by ATP-citrate lyase (ACLY)-mediated catalysis into acetyl-CoA and oxaloacetate (Lee et al., 2014). ACLY is an important enzyme regulating lipid and cholesterol biosynthesis (Ference et al., 2019). Apart from being regulated by sterol regulatory element-binding protein 1α (SREBP-1α) at the transcriptional level (Sato et al., 2000), ACLY is activated by Akt-dependent phosphorylation of Ser455 during nutrient stress (Migita et al., 2008). Activated ACLY controls the acetylation of nuclear transcription factors and histones to regulate gene expression (Sivanand et al., 2017). Hence, cells possess precise epigenetic mechanisms to adapt to nutrient signals and coordinate gene expression programs (McDonnell et al., 2016). As details of acetylation have gradually been revealed, acetylation and inflammation have remained popular research topics in the field of immunity (Song et al., 2020). Moreover, research focus has also shifted from broad histone acetylation to specific targets aimed at pathogen recognition receptors (PRRs) and transcription factors (Shakespear et al., 2011).
Recently, the functions of glucose oxidation in fueling the production of acetyl-CoA and aggravating inflammation have been revealed (Langston et al., 2019; Lauterbach et al., 2019). Other nutrient sources besides glucose have also been shown to regulate acetylation homeostasis (Falcone and Maddocks, 2020; Soliman and Rosenberger, 2011). McDonnell et al. (2016) and Donohoe et al. (2012) emphasized that lipid metabolism can also provide acetyl-CoA for protein acetylation. However, it is poorly understood whether acetyl-CoA is involved in the regulation of inflammation under HFD conditions. Hence, we investigated whether a lipid surplus could provide acetyl-CoA to alter the acetylation modification of nuclear proteins and subsequently affect inflammation. Although fish are relatively lower vertebrates, they have evolutionarily conserved systems for nutrient- and pathogen-sensing and immune response pathways (Hotamisligil, 2006). In aquatic environments, fish are subjected to many pathogen-associated molecular patterns (PAMPs) (Li et al., 2020), and HFD feeding is extensively used because of the protein-sparing effect (Wang et al., 2016). Hence, fish is an appropriate model for overnutrition and inflammation studies. In the current study, alpha mouse liver 12 (AML12) hepatocytes were also used to verify the conservation of the findings between teleosts and mammals. The present study expands the understanding of inflammation caused by overnutrition from the perspective of epigenetics, which contributes to the search for appropriate therapeutic targets for metabolic diseases.
Results
HFD-induced hepatic inflammatory responses were associated with enhanced acetylation levels of p65
To establish a lipid surplus-induced liver inflammation model in the large yellow croaker, a 10-week HFD feeding experiment was carried out. There was no significant difference in the final body weight between the control group and the HFD group (Figure 1A). However, fish fed an HFD showed greater lipid accumulation in the liver than fish fed a control diet, as revealed by liver triglycerides (TG) content analysis (Figure 1B) and hematoxylin and eosin (H&E) staining of liver sections (Figure 1D). The free fatty acid levels in plasma were also significantly higher in the HFD group (Figure 1B). The elevations in activities of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) in plasma under HFD feeding suggested that excessive lipid accumulation induced liver injury (Figure 1C). In addition, H&E staining of liver sections showed that HFD feeding promoted the infiltration of inflammatory cells (Figure 1D). Correspondingly, compared to the control group, the HFD group exhibited significantly higher mRNA levels of proinflammatory cytokines (Figure 1E), which was consistent with the findings of our previous study (Wang et al., 2016). In particular, HFD feeding significantly enhanced the protein expression of COX-2 and cleaved IL-1β and secreted IL-1β in plasma (Figures 1E and1F). Furthermore, the elevation in p65 nuclear localization indicated that the NF-κB pathway was activated by HFD feeding (Figure 1G). In the nucleus, p65-mediated modulation of gene transcription is regulated by multiple coactivators and corepressors, such as KATs and lysine deacetylases (KDACs) (Chen et al., 2001). In the present study, HFD feeding significantly increased mRNA levels of all tested KATs except for p300/CBP-associated factor (Pcaf) and increased the protein levels of CREB-binding protein (CBP) (Figure 1H). In addition, HFD feeding significantly increased the total KAT activity (Figure 1I). In contrast, the mRNA level of nicotinamide-nucleotide adenylyltransferase 1 (Nmnat1), which is associated with NAD synthesis, was significantly reduced by HFD feeding (Figure 1H). Relatedly, the mRNA and protein expression levels of sirtuin 1 (SIRT1) were significantly decreased as the NAD+/NADH ratio decreased (Figures 1H and1J). The breakdown of the balance between KATs and KDACs under HFD feeding increased the acetylation level of p65 (Figure 1K). To further investigate the effect of acetylation modification on p65 transcriptional activity, dual luciferase experiments and electrophoretic mobility shift assay (EMSA) were conducted to show that site-specific mutation of p65 at Lys63 and Lys147 promoted p65 transcriptional activity by enhancing its DNA binding activity (Data S1 and S2,Figures 1L–1n). Collectively, these results indicated that HFD may have broken down acetylation homeostasis to increase the acetylation level of p65, which enhanced its transcriptional activity and aggravated inflammation.
Figure 1.
HFD-induced hepatic inflammatory responses were associated with enhanced acetylation levels of p65
(A) Final body weight of large yellow croaker after treatment with different diets (n = 12).
(B) Liver TG levels and free fatty acids in plasma were analyzed after treatment with different diets (n = 4).
(C) ALT and AST in plasma were analyzed after treatment with different diets (n = 4).
(D) H&E staining of liver sections from different diet groups (n = 4). Scale bar, 50 μm.
(E) The mRNA expression levels of hepatic inflammation related genes (Il-1β, Tnf-α andCox-2) and secreted IL-1β in the plasma were analyzed after treatment with different diets (n = 4).
(F) COX-2 and cleaved IL-1β in the liver were examined by Western blot analysis after treatment with different diets (n = 3).
(G) Nuclear p65 in the liver was examined by Western blot analysis and quantitated after treatment with different diets (n = 3).
(H) The mRNA expression levels of key hepatic deacetylase and acetyltransferase genes (Nmnat1, Sirt1, Hdac3, Cbp, Gcn5, Pcaf and Tip60) were analyzed (n = 4). CBP and SIRT1 in the liver were examined by Western blot and quantitated after treatment with different diets (n = 3).
(I)The activity of hepatic KATs was analyzed after treatment with different diets (n = 3).
(J) The NAD+/NADH ratio was analyzed after treatment with different diets (n = 3).
(K) The acetylation level of p65 in the liver was examined by Western blot analysis and quantitated after treatment with different diets (n = 3).
(L) Site-specific mutation of p65 at Lys63 and Lys147 altered the acetylation level of p65 in HEK293T cells.
(M) Site-specific mutation of p65 at Lys63 and Lys147 altered the transcriptional activity of p65 in HEK293T cells (n = 3).
(N) Site-specific mutation of p65 at Lys63 and Lys147 altered the DNA binding activity of p65 in HEK293T cells. The results are presented as the mean ± SD and were analyzed using independent t-tests (∗p< 0.05). See also Table S1 and Data S1 and S2.
HFD increased acyl-CoA levels and coordinately acetylated multiple targets in metabolic pathways
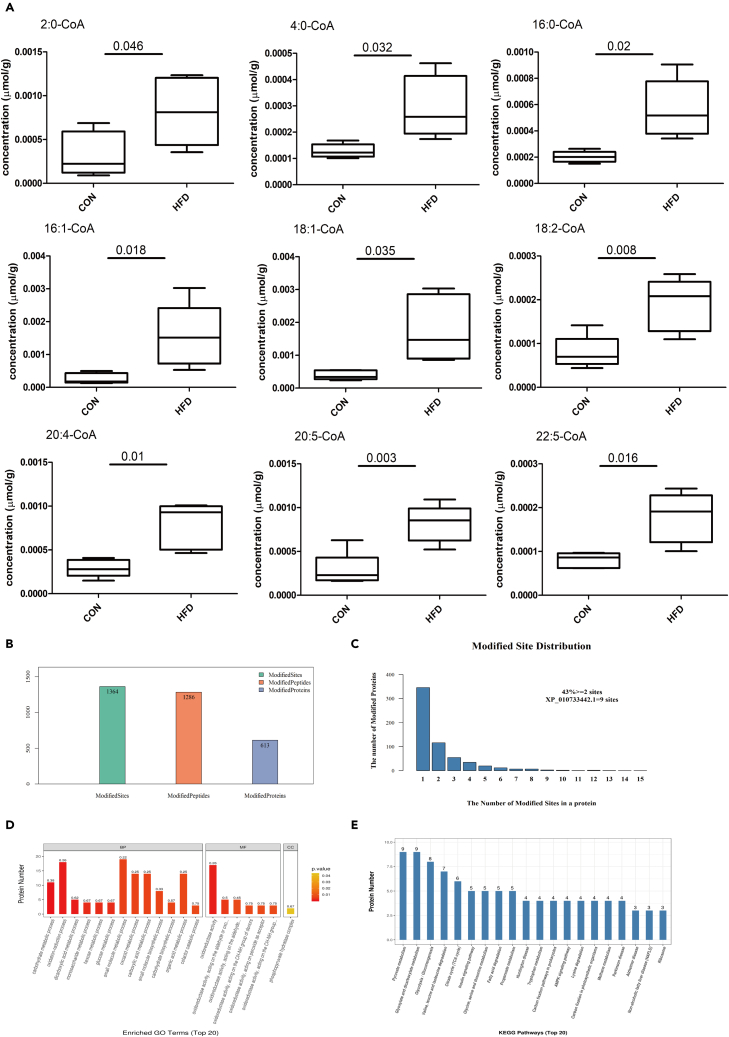
Acetyl-CoA is the direct acetyl donor of acetylation (Menzies et al., 2016), and the elevation in p65 acetylation indicated that HFD may affect acetyl-CoA levels. Therefore, liquid chromatography-MS (LC-MS) was conducted to test the effects of HFD feeding on fatty acyl-CoA levels. We found that the HFD significantly increased the contents of 2:0-CoA, 4:0-CoA, 16:0-CoA, 16:1-CoA, 18:1-CoA, 18:2-CoA, 20:4-CoA, 20:5-CoA and 22:5-CoA in the liver (Figure 2A, Data S3). In order to determine the changes in the lysine acetylation landscape that occurred during the HFD-induced increases in acetyl-CoA abundance, LC-MS/MS was conducted to identify and quantify acetylated lysine (ac-K) peptides. In total, 613 acetylated proteins, 1,286 acetylated peptides, and 1,364 acetylation sites were identified by label-free quantitative proteomics (Figure 2B). Almost half of all proteins were multiacetylated, with 43% having two or more ac-K sites (Figure 2C, Data S4). Acetylation levels at 121 sites among the 1,364 identified ac-K sites were significantly increased or were detected only in the HFD group, whereas only 19 ac-K sites were significantly decreased or were detected only in the control group (Data S5). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were used to further understand the functions and features of differentially expressed acetylated peptides following feeding of different diets. GO analysis showed that differentially expressed acetylated peptides were enriched mainly in carbohydrate metabolic processes, oxidation reduction processes, small molecule metabolic processes, and carboxylic acid metabolic processes (Figure 2D). KEGG pathway analysis showed that most of the differentially expressed acetylated peptides were involved in pyruvate metabolism, glyoxylate/dicarboxylate metabolism, glycolysis/gluconeogenesis, amino acid metabolism, the tricarboxylic acid (TCA) cycle, the insulin signaling pathway and fatty acid degradation (Figure 2E). The high incidence of metabolic enzymes found via LC-MS/MS analysis indicates a key role of acetylation in regulating cellular metabolism (Table S2). Therefore, we demonstrated that HFD feeding was responsible for a striking increase in acetylation of a variety of metabolic enzymes and transcription factors in the whole liver.
Figure 2.
HFD increased acyl-CoA levels and coordinately acetylated multiple targets in metabolic pathways
(A) The levels of acyl-CoA in the liver were measured using LC-MS analysis after treatment with different diets (n = 5).
(B)Number of lysine acetylation sites and proteins identified and quantified by LC-MS/MS of the acetylome.
(C) Distribution of acetylated proteins based on the number of lysine acetylation sites.
(D) GO functional classification of the differentially expressed proteins after treatment with different diets (n = 3).
(E) KEGG pathway-based enrichment analysis of the identified proteins (n = 3). The differences between experimental groups were tested using independent t-tests (the p values of each acyl-CoA were shown above the horizontal line in each graph). See also Figure S1, Table S2, and Datas S3–S5.
Activation of fatty acid oxidation fueled p65 acetylation and inflammatory gene expression under lipid surplus
Acetyl-CoA can be produced from multiple substrates, including glucose, amino acids, fatty acids, and glutamine. In the present study, the elevation in fatty acyl-CoA induced by HFD feeding indicated that mitochondrial fatty acid β-oxidation might have been activated. Indeed, HFD feeding significantly increased fatty acid β-oxidation-related gene expression (Figure 3A). In addition, the HFD also increased the activity of carnitine palmitoyltransferase 1A (CPT1α) (Figure 3A) and protein expressions of long-chain specific acyl-CoA dehydrogenase (LCAD) and medium-chain specific acyl-CoA dehydrogenase (MCAD) (Figure 3B). Hence, we speculated that activation of mitochondrial fatty acid β-oxidation might provide acetyl-CoA to influence acetylation homeostasis. To explore the mechanism by which lipid surplus increased the levels of acetylated p65 (ac-p65) and to clarify the source of acetyl-CoA, we treated fish hepatocytes with palmitate (PA) according to acyl-CoA levels and fatty acid compositions in the experimental diets. Similar to the above observations in vivo, PA increased the mRNA levels of proinflammatory cytokines associated with the increase in ac-p65 (Figures 3C and3F). PA also increased the expression levels of mRNA and proteins related to fatty acid oxidation (Figures 3D and3E). Importantly, inhibition of fatty acid utilization with etomoxir attenuated PA-induced acetylation of p65 and mRNA expression of Cox-2 and Tnf-α (Figures 3F and3G). These results indicated that mitochondrial fatty acid oxidation regulated p65 acetylation and inflammatory gene expression by integrating acetyl-CoA metabolism under lipid surplus.
Figure 3.
Activation of fatty acid oxidation fueled p65 acetylation and inflammatory gene expression under lipid surplus
(A) The mRNA expression levels of genes associated with fatty acid oxidation (Cpt1α, Cpt1β, Pgc1-α and Acat2) and the activity of CPT1α were measured after treatment with different diets (n = 4).
(B) The levels of LCAD and MCAD in the liver were examined by Western blot analysis and quantitated after treatment with different diets (n = 3).
(C) The mRNA expression levels of inflammatory genes (Tnf-α, Cox-2, Il-1β and Il-6) were analyzed under PA treatment (n = 3).
(D) The levels of LCAD and PPARα were examined by Western blot analysis and quantitated after treatment with PA (n = 3).
(E) The mRNA expression levels of genes associated with fatty acid β-oxidation (Pparα, Cpt1α, Cpt1β, Acads and Acat2) were analyzed after 100 μM PA treatment for 12 h (n = 3).
(F) Inhibiting fatty acid oxidation with etomoxir blocked the increase in ac-p65 induced by PA (n = 3).
(G) Inhibiting fatty acid oxidation with etomoxir attenuated the increase in inflammatory gene (Cox-2 and Tnf-α) expression induced by PA (n = 3).
(H) The acetyl-CoA levels were analyzed after 2 mM acetate treatment for 12 h (n = 3).
(I)The acetylation level of p65 was examined by Western blot analysis after 2 mM acetate treatment for 12 h (n = 3).
(J) Nuclear p65 in hepatocytes was examined by Western blot analysis after treatment with 2 mM acetate for 12 h (n = 3).
(K) The mRNA expression levels of inflammatory genes (Tnf-α, Cox-2, Il-1β and Il-6) were analyzed after treatment with acetate for 12 h (n = 3).
(L) The mRNA expression levels of inflammatory genes (Tnf-α and Il-1β) were analyzed after PA and acetate treatment for 12 h (n = 3) . The results are presented as the mean ± SD and were analyzed using independent t-tests (∗p< 0.05, ∗∗∗p< 0.001).
On the basis of the above observations, we wondered whether increases in acetyl-CoA levels independent of lipid oxidation would also activate p65 and cause inflammation. Hence, sodium acetate was added to complete medium, which enabled generation of acetyl-CoA by acetyl-CoA synthetase 2 (ACSS2). Acetate addition significantly increased intracellular acetyl-CoA levels (Figure 3H). However, acetate alone did not promote the nuclear localization and acetylation of p65 (Figures 3I and3J). In addition, acetate had no significant effects on the mRNA expression levels of proinflammatory cytokines (Figure 3K). In contrast, acetate aggravated PA-induced mRNA expression of proinflammatory cytokines (Il-1β and Tnf-α) (Figure 3L). Overall, the findings indicated that acetyl-CoA, a crucial and ubiquitous intermediate metabolite, was not able to induce inflammation alone, but functioned like a second messenger under PA treatment to exacerbate the inflammatory response.
Activation of ACLY supported increases in ac-p65 levels and inflammatory gene expression under lipid surplus
Acetyl-CoA derived from mitochondrial fatty acid β-oxidation cannot penetrate the mitochondrial membrane and enter the cytoplasm freely. However, in the present study, apart from the transcription factor p65, 37 of the 103 differentially expressed proteins in the HFD group were located in mitochondria, while 66 were located outside mitochondria (Table 1). Therefore, we speculated that ACLY might promote the transport of acetyl-CoA from mitochondria to cytosol under lipid surplus. HFD feeding significantly increased the phosphorylation of ACLY at Ser455 (Figure 4A). Correspondingly, PA also significantly increased the phosphorylation of ACLY at Ser455 in vitro (Figure 4B), which indicated that lipid surplus activated ACLY. To further investigate the role of ACLY in lipid surplus-induced inflammation and increases in ac-p65 levels, fish hepatocytes were incubated with the ACLY inhibitor SB-204990 before PA treatment. Inhibition of ACLY decreased the content of acetyl-CoA in hepatocytes (Figure 4C). Moreover, inhibition of ACLY attenuated PA-induced p65 acetylation (Figure 4D) and decreased the mRNA levels of Il-6 and Tnf-α (Figure 4E). ACLY phosphorylation is regulated by Akt (Lee et al., 2014), and treatment with the Akt inhibitor MK-2206 blocked PA-induced mRNA expression of Il-6 and Tnf-α by inhibiting ACLY (Figures 4F and4G). Overall, these findings indicated that ACLY activation governed the citrate-malate-pyruvate shuttle to support p65 acetylation and enhance inflammatory gene expression under lipid surplus.
Table 1.
The subcellular localization distribution of differential proteins
| Subcellular localization | CON | HFD |
|---|---|---|
| Acetyl sites | 94 | 128 |
| Acetyl proteins | 72 | 103 |
| Mitochondrial acetyl sites | 25 | 43 |
| Mitochondrial acetyl proteins | 23 | 37 |
| Out-mitochondrial acetyl sites | 69 | 85 |
| Out-mitochondrial acetyl proteins | 49 | 66 |
Figure 4.
Activation of ACLY supported increases in ac-p65 levels and inflammatory gene expression under lipid surplus
(A) The levels of total and phosphorylated ACLY were examined by Western blot analysis and quantitated after treatment with different diets (n = 3).
(B) The levels of total and phosphorylated ACLY were examined by Western blot analysis and quantitated after treatment with PA in fish hepatocytes (n = 3).
(C) Inhibition of ACLY with SB-204990 decreased the content of acetyl-CoA in fish hepatocytes (n = 3).
(D) Inhibiting ACLY with SB-204990 blocked the increase in ac-p65 induced by PA in fish hepatocytes (n = 3).
(E) Inhibiting ACLY with SB-204990 attenuated the increase in inflammatory gene (Tnf-α andIl-6) expression induced by PA in fish hepatocytes (n = 3).
(F) Inhibiting Akt with MK2206 attenuated the increase in inflammatory gene (Tnf-α andIl-6) expression induced by PA in fish hepatocytes (n = 3).
(G) Inhibiting Akt with MK2206 attenuated PA-induced increase in p-ACLY in fish hepatocytes (n = 3). The results are presented as the mean ± SD and were analyzed using independent t-tests (∗p< 0.05).
CBP and SIRT1 were involved in the regulation of ac-p65 under PA treatment
An increase in the ac-p65 level relies not only on the source of acetyl-CoA, but also on the participation of KATs and KDACs. First, we investigated the mRNA levels of KATs, including Cbp (p300 family), Pcaf, and Gcn5 (GCN5 family). PA significantly increased the mRNA levels of Cbp and Gcn5, while it decreased the mRNA level of Pcaf (Figure 5A). To further explore the effects of KATs on the transcriptional activity of p65, PCAF, CBP, GCN5, and p65 were transfected into HEK293T cells for dual luciferase experiments. As revealed in Figure 5B, PCAF and CBP significantly increased the transcriptional activity of p65. The modification of transcription factors by KATs generally involves direct physical binding. In the present study, CBP and PCAF directly bound to p65 (Figure 5C). In addition, CBP increased the acetylation of p65 (Figure 5D). Moreover, pretreatment of hepatocytes with the CBP inhibitor c646 blocked PA-induced increases in ac-p65 and proinflammatory cytokine expression (Figures 5E and5F). Hence, we speculated that CBP played a vital role in p65 acetylation during PA treatment. Lysine acetylation is a reversible PTM, which suggests that specific KDACs may be responsible for p65 deacetylation. Notably, PA decreased the mRNA levels of nicotinamide phosphoribosyltransferase (Nampt), Nmnat1, and Sirt1, as well as the protein expression of SIRT1 (Figures 5G and5H). Dual luciferase experiments showed that SIRT1 significantly decreased the transcriptional activity of p65 (Figure 5I). Furthermore, SIRT1 decreased the acetylation level of p65 by directly binding with p65 (Figure 5J). Pretreatment of hepatocytes with SIRT1 activator resveratrol attenuated PA-induced increase in proinflammatory cytokine expression (Figure 5K). Collectively, our results suggested that CBP and SIRT1 were involved in the regulation of p65 acetylation and influenced inflammation under PA treatment.
Figure 5.
CBP and SIRT1 were involved in the regulation of ac-p65 under PA treatment
(A) The mRNA expression levels of key hepatic acetyltransferase genes (Cbp, Gcn5 and Pcaf) were analyzed after PA treatment in fish hepatocytes (n = 3).
(B) CBP and PCAF enhanced the transcriptional activity of p65 in HEK293T cells (n = 3).
(C) CBP and PCAF directly bound to p65 in HEK293T cells.
(D) CBP increased the acetylation level of p65 in HEK293T cells.
(E) Inhibiting CBP with c646 blocked the increase in ac-p65 induced by PA in fish hepatocytes (n = 3).
(F) Inhibiting CBP with c646 attenuated the increase in inflammatory gene (Il-1β and Il-6) expression induced by PA in fish hepatocytes (n = 3).
(G) The mRNA expression levels of key hepatic deacetylase genes (Nampt, Nmnat1, Sirt1 and Hdac3) were analyzed after PA treatment in fish hepatocytes (n = 3).
(H) The levels of SIRT1 were examined by Western blot analysis and quantitated after treatment with PA in fish hepatocytes (n = 3).
(I) SIRT1 decreased the transcriptional activity of p65 in HEK293T cells (n = 3).
(J) SIRT1 can directly bind to p65, and SIRT1 decreased the acetylation level of p65 in HEK293T cells.
(K) Activating SIRT1 with resveratrol attenuated the increase in inflammatory gene (Tnf-α andCox-2) expression induced by PA in fish hepatocytes (n = 3). The results are presented as the mean ± SD and were analyzed using independent t-tests (∗p< 0.05) and Tukey’s tests (bars bearing different letters are significantly different among treatments (p< 0.05)).
PA enhanced p65 acetylation to aggravate inflammation in AML12 hepatocytes
To extend our observations to additional cell types, an inflammatory model was established by treating alpha mouse liver 12 (AML12) hepatocytes with 500 μM PA for 24 h (Figure 6A). As shown in Figure 6B, PA significantly increased intracellular acetyl-CoA levels. In addition, PA enhanced the acetylation level of p65 in a dose-dependent manner, which was consistent with its effects in fish. Based on previous studies revealing the enhancement of p65 transcriptional activity by acetylation (Chen et al., 2001), we hypothesized that increased p65 acetylation might aggravate PA-induced inflammation. Similar to the effects in fish, PA treatment enhanced the mRNA expression of genes involved in fatty acid β-oxidation (Figure 6D). Notably, inhibition of fatty acid β-oxidation with etomoxir alleviated PA-induced acetylation of p65 (Figure 6E) and mRNA expression of Il-18 (Figure 6F). These results indicated that fatty acid β-oxidation drove p65 acetylation and exacerbated inflammation under PA treatment, which was conserved between fish and mammals. The role of ACLY during PA stimulation was also tested. The response of ACLY to PA was not as obvious as that in fish. PA significantly enhanced the phosphorylation of ACLY at Ser455 after 12 h of treatment but not after 24 h (Figure 6G). Hence, we hypothesized that ACLY might have played a role in the early stages of the response to PA stimulation. To test the above hypothesis, cells were preincubated with SB-204990 before PA treatment. As shown in Figures 6H and 6I, inhibition of ACLY blocked the increases in p65 acetylation and Il-6 mRNA expression induced by PA. Therefore, the early activation of ACLY was critical for the conversion of acetyl-CoA between mitochondria and the cytosol/nucleus under PA treatment.
Figure 6.
PA enhanced p65 acetylation to intensify inflammation in AML12 hepatocytes
(A) The mRNA expression levels of inflammatory genes (Cox-2, Il-6 and Il-18) were analyzed after treatment with different concentrations of PA for 24 h (n = 3).
(B) The acetyl-CoA levels were analyzed after 500 μM PA treatment for 24 h (n = 3).
(C) The acetylation level of p65 was examined by Western blot analysis and quantitated after treatment with different concentrations of PA for 24 h (n = 3).
(D) The mRNA expression levels of genes associated with fatty acid oxidation (Cpt1α, Acaa2 and Ech1) were analyzed after treatment with different concentrations of PA for 24 h (n = 3).
(E) Inhibition of fatty acid oxidation with etomoxir blocked the increase in ac-p65 induced by PA (n = 3).
(F) Inhibition of fatty acid oxidation with etomoxir relieved the increase in Il-18 mRNA expression induced by PA (n = 3).
(G) The levels of total and phosphorylated ACLY were examined by Western blot analysis and quantitated after treatment with different concentrations of PA for 12 h and 24 h (n = 3).
(H) Inhibition of ACLY with SB-204990 blocked the increase in ac-p65 induced by PA (n = 3).
(I) Inhibition of ACLY with SB-204990 attenuated the increase in Il-6 mRNA expression induced by PA (n = 3).
(J) The mRNA expression levels of KATs (Cbp, Gcn5 and Pcaf) and Sirt1 were analyzed after 500 μM PA treatment for 24 h (n = 3).
(K) Inhibition of GCN5 with MB-3 relieved the increase in Il-18 mRNA expression induced by PA (n = 3).
(L) KDAC inhibitor treatment further increased the mRNA expression levels of inflammatory genes (Il-6 and Il-18) under PA treatment (n = 3). The results are presented as the mean ± SD and were analyzed using independent t-tests (∗p< 0.05) and Tukey’s tests (bars bearing different letters are significantly different among treatments (p< 0.05)).
In addition to acetyl-CoA metabolism, the KATs and KDACs involved in inflammatory regulation under PA treatment were also explored in AML12 hepatocytes. Unlike in fish hepatocytes, PA did not significantly affect the mRNA expression of Cbp in AML12 hepatocytes (Figure 6J). In contrast, PA significantly increased the mRNA expression of Gcn5 (Figure 6J). Moreover, inhibition of GCN5 with MB-3 attenuated PA-induced increases in Il-18 mRNA levels (Figure 6K). Hence, GCN5, rather than CBP relieved PA-induced inflammation in AML12 hepatocytes. Surprisingly, PA did not significantly affect the mRNA expression of Sirt1 (Figure 6J). However, incubation with broad-spectrum KDAC inhibitors significantly increased the mRNA expression of proinflammatory cytokines induced by PA (Figure 6L). Therefore, it was determined that KDACs participated in the regulation of inflammation under PA. However, the specific KDAC responsible for inflammatory regulation needs further study.
Simulation of lipid oxidation with octanoate increased the acetylation of histone H3
To further investigate whether lipid oxidation provides a carbon source for nuclear protein acetylation in the same manner under physiological conditions, octanoate was used to drive lipid oxidation (Currie et al., 2013) without inducing inflammation (Figures S2A–S2C). In the present study, 2 mM octanoate significantly increased the mRNA expression of genes involved in lipid oxidation (Figure 7A). Furthermore, octanoate enhanced the protein expression of MCAD (Figure 7B), which suggested that it increased fatty acid oxidation. In particular, octanoate enhanced histone H3 acetylation in a dose-dependent manner (Figure 7B). Similar to addition of PA, addition of octanoate increased the phosphorylation of ACLY at Ser455 (Figure 7B). To determine whether octanoate-induced increase in acetylated H3 (ac-H3) was dependent on ACLY, SB-204990 was used to inhibit ACLY before octanoate treatment. As shown in Figure 7C, SB-204990 blocked the increase in ac-H3 induced by octanoate.
Figure 7.
Simulating lipid oxidation with octanoate increased the acetylation of histone H3
(A) The mRNA expression levels of key hepatic acetyltransferases (Cbp, Gcn5, Tip60 and Pcaf) and fatty acid oxidation genes (Ehhadh, Acat1, Acat2, Asads and Pparα) were analyzed under octanoate treatment in fish hepatocytes (n = 3).
(B) The levels of total and phosphorylated ACLY, Akt, AMPK, and MCAD and the acetylation of H3 were examined by Western blot analysis and quantitated after treatment with octanoate in fish hepatocytes (n = 3).
(C) Inhibition of ACLY blocked the increase in ac-H3 induced by octanoate in fish hepatocytes (n = 3).
(D) Inhibiting Akt with MK2206 blocked the increase in ac-H3 under octanoate in fish hepatocytes (n = 3).
(E) Activating AMPK with AICAR attenuated the increase in ac-H3 under octanoate in fish hepatocytes (n = 3).
(F) Octanoate reduced SIRT1 protein expression in a dose-dependent manner in fish hepatocytes (n = 3).
(G) Resveratrol attenuated the increase in ac-H3 induced by octanoate in fish hepatocytes (n = 3). The results are presented as the mean ± SD and were analyzed using independent t-tests (∗p< 0.05) and Tukey’s tests (bars bearing different letters are significantly different among treatments (p< 0.05)). See also Figures S2 and S3.
Furthermore, octanoate increased the phosphorylation of Akt at Ser473 (Figure 7B) and decreased the phosphorylation of AMPK at Thr172 (Figure 7E). Thus, we wondered whether Akt and AMPK were related to the H3 acetylation induced by octanoate. In the present study, inhibition of Akt with MK-2206 significantly decreased the phosphorylation of ACLY at Ser455 and attenuated octanoate-induced increase in ac-H3 (Figure 7D). Thus, we speculated that octanoate increased ac-H3 in an Akt/ACLY-dependent manner. Interestingly, the AMPK activator 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) also blocked octanoate-induced increase in ac-H3 without inhibiting the phosphorylation of ACLY at Ser455 (Figure 7E), which suggested that there was another underlying mechanism controlling ac-H3 during octanoate treatment. Recently, AMPK was found to enhance SIRT1 activity by increasing cellular NAD+ levels and modulated the activity of downstream SIRT1 targets (Cantó et al., 2009). In the present study, octanoate dose-dependency decreased the protein expression of SIRT1 (Figure 7F) and AICAR counteracted this decline (Figure 7E). Hence, AICAR might enhance the activity of SIRT1 and reverse octanoate-induced increase in ac-H3. Correspondingly, resveratrol treatment inhibited octanoate-induced increase in ac-H3 (Figure 7G), which further confirmed that SIRT1 played an important role in the regulation of histone H3 acetylation under octanoate treatment. However, treatment with etomoxir to inhibit CPT1α did not block octanoate-induced increase in ac-H3 (Figure S3). This result is in accordance with the idea that octanoate can enter mitochondria without CPT1α. Collectively, our results demonstrate that octanoate increases fatty acid oxidation and increases ac-H3 by activating the Akt/ACLY signaling pathway and inhibiting the AMPK/SIRT1 signaling pathway, which further confirms that mitochondrial fatty acid β-oxidation can provide carbon sources for acetylation modification of nuclear proteins.
Discussion
Aquatic animals are subjected to chronic inflammation due to the aquatic environment and HFD, which seriously restricts the development of aquaculture. Correspondingly, chronic inflammation, as a complication of metabolic syndrome such as obesity and insulin resistance, also severely infects people's health (Chalkiadaki and Guarente, 2012). Although it inspired intensive research seeking to elucidate the cellular and molecular mechanism in mammals (Ogden et al., 2014), little information is available in fish. Recent studies have proposed that intermediate metabolites can not only perform normal coenzyme functions, but also serve as substrates for PTMs to regulate cellular processes (Menzies et al., 2016). Given the discovery of notable signaling roles for intermediary metabolites, we focused on the metabolic flow of acetyl-CoA and aimed to develop nutrition strategies to relieve inflammation and produce green and safe food for humans.
Acetylation modification is an evolutionarily conserved PTM that dynamically regulates metabolism and immunity by affecting enzyme or transcription factor activity (Narita et al., 2019). NF-κB/p65 is a major proinflammatory nuclear transcription factor that can directly increase inflammatory gene expression (Jimenez-Gomez et al., 2013). Emerging evidence has shown that HFD consumption activates the p38 and JNK/MAPK pathways to promote IκB degradation and p65 nuclear translocation (Wang et al., 2016), which was confirmed in the present study. However, degradation of IκB and nuclear translocation of p65 do not maximize the transcriptional activity of p65 (Chen et al., 2005; Ishinaga et al., 2007). Moreover, p65 preferentially undergoes PTMs to enhance the transcriptional activity in nucleus. For example, acetylation at Lys310 can increase the transcriptional activity of p65 in mammals (Chen et al., 2002). However, how nutrient stress affects the acetylation level of p65 remains poorly understood. HFD feeding significantly enhanced the acetylation level of p65 in the present study. LC-MS/MS was conducted to detect possible acetylation sites of p65. A total of nine sites that may undergo acetylation modification (such as Lys57, Lys63, and Lys124) were detected. Single-point mutations at several sites of p65 did not have significant impacts on the levels of acetylation modification. In contrast, mutation of Lys63 and Lys147 to glutamine significantly increased the acetylation level of p65 and enhanced p65 transcriptional activity by increasing DNA binding activity, which further confirmed that an elevation in ac-p65 might aggravate inflammation under lipid surplus. The acetylation of p65 requires acetyl-CoA as an acetyl donor, and the increase in KAT expression and activity in the present study indicated that HFD feeding may increase the content of acetyl-CoA. As expected, LC-MS analysis showed that two short-chain acyl-CoAs (2:0-CoA and 4:0-CoA) and seven long-chain acyl-CoAs (16:0-CoA, 16:1-CoA, 18:1-CoA, 18:2-CoA, 20:4-CoA, 20:5-CoA and 22:5-CoA) displayed significant elevations in the HFD group. Changes in C16 and C18 acyl-CoA levels indicated that HFD feeding might have promoted mitochondrial fatty acid β-oxidation, which was confirmed by increases in the expression of related genes and the enzyme activity of CPT1α associated with LCAD and MCAD protein expression in the HFD group. The activation of fatty acid β-oxidation subsequently led to the production of abundant acetyl-CoA, which in turn affected acetylation homeostasis. Indeed, acetylome analysis showed that HFD feeding significantly increased the acetylation levels of multiple metabolic enzymes. In vitro, inhibition of CPT1α with etomoxir also blocked lipid surplus-induced increases in ac-p65 and proinflammatory cytokine expression. Importantly, an elevation in acetyl-CoA alone derived from acetate independent of lipid surplus did not induce hepatic inflammation. In contrast, acetate aggravated PA-induced inflammation. Hence, acetyl-CoA itself was not an inflammatory factor, but rather contributed to the increase in p65 acetylation and exacerbated HFD-induced inflammation. Collectively, these results indicated that mitochondrial fatty acid β-oxidation provided acetyl-CoA for protein acetylation to integrate metabolism and inflammation under conditions of overnutrition.
In addition to changes in total content, the subcellular fluxes of acetyl-CoA also change strongly based on variations in synthesis and utilization (Pietrocola et al., 2015). In the present study, among the 103 differentially expressed proteins in the HFD group, approximately 34% were in mitochondria, while almost 66% were located outside mitochondria. Acetylation of mitochondrial proteins occurs largely through nonenzymatic mechanisms (Ghanta et al., 2013), which means that physiological and nutritional status can drive these protein modifications by increasing the concentration of mitochondrial acetyl-CoA to expand the local pool of acetyl donors (Davies et al., 2016). The changes in the acetylation levels of extramitochondrial proteins inspired us to investigate how fluctuations in acetyl-CoA content feedback to the cytosol/nucleus and affect inflammatory responses and other biological processes. Mitochondrial membranes are impermeant to the acetyl-CoA molecule (Pietrocola et al., 2015). Cytosolic acetyl-CoA depends mainly on two reactions: citrate export from mitochondria and subsequent ACLY-mediated catalysis (Langston et al., 2019). Extramitochondrial acetyl-CoA serves as a substrate for protein acetylation. Numerous studies have shown that ACLY can sense changes in nutritional status through acetylation or phosphorylation (Feng et al., 2020; Lin et al., 2013). In the present study, lipid surplus increased p-ACLY (Ser455) in vivo and in vitro. Inhibition of ACLY with SB-204990 decreased the content of acetyl-CoA and blocked PA-induced increases in inflammation-related gene expression and ac-p65, which suggested that ACLY played a dominant role in mediating acetyl-CoA conversion to aggravate inflammation under lipid surplus. Indeed, some studies have showed that an HFD decreases ACLY expression in mammals (Carrer et al., 2017). However, Collins et al. (2010) also found that PA treatment increased the mRNA expression of ACLY. The difference in findings is probably due, at least in part, to the differences in species and nutrient conditions. In addition to acetyl-CoA, KATs and KDACs are also important for the acetylation of p65. The present study demonstrated that CBP was crucial for lipid surplus-induced p65 acetylation. CBP was activated and linked with p65 to increase the transcriptional programs of inflammatory genes. Notably, NAD+ levels in cells fluctuate in response to feeding or fasting (Menzies et al., 2016). Hence, the sirtuin family might also regulate the transcriptional programs of inflammatory genes under nutrient stress. Given the nuclear localization of SIRT1, SIRT1 might be responsible for the deacetylation of p65. Through in vivo and in vitro experiments, we found that SIRT1 was deactivated with the decline in NAD+/NADH, which further increased ac-p65 and aggravated inflammation under lipid surplus. Overall, the function of these KATs and KDACs in inflammatory regulation might provide therapeutic targets for the development of anti-inflammatory drugs. For example, vorinostat (SAHA) and romidepsin (FK-228) are available drugs for the treatment of various diseases (Bradner et al., 2010).
The above study in fish revealed that the fluctuation of acetyl-CoA can be an important modulator of inflammation through protein acetylation. To understand the correlated evolution of nutrient stress response from the initial vertebrates to mammals, we used AML12 hepatocytes to explore the role of p65 acetylation in regulating inflammation under lipid surplus. In a lipid surplus-induced inflammation model, PA significantly increased the acetyl-CoA level and enhanced the acetylation of p65 in a dose-dependent manner, which was consistent with results obtained in fish. Considering that p65 acetylation can enhance the transcriptional activity of inflammatory genes, this finding further illustrates that p65 acetylation modification can be targeted to alleviate inflammation caused by lipid surplus. In addition, the pathway that the activation of fatty acid oxidation provides acetyl-CoA for p65 acetylation under PA treatment appears to be conserved across evolution from teleosts to mammals. Notably, although the response of ACLY to lipid surplus was not as obvious as that in fish, inhibition of ACLY with SB-204990 still relieved the increases in ac-p65 and the mRNA expression of proinflammatory cytokines, which suggests that ACLY inhibitors still have broad prospects for use in the treatment of inflammation caused by overnutrition. In contrast to acetyl-CoA metabolism, the KAT/KDAC involved in regulating p65 acetylation under HFD feeding seemed to not be conserved between fish and mammals. The mRNA levels of Cbp and Sirt1 were not influenced by PA in AML12 hepatocytes. In contrast, PA increased Gcn5 mRNA expression, and inhibition of GCN5 relieved PA-induced inflammation. Given the finding that broad-spectrum KDAC inhibitors aggravated PA-induced inflammation, these results expand the therapeutic potential of KATs and KDACs in immune-related applications. Overall, although KATs and KDACs activated by PA were different between fish and mammals, the role of fatty acid oxidation-ACLY-ac-p65 axis in exacerbating lipid surplus-induced inflammation is conserved between fish and mammals to some extent. Cells likely use this acetylation-mediated transcriptional regulation to help deal with physiological or pathological fluctuations in the cellular nutrient status.
The above observations in vivo and in vitro demonstrate that acetyl-CoA-dependent regulation of transcription factor activity is an approach to adapt to proinflammatory stimuli under nutrient stress. To further investigate whether fatty acid oxidation and ACLY coordinated the metabolism of acetyl-CoA and provided acetyl-CoA for acetylation modification of extensive nuclear proteins, octanoate, a medium-chain fatty acid, was selected to simulate fatty acid oxidation. Octanoate dose-dependently increased the acetylation level of histone H3. Consistent with the effects of PA treatment, ACLY played an important role in the regulation of ac-H3 under octanoate treatment. Moreover, Akt, which was able to phosphorylate and activate ACLY (Migita et al., 2008), also regulated octanoate-enhanced ac-H3. Hence, Akt/ACLY dependent acetyl-CoA production participates in the regulation of cell metabolism and immunity by affecting protein acetylation. In addition, octanoate inhibited the AMPK/SIRT1 pathway to enhance the acetylation of histone H3, which expands the knowledge regarding the metabolic link between NAD+ and sirtuin-mediated nuclear processes. Therefore, the availability of acetyl-CoA and NAD+ enables KATs and KDACs to regulate biological processes to adapt to the physiological milieu. McDonnell et al. (2016) used isotopic tracing to reveal that lipids provide acetyl-CoA for histone acetylation in an ACLY independent pathway, which is inconsistent with our present results. Differences between cell lines and culture conditions may account for the contrasting findings.
In conclusion, the present study, for the first time, systematically revealed a potential mechanism that metabolic flux of acetyl-CoA increases p65 acetylation level to aggravate inflammation under lipid surplus in fish (Figure 8). The discovery of inflammatory regulatory roles for these metabolites and cofactors (i.e., acetyl-CoA and NAD+) enhances understanding about how nutrient stress induces metabolic diseases from the perspective of epigenetic modification. The development of nutrition strategies that target metabolic-coupled epigenetics can reduce the use of drugs in aquaculture, which contributes to the production of green and safe food. In addition, regarding that dysregulation of epigenetic modifications is linked to many metabolic diseases, such as obesity and diabetes, more work should be carried out to determine the preventive and therapeutic potential of metabolic substrates and KATs/KDACs for the management of multiple pathologies in vertebrates.
Figure 8.
A work model of how HFD altered the acetylation of p65 to aggravate inflammatory responses
Limitations of study
Our in vivo and in vitro analyses in fish demonstrated that lipid surplus improved fatty acid β-oxidation in mitochondria and activated ACLY to transfer acetyl-CoA from mitochondria to nucleus, which in turn increased ac-p65 and exacerbated inflammation. However, results in AML12 cells indicated that the types of lysine acetyltransferases (KATs) and lysine deacetylases (KDACs) involved in p65 acetylation under palmitate treatment were not conserved. Species limitations affect the translatability of the findings in mammals. Hence, studies aimed at characterizing the precise mechanisms underlying the inflammation regulatory effects of acetylation under lipid surplus in mammals will continue in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal Anti-p65 | Cell Signaling Technology | CAT#8242; RRID:AB_10859369 |
| Rabbit polyclonal Anti-Akt | Cell Signaling Technology | CAT#9272; RRID:AB_329827 |
| Rabbit monoclonal Anti-Akt (phospho Ser473) | Cell Signaling Technology | CAT#4060; RRID:AB_2315049 |
| Rabbit polyclonal Anti-ACLY (phospho Ser455) | Cell Signaling Technology | CAT#4331; RRID: AB_2257987 |
| Rabbit polyclonal Anti-AMPKα | Cell Signaling Technology | CAT#2532; RRID:AB_330331 |
| Rabbit polyclonal Anti-AMPKα (phospho Thr172) | Cell Signaling Technology | CAT#2531; RRID:AB_330330 |
| Rabbit polyclonal Acetyl-Lysine antibody | Cell Signaling Technology | CAT#9441; RRID:AB_331805 |
| Rabbit monoclonal Anti-SIRT1 | Cell Signaling Technology | CAT#9475; RRID:AB_2617130 |
| Rabbit monoclonal Anti-DYKDDDDK Tag | Cell Signaling Technology | CAT#14793; RRID:AB_2572291 |
| Rabbit monoclonal Anti-HA Tag | Cell Signaling Technology | CAT#3724; RRID:AB_1549585 |
| Rabbit polyclonal Anti-Histone H3 | Abcam | CAT#ab1791; RRID: AB_302613 |
| Rabbit monoclonal Anti-ACLY | Abcam | CAT#ab40793 |
| Rabbit monoclonal Anti-LCAD | Abcam | CAT#ab196655 |
| Rabbit monoclonal Anti-MCAD | Abcam | CAT#ab92461; RRID:AB_10563530 |
| Rabbit polyclonal Anti-CBP | Wan Lei Bio | CAT#WL02842 |
| Rabbit polyclonal Anti-PPARα | Absin | CAT#abs117362 |
| Mouse polyclonal Anti-GAPDH | Golden Bridge Biotechnology | CAT#TA-08 |
| Rabbit polyclonal Anti-IL-1β | This paper | N/A |
| Mouse monoclonal Anti-COX-2 | Santa Cruz | CAT#sc-376861 |
| Bacterial and virus strains | ||
| Escherichia coli-DH5α | TransGen Biotech | CAT#CD201-01 |
| Chemicals, peptides, and recombinant proteins | ||
| Palmitic acid | Merck | CAT#P0500; CAS: 57-10-3 |
| Etomoxir | MedChemExpress | CAT#HY-50202 |
| Sodium acetate | Merck | CAT#S2889; CAS: 127-09-3 |
| SB 204990 | MedChemExpress | CAT#HY-16450 |
| MK 2206 | MedChemExpress | CAT#HY-10358 |
| C646 | MedChemExpress | CAT#HY-13823 |
| Resveratrol | MedChemExpress | CAT#HY-16561 |
| KDAC inhibitor | MedChemExpress | CAT#HY-K0030 |
| AICAR | MedChemExpress | CAT#HY-13417 |
| SYBR qPCR Master Mix | Vazyme | CAT#Q711 |
| EZ Trans transfection reagent | LIFE iLAB | CAT#AC04L099 |
| Octanoate | Merck | CAT#C2875; CAS: 124-07-2 |
| Critical commercial assays | ||
| HAT Activity Colorimetric Assay Kit | BioVision | CAT#K332 |
| EZScreen™ NAD+/NADH Colorimetric Assay Kit | BioVision | CAT#K358 |
| Acetyl-CoA Assay Kit | Merck | CAT#MAK039 |
| NE-PER Nuclear and Cytoplasmic Extraction Reagents | Thermo Fisher Scientific | CAT#78835 |
| Dual-Luciferase Reporter Assay System Kit | TransGen Biotech | CAT#FR201 |
| LightShift Chemiluminescent EMSA Kit | Thermo Fisher Scientific | CAT#20148 |
| Mut Express II Fast Mutagenesis Kit V2 | Vazyme | CAT#C214 |
| Deposited data | ||
| Acetylome of large yellow croaker liver | This paper | PXD027084 |
| Experimental models: Cell lines | ||
| Human: HEK293T cell line | (Zhu et al., 2019) | N/A |
| Mouse: AML12 cell line | Procell | CAT#CL-0602 |
| Large yellow croaker: primary hepatocytes | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Juvenile large yellow croaker | NingdeFufa Fishery Co., Ltd. (China) | N/A |
| Oligonucleotides | ||
| Primers for qRT-PCR, see Data S6 | This paper | N/A |
| Primers for recombinant DNA, see Data S6 | This paper | N/A |
| Primers for 5’-biotin-labeled double-stranded oligomers, Fw: 5’-CTAGCGGGAATTTCCGGGA ATTTCCGGGAATTTCCGGGAATTTCC-3’ |
This paper | N/A |
| Primers for 5’-biotin-labeled double-stranded oligomers, Rv: 5’-GGAAATTCCCGGAAATTCC CGGAAATTCCCGGAAATTCCCGCTAG-3′ |
This paper | N/A |
| Recombinant DNA | ||
| pCDNA3.1(+) | Miaoling bio | CAT#P0157 |
| Plasmid: pcDNA3.1(+)-CBP | This paper | N/A |
| Plasmid: pcDNA3.1(+)-p65 | This paper | N/A |
| Plasmid: pcDNA3.1(+)-GCN5 | This paper | N/A |
| Plasmid: pcDNA3.1(+)-PCAF | This paper | N/A |
| Plasmid: pcDNA3.1(+)-SIRT1 | This paper | N/A |
| pGL3-CMV-IL-1β-Luc | (Li et al., 2020) | N/A |
| pGL3-CMV-pNFκB-Luc | (Du et al., 2020) | N/A |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| SPSS 23.0 software | https://www.ibm.com/support/pages/node/581811 | N/A |
| 2-ΔΔCT method | (Livak and Schmittgen, 2001) | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Qinghui Ai (qhai@ouc.edu.cn).
Materials availability
Plasmids generated in this study are available from the lead contact upon reasonable request.
Experimental model and subject details
Animals and diets
Briefly, 4-month-old large yellow croakers of similar size (mean weight 15.87 ± 0.14 g) were obtained from NingdeFufa Fishery Co., Ltd. (China) and reared in floating sea cages under conditions of 26 ± 2°C, 29–32‰ salinity and 6–7 mg/L dissolved oxygen (Yang et al., 2020). The fish were divided into two groups at random and fed the experiment diets. A diet (Fang et al., 2021) (Table S3) containing 12% crude lipid was used as the control (CON) diet because this dietary lipid level is optimal for the growth of the large yellow croaker (Larimichthys crocea) (Yan et al., 2015). While, a diet (Fang et al., 2021) (Table S3) containing 18% crude lipid was used as the high fat diet (HFD). The fish were fed twice a day at 05:00 and 17:00 for 10 weeks. The fatty acid compositions of the two diets (CON and HFD) are listed in Table S4 (Fang et al., 2021). In the study, both sexes were used. At the end of the experiment, the fish were anesthetized with MS222 (1:10, 000; Sigma, USA) prior to sampling. Livers of the experimental fish were collected, frozen in liquid nitrogen, and then stored at −80°C until use.
In the present study, all experimental procedures performed on fish and all animal care procedures were conducted in strict accordance with the Management Rule of Laboratory Animals (Chinese Order No. 676 of the State Council, revised 1 March 2017) (Li et al., 2019).
Cell culture and reagents
Livers of juvenile yellow croakers were removed and placed in sterile phosphate buffer (PBS, Biological Industries, Israel) containing penicillin and streptomycin (cat. no. P1400, Solarbio, China). After washing with Dulbecco’s modified Eagle medium/Ham’s F12 medium (1:1) (DMEM/F12, Biological Industries), the liver tissue was chopped into 1-mm3 slices and digested with 0.25% trypsin (Thermo Fisher Scientific, USA) for 10 min. After neutralization with DMEM/F12 medium containing fetal bovine serum (FBS, Biological Industries), the cell precipitate was suspended in complete medium composed of DMEM/F12 medium supplemented with 15% FBS, 100 U penicillin and 100 μg/mL streptomycin. The cell suspension was inoculated into a six-well culture plate and incubated at 28°C (Zhu et al., 2019).
HEK293T cells were maintained in DMEM with high glucose (Biological Industries) supplemented with 10% FBS and antibiotics within an atmosphere of 5% CO2 at 37°C (Zhu et al., 2019).
AML12 cells were maintained in DMEM/F12 medium (Procell, China) containing 10% FBS, 10 μg/mL insulin, 5.5 μg/mL transferrin, 5 ng/mL selenium, 40 ng/mL dexamethasone and 1% penicillin/streptomycin at 37°C under 5% CO2.
Method details
Histological and biochemical analysis
Paraffin-embedded liver sections were stained with H&E for morphological analysis. Liver triglycerides (TG) were measured using commercially available kits (Applygen Technologies Inc., China). ALT/AST and plasma free fatty acids (FFAs) were quantified with commercially available kits (Nanjing Jiancheng Bioengineering Inc., China).
KAT activity, NAD+ and acetyl-CoA measurements
KAT activity was measured with KAT Activity Colorimetric Assay Kits (cat. no. K332; BioVision, USA). In brief, the kits utilized active nuclear extract (NE) as a positive control and acetyl-CoA as a cofactor. Acetylation of peptide substrate by active KAT released the free form of CoA, which then served as an essential coenzyme for production of reduced nicotinamide adenine dinucleotide (NADH). NADH can easily be detected spectrophotometrically upon reacting with a soluble tetrazolium dye (Yang et al., 2014b). The absorbance of each sample was detected at 440 nm with a microplate reader (SpectraMax i3x).
NAD+/NADH was measured with an EZScreen NAD+/NADH Colorimetric Assay Kit (cat. no. K958; BioVision) according to the manufacturer’s instructions. Extracts were prepared from frozen tissue according to the kit protocol and assayed on two parallel plates, one for total NAD (NADt) and another for NADH. For NADH only, 100 μL of the extract was heated at 60°C for 30 min before the measurement and then cooled to decompose NAD; for NADt, the extract was measured without treatment. The NAD concentration was determined using the provided reference standard and is expressed in nmol of NAD per mg of protein. Oxidized NAD was calculated as [NADt] - [NADH].
The intracellular acetyl-CoA content was determined using an Acetyl-Coenzyme A Assay Kit (MAK039, Sigma) according to the manufacturer’s instructions. Briefly, after deproteinization using 1 M perchloric acid, Acetyl-CoA Quencher and Quench Remover were added to the sample to correct for background created by free coenzyme A and succinyl-CoA. Then, appropriate reaction mix was added to samples. After 30 min of incubation at 37°C, the fluorescence intensity was measured using a plate reader and the following settings: λex = 535 nm, λem = 587 nm.
Acyl-CoA quantification by LC-MS
Extraction of acyl-CoAs was carried out following established protocols (Woldegiorgis et al., 1985) with some modifications. Briefly, 300 μL of extraction buffer containing isopropanol, 50 mM KH2PO4, and 50 mg/mL BSA (25:25:1 v/v/v) acidified with glacial acetic acid was added to sample. Next, 19:0-CoA was added as an internal standard and lipids were extracted by incubation at 4°C for 1 h at 1,500 rpm. Next, 300 μL of petroleum ether was added and the sample was centrifuged at 12,000 rpm for 2 min at 4°C. The upper phase was removed. The samples were extracted two more times with petroleum ether as described above. Finally, 5 μL of saturated ammonium sulfate followed by 600 μL of chloroform: methanol (1:2 v/v) was added to the remaining lower phase. The sample was then incubated on a thermomixer at 450 rpm for 20 min at 25°C and centrifuged at 12,000 rpm for 5 min at 4°C. The clear supernatant was transferred to a fresh tube and subsequently dried in the SpeedVac under OH mode (Genevac). Dry extracts were resuspended in an appropriate volume of methanol:water (9:1 v/v) prior to LC-MS analyses on a Thermo Fisher U3000 DGLC coupled to a SCIEX QTRAP 6500 Plus (Lam et al., 2020).
LC-MS/MS analysis of the acetylome
First, 20 mg of sample was taken, and DL-dithiothreitol (DTT) was added to a final concentration of 10 mM. Subsequently, the sample was placed in a thermomixer (600 rpm, 37°C) for 1.5 h, after which it was taken out and cooled to room temperature. Iodoacetamide (IAA) was added to a final concentration of 50 mM, and the reaction was protected from light for 30 min. Four volumes of 50 mM Tris HCl (pH 8.0) were added, and the urea (UA) concentration was diluted to 2 M. Trypsin was added at a protein: trypsin mass ratio of 50:1, and then the mix was incubated overnight (15–18 h) at 37°C. Trifluoroacetic acid (TFA) was added to a final concentration of 0.1%, and each sample was brought to a pH ≤ 3 by adjusting the volume of 10% TFA. Peptides were desalted using a C18 SPE cartridge and lyophilized (Wiśniewski et al., 2009). Then, samples were reconstituted with 1.4 mL of pre-cooled IAP buffer and were incubated with anti-ac-K antibody beads (PTMScan Acetyl-Lysine Motif (Ac-K) Kit, Cell Signaling Technology, USA) at 4°C for 1.5 h. Subsequently, samples were centrifuged at 2,000 ×g for 30 s, after which the supernatant was discarded. Anti-ac-K antibody beads were washed three times with 1 mL of pre-cooled immunoaffinity purification (IAP) buffer and then washed three times with 1 mL of pre-cooled water. The washed anti-ac-K antibody beads were added to 40 μL of 0.15% TFA, and the mixture was incubated at room temperature for 10 min; 0.15% TFA was added twice. After centrifugation at 2,000 ×g for 30 s, the supernatant was desalted with C18 STAGE Tips. Each fractionated sample was separated using an Easy nLC HPLC liquid phase system at a nanoliter flow rate. Buffer A was a 0.1% formic acid aqueous solution, and buffer B was a 0.1% formic acid/acetonitrile aqueous solution (acetonitrile was 84%). The column was equilibrated with 95% buffer A. The sample was loaded from the autosampler into the loading column (Thermo Scientific Acclaim PepMap100, 100 μm × 2 cm, nanoViper C18) and passed through an analytical column (Thermo Scientific EASY column, 10 cm, ID75 μm, 3 μm, C18-A2) and separated at a flow rate of 300 nL/min. The sample was chromatographed and subjected to MS using a Q Exactive mass spectrometer. The detection mode was positive ion, the parent ion scanning range was 300–1800 m/z, the primary mass spectrometer resolution was 70,000 at 200 m/z, the automatic gain control (AGC) target was 106, the maximum ion time (IT) was 50 ms, and the dynamic exclusion time was 60 s. The mass-to-charge ratios of the polypeptides and polypeptide fragments were collected as follows: 20 fragments were acquired after each full scan, the MS2 activation type was higher-energy collisional dissociation (HCD), the isolation window was 2 m/z, the resolution of the secondary mass spectrum was 17,500 at 200 m/z, the normalized collision energy was 30 eV, and the underfill was 0.1%.
RNA extraction, reverse transcription, and qPCR
Total RNA of liver tissue or hepatocytes was extracted with TRIzol Reagent (Takara, Japan) according to the manufacturer’s protocol. The amount of RNA was determined by a NanoDrop 2000spectrophotometer (Thermo Fisher Scientific). The extracted RNA was reverse transcribed into first-strand cDNA with a PrimeScript RT Reagent Kit (Takara) according to the manufacturer’s instructions. QRT-PCR was performed using SYBR qPCR Master Mix (Vazyme, China) according to the manufacturer’s instructions. Primers were designed according to published sequences in NCBI and are listed in Data S6. To calculate the mRNA levels of genes, the comparative cycle threshold (CT) method (2−ΔΔCT method) was adopted (Livak and Schmittgen, 2001).
Western blot analysis
Total proteins were extracted from livers of the experimental fish and from hepatocytes using RIPA lysis buffer with protease inhibitors and phosphatase inhibitors. Nuclear protein was collected using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) according to the manufacturer’s instructions. Protein concentrations were determined with a BCA Protein Assay Kit (Beyotime Biotechnology, China) according to the manufacturer’s instructions. After standardization, the samples were separated by SDS-PAGE and transferred to 0.45 μm PVDF membranes (Millipore, USA). Membranes were blocked with 5% nonfat dry milk in TBST for 2 h and were then incubated with primary antibodies overnight. Subsequently, the membranes were incubated with secondary antibodies for 1 h and developed with Beyo ECL Plus Reagent (Beyotime Biotechnology) (Jiang et al., 2017). The details of antibodies in the present study were listed in the Key resources table. The densities of protein bands were normalized to the density of internal control and were quantified using NIH Image 1.63 software.
Plasmid constructs
For expression plasmids, coding sequences (CDSs) of p65, CBP, KAT 2A (GCN5), KAT 2B (PCAF) and SIRT1 were cloned from large yellow croaker cDNA by PCR with the primers listed in Data S6. A linearized pcDNA3.1 (+) vector was obtained using QuickCut EcoRI (Takara). A flag fusion protein (DYKDDDDK) was constructed at the C-terminus of p65, and an HA fusion protein (YPYDVPDYA) was constructed at the C-termini of CBP, GCN5, PCAF and SIRT1. Homologous recombination was completed using a ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd., China). For reporter plasmids, the IL-1β promoter and pNFκB-Luc reporter plasmid were donated by colleagues in our laboratory (Du et al., 2020; Li et al., 2020).
MS analysis of p65 acetylation sites
Purified p65-Flag was resolved by SDS-PAGE and was then fixed and stained with Coomassie Brilliant Blue. The gel was rinsed with water several times, and bands of interest were excised and cut into cubes (∼1 × 1 mm). These gel pieces were then transferred to Eppendorf tubes and dehydrated with neat acetonitrile. Afterward, approximately 50 μL of DTT solution (10 mM, in 100 mM ammonium bicarbonate) was added to completely cover the gel pieces. The tubes were incubated at 56°C for 30 min and then cooled to room temperature. Neat acetonitrile was added to dehydrate the gel pieces. All the liquid was then removed. Subsequently, 50 μL of IAA solution (55 mM, in 100 mM ammonium bicarbonate) was added to completely cover gel pieces. The tubes were incubated at 37°C in the dark for 10 min and then cooled to room temperature. Neat acetonitrile was added again to dehydrate the gel pieces. Thereafter, 50% acetonitrile in 100 mM ammonium bicarbonate was then added, and the tubes were vortexed at room temperature until the Coomassie stain was removed. An appropriate volume of trypsin buffer (13 ng/μL, in 10 mM ammonium bicarbonate containing 10% acetonitrile) was added to cover dry gel pieces, and the samples were then incubated at 37°C overnight. Digested peptides were extracted from gel pieces by sequentially adding 0.1% formic acid in 50% acetonitrile, 0.1% formic acid in 80% acetonitrile and 100% acetonitrile. Extraction products were then combined and lyophilized for following steps. Each sample was analyzed by an online nanospray liquid chromatography MS (LC-MS/MS) on a Q Exactive Plus coupled to an EASY-nano-LC 1200 system (Thermo Fisher Scientific).
Mutations at putative acetylation sites
Flag-tagged p65 was mutated using a Mut Express II Fast Mutagenesis Kit V2 (Vazyme Biotech Co., Ltd.). Putative acetylation sites in p65 were mutated to arginine or glutamine. The sites mutated in the present study were based on our LC-MS/MS results. We mutated a single site or conducted two-site and three-site mutations in p65.
Dual-luciferase reporter assays
HEK293T cells were evenly plated in 24-well plates. After overnight culture, the p65 expression vector (control or mutant) and the IL-1β promoter reporter vector were transfected into HEK293T cells at a ratio of 3:1 using Lipofectamine 2000 reagent (Invitrogen, USA). Luciferase activity was detected using a Dual-Luciferase Reporter Assay System Kit (TransGen Biotech Co., Ltd., China) (Zhu et al., 2019). To detect the effects of acetyltransferases on p65 transcriptional activity, p65 and GCN5, CBP, or PCAF were transfected into HEK293T cells at a ratio of 1:1 along with the IL-1β promoter, and luciferase activity was measured after 24 h. To detect the effects of SIRT1 on p65 transcriptional activity, p65 and SIRT1 were transfected into HEK293T cells at a ratio of 1:1 along with a pNFκB-Luc reporter plasmid.
EMSA
p65 and various lysine mutants were transferred into HEK293T cells, and cells were collected after 48 h. Nuclear protein was prepared and collected with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). The protein concentration of each sample was measured by BCA assay (Beyotime Biotechnology), and all samples were then adjusted to the same protein level. The sequences used for 5’-biotin-labeled double-stranded oligomers were as follows: sense sequence, 5’-CTAGCGGGAATTTCCGGGAATTTCCGGGAATTTCCGGGAATTTCC-3’; anti-sense sequence, 5’-GGAAATTCCCGGAAATTCCCGGAAATTCCCGGAAATTCCCGCTAG-3’. Then, DNA-protein interactions were detected with a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) (Du et al., 2020).
Immunoprecipitation and coimmunoprecipitation
For immunoprecipitation (IP), the tissue was lysed with Cell Lysis Buffer for Western Blotting and IP (Beyotime Biotechnology) for 20 min. Then, 40 μL of anti-ac-K antibody agarose beads (Cytoskeleton, Inc., USA) was added to the lysate. The mixture was incubated for 12 h at 4°C. The immunoprecipitates were recovered by centrifugation at 3,500 ×g for 1 min and washed five times with lysis buffer, after which they were analyzed via western blotting.
For co-immunoprecipitation (co-IP), cells transfected with p65-Flag and other acetyltransferases or SIRT1 were harvested at 48 h after transfection and lysed with Cell Lysis Buffer for Western Blotting and IP (Beyotime Biotechnology) for 20 min. Then, the lysate was divided into two equal parts. The protein was enriched by incubation with ANTI-Flag M2 Affinity Gel (Sigma) and mouse IgG-agarose (Sigma), or with Pierce anti-HA agarose (Thermo Fisher Scientific) and mouse IgG-agarose, for 4 h on a 4°C shaker, after which the supernatant was removed by centrifugation at 3,500 ×g for 1 min. The agarose beads were washed five times with lysis buffer and TBST. Finally, the proteins were detached from the agarose beads using a Flag peptide (MedChem Express, USA) or HA peptide (MedChem Express) for use in subsequent experiments (Du et al., 2020).
Quantification and statistical analysis
The data are presented as the mean ± standard deviation (SD). The statistical analyses between two groups were performed with independent t-tests, and comparisons between multiple groups were performed with one-way analysis of variance (ANOVA) in SPSS 23.0 software. P< 0.05 was considered to indicate statistical significance. The number of replicates for each experiment are indicated in the figure legends.
Acknowledgments
This work was supported by the National Science Fund for Distinguished Young Scholars of China [grant number: 31525024], Key Program of National Natural Science Foundation of China [grant number: 31830103], Ten-thousand Talents Program (grant number: 2018-29), and the Agriculture Research System of China [grant number: CARS47-11], Scientific and Technological Innovation of Blue Granary [grant number: 2018YFD0900402].
We are grateful to Hongyuan Yang (The University of New South Wales), Li Xu (Tsinghua University), Wei Yu (Fudan University), Xinxia Wang (Zhejiang University) and Haoxing Zhang (Shenzhen University) for their constructive suggestions on the experimental design and revising article. We thank Wei Xu, Yanjiao Zhang, Xiaojun Xiang, Qiangde Liu and Fan Chen for their experimental assistance. Additionally, we thank Tao Ding and Ning Xu for their help in fish rearing.
Author contributions
Q.C., K.S.M., and Q.H.A. designed the experiments. Q.C., J.L.D., and K.C. performed the experiments. W.F., Z.Q.Z., and Q.C.C. prepared samples for LC/MS-MS. Q.C., and Q.H.A. wrote the paper. All authors discussed the results and improved on the paper.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure diversity in experimental samples through the selection of the cell lines.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103244.
Supplemental information
Data and code availability
-
•
All relevant data are within the manuscript and Supplementary Material. The mass spectrometry proteomics data from this publication have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Ma et al., 2019) with the dataset identifier PXD027084.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alessandrini A., Facci P. Unraveling lipid/protein interaction in model lipid bilayers by atomic force microscopy. J. Mol. Recognit. 2011;24:387–396. doi: 10.1002/jmr.1083. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I., Manning B.D. mTORC1 signaling and the metabolic control of cell growth. Curr.Opin.Cell Biol. 2017;45:72–82. doi: 10.1016/j.ceb.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner J.E., West N., Grachan M.L., Greenberg E.F., Haggarty S.J., Warnow T., Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010;6:238. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S.L., Wellen K.E. Metabolic signaling to the nucleus in cancer. Mol. Cell. 2018;71:398–408. doi: 10.1016/j.molcel.2018.07.015. [DOI] [PubMed] [Google Scholar]
- Cantó C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer A., Parris J.L.D., Trefely S., Henry R.A., Montgomery D.C., Torres A., Viola J.M., Kuo Y.M., Blair I.A., Meier J.L. Impact of a high-fat diet on tissue acyl-CoA and histone acetylation levels. J. Biol. Chem. 2017;292:3312–3322. doi: 10.1074/jbc.M116.750620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkiadaki A., Guarente L. High-fat diet triggers inflammation-induced cleavage of SIRT1 in adipose tissue to promote metabolic dysfunction. Cell Metab. 2012;16:180–188. doi: 10.1016/j.cmet.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Fischle W., Verdin E., Greene W.C. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- Chen L.f., Mu Y., Greene W.C. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.F., Williams S.A., Mu Y., Nakano H., Duerr J.M., Buckbinder L., Greene W.C. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol. Cell.Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J.M., Neville M.J., Hoppa M.B., Frayn K.N. De novo lipogenesis and stearoyl-CoA desaturase are coordinately regulated in the human adipocyte and protect against palmitate-induced cell injury. J. Biol. Chem. 2010;285:6044–6052. doi: 10.1074/jbc.M109.053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E., Schulze A., Zechner R., Walther T.C., Farese R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M.N., Kjalarsdottir L., Thompson J.W., Dubois L.G., Stevens R.D., Ilkayeva O.R., Brosnan M.J., Rolph T.P., Grimsrud P.A., Muoio D.M. The acetyl group buffering action of carnitine acetyltransferase offsets macronutrient-induced lysine acetylation of mitochondrial proteins. Cell Rep. 2016;14:243–254. doi: 10.1016/j.celrep.2015.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D.R., Collins L.B., Wali A., Bigler R., Sun W., Bultman S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Chen Q., Li Y., Xiang X., Xu W., Mai K., Ai Q. Activation of the Farnesoid X receptor (FXR) suppresses linoleic acid-induced inflammation in the large yellow croaker (Larimichthys crocea) J. Nutr. 2020;150:2469–2477. doi: 10.1093/jn/nxaa185. [DOI] [PubMed] [Google Scholar]
- Falcone M., Maddocks O.D.K. The KRAS-BCAA-BCAT2 axis in PDAC development. Nat. Cell Biol. 2020;22:139–140. doi: 10.1038/s41556-020-0467-2. [DOI] [PubMed] [Google Scholar]
- Fang W., Chen Q., Cui K., Chen Q., Li X., Xu N., Mai K., Ai Q. Lipid overload impairs hepatic VLDL secretion via oxidative stress-mediated PKCδ-HNF4α-MTP pathway in large yellow croaker (Larimichthys crocea) Free Radic. Biol. Med. 2021;172:213–225. doi: 10.1016/j.freeradbiomed.2021.06.001. [DOI] [PubMed] [Google Scholar]
- Feng X., Zhang L., Xu S., Shen A.-z. ATP-citrate lyase (ACLY) in lipid metabolism and atherosclerosis: an updated review. Prog.Lipid Res. 2020;77:101006. doi: 10.1016/j.plipres.2019.101006. [DOI] [PubMed] [Google Scholar]
- Ference B.A., Ray K.K., Catapano A.L., Ference T.B., Burgess S., Neff D.R., Oliver-Williams C., Wood A.M., Butterworth A.S., Di Angelantonio E. Mendelian randomization study of ACLY and cardiovascular disease. N. Engl. J. Med. 2019;380:1033–1042. doi: 10.1056/NEJMoa1806747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D., Shaw R.J. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol. Cell. 2017;66:789–800. doi: 10.1016/j.molcel.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanta S., Grossmann R.E., Brenner C. Mitochondrial protein acetylation as a cell-intrinsic, evolutionary driver of fat storage: chemical and metabolic logic of acetyl-lysine modifications. Crit. Rev. Biochem. Mol. Biol. 2013;48:561–574. doi: 10.3109/10409238.2013.838204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Ishinaga H., Jono H., Lim J.H., Kweon S.M., Xu H., Ha U.H., Xu H., Koga T., Yan C., Feng X.H. TGF-β induces p65 acetylation to enhance bacteria-induced NF-κB activation. EMBO J. 2007;26:1150–1162. doi: 10.1038/sj.emboj.7601546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Bian F., Zhou H., Wang X., Wang K., Mai K., He G. Nutrient sensing and metabolic changes after methionine deprivation in primary muscle cells of turbot (Scophthalmus maximus L.) J. Nutr. Biochem. 2017;50:74–82. doi: 10.1016/j.jnutbio.2017.08.015. [DOI] [PubMed] [Google Scholar]
- Jimenez-Gomez Y., Mattison J.A., Pearson K.J., Martin-Montalvo A., Palacios H.H., Sossong A.M., Ward T.M., Younts C.M., Lewis K., Allard J.S. Resveratrol improves adipose insulin signaling and reduces the inflammatory response in adipose tissue of rhesus monkeys on high-fat, high-sugar diet. Cell Metab. 2013;18:533–545. doi: 10.1016/j.cmet.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G., Jr., McKnight S.L. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.M., Zhou T., Li J., Zhang S., Chua G.H., Li B., Shui G. A robust, integrated platform for comprehensive analyses of acyl-coenzyme as and acyl-carnitines revealed chain length-dependent disparity in fatty acyl metabolic fates across Drosophila development. Sci. Bull. 2020;65:1840–1848. doi: 10.1016/j.scib.2020.07.023. [DOI] [PubMed] [Google Scholar]
- Langston P.K., Nambu A., Jung J., Shibata M., Aksoylar H.I., Lei J., Xu P., Doan M.T., Jiang H., MacArthur M.R. Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses. Nat. Immunol. 2019;20:1186–1195. doi: 10.1038/s41590-019-0453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach M.A., Hanke J.E., Serefidou M., Mangan M.S., Kolbe C.-C., Hess T., Rothe M., Kaiser R., Hoss F., Gehlen J. Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP-citrate lyase. Immunity. 2019;51:997–1011.e1017. doi: 10.1016/j.immuni.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Lee J.V., Carrer A., Shah S., Snyder N.W., Wei S., Venneti S., Worth A.J., Yuan Z.-F., Lim H.-W., Liu S. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Cui K., Wu M., Xu D., Mai K., Ai Q. Polyunsaturated fatty acids influence LPS-induced inflammation of fish macrophages through differential modulation of pathogen recognition and p38 MAPK/NF-κB signaling. Front. Immunol. 2020;11:559332. doi: 10.3389/fimmu.2020.559332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Pang Y., Xiang X., Du J., Mai K., Ai Q. Molecular cloning, characterization, and nutritional regulation of Elovl6 in large yellow croaker (Larimichthys crocea) Int. J. Mol. Sci. 2019;20:1801. doi: 10.3390/ijms20071801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Tao R., Gao X., Li T., Zhou X., Guan K.L., Xiong Y., Lei Q.Y. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol. Cell. 2013;51:506–518. doi: 10.1016/j.molcel.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47:1211–1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell E., Crown S.B., Fox D.B., Kitir B., Ilkayeva O.R., Olsen C.A., Grimsrud P.A., Hirschey M.D. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 2016;17:1463–1472. doi: 10.1016/j.celrep.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies K.J., Zhang H., Katsyuba E., Auwerx J. Protein acetylation in metabolism - metabolites and cofactors. Nat. Rev. Endocrinol. 2016;12:43–60. doi: 10.1038/nrendo.2015.181. [DOI] [PubMed] [Google Scholar]
- Migita T., Narita T., Nomura K., Miyagi E., Inazuka F., Matsuura M., Ushijima M., Mashima T., Seimiya H., Satoh Y. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–8554. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- Narita T., Weinert B.T., Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019;20:156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- Ogden C.L., Carroll M.D., Kit B.K., Flegal K.M. Prevalence of childhood and adult obesity in the United States, 2011–2012. Jama. 2014;311:806–814. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrocola F., Galluzzi L., Bravo-San Pedro J.M., Madeo F., Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- Sato R., Okamoto A., Inoue J., Miyamoto W., Sakai Y., Emoto N., Shimano H., Maeda M. Transcriptional regulation of the ATP citrate-lyase gene by sterol regulatory element-binding proteins. J. Biol. Chem. 2000;275:12497–12502. doi: 10.1074/jbc.275.17.12497. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakespear M.R., Halili M.A., Irvine K.M., Fairlie D.P., Sweet M.J. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Sivanand S., Rhoades S., Jiang Q., Lee J.V., Benci J., Zhang J., Yuan S., Viney I., Zhao S., Carrer A. Nuclear acetyl-CoA production by ACLY promotes homologous recombination. Mol. Cell. 2017;67:252–265.e256. doi: 10.1016/j.molcel.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman M.L., Rosenberger T.A. Acetate supplementation increases brain histone acetylation and inhibits histone deacetylase activity and expression. Mol. Cell.Biochem. 2011;352:173–180. doi: 10.1007/s11010-011-0751-3. [DOI] [PubMed] [Google Scholar]
- Song Z.-M., Lin H., Yi X.-M., Guo W., Hu M.-M., Shu H.-B. KAT5 acetylates cGAS to promote innate immune response to DNA virus. Proc. Natl. Acad. Sci. U. S. A. 2020;117:21568–21575. doi: 10.1073/pnas.1922330117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., McCaffery J.M., Irizarry R.A., Boeke J.D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Wang T., Yan J., Xu W., Ai Q., Mai K. Characterization of cyclooxygenase-2 and its induction pathways in response to high lipid diet-induced inflammation in Larmichthys crocea. Sci. Rep. 2016;6:19921. doi: 10.1038/srep19921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.P., Hunter D., Huber R., Lemieux J., Slaymaker S., Vaddi K., Charo I., Leibel R.L., Ferrante A.W., Jr. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J. Clin. Invest. 2006;116:115–124. doi: 10.1172/JCI24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Woldegiorgis G., Spennetta T., Corkey B., Williamson J., Shrago E. Extraction of tissue long-chain acyl-CoA esters and measurement by reverse-phase high-performance liquid chromatography. Anal.Biochem. 1985;150:8–12. doi: 10.1016/0003-2697(85)90434-8. [DOI] [PubMed] [Google Scholar]
- Yan J., Liao K., Wang T., Mai K., Xu W., Ai Q. Dietary lipid levels influence lipid deposition in the liver of large yellow croaker (Larimichthys crocea) by regulating lipoprotein receptors, fatty acid uptake and triacylglycerol synthesis and catabolism at the transcriptional level. PLoS One. 2015;10:e0129937. doi: 10.1371/journal.pone.0129937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Zhou Y., Wu M., Li X., Mai K., Ai Q. ω-6 Polyunsaturated fatty acids (linoleic acid) activate both autophagy and antioxidation in a synergistic feedback loop via TOR-dependent and TOR-independent signaling pathways. Cell Death Dis. 2020;11:1–19. doi: 10.1038/s41419-020-02750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Ko B., Hensley C.T., Jiang L., Wasti A.T., Kim J., Sudderth J., Calvaruso M.A., Lumata L., Mitsche M. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell. 2014;56:414–424. doi: 10.1016/j.molcel.2014.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Li L., Chen L., Yuan W., Dong L., Zhang Y., Wu H., Wang C. PARP-1 mediates LPS-induced HMGB1 release by macrophages through regulation of HMGB1 acetylation. J. Immunol. 2014;193:6114–6123. doi: 10.4049/jimmunol.1400359. [DOI] [PubMed] [Google Scholar]
- Yu C., Chen Y., Cline G.W., Zhang D., Zong H., Wang Y., Bergeron R., Kim J.K., Cushman S.W., Cooney G.J. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J. Biol. Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
- Zaini M.A., Müller C., de Jong T.V., Ackermann T., Hartleben G., Kortman G., Gührs K.-H., Fusetti F., Krämer O.H., Guryev V. A p300 and SIRT1 regulated acetylation switch of C/EBPα controls mitochondrial function. Cell Rep. 2018;22:497–511. doi: 10.1016/j.celrep.2017.12.061. [DOI] [PubMed] [Google Scholar]
- Zhu S., Xiang X., Xu X., Gao S., Mai K., Ai Q. TIR domain-containing adaptor-inducing interferon-β (TRIF) participates in antiviral immune responses and hepatic lipogenesis of large yellow croaker (Larimichthys crocea) Front. Immunol. 2019;10:2506. doi: 10.3389/fimmu.2019.02506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All relevant data are within the manuscript and Supplementary Material. The mass spectrometry proteomics data from this publication have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (Ma et al., 2019) with the dataset identifier PXD027084.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.