Highlights
-
•
Solid-state NMR spectroscopy probes molecular conformation and dynamics in intact ECM.
-
•
Collagen conformational dynamics has roles in mechanical properties of fibrils and cell adhesion.
-
•
Solid-state NMR spectroscopy has shed new light on the chemical structure of bone mineral.
Keywords: Extracellular matrix structure, Collagen, Multidimensional solid-state NMR spectroscopy, Proline conformation, Bone mineral, Biomineralization
Abstract
Solid-state NMR spectroscopy has played an important role in multidisciplinary studies of the extracellular matrix. Here we review how solid-state NMR has been used to probe collagen molecular conformations, dynamics, post-translational modifications and non-enzymatic chemical changes, and in calcified tissues, the molecular structure of bone mineral and its interface with collagen. We conclude that NMR spectroscopy can deliver vital information that in combination with data from structural imaging techniques, can result in significant new insight into how the extracellular matrix plays its multiple roles.
1. Introduction
This review aims to exemplify how solid-state NMR spectroscopy has been used to answer questions about the molecular structure, dynamics and chemical modifications of mammalian extracellular matrix in structural tissues such as tendon, bone and the vasculature, rather than to be an exhaustive review of the extensive literature in this area. The extracellular matrix is composed of complex arrangements of fibrillar and other collagens, proteoglycans and glycoproteins such as fibronectin. We have chosen to focus the examples we highlight primarily around extracellular matrix collagens because of their centrality in the ECM 3D architecture, and in cell adhesion and migration. Throughout the review, we consider where NMR spectroscopy can bring unique information on ECM as well as the experimental requirements and limitations. The review begins with an overview of the information content of solid-state NMR spectra, and then goes on to discuss the role of NMR in probing molecular structures, dynamics and post-translation modifications of collagens in ECM, and finally explores how solid-state NMR spectroscopy has shed important, new light on the structure of mineral in calcified tissues and on how mineral interfaces with collagen.
2. What is NMR spectroscopy?
Solid state NMR spectroscopy can provide valuable insight into the primary and secondary structures of proteins and is one of the few methods that can inform on molecular dynamics in solid materials. At the simplest level, an NMR spectrum consists of a series of signals from particular nuclei, e.g. 13C, the only NMR-active isotope of carbon. NMR signal frequencies are normally measured as relative differences from a reference signal frequency in parts per million (ppm), so-called chemical shifts [1]. The chemical shifts of NMR signals are dependent in a largely predictable way on the bonding environment of the nucleus, e.g. 13C, that gives rise to the signal (Fig. 1A). So, 13C chemical shifts for proteins are characteristic for each residue type because each residue presents an inherently different local environment for its constituent 13C nuclei. Further, the 13C chemical shifts, for example, for the Gly C in collagens are different to those for Gly C in an alpha helix structure [2], [3], reflecting the different bond geometries associated with the different secondary structures.
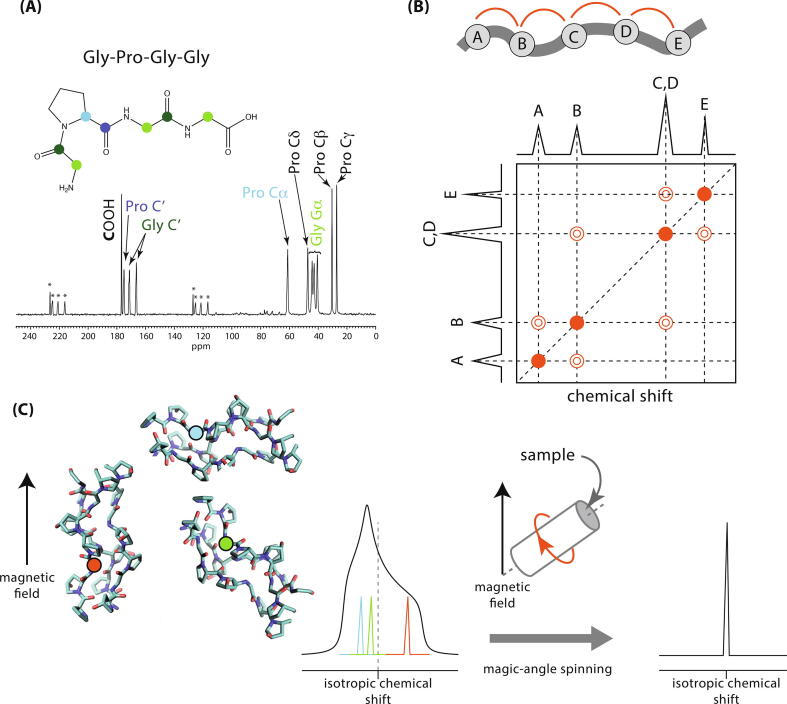
Fig. 1.
(A) A 1D 13C (MAS) NMR spectrum of a lyophilised sample of Gly-Pro-Gly-Gly peptide. The backbone carbons are indicated with coloured circles and their respective NMR signals assigned on the spectrum. Signals marked * are spinning sidebands, necessary artefacts of magic-angle spinning (see (C)). (B) Schematic of a 2D homonuclear correlation spectrum, e.g. 13C - 13C for the molecule at the top with nuclear (atomic) sites A, B, C, D, E. In this hypothetical spectrum, cross-peaks, indicated as contours (open circles), occur between signals from nuclei that are physically close in space, i.e. A-B, B-C, etc. The diagonal signals in the 2D spectrum (filled circles) are typical for this type of 2D spectrum and do not indicate that e.g. site As are close together with other site As. (C) Solid-state NMR spectroscopy typically requires magic-angle spinning (MAS) to remove the effects of anisotropic nuclear spin interactions. The strength of the effect of these interactions on the NMR signal frequency depends on the orientation of the molecule containing the nucleus with respect to the magnetic field applied in the NMR experiment. Thus in a sample where, e.g. there are collagen molecules with multiple orientations, there will be multiple, overlapping signals, resulting in a broad line (exemplified here with the lineshape resulting from shielding anisotropy - interaction of e.g. 13C nuclei with surrounding electrons in the collagen molecules) which limits resolution. Spinning the whole sample at the so-called magic-angle (54.74∘) with respect to the magnetic field, removes the molecular orientation dependence of the chemical shift, resulting in a sharp line at the isotropic chemical shift for each nuclear site. If the spinning rate is slower than width of the NMR signal, so-called spinning sidebands also appear in the spectrum, radiating out from the isotropic signal at the spinning frequency apart.
The NMR-active nuclei of primary interest to matrix biology are 13C (1.1% natural abundance), 15N (0.4% natural abundance) and 1H (100% natural abundance). Structural biologists utilise the dependence of 13C and 15N chemical shifts on amino acid type and secondary structure to generate molecular structural models for proteins from chemical shift data using algorithms trained on experimental data [3], [4], [5]. The limitation on the size of the protein molecule that can be characterized in this way is the spectral resolution and being able to assign e.g. each 13C NMR signal to a particular carbon site in the protein. Resolved, assigned signals are needed for a substantial proportion of the carbon and/or nitrogen sites in the molecule to generate a robust structure. The larger the molecule, the greater the chance of NMR signals from different carbons, nitrogens or hydrogens having similar chemical shifts, and so NMR signals from those sites overlapping. Equally, the difficulty of assigning NMR signals to specific atomic sites significantly increases with the size of the protein.
Multidimensional NMR spectra deal with both the resolution and the assignment problem to a significant extent and moreover, can be designed to give additional, highly useful structural information [3], [5], [6]. An example of a 2D 13C-13C NMR correlation spectrum is shown in Fig. 1B. The projection of the 2D spectrum onto each of the spectral axes in this example is a normal 1D 13C spectrum of signals from each 13C site in the sample as described above. In this case, the 2D plane of the spectrum correlates signals from 13C nuclei that are physically close in space - in practise within approximately 0.8 nm of each other for 13C nuclei. Thus an off-diagonal or cross-peak signal between signals A and B in the 1D projections of Fig. 1B indicate the 13C sites A and B are physically close in space. The intensity of the cross peak typically increases the closer the A and B sites are together, a feature that can be made at least semi-quantitative with proper calibration [5], [7]. The ensuing internuclear distance data is highly useful structural constraint information when building a 3D structural model of a protein [7].
The spectrum in Fig. 1B also demonstrates how 2D spectra improve resolution. Two signals (C and D in Fig. 1B) that overlap in a 1D NMR spectrum have resolved cross-peaks in the 2D spectrum providing that they are close to nuclei with different chemical shifts, i.e. they are correlated with different NMR signals, allowing clear observation that there are indeed two overlapping signals in the 1D spectrum.
There are many options for 2D correlation NMR spectra: 13C NMR signals can be correlated with 15N, 1H, 31P…, 15N signals can be correlated with 1H, etc. [5]. That the spatial proximity of almost any pair of NMR-active nuclei can be determined through a 2D spectral correlation in this way greatly aids the assignment of NMR signals to specific 13C/15N sites in the molecule. For instance so-called N-Ca-CO 3D spectra correlate each peptide backbone amide 15N signal with the 13Cα/13CO of the preceding residue. Therefore providing the amino acid sequence for the protein is known, signal assignment can proceed by a trial assignment of a subset of signals, and working through the signals correlated with those assigned signals to find the signals of the neighbouring residues, and so on [5].
The patterns of signals in a set of 2D spectra from a sample thus provide a powerful fingerprint of the spatial relationship of the carbon/ nitrogen/ hydrogen sites in the molecules in the sample. The resolved chemical shifts of the signals and internuclear distance constraints from 2D NMR spectra provide information on local molecular conformation and protein folding. Many different 2D and higher dimensionality NMR experiments have been designed that allow e.g. different structural lengthscales to be probed or that edit NMR signals according to different conditions, as well as giving increased resolution and aiding spectral assignment. Multidimensional NMR spectroscopy thus offers a plethora of tools for probing molecular structure in matrix biology. “Real life” examples of the use of 2D spectra in ECM studies are shown in Fig. 2.
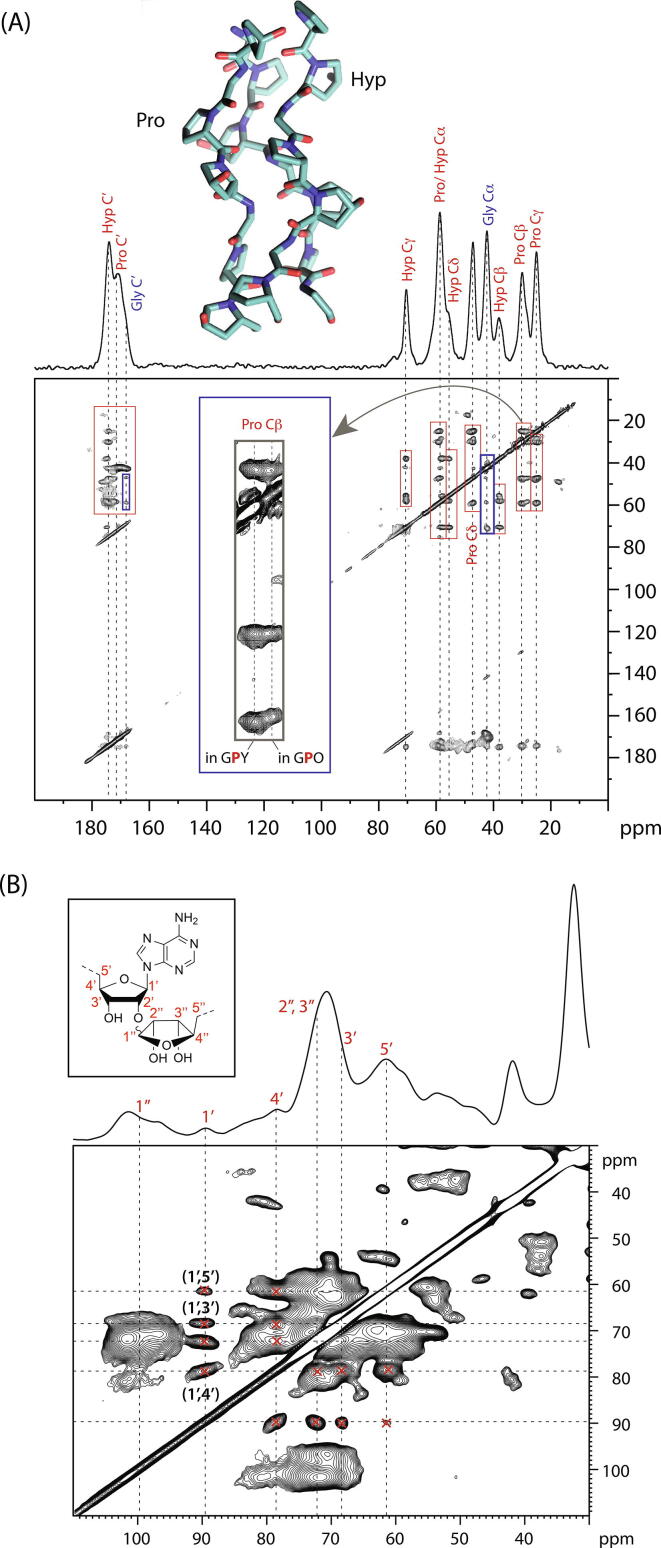
Fig. 2.
(A) A 2D 13C-13C correlation NMR spectrum (proton-driven spin-diffusion) of in vitro ECM from foetal sheep osteoblasts in which U-13C-Pro, Gly were used in the cell culture media, resulting in ECM proteins in which Gly, Pro and Hyp are extensively 13C-labelled. Correlation signals from these (primarily collagen) residues thus appear in this 2D correlation spectrum. Intraresidue signals are indicated in red rectangles and inter-residue correlations in blue rectangles. The inset (grey rectangle) is an expansion of the Pro C signals, showing they can be resolved into those from Pro in GPO and more general GPY triplets. The 1D 13C MAS NMR spectrum of the same sample is shown at the top. (B) 2D 13C-13C correlation NMR spectrum (proton-driven spin-diffusion) of in vitro calcifying (bovine) vascular smooth muscle cell ECM showing assignments of some of the correlation signals from poly(ADP ribose) (inset: structure of one monomer unit of poly(ADP ribose) showing the 13C site labelling scheme). The 1D 13C projection of the spectrum is shown above the 2D spectrum. The poly(ADP ribose) signals are not resolved from other glycan species in this 1D spectrum, although they are well-resolved in the 2D spectrum, showing the power of 2D NMR correlation spectroscopy.
In this review, we will explore the new information that NMR spectroscopy has brought to matrix biology, how it can be fruitfully interfaced with other techniques, and consider where it can be used in the future.
3. Experimental considerations
The nuclei of interest in matrix biology are stable (non-radioactive) isotopes, but the natural abundance of the key NMR-active nuclei for biology are low, at 1.1% for 13C and 0.4% for 15N. This means that enrichment in one or more of these isotopes is usually needed if NMR spectroscopy is to be used for detailed molecular structure studies [5], [8]. How such enrichment can be obtained is one aspect we will address throughout this review.
One powerful strategy is to design the isotope enrichment procedure so that only the molecules of interest are enriched (see Fig. 2 for example). The NMR spectra then contain only signals from those molecules without any need to extract them from the sample. This means that NMR experiments can, and often are, performed on intact biological samples with very little sample preparation needed [9], [10], [11], [12], [13]. This makes NMR spectroscopy of great use for studying the extracellular matrix, where extracting the molecules of interest is often laborious and can result in loss of essential features, such as labile post-translational modifications, and molecular structures that are not representative of in vivo structures. A powerful advantage of solid-state NMR is that one can preserve the native hydration state of the tissues which is a essential for studying molecular structures and dynamics; NMR studies on the effect of the hydration level on ECM structure and mechanics is discussed in Section 5. Samples may be frozen or fixed in a variety of ways, as best preserves the features of the sample being studied. Ex vivo and in vitro tissue samples can be placed directly into cold NMR sample holders, for instance, and chilled throughout the NMR experiment to maintain the structural integrity of the sample without chemical fixing, which would typically dehydrate the sample and distort molecular structures. Moreover, studying intact samples is a very attractive feature compared to other ECM structure/ composition analysis techniques such as LC/ MS and proteomics which require comparatively harsh sample preparation conditions that can lead to preferential or complete loss of some components of the sample.
The NMR experiment involves placing generally a few milligrams or less of a sample into a very large magnetic field (typically 10 Tesla) and directing powerful radiofrequency electromagnetic waves at the sample to excite the nuclear spins of interest. The strong magnetic field means that samples with paramagnetic transition metal ions can cause complications. The radiofrequency electromagnetic waves can cause sample heating, particularly for highly hydrated, salty samples, though newer NMR technology has reduced this problem (with so-called low E-field probes) and samples can also be actively cooled during the NMR experiment.
Experiments on matrix samples, and solid samples in general, require that the sample is spun at rates of tens of kHz, and often over 100 kHz to acquire spectra where resolution of a 1H spectrum is required. This so-called magic-angle spinning (MAS) is routinely applied in solid-state NMR spectroscopy (Figure1C). MAS is required because the nuclei being observed in the NMR experiment experience interactions with other NMR-active nuclei (dipolar coupling) and with the surrounding electrons (shielding) in their respective molecules. Those interactions alter the NMR signal frequency observed for a given nucleus to an extent that depends on the orientation of the molecule containing the nucleus with respect to the NMR magnetic field. This means that the 1H, 13C and 15N signals from, e.g. collagen molecules in differently oriented collagen fibrils in the sample will have different chemical shifts for each different fibril orientation (Figure1C), making the NMR signals very broad. The lineshapes of these broad NMR signals contain potentially useful information on the molecular symmetry at the atomic site, molecular dynamics and in oriented samples, e.g. phospholipid membrane, molecular orientation. However, the widths of these lineshapes are often comparable to the frequency range of the entire NMR spectrum for any given nucleus type, meaning that there is significant overlap of signals from e.g. 13C in different collagen residues in 13C NMR spectra of collagenous ECMs. MAS removes this molecular orientation dependence of the signal frequency and results in a spectrum of so-called isotropic chemical shifts. The linewidths of the signals in an isotropic chemical shift spectrum produced by MAS reflect a distribution of chemical environments (rather than molecular orientations) and/ or the presence of molecular dynamics and/ or residual effects of nuclear interactions (particularly for 1H spectra because 1H - 1H interactions are typically strong causing linewidths of 50 kHz in the absence of MAS). So for instance, the broad isotropic chemical shift 13C signal from collagen Gly Cs in Fig. 2A (1D spectrum at the top of the figure) which has a width at half maximum intensity of 190 Hz compared to 30 Hz for the Gly C signals for the simple (crystalline) Gly-Pro-Gly-Gly peptide in Fig. 1A, reflects the fact that each Gly C in a collagen molecule is in a slightly different chemical environment by virtue of having different residues as neighbours, slightly different bond geometries around it etc. In contrast, the signals for the Gly Cs in the GPGG peptide are well resolved from each other and each Gly Cs 13C signal is from essentially a single carbon environment.
4. Collagen molecular structure by NMR
Our own work in this area began with using the molecular-structure fingerprints that 2D NMR spectra give to compare the molecular conformations between ex vivo and in vitro mammalian extracellualr matrix [14]. Models of ECM need to reproduce the intermolecular interactions that occur in vivo - the ECM 3D nano-architecture relies on the intricate network of intermolecular interactions between ECM proteins (and glycans) and ECM-cell molecular interactions, e.g. integrin-collagen binding interactions, are critical in determining cell functionality. A necessary condition for reproducing the correct intermolecular interactions in an ECM model is that the model’s constituent proteins must have molecular conformations similar to those in vivo. Using a set of 2D 13C-13C and 13C-15N correlation spectra as molecular-structure fingerprints to compare molecular conformations between in vivo and in vitro tissues presented a unique opportunity to verify the extent to which any in vitro cell cultured ECM models in vivo matrix. This strategy necessitated ex vivo tissues in which ECM proteins were highly enriched in 13C and 15N. We achieved this by feeding a pregnant mouse with a diet based on 30% fully enriched 13C, 15N amino acids during the pregnancy and until the infant mice weaned. This generated an enrichment of up to 90% for 13C in essential amino acids in the bone tissue of the pups, for instance [11]. The overall NMR fingerprint we generated here consisted of chemical shift distributions of spatially close 13C and 15N and directly bonded 13C pairs in the most abundant proteins of each tissue, primarily collagens, through 2D correlation 13C-13C and 13C-15N experiments [11], [14], [15].
Applying the same amino acid 13C, 15N labelling to in vitro cell cultures with e.g. foetal sheep osteoblasts (Fig. 2A), allowed us to generate the equivalent 2D NMR correlation spectra for in vitro tissue. Comparing the 2D spectra from the in vivo and in vitro tissues then allowed us to assess directly the similarity in collagen molecular conformations between those tissues and moreover, gave us a rational route to refining in vitro models to generate ECM more similar in terms of molecular conformations to in vivo tissues [14]. For instance, it was straightforward to detect denatured collagens in the 2D NMR spectra; we thus observed that our first in vitro models had significant denatured collagen in them compared to in vivo tissues. Knowing this, we then trialled different ways of growing and preparing the in vitro samples to minimise protein damage and assessed the effectiveness of each strategy by reference to 2D 13C correlation NMR spectra on each in vitro sample produced. The same approach allowed us to identify poly(ADP ribose) in the extracellular matrix of calcifying tissues (Fig. 2B), which is discussed in more detail in Section 7.
De Sa Peixoto and co-workers [16] have investigated the changes in collagen molecular conformation associated with the formation of collagen nanofibrils through observing changes in 13C chemical shift in key collagen amino acid residues between soluble collagen at acidic pH, fibrillar collagen in dense, pure collagen matrices, and denatured collagen, the latter being a reference for 13C chemical shifts of collagen in a random coil conformation. They used natural abundance 13C (unlabelled) collagen and 2D 1H-13C correlation spectra to improve 13C spectral resolution over that in 1D 13C NMR spectra. Changes in the 13C chemical shifts for different types of collagen residues between these different conditions allowed the authors to conclude that there is significant change in both backbone and side chain conformations when collagen molecules aggregate into fibrils. For example, some Arg and Lys residues adopt a conformation where their side chains are more extended away from the triple helix in fibrils than in collagen molecules in (acidic) solution [16]. Given that these residues are charged, this partial extension might facilitate electrostatic interactions between collagen molecules to stabilise the fibril structure [17].
Enhancement of the sensitivity of the NMR experiment can be achieved using dynamic nuclear polarisation (DNP) NMR spectroscopy [18]. DNP NMR consists of transferring the significantly larger polarisation from electron spins (excited by a microwave source) to the NMR active nuclei, usually at very low temperatures (100 K). The DNP experiment therefore requires the presence of paramagnetic centres as the source of electron spins which is typically achieved by adding stable radicals to the sample. The additional complexities of the DNP experiment over more “standard” NMR experiments are compensated by enhancement of the NMR sensitivity by up to several orders of magnitude for 13C for instance [19]. Tiwari et al. [20] used DNP NMR to enhance the signals from aromatic residues in collagen and showed through 1H-13C correlation NMR spectra, putative interactions between collagen Pro/ Hyp and aromatic residues.
5. Collagen molecular dynamics
The dependence of NMR chemical shifts on molecular orientation (see Fig. 1C) means that NMR spectra are sensitive to molecular dynamics, as well as molecular structure. Motion between two different molecular orientations changes the chemical shifts of the nuclear sites involved in the motion. If the motion is very rapid compared to the chemical shift differences for the nuclei in the two molecular orientations, a single signal will observed in the NMR spectrum at the population-weighted average of the chemical shifts for the two orientations. For solution-state samples where molecules typically move rapidly between all possible molecular orientations randomly, the observed chemical shifts are the isotropic chemical shifts, as defined in Section 3 and Fig. 1C, and all molecular orientation dependence of the chemical shift is lost. That is not the case however for the components of the extracellular matrix with more restricted molecular motions, such as collagen fibrils, for instance, and molecular components bound to fibril surfaces. If the inverse of the motion’s correlation time is similar to the chemical shift differences caused by the molecule changing orientation, distinct and predictable changes occur in NMR spectra on static, i.e. non-MAS, samples that allow the angular reorientation and correlation time of the motion to be estimated. A related effect occurs in MAS NMR spectra where molecular motion causes a nucleus to sample different chemical environments, for instance through motion between different molecular conformations.
Torchia and co-workers were among the first to study the dynamics of collagen molecules within native tissues [21], [22], [23], specifically bone. The key in their studies was isotopic labelling to enable them to study the molecular dynamics of specific residue types in collagen molecules. They achieved isotopic labelling through injection of labelled amino acids into in vivo tissue in young animals/ embryos or addition of labelled amino acids to cell culture media in in vitro models. The isotopic enrichment included glycine 13C at C1 [21] or C2 [22], or deuterated (2H) proline and methionine [23]. Their NMR experiments on these tissues led to the determination of motion correlation times for Gly and Pro in fibrillar collagen. Gly and Pro being highly abundant throughout the triple helical region of collagen molecules means that the Gly/ Pro motional correlation times they deduced are for motions that affect a substantial part of the triple helix, e.g. whole molecule reorientation or twisting, or more local motions that are common around Gly/ Pro residues. Their results brought insight into the types of movements and flexibility of the collagen backbone and side chains in crosslinked versus uncrosslinked collagen and calcified versus non-calcified tissues. They concluded that collagen intermolecular cross-linking affects the collagen backbone mobility to a relatively small degree, whereas mineralization of the tissue has a significant immobilisation effect [21], reflected in the greater stiffness of calcified tissues. Collagen mineralization has a less pronounced effect on the motions of the collagen sidechains compared to backbone motion [23], perhaps an indication that most collagen sidechains are not in intimate contact with mineral and that the local motions that collagen sidechains undergo are not strongly coupled to global collagen molecular motions.
These early studies could not determine whether collagen molecules move their spatial positions or whether the molecular motions observed by NMR were reorientations on essentially fixed “lattice” positions. A very interesting NMR study on cartilage showed that collagen molecules are essentially rigid in terms of their spatial positions in the ECM, even despite the high degree of ECM hydration in cartilage, but with significant segmental mobility, the chain backbones undergoing fluctuations with 35∘ amplitudes, and larger for the sidechains () [24].
The motion of Pro residues in collagens is particularly interesting. The Pro sidechain ring can be exo (ring tip pointing away from the peptide backbone) or endo (ring tip pointing towards the peptide backbone), each conformation being necessarily associated with a significantly different residue backbone structure; typically the Pro backbone dihedral angle differs by between endo and exo conformations. XRD studies of small collagen-mimetic peptides suggest that Pro rings in the X-position of collagen Gly-X-Y triplets preferentially adopt an endo conformation and hydroxyproline/ proline rings in the Y-position adopt almost exclusively exo conformations [25]. The Pro and Hyp ring 13C chemical shifts are highly sensitive to the ring conformation, in particular, the 13C chemical shift which differs by 2 ppm between endo and exo conformations [26]. Motion between different chemical sites that have intrinsically different isotropic chemical shifts associated with them means the motion can be detected in spectra from samples undergoing MAS. Rapid motions with correlation times much larger than the inverse of the isotropic NMR frequency difference between the chemical sites involved in the motion results in a single NMR signal at the weighted-average chemical shift. Thus rapid Pro ring flips between endo and exo conformations results in an isotropic 13C signal at the population-weighted average chemical shift between those for the endo and exo conformations. In their study, De Sa Peixoto and collaborators [16] showed that upon fibrillogenesis, the proline C and C undergo increases in their respective 13C chemical shifts by up to 0.6 ppm, suggesting a Pro ring conformational preference more towards the exo conformation for collagen molecules in fibrils compared to those in solution. Conversely, the hydroxyproline, 13C chemical shift does not change much upon fibril formation. This is most likely because the C hydroxyl group is involved in hydrogen bonding and the exo Hyp ring conformation optimises those hydrogen bond interactions, implying that the hydrogen bond interactions are more important energetically in the collagen fibril than optimising the Hyp backbone structure [16].
An intriguing feature of collagens is the relatively high abundance not just of Pro/ Pro-derived residues, but specifically of GPO triplets in their sequences. Well-known for increasing the thermodynamic stability of the collagen molecule triple helix structure [25], we were curious as to whether these triplets had any impact on the fibril properties. De Sa Peixoto et al.’s results [16] imply that the collagen backbone structure changes at Pro residues in fibril formation, even in the absence of the binding partners that collagen molecules and fibrils have in native ECM. We hypothesised that Pro residues may provide important flexibility for the collagen molecular backbone structure to adapt to form stable fibril structures in vivo. The ground state fibril structure in vivo is likely to be influenced by non-collagenous binding partners of collagen molecules as well as collagen-collagen molecular interactions, suggesting that specific local, as well as more global, flexibility may be important in ECM structural integrity and functionality. We used a 2D 13C-13C correlation NMR approach to examine proline ring dynamics and preferred ring conformations in collagen fibrils in intact in vitro and ex vivo tissues [26], [27]. Utilizing the Pro 13C isotropic chemical shift dependence on the Pro ring conformation in parallel with energy landscape modelling of collagen-like triple helices, we found that X-position Pro rings in Gly-Pro–Hyp triplets in native collagen type I have essentially no preference for endo or exo conformations and flip rapidly ( Hz) between endo and exo conformations at biologically-relevant temperatures [27]. Pro endo-exo ring flips have the effect of extending/ compressing the peptide strand, because of the difference in the Pro dihedral angle in the endo and exo ring conformations. Thus if the Pro ring flipping occurs in all three chains of the collagen triple helix, as will be the case in homotrimeric collagens and in heterotrimeric collagens if the GPO sequence is present at the same sequence location in all three chains, then the triple helix is expected to be locally extensible/ compressible at GPO sites. If the GPO sequence is present in only one or two of the triple helix chains (in a heterotrimeric collagens), then the result of GPO Pro endo-exo ring flipping can only be bending of the triple helix. In both scenarios, the GPO Pro is a point of facile and highly specific triple helix flexibility. The alignment of collagen molecules within fibrils is such that GPO sites occur in bands across collagen type I fibrils and, furthermore, those bands are in close proximity to known cell binding sites, raising the intriguing possibility that this GPO flexibility is important in cell adhesion [27]. Furthermore, 90 of the GPO locations in the hole zones of mammalian collagen type I fibrils are “bend only” ones suggesting local, highly controlled backbone bends are important in these regions of the fibril.
Water is one of the most important components of the extracellular matrix undoubtedly having a major influence on protein and glycan molecular dynamics, and so playing roles in both the mechanical and biological properties of the matrix. However, water in biological systems is surprisingly hard to study beyond quantifying its total amount. Solid-state NMR gives insights into the many environments of water in the ECM, the mobility of water molecules in those environments and into how the level of hydration affects the mobility of other ECM molecules, and thus the contribution of those molecules to mechanical properties. Several studies have shown that collagen backbone and sidechain molecular dynamics decrease in amplitude as water is removed from the ECM [24], [9], [28], [29], showing that collagen has crucial interactions with water governing its mechanical properties. Mechanical loads are expected to (transiently) remove water from the ECM, and NMR results showing that ECM dehydration results in decreased amplitudes of collagen molecular motions suggests that mechanical loading can stiffen collagen fibrils solely through change of their hydration state [24].
Different water environments in the ECM are detected in NMR through the respective water 1H signal relaxation times [30]. Two different relaxation times can be readily measured: T1, whose magnitude depends on water tumbling motions on the nanosecond (or faster) timescale and T2, which depends on slower water molecular motion in the ms - s timescale, relevant to water bound to proteins etc. Measuring T1 and T2 for water 1H typically results in multiple values for the relaxation times which can be interpreted as different “pools” of water with different molecular mobilities [31], [32]. Interestingly, the distribution of water in the ECM between bound and “free” bulk states determined by NMR in this way correlates with bone quality, the extent of bound water decreasing in osteopenic trabecular bone [32].
6. Post-translational modification of matrix proteins
6.1. Glycosylation
The study of the sugar species present in the extracellular matrix is important as they play key roles both under physiological conditions and in pathologies. Glycosylation is the most common post-translational modification of proteins and is extensive in the extracellular matrix, where more than 90% of proteins carry this post-translational modification [33]. The most common glycosylations in the ECM are linear polysaccharides linked via N-glycosidic bonds through amino acids such as lysine, and O-glycosidic bonds linked to amino acids with hydroxyl functional groups, such as serine, tyrosine and hydroxylysine. ECM proteoglycans are the most extensively glycosylated proteins in the ECM but fibrillar collagens are also glycosylated, albeit to a lower extent compared to other members of the collagen family [34]. Although the roles of fibrillar collagen glycosylation are yet to be fully elucidated, decoration of collagen molecular and/ or fibril surfaces with sugars might offer resistance to degradation by proteases, mediate fibrillogenesis, support interaction with other matrix proteins and facilitate binding to collagen-specific cell-receptors [35].
The study of protein glycosylation in the extracellular matrix is not straightforward; glycosylation can occur at multiple amino acid side chains, and sometimes different sugar residues are attached to the same site in different copies of a given protein. The presence of glycosylation in proteins can be confirmed using different staining methodologies for proteins resolved on SDS–PAGE, mainly based on oxidation of sugar residues with periodic acid and further reaction with a Schiff reagent, or stains detecting negatively charged glycoproteins and GAGs. These methodologies are limited by non-specific reactions that can take place with easily oxidisable/ highly negatively charged protein segments, and are not sensitive to low levels of glycosylation [36]. LC/MS has become the gold standard for quantifying glycosylation in matrix collagens [37], [38], however, the relatively harsh chemical treatments needed to extract the collagen from the tissue may lead to loss of more labile glycosylation entities. Once extracted from the ECM, the structure of glycans can be elucidated using mass spectroscopy. This process is however laborious because of the need to isolate the protein of interest from the matrix [17], subsequent removal of the glycans from their proteins via enzymatic or chemical treatments, followed by purification and desalting [36].
Solid state NMR can provide a useful complementary approach for the study of collagen glycosylation, because NMR can be applied to intact matrix samples, meaning minimal sample preparation is required and thus there is a high probability of retaining all glycosylation moieties in their native state. Supplementing cell culture media with 13C-enriched glucose concentrates 13C nuclei in glycosylation moieties within the resulting in vitro ECM. Despite the large number of matrix glycosylated proteins, the collagen glycosylation species are typically the most abundant because of the predominance of collagen in the ECM. Thus, the most prominent signals in 13C NMR spectra of the tissues are typically those from collagen glycosylation. The characterization of the glycosylated species can be achieved using 2D 13C-13C correlation experiments as described in Section 2. Armed with knowledge of the glucose metabolic pathways prevalent in the cells producing the ECM, such as Krebs cycle or pentose phosphate pathway, using glucose which is 13C enriched at different carbons, and the information from the resulting 2D NMR spectra for each different glucose 13C labelling pattern, it is possible to assign the signals in the spectra to specific glycosylation moieties. Using this approach, Chow et al. have identified several O-, and N-linked glucosyl, galactosyl and glucosyl galactosyl species in the mineralised matrix deposited by fetal sheep osteoblasts in vitro, for instance, and shown that in bone, collagen glycosylation is in intimate contact with mineral, and closer to mineral than any collagen protein sidechains [14]. It is known from other studies that the extent and type of collagen glycosylation is different between bone and non-calcifying tissues. Combining these results suggests that collagen glycosylation may play an important role in bone mineral nucleation and/or growth [39].
Although solid state NMR is highly advantageous for studying intact tissue samples, it is an inherently low sensitivity technique which makes it difficult to study very sparse post-translational modifications of collagen, even with isotopic labelling [40]. Chow et al. [40] used the enhanced sensitivity of DNP NMR to identify 13C NMR signals from glycosylated, glycated and crosslinked collagen hydroxylysines in mouse skin samples (from mice fed a (U-13C6)-lysine diet) and in vitro vascular smooth muscle cell matrix, grown with (U-13C6, 15N2)-lysine respectively. Hydroxylysine accounts for less than 2% of the lysine found in collagen molecules so observing and resolving these signals from different modifications of collagen hydroxylysine demonstrates the potential power of DNP NMR for studying native tissues [40].
6.2. Glycation
Glycation is a non-enzymatic chemical reaction that takes place spontaneously between reducible sugars and nucleophilic protein side chains, such as terminal amine and guandino groups [41]. The process starts with Schiff base formation, followed by Amadori rearrangement to a keto-imine, and then a series of less well characterised reactions such as oxidations, fragmentations and dehydrations, the endstages of which are a range of Advanced Glycation End products (AGEs). The latter can present either as unnatural cross-links (e.g. glucosepane, pentosidine, imidazolium compounds) or as amino acid monovalent side chain modifications (carboxymethyl/ethyl lysine, imidazolones etc) [41]. Glycation is a slow process, affecting mostly proteins with longer biological half-life, and is often associated with ageing and hyperglycemic conditions such as diabetes [41]. It leads to a range of deleterious effects such as the stiffening and decreased proteolytic turnover of proteins in the matrix [42] and modification of the charge profile of proteins [41], which can be expected to impact intermolecular interactions in the ECM, potentially affecting the 3D architecture of the ECM and crucial functions like cell adhesion [43]. Given the complexity of the reactions that take place and their negative consequences on tissue function, the investigation and characterisation of the distribution of glycation products in the ECM is highly important in the understanding of a range of tissue diseases.
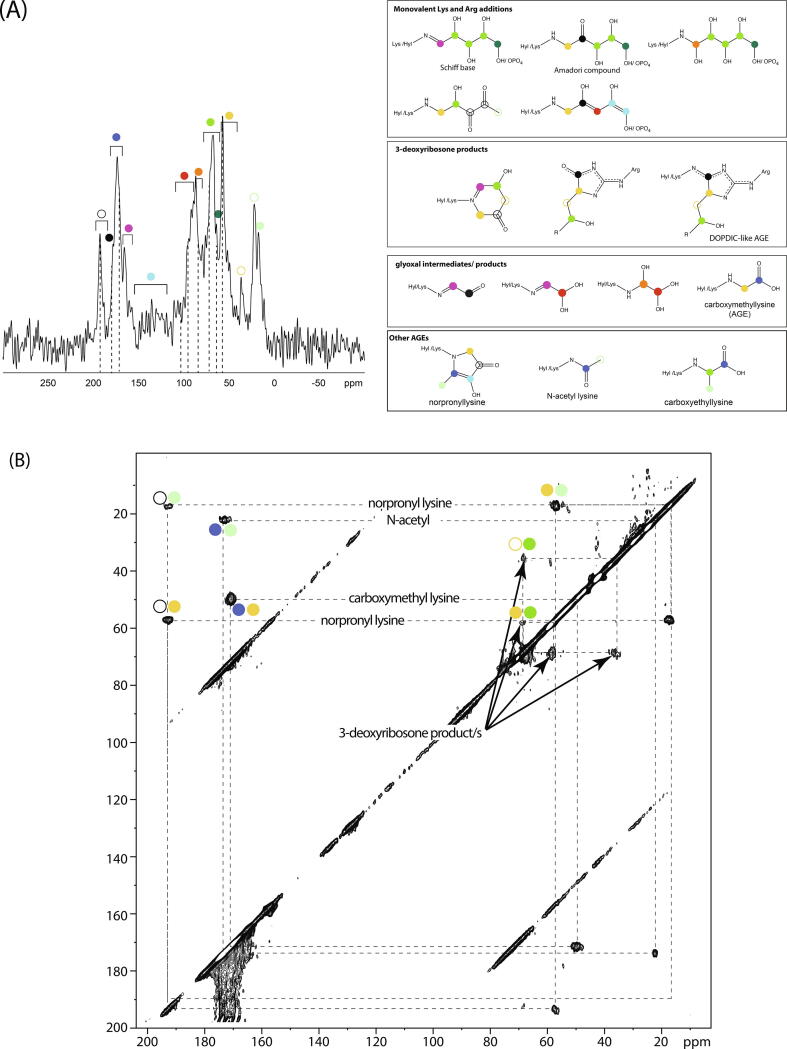
Solid state NMR is a very useful method for the identification of glycation products produced in vitro, both in matrix samples grown by cells, and glycated collagen matrices, where the concentration of glycation products is to a significant extent under the control of the experimenter. Under these conditions, experiments can be designed such that relatively high concentrations of glycation products result and so those products are amenable to characterisation by NMR spectroscopy. Li et al. [44] used 2D 13C NMR to show that the dominant products of ECM glycated with 13C-labelled ribose and ribose-5-phosphate (Fig. 3) are monovalent amino acid side chain modifications and that these are significantly more abundant than AGE cross-links. They hypothesised that these abundant side chain modifications must play a substantive role in the deleterious mechanical and functional consequences of glycation in collagen fibrils [44], [45] and further showed, using TEM, that the side chain modifications cause misalignment of collagen molecules in fibrils. They thus suggested that glycation-associated stiffening of collagen fibrils is substantially caused by misalignment of the collagen molecules in those fibrils. This idea is consistent with the finding that flexible GPO sequences are normally aligned in collagen fibrils [45]. Given the misalignment of collagen molecules with glycation, the GPO sequences are presumably misaligned in glycated fibrils. Alignment of flexible molecular regions means that flexions of those regions in neighbouring molecules across the fibril thickness has the effect of flexing the whole fibril, i.e. the molecular flexibility is conferred across the fibril; in contrast, misalignment of flexible molecular regions inevitably destroys that source of fibrillar flexibility. This study is an example of how integrating NMR spectroscopy with other physical characterization techniques can result in understanding of the ECM on multiple lengthscales, which is essential for a material where both molecular structures e.g. collagen molecule cell binding sites, are inherently coupled to the larger lengthscale nano-micrometre 3D architecture of the ECM.
Fig. 3.
(A) Left: 1D double-quantum-filtered solid-state 13C NMR spectrum of bovine collagen type I fibrils glycated with U-13C-R5P. The spectra contain only signals from 13C bonded to another 13C and so contain only signals from molecular species derived from the respective 13C-labelled sugar. Possible assignments for each signal/ range of signals are indicated with coloured circles; the similarity of chemical structure between possible glycation products means that there is both overlap of signals from different glycation products and several possible assignments for most signals. Dotted lines are to allow easy comparison of chemical shifts of signals for the different glycation sugars. Right: possible glycation products with 13C site chemical shifts expected indicated with coloured circles that correspond to the colour scheme used to designate signals in the spectra on the left. (B) 2D 13C-13C proton-driven spin-diffusion correlation spectrum for U-13C-R5P-glycated collagen type I, showing the assignment of norpronyl-lysine, (carboxymethyl) lysine, N-acetyl and products of Lys-Arg glycation with 3-deoxyribosone-like molecules as the major collagen - R5P glycation products. Products of 3-deoxyribosone-like molecules are expected to include products, such as DOPDIC, with a -CH(N)....CH2-CH(OH)- fragment [46]. The 13CH2 component in such fragments has a distinctive 37 ppm signal [46].
7. Studying biomineralisation processes by NMR
The detailed composition and structure of mineralised tissues, such as bone or calcified arteries, at the molecular lengthscale and the mechanisms behind calcification are still not fully elucidated. The original view that bone mineral is a carbonate-substituted hydroxyapatite, a view derived from XRD and FTIR studies, has been challenged by solid state NMR data. The methodologies used in XRD and FTIR often involve invasive treatment of samples, such as dehydration or removal of the organic matrix, which leads to inevitable changes in the chemical composition, phase and structure of the mineral component. Solid-state NMR spectroscopy on the other hand can be applied directly to tissue samples with little sample preparation required other than steps to preserve the sample [12], [13]. Furthermore, NMR spectroscopy allows separate study of the mineral and organic components of calcified extracellular matrix by the choice of nucleus, without any need to physically separate those components: 31P and 43Ca to study the calcium phosphate mineral component and 13C to study the collagen/ organic matrix [47], [48], [49], [50], [51]. Indeed, the presence of different nuclear species in the mineral and organic components provides the unique opportunity to determine the structural and functional relationships between the two [47], [48], [49], [51], a feature which is strongly enhanced in DNP NMR of bone [52] and utilized, for instance, to follow de novo bone formation in bone implant materials in vivo and in vitro [53], [54], [55]. There have been many interesting studies of bone mineral using solid-state NMR; the scope of this review does not allow discussion of all of them, rather we have collected together the main themes of recent research in this area.
7.1. NMR for the study of inorganic component of mineralised tissues
31P and 1H are the main nuclear species of interest for studying the mineral component of calcified tissues by NMR, as there is a substantial database of 31P and 1H chemical shifts available for calcium phosphates, including ion-substituted materials that may be of relevance in ECM calcification. The isotropic 31P chemical shift spectrum of various mineralised tissues, including bone or teeth, reveals a single peak (3.2 ppm; Fig. 4A) consistent with apatitic orthophosphate [56], [57], [58], [59], but the signal is broader than the equivalent peak observed in crystalline, synthetic hydroxyapatite, indicating biological mineral contains a range of phosphate species and environments that cannot be modelled by hydroxyapatite alone.
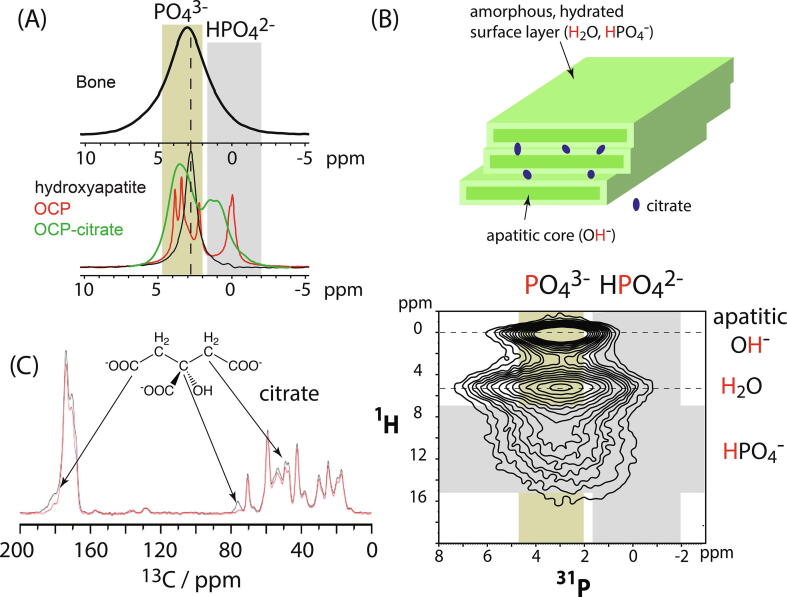
Fig. 4.
(A) 1D 31P MAS NMR spectrum of bone (top) and representative synthetic calcium phosphate phases to indicate the chemical shift ranges expected for orthophosphate (inorganic , olive box) and hydrogen phosphate (, grey box). (B) The current model of bone mineral chemical structure (top) comes largely from solid-state NMR spectroscopy such as the 2D 1H - 31P correlation spectrum of intact sheep cortical bone (bottom), where the correlation signals are assigned to apatitic hydroxyl groups and non-apatitic water and hydrogen phosphate ions. The expected chemical shift ranges for orthophosphate (inorganic , olive box) and hydrogen phosphate (, grey box) are indicated as for (A). From TEM, bone mineral consists of stacks of 2–4 nanoscopic mineral platelets [62], [63]. The same work shows that the mineral platelets schematically depicted here likely derive from sideways-aggregated needle-like structures rather than being homogeneous platelets. Citrate anions are hypothesised to bridge between the mineral platelets [83]. (C) A 13C31P REDOR spectrum of sheep bone. Signals in the REDOR spectrum (red) with lower intensity than those in the reference specrtum (black) are from carbons close in space to phosphorus. The strongest reductions in signal intensity are from signals assigned to the various carbons of citrate, showing that citrate is physically close to bone mineral phosphate.
TEM studies have shown bone mineral to consist of stacks of 2–4 platelet-type structures with dimensions of a few tens of nanometers and 2–5 nm thick [60], [61], [62], [63]. TEM tomography has most recently shown that the platelet structures are in fact comprised of sideways-aggregated needle-like structures resulting in a “picket-fence”-like morphology [62], [63]. NMR spectroscopy of native bone and model mineral phases have been instrumental in explaining the chemical structures that lead to these mineral morphologies [64], [65], [61]. 2D 1H-31P correlation spectra [64], [65], [61], [66], [67] have shown that in addition to more crystalline apatitic structures, bone mineral contains substantial quantities of highly-hydrated, disordered non-apatitic calcium phosphate structures, containing hydrogen phosphate as well as orthophosphate. If the bone sample is soaked in D2O, the 1H signal from the water in the hydrated, disordered, non-apatitic structures in bone mineral disappears [61], indicating that this water in can be readily exchanged. This along with many previous solid-state NMR studies of bone mineral has led to a chemical model of bone mineral [64], [61], [68], [69] in which the platelet-type structures have a core of more crystalline hydroxyapatite (likely partially substituted at all ionic sites) and a surface layer of highly-hydrated, amorphous calcium phosphate (Fig. 4B). The presence of this hydrated amorphous mineral layer in the mineral platelets is important for the parallel organization of the platelets in bone, an arrangement which is not present upon dehydration of bone or in samples of biomimetic nanocrystals of hydroxyapatite [51], [61]. A useful method has been proposed to interpret 2D 1H-31P NMR spectra of bone in terms of contributions from complex synthetic calcium phosphate phases, in particular to distinguish the presence of amorphous mineral that is integral to the mature bone mineral structure from transient amorphous calcium phosphate (ACP) precursors of bone mineral [70]; such approaches are essential to progress understanding of the molecular structure of bone mineral and its development in vivo.
The chemical structure of bone mineral is complicated by the presence of metabolic anions from cell respiration in its structure: carbonate (6–8 wt%) [71], citrate (2–5 wt%) [72], [73], [74], [75] and lactate (variable quantities) [72], [73], [76] are the most abundant of these. Several solid-state NMR studies have led to the proposal that carbonate is present both as substitutions within the lattice of the apatitic component of bone mineral and in the amorphous mineral regions around each mineral platelet [61], [77]. Release of carbonate from mineral platelet surfaces layers is hypothesised to buffer rising plasma pH (acidosis) in turtles in anoxic conditions [78]. By contrast, the turtle skeleton takes up lactate in anoxic conditions with both carbonate and lactate levels in bone mineral re-establishing to expected levels once the animals return to normoxic conditions [76]. The mammalian foetal skeleton is the other notable occurrence of high lactate content in bone mineral, but as with the turtle skeleton, decreases in concentration in the normoxic conditions that prevail after birth [79]. These observations collectively suggest that the carbonate and lactate components of bone mineral are at least in part derived from the extracellular fluid and its contact with blood plasma. A connection of these mineral components to whole-body respiration and metabolism may explain their variable concentration in bone mineral and changes of their concentration in bone mineral with age as a progressively smaller fraction of bone mineral is in equilibrium with body fluids [80]. On the other hand, osteoblasts are professional citrate-producing cells suggesting that the presence of citrate in bone mineral may be actively regulated by cells and therefore that citrate is an inherent, essential part of bone mineral molecular structure, morphology or mineral crystal organization [81], [82]. A huge step in understanding of bone mineral occurred when citrate was shown by NMR [47] to be closely associated with mineral and subsequently, citrate was found to be a common component of both physiological and pathological mammalian calcifications [50]. Because of the large size of citrate, this anion cannot fit in the crystal lattice of hydroxyapatite as carbonate anions can. Instead, Davies et al. [83] proposed the synthetic phase, octacalcium phosphate citrate (OCP-citrate), as a model for how citrate is incorporated into bone mineral. They showed that in OCP-citrate, the citrate anion resides in the disordered hydrated layer of OCP, bridging between the apatitic layers of the OCP crystal structure. 17O, 43Ca and 2D 1H-31P NMR spectra of native bone showed that the citrate-containing disordered hydrated layer in OCP-citrate is highly similar to the disordered hydrated layers in bone mineral [83], [84].
43Ca NMR of native bone samples is challenging [49], [85], [86] because of the low natural abundance of 43Ca, and the additional complication of the 43Ca nucleus possessing an electric quadrupole moment which has the effect of very significantly complicating the NMR spectrum even under MAS. The 43Ca NMR spectrum of bone shows a broad signal which has some spectral overlap with the 43Ca spectrum of 10% carbonated hydroxyapatite but comparatively little with the 43Ca of pure hydroxyapatite or apatites with lower carbonate content [86]. Furthermore, the 43Ca NMR signals from cortical bone indicates larger 43Ca quadrupolar coupling constants than those of synthetic apatites even those with carbonate substitution [86]. Thus the calcium environments in bone mineral cannot be fully modelled by carbonate-substituted or pure hydroxyapatite. Given that the presence of apatitic components to bone mineral is undisputed, it would seem to be the amorphous structural regions that fail to be fully modelled by carbonate-substituted hydroxyapatite. Carbonate-substituted hydroxyapatite contains amorphous structural regions [61], but clearly these alone cannot model the amorphous structural regions of bone mineral. Interestingly, 43Ca NMR spectra of OCP derivatives [84] have signals centred at lower frequencies than apatitic materials and this is the spectral region where 43Ca NMR spectra of bone mineral have signal intensity that is not replicated in 43Ca NMR spectra of synthetic apatitic materials. This observation supports the presence of structures in bone mineral - presumably in the amorphous structural regions - that can be modelled by disordered OCP-like derivatives, disordered by the addition of metabolic anions such as citrate. The amorphous structural regions of bone mineral are intriguing from an NMR perspective too. A dominant component of the amorphous regions is undoubtedly anions. These anions are identified by 2D 1H-31P NMR spectra of bone by a characteristic 1H signal; the acidic 1H of groups is typically in the region 7–15 ppm [87] which is high for a 1H signal, so a 1H signal in this spectral region can be unambiguously assigned. 2D 1H-31P NMR spectra of bone show a very broad 1H signal between 7 and 15 ppm (Fig. 4B and see e.g. reference [66]), the only plausible assignment of which would seem to be to anions. The 31P chemical shift of anions is typically between 2 ppm and −1 ppm [88]. Curiously however, the dominant 31P chemical shift of the 2D 1H-31P NMR signal assigned to in bone samples is essentially that of hydroxyapatite 31P, around 3 ppm, although the signal is very broad and does also cover the chemical shift range more typically expected for [66], including that of stoichiometric compounds containing such as OCP and brushite, as well as OCP-citrate (Fig. 4B). Moreover, 2D 1H-31P NMR spectra on fresh bone [12] show multiple signals in the region of the spectrum which broaden and reduce in intensity (in favour of the apatitic 1H-31P signal) as the sample is dehydrated. These data collectively suggest a wide range of environments in bone mineral which are highly dependent on the degree of hydration, including some environments not characteristic of stoichiometric calcium phosphate phases. The conundrum on the environment of anions in bone mineral is the subject of a recent, excellent critical review by Eden [89]. Consideration of how metabolic anions influence the structure of the amorphous, -containing regions of bone mineral must be high on the agenda for further insight into the structure of bone mineral, as indeed must a complete catalogue of all the organic anions involved in bone mineral, as carbonate, citrate and lactate, whilst probably the dominant species, are unlikely to be an exhaustive list.
7.2. The bone mineral-organic interface
A key question in bone biology is what are the molecules that control the mineral nucleation? Without a hypothesis, it is difficult to answer such questions. One advantage of NMR spectroscopy is that no prior assumptions need to be made about the composition of the sample in order to observe its components by NMR. All 13C–containing molecules can be observed in a 13C NMR spectrum of a sample, for instance. This feature meant that we saw characteristic signals from an unexpected molecule, poly(ADP-ribose) (PAR) in 2D 13C-13C correlation spectra from ECM produced by in vitro calcifying osteoblasts [14] and vascular smooth muscle cells (Fig. 2B) [90]. Further in vitro experiments showed PAR associates with Ca2+ ions to form spherical electron-dense structures that bind specifically to the gap zone of collagen fibrils [90], where nucleation of bone mineral is believed to occur [91]. These observations led to the hypothesis that poly(ADP-ribose) gathers calcium ions from which mineral nucleates once the Ca-laden poly(ADP-ribose) is bound within the extracellular matrix [90].
The 13C{31P} Rotational Echo DOuble Resonance (REDOR) NMR experiment can be used to probe the organic molecules close to collagen (Fig. 4C). The REDOR experiment results in a reference (normal) 1D 13C NMR spectrum of the sample and a REDOR 13C NMR spectrum. 13C signals with lower intensity in the REDOR spectrum compared with the reference spectrum are from carbon sites close in space (within 0.8–1 nm typically) to phosphorus nuclei. The REDOR NMR experiment was what first identified citrate anions to be present in bone mineral [47] (Fig. 4C). The REDOR experiment has also been used to understand how the collagen-mineral interface changes with the hydration level of the bone ECM [12].
The role of non-collagenous proteins in bone mineralisation can also be probed using solid state NMR, to complement data from other biophysical techniques. NMR studies of model systems with nanoscopic hydroxyapatite and osteocalcin [92] and osteopontin [93] have given useful hypotheses about how these osteogenic proteins may interact with bone mineral particles and influence mineral morphology. An ex vivo approach was used to gain insight into the role of osteopontin and osteocalcin in the mechanical properties of bone [94]. Comparison of 1D 13C and 2D 1H-13C correlation NMR spectra between bone samples from osteocalcin/ osteopontin knockout mice (OC-OPN -/-, -/-) with those from wild type animals showed no significant changes in the molecular conformations/ composition of the organic part of the ECM. However, REDOR NMR identified some alterations at the interface between mineral and organic matrix, specifically increased exposure of collagen Arg and Lys residues to mineral. The overall similarity in the mineral and the organic matrix chemical structures between wild type and knockout animal bone tissue suggests that the change in mechanical properties, such as decreased toughness found in the knockout animals, is not related to any structural changes in the mineral or collagen. Instead OPN and OC are likely to contribute to the toughness of the tissue through their involvement in the collagen-mineral interface [94].
8. Conclusions
Solid-state NMR spectroscopy has played an important role in providing molecular structure and dynamics information for the ECM that complements the insight from ultrastructural methods such as TEM and AFM, leading to a far more comprehensive picture of how the ECM is structured, how delivers its mechanical and signalling properties and how its function may be altered. With the continuing development of high-resolution cryo-EM methods, we can expect to see further profitable integration of NMR and ultrastructural methods. For instance, NMR can determine the conformational dynamics. Mapping this information onto molecular structures determined from cryo-EM then gives a hypothesis about how the particular protein plays its role in vivo. Now that solid-state NMR is beginning to mature its view of collagen structures and dynamics in native ECM, there is a new opportunity to study how those structures and dynamics are altered in disease. One obvious application here is to use NMR to understand how cancer cells alter the surrounding ECM structure to signal to other cells and to migrate. The ongoing development of higher NMR magnetic field strengths and higher sensitivity in NMR, such as DNP NMR and the development of so-called cryoMAS NMR technology will facilitate these developments, and very likely bring about fundamental new insight into matrix structure and biology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
AM was funded by a UK BBSRC Doctoral Training Program PhD studentship.
References
- 1.Bernd Reif, Sharon E. Ashbrook, Lyndon Emsley, and Mei Hong. Solid-state NMR spectroscopy. Nature Reviews Methods Primers, 1(1), 2021. [DOI] [PMC free article] [PubMed]
- 2.Wishart D.S., Sykes B.D., Richards F.M. The Chemical shift index. A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31(1992):1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 3.K J Fritzsching, Y Yang, K Schmidt-Rohr, and Mei Hong. Practical use of chemical shift databases for protein solid-state NMR: 2D chemical shift maps and amino-acid assignment with secondary-structure information. Journal of Biomolecular NMR, 56(2):155–167, 2013. [DOI] [PMC free article] [PubMed]
- 4.Andrea Cavalli, Xavier Salvatella, Christopher M Dobson, and Michele Vendruscolo. Protein structure determination from NMR chemical shifts. Proceedings of the National Academy of Sciences of the United States of America, 104(23):9615–9620, 2007. [DOI] [PMC free article] [PubMed]
- 5.Böckmann Anja. Structural and dynamic studies of proteins by high-resolution solid-state NMR. C. R. Chim. Mar 2006;9(3–4):381–392. [Google Scholar]
- 6.Seidel Karsten, Lange Adam, Becker Stefan, Hughes Colan E, Heise Henrike, Baldus Marc. Protein solid-state NMR resonance assignments from (13C, 13C) correlation spectroscopy. Phys. Chem. Chem. Phys. 2004;6:5090–5093. [Google Scholar]
- 7.Manolikas Theofanis, Herrmann Torsten, Meier Beat H. Protein structure determination from 13C spin-diffusion solid-state NMR spectroscopy. J. Am. Chem. Soc. 2008;130(12):3959–3966. doi: 10.1021/ja078039s. [DOI] [PubMed] [Google Scholar]
- 8.L.Y. Lian and D.A. Middleton. Labelling approaches for protein structural studies by solution-state and solid-state NMR. Progress in Nuclear Magnetic Resonance Spectroscopy, 39(3):171–190, 2001.
- 9.Detlef Reichert, Ovidiu Pascui, Eduardo R DeAzevedo, Tito J Bonagamba, Klaus Arnold, and Daniel Huster. A solid-state NMR study of the fast and slow dynamics of collagen fibrils at varying hydration levels. Magnetic resonance in chemistry, 42(2):276–84, Feb 2004. [DOI] [PubMed]
- 10.Daniel Huster, Jürgen Schiller, and Klaus Arnold. Comparison of collagen dynamics in articular cartilage and isolated fibrils by solid-state NMR spectroscopy. Magnetic resonance in medicine: official journal of the Society of Magnetic Resonance in Medicine/ Society of Magnetic Resonance in Medicine, 48(4):624–632, Oct 2002. [DOI] [PubMed]
- 11.Veronica Wai Ching Wong, David G Reid, Wing Ying Chow, Rakesh Rajan, Maggie Green, Roger A Brooks, and Melinda J Duer. Preparation of highly and generally enriched mammalian tissues for solid state NMR. Journal of Biomolecular NMR, 63(2):119–123, 2015. [DOI] [PubMed]
- 12.Rai and Neeraj Sinha Ratan Kumar, Sinha Neeraj. Dehydration-Induced Structural Changes in the Collagen-Hydroxyapatite Interface in Bone by High-Resolution Solid-State NMR Spectroscopy. J. Phys. Chem. C. 2011;115:14219–14227. [Google Scholar]
- 13.Nikel Ondrej, Laurencin Danielle, Bonhomme Christian, Sroga Grażyna E., Besdo Silke. Anna Lorenz, and Deepak Vashishth. Solid state NMR investigation of intact human bone quality: balancing issues and insight into the structure at the organic-mineral interface. J. Phys. Chem. C, Nanomaterials Interfaces. 2012;116(10):6320–6331. doi: 10.1021/jp2125312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ying Chow W., Rajan Rakesh, Muller Karin H., Reid David G., Skepper Jeremy N., Wong Wai Ching, Brooks Roger A., Green Maggie, Bihan Dominique, Farndale Richard W., Slatter David A., Shanahan Catherine M., Duer Melinda J. NMR spectroscopy of native and in vitro tissues implicates poly(ADP ribose) in biomineralization. Science (New York N.Y.) 2014;344:742–746. doi: 10.1126/science.1248167. [DOI] [PubMed] [Google Scholar]
- 15.Wai Ching Veronica Wong, Aurimas Narkevicius, Wing Ying Chow, David G. Reid, Rakesh Rajan, Roger A. Brooks, Maggie Green, and Melinda J. Duer. Solid state NMR of isotope labelled murine fur: a powerful tool to study atomic level keratin structure and treatment effects. Journal of Biomolecular NMR, 66(2):93–98, 2016. [DOI] [PMC free article] [PubMed]
- 16.De Sa Paulo, Peixoto Guillaume Laurent, Azaïs Thierry, Mosser Gervaise. Solid-state nmr study reveals collagen i structural modifications of amino acid side chains upon fibrillogenesis. J. Biol. Chem. 2013;288(11):7528–7535. doi: 10.1074/jbc.M112.390146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberga Ieva, Li Rui, Duer Melinda J. Collagen structure–function relationships from solid-state nmr spectroscopy. Acc. Chem. Res. 2018;51(7):1621–1629. doi: 10.1021/acs.accounts.8b00092. [DOI] [PubMed] [Google Scholar]
- 18.Jaudzems Kristaps, Polenova Tatyana, Pintacuda Guido, Oschkinat Hartmut, Lesage Anne. DNP NMR of biomolecular assemblies. J. Struct. Biol. 2019;206(1):90–98. doi: 10.1016/j.jsb.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Aany Sofia Lilly Thankamony, Johannes J Wittmann, Monu Kaushik, and Björn Corzilius. Dynamic nuclear polarization for sensitivity enhancement in modern solid-state nmr. Progress in Nuclear Magnetic Resonance Spectroscopy, 102:120–195, 2017. [DOI] [PubMed]
- 20.Tiwari Nidhi, Wi Sungsool, Mentink-Vigier Frederic, Sinha Neeraj. Mechanistic Insights into the Structural Stability of Collagen-Containing Biomaterials Such as Bones and Cartilage. J. Phys. Chem. B. 2021;125(18):4757–4766. doi: 10.1021/acs.jpcb.1c01431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkar S.K., Sullivan C.E., Torchia D.A. Solid state 13c nmr study of collagen molecular dynamics in hard and soft tissues. J. Biol. Chem. 1983;258(16):9762–9767. [PubMed] [Google Scholar]
- 22.Sarkar Susanta K, Sullivan Catherine E, Torchia Dennis A. Nanosecond fluctuations of the molecular backbone of collagen in hard and soft tissues: a carbon-13 nuclear magnetic resonance relaxation study. Biochemistry. 1985;24(9):2348–2354. doi: 10.1021/bi00330a033. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar Susanta K, Yukio Hiyama C.H., Niu PE Young, Gerig J.T., Torchia D.A. Molecular dynamics of collagen side chains in hard and soft tissues. a multinuclear magnetic resonance study. Biochemistry. 1987;26(21):6793–6800. doi: 10.1021/bi00395a032. [DOI] [PubMed] [Google Scholar]
- 24.Zernia Göran, Huster Daniel. Collagen dynamics in articular cartilage under osmotic pressure. NMR Biomed. 2006;19(8):1010–1019. doi: 10.1002/nbm.1061. [DOI] [PubMed] [Google Scholar]
- 25.Matthew D. Shoulders and Ronald T. Raines. Collagen Structure and Stability. Annual Review of Biochemistry, 78(1):929–958, Jan 2009. [DOI] [PMC free article] [PubMed]
- 26.Ying Chow W., Bihan Dominique, Forman Chris J., Slatter David A., Reid David G., Wales David J., Farndale Richard W., Duer Melinda J. Hydroxyproline ring pucker causes frustration of helix parameters in the collagen triple helix. Scientific Reports. 2015;5(1):1–11. doi: 10.1038/srep12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wing Ying Chow, Chris J. Forman, Dominique Bihan, Anna M. Puszkarska, Rakesh Rajan, David G. Reid, David A. Slatter, Lucy J. Colwell, David J. Wales, Richard W. Farndale, and Melinda J. Duer. Proline provides site-specific flexibility for in vivo collagen. Scientific Reports, 8:13809, 2018. [DOI] [PMC free article] [PubMed]
- 28.Peizhi Zhu, Jiadi Xu, Nadder Sahar, Michael D Morris, David H Kohn, and Ayyalusamy Ramamoorthy. Time-resolved dehydration-induced structural changes in an intact bovine cortical bone revealed by solid-state NMR spectroscopy. Journal of the American Chemical Society, 131(47):17064–17065, Dec 2009. [DOI] [PMC free article] [PubMed]
- 29.Jiadi Xu., Zhu Peizhi, Morris Michael D., Ramamoorthy Ayyalusamy. Solid-State NMR Spectroscopy Provides Atomic-Level Insights Into the Dehydration of Cartilage. The journal of physical chemistry. B. 2011;115(33):9948–9954. doi: 10.1021/jp205663z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clare M Lewandowski. No Title No Title. The effects of brief mindfulness intervention on acute pain experience: An examination of individual difference, 1:20958, 2015.
- 31.Nidhi Tiwari, Rama Nand Rai, and Neeraj Sinha. Water-lipid interactions in native bone by high-resolution solid-state NMR spectroscopy. Solid State Nuclear Magnetic Resonance, 107(April):101666, 2020. [DOI] [PubMed]
- 32.Ratan Kumar Rai, Tarun Barbhuyan, Chandan Singh, Monika Mittal, Mohd Parvez Khan, Neeraj Sinha, and Naibedya Chattopadhyay. Total water, phosphorus relaxation and inter-atomic organic to inorganic interface are new determinants of trabecular bone integrity. PloS one, 8(12):e83478, 2013. [DOI] [PMC free article] [PubMed]
- 33.Lynch M., Barallobre-Barreiro J., Jahangiri M., Mayr Manuel. Vascular proteomics in metabolic and cardiovascular diseases. J. Intern. Med. 2016;280(4):325–338. doi: 10.1111/joim.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masahiko Terajima, Irina Perdivara, Marnisa Sricholpech, Yoshizumi Deguchi, Nancy Pleshko, Kenneth B Tomer, and Mitsuo Yamauchi. Glycosylation and cross-linking in bone type i collagen. Journal of Biological Chemistry, 289(33):22636–22647, 2014. [DOI] [PMC free article] [PubMed]
- 35.Perdivara Irina, Yamauchi Mitsuo, Tomer Kenneth B. Molecular characterization of collagen hydroxylysine o-glycosylation by mass spectrometry: current status. Aust. J. Chem. 2013;66(7):760–769. doi: 10.1071/CH13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth Ziv, Yehezkel Galit, Khalaila Isam. Identification and quantification of protein glycosylation. Int. J. Carbohydrate Chem. 2012;2012 [Google Scholar]
- 37.Melanie Stammers, Izabella S Niewczas, Anne Segonds-Pichon, and Jonathan Clark. Mechanical stretching changes cross-linking and glycation levels in the collagen of mouse tail tendon. Journal of Biological Chemistry, 295:jbc.RA119.012067, 2020. [DOI] [PMC free article] [PubMed]
- 38.Hudson David M., Archer Marilyn, King Karen B., Eyre David R. Glycation of type I collagen selectively targets the same helical domain lysine sites as lysyl oxidase-mediated cross-linking. J. Biol. Chem. 2018;293(40):15620–15627. doi: 10.1074/jbc.RA118.004829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsuo Yamauchi and Marnisa Sricholpech. Lysine post-translational modifications of collagen. Essays in biochemistry, 52:113–133, 2012. [DOI] [PMC free article] [PubMed]
- 40.Wing Ying Chow, Rui Li, Ieva Goldberga, David G. Reid, Rakesh Rajan, Jonathan Clark, Hartmut Oschkinat, Robert Hayward, Catherine M Shanahan, and Melinda J Duer. Essential but sparse collagen hydroxylysyl post-translational modifications detected by DNP NMR. Chemical Communications, 54(89):12570–12573, 2018. [DOI] [PubMed]
- 41.Allen J Bailey, Robert Gordon Paul, and Lynda Knott. Mechanisms of maturation and ageing of collagen. Mechanisms of ageing and development, 106(1–2):1–56, 1998. [DOI] [PubMed]
- 42.Sell David R., Biemel Klaus M., Reihl Oliver, Lederer Markus O., Strauch Christopher M., Monnier Vincent M. Glucosepane is a major protein cross-link of the senescent human extracellular matrix: Relationship with diabetes. J. Biol. Chem. 2005;280(13):12310–12315. doi: 10.1074/jbc.M500733200. [DOI] [PubMed] [Google Scholar]
- 43.McCarthy Antonio Desmond, Uemura Toshimasa, Etcheverry Susana Beatriz, Cortizo Ana María. Advanced glycation endproducts interfere with integrin-mediated osteoblastic attachment to a type-I collagen matrix. Int. J. Biochem. Cell Biol. 2004;36(5):840–848. doi: 10.1016/j.biocel.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Li R., Rajan R., Wong W.C.V., Reid D.G., Duer M.J., Somovilla V.J., Martinez-Saez N., Bernardes G.J.L., Hayward R., Shanahan C.M. In situ characterization of advanced glycation end products (AGEs) in collagen and model extracellular matrix by solid state NMR. Chem. Commun. 2017;53(100):13316–13319. doi: 10.1039/c7cc06624d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sneha Bansode, Uliana Bashtanova, Rui Li, Jonathan Clark, Karin H Müller, Anna Puszkarska, Ieva Goldberga, Holly H Chetwood, David G Reid, Lucy J Colwell, Jeremy N Skepper, Catherine M Shanahan, Georg Schitter, Patrick Mesquida, and Melinda J Duer. Glycation changes molecular organization and charge distribution in type I collagen fibrils. Scientific Reports, 10(1):1–13, 2020. [DOI] [PMC free article] [PubMed]
- 46.Biemel Klaus M, Reihl Oliver, Lederer Markus O. Formation Pathways for Lysine-Arginine Cross-links Derived from Hexoses and Pentoses by Maillard Processes. J. Biol. Chem. 2001;276(26):23405–23412. doi: 10.1074/jbc.M102035200. [DOI] [PubMed] [Google Scholar]
- 47.Y-Y Hu, A Rawal, and K Schmidt-Rohr. Strongly bound citrate stabilizes the apatite nanocrystals in bone. Proceedings of the National Academy of Sciences of the USA, 107(52):22425–22429, 2010. [DOI] [PMC free article] [PubMed]
- 48.Wise Erica R, Sergey Maltsev M., Davies Elisabeth, Duer Melinda J, Jaeger Christian, Loveridge Nigel, Murray Rachel C., Reid David G. The organic- mineral interface in bone is predominantly polysaccharide. Chem. Mater. 2007;19(21):5055–5057. [Google Scholar]
- 49.Danielle Laurencin, Gilles Guerrero, Julien Amalric, Christian Bonhomme, Christel Gervais, Mark E Smith, and P Hubert Mutin. Advanced solid-state NMR techniques for the investigation of the organic-mineral interfaces in biomaterials. Mater. Res. Soc. Symp. Proc., 1236:2–7, 2010.
- 50.D G Reid, M J Duer, G E Jackson, R C Murray, A L Rodgers, and C M Shanahan. Citrate occurs widely in healthy and pathological apatitic biomineral: Mineralized articular cartilage, and intimal atherosclerotic plaque and apatitic kidney stones. Calcified Tissue International, 93(3), 2013. [DOI] [PubMed]
- 51.Duer Melinda J. The contribution of solid-state NMR spectroscopy to understanding biomineralization: Atomic and molecular structure of bone. J. Magn. Reson. 2015;253:98–110. doi: 10.1016/j.jmr.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 52.Thierry Azaïs, Stanislas Von Euw, Widad Ajili, Stéphanie Auzoux-Bordenave, Philippe Bertani, David Gajan, Lyndon Emsley, Nadine Nassif, and Anne Lesage. Structural description of surfaces and interfaces in biominerals by DNP SENS. Solid State Nuclear Magnetic Resonance, 102(February):2–11, 2019. [DOI] [PubMed]
- 53.Marchandise Xavier, Belgrand Patrick, Legrand André-Pierre. Solid-State 31P NMR Spectroscopy of Bone and Bone Substitutes. Magn. Res. Med. 1992;28:1–8. doi: 10.1002/mrm.1910280102. [DOI] [PubMed] [Google Scholar]
- 54.Anja Penk, Yvonne Förster, Holger A. Scheidt, Ariane Nimptsch, Michael C. Hacker, Michaela Schulz-Siegmund, Peter Ahnert, Jürgen Schiller, Stefan Rammelt, and Daniel Huster. The pore size of PLGA bone implants determines the de novo formation of bone tissue in tibial head defects in rats. Magnetic Resonance in Medicine, 70(4):925–935, 2013. [DOI] [PubMed]
- 55.J Schulz, M Pretzsch, I Khalaf, a Deiwick, H a Scheidt, G Salis-Soglio, a Bader, and D Huster. Quantitative monitoring of extracellular matrix production in bone implants by 13C and 31P solid-state nuclear magnetic resonance spectroscopy. Calcified tissue international, 80(4):275–285, May 2007. [DOI] [PubMed]
- 56.a H Roufosse, W P Aue, J E Roberts, M J Glimcher, and R G Griffin. Investigation of the mineral phases of bone by solid-state phosphorus-31 magic angle sample spinning nuclear magnetic resonance. Biochemistry, 23(25):6115–6120, dec 1984. [DOI] [PubMed]
- 57.Roberts J.E., Bonar L.C., Griffin R.G., Glimcher M.J. Characterization of Very Young Mineral Phases of Bone by Solid State 31Phosphorus Magic Angle Sample Spinning Nuclear Magnetic Resonance and X-Ray Diffraction. Calcif. Tissue Int. 1992;50:42–48. doi: 10.1007/BF00297296. [DOI] [PubMed] [Google Scholar]
- 58.Yaotang Wu, Jerome L Ackerman, Hyun-Man Kim, Christian Rey, Allal Barroug, and Melvin J Glimcher. Nuclear magnetic resonance spin-spin relaxation of the crystals of bone, dental enamel, and synthetic hydroxyapatites. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research, 17(3):472–80, Mar 2002. [DOI] [PubMed]
- 59.Wu Y., Ackerman J.L., Strawich E.S., Rey C., Kim H.-M., Glimcher M.J. Phosphate ions in bone: identification of a calcium-organic phosphate complex by 31P solid-state NMR spectroscopy at early stages of mineralization. Calcified Tissue Int. 2003;72(5):610–626. doi: 10.1007/s00223-002-1068-8. [DOI] [PubMed] [Google Scholar]
- 60.Elizabeth A McNally, Henry P Schwarcz, Gianluigi A Botton, and A Larry Arsenault. A model for the ultrastructure of bone based on electron microscopy of ion-milled sections. PLOS One, 7(1):e29258, 2012. [DOI] [PMC free article] [PubMed]
- 61.Yan Wang, Stanislas Von Euw, Francisco M Fernandes, Sophie Cassaignon, Mohamed Selmane, Guillaume Laurent, Gérard Pehau-Arnaudet, Cristina Coelho, Laure Bonhomme-Coury, Marie-Madeleine Giraud-Guille, et al. Water-mediated structuring of bone apatite. Nature Materials, 12(12):1144–1153, 2013. [DOI] [PubMed]
- 62.Natalie Reznikov, Matthew Bilton, Leonardo Lari, Molly M. Stevens, and Roland Kröger. Fractal-like hierarchical organization of bone begins at the nanoscale. Science, 360(6388), 2018. [DOI] [PMC free article] [PubMed]
- 63.Yi Fei Xu, Fabio Nudelman, E. Deniz Eren, Maarten J.M. Wirix, Bram Cantaert, Wouter H. Nijhuis, Daniel Hermida-Merino, Giuseppe Portale, Paul H.H. Bomans, Christian Ottmann, Heiner Friedrich, Wim Bras, Anat Akiva, Joseph P.R.O. Orgel, Fiona C. Meldrum, and Nico Sommerdijk. Intermolecular channels direct crystal orientation in mineralized collagen. Nature Communications, 11(1):1–12, 2020. [DOI] [PMC free article] [PubMed]
- 64.Erin E Wilson, Ayorinde Awonusi, Michael D Morris, David H Kohn, Mary M J Tecklenburg, and Larry W Beck. Three structural roles for water in bone observed by solid-state NMR. Biophysical journal, 90(10):3722–3731, May 2006. [DOI] [PMC free article] [PubMed]
- 65.S. Maltsev, M.J. Duer, R.C. Murray, and C. Jaeger. A solid-state NMR comparison of the mineral structure in bone from diseased joints in the horse. Journal of Materials Science, 42(21), 2007.
- 66.Stanislas Von Euw, Yan Wang, Guillaume Laurent, Christophe Drouet, Florence Babonneau, Nadine Nassif, and Thierry Azaïs. Bone mineral: new insights into its chemical composition. Scientific Reports, 9(1):1–11, 2019. [DOI] [PMC free article] [PubMed]
- 67.Ratan Kumar Rai and Neeraj Sinha Dehydration-Induced Structural Changes in the CollagenÀHydroxyapatite Interface in Bone by High-Resolution Solid-State NMR Spectroscopy. 2011;115:14219–14227. [Google Scholar]
- 68.Sergey Maltsev, Melinda J. Duer, Rachel C. Murray, and Christian Jaeger. A solid-state NMR comparison of the mineral structure in bone from diseased joints in the horse. Journal of Materials Science, 42(21):8804–8810, Jul 2007.
- 69.Duer Melinda J, Frisciç Tomislav, Murray Rachel C, Reid David G, Wise Erica R. The mineral phase of calcified cartilage: its molecular structure and interface with the organic matrix. Biophys. J . 2009;96(3372–8):4. doi: 10.1016/j.bpj.2008.12.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stanislas Von Euw, Widad Ajili, Tsou Hsi Camille Chan-Chang, Annette Delices, Guillaume Laurent, Florence Babonneau, Nadine Nassif, and Thierry Azaïs. Amorphous surface layer versus transient amorphous precursor phase in bone – A case study investigated by solid-state NMR spectroscopy. Acta Biomaterialia, 59:351–360, 2017. [DOI] [PubMed]
- 71.Sergey V. Dorozhkin. Calcium Orthophosphates in Nature, Biology and Medicine. Materials, 2(2):399–498, Apr 2009.
- 72.Borle A.B., Nichols N., Nichols G.N. Metabolic Studies of Bone in Vitro. J. Biol. Chem. 1960;235(4):1211–1214. [PubMed] [Google Scholar]
- 73.Neuman W.F., Neuman M.W., Brommage R. Aerobic glycolysis in bone: Lactate production and gradients in calvaria. Am. J. Physiol. - Cell Physiol. 1978;3(1):C41–C50. doi: 10.1152/ajpcell.1978.234.1.C41. [DOI] [PubMed] [Google Scholar]
- 74.Kanz Fabian, Reiter Christian, Risser Daniele U. Citrate content of bone for time since death estimation: Results from burials with different physical characteristics and known PMI. J. Forensic Sci. 2014;59(3):613–620. doi: 10.1111/1556-4029.12341. [DOI] [PubMed] [Google Scholar]
- 75.John P Kemp, Adrian Sayers, William D Fraser, George Davey Smith, Mika Ala–Korpela, David M Evans, and Jonathan H Tobias. A Metabolic Screen in Adolescents Reveals an Association Between Circulating Citrate and Cortical Bone Mineral Density. Journal of Bone and Mineral Research, page e3697, 2019. [DOI] [PubMed]
- 76.Elizabeth C. Davis and Donald C. Jackson. Lactate uptake by skeletal bone in anoxic turtles, Trachemys scripta. Comparative Biochemistry and Physiology - A Molecular and Integrative Physiology, 146(3):299–304, 2007. [DOI] [PubMed]
- 77.Ozlen F. Yasar, Wei Chih Liao, Renny Mathew, Yang Yu, Baltzar Stevensson, Yihong Liu, Zhijian Shen, and Mattias Edén. The Carbonate and Sodium Environments in Precipitated and Biomimetic Calcium Hydroxy-Carbonate Apatite Contrasted with Bone Mineral: Structural Insights from Solid-State NMR. Journal of Physical Chemistry C, 125(19):10572–10592, 2021.
- 78.Jackson Donald C., Goldberger Zachary, Visuri Susanna, Armstrong Ryan N. Ionic exchanges of turtle shell in vitro and their relevance to shell function in the anoxic turtle. J. Exp. Biol. 1999;202(5):513–520. doi: 10.1242/jeb.202.5.513. [DOI] [PubMed] [Google Scholar]
- 79.P Cartier. The mineral constituents of calcified tissues. 5. Organic anions of bone. Bulletin de la Societe de Chimie Biologique, 33:161–169, 1951. [PubMed]
- 80.Neuman W.F.F., Neuman M.W.W. The Nature of the Mineral Phase of Bone. Chem. Rev. 1953;53(1):1–45. [Google Scholar]
- 81.Leslie C Costello, Renty B Franklin, Mark a Reynolds, and Meena Chellaiah. The Important Role of Osteoblasts and Citrate Production in Bone Formation: Osteoblast Citration as a New Concept for an Old Relationship. The Open bone journal, 4(Lcc):27–34, 2012. [DOI] [PMC free article] [PubMed]
- 82.Renty B Franklin, Meena Chellaiah, Jing Zou, Mark a Reynolds, and Leslie C Costello. Evidence that osteoblasts are specialized citrate-producing cells that provide the citrate for incorporation into the structure of bone. The open bone journal, 6:1–7, 2014. [DOI] [PMC free article] [PubMed]
- 83.Erika Davies, Karin H Müller, Wai Ching Wong, Chris J Pickard, David G Reid, Jeremy N Skepper, and Melinda J Duer. Citrate bridges between mineral platelets in bone. Proceedings of the National Academy of Sciences of the USA, 111(14):E1354–E1363, 2014. [DOI] [PMC free article] [PubMed]
- 84.Danielle Laurencin, Yang Li, Melinda J. Duer, Dinu Iuga, Christel Gervais, and Christian Bonhomme. A 43Ca nuclear magnetic resonance perspective on octacalcium phosphate and its hybrid derivatives. Magnetic Resonance in Chemistry, (January):1–14, 2021. [DOI] [PubMed]
- 85.Laurencin Danielle, Alan Wong J.V., Hanna Ray Dupree, Smith M.E. A High-Resolution 43Ca Solid-State NMR Study of the Calcium Sites of Hydroxyapatite. J. Am. Chem. Soc. 2008;130(8):2412–2413. doi: 10.1021/ja710557t. [DOI] [PubMed] [Google Scholar]
- 86.Jiadi Xu, Peizhi Zhu, Zhehong Gan, Nadder Sahar, Mary Tecklenburg, Michael D Morris, David H Kohn, and Ayyalusamy Ramamoorthy. Natural-abundance 43ca solid-state nmr spectroscopy of bone. Journal of the American Chemical Society, 132(33):11504–11509, 2010. [DOI] [PMC free article] [PubMed]
- 87.James P Yesinowski and Hellmut Eckert. Hydrogen environments in calcium phosphates: proton MAS NMR at high spinning speeds. Journal of the American Chemical Society, 109(21):6274–6282, Oct 1987.
- 88.J L L Miquel, L Facchini, a.P. Legrand, C Rey, J Lemaitre, A P Legrand, C Rey, J Lemaitre, a.P. Legrand, C Rey, J Lemaitre, A P Legrand, C Rey, and J Lemaitre. Solid state NMR to study calcium phosphate ceramics. Colloids and Surfaces, 45:427–433, 1990.
- 89.Mattias Edén. Structure and formation of amorphous calcium phosphate and its role as surface layer of nanocrystalline apatite: Implications for bone mineralization. Materialia, 17(April), 2021.
- 90.Karin H Müller, Robert Hayward, Rakesh Rajan, Meredith Whitehead, Andrew M Cobb, Sadia Ahmad, Mengxi Sun, Ieva Goldberga, Rui Li, Uliana Bashtanova, et al. Poly (adp-ribose) links the dna damage response and biomineralization. Cell Reports, 27(11):3124–3138, 2019. [DOI] [PMC free article] [PubMed]
- 91.Nudelman Fabio, Lausch Alexander J., Sommerdijk Nico A.J.M., Eli D. Sone. In vitro models of collagen biomineralization. J. Struct. Biol. 2013;183(2):258–269. doi: 10.1016/j.jsb.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 92.Iline-Vul Taly, Kulpanovich Alexey, Nadav-Tsubery Merav, Semionov Artyom, Keinan-Adamsky Keren, Goobes Gil. How does osteocalcin lacking γ-glutamic groups affect biomimetic apatite formation and what can we say about its structure in mineral-bound form? J. Struct. Biol. 2019;207(2):104–114. doi: 10.1016/j.jsb.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 93.Taly Iline-Vul, Raju Nanda, Borja Mateos, Shani Hazan, Irina Matlahov, Ilana Perelshtein, Keren Keinan-Adamsky, Gerhard Althoff-Ospelt, Robert Konrat, and Gil Goobes. Osteopontin regulates biomimetic calcium phosphate crystallization from disordered mineral layers covering apatite crystallites. Scientific Reports, 10(1):1–16, 2020. [DOI] [PMC free article] [PubMed]
- 94.Nikel Ondrej, Laurencin Danielle, McCallum Scott A., Gundberg Caren M, Vashishth Deepak. Nmr investigation of the role of osteocalcin and osteopontin at the organic–inorganic interface in bone. Langmuir. 2013;29(45):13873–13882. doi: 10.1021/la403203w. [DOI] [PMC free article] [PubMed] [Google Scholar]