Summary
Most non-Hodgkin's lymphomas grow exclusively in the lymph node compartment protected by the tumor microenvironment. To better understand the cellular heterogeneity and the complex interaction between malignant and non-malignant cells, experiments with primary, patient-derived samples are often indispensable. Here, we describe a time-efficient but gentle protocol to process human lymph node samples. This protocol avoids enzymatic or mechanical stress and was optimized for the purpose of generating single-cell suspension suitable for delicate assays, such as single-cell RNA sequencing.
For complete details on the use and execution of this protocol, please refer to Roider et al. (2020).
Subject areas: Cancer, Cell isolation, Flow Cytometry/Mass Cytometry, Health Sciences, Single Cell
Graphical abstract
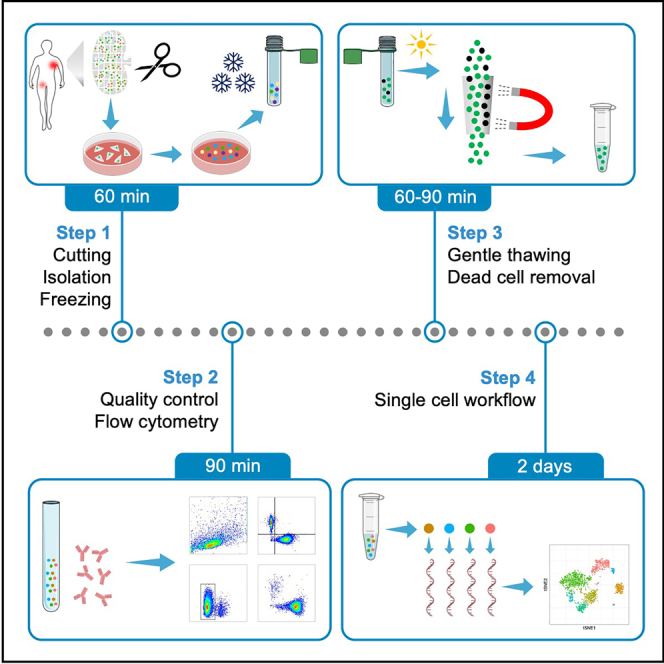
Highlights
-
•
Protocol to isolate and freeze lymphocyte suspensions from human lymph node samples
-
•
Avoiding enzymatic stress to preserve gene expression and epitope profiles
-
•
Optimized thawing and dead cell removal to meet the standards of single-cell sequencing
Most non-Hodgkin's lymphomas grow exclusively in the lymph node compartment protected by the tumor microenvironment. To better understand the cellular heterogeneity and the complex interaction between malignant and non-malignant cells, experiments with primary, patient-derived samples are often indispensable. Here, we describe a time-efficient but gentle protocol to process human lymph node samples. This protocol avoids enzymatic or mechanical stress and was optimized for the purpose of generating single-cell suspension suitable for delicate assays, such as single-cell RNA sequencing.
Before you begin
Introduction
Biobanking of lymph node-derived viable cells is not part of a standard routine in most clinical institutions. However, the majority of T and B cell non-Hodgkin lymphomas (NHL) grow exclusively in the lymph node compartment. Despite the large number of available cell line models (Quentmeier et al., 2019), primary cells are indispensable, especially for studies investigating tumor heterogeneity or tumor microenvironment.
Here, we describe a time-efficient but gentle protocol to isolate lymphocytes from human lymph node samples. Many protocols require enzymatic digestion to prepare single cell suspensions from tissues; however, enzymatic stress not only affects the expression of surface proteins (Reichard and Asosingh, 2019) but can also drastically alter single cell gene expression profiles (Mattei et al., 2020). Our protocol deliberately avoids enzymatic digestion or major mechanical stress and therefore provides an optimized procedure to prepare single cell suspensions from lymph node samples. Before freezing, quality of lymph-node-derived cells is assessed by flow cytometry. We further optimized the thawing procedure and included magnetic removal of dead cells so that thawed samples meet the standards for delicate assays, such as single cell RNA sequencing. Single cell suspensions processed and prepared this way can also be used for other kinds of assays, e.g., drug response profiling (Roider et al., 2020, Roider et al., 2021). Moreover, this protocol can be used to collect lymph node-derived exosomes (Bordas et al., 2020).
General preparations
As lymph node excisions represent relatively invasive diagnostic procedures, ethical approval adhering to regional and institutional regulations is of outstanding importance. Informed consent must be collected of all patients before surgery. Patients whose tissue is being processed must test negatively for hepatitis B, C and HI virus. Excised lymph node tissue must be processed in accordance with regional and institutional safety regulations.
Biobanking of viable lymph node tissue requires specific infrastructure to ensure a standardized workflow and consistently high quality of lymph node-derived cells. Surgeons and perioperative nurses must be instructed in details how to handle lymph node tissue for this specific purpose. We recommend splitting of lymph node tissue immediately after excision. One piece is handled according to the standard diagnostic routine, whereas the second piece is wrapped in a saline-soaked compress and then placed in a sterile container. Subsequent processing of lymph node samples should be performed as soon as possible.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-human CD8 | BioLegend | RRID:AB_314110 |
| BV421 anti-human CD19 | BioLegend | RRID:AB_11142678 |
| BV605 anti-human CD20 | BioLegend | RRID:AB_2563398 |
| PE anti-human Ig light chain κ | BioLegend | RRID:AB_493613 |
| PE/Dazzle 597 anti-human Ig light chain λ | BioLegend | RRID:AB_2687261 |
| PerCP/Cy5.5 anti-human CD3 | BioLegend | RRID:AB_2561628 |
| PE/Cy7 anti-human CD5 | BioLegend | RRID:AB_2275812 |
| AF700 anti-human CD4 | BioLegend | RRID:AB_493743 |
| Viability Dye eFluor506 | Thermo Fisher Scientific | 65-0866-18 |
| Chemicals, peptides, and recombinant proteins | ||
| Fetal bovine serum (FBS) | Gibco | 10500-064 |
| Penicillin/Streptomycin | Gibco | 15140-122 |
| EDTA | Sigma Aldrich | E8008 |
| RPMI medium | Gibco | 21875-034 |
| Trypan blue solution 0.4% | Gibco | 11538886 |
| DMSO | Serva | 20385.01 |
| Critical commercial assays | ||
| MS Columns | Miltenyi | 130-042-201 |
| Dead Cell Removal Kit | Miltenyi | 130-090-101 |
| Software and algorithms | ||
| FlowJo | BD Biosciences | RRID:SCR_008520 |
| FACSDiva | BD Biosciences | RRID:SCR_001456 |
| Other | ||
| Sterilizable forceps | n/a | n/a |
| Sterilizable scissors | n/a | n/a |
| Sterilizable mortar | n/a | n/a |
| Strainer 100 μm | Greiner Bio-One | 542000 |
| Strainer 70 μm | Greiner Bio-One | 542070 |
| Pasteur pipette | LP Italiana | 135030 |
| Centrifuge | Thermo Scientific | Heraeus Megafuge 16R |
| Cryo vials | Greiner Bio-One | 122263 |
| MACS Multistand | Miltenyi | 130-042-303 |
| OctoMACS Separator | Miltenyi | 130-042-108 |
Materials and equipment
Cell culture medium
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI (1X) | n/a | 445 mL |
| Fetal bovine serum (FBS) | 10% | 50 mL |
| Penicillin 10,000 units/mL Streptomycin 10 mg/mL |
100 units/mL 0.1 mg/mL |
5 mL |
| Total | n/a | 500 mL |
Store at 4°C for three months, prepare and use under sterile conditions.
Washing buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Phosphate-buffered saline (PBS) 1× | n/a | 492.5 mL |
| Fetal bovine serum (FBS) | 1% | 5 mL |
| EDTA | 0.5% | 2.5 mL |
| Total | n/a | 500 mL |
Store at 4°C for three months.
Freezing medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Fetal bovine serum (FBS) | 80% | 40 mL |
| Dimethyl sulfoxide (DMSO) | 20% | 10 mL |
| Total | n/a | 50 mL |
Store at 4°C for three months, prepare and use under sterile conditions.
EDTA-free washing buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS 1× | n/a | 47.5 mL |
| Fetal bovine serum (FBS) | 5% | 2.5 mL |
| Total | n/a | 50 mL |
Store at 4°C for three months.
EDTA-containing cell culture medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Cell culture medium | n/a | 50 mL |
| EDTA | 0.5% | 0.25 mL |
| Total | n/a | ≈ 50 mL |
Store at 4°C for three months, prepare and use under sterile conditions.
1X Binding buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Binding buffer 20× | n/a | 2.5 mL |
| Water (sterile, distilled) | n/a | 47.5 mL |
| Total | n/a | 50 mL |
Prepare freshly for each experiment.
Alternatives: You may replace cell culture medium and washing buffer by solutions you are most experienced with. However, we recommend the usage of EDTA, as indicated, because it binds calcium and thus prevents clumping of cells.
Step-by-step method details
Preparation of single cell suspension from human lymph nodes
Timing: 60 min
Lymph node tissue is minced to obtain single cell suspension. The following steps are performed under sterile conditions.
-
1.
Transfer lymph node into a sterile mortar (Figure 1A) and add 5 mL cell culture medium.
-
2.
Remove excessive fat and connective tissue (Figure 1B) using dissecting scissors or a scalpel.
-
3.
Cut lymph node into small pieces of approx. 1–2 mm size using dissecting scissors and forceps (Figure 1C).
Note: The smaller the pieces, the higher the cell yield. However, for larger lymph node pieces, step 3 can be very time-consuming and defines the overall duration of this protocol. If you rely on fast processing rather than high cell yield, you can drastically shorten this step by cutting the lymph node into fewer pieces. We do not recommend using a pestle because it generates more debris.
-
4.
Using a 5 mL serological pipette, thoroughly pipette up and down to gently wash out the lymphocytes.
-
5.
Filter lymphocyte-containing medium through a 100 μm strainer into a sterile 50 mL tube (Figure 1C).
-
6.
Add 5 mL cell culture medium and repeat steps 3–5 once or twice depending on the size of the lymph node. The strainer can be reused unless it becomes clogged.
-
7.
Rinse cell strainer with fresh cell culture medium to fill up the tube to 50 mL.
-
8.
Centrifuge tube at 400 g for 5 min at 18°C–25°C.
-
9.
Carefully decant supernatant (Figure 1D).
Optional: At this step, lymph node-derived exosomes can be collected. To do so, the use of exosome-free FBS and cell culture medium is required in all preceding steps (Bordas et al., 2020).
-
10.
Gently resuspend the pellet in 10 mL cell culture medium and pass cell suspension through 70 μm strainer.
-
11.
Centrifuge at 400 g for 5 min at 18°C–25°C.
-
12.
Resuspend cells in cell culture medium. In case of clumps, repeat steps 10–12.
-
13.
Count cells and set 1 × 106 cells aside for quality control by flow cytometry (see below).
Optional: After this step, cells can be used freshly for any subsequent assays.
Note: For single cell (RNA) sequencing non-frozen cells might generally be the better option. However, single cell (RNA) library preparation is an elaborate process and rather inefficient if performed for only one sample at a time. In addition, batch effects could mask interpatient differences. Therefore, we freeze all samples before use.
-
14.
Fill 2 mL cryotubes with 700 μL of cold freezing medium.
-
15.
Dilute cell suspensions to desired cell concentration using cell culture medium, for instance 5 × 106 cells per 700 μL medium.
-
16.
Add 700 μL cell suspension drop-wise to cryotubes and gently invert tubes a few times.
-
17.
Place cryotubes quickly into a slow freezing container and store at −80°C for ≥ 24 h. Long-term storage should be below −150°C.
Note: Depending on the total yield of cells, we freeze 5 × 106, 1 × 107, 2 × 107 and 5 × 107 cells per vial. Aliquot sizes can be adjusted depending on the planned set of experiments. Please see the section expected outcomes for cell yield of an average lymph node sample.
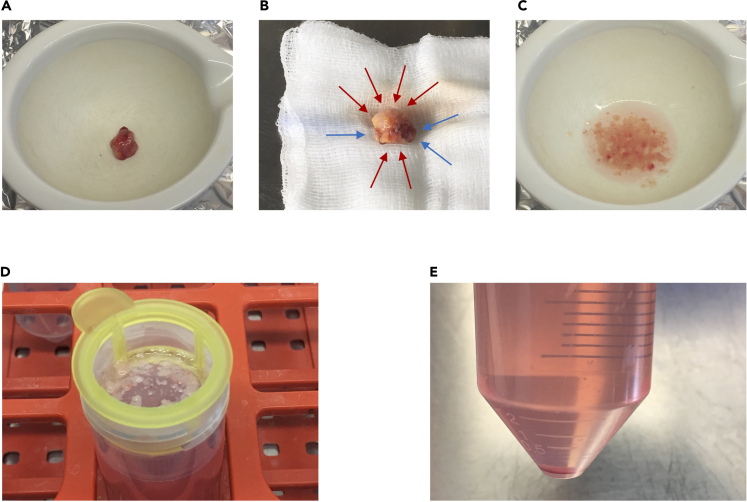
Figure 1.
Key steps of lymph node processing
(A and B) Two different representative lymph nodes in a mortar (A) or on a saline-soaked compress (B) immediately after excision. B) Lymph node or excessive fat tissue is highlighted with blue or green arrows, respectively.
(C) Lymph node in cell culture medium after being cut into small pieces.
(D) Lymph node pieces in a cell strainer. Tube filled with approximately 50 mL medium containing lymph node-derived cells.
(E) Pellet of lymph node-derived cells after centrifugation. Reddish stain indicates slight contamination with erythrocytes.
Output control using flow cytometry
Timing: 60–90 min
In order to check and document the cellular output, lymph node-derived cells are stained for viability, as well as basic B and T cell markers and analyzed on a flow cytometer. This step is crucial as the frequency of B and T cells varies significantly across patient samples.
-
18.
Transfer leftover 1 × 106 cells (from step 13) into a 5 mL flow cytometry tube.
-
19.
Add 1 mL of washing buffer and centrifuge cells at 400 g for 5 min at 18°C–25°C.
-
20.
Decant supernatant and resuspend pellet in remaining buffer (ideally 50–100 μL). Then, add the antibodies, as shown in Table 1.
Note: Staining for kappa and lambda light chain enables quick detection of light chain-restricted populations proving the diagnosis of B cell lymphoma (Horna et al., 2011). The same is true for co-expression of CD5 and CD19 (Hallek et al., 2008). Quantification of B and CD4+/CD8+ T cells might help to plan future experiments. The staining panel can be adjusted according to the titration or availability of antibodies and the configuration of flow cytometer at your means. To detect the above mentioned fluorophores, the flow cytometer must be equipped with suitable lasers (as shown) and filters.
-
21.
Briefly vortex and incubate at 4°C protected from light for 30 min.
-
22.
Add 1 mL of washing buffer, centrifuge cells at 400 g for 5 min and discard supernatant.
-
23.
Repeat for a total of three washes.
-
24.
Resuspend cells in 200 μL washing buffer.
-
25.
Put cells on ice and analyze them on a flow cytometer within 24 h. Please see Figure 2C for suggested gating strategy.
Table 1.
Suggested flow cytometry panel
| Antibody/Target | Conjugate/Dye | Recommended amount | Excitation laser |
|---|---|---|---|
| Anti-human CD19 | BV421 | 0.5 μL | Blue (405 nm) |
| Viability | eFluor 506 | 0.2 μL | Blue (405 nm) |
| Anti-human CD8 | FITC | 0.5 μL | Green (488 nm) |
| Anti-human CD3 | PerCP/Cy5.5 | 1.0 μL | Green (488 nm) |
| Anti-human Ig light chain κ | PE | 1.0 μL | Yellow (561 nm) |
| anti-human Ig light chain λ | PE/Dazzle | 1.0 μL | Yellow (561 nm) |
| Anti-human CD5 | PE/Cy7 | 1.0 μL | Yellow (561 nm) |
| Anti-human CD4 | AF700 | 1.0 μL | Red (633 nm) |
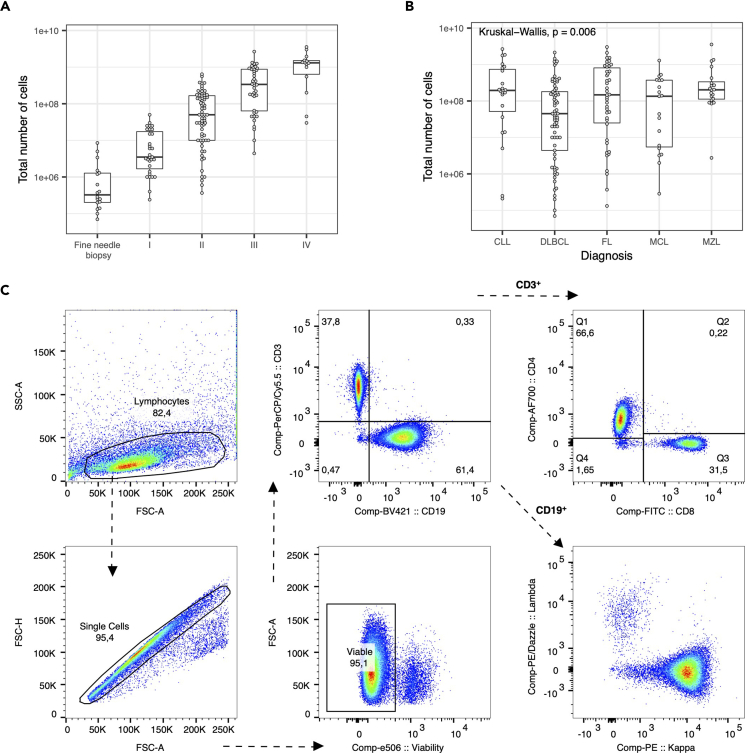
Figure 2.
Quality control of lymph node processing
(A) Lymph nodes were processed as described in the protocol and roughly grouped into four categories based on their size: fine needle biopsies, I (small piece), II (medium size piece), III (big piece) or IV (very big piece). The total number of harvested cells was compared to estimated size of the obtained lymph node piece. Box plots show minimum, first quartile, median, second quartile, and maximum.
(B) The total number of harvested cells across different lymphoma entities. Box plots show minimum, first quartile, median, second quartile, and maximum.
(C) Representative pseudocolor plots showing the usual output of harvested lymph node-derived lymphocytes.
Thawing lymph node-derived cells and dead-cell removal
Timing: 60–90 min
Lymph node-derived cells are thawed using EDTA to prevent clumping of cells prior to removal of dead cells and library preparation for single cell sequencing. Until library preparation, the following steps are performed under sterile conditions.
-
26.
Pre-warm water bath and EDTA-containing cell culture medium to 37°C.
-
27.
Prepare 50 mL tubes with 10 mL of EDTA-containing cell culture medium.
Note: EDTA reduces the formation of cell clumps, and thereby increases the yield of viable cells after thawing.
-
28.
Thaw frozen aliquots in pre-warmed water bath and quickly transfer cell suspension dropwise into the prepared tubes.
-
29.
Centrifuge at 200 g for 5 min at 18°C–25°C and discard supernatant.
-
30.
Resuspend cells in EDTA-free washing buffer.
Note: This step aims to remove EDTA again from the cell suspension, as calcium is required for binding to the column of the dead-cell removal kit.
-
31.
Filter cell suspensions through 75 μm strainer.
-
32.
Count cells, for instance manually by means of trypan blue and a hemocytometer.
-
33.
Centrifuge at 200 g for 5 min at 18°C–25°C and discard supernatant.
-
34.
To magnetically label the dead cells, resuspend the cell pellet in 100 μL magnetic beads per 1 × 107 cells. For fewer than 1 × 107 cells, use 100 μL.
-
35.
Incubate for 15 min protected from light at 18°C–25°C.
-
36.
Meanwhile, prepare magnetic Multistand, OctoMACS™ separator, and one MS columns per sample (Figure 3A).
Note: For single cell RNA sequencing, we strongly recommend the OctoMACS™ separator, as sequential handling of cells might confound gene expression profiles.
-
37.
Wash each column with 500 μL 1X binding buffer (Figure 3B) and wait until the liquid has passed the column. Put a tissue beneath the columns to soak up the flow-through.
-
38.
Place 15 mL tubes under each column to collect the flow-through (Figure 3C).
-
39.
Add 1 mL 1X binding buffer to each sample and apply each sample onto one column (Figure 3D).
-
40.
Wait until the liquid has passed the column before rinsing each column with 500 μL 1X binding buffer. Wait until the liquid has passed the column.
-
41.
Perform a total of 4 washes in this way.
-
42.
Centrifuge the collected flow-through at 200 g for 5 min at 18°C–25°C and discard supernatant.
-
43.
Resuspend cells in washing buffer and from here on handle them at 4°C.
-
44.
Count cells and assess viability, for instance by means of trypan blue and a hemocytometer.
Note: In 10%–15% of all samples that we processed by this protocol, viability did not exceed 85%. In these cases, we reevaluate continuation of the protocol. To avoid unexpected termination of the planned follow-up experiment, we usually thaw and process an additional sample as back-up. It should generally be considered that when thawing a vial of a vial of 1 × 107 cells, only a median of 3.5 × 106 viable cells can be recovered. Please see the troubleshooting section on how to optimize conditions if you encounter low viability or exorbitant loss of cells.
Optional: At this step, cells can be labelled with barcoded antibodies. We do not recommend staining before removal of dead cells.
-
45.
Continue with library preparation for single cell sequencing according to the manufacturer’s protocol.
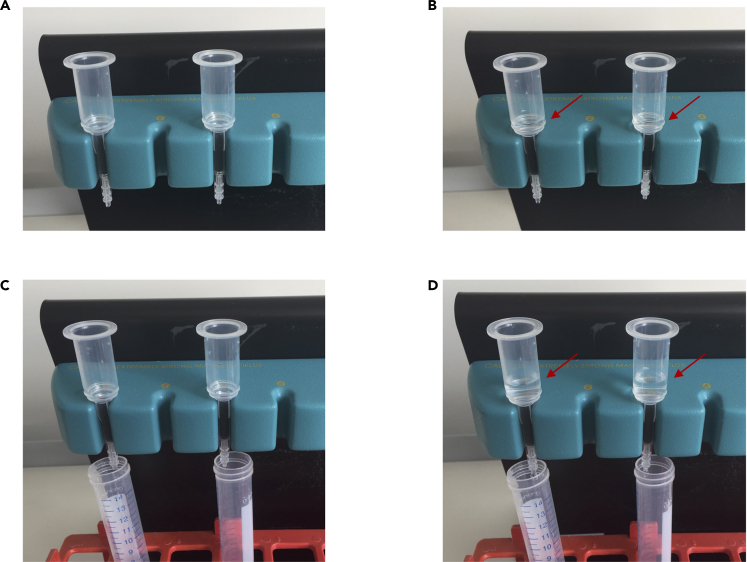
Figure 3.
Dead cell removal
(A) Magnetic multistand with OctoMACS™ separator and two MS columns.
(B) Columns are rinsed with 500 μL binding buffer (red arrow).
(C) To collect flow-through 15 mL tubes are placed beneath each column.
(D) 1000 μL binding buffer was add to each sample. Then, each sample (red arrow) is applied onto one column.
Expected outcomes
This protocol describes how to prepare single cell suspensions from human lymph nodes without enzymatic digestions. The expected yield of cells strongly depends on the size of the processed lymph node (Figure 2A). Comparing the different B cell lymphoma entities, the lowest yield of cells is obtained from diffuse large B cell lymphoma (Figure 2B). Across several hundred lymph node samples, we harvested a median number of 9 × 107 cells per sample. Lymph node samples that we receive have an approximate size of 1.5–2.5 cm3 and an average weight of 1.5–2.5 g. Flow cytometric analysis of isolated cells is indispensable to evaluate the quality and viability of isolated cells. Figure 2C illustrates representative pseudocolor plots of lymph node-derived lymphocytes of a patient with mantle cell lymphoma.
Limitations
This protocol was optimized to isolate lymphocytes as quickly as possible while reducing the level of cellular stress. For this reason, it includes neither enzymatic digestion nor harsh mechanical dissociation. This protocol was not designed for isolation or analysis of lymph node-derived myeloid cells. Among the isolated cells, the proportion of myeloid cells is below 5%. However, we have not systematically investigated whether this proportion is comparable to the proportion of myeloid cells in vivo or after enzymatic digestions of human lymph nodes.
This protocol is neither suitable to isolate or investigate lymph node-derived stroma cells, for which enzymatic digestion is required (Takeda and Jalkanen, 2020).
Another limitation is that fine needle biopsies, e.g., computer tomography-guided biopsies, might yield very low numbers of cells in the range of 1 × 106 (Figure 2A).
Troubleshooting
Problem 1
Low cell yield during initial preparation (step 13)
Potential solution
The number of isolated cells highly depends on the amount of lymph node tissue provided. In particular, ultrasound- or computer tomography-guided biopsies are only 1 mm thick and consequently yield very low numbers of cells.
Low yield of cells can also occur when lymph node tissue is mistaken for conjunctive tissue and then removed during the initial preparation (step 2). Differentiation of the tissue types (Figure 1B) by eye requires some experience. Alternatively, ensure that the excised lymph node tissue is already cleaned by surgeons or pathologists.
Problem 2
Low cell viability after initial preparation (step 13)
Potential solution
There are multiple factors affecting the viability of lymph node-derived cells. To achieve the best possible results, reduce the timespan from excision to processing and freezing in the lab, e.g., by optimizing the transfer from operating room to laboratory. For susceptible assays, such as single cell (RNA) sequencing, we recommend to only use samples that were processed on the day of excision. However, it is possible to store lymph node samples for 12–16 h at 4°C if the harvested cells are to be used in protein, DNA, or flow cytometric assays.
Problem 3
Red cell pellet during initial preparation (step 12)
Potential solution
Similar to peripheral blood mononuclear cells, lymph node-derived lymphocytes can be contaminated by erythrocytes, which can potentially affect downstream experiments and results (Figure 1E). Potential solutions include gradient centrifugation of isolated cell suspensions or lysis of erythrocytes using a hypotonic solution. However, we discourage you from additional steps before susceptible assays, such as single cell (RNA) sequencing. More precisely, heterogeneous sample treatment can drastically confound gene expression profiles and introduce difficult-to-correct batch effects. Record any kind of deviation from the standard protocol and avoid using such samples for delicate assays.
Problem 4
Black/dark cell pellet during initial preparation (step 12)
Potential solution
Black or dark cell pellets may be the result of tattoos in the draining region of the lymph node. As little is known about how the pigments may affect freezing of the cells and downstream experiments, isolated cells should be discarded.
Problem 5
Low cell yield or viability after removal of dead cells (step 44)
Potential solution
Ensure that cells were carefully resuspended before applying them to the column. Any clumps might drastically reduce the cellular output. In addition, low starting viability can also lead to low output or viability after dead cell removal.
Resource availability
Lead contact
Further information and requests for resources or reagents should be directed to the lead contact, Tobias Roider (tobias.roider@embl.de).
Materials availability
This study did not generate new unique materials or reagents.
Acknowledgments
T.R. was supported by a physician scientist fellowship of the Medical Faculty of University Heidelberg. S.D. was supported by a grant of the Hairy Cell Leukemia Foundation, the Heidelberg Research Centre for Molecular Medicine (HRCMM), and an e:med BMBF junior group grant and Deutsche Forschungsgemeinschaft through the SFB873 project B7. We thank Carolin Kolb, Angela Lenze, and Mareike Knoll (University Hospital Heidelberg) for their excellent technical assistance.
Author contributions
T.R. performed experiments; T.R. and S.D. designed and supervised the research; T.R., B.J.B., and S.D. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Data and code availability
The study did not generate/analyze data sets or codes.
References
- Bordas M., Genard G., Ohl S., Nessling M., Richter K., Roider T., Dietrich S., Maass K.K., Seiffert M. Optimized protocol for isolation of small extracellular vesicles from human and murine lymphoid tissues. Int. J. Mol. Sci. 2020;21:5586. doi: 10.3390/ijms21155586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallek M., Cheson B.D., Catovsky D., Caligaris-Cappio F., Dighiero G., Dohner H., Hillmen P., Keating M.J., Montserrat E., Rai K.R. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National cancer Institute-Working group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horna P., Olteanu H., Kroft S.H., Harrington A.M. Flow cytometric analysis of surface light chain expression patterns in B-cell lymphomas using monoclonal and polyclonal antibodies. Am. J. Clin. Pathol. 2011;136:954–959. doi: 10.1309/AJCP3C2QZZBPTMLB. [DOI] [PubMed] [Google Scholar]
- Mattei D., Ivanov A., van Oostrum M., Pantelyushin S., Richetto J., Mueller F., Beffinger M., Schellhammer L., Vom Berg J., Wollscheid B. Enzymatic dissociation induces transcriptional and proteotype bias in brain cell populations. Int. J. Mol. Sci. 2020;21:7944. doi: 10.3390/ijms21217944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentmeier H., Pommerenke C., Dirks W.G., Eberth S., Koeppel M., MacLeod R.A.F., Nagel S., Steube K., Uphoff C.C., Drexler H.G. The LL-100 panel: 100 cell lines for blood cancer studies. Sci. Rep. 2019;9:8218. doi: 10.1038/s41598-019-44491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard A., Asosingh K. Best practices for preparing a single cell suspension from solid tissues for flow cytometry. Cytometry A. 2019;95:219–226. doi: 10.1002/cyto.a.23690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roider T., Brinkmann B.J., Kim V., Knoll M., Kolb C., Roessner P.M., Bordas M., Dreger P., Mueller-Tidow C., Huber W. Autologous culture model of nodal B-cell lymphoma identifies ex vivo determinants of response to bispecific antibodies. Blood Advances. 2021 doi: 10.1182/bloodadvances.2021005400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roider T., Seufert J., Uvarovskii A., Frauhammer F., Bordas M., Abedpour N., Stolarczyk M., Mallm J.P., Herbst S.A., Bruch P.M. Dissecting intratumour heterogeneity of nodal B-cell lymphomas at the transcriptional, genetic and drug-response levels. Nat. Cell Biol. 2020;22:896–906. doi: 10.1038/s41556-020-0532-x. [DOI] [PubMed] [Google Scholar]
- Takeda A., Jalkanen S. Single-cell transcriptomics of human lymph node stroma. STAR Protoc. 2020;1:100021. doi: 10.1016/j.xpro.2020.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The study did not generate/analyze data sets or codes.