Summary
Oral mucosal tissue is composed of several cell types that are difficult to dissociate while maintaining high cell viability. We describe a protocol for the preparation and dissociation of human buccal and gingival oral mucosal tissue to a high-viability single-cell suspension composed of heterogeneous cell types. This heterogeneous cell suspension can subsequently be used for cytometric analyses or to generate single-cell RNA sequencing libraries.
For complete details on the use and execution of this protocol, please refer to Williams et al. (2021).
Subject areas: Cell Biology, Cell isolation, Single Cell, Health Sciences, Immunology
Graphical abstract
Highlights
-
•
Dissociation of oral mucosal tissue and preparation of a single-cell suspension
-
•
Confirmation of highly viable heterogenous single cells after isolation
-
•
Cell suspensions suitable for single-cell RNA sequencing and cytometric analysis
-
•
Adapted for human gingival and buccal mucosa tissues
Oral mucosal tissue is composed of several cell types that are difficult to dissociate while maintaining high cell viability. We describe a protocol for the preparation and dissociation of human buccal and gingival oral mucosal tissue to a high-viability single-cell suspension composed of heterogeneous cell types. This heterogeneous cell suspension can subsequently be used for cytometric analyses or to generate single-cell RNA sequencing libraries.
Before you begin
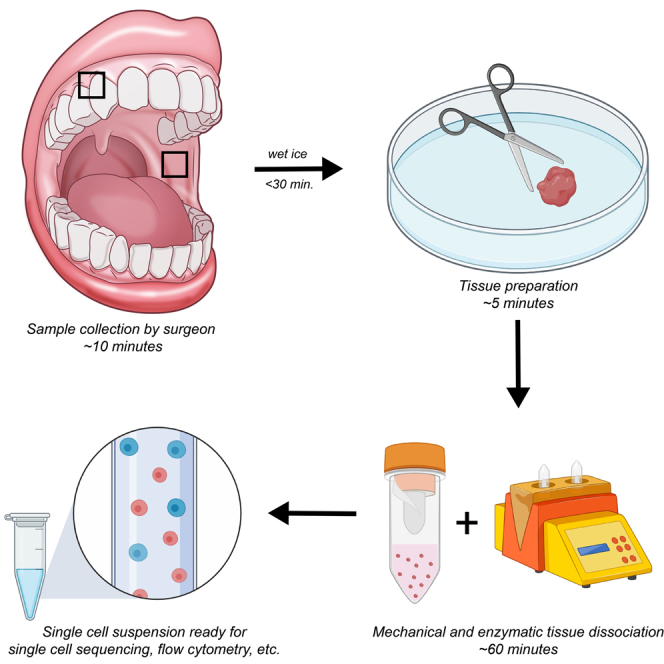
The protocol below describes specific steps for dissociation of human oral mucosal tissue to a high-viability single-cell suspension suitable for downstream assays such as flow cytometry and single cell sequencing. This protocol was modified from Derycke et al. (2012).
-
1.
Human Biopsies should be obtained under an IRB (Institutional Review Board)-approved clinical protocol, with informed consent from human subjects. Protocol is based on a single oral biopsy for downstream analysis.
-
2.
Oral tissue samples should be transferred to the laboratory on wet ice within 30 min from surgery.
-
3.
Ensure adequate supply of stock solutions as described in materials and equipment.
-
4.
Perform all preparation steps for tissue samples and stock solutions in a biosafety cabinet.
-
5.
Pre-heat an incubator shaker to 37°C.
-
6.
Equilibrate a centrifuge with 50cc tube rotor to room temperature.
-
7.
Make digestion buffer and tissue culture medium. (10 min; refer to materials and equipment for recipe tables.)
-
8.
Make RBC lysis and re-equilibration solutions. (15 min; refer to materials and equipment for recipe tables.)
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Oral gingiva (age range: 21–61 yrs; both male and female) | Human Biopsy tissue (NIH Clinical Center) | Table S1 in Williams et al. (2021) |
| Buccal mucosa (age range: 23–33 yrs; both male and female) | Human Biopsy tissue (NIH Clinical Center) | Table S1 in Williams et al. (2021) |
| Chemicals, peptides, and recombinant proteins | ||
| DNase I | Sigma | DN25-1G |
| Collagenase II | Worthington Biochemical Corp. | LS004176 |
| RPMI 1640 | Lonza/Biowhittaker | 12–167F |
| Fetal Bovine Serum (FBS) | Gemini | 100–106 |
| PBS | Gibco | 10010–023 |
| Germicidal bleach | Clorox | N/A |
| 10× PBS | Fisher Scientific | BP3994 |
| 7.5% BSA | Sigma | A8412-100mL |
| Other | ||
| Scissors | Fine Science Tools | 14058–09 |
| Forceps | Roboz | RS-8254 |
| Gentlemacs Dissociator | Miltenyi | 130-093-235 |
| Gentlemacs C tube | Miltenyi | 130-096-334 |
| 70 micron cell strainer | Falcon | 352350 |
| 500 mL Sterile Filter | Millipore | S2GPU05RE |
| Cellometer | Nexcelom | Auto 2000 |
| Petri dish | Falcon | 351029 |
| 50cc centrifuge tube | Falcon | 352098 |
| Centrifuge | Beckman Coulter | Allegra X-14R |
| 3 mL syringe | BD | 309585 |
| AOPI | Nexcelom | CS2-0106-5ML |
| Cell counting chamber | Nexcelom | CHT4-PD100 |
| Incubator/shaker | New Brunswick | M1282-0004 |
| 2 mL Sarstedt tube | Sarstedt | 72.693.005 |
Materials and equipment
Tissue culture media
| 50 mL RPMI 1640 + 1 mL of FBS for final of 2% FBS in 50cc tube. |
Stock solution can be stored for 2 months at 4°C.
Digestion media
| Reagent | Final concentration | Amount |
|---|---|---|
| Tissue culture media | – | 10 mL |
| Collagenase II | 2 mg/mL | 20 mg |
| DNase I | 0.04 mg/mL | 0.4 mg |
| Total | – | 10 mL |
Rock tube back and forth to mix.
Make day of experiment and keep on wet ice until use.
Hypotonic lysis solution
| 6.8 mL 10× PBS in 300 mL water. |
Swirl to mix, then sterile filter.
Store 4C° for 6 months.
Re-equilibration buffer
| 108.1 mL 10× PBS in 500 mL water. |
Swirl to mix, then sterile filter.
Store 4°C for 6 months.
Step-by-step method details
Sample collection
Timing: 30 min
Oral mucosal biopsies (buccal mucosa 3–4 mm punch and/or gingival collar single sample ) are obtained by qualified clinicians under local anesthesia. Collected tissue samples are placed in a 2 mL Sarstedt tube containing 1 mL of RPMI 1640 (no FBS) on wet ice and transferred quickly (within 15–30 min) to the laboratory for processing.
Initial tissue preparation
Timing: 5 min
The purpose of this step is to mince the tissue into smaller pieces that can be further digested enzymatically. Tissue is transported to the lab on ice, but dissection and subsequent steps are performed at room temperature (20°C–24°C) unless otherwise stated.
-
1.Rinse tissue with PBS to remove remaining blood.
-
a.Place tissue in bottom of petri dish, put 1–2 mL of PBS on top of it.
-
b.With forceps, gently agitate tissue in PBS.
-
c.Transfer the tissue with forceps to top of petri dish and add 200ul tissue culture media to prevent tissues from drying out.
-
a.
-
2.
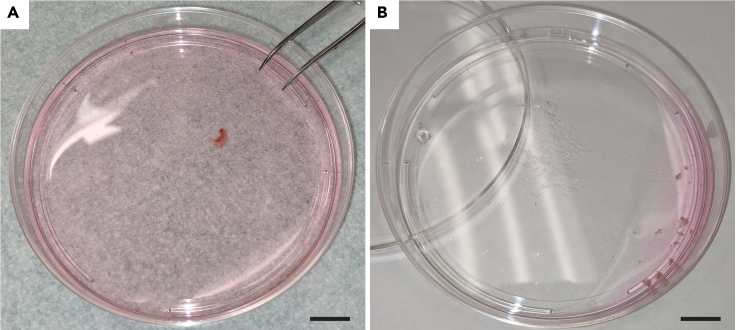
Mince tissue in petri dish with tissue culture media by using forceps to hold the tissue and scissors to cut. Keep cutting until tissue is minced into pieces <1 mm in all dimensions. (Figures 1A and 1B)
-
3.Transfer minced tissue to a gentleMACS C tube containing 10 mL of tissue culture media.
-
a.Minced pieces can be transferred using a 1000ul pipette tip that has the narrow end cut off. (Figure 2)
-
b.Remove all visible tissue, then rinse petri dish with 1 mL tissue culture media. Collect media and transfer to C tube.
-
a.
Figure 1.
Gingiva dissections
(A) Gingiva in petri dish containing tissue culture media (scale bar 1 cm).
(B) Minced tissue prior to gentleMACS
Figure 2.
Aspirate minced tissue using a 1000ul tip with the narrow end cut off
Mechanical and enzymatic tissue dissociation
Timing: 60 minfor steps 4–13
-
4.
Attach C tube containing tissue upside down to gentleMACS dissociator, then run program m_lung_01.01 (8 s at 168 rounds per run no heat). (Figure 3)
CRITICAL: During the dissociation process, some tissue may get stuck to the plastic blades in the C tube. If this occurs, use forceps to transfer the tissue to the liquid prior to centrifugation.
Note: All centrifugation steps are carried out at room temperature.
-
5.
Centrifuge C tube at 300 × g for 5 min.
-
6.
Dispose of the supernatant into a 10% bleach solution and add 10 mL of digestion buffer to C tube containing tissue.
-
7.
Place tube in Innova 44 incubator shaker (or equivalent) at 37°C 220 RPM for 45 min.
-
8.
Attach C tube upside down to gentleMACS dissociator and run program m_lung_02.01 (38 s 2083 rounds per run no heat).
-
9.
Centrifuge C tube at 300 × g for 5 min.
-
10.
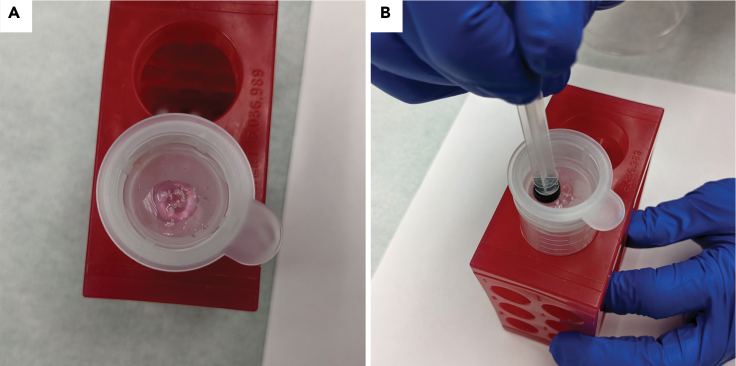
Resuspend sample in 0.5 mL tissue culture media by pipetting and decant contents through a 70 μm cell strainer fitted onto a 50cc tube. Use the rubber end of a 3 mL syringe plunger to press any remaining tissue through the filter. (Figures 4A and 4B)
-
11.
Rinse C tube with remaining tissue culture media and pour through filter.
-
12.
Centrifuge 300 × g for 5 min.
-
13.Dispose of supernatant into a 10% bleach solution.Optional: Timing 10-30 min.Perform red blood cell lysis if necessary for downstream application. Lyse red blood cells by resuspending cells in 5 mL of hypotonic lysis solution and rick the tube back and forth for 60 s. Add 5 mL of re-equilibration buffer, then top off tube with PBS. Centrifuge 5 min at 300 x g. Repeat lysis steps if RBC are still present.
-
14.
Resuspend sample in 100 μL of 0.01% BSA in PBS or other resuspension solution according to the single-cell technology protocol and transfer to a 1.5 mL microcentrifuge tube for single-cell library preparation. Resuspend in 100 μL PBS for flow cytometry, or other buffer depending on downstream application. Keep sample on wet ice and proceed as quickly as possible with subsequent processing as cells will begin to attach to each other. For scRNA-seq, filter the sample immediately before loading the cells into the instrument for the single cell protocol.
Figure 3.
Image of gentleMACS with C tube attached
Figure 4.
Application of minced tissue to cell strainer
(A) Decant contents into a 70 μm cell strainer.
(B) Press any remaining tissue pieces through cell strainer with rubber end of a syringe.
Cell counting and viability
Timing: 5 min
-
15.
On a small piece of parafilm, mix 10 μL of cell suspension and 10 μL of AOPI.
-
16.
Load 20 μL onto slide and count using Auto 2000. Adjust volume to obtain the required cell recovery target.
Expected outcomes
This protocol allows for successful isolation of all expected major oral mucosal cell types (Williams et al., 2021).
Successful dissociation/digestion should yield a viability of 75% or greater for optimal downstream application results. The cell number is dependent upon the size and health status of the tissue. 3–4 mm oral biopsies of healthy tissues will typically yield >10K cells (10–40K, median 14K). For single cell sequencing the cDNA should yield a distinct peak between 300 bp and 10 kb.
Limitations
This protocol was only tested on buccal mucosa and gingiva and may need to be adapted for other mucosal sites.
All tissues tested were processed within 30 min of biopsy procedure, the protocol may yield different results if tissues are not processed in a timely manner.
Red blood cell lysis may lead to a significant reduction in cell viability and/or count.
Use of a manual method utilizing the same enzyme yields lower cell count and viability.
Troubleshooting
Problem 1
Low cell viability after dissociation (<75%) (step 17 )
Potential solution
Timely processing of tissue after biopsy: Complete the tissue processing procedure as soon as possible following biopsy collection. Work with the surgeon to ensure the tissue is placed in the collection tube on wet ice and transferred immediately to the area where tissue dissociation will occur.
Adjustment of Red Blood Lysis steps: the protocol for red blood cell lysis can reduce cell viability. When preparing the tissue for mincing, wash away as much blood as possible with PBS to minimize the number of red blood cells in the final cell suspension.
Make all solutions fresh just prior to use.
Problem 2
Low cell count (step 17)
Potential solution
Ensure incubator is set at 37°C prior to enzymatic digestion.
Ensure collagenase II is stored properly at 4°C and the tissue is completely minced prior to enzymatic and mechanical digestion.
Make all solutions fresh just prior to use.
The size of the tissue biopsy should be at least 3 mm in diameter (for either single cell library prep or flow cytometry). Healthy tissue biopsies will yield far fewer cells than diseased tissue biopsies of the same size. Adjust biopsy size when possible (>3 mm punch biopsy used in this protocol).
Cells may stick to plasticware. Consider using plastic made of polypropylene, which minimizes cell binding to the side of tubes.
Problem 3
Clumps of cells, not single cells (step 17 )
Potential solution
Ensure DNase I has been added to digestion media.
After mechanical and enzymatic dissociation, thoroughly press any remaining tissue through the 70 μm cell strainer.Filter the sample immediately before loading onto the 10× controller.
Problem 4
High red blood cell number in subsequent sequencing analysis (step 14 )
Potential solution
For future biopsy preparation, consider the following:
Prior to mincing the tissue, attempt to separate as much blood from the tissue as possible. Remove any large coagulates and if necessary, rinse tissue in several exchanges of PBS.
Reduce the number of red blood cells by using the optional red blood cell lysis step.
Problem 5
High mitochondrial gene expression in subsequent sequencing analysis (step 15)
Potential solution
High mitochondrial gene expression can be attributed to cells undergoing apoptosis and is used as a metric in assessing quality of single cell libraries. For future biopsy preparation, ensure that biopsies are processed in a timely manner, as cells within tissues start undergoing apoptosis quickly after biopsy procedure is completed.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Niki Moutsopoulos (nmoutsop@mail.nih.gov).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This study was funded in part by the intramural program of NIH/NIDCR (NMM). Figure illustrations were created in part with Biorender.com.
Author contributions
Conceptualization, T.G.-W. and N.M.M.; methodology, T.G.-W., D.W.W., and N.M.M.; investigation, T.G.-W. and D.W.W.; writing – original draft, T.G.-W.; writing – review and editing, D.W.W. and N.M.M.; funding acquisition and supervision, N.M.M.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Teresa Greenwell-Wild, Email: twild@nih.gov.
Niki Maria Moutsopoulos, Email: nmoutsop@mail.nih.gov.
Data and code availability
This study did not generate new data or code.
References
- Derycke L., Zhang N., Holtappels G., Dutre T., Bachert C. IL-17A as a regulator of neutrophil survival in nasal polyp disease of patients with and without cystic fibrosis. J. Cyst. Fibros. 2012;11:193–200. doi: 10.1016/j.jcf.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Williams D.W., Greenwell-Wild T., Brenchley L., Dutzan N., Overmiller A., Sawaya A.P., Webb S., Martin D., NIDCD/NIDCR Genomics and Computational Biology Core, Hajishengallis G. Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell. 2021;184:4090–4104.e15. doi: 10.1016/j.cell.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new data or code.