Abstract
The NK-2 homeobox genes have been shown to play critical roles in the development of specific organs and tissues. Nkx2.6 is a member of the NK-2 homeobox gene family and is most closely related to the Drosophila tinman gene. Nkx2.6 is expressed in the caudal pharyngeal pouches, the caudal heart progenitors, the sinus venosus, and the outflow tract of the heart and in a short segment of the gut at early stages of embryogenesis. To investigate the function of Nkx2.6 in vivo, we generated mice with null mutations of Nkx2.6 by the gene targeting technique. Homozygous Nkx2.6 mutant mice were viable and fertile. There were no obvious abnormalities in the caudal pharyngeal pouch derivatives (the thymus, parathyroid glands, and thyroid gland), heart, and gut. Expression of Nkx2.6 overlaps that of Nkx2.5 in the pharynx and heart and that of Nkx2.3 in the pharynx. Interestingly, in mutant embryos homozygous for Nkx2.6, Nkx2.5 expression extended to the lateral side of the pharynx, suggesting a compensatory function of Nkx2.5 in the mutant pharyngeal pouches.
The NK-2 family genes are a divergent class of homeobox transcription factors. Four NK genes, NK-1 to -4, were first isolated in Drosophila (10), and later these genes were classified into two groups, the NK-1 and NK-2 families (4). The products of the NK-2 family genes have a characteristic tyrosine at position 54 of the homeodomain, and the products of most members also contain two other conserved domains, the TN domain, near the amino terminus, and the NK-2-specific domain, just carboxy terminal to the homeodomain (6). The NK-2 gene family, containing Drosophila NK-2, -3, and -4, now comprises human, mouse, rat, chicken, Xenopus zebra fish, Caenorhabditis elegans, leech, and planaria genes (5, 6, 17).
So far, nine different NK-2 genes have been isolated in mice. Nkx2.1, also known as TTF1 or T/ebp, is expressed in the thyroid anlage, the fetal bronchial epithelium, and the restricted region of the forebrain (13). It was shown by gene targeting that this gene was required for formation of the lung, thyroid gland, pituitary gland, and ventral forebrain (11). Nkx2.2 is expressed in the ventral forebrain at early stages of development (21), and later on Nkx2.2 expression is observed in the pancreatic bud and β cells in the pancreas (24). Homozygous Nkx2.2 mutant mice showed abnormal ventral neuronal patterning at early stages. At a neonatal stage, these mice developed marked hyperglycemia due to a lack of β cells in the pancreas (24). Nkx2.3 is expressed in gut mesenchyme, the epithelium of branchial arches, and the spleen during development (19). Disruption of Nkx2.3 by gene targeting caused abnormal development of the small intestine and spleen (20). Nkx2.5 (also called Csx) starts to be expressed in the cardiac crescent and continues to be expressed in the myocardium during development and in the adult (12, 14). Null mutation of Nkx2.5 arrested heart formation at the looping stage (15, 25), and heterozygous mutations of human Nkx2.5 were shown to cause familial atrial septal defect and atrioventricular conduction delays (23).
Nkx2.6 (also called Tix) is a member of the NK-2 gene family and is closely related to the Drosophila tinman gene in terms of the homeodomain sequence and the genomic organization (1, 6, 26). Nkx2.6 is expressed in the caudal pharyngeal pouches, restricted regions of the gut endoderm, the sinus venosus, and the outflow tract of the developing heart (2, 18, 26). To examine the function of Nkx2.6 in vivo, we generated mice with a null mutation of Nkx2.6 by gene targeting. Surprisingly, homozygous mutant mice showed no obvious defects, raising the possibility of redundant functions of NK-2 genes in these regions.
MATERIALS AND METHODS
Gene targeting.
An Nkx2.6 genomic clone was isolated from a mouse 129Sv genomic library using a genomic fragment containing exon 2 of Nkx2.6 (26). A 5.0-kb upstream fragment containing 5′ flanking sequence, the first exon, and part of the intron and a 2.5-kb downstream fragment were ligated into pPNT (27). R1 embryonic stem (ES) cells (16) were cultured on mouse embryonic fibroblast feeder layers in high-glucose Dulbecco's modified Eagle medium containing 15% fetal calf serum and 103 U of leukemia inhibitory factor per ml. ES cells were electroporated with 30 μg of the targeting vector and were cultured on neomycin-resistant mouse embryonic fibroblast feeders with 300 μg of G418 per ml and 2 μM ganciclovir for 7 days. Forty-three drug-resistant colonies were picked up and genotyped by Southern blotting. Four correctly targeted clones were obtained, and two of them were injected into blastocysts from C57BL/6J mice. Male chimeric mice were bred with female C57BL/6J mice to test for germ line transmission.
Genotyping of progeny.
DNA was isolated from yolk sacs or tail biopsy specimens of weaned mice. PCR was performed to genotype embryos and mice. Results of PCR assays were confirmed by Southern blot analysis. The primers used for detection of the wild-type allele were 5′-GCATCCGTGTTTGTGAAGTGTG-3′ and 5′-TGTGGCTTTTGTACCCTCCAGAG-3′. Primers 5′-GCATCCGTGTTTGTGAAGTGTG-3′ and 5′-TTCCTGACTAGGGGAGGAGTAGAAG-3′ were used to detect the targeted allele.
Histological analysis.
Tissues were fixed with 4% paraformaldehyde at 4°C overnight, dehydrated through graded ethanol and xylene, and embedded in paraffin wax. In situ hybridization was performed as described previously (25). Whole-mount in situ hybridization was performed using a digoxigenin-labeled RNA probe as described previously (7). The Nkx2.5 and Nkx2.6 probes comprised nucleotides 313 to 648 (14) and 146 to 360 (2), respectively. They were subcloned into pBluescript (Stratagene, La Jolla, Calif.) and transcribed using T7 RNA polymerase (Promega, Madison, Wis.). Sense riboprobes for Nkx2.5 and Nkx2.6 gave no signals (data not shown). The entire coding region of Pax9, kindly provided by H. Peters (Brigham and Women's Hospital, Boston, Mass.), was subcloned into pBluescript and transcribed using T7 RNA polymerase. Probes for the atrial natriuretic factor (ANF), brain natriuretic peptide (BNP), and myosin light chain 2V (MLC2V) were described previously (25).
RESULTS
Generation of Nkx2.6 mutant mice.
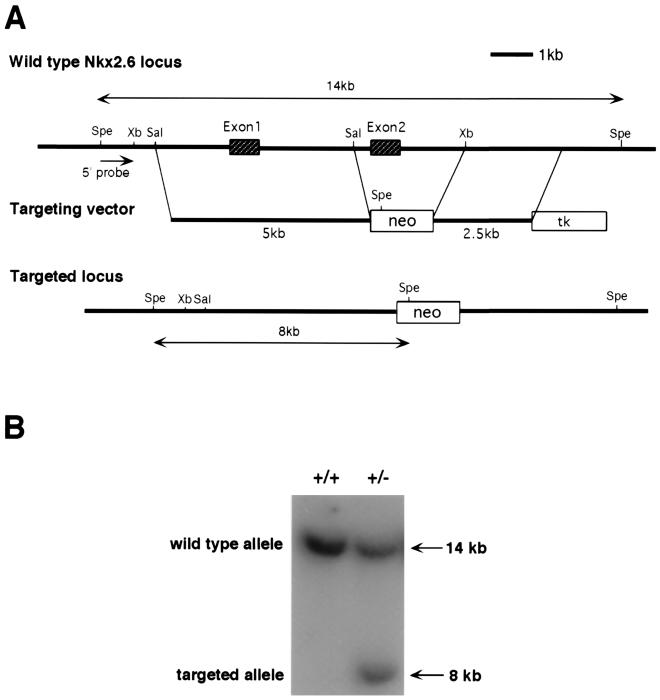
To inactivate Nkx2.6, we constructed a replacement vector that would result in deletion of the entire second exon, encoding the homeodomain and NK-2-specific domain (Fig. 1A). After electroporation and drug selection, 43 drug-resistant colonies were obtained. Southern blot analysis identified four independent clones that had been correctly targeted at the Nkx2.6 locus (Fig. 1B). All of the four clones contained a single integration of the targeting vector, as demonstrated by Southern blotting with a neo probe (data not shown). Two clones with homologous recombination were injected into blastocysts from C57BL/6J mice, and both of them transmitted the targeted allele through the germ line. The two lines of mutant mice showed the same phenotype.
FIG. 1.
Gene targeting of Nkx2.6. (A) The Nkx2.6 genomic locus and the structure of the targeting vector are shown. Homologous recombination resulted in the replacement of the second exon, which encodes the homeodomain and the NK-2-specific domain. Spe, SpeI; Xb, XbaI; Sal, SalI. (B) Genotyping of ES cell clones. Genomic DNA was digested with SpeI and analyzed by Southern blotting using the 5′ probe (SpeI-XbaI fragment).
Analysis of Nkx2.6 mutant mice.
Heterozygous Nkx2.6 mutant mice exhibited no obvious defects and were fertile. In crosses of heterozygous mutant mice, mice with all three possible genotypes were born with normal Mendelian frequency (ratio of +/+ to +/− to −/− = 61:111:59), indicating that disruption of Nkx2.6 did not cause embryonic lethality.
It was reported that Nkx2.6 transcripts were detected in embryos between embryonic day 8.0 (E8.0) and E11.5, but not in later-stage embryos (2, 18). Thus, we examined Nkx2.6 expression at the neonatal stage by in situ hybridization using serial sections of the whole neonates. However, we could not detect transcripts for Nkx2.6 in any organs or tissues of neonates (data not shown). We also examined expression of Nkx2.6 by in situ hybridization in adult organs, such as the brain, heart, lung, liver, pancreas, esophagus, stomach, intestine, kidney, and adrenal gland, and in adult skeletal muscle and aorta. However, we could not observe Nkx2.6 expression in any adult organs or tissues examined (data not shown). Therefore, we focused on the analysis of the tissues and organs where Nkx2.6 is expressed at early stages of embryogenesis.
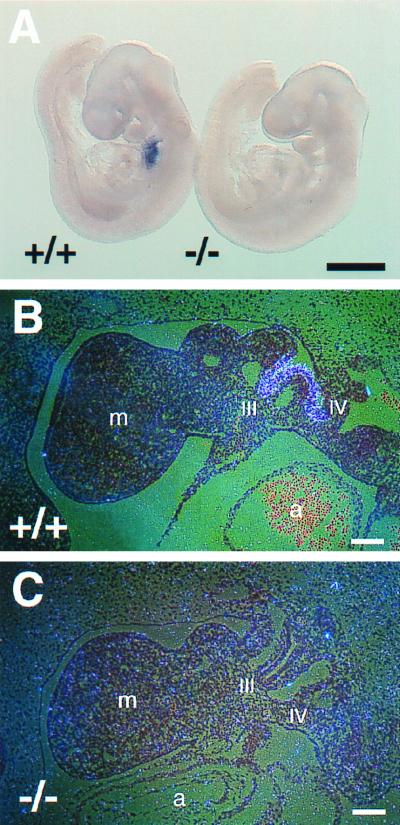
We first confirmed the absence of Nkx2.6 transcripts in homozygous mutant (Nkx2.6−/−) embryos by in situ hybridization using a probe containing the first and second exons of Nkx2.6. In wild-type embryos, Nkx2.6 expression was clearly detected in the pharyngeal pouches (Fig. 2A and B). In contrast, no transcripts could be detected in Nkx2.6−/− embryos (Fig. 2A and C), indicating that we created mice with a null mutation of Nkx2.6.
FIG. 2.
Absence of Nkx2.6 transcripts in homozygous mutant embryos. (A) Wild-type (+/+) and homozygous mutant (−/−) embryos at E9.5 were analyzed by whole-mount in situ hybridization using an Nkx2.6 probe containing the first and second exons. Note the complete absence of Nkx2.6 transcripts in the pharynx of the homozygous mutant embryo. Scale bar = 0.5 mm. (B and C) Tissue sections of wild-type (+/+) and homozygous mutant (−/−) embryos at E9.5 were hybridized with the Nkx2.6 probe. In the wild-type embryo, Nkx2.6 is strongly expressed in the third and fourth pharyngeal pouches. In contrast, no signals were detected in the homozygous mutant embryo. m, mandibular component of the first pharyngeal arch; III and IV, third and fourth pharyngeal pouches; a, atrium. Scale bars = 100 μm.
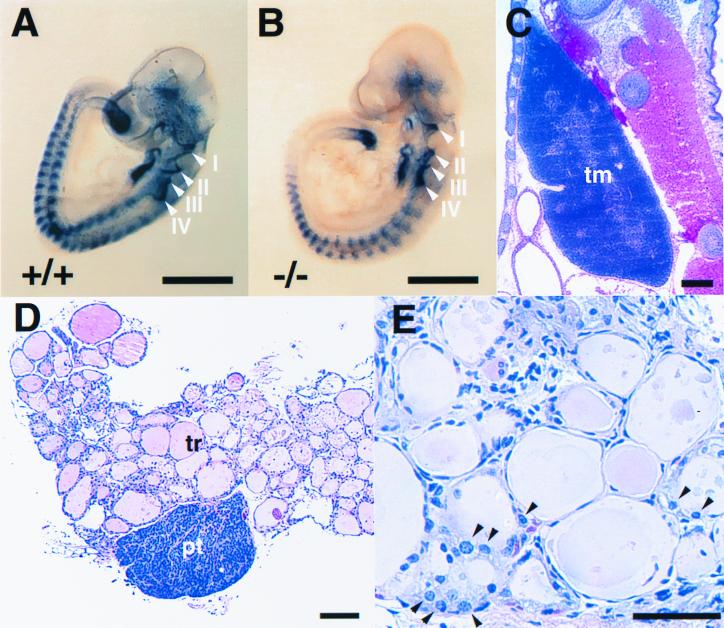
We then performed whole-mount in situ hybridization of wild-type and Nkx2.6−/− embryos using a Pax9 probe to examine pharyngeal pouch formation in mutant embryos. All four pharyngeal pouches (I to IV) were present in Nkx2.6−/− embryos (Fig. 3B), as they were in wild-type littermates (Fig. 3A). We next examined formation of caudal pharyngeal pouch derivatives. We could observe normal formation of the thymus and parathyroid gland (Fig. 3C and D). The numbers (means ± standard deviations) of the chief cells in the parathyroid gland were comparable in Nkx2.6−/− and wild-type mice (1,522 ± 168 and 1,536 ± 106, respectively, per 400× microscopic field, six sections from two mice of each group). The parafollicular cells of the thyroid gland also derive from caudal pharyngeal pouches. Histological analysis showed that parafollicular cells normally exist in the thyroid gland of Nkx2.6−/− mice (Fig. 3E). There were no significant differences in the ratio of parafollicular cells to follicle epithelial cells between Nkx2.6−/− and wild-type mice (0.26 ± 0.05 and 0.27 ± 0.04, respectively, six sections from three mice of each group). Moreover, the serum calcium levels were comparable in Nkx2.6−/− and wild-type mice (9.2 ± 0.3 and 9.5 ± 0.5 mg/dl, respectively, five mice from each group).
FIG. 3.
Caudal pharyngeal pouch derivatives normally formed in homozygous mutant mice. (A and B) Whole-mount in situ hybridization of wild-type (+/+) and homozygous mutant (−/−) embryos using a Pax9 probe. The arrowheads show the pharyngeal pouches (I to IV). Scale bars = 0.5 mm. (C) Normal formation of the thymus in a homozygous mutant neonate. Hematoxylin and eosin (H&E) staining was used. tm, thymus. Scale bar = 300 μm. (D) Normal histology of the parathyroid gland in a homozygous mutant mouse (5 months old). H&E staining was used. tr, thyroid gland; pt, parathyroid gland. Scale bar = 100 μm. (E) Parafollicular cells in the mutant thyroid gland (5 months old), indicated by arrowheads. H&E staining was used. Scale bar = 50 μm.
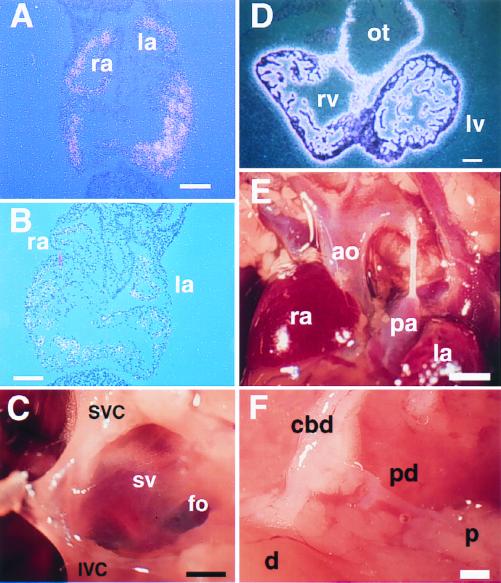
We next examined the heart and gut in Nkx2.6−/− mice. Since Nkx2.6 expression can be detected in caudal heart progenitors at E8.0, we examined expression of ANF and BNP in the mutant atrium. However, both ANF and BNP were normally expressed in the atrium in Nkx2.6−/− embryos (Fig. 4A and B). Nkx2.6 is also expressed in the sinus venosus and in the myocardium of the outflow tract. No abnormalities were detected in either the superior and inferior venae cavae (Fig. 4C), the sinus venarum cavum of the right atrium (Fig. 4D), or the coronary sinus (data not shown), which are all derived from the sinus venosus. We also examined expression of MLC2V, which marks the myocardium of the ventricle and the outflow tract during development, in Nkx2.6−/− embryos. However, we could observe normal expression of MLC2V in the myocardium of the outflow tract in Nkx2.6−/− embryos (Fig. 4E). There were no abnormalities in the outflow tract region, including the pulmonary artery and the aorta (Fig. 4F), in adult Nkx2.6−/− mice, either. In the embryonic gut, Nkx2.6 is expressed in a segment at the foregut-midgut junction spanning the proximal parts of the common bile duct and pancreatic ducts (2). However, there were no anatomical defects in these regions. Moreover, there were no significant differences in the serum bilirubin concentration between Nkx2.6−/− and wild-type mice (0.2 ± 0.1 and 0.2 ± 0.1 mg/dl, respectively, six mice from each group).
FIG. 4.
Normal gene expression and morphology of the heart and gut in homozygous mutant mice. (A and B) Normal expression of ANF (A) and BNP (B) in the atria of Nkx2.6−/− mice. In situ hybridization revealed normal expression of ANF and BNP in both the atria and the ventricles of Nkx2.6−/− embryos (E10.5). ra, right atrium; la, left atrium. Scale bars = 100 μm. (C) Sinus venosus derivatives in a homozygous mutant mouse (6 months old). SVC, superior vena cava; IVC, inferior vena cava; sv, sinus venarum cavum; fo, fossa ovalis. Scale bar = 0.5 mm. (D) Normal expression of MLC2V in the ventricle and outflow tract in Nkx2.6−/− mice. rv, right ventricle; lv, left ventricle; ot, outflow tract. Scale bar = 100 μm. (E) Normal morphology of the outflow tract in a homozygous mutant mouse (6 months old). ra, right atrium; ao, aorta; pa, pulmonary artery; la, left atrium. Scale bar = 1 mm. (F) Normal formation of the common bile duct (cbd) and the pancreatic duct (pd) in a homozygous mutant mouse (6 months old). A part of the pancreatic tissue was removed to show the pancreatic duct. d, duodenum; p, pancreas. Scale bar = 0.5 mm.
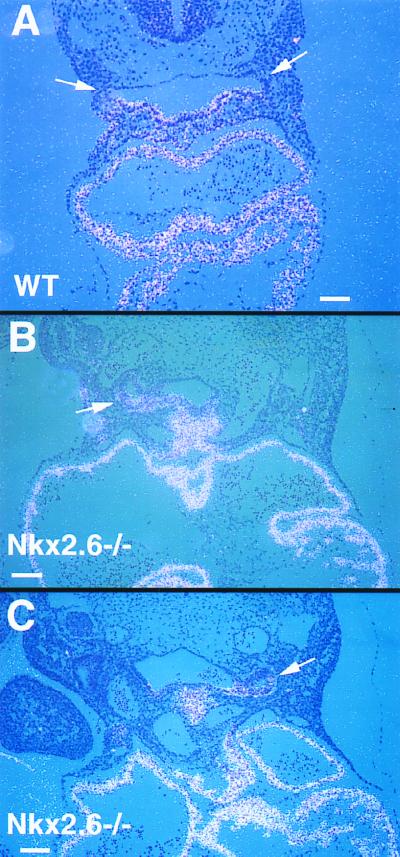
Since Nkx2.3 and Nkx2.5 are expressed in the pharyngeal region similarly to Nkx2.6, we tested the hypothesis that alterations in Nkx2.3 or Nkx2.5 expression may compensate for a loss of Nkx2.6 expression in the pharynx in Nkx2.6−/− mice. Normally, Nkx2.5 expression is restricted to the ventral side of the pharynx, the pharyngeal floor (Fig. 5A). Interestingly, however, in Nkx2.6−/− embryos Nkx2.5 expression extended to the lateral side of the pharynx, where Nkx2.6 is normally expressed (Fig. 5B and C). These results suggested that ectopic expression of Nkx2.5 might compensate for a loss of Nkx2.6 functions in the mutant pharynx. There were no differences in expression of Nkx2.3 between Nkx2.6−/− and wild-type mice (data not shown).
FIG. 5.
Expression of Nkx2.5 in Nkx2.6−/− embryos. (A) Expression of Nkx2.5 in a wild-type (WT) embryo at E9.5. Nkx2.5 is expressed in the pharyngeal floor. Note the absence of Nkx2.5 transcripts in the pharyngeal pouches (arrows). Scale bar = 100 μm. (B and C) Expression of Nkx2.5 in Nkx2.6−/− embryos at E9.5. Note the ectopic expression of Nkx2.5 in pharyngeal pouches (arrows). Scale bars = 100 μm.
DISCUSSION
In spite of the intensive analysis, we could not detect any obvious defects in Nkx2.6−/− mice. So far, we have observed Nkx2.6−/− mice for up to 2 years, but they showed no differences, including in life span. The partially overlapping expression patterns of the NK-2 genes may have redundant functions in specific organs and tissues. It is well known that expression of Hox genes partially overlaps along the anterior-posterior axis. This expression pattern led to the concept of a “Hox code,” which means that a particular combination of Hox genes may play critical roles in specification and formation of particular tissues and organs (8, 9). Thus, the partially overlapping expression patterns of the NK-2 genes raises the possibility of an “Nkx code” (22). Moreover, as shown in this study, alterations in expression patterns of another NK-2 gene may also work as a compensatory mechanism. Ectopic expression of Nkx2.5 may make up for a loss of Nkx2.6 in the mutant pharyngeal pouches.
Interestingly, in gene targeting studies of the NK-2 family genes, not all sites of expression of the NK-2 genes showed phenotypic alterations. For example, homozygous Nkx2.3 or Nkx2.5 mutant mice showed no obvious abnormalities in the pharynx (15, 20, 25). The gut phenotype of Nkx2.3 mutant mice was confined to the small intestine (20). Furthermore, disruption of Nkx2.5 did not abolish the cardiac cell lineage (25), although tinman mutant flies have no cardiac precursor cells (3). It would be interesting to examine the effects of double and triple inactivation of Nkx2.6, Nkx2.3, and Nkx2.5 in vivo.
ACKNOWLEDGMENTS
We thank I. Komuro and H. Inagaki for the initial isolation of an Nkx2.6 genomic clone.
This work was supported by a grant from NIH to S.I. M.T. is a Paul Dudley White Fellow of the American Heart Association, Massachusetts affiliate.
REFERENCES
- 1.Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 2.Biben C, Hatzistavrou T, Harvey R P. Expression of NK-2 class homeobox gene Nkx2-6 in foregut endoderm and heart. Mech Dev. 1998;73:125–127. doi: 10.1016/s0925-4773(98)00037-9. [DOI] [PubMed] [Google Scholar]
- 3.Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 4.Burglin T R. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the homeobox genes. Oxford, England: Oxford University Press; 1993. pp. 25–71. [Google Scholar]
- 5.Garcia-Fernandez J, Baguna J, Salo E. Planarian homeobox genes: cloning, sequence analysis, and expression. Proc Natl Acad Sci USA. 1991;88:7338–7342. doi: 10.1073/pnas.88.16.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey R P. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- 7.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the mouse embryo: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 352–367. [Google Scholar]
- 8.Hunt P, Krumlauf R. Deciphering the Hox code: clues to patterning branchial regions of the head. Cell. 1991;66:1075–1078. doi: 10.1016/0092-8674(91)90029-x. [DOI] [PubMed] [Google Scholar]
- 9.Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y, Nirenberg M. Drosophila NK-homeobox genes. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox C H, Ward J M, Gonzalez F J. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 12.Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- 14.Lints T J, Parsons L M, Hartley L, Lyons I, Harvey R P. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- 15.Lyons I, Parsons L M, Hartley L, Li R, Andrews J E, Robb L, Harvey R P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 16.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nardelli-Haefliger D, Shankland M. Lox10, a member of the NK-2 homeobox gene class, is expressed in a segmental pattern in the endoderm and in the cephalic nervous system of the leech Helobdella. Development. 1993;118:877–892. doi: 10.1242/dev.118.3.877. [DOI] [PubMed] [Google Scholar]
- 18.Nikolova M, Chen X, Lufkin T. Nkx2.6 expression is transiently and specifically restricted to the branchial region of pharyngeal-stage mouse embryos. Mech Dev. 1997;69:215–218. doi: 10.1016/s0925-4773(97)00174-3. [DOI] [PubMed] [Google Scholar]
- 19.Pabst O, Schneider A, Brand T, Arnold H H. The mouse Nkx2-3 homeodomain gene is expressed in gut mesenchyme during pre- and postnatal mouse development. Dev Dyn. 1997;209:29–35. doi: 10.1002/(SICI)1097-0177(199705)209:1<29::AID-AJA3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 20.Pabst O, Zweigerdt R, Arnold H H. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 21.Price M, Lazzaro D, Pohl T, Mattei M G, Ruther U, Olivo J C, Duboule D, Di Lauro R. Regional expression of the homeobox gene Nkx-2.2 in the developing mammalian forebrain. Neuron. 1992;8:241–255. doi: 10.1016/0896-6273(92)90291-k. [DOI] [PubMed] [Google Scholar]
- 22.Reecy J M, Yamada M, Cummings K, Sosic D, Chen C Y, Eichele G, Olson E N, Schwartz R J. Chicken Nkx-2.8: a novel homeobox gene expressed in early heart progenitor cells and pharyngeal pouch-2 and -3 endoderm. Dev Biol. 1997;188:295–311. doi: 10.1006/dbio.1997.8641. [DOI] [PubMed] [Google Scholar]
- 23.Schott J J, Benson D W, Basson C T, Pease W, Silberbach G M, Moak J P, Maron B J, Seidman C E, Seidman J G. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- 24.Sussel L, Kalamaras J, Hartigan O C D J, Meneses J J, Pedersen R A, Rubenstein J L, German M S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M, Kasahara H, Bartunkova S, Schinke M, Komuro I, Inagaki H, Lee Y, Lyons G E, Izumo S. Vertebrate homologs of tinman and bagpipe: roles of the homeobox genes in cardiovascular development. Dev Genet. 1998;22:239–249. doi: 10.1002/(SICI)1520-6408(1998)22:3<239::AID-DVG6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]